Abstract
Genome-wide association studies (GWASs) have identified hundreds of genetic risk variants for human cancers. However, target genes for the majority of risk loci remain largely unexplored. It is also unclear whether GWAS risk-loci-associated genes contribute to mutational signatures and tumor mutational burden (TMB) in cancer tissues. We systematically conducted cis-expression quantitative trait loci (cis-eQTL) analyses for 294 GWAS-identified variants for six major types of cancer—colorectal, lung, ovary, prostate, pancreas, and melanoma—by using transcriptome data from the Genotype-Tissue Expression (GTEx) Project, the Cancer Genome Atlas (TCGA), and other public data sources. By using integrative analysis strategies, we identified 270 candidate target genes, including 99 with previously unreported associations, for six cancer types. By analyzing functional genomic data, our results indicate that 180 genes (66.7% of 270) had evidence of cis-regulation by putative functional variants via proximal promoter or distal enhancer-promoter interactions. Together with our previously reported associations for breast cancer risk, our results show that 24 genes are shared by at least two cancer types, including four genes for both breast and ovarian cancer. By integrating mutation data from TCGA, we found that expression levels of 33 and 66 putative susceptibility genes were associated with specific mutational signatures and TMB of cancer-driver genes, respectively, at a Bonferroni-corrected p < 0.05. Together, these findings provide further insight into our understanding of how genetic risk variants might contribute to carcinogenesis through the regulation of susceptibility genes that are related to the biogenesis of somatic mutations.
Keywords: human cancers, GWAS-identified variants, cis-eQTL, gene expression, susceptibility genes, functional variants, cancer driver genes, mutational signature, tumor mutational burden
Introduction
Genome-wide association studies (GWASs) have identified hundreds of genetic risk variants for human cancers; the majority of these risk variants are for three common cancer types: breast, colorectal, and prostate.1, 2, 3, 4, 5, 6 Approximately 90% of these GWAS-identified single nucleotide polymorphisms, or index SNPs, reside in noncoding regions such as intergenic or intronic regions. However, the target genes and biological mechanisms driving cancer susceptibility remain unclear for many of these variants. In recent years, by using functional epigenetic data from the Encyclopedia of DNA Elements (ENCODE) and the Roadmap Epigenomics project (Roadmap), several studies have shown that index SNPs or their correlated variants in strong linkage disequilibrium (LD) are enriched with cis-regulatory elements, including histone markers, DNase I hypersensitive sites, and transcription factor (TF) binding motifs.7, 8, 9 These findings, together with previous fine-mapping and expression quantitative trait loci (eQTL) studies, indicate that the majority of noncoding index SNPs, or their correlated variants, contribute to cancer pathogenesis through roles in the gene regulation of nearby genes.7, 8, 10, 11, 12, 13, 14
Previous fine-mapping and eQTL analyses, including our own work in breast and colorectal cancers, have revealed a number of candidate cancer-susceptibility genes that are regulated by index SNPs or their correlated variants.12, 14, 15, 16, 17, 18, 19, 20, 21 However, target genes for the majority of index SNPs identified for several other cancer types—melanoma, ovarian, and pancreatic cancers—have remained largely unexplored. In addition, many index SNPs have been recently identified for prostate and colorectal cancers,5, 22, 23 and existing eQTL analyses are often limited by single transcriptome datasets. Thus, the systematic characterization of previously reported index SNPs and the exploration of candidate target genes with multiple transcriptome datasets, such as The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) Project, might provide further understanding of the biological mechanisms that contribute to cancer development.
Recently, variants have been annotated in functional regions genome-wide through the use of various emerging functional genomic resources from the ENCODE,24 Roadmap25, 26 and FANTOM5.27 Potential functional variants can be examined by their locations in transcription factor motifs,9, 28, 29 histone modifications, DNase I hypersensitive sites, and Chromatin Immunoprecipitation Sequencing (ChIP-seq) binding sites. In particular, the advance of chromatin interaction technology including Hi-throughput Genome-wide Chromosome Conformation Capture (Hi-C), Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET), and Integrated Method for Predicting Enhancer Targets (IM-PET) has produced large amounts of chromatin-chromatin interaction data in various normal and cancer cell lines.30, 31, 32, 33 These data are valuable resources for linking particular functional variants to target genes. However, work to systemically link this new information to functional variants and target genes in cancer remains largely unexplored.
Somatic mutations are one of the most common causes of human carcinogenesis.34 However, it is unclear whether cancer-susceptibility genes are associated with somatic mutations, by which they could influence cancer risk and prognosis. The somatic mutation catalogs of base substitutions can be characterized into distinct mutational signatures.35 A total of 30 reference signatures were characterized with mutation data from TCGA in the COSMIC database; some of these have been reported to be a result of either the activity or inactivity of specific cancer-driver genes.36, 37, 38, 39, 40, 41 For example, signature 3 is strongly associated with functional loss of either BRCA1 (MIM: 113705) or BRCA2 (MIM: 600185) in breast cancer. Signatures 2 and 13 are attributed to the activity of APOBEC cytidine deaminases, especially APOBEC3A (MIM: 607109) and APOBEC3B (MIM: 607110), across multiple cancer types. Signatures 10 and 11 are associated with deleterious mutations in the DNA repair POLE (MIM: 174762) and the MGMT (MIM: 156569) genes, respectively.
In this study, we systemically characterized a total of 294 index SNPs identified from GWASs for six major types of cancer—colorectal, lung (lung squamous cell carcinoma and lung adenocarcinoma), ovary, prostate, pancreas, and melanoma—among European populations. We searched for target genes (as putative cancer-susceptibility genes) for index SNPs by meta-analyses of eQTL data from multiple transcriptome datasets (TCGA, GTEx, and other datasets, such as Colonomics for colorectal cancer). Furthermore, we systemically evaluated associations between the expression of putative cancer-susceptibility genes with mutational signatures and tumor mutational burden (TMB) of cancer-driven genes. Our findings provide further insight into how genetic risk variants might contribute to carcinogenesis by affecting the biogenesis of somatic mutations though regulation of putative susceptibility genes.
Material and Methods
Data Resources
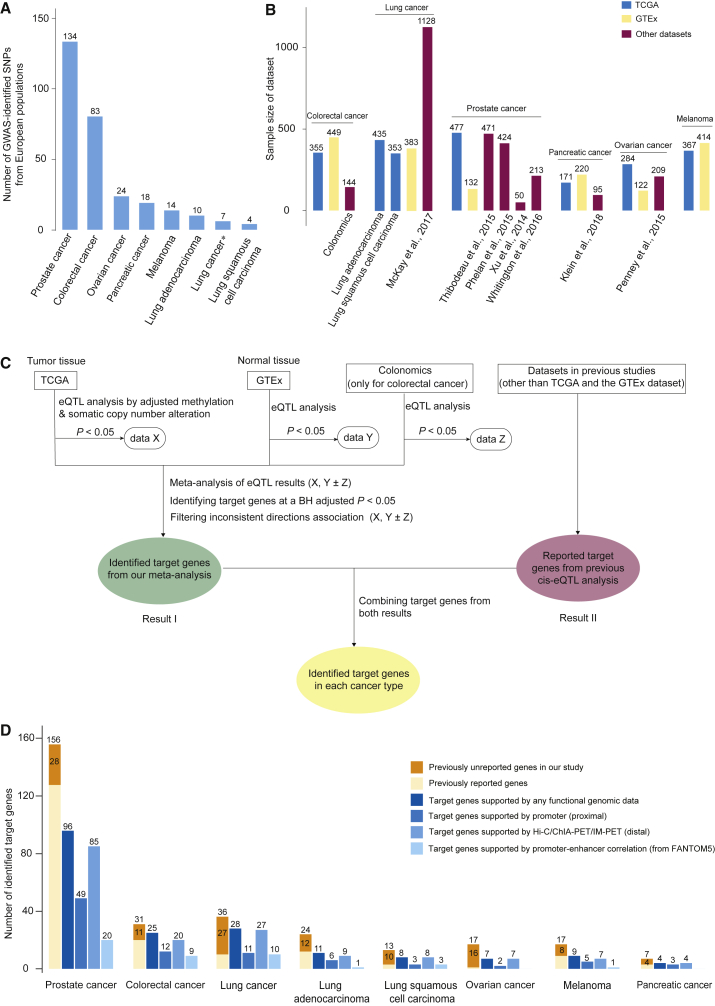
We included in this study a total of 409 GWAS-identified SNPs at p < 5.0 × 10−8 that are from the NHGRI-EBI GWAS catalog and that are associated with six major cancer types: colorectal, lung (lung squamous cell carcinoma and lung adenocarcinoma), ovary, prostate, pancreas, and melanoma in European populations. We filtered index SNPs by LD (distance within 2 Mb, R2 > 0.1), selecting those that had the strongest associations with risk for each cancer. A total of 294 index SNPs remained for further analysis (Figure 1A and Table S1) for: colorectal cancer (n = 83), lung cancer (specific for lung adenocarcinoma: n = 10; specific for lung squamous cell carcinoma: n = 4; common to both lung adenocarcinoma and squamous cell carcinoma: n = 7), ovary (n = 24), prostate (n = 134), pancreas (n = 18), and melanoma (n = 14).
Figure 1.
Identification of Candidate Target Genes for GWAS-Identified SNPs in Six Cancer Types
(A) A histogram showing the number of characterized GWAS-identified SNPs in this study across six cancer types. The ∗ refers to SNPs commonly identified for both lung adenocarcinoma and squamous cell carcinoma (this note applies to other legends in this figure).
(B) A histogram showing sample size for each dataset across six cancer types. The dataset from TCGA is depicted in blue, the dataset from the GTEx is depicted in yellow, and datasets other than TCGA and the GTEx are depicted in red.
(C) A flow chart illustrating the identification of target genes for GWAS-identified SNPs on the basis of cis-eQTL analysis, using data from both TCGA and the GTEx datasets across six cancer types. The rounded rectangle indicates the eQTL target genes identified by TCGA and the GTEx. The green box indicates the target genes that are identified using BH-corrected p < 0.05 from a meta-analysis of eQTL results from TCGA and the GTEx. The data from the Colonomics was also included for colorectal cancer. The red box refers to previously reported target genes. The yellow box refers to target genes after combining results from both our meta-analysis and previous eQTL analysis.
(D) A histogram showing the number of target genes identified and those supported by additional evidence from functional genomic data. The previously unreported target genes in our study are highlighted with deep yellow. Previously reported target genes are depicted in light yellow. The shades of blue from left to right refer to target genes supported by evidence of additional functional genomic data including promoter (proximal), chromatin-chromatin interaction data (distal), and promoter-enhancer correlation data (from FANTOM5).
We downloaded RNA-seq V2 data (level 3), DNA methylation data (level 3, Infinium HumanMethyaltion27K for ovarian cancer and Infinium HumanMethylation450K for the other cancers), and somatic copy number alteration data (level 3) from TCGA from cBioPortal. For gene expression data, normalized gene expression values were further transformed across samples by an inverse normalizing transformation method. We also downloaded SNP data (level 3), genotyped on Affymetrix SNP 6.0, from TCGA. We imputed additional genetic data from genotype data by using the mixed populations of the 1000 Genomes Project Phase 3 and the Minimac tool,42, 43 implemented on the Michigan Imputation Server. Only common SNPs (minor allele frequency [MAF] > 0.05) with high imputation quality (R2 > 0.3) were further evaluated. If the index SNP failed to meet these criteria, we used a surrogate SNP in strong LD (R2 > 0.8). For cis-eQTL analyses, we used samples with matched gene expression, DNA methylation, somatic copy number alteration, and SNP data from European populations; samples numbered: colorectal cancer (n = 355), lung adenocarcinoma (n = 435), lung squamous cell carcinoma (n = 353), ovarian cancer (n = 284), prostate cancer (n = 477), pancreatic cancer (n = 171), and melanoma (n = 367). For somatic mutational signatures, we used 30 reference signatures from the COSMIC database, and we characterized each TCGA sample with mSignatureDB. We also downloaded somatic mutations identified from whole-exome sequencing data for each TCGA sample from the genomic data commons (GDC).
cis-eQTL Analysis
For data from TCGA, we conducted linear regression analyses that included adjustment for DNA methylation, somatic copy number alteration, and the top five principal components for genetic ancestry in order to evaluate associations between the genotypes of index SNPs and expression levels of nearby genes (±1 Mb).
We extracted cis-eQTL results for index SNPs and nearby genes from the most recent GTEx database (v.7)44 on the basis of normal tissues from transverse colorectal (n = 246), lung (n = 383), ovary (n = 122), prostate (n = 132), pancreas (n = 220), and skin (n = 414). For colorectal cancer, we performed an additional cis-eQTL analysis with data from healthy colonic mucosa (n = 47) and normal mucosa adjacent to colon tumor tissues (n = 97) from the Colonomics project.16, 21
We systematically searched for cis-eQTL results from prior studies on six cancers, and, when they could be identified, we included transcriptome datasets other than TCGA and GTEx. Specifically, we included eQTL results from four lung cancer datasets, Laval (N = 409), UBC (N = 287), Groningen (N = 342), and NCI (N = 90);19 four datasets for prostate cancer, Mayo (N = 471),18 PHS/HPFS (N = 264),45 Weill Cornell Medical College (N = 50),46 and Stockholm and Cambridge (N = 213);47 and one dataset each for pancreatic cancer, LTG (N = 95),48 and ovarian cancer, Mayo dataset (N = 209)49 (Figure 1B).
Identification of Putative Cancer-Susceptibility Genes
To identify cancer-susceptibility genes, we combined regression results for eQTL from TCGA and GTEx for each cancer type, as well as from the Colonomics project for colorectal cancer, by using a meta-analysis method based on association direction and p values.50 The combined p values were further adjusted with the Benjamini-Hochberg (BH) procedure. The BH-adjusted p < 0.05 threshold was applied to identify eQTL target genes for each cancer type. In addition, the candidate target genes were filtered to remove those with inconsistent directions of nominally significant association across TCGA and GTEx data (Figure 1C). The eQTL target genes identified in previous literature were also included for us to characterize as putative cancer-susceptibility genes. In addition, an unpublished fine-mapping study recently reported a total of 178 high-confidence target genes from 150 susceptibility regions in breast cancer51 by using the integrated expression quantitative traits and in-silico prediction of GWAS targets (INQUISIT) pipeline.2, 51 We also evaluated the associations of these genes with mutational signatures and TMB of cancer-driver genes (see Statistical Analysis) and included the results in the Discussion.
Pathway Enrichment Analysis
By using the Ingenuity Pathway Analysis (IPA) tool, we examined the functional enrichment in the gene function category and biological pathways for the identified putative cancer-susceptibility genes. We presented the most significant gene function categories and biological pathways.
Functional Annotation of Variants in Strong LD with Index SNPs
Functional annotation was evaluated via epigenetic data, including DNase I hypersensitive sites and TF ChIP-seq binding peaks, from both ENCODE and Roadmap. We evaluated variants for potential functional significance by using chromHMM annotation in all available ENCODE and Roadmap cell lines. For each SNP or indel, we investigated whether it mapped to functional regions, including promoters or enhancers, as annotated from ChromHMM based on the HaploReg v4 database.52 Additional functional prediction scores for each variant from multiple bioinformatic tools (e.g., CADD,53 RegulomeDB,54 and Funseq229) were also evaluated with the WGS Annotator.55
Chromatin-Chromatin Interaction Data Analysis
Experimentally-derived chromatin interactions generated by Hi-C, ChIA-PET, and IM-PET were collected from the 4DGenome.56 To analyze chromatin-chromatin interactions with promoter regions of putative susceptibility genes, we first used data from European populations from the 1000 Genomes project to identify functional variants in strong LD (R2 > 0.8) for index SNPs. We then examined the flanking regions of functional variants (±250 bp) and the flanking regions of gene transcription start sites (TSS; ± 2 kb) to assess potential chromatin-chromatin interactions.
Mutational Signatures and TMB of Cancer-Driver Genes
Mutational signatures have previously been characterized in TCGA samples.35, 57 We analyzed the relative contribution of mutational signatures to overall TMB by assigning contribution values ranging from 0 to 1 for each TCGA sample across seven cancer types (the six types above plus breast cancer). To determine the TMB of cancer-driver genes, we analyzed a total of 299 genes that were recently identified in a PanCancer and PanSoftware analysis of TCGA data.58 We calculated the TMB of cancer-driver genes by calculating the sum of genes harboring at least one missense, deleterious, or disruptive mutation for each sample; specifically, we summed the number of mutations from frame_shift_del, frame_shift_ins, in_frame_del, in_frame_ins, missense_mutation, nonsense_mutation, nonstop_mutation, splice_site, and translation_start_site.
Statistical Analysis
The associations between mutational signatures and gene expression levels in European populations were analyzed with semi-parametric ordinal regression models, which are tailored to fit the severely right-skewed distribution of mutational signatures; these analyses were implemented with the ‘orm’ function from the ‘rms’ library in R.59 The associations between the over-dispersed count of TMB of cancer-driver genes and the expression levels of putative cancer-susceptibility genes in European populations were analyzed via negative binomial regression; these regressions were implemented with the ‘glm.nb’ function from the ‘MASS’ library in R. To evaluate the enrichment of significant associations for identified susceptibility genes with mutational signatures and TMB of cancer-driver genes for each cancer type, we used Fisher’s exact test to compare the number of significant associations identified from the target genes with those from all protein-coding genes across the genome.
Results
Identification of Candidate Target Genes for Index SNPs in Six Cancer Types
We characterized a total of 294 index SNPs that were identified in European populations for the risk of six types of cancer from the GWAS catalog (Figure 1A and Table S1; see Material and Methods). To identify target genes for these index SNPs, we used integrative analysis strategies that included target genes identified from a meta-analysis of cis-eQTL analysis that used multiple transcriptome datasets from TCGA, GTEx, and other available data (e.g., Colonomics for colorectal cancer; Figure 1B), as well as genes previously reported in the literature (Figure 1C; see Material and Methods).
For colorectal cancer, results from a meta-analysis of eQTL results from TCGA, GTEx, and Colonomics revealed a total of 31 target genes for 14 index SNPs at BH-corrected p < 0.05 (Table S2; see Material and Methods). Of these target genes, 20 have been reported in previous studies.16, 21, 60 The remaining 11 target genes, including eight identified from five recently-reported index SNPs, have not been previously linked to colorectal cancer risk (Figure 1D and Table S2).
For prostate cancer, of the 134 SNPs investigated, results from the integrative analyses revealed a total of 156 target genes for 81 index SNPs (Figure 1D and Table S2). Of these, our meta-analysis identified a total of 74 target genes for 39 index SNPs at BH-corrected p < 0.05; of these, 28 genes have not been previously linked to prostate cancer risk (Figure 1D and Table S2).
For lung cancer, results from the integrative analyses revealed a total of 64 target genes for lung cancer; this included 13 genes for lung squamous cell carcinoma (four index SNPs), 24 genes for lung adenocarcinoma (10 index SNPs), and 36 genes for both lung adenocarcinoma and lung squamous cell carcinoma (six index SNPs). Of them, our meta-analysis identified a total of 31 genes for both (three index SNPs), 16 for lung adenocarcinoma (seven index SNPs), and 11 for lung squamous cell carcinoma (two index SNPs). Of these, a total of 40 genes have not been previously linked to lung cancer risk; these genes include 12 for adenocarcinoma, 10 for squamous cell carcinoma, and 27 for both lung adenocarcinoma and lung squamous cell carcinoma (Figure 1D and Table S2).
For ovarian cancer, results from the integrative analyses revealed a total of 17 genes for four index SNPs (Figure 1D and Table S2). A total of 31 genetic loci have been identified by previous GWASs. However, a subset of them (a total of 12 loci) have been identified in eQTL analysis that used datasets other than TCGA and the GTEx, and only OBFC1 (MIM: 613128)was identified for index SNP dbSNP: rs7902587.49 Here, our analysis revealed a total of 16 previously unreported genes for three index SNPs associated with ovarian cancer risk.
For pancreatic cancer, results from the integrative analyses revealed a total of seven target genes for six index SNPs. Six genes were identified from our meta-analysis (Figure 1D and Table S2). Of them, our analysis revealed four previously unreported genes, PVT1 (MIM: 165140), XBP1 (MIM: 194355), ABO (MIM: 110300), and PDX1 (MIM: 600733), and the remaining three genes, KLHL17, NOC2L (MIM: 610770), and HNF4G (MIM: 605966), have been previously reported.48
For melanoma, we identified a total of 17 target genes for nine index SNPs from our meta-analysis (Figure 1D and Table S2). Of them, our analysis revealed eight previously unreported genes, and the remaining nine genes, CASP8 (MIM: 601763), KDELC2, ASIP (MIM: 600201), ANKRD54 (MIM: 613383), OCA2 (MIM: 611409), CDK10 (MIM: 603464), ALS2CR12, CHMP1A (MIM: 164010), and DBNDD1, have been previously reported.61
Overall, results from our integrative analyses revealed a total of 270 target genes (based on 134 index SNPs) as putative susceptibility genes for six types of cancer (Table 1). Of these, a total of 99 genes (36.7%) had not been previously associated with cancer risk.
Table 1.
Summary of the Identified Putative Susceptibility Genes Associated with Index SNP, Mutational Signature, and TMB of Cancer-Driver Genes for Each Cancer Type
| Cancer Type | Number of Index SNPs | Number of Putative Susceptibility Genes |
Number of Putative Susceptibility Genes |
|
|---|---|---|---|---|
| Mutational Signature | TMB of Cancer-Driver Genes | |||
| Breast cancera | 51 | 101 | 24 | 24 |
| Colorectal cancer | 14 | 31 | 4 | 10 |
| Lung cancer | 20 | 64 | 1 | 8 |
| Prostate cancer | 81 | 156 | 2 | 23 |
| Pancreatic cancer | 6 | 7 | 0 | 1 |
| Ovarian cancer | 4 | 17 | 0 | 1 |
| Melanoma | 9 | 17 | 2 | 0 |
| Total | 185 | 393 (365b) | 33 | 67 (66b) |
Refers to putative breast-cancer-susceptibility genes reported from Guo et al., 2018.20
Refers to the number of unique putative cancer-susceptibility genes.
Target Genes Supported by Functional Genomic Analysis
To search for evidence of regulatory mechanisms underlying the identified target genes for index SNPs, we performed extensive functional annotation analysis to identify candidate functional variants in strong LD (see Material and Methods). We evaluated the functionalities of a total of 2,981 variants in strong LD (R2 > 0.8) with the index SNPs. Of them, we analyzed 2,023 putative functional variants, which showed evidence of the epigenetic signals from the data analysis of ENCODE and/or Roadmap (Table S3; see Material and Methods). Specifically, a total of 722 variants were mapped to promoter regions, whereas the remaining 1,301 variants were mapped to enhancer regions. Functional significance for a majority of these variants was further supported by evaluating the annotation from the CADD and other functional prediction tools (Table S3; see Material and Methods).
To search for direct evidence that variants regulate the putative target genes identified from our eQTL analysis, we first examined whether the putative functional variants were positioned in proximal promoter regions because such variants would most likely play a regulatory role in their closest genes. We found a total of 57 target genes that were the closest genes for these putative functional variants (Figure 1D and Table S3). Then we examined whether putative functional variants were located in enhancer regions. We further analyzed chromatin-chromatin interaction data to examine whether the target genes could be regulated by these variants via long-distance promoter-enhancer interactions (see Material and Methods). We collected and analyzed chromatin-chromatin interaction data generated from multiple normal and cancer cell lines (see Material and Methods). We found a total of 104 genes with evidence of distal regulation by putative functional variants via promoter-enhancer interactions (Figure 1D and Table S3). By using promoter and enhancer data from FANTOM5, we observed an additional 13 genes with evidence of distal regulation by putative functional variants (Figure 1D and Table S3).
Taken together, a total of 180 genes (66.7%) showed evidence of regulations by putative functional variants via proximal promoter or distal enhancer-promoter interactions, providing an additional layer of evidence to support the identified target genes for index SNPs (Table S3).
Common Candidate Target Genes in Multiple Cancer Types
To investigate whether the putative target genes identified were common across different cancer types, we analyzed a total of 365 target genes, including 270 candidate target genes for the six cancers above and 101 genes from our previous cis-eQTL analysis for breast cancer.20 Our results revealed that 24 target genes were commonly implicated in cancers of breast and ovarian (n = 4); breast and lung (n = 1); breast and melanoma (n = 1); breast, prostate, and melanoma (n = 1); colorectal and prostate (n = 4); lung and prostate (n = 9); and colorectal, lung, and prostate (n = 4) cancers (Figures 2A and 2B). Notably, 23 of these genes are located in three regions: 17q21.3, 6p22.1–6p21.33–6p21.32, and 2q33.1 (Figures 2B and 2C).
Figure 2.
Putative Cancer-Susceptibility Genes Commonly Implicated in Multiple Cancer Types
(A) A histogram showing the number of target genes commonly implicated in multiple cancer types. The ∗ refers to genes for lung adenocarcinoma and/or lung squamous cell carcinoma (this note applies to all other following figure legends).
(B) A heatmap showing target genes commonly observed from different cancer types. The arrow refers to a putative oncogene or putative tumor suppressor gene inferred by associations between expression levels of these genes and risk alleles of index SNPs from GWAS.
(C) A total of 23 target genes commonly observed in different cancer types are located in the three regions: 17q21.3, 6p22.1-6p21.33-6p21.32, and 2q33.1. Lines with different colors refer to different cancer types. LD values (based on data from European populations from the 1000 Genomes project) are presented for two index SNPs linked by a dashed curve.
At the locus 17q21.3, our results showed that four genes, LRRC37A (MIM: 616555), MAPK8IP1P2, KANSL1-AS1, and LRRC37A4P, were commonly observed for both breast and ovarian cancer. Risk alleles of dbSNP: rs2532263, rs17631303, and rs183211 were associated with increased expression of all of the genes except LRRC37A4P for both breast and ovarian cancers (Figures 2B and 2C).
At the loci 6p22.1–6p21.33–6p21.32, our results showed that 17 genes were commonly implicated in cancers of colorectal and prostate; lung and prostate; and colorectal, lung, and prostate. Similar to the above observation, decreased expression levels of HLA-DQB1 (MIM: 604305) and HLA-DQA1 (MIM: 146880) and increased expression level of HLA-DQA2 (MIM: 613503) were consistently associated with risk alleles of different index SNPs: dbSNP: rs9271695 for colorectal cancer and dbSNP: rs3096702 and rs115306967 for prostate cancer (Figures 2B and 2C). A decreased expression level of NOTCH4 (MIM: 164951) was consistently associated with risk alleles of different index SNPs: dbSNP: rs3117582 for lung cancer and dbSNP: rs3096702 for prostate cancer. In contrast, we observed that the expression levels of the remaining 12 genes were inconsistently associated with risk alleles of different index SNPs from different cancer types (index SNPs dbSNP: rs9271695 for colorectal cancer, rs4324798 and rs3117582 for lung cancer, rs115457135, rs12665339, rs130067, rs3096702, and rs115306967 for prostate cancer) (Figures 2B and 2C).
At locus 2q33.1, our results showed that CASP8 and ALS2CR12 were commonly implicated in cancers of breast and melanoma and breast, prostate, and melanoma, respectively. A decreased expression level of CASP8 was associated with risk alleles of different index SNPs: dbSNP: rs2110693 and rs3769821 for breast cancer and dbSNP: rs13016963 for melanoma (Figures 2B and 2C). Similarly, the increased expression level of ALS2CR12 was associated with risk alleles of different index SNPs: dbSNP: rs3769821 for breast cancer and dbSNP: rs13016963 for melanoma. In contrast, an opposite pattern was observed for the expression level of ALS2CR12, associated with the index SNP for the prostate (Figures 2B and 2C).
We further inferred potential oncogenes and tumor suppressor genes on the basis of positive or negative associations, respectively, between risk alleles of an index SNP (from GWASs) and gene expression. Our results indicated five putative oncogenes and five putative suppressor genes that were associated with risk across different cancers, as well as an additional 14 genes that might play distinct oncogene or tumor suppressor roles among different cancer types (Figure 2B).
Putative Susceptibility Genes Associated with Mutational Signatures
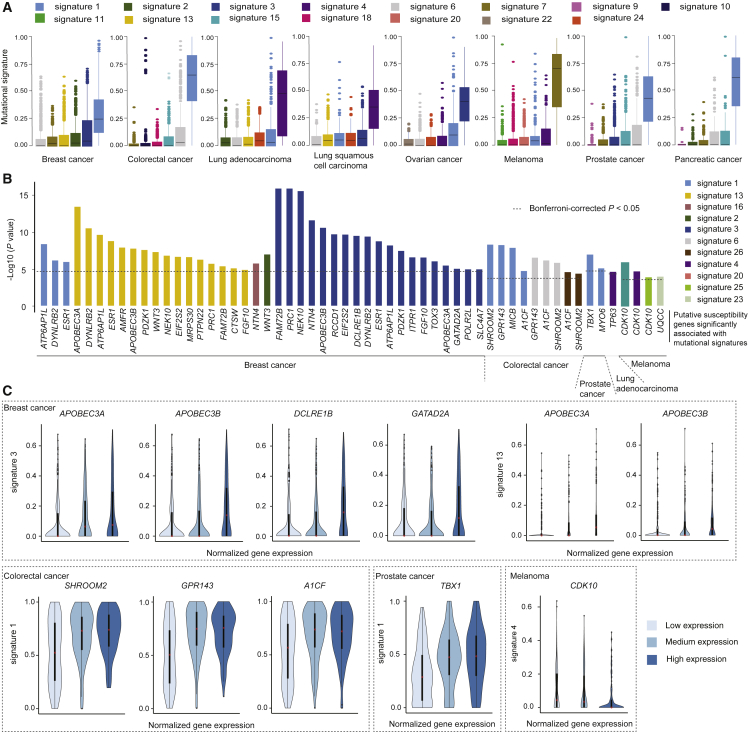
To investigate whether the putative susceptibility genes identified might affect the biogenesis of somatic mutations, we characterized the top mutational signatures that substantially contributed to TMB for each cancer type. In line with previous studies,62, 63, 64 our results showed that the highest proportions of TMB were characterized by signature 1 (with deamination at NpCpGs in breast, colorectal, prostate, and pancreatic cancer), signature 4 (tobacco-smoking-associated signature in lung adenocarcinoma and lung squamous cell carcinoma), signature 3 (BRCA1/2 alteration signature in breast and ovarian cancer), and signature 7 (ultraviolet-light-exposure-associated signature in melanoma) (Figure 3A).
Figure 3.
Putative Susceptibility Genes Associated with Specific Somatic Mutational Signatures
(A) Top mutational signatures contributing to TMB for each cancer type. Each color refers to a specific mutational signature.
(B) Bar plots showing the significance of putative susceptibility genes associated with mutational signatures at nominal p < 0.05 identified for four cancer types. The dashed lines indicate a cutoff of Bonferroni-corrected p < 0.05 for each cancer type.
(C) Violin plots of samples separated by low, median, and high expression levels of the highlighted genes (see Results); the genes were associated with specific mutational signatures at Bonferroni-corrected p < 0.05 for four cancer types. The upper dashed box shows the associations between represented genes and signature 3, as well as signature 13 in breast cancer. Lower diagram: in the dashed boxes from the left to the right, the genes SHROOM2, GPR143, and AICF in colorectal cancer, TBX1 in prostate cancer, and CDK10 in melanoma are presented.
We next evaluated associations between putative cancer-susceptibility-gene expression with each mutational signature for each cancer type (see Material and Methods). Of the 365 genes evaluated, 285 (78.1%) were associated with at least one mutational signature across the seven cancer types, at nominal p < 0.05 (Table S4). By using a more stringent threshold, we identified a total of 33 genes with significant associations with specific mutational signatures across five cancer types (Bonferroni-corrected p < 0.05). Of these, the majority (n = 24, 72.7%) were for breast cancer; additional genes included four (SHROOM2 [MIM: 300103], GPR143 [MIM: 300808], MICB [MIM: 602436], and A1CF [MIM: 618199]) for colorectal cancer, two (CDK10 and UQCC [MIM: 611797]) for melanoma, two (TBX1 [MIM: 602054] and MYO6 [MIM: 600970]) for prostate cancer, and one (TP63 [MIM: 603273]) for lung adenocarcinoma (Figure 3B, Tables 1 and 2). Specifically, high expression levels of APOBEC3A and APOBEC3B were associated with increased signatures 3 and 13, and high expression levels of two other DNA repair related genes, DCLRE1B (MIM: 609683) and GATAD2A (MIM: 614997), were associated with an increased signature 3 in breast cancer. Similarly, we observed that associations were also observed for SHROOM2, GRP143, and A1CF with signature 1 in colorectal cancer, for TBX1 with signature 1 in prostate cancer, and for CDK10 with signature 4 in melanoma (Figure 3C).
Table 2.
Associations Between Putative Susceptibility Gene Expression Levels and Mutational Signatures for Five Cancer Types (Bonferroni-Corrected p < 0.05)
| Gene | Beta | P | Bonferroni-Corrected P |
|---|---|---|---|
| Breast Cancer (Signature 3)∗ | |||
| FAM72B | 0.54 | <2.20 × 10−16 | 5.46 × 10−13 |
| PRC1 | 0.55 | <2.20 × 10−16 | 5.46 × 10−13 |
| NEK10 | −0.48 | 2.22 × 10−16 | 6.06 × 10−13 |
| NTN4 | −0.39 | 1.89 × 10−12 | 5.16 × 10−9 |
| APOBEC3B | 0.44 | 1.92 × 10−11 | 5.24 × 10−8 |
| RCCD1 | 0.38 | 1.46 × 10−10 | 3.99 × 10−7 |
| EIF2S2 | 0.37 | 1.58 × 10−10 | 4.31 × 10−7 |
| DCLRE1B | 0.35 | 2.34 × 10−10 | 6.39 × 10−7 |
| DYNLRB2 | −0.37 | 2.93 × 10−10 | 8.00 × 10−7 |
| ESR1 | −0.38 | 1.28 × 10−9 | 3.49 × 10−6 |
| ATP6AP1L | −0.34 | 4.59 × 10−9 | 1.25 × 10−5 |
| PDZK1 | −0.30 | 2.52 × 10−8 | 6.88 × 10−5 |
| ITPR1 | −0.33 | 1.83 × 10−7 | 5.00 × 10−4 |
| FGF10 | −0.32 | 1.93 × 10−7 | 5.27 × 10−4 |
| TOX3 | −0.28 | 6.31 × 10−7 | 1.72 × 10−3 |
| APOBEC3A | 0.26 | 2.35 × 10−6 | 6.42 × 10−3 |
| GATAD2A | 0.29 | 6.35 × 10−6 | 0.017 |
| POLR2L | −0.25 | 7.58 × 10−6 | 0.021 |
| SLC4A7 | −0.26 | 7.61 × 10−6 | 0.021 |
| Breast Cancer (Signature 13) | |||
| APOBEC3A | 0.48 | 2.84 × 10−14 | 7.75 × 10−11 |
| DYNLRB2 | −0.41 | 2.26 × 10−11 | 6.17 × 10−8 |
| ATP6AP1L | −0.39 | 1.73 × 10−10 | 4.72 × 10−7 |
| ESR1 | −0.37 | 1.19 × 10−9 | 3.25 × 10−6 |
| AMFR | −0.35 | 8.97 × 10−9 | 2.45 × 10−5 |
| APOBEC3B | 0.40 | 1.24 × 10−8 | 3.39 × 10−5 |
| PDZK1 | −0.34 | 1.85 × 10−8 | 5.05 × 10−5 |
| WNT3 | −0.34 | 3.62 × 10−8 | 9.88 × 10−5 |
| NEK10 | −0.32 | 1.10 × 10−7 | 3.00 × 10−4 |
| EIF2S2 | 0.33 | 1.59 × 10−7 | 4.34 × 10−4 |
| MRPS30 | −0.31 | 1.71 × 10−7 | 4.67 × 10−4 |
| PTPN22 | 0.32 | 3.82 × 10−7 | 1.04 × 10−3 |
| PRC1 | 0.33 | 1.35 × 10−6 | 3.69 × 10−3 |
| FAM72B | 0.32 | 2.96 × 10−6 | 8.08 × 10−3 |
| CTSW | 0.28 | 5.56 × 10−6 | 0.015 |
| FGF10 | −0.28 | 8.95 × 10−6 | 0.024 |
| Breast Cancer (Signature 1) | |||
| ATP6AP1L | 0.05 | 3.05 × 10−9 | 8.33 × 10−6 |
| DYNLRB2 | 0.04 | 4.84 × 10−7 | 1.32 × 10−3 |
| ESR1 | 0.04 | 7.63 × 10−7 | 2.08 × 10−3 |
| Breast Cancer (Signature 2) | |||
| WNT3 | −0.29 | 7.48 × 10−8 | 2.04 × 10−4 |
| Breast Cancer (Signature 16) | |||
| NTN4 | 0.60 | 1.26 × 10−6 | 3.44 × 10−3 |
| Colorectal Cancer (Signature 1) | |||
| SHROOM2 | 0.09 | 3.60 × 10−9 | 2.38 × 10−6 |
| GPR143 | 0.09 | 4.07 × 10−9 | 2.69 × 10−6 |
| MICB | −0.09 | 6.26 × 10−9 | 4.13 × 10−6 |
| A1CF | 0.08 | 1.18 × 10−5 | 7.80 × 10−3 |
| Colorectal Cancer (Signature 26) | |||
| A1CF | −1.39 | 4.12 × 10−5 | 0.027 |
| SHROOM2 | −1.12 | 4.77 × 10−5 | 0.032 |
| Colorectal Cancer (Signature 6) | |||
| GPR143 | −0.47 | 2.05 × 10−7 | 1.35 × 10−4 |
| A1CF | −0.52 | 2.75 × 10−7 | 1.81 × 10−4 |
| SHROOM2 | −0.42 | 1.02 × 10−6 | 6.73 × 10−4 |
| Lung Adenocarcinoma (Signature 4) | |||
| TP63 | −0.23495 | 1.94 × 10−5 | 2.56 × 10−2 |
| Prostate Cancer (Signature 1) | |||
| TBX1 | 0.07 | 7.18 × 10−8 | 3.43 × 10−4 |
| MYO6 | 0.07 | 2.68 × 10−6 | 1.28 × 10−2 |
| Melanoma (Signature 20) | |||
| CDK10 | −0.40 | 1.09 × 10−6 | 4.85 × 10−4 |
| Melanoma (Signature 4) | |||
| CDK10 | −0.31 | 1.43 × 10−5 | 6.36 × 10−3 |
| Melanoma (Signature 25) | |||
| CDK10 | 0.72 | 8.92 × 10−5 | 0.040 |
| Melanoma (Signature 23) | |||
| UQCC | 0.32 | 7.77 × 10−5 | 0.035 |
The mutational signatures for each cancer type were derived from TCGA samples.
Putative Susceptibility Genes Associated with TMB of Cancer-Driver Genes
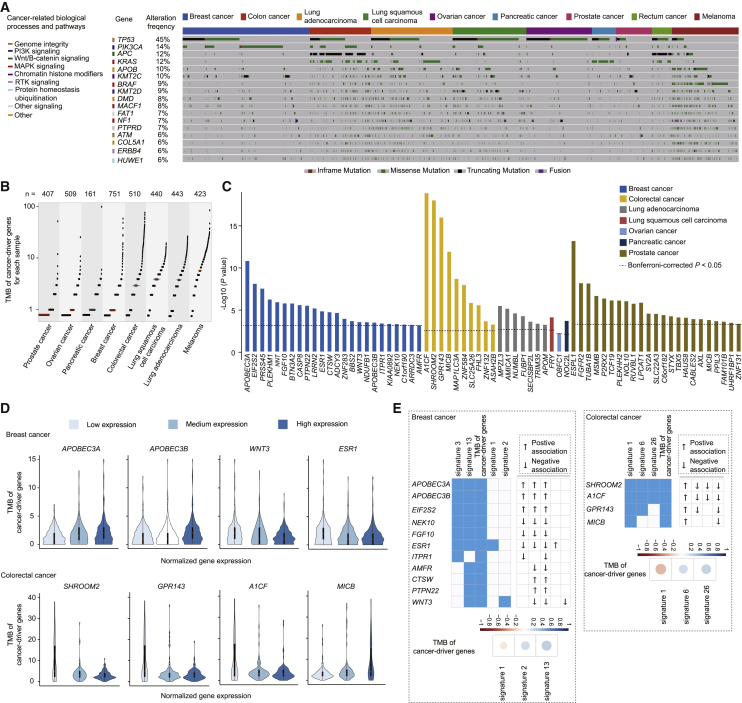
To investigate whether putative cancer-susceptibility genes might affect TMB of cancer-driver genes, we first characterized the mutation spectrum of 299 known cancer-driver genes for each cancer type; as expected, these genes were frequently mutated across the seven cancers evaluated (Figures 4A and 4B). When we evaluated associations between the expression levels of identified target genes and TMB of cancer-driver genes, 139 genes were associated across seven cancer types at nominal p < 0.05 (Table S5). At a Bonferroni-corrected significance threshold, 66 genes were associated with TMB of cancer-driver genes among six types of cancer (Figure 4C and Tables 1 and 3); this included 24 genes in breast cancer, 23 genes in prostate cancer, 10 genes in colorectal cancer, seven genes in lung adenocarcinoma, one gene (FRY [MIM: 614818]) in lung squamous cell carcinoma, one gene (OBFC1) in ovarian cancer, and one gene (NOC2L) in pancreatic cancer. Specifically, high expression levels of APOBEC3A and APOBEC3B were associated with increased TMB of cancer-driver genes, whereas low expression levels of another two genes, WNT3 and ESR1 (MIM: 133430), were associated with increased TMB of cancer-driver genes in breast cancer (Figure 4D). Similarly, we observed that high expression levels of MICB were associated with increased TMB of cancer-driver genes, whereas low expression levels of another three genes, SHROOM2, GRP143, and A1CF, were associated with increased TMB of cancer-driver genes in colorectal cancer (Figure 4D).
Figure 4.
Putative Susceptibility Genes Associated with TMB of Cancer-Driver Genes
(A) Mutation spectrum of cancer-driver genes with high alteration frequency (≥6%) in each sample across cancer types. The top boxes with different colors indicate samples of different cancer types. The lines in different colors indicate the mutation of each driver gene in each sample. The carcinogenesis signaling pathways for these driver genes are indicated with different colors on the left.
(B) Scatterplots for TMB of cancer-driver genes for each sample across seven cancer types. Each dot represents each sample and “n” refers to sample size. The red line indicates the median of TMB of cancer-driver genes for each cancer type.
(C) Bar plots showing the statistical significance between expression levels of putative susceptibility genes and TMB of cancer-driver genes across seven cancer types. The dashed lines indicate the genes with statistical associations at Bonferroni-corrected p < 0.05 in each cancer type.
(D) Violin plots of samples separated by low, median, and high expression levels of the highlighted genes (see Results); the genes were associated with TMB of cancer-driver genes at Bonferroni-correction p < 0.05. The upper plots show the associations in breast cancer, the lower plots show the associations in colorectal cancer.
(E) Heatmap plots showing the putative susceptibility genes associated with both TMB of cancer-driver genes and mutational signatures in breast and colorectal cancer. The ↑ and ↓ refer to a positive and negative association, respectively. The lower plots show the correlation coefficients between TMB of cancer-driver genes and mutational signatures in breast and colorectal cancer, respectively.
Table 3.
Associations Between Putative Susceptibility Gene Expression Levels and TMB of Cancer-Driver Genes for Six Cancer Types (Bonferroni-Corrected p < 0.05).
| Gene | Beta | P | Bonferroni-Corrected P |
|---|---|---|---|
| Breast Cancer | |||
| APOBEC3A | 0.32 | 1.23 × 10−11 | 1.12 × 10−9 |
| EIF2S2 | 0.28 | 6.24 × 10−9 | 5.68 × 10−7 |
| PRSS45 | −0.29 | 2.35 × 10−8 | 2.14 × 10−6 |
| PLEKHM1 | −0.25 | 4.59 × 10−7 | 4.18 × 10−5 |
| NNT | −0.23 | 9.59 × 10−7 | 8.73 × 10−5 |
| FGF10 | −0.24 | 1.32 × 10−6 | 1.20 × 10−4 |
| BTN3A2 | 0.22 | 1.37 × 10−6 | 1.25 × 10−4 |
| CASP8 | 0.22 | 2.12 × 10−6 | 1.93 × 10−4 |
| PTPN22 | 0.22 | 2.50 × 10−6 | 2.28 × 10−4 |
| LRRN2 | −0.21 | 5.27 × 10−6 | 4.80 × 10−4 |
| ESR1 | −0.22 | 1.15 × 10−5 | 1.05 × 10−3 |
| CTSW | 0.20 | 1.48 × 10−5 | 1.35 × 10−3 |
| ADCY3 | −0.21 | 1.79 × 10−5 | 1.63 × 10−3 |
| ZNF283 | 0.18 | 9.02 × 10−5 | 8.21 × 10−3 |
| BBS2 | −0.19 | 1.67 × 10−4 | 0.015 |
| WNT3 | −0.17 | 2.24 × 10−4 | 0.020 |
| NDUFB1 | 0.17 | 2.27 × 10−4 | 0.021 |
| APOBEC3B | 0.19 | 2.52 × 10−4 | 0.023 |
| ITPR1 | −0.18 | 3.22 × 10−4 | 0.030 |
| KIAA0892 | −0.17 | 3.60 × 10−4 | 0.033 |
| NEK10 | −0.17 | 3.78 × 10−4 | 0.034 |
| C1orf190 | −0.18 | 4.35 × 10−4 | 0.040 |
| ARRDC3 | 0.18 | 4.77 × 10−4 | 0.043 |
| AMFR | −0.16 | 5.29 × 10−4 | 0.048 |
| Colorectal Cancer | |||
| A1CF | −0.58 | 7.07 × 10−20 | 1.56 × 10−18 |
| SHROOM2 | −0.53 | 7.92 × 10−19 | 1.74 × 10−17 |
| GPR143 | −0.53 | 8.48 × 10−17 | 1.87 × 10−15 |
| MICB | 0.47 | 2.71 × 10−12 | 5.96 × 10−11 |
| MAP1LC3A | −0.37 | 1.63 × 10−9 | 3.58 × 10−8 |
| ZNF584 | 0.38 | 8.62 × 10−9 | 1.90 × 10−7 |
| SLC25A26 | −0.32 | 1.18 × 10−6 | 2.60 × 10−5 |
| FHL3 | 0.32 | 2.24 × 10−6 | 4.93 × 10−5 |
| ZNF132 | 0.26 | 1.75 × 10−4 | 3.84 × 10−3 |
| ASAH2B | 0.21 | 1.82 × 10−3 | 0.040 |
| Lung Adenocarcinoma | |||
| MPZL3 | 0.20 | 2.50 × 10−6 | 1.10 × 10−4 |
| AMICA1 | −0.23 | 5.41 × 10−6 | 1.19 × 10−4 |
| NUMBL | 0.19 | 2.49 × 10−5 | 3.65 × 10−4 |
| FUBP1 | 0.19 | 4.35 × 10−5 | 4.79 × 10−4 |
| SECISBP2L | −0.18 | 1.94 × 10−4 | 1.71 × 10−3 |
| TRIM35 | −0.17 | 3.66 × 10−4 | 2.68 × 10−3 |
| APOM | −0.14 | 4.63 × 10−4 | 2.91 × 10−3 |
| Lung Squamous Cell Carcinoma | |||
| FRY | −0.16 | 5.97 × 10−5 | 1.67 × 10−3 |
| Pancreatic Cancer | |||
| NOC2L | −0.45 | 1.59 × 10−4 | 1.11 × 10−3 |
| Prostate Cancer | |||
| ESPL1 | 0.60 | 5.27 × 10−14 | 8.38 × 10−12 |
| FGFR2 | −0.48 | 5.31 × 10−9 | 3.05 × 10−7 |
| TUBA1B | 0.44 | 5.76 × 10−9 | 3.05 × 10−7 |
| MSMB | −0.43 | 1.61 × 10−7 | 6.40 × 10−6 |
| P2RX2 | −0.40 | 3.50 × 10−7 | 1.11 × 10−5 |
| TCF19 | 0.38 | 5.96 × 10−7 | 1.29 × 10−5 |
| PLEKHH2 | −0.41 | 6.10 × 10−7 | 1.29 × 10−5 |
| NOL10 | 0.39 | 6.48 × 10−7 | 1.29 × 10−5 |
| RUVBL1 | 0.41 | 8.63 × 10−7 | 1.52 × 10−5 |
| LPCAT1 | 0.40 | 1.10 × 10−6 | 1.75 × 10−5 |
| SV2A | −0.34 | 2.21 × 10−5 | 3.10 × 10−4 |
| SLC22A3 | −0.34 | 2.34 × 10−5 | 3.10 × 10−4 |
| C6orf182 | 0.31 | 3.27 × 10−5 | 4.00 × 10−4 |
| STYX | 0.31 | 4.47 × 10−5 | 5.08 × 10−4 |
| TBX5 | −0.34 | 6.10 × 10−5 | 6.47 × 10−4 |
| HAUS8 | 0.30 | 6.73 × 10−5 | 6.69 × 10−4 |
| CABLES2 | 0.31 | 9.73 × 10−5 | 8.67 × 10−4 |
| AXL | −0.31 | 9.81 × 10−5 | 8.67 × 10−4 |
| MICB | 0.28 | 5.76 × 10−9 | 1.24 × 10−3 |
| PPIL3 | 0.30 | 1.69 × 10−4 | 1.34 × 10−3 |
| FAM101B | −0.31 | 1.95 × 10−4 | 1.48 × 10−3 |
| UHRF1BP1 | 0.28 | 2.68 × 10−4 | 1.94 × 10−3 |
| ZNF131 | 0.28 | 3.56 × 10−4 | 2.46 × 10−3 |
| Ovarian Cancer | |||
| OBFC1 | −0.25 | 4.46 × 10−3 | 0.045 |
Finally, when we examined correlations between TMB of cancer-driver genes and mutational signatures, correlations varied among cancer types (Table S6). In particular, mutational signatures that were associated with putative cancer-susceptibility genes (e.g., signatures 13 and 1, for breast and colorectal cancer, respectively) were strongly correlated with TMB of cancer-driver genes. Specifically, signature 13 was positively correlated and signature 1 was negatively correlated with TMB of cancer-driver genes, respectively (Figure 4E).
Discussion
We systematically evaluated transcriptome data from TCGA, GTEx, and other publicly available data sources and identified putative susceptibility genes for GWAS-identified SNPs for six major cancer types. By using an integrative analysis approach, we characterized a total of 270 candidate genes, including 99 that had not been previously associated with cancer risk. Of the candidate target genes, 180 (66.7%) showed evidence of cis-regulation by putative functional variants via proximal promoter or distal enhancer-promoter interactions. Furthermore, our results showed that a total of 33 and 66 putative susceptibility genes were associated with specific mutational signatures and TBM of cancer-driver genes, respectively. These findings provide additional putative susceptibility genes and further insight into understanding how genetic risk variants might contribute to carcinogenesis, mediated by regulation of susceptibility genes that might affect the biogenesis of somatic mutations.
Our identification of a total of 270 putative cancer-susceptibility genes is supported by several lines of additional evidence. First, 110 (40.7%) target genes showed significance at nominal p < 0.05 in both the GTEx and TCGA datasets. Second, a substantial proportion of target genes (66.7% of 270) had evidence of cis-regulation by putative functional variants via proximal promoter or distal enhancer-promoter interactions. Third, our results showed that ten genes commonly observed in different cancer types were found to be consistently associated with risk alleles of different index SNPs, and an additional 14 genes were also observed among different cancer types, although they had inconsistent associations with index SNPs in different cancer types (Figures 2B and 2C). Finally, a functional enrichment analysis of these genes done with Ingenuity Pathway Analysis (IPA) revealed that the most significantly enriched molecular and cellular function was involved in cancer-related functions (p < 0.05 for the enrichment analysis). In particular, many genes identified by our analysis, including WNT3, SLC22A3 (MIM: 604842), PCAT19 (MIM: 618192), CEACAM21 (MIM: 618191), and CABLES2, have been verified by in vitro or in vivo experiments to be involved in cell growth, proliferation, and apoptosis.65, 66, 67, 68, 69, 70, 71 It should be noted that a total of 16 identified eQTL target genes, five in lung adenocarcinoma and 11 in prostate cancer, showed inconsistent direction of association in previous literature18, 19 (Table S2).
Our results showed that 23 genes, located in three regions (17q21.3, 6p22.1–6p21.33–6p21.32, and 2q33.1) were commonly implicated in at least two cancer types. In line with findings from previous studies, the 17q21.3 locus was associated with risk of both breast and ovarian cancer, and the 6p22.1–6p21.33–6p21.32 loci have been reported to be associated with risk of both lung and prostate cancers.72 The 2q33.1 locus has also been reported to be associated with risk of melanoma, breast, and prostate cancers.73 Together with these findings, our study revealed putative cancer-susceptibility genes in these loci, providing evidence of regulatory mechanisms underlying cancer pleiotropy for shared cancer risk.
Our approach of combining regression results of eQTL analyses from normal tissues and tumor tissues might be questioned. To address this concern, eQTL regression models for tumor tissues from TCGA were adjusted for DNA methylation and somatic copy number alterations to account for any potential influence of somatic alterations in tumor tissue. Even though it might be inappropriate to combine effect sizes from tumor and normal tissues, we reasonably combined the covariate-adjusted p values from TCGA with p values from GTEx following meta-analysis methods based on directions of association.
It should be noted that a particular index SNP might be a surrogate for multiple variants for cancer risk in the locus. The target genes for those potential causal variants, which are in weak LD with index SNPs, might not be identified by cis-eQTL analysis. On the other hand, index SNPs could be statistically excluded as candidate causative variants in some GWAS-identified loci. Nevertheless, the identification of target genes on the basis of the index SNPs is still reliable because most statistically causative variants are still expected to be in strong LD with them.
Our findings showed that many putative cancer-susceptibility genes were associated with specific mutational signatures and TMB of cancer-driver genes. Of the 180 genes with evidence of cis-regulation by putative functional variants via proximal promoter or distal enhancer-promoter interactions, a total of 21 genes showed associations with specific mutational signatures and TMB of cancer-driver genes; these genes included seven for colorectal cancer, one for lung adenocarcinoma, one for pancreatic cancer, and 12 for prostate cancer (Table S7). In breast cancer, an unpublished fine-mapping study has reported a total of 178 high-confidence target genes from 150 susceptibility regions.51 We additionally evaluated the associations of these genes with mutational signatures and TMB of cancer-driver genes (see Material and Methods). We observed 55 genes associated with mutational signatures and 29 with TMB of cancer-driver genes by using a Bonferroni-corrected significance threshold of p < 0.05 (Table S8). Of them, 13 genes showed associations with both specific mutational signatures and TMB of cancer-driver genes. In comparison with the results from the 101 eQTL target genes reported in our previous study,20 a total of 10 genes associated with either mutational signatures or TMB of cancer-driver genes were commonly observed (Figure S1). Additionally, we also evaluated associations between the expression of all protein-coding genes and each mutational signature and TMB of cancer-driver genes for each cancer type. Our results suggested that the proportion of cancer-susceptibility genes associated with specific mutational signatures and TMB of cancer-driver genes was significantly higher than the proportion of all protein-coding genes that were thus associated; this result was found in a majority of cancer types (including breast, colorectal and prostate cancers, and melanoma; Fisher’s exact test, p < 0.05 for the enrichment analysis; Figures S2 and S3; see Material and Methods).
It should be noted that the biological mechanisms of how these cancer-susceptibility genes might affect mutational signatures in different cancer types remains unclear. In line with previous studies, we found that APOBEC3A and APOBEC3B were associated with mutational signatures 3 and 13 for breast cancer.37, 38, 74, 75 By using gene expression data in breast cancer tumor tissues from TCGA, we observed that a majority of genes identified were respectively associated with mutational signatures and also correlated with APOBEC3A and APOBEC3B (p < 0.05; Figure S4). This might indicate that these putative cancer-susceptibility genes affect mutational signatures through a common APOBEC3 pathway. Several DNA repair genes, such as DCLRE1B and GATAD2A, were observed to be associated with mutational signatures. DCLRE1B, encoding a 5ʹ–3ʹ exonuclease, has been reported to play a vital role in DNA double-strand break (DSB) repair by affecting efficient localization of BRCA1 and the MRE11/RAD51/NBS1 complex.76 GATAD2A, encoding a subunit of the nucleosome remodeling and histone deacetylase (NuRD) complex, plays an important role in recruiting NuRD to the sites of damaged chromatin for repair of DSBs by homologous recombination.35, 77, 78, 79 Interestingly, we also found that a low expression level of ESR1 was associated with an increase of TMB of cancer-driver genes. This might be supported by observation of the expression correlations between ESR1 and the DNA-repair gene DLCLRE1B and both APOBEC3A, and APOBEC3B (Figure S4). On the other hand, in previous studies from ChIP-seq analyses, thousands of candidate target genes, including many DNA-repair genes, have been identified for ESR1, adding other possibilities of regulating the related DNA-repair genes and pathways, consequently affecting the TMB of cancer-driver genes.80, 81, 82
In conclusion, we evaluated a total of 270 putative susceptibility genes, including 99 with previously unreported associations, for six major cancer types. Our results indicate that many GWAS-identified variants might influence cancer risk through cis-regulation through proximal promoter or distal enhancer-promoter interactions with putative functional variants. In addition, our results indicate that a substantial proportion of these genes were associated with specific mutational signatures and TMB of cancer-driver genes. Together, our findings provide further insight into understanding the role of genetic variants and how regulation of target susceptibility genes might affect the biogenesis of somatic mutations in multiple types of cancer.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank TCGA, GTEx, ENCODE, Roadmap, 4DGenome, and FANTOM5 for generating valuable data resources for this study. We also thank Marshal Younger for assistance with editing and manuscript preparation. This work was supported by the US National Institutes of Health grant 1R37CA227130-01A1 and the research development fund from Vanderbilt University Medical Center to X.G.. The data analyses were conducted using the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University.
Published: August 8, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.07.006.
Web Resources
4DGenome, https://4dgenome.research.chop.edu/
cBioPortal for Cancer Genomics, http://www.cbioportal.org/
Colonomics, https://www.colonomics.org/
ENCODE, https://www.encodeproject.org/
FANTOM5, http://fantom.gsc.riken.jp/5/
GWAS Catalog, https://www.ebi.ac.uk/gwas/
GTEx Portal, https://www.gtexportal.org/home/
Michigan Imputation Server, https://imputationserver.sph.umich.edu
mSignatureDB, http://tardis.cgu.edu.tw/msignaturedb
OMIM, https://www.omim.org/
TCGA Portal, https://portal.gdc.cancer.gov/
WGS Annotator, https://sites.google.com/site/jpopgen/wgsa
Supplemental Data
References
- 1.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S., Lemaçon A., Soucy P., Glubb D., Rostamianfar A., NBCS Collaborators; ABCTB Investigators; ConFab/AOCS Investigators Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benafif S., Kote-Jarai, and Z., Eeles R.A., PRACTICAL Consortium A review of prostate cancer genome-wide association studies (GWAS) Cancer Epidemiol. Biomarkers Prev. 2018;27:845–857. doi: 10.1158/1055-9965.EPI-16-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossé Y., Amos C.I. A Decade of GWAS results in lung cancer. Cancer Epidemiol. Biomarkers Prev. 2018;27:363–379. doi: 10.1158/1055-9965.EPI-16-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huyghe J.R., Bien S.A., Harrison T.A., Kang H.M., Chen S., Schmit S.L., Conti D.V., Qu C., Jeon J., Edlund C.K. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2019;51:76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y., Kweon S.S., Tanikawa C., Jia W.H., Xiang Y.B., Cai Q., Zeng C., Schmit S.L., Shin A., Matsuo K. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019;156:1455–1466. doi: 10.1053/j.gastro.2018.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaub M.A., Boyle A.P., Kundaje A., Batzoglou S., Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu X.J., Lu Z.J., Kong Y., Lam H.Y., Gerstein M.B. Analysis of genomic variation in non-coding elements using population-scale sequencing data from the 1000 Genomes Project. Nucleic Acids Res. 2011;39:7058–7076. doi: 10.1093/nar/gkr342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C.Y., Chang I.S., Hsiung C.A., Wasserman W.W. On the identification of potential regulatory variants within genome wide association candidate SNP sets. BMC Med. Genomics. 2014;7:34. doi: 10.1186/1755-8794-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X., Long J., Zeng C., Michailidou K., Ghoussaini M., Bolla M.K., Wang Q., Milne R.L., Shu X.O., Cai Q., kConFab Investigators Fine-scale mapping of the 4q24 locus identifies two independent loci associated with breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2015;24:1680–1691. doi: 10.1158/1055-9965.EPI-15-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghoussaini M., French J.D., Michailidou K., Nord S., Beesley J., Canisus S., Hillman K.M., Kaufmann S., Sivakumaran H., Moradi Marjaneh M., kConFab/AOCS Investigators; NBCS Collaborators Evidence that the 5p12 variant rs10941679 confers susceptibility to estrogen-receptor-positive breast cancer through FGF10 and MRPS30 regulation. Am. J. Hum. Genet. 2016;99:903–911. doi: 10.1016/j.ajhg.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dadaev T., Saunders E.J., Newcombe P.J., Anokian E., Leongamornlert D.A., Brook M.N., Cieza-Borrella C., Mijuskovic M., Wakerell S., Olama A.A.A., PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat. Commun. 2018;9:2256. doi: 10.1038/s41467-018-04109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen J.D., Lamontagne M., Couture C., Conti M., Paré P.D., Sin D.D., Hogg J.C., Nickle D., Postma D.S., Timens W. Susceptibility loci for lung cancer are associated with mRNA levels of nearby genes in the lung. Carcinogenesis. 2014;35:2653–2659. doi: 10.1093/carcin/bgu184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q., Seo J.H., Stranger B., McKenna A., Pe’er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Closa A., Cordero D., Sanz-Pamplona R., Solé X., Crous-Bou M., Paré-Brunet L., Berenguer A., Guino E., Lopez-Doriga A., Guardiola J. Identification of candidate susceptibility genes for colorectal cancer through eQTL analysis. Carcinogenesis. 2014;35:2039–2046. doi: 10.1093/carcin/bgu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q., Stram A., Chen C., Kar S., Gayther S., Pharoah P., Haiman C., Stranger B., Kraft P., Freedman M.L. Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum. Mol. Genet. 2014;23:5294–5302. doi: 10.1093/hmg/ddu228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thibodeau S.N., French A.J., McDonnell S.K., Cheville J., Middha S., Tillmans L., Riska S., Baheti S., Larson M.C., Fogarty Z. Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nat. Commun. 2015;6:8653. doi: 10.1038/ncomms9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKay J.D., Hung R.J., Han Y., Zong X., Carreras-Torres R., Christiani D.C., Caporaso N.E., Johansson M., Xiao X., Li Y., SpiroMeta Consortium Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017;49:1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X., Lin W., Bao J., Cai Q., Pan X., Bai M., Yuan Y., Shi J., Sun Y., Han M.R. A comprehensive cis-eQTL analysis revealed target genes in breast cancer susceptibility loci identified in genome-wide association studies. Am. J. Hum. Genet. 2018;102:890–903. doi: 10.1016/j.ajhg.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno V., Alonso M.H., Closa A., Vallés X., Diez-Villanueva A., Valle L., Castellví-Bel S., Sanz-Pamplona R., Lopez-Doriga A., Cordero D., Solé X. Colon-specific eQTL analysis to inform on functional SNPs. Br. J. Cancer. 2018;119:971–977. doi: 10.1038/s41416-018-0018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmit S.L., Edlund C.K., Schumacher F.R., Gong J., Harrison T.A., Huyghe J.R., Qu C., Melas M., Van Den Berg D.J., Wang H. Novel common genetic susceptibility loci for colorectal cancer. J. Natl. Cancer Inst. 2019;111:146–157. doi: 10.1093/jnci/djy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., Profile Study; Australian Prostate Cancer BioResource (APCB); IMPACT Study; Canary PASS Investigators; Breast and Prostate Cancer Cohort Consortium (BPC3); PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium; Cancer of the Prostate in Sweden (CAPS); Prostate Cancer Genome-wide Association Study of Uncommon Susceptibility Loci (PEGASUS); Genetic Associations and Mechanisms in Oncology (GAME-ON)/Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) Consortium Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium T.E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chadwick L.H. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Touzet H., Varré J.S. Efficient and accurate P-value computation for Position Weight Matrices. Algorithms Mol. Biol. 2007;2:15. doi: 10.1186/1748-7188-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y., Liu Z., Lou S., Bedford J., Mu X.J., Yip K.Y., Khurana E., Gerstein M. FunSeq2: A framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014;15:480. doi: 10.1186/s13059-014-0480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jäger R., Migliorini G., Henrion M., Kandaswamy R., Speedy H.E., Heindl A., Whiffin N., Carnicer M.J., Broome L., Dryden N. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat. Commun. 2015;6:6178. doi: 10.1038/ncomms7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L., Wingett S.W., Andrews S., Grey W., Ewels P.A. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 34.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns M.B., Temiz N.A., Harris R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J., Mouw K.W., Polak P., Braunstein L.Z., Kamburov A., Kwiatkowski D.J., Rosenberg J.E., Van Allen E.M., D’Andrea A., Getz G. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knijnenburg T.A., Wang L., Zimmermann M.T., Chambwe N., Gao G.F., Cherniack A.D., Fan H., Shen H., Way G.P., Greene C.S. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23:239–254.e6. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchsberger C., Abecasis G.R., Hinds D.A. minimac2: Faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 45.Penney K.L., Sinnott J.A., Tyekucheva S., Gerke T., Shui I.M., Kraft P., Sesso H.D., Freedman M.L., Loda M., Mucci L.A., Stampfer M.J. Association of prostate cancer risk variants with gene expression in normal and tumor tissue. Cancer Epidemiol. Biomarkers Prev. 2015;24:255–260. doi: 10.1158/1055-9965.EPI-14-0694-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X., Hussain W.M., Vijai J., Offit K., Rubin M.A., Demichelis F., Klein R.J. Variants at IRX4 as prostate cancer expression quantitative trait loci. Eur. J. Hum. Genet. 2014;22:558–563. doi: 10.1038/ejhg.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitington T., Gao P., Song W., Ross-Adams H., Lamb A.D., Yang Y., Svezia I., Klevebring D., Mills I.G., Karlsson R. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat. Genet. 2016;48:387–397. doi: 10.1038/ng.3523. [DOI] [PubMed] [Google Scholar]
- 48.Klein A.P., Wolpin B.M., Risch H.A., Stolzenberg-Solomon R.Z., Mocci E., Zhang M., Canzian F., Childs E.J., Hoskins J.W., Jermusyk A. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat. Commun. 2018;9:556. doi: 10.1038/s41467-018-02942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phelan C.M., Kuchenbaecker K.B., Tyrer J.P., Kar S.P., Lawrenson K., Winham S.J., Dennis J., Pirie A., Riggan M.J., Chornokur G., AOCS study group. EMBRACE Study; GEMO Study Collaborators; HEBON Study; KConFab Investigators. OPAL study group Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017;49:680–691. doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borenstein M., Hedges L.V., Higgins J.P.T., Rothstein H.R. Wiley; 2009. Introduction to Meta-Analysis. [Google Scholar]
- 51.Fachal L., Aschard H., Beesley J., Barnes D.R., Allen J., Kar S., Pooley K.A., Dennis J., Michailidou K., Turman C. Fine-mapping of 150 breast cancer risk regions identifies 178 high confidence target genes. bioRxiv. 2019:521054. doi: 10.1038/s41588-019-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward L.D., Kellis M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X., White S., Peng B., Johnson A.D., Brody J.A., Li A.H., Huang Z., Carroll A., Wei P., Gibbs R. WGSA: An annotation pipeline for human genome sequencing studies. J. Med. Genet. 2016;53:111–112. doi: 10.1136/jmedgenet-2015-103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng L., He B., Wang J., Tan K. 4DGenome: A comprehensive database of chromatin interactions. Bioinformatics. 2015;31:2560–2564. doi: 10.1093/bioinformatics/btv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang P.J., Chiu L.Y., Lee C.C., Yeh Y.M., Huang K.Y., Chiu C.H., Tang P. mSignatureDB: A database for deciphering mutational signatures in human cancers. Nucleic Acids Res. 2018;46(D1):D964–D970. doi: 10.1093/nar/gkx1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M.C., Kim J., Reardon B. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harrell F.E. Second Edition. Springer Ser Stat; 2015. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis; pp. 1–582. [Google Scholar]
- 60.Loo L.W.M., Lemire M., Le Marchand L. In silico pathway analysis and tissue specific cis-eQTL for colorectal cancer GWAS risk variants. BMC Genomics. 2017;18:381. doi: 10.1186/s12864-017-3750-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nica A.C., Parts L., Glass D., Nisbet J., Barrett A., Sekowska M., Travers M., Potter S., Grundberg E., Small K., MuTHER Consortium The architecture of gene regulatory variation across multiple human tissues: The MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martincorena I., Campbell P.J. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 63.Alexandrov L.B., Ju Y.S., Haase K., Van Loo P., Martincorena I., Nik-Zainal S., Totoki Y., Fujimoto A., Nakagawa H., Shibata T. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354:618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nik-Zainal S., Morganella S. Mutational signatures in breast cancer: The problem at the DNA level. Clin. Cancer Res. 2017;23:2617–2629. doi: 10.1158/1078-0432.CCR-16-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y., Liu P., Wen W., James M.A., Wang Y., Bailey-Wilson J.E., Amos C.I., Pinney S.M., Yang P., de Andrade M. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24-25.1. Cancer Res. 2009;69:7844–7850. doi: 10.1158/0008-5472.CAN-09-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grisanzio C., Werner L., Takeda D., Awoyemi B.C., Pomerantz M.M., Yamada H., Sooriakumaran P., Robinson B.D., Leung R., Schinzel A.C. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc. Natl. Acad. Sci. USA. 2012;109:11252–11257. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao P., Xia J.H., Sipeky C., Dong X.M., Zhang Q., Yang Y., Zhang P., Cruz S.P., Zhang K., Zhu J. Biology and clinical implications of the 19q13 aggressive prostate cancer susceptibility locus. Cell. 2018;174:576–589.e18. doi: 10.1016/j.cell.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ren A., Sun S., Li S., Chen T., Shu Y., Du M., Zhu L. Genetic variants in SLC22A3 contribute to the susceptibility to colorectal cancer. Int. J. Cancer. 2019;145:154–163. doi: 10.1002/ijc.32079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu J.H., Zhong X.Y., Zhang W.G., Wang Z.D., Dong Q., Tai S., Li H., Cui Y.F. CDK10 functions as a tumor suppressor gene and regulates survivability of biliary tract cancer cells. Oncol. Rep. 2012;27:1266–1276. doi: 10.3892/or.2011.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu L., Qin Y.R., Ming X.Y., Zuo X.B., Diao Y.W., Zhang L.Y., Ai J., Liu B.L., Huang T.X., Cao T.T. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc. Natl. Acad. Sci. USA. 2017;114:E4631–E4640. doi: 10.1073/pnas.1703178114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weiswald L.B., Hasan M.R., Wong J.C.T., Pasiliao C.C., Rahman M., Ren J., Yin Y., Gusscott S., Vacher S., Weng A.P. Inactivation of the kinase domain of CDK10 prevents tumor growth in a preclinical model of colorectal cancer, and is accompanied by downregulation of Bcl-2. Mol. Cancer Ther. 2017;16:2292–2303. doi: 10.1158/1535-7163.MCT-16-0666. [DOI] [PubMed] [Google Scholar]
- 72.Jiang X., Finucane H.K., Schumacher F.R., Schmit S.L., Tyrer J.P., Han Y., Michailidou K., Lesseur C., Kuchenbaecker K.B., Dennis J. Shared heritability and functional enrichment across six solid cancers. Nat. Commun. 2019;10:431. doi: 10.1038/s41467-018-08054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fehringer G., Kraft P., Pharoah P.D., Eeles R.A., Chatterjee N., Schumacher F.R., Schildkraut J.M., Lindström S., Brennan P., Bickeböller H., Ovarian Cancer Association Consortium (OCAC); PRACTICAL Consortium; Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON); Colorectal Transdisciplinary (CORECT) Study; African American Breast Cancer Consortium (AABC) and African Ancestry Prostate Cancer Consortium (AAPC) Cross-cancer genome-wide analysis of lung, ovary, breast, prostate, and colorectal cancer reveals novel pleiotropic associations. Cancer Res. 2016;76:5103–5114. doi: 10.1158/0008-5472.CAN-15-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor B.J., Nik-Zainal S., Wu Y.L., Stebbings L.A., Raine K., Campbell P.J., Rada C., Stratton M.R., Neuberger M.S. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mason J.M., Das I., Arlt M., Patel N., Kraftson S., Glover T.W., Sekiguchi J.M. The SNM1B/APOLLO DNA nuclease functions in resolution of replication stress and maintenance of common fragile site stability. Hum. Mol. Genet. 2013;22:4901–4913. doi: 10.1093/hmg/ddt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gong F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., Cammarata M., Perez M., Agarwal P., Brodbelt J.S. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spruijt C.G., Luijsterburg M.S., Menafra R., Lindeboom R.G., Jansen P.W., Edupuganti R.R., Baltissen M.P., Wiegant W.W., Voelker-Albert M.C., Matarese F. ZMYND8 co-localizes with NuRD on target genes and regulates poly(ADP-ribose)-dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Rep. 2016;17:783–798. doi: 10.1016/j.celrep.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 79.Gong F., Clouaire T., Aguirrebengoa M., Legube G., Miller K.M. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 2017;216:1959–1974. doi: 10.1083/jcb.201611135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 81.Welboren W.J., van Driel M.A., Janssen-Megens E.M., van Heeringen S.J., Sweep F.C., Span P.N., Stunnenberg H.G. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fletcher M.N., Castro M.A., Wang X., de Santiago I., O’Reilly M., Chin S.F., Rueda O.M., Caldas C., Ponder B.A., Markowetz F., Meyer K.B. Master regulators of FGFR2 signalling and breast cancer risk. Nat. Commun. 2013;4:2464. doi: 10.1038/ncomms3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.