Significance
Mutations are often assumed to be largely detrimental to fitness, but they may also be beneficial, and mutations with large phenotypic effects can persist in nature. One explanation for these observations is that mutations may be beneficial in specific environments because these conditions shift trait expression toward higher fitness. This hypothesis is rarely tested due to the difficulty of replicating mutants in multiple natural environments and measuring their phenotypes. We did so by planting Arabidopsis thaliana genotypes with large-effect flowering time mutations in field sites across the species’ European climate range. We quantified the adaptive value of mutant traits, finding that certain mutations increased fitness in some environments but not in others.
Keywords: fitness landscape, flowering time, branching, natural selection, mutation
Abstract
Contrary to previous assumptions that most mutations are deleterious, there is increasing evidence for persistence of large-effect mutations in natural populations. A possible explanation for these observations is that mutant phenotypes and fitness may depend upon the specific environmental conditions to which a mutant is exposed. Here, we tested this hypothesis by growing large-effect flowering time mutants of Arabidopsis thaliana in multiple field sites and seasons to quantify their fitness effects in realistic natural conditions. By constructing environment-specific fitness landscapes based on flowering time and branching architecture, we observed that a subset of mutations increased fitness, but only in specific environments. These mutations increased fitness via different paths: through shifting flowering time, branching, or both. Branching was under stronger selection, but flowering time was more genetically variable, pointing to the importance of indirect selection on mutations through their pleiotropic effects on multiple phenotypes. Finally, mutations in hub genes with greater connectedness in their regulatory networks had greater effects on both phenotypes and fitness. Together, these findings indicate that large-effect mutations may persist in populations because they influence traits that are adaptive only under specific environmental conditions. Understanding their evolutionary dynamics therefore requires measuring their effects in multiple natural environments.
Throughout much of the 20th century, mutations were assumed to be largely deleterious, diminishing the adaptive value of a trait in a particular environment (1–3). However, more recently, a robust empirical body of work has shown that the frequency of beneficial mutations considerably exceeds previous assumptions (4–10). Many mutations, even of large effect, are known to have become resident in populations and persist as segregating alleles (11–16). The few experiments testing mutations’ fitness effects in nature have revealed complex patterns among environments, with effects in one environment poorly predicting them in another (6, 17–19). A major reason for this complexity may be that environments affect trait expression, but few, if any, studies have gauged the mutation–phenotype relationship in multiple natural environments. Rather than examining unbiased mutation spectra consisting of many neutral mutations present in mutation accumulation lines (19), one way to test this is to examine targeted mutations known to affect ecologically important phenotypes. Here, we do this by constructing genotype–phenotype–fitness landscapes of Arabidopsis thaliana mutants harboring large-effect mutations targeted to known flowering-time genes in common gardens across its European climate range.
Fitness landscapes were introduced as a metaphor to conceptualize how populations evolve through states of low fitness to high fitness (20) and have since been extended and formalized to express mathematically the relationships among genotypes, traits, and fitness (21–23). In practice, empirical fitness landscapes often examine only one-half of the genotype–phenotype–fitness relationship. In the phenotype–fitness approach, landscapes relate trait values to fitness so that a peak represents a combination of trait values that is highly fit. However, these landscapes are usually constructed without reference to specific genes that underlie these traits (24–36). Genotype–fitness landscapes, on the other hand, relate combinations of alleles to fitness so that a peak represents a highly fit genotype. While powerful for understanding evolution, measurements of genotype–fitness landscapes have been rare in multicellular organisms, and most do not consider higher order traits directly related to fitness (20, 37–50).
A major goal in evolutionary biology is to synthesize these approaches in a complete genotype–phenotype–fitness landscape. However, in eukaryotic organisms, combining phenotype–fitness and genotype–fitness landscapes is frequently limited by either a lack of knowledge of adaptive traits or of genes that control them. The genetic model species A. thaliana provides a valuable system to tackle this challenge: The genetic control of ecologically important phenotypic traits is well studied (51–54), and field experiments allow the measurement of phenotypic selection in natural environments (55–61).
Both flowering time and branching architecture are potentially important adaptive traits in A. thaliana and other annual plants. The timing of flowering determines the critical transition from vegetative to reproductive growth; aligning this transition with favorable environmental conditions is critical to successful reproduction. Flowering time varies clinally across the species’ range (53, 56, 60, 62–66), evidence for adaptive differentiation in this trait across different environments. This clinal differentiation is due to natural variation in responses to environmental signals like temperature and day length (66–69). Flowering time is closely linked to branching architecture through development. Prior to flowering, Arabidopsis plants produce rosette leaves and rarely secondary branches. However, after commitment to flowering, side branches are produced from axillary meristems, and these side branches also produce flowers (70, 71). The number of axillary meristems and branches available to produce seeds depends upon flowering time since prolonged vegetative growth due to delayed flowering allows a plant to develop more leaves and axillary meristems (72–74). Thus fecundity is at least partially a function of flowering time-dependent branching architecture (75). Importantly, the hormone gibberellin (GA) partially decouples branching from fecundity since it promotes branching but simultaneously inhibits flower formation (76). This decoupling enables branching and reproductive phenology to act as partially independent developmental modules that may be independently selected upon but that are still partially united. Thus, plant fitness depends on a balance between branching and the timing of flowering to determine fitness in morphological and phenological space. Since both flowering time and gibberellin flux depend upon growing conditions (77–79), this balance is likely to shift among environments.
The genotype–phenotype map is further complicated by the complex and nonlinear relationships among genes, especially for quantitative traits like branching and flowering time with highly polygenic bases. Genes influencing these traits interact through multilevel networks. Some are hubs that influence a large number of other genes, but others are subject to multiple inputs. These highly connected genes within regulatory networks may allow mutations in them to be buffered against producing pernicious phenotypes (80–82). The degree of this buffering is a function of both a gene’s network connectedness and redundancy permitting increases or decreases in these factors, respectively, to lead to more severe deviation in phenotype when mutated (80, 83–86). Together, this implies that potentially adaptive mutations may be masked due to network topography, robustness, and redundancy. Thus, adaptive zones in fitness landscapes may be accessible only through mutations in large-effect hub genes. On the other hand, such robustness is also a function of the environment so that mutations buffered in one environment may be exposed in another (87–90). In this way, genetic network position can interact with environment to shift mutants in fitness landscapes.
Although the accumulation of many small-effect mutations can collectively alter phenotypes, we focus on individual large-effect allelic variants induced by chemical and radiation mutagenesis. Single, large-effect mutations represent a crucial evolutionary step by which a lineage can explore more distant regions of the fitness landscape than it can by single small-effect mutations (91). We define “large-effect” as mutations that cause specific, replicable, and major changes in flowering time and have been extensively experimentally validated (Table 1 and SI Appendix, Table S1). The majority of our mutations were loss-of-function, ascertained at the gene expression and protein levels (SI Appendix, Table S1). This is realistic for alleles segregating in natural populations of A. thaliana that underlie adaptive phenotypic variation in metabolic, defensive, morphometric, and phenological traits (14–16, 92–100). We also included a single instance of gain-of-function through introgression of a strong functional allele to replace a nonfunctional variant of the gene FRIGIDA in the reference ecotype (101). We do not speculate about the precise nature of DNA sequence-level change since there are many potential paths to large effect, but rather we seek to understand how large-effect mutations interact with the environment to influence selection on phenotypes. In order to disentangle the effects of correlated environmental signals like day length and temperature, we selected field sites in a latitudinal gradient, in which day length and temperature covary; a longitudinal gradient, in which temperature varies independent of day length; and a seasonal gradient, in which plantings were repeated within a site for several seasons (SI Appendix, Fig. S1 A and C) (55, 58, 102, 103). We report morphometric, phenological, and fecundity data from this common garden experiment to answer 3 main questions:
-
1)
How do large-effect mutations in flowering time genes affect phenology, morphology, and fitness in different natural environments?
-
2)
How do phenotype–fitness and genotype–phenotype–fitness landscapes change with the environment?
-
3)
How does gene position within a regulatory network modulate mutational effects on the genotype–phenotype–fitness map?
Table 1.
Description of genes mutant in this experiment with their classical pathway designation
| Gene | Pathway | Effect on flowering time |
| FVE* | Autonomous | Pos. regulator that suppresses FLC expression |
| HUA2*,† | Autonomous | Neg. regulator that enhances late flowering in FRI functionals |
| LD* | Autonomous | Pos. regulator that suppresses FLC expression |
| FT* | Integrator | Florigen integrating all pathways to activate floral meristem genes |
| TFL2* | Integrator | Floral meristem identity; maintains high FLC despite brief warmth |
| GAI* | Hormone | Pos. regulator by up-regulating FT and gibberellin synthesis |
| SPY* | Hormone | Neg. regulator downstream of GAI that suppresses gibberellin |
| CO* | Photoperiod | Sensor of inductive long days, downstream of PHYs |
| CRY1* | Photoperiod | Pos. regulator of CO; perceives blue light of long days |
| GI* | Photoperiod | Integrates circadian clock information to perceive long days |
| PHYA* | Photoperiod | Pos. regulator of CO; perceives red light of long days |
| PHYB* | Photoperiod | Neg. regulator of CO; perceives red:far-red ratio of light of long day |
| PHYD* | Photoperiod | Neg. regulator of CO; obligately interacts with PHYB |
| PHYE* | Photoperiod | Neg. regulator of CO; obligately interacts with PHYB |
| FRL† | Vernalization | Constituent of FRI activation complex; up-regulates FLC |
| FRI‡ | Vernalization | Activates FLC, conferring vernalization requirement |
| FLC† | Vernalization | FT repressor that is epigenetically down-regulated by vernalization |
| VIN3† | Vernalization | Required for vernalization response in FRI:FLC functionals |
Pathways are not exclusive since some genes act in multiple pathways simultaneously. Some mutant genotypes harbored multiple mutations. For example, phyabde is a quadruple mutant with LoF in PHYA, PHYB, PHYD, and PHYE. Neg., negative; Pos., positive.
Mutations in this gene were not combined with any natural allele.
Mutations in this gene were induced in a background with a functional FRI allele introgressed from the Sf-2 ecotype.
Natural alleles only by introgressing a functional Sf-2 version without induced mutation.
Results
Here, we analyze subsets of the genotype–phenotype–fitness map across environments and then combine these components. First, we examine the genotype–phenotype landscape by quantifying genotype-phenology and genotype-branching relationships in each environment. Second, we examine the phenotype–fitness landscape by quantifying phenology-fitness and branching-fitness relationships in each environment. Finally, we synthesize these across environments to describe the environmental dependence of mutational effects on phenology, branching, and fitness. Large-effect mutations in environmental signaling pathways are especially useful in these environmental comparisons because plants harboring these mutations are unable to respond to specific environmental inputs. By comparing their phenotypes to those of the same background genotype without the mutation, these mutants can be used as phytometers to gauge the relative importance of specific environmental cues for determining phenology and branching (103).
Phenology.
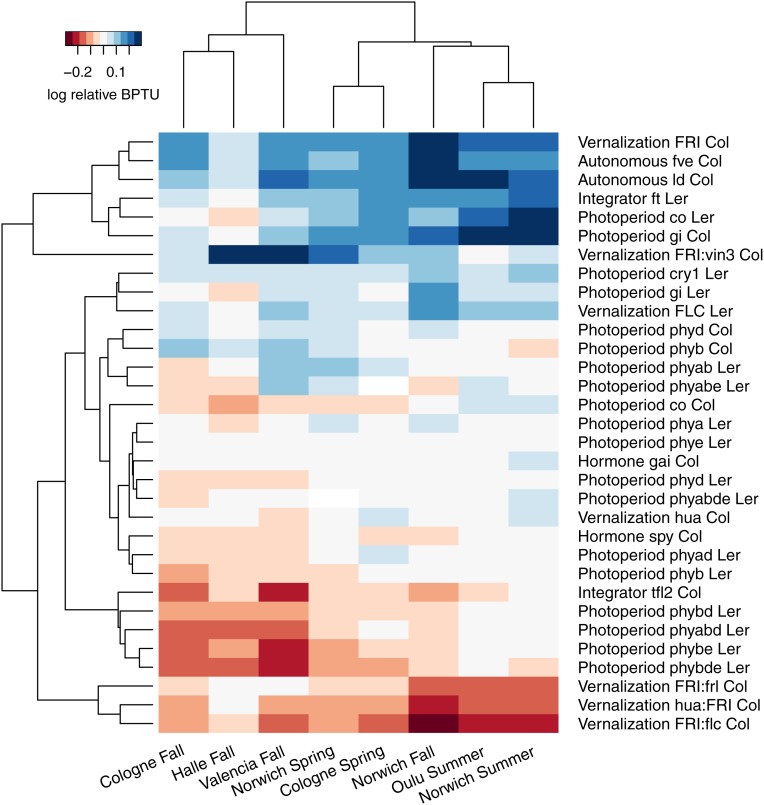
To describe how these mutations affected phenological traits relative to ecotype background across 8 natural environments, we used a 2-part hierarchical clustering algorithm to uncover environmental and mutant trends (Fig. 1). A linear model detected significant heterogeneity in mutant bolting times measured in photothermal units (BPTUs) among plantings, among genotypes, and among multifactorial combinations of planting and genotype (SI Appendix, Table S2). In particular, we identified genotype × environment interactions, with the majority of mutants showing magnitude differences in mutant effects in different environments and several showing both magnitude and directional differences (SI Appendix, Table S3). Together, these results quantify the environmental contingency of genetic effects in determining bolting time among these different environments.
Fig. 1.
Heat map of least-square means of accumulated photothermal units to bolting (BPTUs) in mutants relative to ecotype background on a log2 scale, centered within plantings, visualizing two-step hierarchical cluster by a Euclidian distance, average-based algorithm for both genotype (rows) and planting (columns). All mutant pathways are represented, although not all genotypes, since some were not planted in all 8 sites and seasons. The first word of the row identifiers shows which pathway was manipulated in a mutant, as defined by FLOR-ID (51). Lowercase gene names indicate diminished function alleles, and uppercase, functional. Colons between pathways or genes indicate multiple genetic manipulations within a line, not gene fusions. Col and Ler indicate each line’s ecotype background, Col-0 and Ler-1, respectively. Genotypes with an induced mutation combined with a functional FRIGIDA (denoted by “FRI”) were relativized against FRI Col instead of Col-0.
Specifically, we identified 3 distinct mutant classes: late-bolting, early-bolting, and sign plasticity clusters (Fig. 1). The late-bolting cluster was dominated by mutations in the vernalization and autonomous pathways known to enhance the expression of the floral repressor FLOWERING LOCUS C (FLC) or to impair its down-regulation (i.e., FRIGIDA:vernalization insensitive 3 [FRI:vin3], FRI, fve, ld), as well as mutants impaired in photoperiod response and signal integration (i.e., ft, cry1). The early-bolting cluster showed a mix of pathways. Many upstream photoperiod mutants accelerated bolting, the most consistent of which were deficient in PHYTOCHROME B (PHYB) in the Ler background. The hormone pathway mutant spy showed accelerated phenology, and gibberellic acid insensitive (gai) did not. Acceleration of bolting in the spy mutant was expected because functional SPY is a repressor of inductive gibberellins (104). The hormone pathway’s importance for accelerated flowering has often been emphasized in short days (105), but spy also accelerated flowering in summer and spring plantings that mainly experienced long days. The vernalization pathway mutations fri, hua, and flc accelerated bolting in the Col-FRI background. These loss-of-function mutations all reduce expression of FLC, allowing flowering without vernalization. Finally, the sign plasticity group consisted of genotypes that accelerated flowering in some environments and delayed flowering in others. Genotypes showing this sign plasticity most strongly included 4 mutants deficient in phytochromes (phyb, phyabe, phyad, phyabe) which are involved in photoperiod and ambient temperature sensing (106–108) as well as the photoperiod integrators gi and co (SI Appendix, Table S3). A detailed discussion of gene-specific results is presented in the SI Appendix, Supplemental Results.
Branching.
Flowering time mutants pleiotropically expanded variation in branching in both ecotypic backgrounds, and, similarly to bolting time, mutant branching varied among seasons, sites, and genotypes (SI Appendix, Fig. S5). Most late-bolting mutants, especially in the vernalization pathway, produced more branches, and most early-bolting mutants in the photoperiod and hormone pathways produced fewer branches than their parental ecotypes. Specifically, gai had fewer branches than its ecotype background, and the spy mutant consistently had the fewest branches across plantings (SI Appendix, Fig. S6). Several phytochrome mutants had many fewer branches than either the parental ecotypes or most other mutants, consistent with the constitutive shade avoidance phenotype expected with loss of photoreceptor function. Combined with their bolting time effects, it is also possible that these mutants had fewer branches because they flowered rapidly and therefore developed fewer nodes from which branches could emerge. Several mutants also exhibited sign plasticity for branch production, including the phytochrome mutants phya, phyd, phyad, and phybe, and the vernalization mutant FRI:vin3. Flowering time mutants generated in the Col-0 background had more branches than those in Ler-1 in most plantings (SI Appendix, Fig. S6). This is likely because Ler-1 harbors a loss-of-function allele of ERECTA, a positive regulator of aerial organogenesis (109), and has fewer branches than Col-0.
Univariate Selection.
The fitness effects of flowering time mutations differed in sign and magnitude across the 5 plantings for which fecundity data were collected: Norwich fall, Norwich spring, Norwich summer, Halle fall, and Valencia fall (SI Appendix, Fig. S7). In particular, nearly half of all mutant genotypes switched between positive or negative relative fitness among environments (SI Appendix, Fig. S7). We quantified phenotype–fitness landscapes separately for the phenology and morphometric traits for each planting. Univariate directional selection for earlier bolting and flowering occurred in 4 plantings while delayed flowering was favored in the Norwich summer planting (Table 2). Furthermore, selection for phenology measured in calendar time was stronger than for photothermal time (SI Appendix, Table S4), which we used to scale environmentally dependent phenologies across sites (SI Appendix, Supplemental Methods). We did not detect significant stabilizing selection in most of the plantings. Significant negative quadratic selection was detected for calendar days to bolting in Valencia, which indicated monotonic selection toward a theoretical maximum on the selection surface for an extremely early bolting time that did not fall within the range expressed by any genotype (Table 2). Directional selection consistently favored increased branching in all sites, with no evidence of stabilizing selection on branching traits (Table 2 and SI Appendix, Table S4).
Table 2.
Univariate selection coefficients for A. thaliana traits in 5 field environments, analogous to partial derivatives of polynomial regression techniques
| Planting | DTB | BPTU | Branching | |||||||||
| β | SE | γ | SE | β | SE | γ | SE | β | SE | γ | SE | |
| Halle fall | −0.144 | 0.023 | −0.517* | 0.062 | −0.083 | 0.07 | −4.222 | 3.180 | 0.766* | 0.067 | 0.174 | 0.380 |
| Norwich fall | 0.204 | 0.13 | 0.021 | 0.08 | 0.192 | 0.092 | 10.014 | 11.045 | 1.136* | 0.157 | 0.620 | 0.846 |
| Norwich spring | −0.350* | 0.087 | −0.001 | 0.043 | −0.270* | 0.050 | 0.000 | 8.310 | 0.789* | 0.044 | −0.106 | 0.165 |
| Norwich summer | 0.151* | 0.040 | −0.071 | 0.038 | 0.159* | 0.042 | −6.998 | 6.013 | 0.530* | 0.041 | 0.085 | 0.169 |
| Valencia fall | −0.214* | 0.060 | −0.230* | 0.029 | −0.180* | 0.057 | −17.385 | 15.921 | 0.740* | 0.082 | 0.044 | 0.323 |
β, analogous to the directional selection coefficient; BPTU, accumulated photothermal units to bolting; Branching, total branch number; DTB, calendar days to bolting; γ, analogous to the stabilizing or disruptive coefficient; SE, numerically approximated SE for the term immediately to the left.
Bolded estimates with asterisks represent significance from Bonferroni-corrected P values <0.01.
Multivariate Selection.
Taken together, univariate selection gradients indicated that fast-developing and highly branched phenotypes were generally favored. One possible genetic mechanism for reaching this optimal fitness zone is gibberellin’s simultaneous promotion of a more branched architecture and fast development; however, high gibberellin activity also suppresses flower formation early in development, which could decrease fecundity. To determine how bivariate selection on phenology and branching occurred, we modeled both in a multivariate generalized additive model (GAM) (Table 3). This showed that directional selection for greater branching was strong in all sites, but direct selection for phenology was significant in only 2 sites: Later flowering was favored in Norwich summer, and earlier flowering in Halle fall. No γ terms, which denote stabilizing, disruptive, or correlative selection, were significant. Together, this implicates the role of indirect selection on phenology while selection operated more strongly on branching (110). However, consistent with previous findings of high heritability for phenology traits and low heritability for fitness estimates (111), the ratio of genetic variation to phenotypic variation of morphometric and fitness traits (0.27 to 0.5) was much lower than phenological traits (>0.7) (SI Appendix, Table S5). Since these mutants were selected for their effects on flowering time, phenology’s stronger genetic basis was expected but allows us to gauge the relative contribution of these underlying mutations in determining phenotype and fitness values.
Table 3.
Bivariate selection gradient analysis
| Term | Halle fall | Norwich fall | Norwich spring | Norwich summer | Valencia fall | |||||
| Est. | SE | Est. | SE | Est. | SE | Est. | SE | Est. | SE | |
| βBPTU | −0.13* | 0.10 | 0.33 | 0.17 | −0.01 | 0.08 | 0.20* | 0.07 | −0.02 | 0.09 |
| βbranch | 0.72* | 0.05 | 1.15* | 0.11 | 0.77* | 0.09 | 0.53* | 0.04 | 0.71* | 0.08 |
| γBPTU | −9.10 | 120 | 5.84 | 146 | −30.2 | 90.8 | −60.1 | 100 | 82.2 | 114 |
| γbranch | 0.06 | 0.31 | 0.90 | 1.11 | −0.00 | 0.18 | 0.06 | 0.16 | 0.18 | 0.22 |
| γBPTU,branch | −0.11 | 0.14 | 0.28 | 0.31 | 0.00 | 0.09 | 0.10 | 0.06 | −0.07 | 0.19 |
β, analogous to the directional selection coefficient; BPTU, accumulated photothermal units to bolting; branch, total number of branches; Est., estimate of the coefficient; γ, analogous to the stabilizing or disruptive coefficient; SE, numerically approximated SE.
Bolded estimates with asterisks represent significance from Bonferroni-corrected P values <0.05.
Genotype–Phenotype–Fitness Landscapes.
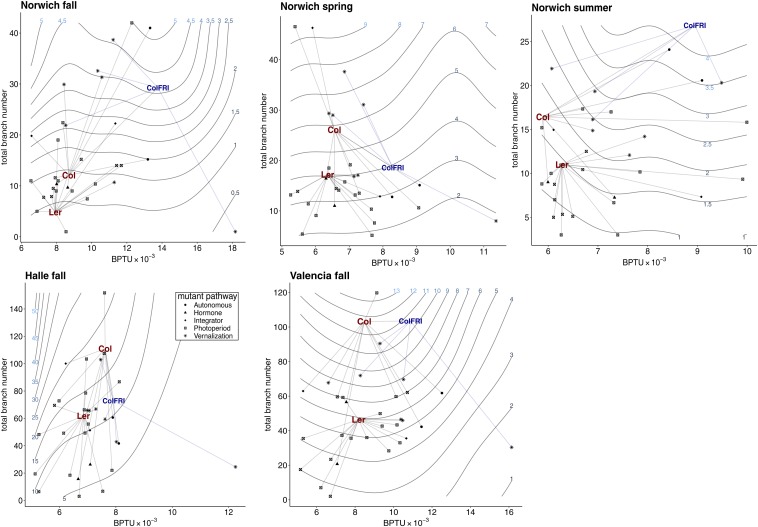
Modeling genotype means in 2D fitness landscapes revealed remarkable variation in the genotype–fitness map among environments (Fig. 2). Ecotype background fitness relative to mutant fitness changed dramatically among environments. Although generally more fit than mutants, our reference ecotypes (Col-0 and Ler-1) showed lower relative fitness than specific mutants in certain environments. For example, in Valencia and Halle, the Col-0 ecotype was more fit than all of its mutants except one, but, in Norwich fall, both ecotypes were less fit than the majority of their mutants. Together, these patterns show that mutants achieved higher fitness through alternate phenotypic paths. For example, some mutants were invariant in phenology in certain environments but were more highly branched whereas others bolted in the direction of higher fitness but were invariant in branching. The mutations underlying these phenotypic shifts were adaptive in conditions that shifted phenotypic expression toward fitness peaks.
Fig. 2.
Fitness landscapes from generalized additive models for accumulated photothermal units bolting and total branch number. Contour line labels show fitness in seed proxy units. BPTU refers to accumulated photothermal units to bolting. Points show line averages where lines are genotypes bulked under the same maternal conditions. “Col” refers to the Col-0 ecotype average; “Ler” to the Ler-1 ecotype average; “ColFRI” refers to the genotype with a functional version of FRI introgressed from the Sf-2 ecotype. Grey vector lines represent mutations induced in the Col and Ler ecotypes, and blue vector lines represent mutations induced in the FRI (Col) genotype.
Network Effect of Mutational Perturbation.
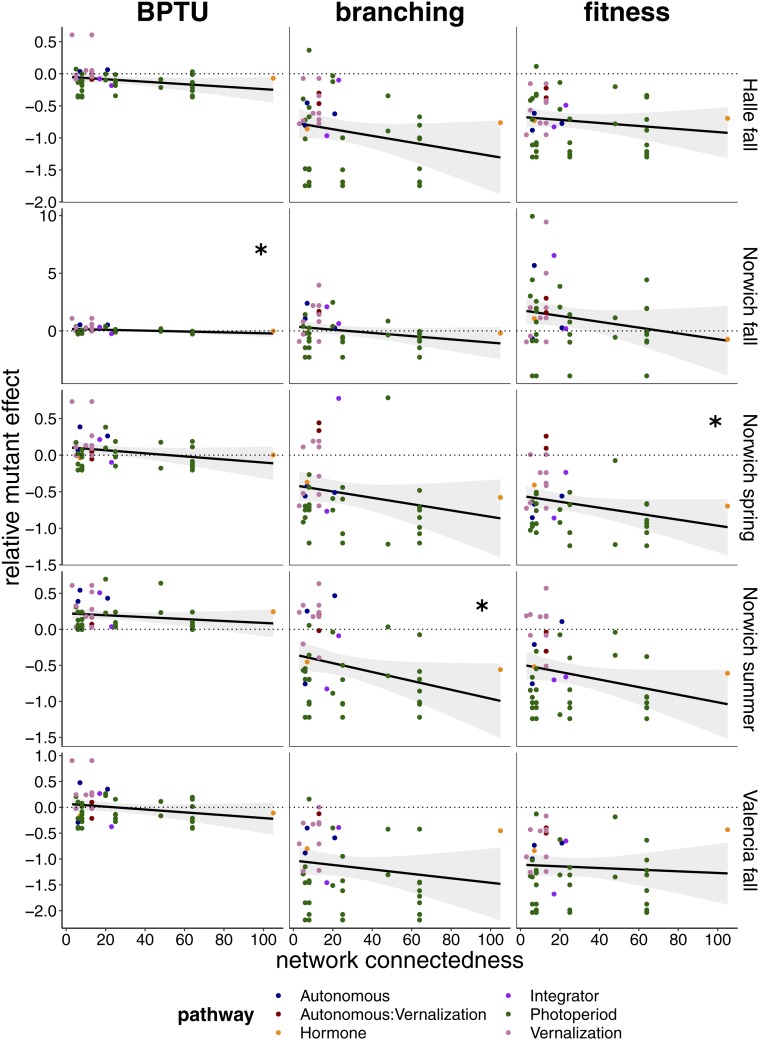
We were interested in how gene network connectivity affected mutational perturbations on phenotypes independent of the pathway in which a mutation was induced. To answer this question, we first assessed how connected the genes in a mutant line were, summing these connections for multiple mutants. We then regressed this metric of connectivity against relativized phenotypic perturbation. Although only 3 of 15 regressions were significant after Bonferroni correction, all showed negative relationships between connectivity and phenotypic perturbation across all 3 traits and all 5 environments (Fig. 3). Overall, the magnitude of bolting time change was considerably dampened relative to branching or fitness. This indicates that bolting time is relatively robust to genetic perturbation in flowering time networks, but that branching and fitness are much less robust. In particular, mutations in more highly connected genes led to consistent and, in at least one environment, significant decreases in fitness.
Fig. 3.
Relationship between network connectedness and mutant phenotypic shift relative to ecotype background. BPTU is for accumulated photothermal units to bolting; branching is the total number of branches; and fitness is seed proxy number. Shaded areas show 95% confidence intervals; panels with asterisks denote significant linear regressions (P < 0.05) after Bonferroni correction.
Discussion
A major goal in evolutionary biology is to understand the relationship among mutations, traits, and fitness in natural environments. Directly inferring these relationships is frequently constrained by limited knowledge of genetic mechanisms in ecological systems and by the difficulty of testing mutants in multiple natural environments. However, by leveraging the well-characterized genetic architecture of flowering time and branching in the model organism A. thaliana, we investigated how mutations in multiple flowering time pathways altered the distribution of A. thaliana genotypes across fitness landscapes, the traits that composed these landscapes, and how they depended upon the environment a plant experienced.
Phenology, branching, and fitness all expressed both magnitude and sign plasticity to growth environment (Fig. 2 and SI Appendix, Figs. S5 and S7). The largest number of lines showed clear sign plasticity for fitness and the fewest for phenology, evidence that fitness was more environmentally labile than phenology. Overall, mutants were more frequently less fit than their parental backgrounds. However, for several mutants, we identified specific natural environments in which they actually had greater fitness. There are 2 explanations for this finding of greater mutant fitness only in certain environments: 1) Mutations’ phenotypic effects might differ between environments, and 2) the underlying selection surface might change. Here, we detected evidence for both mechanisms since certain environments selected for different directions and magnitudes of trait values (evidence of changing selection surfaces) and mutants’ phenological and branching effects differed among plantings (evidence of different mutant effects). Thus, we explain our patterns of conditional fitness advantage in terms of the dramatically different positions in phenotypic and fitness space that the mutants occupied upon exposure to new environments, as well as of those environments’ different selection regimes. This finding points to the importance of empirical tests of fitness in the field. While phenotypic effects may be predictable under limited laboratory conditions, the ramification of genetic perturbation on fitness in nature, mediated by environmentally influenced phenotypes, is highly environmentally contingent.
Mutation Effects on Traits.
Natural environments are far more complex than the controlled laboratory conditions in which most mutations are assessed so we might expect the unexpected when observing mutants in the wild. For example, for A. thaliana flowering time, recent empirical work showed that realistic daily amplitudes in temperature considerably decreased the delay caused by introgressing a strong FRI allele to a nonfunctional background compared to the extreme delay that occurred in constant laboratory temperatures (112). Many of our experimental mutants altered flowering in the direction we expected, but others showed complex and unexpected patterns and were likely responding to multivariate cues in their natural environments that have not yet been tested for these genes. For example, the autonomous pathway mutants lumindependens (ld) and fve were constitutively late-bolting (Figs. 1 and 2). LD and FVE down-regulate the floral repressor FLC in an environmentally insensitive manner (113–115) so we expected mutations in those genes to delay flowering in all sites. However, HUA2 is also an activator of FLC; the hua2 mutant in the low FLC ecotype background Col caused acceleration but, in the high FLC Col FRI background, switched between slight delays in Cologne, Halle, and Valencia falls and acceleration in the other plantings. We speculate that this unexpected sign plasticity may result from either 1) the ecotype’s unmutagenized background allele being responsive to an uncharacterized or multiple environmental inputs or 2) the ecotype background allele epistatically regulates an environmentally responsive module of the flowering time network. The former possibility is especially relevant for complex environments since flowering time genes have been shown to respond to a diverse set of environmental signals. For example, CO has classically been considered a photoperiod pathway gene whose primary role for flowering time is the perception of long days (114, 116, 117). However, recent work has shown that, in addition to this role, CO also integrates ambient temperature information to promote flowering (107, 118–120). The co mutant in the Ler background was delayed in the long-day plantings of summer and spring, which was likely influenced by a combination of the diminished perception of inductive photoperiods and high ambient temperatures. Remarkably, co in the Col background reversed this trend by accelerating bolting in most plantings, a dramatic demonstration of epistasis with genetic background. Thus, the effect of a mutation might change among environments as the relative importance of its roles as a photoperiod- and temperature-sensor also changes.
Fitness Landscapes.
Environmental variation in the phenotypic effects of mutations does not completely explain why the relative fitness among mutants varies across environments. Variation in the strength and direction of multivariate selection was also critically important. This is exemplified by the autonomous mutant ld, which was consistently late-bolting but switched between relative fitness increases and decreases among environments (SI Appendix, Fig. S7). Thus, the topography of the selection surface shifted so that, although broad phenotypic trends remained constant, relative fitness among mutants changed. Fitness is the most composite phenotype expressed by an organism, integrating all preceding traits and developmental contingencies (111, 121). Thus, it is expected that growing conditions could perturb critical junctures during a plant’s developmental program, resulting in different fitness outcomes.
Although these mutants were chosen to expand phenological variation, they also pleiotropically expanded morphological variation in branching. Branching proved to be under stronger selection than phenology (Table 3) although both appeared to be under selection when analyzed in a univariate framework (Table 2). Predictions of responses to selection often center on highly heritable traits like phenology; however, in this experiment, phenology was under weaker selection than branching, which was much less heritable. Since a trait’s response to selection is a function of both its heritability and the strength of selection, phenology may show a marked response to selection due to its greater genetic basis, but this may represent an indirect evolutionary response to stronger selection on branching. Together, this points to the importance of considering the pleiotropy of mutations’ phenotypic effects since they may affect suites of traits with different selective potentials.
Gene Regulatory Networks.
Flowering time genes have been conceptually organized into distinct pathways such as the photoperiod and vernalization pathways (51, 122, 123). This conceptual organization is a consequence of the cue-specific manner in which the functional effects of flowering time genes are initially characterized (124, 125). However, as the interactome has been elucidated by protein–protein and genetic interaction screens, the complexity of gene regulatory networks has grown (126, 127). Indeed, recent work has shown that genes often act outside of their canonical pathways to respond to multiple environmental variables (51). For example, several “photoperiod genes” have also been shown to act as ambient temperature thermosensors (106, 119, 128). Here, we challenged mutants with natural environments that integrated multiple environmental influences. When examining phenotype deviations due to mutations, we found evidence that, indeed, more highly connected genes caused greater changes in phenotypes and greater reductions in fitness, indicating that their connections within the regulatory network were nonredundant (Fig. 3). However, this relationship was not significant for all environments or traits. This indicates that some environments exposed a gene’s connectedness more than others, possibly due to responses to different multivariate environmental inputs.
Conditionally Adaptive Mutations in Genotype–Phenotype–Fitness Landscapes.
Our results point to genotype × environment interaction as an important explanation for recent findings of nonnegligible frequencies of large-effect mutations segregating in natural populations of many species (14–16, 92–100). Contrary to the predictions of mutation-selection balance, which emphasizes inefficient negative selection to explain why mutations linger in a population (129, 130), we found evidence for balancing selection by environmentally dependent fitness. In several cases, the same flowering time pathway mutation had opposite effects on fitness in different environments, and environmentally dependent positive or negative selection acted upon conditionally adaptive traits that ramified from these specific mutations. As these targeted flowering time mutations were grown in different environments, some formed a path to high fitness through phenology, some through branching, and some through a combination of both. Phenological deviations in mutants were more predictable than branching or fitness but still differed among environments. The branching path was under stronger selection, but the phenological path had a stronger genetic basis. Thus, adaptation to natural selection on large-effect mutations is a contingent process that must be measured across multiple settings to gauge the pleiotropic adaptive value of underlying mutations.
Methods
Plant Materials.
In 2006 and 2007, the mutants were planted as part of a larger field experiment in common gardens in 5 locations (Norwich, United Kingdom; Halle, Germany; Cologne, Germany; Valencia, Spain; and Oulu, Finland) and several seasons (summer, spring, and fall) (55, 102, 103, 106, 131). Mutant genes, their primary flowering time pathway designation, and relevant environmental or genetic interactions are summarized in Table 1. All mutants were nontransgenic, induced by radiation or chemical mutagenesis, identified by screens for major effects on flowering tie and experimentally confirmed (SI Appendix, Table S1). The single exception was the introgression of a functional FRI allele from the Sf-2 ecotype to complement loss-of-function alleles of this gene in Col-0; this allowed tests of the effect of induced mutations in backgrounds with a functional vernalization pathway (101). Seeds were bulked and stratified under common conditions (103). Germination occurred under natural light conditions in unheated greenhouses at each field site, and seedlings were transplanted to the field synchronously with germination flushes observed in natural A. thaliana populations near each field site. Between 15 and 20 replicates for each genotype were transplanted in a randomized block design. See SI Appendix, Supplemental Methods for further details.
Phenotypic and Fitness Measurements.
We report 12 morphometric, phenological, and fitness traits (SI Appendix, Table S1). Together with branching’s partial independence as a developmental module and its mechanistic interactions with flowering time and fitness (see Introduction), we focused on branching as a morphological trait; bolting time as a phenological trait; and fecundity as a fitness metric. Days to bolting was the number of days from transplant into the field until an inflorescence shoot was visible at the center of a rosette. To convert these measurements to photothermal time, we scaled by temperature greater than 3 °C during daylight, resulting in photothermal units to bolting (BPTUs) as in Wilczek et al. (103). Total branch number was the total number of primary, secondary, and cauline branches longer than 1 cm. We estimated fitness as the number of seeds produced by a plant in seed proxy units. These were calculated as a plant’s silique number multiplied by an average-length silique’s distance from its valve’s base to apex. Seed set is a direct measurement of fitness, and, since seeds are minute and numerous in each silique, silique length is a practical and reliable proxy for the number of seeds (132, 133). Fitness was scored as 0 for plants that died before producing siliques.
Statistical Methods.
All analyses were performed in R version 3.0.2 (134). To estimate the ratio of genetic variation to phenotypic variation for a trait, we used ANOVA procedures to calculate broad-sense heritability. In order to identify phenotype classes responding to genetic perturbation and to growth environment, we hierarchically clustered phenotypes by genotype and planting, using average, Euclidian-based clustering. To scale phenotypes for visualization in heat maps, we relativized mutant least-square means against within-planting least-square means of 1) Col-0 if the mutant was generated in a Col-0 background; 2) Ler-1 if in a Ler-1 background; or 3) Col-FRI if the mutation was in the Col-0 background with an introgressed functional Sf-2 FRI allele. Relativized traits were log2 transformed for decrease–increase symmetry, centering within plantings. The identified clusters were robust to clustering method. For phenological traits, calendar and photothermal time clusters were similar (SI Appendix, Figs. S2–S4).
To construct fitness landscapes for the 5 plantings for which fitness and growth metrics were taken, we first fit a generalized additive model (GAM) under a Gaussian distribution for each planting. To test for correlated selection on these traits, we included the interaction term BPTU × total branches. Since we wished to simultaneously estimate the magnitude, direction, and curvature of selection gradients, as well as to perform hypothesis testing on whether this selection was significant, we assessed the significance of GAM coefficient estimates with case bootstrapping procedures (32, 33, 135). This analysis combined the biological interpretability of traditional regression techniques with splines’ ability to explain complex surfaces, producing GAM coefficients that are analogous to partial derivatives of the directional selection coefficient (β) and stabilizing selection coefficient (γ) (136). P values were adjusted for multiple testing by Bonferroni corrections.
In order to quantify the network position of the mutant genes, we manually curated nonredundant genetic and physical interactions deposited in the gene network databases BioGRID 3.5.167 (137) and IntAct (European Molecular Biology Laboratory–European Bioinformatics Institute) (138). We counted each mutant gene’s experimentally validated physical or genetic interaction as a connection, and the sum of these connections as its connectedness. We considered these interactions to be edges in our network analysis, and the mutant genes as nodes. In this analysis, we regressed connectedness against relative phenotype deviation from ecotype background, reporting Bonferroni-corrected results.
Supplementary Material
Acknowledgments
We thank M. Blazquez, G. Coupland, C. Dean, M. Hoffman, M. Koornneef, and O. Savolainen for hosting field plantings, as well as L. Albertson, J. F. Egan, L. Martin, C. Muir, R. Petipas, S. Sim, A. Walker, J. Anderson, D. Eaton, B. Moyers, R. Schaeffer, and C. Lopez-Gallego for collecting field data. We also thank B. Robertson, M. Gosling, T. Kauppila, and H. Eissner for assistance in establishing field sites, and E. Albright, Y. Cheng, C. Cooper, M. F. Cooper, M. Dinneen, E. Hsieh, B. Laginhas, W. Jackson, C. Lung, S. Makava, W. Miller, C. M. Sales, L. Sigalla, S. Stein, J. Wade, A. Wilde, A. Williard, and H. Williard for counting siliques for fitness estimates. Jeffrey Ross-Ibarra provided helpful comments on the manuscript. This work was supported by National Science Foundation Grants EF-0425759, DEB-1754102, and GRFP-2014157993.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902731116/-/DCSupplemental.
References
- 1.Fisher R., The Genetical Theory of Natural Selection (Clarendon Press, Oxford, 1930). [Google Scholar]
- 2.Lynch M., et al. , Perspective: Spontaneous deleterious mutation. Evolution 53, 645–663 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Eyre-Walker A., Keightley P. D., The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8, 610–618 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Hall D. W., Mahmoudizad R., Hurd A. W., Joseph S. B., Spontaneous mutations in diploid Saccharomyces cerevisiae: Another thousand cell generations. Genet. Res. 90, 229–241 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Rutter M. T., et al. , Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution 66, 2335–2339 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Rutter M. T., Shaw F. H., Fenster C. B., Spontaneous mutation parameters for Arabidopsis thaliana measured in the wild. Evolution 64, 1825–1835 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Shaw F. H., Geyer C. J., Shaw R. G., A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution 56, 453–463 (2002). [DOI] [PubMed] [Google Scholar]
- 8.García-Dorado A., Monedero J. L., López-Fanjul C., The mutation rate and the distribution of mutational effects of viability and fitness in Drosophila melanogaster. Genetica 102, 255–265 (1998). [PubMed] [Google Scholar]
- 9.Perfeito L., Fernandes L., Mota C., Gordo I., Adaptive mutations in bacteria: High rate and small effects. Science 317, 813–815 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Kassen R., Bataillon T., Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38, 484–488 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Sharp N. P., Agrawal A. F., An experimental test of the mutation-selection balance model for the maintenance of genetic variance in fitness components. Proc. Biol. Sci. 285, 20181864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houle D., Comparing evolvability and variability of quantitative traits. Genetics 130, 195–204 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlesworth B., Causes of natural variation in fitness: Evidence from studies of Drosophila populations. Proc. Natl. Acad. Sci. U.S.A. 112, 1662–1669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Exposito-Alonso M., et al. , The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 14, e1007155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toomajian C., et al. , A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4, e137 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johanson U., et al. , Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Roles A. J., Conner J. K., Fitness effects of mutation accumulation in a natural outbred population of wild radish (Raphanus raphanistrum): Comparison of field and greenhouse environments. Evolution 62, 1066–1075 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Roles A. J., Rutter M. T., Dworkin I., Fenster C. B., Conner J. K., Field measurements of genotype by environment interaction for fitness caused by spontaneous mutations in Arabidopsis thaliana. Evolution 70, 1039–1050 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Halligan D. L., Keightley P. D., Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40, 151–172 (2009). [Google Scholar]
- 20.Wright S., “The roles of mutation, inbreeding, crossbreeding and selection in evolution” in Proceedings of the Sixth International Congress on Genetics, Jones D. F., Ed. (Brooklyn Botanic Garden, Brooklyn, NY, 1932), vol. 1, pp. 356–366. [Google Scholar]
- 21.Gavrilets S., Fitness Landscapes and the Origin of Species (Princeton University Press, Princeton, NJ, 2004). [Google Scholar]
- 22.Orr H. A., Fitness and its role in evolutionary genetics. Nat. Rev. Genet. 10, 531–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schluter D., The Ecology of Adaptive Radiation (Oxford University Press, Oxford, 2000). [Google Scholar]
- 24.Siepielski A. M., et al. , The spatial patterns of directional phenotypic selection. Ecol. Lett. 16, 1382–1392 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Arnold S. J., Performance surfaces and adaptive landscapes. Integr. Comp. Biol. 43, 367–375 (2003). [DOI] [PubMed] [Google Scholar]
- 26.de Visser J. A. G. M., Krug J., Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15, 480–490 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Simpson G. G., Tempo and Mode in Evolution (Columbia Univeristy Press, New York, NY, 1944). [Google Scholar]
- 28.Brodie E. D. 3rd, Moore A. J., Janzen F. J., Visualizing and quantifying natural selection. Trends Ecol. Evol. 10, 313–318 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Kingsolver J. G., et al. , The strength of phenotypic selection in natural populations. Am. Nat. 157, 245–261 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski J., Plotkin J. B., Inferring fitness landscapes by regression produces biased estimates of epistasis. Proc. Natl. Acad. Sci. U.S.A. 111, E2301–E2309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw R. G., Geyer C. J., Inferring fitness landscapes. Evolution 64, 2510–2520 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Lande R., Arnold S. J., The measurement of selection on correlated characters. Evolution 37, 1210–1226 (1983). [DOI] [PubMed] [Google Scholar]
- 33.Morrissey M. B., Sakrejda K., Unification of regression-based methods for the analysis of natural selection. Evolution 67, 2094–2100 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Weinig C., Stinchcombe J. R., Schmitt J., QTL architecture of resistance and tolerance traits in Arabidopsis thaliana in natural environments. Mol. Ecol. 12, 1153–1163 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Erickson D. L., Fenster C. B., Stenøien H. K., Price D., Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 13, 2505–2522 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Stinchcombe J. R., Weinig C., Heath K. D., Brock M. T., Schmitt J., Polymorphic genes of major effect: Consequences for variation, selection and evolution in Arabidopsis thaliana. Genetics 182, 911–922 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilke C. O., Adami C., Interaction between directional epistasis and average mutational effects. Proc. Biol. Sci. 268, 1469–1474 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinkley T., et al. , A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nat. Genet. 43, 487–489 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Hall B. G., Predicting evolution by in vitro evolution requires determining evolutionary pathways. Antimicrob. Agents Chemother. 46, 3035–3038 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitt J. N., Ferré-D’Amaré A. R., Rapid construction of empirical RNA fitness landscapes. Science 330, 376–379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouyos R. D., et al. , Exploring the complexity of the HIV-1 fitness landscape. PLoS Genet. 8, e1002551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiménez J. I., Xulvi-Brunet R., Campbell G. W., Turk-MacLeod R., Chen I. A., Comprehensive experimental fitness landscape and evolutionary network for small RNA. Proc. Natl. Acad. Sci. U.S.A. 110, 14984–14989 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnikov A., Rogov P., Wang L., Gnirke A., Mikkelsen T. S., Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 42, e112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C., Qian W., Maclean C. J., Zhang J., The fitness landscape of a tRNA gene. Science 352, 837–840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski J., Nemenman I., Genotype to phenotype mapping and the fitness landscape of the E. coli lac promoter. PLoS One 8, e61570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu N. C., Dai L., Olson C. A., Lloyd-Smith J. O., Sun R., Adaptation in protein fitness landscapes is facilitated by indirect paths. eLife 5, e16965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X., Liu L., EVOLUTION. Toward a prospective molecular evolution. Science 352, 769–770 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Puchta O., et al. , Network of epistatic interactions within a yeast snoRNA. Science 352, 840–844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hietpas R., Roscoe B., Jiang L., Bolon D. N. A., Fitness analyses of all possible point mutations for regions of genes in yeast. Nat. Protoc. 7, 1382–1396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salverda M. L. M., Koomen J., Koopmanschap B., Zwart M. P., de Visser J. A. G. M., Adaptive benefits from small mutation supplies in an antibiotic resistance enzyme. Proc. Natl. Acad. Sci. U.S.A. 114, 12773–12778 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouché F., Lobet G., Tocquin P., Périlleux C., FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44, D1167–D1171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bentsink L., et al. , Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 4264–4269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atwell S., et al. , Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergelson J., Roux F., Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat. Rev. Genet. 11, 867–879 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Fournier-Level A., et al. , Paths to selection on life history loci in different natural environments across the native range of Arabidopsis thaliana. Mol. Ecol. 22, 3552–3566 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Korves T. M., et al. , Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am. Nat. 169, E141–E157 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Donohue K., et al. , The evolutionary ecology of seed germination of Arabidopsis thaliana: Variable natural selection on germination timing. Evolution 59, 758–770 (2005). [PubMed] [Google Scholar]
- 58.Wilczek A. M., Cooper M. D., Korves T. M., Schmitt J., Lagging adaptation to warming climate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 111, 7906–7913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Exposito-Alonso M., Brennan A. C., Alonso-Blanco C., Picó F. X., Spatio-temporal variation in fitness responses to contrasting environments in Arabidopsis thaliana. Evolution 72, 1570–1586 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Méndez-Vigo B., Gomaa N. H., Alonso-Blanco C., Picó F. X., Among- and within-population variation in flowering time of Iberian Arabidopsis thaliana estimated in field and glasshouse conditions. New Phytol. 197, 1332–1343 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Frachon L., et al. , Intermediate degrees of synergistic pleiotropy drive adaptive evolution in ecological time. Nat. Ecol. Evol. 1, 1551–1561 (2017). Erratum in: Nat. Ecol. Evol.2, 194 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Lempe J., et al. , Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1, 109–118 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brachi B., et al. , Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6, e1000940 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brachi B., et al. , Investigation of the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Mol. Ecol. 22, 4222–4240 (2013). [DOI] [PubMed] [Google Scholar]
- 65.The 1001 Genomes Consortium , 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stinchcombe J. R., et al. , A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. U.S.A. 101, 4712–4717 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Debieu M., et al. , Co-variation between seed dormancy, growth rate and flowering time changes with latitude in Arabidopsis thaliana. PLoS One 8, e61075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samis K. E., et al. , Longitudinal trends in climate drive flowering time clines in North American Arabidopsis thaliana. Ecol. Evol. 2, 1162–1180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vidigal D. S., et al. , Altitudinal and climatic associations of seed dormancy and flowering traits evidence adaptation of annual life cycle timing in Arabidopsis thaliana. Plant Cell Environ. 39, 1737–1748 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Ratcliffe O. J., Bradley D. J., Coen E. S., Separation of shoot and floral identity in Arabidopsis. Development 126, 1109–1120 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Hempel F. D., et al. , Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124, 3845–3853 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Grbić V., Bleecker A. B., Axillary meristem development in Arabidopsis thaliana. Plant J. 21, 215–223 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Beveridge C. A., Weller J. L., Singer S. R., Hofer J. M. I., Axillary meristem development. Budding relationships between networks controlling flowering, branching, and photoperiod responsiveness. Plant Physiol. 131, 927–934 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Umehara M., et al. , Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Bowman J. L., Alvarez J., Weigel D., Meyerowitz E. M., Smyth D. R., Control of flower development in Arabidopsis thaliana by Apetala1 and interacting genes. Development 119, 721–743 (1993). [Google Scholar]
- 76.Yamaguchi N., et al. , Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344, 638–641 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Porri A., Torti S., Romera-Branchat M., Coupland G., Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139, 2198–2209 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Arana M. V., Marín-de la Rosa N., Maloof J. N., Blázquez M. A., Alabadí D., Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 9292–9297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai M. Y., et al. , Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lempe J., Lachowiec J., Sullivan A. M., Queitsch C., Molecular mechanisms of robustness in plants. Curr. Opin. Plant Biol. 16, 62–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu J., et al. , System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat. Genet. 41, 166–167 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Mestek Boukhibar L., Barkoulas M., The developmental genetics of biological robustness. Ann. Bot. 117, 699–707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lachowiec J., Queitsch C., Kliebenstein D. J., Molecular mechanisms governing differential robustness of development and environmental responses in plants. Ann. Bot. 117, 795–809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levy S. F., Siegal M. L., Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 6, e264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanada K., et al. , Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis. Genome Biol. Evol. 1, 409–414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bauer C. R., Li S., Siegal M. L., Essential gene disruptions reveal complex relationships between phenotypic robustness, pleiotropy, and fitness. Mol. Syst. Biol. 11, 773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waddington C. H., Canalization of development and the inheritance of acquired characters. Nature 150, 563–565 (1942). [Google Scholar]
- 88.Abley K., Locke J. C. W., Leyser H. M. O., Developmental mechanisms underlying variable, invariant and plastic phenotypes. Ann. Bot. 117, 733–748 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Waddington C. H., Genetic assimilation. Adv. Genet. 10, 257–293 (1961). [DOI] [PubMed] [Google Scholar]
- 90.Masel J., Siegal M. L., Robustness: Mechanisms and consequences. Trends Genet. 25, 395–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Elena S. F., Cooper V. S., Lenski R. E., Punctuated evolution caused by selection of rare beneficial mutations. Science 272, 1802–1804 (1996). [DOI] [PubMed] [Google Scholar]
- 92.Lloyd J., Meinke D., A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol. 158, 1115–1129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meinke D. W., A survey of dominant mutations in Arabidopsis thaliana. Trends Plant Sci. 18, 84–91 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Barboza L., et al. , Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proc. Natl. Acad. Sci. U.S.A. 110, 15818–15823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mouchel C. F., Briggs G. C., Hardtke C. S., Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 18, 700–714 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grant M. R., et al. , Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 15843–15848 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiang Y., et al. , Sequence polymorphisms at the REDUCED DORMANCY5 pseudophosphatase underlie natural variation in Arabidopsis dormancy. Plant Physiol. 171, 2659–2670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Monroe J. G., et al. , Drought adaptation in Arabidopsis thaliana by extensive genetic loss-of-function. eLife 7, e41038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Le Corre V., Variation at two flowering time genes within and among populations of Arabidopsis thaliana: Comparison with markers and traits. Mol. Ecol. 14, 4181–4192 (2005). [DOI] [PubMed] [Google Scholar]
- 100.Xu Y.-C., et al. , Adaptation and phenotypic diversification in Arabidopsis through loss-of-function mutations in protein-coding genes. Plant Cell 31, 1012–1025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee I., Amasino R. M., Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fournier-Level A., et al. , A map of local adaptation in Arabidopsis thaliana. Science 334, 86–89 (2011). [DOI] [PubMed] [Google Scholar]
- 103.Wilczek A. M., et al. , Effects of genetic perturbation on seasonal life history plasticity. Science 323, 930–934 (2009). [DOI] [PubMed] [Google Scholar]
- 104.Silverstone A. L., et al. , Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143, 987–1000 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Conti L., Hormonal control of the floral transition: Can one catch them all? Dev. Biol. 430, 288–301 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Chew Y. H., et al. , An augmented Arabidopsis phenology model reveals seasonal temperature control of flowering time. New Phytol. 194, 654–665 (2012). [DOI] [PubMed] [Google Scholar]
- 107.Legris M., et al. , Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Qiu Y., Li M., Kim R. J. A., Moore C. M., Chen M., Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nat. Commun. 10, 140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shpak E. D., Berthiaume C. T., Hill E. J., Torii K. U., Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131, 1491–1501 (2004). [DOI] [PubMed] [Google Scholar]
- 110.Johnson T., Barton N., Theoretical models of selection and mutation on quantitative traits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1411–1425 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Merilä J., Sheldon B. C., Genetic architecture of fitness and nonfitness traits: Empirical patterns and development of ideas. Heredity 83, 103–109 (1999). [DOI] [PubMed] [Google Scholar]
- 112.Burghardt L. T., et al. , Fluctuating, warm temperatures decrease the effect of a key floral repressor on flowering time in Arabidopsis thaliana. New Phytol. 210, 564–576 (2016). [DOI] [PubMed] [Google Scholar]
- 113.Simpson G. G., The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7, 570–574 (2004). [DOI] [PubMed] [Google Scholar]
- 114.Simpson G. G., Dean C., Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289 (2002). [DOI] [PubMed] [Google Scholar]
- 115.Pazhouhandeh M., Molinier J., Berr A., Genschik P., MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 3430–3435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Franklin K. A., Quail P. H., Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Halliday K. J., Koornneef M., Whitelam G. C., Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol. 104, 1311–1315 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Halliday K. J., Salter M. G., Thingnaes E., Whitelam G. C., Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885 (2003). [DOI] [PubMed] [Google Scholar]
- 119.Fernández V., Takahashi Y., Le Gourrierec J., Coupland G., Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 86, 426–440 (2016). [DOI] [PubMed] [Google Scholar]
- 120.Kinmonth-Schultz H. A., et al. , Cool night-time temperatures induce the expression of CONSTANS and FLOWERING LOCUS T to regulate flowering in Arabidopsis. New Phytol. 211, 208–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Laughlin D. C., Messier J., Fitness of multidimensional phenotypes in dynamic adaptive landscapes. Trends Ecol. Evol. 30, 487–496 (2015). [DOI] [PubMed] [Google Scholar]
- 122.Pajoro A., et al. , The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: A two-decade history. J. Exp. Bot. 65, 4731–4745 (2014). [DOI] [PubMed] [Google Scholar]
- 123.Fornara F., de Montaigu A., Coupland G., SnapShot: Control of flowering in Arabidopsis. Cell 141, 550, 550.e1–550e2 (2010). [DOI] [PubMed] [Google Scholar]
- 124.Romera-Branchat M., Andrés F., Coupland G., Flowering responses to seasonal cues: What’s new? Curr. Opin. Plant Biol. 21, 120–127 (2014). [DOI] [PubMed] [Google Scholar]
- 125.Andrés F., Coupland G., The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 (2012). [DOI] [PubMed] [Google Scholar]
- 126.Fraser D. P., Hayes S., Franklin K. A., Photoreceptor crosstalk in shade avoidance. Curr. Opin. Plant Biol. 33, 1–7 (2016). [DOI] [PubMed] [Google Scholar]
- 127.Jung J. H., Ju Y., Seo P. J., Lee J. H., Park C. M., The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69, 577–588 (2012). [DOI] [PubMed] [Google Scholar]
- 128.Jung J. H., et al. , Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016). [DOI] [PubMed] [Google Scholar]
- 129.Zhang X. S., Hill W. G., Genetic variability under mutation selection balance. Trends Ecol. Evol. 20, 468–470 (2005). [DOI] [PubMed] [Google Scholar]
- 130.Nielsen R., Slatkin M., An Introduction to Population Genetics: Theory and Applications (Sinauer Associates, Oxford University Press, Sunderland, MA, 2013). [Google Scholar]
- 131.Wilczek A. M., et al. , Genetic and physiological bases for phenological responses to current and predicted climates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 3129–3147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Horton M. W., et al. , Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44, 212–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Alonso-Blanco C., Blankestijn-de Vries H., Hanhart C. J., Koornneef M., Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96, 4710–4717 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.R Core Development Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2015). [Google Scholar]
- 135.Diederich A., Generalized additive models. An introduction with R. J. Math. Psychol. 51, 339 (2007). [Google Scholar]
- 136.Lynch M., Walsh B., Genetics and Analysis of Quantitative Traits (Sinauer Associates, Inc., Sunderland, MA, 1998). [Google Scholar]
- 137.Chatr-Aryamontri A., et al. , The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Orchard S., et al. , The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.