Abstract
Deciphering the environmental contexts at which genetic effects are most prominent is central for making full use of GWAS results in follow-up experiment design and treatment development. However, measuring a large number of environmental factors at high granularity might not always be feasible. Instead, here we propose extracting cellular embedding of environmental factors from gene expression data by using latent variable (LV) analysis and taking these LVs as environmental proxies in detecting gene-by-environment (GxE) interaction effects on gene expression, i.e., GxE expression quantitative trait loci (eQTLs). Applying this approach to two largest brain eQTL datasets (n = 1,100), we show that LVs and GxE eQTLs in one dataset replicate well in the other dataset. Combining the two samples via meta-analysis, 895 GxE eQTLs are identified. On average, GxE effect explains an additional ∼4% variation in expression of each gene that displays a GxE effect. Ten of these 52 genes are associated with cell-type-specific eQTLs, and the remaining genes are multi-functional. Furthermore, after substituting LVs with expression of transcription factors (TF), we found 91 TF-specific eQTLs, which demonstrates an important use of our brain GxE eQTLs.
Keywords: gene by environment interactions, eQTL, cell-type specificity, context-specific genotype effects, gene expression, cellular embedding of environment
Introduction
Large-scale genome-wide association studies (GWASs) have identified numerous genetic risk loci for complex neurological and psychiatric disorders.1, 2, 3 However, the majority of disease-associated loci are non-coding and likely regulatory.4 Inferring their downstream impact on molecular mechanisms thus requires additional data, such as expression quantitative trait loci (eQTLs) datasets.5 Although many GWAS variants are shown to affect expression of nearby genes, the contexts under which effects are most prominent are largely unknown. Toward this end, studies have shown that certain eQTLs are more pronounced in specific cell types,6, 7 which helps prioritize cell targets for follow-up experiments and treatment development. Other studies have identified response QTLs, where response to certain exposures is dependent on genotype.8, 9, 10 More broadly, gene-by-environment (GxE) eQTL studies have identified genetic variants that affect gene expression in sex-, age-, cellular-environment-, and developmental-stage-specific manners.11, 12, 13, 14 The small number of eQTL studies that measure environmental factors, such as those related to lifestyles (e.g., smoking, drinking, and exercise) and physical environment (e.g., air pollution), have provided a rich resource for identifying GxE eQTLs.15, 16
A central challenge for identifying GxE eQTLs is the scarcity of large datasets with both gene expression data and environmental variables from the same individuals. One approach for addressing this challenge is to infer the cellular embedding of environmental factors from gene expression data.17, 18, 19, 20 A powerful tool for such inference is latent variable (LV) analysis.21, 22, 23, 24, 25 LVs inferred from gene expression data often reflect common environmental variables, such as age, sex, smoking, and drug intake.26 LVs have also been shown to correlate with the proportion of constituent cell types in bulk tissue samples and tissue-specific activation of various gene pathways.27 In fact, LVs might capture cellular embedding of environmental factors at a granularity that is currently not possible to directly measure, hence enabling identification of novel GxE eQTLs.
To test the possibility of identifying GxE eQTLs with expression-based LVs as environmental factors in the brain, we assembled the two largest eQTL datasets from dorsolateral prefrontal cortex (DLPFC, n = 1,100) and applied a biologically informed latent variable analysis25 to derive a large number of LVs (q = 135). Using these LVs, we found 895 GxE eQTLs, corresponding to 52 unique genes, at a dependent false discovery rate (FDR)28 threshold of 0.1 (Figure 1). On average, modeling the interaction between genotype and LV explained an additional ∼4% variation in expression of each gene that exhibits a GxE effect. Ten of these 52 genes are associated with cell-type-specific eQTLs. The remaining genes are multi-functional, which is consistent with how their expression levels are expected to be differentially regulated by context. Hypothesizing that certain LVs might reflect the effects of transcription factors (TFs), we substituted the LV in each identified GxE eQTL with its significantly correlated TFs, and tested for interaction effect. We found ninety-one TF-specific eQTLs, corresponding to four unique genes and three unique TFs, which demonstrates an important utility of our GxE eQTLs.
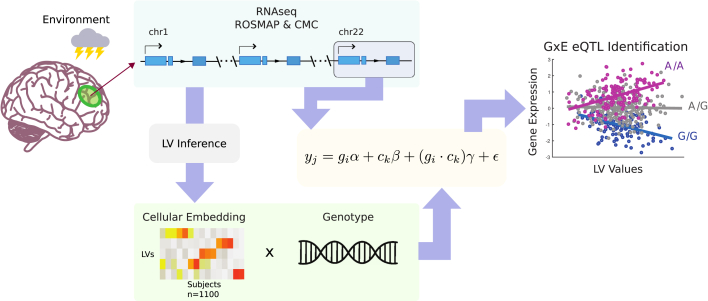
Figure 1.
Graphical Summary of GxE Analysis
Cellular embedding of environmental factors is first extracted from whole genome RNA-sequencing data via PLIER. For each gene, multiple regression is then applied so that the interaction effect between a LV and each cis SNP of the given gene can be tested. Significant interaction is declared at a dependent FDR threshold of 0.1.
Methods
RNA-Sequencing and Genotype Data
In this work, we used data from the ROSMAP29 and CMC30 studies, which have been approved by their respective institutional review boards. Five hundred eight and 592 individuals have both genotype and RNA-sequencing data from the DLPFC in the ROSMAP and CMC studies, respectively. The data preprocessing pipelines are exactly the same as previously described,30, 31 except the top 10 principal components (PCs) were removed from the gene expression data after LV extraction. 13,484 and 13,078 highly expressed genes were retained after QC for the ROSMAP and CMC expression datasets, respectively, and 10,961 genes were shared between the two datasets.
LV Extraction from Gene Expression Data
Given that environmental effects are reflected in the transcriptome,17, 18, 19, 20 inferring environmental proxies from gene expression data should capture an individual’s exposures. Conventional LV inference methods, such as non-negative matrix factorization,32 principle-component analysis (PCA),21 and their variants,23, 33 enable extraction of LVs in an unsupervised manner. Most LV inference methods can be formulated as the following optimization problem:
| (Equation 1) |
where X is a n × p gene expression data matrix of n subjects and p genes that has already been normalized to account for known technical confounding factors, such as batch. L is a n × q matrix containing q LVs, and Z is the corresponding q × p loading matrix. Ω(L, Z) is a regularization function, e.g., ZiZj and ||Zi||2 = 1 for all i, j in the case of PCA, where Zi is the ith row of Z. These methods summarize the variations common across features in X into a small number of LVs. In practice, some of the inferred LVs are often found to correlate with biologically relevant factors, such as age and sex, despite the fact that no mathematical mechanism in Equation 1 imposes such a property. However, some of the LVs might correlate with hidden technical confounding factors.
To encourage inference of environmentally relevant LVs and reduce those that capture technical confounding factors, one can impose additional constraints on Equation 1. One strategy, implemented in PLIER,25 imposes constraints to encourage Z to be a combination of known gene sets and pathways:34, 35, 36, 37
| (Equation 2) |
where G is a h × g binary matrix with Gij = 1 if gene j belongs to a known gene set or pathway i. B is a q × h non-negative weight matrix encouraged to be sparse via incorporation of ||B||1 so that each row of Z would be constructed from only a small number of gene sets and pathways, which eases interpretation of the LVs. Equation 2 can be solved with block coordination descent, and PLIER provides heuristics for setting the tuning parameters λ1, λ2, λ3, and q.
In this work, we focused on using PLIER to extract LVs. We first applied PLIER separately to the ROSMAP and CMC gene expression data after removing technical confounding factors but without regressing out expression PCs. The reason for not regressing out PCs is that they often capture broad patterns related to non-genetic factors, which are indeed the type of variation we like to capture. As for parameter selection, PLIER is shown to be robust for a wide range of parameter combinations around the default values.25 Hence, we opted to use the default parameter setting. In brief, λ1 and λ2 are based on the singular value of X. λ3 is set such that the fraction of LVs associated with prior pathway information is 0.7, and the statistical significance of the pathway associations (rows of B) are assessed via a pseudo-cross-validation procedure for labeling LVs with specific gene sets and pathways. q is set on the basis of the “elbow” of the eigenspectrum of X. To enable subsequent GxE meta-analysis, we concatenated the ROSMAP and CMC gene expression data after standardization and applied PLIER to generate a LV set common to the two datasets. We note that 10 LVs (LV12, LV20, LV26, LV32, LV88, LV113, LV114, LV115, LV116, and LV133) have values close to 0, which PLIER correctly assigned to no known gene sets or pathways, and no significant GxE eQTLs are associated with these LVs.
To further aid interpretation of the LVs, we correlated the (ROSMAP portion of the) LVs to phenotypic and demographic variables of the ROSMAP samples as well as gene-expression-based estimates of cell-type proportions.35 The phenotypic and demographic variables included those related to cognition, clinical, personality, age, sex, alcohol, smoking, self-reported thyroid diseases, and pathology.
LV Replication
To assess LV replication, we computed the correlation between gene loadings of all LV pairs across the ROSMAP and CMC datasets and matched the LVs by using Hungarian clustering.38 The correlation between gene loadings of matched LVs was used as the replication metric. To establish a baseline, we extracted LVs from the blood-based DGN expression dataset26 and examined the correlation between matched LVs derived from the two brain datasets versus this blood dataset. We also assessed how well the common LV set reflects the LVs derived from each brain-based dataset by computing the correlation between LVs across the concatenated and individual datasets; we applied Hungarian clustering to match the LVs and again used the correlation between matched LVs as the evaluation metric.
Modeling GxE Effects
We modeled the expression levels of each gene j as a function of SNP i’s genotype, LV k, and their interaction:
| (Equation 3) |
where the n × 1 vector, yj, contains the expression levels of gene j from n individuals and where known confounding factors in addition to the top 10 PCs of expression were removed (here, we regressed out the top 10 PCs to better capture the genetic and GxE component of expression17). We note that yj is different from X in Equation 2 in that X has only known technical confounding factors removed. The n × 1 vector, gi, contains the genotype values of cis SNPs that are within 1Mb from the transcription starting site (TSS) of gene j and part of the previously found brain xQTL SNP set.31 The rationale for restricting our analysis to the xQTL SNP set is that cell-type-specific eQTL SNPs typically display significant main effects.20 We thus only analyzed SNPs exhibiting main effects on molecular traits31 to focus on SNPs that are more likely to display GxE effects while reducing the multiple testing burden. The n × 1 vector, ck, corresponds to LV k, derived by concatenation of the ROSMAP and CMC gene expression data and application of PLIER. To reduce false GxE detections, we also applied a number of filters in addition to standard QC. In particular, outliers in LVs and gene expression can easily result in false GxE detections, especially if the outliers happened to only belong to one genotype but not the others. Another problematic scenario is when a SNP has an acutely smaller number of samples for one genotype than for the other genotypes, which also tends to result in false GxE detections. Therefore, we restricted our GxE analysis to SNPs with all three genotypes, each of which has sample size > 5% of the total sample. We also excluded subjects with LV values or expression levels beyond 3 standard deviations from the median. We first applied the above procedures separately to the ROSMAP and CMC datasets and subsequently combined the results by using meta-analysis to increase statistical power. Significant GxE eQTLs were declared a dependent FDR threshold28 of 0.1, correcting for all LVs examined.
GxE Replication
To assess replicability, we computed the π1 statistics39 to estimate the proportion of GxE eQTLs that were in ROSMAP and were also significant in CMC. To declare significance, we generated an empirical null distribution by computing π1 for 104 random p value subsets of size m, where m is the number of GxE eQTLs. Only p values of associations that did not overlap with the GxE eQTLs were used for null estimation. We note that the modest number of detected GxE eQTLs limited the accuracy of the empirical p value distribution for π1 estimation. The magnitude of the estimated π1 should thus be interpreted with caution, and statistical testing of π1 is needed.
Mapping Transcription Factors to Their Targeted Genes with GxE eQTLs
We hypothesized that some of the LVs would capture effects of TFs, hence the GxE eQTL genes could potentially be their targets. To test this hypothesis, we first used a stringent criterion to assign TFs to LVs. Specifically, we took the list of 1,734 TFs encoded in human genome from the Catalog of Inferred Sequence Binding Preferences (CIS-BP),40 intersected this list with the highly expressed genes in the ROSMAP samples (892 genes), and used the expression of the intersected genes as the representation of the TFs. We then modeled the expression of each intersected gene (without PC removal) as a linear combination of all LVs by using multiple regression and applied stability selection41 to identify the significant LVs for each TF. To perform stability selection, we generated 10,000 bootstrap samples, applied multiple regression to each bootstrap sample, and identified significant regression coefficients at an α of 0.05 with Bonferroni correction. LVs with a selection frequency of 1, i.e., those that passed the Bonferroni-corrected threshold for all 10,000 bootstraps, were declared as significant for a given TF. We note that including all LVs into multiple regression, as opposed to correlating each LV separately with each TF, semi-partials out the variations of other LVs and thus highlights the unique aspect of each LV. Also, we opted to use gene expression without PC removal as TF representation because LVs were extracted from gene expression data without PC removal. After we assigned TFs to LVs, for each identified GxE eQTL, we used the gene expression representation of the TFs in place of their corresponding LVs and tested for interaction effect. Significant interaction was declared at 0.05 with Bonferroni correction for the number of GxE SNP-TF pairs tested.
Results
Deriving and Interpreting Latent Variables
To derive biologically informed LVs, we applied a variant of factor analysis called PLIER25 on the ROSMAP and CMC gene expression data. PLIER introduces a regularization term to factor analysis and thereby encourages factor loadings to be consistent with known gene sets and pathways.34, 35, 36, 37 This modification tends to yield LVs that are biologically interpretable and more robust across datasets because the resulting LVs are less likely to represent data-specific technical factors.
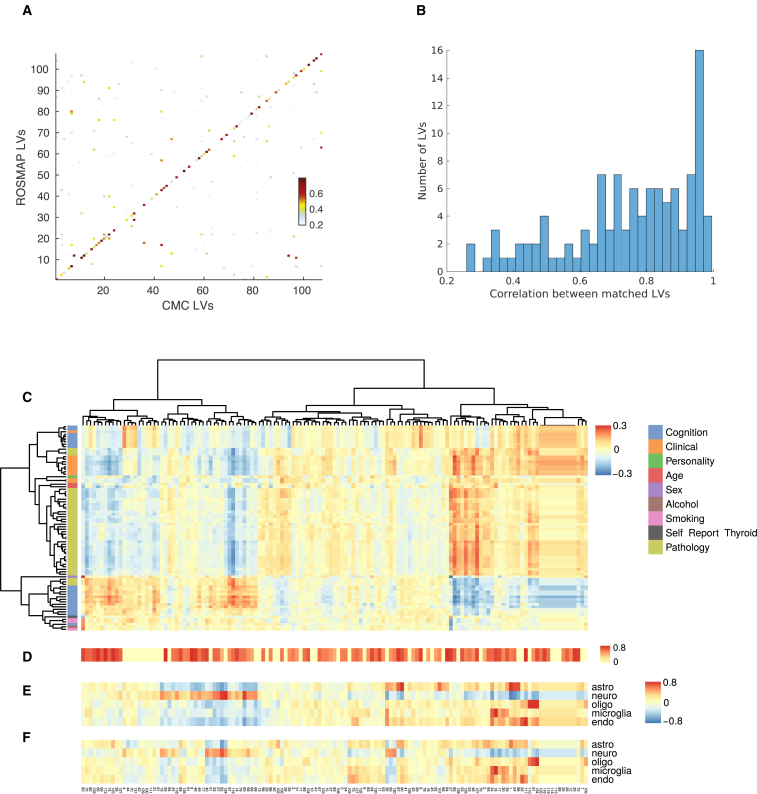
To assess LV replicability, we first applied PLIER (with the default parameter setting) separately to the ROSMAP and CMC gene expression datasets to derive 111 LVs and 107 LVs, respectively. We then computed the correlation between gene loadings of matched LVs, which we used as the LV replication metric (see Methods). To establish a baseline, we used the DGN expression dataset,26 which was derived from blood samples of 902 individuals, and extracted 109 LVs with PLIER’s default setting. We then examined the correlation between matched LVs derived from the two brain datasets versus this blood dataset. The correlation between matched LVs of ROSMAP and those of CMC (both from brain tissue) is 0.3706 ± 0.2553, which is significantly higher than ROSMAP versus DGN (0.1653 ± 0.1578) and CMC versus DGN (0.1648 ± 0.1619), as determined by the Wilcoxon rank sum test, with p < 10-9 for both cases. Our results thus suggest that LVs from the two brain datasets are reasonably replicable, which is encouraging given the substantial differences in the underlying populations. Also, each LV derived from one dataset highly correlates with only a single LV derived from the other dataset for the majority of the LVs (Figure 2A). To facilitate meta-analysis for increasing statistical power in GxE eQTL detection, we further applied PLIER to the entire sample (n = 1,100) to generate a common LV set, which resulted in 135 LVs (see Methods). All subsequent GxE analyses were based on this LV set. Among the 135 LVs, 49% displayed a correlation of >0.8 with LVs derived from the ROSMAP dataset alone (Figure 2B).
Figure 2.
LV Replication and Characterization
Let LV_R and LV_C denote LVs derived from the ROSMAP data alone and the CMC data alone, respectively. Let LV_RC denote LVs derived from the concatenation of ROSMAP and CMC data, and let the ROSMAP and CMC components of LV_RC be denoted as LV_RCr and LV_RCc, respectively.
(A) Correlation of gene loadings between LV_R and LV_C. Each LV_R is matched with its best corresponding LV_C via Hungarian clustering. LVs are arranged along the rows and columns so that the diagonal elements correspond to correlations between matched LVs.
(B) Correlation between LV_R and LV_RCr. 49% of the matched LVs have correlation >0.8.
(C) Spearman’s correlation between LV_RCr and phenotypes across nine categories present in the ROSMAP cohort are shown as heatmaps with LV_RC hierarchically clustered. Only phenotypes significantly correlated to any LV_RCr are displayed, and significance is declared at a dependent FDR threshold of 0.05. The correlation range is clipped to -0.3 to 0.3 for clarity.
(D) Pathway enrichment of LV_RC is summarized in terms of area under the curve (AUC).24 AUC of the most enriched pathway is displayed. Certain LVs (in yellow) are not enriched for any particular pathway.
(E) Spearman’s correlation between LV_RCr and expression-based cell-type proportion estimates.32 Correlation range is clipped to -0.8 to 0.8.
(F) Spearman’s correlation between LV_RCc and expression-based cell-type proportion estimates.32 The correlation range is clipped to -0.8 to 0.8.
Each LV was associated with a prior weight vector that indicates the biological processes it captures. Overall, 70 (51%) of the 135 LVs were significantly associated with known gene sets and pathways (Table S1, Figure 2D). These include oxidative and stress-response pathways, specific immune activation pathways (such as NFKB and IFN pathways), and mitochondrial processes. 16 LVs were associated with five major brain cell types (neurons, endothelial cells, microglia, astrocytes, and oligodendrocytes) on the basis of the LV prior weights (Table S1). These 16 LVs themselves highly correlate with expression-based cell-type markers35 (Figures 2E and 2F), which confirms PLIER’s annotation.
In addition to annotating LVs with known gene sets and pathways with PLIER, we also associated phenotype and demographical variables to LVs by using correlation analysis (Table S2). The phenotypic and demographic variables were assembled into nine categories on the basis of expert knowledge. Categories include cognition, clinical, personality, age, sex, alcohol, smoking, self-reported thyroid diseases, and pathology. Several LVs are associated with these categories; for example, LV27 is associated with smoking, LV56 with age, and LV60 with sex (Figure 2C).
Identifying GxE eQTLs with Latent Variables
To identify GxE eQTLs, we modeled the expression levels of each gene j, yj, as a function of SNP i’s genotype, gi, LV k, ck, and their interaction:
| (Equation 4) |
For each gene, we assessed each cis SNP within 1 Mb of the TSS. Considering how SNPs affecting gene expression in a cell-type-specific manner typically display strong main effects,20 we opted to restrict our GxE analysis to xQTL SNPs, i.e., SNPs that affect molecular traits. Specifically, we restricted analysis to SNPs that affect gene expression (eQTLs), DNA methylation (mQTLs), or histone acetylation (haQTLs) in the DLPFC as found in our previous work;31 this resulted in 702,103 tested SNPs. LVs in (Equation 4) correspond to those derived by concatenation of the ROSMAP and CMC gene expression datasets (after per-dataset standardization) and application of PLIER. Known confounds and the top 10 expression PCs were regressed out from yj, and outlier subjects were removed so that false GxE eQTL detection would be reduced (see Methods). Significant interaction was declared at a dependent FDR threshold28 of 0.1.
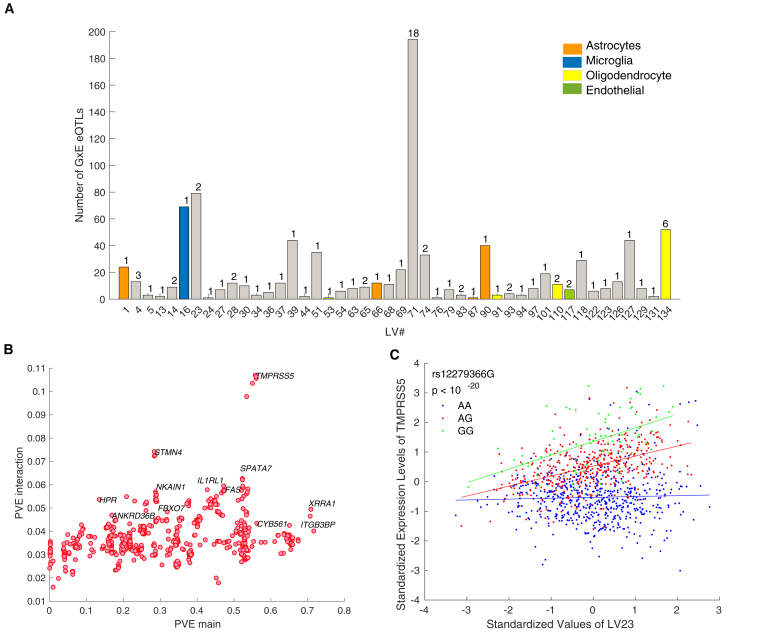
The GxE eQTLs derived from the ROSMAP data alone replicated well in the CMC data with a replication π1 of 0.7 (p = 0.01). This replication rate is larger than those for trans eQTLs but is smaller than the replication rate of cis eQTLs on the same tissue type.5 231 GxE eQTLs corresponding to 10 unique genes were detected. When we doubled the sample size by applying meta-analysis to the ROSMAP and CMC samples, we detected 895 GxE eQTLs corresponding to 52 unique genes (Table S3, Figure 3A), which is a ∼4 × increase in detection rate. The substantial increase in detection rate suggests that the current sample size (n = 1,100) is just large enough to start detecting interaction effects, i.e., detection is far from plateauing, which is consistent with previous sub-sampling analysis.20
Figure 3.
GxE eQTL Characterization
(A) The number of GxE eQTLs detected by each LV. Only LVs associated with ≥1 GxE eQTLs are displayed. The number of unique GxE genes is indicated on top of each bar. The colored bars correspond to LVs representing cell-type proportions.
(B) The percentage variance explained (PVE) by the main effect of a SNP versus the effect of interaction between SNP and LV. Interaction effect explains an additional ∼4% variance in expression of each GxE gene on average.
(C) Gene expression of TMPRSS5 versus LV23 with respect to the genotype of rs12279366. The interaction effect between rs12279366 and LV23 corresponds to the highest amount of additional variance explained in gene expression.
On average, modeling LV-genotype interaction explains an additional ∼4% variance in the expression of each gene that shows a significant interaction effect (Figure 3B), which is consistent with previous reports.17, 42 In a few genes, an additional 7%–10% variation in gene expression is explained by the interaction effect. For example, 53% variation in expression level of TMPRSS5 (MIM: 606751) is explained by genotype (rs12279366) alone, and the interaction between LV23 and rs12279366 explains an additional 10% variation in gene expression (Figure 3C).
Ten GxE eQTL genes were found for LVs that represent cell types (Figure 3A). These cell-type-specific eQTLs facilitate validation with external data. For instance, the genetic regulation of five genes is significantly modified by LV134 (a LV that reflects oligodendrocyte proportions). Among these genes are STMN4, NKAIN1 (MIM: 612871), and FAM221A, which are mainly expressed in oligodendrocytes.35 Other examples include an astrocyte-specific genetic regulation of FAS (MIM: 134637).35
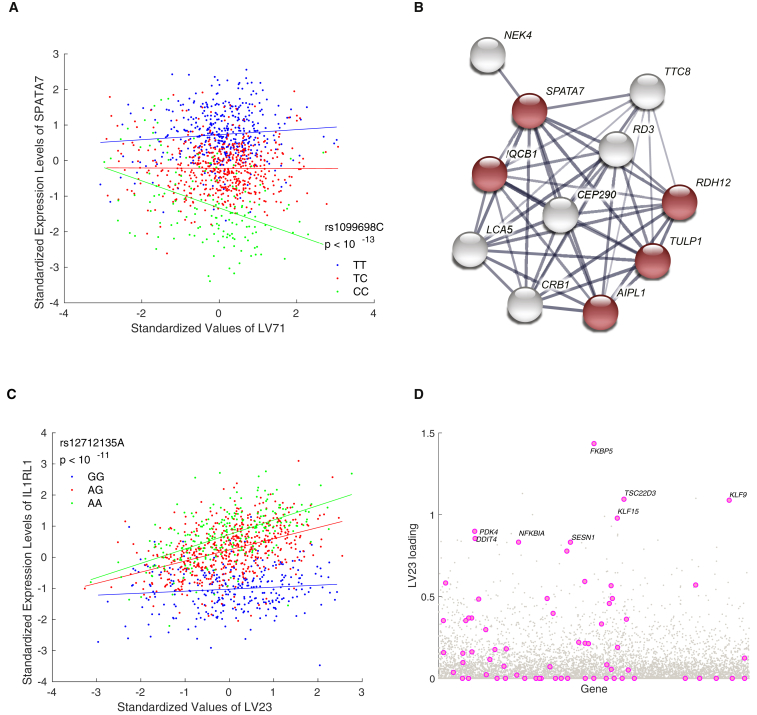
In addition to cell-type-specific eQTLs, we identified several GxE eQTLs that point to a context-specific impact of cis SNPs on well-known disease genes. For these SNPs, the associated LVs can yield insights into condition-specific regulation of the implicated genes, which provides promising directions in experimental conditions and stimulations for follow-up experiments. For instance, we found significant effects of interaction between cis SNPs for 18 genes and LV71 (enriched for genes annotated in the retinol metabolism pathway, p < 10-5). One such gene is SPATA7 (MIM: 609868) (Figure 4A), which is known to cause childhood-onset severe retinal dystrophy.43 Although SPATA7 is expressed in various brain regions and plays an important role in the retina, its specific function is unclear. Our results indicate that genetic regulation of SPATA7 is sensitive to activation of retinol metabolism. Fittingly, four of 10 functional interaction partners of SPATA7 are enriched for retinol metabolism, according to results obtained with STRINGdb,44 p < 10-7 (Figure 4B). Another example is a ITGB3BP (MIM: 605494) eQTL SNP that interacts with LV71. ITGB3BP is a multi-functional gene involved in the modulation of several critical pathways, including retinoid X receptor, NF-kappaB-dependent signaling, caspase signaling, and mitotic progression. Given the importance of ITGB3BP in these pathways, it is plausible that the genetic effects on these pathways might be partly regulated by retinoic acid activity.
Figure 4.
Examples of GxE eQTL Genes
(A) Expression levels of SPATA7 versus LV71 with respect to the genotype of rs10998698. LV71 is enriched for “retinol metabolism” genes.
(B) A SPATA7 functional interaction network obtained from the STRING database. Genes annotated to “retinol metabolism” are shown in red.
(C) Expression levels of IL1RL1 versus LV23 with respect to the genotype of rs12712135.
(D) Gene loading score for LV23; genes annotated to “repeatable Glucocorticoids response genes”40 are highlighted in pink.
A further example is the interaction between SNPs near IL1RL1 (MIM: 601203) and LV23 (Figure 4C). IL1RL1 is a member of Toll-like receptor superfamily, which has been associated with cardiovascular disease45 as well as allergy and immune disorders.46 Interestingly, we found that PLIER associated LV23 mainly with a gene set that is upregulated in heart tissue of patients with heart failure after the implantation of assistive devices,47 p < 10-5. After closer examinations of genes with higher weights for LV23, we also found a strong enrichment for genes induced by glucocorticoids specifically in the brain48 (p < 10-10 hypergeometric test, Figure 4D). Indeed, FKBP5 (MIM: 602623) has the largest weight for LV23, and is known for regulating glucocorticoid receptor sensitivity. Thus, the GxE analysis predicts that the eQTL SNP rs12712135 has a glucocorticoid-dependent effect on IL1RL1 expression.
Hypothesizing that LVs might reflect the effects of TFs, we used the detected GxE eQTLs to identify gene targets of TFs.40 Specifically, we first assigned TFs to LVs by using a bootstrap procedure41 (see Methods). We then replaced the LV in each detected GxE eQTL with its corresponding TFs, and tested for an interaction effect (Table S4). Ninety-one TF-specific eQTLs, corresponding to four unique genes and three unique TFs, were found at a Bonferroni-corrected threshold of 0.05. Interestingly, KLF15 (MIM: 606465) was found to be the top TF for LV23 (correlation of 0.7346, p < 10-86) and shows a significant interaction effect with rs12712135 on the expression of IL1RL1, p < 10-6. This finding, in combination with prior evidence for induction of KLP15 by the glucocorticoid response,49 provides further support for our finding of a glucocorticoid-dependent effect on IL1RL1 expression.
Although we found several LVs that strongly correlate with common environmental factors, such as age and sex, we did not find GxE eQTLs for these LVs. Finally, we overlapped the set of GxE eQTL SNPs with three well-powered, brain-relevant GWASs (Schizophrenia [MIM: 181500],50 MDD [MIM: 608516],51 and AD [MIM: 104300]52). Given the small number of independent loci obtained from our GxE analysis, we did not find enrichment for disease SNPs. One locus near PPM1M (MIM: 608979) overlapped with a Schizophrenia-associated region, which showed an interaction effect with LV71 (a LV that is enriched for genes annotated in the retinol metabolism pathway). PPM1M is a protein phosphatase that is preferentially expressed in a few tissues, including brain tissue. Although little is known about PPM1M’s function in the brain, an early study linked its function to neurite growth. Fittingly, retinol metabolism is a critical pathway for neurite outgrowth and plays an important role in pathogenesis of Schizophrenia.53
Discussion and Conclusion
In this study, we investigated how SNPs influence gene expression in DLPFC through their interaction with LVs that reflect environmental conditions. Our approach was motivated by the observation that broad variability in gene expression across individuals, as summarized by LVs, often reflects cellular and environmental factors. We thus sought to represent a large set of environmental variables with LVs whose impact is embedded at the cellular level, and we used these LVs to identify GxE eQTLs. To this end, we applied a biologically informed latent variable analysis to infer 135 LVs from the two largest brain eQTL datasets (n = 1,100) and showed that the majority of these LVs are highly reproducible across datasets. We then used these LVs in a standard statistical interaction model to identify interaction effects between LVs and genotype, as manifested on gene expression levels. At a dependent FDR threshold of 0.1, we identified 52 genes whose expression levels were impacted by an interaction effect between genotype and LVs. On average, the interaction term explains an additional ∼4% variation in expression levels for genes exhibiting GxE effects. We observed that ∼20% of the GxE eQTLs correspond to cell-type-specific eQTLs. Other GxE eQTLs are mostly associated with multi-functional genes, such as ITGB3BP, where the impact of specific regulatory variants depends on the cellular context.
Our study builds upon a previous work on identifying context-specific eQTLs;17 in that work, different contexts were defined by individual proxy genes. We chose to use LVs, as opposed to single proxy genes, for three reasons. First, our preliminary experiments showed that LVs can more accurately represent a latent context and hence improve the statistical power for identifying interaction effects. Specifically, we compared the discovery rate for five proxy genes that are typically used to represent five major cell types (ENO2 [MIM: 131360] for neurons, OLIG2 [MIM: 606386] for oligodendrocytes, CD34 [MIM: 142230] for endothelial cells, CD68 [MIM: 153634] for microglia, and GFAP [MIM: 137780] for astrocytes) against LVs that represent cell types with the ROSMAP data. At the same dependent FDR threshold of 0.1, we found 75 cell-type-specific eQTLs with LVs, whereas we found only five cell-type-specific eQTLs with single proxy genes (Figure S1). Second, LVs are typically associated with tens to hundreds of genes, providing more information for interpreting the specific pathways and/or cellular context that they represent. Third, because LVs are constructed by aggregation of signals that are common across a specific set of genes, genetic components of expression that are disparate across these genes would be averaged out. Thus, LVs presumably provide a “cleaner” representation of environmental factors than single proxy genes, which inherently have the genetic component of gene expression intact.
The discovery rate of GxE eQTLs greatly depends on the sample size. By doubling the sample size from ∼500 to ∼1,000, we observed an approximately 4× increase. Although already the largest sample for brain tissue, the discovery rate for GxE eQTLs in this study is rather low, which most likely implies that much larger sample sizes are needed to fully recover the range of eQTLs that are context dependent. Recent multivariate models that combine multiple environments and genotypes might also help in improving statistical power.42
Considering that cell-type-specific eQTL SNPs typically exhibit strong main effects on gene expression,20 we restricted the GxE analysis to xQTL SNPs,31 i.e., SNPs shown to affect molecular traits. This SNP selection hones in on SNPs that are more likely to display GxE effects while reducing the multiple testing burden. Also, by including mQTL and haQTL SNPs, we permitted the possibility of finding non-eQTL SNPs with GxE effects. Indeed, the majority of detected GxE eQTLs are eQTLs also. Hence, the GxE analysis is providing only a few new eQTL discoveries (Table S3). To further test this observation, we compared restricting the analysis to xQTL SNPs to using all SNPs within 1 Mb of the TSS of each gene. At α = 0.1 with Bonferroni correction, 222 GxE eQTLs were found both when xQTL SNPs were used and when all SNPs were used. Fifty-three GxE eQTLs were found only when xQTL SNPs were used, and 31 GxE eQTLs were found only when all SNPs were used. This high overlap in GxE eQTLs provides additional evidence that GxE eQTL SNPs typically display strong main effects on gene expression. Also, matching our expectation, restricting the SNPs to xQTL SNPs increases detection sensitivity. Importantly, we note that the actual value of the GxE analysis is the identification of environmental conditions for which the effects of the eQTL SNPs are more pronounced. For instance, the GxE eQTL SNPs near CD53 (MIM: 151525) (a gene primarily expressed in microglia) display significant main effects, but in addition, the GxE analysis predicts that the impact of these SNPs on CD53 expression is much greater in microglia cells. Also, we showed that the detected GxE eQTLs can be used for finding potential gene targets of TFs. For example, SPL1 was found to modify the genetic effects on CD53 expression, which aligns with how CD53 is a target of SPL1 in mice.54
In summary, we investigated GxE eQTLs in the brain by inferring LVs from gene expression data and using these LVs to represent cellular context. Our investigation identified 52 unique genes, whose eQTLs showed context dependency. The identified GxE eQTLs provide insights into cell-type-specificity and gene function.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank Maria Chikina for helpful comments. This work has been partly supported by National Institutes of Health grant P330AG10161, U01 (D.L. and D.B.) and by a Natural Sciences and Engineering Research Council of Canada Discovery Grant (S.M.).
Published: August 22, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.07.016.
Web Resources
CMC study, https://www.synapse.org/#!Synapse:syn2759792
Mostafavi lab (all results freely available), http://saramostafavi.github.io/software.html
ROSMAP study, https://www.radc.rush.edu
Supplemental Data
References
- 1.Hyman S.E. The daunting polygenicity of mental illness: Making a new map. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20170031. doi: 10.1098/rstb.2017.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallagher M.D., Chen-Plotkin A.S. The post-GWAS era: From association to function. Am. J. Hum. Genet. 2018;102:717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gratten J., Wray N.R., Keller M.C., Visscher P.M. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat. Neurosci. 2014;17:782–790. doi: 10.1038/nn.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F., Lupski J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015;24(R1):R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GTEx Consortium. Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raj T., Rothamel K., Mostafavi S., Ye C., Lee M.N., Replogle J.M., Feng T., Lee M., Asinovski N., Frohlich I. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairfax B.P., Makino S., Radhakrishnan J., Plant K., Leslie S., Dilthey A., Ellis P., Langford C., Vannberg F.O., Knight J.C. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowles D.A., Burrows C.K., Blischak J.D., Patterson K.M., Serie D.J., Norton N., Ober C., Pritchard J.K., Gilad Y. Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes. eLife. 2018;7:e33480. doi: 10.7554/eLife.33480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M.N., Ye C., Villani A.C., Raj T., Li W., Eisenhaure T.M., Imboywa S.H., Chipendo P.I., Ran F.A., Slowikowski K. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye C.J., Feng T., Kwon H.K., Raj T., Wilson M.T., Asinovski N., McCabe C., Lee M.H., Frohlich I., Paik H.I. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor D.L., Knowles D.A., Scott L.J., Ramirez A.H., Casale F.P., Wolford B.N., Guan L., Varshney A., Albanus R.D.O., Parker S.C.J. Interactions between genetic variation and cellular environment in skeletal muscle gene expression. PLoS ONE. 2018;13:e0195788. doi: 10.1371/journal.pone.0195788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao C., Joehanes R., Johnson A.D., Huan T., Esko T., Ying S., Freedman J.E., Murabito J., Lunetta K.L., Metspalu A. Sex- and age-interacting eQTLs in human complex diseases. Hum. Mol. Genet. 2014;23:1947–1956. doi: 10.1093/hmg/ddt582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kukurba K.R., Parsana P., Balliu B., Smith K.S., Zappala Z., Knowles D.A., Favé M.J., Davis J.R., Li X., Zhu X. Impact of the X Chromosome and sex on regulatory variation. Genome Res. 2016;26:768–777. doi: 10.1101/gr.197897.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannon E., Spiers H., Viana J., Pidsley R., Burrage J., Murphy T.M., Troakes C., Turecki G., O’Donovan M.C., Schalkwyk L.C. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 2016;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knowles D.A., Davis J.R., Edgington H., Raj A., Favé M.J., Zhu X., Potash J.B., Weissman M.M., Shi J., Levinson D.F. Allele-specific expression reveals interactions between genetic variation and environment. Nat. Methods. 2017;14:699–702. doi: 10.1038/nmeth.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favé M.-J., Lamaze F.C., Soave D., Hodgkinson A., Gauvin H., Bruat V., Grenier J.-C., Gbeha E., Skead K., Smargiassi A. Gene-by-environment interactions in urban populations modulate risk phenotypes. Nat. Commun. 2018;9:827. doi: 10.1038/s41467-018-03202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhernakova D.V., Deelen P., Vermaat M., van Iterson M., van Galen M., Arindrarto W., van ’t Hof P., Mei H., van Dijk F., Westra H.J. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 18.Tung J., Gilad Y. Social environmental effects on gene regulation. Cell. Mol. Life Sci. 2013;70:4323–4339. doi: 10.1007/s00018-013-1357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi J.K., Kim S.C. Environmental effects on gene expression phenotype have regional biases in the human genome. Genetics. 2007;175:1607–1613. doi: 10.1534/genetics.106.069047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westra H.J., Arends D., Esko T., Peters M.J., Schurmann C., Schramm K., Kettunen J., Yaghootkar H., Fairfax B.P., Andiappan A.K. Cell Specific eQTL Analysis without Sorting Cells. PLoS Genet. 2015;11:e1005223. doi: 10.1371/journal.pgen.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stegle O., Parts L., Durbin R., Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mostafavi S., Battle A., Zhu X., Urban A.E., Levinson D., Montgomery S.B., Koller D. Normalizing RNA-sequencing data by modeling hidden covariates with prior knowledge. PLoS ONE. 2013;8:e68141. doi: 10.1371/journal.pone.0068141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao W., Zaslavsky E., Hartmann B.M., Sealfon S.C., Chikina M. Pathway-level information extractor (PLIER) for gene expression data. Nat. Methods. 2019;16:607–610. doi: 10.1038/s41592-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battle A., Mostafavi S., Zhu X., Potash J.B., Weissman M.M., McCormick C., Haudenschild C.D., Beckman K.B., Shi J., Mei R. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parts L., Stegle O., Winn J., Durbin R. Joint genetic analysis of gene expression data with inferred cellular phenotypes. PLoS Genet. 2011;7:e1001276. doi: 10.1371/journal.pgen.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 29.Bennett D.A., Buchman A.S., Boyle P.A., Barnes L.L., Wilson R.S., Schneider J.A. Religious orders study and rush memory and aging project. J. Alzheimers Dis. 2018;64(s1):S161–S189. doi: 10.3233/JAD-179939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng B., White C.C., Klein H.U., Sieberts S.K., McCabe C., Patrick E., Xu J., Yu L., Gaiteri C., Bennett D.A. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fusi N., Stegle O., Lawrence N.D. Joint modelling of confounding factors and prominent genetic regulators provides increased accuracy in genetical genomics studies. PLoS Comput. Biol. 2012;8:e1002330. doi: 10.1371/journal.pcbi.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbas A.R., Baldwin D., Ma Y., Ouyang W., Gurney A., Martin F., Fong S., van Lookeren Campagne M., Godowski P., Williams P.M. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 35.Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhn H.W. The Hungarian method for the assignment problem. Naval Research Logistics Quarterly. 1955;2:83–97. [Google Scholar]
- 39.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meinshausen N., Bühlmann P. Stability selection. J. R. Stat. Soc. Series B Stat. Methodol. 2010;72:417–473. [Google Scholar]
- 42.Moore R., Casale F.P., Jan Bonder M., Horta D., BIOS Consortium. Franke L., Barroso I., Stegle O. A linear mixed-model approach to study multivariate gene-environment interactions. Nat. Genet. 2019;51:180–186. doi: 10.1038/s41588-018-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackay D.S., Ocaka L.A., Borman A.D., Sergouniotis P.I., Henderson R.H., Moradi P., Robson A.G., Thompson D.A., Webster A.R., Moore A.T. Screening of SPATA7 in patients with Leber congenital amaurosis and severe childhood-onset retinal dystrophy reveals disease-causing mutations. Invest. Ophthalmol. Vis. Sci. 2011;52:3032–3038. doi: 10.1167/iovs.10-7025. [DOI] [PubMed] [Google Scholar]
- 44.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho J.E., Chen W.-Y., Chen M.-H., Larson M.G., McCabe E.L., Cheng S., Ghorbani A., Coglianese E., Emilsson V., Johnson A.D., CARDIoGRAM Consortium. CHARGE Inflammation Working Group. CHARGE Heart Failure Working Group Common genetic variation at the IL1RL1 locus regulates IL-33/ST2 signaling. J. Clin. Invest. 2013;123:4208–4218. doi: 10.1172/JCI67119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akhabir L., Sandford A. Genetics of interleukin 1 receptor-like 1 in immune and inflammatory diseases. Curr. Genomics. 2010;11:591–606. doi: 10.2174/138920210793360907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y., Park S., Li Y., Missov E., Hou M., Han X., Hall J.L., Miller L.W., Bache R.J. Alterations of gene expression in failing myocardium following left ventricular assist device support. Physiol. Genomics. 2003;14:251–260. doi: 10.1152/physiolgenomics.00022.2003. [DOI] [PubMed] [Google Scholar]
- 48.Juszczak G.R., Stankiewicz A.M. Glucocorticoids, genes and brain function. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;82:136–168. doi: 10.1016/j.pnpbp.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Sasse S.K., Mailloux C.M., Barczak A.J., Wang Q., Altonsy M.O., Jain M.K., Haldar S.M., Gerber A.N. The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol. Cell. Biol. 2013;33:2104–2115. doi: 10.1128/MCB.01474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M.F. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marioni R.E., Harris S.E., Zhang Q., McRae A.F., Hagenaars S.P., Hill W.D., Davies G., Ritchie C.W., Gale C.R., Starr J.M. GWAS on family history of Alzheimer’s disease. Transl. Psychiatry. 2018;8:99. doi: 10.1038/s41398-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lane M.A., Bailey S.J. Role of retinoid signalling in the adult brain. Prog. Neurobiol. 2005;75:275–293. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Satoh J., Asahina N., Kitano S., Kino Y. A comprehensive profile of ChIP-Seq-based PU.1/Spi1 target genes in microglia. Gene Regul. Syst. Bio. 2014;8:127–139. doi: 10.4137/GRSB.S19711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.