Abstract
Caspases are an important protease family that coordinate inflammation and programmed cell death. Two closely related caspases, caspase-3 and caspase-7, exhibit largely overlapping substrate specificities. Assessing their proteolytic activities individually has therefore proven extremely challenging. Here, we constructed an outer membrane protein G (OmpG) nanopore with a caspase substrate sequence DEVDG grafted into one of the OmpG loops. Cleavage of the substrate sequence in the nanopore by caspase-7 generated a characteristic signal in the current recording of the OmpG nanopore that allowed the determination of the activity of caspase-7 in Escherichia coli cell lysates. Our approach may provide a framework for the activity-based profiling of proteases that share highly similar substrate specificity spectrums.
Introduction
Caspases are a family of cysteine proteases with specificity for cleaving principally after aspartate residues, which play key roles in apoptosis (1, 2) and inflammation (3). Caspases can be divided into two subgroups based on their predominant functional roles (4, 5, 6). Caspase (casp)-1, -4, -5, -11, and -12 are mainly involved in inflammation, whereas casp-3, -6, -7, -8, and -9, are implicated in apoptosis. During apoptosis, the initiator caspases, casp-8 and -9, activate downstream executioner caspases, casp-3, -6, and -7, which carry out selective proteolysis in the cell. All caspase family members have a catalytic cysteine-histidine dyad at the active site, and cleavage only occurs after certain aspartate (or in rare instances, glutamate or phosphoserine (7)) residues in the substrate.
The aberrant caspase activity is associated with the progression of several diseases, such as neurodegenerative disorders (8, 9, 10), cancer (11, 12), and sepsis (13). The diverse functions for caspases in human health and disease provide promising leads toward understanding important biological phenomena as well as avenues for discovering disease therapeutics. Caspases have high levels of sequence homology and overlapping substrate specificities. For example, both casp-3 and -7 can effectively cleave DEVD↓G; casp-2, -3, and -7 potentially cleave VDVAD↓G; and casp-1 and casp-9 favor WEHD↓G (14, 15). Among the caspase family members, casp-3 and casp-7 are the most closely related to each other, with an overall sequence identity of 56% and sequence similarity of 73% (16). They exhibit almost identical substrate specificity, albeit with different cleavage efficiencies (14, 17, 18, 19, 20, 21). However, recent data suggest that casp-3 and casp-7 may have distinct functions because cells deficient in these caspases showed differences in the regulation of reactive oxygen species production during intrinsic cell death (22). To define their distinct role in programmed cell death, probes that can specifically address their activity are needed. Both casp-3 and casp-7 are commonly studied using inhibitors and probes that rely on the canonical DEVD tetrapeptide recognition sequence (14, 21). Unfortunately, the DEVD-based probes promiscuously bind to casp-3, -6, -7, and -8, preventing the investigation of the activity of each isoform in native cellular mixtures (19, 23, 24). Despite the availability of the antibody-coupled enzyme-linked immunosorbent assay to analyze the abundance of the isoforms, it does not directly reflect the activity of these enzymes. Recently, a Förster resonance energy transfer (FRET) substrate that exhibited 1000-fold enhanced selectivity for the active casp-3 over other caspases in apoptotic cells has been successfully developed (25). Although, so far, a method to selectively detect active casp-7 in complex biological systems is still not available.
Nanopore sensing is a single-molecule detection method that measures the ionic current signal induced by the interaction of analytes with a nanopore at an applied potential (26, 27, 28, 29). Recently, protein nanopores have been developed as powerful tools for fundamental studies in chemistry and biology (30, 31) as well as biotechnology applications such as biosensing (32) and DNA and RNA sequencing (30, 33, 34, 35, 36, 37, 38, 39). Nanopores can also detect protein analytes (26, 40) at the single-molecule level by coupling with high-affinity binding sites such as aptamers (41, 42, 43), inhibitors (44), ligands (45, 46), and peptide sequences (47, 48). Of particular interest here, nanopore sensors have been successfully used to analyze enzyme activities (49, 50). Several enzymes of the protease family, such as HIV-1 protease (51), trypsin (52), and renin protease (49), have been detected by measuring the translocation of enzymatically cleaved peptides through a nanopore (49, 51, 52). Peptides derived from cleaved substrates caused different ionic current blockages compared to uncleaved protein substrates (49, 52, 53, 54, 55). A weakness of this approach is that proteases that cleave the same peptide sequence cannot be unambiguously distinguished. Also, the presence of nonspecific proteases with broad cleavage specificity (e.g., trypsin, Arg-C, Glu-C, or proteinase K) could cleave target peptide sequences and thus cause false-positive results. Moreover, direct protease sensing using nanopores has not been achieved in complex biological samples. This is because some highly abundant small molecules in complex samples could block the nanopore and thereby produce false-positive signals that interfere with the identification of the target peptide products (49). To address this problem, it was possible to reduce sample complexity by applying a precleaning column before nanopore analysis (49). However, this approach does not eliminate the interference of other peptidases.
In addition to measurement of the enzymatic peptide product, proteases can also be detected by their direct interactions with nanopores. For example, specific binding and detection of thrombin were achieved with an α-hemolysin (α-HL) pore modified with an aptamer attached near the entrance of the nanopore (42). In addition, when a tobacco etch viral protease cleavage site was inserted at one end of α-hemolysin, the cleavage of tobacco etch virus caused a noticeable change in the current signal (56).
Recently, we demonstrated the discrimination of protein homologs in a complex mixture using an outer membrane protein G (OmpG)-based nanopore (57). OmpG from Escherichia coli (E. coli) is a monomeric β-barrel protein (58, 59, 60) housing seven long extracellular loops (61). In this study, we show that an OmpG nanopore with a substrate peptide DEVD sequence inserted into loop 6 can directly detect and discriminate homologs casp-3 and casp-7 in diluted bacterial cell lysates. Our results demonstrate the ability of the OmpG nanopore sensor to resolve protein homologs in untreated biological samples.
Materials and Methods
Cloning, expression, and purification of OmpGcasp
To minimize the gating frequency of OmpG constructs, we first engineered wild-type (WT) OmpG with three mutations, H231A, H261A, and D215 deletion (62, 63). This OmpG construct is referred to as OmpG2HD. Two peptide sequences (–GDEVDG– and –GEDDVG–) were respectively introduced to replace the aspartic acid 224 by mutagenesis PCR taking the plasmid pT7-OmpG2HD as the template. The mutant constructs are called OmpGcasp and OmpGEDDV.
The pT7-OmpGcasp, pT7-OmpGEDDV, and pT7-OmpG2HD were transformed into BL21 (DE3) cells and were grown in lysogeny broth medium at 37°C until the OD600 reached 0.5–0.6 and induced with 0.5 mM isopropyl β–D-1-thiogalactopyranoside (IPTG). Cells were harvested 3 h later and lysed in lysis buffer 50 mM Tris (pH 8.0), 200 μg/mL lysozyme, and 1 mM EDTA with sonication. The lysate was centrifuged at 19,000 × g for 30 min before washing the inclusion body once with 30 mL 50 mM Tris (pH 8.0) and 1.5 M urea. Then the OmpG-containing inclusion body was dissolved in 30 mL 50 mM Tris (pH 8.0) and 8 M urea and passed through a 0.45 μm filter before fast protein liquid chromatography purification. All OmpG proteins were purified using a 5 mL QFF-anionic exchange column (GE Healthcare, Chicago, IL) and eluted in buffer 50 mM Tris (pH 8.0), 8 M urea, and 500 mM NaCl by applying a gradient.
To refold OmpGcasp, OmpGEDDV, and OmpG2HD, the purified proteins were diluted with the refolding buffer 50 mM Tris (pH 9.0) and 3.25% octyl-glucoside until the final concentration of urea reached 3.0 M. Samples were then incubated at 37°C for 3 days. The refolded sample was flash frozen with 20% glycerol in liquid nitrogen and stored at −80°C until further use.
Expression and purification of caspase-3, -6, -7, -8, and -9
Caspase-3 was expressed as the full-length WT protein, which undergoes complete self-activation during induction. Caspase-3 WT DNA was transformed into BL21 (DE3) E. coli cells and grown in 2xYT media in the presence of ampicillin until they reached an OD600 of 0.6. Expression was induced by the addition of 1 mM IPTG, and the temperature was lowered to 30°C for 3 h. In this research, we used caspase-6 D23A D179A constitutively two chain (CTC), in which the large subunit contains both prodomains due to D23A mutation and linker due to D179A mutation (64). This construct was grown in 2xYT media until OD600 reached 0.6, and expression was induced by the addition of 1 mM IPTG when the temperature was lowered to 20°C for 18 h. Caspase-7 CTC ΔN ΔISL WT DNA encodes for independent translation of the large and small subunits lacking both the prodomain and the intersubunit linker to ensure homogeneous protein preparations (65). Caspase-7 CTC ΔN ΔISL WT DNA was transformed into BL21 (DE3) E. coli cells and spread onto lysogeny broth agar plates containing 100 μg/mL ampicillin. The cells were grown at 37°C in 2xYT media with ampicillin until OD600 reached 0.6. Expression was induced by adding 1 mM IPTG and lowering the temperature to 18°C for 12 h. All cells were stored as pellets at −80°C for further purification.
Caspase-3, -6, and -7 were purified in an identical manner. The cells were harvested, resuspended in a lysis buffer containing 50 mM sodium phosphate (pH 8), 300 mM NaCl, and 2 mM imidazole and lysed in a microfluidizer. Lysates were clarified by centrifuging at 27,000 × g, and the supernatant was loaded onto a 5 mL HiTrap Ni-affinity column preequilibrated with lysis buffer. The column was washed with a buffer containing 50 mM sodium phosphate, pH 8, 300 mM NaCl and 50 mM imidazole. The proteins were eluted with a buffer of 50 mM sodium phosphate, pH 8, 300 mM NaCl and 300 mM imidazole and diluted 6-fold in buffer A containing 20 mM Tris (pH 8.5) and 2 mM dithiothreitol (DTT) before loading onto a 5 mL HiTrap Q column. A linear salt gradient was used to elute the proteins at 120 mM NaCl.
Caspase-8 WT ΔDED (death effector domain) protein (66, 67) was expressed in BL21 (DE3) E. coli cells using 2xYT media. Cells were grown at 37°C until they reached an OD6000 of 0.6. The temperature was dropped to 25°C and the cells were induced by the addition of 1 mM IPTG for 3 h. Cells were harvested by centrifugation at 5000 × g and stored as pellets at −80°C. Pellets were resuspended in a buffer containing 50 mM sodium phosphate pH 8.0, 500 mM NaCl, and 2 mM imidazole. Cells were lysed by a microfluidizer and subsequently centrifuged at 27,000 × g to remove cellular debris. The supernatant was passed over a nickel column (5 mL HiTrap Ni-affinity column; GE Healthcare), washed until the absorbance reached baseline, and eluted with a buffer containing 50 mM sodium phosphate, 300 mM NaCl, and 300 mM imidazole. The elution was collected and diluted sixfold into a buffer containing 20 mM Tris (pH 8.5) and 2 mM DTT. The nickel elution fraction was loaded onto a Q column (5 mL HiTrap Q column; GE Healthcare) and eluted by a linear NaCl gradient.
Caspase-9 WT protein was expressed in BL21 (DE3) E. coli cells using 2xYT media and grown at 37°C until they reached an OD600 of 0.9. The temperature was dropped to 15°C, and the cells were induced by the addition of 1 mM IPTG for 3 h. Cells were harvested by centrifugation at 5000 × g and stored as pellets at −80°C. Pellets were resuspended in a buffer containing 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, and 2 mM imidazole. Cells were lysed by a microfluidizer and subsequently centrifuged at 27,000 × g to remove cellular debris. The supernatant was loaded onto a nickel column, washed, and eluted with an imidazole gradient extending from 2 to 100 mM. Elution fractions were assessed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), and those containing caspase-9 were pooled and diluted 10-fold into a buffer containing 20 mM Tris (pH 8.5) and 5 mM DTT. The diluted caspase-9 was loaded onto a Q column and eluted by a linear NaCl gradient.
The purity of the prepared caspase proteins was assessed by using SDS-PAGE and stored at −80°C (Fig. S1).
Cleavage assay of OmpG proteins by different proteases
Each protease was added to either the refolded or denatured OmpG with the mole ratio being 1:20. The mixture was diluted with the same volume of buffer (150 mM NaCl and 50 mM HEPES (pH = 7.5), followed by a 0.5–4 h incubation at room temperature (RT ∼23°C). After incubation, all samples were heated at 95°C for 15 min with SDS-PAGE sample buffer, before loading to SDS-PAGE gel.
Preparation of lysate samples
After the cells expressing either caspase-3 or caspase-7 were harvested, the cell pellets were resuspended in phosphate buffered saline and lysed using sonication. Lysates were clarified by centrifugation at 27,000 × g. The identical experiment was performed without the addition of IPTG to act as a negative control for caspase expression.
Single-channel recording of OmpG proteins
Single-channel recording of OmpGcasp was similar to the previous study (45). Briefly, experiments were performed in a device containing two chambers separated by a 25-μm-thick polytetrafluoroethylene film (Goodfellow, Huntingdon, United Kingdom). The film contains an aperture of ∼100 μm diameter located at the center of the film made with an electric spark. A hexadecane/pentane (10% v/v) solution was first added to cover both sides of the aperture. Each chamber was then filled with buffer containing 300 mM KCl, 50 mM HEPES (pH 7.5). 1, 2-diphytanoyl-sn-glycero-3-phosphocholine dissolved in pentane (10 mg/mL) was dropped on the surface of the buffer in both chambers. After the pentane evaporated, the lipid bilayer was formed by pipetting the solution in both chambers several times. An Ag/AgCl electrode was immersed in each chamber with the cis side grounded. OmpG proteins (∼2–3 nM, final concentration) were also added to the cis chamber. To promote OmpG insertion, ≥ +250 mV was usually applied.
After a single OmpGcasp pore inserted into the planar lipid bilayer, the applied voltage was decreased to 30 mV for recording. OmpG proteins inserted in the bilayer bidirectionally with its extracellular loops located at either cis or trans side. After 5–10 min recording in both positive and negative voltages, the orientation of the OmpG pore in the lipid bilayer was determined by analyzing the asymmetrical gating pattern at both potentials (68).
Target proteases (like casp-3, -6, -7, -8, -9, and proteinase K) were added to the chamber where the extracellular loops of OmpGcasp were located; then, the solution in that chamber was stirred for around 15 s. We define a positive potential when the potential of the chamber where the extracellular loops were exposed was positive. The current was amplified with an Axopatch 200B integrating patch-clamp amplifier (Axon Instruments, Foster City, CA). Signals were filtered with a Bessel filter at 2 kHz (unless otherwise stated) and then acquired by a computer (sampling at 100 μs) after digitization with a Digidata 1440A/D board (Axon Instruments).
Results and Discussion
Design of OmpGcasp to detect active caspase-7
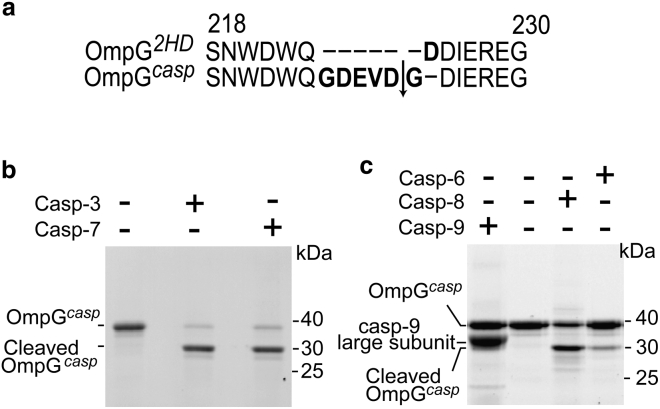
To create the OmpG nanopore for detection of caspases, we inserted a caspase recognition sequence preceded by a Gly residue (–GDEVD↓G–) into loop 6 of an OmpG2HD construct. OmpG2HD denotes an OmpG construct containing three mutations (H231A, H261A, and D215 deletion) (Fig. 1 a). The resultant nanopore construct, OmpGcasp, exhibited refolding efficiency similar to that of OmpG2HD (∼95% refolding efficiency (Fig. S2)). To test whether the casp-7 could cleave OmpGcasp, we conducted cleavage assays with casp-7 and other caspases, which also cleave DEVD-containing substrates, including casp-3, -6, -8, and -9 (14, 15, 19, 69) (Fig. 1, b and c). OmpGcasp was incubated with casp-3 and -7 at a 20:1 molar ratio (30 min at 23°C) and analyzed by SDS-PAGE. We observed that OmpGcasp was efficiently cleaved by both casp-3 and casp-7, with around 90% efficiency for both caspases (Fig. 1 b). Because their reported proteolytic activity with –DEVD– substrates were lower than casp-3 and -7 (19, 23, 25), casp-6, -8, and -9 were incubated with OmpGcasp separately at a 1:1 molar ratio (1 h at 23°C). It was shown that OmpGcasp could be cleaved by casp-6 and -8 with around 8% and 55% efficiency (analyzed by ImageJ), respectively, whereas casp-9 showed some negligible cleavage (Fig. 1 c). This result is consistent with previous findings that DEVD-based chemical probes were processed by casp-3, -6, -7, -8, and -9 albeit with different efficiency (i.e., the kcat/Km of casp-6, -7, -8, and -9 were 0.2, 30.7, 1.5, and 0.03% compared to casp-3 (19, 23, 25)).
Figure 1.
Design of OmpGcasp nanopore to detect casp-7. (a) Loop 6 sequence alignment of OmpG2HD and OmpGcasp. The arrow represents the theoretical caspase cleavage site. (b) SDS-PAGE analysis of the cleavage of OmpGcasp by casp-3 and casp-7. The mole ratio of OmpG to caspase was 20:1, and the mixture was incubated for 30 min at 23°C. (c) SDS-PAGE analysis of the cleavage of OmpGcasp by casp-6, casp-8, and casp-9. The mole ratio of OmpG to caspase was 1:1, and the mixture was incubated for 1 h at 23°C. Buffer used in cleavage assay is 150 mM NaCl and 50 mM HEPES buffer (pH 7.5). The shadow bands in (c) may be caused by degraded proteins and the impurities in the caspase samples. The same amount of OmpGcasp was loaded in each lane. The cleavage efficiency was calculated by comparison of the intensity of the intact OmpGcasp bands that were analyzed by ImageJ.
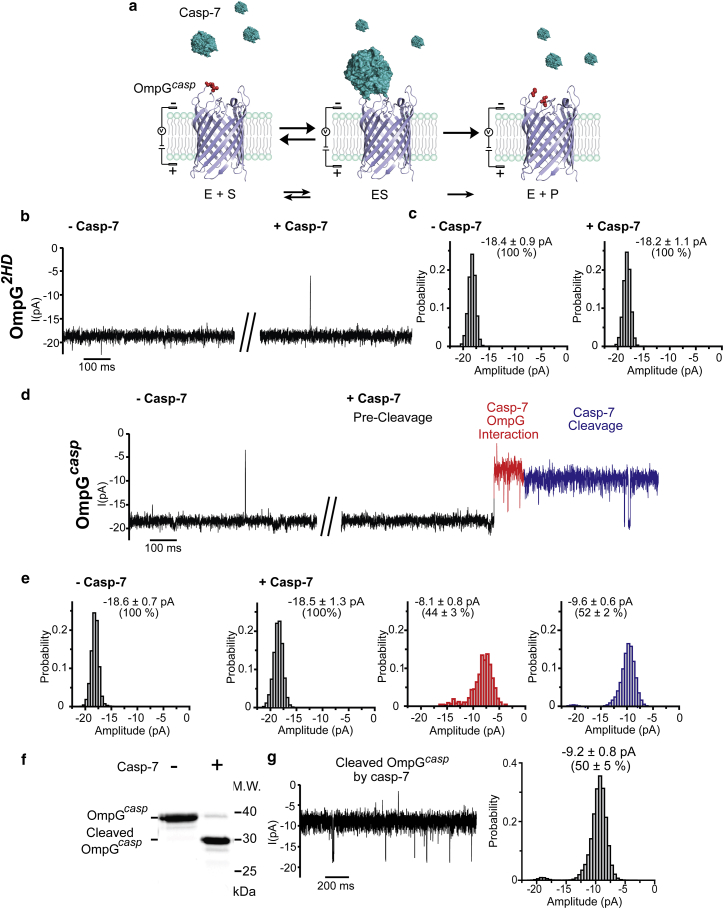
Because our previous study revealed that the OmpG nanopore was able to resolve protein homologs (70), we further tested whether the OmpGcasp nanopore is also capable of detecting any specific interactions with casp-7 using single-channel recording (Fig. 2 a), which could be used to discriminate its presence from other caspases. In the absence of casp-7, the current trace of OmpGcasp was indistinguishable from that of the OmpG2HD, indicating the inserted peptide had little effect on the gating behavior of the OmpG nanopore (Fig. 2, b and d, left panels; Fig. S3). After adding the casp-7 (at a final concentration of 0.2 μM) into the recording chamber where the OmpG loops were located, we observed no change in the open current of the OmpG2HD (Fig. 2, b and c, right panels), whereas the current of OmpGcasp decreased to ∼52%, from −18.5 ± 1.3 to −9.6 ± 0.6 pA (n = 36) (Fig. 2, d and e). This signal was irreversible (Fig. S4), implying it was due to casp-7 cleaving the loop. Interestingly, before the postcleavage signal of the 52% current is observed, a unique current signature around −8.1 pA, 44% of the open current, was observed from every independent pore (more than 150 pores in total) after adding casp-7 (Fig. 2, d and e). We termed the characteristic 44% current signal as “casp-7-OmpG interaction.” We suspect that the interactions between the OmpG loops and casp-7 are weak or extremely transient because we did not observe any reversible caspase binding states before cleavage of OmpGcasp (Fig. 2, d and e). However, the casp-7-OmpG interaction may be enhanced after casp-7 and OmpGcasp form a relatively stable conformation, resulting in the 44% decrease in the open pore current before the postcleavage signal. We suspect that this signal may represent the tetrahedral intermediate state of the proteolytic event.
Figure 2.
Cleavage signals of OmpGcasp generated by casp-7. (a) A schematic of caspase detection by OmpGcasp nanopore. (b) Representative current recording trace and (c) the events amplitude histogram of OmpG2HD before and after adding casp-7. (d) Representative single-channel recording trace and (e) the corresponding all-events histogram of OmpGcasp before and after adding casp-7. The postcleavage event is highlighted in blue, and the casp-7-OmpG interaction is in red. The histogram was fitted with a Gaussian function to derive the current amplitude. The average current was calculated from at least four individual pores, and the errors represent the SD. The percentage of the current was normalized against the current amplitude of the precleaved OmpGcasp pore that was set as 100%. (f) SDS-PAGE analysis of cleaved OmpGcasp by casp-7. Gel analysis by ImageJ showed that 95% of OmpGcasp was cleaved by casp-7. (g) The representative current recording and the events amplitude histogram of cleaved OmpGcasp by casp-7. Cleaved OmpGcasp was prepared by incubating OmpGcasp proteins with casp-7 (1:1 mol ratio) for 1 h at 23°C in 150 mM NaCl and 50 mM HEPES buffer (pH 7.5). To see this figure in color, go online.
To confirm that the 52% decreased current is due to casp-7 cleavage, we preincubated the OmpGcasp proteins with casp-7 for an hour until ∼95% of the OmpGcasp proteins were cleaved as confirmed by SDS-PAGE (Fig. 2 f). The current traces of the precleaved OmpGcasp were recorded by single-channel recording. We observed that the precleaved pores had an averaged current of −9.2 ± 0.8 pA (n = 4), similar to the pores that were cleaved in situ (Fig. 2 g). Because the cleavage events resulted in a reduction in the open current of OmpGcasp, we surmise that this is because one or both of the newly generated cleaved peptides of loop 6 have persistently collapsed into the pore lumen. The two cleaved peptides of loop 6 are still flexible, so they may briefly flip in and out of the pore lumen, generating the current spikes as shown in Fig. 2, d and g. In addition, we tested the OmpGcasp nanopore with an inactive casp-7 mutant H144A protein in which the catalytic residue H144 was substituted by an alanine (Fig. S5). In the presence of casp-7 H144A, the OmpGcasp nanopore did not generate any noticeable change in the current recording. Furthermore, we also created an OmpG construct containing –GEDDVG– peptide, termed OmpGEDDV as a control. OmpGEDDV has the same amino acid composition as the OmpGcasp, but its peptide –GEDDVG– cannot be cleaved by casp-7 in solution as shown by the SDS-PAGE analysis (Fig. S6). Adding casp-7 to the OmpGEDDV nanopore did not induce any current change similar to the 52% decrease in the OmpGcasp nanopore (Fig. S6). Taken together, these data indicate that the significant change in the current of OmpGcasp is caused by the cleavage of the substrate peptide DEVD by casp-7.
Selectivity of OmpGcasp over other proteases
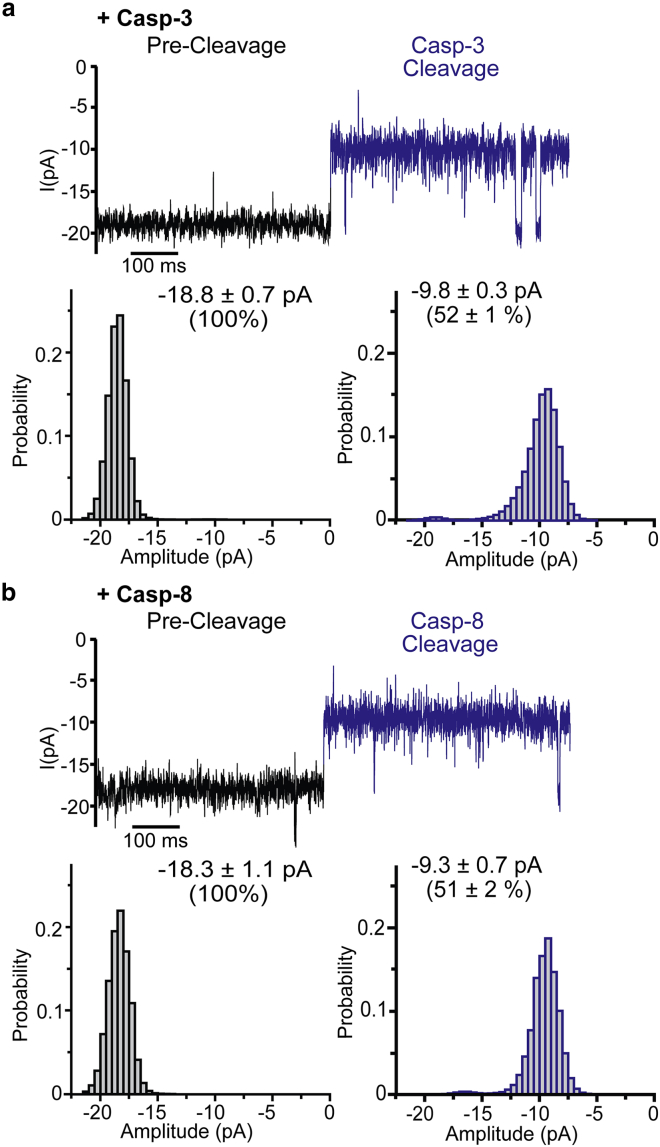
Because the cleavage assay showed that OmpGcasp can be cleaved by other caspases with different efficiency (Fig. 1, b and c), we then tested whether OmpGcasp could differentiate any different cleavage and/or interaction signals between OmpGcasp and other caspases in situ. After the addition of casp-3 (a final concentration of 0.2 μM), an ∼52% decrease in the open pore current was also observed, from −18.8 ± 0.7 to −9.8 ±0.3 pA (n = 10) (Fig. 3 a), like casp-7 cleavage. Similarly, casp-8 (final concentration of 0.7 μM) generated an irreversible signal after cleavage, the open pore current changed from −18.3 ± 1.1 to −9.3 ± 0.7 pA (n = 3) (Fig. 3 b). Interestingly, neither casp-3 nor casp-8 generated the unique 44% current level that is associated with casp-7-OmpG interaction. The difference in their signals may be contributed by the slight difference between the surface charges of casp-3, -7, and -8 (Fig. S7), as our previous study revealed that OmpG’s loops are involved in interacting and sampling the surface of target proteins via electrostatic interactions (70, 71). This feature is responsible for OmpG’s high sensitivity for resolving protein homologs with different surface charges. Although further studies are required to decipher the precise mechanisms of the interactions between OmpGcasp and caspases, we have shown that this nanopore has the ability to distinguish casp-7 from other caspases despite their similar postcleavage signals.
Figure 3.
Cleavage signals of OmpGcasp generated by caspase-3 and -8. (a and b) Representative single-channel recording traces and the corresponding all-events histogram of events generated by (a) casp-3 and (b) casp-8. The average current was calculated from at least four individual pores, and the errors represent the SD. The final concentration of casp-3 was 0.2 μM whereas that of casp-8 was 0.7 μM. The postcleavage event is highlighted in blue. The current amplitude of precleaved OmpGcasp was normalized to 100%. To see this figure in color, go online.
Although casp-6 showed a weak cleavage activity from the cleavage assay at 1:1 molar ratio with OmpGcasp after 1 h at 23°C (Fig. 1 c), we could not observe any cleavage signal after adding 1.3 μM casp-6 (final concentration) after more than 16 h of recording (n = 3). Casp-9 also did not cleave OmpGcasp in either the cleavage assay (Fig. 1 c) or the single-channel recording even after OmpGcasp was recorded for more than 16 h in the presence of 10 μM casp-9 (n = 3). These results are in line with the previous studies that casp-6 and -9 show poor catalytic activity with the –DEVD– substrate (19, 23). In short, the OmpGcasp nanopore exhibits a great selectivity to analyze the casp-7 activity.
Selective detection of casp-7 over other common proteases
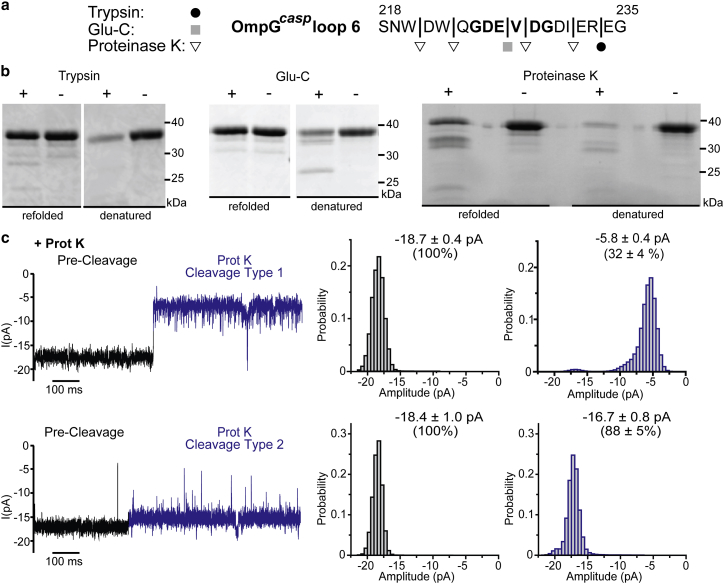
Because of the ubiquitous expression of several classes of proteases, biological samples usually contain complex mixtures of these proteolytic enzymes. To test whether naturally occurring protease sites in OmpGcasp were accessible for unspecific protease cleavages that may generate false-positive signals, we first incubated OmpGcasp with trypsin, Glu-C, or proteinase K (prot K). These three enzymes are serine proteases for which compatible cleavage site(s) on OmpG’s loop 6 and other loops are predicted based on sequence analysis (Figs. 4 a and S8 a). Of these three proteases, only prot K cleaved refolded OmpGcasp in solution (Fig. 4 b). We tested the cleavage efficiency of trypsin and Glu-C with denatured OmpGcasp and found that both trypsin and Glu-C successfully cleaved the denatured OmpG sample (Fig. 4 b). Because the refolding of OmpGcasp is around 95% (Fig. S2), trypsin could cleave the small amount of unfolded/misfolded OmpG nanopore in the sample, which visualized as low molecular-weight bands of cleavage products (less than 5%, analyzed by ImageJ). These results suggest that the relevant cleavage sites in folded OmpGcasp are not accessible to trypsin and Glu-C, presumably because of burial in the barrel or steric protection within the detergent micelle.
Figure 4.
Cleavage assay of OmpGcasp with trypsin, Glu-C, and proteinase K (prot K). (a) Theoretical cleavage sites of trypsin (depicted as black circles), Glu-C (depicted as gray squares), and prot K (depicted as white inverted triangles) in OmpGcasp loop 6. Other theoretical cleavage sites of three proteases in OmpG’s loops are shown in Fig. S8a. (b) SDS-PAGE analysis of the cleavage of OmpG by Glu-C, trypsin, and prot K as visualized by SDS-PAGE. Each protease was added to either the refolded or denatured OmpGcasp with the mole ratio being 1:20. The mixture was diluted with the same volume of buffer (150 mM NaCl 50 mM HEPES (pH = 7.5)), followed by a 4-hr incubation at room temperature (RT ∼23°C). After incubation, all samples were heated at 95°C for 15 min with SDS-PAGE sample buffer, before loading to 12% SDS-PAGE gel. Denatured OmpG protein was prepared by first heating refolded OmpGcasp at 95°C for 15 min with around 3–4 M urea (final concentration). After cooling to RT, the samples were used for cleavage assays. (c) Representative single-channel recording traces and the corresponding all-events histogram of events generated by prot K (the final concentration was 0.2 μM). The postcleavage event is highlighted in blue. The average current was calculated from at least three individual pores, and the errors represent the SD. The percentages of the current were normalized against the current amplitude of the precleaved OmpGcasp pore that was set as 100%. To see this figure in color, go online.
We then investigated how the current signal of OmpGcasp was affected after cleavage by prot K protease. Upon treatment with prot K, we observed two types of prot K cleavage. The first type showed the decreasing in the open current of OmpGcasp from −18.5 ± 0.7 to −5.8 ± 0.4 pA (n = 17), a 32% open current (Fig. 4 c, first row). A 12% decrease in the open pore current was observed in the type 2 prot K cleavage signal (from −18.4 ± 1.0 to −16.7 ± 0.8 pA (n = 3)) (Fig. 4 c, second row). In comparison, cleavage with casp-3 and casp-7 resulted in a 52% open current (Fig. 2, a and b). Because there are four potential proteinase K cleavage sites on loop 6 (Fig. 4 a) as well as several cleavage sites on other loops (Fig. S8 a), we anticipated multiple types of postcleavage signals for prot K. Surprisingly, we observed reproducibly only two types of prot K cleavage current signal with the type 1 signal occurring five times more frequently than that of the type 2 (Fig. 4 c). This is likely due to the prot K’s preference for certain cleavage sites within the loops of OmpGcasp. Notably, the cleavage signals of prot K are significantly different from the cleavage signal generated by casp-3, -7, and -8 (Figs. 2 d and 3). This result suggests that the ionic current generated by OmpGcasp is sensitive to the cleaved peptide products produced by different proteases.
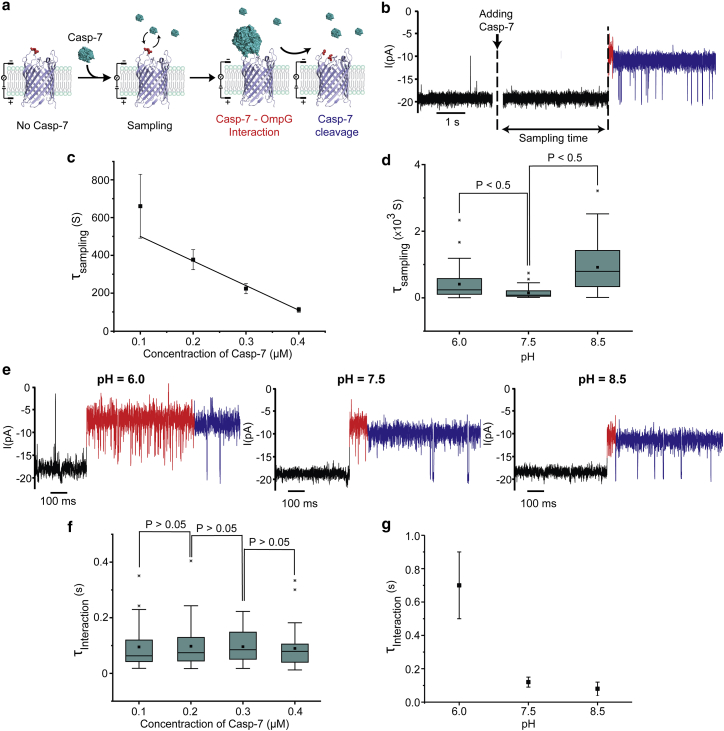
Quantification of casp-7 by the sampling time
To quantify casp-7 concentration, we first explored the effect of the concentration of casp-7 on the time from addition of casp-7 to the occurrence of the interaction signal, termed as the sampling time τsampling (Figs. 5, a and b and S9 a). When the casp-7 concentration was increased from 0.1 to 0.4 μM, the sampling time gradually decreases linearly with the casp-7 concentration (Fig. 5 c). Our results suggest that the sampling time is directly correlated to the activity of casp-7, which can be used for profiling the concentration of active proteases by the OmpG nanopore.
Figure 5.
Quantitative analysis of casp-7 using OmpGcasp. (a) A schematic of the hypothesized cleavage process after adding casp-7 to OmpGcasp. (b) Representative single-channel recording trace of OmpGcasp before and after adding casp-7. The cleavage event is highlighted in blue, and the casp-7–OmpG interaction is in red. The sampling time was measured from adding casp-7 until the interaction signal was observed. (c) Concentration dependence of τsampling. Linear regression has the coefficient of determination (R2) of 0.97. (d) A box-and-whisker plot shows the sampling time in different pH with 0.4 μM casp-7. (e) Representative single-channel recording trace of OmpGcasp after adding casp-7 at different pH values. (f) A box-and-whisker plot shows the casp-7-OmpG interaction time (τinteraction) in different concentrations of casp-7. (g) pH dependence of τinteraction with 0.4 μM casp-7. All the data presented here are from at least 30 independent pores. Error bars represent the SDs from the measurements. In the box-and-whisker plot, the horizontal line in the middle of each box represents the median, whereas the box limits represent the 25th and 75th percentiles. The whiskers represent the minimum and maximum values. The two-tailed p-value from unpaired t-test between two data sets was compared with the significant level (α = 0.05). To see this figure in color, go online.
Because the activity of proteases are dependent on pH (72, 73, 74, 75), we then investigated whether the sampling time would change in different pH. The sampling time was measured at three different pH values, 6.0, 7.5, and 8.5, with 0.4 μM casp-7 (n ≥ 30). The unpaired t-test showed the mean values of sampling time in three pH conditions were significantly different (Table S1). The shortest sampling time was observed at pH 7.5 (Fig. 5 d), consistent with the previous finding that casp-7 is the most active at this pH (72). This result further confirms that the sampling time correlates with the enzymatic activity of caspase-7.
In addition, we investigated the effect of casp-7 concentration and pH on the dwell time τinteraction of the casp-7-OmpG interaction signal (44% current level) (Figs. 5 e and S9 b). We first observed that the average τinteraction is concentration independent (around 0.1 s) at pH = 7.5 at three casp-7 concentrations (Fig. 5 f) because the unpaired t-test showed that the difference of dwell times of casp-7-OmpG interaction between three conditions is not statistically significant (p-value > 0.05) (Table S2). Interestingly, when pH changed from 6.0 to 8.5, the interaction time significantly decreased (Fig. 5 g). We surmise that the casp-7-OmpG interaction current state may represent an intermediate state in the catalytic cycle of the caspase-7. The decreasing in τinteraction in elevated pH may be due to the fact that the higher pH promotes the deprotonation of the catalytic cysteine residue (72, 76, 77), which participates in the nucleophilic attack on the peptide carbonyl bond. Thus, the dwell time of the unique interaction signal is only dependent on how long the intermediate complex of casp-7 and OmpGcasp exists before cleavage. It is unclear why casp-7 can temporarily be trapped in an intermediate state with OmpGcasp before the release of the product whereas other caspases were unable to trigger a similar signal. Previously, we have observed that the electrostatic interaction between the OmpG sensor and the analyte proteins determines the selectivity of the OmpG in differentiating the protein homologs and isoforms (45, 70, 71). Thus, we surmise that the surface charge distribution of OmpG loops and the casp-7 active site, as well as the sequence of loop 6 may play a role in facilitating the formation of the casp-7-OmpGcasp complex. Further studies, such as altering the residues at the OmpG surface or near the active site of casp-7 and exploring the isotope effect in deuterium buffers are required to understand the origin and nature of the casp-7-OmpG interaction signal (e.g., the noncovalent casp-7-OmpG intermediate or the tetrahedral intermediates). Nevertheless, our findings suggest the potential use of OmpG for profiling protease activity as well as studying the catalytic mechanisms.
Direct detection of caspases in bacterial lysates
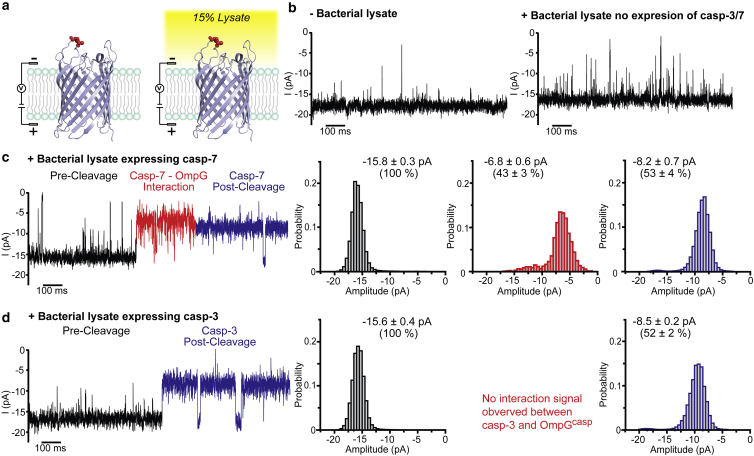
Previous studies have applied nanopores to sense analytes in samples containing serum (47, 57). To determine if the OmpGcasp nanopore could be used to analyze “real-world” samples with minimal sample preparation, we tested whether OmpGcasp could distinguish casp-3 and casp-7 activity directly in E. coli cell lysates (Fig. S10). E. coli has many endogenous proteases, such as Lon, ClpXP, and HflB (78, 79). We first tested the bacterial lysate without the caspase expression with our OmpGcasp to verify if those endogenous proteases would interfere with the caspase detection. We found that the bilayer became unstable in the presence of ≥30% lysate, so 15% lysate was used in this study (Fig. 6 a). The addition of 15% (v/v) E. coli lysate lacking caspase expression slightly increased the gating signal in the OmpGcasp nanopores. A slight decrease in the open current, from −18.5 ± 0.7 to −15.6 ± 0.4 pA (Fig. 6 b) was also observed. This is because the conductivity of the bacterial lysate (in phosphate buffered saline, ∼18 mS/cm) was lower than that of the recording buffer (300 mM KCl, 50 mM HEPES, ∼40 mS/cm). The increased OmpGcasp gating signals likely resulted from interactions between OmpGcasp and cellular components, such as proteins, nucleic acids, or small molecules in the cell lysate. In short, no similar signal to casp-3/7 postcleavage was observed after more than 40 min of recording with E. coli lysate without caspase expression (n = 5) (Fig. S11).
Figure 6.
Direct detection of two caspases in bacterial cell lysate. (a) A schematic of OmpG nanopore in absence and presence of lysate. (b) Representative current traces of OmpGcasp pore in the absence and presence of lysates. 160 μL lysate was added to the loop-containing chamber for a final concentration of 15% (v/v). (c and d) Representative single-channel current recording traces and the corresponding all-events histogram of the events generated by (c) casp-3 and (d) casp-7. The postcleavage event is in blue, and the casp-7-OmpG interaction is in red. The average current was calculated from at least four individual pores, and the errors represent the SD. The current percentages of the events were normalized against the current amplitude of the precleaved OmpGcasp pore that was set as 100%. To see this figure in color, go online.
The cell lysates expressing casp-3 or casp-7 were then added to the OmpGcasp nanopore. The addition of the lysates to the recording chamber induced a reduction in open pore current by 52% (Fig. 6, c and d) after less than 10 min (n = 4), which is similar to the postcleavage signal from purified proteins (Fig. 2, a and b). Importantly, a casp-7-OmpG interaction signal was readily visible (Fig. 6 d) with the average τinteraction 0.12 ± 0.10 s (n = 4). Although the current traces for caspase cleavage in the presence of lysate were slightly noisier than those in the absence of lysate, the characteristic signals of the interaction and cleavage of both caspases were well preserved, which allowed the unambiguous detection and differentiating casp-7 from casp-3 in a complex mixture.
Conclusion
In this study, we have developed a real-time, label-free method based on the OmpG nanopore for selectively detecting the caspase-7 activity directly in 15% (v/v) E. coli cell lysates. The OmpG nanopore with an inserted peptide sequence for protein detection has several advantages. First, it simplifies the production of OmpG sensors with the 100% yield. The previous OmpG-biotin nanopore, in comparison, requires a postpurification chemical modification to attach a ligand, in which the 100% labeling yield was difficult to achieve. Second, the OmpGcasp nanopore shows high selectivity for casp-7 over common interfering proteases such as proteinase K, trypsin, and Glu-C. Although the DEVDG sequence is the most efficient substrate sequence for both casp-3 and casp-7 (15), our OmpG nanopore with the DEVDG incorporated sequence is able to selectively detect casp-7 from its homolog, casp-3. Our OmpGcasp nanopore also succeeds to distinguish casp-7 from some other caspases, which may inefficiently cleave DEVDG, including casp-6, -8, and -9. Because other caspases differ from casp-7 with respect to the substrate specificity (i.e., casp-2 for VDVADG, casp-9 for WEHDG, and casp-10 for LEHDG) (15). Thus, in practice, the OmpGcasp approach can be used to identify the casp-7 activity in the presence of the peptide inhibitors that are preferentially cleaved by other caspases family members (e.g., VDVADG, WEHDG, and LEHDG to mask the potential interferences from casp-2, casp-9, and casp-10).
Our study also highlighted the feasibility of directly discriminating two highly related proteases in complex mixtures, like diluted bacterial cell lysates. To our knowledge, our OmpGcasp nanopore is the first tool for specific detection of casp-7 activity. In addition, no other nanopore system has shown this ability to directly distinguish two highly homologous proteases in diluted cell lysate. Moreover, our nanopore design can quantify the activity of caspases by measuring the sampling time. With its ability to specifically identify and quantify proteins in diluted cell lysates, the OmpG nanopore shows the potential to be used as a fast and powerful detection platform for proteomics research as well as disease diagnostics (80, 81).
With our current sensing scheme, in which only one OmpG pore is recorded at a time, quick quantification of caspase activity is not feasible. By integrating OmpG into a nanopore array, such as the MinION platform from Oxford Nanopore Technology (Oxford, United Kingdom), in which several hundred protein pores can be recorded simultaneously, the kinetics of proteolytic activity could be rapidly and accurately estimated (82, 83). Similarly, we were not able to directly analyze the casp-7 activity in the mixture of casp-3 and casp-7 under the current instrument setting. This limitation could also be circumvented by using a nanopore array. Previous study has shown that enzymatic efficiency of casp-3 is around threefold higher than that of casp-7 (17, 25). Thus, with nanopore arrays, ∼25% of pores are expected be cleaved by casp-7 when casp-3 and casp-7 are equally present in the sample. When the concentration of casp-3 becomes dominant than casp-7, the activity of casp-3 will likely mask the casp-7. In such a case, casp-3-specific inhibitors (19, 84) could be added to the sample until the casp-7 signal becomes significant for quantification. In addition, future work can be done to tune the OmpGcasp nanopore to achieve a higher cleavage efficiency for casp-7 over casp-3. In this way, an OmpGcasp array could be used for profiling casp-7 activity directly from real-world samples such as serum or cell lysate. In the future, this approach may be extended to detect other proteolytic enzymes of equally high medical relevance, such as HIV protease, Hepatitis C protease, or the flavivirus proteases.
Author Contributions
B.P. designed and performed the experiments, analyzed data, and wrote the manuscript. X.L. participated in the single-channel experiment. S.J.E., M.E.H., and J.A.H. provided caspase samples and the expertise to work with caspases. M.A.F. initiated the study. M.C. conceived, designed, and coordinated the study and wrote the manuscript. All authors edited the manuscript and gave final approval for publication.
Acknowledgments
B.P. was supported in part by a fellowship from the University of Massachusetts as part of the Chemistry-Biology Interface training grant (National Institutes of Health T32 GM008515). This research was supported by the US National Science Foundation grant CBET 1511367 (to J.A.H.) and the US National Institutes of Health grant R01-GM115442 (to M.C.).
Editor: Thomas Perkins.
Footnotes
Scott J. Eron’s present address is C4 Therapeutics, Cambridge, Massachusetts.
Maureen E. Hill’s present address is FogPharma, Cambridge, Massachusetts.
Monifa A. Fahie’s present address is Department of Biology, Brandeis University, Waltham, Massachusetts.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.07.045.
Supporting Material
References
- 1.Kuida K., Zheng T.S., Flavell R.A. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 2.Yuan J., Shaham S., Horvitz H.R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 β-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 3.Ghayur T., Banerjee S., Allen H. Caspase-1 processes IFN-γ-inducing factor and regulates LPS-induced IFN-γ production. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 4.Martinon F., Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Harvey N.L., Kumar S. The role of caspases in apoptosis. Adv. Biochem. Eng. Biotechnol. 1998;62:107–128. doi: 10.1007/BFb0102307. [DOI] [PubMed] [Google Scholar]
- 6.McIlwain D.R., Berger T., Mak T.W. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seaman J.E., Julien O., Wells J.A. Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues. Cell Death Differ. 2016;23:1717–1726. doi: 10.1038/cdd.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Amelio M., Sheng M., Cecconi F. Caspase-3 in the central nervous system: beyond apoptosis. Trends Neurosci. 2012;35:700–709. doi: 10.1016/j.tins.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Graham R.K., Deng Y., Hayden M.R. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Leyva M.J., Degiacomo F., Ellman J.A. Identification and evaluation of small molecule pan-caspase inhibitors in Huntington’s disease models. Chem. Biol. 2010;17:1189–1200. doi: 10.1016/j.chembiol.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putt K.S., Chen G.W., Hergenrother P.J. Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nat. Chem. Biol. 2006;2:543–550. doi: 10.1038/nchembio814. [DOI] [PubMed] [Google Scholar]
- 12.Olsson M., Zhivotovsky B. Caspases and cancer. Cell Death Differ. 2011;18:1441–1449. doi: 10.1038/cdd.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotchkiss R.S., Nicholson D.W. Apoptosis and caspases regulate death and inflammation in sepsis. Nat. Rev. Immunol. 2006;6:813–822. doi: 10.1038/nri1943. [DOI] [PubMed] [Google Scholar]
- 14.Talanian R.V., Quinlan C., Wong W.W. Substrate specificities of caspase family proteases. J. Biol. Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 15.Julien O., Wells J.A. Caspases and their substrates. Cell Death Differ. 2017;24:1380–1389. doi: 10.1038/cdd.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh J.G., Cullen S.P., Martin S.J. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl. Acad. Sci. USA. 2008;105:12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McStay G.P., Salvesen G.S., Green D.R. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 18.Hill M.E., MacPherson D.J., Hardy J.A. Reprogramming caspase-7 specificity by regio-specific mutations and selection provides alternate solutions for substrate recognition. ACS Chem. Biol. 2016;11:1603–1612. doi: 10.1021/acschembio.5b00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers C.J., González-Páez G.E., Wolan D.W. Selective detection and inhibition of active caspase-3 in cells with optimized peptides. J. Am. Chem. Soc. 2013;135:12869–12876. doi: 10.1021/ja406399r. [DOI] [PubMed] [Google Scholar]
- 20.Agard N.J., Mahrus S., Wells J.A. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc. Natl. Acad. Sci. USA. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornberry N.A., Rano T.A., Nicholson D.W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 22.Brentnall M., Rodriguez-Menocal L., Boise L.H. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013;14:32–40. doi: 10.1186/1471-2121-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger A.B., Sexton K.B., Bogyo M. Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res. 2006;16:961–963. doi: 10.1038/sj.cr.7310112. [DOI] [PubMed] [Google Scholar]
- 24.Vickers C.J., González-Páez G.E., Wolan D.W. Selective detection of caspase-3 versus caspase-7 using activity-based probes with key unnatural amino acids. ACS Chem. Biol. 2013;8:1558–1566. doi: 10.1021/cb400209w. [DOI] [PubMed] [Google Scholar]
- 25.Vickers C.J., González-Páez G.E., Wolan D.W. Discovery of a highly selective caspase-3 substrate for imaging live cells. ACS Chem. Biol. 2014;9:2199–2203. doi: 10.1021/cb500586p. [DOI] [PubMed] [Google Scholar]
- 26.Shi W., Friedman A.K., Baker L.A. Nanopore sensing. Anal. Chem. 2017;89:157–188. doi: 10.1021/acs.analchem.6b04260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu L.Q., Shim J.W. Single molecule sensing by nanopores and nanopore devices. Analyst. 2010;135:441–451. doi: 10.1039/b907735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howorka S., Siwy Z. Nanopore analytics: sensing of single molecules. Chem. Soc. Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 29.Ma L., Cockroft S.L. Biological nanopores for single-molecule biophysics. ChemBioChem. 2010;11:25–34. doi: 10.1002/cbic.200900526. [DOI] [PubMed] [Google Scholar]
- 30.Kasianowicz J.J., Robertson J.W., Stanford V.M. Nanoscopic porous sensors. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 2008;1:737–766. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 31.Majd S., Yusko E.C., Mayer M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010;21:439–476. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayley H., Cremer P.S. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 33.Kasianowicz J.J., Brandin E., Deamer D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branton D., Deamer D.W., Schloss J.A. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wanunu M. Nanopores: a journey towards DNA sequencing. Phys. Life Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Jong L.C., Cree S., Walker L.C., kConFab Investigators Nanopore sequencing of full-length BRCA1 mRNA transcripts reveals co-occurrence of known exon skipping events. Breast Cancer Res. 2017;19:127–136. doi: 10.1186/s13058-017-0919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain M., Koren S., Loose M. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stranges P.B., Palla M., Church G.M. Design and characterization of a nanopore-coupled polymerase for single-molecule DNA sequencing by synthesis on an electrode array. Proc. Natl. Acad. Sci. USA. 2016;113:E6749–E6756. doi: 10.1073/pnas.1608271113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuller C.W., Kumar S., Ju J. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc. Natl. Acad. Sci. USA. 2016;113:5233–5238. doi: 10.1073/pnas.1601782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammad M.M., Movileanu L. Protein sensing with engineered protein nanopores. Methods Mol. Biol. 2012;870:21–37. doi: 10.1007/978-1-61779-773-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Meervelt V., Soskine M., Maglia G. Detection of two isomeric binding configurations in a protein-aptamer complex with a biological nanopore. ACS Nano. 2014;8:12826–12835. doi: 10.1021/nn506077e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rotem D., Jayasinghe L., Bayley H. Protein detection by nanopores equipped with aptamers. J. Am. Chem. Soc. 2012;134:2781–2787. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohammad M.M., Iyer R., Movileanu L. Engineering a rigid protein tunnel for biomolecular detection. J. Am. Chem. Soc. 2012;134:9521–9531. doi: 10.1021/ja3043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie H., Braha O., Bayley H. Single-molecule observation of the catalytic subunit of cAMP-dependent protein kinase binding to an inhibitor peptide. Chem. Biol. 2005;12:109–120. doi: 10.1016/j.chembiol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 45.Fahie M., Chisholm C., Chen M. Resolved single-molecule detection of individual species within a mixture of anti-biotin antibodies using an engineered monomeric nanopore. ACS Nano. 2015;9:1089–1098. doi: 10.1021/nn506606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soskine M., Biesemans A., Maglia G. Single-molecule analyte recognition with ClyA nanopores equipped with internal protein adaptors. J. Am. Chem. Soc. 2015;137:5793–5797. doi: 10.1021/jacs.5b01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., Haque F., Guo P. Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano. 2013;7:9814–9822. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheley S., Xie H., Bayley H. A genetically encoded pore for the stochastic detection of a protein kinase. ChemBioChem. 2006;7:1923–1927. doi: 10.1002/cbic.200600274. [DOI] [PubMed] [Google Scholar]
- 49.Kukwikila M., Howorka S. Nanopore-based electrical and label-free sensing of enzyme activity in blood serum. Anal. Chem. 2015;87:9149–9154. doi: 10.1021/acs.analchem.5b01764. [DOI] [PubMed] [Google Scholar]
- 50.Willems K., Van Meervelt V., Maglia G. Single-molecule nanopore enzymology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0230. 20160230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L., Han Y., Guan X. Real-time label-free measurement of HIV-1 protease activity by nanopore analysis. Biosens. Bioelectron. 2014;62:158–162. doi: 10.1016/j.bios.2014.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou S., Wang L., Guan X. Label-free nanopore single-molecule measurement of trypsin activity. ACS Sens. 2016;1:607–613. doi: 10.1021/acssensors.6b00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kukwikila M., Howorka S. Electrically sensing protease activity with nanopores. J. Phys. Condens. Matter. 2010;22:454103. doi: 10.1088/0953-8984/22/45/454103. [DOI] [PubMed] [Google Scholar]
- 54.Zhao Q., de Zoysa R.S., Guan X. Real-time monitoring of peptide cleavage using a nanopore probe. J. Am. Chem. Soc. 2009;131:6324–6325. doi: 10.1021/ja9004893. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Montana V., Gu L.Q. Nanopore sensing of botulinum toxin type B by discriminating an enzymatically cleaved Peptide from a synaptic protein synaptobrevin 2 derivative. ACS Appl. Mater. Interfaces. 2015;7:184–192. doi: 10.1021/am5056596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrington L., Cheley S., Bayley H. Stochastic detection of Pim protein kinases reveals electrostatically enhanced association of a peptide substrate. Proc. Natl. Acad. Sci. USA. 2013;110:E4417–E4426. doi: 10.1073/pnas.1312739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fahie M.A., Yang B., Chen M. Selective detection of protein homologues in serum using an OmpG nanopore. Anal. Chem. 2015;87:11143–11149. doi: 10.1021/acs.analchem.5b03350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yildiz O., Vinothkumar K.R., Kühlbrandt W. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 2006;25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang B., Tamm L.K. Structure of outer membrane protein G by solution NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subbarao G.V., van den Berg B. Crystal structure of the monomeric porin OmpG. J. Mol. Biol. 2006;360:750–759. doi: 10.1016/j.jmb.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 61.Conlan S., Zhang Y., Bayley H. Biochemical and biophysical characterization of OmpG: a monomeric porin. Biochemistry. 2000;39:11845–11854. doi: 10.1021/bi001065h. [DOI] [PubMed] [Google Scholar]
- 62.Korkmaz-Ozkan F., Köster S., Yildiz O. Correlation between the OmpG secondary structure and its pH-dependent alterations monitored by FTIR. J. Mol. Biol. 2010;401:56–67. doi: 10.1016/j.jmb.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 63.Chen M., Khalid S., Bayley H. Outer membrane protein G: engineering a quiet pore for biosensing. Proc. Natl. Acad. Sci. USA. 2008;105:6272–6277. doi: 10.1073/pnas.0711561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaidya S., Velázquez-Delgado E.M., Hardy J.A. Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J. Mol. Biol. 2011;406:75–91. doi: 10.1016/j.jmb.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Witkowski W.A., Hardy J.A. A designed redox-controlled caspase. Protein Sci. 2011;20:1421–1431. doi: 10.1002/pro.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eron S.J., MacPherson D.J., Hardy J.A. Multiple mechanisms of zinc-mediated inhibition for the apoptotic caspases-3, -6, -7, and -8. ACS Chem. Biol. 2018;13:1279–1290. doi: 10.1021/acschembio.8b00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maelfait J., Beyaert R. Non-apoptotic functions of caspase-8. Biochem. Pharmacol. 2008;76:1365–1373. doi: 10.1016/j.bcp.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 68.Chen M., Li Q.H., Bayley H. Orientation of the monomeric porin OmpG in planar lipid bilayers. ChemBioChem. 2008;9:3029–3036. doi: 10.1002/cbic.200800444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pop C., Salvesen G.S. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fahie M.A., Chen M. Electrostatic interactions between OmpG nanopore and analyte protein surface can distinguish between glycosylated isoforms. J. Phys. Chem. B. 2015;119:10198–10206. doi: 10.1021/acs.jpcb.5b06435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fahie M.A., Yang B., Chen M. Tuning the selectivity and sensitivity of an OmpG nanopore sensor by adjusting ligand tether length. ACS Sens. 2016;1:614–622. doi: 10.1021/acssensors.6b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stennicke H.R., Salvesen G.S. Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 73.Vernet T., Tessier D.C., Ménard R. Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 1995;270:16645–16652. doi: 10.1074/jbc.270.28.16645. [DOI] [PubMed] [Google Scholar]
- 74.Rezanejad H., Karbalaei-Heidari H.R., Yousefi R. Microsciadin, a new milk-clotting cysteine protease from an endemic species, Euphorbia microsciadia. Biomacromol. J. 2015;1:93–103. [Google Scholar]
- 75.Hsiao N.W., Chen Y., Kao C.H. Purification and characterization of an aspartic protease from the Rhizopus oryzae protease extract, peptidase R. Electron. J. Biotechnol. 2014;17:89–94. [Google Scholar]
- 76.Fuentes-Prior P., Salvesen G.S. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elsässer B., Zauner F.B., Brandstetter H. Distinct roles of catalytic cysteine and histidine in the protease and ligase mechanisms of human legumain as revealed by DFT-based QM/MM simulations. ACS Catal. 2017;7:5585–5593. doi: 10.1021/acscatal.7b01505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gottesman S. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 79.Lazdunski A.M. Peptidases and proteases of Escherichia coli and Salmonella typhimurium. FEMS Microbiol. Rev. 1989;5:265–276. doi: 10.1016/0168-6445(89)90035-1. [DOI] [PubMed] [Google Scholar]
- 80.Nomura D.K., Dix M.M., Cravatt B.F. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao J., Patwa T.H., Simeone D.M. Protein biomarkers in cancer: natural glycoprotein microarray approaches. Curr. Opin. Mol. Ther. 2008;10:602–610. [PMC free article] [PubMed] [Google Scholar]
- 82.Feng Y., Zhang Y., Du C. Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics. 2015;13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garalde D.R., Snell E.A., Turner D.J. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- 84.Schroeder T., Barandun J., Grütter M.G. Specific inhibition of caspase-3 by a competitive DARPin: molecular mimicry between native and designed inhibitors. Structure. 2013;21:277–289. doi: 10.1016/j.str.2012.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.