Significance
Our new method uses only epigenomic patterns to classify the expression potential of annotated genes and identifies pseudogenes that are difficult to classify based solely on sequence. Genes were divided into those with protein expression, those with mRNA expression, and those that are silent. A large fraction of annotated genes are constitutively silent in one lineage but can be transcribed in others. We refer to the species-wide set of transcribed genes as the expressome and show that it is much larger than the expressible gene set in any individual. Additionally, we find that DNA methylation patterns within the gene body can differentiate between genes that express proteins and genes that express only RNAs.
Keywords: machine learning, genome annotation, maize, epigenomics, proteomics
Abstract
Accurate annotation of plant genomes remains complex due to the presence of many pseudogenes arising from whole-genome duplication-generated redundancy or the capture and movement of gene fragments by transposable elements. Machine learning on genome-wide epigenetic marks, informed by transcriptomic and proteomic training data, could be used to improve annotations through classification of all putative protein-coding genes as either constitutively silent or able to be expressed. Expressed genes were subclassified as able to express both mRNAs and proteins or only RNAs, and CG gene body methylation was associated only with the former subclass. More than 60,000 protein-coding genes have been annotated in the reference genome of maize inbred B73. About two-thirds of these genes are transcribed and are designated the filtered gene set (FGS). Classification of genes by our trained random forest algorithm was accurate and relied only on histone modifications or DNA methylation patterns within the gene body; promoter methylation was unimportant. Other inbred lines are known to transcribe significantly different sets of genes, indicating that the FGS is specific to B73. We accurately classified the sets of transcribed genes in additional inbred lines, arising from inbred-specific DNA methylation patterns. This approach highlights the potential of using chromatin information to improve annotations of functional genes.
In maize, proteins are observed only from a subset of transcribed genes: 87% of genes observed to make proteins have syntenic orthologs in sorghum, even though syntenic genes account for only 23% of transcribed genes (1). This observation explains why nearly all genes with known functions are syntenic (2), and it raises a new question: How can the cell distinguish between syntenic and species-specific genes such that both are transcribed but only the former expresses proteins?
To begin to answer this question, we present a machine-learning-based approach that provides genome-wide classifications of annotated protein-coding genes as expressible or constitutively silent based on patterns of DNA methylation or histone modifications. The classifiers are additionally able to distinguish between genes that can express proteins and genes that can only express RNAs. Our findings address a long-standing challenge in genome biology to discover the expressible gene set (EGS) which comprises all protein-coding genes with the potential to be expressed in an individual. Efforts to identify the EGS have been based on surveys of expression and on comparative genomics. While these criteria have been useful, surveys only sample some of the conditions, cell types, and genetic diversity that affect gene expression, leaving some functional genes without evidence for expression. New methods are needed to identify the EGS. It is equally important to identify the silent gene set because these genes may or may not be expressed in other individuals with different genetic backgrounds and epigenetic marks. Our EGS for protein expression was contained within the larger EGS for mRNA expression. Collectively, the EGS from all individuals constitutes the expressome: all protein-coding genes in a species with the potential to be expressed as proteins or only as RNAs.
Most researchers study the predicted genes that are derived from whole-genome annotations. These annotation approaches can be complicated by the presence of sequences with homology to protein coding genes that may not be functional genes. These false gene annotations can result from silenced paralogs following either whole-genome duplications or tandem duplications, or they may arise from capture of gene fragments by transposable elements. Here we show that the analysis of DNA methylation patterns can help identify annotated genes that are not likely to be expressed or can be expressed only as RNAs.
We found that the most significant genome features used by our random forest classifiers are well-known patterns of DNA methylation and histone modification, indicating that these patterns may play roles in establishing permissions for gene expression. Genes that expressed both mRNAs and proteins had DNA methylation patterns that were distinct from genes that only expressed RNAs. Silent gene patterns differed from both expressible classes. Our models matched or outperformed expert curation for the ability to differentiate between expressed and silent genes. Extension of our method to other inbred lines with differential DNA methylation patterns demonstrated that the EGS differs between inbreds by thousands of genes that correlate with variations in the epigenome. A small but significant difference in the EGS was observed between organs. Discovery of the EGS for individuals and of the expressome for a species will contribute to understanding and fully utilizing the genetic potential of organisms. Our characterization of the EGS for the widely used maize inbred, B73, provides a first step toward discovery of the expressome for the world’s most valuable crop.
Results and Discussion
Genic DNA Methylation Can Classify Expression Potential.
Syntenic genes are hypomethylated relative to their nonsyntenic counterparts, suggesting that epigenomic features may enable robust gene classifications (3, 4). To further explore the relationship between DNA methylation and gene expression, we used the random forest algorithm (5) to build classifiers for all genes of the maize inbred line B73. Two classifiers were built based solely on genic DNA methylation features. Both used a combination of proteome and transcriptome data for training from 23 different tissues or times of development (1). For the expressible protein classifier (EPC), the silent class consisted of annotated genes with no observed mRNAs or proteins (NR_NP). The expressible class consisted of genes with high levels of mRNAs [fragments per kilobase per million reads (FPKM) > 1, defined by (1)] and observed proteins (HR_OP). The training classes of the second classifier (expressible mRNA classifier, ERC) were defined using all genes with no detectable mRNAs (NR) vs. all genes with high mRNA levels (HR), and it did not use protein data (Fig. 1).
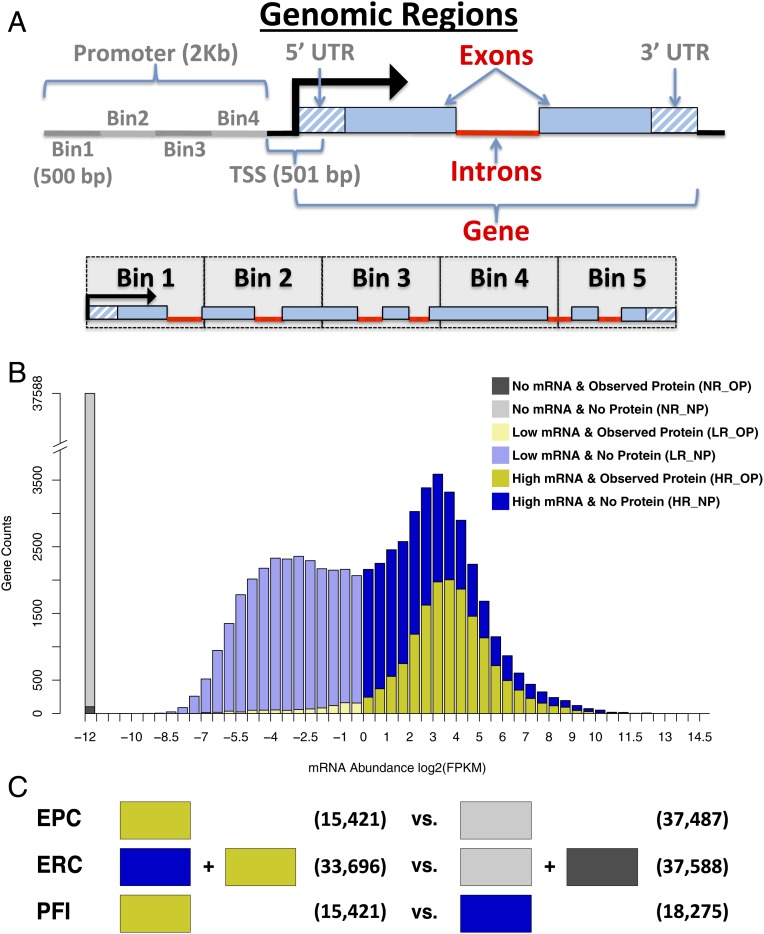
Fig. 1.
Overview of model features and training set definitions. (A) The various genomic regions where DNA methylation levels were quantified and used as features for classification. Features with gray labels were discarded after initial testing. Each gene was also split into 5 equivalent regions, called bins, and features were quantified separately in each bin. (B) The distribution of detected mRNA abundance is bimodal. The 2 mRNA populations can be roughly separated using an FPKM of 1. Here the nondetected mRNA (No mRNA) is represented as a separate population and given an artificial value of −12. Each population can be further refined into observed vs. nonobserved protein (No Protein) to yield 6 different groups of genes indicated by the different colors. LR_OP refers to all annotated genes that were observed to express low levels of mRNAs and detectable levels of proteins. (C) Three separate random forest models were built. Colored blocks correspond to the gene sets (from B) used for each training class. Blocks on the left indicate the positive (true) training instances vs. blocks on the right that indicate the negative (false) training instances. Numbers in parentheses indicate the number of genes in each training class.
Several DNA methylation features were tested. The importance of features was determined using the mean decrease in accuracy upon random permutation of each individual variable (5). Three methylation sequence contexts (CHG, CG, and CHH) were quantified separately and summarized within gene regions (Fig. 1A), including the promoter (2 kilobases upstream of the transcription start site [TSS], split into 4 bins), the TSS, 5′ UTR, 3′ UTR, introns, exons, and a summed value encompassing the gene model. The summarized random forest feature importance is shown for each methylation context and genomic region (SI Appendix, Fig. S1). Based on these scores, multiple features with low importance were deleted from the models. Features retained were CHG and CG methylation in exons and introns, plus the aggregation of all retained features (labeled “Gene”). To account for variable distributions of CG and CHG methylation along the gene body (4, 6, 7), we divided each gene into 5 equal-proportion bins; methylation features were summarized separately for each bin (Fig. 1A and SI Appendix, Figs. S2 and S3). Classification accuracies were determined using random forest out-of-bag cross-validation on the training set genes (Fig. 2A and SI Appendix, Fig. S4A). Both classifiers had high accuracy with areas under the curve (AUCs) of 0.94 or higher for receiver operating characteristic (ROC) curves and precision vs. recall (PR) curves. The EPC achieved a near-perfect AUC of 0.99.
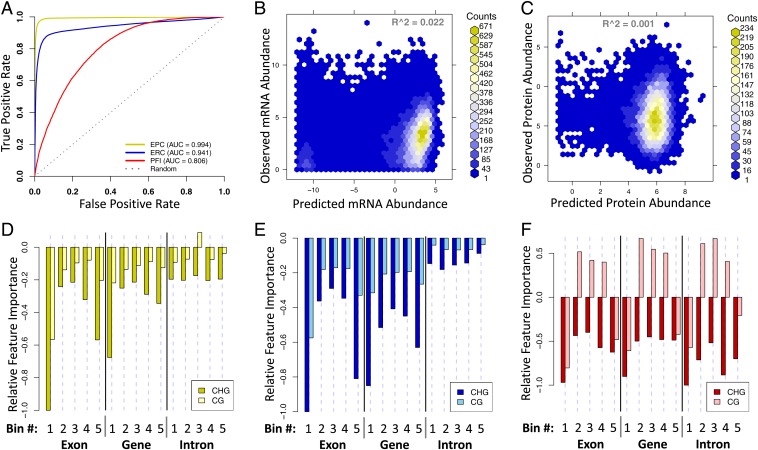
Fig. 2.
Results for random forest models. (A) Receiver operating characteristic (ROC) curves showing classification accuracy of the EPC, ERC, and PFI models. (B and C) Binned scatterplot showing prediction accuracy for quantitative abundance models considering only genes with observed expression for mRNA abundance (B) and protein abundance (C). (D–F) Signed feature importance measures for 3 different models. The values reflect the random forest “mean decrease in accuracy” measure of feature importance. The sign is based on the relationship of the feature values to the training class assignments. Positive values indicate a positive correlation between the feature and either protein observation (EPC and PFI) or high mRNA (ERC).
CHG and CG Methylation Are Negatively Associated with Expression.
To determine whether silent genes were associated with high or low methylation levels, each feature was given a positive or negative sign (Fig. 2 D and E and SI Appendix, Materials and Methods and Fig. S5). The structures of the EPC and ERC models were very similar based on feature importance (Fig. 2 D and E and SI Appendix, Fig. S4D). Methylation of CHG and CG at the 5′ ends of genes (bin 1) was most strongly associated with silent genes; CHG methylation at the 3′ ends of genes (bin 5) was also significant. In vitro methylation of CG sites in the 5′ region of the gene is a potent inhibitor of transgene expression (8).
Gene Classification Is Not Associated with Patterns or Levels of Expression.
To test whether genic DNA methylation is associated with the patterns or levels of expression, random forest models were run using the same methylation training data but replacing the binary class vector with quantitative mRNA and protein abundance for the ERC and EPC, respectively. This produced the protein expression-level predictor and the mRNA expression-level predictor; both failed to accurately predict expression levels. When examining the full set of predictions (SI Appendix, Fig. S4 B and C), we observed good R2 values but observed low R2 values when considering only genes with detectable expression (Fig. 2 B and C). Therefore, the good R2 values observed on the full set of predictions may be mostly due to the ability of the model to discriminate between observed and nonobserved products of expression.
Genome-Wide Classifications.
The ERC and EPC were used to reclassify all protein-coding genes based solely on DNA methylation patterns, including the 98,296 members of the working gene set for which we had methylation coverage. The ERC classified 41,056 genes as able to express mRNAs, but only 32,979 genes were classified by the EPC as able to express proteins; 55% of the EPC expressible genes were absent from the training set (SI Appendix, Fig. S6A). This highlights the power of classification models to learn from a high-confidence subset of genes and then provide accurate genome-wide classifications. Comparison of results from the ERC and EPC identified 2 groups of genes that are expressed as RNAs only (8,078) or as mRNAs plus proteins (32,978) (Dataset S1).
We compared our classifications to the most recent (RefGen version 4 [v4]) and the previous (RefGen v2) curated classifications. Maize RefGen v2 was the last version where a full gene set was annotated (5a working gene set [WGS]), yielding over 110,000 gene models at distinct loci. The maize filtered gene set (FGS) is a subset of high-confidence protein-coding genes from the RefGen v2 WGS (see SI Appendix, Materials and Methods for description). For the newest assembly, RefGen v4 (9), only a filtered gene set has been annotated. We cross-referenced the RefGen v4 FGS to the RefGen v2 accessions (Dataset S1) so that our classifications could be compared to both versions of the maize genome. Only ∼77% of the RefGen v4 FGS can be converted to RefGen v2 accessions, which constrained our ability to make comparisons.
Our ERC differs from the curated FGS by 17,684 genes and 18,019 genes for RefGen v2 and RefGen v4, respectively (SI Appendix, Fig. S7A). We classified the remaining genes (57,239) as silent in B73. However, epialleles of silent genes can be transcribed in other genetic backgrounds as described below.
The EPC and ERC classified as silent 33 and 23% of the RefGen v2 FGS and 16 and 12% of the RefGen v4 FGS (SI Appendix, Fig. S7A); these proportions rose to 66 and 58% for all potential genes (RefGen v2 WGS). The Maize Genetics and Genomics Database (MaizeGDB) curation project (10) annotates a biotype to each gene model. The biotype can be used to filter out all likely transposable elements (TEs) and pseudogenes to yield 63,331 probable protein-coding genes in the WGS. Of these, 60,295 have coverage in our DNA methylation data. Using the higher-confidence EPC classifier, 48% were classified as silent (SI Appendix, Fig. S8), and nearly all of the TEs (97%) and pseudogenes (94%) in the WGS were classified as silent.
The Accuracy of Random Forest Models Matches or Exceeds That of Expert Curation.
We compared the abilities of the curated RefGen v2 and v4 filtered gene sets and the EPC and ERC classifiers to identify the set of expressible genes in B73. ROC and PR curves were created to evaluate each set (SI Appendix, Fig. S7 F and G). Each set represents a 2-category classification (expressible or silent). The ROC curve (SI Appendix, Fig. S7F) shows that the EPC classifier achieves the highest accuracy (solid yellow line). The RefGen v2 set achieves a similar true-positive rate but with a higher false-positive rate, meaning that the RefGen v2 FGS, being the largest set, includes nearly all observed proteins but has more false positives. Precision (SI Appendix, Fig. S7G) represents the proportion of the corresponding set that is correctly called and should be insensitive to incomplete data assuming this subset is random. Looking at precision, we see the EPC and ERC (solid lines) outperform the RefGen v2 FGS (dashed lines). The RefGen v4 FGS performed well, with higher precision than the ERC for expressible mRNAs, but the EPC is still the top performer, indicating that the addition of protein data substantially improves classifications. The observed graphs of bimodal mRNA abundance could be more or less reconstructed from the 4 classified gene sets by plotting the average mRNA abundance for each silent and expressed gene (SI Appendix, Fig. S7 B–E). This indicates that DNA methylation patterns are sufficient to explain the observed bimodal component in the distributions of gene expression.
Hypermethylation of Transposable Elements Does Not Affect the Prediction Accuracy of Protein Coding Genes.
The correlation between DNA methylation and gene expression was established first in studies of the maize autonomous TEs Ac, Spm, and MuDR. Using combinations of restriction enzymes, investigators found that the methylation of TEs was associated with their ability to transpose and to cause transposition of additional members of their TE family (11). TE methylation was negatively associated with mRNA and protein expression and with cycling between active and inactive states. High levels of CHG and CG methylation repress expression of TEs and repetitive elements (4, 7, 12, 13). Subsequent work has shown that plant TEs are silenced by RNA-dependent DNA methylation in the CHH context (14). Because these elements are so abundant in the genome, many gene models in the WGS are TEs that have escaped sequence masking. Of the 110,028 RefGen v2 gene models, 29,082 have been categorized as likely TEs. In addition, we identified 7,612 gene models that have a high basic local alignment search tool (BLAST) hit to one or more reference TE sequences in the maize TE database (15) and may be protein-coding genes with TEs inserted into their gene body. To determine the extent to which these previously characterized, highly methylated elements are affecting our classifiers and conclusions, we rebuilt all of the classification models after filtering out all 36,694 TEs and TE-containing gene models. The new classifier is nearly identical to the original both in classification accuracy (SI Appendix, Fig. S9 A and B) and in feature importance (SI Appendix, Fig. S9 C–E). We have left the TEs in the final models because our goal is to examine the relationship between genic methylation and expression potential. A subset of these 36,694 TEs and TE-containing genes was observed as proteins (2,423) or as highly expressed RNAs (5,065).
Inbred-Specific Expressible Gene Sets.
To determine whether inbred-specific DNA methylation is associated with an inbred-specific EGS, the ERC was remade using data from multiple maize inbreds. The third leaf from the genetically diverse inbreds Mo17, CML322, Oh43, Tx303, and B73 was used to produce DNA methylation data (16) and RNA sequencing (RNA-seq) data (17). The methylation data were processed by quantifying weighted methylation levels for consecutive 100 base pair (bp) tiles along each chromosome. A new classifier, ERC-2, was constructed using the same class definitions as the ERC (summarized expression from many tissues). The model was trained using these class definitions plus the 100 bp tile DNA methylation data from the third leaf B73 sample. Genes of the remaining 4 inbreds were classified using their DNA methylation and the ERC-2 model (Fig. 3 A, C, and E).
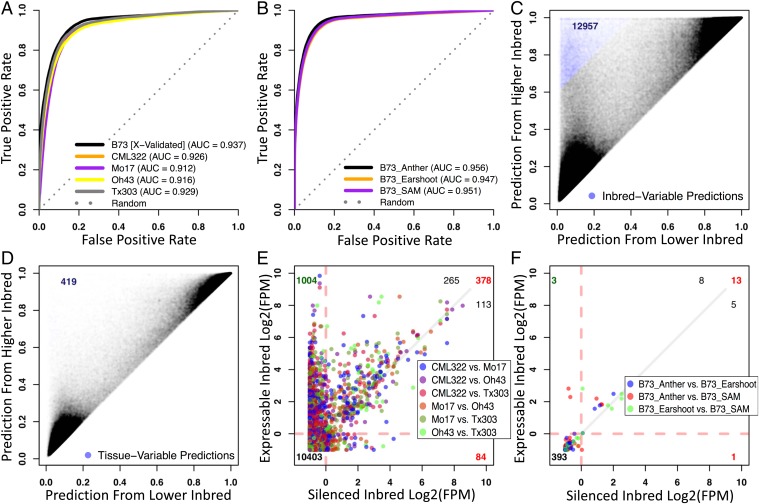
Fig. 3.
A new version of the ERC was generated (called ERC-2) using the same training classes defined for ERC but with WGBS data from B73 third leaf tissue that was summarized in 100 bp windows along the genome. This ERC-2 was then used to classify 2 test data sets of similar WGBS data. The first set (A, C, and E) was sampled from the third leaf of 4 diverse maize inbred lines (CML322, Mo17, Oh43, and Tx303). The second set (B, D, and F) was sampled from 3 additional B73 tissues (anther, ear shoot, and shoot apical meristem). In addition, each of these test samples has corresponding transcript profiling via RNA-seq available. (A) Receiver operating characteristic (ROC) curve showing prediction accuracies achieved by ERC-2 model on the B73 training genotype, using cross validation and when the ERC-2 model is tested with new methylation data from different maize inbreds. (B) Receiver operating characteristic (ROC) curve showing prediction accuracies achieved by the ERC-2 model on 3 additional B73 tissues. (C and D) Scatterplots showing the prediction scores between pairwise comparisons of all 4 test inbreds (C) or all 3 test tissues of B73 (D). Each point represents one gene for one test inbred–to–test inbred or test tissue–to–test tissue comparison. Upper left (blue) represent genes that are classified differently in one sample compared to another. (E and F) Scatterplots showing comparison of mRNA abundance in test sample pairs for differentially classified genes (blue dots in C and D). The numbers in the corners represent gene counts in each quadrant (quadrants are defined using cutoffs at log2[FPM] = 0). Quadrant 1 is further split into 2 via a diagonal gray line, with black numbers representing corresponding gene counts.
The ERC-2 was also used to determine whether developmentally regulated differences in genic methylation are associated with tissue-specific gene expression potentials (Fig. 3 B, D, and F). Three previously published B73 tissue data sets were examined (anther, developing ear, and shoot apical meristem [SAM]). For each sample, both DNA methylation and RNA-seq data were collected (18). We observed much greater variability in classification scores between inbreds than we did between tissues (Fig. 3 C and D; blue dots). On average, 2,160 genes have differential classifications between 2 inbreds while only 140 genes have differential classifications between 2 tissues.
The ERC-2 performed well for all of the inbred lines (Fig. 3A) and the tested tissues (Fig. 3B), with areas under ROC curves of 0.9 or greater. The test samples were compared to each other in a pairwise fashion to give 6 comparisons among the 4 inbreds and 3 comparisons among the 3 tissues. The ERC-2 classification scores were plotted for each comparison, with the lower score plotted on the x axis and the higher score plotted on the y axis (Fig. 3 C and D). As expected, most genes receive the same classification for the 2 samples in question (98 and 99.9% for inbred and tissue comparisons, respectively), indicating that most genes possess similar methylation patterns (Fig. 3 C and D; gray dots). On average, 2,160 and 140 genes have a differential classification between 2 inbreds and tissues, respectively, resulting from differential DNA methylation (Fig. 3 C and D; blue dots), causing them to be classified as silent in one sample type and expressible in another. We designated differential classifications as a difference in score greater than 0.6. We plotted observed mRNA abundance for the differentially classified genes (Fig. 3E). As with the prediction scores, each gene was plotted with the expression of the lower-predicted sample on the x axis and the higher-predicted sample on the y axis. We used fragments per million (FPM) of 1 as a cutoff below which genes were considered not expressed. A total of 1,466 genes was expressed in at least one inbred. Of these, 1,004 (∼68%) were not expressed when classified as silent, and 1,269 (∼87%) had lower expression when classified as silent. This is similar to the average accuracy of the expression models (AUC = 0.92; Fig. 3A). We compared the different tissues, but only 17 genes were differentially expressed with 65% above the diagonal in Fig. 3F. These results indicate that the set of transcribed genes varies significantly between genotypes and much less so between tissues, consistent with previous reports on differential DNA methylation (19).
Many of the silenced genes may arise from recently copied gene fragments that have been captured inside of TEs (20). We observed that as new inbreds are added to the analysis, the transcribed gene set of the species is expanded (SI Appendix, Fig. S10A). Thus, maize appears to have a panexpressible gene set of significantly more genes than are transcribed in any individual inbred. The inherited epigenomic patterns that we have associated with permission for expression, along with cis and trans transcriptional regulation, give rise to the pantranscriptome where the phenomenon of inbred-specific expression has been characterized (21). The panexpressible gene set is distinct from the pangenome which arises from structural variation (9). Differential DNA methylation exhibits relatively stable transgenerational inheritance (17), and therefore, our models predict that hybrids will express a set of genes that is the sum of the sets expressed by the 2 inbred parents. This prediction has been confirmed (22). While DNA methylation is generally stable, it likely has a spontaneous rate of change greater than DNA sequence yet low enough to maintain a long-term selection response (23). Therefore, spontaneous mutations in DNA methylation will occasionally cause expressible genes to become silent and silent genes to become expressible.
Silenced Genes Have Distinct Attributes Compared to Expressible Genes.
The ERC-2 results allow us to compare attributes of expressible genes to those of silenced genes to characterize the large set of silenced genes. The ERC-2 classifications predict that 32,333 genes can be transcribed across all 5 inbreds. Of these, 22,101 have syntenic orthologs with sorghum, and 10,232 are nonsyntenic. We refer to these groups as “all inbreds syntenic” and “all inbreds nonsyntenic.” In addition, 18,289 genes are transcribed in some subset of the 5 inbreds. We will call this the “any inbred” group. Finally, 50,103 genes are predicted to be silenced in all inbreds, the “no inbreds” group (SI Appendix, Fig. S10 and Datasets S3 and S19). All potential TEs were discarded from all 4 groups for this analysis (SI Appendix, Fig. S10B). Categorical enrichment was carried out on each group (Dataset S3, Tabs 2–5). Interestingly, we see significant enrichment for “biotic stress” and “secondary metabolism” categories in the all inbreds nonsyntenic group, indicating more recent selection for these functions in maize. We found half (51%) of the no inbred group to be potential TEs, so they were removed, leaving 24,433 genes that are silenced in all examined inbreds but have been annotated to potentially encode proteins. Compared to genes that are expressible across all inbreds, non-TE genes in the no inbred group tend to be shorter, contain fewer introns, contain fewer known protein domains, and are less conserved in sorghum (SI Appendix, Fig. S10 C–F). Many of these silent genes (32%) express RNAs in B73 at very low levels (SI Appendix, Fig. S10 H and I). Interestingly, 387 of them have detectable proteins expressed at moderate levels (SI Appendix, Fig. S10 H and J). We clustered genes based on protein sequence similarity. All inbreds genes tend to be in larger clusters (SI Appendix, Fig. S10G). The network clusters tend to form around protein domains and therefore represent known gene families (SI Appendix, Fig. S10K); we displayed only the 15 largest clusters. Many of the silent genes cluster with known protein domains. Several large gene families comprise mostly silent genes, and these sequences have no known functions (“unknown” cluster in SI Appendix, Fig. S10K).
Of the 50,103 silent (no inbreds) genes, 25,670 are likely to be TEs. Of the remaining non-TE genes, 15,638 can be clustered with other genes based on protein sequence similarity. Within this sequence similarity network, ∼54% of the connections are between non-TE silent genes and expressible genes (SI Appendix, Fig. S10L). These silent genes may be recently copied gene fragments that have been captured by TEs (20). The remaining 46% of the edges are with other silent genes, forming groups of what appear to be protein domains of unknown function. This leaves 8,795 silent genes with no significant sequence similarity to other maize genes.
CG Gene Body Methylation Is Associated with Protein Expression.
The protein-specific feature illuminator (PFI) was built to find genic methylation patterns that distinguish between genes which express high RNA levels without proteins and genes that have high mRNA levels plus observed proteins (HR_NP vs. HR_OP). Of the 33,696 genes with observed RNAs in the HR, less than half (15,421) had observed proteins. The PFI was able to differentiate between the HR_NP and HR_OP with good accuracy (Fig. 2A and SI Appendix, Fig. S4A), achieving an area under the ROC curve of 0.8. Comparison of the feature importance between the PFI and the EPC/ERC showed that most of the important features were shared. However, there was a key difference. We observed a change in sign for midgene CG methylation (bins 2 to 4), indicating an association between protein expression and high CG methylation in the middle of genes (Fig. 2F). This pattern was previously described as gene body methylation (gbM); it is specifically defined as CG hypermethylation that occurs in the middle of the gene while both the 5′ and 3′ ends of the gene body remain hypomethylated (24, 25).
We examined the association between gbM and protein expression. Genes with less than 50% methylation in bins 1 and 5 plus greater than 50% methylation in at least one of bins 2 to 4 were defined as having gbM. Of these 9,071 genes, 59% had observed proteins, which is 3.5 times more than expected by chance (P value = 0, based on a hypergeometric test using the upper tail) (SI Appendix, Fig. S11A). High RNA and no protein genes showed a lesser enrichment of 1.4-fold (P value = 1e-62), while low mRNA and silent genes were underenriched at 0.4 and 0.1-fold, respectively (P values = 0 for both, based on a hypergeometric test using the lower tail). Of the 9,071 gbM genes, 88% were classified by the EPC as able to express proteins (SI Appendix, Fig. S11B). The results illustrate the increased sensitivity and accuracy of machine learning for gene classifications compared to a prospective approach using known DNA methylation patterns.
Gene body methylation has been described for both plants and animals. The occurrence of gbM in plants appears to be specific to angiosperms (26). However, within angiosperms, there are several reports of species that do not have gbM (26, 27). Although functions of gbM remain unknown, it is associated with constitutive mRNA expression, and these mRNAs also tend to have relatively high abundance (24, 25). One hypothesis is that gbM acts to block TE insertion (7) and therefore prevents mutagenesis of expressed genes. Our finding that gbM is associated with protein expression was unexpected, and it will be interesting to see whether this is true in other species.
Intronic regions have the highest feature importance in the PFI model (Fig. 2F). Of the 110,028 genes in the WGS, only 55,558 (50%) contain introns; 70% of genes in the FGS contain introns. Of the genes with observed proteins, 88% contain one or more introns, while only 58% of genes with high RNA and no protein contain one or more introns (SI Appendix, Fig. S12A). Therefore, the presence of introns helps to distinguish protein-expressing genes from the other members of the highly transcribed set. The link between introns and gene expression was discovered using transgenes in maize (28). We observed that 93% of genes with gbM contain introns (SI Appendix, Fig. S12A) and methylation is localized to the middle of intron-containing genes (SI Appendix, Fig. S12B) in contrast to genes without introns (SI Appendix, Fig. S12C); this is consistent with the hypothesis that gbM plays a role in RNA splicing (7, 29, 30). CG methylation specifically at intron–exon junctions may play a role in splicing (31).
Gene synteny combined with gene length and transcript expression was previously used to curate high-confidence and low-confidence gene models in sorghum (32). More than 73% of high-confidence gene models were found to be associated with CHG hypomethylation in the gene body, whereas the gene ends displayed CG hypomethylation plus hypermethylation in the midbody. Curated models were used to train a J48 decision tree classifier to recognize high- and low-confidence gene models based on expression, synteny, and DNA methylation patterns; all 3 data types were required for the decision tree classifier to perform fully.
Histone Modifications Are Positively Associated with Expression.
We compared genes classified by the EPC and ERC models to published patterns of histone modifications (33). Genes classified as expressible had high levels of H3K36me3, H3K9Ac, and H3K4me3, whereas silent genes had low levels (SI Appendix, Fig. S13 A and B). These modifications are associated with transcriptional activation (34). To determine whether the distribution of these modifications could classify genes according to their expressibility, we trained new EPC and ERC models using histone modifications as features instead of DNA methylation. The histone-based models performed well, with only slightly less accuracy than using DNA methylation (SI Appendix, Fig. S13 C and D). The addition of histone features to the EPC and ERC did not improve the accuracy of the models. The important features of the histone models were high levels of H3K36me3 and H3K4me3, especially at the 5′ ends of expressible genes, and high levels of H3K9Ac in the gene midbody (SI Appendix, Fig. S13 G and H). Using similar models, H3K36me3 was previously identified as the most important feature used to identify gene bodies in human embryonic stem cells (35). Histone modifications did not predict the time, place, or levels of expression (SI Appendix, Fig. S13 E and F).
Our findings may shed light on mechanisms of evolution and domestication. DNA methylation and histone features that were important in our classifiers may be part of a mechanism for selectively silencing genes that arise through gene or whole-genome duplication (36). This may enable retention of new genes without immediate or extreme phenotypic effects. New genes may be purged from the genome or persist as a reservoir of adaptive potential that can be tapped through spontaneous, heritable changes in DNA methylation or histone marks.
Supplementary Material
Acknowledgments
This work was supported by NSF Grant IOS-1546899 (to S.P.B.).
Footnotes
Conflict of interest statement: N.M.S. and V.W. are coauthors on a 2016 Review article.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813645116/-/DCSupplemental.
References
- 1.Walley J. W., et al. , Integration of omic networks in a developmental atlas of maize. Science 353, 814–818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnable J. C., Freeling M., Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One 6, e17855 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichten S. R., et al. , Heritable epigenetic variation among maize inbreds. PLoS Genet. 7, e1002372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West P. T., et al. , Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9, e105267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 6.Gent J. I., et al. , CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23, 628–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regulski M., et al. , The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23, 1651–1662 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohn T., Corsten S., Rieke S., Müller M., Rothnie H., Methylation of coding region alone inhibits gene expression in plant protoplasts. Proc. Natl. Acad. Sci. U.S.A. 93, 8334–8339 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao Y., et al. , Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portwood II J. L., et al. , MaizeGDB 2018: The maize multi-genome genetics and genomics database. Nucleic Acids Res. 47, D1146–D1154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler V. L., Walbot V., DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. U.S.A. 83, 1767–1771 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slotkin R. K., Martienssen R., Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Zemach A., McDaniel I. E., Silva P., Zilberman D., Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Gallego-Bartolomé J., et al. , Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis. Cell 176, 1068–1082.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wessler S. R., et al. , Maize transposable element database. https://figshare.com/articles/MaizeTE_Seqs_12-Feb-2015_fasta_fa/9172439/1. Accessed 6 August 2019.
- 16.Li Q., et al. , Examining the causes and consequences of context-specific differential DNA methylation in maize. Plant Physiol. 168, 1262–1274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichten S. R., et al. , Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25, 2783–2797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q., et al. , RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. U.S.A. 112, 14728–14733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichten S. R., Vaughn M. W., Hermanson P. J., Springer N. M., Variation in DNA methylation patterns is more common among maize inbreds than among tissues. Plant Genome 6, 1–10 (2013). [Google Scholar]
- 20.Anderson S. N., et al. , Transposable elements contribute to dynamic genome content in maize. bioRxiv:10.1101/547398 (12 February 2019). [DOI] [PubMed]
- 21.Hirsch C. N., et al. , Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26, 121–135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldauf J. A., et al. , Single-parent expression is a general mechanism driving extensive complementation of non-syntenic genes in maize hybrids. Curr. Biol. 28, 431–437.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 23.van der Graaf A., et al. , Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. U.S.A. 112, 6676–6681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., et al. , Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Zilberman D., Gehring M., Tran R. K., Ballinger T., Henikoff S., Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Bewick A. J., et al. , On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 113, 9111–9116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niederhuth C. E., et al. , Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17, 194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callis J., Fromm M., Walbot V., Introns increase gene expression in cultured maize cells. Genes Dev. 1, 1183–1200 (1987). [DOI] [PubMed] [Google Scholar]
- 29.Wang B., et al. , Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 7, 11708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lev Maor G., Yearim A., Ast G., The alternative role of DNA methylation in splicing regulation. Trends Genet. 31, 274–280 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Wang X., et al. , DNA methylation affects gene alternative splicing in plants: An example from rice. Mol. Plant 9, 305–307 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Olson A., et al. , Expanding and vetting Sorghum bicolor gene annotations through transcriptome and methylome sequencing. Plant Genome 7, 10.3835/plantgenome2013.08.0025 (2014). [DOI] [Google Scholar]
- 33.He G., et al. , Conservation and divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol. 14, R57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger S. L., The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Rajagopal N., et al. , Distinct and predictive histone lysine acetylation patterns at promoters, enhancers, and gene bodies. G3 (Bethesda) 4, 2051–2063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panchy N., Lehti-Shiu M., Shiu S. H., Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.