Microscopic imaging methods facilitate single-molecule nanopore sensing without the need of electronic connections.
Abstract
A wide variety of single molecules can be identified by nanopore sensing. However, all reported nanopore sensing applications result from the same measurement configuration adapted from electrophysiology. Although urgently needed in commercial nanopore sequencing, parallel electrophysiology recording is limited in its cost and its throughput due to the introduced complexities from electronic integration. We present the first electrode-free nanopore sensing method defined as DiffusiOptoPhysiology (DOP), in which single-molecule events are monitored optically without any electrical connections. Single-molecule sensing of small molecules, macromolecules, and biomacromolecules was subsequently demonstrated. As a further extension, a fingertip-sized, multiplexed chip with single-molecule sensing capabilities has been introduced, which suggests a new concept of clinical diagnosis using disposable nanopore sensors. DOP, which is universally compatible with all types of channels and a variety of fluorescence imaging platforms, may benefit diverse areas such as nanopore sequencing, drug screening, and channel protein investigations.
INTRODUCTION
Natural cross-membrane transport is assisted by different membrane transport proteins (1). The transported solutes such as small ions (2), water (3), sugar (4), or even genetic materials (5) are critical for the regulation of different cell activities. Although the detailed transport mechanism varies (6), the fact that single-molecule identities could be reported during channel translocation forms the fundamental basis of nanopore sensing as a biomimetic approach (7). As reported, nanopore sensing has been carried out from a planar lipid membrane (8), a droplet interface bilayer (DIB) (9, 10), a hydrogel interface bilayer (11, 12), synthetic solid-state membranes (13), a glass nanopipette (14), or a cell membrane (15). However, the core setup, which was adapted from electrophysiology, has remained unchanged since its first appearance (8, 16, 17).
During electrophysiology, the Ag/AgCl electrode pair serves to apply a transmembrane electrical potential, which drives a sustained electromigration of ions and charged analytes. It also serves to record the ionic current fluctuations for single-molecule identification (Fig. 1A). Without the electrodes, although thermal diffusion of ions across a nanopore exists in both directions, the net flow of ions and the electric field within the whole electrolyte-containing space is strictly zero due to the rule of electroneutrality (Fig. 1B) (18), when the ion selectivity of nanopores is too weak to be considered.
Fig. 1. DiffusiOptoPhysiology and its application in trimethyl-β-cyclodextrin sensing.
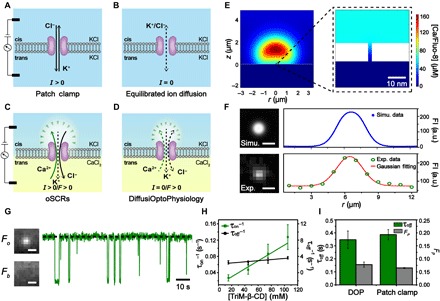
(A to D) Schematics of ion transport through a nanopore in different measurement platforms. Electromigration and diffusion of ions are indicated by solid and dashed lines, respectively. (A) During electrophysiology recording, electrophoretic motion of K+ and Cl− through a nanopore is observed when a transmembrane potential is applied via a pair of Ag/AgCl electrodes. (B) In the absence of electrodes, although thermal motion of ions across the nanopore exists in both directions, no net flow of ion transport should happen according to the rule of electroneutrality. (C) During optical single-channel recordings (oSCRs), directional motion of Ca2+, which is electrophoretically driven through a nanopore, establishes a steep Ca2+ concentration gradient. Upon binding with Fluo-8 in cis, the Fluo-8/Ca2+ complex around the pore vicinity emits strong fluorescence. (D) During DiffusiOptoPhysiology (DOP), a mild Ca2+ concentration gradient could be established around the pore vicinity due to the thermal motion of ions. Upon binding with Fluo-8, a weaker fluorescence emission than (C) is expected. (E) A cross-sectional view of the spatial distribution of the Fluo-8/Ca2+ complex around the pore. Dashed box: The zoomed-in view of the immediate vicinity area near the nanopore. (F) Top left: Corresponding image result from computer simulation. Top right: The simulated fluorescence intensity (FI) profile follows a Gaussian distribution. Bottom left: A representative frame acquired from DOP recording for a single wild-type (WT) α-hemolysin (α-HL) nanopore. Bottom right: The corresponding fluorescence intensity profile also follows a Gaussian distribution. Scale bars, 4 μm. a.u., arbitrary units. (G) Single-molecule sensing of trimethyl-β-cyclodextrin (TriM-β-CD) (75 mM) with an α-HL nanopore during DOP recording. Scale bars, 4 μm. (H) Plot of the reciprocals of the mean interevent intervals (1/τon) and mean dwell time (1/τoff) versus TriM-β-CD concentration. The mean and SD come from three independent experiments for each condition (n = 3). (I) Statistics of τoff and FP results acquired from DOP and electrophysiology recording at +20 mV, respectively. The DOP recordings (F to I) were performed with 1.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 0.75 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. The electrophysiology recordings were performed with 1.5 M KCl and 10 mM HEPES (pH 7.0) in both sides of the membrane. TriM-β-CD was added to the cis side with a final concentration of 4 mM.
Electrophysiology measurements, which provide a superior temporal (~10 μs) and amplitude resolution (<0.1 pA), satisfy the need of single-channel recording–based applications but are disadvantageous in the throughput (19). Although urgently needed in nanopore sequencing and drug screening, simultaneous readout from 1 million channels has not been achieved without a substantial sacrifice in the cost or the size of the device (19, 20). Such an urgent need thus stimulated us to reconsider a simplified strategy for high-throughput channel recordings, which may be further acquired from biomimicry.
Bacterial phage T4 injects its genomic DNA through channel proteins when intact with host cells (21). Staphylococcus aureus α-hemolysin (α-HL) leads to hemolysis of target cells due to passive leakage of nutrients through inserted channels (22). These spontaneous molecular transport processes, as acquired from natural evolution, remind us that external electronics are not indispensable for molecular transport. The challenge remaining is how nanopore sensing signals could be acquired without electrical connections.
Adapted from optical patch clamping (23, 24), optical single-channel recording (oSCR) (25–29) has demonstrated an alternative strategy, which optically monitors Ca2+ fluxes through individual nanopores embedded in a DIB (Fig. 1C). Although oSCR is advantageous in high-throughput measurements, a pair of electrodes was still used to electrophoretically drive a sustained flow of Ca2+ through nanopores (29). Manual insertion of electrodes into aqueous droplets, which requires delicate micromanipulation skills and may lead to a high risk of bilayer rupture (29), has hampered its wide use in academic studies and industrial applications.
DiffusiOptoPhysiology (DOP), which is a simplified form of oSCR created by omitting all electrical connections, optically monitors fluorescence emission resulting from diffusive binding of Ca2+ with its indicator dye Fluo-8 through a nanopore sensor (Fig. 1D). Direct sensing of small molecules, macromolecules, and biomacromolecules was subsequently demonstrated from direct fluorescence readout. DOP enables parallel measurements from thousands of nanopores with highly accessible and biocompatible materials and costs <$1 in consumables for a single use. New concepts of clinical diagnosis may thus be developed using disposable chips equipped with nanotechnology sensors. The reduced technical barriers concerning the cost, facilities, and skills enable facile accomplishment of a nanopore measurement by any researcher and requires negligible training. Diverse research areas such as high-throughput drug screening or fundamental investigations of ion channels may thus benefit.
RESULTS
Single-molecule sensing of trimethyl-β-cyclodextrin by DOP: A proof of concept demonstration
According to Fig. 1D, the basic configuration of DOP recording includes asymmetric electrolyte buffers separated by a semipermeable membrane with inserted nanopores. The compartment that is filled with KCl, Fluo-8, and EDTA is defined as the cis side, and the compartment that is filled with CaCl2 is defined as the trans side. A biological nanopore, which is the only conducting path between cis and trans sides, promotes binding of Ca2+ and Fluo-8 via thermodynamic diffusion through channel transport, which is driven by the chemical gradient. FluoCa, which is the bound form of Ca2+ and Fluo-8, emits fluorescence around each nanopore to report the opening state of the sensor.
Theoretically, a finite element method (FEM) simulation was established, which was adapted from the Poisson-Nernst-Planck-Stokes model (30) (section S1 and fig. S1). To mimic experimental operations, simulation parameters such as different combinations of reagent concentrations could be adjusted by setting different boundary conditions. Tentatively, the simulation was performed by setting the boundary condition in cis as 1.5 M KCl, 40 μM Fluo-8, and 400 μM EDTA and the boundary condition in trans as 0.75 M CaCl2 along with a cylindrical channel geometry of 2 nm in diameter. From the result, a concentration gradient of FluoCa was established immediately above the nanopore (Fig. 1E). Resulting from simultaneous emission from FluoCa, a strong fluorescence intensity contrast is expected on top of the nanopore. The intensity profile, which is generated as a mimic of total internal reflection fluorescence (TIRF) imaging (section S1), follows a Gaussian distribution with a full width at half maximum (FWHM) of 2.670 μm (Fig. 1F, top).
By omitting the need for an electrode arrangement, we carried out experiments on a miniaturized device (10 mm by 10 mm by 1 mm) manufactured from bulk polymethyl methacrylate (PMMA) (fig. S2). A DIB was established between an aqueous droplet and a 100-nm-thick hydrogel sheet (fig. S3). The aqueous droplet was composed of 1.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0). The hydrogel sheet was composed of 0.75 M CaCl2 and 10 mM HEPES (pH 7.0) with 2.5% (v/w) low-melt agarose. Wild-type (WT) α-HL, which was placed in the aqueous droplet, spontaneously inserts into the DIB and appeared as a bright fluorescent spot during TIRF imaging (Fig. 1F, bottom). The fluorescence intensity profile of a representative frame from TIRF imaging follows an approximate Gaussian distribution with an FWHM of 2.583 μm (Fig. 1F, bottom), which resembles that from the simulation.
Trimethyl-β-cyclodextrin (TriM-β-CD), which interacts with the restriction of a WT α-HL nanopore, generates long-residing and deep pore blockade events during electrophysiology recordings (31). This ease of observation enables TriM-β-CD as a representative small-molecule analyte for a proof-of-concept demonstration of single-molecule sensing using DOP. To maintain a stable analyte concentration during DOP recording, we added TriM-β-CD in cis with a final concentration of 75 mM. TriM-β-CD binding from cis was verified by corresponding electrophysiology measurements (fig. S4). During DOP recording, stochastic binding of TriM-β-CD with α-HL, which results in a restricted Ca2+ flux through the channel, generates a highly distinguishable image contrast between the open (Fo) and the blocked (Fb) states of an α-HL nanopore (Fig. 1G). Successive pore blockades were observed from the corresponding fluorescence traces (Fig. 1G), which were extracted from continuously recorded image series (section S2). To perform quantitative comparisons between different trials, we calibrated and normalized all fluorescence traces before the analysis (fig. S5 and S6).
Single-molecule sensing events from a normalized fluorescence trace were characterized by the event dwell time (toff), the interevent duration (ton), and the percentage blockade depth (%Fb). A histogram of toff and ton displays an exponential distribution, which could be fit and characterized by their mean time constants τoff and τon, respectively (fig. S7). By varying the TriM-β-CD concentration in cis, the reciprocal of dwell time (1/τoff) remains constant, whereas the reciprocal of interevent interval (1/τon) correlates linearly with the TriM-β-CD concentration in cis (Fig. 1H and table S1).
From DOP recording, a mean τoff value of 0.347 ± 0.067 s and a mean FP value of 0.078 ± 0.010 were recorded (n = 3). Here, FP is defined as the mean blockade depth, which was derived from %Fb values from each trial of DOP recording (fig. S7). Corresponding electrophysiology results acquired with a +20-mV potential bias generated a τoff value of 0.386 ± 0.392 s and an IP value of 0.065 ± 0.002. Here, IP is defined as the mean blockade depth %Ib from each electrophysiology recording trial. Three independent trials were performed for each set of measurement conditions to form the statistics. This resemblance of results thus confirmed the feasibility of single-molecule sensing by DOP (Fig. 1I).
Although not demonstrated, single-molecule sensing of other small molecules, such as sugar (32, 33), ions (34), nucleotides (35), neurotransmitters (36), and amino acids (37) could, in principle, be similarly performed by DOP recording with an added advantage to the throughput. However, the strength of fluorescence emission and the analyte binding efficiency could be further improved to match those in electrophysiology.
Enhanced DOP sensing by directional osmosis
During routine electrophysiology recording, the applied electrochemical gradient is critical to drive a sustained flow of charged particles, such as ions and analytes. Intuitively, to drive a directed flow of analytes into the nanopore sensor without the use of electrodes, it would appear that an asymmetry of another form has to be introduced.
The DIB, which is a self-assembled membrane composed of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) lipid, is selectively permeable to water molecules rather than ions (38, 39). When a difference of osmolarity concentration (Csolute = iMsolute) exists across the DIB, an osmotic pressure is established according to ∆π = (Csolute, cis − Csolute, trans)RT, where i is the dimensionless van’t Hoff index that addresses the number of dissociated ions from each solute molecule, Msolute is the molar concentration of the solute, R is the ideal gas constant, and T is the temperature in Kelvin (40). Here, the positive direction of the osmotic pressure is defined to be from trans to cis. This osmotic pressure subsequently drives an oriented flow of water, ions, and analytes through a biological nanopore, which is inserted in the membrane (27). As a result, an enhanced translocation efficiency for analyte should be achieved with this introduced asymmetry.
To verify this hypothesis experimentally, we carried out a series of DOP recordings in DIBs (Fig. 2A) with varying KCl concentrations (1.0 to 2.5 M) in cis, while the CaCl2 concentration in trans was kept constant at 0.75 M. Tentatively, α-HL and TriM-β-CD were selected again as the model sensor and analyte, respectively, and the concentration of TriM-β-CD in cis was fixed at 15 mM. From representative DOP recordings, we observed an enhanced capture rate of TriM-β-CD from time-extended fluorescence traces, when the KCl concentration in cis was decreased from 2.5 to 1.0 M (Fig. 2A). By evaluating the 1/τon values for events from independent measurements, a systematic decrease of 1/τon was observed in accordance with the decrease of osmotic pressure (Fig. 2B and table S2), which means that a higher rate of event detection was observed with the assistance from a directed osmotic flow.
Fig. 2. Enhanced sensing performance with adjusted osmosis during DOP recording.
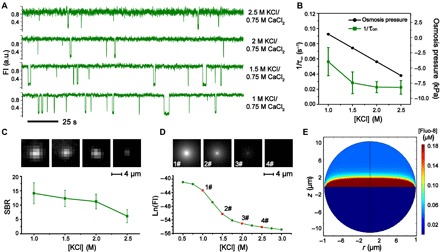
(A) Fluorescence traces acquired from DOP recording with different electrolyte combinations. DOP recordings were performed with α-HL nanopores. TriM-β-CD was added to cis with a final concentration of 15 mM. The interevent interval τon decreases when a larger osmosis gradient between cis and trans was established. (B) Plot of the osmotic pressure and 1/τon as a function of the KCl concentration in cis. The mean and SD are derived from three sets of independent experiments (n = 3). (C) The SBR analysis of fluorescent imaging results with different KCl concentrations in cis. Top: Representative images acquired from DOP recordings. The images from left to right were acquired by DOP recording with [KCl] in cis as 1.0, 1.5, 2.0, and 2.5 M, respectively. The value of SBR decreases when KCl concentration in cis increases. The mean and SD comes from five independent experiments (n = 5). Experiments in (A) to (C) were performed with 1 to 2.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 0.75 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. (D) Plot of simulated fluorescence intensity as a function of the KCl concentration in cis. Top: Corresponding two-dimensional (2D) profiles of fluorescence intensities from the simulation, which resembles the results acquired from DOP in (C). The simulations were performed with 1 to 2.5 M KCl in cis and 0.75 M CaCl2 in trans. (E) A cross-sectional view of the spatial distribution of Fluo-8 in the simulation space. The boundary condition of the simulation was set as 1.0 M KCl in cis and 0.75 M CaCl2 in trans. Sustained osmotic flow from cis to trans gives rise to an enriched distribution of Fluo-8 in the vicinity of the DIB.
It was also discovered that a notably improved fluorescence image contrast was observed from DOP recordings when an osmotic flow from cis to trans exists. This phenomenon could be observed from the reduced thermal noises in the fluorescence traces with measurement conditions of lower KCl concentrations (Fig. 2A and fig. S8). Here, the high thermal noise observed from the fluorescence trace is a consequence of reduced photon counts during imaging.
To further investigate why the fluorescence intensity from DOP recording could be modulated by osmosis (Fig. 2C and table S3), we carried out a different set of experiments with 1 to 2.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 0.75 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. To avoid interference from analyte binding, we omitted TriM-β-CD. To avoid interference from uneven TIRF illumination or laser power fluctuations when evaluating the brightness of the fluorescence, we introduced a signal-to-background ratio (SBR) value to compare quantitatively different trials of DOP recordings (section S3). From representative image frames and the corresponding SBR values, the brightness of the fluorescence spot was enhanced when a larger osmotic pressure from cis to trans was introduced. Five independent measurements were included for each condition to generate the statistics (Fig. 2C).
This phenomenon could also be observed with the corresponding FEM simulations, which were carried out by setting the boundary condition in cis as 1.0 to 2.5 M KCl, 40 μM Fluo-8, and 400 μM EDTA and the boundary condition in trans as 0.75 M CaCl2 (Fig. 2D). By plotting the Fluo-8 distribution within the simulation space, it can be noticed that a concentrated Fluo-8 distribution was established near the cis side of the membrane when a directed osmotic flow from cis to trans exists (Fig. 2E). This happens because Fluo-8, which is impermeable to the lipid membrane, is enriched via the osmotic flow and, consequently, the fluorescence intensity is enhanced (fig. S9). Although the simulation was carried out with a simplified cylindrical pore geometry with an electrical neutral pore inner surface, an osmotic flow speed of a few micrometers per second was estimated (fig. S10), which explains the enhanced capture rate of TriM-β-CD during DOP recordings (Fig. 2B).
Further SBR optimization with enlarged Ca2+ flux
However, Fluo-8 enrichment by osmosis should not happen in a solid-state nanopore device in which the membrane does not have a semipermeability feature. Alternatively, the SBR from DOP recording could be improved by the introduction of more Ca2+ flux through nanopores. An immediate solution following this strategy is to increase the [CaCl2] in trans, which directly raises the chemical gradient of [Ca2+] across the membrane. To confirm this hypothesis, we carried out a series of DOP recordings by gradually up-regulating the CaCl2 concentration in trans. To avoid the interference from osmosis, the KCl concentration in cis was adjusted accordingly so that the osmolarity concentration in cis and trans were isotonic throughout.
Experimentally, a DIB was established with 0.75, 1.5, or 2.25 M KCl; 400 μM EDTA; 40 μM Fluo-8; and 10 mM HEPES (pH 7.0) in cis and 0.5, 1, or 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. Representative image frames show a systematic enlargement of the fluorescence spot size when acquired with a combination of electrolyte buffer where the [CaCl2] in trans was higher (Fig. 3A). Corresponding two-dimensional (2D) Gaussian fittings (Fig. 3A), which were color coded according to the fitting amplitude, give a more straightforward comparison of the fluorescence intensity acquired under these conditions. A quantitative measure of the SBR and the FWHM from DOP recordings acquired with these electrolyte combinations is demonstrated in Fig. 3B, in which both the FWHM and the SBR (table S4) increase when the osmolarity concentration in both sides of the membrane is up-regulated. A total of 12 independent measurements were included for each condition to produce the statistics.
Fig. 3. Enhanced SBR with increased Ca2+ flux during DOP recording.
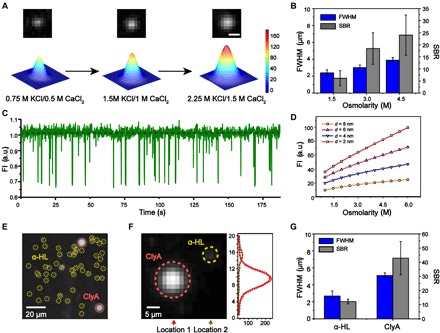
(A) Imaging results (top) and the corresponding 2D Gaussian fittings (bottom) acquired from DOP recording. The CaCl2 concentration in trans was increased when the KCl concentration in cis was adjusted so that the osmolarity concentrations were kept isotonic. The fluorescence spot, which corresponds to Ca2+ flux through a WT α-HL nanopore, becomes brighter with an increased Ca2+ flux. Scale bar, 4 μm. (B) The FWHM and SBR of the fluorescence imaging signals with different electrolyte osmolarity concentrations (n = 12). The DOP recordings in (A) and (B) were carried out with 0.75 to 2.25 M KCl, 400 μM EDTA, 40 μM Fluo-8, 10 mM HEPES, pH 7.0 in cis and 0.5–1.5 M CaCl2, 10 mM HEPES, pH 7.0 in trans. (C) A representative fluorescence trace shows PEG 1500 translocation signals through a WT α-HL nanopore, as acquired by DOP recording. PEG 1500 was added to the agarose substrate reaching a final concentration of 20 mM. The DOP recording was carried out with 2.25 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. (D) Simulated total fluorescence intensity as a function of osmolarity concentration, shown for four different pore sizes with a diameter of 2, 4, 6, and 8 nm, respectively. The electrolyte concentrations were kept isotonic to avoid the interference of osmosis in this demonstration. (E) Simultaneously imaging of WT α-HL and ClyA-RR nanopores in the same DIB. Because of a larger channel conductance, ClyA-RR appears as a larger and brighter spot in comparison with WT α-HL in the same field of view (yellow dashed circles, WT α-HL; red dashed circles, ClyA-RR). Scale bar, 20 μm. (F) Left: Simultaneous imaging of an α-HL and a ClyA-RR. Right: The fluorescence intensity profile along vertical lines as marked by location 1 and 2, respectively. The fluorescence intensity profile is fitted with a Gaussian distribution. Scale bar, 5 μm. (G) The FWHM and SBR of the fluorescence imaging signals of WT α-HL and ClyA-RR (n = 5). DOP recordings as demonstrated in (E) and (F) were carried out with 1.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans.
Polyethylene glycol (PEG), which is a macromolecule that is electrically neutral when dissolved in a buffer of neutral pH, has been demonstrated to translocate through α-HL nanopores during electrophysiology recordings (41). It was reported that the capture rate was enhanced, and the event residing time was extended when measured with an electrolyte buffer at higher salt concentrations (42). As a demonstration, PEG 1500 was selected as the model analyte for single-molecule sensing of macromolecules by DOP.
Experimentally, the DIB was established with 2.25 M KCl, 400 μM EDTA, 40 μM Fluo-8, 20 mM PEG 1500, and 10 mM HEPES (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. Upon the addition of 20 mM PEG 1500 in cis, an abundance of spiky translocation events immediately appeared from the extracted fluorescence traces (Fig. 3C). The resemblance of these pore translocation features to reported electrophysiology data confirms that PEG 1500 could be sensed by DOP recording, similar to that demonstrated with TriM-β-CD.
However, the solubility of the analyte is normally decreased in an electrolyte buffer of high salt concentrations (42). Furthermore, the salt concentration is also limited by the maximum solubility of electrolytes in water (CaCl2, 6.767 M; KCl, 3.408 M, at 20°C). To produce more Ca2+ flux without reaching this limit, a nanopore sensor with a larger aperture could be introduced during DOP recording, and this is confirmed by the corresponding FEM studies (section S1 and Fig. 3D). According to reported crystallography results, the restriction of a Cytolysin A (ClyA) nanopore, which measures 3.8 nm in diameter, is 2.7 times the diameter of an α-HL (43). Acknowledging its large channel opening, ClyA and its variants were developed to sense large biomacromolecules such as double-stranded DNA (dsDNA) or small proteins (44, 45). ClyA-RR, which was reported to be a charge-optimized mutant and could effectively translocate dsDNA during electrophysiology recording (44), was selected for DOP recording (see Materials and Methods and fig. S11). Although not demonstrated, phi29 connector protein (45) or solid-state nanopores (26) are also good candidates.
Tentatively, the DIB was established with 1.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. To perform a quantitative comparison with nanopores of different channel openings during DOP recording, we placed both dodecameric ClyA-RR nanopores and heptameric α-HL nanopores in the droplet for simultaneous measurements from the same DIB.
When inserted, a ClyA-RR nanopore appears as a huge and dazzling fluorescence spot, whereas an α-HL nanopore appears small in size with a dim intensity (Fig. 3, E and F, and movie S1). The FWHM and SBR as derived from DOP recordings with ClyA-RR outperform those from α-HL with the introduction of more Ca2+ flux across the membrane (Fig. 3G and table S5). Five independent measurements were performed to generate the statistics. The observed fluorescence enhancement of ClyA-RR in contrast to α-HL results from a much larger pore conductance, which is 10 times more conductive than that of α-HL (fig. S11).
Although numerous efforts have been made to counteract the electrophoretic force during DNA translocation (46), the electrophoretic force, which efficiently untangles coiled DNA during translocation (47), was still considered indispensable during DNA sensing. However, the long persistence length of dsDNA (48) and the wide opening of ClyA nanopore may reduce the entropic barrier for dsDNA translocation (49). Furthermore, the relatively large vestibule of ClyA may also serve to accommodate dsDNA in a form of partial translocation to report the sensing signal of dsDNA during the DOP recording as well.
dsDNA sensing by ClyA nanopores
Experimentally, the DIB was established with 2.25 M KCl, 10 mM HEPES, 400 μM EDTA, 40 μM Fluo-8 (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. dsDNA, which is composed of 78 base pairs (bp) (table S6 and “Materials” section), was optionally dissolved in the aqueous droplet with a final concentration of 2 μM (Fig. 4A).
Fig. 4. dsDNA sensing using ClyA-RR nanopores.
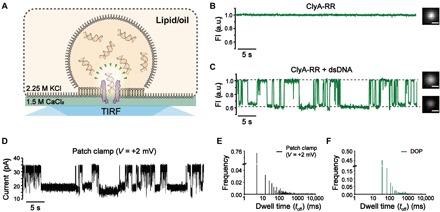
(A) The schematic diagram of dsDNA sensing using ClyA-RR during DOP recording. (B) DOP imaging of a ClyA-RR nanopore and the corresponding fluorescence trace. No dsDNA was added in the droplet. (C) DOP imaging of ClyA-RR nanopore and the corresponding fluorescence trace when dsDNA was added in the droplet with a final concentration of 2 μM. Successive deep and long-residing fluorescence blockades were observed. Scale bar, 5 μm. The DOP recordings in (B) and (C) were performed with 2.25 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. dsDNA, which is of 78 bp, was add to cis with a final concentration of 2 μM. (D) Electrophysiology recording of dsDNA events recorded with ClyA-RR nanopore at +2 mV. With voltages as low as +2 mV, current blockages were still observable. The electrophysiology recordings were performed with 2.25 M KCl and 10 mM HEPES (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. dsDNA, which is of 78 bp, was add to cis with a final concentration of 2 μM. (E) Histogram of the dwell time for dsDNA events extracted from electrophysiology recordings. (F) Histogram of the dwell time for dsDNA events extracted from DOP recordings. Because of a limited acquisition time (30 ms) of the electron-multiplying charge-coupled device in a large field of view, fast dsDNA translocations cannot be fully resolved. Data from DOP recordings (shown in olive color) were acquired with a frame rate of 30 ms. The electrophysiology trace (shown in black) were recorded with a +2-mV applied bias, sampled at 25 kHz, and low-pass–filtered at 1 kHz.
With no dsDNA added into the droplet, the representative fluorescence trace from a ClyA nanopore appears stably open with no spontaneous gating activity (Fig. 4B). When dsDNA was added to the droplet, successive fluorescence blockades spontaneously appeared during the DOP recording (Fig. 4C and movie S2). Being able to perform parallel measurements by DOP, dsDNA sensing from multiple ClyA-RR pores could be monitored simultaneously (movie S3). The observed blockades from DOP recording showed a mean FP of 0.682 ± 0.026 and a mean τoff of 0.224 ± 0.148 s (n = 3; table S7). These results suggest that dsDNA interaction with ClyA-RR was happening without an applied electrochemical gradient. The long residing time may result from the omitted electrophoretic force during the measurements or it may be because the dsDNA was trapped within the huge vestibule structure of ClyA. DOP recordings were further carried out with a gradient of final concentration of dsDNA in the droplet. We noticed that the 1/τon correlates linearly with the dsDNA concentration in cis (fig. S12), which has evidenced that the events observed results from the added dsDNA.
To further verify this phenomenon via routine electrophysiology recordings, we established a planar lipid membrane with 2.25 M KCl and 10 mM HEPES (pH 7.0) in cis and 1.5 M CaCl2 and 10 mM HEPES (pH 7.0) in trans. The 78-bp dsDNA was added in cis with a final concentration of 2 μM. A +2-mV bias was applied to mimic DOP recording, in which the transmembrane electric potential was strictly zero. Figure 4D shows representative electrophysiology traces recorded with a +2-mV transmembrane potential. Similar DNA events as observed from DOP recordings were monitored, which verified our hypothesis that dsDNA could interact with ClyA and generate detectable sensing signals when optically monitored without electrodes. The IP value, which was 0.645 ± 0.003 at +2 mV, was also qualitatively consistent with the blockades observed from DOP recordings (fig. S13). The dwell time was widely distributed from 1 to 105 ms during electrophysiology recordings, whereas because of the limited acquisition bandwidth from DOP recording, fast events below 30 ms are not detectable optically (Fig. 4, E and F).
The resemblance in the % blockade depth of the dsDNA events from DOP and electrophysiology from our measurements with that from reported literature (44) has suggested that the dsDNA might translocate through the nanopore. With an isotonic electrolyte buffer combination across the DIB and without an applied electrophoretic force, the observation of dsDNA blockade events from ClyA-RR nanopores in a DOP platform has suggested that the nature of translocation might be diffusion limited. We believe that the large channel opening of ClyA-RR has considerably reduced the entropic barrier that a piece of dsDNA needs to overcome to translocate through the pore.
To further evidence dsDNA translocation through a ClyA-RR nanopore during a DOP measurement, a follow-up investigation that uses fluorescently labeled DNA as the analyte may be carried out in a dual-color fluorescence microscopy assay. However, the fact that the observed events in Fig. 4C and fig. S12 result from dsDNA interaction with ClyA-RR provide concrete evidence for single-molecule sensing of dsDNA by DOP.
Multiplex DOP recording and future prospects
By omitting the need for an electrode arrangement, DOP enables a much more compact device size while the advantages of low cost (<1$ for disposable devices for each measurement) and high throughput are retained. This configuration is suitable for fabricating disposable nanopore chips for clinical diagnosis, where cross contamination is strictly prohibited.
However, DOP measurements from a single DIB were restricted to one combination of pore and analytes. By omitting the need for electrode accommodation, DOP enables multiplex recording from different DIBs with an extremely simple configuration and a greatly reduced measurement volume, in which different analytes could be physically separated into various water in oil compartments.
As a proof of concept, microdroplets (~30 pl) containing ClyA-RR nanopores were generated and pipetted into a measurement reservoir filled with lipid/oil solution (Fig. 5A). In preparation of aqueous droplets, ClyA-RR nanopores were added into the aqueous buffer, which was composed of 1.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0). Aqueous droplets (0.5 μl) were then pipetted into the lipid/oil solution [5 mg of DPhPC dissolved in a 2-ml 1:1 (v/v) mixture oil of hexadecane and silicone oil]. After vortex oscillation, small droplets of varied sizes were generated. Last, droplets with ~30 pl volume were selected for downstream DOP demonstration. Although not monodispersed in size, many independent DIBs could spontaneously form with extreme ease for subsequent DOP recording (Fig. 5B). In a DIB that was ~40 μm in diameter, a single inserted ClyA nanopore was unambiguously observed as a bright fluorescent spot (Fig. 5C and movie S4), which lasts for ~10 min before the EDTA in the droplet was depleted by Ca2+ binding. This corresponds to an effective measurement density of 103 independent DIBs per square millimeter, such as in a highly ordered DIB array that could be formed with the assistance from microfluidics setups (50). However, this high measurement density could not be easily achieved by oSCR or electrophysiology due to the complexities from the integration of the electronics.
Fig. 5. Multiplex DOP recording with mini-DIBs.
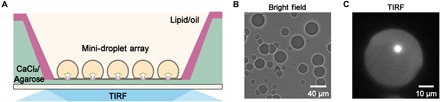
(A) Schematics of a mini-DIB array for multiplex DOP recordings. (B) A bright-field image of the mini-DIB array. (C) A frame of ClyA-RR nanopore inserted in a mini DIB. The diameter of the DIB is ~40 μm. This mini-DIB was established with 1.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0) in cis and 1.5 M KCl and 10 mM HEPES (pH 7.0) in trans.
Although with the advantage of being simple to carry out, electrode-free, economic in consumables, low in measurement volume, and high in throughput, DOP is not without limitations. As a fluorescence imaging technique, the temporal resolution of DOP is generally limited to ~10 ms per frame when recorded in the full field of view (135 μm by 135 μm). The ultimate temporal resolution of DOP may also be limited by many other parameters such as the time of chemical reactions and the photon capture efficiency of the camera; however, these were not observed with the current 10-ms acquisition rate. The slow acquisition speed has limited DOP recordings to analytes that report long-residing pore blockade events. Fluorescence readout from a reduced amount of image pixels could immediately boost the speed of acquisition. With the improved addressability available from a highly ordered array of nanopores, high speed DOP recording could be potentially carried out by multipoint confocal readout (28) or by spinning disk confocal imaging (51).
In the absence of an electric field, the limit of detection during DOP was generally higher than that from electrophysiology or oSCR. However, with the omission of electrode accommodation, DOP reports the smallest measurement volume (down to ~30 pl) from which a single-channel activity can still be probed. This volume is ~1/106 of that from conventional electrophysiology recordings (52) and ~1/104 of that from reported oSCR recordings (25), respectively. This pronounced reduction of the measurement volume is advantageous in reducing the consumption of sample in each measurement. On the other side, the omission of electrodes has disabled the convenience to control the motion direction of analytes during recording. With the gained advantages of low sample consumption and multiplex sensing, the small measurement volume also limits the acquisition time of a DOP assay to ~30 to 45 min. However, within this limited measurement time, a desired amount of sensing events can normally be acquired because of its high-throughput nature.
Although demonstrated with an expensive microscopic setup as a proof of concept, the strong fluorescence emitted from nanopores during DOP recordings and its ease in the configuration has suggested future developments of DOP recordings with low-cost hardware in the form of a handheld instrument. With the above listed reasons, DOP should be considered as a complement instead of a replacement to existing single-channel recording techniques.
DISCUSSION
In summary, we have demonstrated how DOP, which was inspired from natural passive channel transport, could be used as a nanopore sensing platform. Although the fluorescence emission during DOP recording was triggered by passive diffusion and subsequent binding of Ca2+ and Fluo-8, the fluorescence intensity was demonstrated to be strong enough to serve a wide variety of single-molecule sensing applications. Upon combined optimization from electrolytes and channel sizes, this technique enables high-throughput nanopore measurements while the sensing performance remains comparable to conventional electrophysiology recording or oSCR. Although demonstrated with TIRF microscopy, DOP is, in principle, compatible with any fluorescence platforms such as confocal or epifluorescence microscopy. Without the need for space to accommodate electrodes, the measurement volume of DOP was further reduced to ~30 pl, which has pronoucedly reduced the sample consumption and may be suitable for measurements of analytes with an extremely low abundance. DOP recording with an array of microdroplets also enables multiplex measurements from independent compartments, which could be easily established from water in oil separations. Integration of nanotechnology sensors into a chip, without electronics has greatly reduced the cost and size of the consumable devices. This scheme can be easily carried out by researchers with no experience of nanopore sensing and may inspire future clinical applications using disposable nanopore chips.
MATERIALS AND METHODS
Detailed methods are available in the Supplementary Materials.
Materials
Hexadecane, silicone oil AR20, pentane, EDTA, Triton X-100, and PEG 1500 were obtained from Sigma-Aldrich. Potassium chloride, calcium chloride, magnesium chloride, and sodium chloride were from Aladdin. Dioxane-free isopropyl-β-d-thiogalacto-pyranoside, dodecyl β-d-maltopyranoside (DDM), kanamycin sulfate, trimethylamine methane (tris), and imidazole were from Solarbio. Low–melting point agarose and wide-range DNA ladder (20 to 500 bp) were from Takara. Precision plus protein marker and 4 to 15% polyacrylamide gels were from Bio-Rad. Ethanol and acetone were from Sinopharm. Fluo-8H sodium salt (Fluo-8) was from AAT Bioquest. DPhPC was from Avanti Polar Lipids. HEPES was from Shanghai Yuanye Bio-Technology. Escherichia coli strain BL21 (DE3) was from BioMed. TriM-β-CD was from Tokyo Chemical Industry (Shanghai). LB was from Hopebio. All the items listed above were used as received.
KCl buffer [1 to 2.5 M KCl and 10 mM HEPES (pH 7.0)] and CaCl2 buffer [0.5 to 1.5 M CaCl2 and 10 mM HEPES (pH 7.0)] were membrane-filtered (0.2 μm cellulose acetate; Nalgene). For simplicity, 1 to 2.5 M KCl buffer stands for 1 to 2.5 M KCl and 10 mM HEPES (pH 7.0). CaCl2 buffer (0.5 to 1.5 M) stands for 0.5 to 1.5 M CaCl2 and 10 mM HEPES (pH 7.0), if not otherwise stated. The KCl buffer was treated with Chelex 100 resin (Bio-Rad) overnight before use to remove contaminating divalent ions.
High-performance liquid chromatography–purified DNA (table S6) was dissolved in deoxyribonuclease/ribonuclease-free water before use. To form dsDNA, complementary single-stranded DNAs were further dissolved in 1.5 M KCl buffer [(1.5 M KCl and 10 mM HEPES (pH 7.0)] heated to 95°C and gradually cooled down (−5°C/min) to room temperature (25°C) on a polymerase chain reaction thermal cycler (ABI 2720). The protein nanopores used in this paper were α-HL WT and ClyA-RR (fig. S11), which were expressed in E. coli and purified on the basis of published protocols (27, 44).
ClyA-RR preparation
The gene coding for monomeric ClyA-RR (D64R/C87A/L99Q/E103G/S110R/F166Y/I203V/C285S/K294R/H307Y) protein was custom-synthesized and constructed in a pET 30a(+) plasmid (GenScript, NJ). A hexahistidine tag was introduced at the C terminus of the protein for later chromatographic purifications. The plasmid was transformed into E. coli BL21 (DE3) competent cells and cultured in the LB agar plate with kanamycin (50 μg/ml) for 18 hours. A single colony was inoculated into the LB medium containing kanamycin (50 μg/ml) and incubated at 37°C until the optical density at 600 nm reached 4.0. Protein expression was inducted by adding isopropyl β-d-thiogalactoside to the LB medium, reaching a final concentration of 1 mM. The culture medium was further shaken (200 rpm) for 16 hours at 15°C. The cells were then harvested by centrifugation (4000 rpm at 4°C for 20 min). The pellets were collected and resuspended in the lysate buffer [150 mM NaCl, 50 mM tris-HCl, and 10% glycerol (pH 8.0)], lysed by sonication (15 min), and centrifuged (14,000 rpm at 4°C for 40 min) to remove intact cells. After syringe filtration, the supernatant was loaded onto a nickel affinity column (HisTrap HP, GE Healthcare). After washing the column with wash buffer A [150 mM NaCl, 50 mM tris-HCl, 10% glycerol, and 20 mM imidazole (pH 8.0)], the target protein was eluted in sequence using three wash buffers [buffer B: 500 mM NaCl, 15 mM tris-HCl, 10% glycerol, and 300 mM imidazole (pH 8.0); buffer C: 500 mM NaCl, 15 mM tris-HCl, 10% glycerol, and 50 mM imidazole (pH 8.0); buffer D: 500 mM NaCl, 15 mM tris-HCl, 10% glycerol, and 20 mM imidazole (pH 8.0)]. The elution fraction that contains ClyA-RR monomers were determined using SDS–polyacrylamide gel electrophoresis (PAGE) (fig. S11) and stored in the buffer of 270 mM NaCl, 50 mM tris-HCl, 10% glycerol, and 0.2% Triton X-100 (pH 8.0) at −80°C.
According to previous studies (44), 0.25% (w/v) β-dodecylmaltoside (DDM) was added to promote the pore oligomerization. After incubation at 25°C for 15 min, the result of pore oligomerization was characterized by blue native PAGE (Bio-Rad) using 4 to 15% polyacrylamide gels (fig. S11). The gel showed that the monomers had been self-assembled into oligomers before the addition of DDM. However, to follow previous studies strictly (44), ClyA-RR dodecamers with DDM addition were still used for follow-up measurements. Here, the bands that correspond to dodecameric ClyA-RR were cut from the gel and soaked in 150 mM NaCl and 15 mM tris-HCl (pH 7.5) supplemented with 0.2% DDM and 10 mM EDTA for 3 hours. The supernatant, which contains dodecameric proteins diffused out of the gel, was collected by centrifugation (20,000g at 4°C for 20 min). The collected dodecameric ClyA-RR proteins were either used immediately for subsequent experiments or stored at 4°C for up to 14 days.
DIB formation
A detailed description of how a droplet/hydrogel bilayer was created was previously reported (27). Briefly, oxygen plasma–treated coverslips (24 mm by 40 mm) were spin-coated (3000 rpm for 30 s) with 200 μl of molten agarose (0.75% w/v in Milli-Q water). The coverslip was stuck to a PMMA device by filling the microfluidic channels within the device with molten agarose [2.5% (w/v) in CaCl2 buffer]. The lipid/oil solution was prepared by dissolving 5-mg dried film of DPhPC lipids in a 2-ml mixture of hexadecane and silicone oil with a 1:1 volume ratio. A lipid monolayer was formed on the agarose-coated glass coverslip when immersed in the lipid/oil solution. In preparation of aqueous droplets, protein nanopores and other analytes could be added into the aqueous buffer, which is composed of 1 to 2.5 M KCl, 400 μM EDTA, 40 μM Fluo-8, and 10 mM HEPES (pH 7.0). Aqueous droplets of various volumes were pipetted into the lipid/oil solution for incubation. After 5 min, a self-assembled lipid monolayer at the water-oil interface could form. When this droplet and the agarose substrate were brought together in the lipid/oil solution, a stable bilayer (DIB) could form spontaneously.
TIRF imaging and optical recordings
The DIB was imaged using an inverted microscope (Eclipse Ti-U, Nikon) equipped with a 60× oil immersion TIRF objective (numerical aperture = 1.49; Plan Apo, Nikon). The fluorescence was excited with a 473-nm diode-pumped solid-state laser (100 mW; Changchun New Industries Optoelectronics Technology). The image was acquired with an electron-multiplying charge-coupled device camera (iXon3 897, Andor). The exposure time was set to 3 to 30 ms. The maximum field of view was 135 μm by 135 μm.
Electrophysiology recordings
Electrophysiology recordings were performed as previously reported (7, 53, 54). The electrophysiology trace was acquired with 25-kHz sampling rate, low-pass–filtered at 1 kHz (Axopatch 200B, Molecular Devices), digitized, and recorded using the Digidata 1550A digitizer (Molecular Devices). Subsequent data analysis was performed with Clampfit 10.7 (Molecular Devices).
Supplementary Material
Acknowledgments
We would like to thank G. Maglia (University of Groningen), G. Huang (University of Groningen), and Z. Zeng (University of Pittsburgh) for useful discussions with ClyA nanopore expression; M. Cheetham (University of Cambridge) for useful discussions with the Comsol modeling; and J. Pan (Nanjing University) for device photography. Funding: This work is supported by the National Natural Science Foundation of China (grant nos. 21327902, 21675083, and 91753108), the Fundamental Research Funds for the Central Universities (grant nos. 020514380142 and 020514380174), the State Key Laboratory of Analytical Chemistry for Life Science (grant nos. 5431ZZXM1804 and 5431ZZXM1902), the Excellent Research Program of Nanjing University (grant no. ZYJH004), the 1000 Plan Youth Talent Program of China, the Program for High-Level Entrepreneurial and Innovative Talents Introduction of Jiangsu Province, and the Technology Innovation Fund Program of Nanjing University. Author contributions: S.H. conceived the project, designed the experiments, and constructed the Comsol model. Yuqin Wang performed the measurements. P.Z. set up the instruments. Yu Wang and S.Y. prepared all protein nanopores. Yuqin Wang and X.D. conducted the electrophysiology measurements with TriM-β-CD and PEG 1500. S.H. and Yuqin Wang analyzed the data and wrote the paper. S.H. and H.-Y.C. supervised the project. Competing interests: S.H. and Yuqin Wang are inventors on a filed PCT patent application related to this work (no. PCT/CN2019/089050, 29 May 2019). The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Raw data can be requested from S.H. (shuo.huang@nju.edu.cn) or H.-Y.C. (hychen@nju.edu.cn).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaar3309/DC1
Section S1. The FEM simulation
Section S2. 2D Gaussian fitting
Section S3. SBR evaluation
Table S1. 1/τon and 1/τoff of TriM-β-CD with different [TriM-β-CD].
Table S2. 1/τon of TriM-β-CD with different [KCl] in cis.
Table S3. FWHM and SBR with different [KCl].
Table S4. FWHM and SBR with different [CaCl2].
Table S5. FWHM and SBR of α-HL and ClyA-RR nanopores.
Table S6. Nucleic acid abbreviations and sequences.
Table S7. Blockade level of dsDNA events.
Fig. S1. FEM model geometry.
Fig. S2. The DIB device.
Fig. S3. The schematic diagram of the setup.
Fig. S4. Cyclodextrin binding kinetics.
Fig. S5. Definition of signal and background during oSCR.
Fig. S6. Demonstration of fluorescence trace normalization.
Fig. S7. Event statistics derivation.
Fig. S8. Baseline comparison during TriM-β-CD sensing.
Fig. S9. FEM modeling of Fluo-8 distribution.
Fig. S10. FEM modeling of the osmotic flow.
Fig. S11. The preparation and characterization of ClyA-RR.
Fig. S12. Observing dsDNA events with different dsDNA concentrations.
Fig. S13. Statistics of dsDNA events acquired from ClyA-RR.
Movie S1. Simultaneous imaging of α-HL and ClyA.
Movie S2. dsDNA sensing by DOP.
Movie S3. Parallel dsDNA sensing by DOP.
Movie S4. A single ClyA-RR nanopore inserted in a miniaturized DIB.
REFERENCES AND NOTES
- 1.Andre B., An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11, 1575–1611 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Gouaux E., MacKinnon R., Principles of selective ion transport in channels and pumps. Science 310, 1461–1465 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Nielsen S., Nagelhus E. A., Amiry-Moghaddam M., Bourque C., Agre P., Ottersen O. P., Specialized membrane domains for water transport in glial cells: High-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J. Neurosci. 17, 171–180 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood I. S., Trayhurn P., Glucose transporters (GLUT and SGLT): Expanded families of sugar transport proteins. Br. J. Nutr. 89, 3–9 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Choi K. H., McPartland J., Kaganman I., Bowman V. D., Rothman-Denes L. B., Rossmann M. G., Insight into DNA and protein transport in double-stranded DNA viruses: The structure of bacteriophage N4. J. Mol. Biol. 378, 726–736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressloff P. C., Newby J. M., Stochastic models of intracellular transport. Rev. Mod. Phys. 85, 135–196 (2013). [Google Scholar]
- 7.Huang S., Nanopore-based sensing devices and applications to genome sequencing: A brief history and the missing pieces. Chin. Sci. Bull. 59, 4918–4928 (2014). [Google Scholar]
- 8.Bezrukov S. M., Kasianowicz J. J., Current noise reveals protonation kinetics and number of ionizable sites in an open protein ion channel. Phys. Rev. Lett. 70, 2352–2355 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Malmstadt N., Nash M. A., Purnell R. F., Schmidt J. J., Automated formation of lipid-bilayer membranes in a microfluidic device. Nano Lett. 6, 1961–1965 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Funakoshi K., Suzuki H., Takeuchi S., Lipid bilayer formation by contacting monolayers in a microfluidic device for membrane protein analysis. Anal. Chem. 78, 8169–8174 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Ide T., Yanagida T., An artificial lipid bilayer formed on an agarose-coated glass for simultaneous electrical and optical measurement of single ion channels. Biochem. Biophys. Res. Commun. 265, 595–599 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Jeon T.-J., Malmstadt N., Schmidt J. J., Hydrogel-encapsulated lipid membranes. J. Am. Chem. Soc. 128, 42–43 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Li J., Stein D., McMullan C., Branton D., Aziz M. J., Golovchenko J. A., Ion-beam sculpting at nanometre length scales. Nature 412, 166–169 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Zhang B., Galusha J., Shiozawa P. G., Wang G., Bergren A. J., Jones R. M., White R. J., Ervin E. N., Cauley C. C., White H. S., Bench-top method for fabricating glass-sealed nanodisk electrodes, glass nanopore electrodes, and glass nanopore membranes of controlled size. Anal. Chem. 79, 4778–4787 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Geng J., Kim K., Zhang J., Escalada A., Tunuguntla R., Comolli L. R., Allen F. I., Shnyrova A. V., Cho K. R., Munoz D., Wang Y. M., Grigoropoulos C. P., Ajo-Franklin C. M., Frolov V. A., Noy A., Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature 514, 612–615 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Bezrukov S. M., Vodyanoy I., Parsegian V. A., Counting polymers moving through a single ion channel. Nature 370, 279–281 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Keller S. L., Bezrukov S. M., Gruner S. M., Tate M. W., Vodyanoy I., Parsegian V. A., Probability of alamethicin conductance states varies with nonlamellar tendency of bilayer phospholipids. Biophys. J. 65, 23–27 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou L. Y. T., Ming K., Chan W. C. W., Strategies for the intracellular delivery of nanoparticles. Chem. Soc. Rev. 40, 233–245 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Dunlop J., Bowlby M., Peri R., Vasilyev D., Arias R., High-throughput electrophysiology: An emerging paradigm for ion-channel screening and physiology. Nat. Rev. Drug Discov. 7, 358–368 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Jain M., Koren S., Miga K. H., Quick J., Rand A. C., Sasani T. A., Tyson J. R., Beggs A. D., Dilthey A. T., Fiddes I. T., Malla S., Marriott H., Nieto T., O'Grady J., Olsen H. E., Pedersen B. S., Rhie A., Richardson H., Quinlan A. R., Snutch T. P., Tee L., Paten B., Phillippy A. M., Simpson J. T., Loman N. J., Loose M., Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letellier L., Plançon L., Bonhivers M., Boulanger P., Phage DNA transport across membranes. Res. Microbiol. 150, 499–505 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Song L., Hobaugh M. R., Shustak C., Cheley S., Bayley H., Gouaux J. E., Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1865 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Parker I., Yao Y., Ca2+ transients associated with openings of inositol trisphosphate-gated channels in Xenopus oocytes. J. Physiol. 491, 663–668 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipp P., Niggli E., Submicroscopic calcium signals as fundamental events of excitation—Contraction coupling in guinea-pig cardiac myocytes. J. Physiol. 492, 31–38 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S., Romero-Ruiz M., Castell O. K., Bayley H., Wallace M. I., High-throughput optical sensing of nucleic acids in a nanopore array. Nat. Nanotechnol. 10, 986–991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivankin A., Henley R. Y., Larkin J., Carson S., Toscano M. L., Wanunu M., Label-free optical detection of biomolecular translocation through nanopore arrays. ACS Nano 10, 10774–10781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Yan S., Zhang P., Zeng Z., Zhao D., Wang J., Chen H., Huang S., Osmosis-driven motion-type modulation of biological nanopores for parallel optical nucleic acid sensing. ACS Appl. Mater. Interfaces 10, 7788–7797 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Anderson B. N., Assad O. N., Gilboa T., Squires A. H., Bar D., Meller A., Probing solid-state nanopores with light for the detection of unlabeled analytes. ACS Nano 8, 11836–11845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heron A. J., Thompson J. R., Cronin B., Bayley H., Wallace M. I., Simultaneous measurement of ionic current and fluorescence from single protein pores. J. Am. Chem. Soc. 131, 1652–1653 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Shuai J., Parker I., Optical single-channel recording by imaging Ca2+ flux through individual ion channels: Theoretical considerations and limits to resolution. Cell Calcium 37, 283–299 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Gu L.-Q., Braha O., Conlan S., Cheley S., Bayley H., Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature 398, 686–690 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Fennouri A., Daniel R., Pastoriza-Gallego M., Auvray L., Pelta J., Bacri L., Kinetics of enzymatic degradation of high molecular weight polysaccharides through a nanopore: Experiments and data-modeling. Anal. Chem. 85, 8488–8492 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Bayley H., Cremer P. S., Stochastic sensors inspired by biology. Nature 413, 226–230 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Braha O., Gu L.-Q., Zhou L., Lu X., Cheley S., Bayley H., Simultaneous stochastic sensing of divalent metal ions. Nat. Biotechnol. 18, 1005–1007 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Clarke J., Wu H.-C., Jayasinghe L., Patel A., Reid S., Bayley H., Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Boersma A. J., Brain K. L., Bayley H., Real-time stochastic detection of multiple neurotransmitters with a protein nanopore. ACS Nano 6, 5304–5308 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Boersma A. J., Bayley H., Continuous stochastic detection of amino acid enantiomers with a protein nanopore. Angew. Chem. Int. Ed. Engl. 51, 9606–9609 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Sacerdote M. G., Szostak J. W., Semipermeable lipid bilayers exhibit diastereoselectivity favoring ribose. Proc. Natl. Acad. Sci. U.S.A. 102, 6004–6008 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishigami T., Matsuyama H., Numerical modeling of concentration polarization in spacer-filled channel with permeation across reverse osmosis membrane. Ind. Eng. Chem. Res. 54, 1665–1674 (2015). [Google Scholar]
- 40.Lewis G. N., The osmotic pressure of concentrated solutions, and the laws of the perfect solution. J. Am. Chem. Soc. 30, 668–683 (1908). [Google Scholar]
- 41.Robertson J. W. F., Rodrigues C. G., Stanford V. M., Rubinson K. A., Krasilnikov O. V., Kasianowicz J. J., Single-molecule mass spectrometry in solution using a solitary nanopore. Proc. Natl. Acad. Sci. U.S.A. 104, 8207–8211 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues C. G., Machado D. C., Chevtchenko S. F., Krasilnikov O. V., Mechanism of KCl enhancement in detection of nonionic polymers by nanopore sensors. Biophys. J. 95, 5186–5192 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soskine M., Biesemans A., Moeyaert B., Cheley S., Bayley H., Maglia G., An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 12, 4895–4900 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franceschini L., Brouns T., Willems K., Carlon E., Maglia G., DNA translocation through nanopores at physiological ionic strengths requires precise nanoscale engineering. ACS Nano 10, 8394–8402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wendell D., Jing P., Geng J., Subramaniam V., Lee T. J., Montemagno C., Guo P., Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat. Nanotechnol. 4, 765–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manrao E. A., Derrington I. M., Laszlo A. H., Langford K. W., Hopper M. K., Gillgren N., Pavlenok M., Niederweis M., Gundlach J. H., Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasianowicz J. J., Brandin E., Branton D., Deamer D. W., Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U.S.A. 93, 13770–13773 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tinland B., Pluen A., Sturm J., Weill G., Persistence length of single-stranded DNA. Macromolecules 30, 5763–5765 (1997). [Google Scholar]
- 49.Nomidis S. K., Hooyberghs J., Maglia G., Carlon E., DNA capture into the ClyA nanopore: Diffusion-limited versus reaction-limited processes. J. Phys. Condens. Matter 30, 304001 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Chen Z., Liao P., Zhang F., Jiang M., Zhu Y., Huang Y., Centrifugal micro-channel array droplet generation for highly parallel digital PCR. Lab Chip 17, 235–240 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Nakano A., Spinning-disk confocal microscopy—A cutting-edge tool for imaging of membrane traffic. Cell Struct. Funct. 27, 349–355 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Laszlo A. H., Derrington I. M., Ross B. C., Brinkerhoff H., Adey A., Nova I. C., Craig J. M., Langford K. W., Samson J. M., Daza R., Doering K., Shendure J., Gundlach J. H., Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 32, 829–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Patil K. M., Yan S., Zhang P., Guo W., Wang Y., Chen H.-Y., Gillingham D., Huang S., Nanopore sequencing accurately identifies the mutagenic DNA lesion O6-carboxymethyl guanine and reveals its behavior in replication. Angew. Chem. Int. Ed. 58, 8432–8436 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Yan S., Li X., Zhang P., Wang Y., Chen H.-Y., Huang S., Yu H., Direct sequencing of 2'-deoxy-2'-fluoroarabinonucleic acid (FANA) using nanopore-induced phase-shift sequencing (NIPSS). Chem. Sci. 10, 3110–3117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaar3309/DC1
Section S1. The FEM simulation
Section S2. 2D Gaussian fitting
Section S3. SBR evaluation
Table S1. 1/τon and 1/τoff of TriM-β-CD with different [TriM-β-CD].
Table S2. 1/τon of TriM-β-CD with different [KCl] in cis.
Table S3. FWHM and SBR with different [KCl].
Table S4. FWHM and SBR with different [CaCl2].
Table S5. FWHM and SBR of α-HL and ClyA-RR nanopores.
Table S6. Nucleic acid abbreviations and sequences.
Table S7. Blockade level of dsDNA events.
Fig. S1. FEM model geometry.
Fig. S2. The DIB device.
Fig. S3. The schematic diagram of the setup.
Fig. S4. Cyclodextrin binding kinetics.
Fig. S5. Definition of signal and background during oSCR.
Fig. S6. Demonstration of fluorescence trace normalization.
Fig. S7. Event statistics derivation.
Fig. S8. Baseline comparison during TriM-β-CD sensing.
Fig. S9. FEM modeling of Fluo-8 distribution.
Fig. S10. FEM modeling of the osmotic flow.
Fig. S11. The preparation and characterization of ClyA-RR.
Fig. S12. Observing dsDNA events with different dsDNA concentrations.
Fig. S13. Statistics of dsDNA events acquired from ClyA-RR.
Movie S1. Simultaneous imaging of α-HL and ClyA.
Movie S2. dsDNA sensing by DOP.
Movie S3. Parallel dsDNA sensing by DOP.
Movie S4. A single ClyA-RR nanopore inserted in a miniaturized DIB.


