Abstract
Antimicrobial resistance (AMR) is one of the greatest public health challenges we are currently facing. To develop effective interventions against this, it is essential to understand the processes behind the spread of AMR. These are partly dependent on the dynamics of horizontal transfer of resistance genes between bacteria, which can occur by conjugation (direct contact), transformation (uptake from the environment) or transduction (mediated by bacteriophages). Mathematical modelling is a powerful tool to investigate the dynamics of AMR; however, the extent of its use to study the horizontal transfer of AMR genes is currently unclear. In this systematic review, we searched for mathematical modelling studies that focused on horizontal transfer of AMR genes. We compared their aims and methods using a list of predetermined criteria and used our results to assess the current state of this research field. Of the 43 studies we identified, most focused on the transfer of single genes by conjugation in Escherichia coli in culture and its impact on the bacterial evolutionary dynamics. Our findings highlight the existence of an important research gap in the dynamics of transformation and transduction and the overall public health implications of horizontal transfer of AMR genes. To further develop this field and improve our ability to control AMR, it is essential that we clarify the structural complexity required to study the dynamics of horizontal gene transfer, which will require cooperation between microbiologists and modellers.
Keywords: antimicrobial resistance, horizontal gene transfer, mathematical modelling, epidemiology, microbiology
1. Introduction
Antimicrobial resistance (AMR) is undeniably one of the greatest global public health challenges we are currently facing [1]. The recent discoveries on the spread of resistance genes for key antimicrobials such as NDM-1 for carbapenem resistance [2–4] suggest that to tackle this challenge, instead of only studying the spread of resistant bacteria, we must understand the processes by which individual resistance genes spread. The first is ‘vertical gene transfer’, where genes are passed from parent to progeny during bacterial replication. The second, which is our focus here, is ‘horizontal gene transfer’ (HGT). This allows bacteria to acquire genetic material, including AMR genes, from their environment or other bacteria [5–7]. There are three mechanisms of HGT. First, ‘transformation’ is the capacity of bacteria to intake genetic material from their environment. Second, ‘conjugation’ occurs when two bacteria come into contact with each other and form a conjugative bridge, enabling direct exchange of genetic material. Finally, ‘transduction’ occurs when a bacteriophage (a virus that can infect bacteria) replicates and packages a bacterial gene instead of its own genetic material and then acts as a vector and transfers this gene into another bacterium.
The consequences of HGT of AMR in a bacterial population are varied and can change depending on the setting where this process occurs. First, HGT can often be at the origin of new combinations of resistances to multiple antimicrobials in single bacteria strains [8]. This is amplified by the fact that HGT can occur both intraspecies and interspecies [9], therefore allowing for mixing between many different gene pools. Fortunately, these resistance mechanisms often impose a fitness cost that reduces the competitiveness of bacteria with AMR genes in settings where antibiotics are absent [10], thereby limiting the increase in the prevalence of these bacteria in the environment. Studying HGT of AMR can be further complicated by differences in transfer rates and importance of transfer mechanisms between bacterial species [11], with transformation, for example, being rare for Staphylococcus aureus [12] but common for Neisseria gonorrhoea [13], and by differences between rates estimated in vitro and in vivo, as was seen with transduction in Staphylococcus aureus [14] and conjugation in Klebsiella pneumoniae and Escherichia coli [15]. Finally, HGT dynamics appear to vary depending on the presence or absence of antibiotics in the surrounding environment [16–20], therefore requiring studies to be conducted in multiple settings to fully capture this process.
It is essential to fully understand HGT of AMR since it can impact the overall transmission of AMR, and therefore the predicted effect of interventions against bacterial infections, to varying degrees depending on the setting. A most striking example of this is phage therapy, where bacteriophages are proposed as antimicrobials. A risk is that therapeutic phages could perform transduction and increase the proportion of bacteria in the patient which carry a resistance gene. In that case, if the phage therapy treatment fails to clear all the bacteria, this could leave the patient at a higher risk of antimicrobial-resistant bacteria infection [21,22]. In addition to the aforementioned differences between bacterial species, HGT mechanisms themselves are biologically complex. For example, the capacity to form a conjugative bridge generally requires the presence of a specific set of ‘tra’ genes [23]. These can themselves be transferred, leading to an increase through time in the prevalence of bacteria that can perform conjugation. Transformation gene expression is extremely variable depending on the environmental conditions that bacteria are exposed to [6], and therefore we cannot realistically assume that bacteria are able to perform transformation at all times. Finally, some phages can undergo either a ‘lytic cycle’, where they immediately replicate upon infecting a bacterium, or a ‘lysogenic cycle’, where they first integrate into the bacterial genome for a variable duration [12]. Consequently, transduction dynamics can be further complicated by the characteristics of the phage life cycle.
Therefore, HGT is complex in its dynamics, and studying these requires appropriate tools. Mathematical modelling is often used to study infectious disease processes [24]. It provides a simulation environment that can be informed by real-life data, in which dynamics can be disentangled and easily studied. Mathematical models can be split into ‘deterministic models', which always generate the same results for a given set of parameter values [24], and ‘stochastic models’, which generate variability in their results using random events [24]. Mathematical modelling is already being used to study AMR dynamics and their public health implications [25,26]. For example, it has been employed to study within-host bacterial dynamics (i.e. the bacterial processes that occur during colonization or infection of a host) and derive conclusions on patterns of AMR seen in the host population [27]. Consequently, it can provide novel insight into optimal strategies to combat AMR spread by analysing the effect that these have on the transmission dynamics [28]. However, existing models may not always capture the relevant and complex microbiological dynamics of HGT. In this systematic review, we aimed to find modelling studies that focus on HGT of AMR to record their methods and research questions and, hence, to identify potential research gaps and areas for improvement in this field.
2. Methods
The methodology of our systematic review follows the recommended PRISMA guidelines [29].
2.1. Inclusion criteria
In order to be included in this review, studies had to fulfil all of the following criteria:
-
(1)
Study the horizontal transfer of genes between bacteria.
-
(2)
The genes studied must explicitly be identified as genes encoding AMR.
-
(3)
Use at least one dynamic population model. A model is ‘dynamic’ if it tracks the changes in the number of bacteria belonging to various populations (e.g. antibiotic-resistant and -susceptible bacteria) over time.
2.2. Screening process
The entire screening process is summarized in figure 1. We searched two databases on 26 April 2019 using the following terms:
-
—
PubMed search: ‘(antimicrobial OR antibacterial OR antibiotic) resist* AND (horizontal transfer OR mobile genetic element OR plasmid OR transformation OR conjugation OR transduction OR phage) AND (math* OR dynamic*) model*’, 171 results.
-
—
Web of Science search: ‘TS = ((antimicrobial OR antibacterial OR antibiotic) resist* AND (horizontal transfer OR mobile genetic element OR plasmid OR transformation OR conjugation OR transduction OR phage) AND (math* OR dynamic*) model*), 185 results.
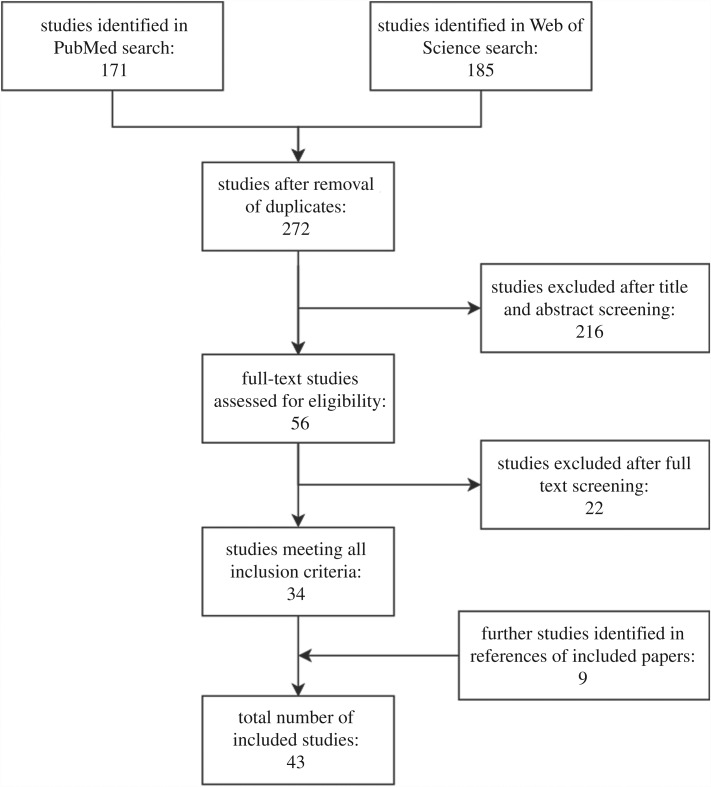
Figure 1.
PRISMA flow diagram of the search and exclusion process.
After removal of duplicates, these combined searches yielded a list of 272 studies. Both Q.J.L. and G.M.K. independently screened the titles and abstracts of all 272 studies. Fifty-four studies were retained by both authors, and two more were discussed and retained after an additional screen of the methods due to uncertainty, leading to a total of 56 studies retained after the first screening step.
The full texts of these 56 studies were then screened by Q.J.L., leading to 34 studies being retained as relevant for this review. Finally, by screening the reference lists in these 34 studies, 9 more were included, leading to a total of 43 studies to discuss in this review.
2.3. Information extracted from the included studies
To maximize comparability between studies, we devised a list of 11 elements to extract from every study. These are summarized and explained in table 1.
Table 1.
Elements recorded from all included studies. Where no ‘possible values’ are given in the table, this indicates that the values were not restricted to a predetermined list.
| recorded element | signification | possible values |
|---|---|---|
| transfer mechanism | biological mechanism of horizontal gene transfer modelled | ‘conjugation’ or ‘transformation’ or ‘transduction’ |
| bacteria | any species of bacteria explicitly modelled | — |
| aim of the study | whether the study looked at gene transfer to understand evolutionary trends seen in the bacterial population or to understand its impact on public health, or both | ‘evolutionary’ or ‘public health’ or ‘both’ |
| bacterial environment | any environment that contained bacteria in the model | — |
| antibiotic effect considered | whether one or more antibiotic(s) were present in the model(s) | ‘yes’ or ‘no’ |
| multiple resistances considered | whether the model(s) tracked multiple resistance genes that could be transferred separately | ‘yes’ or ‘no’ |
| fitness cost of resistance considered | whether the model(s) included a fitness cost for bacteria carrying a resistance gene | ‘yes’ or ‘no’ |
| source of model parameters | whether the study also generated its own experimental data to support its parameter values, or chose values informed by previous studies (which could be experimental studies or not), or assumed values | ‘experimental’ and/or ‘external’ and/or ‘assumed’ |
| type of model | whether the structure of the model(s) was deterministic or stochastic, or both (if the study presented more than one model) | ‘deterministic’ or ‘stochastic’ or ‘both’ |
| type of parameter values | if the model(s) structure was ‘deterministic’, whether the parameter values were constant or were sampled from distributions before each model run | ‘constant’ or ‘sampled’ |
| sensitivity analysis performed | whether the study performed any type of sensitivity analysis of the effect of model parameter values on the results | ‘yes’ or ‘no’ |
Note that in our analysis, ‘Type of parameter values' and ‘Sensitivity analysis performed’ are two independent criteria. Therefore, we can report that a study only uses ‘Constant’ parameter values, yet still performs a sensitivity analysis. If a study is reported to have ‘Sampled’ parameters, this means that the values of the parameters vary for each model run and that this is represented in the main results, with figures showing the model output with ranges instead of single lines for example. If a sensitivity analysis was performed, this means that the authors report conducting such a procedure to support their findings (e.g. to argue that their choice of ‘Constant’ parameter values is a reasonable assumption and does not significantly affect their results).
3. Results
The table showing all of the recorded elements from the 43 included studies can be found in the electronic supplementary material of this paper.
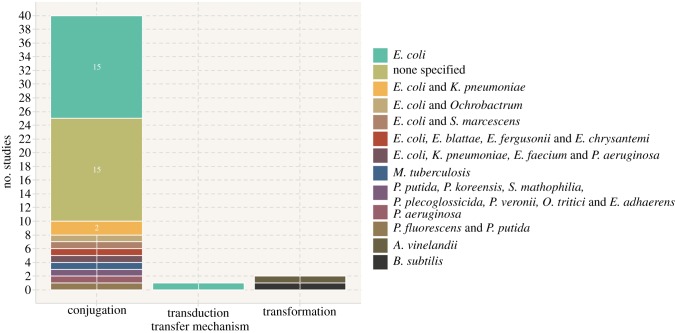
First, when looking at the transfer mechanism modelled by these studies, we observe that almost all exclusively focused on conjugation (40 of 43) [30–69] (figure 2). Of the remaining three, two focused on transformation [70,71] and one on transduction [72]. Additionally, more than a third of the studies (16/43) chose exclusively Escherichia coli as the bacteria in which to model the transfer processes [30,34,36,41–46,52,53,59,64,66,68,72] (figure 2). It is also worth noting that another one-third of the studies (15/43) did not model a specific organism and instead indicate that they are looking at bacteria in general [31,32,37,38,48,51,54,56–58,61,62,65,67,69]. Finally, while eight studies applied their model to more than one bacterial species [33,35,39,40,47,49,60,63], only four of these modelled two strains of bacteria simultaneously and captured interspecies transfer of resistance genes [39,49,60,63].
Figure 2.
Transfer mechanisms and bacterial species modelled in the 43 studies included in our review. (Online version in colour.)
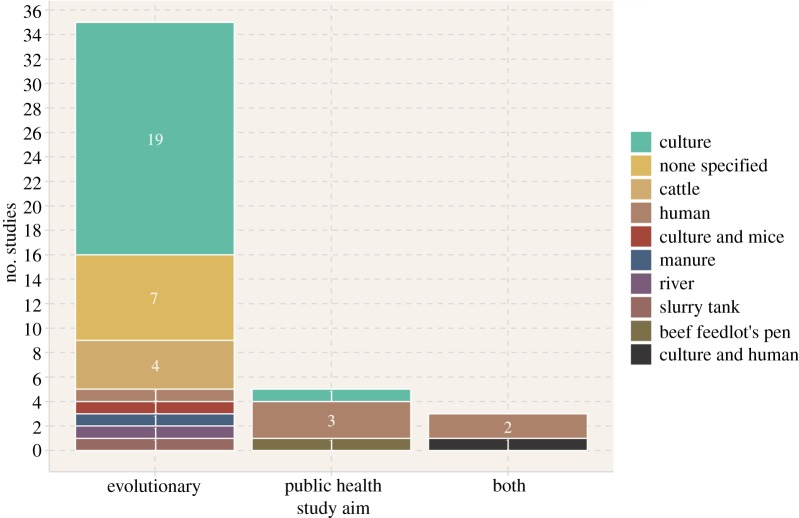
In terms of the aims of these studies, all except eight studies [32,55,58,60,63–65,69] used modelling approaches exclusively to improve the understanding of bacterial evolutionary dynamics (figure 3). This covered questions such as how the prevalence of resistance genes in the bacterial population changes over time (as in [34], for example), or how the rise of multidrug-resistant bacteria varied under different environmental conditions (as in [30], for example). Inversely, the remaining eight studies [32,55,58,60,63–65,69] attempted to place at least some of their results in a public health setting by, for example, quantifying the impact of transfer on the incidence of multidrug-resistant bacteria infection in humans [32,69]. In accordance with this previous point, almost half of the studies (20/43) modelled bacteria exclusively in culture [33–42,47,49,50,52,53,58,59,66,70,71], and only seven modelled bacteria in humans [30,32,55,60,63,65,69] (figure 3). In the remaining studies, seven did not specify an environment for their bacteria [31,48,56,57,61,62,67].
Figure 3.
Aims and environments modelled in the 43 studies included in our review. (Online version in colour.)
Almost all of the studies included a bacterial fitness cost for the carriage of a resistance gene in their models (table 2), except for six [32,42,48,63,66,71]. On the other hand, despite the fact that in reality bacteria can acquire multiple AMR genes independently (i.e. the acquisition of each gene is a separate HGT event), only four studies included this feature [30,32,60,69] (table 2). Finally, it is important to note that almost half of the studies did not model the presence of antibiotics and therefore did not consider the effect of antibiotics on transfer rates [33–36,39–42,47,52,53,59,63,66,68,71,72] (table 2).
Table 2.
Summary of the presence or absence of model characteristics in the 43 studies we reviewed.
| include antibiotic effect | include multiple AMR genes | include fitness cost | include sensitivity analysis | |
|---|---|---|---|---|
| yes | 26 | 4 | 37 | 29 |
| no | 17 | 39 | 6 | 14 |
Almost half of these modelling studies (19/43) included their own experimental work to generate data and estimate at least some parameter values for their models [33–36,39–42,47,49,51–54,59,66,68,70,71] (figure 4). On the other hand, more than half (23/43) chose to assume the values of at least some of their parameters, without explicitly citing any sources to support their choices, and a quarter (12/43) assumed the values of all of their parameters [31,32,37,38,65,67]. Finally, a third (15/43) used previous studies to obtain at least some of their parameter values. For these, except for three studies (two of which were each the direct follow-up of another one on the same topic [44,50], and one an analysis of data collected during an outbreak [63]), more than one previous study was taken to estimate the value of parameters, with a median number of studies of 8 and a maximum of 42.
Figure 4.
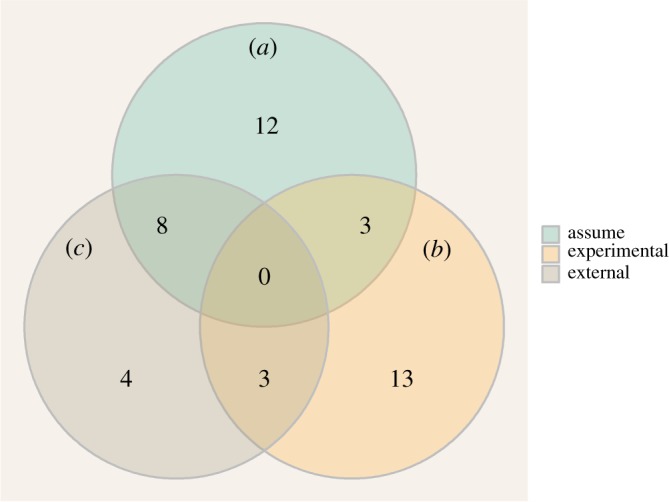
Sources of parameter values in the 43 studies included in our review. ‘Assume’ ((a), green): no clear reference is given to support the choice of parameter value; ‘Experimental’ ((b), orange): the study generated its own experimental data to support the choice of parameter value; ‘External’ ((c) brown): the study references a previous study to support the choice of parameter value. Studies in an overlap region used each of the corresponding methods at least once to estimate the value of their parameters. (Online version in colour.)
Finally, more than three quarters of the studies (33/43) exclusively relied on deterministic models to obtain their results [30,32,34,36–40,42,43,45–51,53–56,58,59,61,63–69,71,72]. All of these deterministic models were composed of a set of ordinary differential equations to track the different subpopulations (susceptible bacteria, resistant bacteria, etc.) through time. As for the ten studies that relied on stochastic models [31,33,35,41,44,52,57,60,62,70], most of these were agent-based models, where the bacteria were tracked individually [31,33,41,52,57,60], while the remaining ones either used stochastic differential equations [44,62,70] or difference equations [35]. Of the studies that exclusively used deterministic models, only eight acknowledge variability in the parameter values by running their model multiple times and sampling parameters from distributions instead of assuming them to be constant [32,38,43,46,56,64,65,72]. Nevertheless, most studies performed sensitivity analyses of the effect of their parameter values on their model results (table 2). Overall, nine studies still relied solely on a deterministic model without either sampling their parameter values or performing sensitivity analyses [30,36,40,42,48,54,55,58,68]. We also noted that except for the one study on transduction [72], all the studies modelled transfer as a mass action process. This assumes that the number of transfer events is determined by multiplying the number of bacteria that can receive the gene, the number of bacteria that can transfer the gene and the rate at which transfer occurs. Therefore, this is generally written as some form of β × S × R/N, where β is a rate of transfer, S is the number of bacteria that can receive the resistance gene, R is the number of bacteria that can provide the resistance gene and N is the total bacterial population in the system.
4. Discussion
We used a systematic literature review of mathematical models of HGT to determine our current understanding of the dynamics of HGT of AMR. The first main observation from our results is that the majority of studies assessed only focus on HGT by conjugation (40 of 43). The likely reason for this is the simplicity of conjugation dynamics. Effectively, these are comparable to infections transmitted upon contact, such as influenza, where established modelling exists using mass action dynamics [24]. Consequently, modelling conjugation does not require much complexity to be added to these models. However, we know that transformation and transduction also contribute to HGT [7,14], and the lack of studies on these mechanisms is worrying.
Conjugation, transformation and transduction fundamentally differ in their biology, making it essential to study each of them in their own modelling framework; it is unknown whether models of conjugation could be directly applied to transformation and transduction. When looking at the studies that attempted to model these two processes, we first see that the one that focused on transduction [72] attempted to place it in a complex setting, with the phage able to undergo both lytic and lysogenic cycles, and the possibility for some bacteria to be resistant to phage infection. Transduction is represented as a multistep process in this model, as opposed to relying on a single rate. The phage must first successfully infect a bacterium and then pick up a resistance gene, before successfully transferring this gene to a different bacterium. This model aims to accurately represent most of the biological complexity of transduction, which necessarily requires many assumptions regarding parameter values. Further study of this trade-off would be greatly beneficial; it is currently unclear whether this complexity is required, at the cost of more assumptions, or if the process of transduction could be simplified and modelled using fewer parameters, which could be estimated from the experimental data. The two studies that focused on transformation [70,71] applied similar mass action dynamics to this process to that which can be seen in models of conjugation. However, this approach assumes that the number of resistance genes available in the environment is equivalent to the number of bacteria carrying these genes. This is questionable, as we would only expect these genes to be available in the environment after the bacteria die and release their genetic material; although it is possible for bacteria to actively release their genetic material while still alive, the extent of this phenomenon is unclear [6]. Further exploration of this assumption and perhaps redesigns of model structures for transformation would be of value.
E. coli is the most commonly studied model organism for bacteria in general [73]. Its rapid growth and consistent behaviour in in vitro settings make it amenable to experimental work, including transfer studies, and therefore its overwhelming presence as the organism of choice for the studies modelling HGT of AMR genes is not a surprise. However, HGT is known to occur with varying rates in multiple bacterial species, and consequently it is unlikely that the rates of transfer estimated by looking at E. coli are equally applicable to other bacterial species [7]. In addition, HGT of AMR is a process that can also occur between bacterial species [9,11], while most models here exclusively focused on E. coli alone. Some resistances in bacterial species are in fact thought to have been originally acquired following a gene transfer event with another species, such as the mecA resistance gene in Staphylococcus aureus acquired from Staphylococcus fleurettii [74].
Despite the fact that the carriage of an AMR gene often imposes a reduction in the growth rate of the bacteria [10], a few studies did not model this (6/43), but only one argued that this element could be ignored after fitting their model to the experimental data [66]. However, this was once more only based on observations in vitro, which are likely to differ from the in vivo reality. Including a fitness cost, while requiring the estimation of an additional parameter, does not add any particular complexity to the model structure itself, effectively only requiring a reduced growth rate value for the bacteria carrying AMR genes as opposed to bacteria susceptible to the modelled antibiotic (as can be seen in [68], for example), and should therefore be included at least for sensitivity analyses. In addition, although it is understandable that the first models of HGT of AMR should focus on tracking single genes to understand the basic dynamics of this process, in reality, many bacteria carry multiple AMR genes that can be transferred independently [8]. However, we only identified four studies in our review which included more than one independent AMR gene in their model [30,32,60,69]. Thirteen studies did model the transfer of multiple linked genes [34,35,40–42,47,49,53,55,59,66,68,70]; however, in these cases, a single HGT event causes the transfer of all of these genes, and therefore, there is little difference between the model structures of these 13 studies and those of other studies that modelled the transfer of single genes.
Many studies did not allow for the presence of an antibiotic in their model. However, antibiotics are likely to modify HGT dynamics by directly affecting transfer rates, as well as the survival of bacteria not carrying the AMR gene [16–20]. The former has been shown to occur for transduction in S. aureus, where the addition of antibiotics induced a higher proportion of transducing phage compared with lytic phage [75]. On the other hand, some studies correctly argue that it is equally important to understand the dynamics of HGT in the absence of antibiotics. Effectively, it is common for bacterial populations to rapidly transition between being exposed to antibiotics or not, with the most obvious example being individuals transiently consuming antibiotics. Consequently, understanding the dynamics of HGT of AMR both in the presence and in the absence of antibiotics is essential.
HGT of AMR has been studied in laboratory settings; consequently data around which models can be built have been generated and are available [7,76]. However, we note that, to the best of our knowledge, most data appear to focus on conjugation in in vitro settings. The availability of the experimental data on HGT of AMR by transformation or transduction, and on any of the three HGT mechanisms in more complex settings (such as in vivo), is unclear. This should be investigated in future work to further refine the recommendations we make here and identify where more data are needed to support the development of mathematical models. This is essential to understand which of the gaps we identify are due to theory outpacing data collection and which are due to underutilization of the available data. In any case, using these external data sources for purposes they were not originally designed for can require assumptions to be made in the model structure and parameters. In addition, it is essential to bear in mind how these data were originally collected since, for example, combining sources that look at bacteria in multiple environments to derive parameters in a single environment-specific model is far from ideal. On the other hand, the fact that a quarter of the studies we reviewed (12/43) assumed all of their parameter values is worrying. While the purpose of some of these studies was to exclusively test a range of parameter values to identify conditions for a specific event to occur (e.g. AMR prevalence increases), the absence of any clear sources for the limits of these ranges is questionable. Looking at studies that determined their parameter values experimentally, we see that some of these also assume values such as the initial proportion of bacteria capable of performing transformation and the rate at which they can gain this ability [70], the bacterial growth rate and the conjugation rate [40] or the fitness cost of carrying an AMR gene and the rate at which such genes are lost by the bacteria [34]. Informing models with data is essential to ensure that they are accurate representations of reality; therefore, as stated above, we believe that further work is required to review the availability of data on HGT of AMR and the methods that could be used to generate them when they are currently missing.
Regarding model structures, the majority of studies relied on deterministic models. To allow variability in the dynamics and therefore increased realism, studies more often chose to sample their parameter values, run their deterministic model and repeat this process a number of times (as can be seen in [32,38,43,46,56,64,65,72]), a simpler alternative to developing new stochastic models. Acknowledging stochasticity when looking at HGT is essential; HGT rates are typically low (estimates from studies in our review include for example 5.1 × 10−15 (cells/ml)−1 h−1 for conjugation [49] and 10−16 (cells/ml)−1 h−1 for transformation [70]). These are therefore models of rare events which, by chance, might not always occur as expected, a feature that deterministic models fail to capture [24]. Sensitivity analysis is extremely important in any case since a small change in parameter value can lead to a greater change in the results. Despite this, nine studies exclusively relied on a deterministic model without sampling parameters or performing sensitivity analyses [30,36,40,42,48,54,55,58,68]. Interestingly, five of these nine studies also generated their own parameter values experimentally [36,40,42,54,68]. Although they capture variation when measuring the parameters experimentally, often providing distributions for their values, they then only retain fixed-point estimates for their corresponding model parameter values instead of sampling them from these distributions and only use these fixed estimates to derive their conclusions. Acknowledging variability in microbiological observations by specifying distributions rather than point estimates is essential, and this must be represented in the corresponding mathematical models.
This also raises the question of how to best represent these microbiological events in mathematical models. Effectively, almost all of the models here describe transfer as a mass action process (42/43). However, as stated above, this approach is acceptable for conjugation, but might not fully apply to transformation, where transfer depends on the density of DNA in the surrounding environment rather than the number of bacteria, and transduction, which follows vector-like dynamics with the phage acting as carriers of resistance genes between bacteria. Therefore, transformation dynamics might be better represented by models of environmental transmission of infections (such as [77]) and transduction by models of vector-borne diseases (such as [78]), as opposed to mass action models. The degree of modelling complexity required to accurately represent HGT is therefore unclear. This is also true for models designed to understand the public health implications of HGT of AMR, for which the level of detail required to represent within-host dynamics must be clarified. In addition, since transfer dynamics have thus far been mostly studied in bacterial culture, mostly ‘short’ time frames have been explored (hours or days), with long-term dynamics remaining unclear despite our knowledge that even resistant bacteria can colonize us for weeks or months [79–81]. To best guide our public health policies with mathematical modelling, we must first clarify the complexity of the process we are actually attempting to model and the time required to fully capture its in vivo dynamics.
This is the first attempt at providing an overview of existing mathematical modelling work on HGT of AMR. Our systematic review methods, with two individuals separately screening the titles and abstracts of candidate studies, allowed us to identify and bring together key studies on this topic. By using our list of comparison elements, we extracted and contrasted essential information between studies, overall allowing us to obtain a broad overview of the field and identify research gaps. However, our approach also has some limitations. First, it was necessary for us to specify ‘(math* OR dynamic*) model*’ rather than just ‘model*’ in the search, since otherwise it would have returned results on experimental models (e.g. mice) as opposed to mathematical models. Effectively, repeating our search with ‘model*’ instead of ‘(math* OR dynamic*) model*’ yields 2360 and 1560 results on PubMed and Web of Science, respectively, as opposed to our 171 and 185 results. However, the consequence of our choice was that nine relevant studies were missed in the search and were only identified by screening the references of already included studies. These nine studies were missed in the original literature search due to the absence of at least one of the search terms, with some studies for example referring to their models as ‘mass action models’ instead of ‘mathematical models’. In addition, we only searched for studies that modelled transfer of AMR genes, as opposed to HGT of any gene. This is first due to our specific research interest; horizontal transfer of AMR genes is an especially strong evolutionary driver for bacteria populations, compared with transfer of other genes. This is because AMR genes can be strongly selected for by environmental factors, such as the presence of antibiotics, while many other genes are often not subject to such selection pressures. In addition, AMR genes can be selected in more settings compared with other genes; for example, genes involved in immune evasion will be selected only during infection of the host, while AMR genes can also be selected for during asymptomatic colonization. The consequences of HGT of AMR in the bacterial population can therefore be greater than for other genes, which is why we believe that it is important to study this process. Second, repeating the search without ‘(antimicrobial OR antibacterial OR antibiotic) resist*’ yields 12 236 and 38 148 results on PubMed and Web of Science, respectively, which would be too many to cover in a single systematic review. Nevertheless, this suggests that there are other studies that model HGT more broadly. These could be a source of methodologies that could be applied to further develop the specific field of HGT of AMR modelling. In terms of the elements gathered from the studies to compare them, we were unable to extract any meaningful quantitative data (e.g. estimated gene transfer rates) common to all studies due to the high variability of study designs. This variability also prevented us from identifying common measures of study quality we could report aside from the presence or absence of sensitivity analysis.
Studying the effect of HGT of AMR on bacterial evolutionary dynamics is a necessary first step to understand the overall importance of this process. This has been the focus of the majority of the studies identified in this review; however, the public health implications remain vastly unknown. This is related to the observation that the majority of studies model bacteria in an in vitro setting; to understand the public health impact of HGT of AMR, it is essential to expand this to include other bacterial environments such as within humans and animals. In addition, important differences have been identified between transfer rates estimated in vitro and in vivo, with in vivo transduction rates in S. aureus and conjugation rates in K. pneumoniae and E. coli for example being much higher than expected [14,15]. This difference in dynamics is attributable to the fact that in vitro conditions fail to capture essential biological mechanisms influencing bacteria and therefore HGT [6,10]. Studying HGT in vitro allows for a controlled environment to understand the basic dynamics of this process and the factors that might influence them (e.g. antibiotic exposure) and consequently offers a starting point to inform in vivo models. Therefore, we recommend that future modelling studies should build upon the work of existing in vitro studies to evaluate HGT of AMR in more complex scenarios, using parameter estimates from in vitro studies as a baseline and refining them using the data generated with in vivo model organisms such as mice [68]. Owing to the added complexity (e.g. immune system, simultaneous within-host and between-hosts dynamics, rapidly varying host exposure to antibiotics and therefore selection pressure on the bacteria), this will require major extensions to existing models. However, we believe that this is necessary to truly assess the potential consequences of HGT of AMR on human well-being.
This systematic review allowed us to identify key research gaps on the dynamics of HGT of AMR. First, we recommend that future studies should focus on developing models of transformation and transduction to determine the required complexity to represent these dynamics. Since these mechanisms fundamentally differ in their biological characteristics, this will likely require substantial, novel modelling work as opposed to the extension of existing models of conjugation. In parallel, since the basic dynamics of conjugation are already reasonably well understood, future studies on this mechanism should focus on other bacterial species than E. coli, preferably in a setting where interspecific HGT and the movement of multiple, separate AMR genes can be observed. This should be achievable simply by re-parametrization or minor extension of existing models; the greatest challenge would be to generate new data on HGT in these currently unexplored settings. The optimal solution to address these research questions would be to design frameworks to study HGT of AMR that encompass both laboratory and modelling work; this would ensure that the data collected are appropriate for the modelling needs and that the actual model is a good representation of the situation measured in the laboratory. Therefore, we believe that to fully understand the complexity of both the biology and the dynamics of HGT, collaboration of both microbiologists and mathematical modellers would be the best strategy for future research on this topic and that studies should attempt to generate both their own data and models to reduce the assumptions they require.
While exclusively microbiological approaches have successfully been used to identify when HGT occurs, combining these with modelling has allowed us to estimate rates at which these events occur and to disentangle the finer temporal dynamics of this process. For example, some studies we identified in our review, which combined microbiology and modelling work, answered questions such as how changing the exposure of bacteria to antibiotics influences the HGT rates [49], how a bacterium interacts in space with its neighbours to perform HGT [31] or how to adjust shaking speed to maximize contact between bacteria, and thus the rate of HGT, in a liquid culture [66]. Modelling also allows faster exploration of situations that could be harder to test using only microbiological methods, since an experiment where the bacteria need to grow for 24 h in the laboratory could be completed in a few seconds using a mathematical model. Crucially, this requires the model to be an accurate representation of reality, which in turn requires it to be informed by the microbiological data to begin with. Therefore, our conclusion here is not that either one of modelling or microbiology is superior to the other, but that both approaches complement each other. Consequently, we believe that close cooperation between these two fields would allow us to greatly improve our understanding of complex microbiological processes, such as HGT of AMR.
5. Conclusion
In this systematic review, we aimed to assess the current state of mathematical modelling as a tool to improve our understanding of HGT of AMR. From the 43 studies identified, we found that the majority focused on conjugation in E. coli, exploring evolutionary dynamics of HGT in culture. While this provides a solid base for a key method of HGT, future work must also consider HGT by transformation and transduction, which are also essential drivers of HGT in bacteria. Importantly for public health implications, only one bacterial species was considered in most models when we know that interspecies transfer is responsible for many of our epidemic AMR clones, and much of the work was fitted to data in the absence of antibiotic exposure. Crucially, to answer these questions, we must first clarify the level of modelling complexity required to accurately represent HGT dynamics, as well as the availability and capacity to generate the experimental data on these processes. This complex topic requires close collaboration between mathematical modellers and microbiologists in order to determine the full impact of these processes on our ability to control the public health threat posed by AMR.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Katherine Atkins for helpful discussions on the search strategy for this systematic review.
Data accessibility
This article has no additional data.
Authors' contributions
All authors jointly developed the search strategy. Q.J.L. and G.M.K. independently screened the titles and abstracts of the identified studies. Q.J.L. then evaluated the full texts of the included studies and wrote the first draft of the manuscript. All authors subsequently edited the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Medical Research Council (grant no. MR/P014658/1) and by a Medical Research Council London Intercollegiate Doctoral Training Program studentship (grant no. MR/N013638/1).
References
- 1.World Health Organisation. 2015. Global action plan on antimicrobial resistance. Geneva, Switzerland: WHO. [Google Scholar]
- 2.Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. ( 10.1016/S1473-3099(10)70143-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodford N, Johnson AP. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 62, 499–513. ( 10.1099/jmm.0.052555-0) [DOI] [PubMed] [Google Scholar]
- 4.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front. Microbiol. 3, 110 ( 10.3389/fmicb.2012.00110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. ( 10.1038/35012500) [DOI] [PubMed] [Google Scholar]
- 6.Thomas CM, Nielsen KM. 2005. Mechanisms of and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. ( 10.1038/nrmicro1234) [DOI] [PubMed] [Google Scholar]
- 7.von Wintersdorff CJH, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PHM, Wolffs PFG. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173 ( 10.3389/fmicb.2016.00173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanwar J, Das S, Fatima Z, Hameed S. 2014. Multidrug resistance: an emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014, 541340 ( 10.1155/2014/541340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naidoo J. 1984. Interspecific co-transfer of antibiotic resistance plasmids in staphylococci in vivo. J. Hyg., Camb. 93, 59–66. ( 10.1017/s0022172400060939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melnyk AH, Wong A, Kassen R. 2015. The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283. ( 10.1111/eva.12196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe T, Fukasawa T. 1961. Episome-mediated transfer of drug resistance in Enterobacteriaceae I. Transfer of resistance factors by conjugation J. Bacteriol. 81, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindsay JA. 2014. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 304, 103–109. ( 10.1016/J.IJMM.2013.11.010) [DOI] [PubMed] [Google Scholar]
- 13.Hamilton HL, Dillard JP. 2006. Natural transformation of Neisseria gonorrhoeae : from DNA donation to homologous recombination. Mol. Microbiol. 59, 376–385. ( 10.1111/j.1365-2958.2005.04964.x) [DOI] [PubMed] [Google Scholar]
- 14.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 6, 2697–2708. ( 10.1093/gbe/evu214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottig S, Gruber TM, Stecher B, Wichelhaus TA, Kempf VAJ. 2015. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin. Infect. Dis. 60, 1808–1815. ( 10.1093/cid/civ191) [DOI] [PubMed] [Google Scholar]
- 16.Velkov VW. 1999. How environmental factors regulate mutagenesis and gene transfer in microorganisms. J. Biosci. 24, 529–559. ( 10.1007/BF02942664) [DOI] [Google Scholar]
- 17.Hastings PJ, Rosenberg SM, Slack A. 2004. Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 12, 401–404. ( 10.1016/J.TIM.2004.07.003) [DOI] [PubMed] [Google Scholar]
- 18.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. ( 10.1038/nature02241) [DOI] [PubMed] [Google Scholar]
- 19.Maiques E, Ubeda C, Campoy S, Salvador N, Lasa I, Novick RP, Barbé J, Penadés JR. 2006. Beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 188, 2726–2729. ( 10.1128/JB.188.7.2726-2729.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys J-P. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313, 89–92. ( 10.1126/SCIENCE.1127912) [DOI] [PubMed] [Google Scholar]
- 21.Jassim SAA, Limoges RG. 2017. Bacteriophage and antimicrobial resistance. In Bacteriophages: practical applications for nature's biocontrol, pp. 19–57. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 22.Verheust C, Pauwels K, Mahillon J, Helinski DR, Herman P. 2010. Contained use of bacteriophages: risk assessment and biosafety recommendations. Appl. Biosaf. 15, 32–44. ( 10.1177/153567601001500106) [DOI] [Google Scholar]
- 23.Drlica K, Gennaro ML. 2001. Plasmids. In Encyclopedia of genetics, pp. 1485–1490. New York, NY: Academic Press. [Google Scholar]
- 24.Anderson RM, May RM. 1991. Infectious diseases of humans : dynamics and control. Oxford, UK: Oxford University Press. [Google Scholar]
- 25.Opatowski L, Guillemot D, Boëlle P-Y, Temime L. 2011. Contribution of mathematical modeling to the fight against bacterial antibiotic resistance. Curr. Opin. Infect. Dis. 24, 279–287. ( 10.1097/QCO.0b013e3283462362) [DOI] [PubMed] [Google Scholar]
- 26.van Kleef E, Robotham JV, Jit M, Deeny SR, Edmunds WJ. 2013. Modelling the transmission of healthcare associated infections: a systematic review. BMC Infect. Dis. 13, 294 ( 10.1186/1471-2334-13-294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies NG, Flasche S, Jit M, Atkins KE. 2019. Within-host dynamics shape antibiotic resistance in commensal bacteria. Nat. Ecol. Evol. 3, 440–449. ( 10.1038/s41559-018-0786-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Kleef E, Luangasanatip N, Bonten MJ, Cooper BS. 2017. Why sensitive bacteria are resistant to hospital infection control. Wellcome Open Res. 2, 16 ( 10.12688/wellcomeopenres.11033.2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberati A, et al. 2009. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 339, b2700 ( 10.1136/bmj.b2700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Agata EMC, Dupont-Rouzeyrol M, Magal P, Olivier D, Ruan S. 2008. The impact of different antibiotic regimens on the emergence of antimicrobial-resistant bacteria. PLoS ONE 3, e4036 ( 10.1371/journal.pone.0004036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehring R, Schumm P, Youssef M, Scoglio C. 2010. A network-based approach for resistance transmission in bacterial populations. J. Theor. Biol. 262, 97–106. ( 10.1016/j.jtbi.2009.09.002) [DOI] [PubMed] [Google Scholar]
- 32.Obolski U, Hadany L. 2012. Implications of stress-induced genetic variation for minimizing multidrug resistance in bacteria. BMC Med. 10, 89 ( 10.1186/1741-7015-10-89) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krone SM, Lu R, Fox R, Suzuki H, Top EM. 2007. Modelling the spatial dynamics of plasmid transfer and persistence. Microbiology 153, 2803–2816. ( 10.1099/mic.0.2006/004531-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundquist PD, Levin BR. 1986. Transitory derepression and the maintenance of conjugative plasmids. Genetics 113, 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ponciano JM, De Gelder L, Top EM, Joyce P. 2007. The population biology of bacterial plasmids: a hidden Markov model approach. Genetics 176, 957–968. ( 10.1534/genetics.106.061937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer EA, Dierikx CM, van Essen-Zandbergen A, van Roermund HJ, Mevius DJ, Stegeman A, Klinkenberg D. 2014. The IncI1 plasmid carrying the blaCTX-M-1 gene persists in in vitro culture of a Escherichia coli strain from broilers. BMC Microbiol. 14, 77 ( 10.1186/1471-2180-14-77) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svara F, Rankin DJ. 2011. The evolution of plasmid-carried antibiotic resistance. BMC Evol. Biol. 11, 130 ( 10.1186/1471-2148-11-130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willms AR, Roughan PD, Heinemann JA. 2006. Static recipient cells as reservoirs of antibiotic resistance during antibiotic therapy. Theor. Popul. Biol. 70, 436–451. ( 10.1016/j.tpb.2006.04.001) [DOI] [PubMed] [Google Scholar]
- 39.Hall JPJ, Wood AJ, Harrison E, Brockhurst MA. 2016. Source-sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc. Natl Acad. Sci. USA 113, 8260–8265. ( 10.1073/pnas.1600974113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dionisio F, Matic I, Radman M, Rodrigues OR, Taddei F. 2002. Plasmids spread very fast in heterogeneous bacterial communities. Genetics 162, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong X, Droesch J, Fox R, Top EM, Krone SM. 2012. On the meaning and estimation of plasmid transfer rates for surface-associated and well-mixed bacterial populations. J. Theor. Biol. 294, 144–152. ( 10.1016/J.JTBI.2011.10.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Z, et al. 2015. Effects of nano-TiO2 on antibiotic resistance transfer mediated by RP4 plasmid. Nanotoxicology 9, 895–904. ( 10.3109/17435390.2014.991429) [DOI] [PubMed] [Google Scholar]
- 43.Cazer CL, Ducrot L, Volkova VV, Gröhn YT. 2017. Monte Carlo simulations suggest current chlortetracycline drug-residue based withdrawal periods would not control antimicrobial resistance dissemination from feedlot to slaughterhouse. Front. Microbiol. 8, 1753 ( 10.3389/fmicb.2017.01753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volkova VV, Lu Z, Lanzas C, Scott HM, Gröhn YT. 2013. Modelling dynamics of plasmid-gene mediated antimicrobial resistance in enteric bacteria using stochastic differential equations. Sci. Rep. 3, 2463 ( 10.1038/srep02463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker M, Hobman JL, Dodd CER, Ramsden SJ, Stekel DJ. 2016. Mathematical modelling of antimicrobial resistance in agricultural waste highlights importance of gene transfer rate. FEMS Microbiol. Ecol. 92, fiw040 ( 10.1093/femsec/fiw040) [DOI] [PubMed] [Google Scholar]
- 46.Volkova VV, Lanzas C, Lu Z, Gröhn YT. 2012. Mathematical model of plasmid-mediated resistance to ceftiofur in commensal enteric Escherichia coli of cattle. PLoS ONE 7, e36738 ( 10.1371/journal.pone.0036738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kneis D, Hiltunen T, Hess S. 2019. A high-throughput approach to the culture-based estimation of plasmid transfer rates. Plasmid 101, 28–34. ( 10.1016/j.plasmid.2018.12.003) [DOI] [PubMed] [Google Scholar]
- 48.Knopoff DA, Sanchez Sanso JM. 2017. A kinetic model for horizontal transfer and bacterial antibiotic resistance. Int. J. Biomath. 10, 1750051 ( 10.1142/S1793524517500516) [DOI] [Google Scholar]
- 49.Lopatkin AJ, Huang S, Smith RP, Srimani JK, Sysoeva TA, Bewick S, Karig DK, You L. 2016. Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1, 16044 ( 10.1038/nmicrobiol.2016.44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peña-Miller R, Rodríguez-González R, MacLean RC, San Millan A. 2015. Evaluating the effect of horizontal transmission on the stability of plasmids under different selection regimes. Mob. Genet. Elements 5, 29–33. ( 10.1080/2159256X.2015.1045115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heuer H, Focks A, Lamshoeft M, Smalla K, Matthies M, Spiteller M. 2008. Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol. Biochem. 40, 1892–1900. ( 10.1016/j.soilbio.2008.03.014) [DOI] [Google Scholar]
- 52.Freese PD, Korolev KS, Jimenez JI, Chen IA. 2014. Genetic drift suppresses bacterial conjugation in spatially structured populations. Biophys. J. 106, 944–954. ( 10.1016/j.bpj.2014.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simonsen L, Gordon DM, Stewart FM, Levin BR. 1990. Estimating the rate of plasmid transfer: an end-point method. J. Gen. Microbiol. 136, 2319–2325. ( 10.1099/00221287-136-11-2319) [DOI] [PubMed] [Google Scholar]
- 54.Gothwal R, Thatikonda S. 2018. Mathematical model for the transport of fluoroquinolone and its resistant bacteria in aquatic environment. Environ. Sci. Pollut. Res. Int. 25, 20 439–20 452. ( 10.1007/s11356-017-9848-x) [DOI] [PubMed] [Google Scholar]
- 55.Ibargueen-Mondragon E, Romero-Leiton JP, Esteva L, Mariela Burbano-Rosero E. 2016. Mathematical modeling of bacterial resistance to antibiotics by mutations and plasmids. J. Biol. Syst. 24, 129–146. ( 10.1142/S0218339016500078) [DOI] [Google Scholar]
- 56.Zwanzig M, Harrison E, Brockhurst MA, Hall JPJ, Berendonk TU, Berger U. 2019. Mobile compensatory mutations promote plasmid survival. mSystems 4, e00186-18 ( 10.1128/mSystems.00186-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connelly BD, Zaman L, McKinley PK, Ofria C. 2011. Modeling the evolutionary dynamics of plasmids in spatial populations. In GECCO-2011: Proc. 13th Annual Genetic and Evolutionary Computation Conf., Dublin, Ireland, 12–16 July 2011, pp. 227–233. New York, NY: ACM; ( 10.1145/2001576.2001608) [DOI] [Google Scholar]
- 58.Khan A, Imran M. 2018. Optimal dosing strategies against susceptible and resistant bacteria. J. Biol. Syst. 26, 41–58. ( 10.1142/S0218339018500031) [DOI] [Google Scholar]
- 59.Malwade A, Nguyen A, Sadat-Mousavi P, Ingalls BP. 2017. Predictive modeling of a batch filter mating process. Front. Microbiol. 8, 461 ( 10.3389/fmicb.2017.00461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campos M, et al. 2019. Simulating multilevel dynamics of antimicrobial resistance in a membrane computing model. MBio 10, e02460-18 ( 10.1128/mBio.02460-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu S, Yang J, Yin C, Zhao X. 2018. The dominance of bacterial genotypes leads to susceptibility variations under sublethal antibiotic pressure. Future Microbiol. 13, 165–185. ( 10.2217/fmb-2017-0070) [DOI] [PubMed] [Google Scholar]
- 62.Raz Y, Tannenbaum E. 2010. The influence of horizontal gene transfer on the mean fitness of unicellular populations in static environments. Genetics 185, 327–337. ( 10.1534/genetics.109.113613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haverkate MR, Dautzenberg MJD, Ossewaarde TJM, van der Zee A, den Hollander JG, Troelstra A, Bonten MJM, Bootsma MCJ. 2015. Within-host and population transmission of bla(OXA-48) in K. pneumoniae and E. coli. PLoS ONE 10, e0140960 ( 10.1371/journal.pone.0140960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volkova VV, Lu Z, Lanzas C, Grohn YT. 2013. Evaluating targets for control of plasmid-mediated antimicrobial resistance in enteric commensals of beef cattle: a modelling approach. Epidemiol. Infect. 141, 2294–2312. ( 10.1017/S0950268812002993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webb GF, D'Agata EMC, Magal P, Ruan S. 2005. A model of antibiotic-resistant bacterial epidemics in hospitals. Proc. Natl Acad. Sci. USA 102, 13 343–13 348. ( 10.1073/pnas.0504053102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong X, Krol JEJE, Top EM, Krone SM. 2010. Accounting for mating pair formation in plasmid population dynamics. J. Theor. Biol. 262, 711–719. ( 10.1016/j.jtbi.2009.10.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart FM, Levin BR. 1977. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87, 209–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freter R, Freter RR, Brickner H. 1983. Experimental and mathematical models of Escherichia coli plasmid transfer in vitro and in vivo. Infect. Immun. 39, 60–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gomes ALC, Galagan JE, Segrè D. 2013. Resource competition may lead to effective treatment of antibiotic resistant infections. PLoS ONE 8, e80775 ( 10.1371/journal.pone.0080775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnsen PJ, Dubnau D, Levin BR. 2009. Episodic selection and the maintenance of competence and natural transformation in Bacillus subtilis. Genetics 181, 1521–1533. ( 10.1534/genetics.108.099523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu N, Massoudieh A, Liang X, Kamai T, Zilles JL, Nguyen TH, Ginn TR. 2015. A kinetic model of gene transfer via natural transformation of Azotobacter vinelandii. Environ. Sci. Res. Technol. 1, 363–374. ( 10.1039/c5ew00023h) [DOI] [Google Scholar]
- 72.Volkova VV, Lu Z, Besser T, Gröhn YT. 2014. Modeling the infection dynamics of bacteriophages in enteric Escherichia coli: estimating the contribution of transduction to antimicrobial gene spread. Appl. Environ. Microbiol. 80, 4350–4362. ( 10.1128/AEM.00446-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cooper GM, Hausman RE. 2015. The cell: a molecular approach, 7th edn See https://www.sinauer.com/media/wysiwyg/samples/TheCell7e_Brochure.pdf. [Google Scholar]
- 74.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. 2010. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 54, 4352–4359. ( 10.1128/AAC.00356-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stanczak-Mrozek KI, Laing KG, Lindsay JA. 2017. Resistance gene transfer: induction of transducing phage by sub-inhibitory concentrations of antimicrobials is not correlated to induction of lytic phage. J. Antimicrob. Chemother. 72, 1624–1631. ( 10.1093/JAC/DKX056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lerminiaux NA, Cameron ADS. 2019. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. ( 10.1139/cjm-2018-0275) [DOI] [PubMed] [Google Scholar]
- 77.Breban R, Drake JM, Stallknecht DE, Rohani P. 2009. The role of environmental transmission in recurrent avian influenza epidemics. PLoS Comput. Biol. 5, e1000346 ( 10.1371/journal.pcbi.1000346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Day T. 2002. Virulence evolution via host exploitation and toxin production in spore-producing pathogens. Ecol. Lett. 5, 471–476. ( 10.1046/j.1461-0248.2002.00342.x) [DOI] [Google Scholar]
- 79.Haverkate MR, Derde LPG, Brun-Buisson C, Bonten MJM, Bootsma MCJ. 2014. Duration of colonization with antimicrobial-resistant bacteria after ICU discharge. Intensive Care Med. 40, 564–571. ( 10.1007/s00134-014-3225-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haverkate MR, Weiner S, Lolans K, Moore NM, Weinstein RA, Bonten MJM, Hayden MK, Bootsma MCJ. 2016. Duration of colonization with Klebsiella pneumoniae carbapenemase-producing bacteria at long-term acute care hospitals in Chicago, Illinois. Open Forum Infect. Dis. 3, ofw178 ( 10.1093/ofid/ofw178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O'Fallon E, Gautam S, D'Agata EMC. 2009. Colonization with multidrug-resistant gram-negative bacteria: prolonged duration and frequent cocolonization. Clin. Infect. Dis. 48, 1375–1381. ( 10.1086/598194) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.