Abstract
DHX9 has numerous functions regulating transcription, translation, RNA processing and transport, and DNA replication and maintenance of genomic stability. It is involved in human cancers as either an oncogene or tumor suppressor. However, its role in the progression of lung cancer and underlying mechanisms remains unclear. In this study, we demonstrated that DHX9 is overexpressed in human lung cancer tissues and serum. Also, a favorable prognosis of lung adenocarcinoma is predicted when DHX9 is at a high level. DHX9 knockdown promoted cell proliferation, migration, and invasion and inhibited apoptosis progression in A549 cells. Moreover, DHX9 knockdown led to a significant decrease of E-cadherin expression, an increase of vimentin and snail, and a significant increase in the phosphorylation of STAT3 in A549 cells. In summary, our studies identified a novel role of DHX9 in driving tumor growth and epithelial-mesenchymal transition progress of A549 cells. We propose that the STAT3 pathway may be implicated in the DHX9-related epithelial-mesenchymal transition of lung adenocarcinoma. Therefore, DHX9 may be a prognostic marker or potential therapeutic target for lung adenocarcinoma.
Keywords: Lung adenocarcinoma, DHX9, EMT, STAT3
Introduction
DHX9, also known as RNA helicase A (RHA), nuclear DNA helicase II (NDH II), or leukophysin (LKP), is an NTP-dependent RNA helicase belonging to the DExH-box family of helicase proteins. DHX9 can unwind double-stranded RNA and DNA in a 3’ to 5’ direction [1] and generate aberrant polynucleotide structures [2]. With multiple domains, DHX9 plays key roles at multiple levels of gene regulation, including transcription [3,4], translation [5,6], RNA processing and transport [7,8], microRNA processing [9], DNA replication, and maintenance of genomic stability [2,10]. In view of its indispensable position in cellular functions, researchers are increasingly concerned about its relationship with human diseases such as cancers or viral infections [11].
Evidence suggests that DHX9 plays different roles in different types of human cancers. DHX9 has been shown to be located downstream of SOX4 in prostate cancer [12]. Overexpression of truncated DHX9 peptides has been shown to result in loss of BRCA1 functions such as DNA repair in breast cancer [13]. A study demonstrated that depletion of DHX9 hinders osteosarcoma cell proliferation [14]. Gene expression profiles exhibit overexpression of DHX9 in osteosarcoma cells with high metastatic ability [15]. In addition, mutation of DHX9 in nonsmokers with lung adenocarcinoma indicates a poor prognosis [16]. Studies have shown that DHX9 may be an oncogene, but some studies still indicate that DHX9 may act as a tumor suppressor. DHX9 binds directly to the p16 promoter, inducing upregulation of p16 [17]; loss of p16 function is an early event in tumor progression [18]. Also, data show that KIF1Bβ interacts with DHX9, causing nuclear accumulation of DHX9 followed by apoptosis of cells [19].
Epithelial-mesenchymal transition (EMT), a crucial mechanism in tumor metastasis, is characterized by the loss of epithelial phenotype and gain of mesenchymal phenotype [20]. TGF-β [21], JAK/STAT3 [22], PI3K/AKT [23], MAPK/ERK [24], Wnt [25], and NF-κB [26] are implicated in the mechanisms of EMT in different types of human cancer. Data have shown that phosphorylation of STAT3 on Tyr705, not Ser727, drives a transcriptional program that converts an epithelial morphology to a migratory mesenchymal one [22].
Previously, we identified DHX9 as an overexpressed protein in lung adenocarcinoma [27] and tested the level of DHX9 with 64 serum samples and 17 tissue samples of lung cancer patients [28]. In this study, we examined the expression of DHX9 in lung cancer tissues and serum with more samples and are the first to report that DHX9 is an essential factor in tumor proliferation, invasion, migration, and EMT in A549 cells, probably through regulation of the phosphorylation of STAT3. These data strongly suggest that DHX9 is an important suppressor in lung adenocarcinoma.
Materials and methods
Patients and specimens
From 2014 to 2015, 123 lung cancer serum specimens and 47 noncancerous (Table 1) serum specimens were obtained from the Respiratory Department of the Second Affiliated Hospital of Xi’an Jiaotong University. The diagnoses were based on clinical and histological examinations using bronchoscopy or lung biopsy. Venous blood samples (2 to 5 ml) from each patient were collected and centrifuged at 1500 rpm for 10 minutes. The supernatant was collected and stored in a 100 μl EP tube at -80°C. No patients had autoimmune diseases, cirrhosis, or other metabolic diseases. All lung cancer patients were newly diagnosed, and serum samples were collected before treatment. This study, as well as follow-up studies, was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University, and written informed consents was obtained from all participants.
Table 1.
Serum DHX9 level in different pathological types of lung cancer
| Total (n=170) | DHX9 (ng/ml) | P value | |
|---|---|---|---|
| Control | 47 | 8.02 ± 0.44 | |
| Lung cancer | 123 | 10.40 ± 0.72 | 0.007** |
| Lung adenocarcinoma | 45 | 10.12 ± 0.72 | 0.012* |
| Lung squamous cell carcinoma | 46 | 9.15 ± 0.55 | 0.122 |
P < 0.05;
P < 0.01.
Cell culture
Human lung adenocarcinoma cell line A549 was incubated in RPMI-1640 (Hyclone, USA) containing 10% fetal bovine serum supplemented with 1% penicillin-streptomycin solution (Hyclone, USA) at 37°C in a 5% CO2 incubator. The cell line was purchased from the Cell Bank at the Chinese Academy of Science (Shanghai, China) in 2015. The cells were authenticated using short tandem repeat (STR) markers.
Lentivirus vector construction: lentivirus-mediated small hairpin RNA (lenti-shRNA) targeting DHX9
The sequences of the DHX9 gene for shRNA (Jiman, Shanghai, China) targeting are shown below: DHX9-shRNA 15’-GAAGGATTACTACTCAAGAAA-3’; DHX9-shRNA 25’-GGGCTATATCCATCGAAATTT-3’; DHX9-shRNA 35’-ACGACAATGGAAGCGGATATA-3’; DHX9-shRNA 45’-TTAAGGAAACCAAGCATATAG-3’.
The shRNA targeting sequence 5’-TTCTCCGAACGTGTCACGT-3’ was used as a negative control (NC) to monitor the responses caused by shRNA. Lentivirus vectors pGMLV-SC5 RNAi containing DHX9 shRNA were used to infect A549 cells, then reverse transcription-polymerase chain reaction and Western blot were used to assess the expression level of stable DHX9.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cells using RNAfast200 (Shaanxi Xianfeng Biotechnology, Shaanxi, China) according to the manufacturer’s instructions. Complementary DNA was synthesized by reverse transcription reactions according to SYBR Premix Ex Taq II (TAKARA BIO INC). Then real-time PCR was performed in triplicate with PrimeScript™RT Master Mix (Takara Bio, Inc.) in the ABI Step-One system. Primers were designed by Sangon Biotech Co. Ltd. (Shanghai, China) or AuGCT Biotech (Beijing, China). The primers used in real-time PCR were as follows: Forward primers: DHX9: 5’-CTGTGGCTACAGCGTTCGAT-3’, E-cadherin: 5’-GAACGCATTGCCACATACAC-3’, Vimentin: 5’-GAAATTGCAGGAGGAGATGC-3’, Snail: 5’-CGAGTGG TTCTTCTGCGCTA-3’, GAPDH: 5’-GTCTCCTCTGACTTCAACAGCG-3’. Reverse primers: DHX9: 5’-GATTCCTCGAATGCCTGCTTC-3’, E-cadherin: 5’-GAATTCGGGCTTGTTGTCAT-3’, Vimentin: 5’-ATTCCACTTTGCGTTCAAGG-3’, Snail: 5’-CTGCTGGAAGGTAAACTCTGGA-3’, GAPDH: 5’-ACCACCCTGTTGCTGTAGCCAA-3’. GAPDH was used as a control. The relative gene expression levels of target mRNA were calculated based on the CT method (2-ΔΔCt).
Western blotting
Total cellular proteins were extracted using RIPA lysis buffer (Xi’an HEART Biotech). Protein (40 μg) was separated through 10% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. The membranes were blocked in 5% (w/v) nonfat dry milk powder in 0.1% Tris buffered saline/Tween 20 (TBST) and then probed with different antibodies overnight at 4°C. Antibodies against DHX9, NF-κB, β-actin, and ERK were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Antibodies against E-cadherin, vimentin, snail, AKT, p-AKT (Ser473), p-STAT3 (Tyr705), and p-ERK (Thr202/Thr204) were purchased from Cell Signaling Technology (Danvers, MA, USA), and antibodies against STAT3, GAPDH, and β-catenin were purchased from Wanlei Biotechnology (Shenyang, China). The blot was then detected with HRP-conjugated secondary antibodies (Cell Signaling Technology). Protein band signals were visualized using ECL chemiluminescence (Thermo Company, USA). The band densities were analyzed using Image Pro Plus software (Media Cybernetics, Inc., Bethesda, MD).
ELISA: detecting the level of DHX9 in serum
The expression level of DHX9 in serum was detected using a DHX9 ELISA kit (Elabscience Biotechnology, Wuhan, China) according to the manufacturer’s instructions.
Immunohistochemistry (IHC)
A total of 61 formalin-fixed, paraffin-embedded (FFPE) tissues (45 primary cancerous, 16 adjacent noncancerous tissues) were collected at the Second Affiliated Hospital of Xi’an Jiaotong University and Shaanxi Provincial Cancer Hospital from 2014 to 2016; cancer was diagnosed in these samples based on histological examinations using bronchoscopy, lung biopsy, or surgery. IHC was performed to detect the protein expression of DHX9. Briefly, FFPE sections were deparaffinized with xylene, rehydrated in a series of graded ethanol, and heated with citrate buffer in a microwave oven for antigen retrieval. After endogenous peroxidase blocking in 0.5% H2O2 and unspecific binding site blocking with normal goat serum for 15 minutes, the sections were incubated with anti-DHX9 (1:500, Abgent, San Diego, CA, USA) at 4°C overnight. After incubating with biotinylated goat anti-mouse HRP-secondary antibody (Boster, Wuhan, China), the sections were stained with DAB and counterstained with hematoxylin. A semi-quantitative scoring system was applied to evaluate the intensity of staining according to the positive staining cells ratio: 0% to 10% (-), 11% to 30% (+), 31% to 70% (++), 71% to 100% (+++).
Cell proliferation assay
Cell proliferation ability was assessed using MTT assays. Briefly, 4 × 103 cells were seeded in 200 microliters of complete medium per well in 96-well plates. After incubation for 24, 48, and 72 hours, cells were further incubated with 5 mg/ml, 20 microliters of MTT (Beyotime, Shanghai, China) for another four hours. The absorbance at 490 nm was measured on a spectrophotometer (Bio Tek, USA). MTT assays were performed with six replicates.
Cell cycle analysis
Cells (1 × 106) were fixed in 70% ethanol at 4°C for 24 hours and stained with propidium iodide (PI). After 30 minutes of incubation at 37°C, cells were analyzed using flow cytometry (Becton Dickinson FACSCalibur).
Apoptosis assay
Cells were centrifuged at 800 rpm for five minutes and suspended in 250 microliters of Binding Buffer. Cells (100 μl) were incubated at room temperature for five minutes with 5 μl of Annexin V/Alexa Fluor647 followed by addition of 20 μg/ml, 10 μl of PI, and then analyzed using flow cytometry (Becton Dickinson FACSCalibur).
“Wound” healing assay
To assess cell migration ability, scratches were made with 10-microliter pipette tips when cells grew to 100% confluence in six-well culture plates and the movement of cells into scraped areas was observed. Images were captured at zero, 24, and 48 hours. Three images were taken per well at a set time point, and the area of the scratch was measured with Image Pro Plus software (Media Cybernetics, Inc., Bethesda, MD) in each image. Cell migration ability was calculated by subtracting the wound area at each time point from the wound area at the zero-hour time point. The wound healing assay was performed in triplicate.
Transwell migration and invasion assays
Transwell migration assays were performed with a 24-well transwell plate (Corning, Inc., New York, NY, USA) according to the manufacturer’s instructions. For the matrigel invasion assay, the upper chambers of a 24-well transwell plate were precoated with 100 μl of 10% Matrigel (BD Biosciences, Franklin, New Jersey, USA) in RPMI-1640 for four hours. Medium with 10% FBS was added to the lower chambers, and aliquots of (1-2) × 104 cells in 200 μl of FBS-free RPMI-1640 were seeded into the upper chambers. After the 24-hour incubation, migrated and invaded cells were fixed and stained with 0.1% crystal violet, counted under a microscope in five random fields per chamber, and photographed. Data were summarized from three independent experiments.
Prognostic analyses
The KM Plotter Online Tool (http://kmplot.com) [29] was used to evaluate the relationship between DHX9 and patient clinical outcomes in non-small cell lung cancer.
Statistical analysis
Statistical analysis was performed using SPSS 20.0 (SPSS, Inc., Chicago, IL). Data were presented as mean ± standard deviation from at least three experiments. Statistical comparisons were based on Student’s t-test or analysis of variance (ANOVA). A P value < 0.05 was considered statistically significant.
Results
DHX9 was highly expressed in serum of human lung cancer patients
Our group has tested the serum level of DHX9 in 64 lung cancer samples previously [28]. To verify the results, we detected the DHX9 protein level in the serum of 123 lung cancer patients and 47 non-cancer patients via ELISA. In line with our previous results, compared with controls, higher levels of DHX9 were detected in the serum of patients with lung cancer (10.40 ± 0.72 vs. 8.02 ± 0.44, P = 0.007, Figure 1A). The serum DHX9 level of lung adenocarcinoma was higher than that of lung squamous cell carcinoma (10.12 ± 0.72 vs. 9.15 ± 0.55, Figure 1A and Table 1). Meanwhile, the serum DHX9 level of lung squamous cell carcinoma was slightly elevated compared with controls, but the difference was not statistically significant (9.15 ± 0.55 vs. 8.02 ± 0.44, P = 0.122, Figure 1A). We observed no statistical correlation of serum DHX9 level with clinicopathological characteristics (data not shown).
Figure 1.
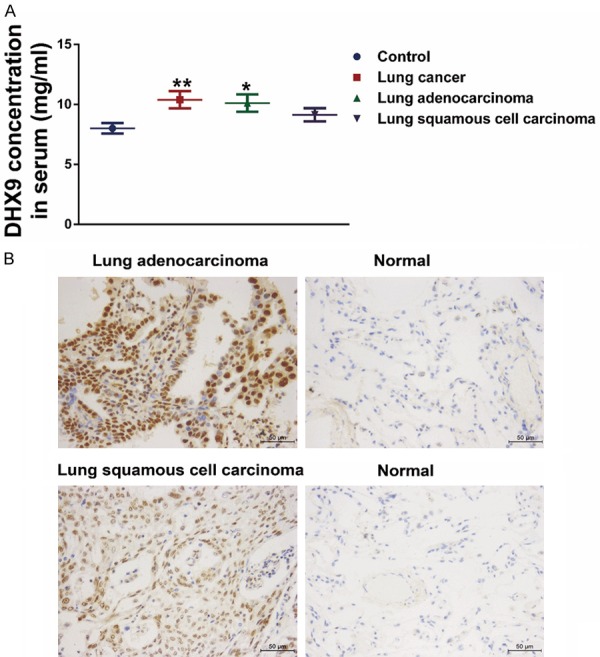
DHX9 was upregulated in lung cancer serum and tissues compared with controls. A. Serum level of DHX9 was overexpressed in lung cancer patients compared with those with benign lung diseases. Subgroup analysis showed that the level of lung adenocarcinoma was higher than that of controls, while the level of the lung squamous cell carcinoma group was not statistically different from the controls. *P < 0.05, **P < 0.01. B. Representative images of DHX9 IHC staining in lung tumor tissues and normal lung tissues. IHC showed that DHX9 was highly expressed in lung tumor tissues, and the expression level in lung adenocarcinoma tissues was higher than that of lung squamous cell carcinoma tissues.
DHX9 was highly expressed in tumor tissues of human lung cancer
Compared with our previous study with 17 lung cancer samples [28], we further detected DHX9 protein levels in tumor tissues through IHC based on 45 primary lung tumor tissues and 16 non-tumor lung tissues. The results showed that DHX9 was strongly stained in lung tumor tissues. DHX9 staining was located both in the nucleus and the cytoplasm of tumor cells, and nuclear staining was stronger than the corresponding cytoplasmic staining (Figure 1B). Moreover, the DHX9 expression in lung adenocarcinoma tissues was significantly higher than that of lung squamous cell carcinoma tissues (P = 0.034) (Figure 1B and Table 2). Based on the IHC results, the patients were designated to one of four groups: DHX9-negative (-: positive cell ratio was 0% to 10%), DHX9 weakly positive (+: positive cell ratio was 11% to 30%), DHX9-positive (++: positive cell ratio was 31% to 70%), and DHX9 strongly positive (+++: positive cell ratio was 71% to 100%) (Figure S1). Accordingly, the summary results of IHC showed that DHX9 was highly expressed in lung cancer tissues compared with controls (Table 2). DHX9 expression level was correlated with histological type, and there were no statistical associations of DHX9 level with other clinical characteristics (Table 2).
Table 2.
The relationships between DHX9 levels and clinicopathological characteristics in lung cancer tissues
| Parameters | Total | Expression levels of DHX9 | P value | |||
|---|---|---|---|---|---|---|
|
| ||||||
| - | + | ++ | +++ | |||
| All case | 45 | 7 | 5 | 16 | 17 | |
| Age | ||||||
| < 60 | 24 | 6 | 1 | 10 | 7 | |
| ≥ 60 | 21 | 1 | 4 | 6 | 10 | 0.194 |
| Gender | ||||||
| Male | 31 | 3 | 5 | 10 | 13 | |
| Female | 14 | 4 | 0 | 6 | 4 | 0.365 |
| Histology | ||||||
| Control | 16 | 13 | 3 | 0 | 0 | |
| Lung cancer | 45 | 7 | 5 | 16 | 17 | 0.000*** |
| LSCC | 12 | 3 | 3 | 4 | 2 | |
| Lung adenocarcinoma | 33 | 4 | 2 | 12 | 15 | 0.034* |
| Lymphatic invasion | ||||||
| N0 | 24 | 5 | 2 | 8 | 9 | |
| N+ | 21 | 2 | 3 | 8 | 8 | 0.719 |
| TNM stage | ||||||
| I+II | 33 | 7 | 2 | 12 | 12 | |
| III+IV | 12 | 0 | 3 | 2 | 5 | 0.467 |
P < 0.05;
P < 0.001.
DHX9 was associated with favorable prognosis in lung adenocarcinoma
To further study the role of DHX9 on lung adenocarcinoma, effects of DHX9 on prognosis were predicted by the online tool KM Plotter (http://kmplot.com). Significant differences in overall survival (OS) and progression-free survival (PFS) were observed between the high-DHX9 and the low-DHX9 group in lung adenocarcinoma (Figure 2A). Lung adenocarcinoma patients with high DHX9 have favorable OS and PFS compared with those with low DHX9 (HR: 0.56, CI: 0.44-0.71, log-rank P = 1.7e-06 and HR: 0.73, CI: 0.53-1, log-rank P = 0.048, for OS and PFS, respectively). However, the expression level of DHX9 had no significant effect on the prognosis of typical patients with non-small cell lung cancer (Figure 2B), and further analysis showed that DHX9 had no significant effect on the prognosis of patients with squamous cell carcinoma (data not shown).
Figure 2.
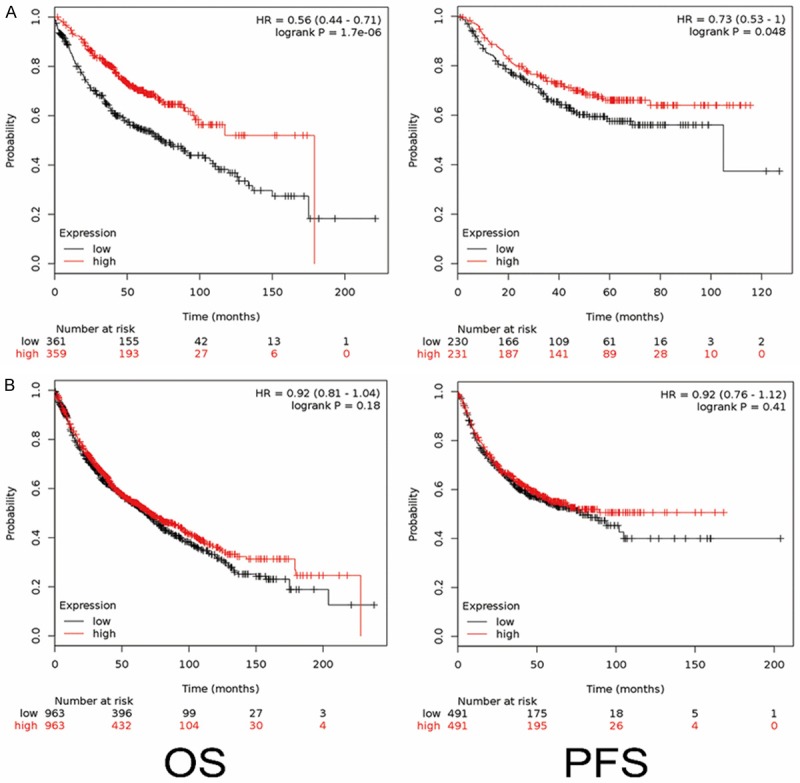
DHX9 was associated with good prognosis in lung adenocarcinoma. A. A high level of DHX9 in lung adenocarcinoma predicted good overall survival compared with the group with low levels, which was evaluated using the KM Plotter online tool based on the mRNA level of 720 patients (left line). Data showed that a high level of DHX9 in lung adenocarcinoma predicted good progression-free survival, according to the mRNA level of 461 patients (right line). B. DHX9 was not associated with overall survival and progression-free survival in non-small cell lung cancer, which was evaluated using the KM Plotter online tool based on the mRNA level of 1926 patients for overall survival study and 982 patients for progression-free survival study.
DHX9 knockdown significantly promoted cell proliferation, migration, and invasion in A549 cells
To investigate the effects of DHX9 on malignant growth potential, we knocked down DHX9 of lung adenocarcinoma cell line A549 using lenti-RNAs, as in Figure 3A, and SH1 cells were used for further investigation [28]. We then detected the knockdown efficiency using RT-PCR and Western blot, respectively. SH1 has a significantly silencing effect of DHX9 (Figure 3B-D). The CON, NC, and SH1 cells were used in the following experiment. MTT assay showed that DHX9 knockdown significantly promoted cell proliferation in A549 cells at 24, 48, and 72 hours (Figure 4C).
Figure 3.
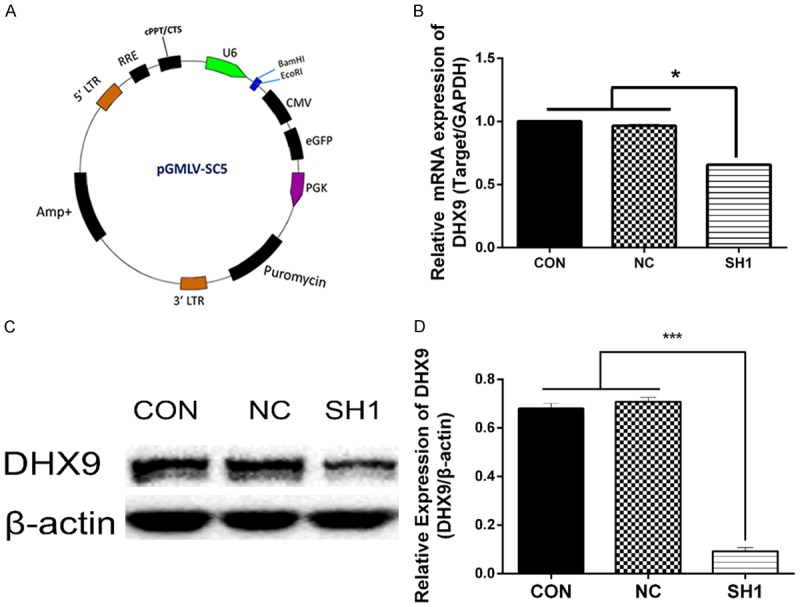
DHX9 was significantly reduced by lenti-shRNA1 (SH1) in lung adenocarcinoma cell line A549. A. The plasmid for the construction of lentivirus mediated DHX9 shRNA. B. qPCR showed that the mRNA of DHX9 in A549 was decreased by SH1 compared with NC and CON. *P < 0.05. C, D. Western blot showed that DHX9 was decreased by SH1 compared with NC and CON. CON, control group without any infection; NC, infected with negative lentivirus; SH1, infected with lenti-shRNA1.
Figure 4.
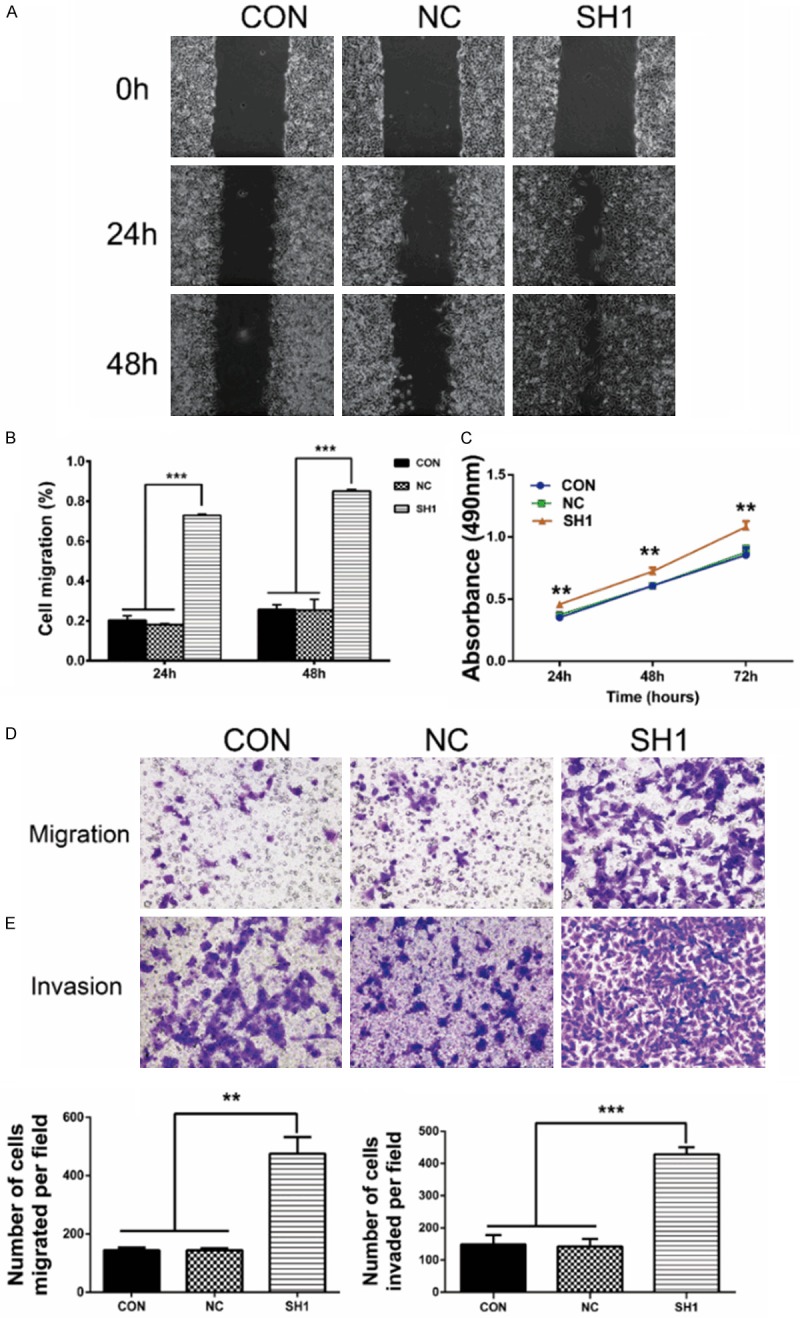
DHX9 knockdown promoted the proliferation, migration, and invasion of lung adenocarcinoma. A. Cell migration monitored by “wound” healing assay at zero, 24, and 48 hours. Pictures showed that SH1 cells migrated more to the scratches at 24 and 48 hours than the control and negative control. B. Wound area was calculated using Image-Pro Plus. SH1 cells covered 73% of the gap at 24 hours and 85% at 48 hours, and NC cells covered 18% at 24 hours and 25% at 48 hours. C. Cells were seeded in 96-well plates. After incubation for 24, 48, and 72 hours, cells were treated with MTT for another four hours. The absorbance at 490 nm was measured. **P < 0.01, when compared with the control group. D, E. Transwell migration and invasion assays on the indicated cells. The migration assay showed that the migrated cells were 475.40 ± 57.05 for SH1 and 144.10 ± 5.77 for NC. The invasion assay showed that the invaded cells were 428.00 ± 11.07 for SH1 and 142.30 ± 11.74 for NC. DHX9 knockdown promoted cell migration and invasion of LAC. **P < 0.01, ***P < 0.001.
DHX9 has been reported to be an important factor in cancer metastasis [15]. Therefore, we subsequently examined the effects of DHX9 on cell migration and invasion in A549 cells. In the “wound” healing assay (Figure 4A, 4B), SH1 cells covered almost 73% of the scratch at 24 hours and at least 85% at 48 hours. In contrast, the scratch was covered only by 18% at 24 hours and 25% at 48 hours in the NC group. In addition, the transwell migration assay showed that SH1 cells migrated faster than control cells. Meanwhile, DHX9 knockdown facilitated the invasive abilities of A549 cells through the matrigel membrane (Figure 4D, 4E).
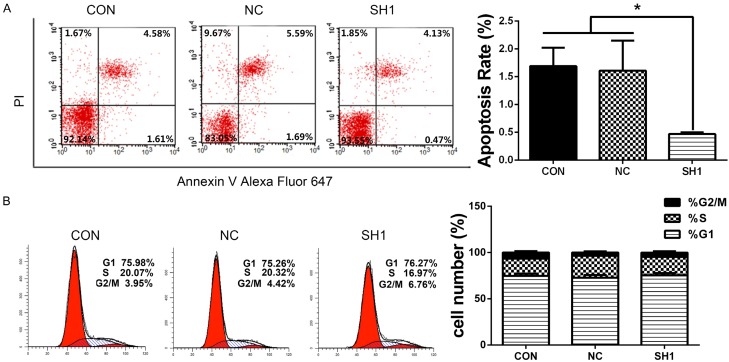
DHX9 knockdown suppressed cell apoptosis and had no effect on cell cycle
Effects of DHX9 knockdown on apoptosis of LAC were evaluated using flow cytometry. The results showed that DHX9 knockdown inhibited apoptosis of LAC (Figure 5A). Cell cycle analysis showed that DHX9 had no effect on the cell cycle of LAC (Figure 5B).
Figure 5.
DHX9 knockdown inhibited tumor apoptosis of lung adenocarcinoma. A. Apoptotic cells were stained with PI/Annexin Alexa Fluor 647, and cells were analyzed using flow cytometry. Quantification values are shown at the right. The early apoptosis rate of SH1 was lower than that of CON and NC. B. Representative images of cell cycle analysis of LAC (CON, NC, SH1) (left panel); the percentages of different cell phases were quantified and presented as mean ± SD (right panel). Data showed no statistical difference between SH1 and NC. *P < 0.05.
DHX9 knockdown promoted EMT and the phosphorylation of STAT3
Epithelial mesenchymal transition (EMT) is a major mechanism of invasion and migration of tumor cells. We further detected the levels of EMT markers, including the epithelial marker E-cadherin and the mesenchymal markers vimentin and snail in DHX9 knockdown cells. As shown in Figure 6A, E-cadherin mRNA expression dramatically decreased, while the mRNA levels of vimentin and snail significantly increased in DHX9 knockdown LAC. We further verified these trends at the protein level using Western blot (Figure 6B). These results indicate that DHX9 may deregulate EMT and thus affect cell migration and invasion in LAC.
Figure 6.
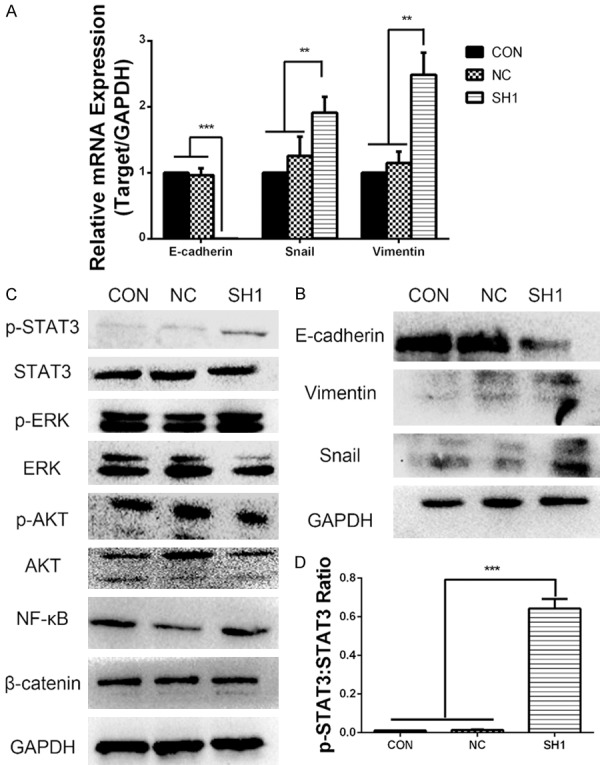
DHX9 knockdown promoted the development of EMT and the phosphorylation of STAT3 in LAC. (A) The mRNA level of E-cadherin decreased, and that of vimentin and snail increased in DHX9 knockdown cells compared with control cells, which were analyzed using real-time PCR. GAPDH was used as a control. (B) The protein levels of E-cadherin, vimentin, and snail were detected using Western blot. The protein level of E-cadherin decreased, and that of vimentin and snail increased while DHX9 was knocked down. GAPDH was used as a control. (C) Protein expression of p-STAT3, STAT3, p-ERK1/2, ERK1/2, p-AKT, AKT, NF-κB, β-catenin, and GAPDH were determined using Western blot in LAC (CON, NC, SH1). DHX9 knockdown promoted the phosphorylation of STAT3 and had no effect on other pathway markers. (D) Densitometric analysis showing the relative p-STAT3:STAT3 ratio in CON, NC, and SH1 cells from (C). Results showed that the STAT3 phosphorylation level of SH1 was significantly higher than that of the control group. **P < 0.01, ***P < 0.001.
Given the evident effects of DHX9 on tumor proliferation and metastasis of LAC, signaling pathways involved in tumorigenesis that might be regulated by DHX9 were evaluated using Western blot to further explore the molecular mechanism. As shown in Figure 6C, the expression of signal transducer and activator of transcription 3 (STAT3), p-STAT3; extracellular signal-regulated kinase (ERK1/2), p-ERK1/2; protein kinase B (AKT), p-AKT; and the levels of NF-κB and β-catenin were detected using Western blot. Results indicated that p-STAT3 level increased in DHX9 knockdown LAC (Figure 6C, 6D). These data showed that knockdown of DHX9 has an effect on the phosphorylation of STAT3.
In summary, knockdown of DHX9 promoted A549 cell proliferation, migration, invasion, and EMT, and DHX9 regulated the phosphorylation of STAT3. Previous study showed that phosphorylation of STAT3 can drive the EMT [22]. We propose that DHX9 influences the biological behavior of A549 cells, perhaps in a DHX9/STAT3/EMT manner.
Discussion
Tumorigenesis is a complex process that begins with genetic mutations and is accompanied by a series of disorders in gene expression, including oncogene activation and tumor suppressor inactivation. The failure of tumor treatment is mainly due to metastasis and recurrence, resulting in organ dysfunction and eventually leading to death. Therefore, a better understanding of the mechanisms involved in lung cancer metastasis may provide a foundation for new treatments. Previous studies have shown that DHX9 plays a central role in cellular processes, especially the regulation of gene expression [3-5,11]. Furthermore, DHX9 is one of the most important mutated genes in lung adenocarcinoma [16]. Whether DHX9 exerts its effects in tumorigenesis and development of lung adenocarcinoma is still unknown.
Accumulating evidence shows that DHX9 is involved in different types of human cancers and may act as either tumor suppressor or oncogene. DHX9 interacts with EGFR [30], BRCA1 [13], and NF-κB [15] and thereby functions as an oncogene in different types of human cancer. In contrast, DHX9 was found to be a tumor suppressor by regulating expression of p53 [31], p16 [17], and KIF1Bβ [19]. Data from previous work suggest that DHX9 may have complex roles in different types of cancer in a variety of ways, possibly correlated with cellular context and/or levels or activity of its interacting partners. By comparing protein expression profiles between lung adenocarcinoma and its paired adjacent normal lung tissue using iTRAQ and 2D-LC-MS/MS, we identified 568 differentially expressed proteins. In particular, DHX9 was overexpressed in lung adenocarcinoma [27]. Consistent with our previous data, DHX9 was upregulated in both the tissues and serum of lung cancer patients. Moreover, compared with LSCC, the level of DHX9 in lung adenocarcinoma was more obvious (Figure 1). In addition, using an online tool (http://kmplot.com), we revealed that a high level of DHX9 in lung adenocarcinoma predicts a favorable prognosis (Figure 2). The dysregulation of DHX9 expression in lung adenocarcinoma implies that it may play an important role in the tumorigenesis and metastasis of lung adenocarcinoma and may be a prognostic marker. This possibility was further confirmed through a series of experiments. Knockdown of DHX9 promoted cell proliferation, migration, and invasion of LAC in vitro. Although DHX9 suppression had no significant effects on the cell cycle, we found that early cell apoptosis was inhibited by knockdown of DHX9 (Figures 4, 5).
EMT promotes cancer cell local infiltration, which is the first step in metastasis [20]. In our study, we found that suppression of DHX9 was coupled with a significant decrease in epithelial marker E-cadherin and an increase of the mesenchymal markers vimentin and snail (Figure 6A, 6B), suggesting that DHX9 may be a regulator in the complex progression of EMT in lung adenocarcinoma. Loss of E-cadherin is a critical marker of EMT, resulting in weak cell-cell adhesion followed by invasion and migration of tumor cells [20]. However, due to the complicated roles of EMT in different types of cancer, it is difficult to explore the underlying mechanism by which DHX9 regulates the EMT process and influences the migration and invasion of LAC. To better understand the mechanism, we then detected the potential signaling pathways by Western blot, including STAT3 and p-STAT3, ERK1/2 and p-ERK1/2, AKT and p-AKT, NF-κB, and β-catenin and identified the significantly altered pathway JAK/STAT3. The levels of p-AKT, p-ERK1/2, β-catenin, and NF-κB did not obviously change after knockdown of DHX9 (Figure 6C, 6D), which may be correlated with cancer types, complex cellular context, or cellular localization.
STAT3 is an important activating factor involved in the regulation of immune response and tumor microenvironment. Activation of STAT3 promotes tumor cell proliferation, invasion, and survival; inhibits tumor immunity; and mediates tumor-associated inflammation [32]. Our in vitro studies suggested that the STAT3 signaling pathway was involved in DHX9-mediated tumor proliferation and metastasis. Knockdown of DHX9 dramatically increased the phosphorylation of STAT3. Similar to findings of a previous study, the JAK/STAT3 pathway is one of the most important pathways associated with the oncogenesis of lung adenocarcinoma [3]. In addition, overactivated STAT3 was reported to promote the EMT process [33]. STAT3 pathway-related EMT are involved in the progression of gastric, esophageal, and prostate cancer [34-36]. Our study demonstrated a novel connection between DHX9 and STAT3. Knockdown of DHX9 activated STAT3 and promoted EMT, leading us to believe that DHX9 may mediate the EMT via STAT3. Together, these findings stress the importance of DHX9 in tumor proliferation and metastasis and highlight its interconnection with STAT3 and its potential as a prognostic marker and therapeutic target to improve the clinical outcomes of lung adenocarcinoma.
In conclusion, our study showed that DHX9 is overexpressed in human lung cancer tissues and serum, and there is a strong correlation between DHX9 and tumor proliferation, migration, invasion, and EMT of LAC. Moreover, we revealed that knockdown of DHX9 promotes tumor progression and EMT of LAC, at least partly through activation of STAT3. Thus, DHX9 may act as a tumor suppressor in lung adenocarcinoma. These findings enhance our understanding of the mechanism that drives the progression of lung adenocarcinoma. Our results highlight the potential value of DHX9 as a promising target for treatment and a biomarker for diagnosis of lung adenocarcinoma.
Acknowledgements
This work was supported by the Science and Technology Funds of Shaanxi Province (2017JM8159) and the National Natural Science Foundation of China (81672300).
Disclosure of conflict of interest
None.
Abbreviations
- DHX9
DEAH box helicase 9
- NDH II
nuclear DNA helicase II
- LKP
leukophysin
- LAC
lung adenocarcinoma cells
- STAT3
signal transducer and activator of transcription 3
- NSCLC
non-small cell lung cancer
- LSCC
lung squamous cell carcinoma
- EMT
epithelial mesenchymal transition
- SOX4
SRY-Box 4
- BRCA1
Breast Cancer 1
- KIF1Bβ
kinesulin family member 1B
- OS
overall survival
- PFS
progression-free survival
- AKT
protein kinase B
- ERK
extracellular signal-regulated kinase
- lenti-shRNA
lentivirus-mediated small hairpin RNA
- IHC
immunohistochemistry
Supporting Information
References
- 1.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 2.Jain A, Bacolla A, Del Mundo IM, Zhao J, Wang G, Vasquez KM. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013;41:10345–10357. doi: 10.1093/nar/gkt804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 4.Tetsuka T, Uranishi H, Sanda T, Asamitsu K, Yang JP, Wong-Staal F, Okamoto T. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. Eur J Biochem. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 5.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 6.Manojlovic Z, Stefanovic B. A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs. RNA. 2012;18:321–334. doi: 10.1261/rna.030288.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bratt E, Ohman M. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA. 2003;9:309–318. doi: 10.1261/rna.2750803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang H, Gaietta GM, Fischer WH, Ellisman MH, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 9.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain A, Bacolla A, Chakraborty P, Grosse F, Vasquez KM. Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–6999. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016;7:42716–42739. doi: 10.18632/oncotarget.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22:983–991. doi: 10.1038/sj.onc.1206195. [DOI] [PubMed] [Google Scholar]
- 14.Cheng D, Zhang H, Yuan J, Li S, Yang Q, Fan C. Minichromosome maintenance protein 2 and 3 promote osteosarcoma progression via DHX9 and predict poor patient prognosis. Oncotarget. 2017;8:26380–26393. doi: 10.18632/oncotarget.15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zucchini C, Rocchi A, Manara MC, De Sanctis P, Capanni C, Bianchini M, Carinci P, Scotlandi K, Valvassori L. Apoptotic genes as potential markers of metastatic phenotype in human osteosarcoma cell lines. Int J Oncol. 2008;32:17–31. [PubMed] [Google Scholar]
- 16.Sun Z, Wang L, Eckloff BW, Deng B, Wang Y, Wampfler JA, Jang J, Wieben ED, Jen J, You M, Yang P. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics. 2014;7:32. doi: 10.1186/1755-8794-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myohanen S, Baylin SB. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- 18.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen ZX, Wallis K, Fell SM, Sobrado VR, Hemmer MC, Ramskold D, Hellman U, Sandberg R, Kenchappa RS, Martinson T, Johnsen JI, Kogner P, Schlisio S. RNA helicase A is a downstream mediator of KIF1Bbeta tumor-suppressor function in neuroblastoma. Cancer Discov. 2014;4:434–451. doi: 10.1158/2159-8290.CD-13-0362. [DOI] [PubMed] [Google Scholar]
- 20.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, MacBeath G. A noncanonical frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell. 2014;159:844–856. doi: 10.1016/j.cell.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr. 2015;9:317–324. doi: 10.1080/19336918.2015.1016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang J, Qian Y, Ma Y, Wang F, Li H, Cheng C, Zhang L, Jia Z, Li Y, Yang B, Xu E, Wang J, Yang J, Bi Y, Chang L, Wang Y, Zhang Y, Song B, Li G, Shi R, Liu J, Zhang M, Cheng X, Cui Y. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017;397:83–93. doi: 10.1016/j.canlet.2017.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 26.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Li W, Hou Y, Niu Z, Zhong Y, Zhang Y, Yang S. Comparative membrane proteomic analysis between lung adenocarcinoma and normal tissue by iTRAQ labeling mass spectrometry. Am J Transl Res. 2014;6:267–280. [PMC free article] [PubMed] [Google Scholar]
- 28.Cao S, Sun R, Wang W, Meng X, Zhang Y, Zhang N, Yang S. RNA helicase DHX9 may be a therapeutic target in lung cancer and inhibited by enoxacin. Am J Transl Res. 2017;9:674–682. [PMC free article] [PubMed] [Google Scholar]
- 29.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, Chang WC, Wang Y, Hsu MC, Yu YL, Huang TH, Ding Q, Chen CH, Tsai CH, Hung MC. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2010;107:16125–16130. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halaby MJ, Harris BR, Miskimins WK, Cleary MP, Yang DQ. Deregulation of internal ribosome entry site-mediated p53 translation in cancer cells with defective p53 response to DNA damage. Mol Cell Biol. 2015;35:4006–4017. doi: 10.1128/MCB.00365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang WP, Sun Y, Lu Q, Zhao JB, Wang XJ, Chen Z, Ni YF, Wang JZ, Han Y, Zhang ZP, Yan XL, Li XF. Gankyrin promotes epithelial-mesenchymal transition and metastasis in NSCLC through forming a closed circle with IL-6/ STAT3 and TGF-beta/SMAD3 signaling pathway. Oncotarget. 2017;8:5909–5923. doi: 10.18632/oncotarget.13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Tang N, Wang C, Xiao L, Yu M, Zhao L, Cai H, Han L, Xie C, Zhang Y. TNF-alpha-inducing protein of helicobacter pylori induces epithelial-mesenchymal transition (EMT) in gastric cancer cells through activation of IL-6/STAT3 signaling pathway. Biochem Biophys Res Commun. 2017;484:311–317. doi: 10.1016/j.bbrc.2017.01.110. [DOI] [PubMed] [Google Scholar]
- 35.Zang C, Liu X, Li B, He Y, Jing S, He Y, Wu W, Zhang B, Ma S, Dai W, Li S, Peng Z. IL-6/STAT3/TWIST inhibition reverses ionizing radiation-induced EMT and radioresistance in esophageal squamous carcinoma. Oncotarget. 2017;8:11228–11238. doi: 10.18632/oncotarget.14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong D, Liu Q, Liu G, Xu J, Lan W, Jiang Y, Xiao H, Zhang D, Jiang J. Metformin inhibits castration-induced EMT in prostate cancer by repressing COX2/PGE2/STAT3 axis. Cancer Lett. 2017;389:23–32. doi: 10.1016/j.canlet.2016.12.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.