Abstract
Encapsulation of proteins within lipid inverse bicontinuous cubic phases (Q2) has been widely studied for many applications, such as protein crystallization or drug delivery of proteins for food and pharmaceutical purposes. However, the use of the lipid sponge (L3) phase for encapsulation of proteins has not yet been well explored. Here, we have employed a lipid system that forms highly swollen sponge phases to entrap aspartic protease (34 kDa), an enzyme used for food processing, e.g., to control the cheese-ripening process. Small-angle x-ray scattering showed that although the L3 phase was maintained at low enzyme concentrations (≤15 mg/mL), higher concentration induces a transition to more curved structures, i.e., transition from L3 to inverse bicontinuous cubic (Q2) phase. The Raman spectroscopy data showed minor conformational changes assigned to the lipid molecules that confirm the lipid-protein interactions. However, the peaks assigned to the protein showed that the structure was not significantly affected. This was consistent with the higher activity presented by the encapsulated aspartic protease compared to the free enzyme stored at the same temperature. Finally, the encapsulation efficiency of aspartic protease in lipid sponge-like nanoparticles was 81% as examined by size-exclusion chromatography. Based on these results, we discuss the large potential of lipid sponge phases as carriers for proteins.
Significance
We show for the first time, to our knowledge, the use of highly swollen lipid sponge (L3) phase for highly efficient (>80%) encapsulation of an active enzyme, aspartic protease, clearly their potential as enzyme carriers. Small-angle x-ray scattering showed that although the L3 phase was maintained at low enzyme concentrations, higher concentration induces a transition to more curved structures (i.e., transition from L3 to inverse bicontinuous cubic (Q2) phase). The Raman spectroscopy data showed minor conformational changes assigned to the lipid molecules that confirm the lipid-protein interactions. The encapsulated aspartic protease was found to maintain its high activity over a longer period of time compared to the free enzyme stored at the same temperature.
Introduction
Lipid inverse bicontinuous cubic phases (Q2) have been extensively investigated for protein crystallization (1, 2, 3), as matrices for encapsulation and drug delivery of peptides or proteins (4, 5, 6, 7, 8, 9), and for biosensing (10, 11). Protein encapsulation is required to improve/maintain the protein stability and thus, keep their function during storage for longer time. In addition, it also allows better control of the function and delivery of the protein. In general, lipid liquid crystals (LLCs) are able to entrap both hydrophilic and hydrophobic biomolecules because of the amphiphilic character of lipids. Inverse bicontinuous phases present higher encapsulation efficiency compared to other LLCs (9) because of their large water pores and their large surface/volume ratio.
Sun et al. reported that both the topology of the mesophase and the water channel dimensions were affecting the activity of the enzyme horseradish peroxidase (∼6 nm) (12, 13). They found that the enzymatic activity was limited when the protein size was larger than the pore size but increased when expanding the water channels (12). In another study, they compared three different LLCs with the same pore size and conclude that the Q2 phase with space group Pn3m (Q224) showed improved horseradish peroxidase activity compared to the cubic phase with Ia3d spacing group (Q230), followed by the inverse hexagonal phase (13). These results suggest that the Q224 is more favorable for entrapment of proteins without disrupting the function but also that the water pores should preferably be larger than the protein size.
Because large pore size is highly relevant for protein encapsulation, swollen bicontinuous phases are needed. Recent studies by Tyler et al. (14) and Zabara et al. (15) demonstrated that inverse bicontinuous cubic phases with large water pore diameters can be achieved (24.9 and 22.6 nm, respectively). In both studies, the lipid formulation was prepared using organic solvents. As we aim for applications within the food and pharmaceutical areas, it is desirable to avoid the use of organic solvents. There is also a larger risk for conformational changes and loss of enzyme activity if organic solvents are used in the formulation process. Another type of inverse bicontinuous phase that can be used for this purpose is the lipid sponge phase, L3. The L3 phase is a disordered Q2 phase that possesses a more flexible structure, and thus, as demonstrated in this study, it can accommodate larger biomolecules as well as larger concentrations of them.
We have previously reported (16) on a lipid system that can form sponge phases with water channels up to 13 nm of diameter at 25°C and without the use of organic solvents. These findings imply that these phases could entrap biomolecules up to 13 nm or even larger if they penetrate into the lipid bilayer. This lipid system was composed of diglycerolmonooleate (DGMO); a mixture of mono-, di- and triglycerides called Capmul GMO-50 (GMO-50) in which the main component is glycerol monoolein; and polysorbate 80 (P80). These lipid sponge phases (and other LLCs) can easily be fragmented in excess water to form nanoparticles with sponge phase structure with the same aqueous pore dimensions. The dispersions of sponge-like nanoparticles (L3-NPs) are more fluid than the corresponding viscous bulk phases. This makes them more suitable for different applications that require continuous processing, like dispensing or injection. Here, we report a study of the inclusion of aspartic protease (EC 3.4.23.22) within this lipid sponge phase matrix, both as a bulk phase and as dispersions in excess water. Importantly, the L3-NPs used here were prepared without the use of high-energy processes (e.g., sonication) that might disrupt the protein structure.
Aspartic protease (Asp) is a monomeric enzyme of 34 kDa (unit cell dimensions of 4.27 × 7.46 × 4.25 nm (17)) that catalyzes the hydrolysis of peptide bonds. It is used in cheese production because it acts as a milk clotting enzyme and is also an important component of the cheese-ripening process. The encapsulation of this protein has been investigated to control enzyme activity to decrease the ripening time and thus reduce cheese production costs. Aspartic protease encapsulation could provide good distribution of the enzyme in the milk and prevent interaction with milk caseins before the curd is formed. It has been shown that free aspartic protease solutions added in the milk before the curd is formed result in weak curd formation, i.e., low cheese yields. Addition of aspartic protease after the curd is formed lead to an inhomogeneous distribution of the enzyme within the cheese product (18). Encapsulation of microbial protease in liposomes was previously explored by Kirby et al. (19), who demonstrated an encapsulation efficiency of 40%. Moreover, they showed that liposomes protected the enzyme from interacting with milk in early stages and that most of the protease was found in the curd, resulting in firmer cheeses.
In this work, we have investigated the used of lipid sponge (and cubic) phases as matrices for aspartic protease encapsulation to improve the enzyme storage stability. It has been shown that these bicontinuous type of phases have higher encapsulation efficiency compared to the liposomes ((20, 21, 22) and references therein). For this purpose, our study focused on three aspects: 1) the changes in the lipid structure caused by aspartic protease, 2) the influence of the lipid matrix on the structure and function of aspartic protease, and 3) the ability of lipid-sponge-like NPs to entrap aspartic protease in terms of encapsulation efficiency.
Structural changes in the lipid sponge phase by addition of aspartic protease and glycerol were investigated by small-angle x-ray scattering (SAXS) and Raman spectroscopy. The influence of glycerol was important here because the enzyme used for food processing is initially preserved in 50% glycerol solution. Raman spectroscopy gave insight into structural alterations of the protein as well as into the lipid interactions once the protein is included in the lipid matrix. Finally, the function of encapsulated aspartic protease was analyzed by using the milk clotting activity test and the encapsulation efficiency by separation of the free enzyme from the encapsulated enzyme using size-exclusion chromatography (SEC).
Materials and Methods
Materials
A mixture of glycerides denoted as Capmul GMO-50 (lot no. 100616-8) was provided by Abitec (Janesville, WI). Its composition was 54.7% monoglycerides, 15–35% diglycerides and 2–10% triglycerides with the following fatty acid composition: 84.6% oleic (C18:1), 6.8% linoleic, 0.8% linolenic, and 6.2% saturated acids. DGMO with 88% of diglycerol monoester and 4.9% free glycerol and polyglycerols was received from Danisco A/S (Brabrand, Denmark). The main fatty acid component was oleic acid, constituting 90.7%, followed by linoleic (4.2%), saturated (2.9%), eicosenoic (1.2%), and linolenic (0.8%). Polyoxyethylene (20) sorbitan monooleate (P80) was supplied by Croda (Chocques, France). Milli-Q purified water (H2O, 18 MΩ · cm) was used for all experiments and heavy water (D2O; Sigma-Aldrich, St. Louis, MO) was employed as well for Raman measurements. The aspartic protease solution (Thermolase 625; https://www.chr-hansen.com/en/food-cultures-and-enzymes/cheese/cards/product-cards/thermolase) was kindly provided by Chr. Hansen A/S, Hoersholm, Denmark. All the other chemicals were of analytical grade and were used as received.
Sample preparation
Aspartic protease solution
Two different enzyme samples were used in this study: the original solution, Thermolase, which contains around 50% glycerol; and freeze-dried aspartic protease (Asp). The latter was obtained by dialyzing Thermolase against an aqueous buffer to remove the glycerol preservative as well as control the electrolytes present in the solution. Removal of glycerol was performed using a dialysis bag with MWCO 6–8 kDa (Spectra/Por1). The dry membrane was first soaked in milli-Q water and 10 mM EDTA at room temperature for 30 min to remove the preservative and heavy metals. The membrane was then rinsed thoroughly with milli-Q water. The dialysis bag (around 50 cm) was filled with 100–150 mL of Thermolase, and dialysis was carried out against 10 mM phosphate buffer at pH 7 during 4 days at 4°C, with two to three changes of outside buffer per day. During this process, the dialysis bag was placed in a cylinder of 2 L that was filled with buffer. The conductivity of the buffer after hours of dialysis was monitored until it was the same as the original buffer. The dialyzed solution was freeze-dried (DW6-85 Heto-Holten Lab Equipment, Allerّd, Denmark) to obtain a powder that was solubilized into water or heavy water before its use. Different concentrations of aspartic protease were prepared (10, 25, 50, 75, and 100 mg/mL). The concentration was checked by measuring the absorbance at 280 nm and/or using the bicinchoninic acid assay (23).
Lipid-enzyme/glycerol bulk phases
First, two lipid stock solutions were prepared by mixing DGMO, melted GMO-50, and P80 at appropriate amounts. The first one was prepared with a DGMO/GMO-50 ratio of 40:60, whereas the second one was 55:45. The lipid/P80 ratio was always kept at 70:30. The stock solutions were left in a roller mixer for 24 h to homogenize. Then, the lipid + P80 blend was mixed with different amounts of an aqueous solution (55–70 wt %). These samples were then mixed in a roller mixer for 24–72 h until they were completely homogeneous. High-viscosity samples were also centrifuged at 371 × g, after which the vials were turned upside down and then again centrifuged. The centrifugation process with changing orientation of vials every second time was then repeated several times to ensure proper mixing. This process was repeated every day until equilibration was reached and the samples appeared homogenous. Samples were left to equilibrate for at least 1 week before being analyzed.
The aqueous solution was composed by mixtures of water/heavy water with Thermolase solution, glycerol, or Asp. Samples with Thermolase were composed of different Thermolase solution/water ratios (80:20, 50:50, and 20:80) to obtain the desired protein concentrations, for which the final protein concentration varied from 3 to 15 mg/mL depending on aqueous phase content. For the “blank,” this aqueous solution was composed of the corresponding 50% glycerol solution/water ratios (80:20, 50:50, and 20:80). In the case of freeze-dried Asp, enzyme solutions at different protein concentrations (10, 25, 50, and 100 mg/mL) were employed, and only formulations at 40:60 wt % lipid + P80/aqueous solution were investigated. Because only samples with 60 wt % of aqueous phases were studied, the final Asp concentration was 6, 15, 30, and 60 mg/mL.
Nanoparticle dispersions
Lipid sponge NPs without enzyme were prepared as a control sample by adding drop by drop 4 wt % of the lipid stock solution to a vial with 96 wt % of water. In the case of nanoparticle dispersions with included aspartic protease, 10 wt % of lipid-Asp bulk phase (with 15 mg/mL Asp) was added to a vial with 90 wt % of water. The vials were sealed, vigorously shaken by hand, and mixed horizontally in an orbital shaker for 24 h at 300 rpm. The final lipid + P80 amount for both samples was equivalent (4 wt %), and the final aspartic protease concentration was ∼1.5 mg/mL.
SAXS
Samples with Thermolase and glycerol were analyzed at the I911-4 SAXS beamline at MAX-Lab (Lund University, Sweden). The scattered intensity was recorded at three temperatures (25, 35, and 45 ± 0.1°C) with a wavelength of 0.91 Å−1 on a Pilatus 1 M 165 mm CCD detector (Paul Scherrer Institute, Villigen, Switzerland). Samples were mounted between Kapton windows in a steel sample holder and were measured for 60 s at a sample-to-detector distance of 1913 mm. The two-dimensional (2D) SAXS patterns were analyzed using the Fit2D software. The detector position of the diffraction pattern from silver behenate was used to calibrate the sample-to-detector distance.
Sample with Asp were measured using the SAXSLab Ganesha 300XL instrument (SAXSLAB ApS, Skovlunde, Denmark). The instrument was equipped with a 2D 300 K Pilatus detector from Dectris (Baden, Switzerland) and a GeniX 3D x-ray source (Xenocs, Sassenage, France). The x-ray wavelength was 1.54 Å−1, and the data was collected in the q range of 0.12−7.6 nm−1. Samples were sealed at room temperature between two thin mica windows in a metallic block. The measuring time was 30 min while the samples were kept at 25 ± 0.2°C by means of water circulating through the sample holder from an external recirculating water bath. The 2D scattering pattern was radially averaged using SAXSGui software to obtain I(q).
Raman measurements
Vibrational spectroscopy measurements of aspartic protease encapsulated in a lipid matrix were carried out using a Fourier transform Raman (FT-Raman) spectrometer MultiRAM (Bruker Optik, Ettlingen, Germany) equipped with a liquid-nitrogen-cooled Ge diode detector. The excitation radiation of the Nd:YAG laser (1064 nm) was adjusted to 200 mW. A gold-plated 90° angle mirror objective (focal length 33 mm) was used for sample irradiation and collection of the light scattered by the sample. The diameter of the laser spot at the sample was 100 μm. Spectral resolution was set to 4 cm−1, and the aperture was 4 mm. All the spectra were collected by averaging 24,000 sample scans.
Frequencies and intensities of the bands were determined by fitting experimental contour with Gaussian-Lorentzian form components using GRAMS/AI 8.0 (Thermo Fisher Scientific, Waltham, MA) software.
Relative milk clotting activity test
Aspartic protease activity test was based on the relative milk clotting activity test (24). The procedure involves adding 200 μL of diluted rennet enzyme (either the reference or the sample) to a tube with 10 mL of reconstituted skim milk at pH 6.5 and 32°C that is continuously rotating at a specific angle in a water bath. The milk clotting time (T) is recorded as the time from the addition of the enzyme (either reference standard or sample) until the formation of small visible flakes appears in the milk. The total milk clotting activity (strength) can then be extracted, and it is expressed as international milk clotting units (IMCUs) per mL or mg:
| (1) |
where SStandard is the strength of the reference standards, T is the milk clotting time in seconds, and D is the dilution factor. Liquid calf rennet solution with initial activity of 100 IMCUs/mL was used as reference standard for these experiments. To obtain reliable results, the difference in milk clotting time between the reference standard and the sample should be below 50 s. The reference standard was diluted to 3 IMCUs/mL, and the unknown sample was diluted, taking into account the protein concentration and the initial concentration and strength of the Thermolase solution (∼625 IMCUs/mL). After each dilution step, samples were mixed with a vortex. The dilution media (84 mM acetate buffer at pH 5.5) was used for these dilutions, and the analysis was carried out in triplicate for each sample (24). The samples analyzed consisted of free enzyme solution kept at 4°C and room temperature, encapsulated Asp in bulk and in lipid nanoparticles (also kept at room temperature). The initial free enzyme solution concentration was the same for all the samples, i.e., had the sample Asp concentration. The activity test was performed over time (day 0, 3, and 6), where the starting point was 1 week after sample preparation because of equilibration of LLC phases. Several identical bulk phase samples with equivalent composition were prepared. Each sample was only used once for each time point when measuring the change in activity with time to avoid structural changes due to removal of sample for the activity test.
SEC
SEC was performed in an Äkta pure instrument (General Electric Healthcare Life Sciences, Chicago, IL) with an ultraviolet monitor that records the absorbance at 280 nm. The column Superose 6 increase (General Electric Healthcare), with a column volume of 24 mL, was employed for these experiments. A 100 mM phosphate buffer at pH 7 and with 100 mM NaCl, filtered through 0.2 μm pore size filter paper, was used as eluent. The flow rate of the system was set to 0.5 mL/min, and the sample volume was 100 μL.
The evaluation of the data was performed by estimating the area under the curve (AUC) of the main peaks as well as the peak position and shape. The lipid nanoparticle peak and the free Asp peak were considered separately. The AUC was estimated by first defining a baseline by selecting a few anchor points and then fitting a cubic function to them. This baseline was then subtracted from the chromatogram. In the case of encapsulated Asp, the unloaded L3-NPs curve was subtracted from the sample chromatogram before baseline correction to evaluate the protein peak (Fig. S1). This step was very important to accurately determine the area under the free enzyme peak, as the lipid nanoparticles left a higher background than the original. To find the AUC of the main Asp peak, the peaks observed between 16 and 24 mL were fitted with a bi-Gaussian function and then each of them were integrated to obtain the area (Fig. S2). For the nanoparticle’s peak, the AUC was then found by integrating the peak, which also included part of the tail (Fig. S1).
Dynamic light scattering (DLS) and cryogenic transmission electron microscopy (cryo-TEM) experimental methods can be found in the Supporting Materials and Methods.
Results and Discussion
Influence of aspartic protease on the lipid sponge phase structure
We have previously reported that the DGMO/GMO-50/P80/water system (16) could form sponge phases with large cell dimensions, which makes this system promising for encapsulation of proteins. The lipid sponge phase is a more flexible structure compared to other LLC phases and hence facilitates the accommodation of macromolecules within the bicontinuous structure. In this section, we discuss the results from the investigated inclusion of aspartic protease into this lipid sponge phase. We have first studied the influence of aspartic protease on the lipid sponge phase structure because changing the LLC phase might also alter the cell dimensions and thus the capacity for this system to entrap large biomolecules. Two aspartic protease solutions were here examined: Thermolase solution, which is an aspartic protease solution with 50% glycerol, and freeze-dried “pure” aspartic protease dispersed in water. Because the initial solution of Thermolase contains large amounts of glycerol, the effect of glycerol on the lipid structure was investigated as well.
Effect of Thermolase (and glycerol)
Lipid/P80 mixtures that included Thermolase (aspartic protease formulated in 50% glycerol) and glycerol were investigated to understand how these mixtures affect the lipid phase behavior. L3 phases with water pores of 9 nm in diameter are formed at 25°C in the neat lipid DGMO/GMO-50/P80/water system, at DGMO/GMO-50 of 40:60, when the lipid/P80 was 70:30 and the water content 60 wt % water (16). Therefore, this composition was chosen as a starting point to investigate the structural changes caused by aspartic protease and/or glycerol. The aqueous phase was composed of mixtures of water with Thermolase formulation or glycerol. Four different hydrations were studied (55–70 wt % aqueous phase) at three different temperatures (25, 35, and 45°C). In this study, we used a mixed lipid system to obtain the desired phase. If the system is at true equilibrium, the lipids should be molecularly mixed for a sample in the one-phase region. It is indeed true that the use of solvent can speed up the time needed to reach equilibrium. In our case, the high concentration of the surfactant, Tween 80, is likely to help mix the lipids. During the course of this study, we prepared many different samples of the same compositions on various occasions, and we always attained the same structure in the samples. This indicated that we are at or close to the equilibrium.
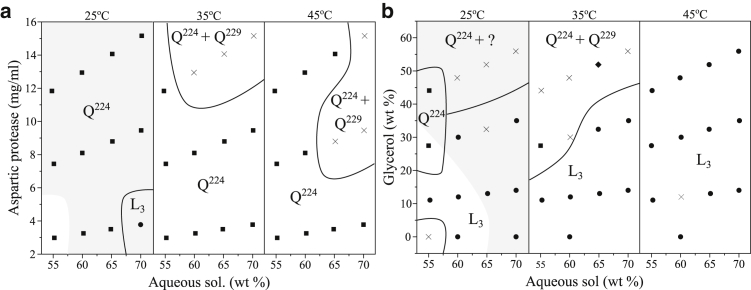
Fig. 1 shows the phase diagrams obtained from the SAXS data. The first things that can be noticed are the differences between the samples with Thermolase formulation and only glycerol aqueous solution. Bulk phases with Thermolase mainly form inverse bicontinuous cubic phases with space group Pn3m for all the formulations and temperatures investigated. This phase was characterized by the presence of at least five Bragg peaks following the ratio √2, √3, √4, √6, √8, and/or √9 (Fig. S3). In addition, samples at 25°C were very viscous and did not present birefringence when observed between crossed polarized light. However, most of the samples contained water in excess (Fig. 1 a), even if the excess was minute at the two lowest water contents.
Figure 1.
Phase diagrams of bulk phases with 40:60 DGMO/GMO-50 ratio and (a) Thermolase (aspartic protease + glycerol) and (b) glycerol. The phase diagrams were constructed as a function of aqueous solution content, aspartic protease, or glycerol final concentration and temperature as indicated on the graphs. Inverse bicontinuous cubic phase with space group Pn3m (Q224) are depicted as filled squares and the co-existence between Pn3m (Q224) and Im3m (Q229) as crosses. L3 phase is shown as solid circles and multiple phase region as crosses. Water excess area is indicated in gray. Lines were drawn to visually guide the eye and do not represent absolute phase boundaries.
In contrast, the glycerol bulk phases formed mostly sponge phases at low glycerol content. At the highest glycerol concentration studied, inverse bicontinuous phases with Pn3m and Im3m spacing group were observed at 25 and 35°C, whereas at 45°C, the L3 phase was predominant. This is quite interesting because structures with higher negative curvature are usually formed with increasing temperature. This behavior could be related to the hydration effect of glycerol, as will be discussed later. The Im3m space group was identified by the observed six Bragg reflections (√2, √4, √6, √10, √12, and √14). The L3 phase scattering curves features two diffuse peaks characteristic of this phase (Fig. S3; (16, 25, 26, 27)). The sponge phase sample appears isotropic when observed between cross-polarizers but is less viscous than the cubic phases. As observed from Fig. 1 b, most of the samples at 25°C contain excess water except at low glycerol concentrations and low water content.
From Fig. S3, it should also be noted that the q-values shift toward higher values as the Thermolase or glycerol concentration is increased. This was a general trend observed by all the formulations and indicates a reduction of the repeat distances with more Thermolase or glycerol. Fig. S4 reflects this behavior very well because the lattice parameters are smaller for the highest concentrations. These results suggest that both aspartic protease and glycerol alter the LLC structure, probably by partition to a larger extend into the lipid bilayer. As a result, bulk phases with Thermolase, which contains both aspartic protease and glycerol, present a larger change in phase behavior compared to bulk phases with only glycerol aqueous solution. This could be explained because 1) glycerol is a less polar solvent and 2) aspartic protease is an amphiphilic molecule, which facilitates their location within the lipid bilayer, as indicated by the decrease in lattice parameter.
The unit cell dimensions for these bulk phases at 40:60 DGMO/GMO-50 were compared to the ones with only water as the aqueous phase (16). At this lipid composition, the sponge phase in 60 wt % water presented a d-spacing of 12.5 nm (q ≈ 0.5 nm−1), and no excess water could be observed by visual inspection. Most of the samples investigated here, especially at medium and high glycerol and protein concentrations, showed the first Bragg peak at higher q-values than without glycerol. The smaller d-spacing, and hence a reduction in the cell dimensions, means a decrease in the water pore dimensions. Consequently, visual observation shows water excess of most of the samples at room temperature (see Fig. 1), that is, glycerol induces a dehydration of the liquid crystalline phases. Richardson et al. (28) suggested that glycerol prevents dehydration of lipid cubic phases by replacing the water from the water channels with a water-glycerol mixture. As a result, lipid phases with less negative curvature were formed. Their study was performed by using thin lipid films of phytantriol/glycerol and monoolein/glycerol mixtures, in which the hydration was changed by changing the relative humidity (water vapor). In our experiment, we have a more complex lipid mixture (DGMO/GMO-50/P80/glycerol) in bulk, and the hydration came from excess aqueous solvent (liquid). These differences may explain why we did not see any hydration effect, even at glycerol concentrations larger than the ones studied by Richardson et al. (28).
Another important aspect here is that we do see a change in lattice parameter as function of glycerol content, indicating that glycerol is strongly interacting with the bilayer or embedded into it. This emphasizes the differences between the lipid system studied here and the one investigated by Richardson et al. (28), in which they did not see a significant change at concentrations up to 40 wt % glycerol. However, the hydration effect observed by Richardson et al. could explain why there is a decrease in negative curvature when increasing temperature at large glycerol concentrations. Nevertheless, as a general trend, the lattice parameter decreased with increment of temperature as expected.
Phase behavior changes induced by Thermolase and glycerol were also observed at the DGMO/GMO-50 ratio of 55/45 and 55–70 wt % aqueous content, but at these compositions, mostly lamellar (Lα) phases are formed (16). Document S1. Supporting Materials and Methods and Figs. S1–S12, Document S2. Article plus Supporting Material shows that L3 phases are formed at medium and highest Thermolase concentration, whereas L3 coexists with Lα at the lowest concentration. In the case of glycerol bulk phases (Fig. S5 b), Lα and L3 mixtures are most predominant, except at the highest glycerol content at which L3 phases are formed. This confirms that both protein and glycerol have an impact on the LLC structure and promote the formation of more curved structures.
In the samples subject for further studies, glycerol was removed from the formulation to be able to reveal the effect of the enzyme alone on the lipid phase behavior. Furthermore, we also expected that the less swollen structures in presence of glycerol would make more difficult the inclusion of proteins.
Effect of freeze-dried aspartic protease
Formulations with freeze-dried aspartic protease were examined to understand better the influence of this enzyme on the lipid sponge phase structure. This also allowed us to load the lipid LLCs with higher enzyme concentration, not possible with the Thermolase formulation. Here, we focused only on bulk phases with 40:60 DGMO/GMO-50 ratio (always in the presence of P80) and 60 wt % of aqueous solution, i.e., with dialyzed and freeze-dried aspartic protease dissolved in water.
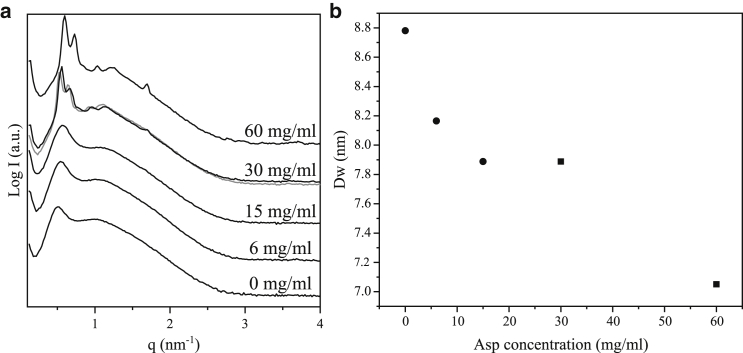
Fig. 2 a shows SAXS curves of these bulk phases as function of aspartic protease final concentration. All the samples are in H2O, except the gray scattering curve at 30 mg/mL that is recorded for the corresponding sample in D2O and is almost identical to curve for similar sample in H2O. At low enzyme concentrations (0–15 mg/mL), two diffuse broad peaks are observed, which are characteristic of the L3 phase. The first peak corresponds to cell-cell dimensions, whereas the second one is related to bilayer distances or correlation length (16, 25, 26, 27). Inverse bicontinuous cubic phase with space group Pn3m was formed at high aspartic protease concentrations (30–60 mg/mL), as apparent from at least four Bragg reflections at spacing ratios of √2, √3, (√4), √6, and √8. In addition, a peak was observed at q = 1.7 nm−1 for both compositions. This peak could be related to the enzyme size, as the d-spacing (= 2π/q) estimated was 3.7 nm, and it was more visible at the highest protein concentration (60 mg/mL). This q-position could also be assigned to the Bragg reflection √18 or √16 (30 and 60 mg/mL, respectively) of the Pn3m space group. However, because the peak position does not change with the enzyme concentration like the other Bragg reflections, we believe it is not related to the LLC structure. Another composition of the same mixture but with a 50:50 DGMO/GMO-50 ratio was also analyzed by SAXS at different aspartic protease concentrations (Fig. S6). At these compositions, coexistence of lamellar and sponge phase was observed. Furthermore, the peak at q = 1.7 nm−1 was also present at 30 and 45 mg/mL of aspartic protease, suggesting that this peak comes from the protein and not from another LLC phase.
Figure 2.
(a) Scattering pattern of bulk phases with included freeze-dried aspartic protease as a function of the final enzyme concentration (0–60 mg/mL). All samples were prepared with DGMO/GMO-50 ratio of 40:60, 70:30 lipid/P80, and 60 wt % of aqueous phase. The scattering pattern of the sample containing 30 mg/mL of aspartic protease in D2O instead of H2O is depicted as a gray line. (b) Water pores diameter (Dw) of sponge phase (circles) and Pn3m bicontinuous cubic phase (squares) as a function of aspartic protease final concentration are shown. All samples were prepared with DGMO/GMO-50 ratio of 40:60, 70:30 lipid/P80, and 60 wt % of aqueous phase.
To check that the size of the enzyme could correspond to the peak observed at q = 1.7 nm−1, DLS of free aspartic protease in solution was performed. The hydrodynamic radius of aspartic protease was determined to be 3.1 and 3.0 nm in 10 mM phosphate buffer and water, respectively (Fig. S7). Therefore, the peak detected at q = 1.7 nm−1 most likely comes from the enzyme, although the enzyme seems to be slightly bigger (3.7 nm) when encapsulated. This might be due to associated lipids or a more extended protein conformation.
Fig. 2 a shows the scattering peaks shift toward higher q-values with more aspartic protease on the lipid matrix. This suggests a decrease in pore dimensions. Using the SAXS data, it was possible to estimate the water channel diameter of the sponge and bicontinuous cubic phase with the Pn3m spacing group (Fig. 2 b). First, the cubic phase water channel radius (rcub) was estimated, and then the L3 water channel radius (rL3), as described in the Supporting Materials and Methods.
Fig. 2 b shows the water channel diameter (Dw = 2r) obtained for bulk phases with freeze-dried aspartic protease. As the enzyme concentration increases, the water pore size is decreased. This indicates that there is an increase in curvature, and thus, the LLC structures eventually changes from inverse bicontinuous sponge to cubic phase. As observed in the case of Thermolase, these results suggest that the aspartic protease is embedded within the lipid bilayer or attached to it in such a way that it affects the LLC structure and reduce the unit cell dimensions. Therefore, it seems that there might strong enzyme-lipid attraction, most probably governed by hydrophobic interaction. This makes aspartic protease disrupt the flexibility of the lipid bilayer until it changes the curvature at high concentrations.
It should be noted that at 15 mg/mL, the sponge phases sample showed a minute amount of water excess in the bottom of the vial. This indicated that at this composition, the LLC phase could not take up more enzyme solution. When further increasing the protein concentration to 30 mg/mL, a change in structure to a cubic phase was observed. However, the water channel dimensions remain quite similar for 15 and 30 mg/mL samples, as shown in Fig. 2 b. This could support the fact that aspartic protease is penetrating into the lipid bilayer and stiffening it.
Comparing these results with the ones obtained with Thermolase (formulation with glycerol) shows that using aspartic protease alone, it is more favorable as relatively swollen L3 phases are formed, which could include up to 15 mg/mL of enzyme. A phase transition was also observed when horse heart cytochrome c was entrapped into phytantriol LLC phase (6). The authors reported that the protein perturbed the inverse bicontinuous cubic structures (Q224 and Q230) transforming into a cubic phase with Im3m space group (Q229). In addition, they observed a decrease in unit cell dimensions with increasing protein concentration, as in this study. This behavior was also observed in other lipid systems (9, 29), although the opposite effect has also been reported (8, 29, 30). Therefore, the phase behavior is highly dependent on the lipid system investigated but also the protein entrapped within it.
Aspartic protease-lipid molecular interactions
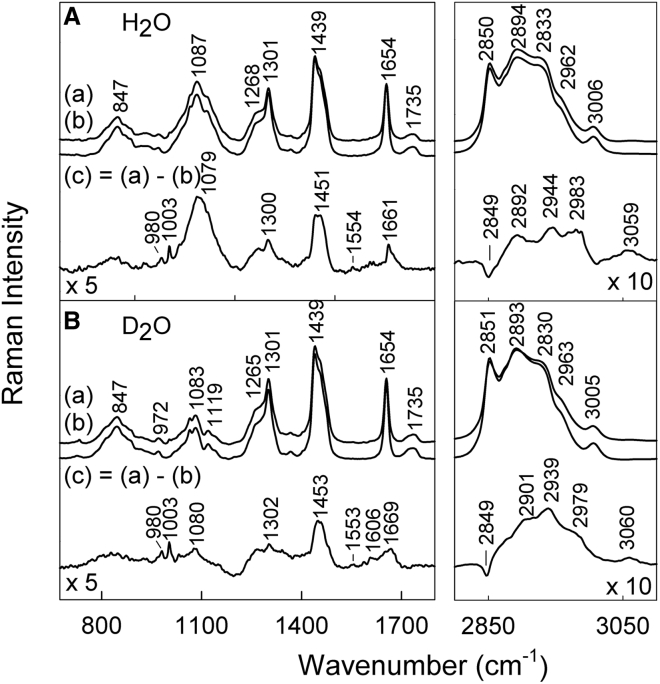
Raman spectroscopy was employed to gain further insight into the lipid-protein interactions. For this purpose, the bulk LLC phase discussed above was prepared from Asp solution with a concentration of 100 mg/mL to give a final sample concentration of 60 mg/mL. The Raman spectra were analyzed and compared to the ones obtained for free Asp at 100 mg/mL. It is noteworthy that these high amounts of enzyme were necessary to be able to detect aspartic protease, and thus, the LLC phase with the highest aspartic protease concentration was chosen for these experiments. In addition, the Raman spectra of the bulk phase with 100 mg/mL Asp were also compared to the lipid LLC with 60 wt % water without the enzyme. As mentioned previously, at this lipid + P80/water composition, L3 phases are formed, but when 100 mg/mL of Asp is included in the aqueous solution, a phase transition to an inverse bicontinuous cubic phase (Q2) occurs. This aspect should be kept in mind during the analyses and interpretation of these data. However, as we showed in a previous study (31), very minor differences were observed between L3 and Q2 phase with the Ia3d spacing group. These differences are not noticeable in the difference spectra in Fig. 3, and thus, the structure of the carrier is not affected.
Figure 3.
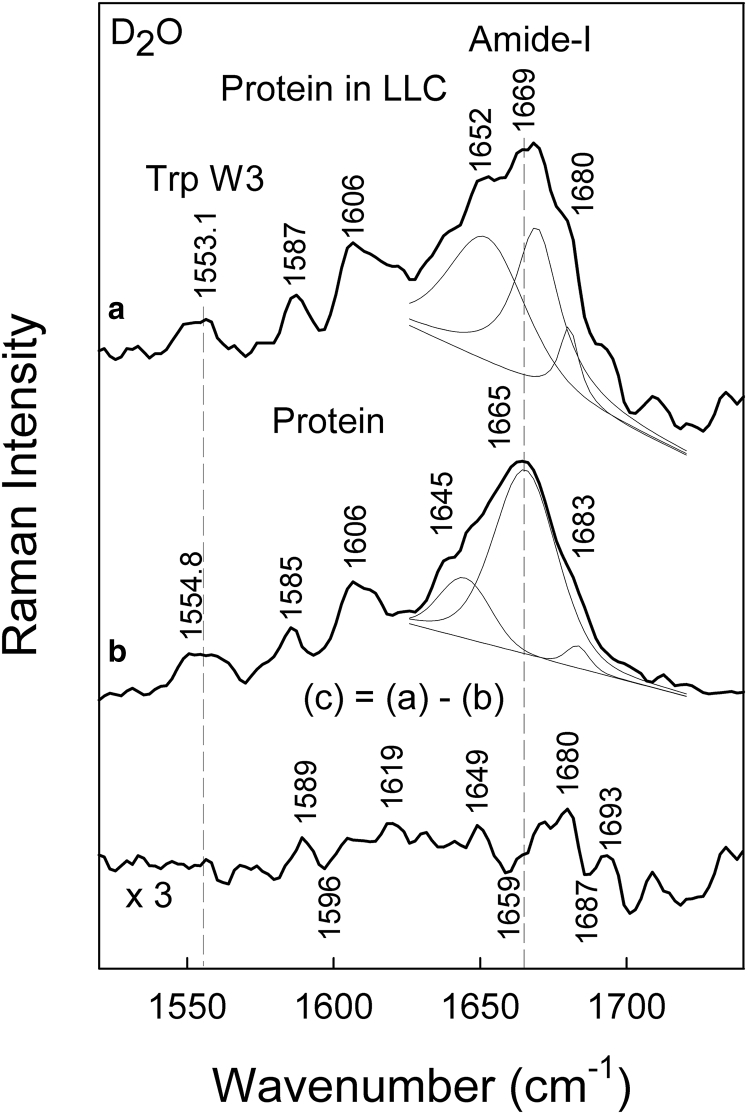
FT-Raman spectra of lipid liquid crystal (LLC)-protein composition in H2O (A) and D2O (B). The spectrum of aspartic protease (60 mg/mL) incorporated in the LLC phase (a), and the lipid matrix reference spectrum without enzyme (b) was used to construct a difference spectra (c). The difference spectra were calculated by taking into account the intensity of the 1654 cm−1 peak. The excitation wavelength was 1064 nm, and 24,000 scans were summed to construct the spectra.
Effect on aspartic protease structure and interactions of the LLC environment
Aspartic protease consists of 330 amino acids, which include 20 phenylalanine (Phe), 13 tyrosine (Tyr), 5 tryptophan (Trp), and 1 histidine (His) residues. The protein secondary structure is reported to be formed by 10% helical and 45% β-sheets (32, 33).
Fig. 3 compares the Raman spectra of the LLC phase with and without aspartic protease in H2O and D2O. Table 1 shows the assignments of protein vibrational bands, which were based on the literature and our previous Raman studies on biomolecules (6, 7, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46). The intensities of protein-related Raman bands were low, and therefore, the difference spectrum (multiplied by 5) is shown to highlight the differences. The presence of Phe residues can be clearly recognized from the high-frequency =C-H stretching band of aromatic residues near 3060 cm−1 and the ring breathing vibrational mode at 1003 cm−1. This suggests that aspartic protease is found within the lipid matrix studied here. However, further analysis should be performed to confirm this. For this purpose, the spectra obtained from preparations with heavy water were further investigated because they do not suffer from laser beam absorption by the second overtone present in H2O preparations. Such absorption may result in heating of the sample and decrease in the signal/noise ratio.
Table 1.
Positions and Tentative Assignments of the Raman Vibrational Modes of Free Aspartic Protease and the Same Protein Incorporated in Liquid Crystalline Lipid Matrix in H2O and D2O
| H2O |
D2O |
Assignment | ||
|---|---|---|---|---|
| Protein in LLC | Protein | Protein in LLC | Protein | |
| 621 | 621 | 621 | 621 | Phe F6b |
| 643 | 642 | 642 | 642 | Tyr |
| 760 | 758 | 757 | Trp W18 | |
| 827 | 827 | 828 | 828 | Tyr Y1 FR |
| 851 | 852 | 848 | 850 | Tyr Y1 FR |
| 876 | 875 | 874 | 873 | Trp W17 |
| 935 | 936 | ν(N-Cα-C) | ||
| 1003 | 1004 | 1003 | 1003 | Phe F12 |
| 1032 | 1032 | 1032 | 1031 | Phe F18a |
| 1057 | 1057 | 1058 | ν(C−C) | |
| 1082 | 1080 | 1083 | ν(C−C) | |
| 1119 | 1126 | 1126 | ν(C−C), ν(C−N) | |
| 1150 | 1158 | ν(C−C), ν(C−N) | ||
| 1206 | 1207 | 1208 | Phe F7a, Tyr | |
| 1271 | 1269 | 1263 | 1263 | Tyr |
| 1302 | 1296 | |||
| 1342 | 1339 | 1341 | 1337 | Trp FR W7 |
| 1359 | 1366 | 1359 | Trp FR W7 | |
| 1401 | ||||
| 1459 | 1459 | 1453 | 1461 | δ(CH2,CH3) |
| 1554 | 1553 | 1553 | 1555 | Trp W3 |
| 1583 | 1586 | 1587 | 1585 | Phe F8b |
| 1605 | 1604 | 1606 | 1606 | Phe F8a |
| 1616 | 1615 | 1615 | Tyr Y8a | |
| 1668 | 1667 | 1669 | 1665 | amide-I |
| 2892 | 2887 | 2871 | 2887 | ν(CH2) |
| 2910 | 2901 | |||
| 2944 | 2939 | 2939 | 2938 | νas(CH3) |
| 2983 | 2971 | 2979 | 2978 | νas(CH3) |
| 3059 | 3062 | 3060 | 3061 | ν(=C−H) aromatic |
δ, deformation; ν, stretching; FR, Fermi resonance.
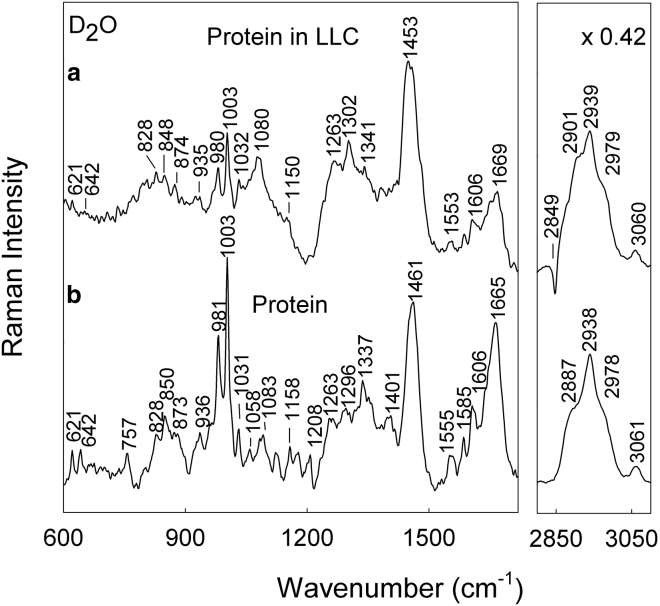
FT-Raman spectra of aspartic protease in D2O solution and included in the LLC phase are presented in Fig. 4. The neat lipid phase spectrum was subtracted from the spectra of the enzyme in the LLC to emphasize the enzyme spectra and be able to more easily compare it to the free enzyme. Fig. 4 a depicts the difference spectrum, which shows a broad dip near 1200 cm−1 due to deformation δ(DOD) vibration of D2O molecules. The characteristic bands of aromatic amino acid residues were found for Phe (621, 1003, 1031, 1585 1606, and 3060 cm−1), Tyr (642, 828, 848, 1208, 1263, and 1615 cm−1), and Trp (757, 873, 1337, 1359, and 1555 cm−1) in heavy water. In addition, deformation motion of methyl δ(CH3) and methylene δ(CH2) groups were observed at 1453−1461 cm−1, and the band near 936 cm−1 was assigned to the backbone N-Cα-C stretch of α-helix structure. The vibrational mode of the amide-I band appears near 1669 cm−1 and will be analyzed later in more detail. All of these features indicate that aspartic protease is found within the lipid Q2 structure. Further details on the Raman band assignments can be found in Table 1.
Figure 4.
Comparison of FT-Raman spectra of (a) aspartic protease incorporated into the LLC phase, in which the lipid phase spectrum was subtracted, and (b) isolated protein in D2O. The excitation wavelength was 1064 nm, and 24,000 scans were co-added.
Several features in the Raman spectra prove the presence of the aspartic protease within the investigated lipid system. The corresponding intensity ratio I1720/I1740 drops from 1.90 to 1.05, indicating a reduced number of carbonyls participating in hydrogen bonding with heavy water. The amide-I spectral band exhibits a minute increase in weakly hydrogen-bonded secondary structure elements when the protein is entrapped. In addition, the spectral marker W3 of the Trp indole ring rotation is found to shift by 1.7 cm−1 to lower wavenumbers (Fig. 5). In conclusion, the lipid-protein interaction induces small spectral changes with introduction of aspartic protease into the quaternary DGMO/GMO-50/P80/D2O system, which reflects only slight structural rearrangements at a molecular level in both main constituents. Even so, these minor alterations cause rather drastic changes in the self-assembly of the lipids, as demonstrated by SAXS. Specific Raman marker bands were used to provide structural information on particular sites of the protein (39, 41, 42). Aspartic protease contains 13 Tyr residues at the following positions: 17, 59, 89, 100, 173, 179, 189, 195, 212, 236, 245, 261, and 287. Raman spectroscopy is able to provide important information on the hydrogen-bonding interaction of the phenolic OH group present in the Tyr residue (36, 38, 41). The Tyr side chain exhibits an intense doublet at 830/850 cm−1, associated with the Fermi resonance between the ring breathing mode Y1 and overtone of an out-of-plane deformation vibration (Y16a) (36). The intensity ratio of the Tyr Fermi doublet I850/I830 (Fig. S8 a) is an indicator for the hydrogen bond state of the tyrosine phenoxyl groups (36, 39). The ratio is found to be at 1.30 for protein in heavy water, and it drastically decreases down to 0.74 upon the protein insertion into the LLC phase. The decrease of the I850/I830 ratio indicates elevated proton donor strength of tyrosine residues upon encapsulation into the LLC matrix.
Figure 5.
Comparison of FT-Raman spectra of (a) LLC-incorporated aspartic protease (lipid phase is subtracted) and (b) isolated protein in D2O. The difference spectrum is also included (c). The wavenumber axis is limited (1500–1750 cm−1). Spectral intensity was normalized according to the Phe ring vibration at 1003 cm−1. The excitation wavelength was 1064 nm, and 24,000 scans were co-added. The bands are decomposed into fundamental Gaussian-Lorentzian shape constituents.
The most interesting spectral region is related to the secondary-structure-sensitive amide-I vibrational mode. This band primarily corresponds to C=O stretching vibration (83%), with some contribution from out-of-phase C-N stretching, C-C-N deformation, and N-H in-plane bending vibrations (37). One can see that the position of the main peak of amide-I mode coincides for both the spectra (Fig. 5). However, the difference spectrum shows slight increase in intensity of high-frequency component near 1680 cm−1 for protein incorporated into the Q2 phase. This can be explained as a slight increase in weakly hydrogen-bonded disordered parts of the secondary structure (39, 41). However, in general, Raman spectra show that the secondary structure of protein in the Q2 phase remains very similar to that observed in solution phase.
The band near 1553−1555 cm−1 is related with W3 mode of Trp residues. The aspartic protease contains five Trp residues. The W3 mode is associated mainly with the stretching vibration of the C2=C3 bond of the aromatic ring (Fig. S8 b) and provides information on the changes of torsional angle about the C2=C3-Cβ-Cα moiety (42). Its frequency is sensitive to the torsion angle χ (7, 41), in accordance with the following relation (42):
| (3) |
The entrapment of protein into the Q2 phase results in red shift of W3 mode by 1.7 cm−1, indicating conformational changes of aromatic Trp rings in the LLC phase, as high as an average rotation of ∼5°. This might be an effect of the more apolar environment in the lipid matrix.
Effect on the LLC structure by the presence of aspartic protease
The difference spectra of the protein within the lipid Q2 phase in Fig. 4 a also shows some lipid features. Encapsulation of protein in LLC causes the intensity drop of methylene stretching motion of lipid molecules, visible as a sharp dip at 2849 cm−1. The frequency of this feature corresponds to vibration of highly ordered alkyl chain segments (47), which is related to the highly organized lipid inverse bicontinuous structure in which aspartic protease is entrapped. However, the reduction in intensity indicates the presence of less ordered hydrocarbon chains, probably due to aspartic protease being embedded into or interacting strongly with the lipid bilayer and disrupting these organization at a molecular level.
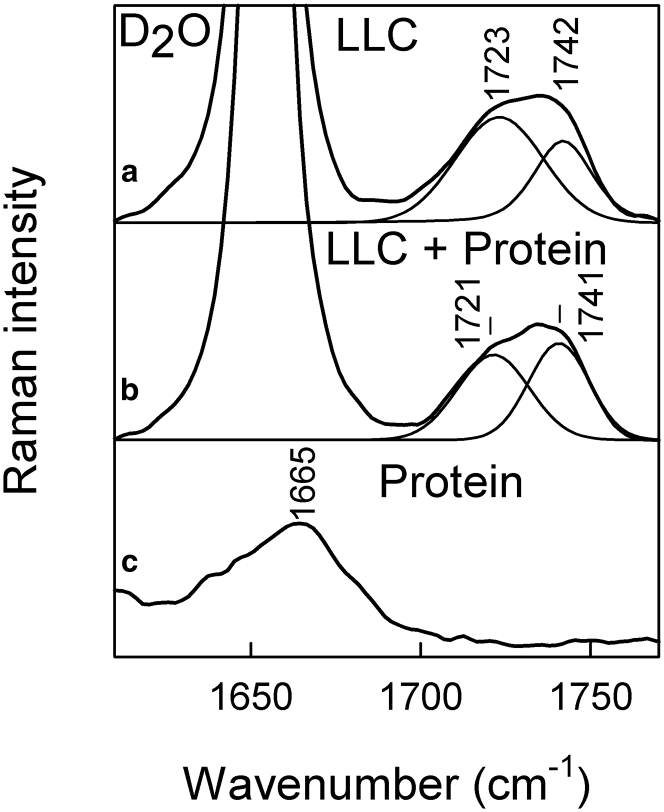
The protein-induced changes in the structure of lipid matrix was further probed by analyzing the C=O stretching vibration region (Fig. 6). To eliminate the influence of water-bending mode δ(HOH) near 1640 cm−1, the Raman spectra were recorded in D2O solvent. The broad C=O stretching band of lipid matrix was fitted by mixed Gaussian-Lorentzian form components. The position of carbonyl stretching vibration is prone to shift to lower wavenumbers when a hydrogen bond is formed (43). As one can see from the Gaussian-Lorentzian υ(C=O) band fit of the lipid matrix alone (Fig. 6 a), the lower energy species are predominant, and the intensity ratio I1720/I1740 is equal to 1.90. Incorporation of protein perturbs the lipid matrix, which reduces the number of water-bound carbonyls and decreases the ratio to 1.05. In a previous study, similar lowering of the ratio from 2.4 to 2.0 and to 1.0 was observed for transition between sponge (60 wt % D2O) to inverse bicontinuous cubic phases with spacing group Ia3d (40 wt % D2O), and to lamellar (10 wt % D2O) phases of the same lipid composition LLC (31). Similar spectral changes were observed in H2O solvent (Fig. S9). Thus, loading of LLCs with protein affects the packing of lipid molecules, leading to rearrangements on the supramolecular scale. This agrees quite well with the SAXS data, as at this composition, the addition of aspartic protease induces phase transition from sponge to cubic phase. It is worth noting that protein is featureless in 1700−1770 cm−1 region (Fig. 6 c), and thus, it does not impair the spectral component analysis of LLC matrix. Unfortunately, other spectral regions contain contributions from both the lipid matrix and protein molecules, and hence, further analysis of LLC response to addition of protein is limited.
Figure 6.
FT-Raman data of (a) LLC, (b) LLC with incorporated protein aspartic protease, and (c) isolated protein in D2O. Spectra are limited to carbonyl stretching υ(C=O) region. The bands are decomposed into Gaussian-Lorentzian shape constituents.
Spectroscopic data suggest only small structural changes for both lipids and protein when the aspartic protease is introduced in the LLC. Firstly, for the lipid ensemble, the aliphatic chain methylene symmetric stretching motion νs(CH2) associated with clusters of well-ordered chains is reduced in intensity. This indicates that the protein is introduced or attached somehow into the lipid bilayer in such a way that, to a certain degree, it disrupts the acyl chain organization at the molecular level. In addition, changes in the band shape near 1735 cm−1 suggest reordering of solvent molecules next to the carbonyl groups. This implies that the protein hampers direct contact between C=O and D2O. Both increase of acyl chain disorder and dehydration in the headgroup region in the presence of aspartic protease is expected to increase the curvature of the LLC structure and thus explains the observed phase transition from L3 to Q2 phase.
Influence of the lipid matrix in aspartic protease activity
The milk clotting activity test was performed to measure the activity of the entrapped Asp compared to the free enzyme, i.e., to assess the retention of the enzyme function by the encapsulation. For this purpose, four different types of samples were analyzed: free Asp solution kept at 4°C and at room temperature and encapsulated Asp in bulk phase and in lipid nanoparticles, both kept at room temperature. The milk clotting strength of each sample was estimated by comparing the clotting time to a reference standard, as described in the experimental section. Then, the activity strength of samples kept at room temperature (free enzyme, entrapped in L3 bulk phase or L3-NPs) was divided by the activity strength of the free Asp stored at 4°C to evaluate the activity retention according to Eq. 4:
| (4) |
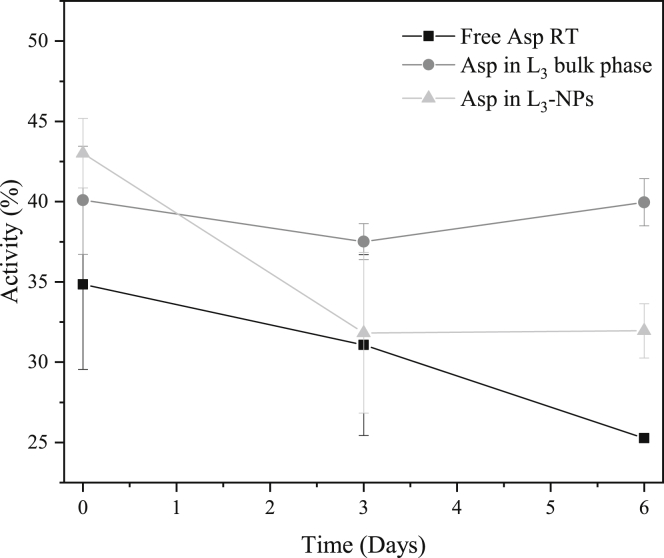
Equation 4 assumes that the free enzyme stored at 4°C maintained 100% of its activity. Here, we note that Asp preserved the activity during storage at 4°C to the same extent as the Thermolase formulation (data not shown). Fig. 7 reveals that encapsulated Asp is more active than the free enzyme stored at the same temperature conditions (25°C) after 1 week of equilibration time. Although the free aspartic protease is losing the activity over time (before equilibration, it was 100%), L3 bulk phases protect the enzyme from degradation as observed by the nearly constant activity value. It is worth mentioning that the bulk phases are dispersed in the buffer to the desired enzyme concentration and this sample is then used for the activity test carried.
Figure 7.
Activity of free Asp (black squares), Asp entrapped in L3 bulk phase (dark gray solid circles), and in L3-NPs (light gray triangles) as a function of time. Note that day 0 is 1 week after the free enzyme solution and bulk phase samples were prepared because bulk phases require some equilibration time. L3 NPs were prepared from a bulk phase a day before the first measurement. All samples were stored at room temperature.
In the case of aspartic protease included in L3-NPs, there is an initial drop of activity, which then is maintained and also presents a higher value than the free enzyme. Therefore, these data indicate that the lipid vehicle preserves the enzyme catalytic function because the free enzyme kept at the same temperature showed lower activity. These data support the conclusion from Raman spectroscopy that the protein structure is not significantly affected by the entrapment process, and hence, its active side is intact.
It is noteworthy that aspartic protease is not totally protected by the lipid matrix, as more than 50% of the activity strength was lost. This reduction could be mainly attributed to fact that the substrate (milk casein) is larger than the pore size of the L3 phases and hence could not enter into the LLC phase. Milk casein micelles are reported to be polydisperse spherical particles with an average diameter of 200 nm, although the size might vary from 50 to 600 nm ((48) and reference therein). Because the water pore diameter of the L3 phase with Asp was ∼8 nm (Fig. 2 b), only casein monomers can enter the pores. In addition, the aspartic protease found on the lipid particle surface or released into solution would be participating in clotting the milk. An interesting point here is that the milk clotting time was highly affected by the dilution of sample used, which was adjusted to obtain a time closer to the reference standard (<50 s). This could support the fact that the milk clotting strength observed is coming from free aspartic protease or the enzyme found in lipid-aqueous interfaces. What is clear, however, is that even if the active enzyme is mostly free, the lipid matrix somehow protects it. This is probably due to protein-lipid interactions, verified by Raman spectroscopy, that stabilize the protein structure. As a result, aspartic protease does not lose its active conformation due to unfolding. These results are in line with van’t Hag et al. (30) or Sun et al. (12), who demonstrated that the enzyme activity was retained after encapsulation within lipid inverse bicontinuous cubic phases.
Further investigation should be carried out to confirm that the loss in activity after encapsulation (compared to the free enzyme stored at 4°C) is due to the substrate (casein) not being able to enter the sponge phase pores. One of the possibilities could be to tune the environmental conditions (temperature, pH, ionic strength) so that the lipid L3 matrix is fragmented or changed in structure and then release the aspartic protease when necessary. This would actually be of great interest because the lipid L3 phase could confer protection when the formulation is added to milk and then release the enzyme in later stages of the cheese processing to form curd.
L3-NPs with encapsulated aspartic protease
DLS and cryo-TEM were employed to analyze the particle size and confirm the L3 structure of nanoparticles with entrapped Asp. As can be seen from Table 2, the particle size and polydispersity are quite similar with and without the presence of Asp. The main difference in size here might arise from small variations in composition of DGMO/GMO-50/P80. To confirm that the sponge structure was still obtained when Asp was encapsulated in agreement with the bulk phase, a representative cryo-TEM image is shown in Fig. S10. Nanoparticles with a well-formed inner sponge phase structure were detected with coexistence of some particle where the structure was less developed. Small-angle neutron scattering confirmed that the main structure was the sponge phase, as the two broad peaks of this phase were clearly observed (data not shown).
Table 2.
Size and Size Distribution of 0.5 wt % of L3-NPs with and without Encapsulated Aspartic Protease
| Parameter | L3-NPs with Asp | L3-NPs |
|---|---|---|
| Diameter (nm) | 154.5 ± 0.5 | 168.5 ± 0.5 |
| Polydispersity | 0.12 ± 0.03 | 0.15 ± 0.02 |
Free versus entrapped aspartic protease
SEC coupled to an ultraviolet detector was employed to separate the free aspartic protease from the encapsulated one but also to evaluate interactions between the lipid and proteins. In addition, the freeze-dried aspartic protease was compared to the Thermolase formulation to confirm that the protein structure was not considerably affected by the dialysis and freeze-drying process.
Fig. S11 shows the comparison between Thermolase and dialyzed + freeze-dried aspartic protease. The main peak, which corresponds to the aspartic protease monomer, is shifted from 18.62 mL for Thermolase to 18.38 mL for Asp at the same protein concentration. This indicates that Asp presents a larger size or slight aggregation, which could be caused by the sample preparation process or differences in terms of the buffer conditions that affects the interaction in the size-exclusion column material. What is important here is that the enzyme is still active, and it is not significantly affected by the sample preparation process.
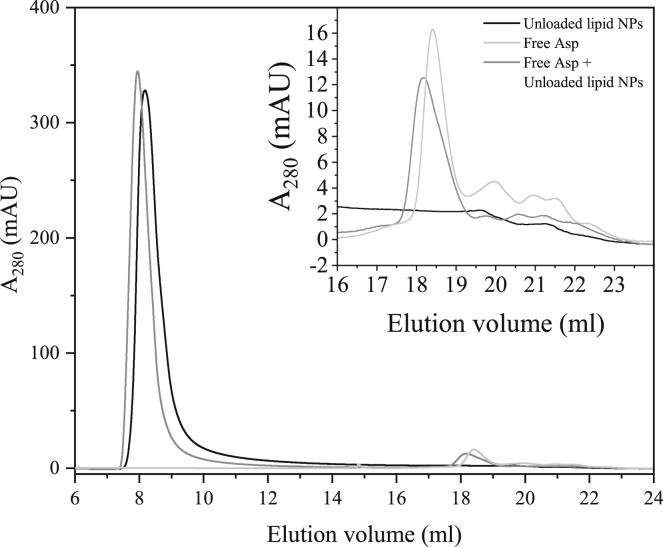
A mixture of unloaded L3-NPs and free Asp was also investigated by SEC to check for possible protein-lipid interactions that would affect the results of entrapped Asp. Additionally, this sample was compared to the SEC results obtained for each component separately to find the elution volume at which the L3-NPs and Asp appear (Fig. 8). L3-NPs were eluted just after the void volume (∼7.2 mL), and the peak maximum was at 8.18 mL. In contrast, 0.75 mg/mL of Asp was eluted at 18.41 mL, which is quite close to the value reported for 1.5 mg/mL. This indicates that the NPs and the aspartic protease can be well separated by SEC using this column.
Figure 8.
SEC results illustrated as absorbance at 280 nm as a function of elution volume of 20 mg/mL unloaded lipid nanoparticles (NPs) (black), 0.75 mg/mL aspartic protease (light gray), and a mixture of both previous samples (dark gray) in which the concentration of lipid NPs and enzyme was the same as the separated components. Embedded graph shows a magnification of the 16–24 mL region.
The AUC as well as the position of the peaks and their shape were here considered to compare these three data sets. Fig. 8 shows that when the L3-NPs and free aspartic protease are mixed, the peaks are shifted toward lower volumes, which indicates that both the nanoparticles and the free Asp are eluted earlier than if they were alone, hence suggesting a slightly larger size. This is also shown in Table 3, in which the peak position and AUC values obtained for the three different samples are indicated.
Table 3.
Peak Position, i.e. Elution Volume, and AUC Obtained for the Lipid NPs and Free Asp Peaks as a Result of SEC Unloaded Lipid Nanoparticles, 0.75 mg/ml Aspartic Protease, and a Mixture of the Two
| Sample | L3-NPs |
Aspartic Protease |
||
|---|---|---|---|---|
| Peak Maximum (mL) | AUC (mAU · mL) | Peak Maximum (mL) | AUC (mAU · mL) | |
| 0.75 mg/mL free Asp | – | – | 18.41 | 9.15 |
| 20 mg/mL unloaded L3-NPs | 8.18 | 300.05 | – | – |
| Mixture of free Asp and unloaded L3-NPs | 7.94 | 265.73 | 18.16 | 10.80 |
Another feature that can be noticed from Fig. 8 is the change in shape of both peaks, especially for the free enzyme. The main peak of aspartic protease is lower and wider than the free enzyme alone, although the AUC is quite similar. This could suggest that some free lipids interact with aspartic protease and might even attach to the enzyme forming larger objects. In contrast, the L3-NP peak is better defined because the peak is narrower and has a shorter tail. The peak corresponding to the NPs also appears at a slightly lower volume, suggesting that they are slightly larger. This might be an effect of aspartic protease being attached to the L3-NP surface. This is supported by Raman spectroscopy that revealed the presence of lipid-aspartic protease interactions.
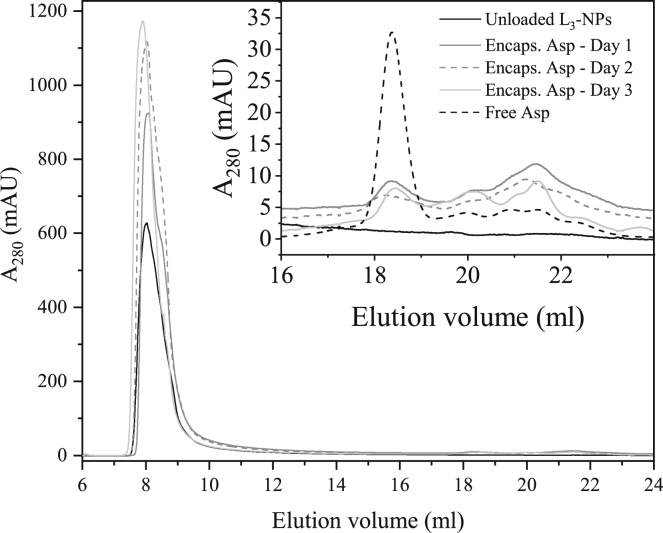
Once the peak position of each separate component was defined, we proceeded to analyze the encapsulated Asp (i.e., 1.5 mg/mL of aspartic protease included in lipid sponge phase NPs). The same sample was analyzed during three consecutive days to trace changes over time (e.g., to observe if the protein is released from the L3-NPs). In addition, a 5-month-old sample was analyzed, although it was from a different batch. Figs. 9 and S12 display the SEC results for this sample compared to the unloaded L3-NPs and free Asp at the same lipid or protein composition (1.5 mg/mL).
Figure 9.
Size-exclusion chromatography (SEC) results of 40 mg/mL of L3-NPs without enzyme (black) and encapsulated Asp analyzed in three consecutive days (dark gray, dashed dark gray, and light gray). An embedded plot includes a magnification of the 16–24 mL region of the same samples after subtraction of the unloaded L3-NPs curve and also of the free Asp at the same concentration (1.5 mg/mL), shown as a dashed black line. Encapsulated enzyme samples contain the same lipid amount as the control sample.
Encapsulated Asp samples present a larger absorbance value compared to the L3-NPs alone. These higher absorbance values are probably due to the scattering of the NPs but also from the aspartic protease absorbance because the sample with only lipid NPs has much lower values. Table 4 also summarizes these results in terms of AUC, where the AUCEnc.Asp > AUCL3-NPs. However, it is quite difficult to assess how much Asp is entrapped within the lipid NPs just by looking at this peak, especially because there are some differences between the three experiments of encapsulated Asp probably because of the polydispersity of these NPs, as shown from the DLS data.
Table 4.
Peak Position, i.e. Elution Volume, and AUC Obtained for the Lipid NPs and Free Asp Peaks as a Result of SEC for L3-NPs Without Enzyme and Encapsulated Asp Analysed During Three Consecutive Days
| Sample | L3-NPs |
Aspartic Protease |
||
|---|---|---|---|---|
| Peak Maximum (mL) | AUC (mAU · mL) | Peak Maximum (mL) | AUC (mAU · mL) | |
| 1.5 mg/mL free Asp | – | – | 18.38 | 17.88 |
| 40 mg/mL unloaded L3-NPs | 8.03 | 603.63 | – | – |
| Encapsulated Asp - day 1 | 8.09 | 808.39 | 18.35 | 3.14 |
| Encapsulated Asp - day 2 | 8.03 | 1063.06 | 18.21 | 3.95 |
| Encapsulated Asp - day 3 | 7.90 | 989.70 | 18.40 | 3.21 |
| Encapsulated Asp - 5 months old | 7.74 | 827.86 | – | – |
The Asp peak was then analyzed to determine how much enzyme is entrapped. The inserted graph in Fig. 9 shows that the free aspartic protease was much lower in the encapsulated Asp samples compared to the completely free enzyme. By subtracting the lipid nanoparticle background, it was possible to estimate the AUC of the peak (as described in the experimental section). The AUC for the completely free enzyme was 17.88 mAU·mL, whereas the averaged AUC for the encapsulated Asp was 3.43 mAU·mL. Because the aspartic protease concentration was the same for all the samples, it is possible to estimate how much enzyme is encapsulated by the equation:
| (5) |
where AUCEnc. Asp and AUCFree Asp are the AUC of the free aspartic protease peak in the encapsulated aspartic protease sample and in the completely free aspartic protease, respectively. These findings indicate that 81 ± 3% of aspartic protease is entrapped or strongly bound to the surface of L3-NPs. This therefore agrees with the increase in absorbance for the NP’s peak and confirms that aspartic protease is included in the lipid NPs.
Similar results were obtained for a sample with encapsulated Asp that was stored for 5 months (Fig. S12). In this case, however, the aspartic protease peak was not visible, suggesting that 100% of the added enzyme is entrapped within this lipid matrix. This suggests that there are significant protein-lipid attractive interactions as previously discussed. As a result, Asp diffuses into the L3 structure of the NPs because it is a bicontinuous system connected to the bulk solvent in a similar way as for bicontinuous cubic phases (49, 50, 51). However, in this case, the enzyme is strongly attached to the lipid bilayer, probably because of hydrophobic interactions. This could explain why an increase in Asp concentration causes a change in lipid structure curvature as observed by SAXS.
Conclusions
In this work, we have shown that lipid sponge bulk phases and NPs have large potential as immobilization matrices for proteins. For this purpose, we have investigated the entrapment of aspartic protease (34 kDa), an enzyme commonly used during the cheese-ripening process, to improve its stability and encapsulation efficiency. The results indicate that the enzyme is embedded into the lipid matrix because it causes structural changes at high concentrations and interacts with the lipid as demonstrated by SAXS and Raman spectroscopy.
Here, we have also shown that the encapsulation process does not affect the aspartic protease structure. This was demonstrated by Raman spectroscopy, in which only minor structural changes were observed at a molecular level. In addition, both L3 bulk phases and L3-NPs with included enzyme presented higher activity than the free enzyme stored at the same temperature. Finally, SEC revealed that 81% of aspartic protease was entrapped within the L3-NPs, which makes this lipid system promising as a carrier for large biomolecules like proteins.
Author Contributions
M.Valldeperas conducted most of the experimental work and wrote the draft of the manuscript and made initial data interpretation. M.T., M.Velička, and G.N. performed and analyzed Raman spectroscopy data. G.N. also helped revise the manuscript. S.K.D. contributed with expertise on the size-exclusion experiment and enzyme assay. J.B. contributed with expertise on the lipid phase behavior. T.N. helped develop and revise the manuscript.
Acknowledgments
The authors thank Professor Marité Cárdenas for the support and access to the SEC instrument at the Department of Biomedical Sciences (Malmö University). G.N. gratefully acknowledges the Center of Spectroscopic Characterization of Materials and Electronic/Molecular Processes (SPECTROVERSUM Infrastructure) for use of the Raman spectrometer.
This work was financially supported by the Biopolymer Based Food Delivery Systems project from the European Commision’s Seventh Framework Programme (PITN-GA-2013-606713 (Bibafoods), as well as the Swedish Foundation for Strategic Research (GSn15-0008, National Graduate School in Neutron Scattering - SwedNess).
Footnotes
Editor: Georg Pabst.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.07.031.
Supporting Material
References
- 1.Wöhri A.B., Johansson L.C., Neutze R. A lipidic-sponge phase screen for membrane protein crystallization. Structure. 2008;16:1003–1009. doi: 10.1016/j.str.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Landau E.M., Rosenbusch J.P. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffrey M. A lipid’s eye view of membrane protein crystallization in mesophases. Curr. Opin. Struct. Biol. 2000;10:486–497. doi: 10.1016/s0959-440x(00)00119-6. [DOI] [PubMed] [Google Scholar]
- 4.Angelova A., Angelov B., Couvreur P. Self-assembled multicompartment liquid crystalline lipid carriers for protein, peptide, and nucleic acid drug delivery. Acc. Chem. Res. 2011;44:147–156. doi: 10.1021/ar100120v. [DOI] [PubMed] [Google Scholar]
- 5.Conn C.E., Drummond C.J. Nanostructured bicontinuous cubic lipid self-assembly materials as matrices for protein encapsulation. Soft Matter. 2013;9:3449. [Google Scholar]
- 6.Misiūnas A., Niaura G., Nylander T. Horse heart cytochrome c entrapped into the hydrated liquid-crystalline phases of phytantriol: X-ray diffraction and Raman spectroscopic characterization. J. Colloid Interface Sci. 2012;378:232–240. doi: 10.1016/j.jcis.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Misiūnas A., Talaikytė Z., Nylander T. Thermomyces lanuginosus lipase in the liquid-crystalline phases of aqueous phytantriol: X-ray diffraction and vibrational spectroscopic studies. Biophys. Chem. 2008;134:144–156. doi: 10.1016/j.bpc.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Angelova A., Ollivon M., Bourgaux C. Lipid cubic phases as stable nanochannel network structures for protein biochip development: x-ray diffraction study. Langmuir. 2003;19:6928–6935. [Google Scholar]
- 9.Leslie S.B., Puvvada S., Rudolph A.S. Encapsulation of hemoglobin in a bicontinuous cubic phase lipid. Biochim. Biophys. Acta. 1996;1285:246–254. doi: 10.1016/s0005-2736(96)00169-1. [DOI] [PubMed] [Google Scholar]
- 10.Barauskas J., Razumas V., Nylander T. Entrapment of glucose oxidase into the cubic Q230 and Q224 phases of aqueous monoolein. Colloid Polym. Sci. 2000;116:16–20. [Google Scholar]
- 11.Razumas V., Kanapieniene J., Larsson K. Electrochemical biosensors for glucose, lactate, urea, and creatinine based on enzymes entrapped in a cubic liquid crystalline phase. Anal. Chim. Acta. 1994;289:155–162. [Google Scholar]
- 12.Sun W., Vallooran J.J., Mezzenga R. Controlling enzymatic activity and kinetics in swollen mesophases by physical nano-confinement. Nanoscale. 2014;6:6853–6859. doi: 10.1039/c4nr01394h. [DOI] [PubMed] [Google Scholar]
- 13.Sun W., Vallooran J.J., Mezzenga R. Enzyme kinetics in liquid crystalline mesophases: size matters, but also topology. Langmuir. 2015;31:4558–4565. doi: 10.1021/acs.langmuir.5b00579. [DOI] [PubMed] [Google Scholar]
- 14.Tyler A.I., Barriga H.M., Brooks N.J. Electrostatic swelling of bicontinuous cubic lipid phases. Soft Matter. 2015;11:3279–3286. doi: 10.1039/c5sm00311c. [DOI] [PubMed] [Google Scholar]
- 15.Zabara A., Chong J.T.Y., Mezzenga R. Design of ultra-swollen lipidic mesophases for the crystallization of membrane proteins with large extracellular domains. Nat. Commun. 2018;9:544. doi: 10.1038/s41467-018-02996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valldeperas M., Wiśniewska M., Barauskas J. Sponge phases and nanoparticle dispersions in aqueous mixtures of mono- and diglycerides. Langmuir. 2016;32:8650–8659. doi: 10.1021/acs.langmuir.6b01356. [DOI] [PubMed] [Google Scholar]
- 17.Erskine P.T., Coates L., Cooper J.B. Atomic resolution analysis of the catalytic site of an aspartic proteinase and an unexpected mode of binding by short peptides. Protein Sci. 2003;12:1741–1749. doi: 10.1110/ps.0305203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law B.A., Goodenough P.W. Enzymes in milk and cheese production BT. In: Tucker G.A., Woods L.F.J., editors. Enzymes in Food Processing. Springer; 1995. pp. 114–143. [Google Scholar]
- 19.Kirby C.J., Brooker B.E., Law B.A. Accelerated ripening of cheese using liposome-encapsulated enzyme. Int. J. Food Sci. Technol. 1987;22:355–375. [Google Scholar]
- 20.Rizwan S.B., McBurney W.T., Hook S. Cubosomes containing the adjuvants imiquimod and monophosphoryl lipid A stimulate robust cellular and humoral immune responses. J. Control. Release. 2013;165:16–21. doi: 10.1016/j.jconrel.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Barriga H.M.G., Holme M.N., Stevens M.M. Cubosomes: the next generation of smart lipid nanoparticles? Angew. Chem. Int. Engl. 2019;58:2958–2978. doi: 10.1002/anie.201804067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matougui N., Boge L., Saulnier P. Lipid-based nanoformulations for peptide delivery. Int. J. Pharm. 2016;502:80–97. doi: 10.1016/j.ijpharm.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Walker J.M. The bicinchoninic acid (BCA) assay for protein ouantitation BT. In: Walker J.M., editor. The Protein Protocols Handbook. Humana Press; 1996. pp. 11–14. [Google Scholar]
- 24.International Organization for Standardization. International Organization for Standardization; Geneva, Switzerland: 2007. Milk: Determination of Total Milk-Clotting Activity of Bovine Rennets. [Google Scholar]
- 25.Roux D., Cates M.E., Bellocq A.M. Light scattering from a surfactant “sponge” phase: evidence for a hidden symmetry. Europhys. Lett. 1990;11:229–234. [Google Scholar]
- 26.Lei N., Safinya C.R., Liang K.S. Synchrotron x-ray-scattering studies on the sodium dodecyl sulfate–water–pentanol–dodecaneL3sponge phase. Phys. Rev. E. 1997;56:608–613. [Google Scholar]
- 27.Angelov B., Angelova A., Pedersen J.S. SAXS investigation of a cubic to a sponge (L3) phase transition in self-assembled lipid nanocarriers. Phys. Chem. Chem. Phys. 2011;13:3073–3081. doi: 10.1039/c0cp01029d. [DOI] [PubMed] [Google Scholar]
- 28.Richardson S.J., Staniec P.A., Squires A.M. Glycerol prevents dehydration in lipid cubic phases. Chem. Commun. (Camb.) 2015;51:11386–11389. doi: 10.1039/c5cc03771a. [DOI] [PubMed] [Google Scholar]
- 29.van ’t Hag L., Shen H.H., Conn C.E. Effect of lipid-based nanostructure on protein encapsulation within the membrane bilayer mimetic lipidic cubic phase using transmembrane and lipo-proteins from the beta-barrel assembly machinery. Langmuir. 2016;32:12442–12452. doi: 10.1021/acs.langmuir.6b01800. [DOI] [PubMed] [Google Scholar]
- 30.van ’t Hag L., Anandan A., Conn C.E. Direct demonstration of lipid phosphorylation in the lipid bilayer of the biomimetic bicontinuous cubic phase using the confined enzyme lipid A phosphoethanolamine transferase. Soft Matter. 2017;13:1493–1504. doi: 10.1039/c6sm02487d. [DOI] [PubMed] [Google Scholar]
- 31.Talaikis M., Valldeperas M., Nylander T. On the molecular interactions in lipid bilayer-water assemblies of different curvature. J. Phys. Chem. B. 2019;123:2662–2672. doi: 10.1021/acs.jpcb.8b11387. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 33.Berman H.M., Westbrook J., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Razmutė-Razmė I., Kuodis Z., Niaura G. In-situ surface-enhanced Raman spectroscopic investigation of NH site of indole ring-terminated self-assembled monolayer on gold electrode. Chemija. 2007;18:16–20. [Google Scholar]
- 35.Carey P.R. Raman crystallography and other biochemical applications of Raman microscopy. Annu. Rev. Phys. Chem. 2006;57:527–554. doi: 10.1146/annurev.physchem.57.032905.104521. [DOI] [PubMed] [Google Scholar]
- 36.Siamwiza M.N., Lord R.C., Shimanouchi T. Interpretation of the doublet at 850 and 830 cm-1 in the Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry. 1975;14:4870–4876. doi: 10.1021/bi00693a014. [DOI] [PubMed] [Google Scholar]
- 37.Krimm S., Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. In: Anfinsen C.B., Edsall J.T., editors. Advances in Protein Chemistry. Academic Press; 1986. pp. 181–364. [DOI] [PubMed] [Google Scholar]
- 38.Niaura G. Raman Spectroscopy in analysis of biomolecules. Encycl. Anal. Chem. 2014 Published online June 16, 2014. 10.1002/9780470027318.a0212.pub3. [Google Scholar]
- 39.Wen Z.Q. Raman spectroscopy of protein pharmaceuticals. J. Pharm. Sci. 2007;96:2861–2878. doi: 10.1002/jps.20895. [DOI] [PubMed] [Google Scholar]
- 40.Gaigalas A.K., Reipa V., Niaura G. Stark spectroscopy of tryptamine immobilized on a gold electrode. J. Colloid Interface Sci. 1998;203:299–310. doi: 10.1006/jcis.1998.5509. [DOI] [PubMed] [Google Scholar]
- 41.Kocherbitov V., Latynis J., Niaura G. Hydration of lysozyme studied by Raman spectroscopy. J. Phys. Chem. B. 2013;117:4981–4992. doi: 10.1021/jp4017954. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi H. Raman structural markers of tryptophan and histidine side chains in proteins. Biopolymers. 2003;72:305–317. doi: 10.1002/bip.10440. [DOI] [PubMed] [Google Scholar]
- 43.Blume A., Hübner W., Messner G. Fourier transform infrared spectroscopy of 13C = O-labeled phospholipids hydrogen bonding to carbonyl groups. Biochemistry. 1988;27:8239–8249. doi: 10.1021/bi00421a038. [DOI] [PubMed] [Google Scholar]
- 44.Ignatjev I., Proniewicz E., Niaura G. Effect of potential on temperature-dependent SERS spectra of neuromedin B on Cu electrode. Phys. Chem. Chem. Phys. 2013;15:807–815. doi: 10.1039/c2cp42077e. [DOI] [PubMed] [Google Scholar]
- 45.Blanpain-Avet P., Hédoux A., Delaplace G. Analysis by Raman spectroscopy of the conformational structure of whey proteins constituting fouling deposits during the processing in a heat exchanger. J. Food Eng. 2012;110:86–94. [Google Scholar]
- 46.Hernández B., Pflüger F., Ghomi M. Vibrational analysis of amino acids and short peptides in hydrated media. VIII. Amino acids with aromatic side chains: L-phenylalanine, L-tyrosine, and L-tryptophan. J. Phys. Chem. B. 2010;114:15319–15330. doi: 10.1021/jp106786j. [DOI] [PubMed] [Google Scholar]
- 47.Orendorff C.J., Ducey M.W., Jr., Pemberton J.E. Quantitative correlation of Raman spectral indicators in determining conformational order in alkyl chains. J. Phys. Chem. A. 2002;106:6991–6998. doi: 10.1021/jp014311n. [DOI] [PubMed] [Google Scholar]
- 48.Glantz M., Devold T.G., Paulsson M. Importance of casein micelle size and milk composition for milk gelation. J. Dairy Sci. 2010;93:1444–1451. doi: 10.3168/jds.2009-2856. [DOI] [PubMed] [Google Scholar]
- 49.Maldonado A., Urbach W., Langevin D. Surface self-diffusion in L3 phases. J. Phys. Chem. B. 1997;101:8069–8073. [Google Scholar]
- 50.Ott A., Urbach W., Hoffmann H. Surfactant self-diffusion in L3 phases. Langmuir. 1992;8:345–347. [Google Scholar]
- 51.Evertsson H., Stilbs P., Engström S. NMR self diffusion measurements of the Monooleoylglycerol/Poly ethylene glycol/water L3 phase. Colloids Surf. B Biointerfaces. 2002;26:21–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.