Abstract
Premature senescence, a death escaping pathway for cells experiencing stress, is conducive to aging and cardiovascular diseases. The molecular switch between senescent and apoptotic fate remains, however, poorly recognized. Nrf2 is an important transcription factor orchestrating adaptive response to cellular stress. Here, we show that both human primary endothelial cells (ECs) and murine aortas lacking Nrf2 signaling are senescent but unexpectedly do not encounter damaging oxidative stress. Instead, they exhibit markedly increased S-nitrosation of proteins. A functional role of S-nitrosation is protection of ECs from death by inhibition of NOX4-mediated oxidative damage and redirection of ECs to premature senescence. S-nitrosation and senescence are mediated by Keap1, a direct binding partner of Nrf2, which colocalizes and precipitates with nitric oxide synthase (NOS) and transnitrosating protein GAPDH in ECs devoid of Nrf2. We conclude that the overabundance of this “unrestrained” Keap1 determines the fate of ECs by regulation of S-nitrosation and propose that Keap1/GAPDH/NOS complex may serve as an enzymatic machinery for S-nitrosation in mammalian cells.
Keywords: Keap1, NOX4, Nrf2, Oxidative stress, S-nitrosation, S-nitrosylation
Graphical abstract
Highlights
-
•
Endothelial cells and aortas with disturbed Nrf2 signaling are senescent.
-
•
Senescence onset is accompanied by a profound S-nitrosation of proteins.
-
•
S-nitrosation of NOX4 protects Nrf2-deficient cells from oxidative damage.
-
•
The protein orchestrating S-nitrosation in endothelial cells is Keap1.
-
•
Keap1 interacts with transnitrosating protein GAPDH and nitric oxide synthase (NOS).
1. Introduction
Cellular senescence is a state of permanent cell cycle arrest with concomitantly maintained metabolic activity and viability. Senescence may be caused by DNA damage, response to oncogene activation, tumor-suppressor loss, or cellular stress. There is much evidence that premature senescence contributes to aging and age-related diseases [1]. Such dependency also applies to the vascular system, manifesting as an increased risk of cardiovascular disorders (CVDs) [2]. The endothelium is particularly exposed to adverse signals, allowing it to undergo premature senescence and become dysfunctional, which facilitates the onset of CVD [3].
Previous studies demonstrated that Nrf2, encoded by NFE2L2 (nuclear factor (erythroid-derived 2)-like 2) is one of the key players maintaining a healthy endothelial phenotype [[4], [5], [6], [7]]. Nrf2 is a transcription factor mediating an adaptive response to cellular stress. A number of genes encoding detoxifying enzymes (glutathione S-transferase (GST), NAD(P)H:quinone oxidoreductase (NQO1)), stress-responsive proteins (heme oxygenase-1 (HMOX1)), and enzymes responsible for the elimination of superoxide and hydroperoxides (glutathione peroxidase (GPX), superoxide dismutase (SOD)), are directly regulated by Nrf2. When Nrf2 escapes Keap1-mediated proteolysis, it activates transcription of a large number of protective enzymes through binding to the electrophile response element, EpRE (also known as the antioxidant response element (ARE)) located in their promoters [8]. Not surprisingly, inhibition of Nrf2 activity was shown to make mice susceptible to oxidative damage [[9], [10], [11], [12]]. Changes in basal Nrf2 level may decrease or increase with age [[13], [14], [15]], but more consistently, the ability to activate Nrf2 above the basal level declines with age, which may explain the ineffective Nrf2-dependent antioxidant defense in vascular cells seen in the elderly [16]. Senescence-associated mechanisms for declining Nrf2 activity are, however, unclear. The molecular mechanism of senescence driven by Nrf2 inhibition, especially in endothelial cells (ECs), also remains unidentified. Furthermore, the factors that determine whether a cell undergoes senescence or apoptosis are not established.
One of the key molecules modulating response and function of a vascular system and ECs is nitric oxide (˙NO), which is produced in cells by nitric oxide synthases (NOS). The classical pathway triggered by ˙NO in blood vessels activates soluble guanyl cyclase, producing cGMP, which results in vasodilatation of vascular smooth muscle cells (VSMCs) and modulation of blood pressure [17]. Noteworthy, ˙NO can also influence the cellular response through protein S-nitrosation (SNO) [18], which is an oxidative chemical reaction that generates a nitroso group on a thiol. The chemistry through which this reaction occurs in mammals is still uncertain, though several non-enzymatic mechanisms have been proposed so far [19]. In bacteria, an enzymatic complex of NarGHI, Hcp and GAPDH, or NO synthase, SNO synthase and transnitrosating protein, respectively, has recently been demonstrated to catalyze SNO formation [20]. The SNO posttranslational modification has been shown in many proteins that are involved in the regulation of apoptosis, migration, transport and cell division [21]. A ‘switch-off’ mechanism is a denitrosation, performed mainly by S-nitrosoglutathione reductase (GSNOR) and thioredoxin reductases (TrxRs). Several proteins are constitutively nitrosated in the cell, and denitrosation serves as a mechanism for their activation [22].
Recently we found that loss of Nrf2 transcription factor leads to the induction of p53-dependent premature senescence in primary human aortic endothelial cells (HAECs) [23]. The molecular mechanism that triggers the onset of senescence when Nrf2 signaling is absent and its relevance in vivo remains to be clarified. In this paper, we demonstrate that Keap1, unhampered by an interaction with Nrf2, is involved in protein S-nitrosation in ECs that provides protection from NOX4-mediated oxidative damage and apoptosis while redirecting them to senescence. Moreover, we found that in Nrf2-deficient cells, Keap1 interacts with GAPDH and NOS, which may provide the machinery for S-nitrosation similar to that seen in bacteria [20].
2. Materials and methods
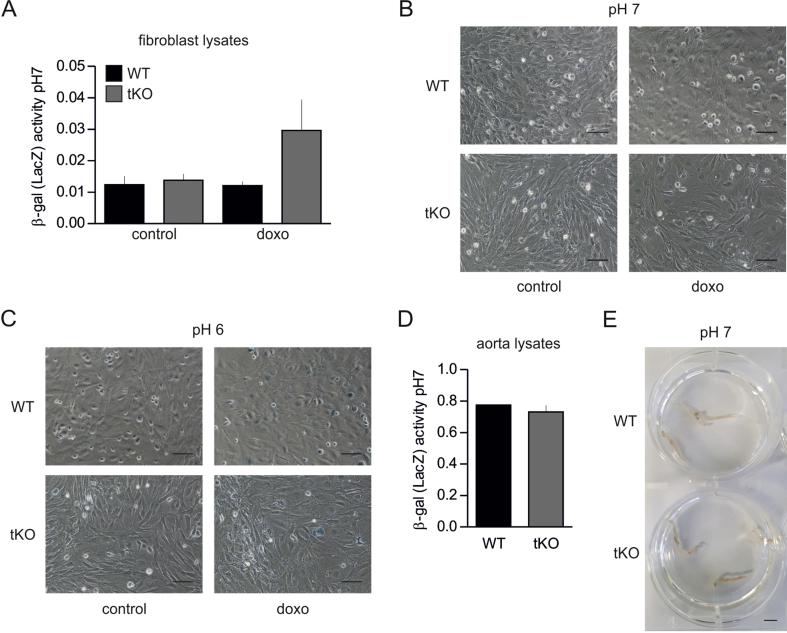
Animals. Nrf2-transcriptional knockout (tKO) C57BL/6 J mice, initially developed by Prof. Masayuki Yamamoto [24], were kindly provided by Prof. Antonio Cuadrado (Universidad Autonoma de Madrid) together with WT mice. 8-week old male and female animals were used in the study. In tKO animals, DNA binding domain of Nrf2 was replaced by LacZ gene [24], which generates a fusion protein N-terminal Nrf2-β-galactosidase [25,26]. As the previous paper shows, this protein can be detected only in response to electrophilic response, but not in control conditions [25]. To make sure that it is possible to distinguish its activity from the senescence-related one, we performed additional experiments on lung fibroblast and aortas isolated from WT and tKO mice. Importantly, the activity of β-gal of LacZ origin can be detected only at pH 7 [27], while senescence-associated-β-gal (SA-β-gal) activity is detected at pH 6 [28,29]. Fibroblasts were additionally stimulated with doxorubicin, known inducer of cellular senescence and Nrf2 activity. Our data shows that β-gal activity at pH 7 is detected above background only in lysates from tKO fibroblasts stimulated with doxorubicin (Fig. S1A). Importantly, it remains undetectable in in situ staining at pH 7 (Fig. S1B), which suggests that detection of LacZ-derived β-gal activity can be only achieved in lysates at pH 7. Accordingly, the blue color of cells stained in situ at pH 6 may come only from SA-β-gal activity (Fig. S1C). In agreement, activity of LacZ-derived β-gal remains below the level of detection in lysates prepared from intact tKO aortas (Fig. S1D) and in in situ staining (Fig. S1E).
Cell culture. Human aortic endothelial cells (HAECs) (Gibco and PromoCell) were grown in Endothelial Basal Medium (EBM-2) (Lonza) supplemented with EGM-2MV SingleQuot Kit Supplements & Growth Factors (Lonza) and 10% fetal bovine serum (FBS) (EURx). HAECs isolated from female donor, age 21 were used in the majority of experiments. Two other preparations, isolated from age 21 male and age 23 male donors were used to verify the observed phenotype. Murine fibroblasts were cultured in DMEM HG (Lonza) containing 10% FBS and penicillin (100 IU/mL)/streptomycin (100 μg/mL) (Lonza). Cells were cultured at 37 °C in a humidified incubator in 5% CO2 atmosphere. The cells used in all experiments were between passages 5 and 14 to exclude the induction of senescence by prolonged cell culture.
Transfection with small interfering RNA. Transfections of HAECs were performed using 50 nM siRNA against human NFE2L2 (Life Technologies s9493 or siNFE2L2seq2 Life Technologies s9492) or scrambled siRNA (Life Technologies 4390846) using Lipofectamine™ 2000 Transfection Reagent (Life Technologies) in Opti-MEM I Reduced Serum medium (Life Technologies). After 24 h medium was changed to EGM-2MV. The experiments were performed 48 h after transfection. Transfection with siRNA against NOX2 (Life Technologies s3788), NOX4 (Life Technologies s27013), NOS2 (Life Technologies s9620), NOS3 (Life Technologies s9623), GAPDH (Life Technologies s5572) and KEAP1 (Life Technologies s18983 or siKEAP1seq2 Life Technologies s18982) was performed concomitantly with silencing of NFE2L2.
Oxidant (commonly referred to as reactive oxygen species) measurement. Oxidants were assessed using 2 methods: MitoTracker Red CM-H2Xros (Life Technologies) and CellRox Deep Red Reagent (Life Technologies). The staining was performed according to the manufacturer's procedure. Data were collected using BD LSR Fortessa. We recognize that the oxidation of these dyes is only a semi-quantitative estimate of increased oxidant generation and not a true measurement of superoxide or any other oxygen-derived species.
Glutathione level measurement. Assessment of glutathione level and GSSG/GSH ratio was performed using Glutathione (GSSG/GSH) Detection Kit (Enzo). The procedure was performed according to manufacturer's protocol.
Thiobarbituric acid reacting substances (TBARS). In brief, cells upon treatment or transfection were washed with PBS and then 20% (w/v) trichloroacetic acid containing 0.8% (w/v) thiobarbituric acid (Sigma) was added to each well. The cells were scratched off to the eppendorf tubes and boiled for 30 min. After cooling to room temperature and centrifugation (350 g, 10 min), the absorbance of the supernatant at 535 nm and for the nonspecific turbidity at 600 nm was subtracted. The amount of MDA equivalents was calculated using the extinction coefficient of MDA-TBA complex which is 1.56 × 105 M−1 cm−1 and the results are expressed as pM MDA/mg protein. We recognize that the TBARS assay is a semiquantitative estimate of the oxidation of molecules, including lipids, that may increase with oxidative stress rather than a true measurement of lipid peroxidation.
Detection of O2•- in the isolated aorta or cells in vitro. Oxidation of dihydroethidium (DHE) to 2-hydroxy-ethidium (2-OH-E+) was used as an indicator of superoxide radical anion (O2•-) production in HAECs and aortas. The freshly harvested aorta was carefully cut out and cleaned from any adherent tissues, opened longitudinally, and placed in wells of a 48-well plate, each well filled with 100 μL of 10 μM DHE in PBS buffer without Ca/Mg ions pH 7.4 and incubated for 45 min at 37 °C in the dark. Then it was immediately frozen in liquid nitrogen in light safe tubes and stored in −80 °C until used. Aorta samples were homogenized in 250 μL of 0.1% Triton X-100 in ice-cold DPBS buffer and then centrifuged for 5 min at 1000 g at 4 °C to obtain supernatant. In order to extract the oxidation products of DHE 200 μL of supernatant was transferred to a fresh tube and mixed with 100 μL of 0.2 M HClO4 in methanol. The samples were incubated on ice for 2 h and subsequently centrifuged for 30 min at 16,600 g at 4 °C. A volume of 120 μL of the supernatant was then transferred to a fresh tube containing 120 μL of 1 M phosphate buffer (pH 2.6). This solution was centrifuged for an additional 15 min at 16,600 g at 4 °C. A volume of 200 μL of the supernatant was transferred to HPLC vials for analysis.
Confluent HAEC cells (85 mm dishes) were incubated for 30 min with 10 μM DHE in cell culture medium without FBS at 37 °C in a fully humidified atmosphere of 5% CO2. All subsequent manipulations were performed on ice. Followed by washing with an ice-cold DPBS, the cells were scrapped in 1 mL of DPBS and centrifuged for 5 min at 1000 g at 4 °C. The supernatant was removed and 150 μL of DPBS containing 0.1% Triton X-100 was added. The cells were lysed using an insulin syringe (10 times) and centrifuged for 5 min at 1000 g at 4 °C. The volume of 2 μL of the supernatant was used for protein determination using BCA assay; the volume of 100 μL of supernatant was transferred into the tube containing 100 μL of 0.2 M HClO4 in MeOH, vortexed (10 s) and placed back on the ice for protein precipitation. After 1–2 h the samples were centrifuged for 30 min at 16,600 g at 4 °C and 100 μL of supernatant was transferred into the tube containing 100 μL of 1 M phosphate buffer pH 2.6, vortexed (5 s) and centrifuged for 15 min at 16,600 g at 4 °C. A volume of 150 μL of the supernatant was transferred into HPLC vials for analysis. Measurement of 2-OH-E+ was performed by HPLC technique with fluorescence detection using Ultimate3000 Dionex system (Thermo, USA). Chromatographic separation was carried out on an analytical column Kinetex C18 (4.6 × 100 mm, 2.6 μm, Phenomenex, USA) with the oven temperature set at 40 °C. The mobile phase consisted of acetonitrile (A) and water (B) both with an addition of 0.1% trifluoroacetic acid with the following linear eluting steps: 0.0 min (A:B, 25/75, v/v) – 0.5 min (A:B, 25/75, v/v) – 8 min (A:B, 35/65, v/v) – 9 min (A:B, 95/5, v/v) – 11 min (A:B, 95/5, v/v) – 12.0 min (A:B, 25/75, v/v) – 14.0 min (A:B, 25/75, v/v). The flow rate was set at 1 mL min−1. A sample volume of 50 μL was injected onto column. The concentration of 2-OH-E+ (corresponding to the concentration of O2•- generated in samples) was calculated from a standard curve and normalized to total protein concentration.
Quantitative RT-PCR. RNA was isolated using RNeasy Mini Kit (Qiagen). Reverse transcription reaction was carried out using High Capacity cDNA Reverse Transcription Kit (Life Technologies). The procedures were performed according to the vendor's instructions. Quantitative RT-PCR was performed in a StepOnePlus real-time PCR system (Applied Biosystems) using SYBR Green PCR Master Mix (Sigma). Sequences of primers are given in Table S1.
EPR measurement of ˙NO in the isolated aorta and in cells in vitro. For measurements of ˙NO, EPR spin-trapping with diethyldithiocarbamic acid sodium salt (DETC) was used. Briefly, Krebs–HEPES buffer (consisting of, in mM: NaCl 99.0; KCl 4.7; MgSO4·7 H2O 1.2; KH2PO4 1.0; CaCl2·H2O 2.5; NaHCO3 25.0; glucose 5.6; and Na-Hepes 20.0) was filtered through a 0.22 μm paper syringe filter and equilibrated to pH 7.4, then deoxygenized by bubbling argon gas for at least 30 min. DETC (3.6 mg) and FeSO4 · 7H2O (2.25 mg) were separately dissolved under argon gas bubbling in two 10 mL volumes of ice-cold Krebs–HEPES buffer and were kept under gas flow on ice until used. One half (upper segment) of freshly harvested aortas was cleaned from adherent tissue, opened longitudinally, and placed in wells of the 48-well plate, each filled with 100 μL Krebs–Hepes and preincubated for 30 min at 37 °C. DETC and FeSO4 · 7 H2O solutions were mixed 1:1 (v/v) to obtain 250 μL Fe(DETC)2 colloid per well (final concentration 285 μM) and immediately added to the aorta in parallel with calcium ionophore A23187 (final concentration 1 μM) to stimulate eNOS and subsequently incubated at 37 °C for 90 min. In some cases, the addition of calcium ionophore was omitted, to show the unstimulated formation of ˙NO–Fe(DETC)2. After incubation, each aorta was removed from the buffer, drained on a piece of Kimwipe for 5 s and the wet mass was measured. Next the aorta was frozen, in liquid nitrogen, into the middle of a 400 μL column of Krebs–HEPES buffer and stored in −80 °C until measured.
Confluent HAEC cells (10 cm dishes) were washed with DPBS and 600 μL of culture medium without FBS was added. 200 μL of freshly prepared Fe(DETC)2 colloid was added into the dishes and the cells were incubated for 60 min at 37 °C in fully humidified atmosphere of 5% CO2, followed by discarding the medium containing the spin trap, collection of the cells by scraping into 1-mL insulin syringe (300 μL per sample) and snap freezing in liquid nitrogen.
EPR spectra were obtained in liquid nitrogen in a finger Dewar using an X-band EPR spectrometer (EMX Plus, Bruker) and were quantified by measuring the total amplitude of the ˙NO–Fe(DETC)2 after correction of baseline. ˙NO production was calculated from a standard curve prepared using MAHMA-NONOate donor and samples were normalized to the weight of the wet aorta.
Intracellular ˙NO. Intracellular ˙NO was measured using fluorescent probe DAF FM (Life Technologies) according to manufacturer's protocol. The data was collected on BD LSR Fortessa cytometer.
Measurement of nitrosylhemoglobin in blood. The concentration of nitrosylhemoglobin was measured by electron paramagnetic resonance (EPR). Blood was drawn from the right heart ventricle to a heparinized syringe (nadroparin, 10 U/mL) and EPR spectra of isolated erythrocytes (snap-frozen isolated erythrocytes (RBCs) obtained from whole blood via centrifugation at 1000g and 4 °C for 5 min) were recorded in liquid nitrogen (77 K) by using an EMX EPR Burker spectrometer operating at a frequency of 9.45 GHz, with the power of 15.98 mW, modulation amplitude equal to 5 G, time constant of 81.92 s, and sweep time of 20.48 s in the range of 200 G. For each sample, 30 individual scans were averaged. NOHb levels were expressed as ˙NO per mg RBCs, were calculated from standard curve samples prepared by incubating washed RBCs with various concentrations of PAPA-NONOate donor at 37 °C for 30 min [30].
Measurement of S-nitrosation by chemical reduction/chemiluminescence. The cells were lysed in PBS with 1% NP-40 (Sigma Aldrich), 1 mM diethylenetriaminepentaacetic acid (Sigma Aldrich) and 0.2 mM neucuproine (SigmaAldrich), centrifuged 10 min, 14,000 g. For detection of SNO, NO was displaced from S–NO bonds by mercuric ion (HgCl2) and measured using the Sievers NOA 280i nitric oxide analyzer, as described by Manning and Schonhoff [31]. Each sample was divided into two equal aliquots and incubated for 5 min at room temperature in the dark with either HgCl2 (final concentration 5 mM) or vehicle (deionized water). All samples were prepared with the addition of 1 mM EDTA to chelate free metals, that might interfere with SNO detection. Next, 50 μL of each sample was introduced into the reaction chamber of the nitric oxide analyzer, filled with 5 mL of glacial acetic acid, 1 mL of KI solution (50 mg per mL water) and antifoaming agent, using an airtight Hamilton syringe. The concentration of ˙NO in each sample was calculated based on a calibration curve. The addition of HgCl2 and/or EDTA did not modify the pH of samples, nor did it influence the calibration curve slope. The increase in the ˙NO content between HgCl2-treated and vehicle-treated aliquots of each sample is the SNO-specific signal, which is expressed as nM per mg protein.
Detection of S-nitrosation by biotin switch assay. The procedure was performed as described in Forrester et al. [32]. For immunofluorescence detection of S-nitrosated proteins streptavidin conjugated to Alexa Fluor 568 or Alexa Fluor 488 (Life Technologies) was used. High-resolution images were taken using a meta laser scanning confocal microscope (LSM-510; Carl Zeiss). Due to the controversy raised on the specificity of biotin switch assay [33,34], quantitative chemiluminescence-based SNO measurement was done and additional controls with DTT, ascorbate and GSNO + cysteine for biotin switch assay were performed.
Detection of S-nitrosation by Western blotting. The amount of total S-nitrosated proteins in cells was detected by Pierce S-nitrosation Western Blot Kit (Life Technologies) according to vendor's procedure. The assay is based on a modified biotin switch assay, in which cysteine-specific iodoTMT (tandem mass tag) is used instead of biotin. The membrane is probed with anti-TMT antibody to detect the S-nitrosated proteins. Due to the controversy raised on the specificity of biotin switch assay [33,34], quantitative chemiluminescence-based SNO measurement was done and additional controls with DTT, ascorbate and GSNO + cysteine for biotin switch assay were performed.
8-isoprostane measurement. 8-isoprostane was measured using OxiSelect™ 8-iso-Prostaglandin F2α ELISA Kit (Cell Biolabs) according to manufacturer's protocol. We recognize that this antibody may also recognize closely related lipid oxidation products.
Neutral comet assay. For measurement of double-strand breaks Trevigen CometAssay (Trevigen) was used according to the manufacturer's protocol. The high-resolution photos were taken using confocal microscope with meta-scanner (LSM-510, Carl Zeiss). Level of DNA damage was assessed using Instem – Comet Assay IV measurement system for the comet assay.
Isolation of fibroblasts. 5-day old pups were sacrificed by decapitation and lungs were collected. After tissue fragmentation, lungs were digested in 4 U/mL dispase (Gibco) for 45 min. After enzyme neutralization, cells were centrifuged and seeded onto plates. After 4 days, the cells were passaged and seeded for the experiment. LacZ-driven β-galactosidase activity was measured using the β-Galactosidase Enzyme Assay with Reporter Lysis Buffer (Promega).
En face immunostaining. For en face immunostaining, the mice were perfused with 1% PFA. Whole aortas were removed, gently cleaned of perivascular fat and fixed for 15 min with 4% paraformaldehyde (PFA). Next, aortas were incubated for 3 h in blocking permeabilizing buffer (0.3% Triton X-100, 5% goat serum, 1% BSA) and then incubated overnight with primary antibody targeting Keap1 (Clone 144, Millipore) in buffer (0.1% Triton X-100, 1% goat serum). Thereafter, the aortas were incubated for 2 h with secondary antibody conjugated with Alexa Fluor 568 (Life Technologies). After washing, the aortas were opened, intima facing up and mounted with Dako Fluorescence Mounting Medium (Dako). High-resolution images were taken using a meta laser scanning confocal microscope (LSM-510; Carl Zeiss).
Immunofluorescence. HAECs were seeded on coverslips. After stimulation, the cells were shortly washed in PBS, fixed for 10 min in 80% methanol and washed 3 times in PBS. Afterwards, the cells were incubated in 0.25% glycine in PBS for 30 min at room temperature and washed 3 times in PBS, then blocked in 3% BSA in PBS for 1 h at room temperature. The cells were probed with primary antibody (anti-Keap1 Millipore, anti-GAPDH Santa Cruz, anti-NOX4 Santa Cruz, anti-iNOS Santa Cruz, anti-phospho-eNOS (Ser1177) Cell Signaling Technology, anti-Nrf2 Santa Cruz) in 3% BSA in PBS, overnight at 4 °C. Next day, cells were 3 times washed in PBS, and incubated with secondary antibodies conjugated with fluorochromes and Hoechst to visualize nuclei. The slides were mounted with Dako Mounting Medium (Dako). High-resolution images were taken using a meta laser scanning confocal microscope (LSM-510; Carl Zeiss).
For the immunofluorescent stainings, the aortas after cleaning from surrounding tissue were put in OCT Tissue Freezing Medium (Leica) and frozen in isopentane at −80 °C. Then, 16 μm slides were cut in cryostat (Leica) and put on poly-l-lysine-covered slides. The slides were fixed for 10 min in 80% methanol and washed 3 times in PBS. Afterwards, the slides were incubated in 0.25% glycine in PBS for 30 min at room temperature and washed 3 times in PBS, then blocked 10% goat serum in PBS for 1 h at room temperature. The aortas were probed with primary antibody against p21 (Santa Cruz) in 1% goat serum in PBS, overnight at 4 °C. Next day, cells were 3 times washed in PBS, and incubated with secondary antibodies conjugated with fluorochrome. The slides were mounted with Dako Mounting Medium (Dako). High-resolution images were taken using a meta laser scanning confocal microscope (LSM-510; Carl Zeiss).
Staining for SA-β-galactosidase activity. Cells or aortas were fixed for 3 min with 4% formaldehyde, washed twice with PBS and incubated overnight at 37 °C with staining solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 150 mM NaCl, 2 mM MgCl2, 1 mg/mL X-gal in citrate buffer pH 6).
Assessment of cell proliferation. Cells were stained for PCNA antigen, as described previously [23].
Transduction with adenoviral vectors AdGFP and AdNFE2L2DN. The transductions with adenoviral vectors AdNFE2L2DN or AdGFP were performed at a multiplicity of infection 50. AdNFE2L2DN vectors code 403–605 aa of Nrf2: domains Neh2 (Keap1 binding), Neh4 and Neh5 (transactivating domains), Neh7 (RXRa binding) and Neh6 (TrCP binding) are missing, but Neh1 (DNA binding) and Neh3 (CHD6 binding) are present. After 24 h of incubation, medium with vectors was removed and fresh EGM-2MV 10% FBS complete medium was added for the next 24 h. The efficiency of transduction was confirmed by detection of GFP expression with the fluorescent microscope and by assessment of mRNA level of Nrf2 target genes.
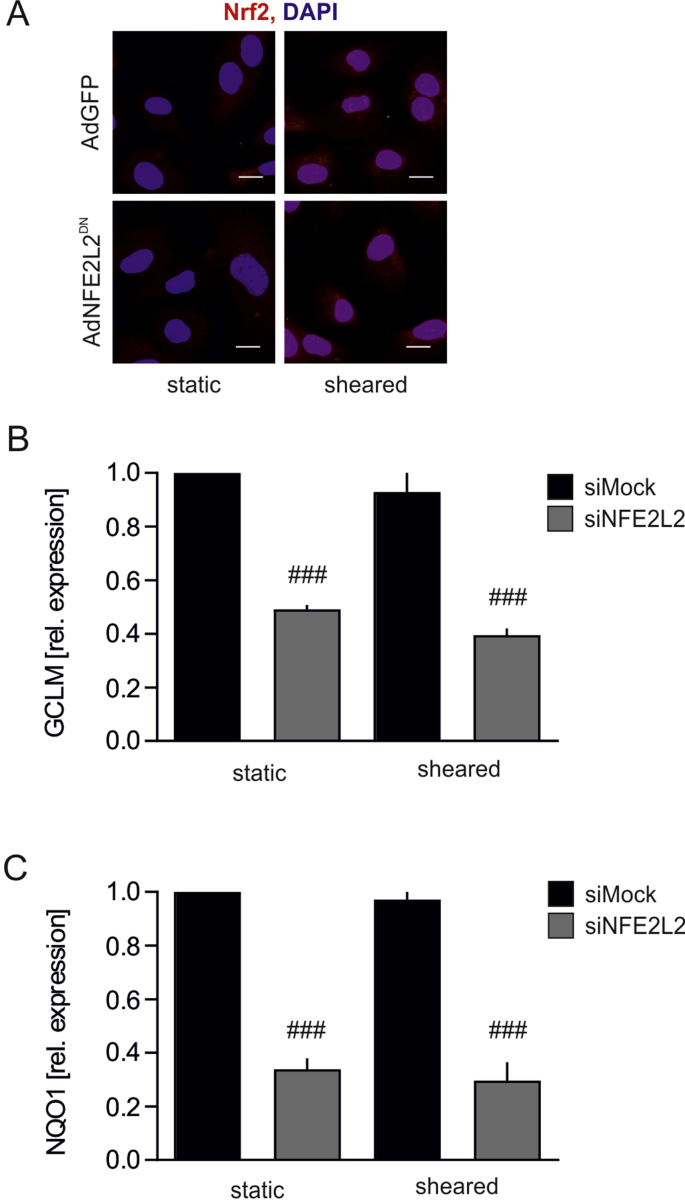
Shear stress conditions. For the shear stress experiments, HAECs were sheared orbitally at 300 rpm for 24 h at 37 °C in a humidified incubator in 5% CO2 atmosphere. The effect of orbital shear stress reflects the one seen in the disturbed and turbulent flow [35]. This sort of shear stress can be seen in the regions of bifurcations, bends and branches. There is an increased number of senescent endothelial cells and accelerated aging at the sites of disturbed flow [36]. Also Nrf2 activity and nuclear translocation are the same in orbital shear stress and disturbed/oscillatory shear stress. In contrast to laminar shear stress (L-flow), in oscillatory (O-flow) conditions Nrf2 does not bind to EpRE, although it translocates into nuclei in both L-flow and O-flow [37]. In our model of shear stress Nrf2 is localized preferentially in the nuclei but remains transcriptionally inactive, as the expression of Nrf2 target genes remained comparable between cells subjected to static and orbital shear stress conditions (Fig. S2).
Measurement of nitrite in media - Griess assay. Collected media were incubated for 30 min 1:1 with a mixture containing 0.2% naphthylethylenediamine dihydrochloride and 2% sulphanilamide in 5% phosphoric acid. Absorbance was measured at 530 nm using Tecan Spectra II Microplate Reader (Tecan).
Western blotting. Total protein was isolated using RIPA buffer with protease inhibitors (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS). Western blotting was performed according to standard procedures. Membranes were probed with antibodies against Nrf2 (Santa Cruz Biotechnology), p21 (Cell Signaling Technology), phospho-eNOS (Ser 1177) (Cell Signaling Technology), eNOS (Cell Signaling Technology), and tubulin (Calbiochem).
Co-immunoprecipitation. The proteins were crosslinked with 500 μM dithiobis succinimidyl propionate (DSP, Thermo Scientific Pierce) for 30 min, RT. The reaction was quenched with 50 mM Tris, pH 7.5. The cells were lysed in RIPA buffer (Pierce), containing protease and phosphatase inhibitors (Roche). The lysates were incubated with 2 μM of rat anti-Keap1 antibody or 2 μM of rat IgG antibody (BD) overnight at 4 °C. The proteins were pulled down using goat anti-rat Magnetic Beads (New England's Biolabs). The eluted proteins were separated by SDS-PAGE. The membranes were probed with anti-GAPDH antibody (Santa Cruz Biotechnology) and anti-phospho(Ser1177)-eNOS antibody (Cell Signaling Technology).
Measurement of nitrate and nitrite in plasma. The concentrations of nitrate and nitrite were measured by HPLC NOx analyzing system ENO-20 (Eicom), as described previously [38].
Identification of S-nitrosated proteins by mass spectrometry analysis. S-nitrosated proteins were pulled down using the biotin switch assay. Precipitated proteins were processed as described previously [23]. Label-Free-Quantification (LFQ) intensity values were calculated using the MaxLFQ algorithm. Data were normalized and analyzed using Scaffold 4.8.2 ProteomeSofware platform. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository.
Stimulation of cells. HAECs were stimulated with 1 mM N-acetyl cysteine (NAC) pH 7.4, 50 μM TEMPOL, 5 mM DTT, 200 μM S-nitrosoglutathione (GSNO) + 50 μM cysteine, 100 μM etoposide, 10 μM antimycin or 5 mM ascorbate in EGM-2MV medium. Stimulation of HAECs with TEMPOL, DTT, antimycin or ascorbate was done for 24 h, with GSNO + cysteine for 15 min, with FeCitrate for 30 min, and with etoposide for 1.5 h. In case of NAC, the cells were prestimulated for 2 h, and then NAC was withdrawn for 6 h. All reagents were purchased from Sigma.
Statistical Analysis. All experiments were performed in duplicates and were repeated at least three times. Data are presented as mean ± SEM. Statistical assessment was done with Student's t-test for two group comparisons or by analysis of variance (ANOVA), followed by a Bonferroni posthoc test for multiple comparisons. Differences were accepted as statistically significant for p < 0.05.
3. Results
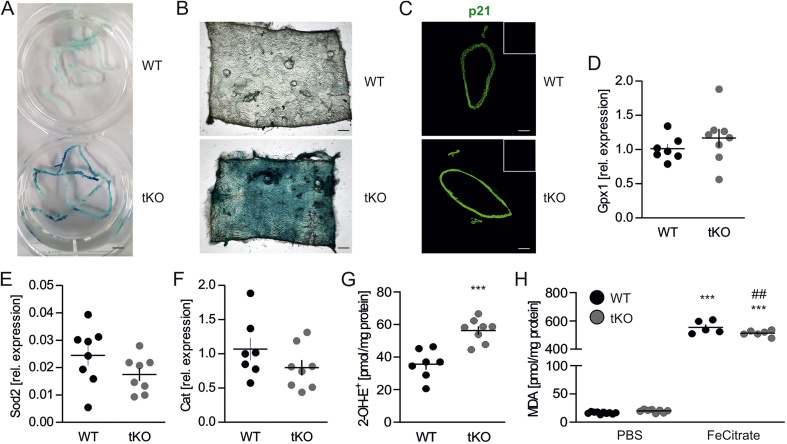
Inhibition of Nrf2 transcriptional activity results in premature senescence, but not oxidative damage, in murine aorta. To verify if disruption of Nrf2 signaling drives senescent phenotype of vascular cells in vivo, aortas of 8-week old wild type (WT) and transcriptional knock out (tKO) mice were stained for senescence-associated β-galactosidase (SA-β-gal) activity. We found that it was profoundly increased in tKO aortas, compared to WT animals, and present throughout the whole length of the vessels (Fig. 1A). En face projection showed the appearance of SA-β-gal positive cells at the luminal surface of the vessels, which were also massively scattered across the whole depth of the aortic wall, including tunica intima and tunica adventitia (Fig. 1B). Moreover, a significant gain of p21, the senescence-mediating protein, was seen in the aortas of tKO mice (Fig. 1C). Considering the key role of Nrf2 signaling in the regulation of oxidative defense [8], and the mechanism of premature senescence induction by oxidative stress [39], oxidative status of WT and tKO aortas was assessed. No differences in glutathione peroxidase (Gpx1), manganese superoxide dismutase (Sod2) and catalase (Cat) expression were detected between the aortic tissue of WT and tKO animals (Figs. 1D–F). The superoxide anion level was elevated by ~1.6 fold in the aortas of tKO mice (Fig. 1G), but TBARS, which are an estimate of oxidative damage, were equal between WT and tKO aortas (Fig. 1H). Significant increase of TBARS was observed for aortas challenged with iron overload, which served as a positive control for the assay (Fig. 1H). These data demonstrate that inhibition of Nrf2 transcriptional activity does not cause damaging oxidative stress in the murine aorta and results in the senescence onset.
Fig. 1.
Senescence, but no oxidative damage is present in Nrf2 tKO aortas.
(A) Assessment of SA-β-gal activity in aortas from WT and Nrf2 tKO mice. Representative pictures. n = 8. Scale bar 4 mm.
(B)En face projection of aortas from WT and Nrf2 tKO mice stained for SA-β-gal activity. Representative pictures. n = 5. Scale bar 0.35 mm.
(C) p21 level in WT and Nrf2 tKO aortas. Representative pictures. n = 6. Scale bar 0.2 mm. Insets – negative control.
(D–F) Expression of Gpx1 (D), Sod2 (E) and Cat (F) in aortas from WT and Nrf2 tKO mice. Relative expression of genes was measured by real-time PCR. EEF2 served as a reference gene. n = 7–8.
(G) Assessment of superoxide anion level in WT and Nrf2 tKO aortas by HPLC measurement of 2-OH-E+, the superoxide-derived oxidation product of DHE. ∗∗∗p < 0.001 vs WT, n = 7–8, two-tailed Student's t-test.
(H) Measurement of TBARS in WT and Nrf2 tKO aortas. The specimens were treated with 5 μM FeCitrate (a positive control) for 30 min. MDA was measured using thiobarbituric acid assay. ∗∗∗p < 0.001 vs PBS, ##p < 0.01 vs WT. n = 5–10, two-way ANOVA + Bonferroni's.
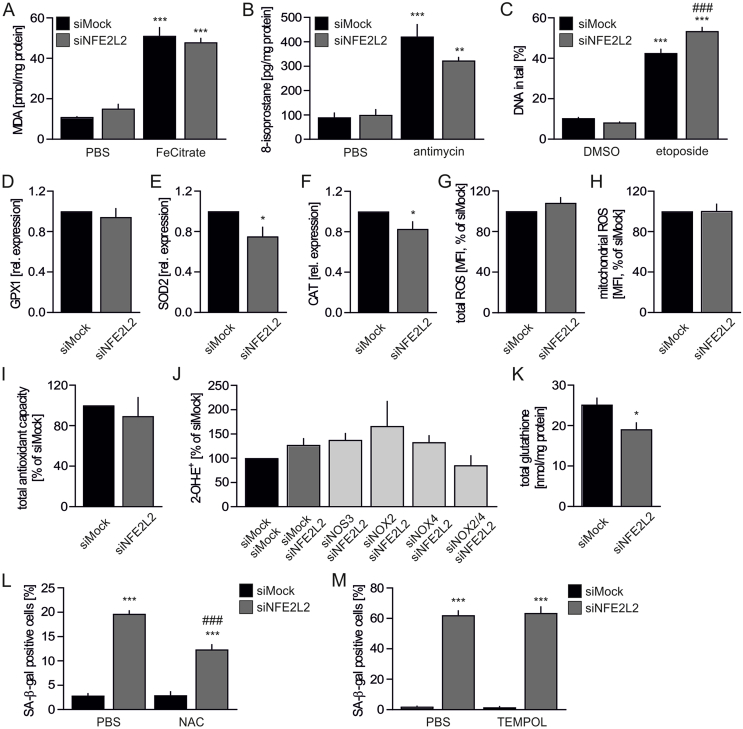
Loss of Nrf2 protein in primary human ECs is not associated with excessive oxidative stress. Similarly to Nrf2 tKO aortas, TBARS remained low in siNFE2L2-transfected HAECs, with the level comparable to siMock cells (Fig. 2A). In accordance, 8-isoprostane, another marker of oxidative damage, was low and equal between siMock and siNFE2L2 cells (Fig. 2B), and level of double-strand DNA breaks (DSB) was also comparable for both groups (Fig. 2C). Antimycin and etoposide treatments were used as positive controls for increased 8-isoprostane and DSB, respectively (Figs. 2B and C). No changes in the expression of GPX1 (Fig. 2D), but significant downregulation of SOD2 (Fig. 2E) and CAT (Fig. 2F) transcripts were observed upon silencing of NFE2L2 in HAECs. Even so, neither total (Fig. 2G) nor mitochondrial (Fig. 2H) oxidation of fluorescent probes, were found to be elevated in Nrf2-deficient cells and their total antioxidant capacity was preserved (Fig. 2I). The HPLC-based analysis showed a tendency of 25% increase in superoxide anion amount in siNFE2L2 HAECs (Fig. 2J). Analysis of potential superoxide sources (uncoupled eNOS, NOX2 and NOX4) revealed no significant differences between inspected groups, however concomitant silencing of NOX2 and NOX4 in Nrf2-deficient cells caused the strongest tendency toward decreased superoxide (Fig. 2J). Total glutathione level was lower in Nrf2-deficient HAECs (Fig. 2K), while the disulfide form of glutathione was undetectable. GSSG was likely exported to account for the decreased total glutathione. Such results may be indicative of a redox status that only slightly shifted towards oxidation in HAECs lacking Nrf2. Thus, Nrf2 deficiency in HAECs does not result in oxidative damage. Finally, we made an attempt to rescue the senescent phenotype of Nrf2-deficient cells (Fig. 2L) by incubation with N-acetylcysteine (NAC), a known glutathione precursor that supplies cysteine, which is normally limiting for glutathione synthesis. NAC triggered only partial attenuation of SA-β-gal activity in HAECs with siRNA-mediated knockdown of Nrf2 (Fig. 2L). Incubation of cells with non-toxic concentration of the superoxide scavenger TEMPOL did not reverse the senescent phenotype of HAECs (Fig. 2M). Collectively, these data show that Nrf2-deficient HAECs do not encounter excessive oxidative stress and damage. Although their microenvironment shows mild disequilibrium towards oxidation, it is not harmful to the cells. Therefore, the major cause of HAEC senescence driven by knockdown of Nrf2 is not linked to the uncontrolled oxidative stress.
Fig. 2.
Loss of Nrf2 in HAECs does not result in oxidative damage.
(A) Measurement of TBARS in siMock and siNFE2L2 HAECs. Cells were treated with 5 μM FeCitrate (a positive control) for 1 h. MDA was measured using thiobarbituric acid assay. n = 3, ∗∗∗p < 0.001 vs PBS, two-way ANOVA + Bonferroni's.
(B) 8-isoprostane level in siMock and siNFE2L2 HAECs measured by ELISA. Cells were treated with 10 μM antimycin (a positive control) for 24 h. n = 3, ∗∗p < 0.01 vs PBS, ∗∗∗p < 0.001 vs PBS, two-way ANOVA + Bonferroni's.
(C) Percentage of DNA in tail in siMock and siNFE2L2 HAECs. Neutral comet assay. Cells were treated with 100 μM etoposide (a positive control) for 1.5 h. n = 115–209, ∗∗∗p < 0.001 vs DMSO, ###p < 0.001 vs siMock, two-way ANOVA + Bonferroni's.
(D–F) Expression of GPX1 (D), SOD2 (E) and CAT (F) in siMock and siNFE2L2 HAECs. Relative expression of genes was measured by real-time PCR. EEF2 served as a reference gene. ∗p < 0.05 vs siMock. n = 17, two-tailed Student's t-test.
(G–H) Total (G) and mitochondrial (H) ROS level in siMock and siNFE2L2 HAECs was assessed by fluorescent probe using flow cytometry. n = 7–9.
(I) Assessment of total antioxidant capacity of siMock and siNFE2L2 HAECs. n = 3.
(J) Assessment of superoxide anion level in siMock/siMock, siMock/siNFE2L2, siNOS3/siNFE2L2, siNOX2/siNFE2L2, siNOX4/siNFE2L2 and siNOX2/4/siNFE2L2 HAECs by HPLC measurement of 2-OH-E+, the superoxide-derived oxidation product of DHE. n = 3–6.
(K) Total glutathione level in siMock and siNFE2L2 HAECs. ∗p < 0.05 vs siMock. n = 4, two-tailed Student's t-test.
(L) Percentage of cells with increased SA-β-gal activity in siMock and siNFE2L2 HAECs treated with N-acetylcysteine (NAC). Cells transfected with control (siMock) or NFE2L2 (siNFE2L2) siRNA were treated for 2 h with 1 mM NAC and left for 6 h for the reversion of signaling. n = 4, ∗∗∗p < 0.001 vs siMock, ###p < 0.001 vs siNFE2L2, two-way ANOVA + Bonferroni's.
(M) Percentage of cells with increased SA-β-gal activity in siMock and siNFE2L2 HAECs treated with TEMPOL. Cells transfected with control (siMock) or NFE2L2 (siNFE2L2) siRNA were treated for 24 h with 50 μM TEMPOL. n = 4, ∗∗∗p < 0.001 vs siMock, two-way ANOVA + Bonferroni's.
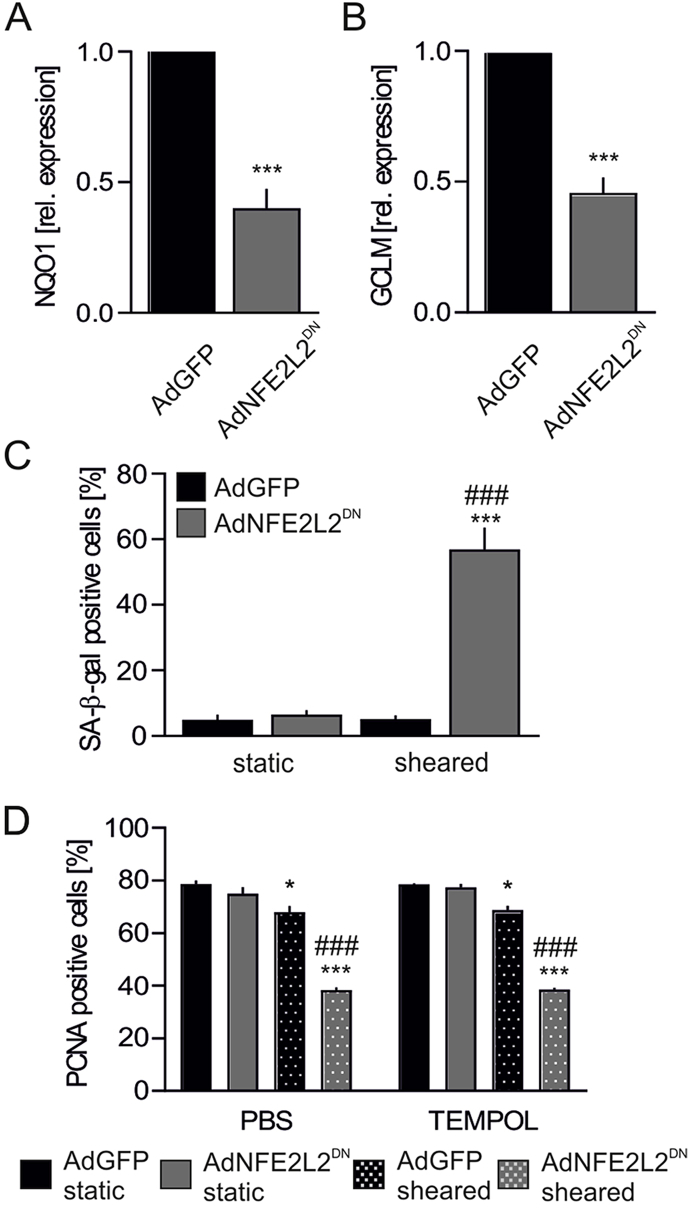
Lack of Nrf2 transcriptional activity induces premature senescence but only in shear stress conditions. The animal model of Nrf2 deficiency used in this study, and broadly used worldwide, is a transcriptional knockout, in which DNA binding domain of Nrf2 had been removed, but the N-terminal fragment of Nrf2, containing Keap1 binding domain Neh2, is preserved [23,24]. On the other hand, in siRNA-based in vitro experimental setting (Fig. 2) the availability of Nrf2 protein is reduced [23]. Moreover, our recent study indicates that Nrf2 activity may reach beyond transactivation of gene expression, as the constant synthesis of Nrf2 occupies Keap1 thereby preventing angiogenic dysfunction in ECs [23]. Therefore, to compare the findings from in vivo setting (Fig. 1) with in vitro experiments (Fig. 2), HAECs were transduced with adenoviral vectors harboring dominant-negative NFE2L2, coding C-terminal fragment of Nrf2 (domains Neh1 and Neh3), which is transcriptionally inactive [23]. The non-toxic titer of AdNFE2L2DN vectors led to ~60% inhibition of Nrf2 transcriptional activity, assessed by the mRNA level of its target genes NQO1 and GCLM (Figs. 3A and B), however it did not result in the onset of senescence (Fig. 3C), unless the cells were subjected to the orbital shear stress conditions (Fig. 3C). The decrease in proliferation marker PCNA was not reversed by treatment of the cells with TEMPOL (Fig. 3D). Taken together, loss of Nrf2 signaling in sheared cells induces senescence onset in vitro in HAECs and in vivo in the murine aorta, but ECs devoid of Nrf2 protein develop a senescent phenotype even in static conditions.
Fig. 3.
Inhibition of Nrf2 transcriptional activity together with subjection to shear stress conditions results in a senescence onset in HAECs.
(A–B) Expression of NQO1 (A) and GCLM (B) in HAECs transduced with dominant negative NFE2L2 at MOI 50. Relative expression of genes was measured by real-time PCR. EEF2 served as a reference gene. ∗∗∗p < 0.001 vs AdGFP. n = 4–5, two-tailed Student's t-test.
(C) Percentage of cells with increased SA-β-gal activity in HAECs transduced with dominant negative NFE2L2 at MOI 50. Cells were subjected for 24 h to shear stress conditions. n = 3, ∗∗∗p < 0.001 vs static, ###p < 0.001 vs AdGFP, two-way ANOVA + Bonferroni's.
(D) PCNA immunofluorescent staining in HAECs transduced with dominant negative NFE2L2 at MOI 50 and treated for 24 h with 50 μM TEMPOL. Cells were subjected for 24 h to shear stress conditions. Quantitative data. n = 3, ∗p < 0.05 vs static, ∗∗∗p < 0.001 vs static, ###p < 0.001 vs AdGFP, two-way ANOVA + Bonferroni's.
Nrf2-dependent senescence onset is associated with potent protein S-nitrosation. Premature cellular senescence can be triggered by markedly increased S-nitrosation of proteins, which impairs multiple cellular processes [3,40,41]. Thus we considered that S-nitrosation might be responsible for the induction of senescent phenotype in endothelial cells with disturbed Nrf2 signaling. Furthermore, we hypothesized that its functional role in these cells is protection from oxidative damage.
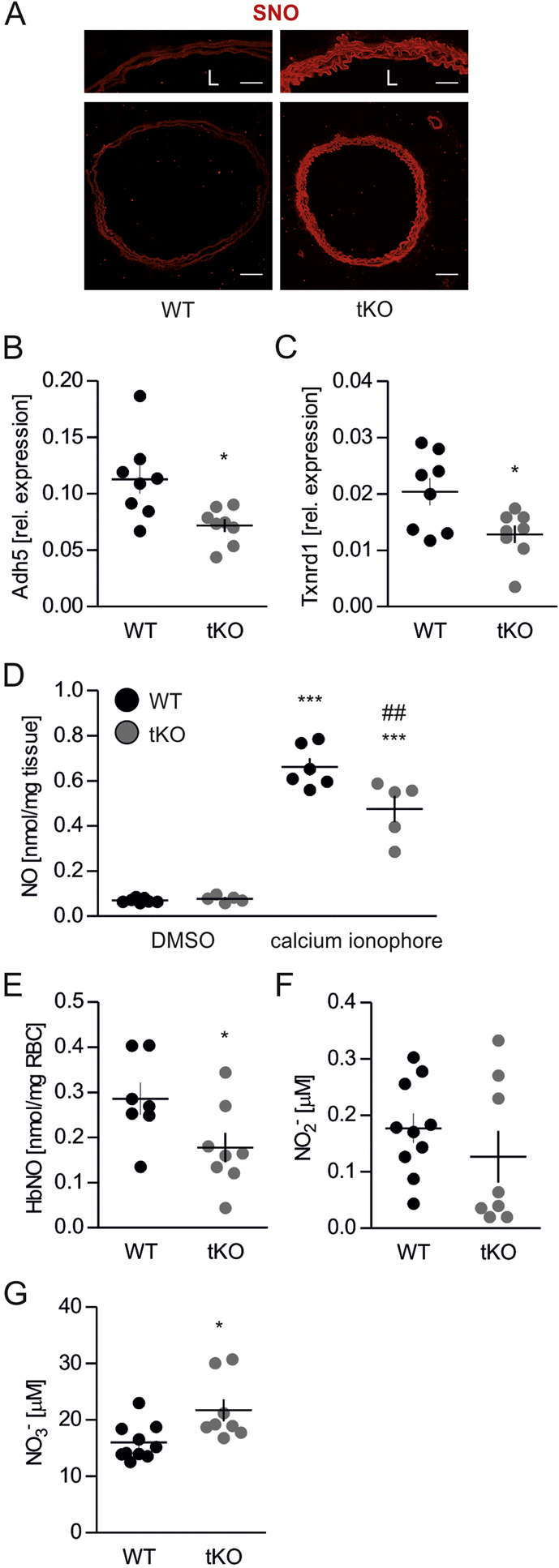
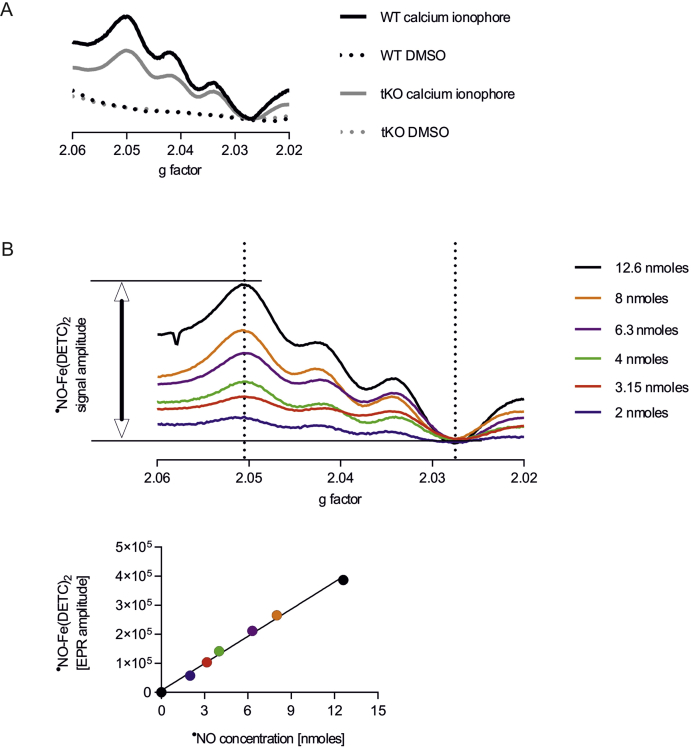
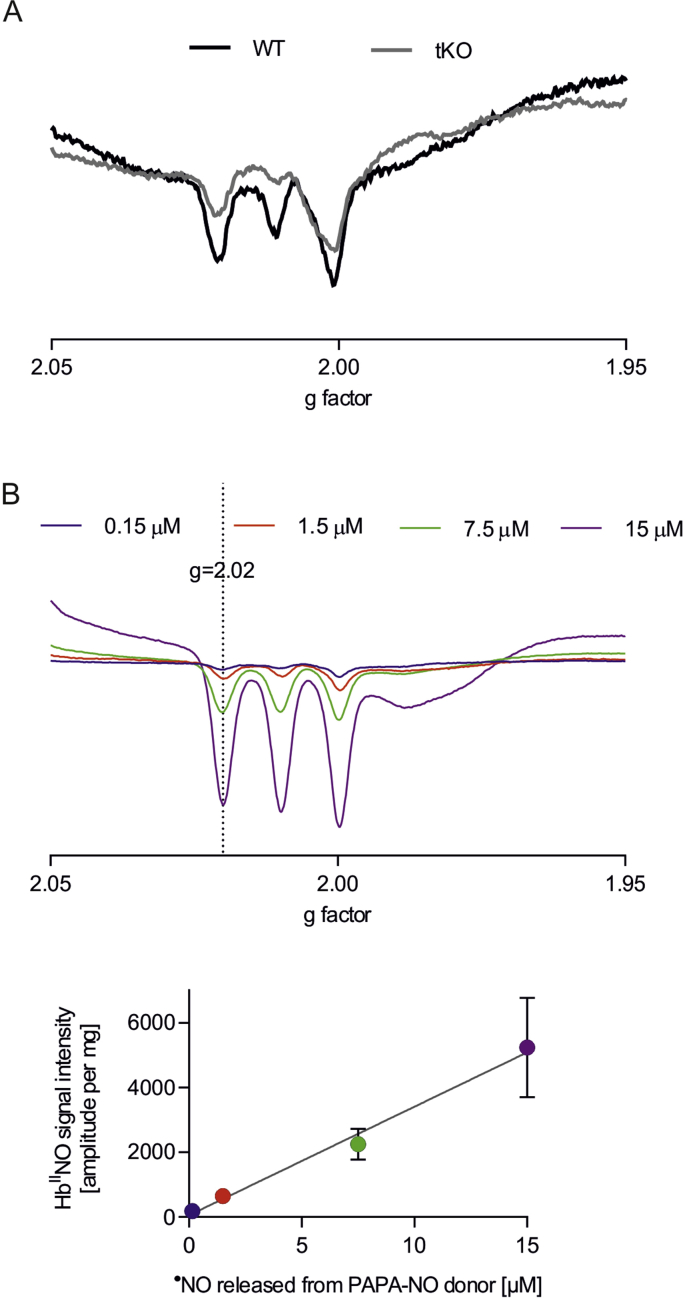
In comparison to WT, tKO aortas exhibited an increased level of S-nitrosation (Fig. 4A), together with the significant downregulation of genes Adh5 (Fig. 4B) and Txnrd1 (Fig. 4C), coding denitrosating enzymes GSNOR and TrxR1, respectively. Analysis of ˙NO metabolism showed lower nitric oxide functional release in the aortic wall upon stimulation with calcium ionophore (Figs. 4D and S6), decreased amount of nitrosylhemoglobin in erythrocytes (Figs. 4E and S7) together with unchanged nitrites (Fig. 4F), but raised nitrates, the products of nitrite oxidation (Fig. 4G), in plasma of Nrf2 tKO mice.
Fig. 4.
Nrf2 tKO aortas exhibit a markedly increased level of S-nitrosation.
(A) Analysis of protein S-nitrosation by immunofluorescent staining in aortas of WT and Nrf2 tKO mice. The biotin switch assay was used to label S-nitrosated protein. Detection was done with Alexa Fluor 568 conjugated streptavidin. Representative pictures. n = 5. Scale bar 175 μm.
(B–C) Expression of Adh5 (B) and Txnrd1 (C) in aortas from WT and Nrf2 tKO mice. Relative expression of genes was measured by real-time PCR. EEF2 served as a reference gene. ∗p < 0.05 vs WT. n = 8, two-tailed Student's t-test.
(D) Level of nitric oxide in aortas of WT and Nrf2 tKO mice. The aortas were stimulated with calcium ionophore for 90 min and NO level was assessed by EPR. ∗∗∗p < 0.001 vs DMSO. ##p < 0.01 vs WT, n = 5–7, two-way ANOVA + Bonferroni's.
(E) Level of nitrosated haemoglobin (HbNO) in the blood of WT and Nrf2 tKO mice assessed by EPR. ∗p < 0.05 vs WT, n = 7–8, two-tailed Student's t-test.
(F–G) Level of nitrite (F) and nitrate (G) in plasma of WT and Nrf2 tKO mice assessed by HPLC. ∗p < 0.05 vs WT, n = 8–10, two-tailed Student's t-test.
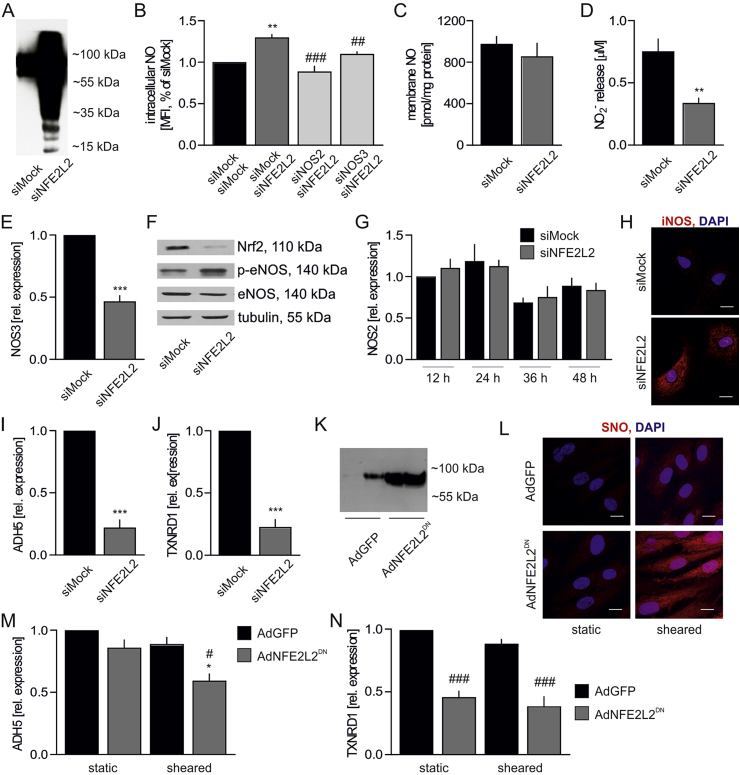
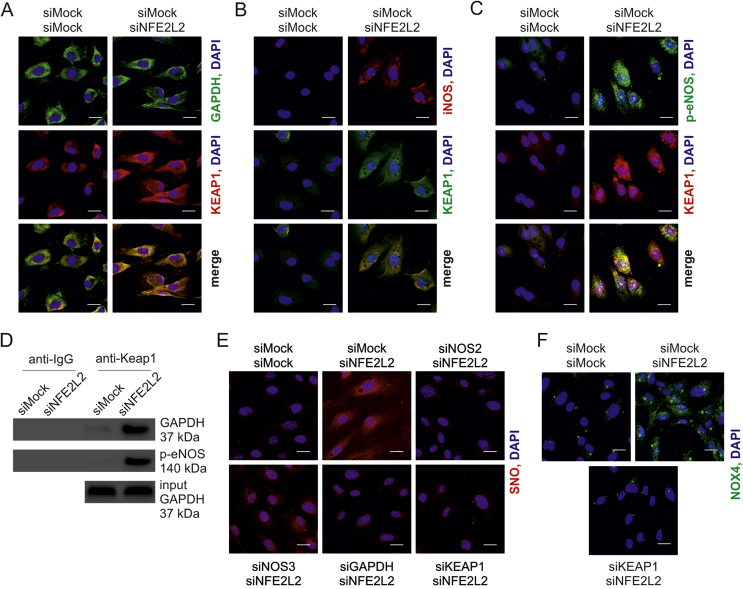
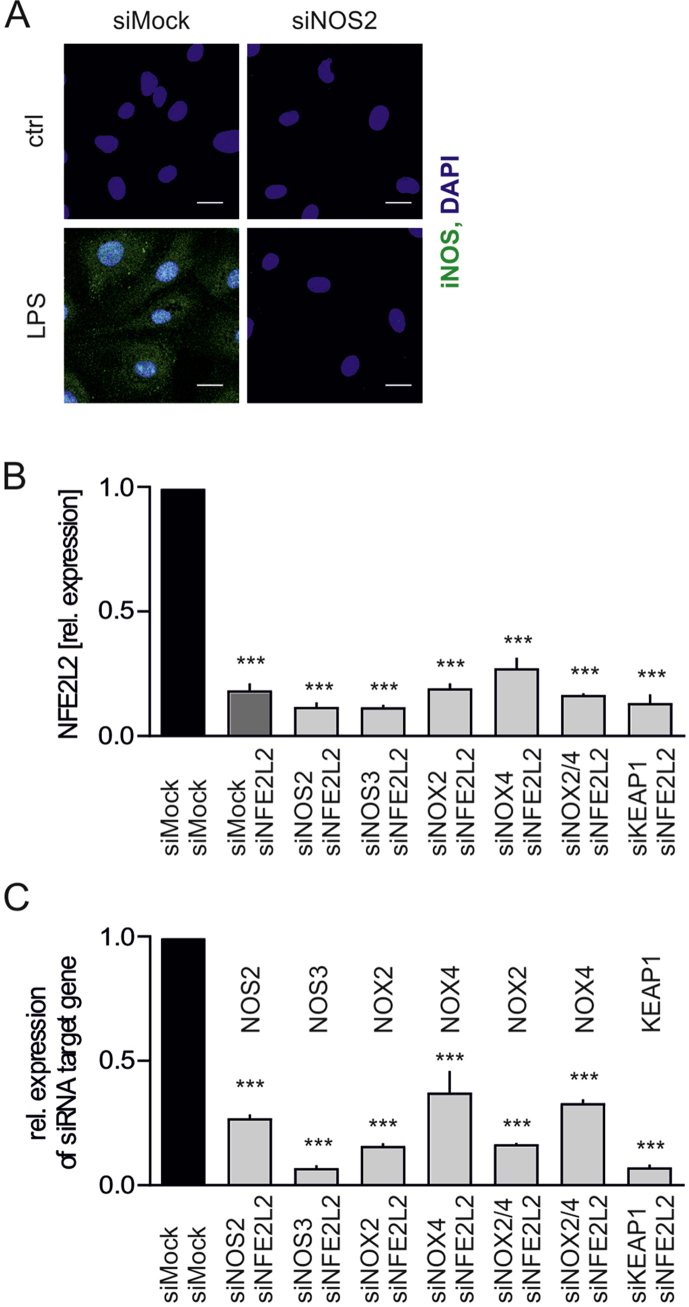
Likewise, silencing of NFE2L2 expression in HAECs resulted in a potent S-nitrosation of proteins (Fig. 5A). Nitrotyrosines, however, were undetectable, which excludes the presence of excessive nitrosative stress in these cells. The intracellular level of ˙NO, measured by DAF FM probe, was raised by ~30% in Nrf2-deficient cells due to eNOS and iNOS activity (Fig. 5B). Cellular ˙NO concentration, measured by EPR with hydrophobic spin trap DETC, was equal between compared groups (Fig. 5C), whereas the level of secreted nitrites, direct products of ˙NO oxidation, was lower in Nrf2-deficient cells (Fig. 5D). Although expression of NOS3, coding endothelial NOS (eNOS), decreased on the mRNA and protein level (Figs. 5E and F), eNOS activating phosphorylation of serine 1177 was strengthened (Fig. 5F). NOS2 gene expression, coding inducible NOS (iNOS) remained unchanged (Fig. 5G), but iNOS protein was strongly elevated in Nrf2-deficient cells (Fig. 5H). The specificity of anti-iNOS antibody was verified (Fig. S3A). Finally, a profound downregulation of ADH5 (Fig. 5I) and TXNRD1 (Fig. 5J) expression was observed in HAECs devoid of Nrf2.
Fig. 5.
Nrf2-dependent senescence onset is associated with potent protein S-nitrosation in HAECs.
(A) Analysis of protein S-nitrosation in siMock and siNFE2L2 transfected HAECs. IodoTMT was used to label S-nitrosated proteins, which were detected by immunoblotting. Representative pictures. n = 6.
(B) Intracellular NO level in siMock/siMock, siMock/siNFE2L2, siNOS2/siNFE2L2 and siNOS3/siNFE2L2 transfected HAECs measured by fluorescent probe DAF FM. n = 4, one-way ANOVA + Bonferroni's.
(C) Membrane trapped nitric oxide level in siMock and siNFE2L2 transfected HAECs assessed by EPR. n = 3.
(D) Concentration of nitrites secreted within 24 h into media by siMock and siNFE2L2 HAECs. The analysis was performed using Griess assay. ∗∗p < 0.01 vs siMock, n = 9, two-tailed Student's test.
(E) Expression of NOS3 in HAECs transfected with siMock or siNFE2L2. Relative expression of gene was measured by real-time PCR. EEF2 served as reference gene. ∗∗∗p < 0.001 vs siMock. n = 11–12, two-tailed Student's t-test.
(F) Level of total and phospho-Ser1177 eNOS in siMock and siNFE2L2 HAECs. Whole-cell lysates were analyzed by immunoblotting. Tubulin served as a loading control. Representative example of 3 experiments.
(G) Time course of the expression of NOS2 in HAECs transfected with siMock and siNFE2L2. Relative expression of genes was measured by real-time PCR. EEF2 served as a reference gene. n = 9.
(H) iNOS immunofluorescent staining in siMock and siNFE2L2 HAECs. Representative pictures of 3 experiments. Scale bar 10 μm.
(I–J) Expression of (I) ADH5 and (J) TXNRD1 in HAECs transfected with siMock and siNFE2L2. Relative expression of genes was measured by real-time PCR. EEF2 served as a reference gene. ∗∗∗p < 0.001 vs siMock, n = 6, two-tailed Student's t-test.
(K) Analysis of protein S-nitrosation in HAECs transduced with dominant negative NFE2L2 at MOI 50. IodoTMT was used to label S-nitrosated proteins, which were detected by immunoblotting. Representative picture of 3 experiments.
(L) Analysis of protein S-nitrosation by biotin switch assay with immunofluorescent detection in HAECs transduced with dominant negative NFE2L2 at MOI 50. Cells were subjected for 24 h to shear stress conditions. Representative picture of 3 experiments. Scale bar 10 μm.
(M–N) Expression of ADH5 (M) and TXNRD1 (N) in HAECs transduced with dominant negative NFE2L2 at MOI 50. Cells were subjected for 24 h to shear stress conditions. Relative expression of genes was measured by real-time PCR. EEF2 served as reference gene. ∗p < 0.001 vs static, #p < 0.05 vs AdGFP, ###p < 0.001 vs AdGFP, n = 3–15, two-way ANOVA + Bonferroni's.
Overexpression of NFE2L2DN in HAECs in static conditions only subtly increased protein S-nitrosation (Figs. 5K and L), which was accompanied by the unchanged level of ADH5 mRNA (Fig. 5M) and a significant decrease of TXNRD1 (Fig. 5N). Culture of HAECs in sheared conditions induced protein S-nitrosation both in GFP- and NFE2L2DN-transduced cells; however, the level of this modification was much stronger in AdNFE2L2DN HAECs (Fig. 5L). Increased S-nitrosation and SA-β-gal activity were seen in sheared AdNFE2L2DN in the entire well. Analysis of gene expression of denitrosating enzymes revealed that stress conditions downregulated ADH5 only in AdNFE2L2DN cells (Fig. 5M), while TXNRD1 mRNA level was lower in AdNFE2L2DN cells in comparison to AdGFP regardless of flow conditions (Fig. 5N).
Collectively, these results demonstrate the presence of exacerbated S-nitrosation in Nrf2-deficient HAECs, NFE2L2DN overexpressing HAECs subjected to orbital shear stress, and in Nrf2 tKO aortas when the onset of senescence is observed. Moreover, intensified S-nitrosation and senescence occur uniquely when the expression of both genes coding denitrosylating enzymes, ADH5 and TXNRD1, is downregulated. Therefore, taking the next step, we aimed to determine the functional significance of S-nitrosation in Nrf2-deficient ECs.
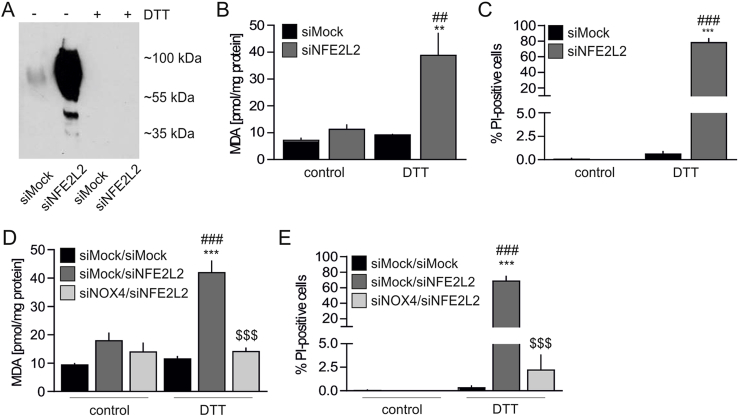
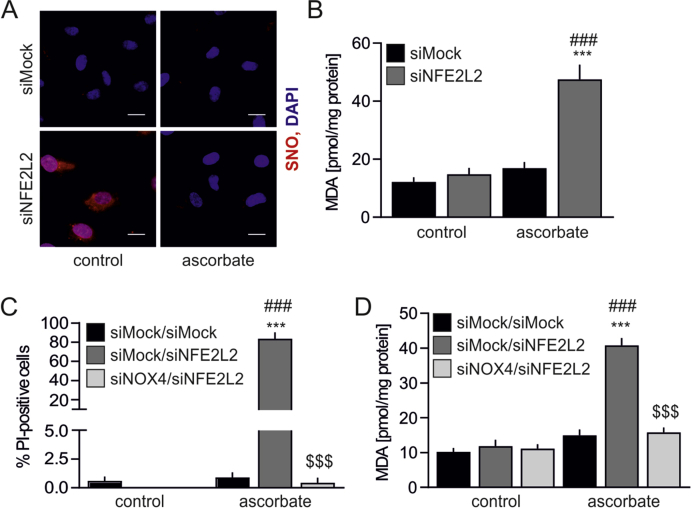
S-nitrosation provides protection from NOX4-mediated oxidative stress and death in Nrf2-deficient cells. The role of S-nitrosation in endothelial physiology remains elusive. It is an essential protein modification to maintain endothelial homeostasis [21]. On the other hand, it contributes to the impairment of various cellular processes, thus leading to senescence onset [42]. To address this issue, the cells were incubated with dithiothreitol (DTT), the reducing agent, which entirely abolished protein S-nitrosation (Fig. 6A). Such treatment led to oxidative damage assessed by an increase in TBARS (Fig. 6B) and sudden Nrf2 deficient cell death (Fig. 6C), which was not the case in siMock HAECs (Figs. 6B and C). This shows that S-nitrosation protects Nrf2-deficient ECs from oxidative injury and death.
Fig. 6.
S-nitrosation provides protection from NOX4-mediated oxidative stress and death in Nrf2-deficient HAECs.
(A) Analysis of protein S-nitrosation in siMock and siNFE2L2 transfected HAECs treated with DTT. Cells were treated with 5 mM DTT for 24 h. TMT was used to label S-nitrosated proteins detected by immunoblotting. Representative picture of 3 experiments.
(B) Measurement of TBARS in HAECs transfected with siMock or siNFE2L2 and treated with 5 mM DTT for 24 h. MDA was measured using thiobarbituric acid assay. n = 3, ∗∗p < 0.01 vs PBS, ##p < 0.01 vs siNFE2L2, two-way ANOVA + Bonferroni's.
(C) Detection of cell death with propidium iodide (PI) staining. Cells were transfected with siMock or siNFE2L2 and treated with 5 mM DTT for 24 h n = 5. ∗∗∗p < 0.001 vs PBS, ###p < 0.001 vs siNFE2L2, two-way ANOVA + Bonferroni's.
(D) Measurement of TBARS. Cells were cotransfected with siMock, siNFE2L2 or siNOX4 and treated with 5 mM DTT for 24 h. MDA was measured using thiobarbituric acid assay. ∗∗∗p < 0.001 vs control, ###p < 0.001 vs siMock/siMock, $$$p < 0.001 vs siMock/siNFE2L2, n = 3, two-way ANOVA + Bonferroni's.
(E) Detection of cell death with propidium iodide (PI) staining. Cells were cotransfected with siMock, siNFE2L2 or siNOX4 and treated with 5 mM DTT for 24 h ∗∗∗p < 0.001 vs control, ###p < 0.001 vs siMock/siMock, $$$p < 0.001 vs siMock/siNFE2L2, n = 5, two-way ANOVA + Bonferroni's.
In the pursuit of the possible mechanism of S-nitrosation based protection of ECs, mass spectrometry analysis was performed to identify modified proteins. Among 28 SNO-proteins, which appeared preferentially in siNFE2L2 HAECs (Table S2), NAPDH oxidase 4 (NOX4), one of the main sources of reactive oxygen species in the cells [43], was found. As S-nitrosation was shown to inhibit NOX4 activity [44], we hypothesized that it provides protection from oxidative damage in HAECs devoid of Nrf2. To address this point, concomitant silencing of NOX4 and NFE2L2 was performed (Figs. S3B and C). Inhibition of NOX4 expression fully prevented denitrosation-induced oxidative damage in HAECs cells lacking Nrf2 (Fig. 6D), which successfully hampered DTT-induced cell death (Fig. 6E). Similarly, stimulation of the cells with another reducing agent, sodium ascorbate, reversed S-nitrosation of proteins (Fig. S4A) and induced oxidative damage in siNFE2L2 HAECs (Fig. S4B), unless the cells were also devoid of NOX4 (Figs. S4C and D). Thus, we identify S-nitrosation of NOX4 as a mechanism for protection of Nrf2-deficient ECs from oxidative damage.
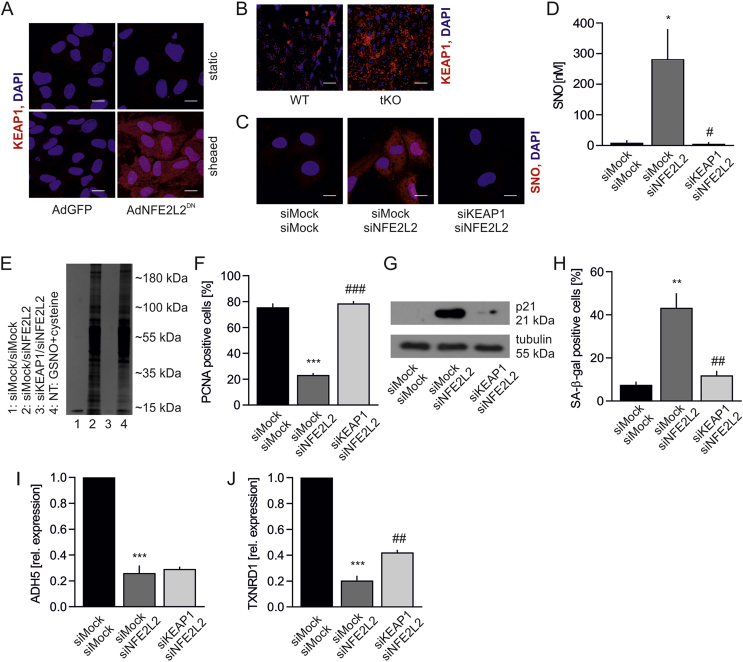
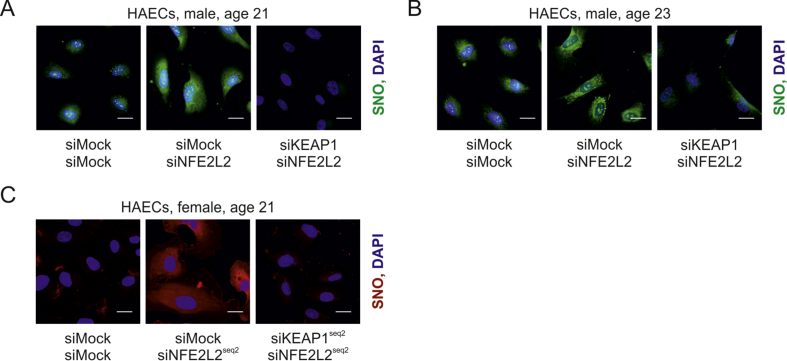
Unrestrained Keap1 triggers protein S-nitrosation and premature senescence in ECs. A prerequisite for the occurrence of increased S-nitrosation and senescence is a lack of Nrf2 protein in static conditions or a loss of Nrf2 transcriptional activity in shear stress conditions both in vitro and in vivo. Our group has recently shown that Keap1 has to be restrained by Nrf2 in ECs to prevent angiogenic dysfunction and that its level is increased, while p62, which Keap1 degradation depends on [45], declines in ECs devoid of Nrf2 [23]. On the other hand, it is known that exposure of ECs to oscillatory shear stress, which effects on Nrf2 are similar to orbital shear stress used in the study (Fig. S2), induces translocation of Nrf2 into the nucleus, although it remains transcriptionally inactive [37]. Such conditions allow Keap1 to function in a different manner than when it is involved in Nrf2 ubiquitination and degradation. Indeed, Keap1 level was increased in sheared HAECs overexpressing dominant-negative NFE2L2 (Fig. 7A) and also on the luminal surface of tKO aortas (Fig. 7B) in comparison to siMock transfected cells and WT mice, respectively. Therefore, we hypothesized that unrestrained and overabundant Keap1 is responsible for S-nitrosation and premature senescence of ECs. Concomitant silencing of NFE2L2 and KEAP1 in HAECs decreased protein S-nitrosation (Fig. 7C), which was confirmed by a direct measurement of SNO using quantitative chemiluminescence assay (Fig. 7D), and by detection of SNO-proteins pull downed with biotin switch assay. GSNO-treated sample served as a positive control (Fig. 7E). Moreover, knockdown of KEAP1 restored cell proliferation (Fig. 7F), lowered p21 level (Fig. 7G), and finally reversed senescent phenotype of Nrf2-deficient cells (Fig. 7H). Keap1-mediated S-nitrosation was also confirmed in two other batches of HAECs isolated from different donors (Figs. S5A and B) and using different siRNA sequences targeting NFE2L2 and KEAP1 (Fig. S5C). Interestingly, silencing of KEAP1 in Nrf2-deficient HAECs did not bring back expression of ADH5 (Fig. 7I), while restoration of TXNRD1 expression was statistically significant, though still low (Fig. 7J). Such a mild restoration of TXNRD1 expression is rather unlikely to mediate the reversal of S-nitrosation in siNFE2L2/siKEAP1 cells.
Fig. 7.
Keap1 controls protein S-nitrosation and senescent phenotype of HAECs.
(A) Keap1 immunofluorescent staining in AdGFP and AdNFE2L2DN HAECs. Cells were subjected for 24 h to shear stress conditions. Representative picture of 3 experiments. Scale bar 10 μm.
(B)En face immunofluorescent staining of WT and tKO aortas for Keap1. Representative picture of 5 experiments. Scale bar 15 μm.
(C) Analysis of protein S-nitrosation by biotin switch assay with immunofluorescent detection in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. Representative picture of 3 experiments. Scale bar 10 μm.
(D) Quantitative analysis of protein S-nitrosation by chemiluminescence in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. n = 3, ∗p < 0.05 vs siMock/siMock, #p < 0.05 vs siMock/siNFE2L2, one-way ANOVA + Bonferroni's.
(E) Analysis of protein S-nitrosation in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. GSNO + cysteine served as a positive control. Cell lysates were subjected to biotin switch assay, pull downed by streptavidin and separated by SDS-PAGE electrophoresis. Proteins were visualized by silver stain. n = 3. NT – non-transfected. Representative picture.
(F) Quantitative analysis of PCNA immunofluorescent staining in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. ∗∗∗p < 0.001 vs siMock/siMock, ###p < 0.001 vs siMock/siNFE2L2. n = 3, one-way ANOVA + Bonferroni's.
(G) p21 level in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. Whole-cell lysates were analyzed by immunoblotting. Tubulin served as a loading control. Representative picture of 3 experiments.
(H) Percentage of cells with increased SA-β-gal activity in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. ∗∗p < 0.01 vs siMock/siMock, ##p < 0.01 vs siMock/siNFE2L2. n = 3, one-way ANOVA + Bonferroni's.
(I–J) Expression of (I) ADH5 and (J) TXNRD1. ∗∗∗p < 0.001 vs siMock/siMock, ##p < 0.01 vs siMock/siNFE2L2. n = 3, one-way ANOVA + Bonferroni's.
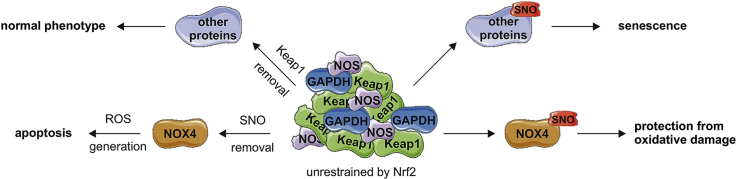
The SNO-proteins formation can be achieved by transnitrosation due to activity of proximal SNO-modified proteins – transnitrosases, which include hemoglobin, thioredoxin, caspase 3, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and protein disulphide isomerase (PDI) [46]. As NO synthase and GAPDH are a part of S-nitrosating enzymatic complex found in bacteria [20], we analyzed colocalization of active eNOS, iNOS and GAPDH with Keap1 in HAECs. GAPDH and iNOS strongly colocalized with Keap1 in siNFE2L2 cells (Figs. 8A and B), and Ser1177-phosphorylated eNOS/Keap1 colocalization was also found in these cells (Fig. 8C). In line with blotting analyses (Fig. 5F) and immunofluorescent staining (Fig. 5H), the levels of activated eNOS and total iNOS were increased in siNFE2L2 HAECs (Figs. 8B and C). GAPDH and phospho-eNOS precipitated with Keap1 (Fig. 8D). We were also trying to check if iNOS precipitates with Keap1, however the antibody against iNOS works well in our hands for immunofluorescence (Fig. S3A), but not for western blotting. Knockdown of every single protein of the complex Keap1/GAPDH/NOS inhibited S-nitrosation (Fig. 8E). The question remained, how denitrosation of proteins in Nrf2 deficient cells redirects them from senescence to apoptosis (Fig. 6), while silencing of Keap1, the protein responsible for the induction of S-nitrosation and senescence in these cells, restores their normal phenotype (Fig. 7). Such reliance implicates that NOX4 can be active only when it is denitrosated and Keap1 is present. As depicted in Fig. 8F, NOX4 protein is abolished in siNFE2L2/KEAP1 cells, which reconciles this discrepancy. Collectively, these data show that the overabundance of unrestrained Keap1 causes protein S-nitrosation. In addition, it determines normal, senescent or apoptotic fate of ECs.
Fig. 8.
Keap1 interacts with GAPDH and NOS in HAECs.
(A) Colocalization between GAPDH (green) and Keap1 (red) in siMock/siMock and siMock/siNFE2L2 HAECs. Representative pictures of 4 experiments. Scale bar 10 μm.
(B) Colocalization between iNOS (red) and Keap1 (green) in siMock/siMock and siMock/siNFE2L2 HAECs. Representative pictures of 3 experiments. Scale bar 10 μm.
(C) Colocalization between phosphorylated (Ser1177) eNOS (green) and Keap1 (red) in siMock/siMock and siMock/siNFE2L2 HAECs. Representative pictures of 3 experiments. Scale bar 10 μm.
(D) Co-immunoprecipitation of Keap1 with phosphorylated (Ser1177) eNOS and GAPDH in siMock/siMock and siMock/siNFE2L2 HAECs. Representative picture of 3 experiments.
(E) Analysis of protein S-nitrosation by biotin switch assay with immunofluorescent detection in siMock/siMock, siMock/siNFE2L2, siNOS2/siNFE2L2, siNOS3/siNFE2L2, siGAPDH/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. Representative picture of 3 experiments. Scale bar 10 μm
(F) NOX4 level in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs. Representative pictures of 3 experiments. Scale bar 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Studies of the past decade suggested that Nrf2 plays a protective role against senescence and aging [47]. These associations are consistent across species, including human, rat, mouse, Drosophila melanogaster and Caenorhabditis elegans (CncC and SKN-1 are Nrf2 homologs in flies and worms, respectively) [48]. Although in the literature no reports show complete loss of Nrf2 in humans, varying levels of its activity, especially in aging, is not without significance. Detrimental effects of lower Nrf2 activity are usually attributed to its influence on antioxidant mechanisms. In the light of our results, the role of Nrf2 in the regulation of Keap1 level and its activity should also be taken into account. Keap1 is degraded by autophagy [45] and one of the Nrf2 target genes is p62, [49] a cargo receptor for autophagosomes. In this manner, Nrf2 activity may affect Keap1 level. The activity of Nrf2 in humans is modulated in many ways. NFE2L2 gene is highly polymorphic, which is associated with risk and progression of numerous diseases including cardiovascular ones [48,50,51]. The electrophile response element (EpRE, also called ARE), a consensus binding sequence for Nrf2, can also be polymorphic. This polymorphism may play a role in the prevention of tauopathies [52,53]. Nrf2 level and its activity may decline with age, which was shown in various cell types, including vascular cells, cardiac cells, epithelial cells and fibroblasts. The reason for a declined Nrf2 signaling remains unidentified, though [48]. Moreover, Nrf2 was shown to be sequestered by progerin and thereby mislocalized subnuclearly, which impairs its transcriptional activity[14]. Finally, senescence-associated miRNA may repress Nrf2 in premature aging [54].
The molecular mechanisms of senescence related to the Nrf2 signaling pathway remains open to question, especially in respect to the cardiovascular system [48]. The overall components of vascular aging have been reviewed, indicating oxidative stress as the major reason for age-related vascular impairment [3]. Here, we present that, paradoxically, the senescent phenotype of endothelial cells and aortas with deregulated Nrf2 signaling is not accompanied by uncontrollable oxidative stress and damage, even though the steady state superoxide anion, a modulator of EC function [55], is slightly increased. In the case of ECs and aortas with Nrf2 impairment, an elevated level of oxidants, particularly H2O2 and other hydroperoxides, which are the predominant redox second messengers [56], can be responsible for the observed weak shift in the redox status of the cell towards more oxidative, but still unharmful, microenvironment. Such conditions may facilitate the covalent attachment of NO+ to cysteine, which requires oxidation of ˙NO in the reactions leading to S-nitrosation [57].
For now, we are limited by the general problem of not understanding how S-nitrosation occurs, though it is likely that Keap1 is a SNO synthase. Human Keap1 is a protein possessing 27 cysteines. How a cysteine of Keap1 differs from the many protein cysteines so that it can be oxidized by H2O2 under physiological conditions remains to be elucidated. Modification of Keap1 cysteines has been suggested to involve the binding of zinc, which would allow the splitting of the O–O bond that cannot be achieved by a thiolate alone [58]. The problem here is that ˙NO must be oxidized to NO+ to react with a thiolate (S−) to form SNO and Zn2+ cannot achieve that. It is possible that a transition metal like Fe3+ could bind to a cysteine in Keap1 in place of Zn2+ and oxidize ˙NO. Or as there was an increase in O2.-, the following could occur if the Keap1 molecule was close enough to the site of O2.- production:
| Keap-S- + O2.- - > Keap-S. + O2 |
| Keap-S. + ˙NO - > Keap-SNO |
where the presence of a Zn2+ bound to the thiolate would enhance its nucleophilicity and facilitate a reaction with O2.- to form a thiyl radical.
Increased S-nitrosation is considered as a fateful posttranslational modification due to its detrimental effect on various cellular processes such as autophagy [42], actin polymerization [59] and protein folding [60]. It is known that this modification may induce the senescent phenotype [42,59], which is also the case in our experimental models. In terms of protecting cells, S-nitrosation of caspases was shown to inhibit their enzymatic activity [61]. S-nitrosation may also have a defensive role as it prevents irreversible protein modifications similar to S-glutathionylation [62,63]. As our data show, S-nitrosation of NOX4 has fundamental significance in endothelial cells, protecting them from apoptosis in the absence of Nrf2, the key cellular redox regulator. Thereby, S-nitrosation determines EC fate, balancing between premature senescence and cellular death. We found that the protein, which is directly controlling S-nitrosation in ECs is Keap1, which broadens the function of Keap1 in the cell beyond the repressor of Nrf2 and redox sensor. We emphasize that the role of Nrf2/Keap1 system in the anti-oxidative defense of ECs is a twin-track approach: a regulation of cytoprotective genes by Nrf2, but also Keap1-mediated S-nitrosation of cysteines.
Until recently, the biology of the modification of macromolecules by ˙NO and the mechanisms associated with this process were poorly understood and thought to be non-enzymatic. As described above, this reaction would require formation of an intermediate NO+ or another nitrosating agent [64], but the kinetics of such mechanisms raises constraints in in vivo conditions [20,65]. Here, we propose that Keap1 creates together with NOS and GAPDH an enzymatic complex, similar to the one found in bacteria [20], which catalyzes the reaction of S-nitrosation in mammalian cells. Although verification of this concept requires further detailed studies, thought-provoking similarities can be found between the bacterial system regulating antioxidative response and S-nitrosation, and mammalian one. The complex of NO synthase (NarGHI), SNO synthase (Hcp) and transnitrosating protein (GAPDH) is responsible for protein S-nitrosation in E. coli [20] and this process is controlled by OxyR [66]. OxyR is the main transcription factor orchestrating the genes involved in a defense system in bacteria. It is a redox-sensitive regulator which is activated by a thiol-disulfide switch model [67]. Thiol groups of OxyR are modified by oxidation or S-nitrosation and turn on the regulons protective from oxidative and nitrosative stress, respectively. Hcp, bacterial SNO synthase, is one of the OxyR target genes and these two may interact [20,66]. Whereas OxyR is the oxidant sensor in bacteria, the Nrf2-Keap1 axis serves as such in mammalian cells [68]. Importantly, deletion of OxyR leads to potent protein S-nitrosation [66], the same effect is found in Nrf2-deficient ECs (Fig. 5A).
As it was previously demonstrated, S-nitrosation of Keap1 in ECs, like oxidative modification or alkylation, allows Nrf2 activation [69]. Here, we demonstrate that Keap1, with its interaction with NOS and GAPDH, resembles the bacterial SNO enzymatic complex, and provides a novel mechanism for NO-based cellular signaling in mammalian cells.
In conclusion, we show that Keap1 governs S-nitrosation in endothelial cells. This serves as an oxidative damage escaping mechanism relying on NOX4 in ECs, and redirects them from death to senescence. Hence, our data unveil a novel non-canonical role of Keap1 in endothelial cell biology.
Disclosures
None.
Acknowledgements
We thank Kamil Kus (Jagiellonian Centre for Experimental Therapeutics, Jagiellonian University) and Katarzyna Sikora (Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University) for technical help. This work was supported by the National Science Centre grants No. 2012/07/B/NZ3/02912 (AGP), and No. 2016/22/E/NZ3/00405 (AGP), and No. 2017/25/B/NZ3/00986 (AJ), as well as the Ministry of Science and Higher Education grant No. 0138/IP1/2015/73 (AGP). Faculty of Biochemistry, Biophysics and Biotechnology and Malopolska Centre of Biotechnology of the Jagiellonian University and Centre the Molecular Biophysics of CNRS in Orleans, France are supported by the International Associated Laboratory (LIA) grant from CNRS and Jagiellonian University. The equipment used was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. HJF is supported by grant ES023864 from the United States National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101304.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Mass spectrometry S-nitrosation profiling in siMock and siNFE2L2 HAECs. Qualitative analysis. n = 4. Mann-Whitney U test.
Sequences of primers used in qPCR.
Fig. S1.
Activity of β-galactosidase of different origin in lung fibroblasts and aortas isolated from WT and Nrf2 tKO newborns.
(A) Activity of β-gal coded by LacZ transgene in pH 7 in fibroblast lysates. n = 3.
(B) Activity of β-gal coded by LacZ transgene in pH 7 in fibroblasts in situ. n = 3. Scale bar = 50 μm.
(C) Activity of SA-β-gal in pH 6 in fibroblasts in situ. n = 3. Scale bar 50 μm.
(D) Activity of β-gal coded by LacZ transgene in pH 7 in aortic lysates. n = 3.
(E) Activity of β-gal coded by LacZ transgene in pH 7 in aortas in situ. n = 3. Scale bar = 4 mm.
Fig. S2.
Effect of orbital shear stress on Nrf2 nuclear localization and its activity in HAECs.
(A) Cellular localization of Nrf2 in AdGFP and AdNFE2L2DN HAECs. Nrf2 immunofluorescent staining. n = 3. Representative pictures. Scale bar 10 μm.
(B–C) Expression of Nrf2 target genes GCLM (B) and NQO1 (C). Relative expression of genes was measured by real-time PCR. EEF2 served as reference gene. n = 3. ###p < 0.001 vs siNFE2L2. Two-way ANOVA + Bonferroni's.
Fig. S3.
Specificity of anti-iNOS antibody and efficiency of target gene silencing by siRNA.
(A) iNOS immunofluorescent staining in HAECs transfected with siMock and siNOS2 and stimulated with LPS for 6 h. Scale bar 10 μm n = 3. Representative pictures.
(B) Expression of NFE2L2 in HAECs transfected with siMock/siMock, siMock/siNFE2L2, siNOS2/siNFE2L2, siNOS3/siNFE2L2, siNOX2/siNFE2L2, siNOX4/siNFE2L2, siNOX2/4/siNFE2L2, siKEAP1/siNFE2L2. Relative expression of gene was measured by real-time PCR. EEF2 served as reference gene. ***p < 0.001 vs siMock/siMock. n = 3–12, two-tailed Student's t-test.
(C) Expression of target genes in HAECs transfected with siMock/siMock, siNOS2/siNFE2L2, siNOS3/siNFE2L2, siNOX2/siNFE2L2, siNOX4/siNFE2L2, siNOX2/4/siNFE2L2, siKEAP1/siNFE2L2. Relative expression of gene was measured by real-time PCR. EEF2 served as reference gene. ***p < 0.001 vs siMock/siMock. n = 3, two-tailed Student's t-test.
Fig. S4.
S-nitrosation provides protection from NOX4-mediated oxidative stress and death in Nrf2-deficient cells.
(A) Analysis of protein S-nitrosation by biotin switch assay and immunofluorescent detection in siMock and siNFE2L2 transfected HAECs treated with 5 mM ascorbate for 24 h. Representative picture of 3 experiments. Scale bar 10 μm.
(B) Measurement of TBARS in HAECs transfected with siMock or siNFE2L2 and treated with 5 mM ascorbate for 24 h. MDA was measured using thiobarbituric acid assay. n = 3, ***p < 0.001 vs PBS, ###p < 0.001 vs siNFE2L2, two-way ANOVA + Bonferroni's.
(C) Detection of cell death with propidium iodide (PI) staining. Cells were cotransfected with siMock, siNFE2L2 or siNOX4 and treated with 5 mM ascorbate for 24 h ***p < 0.001 vs control, ###p < 0.001 vs siMock/siMock, $$$p < 0.001 vs siMock/siNFE2L2, n = 3, two-way ANOVA + Bonferroni's.
(D) Measurement of TBARS. Cells were cotransfected with siMock, siNFE2L2 or siNOX4 and treated with 5 mM ascorbate for 24 h. MDA was measured using thiobarbituric acid assay. ***p < 0.001 vs control, ###p < 0.001 vs siMock/siMock, $$$p < 0.001 vs siMock/siNFE2L2, n = 3, two-way ANOVA + Bonferroni's.
Fig. S5.
Analysis of protein S-nitrosation in other HAEC batches and with additional siRNA sequences.
(A–B) Analysis of protein S-nitrosation by biotin switch assay with immunofluorescent detection in siMock/siMock, siMock/siNFE2L2 and siKEAP1/siNFE2L2 HAECs isolated from (A) 21 years old male and (B) 23 years old male donors.
(C) Analysis of protein S-nitrosation by biotin switch assay with immunofluorescent detection in siMock/siMock, siMock/siNFE2L2seq2 and siKEAP1seq2/siNFE2L2seq2 HAECs isolated from 21 years old female donor. Representative picture of 3 experiments. Scale bar 10 μm.
Fig. S6.
NO EPR spectra of (A) representative samples and (B) standards.
Fig. S7.
HbNO EPR spectra of (A) representative samples and (B) standards.
References
- 1.Campisi J., Fagagna F d'Adda di. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 2.Nilsson P.M. Early vascular aging (EVA): consequences and prevention. Vasc. Health Risk Manag. 2008;4:547–552. doi: 10.2147/vhrm.s1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erusalimsky J.D. Vascular endothelial senescence: from mechanisms to pathophysiology. J Appl Physiol Bethesda Md 1985. 2009;106:326–332. doi: 10.1152/japplphysiol.91353.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zakkar M., Van der Heiden K., Luong L.A., Chaudhury H., Cuhlmann S., Hamdulay S.S., Krams R., Edirisinghe I., Rahman I., Carlsen H., Haskard D.O., Mason J.C., Evans P.C. Activation of Nrf2 in endothelial cells protects arteries from exhibiting a proinflammatory state. Arterioscler. Thromb. Vasc. Biol. 2009;29:1851–1857. doi: 10.1161/ATVBAHA.109.193375. [DOI] [PubMed] [Google Scholar]
- 5.Ungvari Z., Bailey-Downs L., Gautam T., Jimenez R., Losonczy G., Zhang C., Ballabh P., Recchia F.A., Wilkerson D.C., Sonntag W.E., Pearson K., Cabo R de, Csiszar A. Adaptive induction of NF-E2-related factor-2-driven antioxidant genes in endothelial cells in response to hyperglycemia. Am. J. Physiol. Heart Circ. Physiol. 2011;300:H1133–H1140. doi: 10.1152/ajpheart.00402.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jyrkkänen H.-K., Kansanen E., Inkala M., Kivelä A.M., Hurttila H., Heinonen S.E., Goldsteins G., Jauhiainen S., Tiainen S., Makkonen H., Oskolkova O., Afonyushkin T., Koistinaho J., Yamamoto M., Bochkov V.N., Ylä-Herttuala S., Levonen A.-L. Nrf2 regulates antioxidant gene expression evoked by oxidized phospholipids in endothelial cells and murine arteries in vivo. Circ. Res. 2008;103:e1–9. doi: 10.1161/CIRCRESAHA.108.176883. [DOI] [PubMed] [Google Scholar]
- 7.Ungvari Z., Tarantini S., Donato A.J., Galvan V., Csiszar A. Mechanisms of vascular aging. Circ. Res. 2018;123:849–867. doi: 10.1161/CIRCRESAHA.118.311378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Kong A.-N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umemura T., Kuroiwa Y., Kitamura Y., Ishii Y., Kanki K., Kodama Y., Itoh K., Yamamoto M., Nishikawa A., Hirose M. A crucial role of Nrf2 in in vivo defense against oxidative damage by an environmental pollutant, pentachlorophenol. Toxicol. Sci. 2006;90:111–119. doi: 10.1093/toxsci/kfj076. [DOI] [PubMed] [Google Scholar]
- 10.Aoki Y., Sato H., Nishimura N., Takahashi S., Itoh K., Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol. Appl. Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 11.Cho H.-Y., Jedlicka A.E., Reddy S.P.M., Kensler T.W., Yamamoto M., Zhang L.-Y., Kleeberger S.R. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 12.Merchant A.A., Singh A., Matsui W., Biswal S. The redox-sensitive transcription factor Nrf2 regulates murine hematopoietic stem cell survival independently of ROS levels. Blood. 2011;118:6572–6579. doi: 10.1182/blood-2011-05-355362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomobe K., Shinozuka T., Kuroiwa M., Nomura Y. Age-related changes of Nrf2 and phosphorylated GSK-3β in a mouse model of accelerated aging (SAMP8) Arch. Gerontol. Geriatr. 2012;54:e1–7. doi: 10.1016/j.archger.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Kubben N., Zhang W., Wang L., Voss T.C., Yang J., Qu J., Liu G.-H., Misteli T. Repression of the antioxidant NRF2 pathway in premature aging. Cell. 2016;165:1361–1374. doi: 10.1016/j.cell.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., Davies K.J.A., Forman H.J. Oxidative stress response and Nrf2 signaling in aging. Free Radic. Biol. Med. 2015;88:314–336. doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ungvari Z., Bailey-Downs L., Sosnowska D., Gautam T., Koncz P., Losonczy G., Ballabh P., Cabo R de, Sonntag W.E., Csiszar A. Vascular oxidative stress in aging: a homeostatic failure due to dysregulation of NRF2-mediated antioxidant response. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H363–H372. doi: 10.1152/ajpheart.01134.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold W.P., Mittal C.K., Katsuki S., Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3’:5’-cyclic monophosphate levels in various tissue preparations. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamler J.S., Simon D.I., Osborne J.A., Mullins M.E., Jaraki O., Michel T., Singel D.J., Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. U. S. A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Ruiz A., Araújo I.M., Izquierdo-Álvarez A., Hernansanz-Agustín P., Lamas S., Serrador J.M. Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxidants Redox Signal. 2013;19:1220–1235. doi: 10.1089/ars.2012.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seth D., Hess D.T., Hausladen A., Wang L., Wang Y.-J., Stamler J.S. A multiplex enzymatic machinery for cellular protein S-nitrosylation. Mol. Cell. 2018;69:451–464.e6. doi: 10.1016/j.molcel.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwakiri Y.S. Nitrosylation of proteins: a new insight into endothelial cell function regulated by eNOS-derived NO. Nitric oxide. Biol Chem Off J Nitric Oxide Soc. 2011;25:95–101. doi: 10.1016/j.niox.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benhar M., Forrester M.T., Stamler J.S. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 23.Kloska D., Kopacz A., Cysewski D., Aepfelbacher M., Dulak J., Jozkowicz A., Grochot-Przeczek A. Nrf2 sequesters Keap1 preventing podosome disassembly: a quintessential duet moonlights in endothelium. Antioxidants Redox Signal. 2019;30(14):1709–1730. doi: 10.1089/ars.2018.7505. [DOI] [PubMed] [Google Scholar]
- 24.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. An nrf2/small maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 25.Itoh K., Wakabayashi N., Katoh Y., Ishii T., O'Connor T., Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 26.Itoh K., Mimura J., Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxidants Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 27.Bell P., Limberis M., Gao G., Wu D., Bove M.S., Sanmiguel J.C., Wilson J.M. An optimized protocol for detection of E. coli beta-galactosidase in lung tissue following gene transfer. Histochem. Cell Biol. 2005;124:77–85. doi: 10.1007/s00418-005-0793-2. [DOI] [PubMed] [Google Scholar]
- 28.Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishima K., Handa J.T., Aotaki-Keen A., Lutty G.A., Morse L.S., Hjelmeland L.M. Senescence-associated beta-galactosidase histochemistry for the primate eye. Investig. Ophthalmol. Vis. Sci. 1999;40:1590–1593. [PubMed] [Google Scholar]
- 30.Dybas J., Berkowicz P., Proniewski B., Dziedzic-Kocurek K., Stanek J., Baranska M., Chlopicki S., Marzec K.M. Spectroscopy-based characterization of Hb-NO adducts in human red blood cells exposed to NO-donor and endothelium-derived NO. Analyst. 2018;143:4335–4346. doi: 10.1039/c8an00302e. [DOI] [PubMed] [Google Scholar]
- 31.Mannick J.B., Schonhoff C.M. Analysis of protein S-nitrosylation. Curr. Protein Pept. Sci. 2006;46(1):14.6.1–14.6.22. doi: 10.1002/0471140864.ps1406s46. [DOI] [PubMed] [Google Scholar]
- 32.Forrester M.T., Foster M.W., Benhar M., Stamler J.S. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic. Biol. Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogg N., Zielonka J., Kalyanaraman B. Chapter 3 - detection of nitric oxide and peroxynitrite in biological systems: a state-of-the-art review. In: Ignarro L.J., Freeman B.A., editors. Nitric Oxide. third ed. Academic Press; 2017. pp. 23–44. [Google Scholar]
- 34.Gladwin M.T., Wang X., Hogg N. Methodological vexation about thiol oxidation versus S-nitrosation -- a commentary on ‘An ascorbate-dependent artifact that interferes with the interpretation of the biotin-switch assay’. Free Radic. Biol. Med. 2006;41:557–561. doi: 10.1016/j.freeradbiomed.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Dardik A., Chen L., Frattini J., Asada H., Aziz F., Kudo F.A., Sumpio B.E. Differential effects of orbital and laminar shear stress on endothelial cells. J. Vasc. Surg. 2005;41:869–880. doi: 10.1016/j.jvs.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Nazari-Shafti T.Z., Cooke J.P. Telomerase therapy to reverse cardiovascular senescence. Methodist DeBakey Cardiovasc J. 2015;11:172–175. doi: 10.14797/mdcj-11-3-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosoya T., Maruyama A., Kang M.-I., Kawatani Y., Shibata T., Uchida K., Warabi E., Noguchi N., Itoh K., Yamamoto M. Differential responses of the Nrf2-Keap1 system to laminar and oscillatory shear stresses in endothelial cells. J. Biol. Chem. 2005;280:27244–27250. doi: 10.1074/jbc.M502551200. [DOI] [PubMed] [Google Scholar]
- 38.Przyborowski K., Wojewoda M., Sitek B., Zakrzewska A., Kij A., Wandzel K., Zoladz J.A., Chlopicki S. Effects of 1-methylnicotinamide (MNA) on exercise capacity and endothelial response in diabetic mice. PLoS One. 2015;10 doi: 10.1371/journal.pone.0130908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Csiszar A., Pacher P., Kaley G., Ungvari Z. Role of oxidative and nitrosative stress, longevity genes and poly(ADP-ribose) polymerase in cardiovascular dysfunction associated with aging. Curr. Vasc. Pharmacol. 2005;3:285–291. doi: 10.2174/1570161054368616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brodsky S.V., Gealekman O., Chen J., Zhang F., Togashi N., Crabtree M., Gross S.S., Nasjletti A., Goligorsky M.S. Prevention and reversal of premature endothelial cell senescence and vasculopathy in obesity-induced diabetes by ebselen. Circ. Res. 2004;94:377–384. doi: 10.1161/01.RES.0000111802.09964.EF. [DOI] [PubMed] [Google Scholar]
- 41.Lamoke F., Shaw S., Yuan J., Ananth S., Duncan M., Martin P., Bartoli M. Increased oxidative and nitrative stress accelerates aging of the retinal vasculature in the diabetic retina. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montagna C., Rizza S., Maiani E., Piredda L., Filomeni G., Cecconi F. To eat, or NOt to eat: S-nitrosylation signaling in autophagy. FEBS J. 2016;283:3857–3869. doi: 10.1111/febs.13736. [DOI] [PubMed] [Google Scholar]
- 43.Ago T., Kitazono T., Ooboshi H., Iyama T., Han Y.H., Takada J., Wakisaka M., Ibayashi S., Utsumi H., Iida M. Nox4 as the major catalytic component of an endothelial NAD(P)H oxidase. Circulation. 2004;109:227–233. doi: 10.1161/01.CIR.0000105680.92873.70. [DOI] [PubMed] [Google Scholar]
- 44.Selemidis S., Dusting G.J., Peshavariya H., Kemp-Harper B.K., Drummond G.R. Nitric oxide suppresses NADPH oxidase-dependent superoxide production by S-nitrosylation in human endothelial cells. Cardiovasc. Res. 2007;75:349–358. doi: 10.1016/j.cardiores.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 45.Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evangelista A.M., Kohr M.J., Murphy E., S-Nitrosylation Specificity, occupancy, and interaction with other post-translational modifications. Antioxidants Redox Signal. 2013;19:1209–1219. doi: 10.1089/ars.2012.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swamy S.M., Rajasekaran N.S., Thannickal V.J. Nuclear factor-erythroid-2-related factor 2 in aging and lung fibrosis. Am. J. Pathol. 2016;186:1712–1723. doi: 10.1016/j.ajpath.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kloska D., Kopacz A., Piechota-Polanczyk A., Nowak W., Dulak J., Jozkowicz A., Grochot-Przeczek A. Nrf2 in aging - focus on the cardiovascular system. Vasc. Pharmacol. 2019;112:42–53. doi: 10.1016/j.vph.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A., McMahon M., Hayes J.D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho H.-Y., Kwak M.-K., Pi J. Nrf2 in host defense: over the rainbow. Oxid Med Cell Longev. 2013;2013:975839. doi: 10.1155/2013/975839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuadrado A., Manda G., Hassan A., Alcaraz M.J., Barbas C., Daiber A., Ghezzi P., León R., López M.G., Oliva B., Pajares M., Rojo A.I., Robledinos-Antón N., Valverde A.M., Guney E., Schmidt H.H.H.W. Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacol. Rev. 2018;70:348–383. doi: 10.1124/pr.117.014753. [DOI] [PubMed] [Google Scholar]
- 52.Wang X., Campbell M.R., Lacher S.E., Cho H.-Y., Wan M., Crowl C.L., Chorley B.N., Bond G.L., Kleeberger S.R., Slattery M., Bell D.A. A polymorphic antioxidant response element links NRF2/sMAF binding to enhanced MAPT expression and reduced risk of parkinsonian disorders. Cell Rep. 2016;15:830–842. doi: 10.1016/j.celrep.2016.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Tomso D.J., Chorley B.N., Cho H.-Y., Cheung V.G., Kleeberger S.R., Bell D.A. Identification of polymorphic antioxidant response elements (AREs) in the human genome. Hum. Mol. Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuosmanen S.M., Sihvola V., Kansanen E., Kaikkonen M.U., Levonen A.-L. MicroRNAs mediate the senescence-associated decline of NRF2 in endothelial cells. Redox Biol. 2018;18:77–83. doi: 10.1016/j.redox.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai H. Hydrogen peroxide regulation of endothelial function: origins, mechanisms, and consequences. Cardiovasc. Res. 2005;68:26–36. doi: 10.1016/j.cardiores.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 56.Forman H.J., Ursini F., Maiorino M. An overview of mechanisms of redox signaling. J. Mol. Cell. Cardiol. 2014;73:2–9. doi: 10.1016/j.yjmcc.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Giacomo G., Rizza S., Montagna C., Filomeni G. Established principles and emerging concepts on the interplay between mitochondrial physiology and S-(De)nitrosylation: implications in cancer and neurodegeneration. Int J Cell Biol. 2012;2012:361872. doi: 10.1155/2012/361872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forman H.J., Davies M.J., Krämer A.C., Miotto G., Zaccarin M., Zhang H., Ursini F. Protein cysteine oxidation in redox signaling: caveats on sulfenic acid detection and quantification. Arch. Biochem. Biophys. 2017;617:26–37. doi: 10.1016/j.abb.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.García-Ortiz A., Martín-Cofreces N.B., Ibiza S., Ortega Á., Izquierdo-Álvarez A., Trullo A., Victor V.M., Calvo E., Sot B., Martínez-Ruiz A., Vázquez J., Sánchez-Madrid F., Serrador J.M. eNOS S-nitrosylates β-actin on Cys374 and regulates PKC-θ at the immune synapse by impairing actin binding to profilin-1. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura T., Lipton S.A. Emerging roles of S-nitrosylation in protein misfolding and neurodegenerative diseases. Antioxidants Redox Signal. 2008;10:87–101. doi: 10.1089/ars.2007.1858. [DOI] [PubMed] [Google Scholar]
- 61.Melino G., Bernassola F., Knight R.A., Corasaniti M.T., Nistico G., Finazzi-Agro A. S-nitrosylation regulates apoptosis. Nature. 1997;388:432–433. doi: 10.1038/41237. [DOI] [PubMed] [Google Scholar]
- 62.Chen Y.-Y., Chu H.-M., Pan K.-T., Teng C.-H., Wang D.-L., Wang A.H.-J., Khoo K.-H., Meng T.-C. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun J., Steenbergen C., Murphy E., S-nitrosylation NO-related redox signaling to protect against oxidative stress. Antioxidants Redox Signal. 2006;8:1693–1705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kovacs I., Lindermayr C. Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation. Front. Plant Sci. 2013;4 doi: 10.3389/fpls.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nedospasov A., Rafikov R., Beda N., Nudler E. An autocatalytic mechanism of protein nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13543–13548. doi: 10.1073/pnas.250398197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seth D., Hausladen A., Wang Y.-J., Stamler J.S. Endogenous protein S-Nitrosylation in E. coli: regulation by OxyR. Science. 2012;336:470–473. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hillion M., Antelmann H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015;396:415–444. doi: 10.1515/hsz-2015-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D'Autréaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 69.Um H.-C., Jang J.-H., Kim D.-H., Lee C., Surh Y.-J. Nitric oxide activates Nrf2 through S-nitrosylation of Keap1 in PC12 cells. Nitric Oxide Biol Chem. 2011;25:161–168. doi: 10.1016/j.niox.2011.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectrometry S-nitrosation profiling in siMock and siNFE2L2 HAECs. Qualitative analysis. n = 4. Mann-Whitney U test.
Sequences of primers used in qPCR.