Abstract
Aberrant expression of miRNAs has been reported to be involved in the development and progression of glioma. But the function of miR-876-3p in glioma is unknown. We found that miR-876-3p is significantly downregulated in glioma tissues and cell lines. Overexpression of miR-876-3p suppressed glioma cell proliferation, epithelial-mesenchymal transition, migration, and invasion. By prediction combining with luciferase reporter assay, we identified that miR-876-3p could decrease the expression of KIF20A by directly targeting the region of its 3’UTR. Furthermore, we observed that overexpression of miR-876-3p inhibited the expression of KIF20A, thus blocking the protein kinase JAK2/STAT3 pathway. Overexpressed KIF20A reversed miR-876-3p-induced suppression of glioma cell proliferation, migration, and invasion. We also demonstrated the inhibitory effect of miR-876-3p on tumor growth in glioma using an in vivo model. The miR-876-3p/KIF20A-axis mediated JAK2/STAT3 pathway have therapeutic potential in glioma treatment.
Keywords: Glioma, miR-876-3p, proliferation, EMT, KIF20A
Introduction
MiRNAs are a class of short single-strand non-coding RNAs, which length are about 19-25 nucleotides [1-3]. MiRNAs negatively regulate gene expression through gene translational repression by binding to the 3’ untranslated region (UTR) [4,5]. The aberrantly expression of miRNAs occur in various types of human cancers, which can act as tumor suppressors or promoters [6,7].
Kinesin family member 20A (KIF20A) is a member of the kinesin super family-6 [8,9]. Previous studies showed that KIF20A could play a important role in the development and progression of human cancers, including gastric [10], pancreatic cancer [11,12], breast cancer [13] and hepatocellular carcinoma [8]. In addition, KIF20A has been reported to be involved in proliferation, migration, invasion and angiogenesis [11,14]. Furthermore, KIF20A is a tumor-associated antigen involved in the glioma cell growth and cell survival [15,16].
Here, we determine whether miR-876-3p directly represses KIF20A expression, which leads to the subsequent inhibition of glioma cell proliferation, epithelial-mesenchymal transition (EMT) and in vivo tumor growth. In an exploration of the mechanism of miR-876-3p action, we demonstrate that dysregulation of miR-876-3p/KIF20A signalling activates the JAK2/STAT3 pathway.
Materials and methods
Cell lines and human tissue samples
Human glioma cell lines (U251, T98, U87, and LN229) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Normal human astrocytes (NHAs) were obtained from Lonza (Walkersville, MD, USA). Glioma cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% of fetal bovine serum (FBS) in a humidified incubator at 37°C and 5% CO2. NHAs were grown in the astrocyte growth media supplemented with rhEGF, insulin, L-glutamine, GA-1000, ascorbic acid and 5% FBS. MiRNA expression data was obtained from Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/portal.php). Gene expression data was obtained from Cancer Genome Atlas (TCGA) database (http://tcga-data.nci.nih.gov). Nontumorous brain tissues (NBTs) and glioma tissues were obtained from the Department of Neurosurgery, Huai’an Hospital Affiliated to Xuzhou Medical University, Second People’s Hospital of Huai’an City. Of these 30 samples, 6 were NBTs, 12 were low-grade glioma samples (grade II, LGGs) and 12 were high-grade glioma samples (grade III and IV, HGGs). Written informed consent was obtained from all the patients.
RNA isolation and quantitative real-time PCR (qRT-PCR)
qRT-PCR were performed using Mir-XTM miRNA qRT-PCR SYBR® Kit (Takara) for miR-876-3p. The expression of U6 was used as an endogenous control. qRT-PCR was performed using TB GreenTM Fast qPCR Mix (Takara) for KIF20A. The expression of GAPDH was used as an internal control.
Cell transfection
Lentiviruses carrying hsa-miR-876-3p or its negative control were purchased from Genepharma (Shanghai, China). For over-expression of KIF20A, cells were transfected with KIF20A over-expressing recombinant vector (GenePharma, Shanghai, China). Stable cell lines were established by infecting U251 and T98 cells, which were collected for the experiments after 48 h of transfection.
Cell Counting Kit-8 assay
Cells (2 × 103) were seeded to 96-well plate. After incubation for 0, 1, 2, 3 and 4 days, 10 μL of CCK8 reagent was added to each well and cells were cultured for 4 h at 37°C. Then, cell proliferation was quantified by measuring the absorbance at 450 nm.
Colony information assay
Cells transfected with miR-876-3p or its negative control (500/well) were planted in a 6-well plate for about 2 weeks. Then, cells were fixed with ethanol and stained with crystal violet. Colonies with ≥ 50 cells were calculated as one colony.
Wound-healing assay
Cells transfected with the miR-876-3p or its negative control were seeded in 6-well plates. A linear wound was created with a plastic pipette tip. Cells were then cultured in serum-free medium and photographed under a microscope at oh and 48 h.
Transwell invasion assay
2 × 105 cells were seeded in the upper chambers of 24-well Transwell plates precoated with 10 μg of Matrigel (BD Biosciences, Franklin, NJ, USA). DMEM containing 10% FBS was added to the lower chamber. After incubation for 36 h, the cells on the upper chambers were removed. Invading cells on the bottom were fixed and stained with 1% crystal violet and photographed with microscope.
Three-dimensional spheroid assay
Cells were seeded at a 3 × 103 cells/ml density in 96-well ultra-low adherence plates. After 48 hours, these cells were induced to aggregate into a multicellular spheroid, and then matrigel was added into wells. After 48 h, motion of cells was confirmed as fully formed with microscopy.
Dual luciferase reporter assay
The wild-type (wt) 3’-UTR of the KIF20A sequence containing a binding site for miR-876-3p was amplified from a human cDNA library and cloned into the downstream region of the luciferase gene. The mutant (mt) 3’-UTR of KIF20A was created by mutating the seed region of the miR-876-3p-binding site. For reporter assay, cells were co-transfected with miR-876-3p and wild-type or mutant 3’UTR of KIF20A by Lipofectamine 3000. Renilla luciferase served for normalization.
Western blotting assay
Western blot assay was performed as described previously [17]. Antibodies against KIF20A (ab104118), Cyclin D1 (ab16663), CDK2 (ab32147), CDK4 (ab108357), Vimentin (ab195877), Fibronectin (ab2413), E-cadherin (ab40772), N-cadherin (ab18203), STAT3 (ab68153), p-STAT3 (ab76315), Actin (ab8227, Abcam); JAK2 (D2E12), and p-JAK2 (D15E2, Cell Signaling Technology) were used for western blotting assay.
Xenograft assay
U251 (1 × 106) cells stable expressing miR-876-3p or its negative control were subcutaneously injected into the flanks of nude mice of 6 weeks. Tumors volume were examined every 5 days. At 33 days after the injection, mice were killed. Tumor volume (mm3) = 1/2 × (length × width2). The study was approved by the Animal Care and Use Ethics Committee of Department of Neurosurgery, Huai’an Hospital Affiliated to Xuzhou Medical University, Second People’s Hospital of Huai’an City.
Statistical analysis
The results are presented as the means ± SEM. All the statistical analyses were conducted in the GraphPad Prism 5 software. The significance of the differences in variables were calculated using Student’s t test. P values < 0.05 were considered significant.
Results
Expression levels of miR-876-3p in human glioma tissues and cell lines
To investigate miR-876-3p expression in glioma, we initially analyzed its expression by database of CGGA (Grade II: 63; Grade III: 44 and Grade IV: 91). The expression of miR-876-3p was higher in low grade glioma tissues (grade II, LGGs) than that in high grade glioma tissues (grades III and IV, HGGs) (Figure 1A). Secondly, we detected miR-876-3p expression in 30 samples, including 6 NBTs, 12 LGG cases and 12 HGG cases by qRT-PCR. The expression of miR-876-3p was decreased observed in LGGs and HGGs compared with nontumorous brain tissues (NBTs) (Figure 1B). Thirdly, we investigated the expression of miR-876-3p in glioma cell lines (U251, T98, U87, and LN229) and normal human astrocytes (NHAs). Compared with NHAs, miR-876-3p was downregulated in glioma cells (Figure 1C). Then we chose a representative glioma specimen and NBTs for FISH analysis. Compared with NBTs, miR-876-3p was downregulated in glioma (Figure 1D).
Figure 1.
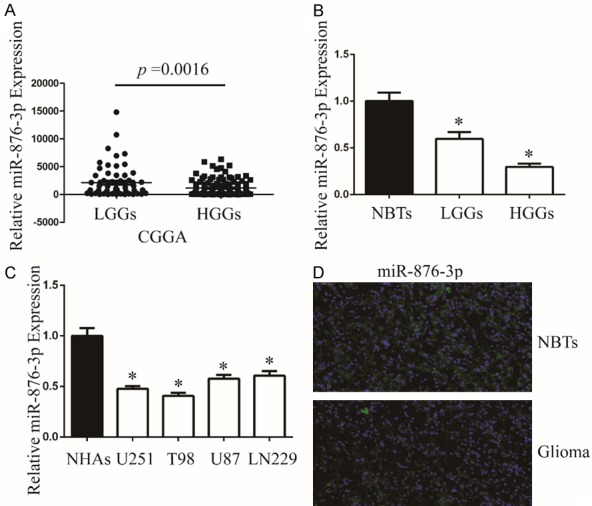
miR-876-3p is downregulated in glioma. A. The expression of miR-876-3p in LGGs and HGGs. *P < 0.05. B. The expression of miR-876-3p in clinical samples (NBTs: 6, LGGs: 12, and HGGs: 12). Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. C. qRT-PCR analysis of miR-876-3p expression in NHAs and glioma cell lines U251, T98, U87, and LN229. Transcript levels were normalized by U6 expression. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. D. The expression of miR-876-3p in NBTs and glioma specimens was assessed by fluorescence in situ hybridization.
MiR-876-3p suppresses glioma cell proliferation
To evaluate the effect of miR-876-3p on malignant biological behaviors in glioma, we stable transfected miR-876-3p or its negative control into glioma cells. The transfection efficiency was assessed by qRT-PCR (Figure 2A). CCK-8, colony formation assays were determined to exam the effects of miR-876-3p on glioma proliferation. Our results showed that overexpression of miR-876-3p significantly impaired the proliferation of both U251 and T98 cells compared its control (Figure 2B-D). To further understand the differences in cell proliferation, flow cytometry was performed to determine the cell cycle distribution. The results showed that overexpression of miR-876-3p cells resulted in G1 arrest (Figure 2E-G). To investigate the molecules of the cell cycle arrest, we measured the expression of cyclin-dependent kinases (CDKs; CDK2 and CDK4) and Cyclin D1 (CCND1) in cells transfected with miR-876-3p or its negative control by western blot (Figures 2H and S1).
Figure 2.
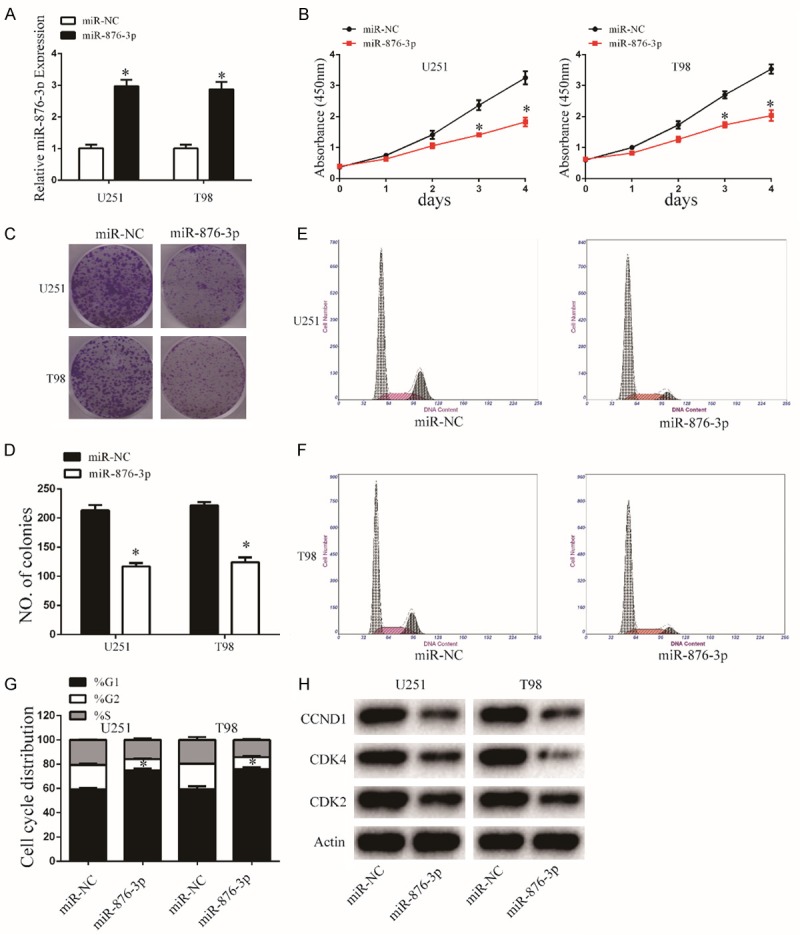
Overexpression of miR-876-3p inhibits glioma cell proliferation. A. The expression level of miR-876-3p in U251 and T98 cells was analyzed by qRT-PCR after transfection. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. B. Overexpression of miR-876-3p on the growth of U251 and T98 cells was examined by CCK8 assays. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. C and D. Cell viability of miR-876-3p was evaluated using the colony formation assay. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. E-G. Overexpression of miR-876-3p effect on cell cycle distribution of U251 and T98 cells. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. H. Western blot analysis indicated the regulation of the cell-cycle-regulatory proteins CDK2, CDK4, and CCND1 protein levels in U251 and T98 cells after stable transfection with miR-876-3p and miR-NC. Actin was used as control.
MiR-876-3p suppresses glioma cell migration, invasion, and EMT
The wound-healing assay showed that miR-876-3p overexpression suppressed cell migration of glioma cell lines (Figure 3A and 3C). Similarly, the invasion ability of glioma cell significantly decreased after tranfected with miR-876-3p in the Transwell and Matrigel-based 3D invasion assays (Figure 3B, 3D and 3E). Next, we investigated the effect of miR-876-3p overexpression on EMT in glioma cells. Expression levels of the mesenchymal markers Vimentin, Fibronectin, and N-cadherin were downregulated, while those of E-cadherin were upregulated in miR-876-3p-overexpressing U251 or T98 cells (Figures 3F and S2). Thus, overexpression of miR-876-3p suppresses glioma cell migration and invasion.
Figure 3.
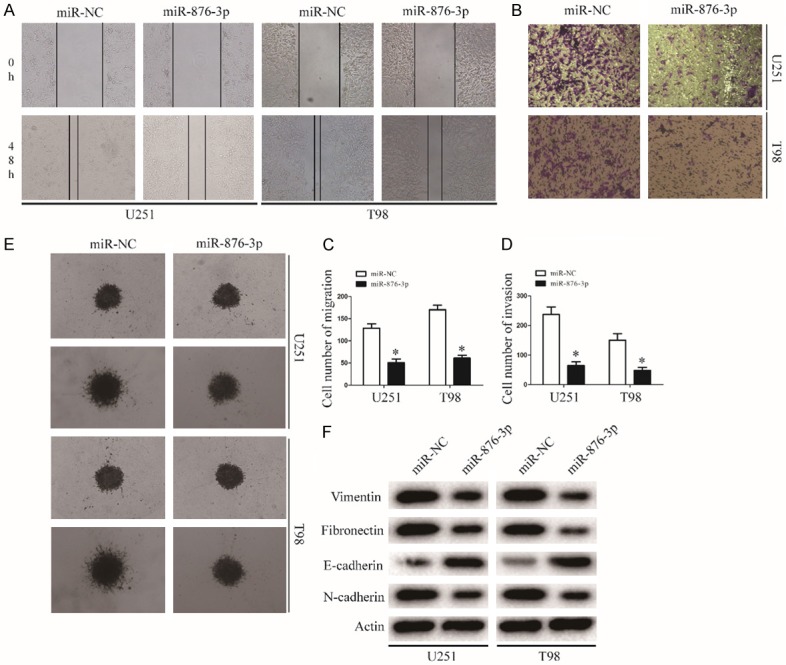
miR-876-3p overexpression blocks invasion and migration of glioma. A and C. Migration of glioma cells was monitored by a wound-healing assay. Representative images of glioma cells transfected with miR-NC or miR-876-3p. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. B and D. Migration of glioma cells was monitored by a Matrigel invasion assay. Representative images of glioma cells transfected with miR-NC or miR-876-3p. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. E. Results of glioma cell invasion were validated by a 3D spheroid migration assay. F. EMT-associated proteins in miR-NC or miR-876-3p transfected U251 or T98 cells were determined by western blotting. Actin was used as the loading control.
KIF20A was a direct target of miR-876-3p
To further investigate the mechanism of miR-876-3p in the proliferation, migration, and invasion of glioma, the TargetScan and miRanda softwares were used to identify potential targets of miR-876-3p. KIF20A was identified as a potential effector of miR-876-3p (Figure 4A). To test whether miR-876-3p directly targets KIF20A, we inserted the target region sequence of the KIF20A 3’-UTR (WT) or the mutated sequence (MUT) into a luciferase reporter vector. As shown in Figure 4B, transfected miR-876-3p resulted in a decrease in the luciferase activity of the construct containing the wt 3’-UTR of KIF20A but had no effect on the mt 3’-UTR reporter. MiR-876-3p overexpression significantly decreased the expression of KIF20A in protein level, thus blocking the protein kinase JAK2/STAT3 pathway (Figures 4E and S3B). Additionally, we examined KIF20A mRNA level in TCGA database. The mRNA level of KIF20A in glioma tissues (GLIMAs) were upregulated compared with those in nontumorous brain tissues (NBTs) (Figure 4C). We examined the protein expression of KIF20A in 24 GLIMAs and 6 NBTs. We found that protein expression of KIF20A was higher expressed in GLIMAs than NBTs (Figures 4D and S3A).
Figure 4.
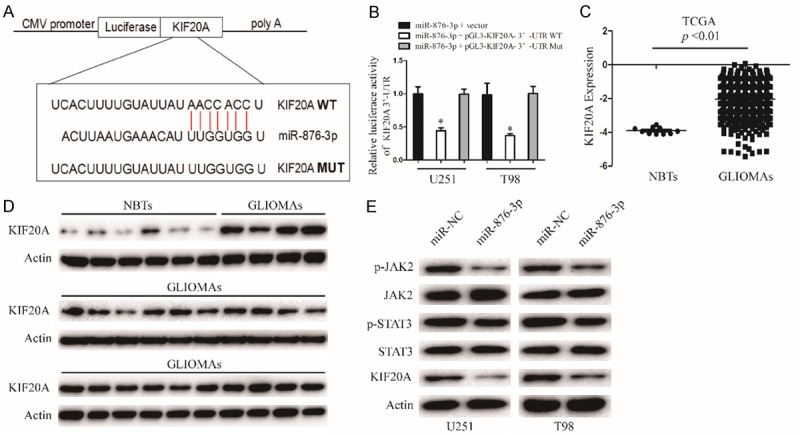
KIF20A is direct target of miR-876-3p in glioma cell. A. The miR-876-3p binding sites in the 3’UTR of KIF20A with the wild type (WT) and mutated (MUT) sequences. B. U251 and T98 cells were co-transfected with miR-876-3p and luciferase reporter constructs containing either pGL3-miR-876-3p-3’-UTR-WT or pGL3-miR-876-3p-3’-UTR-MUT. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05. C. TCGA database showing KIF20A expression in NBTs and GLIOMAs. D. The protein levels of KIF20A in NBTS and glioma specimens were determined by western blotting. E. Western blot analysis of KIF20A, JAK2, p-JAK2, STAT3, and p-STAT3 protein levels in U251 and T98 cells after transfection with miR-NC or miR-876-3p.
KIF20A attenuates the effects of miR-876-3p on glioma cell proliferation, migration, and invasion
To further determine whether miR-876-3p functionally targets KIF20A, we transfected KIF20A into miR-876-3p overexpressing glioma cells. The expression levels of p-JAK2 and p-STAT3 recovered after exogenous introduction of KIF20A (Figures 5A and S4). Furthermore, transfected with KIF20A significantly attenuated the miR-876-3p-induced suppression of glioma cell proliferation, migration, and invasion (Figure 5B-G). These results indicate that KIF20A was a direct target gene through which miR-876-3p suppressed the migration and invasion of glioma cells.
Figure 5.
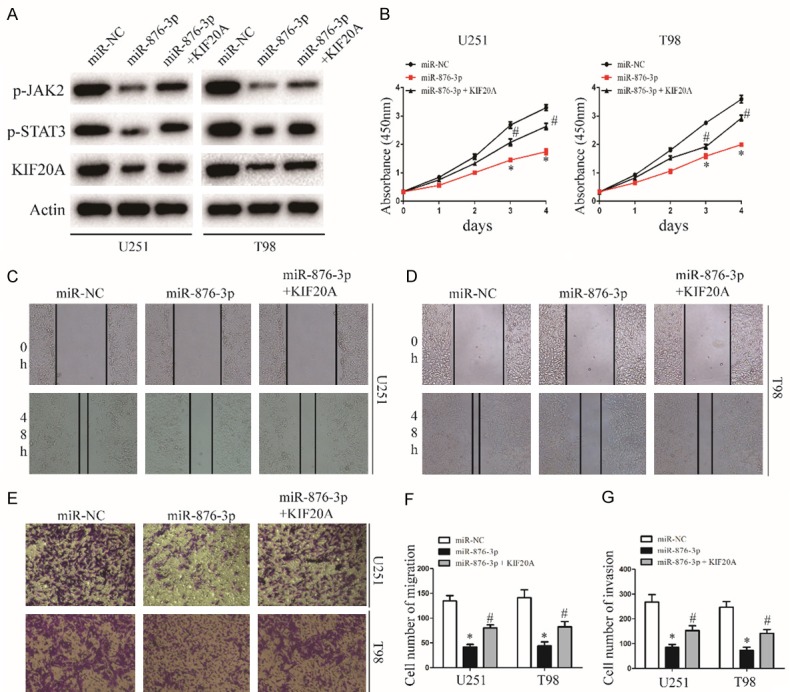
KIF20A reintroduction reverses the inhibitory effect of miR-876-3p. A. Western blot analysis of KIF20A, p-JAK2, and p-STAT3 protein levels in cells transfected with KIF20A in the presence of miR-NC or miR-876-3p. B. Effect of KIF20A reintroduction on miR-NC or miR-628-5p transfected U251 and T98 growth rates as measured by CCK-8 assay. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05, #P < 0.05. C, D and F. Effect of KIF20A reintroduction on miR-NC or miR-876-3p transfected on U251 and T98 migration ability as determined by wound-healing assay. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05, #P < 0.05. E and G. Effect of KIF20A reintroduction on miR-NC or miR-876-3p transfected on U251 and T98 invasion ability as determined by a Matrigel invasion assay. Data were expressed as the mean ± SEM from three independent experiments. *P < 0.05, #P < 0.05.
The function of miR-876-3p is proved in orthotopic and subcutaneous nude mice model
To future prove the function of miR-876-3p, a xenograft model was constructed. U251 cells transfected with miR-876-3p and its negative control were subcutaneously inoculated into nude mice. Overexpression of miR-876-3p inhibited the glioma growth in vivo, as revealed by measuring the tumors volume and tumors weight (Figure 6A-C). In addition, overexpression of miR-876-3p downregulated protein levels of KIF20A, p-JAK2, and p-STAT3, which consistent with the results in vivo (Figures 6D and S5). Altogether, these findings indicate that miR-876-3p suppresses the progression and development of glioma in vivo.
Figure 6.
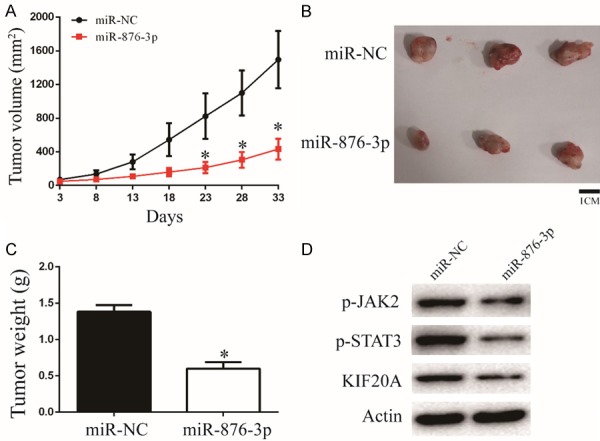
Overexpression of miR-876-3p suppresses glioma growth in vivo. (A) Tumor growth curves after subcutaneous injection of nude mice with U251 cells stably expressing miR-NC or miR-876-3p. Tumor volumes were measured every 5 d from days 3 to 33. (B and C) Analysis of miR-NC and miR-876-3p tumors on day 33 after injection: tumor images (B); tumor weights (C). *P < 0.05. (D) Western blot analysis of KIF20A, p-JAK2, and p-STAT3 protein levels in tumors of miR-NC group or miR-876-3p group.
Discussion
Glioma is the most common tumor in the central nervous system of human, which outcome is poor [18-20]. Previous studies have demonstrated that miRNAs play important roles in glioma initiation and progression as tumor promoter or suppressor [21,22]. Therefore, exploring the molecular and cellular mechanism of glioma-related miRNAs is the key to improve the treatment of glioma. In this study, we detected miR-876-3p expression and the function in glioma for the first time. We determined that miR-876-3p was significantly down-regulated in glioma samples and cells. Then, we revealed the function of miR-876-3p in glioma cell proliferation, cell cycle distribution, migration, invasion and EMT. The results showed that upregulated miR-876-3p suppressed cell growth, migration, invasion, induced G1 cell cycle arrest. These data suggest that miR-876-3p functions as a tumor suppressor in glioma.
KIF20A is a microtubule-associated motor protein, which is a member of the kinesin superfamily-6. Previous studies showed that KIF20A may involve in the development and progression of cancers, including pancreatic cancer [23], hepatocellular carcinoma [8,24], and breast cancer [13]. In addition, KIF20A expression was found to be associated with poor oncologic outcome.
JAK/STAT pathway is one of the most significant and active signalling pathways in cells which plays an important role in many physiological processes, such as cell growth, differentiation, and immune function [25,26]. More and more studies have found the abnormal activation of the JAK/STAT signaling pathway in various tumor tissues. Additionally, previous studies showed that inhibition of JAK2/STAT3 pathway could suppress migratory, invasive potential and EMT in some cancers [27,28].
The functions of microRNA are associated with its target genes. In the further mechanistic study, we first predicted the downstream target gene of miR-876-3p by TargetScan and miRanda softwares. Here, we showed that mRNA and protein expression of KIF20A was upregulated in glioma samples. Additionally, the luciferase assay was conducted to determine the targeting of miR-876-3p to the 3’UTRs of KIF20A. We observed that miR-876-3p-mediated downregulation of KIF20A inactivates the JAK2/STAT3 pathway in glioma cells. Furthermore, overexpression of KIF20A reversed the miR-876-3p-induced inhibition of JAK2/STAT3 pathway and suppression of glioma cell proliferation, migration, and invasion. These results suggested that miR-876-3p reduced glioma cell proliferation, migration, and invasion by targeting KIF20A. Then, we confirmed that overexpression of miR-876-3p inhibits the development and progression of xenograft tumors in vivo.
In summary, miR-876-3p downregulation plays an important role in the progression and malignancy of glioma. miR-876-3p is a critical repressor of KIF20A. Understanding the dysregulation of miR-876-3p/KIF20A stimulated JAK2/STAT3 pathway in glioma is important to improve our knowledge of the biological basis of glioma development and progression and has therapeutic potential treatment in glioma.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen L, Gao H, Liang J, Qiao J, Duan J, Shi H, Zhen T, Li H, Zhang F, Zhu Z, Han A. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am J Cancer Res. 2018;8:2387–2401. [PMC free article] [PubMed] [Google Scholar]
- 2.Huang H, Wang Y, Li Q, Fei X, Ma H, Hu R. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett. 2019;446:81–89. doi: 10.1016/j.canlet.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Jin C, Wang A, Liu L, Wang G, Li G, Han Z. miR-145-5p inhibits tumor occurrence and metastasis through the NF-kappaB signaling pathway by targeting TLR4 in malignant melanoma. J Cell Biochem. 2019 doi: 10.1002/jcb.28388. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Du YM, Wang YB. MiR-637 inhibits proliferation and invasion of hepatoma cells by targeted degradation of AKT1. Eur Rev Med Pharmacol Sci. 2019;23:567–575. doi: 10.26355/eurrev_201901_16869. [DOI] [PubMed] [Google Scholar]
- 5.Dusaulcy R, Handgraaf S, Visentin F, Vesin C, Philippe J, Gosmain Y. miR-132-3p is a positive regulator of alpha-cell mass and is downregulated in obese hyperglycemic mice. Mol Metab. 2019;22:84–95. doi: 10.1016/j.molmet.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan H, Zhang YS. miR-490-3p modulates the progression of prostate cancer through regulating histone deacetylase 2. Eur Rev Med Pharmacol Sci. 2019;23:539–546. doi: 10.26355/eurrev_201901_16866. [DOI] [PubMed] [Google Scholar]
- 7.Lin L, Wang Y. miR-548b-3p regulates proliferation, apoptosis, and mitochondrial function by targeting CIP2A in hepatocellular carcinoma. Biomed Res Int. 2018;2018:7385426. doi: 10.1155/2018/7385426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu M, Huang X, Chen Y, Fu Y, Xu C, Xiang W, Li C, Zhang S, Yu C. Aberrant KIF20A expression might independently predict poor overall survival and recurrence-free survival of hepatocellular carcinoma. IUBMB Life. 2018;70:328–335. doi: 10.1002/iub.1726. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Zhou LL, Li X, Ni J, Chen P, Ma R, Wu J, Feng J. Overexpression of KIF20A confers malignant phenotype of lung adenocarcinoma by promoting cell proliferation and inhibiting apoptosis. Cancer Med. 2018;7:4678–4689. doi: 10.1002/cam4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng Y, Wang W, Hong B, Jiang X, Sun R, Yan Q, Zhang S, Lu M, Wang S, Zhang Z, Lin W, Li Y. Upregulation of KIF20A correlates with poor prognosis in gastric cancer. Cancer Manag Res. 2018;10:6205–6216. doi: 10.2147/CMAR.S176147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stangel D, Erkan M, Buchholz M, Gress T, Michalski C, Raulefs S, Friess H, Kleeff J. Kif20a inhibition reduces migration and invasion of pancreatic cancer cells. J Surg Res. 2015;197:91–100. doi: 10.1016/j.jss.2015.03.070. [DOI] [PubMed] [Google Scholar]
- 12.Taniuchi K, Furihata M, Saibara T. KIF20A-mediated RNA granule transport system promotes the invasiveness of pancreatic cancer cells. Neoplasia. 2014;16:1082–1093. doi: 10.1016/j.neo.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW, Zhao F, Monteiro LJ, Coombes RC, Medema RH, Khoo US, Lam EW. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35:990–1002. doi: 10.1038/onc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai Y, Shibata K, Sakata J, Suzuki S, Utsumi F, Niimi K, Sekiya R, Senga T, Kikkawa F, Kajiyama H. KIF20A expression as a prognostic indicator and its possible involvement in the proliferation of ovarian clearcell carcinoma cells. Oncol Rep. 2018;40:195–205. doi: 10.3892/or.2018.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito K, Ohta S, Kawakami Y, Yoshida K, Toda M. Functional analysis of KIF20A, a potential immunotherapeutic target for glioma. J Neurooncol. 2017;132:63–74. doi: 10.1007/s11060-016-2360-1. [DOI] [PubMed] [Google Scholar]
- 16.Duan J, Huang W, Shi H. Positive expression of KIF20A indicates poor prognosis of glioma patients. Onco Targets Ther. 2016;9:6741–6749. doi: 10.2147/OTT.S115974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Chong Y, Tu Y, Liu N, Yue C, Qi Z, Liu H, Yao Y, Liu H, Gao S, Niu M, Yu R. CRM1/XPO1 is associated with clinical outcome in glioma and represents a therapeutic target by perturbing multiple core pathways. J Hematol Oncol. 2016;9:108. doi: 10.1186/s13045-016-0338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Wang M, Wu J, Han Q, Zhang X. ZNF326 promotes malignant phenotype of glioma by up-regulating HDAC7 expression and activating Wnt pathway. J Exp Clin Cancer Res. 2019;38:40. doi: 10.1186/s13046-019-1031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Xue Y, Ma J, Shao L, Wang D, Zheng J, Liu X, Yang C, He Q, Ruan X, Li Z, Liu Y. SNHG1 promotes malignant biological behaviors of glioma cells via microRNA-154-5p/miR-376b-3p- FOXP2- KDM5B participating positive feedback loop. J Exp Clin Cancer Res. 2019;38:59. doi: 10.1186/s13046-019-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, Xu J, Liu J, Zhang H, Sun C, Wang Q, Shi C, Zhou X, Hua D, Luo W, Bian X, Yu S. The novel chromatin architectural regulator SND1 promotes glioma proliferation and invasion and predicts the prognosis of patients. Neuro Oncol. 2019;21:742–754. doi: 10.1093/neuonc/noz038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Chen X, Zeng K, Xu M, He B, Pan Y, Sun H, Pan B, Xu X, Xu T, Hu X, Wang S. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/beta-catenin signal pathways. Cell Death Dis. 2018;9:1037. doi: 10.1038/s41419-018-1105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayraktar R, Ivan C, Bayraktar E, Kanlikilicer P, Kabil NN, Kahraman N, Mokhlis HA, Karakas D, Rodriguez-Aguayo C, Arslan A, Sheng J, Wong S, Lopez-Berestein G, Calin GA, Ozpolat B. Dual suppressive effect of miR-34a on the FOXM1/eEF2-kinase axis regulates triple-negative breast cancer growth and invasion. Clin Cancer Res. 2018;24:4225–4241. doi: 10.1158/1078-0432.CCR-17-1959. [DOI] [PubMed] [Google Scholar]
- 23.Asahara S, Takeda K, Yamao K, Maguchi H, Yamaue H. Phase I/II clinical trial using HLA-A24-restricted peptide vaccine derived from KIF20A for patients with advanced pancreatic cancer. J Transl Med. 2013;11:291. doi: 10.1186/1479-5876-11-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi C, Huang D, Lu N, Chen D, Zhang M, Yan Y, Deng L, Lu Q, Lu H, Luo S. Aberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosis. Oncotarget. 2016;7:26206–26219. doi: 10.18632/oncotarget.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southworth T, Mason S, Bell A, Ramis I, Calbet M, Domenech A, Prats N, Miralpeix M, Singh D. PI3K, p38 and JAK/STAT signalling in bronchial tissue from patients with asthma following allergen challenge. Biomark Res. 2018;6:14. doi: 10.1186/s40364-018-0128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Pan C, Huang Z, Yuan H, Chen J. WSV181 inhibits JAK/STAT signaling and promotes viral replication in Drosophila. Dev Comp Immunol. 2019;92:20–28. doi: 10.1016/j.dci.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Liu K, Tian T, Zheng Y, Zhou L, Dai C, Wang M, Lin S, Deng Y, Hao Q, Zhai Z, Dai Z. Scutellarin inhibits proliferation and invasion of hepatocellular carcinoma cells via down-regulation of JAK2/STAT3 pathway. J Cell Mol Med. 2019;23:3040–3044. doi: 10.1111/jcmm.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu SC, Huang CM, Bamodu OA, Lin CS, Liu BL, Tzeng YM, Tsai JT, Lee WH, Chen TM. Ovatodiolide suppresses nasopharyngeal cancer by targeting stem cell-like population, inducing apoptosis, inhibiting EMT and dysregulating JAK/STAT signaling pathway. Phytomedicine. 2018;56:269–278. doi: 10.1016/j.phymed.2018.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


