Abstract
Food production in green crops is severely limited by low activity and poor specificity of D-ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) in natural photosynthesis (NPS). This work presents a scientific solution to overcome this problem by immobilizing RuBisCO into a microfluidic reactor, which demonstrates a continuous production of glucose precursor at 13.8 μmol g−1 RuBisCO min−1 from CO2 and ribulose-1,5-bisphosphate. Experiments show that the RuBisCO immobilization significantly enhances enzyme stabilities (7.2 folds in storage stability, 6.7 folds in thermal stability), and also improves the reusability (90.4% activity retained after 5 cycles of reuse and 78.5% after 10 cycles). This work mimics the NPS pathway with scalable microreactors for continuous synthesis of glucose precursor using very small amount of RuBisCO. Although still far from industrial production, this work demonstrates artificial synthesis of basic food materials by replicating the light-independent reactions of NPS, which may hold the key to food crisis relief and future space colonization.
Subject terms: Lab-on-a-chip, Microfluidics, Immobilized enzymes, Biomimetic synthesis
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is a difficult enzyme to work with. Here, the authors covalently immobilized it in a microfluidic reactor to enhance its storage/thermal stabilities and reusability, which enabled the continuous artificial synthesis of glucose precursor.
Introduction
The world is moving rapidly into food crisis due to extreme weather, shortage of farmland, and population explosion. And glucose is the basic material of food produced by green plants using the natural photosynthesis (NPS)1. However, the NPS suffers from low-energy efficiency (~1%) and is greatly limited by soil, climate, water, and labor2,3. It is reported that food demand is expected to increase by 70% by 20504, which makes an extremely urgent demand for a scientific solution beyond NPS for food production. Recently, scientists have figured out an excellent alternative to NPS, which is called artificial photosynthesis (APS), for the energy-rich chemicals production. It mimics the green plants to produce energy-rich materials and fuels but using man-made nanomaterials and engineered photosynthetic reactors. Compared with the NPS, the APS usually presents much higher efficiency (~12%) and simplicity5. Although the APS has attracted substantial research interest, most of efforts have been devoted to the light-dependent reactions for hydrogen generation and water splitting6,7, microbial growth and biofuel production8–12, and photocatalytic cofactor regeneration13–16. The exploration of light-independent reactions for food production is worth more attentions. Nevertheless, it is found that the light-independent reactions, though crucial for glucose production from CO2, are more challenging for the artificial replication, since they involve multi-step enzymatic reactions (Fig. 1a).
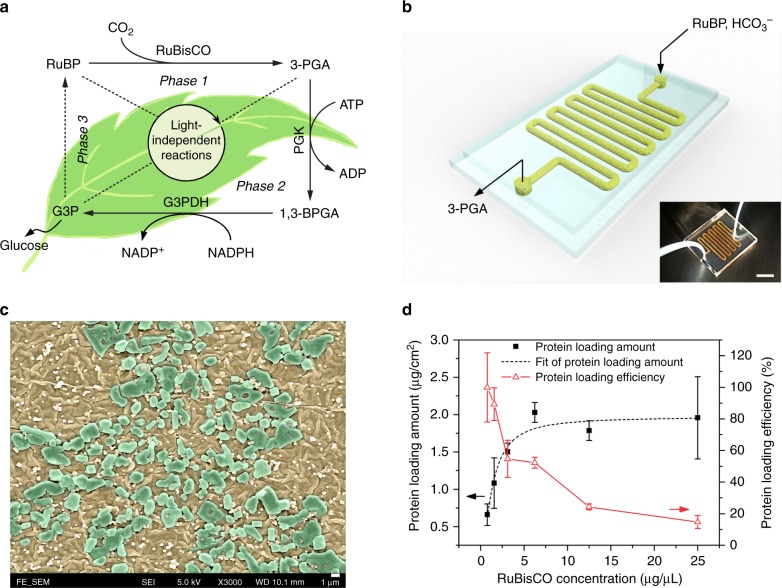
Fig. 1.
Design and characterization of RuBisCO immobilized microfluidic reactors (RIMRs). a Scheme of light-independent reactions of NPS: Phase 1: Carbon fixation starts with ribulose 1,5-bisphosphate (RuBP) and uses the enzyme RuBisCO to fix CO2 into 3-phosphoglycerate (3-PGA); Phase 2: Reduction reaction uses adenosine triphosphate (ATP), nicotinamide adenine dinucleotide phosphate (NADPH) and the enzyme phosphoglycerate kinase (PGK) and glyceraldehyde 3-phosphate dehydrogenase (G3PDH) to reduce 3-PGA into glyceraldehyde 3-phosphate (G3P), two of which can form the end product glucose; Phase 3: RuBP regeneration from G3P using up to 9 steps of enzymatic reactions. b Three-dimensional diagram and the photograph (inset) of the RIMRs, the scale bar of the inset is 1 cm. c SEM image of the inner surfaces of RIMRs. Flat and smooth PDMS becomes rough and is covered by PDA nanoparticles after PDA modification (brownish color). Large blocks are the immobilized RuBisCO (green blocks). The scale bar is 1 μm. d Protein-loading amount and protein-loading efficiency as a function of the concentrations of injected RuBisCO to find the optimal RuBisCO concentration for further experiments. Error bars represent the standard deviations from three independent experiments. Source data are provided as a Source Data file
The first carbon fixation phase in light-independent reactions is catalyzed by the most abundant enzyme in plants, RuBisCO (EC 4.1.1.39). Even so, the low catalytic rate and the poor specificity of RuBisCO seriously limit the agricultural productivity17. Although many studies have been conducted to improve the efficiency of RuBisCO18–23, they are mainly designed for in vivo biological systems24,25. Lots of energy and labor are consumed to balance the metabolic pathways in intact cells. For the industrial cell-free application with the demands of sustainability and scalability, immobilizing and concentrating RuBisCO into man-made structures presents to be a more promising method to enhance the overall catalytic efficiency26–28. Enzyme immobilization has many advantages, such as high enzyme-to-reactant ratio, enhanced enzyme stability, admirable enzyme reusability, weakened feedback inhibition, and possible modulation of catalytic property29–31. Various enzyme immobilization techniques have been thoroughly explored, such as physical adsorption, affinity bonding, covalent binding, and encapsulation. However, the choice of techniques should be very careful, since inappropriate immobilization may cause the reduction of enzyme activity due to conformational change, detachment, and inactivation32. Among all the enzyme immobilization methods, covalent immobilization always offers the strongest bond between enzyme and support33,34. But it usually involves complex linkers and organic solvents. Recently, polydopamine (PDA) becomes rather popular in biomimetic systems for the covalent immobilization of enzymes thanks to their excellent biocompatibility and high efficiency in imparting biological functionality on inorganic materials35–38. Moreover, the PDA functionalized on smooth substrates would form a coarse surface assembled by many particles, which could enlarge the active surface for immobilization37,38.
In plants, the NPS occurs in a natural microfluidic system of microscale chloroplasts and fluids-filled areas39. In laboratory research, scientists usually make use of microfluidic systems to mimic the reactions and behaviors in natural instead of bulk systems. Thereby, the inherent advantages of microfluidics can be well taken, such as low sample consumption, flexible manipulation, large surface-area-to-volume ratio, and self-refreshing surface effect40. In addition, remarkable enhancement of the overall reaction production can also be expected41–43. Moreover, when the enzyme is immobilized into microfluidic reactors, separation and recovery of enzyme can be achieved simultaneously along with the product generation, avoiding the contamination and reducing the enzyme consumption44,45.
On account of the above benefits, this work plans to constitute the NPS system for glucose precursor (3-PGA) production by replicating the first phase reaction of NPS using microfluidic reactors. PDA is employed as the linker to immobilize RuBisCO on the inner surfaces of reactors. The reaction performance of artificial chemosynthesis of glucose precursor in the RuBisCO-immobilized microfluidic reactors (RIMRs) is compared with that in the bulk reaction in terms of the stability, reusability, and long-term 3-PGA production subsequently. This is a proof of concept for constructing the NPS pathway using enzyme-immobilized microfluidic reactors for continuous synthesis of glucose precursor.
Results
Fabrication and characterization of RIMRs
The three-dimensional diagram and the photograph (inset) of the RIMRs is presented in Fig. 1b. It consists of two PDMS layers, and the inner surfaces of microchannels are functionalized by PDA. The abundant catechol groups of PDA layer can react with the amine groups of RuBisCO through the Michael addition or Schiff base reactions35, facilitating the immobilization of RuBisCO (see Supplementary Fig. 1). Figure 1c is the SEM image of the microchannels’ inner surfaces after the RuBisCO immobilization. The PDA-modified PDMS is coarse and rough and is covered with many PDA nanoparticles. The RuBisCO immobilization then leads to the formation of some blocks, proving the successful immobilization (see Supplementary Fig. 3). The successful immobilization of RuBisCO is further confirmed by the measurements of water contact angle, Raman and ATR-FTIR spectrum and fluorescence images of the microfluidic channels (see Supplementary Figs. 4–6).
Protein-loading capacity and kinetic parameters
After the successful immobilization of RuBisCO, the protein-loading capacity of the PDA-modified microfluidic reactors is investigated to determine the optimal RuBisCO concentration for further experiments. As plotted in Fig. 1d (dark solid squares), the amount of protein loaded in the reactors rises with the increase of the injected RuBisCO concentration. It then saturates to about 2.0 μg cm−2 when the RuBisCO concentration reaches about 6.25 μg μL−1. However, the protein-loading efficiency decreases with the increase of RuBisCO concentration (red open triangles in Fig. 1d). This shows that RuBisCO can be easily immobilized onto the PDA layer. Once all the functional groups of the PDA are covalently bonded with RuBisCO, the excessive RuBisCO would be easily rinsed off from the reactor. Consequently, 6.25 μg μL−1 is chosen as the optimal RuBisCO concentration for the following RIMRs experiments. The protein-loading capacity of the PDA-modified microfluidic reactor is estimated to be 2.03 μg cm−2, and the protein-loading efficiency is ~50%. In the following experiments, the RuBisCO amounts used for both the immobilized and the free RuBisCO test are 21.875 μg.
Then, the kinetic parameters of the immobilized RuBisCO in the RIMRs reaction and the free RuBisCO in the bulk reaction are determined (see Table 1). Compared with the kinetic parameters of free RuBisCO, the Km value (the Michaelis–Menten constant) for the RIMRs is relatively higher (0.070 vs. 0.049 mM), inferring a lower affinity of the immobilized enzyme for the reactant. This is a common and intrinsic weakness of immobilized enzymes, owing to the steric hindrance introduced by the coverage of some active site by the support46. In addition, the immobilized enzyme may lose the flexibility to bind the natural ligands during catalysis and the diffusion distance for the reactant to the immobilized enzyme on the microchannel sidewalls becomes relatively long (as compared to the free enzyme)46. From Table 1, the RIMRs also exhibited a lower Vmax (the maximal reaction rate) than that of the free RuBisCO. The activity loss induced by the immobilization procedure is ascribed to the conformational changes of RuBisCO. Such conformational changes can also be observed from the presence of a weak band at about 1600 cm−1 of RuBisCO-PDA-PDMS in Supplementary Fig. 5b. According to the previous work, the mechanism of PDA for the enzyme immobilization is to link the amino groups of enzymes to the catechol groups of PDA35,38. It has been reported that the amino groups of lysine residues at the active sites of RuBisCO play a critical role in catalysis47,48. Therefore, the immobilization of RuBisCO by PDA modification may cause irreversible conformational changes of RuBisCO and thus result in the reduction of its activity.
Table 1.
Kinetic parameters obtained from RIMRs reaction and bulk reactiona
| Reaction type | Vmax (mmol min−1 g−1 RuBisCO) | Km (RuBP) (mM) |
|---|---|---|
| RIMRs reaction | 0.070 ± 0.003 | 0.070 ± 0.012 |
| Bulk reaction | 0.169 ± 0.006 | 0.049 ± 0.008 |
aThe collected production solutions I and II are both 100 μL. RuBP concentrations are 0.01–2 mM for the bulk reaction and 0.025–2 mM for the RIMRs reaction. The concentration of bicarbonate in the reaction buffer is 66 mM. Km and Vmax values are the means ± s.d. of three independent experiments, which is calculated by the GraphPad Prism 7 according to the nonlinear fitting of Michaelis–Menten model. Source data are provided as a Source Data file
It is noted that the activity of free RuBisCO is very low compared with the reported RuBisCO activity before. According to Table 1, the kcat (turnover number) of free RuBisCO here is about 0.19 s−1, which is remarkably smaller than the natural kcat of RuBisCO from Spinach (~3 s−1)49. One reason is likely the irreversible binding of inhibitory contaminants produced during the commercial preparation of RuBP50,51. Another reason for the low activity of RuBisCO here may be the fallover during catalysis, which is caused by the tight binding of inhibitory sugar phosphates to the active site52–54.
Feasibility of RIMRs
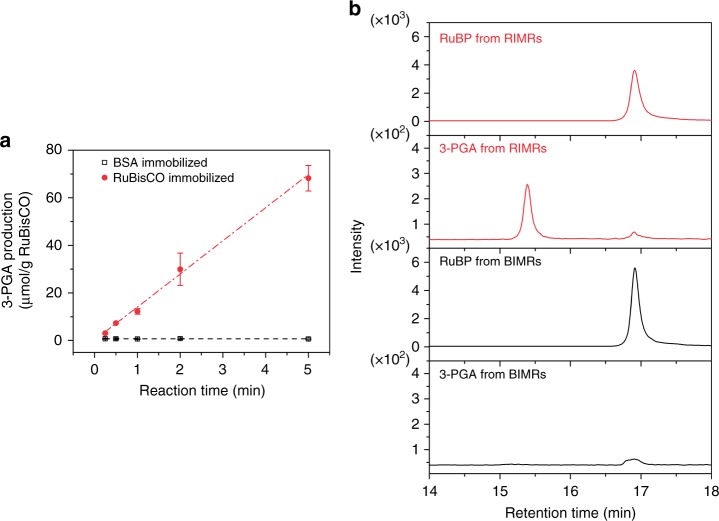
To check the feasibility of the reactors for artificial chemosynthesis of glucose precursor, the prepared RIMRs are used to produce 3-PGA from RuBP and CO2 under different reaction times. In the control experiment that albumin from bovine serum (BSA) is immobilized onto the microfluidic reactor, the BSA-immobilized microfluidic reactors (BIMRs) produces negligible amount of 3-PGA (dark open squares in Fig. 2a), and the production amount does not increase with reaction time either. In contrast, the RIMRs demonstrate a linearly increased production of 3-PGA at the rate of 13.8 μmol g−1 RuBisCO min−1 (red solid circles and red dash-dotted fitting line of Fig. 2a) when the reaction time is 5 min or shorter. Production of 3-PGA by free RuBisCO as a function of time is provided in Supplementary Fig. 9 for comparison.
Fig. 2.
Feasibility of RIMRs. a Production amount of 3-PGA as a function of the reaction time for the RIMRs (solid red circles) and the BIMRs (open dark squares). The red dash-dotted line and dark dashed line are the liner fitting regressions. The slope of liner fitting regression of 3-PGA production in RIMRs is 13.8 μmol g−1 RuBisCO min−1 and that in BIMRs is almost 0. The initially injected RuBisCO and BSA concentrations for immobilization are both 6.25 μg μL−1. The collected production solutions I from RIMRs and BIMRs are 21 μL. RuBP concentration is 0.5 mM. in the reaction buffer is 66 mM. Reaction temperature is 30 °C. b HPLC–MS/MS chromatography of RuBP and 3-PGA in the production solutions I obtained from RIMRs and BIMRs at the reaction time of 5 min. Error bars represent the standard deviations from three independent experiments. Source data are provided as a Source Data file
Apart from the UV–visible spectrometer, a high-performance liquid chromatography–tandem mass spectrometer (HPLC–MS/MS) system is also applied to directly examine the ingredients of 3-PGA and RuBP in the production solution I. The retention times of standard RuBP and 3-PGA are 16.97 and 15.21 min, respectively (see Supplementary Fig. 10a). The production amount of 3-PGA can also be derived from the calibration of the integrated peak areas as a function of its concentrations (see Supplementary Fig. 10b). As shown in Fig. 2b, the peaks with the similar retention time of RuBP and 3-PGA are observed in the production solution I obtained from the RIMRs. The 3-PGA production amount by RIMRs in 5 min is calculated to be about 71.9 μmol g−1 RuBisCO from the chromatography. It is consistent with the 3-PGA production amount of 68.2 ± 5.4 μmol g−1 RuBisCO in 5 min as determined by UV–visible spectrometer. However, no significant 3-PGA amount can be detected from the production solution I of the BIMRs. It is also noted that the RuBP signal from BIMRs is higher than that from RIMRs, indicating the conversion of RuBP to 3-PGA in RIMRs.
In consequence, it is well proved that 3-PGA is successfully synthesized from the RIMRs by implementing the light-independent reactions pathway. Moreover, in the RIMRs reaction, the products are flowed through the reactor and pumped out immediately after reaction. The accumulation of 3-PGA around RuBisCO is reduced and the control of the reaction time can be achieved straightforwardly by changing the flow rate of the reactant mixture injection. This is one of the core features of RIMRs.
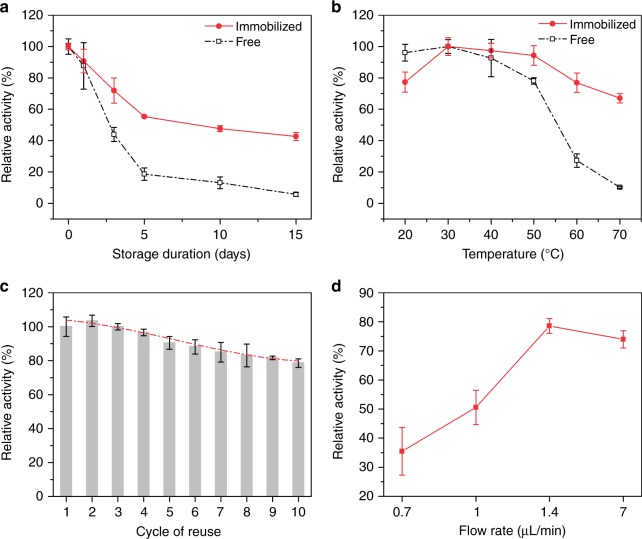
Storage and thermal stabilities of RIMRs
The storage and thermal stabilities of RIMRs are very important to practical applications. Regarding the storage stability (see Fig. 3a), the half-life (the storage time that 50% of the initial activity can be retained after the incubation in reaction buffer at 4 °C) is about 2.6 days for the free RuBisCO, and it is enhanced to be three times (8 days) after the immobilization. In addition, the storage stability of the immobilized RuBisCO is enhanced by 7.2 times compared with the free one (43% vs. 6% of the initial activity retention) after the incubation for 15 days. More characterizations of free RuBisCO under different incubation time including sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) are provided in Supplementary Fig. 11.
Fig. 3.
Stability and reusability of RIMRs. a Storage stability (incubation at 4 °C) of immobilized (red line) and free RuBisCO (dark line). b Thermal stability of immobilized (red line) and free RuBisCO (dark line). All samples are incubated for 10 min before the activity assay. c Reusability of the RIMRs when RuBP is injected at 1.4 μL min−1 (the reaction time is 5 min). Red dash-dotted line is the third-order polynomial fitting, representing that the activity drop trend tends to slow down with the increase of cycles of reuse. d Relative activity as a function of the flow rate of RuBP injection (from 7 to 0.7 μL min−1) for repeated uses. The amount of RuBisCO is 21.875 μg for both the immobilized and free ones in all the experiments. The volume of the collected production solutions for the storage and thermal stability tests are 100 μL. RuBP concentration is 0.5 mM and in the reaction buffer is 66 mM. Error bars represent the standard deviations from three independent experiments. Source data are provided as a Source Data file
Figure 3b shows the influence of temperature on the stability of RuBisCO. The highest activities (Aht) of immobilized and free RuBisCO both appear at 30 °C. And the optimal incubation temperature range (i.e., the temperature range that over 98% of maximum activity is retained after incubation) of RuBisCO is expanded from 24–34 °C to 29–40 °C after the immobilization. It is also worth noting that the immobilized RuBisCO retains 67% of Aht at 70 °C, whereas the free RuBisCO retains only 10%, showing a difference of 6.7 times. When the immobilized and free RuBisCO are incubated at 50 °C for a long time up to 60 min (as shown in Supplementary Fig. 12), the immobilized RuBisCO retains 75% of Aht while the free RuBisCO retains only 46%. These show that the immobilization significantly enhances the resistance of RuBisCO to the thermal inactivation. For direct comparison, Table 2 lists the major performances of the immobilized and free RuBisCO.
Table 2.
Comparison of the RIMRs reaction and the bulk reaction
| Reaction type | Storage stabilitya | Thermal stabilityb | Reusabilityc | Production of 3-PGA (μmol g−1 RuBisCO) |
|---|---|---|---|---|
| RIMRs reaction | 43% | 67% | 90.4% | |
| Bulk reaction | 6% | 10% | – | Saturation at ~360 |
| Enhancement factor | 7.2-fold | 6.7-fold | – | 1.8-folde |
aStorage stability is defined as the remaining relative activity after 15 days
bThermal stability is the remaining relative activity after incubation at 70 °C for 10 min
cReusability is the remaining relative activity after five cycles of reuse at the flow rate of 1.4 μL min−1
dVp is the volume of the reactant mixture (RuBP and in the reaction buffer)
eData is obtained when Vp is 1680 μL
Both the improved storage and thermal stabilities of RuBisCO can be ascribed to the enhanced stability of the enzyme conformation after the covalent immobilization55,56. Generally, the enzyme activity in solution could be greatly affected by the structure change caused by the unfolding of protein amino acid chains. On one hand, the elevated temperature or long-time storage would consequently influence the conformation of RuBisCO, leading to the activity reduction. On the other hand, the immobilization technique can offer multipoint anchors of RuBisCO on the support of the microfluidic reactors. As a result, the enzyme structure and the active sites are properly preserved, thus maintaining the activity of immobilized RuBisCO.
Reusability of RIMRs
In addition to the thermal stability and the storage stability, enzymes are also required to have an outstanding reusability for industrial usage. As shown by the gray bars in Fig. 3c, the relative activity decreases with the increase of cycles of reuse but the trend of activity drop gradually slows down (red dash-dotted line by third-order polynomial fitting). More specifically, 90.4% and 78.5% of initial activity can be retained after five cycles and 10 cycles of reuse, respectively, when the flow rate is kept at 1.4 μL min−1. The activity loss after long-time assay may be attributed to two factors. The first factor is the enzyme inactivation after repeated uses and the inhibition of accumulative 3-PGA and RuBP degradation products in aqueous solution50. Therefore, RuBisCO activity is usually measured in a short assay that is stopped in <1 min57. The second factor is the enzyme detachment due to the flushing by the running flow. When the flow rate is lower down, the flow in the microchannel moves slower but the reaction time and the operation time is extended. Correspondingly, the flushing effect is reduced but the inactivation effect is increased, and their combination affects the reusability of RuBisCO in RIMRs. Based on the measured reusability at varying flow rates at 30 °C (see Fig. 3d and Supplementary Fig. 13), the flow rate of 1.4 μL min−1 yields the highest remaining activity after 10 cycles of reuse. The reusability of RuBisCO in the bulk reaction is hard to determine, since it needs special techniques to filter out and recycle the enzyme. In contrast, it is quite facile in the RIMRs reaction by pumping out the production solution and then injecting the new reactant mixture.
Continuous production of 3-PGA from RIMRs
When the reactant mixture is pumped constantly into the RIMRs, continuous and accumulative production of 3-PGA at maximum efficiency can be achieved using only a small amount of RuBisCO. As shown in Fig. 4, when the volume of collected production solution (Vp) is small, the steric hinderance probably causes the RIMRs reaction to have a lower 3-PGA production than the bulk reaction. Nevertheless, the 3-PGA production by the RIMRs in the continuous mode (reaction temperature: 30 °C, reaction time: 1 min) increases with Vp at a rate of 0.39 μmol g−1 RuBisCO min−1 L−1 (red solid circles in Fig. 4). In contrast, the 3-PGA production in the bulk reaction goes up gradually and tends to saturate at about 360 μmol g−1 RuBisCO when Vp is larger than 600 μL (dark open squares in Fig. 4). The saturation of 3-PGA production in the bulk reaction is attributed to the reduced RuBisCO concentration with the increased Vp when the amount of RuBisCO remains the same. Regarding the RIMRs reaction, the increase of Vp does not affect the RuBisCO concentration thanks to the immediate pumping out of the production solution I and the continuous injection of fresh reactant mixture, which also help to avoid the feedback inhibition. Together with the good reusability of immobilized RuBisCO, the RIMRs enable the accumulative production of 3-PGA using only a small amount of RuBisCO. This is another core merit of RIMRs.
Fig. 4.
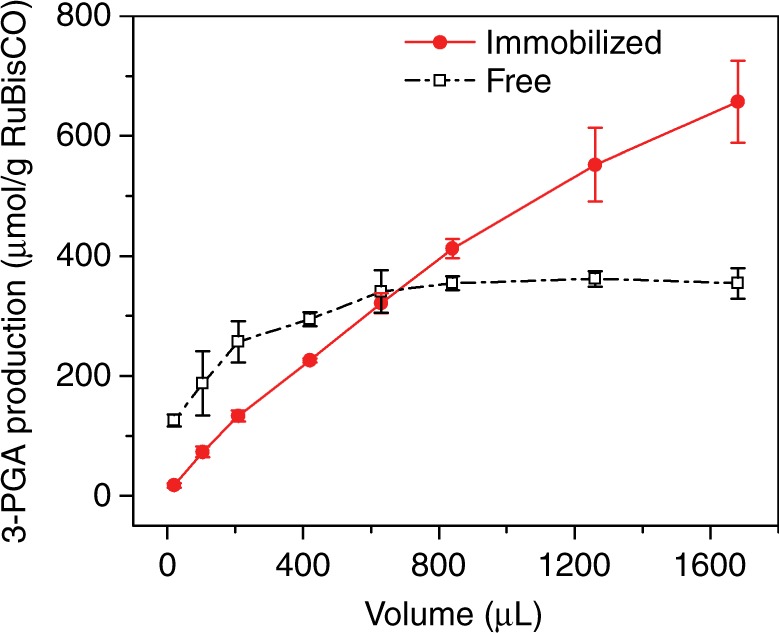
Continuous production of 3-PGA as a function of the volume of collected production solution. The dark open squares represent the 3-PGA production in the bulk reaction with free RuBisCO and the red solid circles represent the 3-PGA production in the RIMRs with the immobilized RuBisCO. The amount of RuBisCO used is 21.875 μg for both the immobilized and free ones. RuBP concentration is 0.5 mM and in the reaction buffer is 66 mM. Reaction temperature is 30 °C. Reaction time is 1 min. Error bars represent the standard deviations from three independent experiments. Source data are provided as a Source Data file
Discussion
In this work, we have immobilized RuBisCO into the PDA-modified microfluidic reactors for continuous production of glucose precursor. As summarized in Table 2, the immobilized RuBisCO showed a 7.2-fold improvement in the storage stability and a 6.7-fold increase in the thermal stability compared with the free RuBisCO. The RIMRs also presented a good reusability with 90.4% of its initial activity retained after five cycles of reuse and 78.5% after 10 cycles. This work lays the foundation to the continuous synthesis of glucose precursor by building up the NPS pathway in the bio-mimic microfluidic reactors. Many advantages of microfluidics for enzymatic reactions are perfectly demonstrated here. Compared with the bulk system, the reaction in microfluidic reactor is easy to control by adjusting the injection of reactant mixture solution. It can also continuously produce 3-PGA by constantly injecting CO2 and RuBP, enabling the accumulative production of 3-PGA with very small amount of RuBisCO in the continuous mode. Moreover, the continuous pumping out of products and injecting in the reactant mixture avoid the feedback inhibition problem and thus maximize the efficiency of RuBisCO enzymatic reaction in a low-cost, convenient, and durable way.
As observed in the experiment, the RuBisCO immobilization would cause some loss of activity due to the detachment and the inactivation of enzyme. In the future, we will try other immobilization methods, like the enzyme encapsulation45,58, with the hope to better retain the activity. Moreover, the immobilization amount of RuBisCO is still small due to the limited space of the microreactor. Some new material such as MOFs59, COFs60, mesoporous nanoparticles61, and carbon nitride62 could be applied to the microreactor to provide larger surface areas in the future. The reactor can also be easily scaled up63–66 to increase the output and more functionalities can be integrated, such as deoxygenation, temperature control of individual reaction, etc. Other enzymes could be incorporated into the microreactor to cascade the reactions for the constitution of full NPS pathway. In a long run, a scientific approach would be available to produce massive basic food materials to relieve the food crisis and to prepare for future space colonization.
Methods
Chemicals and reagents
The reaction buffer (pH 8.0) used for enzyme assay consisted of 0.1 M tris(hydroxymethyl)aminomethane buffer (Tris–HCl, pH 8.0, Beijing Solarbio Technology Co., Ltd., Beijing), 5 mM magnesium chloride hexahydrate (MgCl2·6H2O, AR, Sinopharm Chemical Reagent Co., Ltd., Shanghai), 66 mM potassium bicarbonate (KHCO3, AR, Sinopharm Chemical Reagent Co., Ltd, Shanghai), and 5 mM dl-dithiothreitol (DTT, 99%, Aladdin Industrial Corporation, Shanghai). (3-aminopropyl)triethoxysilane (APTES, 99%) was bought from Aladdin Industrial Corporation, Shanghai. Dopamine hydrochloride (98%, J&K Scientific Ltd., Beijing) was used as 1 mg mL−1 solution in 10 mM Tris–HCl buffer (pH = 8.8, Beijing Solarbio Technology Co., Ltd., Beijing). Reduced nicotinamide adenine dinucleotide disodium salt (NADH Na2, ≥98.0%), adenosine-5′-triphosphate disodium salt trihydrate (ATP Na2, ≥98.0%) and fluorescein isothiocyanate isomer I (FITC, ≥90%, HPLC) were purchased from Beijing Solarbio Technology Co., Ltd. Albumin (98%, from bovine serum) was from J&K Scientific Ltd., Beijing. d-ribulose 1,5-bisphosphate sodium salt hydrate (RuBP, ~90%), d-ribulose 1,5-diphosphate carboxylase (RuBisCO, from spinach partially purified powder, 0.01–0.1 unit/mg solid), d-(−)−3-phosphoglyceric acid disodium salt (3-PGA, ≥93%), glyceraldehyde 3-phosphate dehydrogenase (GAPDH, from rabbit muscle lyophilized powder), 3-phosphoglyceric phosphokinase (PGK, from baker’s yeast (S. cerevisiae), ammonium sulfate suspension, ≥1000 units/mg protein), glycerol 3-phosphate oxidase (G3POX, from Pediococcus sp. lyophilized powder), α-glycerophosphate dehydrogenase (G3PDH, from rabbit muscle, type I, ammonium sulfate suspension), triosephosphate isomerase (TPI, from baker’s yeast (S. cerevisiae), ammonium sulfate suspension) and catalase (from bovine liver powder) were provided by Sigma-Aldrich.
Fabrication of RIMRs
The RIMRs were made in three main steps: (a) preparation of the pristine polydimethylsiloxane (PDMS) microfluidic reactors; (b) modification of PDA; and (c) immobilization of RuBisCO. First, the PDMS microfluidic reactors were fabricated by sealing one molded PDMS layer with another flat PDMS layer on the top. The molded PDMS layer was made by using the standard soft-lithography technique67 with the Sylgard 184 elastomer kit (Dow Corning Corporation) and a SU-8 mold (SU-8 50, MicroChem) fabricated by the photolithography. Then, the pristine PDMS inner surface of the microfluidic reactors were modified by PDA. The strategy for the PDA modification was adopted from Zheng’s group with minor modification38. The detailed procedures are provided in Supplementary Methods. Next, 7 μL of RuBisCO in the reaction buffer was injected into the PDA-PDMS microfluidic reactor by pipette and kept at room temperature for 6 h for the RuBisCO immobilization. After that, the reactor was rinsed by the reaction buffer using a syringe pump at a flow rate of 2.5 μL min−1 for 40 min. Finally, the RIMRs were ready for use.
Confirmation of RuBisCO immobilization
The surface characterization of the inner surfaces of the microfluidic reactors after each step of RuBisCO immobilization was first conducted by scanning electron microscope (SEM, JEOL JSM-6490) for the confirmation of RuBisCO immobilization. More characterization details such as optical microscopic images, water contact angles, Raman spectra, ATR-FTIR spectra and fluorescence images are presented in Supplementary Figs. 2–6.
Determination of protein-loading capacity
The protein-loading capacity of the PDA-modified microfluidic reactors was investigated to select an optimal RuBisCO concentration for the RIMRs fabrication in the following research. Increased concentrations of RuBisCO solutions (1.5625–25 μg μL−1) were injected into the PDA-modified microfluidic reactors. Then, the rinsed-out RuBisCO solutions were carefully collected to determine the protein concentration difference between the initially injected and the rinsed-out RuBisCO solutions. The protein concentration was qualified by the Bradford method68 using the Quick Start Bradford Protein Assay kit (Bio-Rad Pacific Limited.), which were determined by measuring the absorbance at the wavelength of 595 nm using a UV–visible spectrometer (UV-2450, Shimadzu). BSA solutions (0.125–1 mg mL−1) were selected as standards to plot the calibration curve (see Supplementary Fig. 7). The protein-loading amount of the reactor was calculated by
| 1 |
where C0 is the protein concentration of initially injected RuBisCO solution (μg μL−1), C1 is the protein concentration of the rinsed-out RuBisCO solution (μg μL−1), Vi is the volume of initially injected RuBisCO solution (μL), Vw is the volume of washing solution collected from the outlet of the reactor (μL), and Ar is the surface area of the reactor (~367.5 mm2). The maximum protein-loading amount was then regarded as the protein-loading capacity of the PDA-modified microfluidic reactor. The protein-loading efficiency was also obtained by
| .2 |
Then the optimal RuBisCO concentration for further research can be determined referring to the protein-loading capacity and the protein-loading efficiency. All the error bars represent the standard deviations from three repeated experiments.
Assay of RuBisCO activity
The activities of the free and immobilized RuBisCO were determined using an amplification signal assay adapted from the previously reported method57. The general details are provided in Supplementary Methods. In the immobilized RuBisCO activity assay, reactant mixture (66 mM and 0.5 mM RuBP in the reaction buffer) was passed through the RIMRs at the flow rate of 7 μL min−1 (reaction time is 1 min). The production solution I (mixture of RuBisCO, RuBP, and products) was collected from the outlet of the reactors before it was added into the assay mixture for 3-PGA amount determination. The specific composition of the assay mixture can be seen in Supplementary Methods of amplification method for RuBisCO activity assay. For the free RuBisCO activity assay, RuBisCO was incubated with reactant mixture for 1 min before the reaction is stopped by the same volume of 80% ethanol. Then, the production solution II (mixture of RuBisCO, RuBP, , products, and ethanol) was added into the assay mixture to determine the 3-PGA amount. The amount of RuBisCO used here was the same as that immobilized into the microfluidic reactors (21.875 μg). The amount of 3-PGA in the production solutions could be determined from the calibration lines generated by adding different amounts of standard 3-PGA into the assay mixture (see Supplementary Fig. 8). Here the calibration line for production solution I is obtained by using standard 3-PGA dissolved in the reaction buffer, and that for production solution II is prepared by using standard 3-PGA dissolved in the reaction buffer:ethanol (60:40, v/v). The enzyme activity was defined as the production rate of 3-PGA (μmol g−1 RuBisCO min−1). The RuBisCO activities under different concentrations of RuBP were fit to a Michaelis–Menten-type model using the hyperbola regression to derive the Vmax and Km parameters of both free and immobilized RuBisCO.
Feasibility of 3-PGA production using RIMRs
The feasibility of producing 3-PGA with RIMRs was tested by injecting 0.5 mM RuBP and 66 mM in the reaction buffer into the as-prepared RIMRs using a syringe pump at a specific flow rate. The production solution I was collected from the outlet for the 3-PGA amount determination by the amplification signal assay. Different amounts of 3-PGA can be produced when the reactant mixture is injected at different flow rates (from 28 to 1.4 μL min−1), which directly determines the reaction time of the 3-PGA production reaction (see Supplementary Methods for reaction time determination). The produced amount of 3-PGA (μmol g−1 RuBisCO) in a specific reaction time can also be determined by the same standard curve obtained from last section. Control experiments were conducted at the same time with the same amount of BSA that was immobilized in microfluidic reactors. HPLC–MS/MS was also applied to detect the produced 3-PGA from the production solution I according to the method tested before69. The details of HPLC–MS/MS analysis are described in the Supplementary Methods.
Storage and thermal stabilities of RIMRs
To test the storage stability of the RIMRs, several prepared RIMRs were incubated at 4 °C for different days. Then, the immobilized RuBisCO activities of them were measured. The storage stability of free RuBisCO was also tested at the same condition for comparison. The highest RuBisCO activity (Ahs) was normalized to 100% and the relative RuBisCO activities after different storage days were calculated as a percentage of Ahs.
Regarding the thermal stability of the RIMRs, several prepared RIMRs were first incubated in the oven at different temperatures (from 20 to 70 °C) for 10 min and then the RuBisCO activities of each RIMRs were examined. RIMRs were also incubated at 50 °C for up to 60 min to examine their stability at elevated temperature for a prolonged time. The thermal stability of free RuBisCO was also tested at the same condition for comparison. Aht was normalized to 100% and the relative RuBisCO activities at different incubation temperatures were calculated as a percentage of Aht.
Reusability of RIMRs
The reusability of the RIMRs means the capability of the RIMRs being used repeatedly. It was evaluated by conducting the 3-PGA production reaction using one reactor for 10 cycles of reuse. Here, one cycle of reuse refers to the test that 21 μL of reactant mixture solution passes through the RIMRs for 3-PGA production. RuBisCO activity was measured from the collected production solution I for each cycle of reuse. Different flow rates were applied to inject the reactant mixture to investigate the effects of flushing. The relative RuBisCO activities were calculated as a percentage of the initial RuBisCO activity in the first cycle.
Continuous production of 3-PGA from RIMRs
Continuous production of 3-PGA was achieved by constantly injecting the reactant mixture into the RIMRs and thus a large amount of 3-PGA can be produced. As a comparison, 3-PGA production in the bulk reaction with the same amount of free RuBisCO was conducted at the same time. Increasing Vp (from 21 to 1680 μL) was applied to examine the difference of the 3-PGA production ability between the RIMRs reaction and the bulk reaction.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work is supported by National Science Foundation of China (no. 61377068) and Research Grants Council (RGC) of Hong Kong (N_PolyU505/13, 152184/15E, 152127/17E, 152126/18E, and 152219/19E). The authors would like to thank The Hong Kong Polytechnic University for the grants G-YBPR, 4-BCAL, 1-ZE14, 1-ZE27, and 1-ZVGH, Shandong Provincial Natural Science Foundation, China (ZR2016XJ004) and Doctoral cooperation fund of Shandong Academy of Sciences (2017BSHZ007). The technical assistance in mass spectrometry facility of the University Research Facility in Life Sciences (ULS) of the Hong Kong Polytechnic University is also appreciated.
Author contributions
Y.Z. designed the research, performed the experiments and data analysis, and prepared the manuscript. Z.H. and X.H. contributed to UV experiments and manuscript revision. Q.C., L.S. and Y.Y. contributed to designing and fabricating the microfluidic reactors. Q.W., P.-K.S. and Z.Y. contributed to the HPLC–MS/MS experiments. Y.J. guided the biotreatment, Z.L. helped the analysis, W.Y. aided the surface modification, A.J., S.S., and W.Z. supported the spectral assay. X.Z. conceived the idea, supervised the whole project and revised the manuscript. All authors discussed the results and commented on the manuscript.
Data availability
The source data underlying Fig. 1d, Table 1, Figs. 2a, b, 3a–d, 4, and Supplementary Figs. 4–10, 11b, 12, 13 are provided as a Source Data file. All data underlying the findings in this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information: Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at 10.1038/s41467-019-12089-6.
References
- 1.Berg, J. M., Tymoczko, J. L. & Stryer, L. Biochemistry, 7th edn (W. H. Freeman, 2012).
- 2.Archer, M. D. & Barber, J. Molecular to Global Photosynthesis (World Scientific, 2004).
- 3.Barber J, Tran PD. From natural to artificial photosynthesis. J. R. Soc. Interface. 2013;10:20120984. doi: 10.1098/rsif.2012.0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Northoff, E. 2050 A Third More Mouths to Feed (Food and Agriculture Organization of the United Nations, 2016).
- 5.Concepcion JJ, House RL, Papanikolas JM, Meyer TJ. Chemical approaches to artificial photosynthesis. Proc. Natl Acad. Sci. USA. 2012;109:15560–15564. doi: 10.1073/pnas.1212254109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tachibana Y, Vayssieres L, Durrant JR. Artificial photosynthesis for solar water-splitting. Nat. Photonics. 2012;6:511. doi: 10.1038/nphoton.2012.175. [DOI] [Google Scholar]
- 7.Chowdhury FA, Trudeau ML, Guo H, Mi Z. A photochemical diode artificial photosynthesis system for unassisted high efficiency overall pure water splitting. Nat. Commun. 2018;9:1707. doi: 10.1038/s41467-018-04067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Razeghifard, R. Natural and Artificial Photosynthesis: Solar Power as an Energy Source (John Wiley & Sons, 2013).
- 9.Ruffing AM. Engineered cyanobacteria: teaching an old bug new tricks. Bioeng. Bugs. 2011;2:136–149. doi: 10.4161/bbug.2.3.15285. [DOI] [PubMed] [Google Scholar]
- 10.Lan EI, Liao JC. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 2011;13:353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Kunjapur AM, Eldridge RB. Photobioreactor design for commercial biofuel production from microalgae. Ind. Eng. Chem. Res. 2010;49:3516–3526. doi: 10.1021/ie901459u. [DOI] [Google Scholar]
- 12.Magnuson A, et al. Biomimetic and microbial approaches to solar fuel generation. Acc. Chem. Res. 2009;42:1899–1909. doi: 10.1021/ar900127h. [DOI] [PubMed] [Google Scholar]
- 13.Lee JS, Lee SH, Kim JH, Park CB. Artificial photosynthesis on a chip: microfluidic cofactor regeneration and photoenzymatic synthesis under visible light. Lab Chip. 2011;11:2309–2311. doi: 10.1039/c1lc20303g. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Antonietti M. Bio-inspired NADH regeneration by carbon nitride photocatalysis using diatom templates. Energy Environ. Sci. 2013;6:1486–1493. doi: 10.1039/c3ee40696b. [DOI] [Google Scholar]
- 15.Huang X, et al. Microfluidic chip-based one-step fabrication of an artificial photosystem I for photocatalytic cofactor regeneration. RSC Adv. 2016;6:101974–101980. doi: 10.1039/C6RA21390A. [DOI] [Google Scholar]
- 16.Zhao Y, et al. Fully conjugated two-dimensional sp2-carbon covalent organic frameworks as artificial photosystem I with high efficiency. Angew. Chem. Int. Ed. 2019;58:5376–5381. doi: 10.1002/anie.201901194. [DOI] [PubMed] [Google Scholar]
- 17.Ellis RJ. Biochemistry: tackling unintelligent design. Nature. 2010;463:164–165. doi: 10.1038/463164a. [DOI] [PubMed] [Google Scholar]
- 18.Tsai Y-CC, Lapina MC, Bhushan S, Mueller-Cajar O. Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat. Commun. 2015;6:8883. doi: 10.1038/ncomms9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonacci W, et al. Modularity of a carbon-fixing protein organelle. Proc. Natl Acad. Sci. USA. 2012;109:478–483. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin MT, Occhialini A, Andralojc PJ, Parry MAJ, Hanson MR. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014;513:547. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruan C-J, Shao H-B, Teixeira, da Silva JA. A critical review on the improvement of photosynthetic carbon assimilation in C3 plants using genetic engineering. Crit. Rev. Biotechnol. 2012;32:1–21. doi: 10.3109/07388551.2010.533119. [DOI] [PubMed] [Google Scholar]
- 22.Aigner H, et al. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science. 2017;358:1272–1278. doi: 10.1126/science.aap9221. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, et al. Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature. 2019;566:131–135. doi: 10.1038/s41586-019-0880-5. [DOI] [PubMed] [Google Scholar]
- 24.Sidhu GK, Mehrotra R, Mehrotra S. Carbon concentrating mechanisms: in rescue of Rubisco inefficiency. Acta Physiol. Plant. 2014;36:3101–3114. doi: 10.1007/s11738-014-1652-3. [DOI] [Google Scholar]
- 25.Carmo‐Silva E, Scales JC, Madgwick PJ, Parry MA. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 2015;38:1817–1832. doi: 10.1111/pce.12425. [DOI] [PubMed] [Google Scholar]
- 26.Chakrabarti S, Bhattacharya S, Bhattacharya SK. Immobilization of D‐ribulose‐1, 5‐bisphosphate carboxylase/oxygenase: a step toward carbon dioxide fixation bioprocess. Biotechnol. Bioeng. 2003;81:705–711. doi: 10.1002/bit.10515. [DOI] [PubMed] [Google Scholar]
- 27.Satagopan S, Sun Y, Parquette JR, Tabita FR. Synthetic CO2-fixation enzyme cascades immobilized on self-assembled nanostructures that enhance CO2/O2 selectivity of RubisCO. Biotechnol. Biofuels. 2017;10:175. doi: 10.1186/s13068-017-0861-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hery TM, Satagopan S, Northcutt RG, Tabita FR, Sundaresan V-B. Polypyrrole membranes as scaffolds for biomolecule immobilization. Smart Mater. Struct. 2016;25:125033. doi: 10.1088/0964-1726/25/12/125033. [DOI] [Google Scholar]
- 29.Rodrigues RC, Ortiz C, Berenguer-Murcia Á, Torres R, Fernández-Lafuente R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013;42:6290–6307. doi: 10.1039/C2CS35231A. [DOI] [PubMed] [Google Scholar]
- 30.Bornscheuer UT. Immobilizing enzymes: how to create more suitable biocatalysts. Angew. Chem. Int. Ed. 2003;42:3336–3337. doi: 10.1002/anie.200301664. [DOI] [PubMed] [Google Scholar]
- 31.Sheldon RA, van Pelt S. Enzyme immobilisation in biocatalysis: why, what and how. Chem. Soc. Rev. 2013;42:6223–6235. doi: 10.1039/C3CS60075K. [DOI] [PubMed] [Google Scholar]
- 32.Cantone S, et al. Efficient immobilisation of industrial biocatalysts: criteria and constraints for the selection of organic polymeric carriers and immobilisation methods. Chem. Soc. Rev. 2013;42:6262–6276. doi: 10.1039/c3cs35464d. [DOI] [PubMed] [Google Scholar]
- 33.Cao, L. Carrier-bound Immobilized Enzymes: Principles, Application and Design (John Wiley & Sons, 2006).
- 34.Zucca P, Sanjust E. Inorganic materials as supports for covalent enzyme immobilization: methods and mechanisms. Molecules. 2014;19:14139–14194. doi: 10.3390/molecules190914139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Dellatore SM, Miller WM, Messersmith PB. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Rho J, Messersmith PB. Facile conjugation of biomolecules onto surfaces via mussel adhesive protein inspired coatings. Adv. Mater. 2009;21:431–434. doi: 10.1002/adma.200801222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao C, et al. Surface modification of halloysite nanotubes with dopamine for enzyme immobilization. ACS Appl. Mater. Interfaces. 2013;5:10559–10564. doi: 10.1021/am4022973. [DOI] [PubMed] [Google Scholar]
- 38.Cheng G, Hao S-J, Yu X, Zheng S-Y. Nanostructured microfluidic digestion system for rapid high-performance proteolysis. Lab Chip. 2015;15:650–654. doi: 10.1039/C4LC01165A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connor, C. M., Adams, J. U. & Fairman, J. Essentials of Cell Biology (NPG Education, 2010).
- 40.Wang N, Zhang X, Wang Y, Yu W, Chan HLW. Microfluidic reactors for photocatalytic water purification. Lab Chip. 2014;14:1074–1082. doi: 10.1039/C3LC51233A. [DOI] [PubMed] [Google Scholar]
- 41.Ma J, et al. Organic-inorganic hybrid silica monolith based immobilized trypsin reactor with high enzymatic activity. Anal. Chem. 2008;80:2949–2956. doi: 10.1021/ac702343a. [DOI] [PubMed] [Google Scholar]
- 42.Křenková J, Foret F. Immobilized microfluidic enzymatic reactors. Electrophoresis. 2004;25:3550–3563. doi: 10.1002/elps.200406096. [DOI] [PubMed] [Google Scholar]
- 43.Lv Y, Lin Z, Tan T, Svec F. Preparation of reusable bioreactors using reversible immobilization of enzyme on monolithic porous polymer support with attached gold nanoparticles. Biotechnol. Bioeng. 2014;111:50–58. doi: 10.1002/bit.25005. [DOI] [PubMed] [Google Scholar]
- 44.Liu, D. -M., Chen, J. & Shi, Y. -P. Advances on methods and easy separated support materials for enzymes immobilization. Trends Anal. Chem. 102, 332–342 (2018).
- 45.Tan H, et al. Heterogeneous multi-compartmental hydrogel particles as synthetic cells for incompatible tandem reactions. Nat. Commun. 2017;8:663. doi: 10.1038/s41467-017-00757-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Oliveira KB, Mischiatti KL, Fontana JD, de Oliveira BH. Tyrosinase immobilized enzyme reactor: development and evaluation. J. Chromatogr. B. 2014;945:10–16. doi: 10.1016/j.jchromb.2013.11.042. [DOI] [PubMed] [Google Scholar]
- 47.Lorimer GH, Miziorko HM. Carbamate formation on the. epsilon.-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by carbon dioxide and magnesium (2+) Biochemistry. 1980;19:5321–5328. doi: 10.1021/bi00564a027. [DOI] [PubMed] [Google Scholar]
- 48.Hartman F, Stringer C, Milanez S, Lee EH. The active site of Rubisco. Philos. Trans. R. Soc. Lond. B. 1986;313:379–395. doi: 10.1098/rstb.1986.0045. [DOI] [Google Scholar]
- 49.Stitt M, Lunn J, Usadel B. Arabidopsis and primary photosynthetic metabolism—more than the icing on the cake. Plant J. 2010;61:1067–1091. doi: 10.1111/j.1365-313X.2010.04142.x. [DOI] [PubMed] [Google Scholar]
- 50.Kane HJ, Wilkin J-M, Portis AR, John Andrews T. Potent inhibition of ribulose-bisphosphate carboxylase by an oxidized impurity in ribulose-1,5-bisphosphate. Plant Physiol. 1998;117:1059–1069. doi: 10.1104/pp.117.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andralojc PJ, et al. 2-Carboxy-d-arabinitol 1-phosphate (CA1P) phosphatase: evidence for a wider role in plant Rubisco regulation. Biochem. J. 2012;442:733–742. doi: 10.1042/BJ20111443. [DOI] [PubMed] [Google Scholar]
- 52.Edmondson DL, Badger MR, Andrews TJ. A kinetic characterization of slow inactivation of ribulosebisphosphate carboxylase during catalysis. Plant Physiol. 1990;93:1376–1382. doi: 10.1104/pp.93.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edmondson DL, Badger MR, Andrews TJ. Slow inactivation of ribulosebisphosphate carboxylase during catalysis is not due to decarbamylation of the catalytic site. Plant Physiol. 1990;93:1383–1389. doi: 10.1104/pp.93.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edmondson DL, Badger MR, Andrews TJ. Slow inactivation of ribulosebisphosphate carboxylase during catalysis is caused by accumulation of a slow, tight-binding inhibitor at the catalytic site. Plant Physiol. 1990;93:1390–1397. doi: 10.1104/pp.93.4.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarı M, Akgöl S, Karataş M, Denizli A. Reversible immobilization of catalase by metal chelate affinity interaction on magnetic beads. Ind. Eng. Chem. Res. 2006;45:3036–3043. doi: 10.1021/ie0507979. [DOI] [Google Scholar]
- 56.Zhu Y, Kaskel S, Shi J, Wage T, van Pée K-H. Immobilization of Trametes versicolor laccase on magnetically separable mesoporous silica spheres. Chem. Mater. 2007;19:6408–6413. doi: 10.1021/cm071265g. [DOI] [Google Scholar]
- 57.SULPICE R, et al. Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ. 2007;30:1163–1175. doi: 10.1111/j.1365-3040.2007.01679.x. [DOI] [PubMed] [Google Scholar]
- 58.Wang H, et al. Biomimetic enzyme cascade reaction system in microfluidic electrospray microcapsules. Sci. Adv. 2018;4:eaat2816. doi: 10.1126/sciadv.aat2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du Y, et al. MOF-based nanotubes to hollow nanospheres through protein-induced soft-templating pathways. Adv. Sci. 2019;6:1801684. doi: 10.1002/advs.201801684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kandambeth S, et al. Self-templated chemically stable hollow spherical covalent organic framework. Nat. Commun. 2015;6:6786. doi: 10.1038/ncomms7786. [DOI] [PubMed] [Google Scholar]
- 61.Zhang N, et al. Surface-functionalized mesoporous nanoparticles as heterogeneous supports to transfer bifunctional catalysts into organic solvents for tandem. Catal. ACS Appl Nano Mater. 2018;1:6378–6386. doi: 10.1021/acsanm.8b01572. [DOI] [Google Scholar]
- 62.Wang Y, et al. Enzymes immobilized on carbon nitride (C3N4) Adv. Mater. Interfaces. 2019;6:1801664. doi: 10.1002/admi.201801664. [DOI] [Google Scholar]
- 63.Elvira KS, Solvas XCI, Wootton RCR, deMello AJ. The past, present and potential for microfluidic reactor technology in chemical synthesis. Nat. Chem. 2013;5:905–915. doi: 10.1038/nchem.1753. [DOI] [PubMed] [Google Scholar]
- 64.Kockmann N, Gottsponer M, Roberge DM. Scale-up concept of single-channel microreactors from process development to industrial production. Chem. Eng. J. 2011;167:718–726. doi: 10.1016/j.cej.2010.08.089. [DOI] [Google Scholar]
- 65.Amador C, Gavriilidis A, Angeli P. Flow distribution in different microreactor scale-out geometries and the effect of manufacturing tolerances and channel blockage. Chem. Eng. J. 2004;101:379–390. doi: 10.1016/j.cej.2003.11.031. [DOI] [Google Scholar]
- 66.Tonkovich A, et al. Microchannel technology scale-up to commercial capacity. Chem. Eng. Res. Des. 2005;83:634–639. doi: 10.1205/cherd.04354. [DOI] [Google Scholar]
- 67.Duffy DC, McDonald JC, Schueller OJ, Whitesides GM. Rapid prototyping of microfluidic systems in poly (dimethylsiloxane) Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 68.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 69.Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography–tandem mass spectrometry. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data underlying Fig. 1d, Table 1, Figs. 2a, b, 3a–d, 4, and Supplementary Figs. 4–10, 11b, 12, 13 are provided as a Source Data file. All data underlying the findings in this study are available from the corresponding author upon reasonable request.