Abstract
There are numerous challenges to identifying, developing and implementing quantitative techniques for use in clinical radiology, suggesting the need for a common translational pathway. We developed the quantitative neuroradiology initiative (QNI), as a model framework for the technical and clinical validation necessary to embed automated segmentation and other image quantification software into the clinical neuroradiology workflow. We hypothesize that quantification will support reporters with clinically relevant measures contextualized with normative data, increase the precision of longitudinal comparisons, and generate more consistent reporting across levels of radiologists’ experience. The QNI framework comprises the following steps: (1) establishing an area of clinical need and identifying the appropriate proven imaging biomarker(s) for the disease in question; (2) developing a method for automated analysis of these biomarkers, by designing an algorithm and compiling reference data; (3) communicating the results via an intuitive and accessible quantitative report; (4) technically and clinically validating the proposed tool pre-use; (5) integrating the developed analysis pipeline into the clinical reporting workflow; and (6) performing in-use evaluation. We will use current radiology practice in dementia as an example, where radiologists have established visual rating scales to describe the degree and pattern of atrophy they detect. These can be helpful, but are somewhat subjective and coarse classifiers, suffering from floor and ceiling limitations. Meanwhile, several imaging biomarkers relevant to dementia diagnosis and management have been proposed in the literature; some clinically approved radiology software tools exist but in general, these have not undergone rigorous clinical validation in high volume or in tertiary dementia centres. The QNI framework aims to address this need. Quantitative image analysis is developing apace within the research domain. Translating quantitative techniques into the clinical setting presents significant challenges, which must be addressed to meet the increasing demand for accurate, timely and impactful clinical imaging services.
THE QUANTITATIVE NEURORADIOLOGY INITIATIVE FRAMEWORK
Current clinical radiology practice provides an assessment or interpretation of usually qualitatively described facts. These facts are often difficult to accurately define, thereby increasing the potential variability of their interpretation. Establishing agreed facts within a robust reference framework would address this problem and consequentially reduce disagreement in reporting. Quantitative imaging biomarkers (QIBs), defined as objectively measured characteristics derived from in vivo images as indicators of normal biological processes, pathogenic processes, or response to a therapeutic intervention,1 offer considerable potential in this context. QIBs are generally first developed in the context of group-level research studies which may fail to translate to individual patient inference. Research software tools may lack the level of quality management and clinical validation required for routine radiology practice and may not be readily deployed within hospital information systems. We therefore propose a translational pipeline for the clinical adoption of neuroradiological QIBs in the context of the UK health service, taking current neuroradiology practice in dementia as an example. We will review the evidence supporting the potential benefit of quantitative neuroradiology in the diagnosis and management of dementia and present our approach to overcoming practical barriers to clinical adoption.
Quantitative neuroradiology aims to automatically derive from clinical neuroimages robust, objective and validated measures related to disease state, recording these in the form of a visually accessible report to be presented within the routine workflow of the reporting neuroradiologist. We hypothesize that this facility will increase diagnostic confidence by providing objective comparison with normative population data and eliminating inter-rater variation for certain assessments, enabling earlier unequivocal detection of pathological changes, as well as reducing reporting times.
The aims of this approach include increased measurement reproducibility v s visual assessment, reduction of intra- and inter-rater variability, increased confidence in achieving a correct diagnosis,2 increased workflow and patient management efficiency, and detection of features and changes with sensitivity and reproducibility not attainable by qualitative observations. It is also possible in principle to provide estimates of measurement uncertainty, providing enhanced assessment of the QIB reliability.
We have developed the quantitative neuroradiology initiative (QNI) framework to expedite the implementation and adoption of automated image quantification into clinical neuroradiology practice. Our focus is on the design and delivery of clinically relevant, user-friendly, quality-assured reports of regional and global brain characteristics at the individual patient level that are fully integrated into the clinical workflow. We envisage that this will support neuroradiologists with clinically useful measures in the context of normative data, increase sensitivity for detecting longitudinal change, and generate more consistent and confident reporting across levels of radiologists’ experience.
We propose the following steps as essential for eventual adoption into routine practice of specific quantitative neuroradiological tools: (1) establishing an area of clinical need and identifying the appropriate proven imaging biomarker(s) for the disease in question; (2) developing a method for automated analysis of these biomarkers, by designing an algorithm and compiling reference data; (3) communicating the results via an intuitive and accessible quantitative report; (4) technically and clinically validating the proposed tool pre-use; (5) integrating the developed analysis pipeline into the clinical reporting workflow; and (6) performing in-use evaluation. These steps are now described in further detail.
The first two steps consist of identifying an area of clinical need, and its corresponding imaging biomarkers, and an effective quantification algorithm and reference data. This involves demonstrating the basis for accepting the identified imaging parameter as a correlate of a pathological process of interest and considering the potential contribution it could make to patient management. The success of a quantitative tool relies on the use of disease-specific algorithm training datasets and disease-specific normative data to contextualize the individual patient's findings. Each disease or biomarker of interest may require a different normative dataset to accurately contextualise the individual results. A source of high-volume, age-matched, generalizable normative data is needed to allow referenced comparison of the subject's biomarker measurements to a range of normal values. Generalizability can be limited by a multitude of factors, including scanner and scanning parameter differences, patient gender and brain volume, and overlap with normal ageing.
The third phase relates to production of a quantitative report. Data should be presented in a clinically and visually meaningful way, providing the radiologist with clinically useful information that can be integrated into the radiology report. This may facilitate the move towards standardized, structured reporting to minimize variability between reporters.
The fourth phase is technical and clinical validation prior to the pipeline being introduced into clinical use. Technical validation at this stage includes consideration of image acquisition quality with attention to reproducibility, error and artefact which may affect the algorithm's performance. Pre-use clinical validation must also be obtained. We propose that this should consist of a proof of concept "credibility" study and a clinical impact "accuracy" study, which we will expand on in more detail under the wider clinical validation process.
The fifth stage in the proposed framework is pipeline integration into the clinical workflow. Smooth integration will increase acceptance of the technique by radiologists and referring physicians. Some basic requirements for this are compatibility with the data format and transfer, i.e. the Digital Imaging and Communications in Medicine (DICOM) standard, and integration into the hospital Picture Archiving and Communication Systems (PACS). Automated outputs, ideally, should be viewed within the same workstation environment as an additional DICOM series alongside the source images, allowing for rapid and efficient integration of the information into the radiologist’s workflow. Software must be developed under a quality management framework for medical devices, along with consideration to patient data protection and other institutional information governance requirements. Key features of the regulatory framework applicable in Europe are summarised in Box 1.
Box 1. .
Summary of the medical device regulatory framework applicable to software medical devices in the European Union.3,4
The National Institutes of Standards and Technology (NIST, https://www.nist.gov/), when considering the role of QIBs in clinical practice, raised the concern that a robust QIB must be immune to variability of clinical interpretation as well as physical measurement variability across imaging platforms.5 There are several common challenges to the practical adoption of QIBS, and in particular methods for automated brain volume quantification, in the clinical setting, including but not limited to those outlined in Table 1.
Table 1. .
Common technical challenges for QIB deployment in the clinical setting
| Suboptimal acquisition protocols in the clinical setting | Routine clinical MRI protocols may be less sophisticated than those specified for research studies, e.g. in many centres clinical T 1 weighted scans may be performed with two-dimensional acquisitions. Isotropic 3D data, which is more suitable for quantitative analysis, may not be available in routine clinical practice. Inconsistencies in scanning parameters can also cause significant variation in tissue contrast, making, for instance, automated GM/WM delineation for a subregion of interest challenging. |
| Interscanner variability | Image geometric accuracy varies between scanners and vendors, resulting in varying spatial distortions which, if uncorrected, may impact upon regional tissue-volume estimates. Quantification accuracy is predicated on high reproducibility between MRI instruments; this however is not generally a primary design concern in clinical systems, since this rarely affects routine clinical practice based on radiologists’ qualitative visual evaluation. |
| Image artefacts | Robust screening of incoming data to detect artefacts, such as those arising from patient motion and other errors must be established, as these may impede the automated algorithm in performing accurate quantification. Adaptive correction schemes prior to analysis, such as bias field or motion artefact correction, may minimize the number of data sets failing to yield reliable volume estimates for a given measurement strategy. Many software packages are automated, meaning they will produce a numerical result whatever the input data and often do not allow intermediate (e.g. segmentation) steps to be scrutinized. |
| Need for full automation | To move away from time-consuming manual or semi-automated techniques requiring frequent intervention and monitoring, often by highly expert practitioners, methods for clinical application must be fully automated. This also protects the process from inter operator variability. Such automated techniques must be generalizable across the range of MRI services in the health system, including both scanner type and acquisition protocol variations. |
3D, three-dimensional.
Issues regarding technical variation and barriers to generalisability are being tackled in part by the international study Alzheimer's disease neuroimaging initiative (ADNI), which has collected a large dataset from multiple institutions and at several time points, as part of the ADNI research into AD pathophysiology and biomarker development. This ambitious project has made progress in the standardisation of MRI scanning protocols in the research context. Their data are publicly accessible, providing a rich source of normative data (http://adni.loni.usc.edu/). Whilst adoption of uniform imaging protocols across clinical centres is not currently seen as attainable, due to service-predicated differences, and differing scanner platforms, many centres across Europe have now adopted a consistent three-dimensional T 1 weighted volume sequence within their dementia imaging protocol6 reflecting increasing interest in acquisition homogeneity within a healthcare provider across clinical timepoints. If there is adequate imaging protocol uniformity within a clinical service, an added software tool should perform reliably within that setting. This reliability should be established on a site-by-site basis, as addressed by step six of our framework.
The final stage of the six-step QNI framework specifies an in-use evaluation of the pipeline with respect to the key areas of patient management and socioeconomic impact, which we will also expand on below. Validation of automated MRI techniques has tended to occur in the research setting largely referenced to the results of other available methods, using well-curated data sets. Studies exploring the validation of these techniques in clinical practice are still sparse and adopt disparate methods. Clinical validation involves both testing the technical performance of the algorithm with clinical quality data, possibly generated using disparate scanner platforms and protocols and, importantly, capturing the outcomes and experience of the radiologist end-users. The latter is necessary to demonstrate that the tool is beneficial in terms of one or more of efficiency; accuracy; inter- and intrareader agreement and diagnostic confidence.7,8 Importantly, interpretation of QIB reports alongside laboratory biomarkers and clinical examination in the MDT setting would establish the added value of this quantitative information in reaching a diagnosis or monitoring and prognostication. This would also allow assessment of the views of neuroradiologists and MDT members on the usability of the quantitative information in clinical practice, and identify and address potential practical barriers hampering the adoption of the biomarker pipeline. Generalized deployment across centres will require further work towards achieving standardized imaging protocols, or adaptation to the analysis methods to account for protocol variation. Eventually a system-wide assessment of the impact of methods integrated via the QNI framework on radiologists, referring physicians, patients, and the hospital will be needed. This ultimate clinical and healthcare economic validation will likely require long-term multi centre assessment.
Within our proposed framework, clinical validation (steps 4 and 6) has thus far received the least attention in QIB development, but it remains a crucial step for adoption into clinical radiological practice. Since attention given to technical development and establishing reference data far outweighs that given to clinical validation and integration into hospital IT environment, we will now discuss in more detail our proposals for these important challenges. In steps 4 and 6 of our QNI framework we suggest four stages of clinical validation: credibility, accuracy, patient management, and socioeconomic impact, which we will expand below.
Clinical validation pathway (QNI framework steps 4 and 6)
Credibility: validation of the proof of concept or biological validation in real-world data should be conducted. This could involve a pilot study in which the chosen quantitative biomarker tool is applied to clinical MRI scans of known cases of the disease of interest and quality checked. This would be followed by technical validation, checks on image acquisition, post-processing, analysis and report generation. At this stage, a limited clinical validation should be performed by experienced blinded expert radiologists who have not seen the cases, and who should rate them first according to their routine practice, blinded to the QIB report and again taking the report into account. Classical evaluation would be compared to the produced report and their consistency evaluated.
Accuracy: once the credibility of the automated technique has been established, its impact on the clinical reporting process should be examined. The setting for this evaluation should reproduce the radiologist’s normal reporting environment as closely as possible, with the automated report displayed alongside the imaging series. Assessment of radiologists’ accuracy, confidence, and reporting efficiency may all be measured, both with and without the quantitative report being present. The images should be presented in a random unpredictable order and should include a spectrum of pre-selected clinical cases, from clearly pathological to more subtle changes, as well as normal-appearing control scans—where available. The pathology of each case should be established to the best available gold-standard, depending on the condition in question (e.g. cerebrospinal fluid (CSF) analysis and neuropsychiatric profile in the case of Alzheimer’s disease, AD, and frontotemporal dementia, FTD). Including a range of severity in the case mix is valuable in discerning whether the added quantitative information is most impactful where the pathology is subtle or unclear, where for instance fine-grained analysis may perform better than coarse visual rating scales. In the evaluation it may also be useful to include radiologists with a range of expertise, representing the wide range of training and experience levels present in a working radiology department. It is then possible to establish whether the quantitative report increased inter-rater agreement between these groups, in addition to increased accuracy when compared with the gold-standard. Additionally, it may be of secondary interest to include a group of non-clinical image analysts in the assessment, e.g. if the tool is being considered for education and training.
Patient management: based on the experience gathered from the accuracy study, the quantitative tool should be integrated within the hospital’s radiology department and the tool rolled out to specific reporting radiologists. Whether this full integration can be achieved or not will depend on the tool design details achieving compliance with the relevant medical device regulations. This is also an important stage for assessing how easily the tool can be integrated into the reporting workflow, and regular structured feedback from the users should be documented. To determine the impact on patient management, a prospective assessment of patients’ clinical pathways, including speed of diagnosis, need for repeat investigations, and therapeutic decisions if available, can be compared with cases for which quantitative analysis was not available.
Socio-economic impact: definitive socioeconomic validation requires a larger scale, multicentre study investigating resource utilisation, productivity, clinical and population perception, and economic impact over time. This phase of clinical validation is especially challenging due to the a priori requirement to invest in these tools and their supporting infrastructure before an economic impact can be demonstrated. Ultimately,, this is the type of business intelligence required to convince purchasers (hospitals and insurance companies) to bear the costs of the additional software tools and processing hardware.
QNI framework applied to dementia imaging
Dementia is a class of conditions which cause an irreversible progressive decline in cognitive function, affecting an increasing number of people worldwide and presenting a significant challenge for health and social care. It is estimated that dementia will affect more than 115 million people worldwide by 2050.9 Conditions causing dementia are varied, and imaging can help to identify the differences between the commonest: AD (50–75% of cases), vascular dementia, (20%), and FTD (5%).10 AD is characterized by histopathological findings of neurofibrillary tangles and amyloid plaques that cause synaptic and axonal loss and subsequent atrophy in a progressive regional pattern.11 This pattern of atrophy is seen on structural imaging in several recognized patterns or subtypes.12,13
We will now use dementia as an exemplar application to discuss the development of QIBs for clinical neuroradiology, referring directly to each step of our QNI framework.
QNI dementia framework, Steps 1 to 6
Step 1: establishing a clinical need
Structural MRI is the mainstay of conventional neuroradiology in current dementia practice.14 It meets clinical needs in the diagnostic setting to exclude alternative pathologies, and to attempt to differentiate between the dementia pathologies by establishing a specific pattern of atrophy.15
Validated and clinically adopted dementia QIBs could facilitate diagnosis in the early or even prodromal disease phases; provide objective measures of difference from the normal ageing spectrum; exclude differential diagnoses; and support powerful preclinical drug trials.16,17 There is also a potential role for QIBs as prognostic measures, since the relationship between progressive biomarker change and clinical disease trajectory can be investigated.18
Classical AD is typified by early medial temporal lobe atrophy (MTA), followed by lateral temporal, medial and lateral parietal, and frontal involvement, with relative sparing of the occipital lobe and sensory-motor cortex. This pattern on MRI is discriminating, as it is not commonly seen in normal ageing, and ante-mortem MRI findings in AD patients correlate with pathological severity post-mortem.19
Mild cognitive impairment (MCI) is identified as a prodromal stage of AD, with a 10–15% annual conversion rate to AD, especially within the subset with an amnestic presentation.20 Longitudinal MRI has demonstrated that MCI subjects initially have focal, limited areas of cerebral atrophy, mainly in the medial temporal lobes, and this increases in increments to the established AD pattern.21 Recent dementia research has prioritised the identification of potential clinical and imaging biomarkers early in the disease course, as it has been shown that AD signs and symptoms can emerge years if not decades before a clear pathological imaging pattern is established.12 Quantification of atrophy rates, brain volume and morphometry have been useful in prediction of which MCI subjects will progress to AD, with differences from stable MCI subjects detectable well before clinical AD diagnosis.20,22,23
With imaging identified as a potentially powerful diagnostic tool, the introduction of objective methods for quantifying dementia-related changes on MRI, especially MTA assessment in AD, has become a priority for clinicians and researchers. Indeed, the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association, NINCDS-ADRDA, issued updated guidelines which incorporated structural MRI of the medial temporal lobe in the assessment of AD; however, no specific method for objective measurement was defined.24
Current practice: visual rating scores
Visual rating scores, with "cut-offs" defining degrees of abnormality, are a semi-quantitative means aimed to facilitate communication among practitioners. MTA grading was established by Scheltens et al.25 using a discrete 5-point scale for the size of the hippocampal formation as well as the prominence of adjacent CSF spaces.
Visual rating scores are useful, rapid and accessible tools in clinical practice. Their limitations include insensitivity to subtle or early changes, ambiguity in distinguishing pathological change from normal ageing, ceiling and/or flooring effects, and being only coarsely discriminating due to their discrete categorizations 26 . They are used to varying degrees by radiologists across Europe, depending on levels of training and with unknown reproducibility both within and across imaging departments.6 Fully quantitative imaging biomarkers may largely address these issues providing practitioner-independent objectivity at least across a single radiology service.
Step 2: developing a method for automated analysis
Many biomarkers show promise in early research phases but face bottlenecks when it comes to validation and clinical implementation. Frisoni et al proposed a 5-phase framework for development of specific QIBs for prodromal AD, adapted from a framework implemented in oncology screening.27 Despite being one of the most established imaging biomarkers in AD, MTA is still in the early stages of this 5-step pathway.28 Standardized validation procedures for automated segmentation algorithms based on a harmonized manual segmentation protocol is identified as an imminent priority, which will allow for meaningful assessment of algorithm reproducibility. Only then can their clinical validity and utility be evaluated in the memory clinic. The Frisoni framework integrates well with the QNI framework in that it focuses on QIB development and clinical validation, which are key parts of the broader, end-to-end translational implementation pathway detailed by the QNI.
In the research context, numerous studies have compared cerebral atrophy between AD, MCI and healthy control groups using MRI-based regional volume measurement (“volumetric”) approaches rather than visual rating scores. Measurement methods have included manual delineation of anatomical regions of interest, as well as automated or semi-automated volumetry, although with varying protocols for anatomical delineation for segmentation.18 More recent methods allow for the parcellation of brain components into grey matter (GM) white matter (WM) and CSF and for automated voxel level quantification.29,30 Methods for detecting and quantifying white matter vascular disease burden and mitigating the effects of severe WM damage on the success of the volume quantification algorithm, will be particularly important in the dementia and ageing populations.31
Several commercial software solutions for clinically applied global and regional brain volume analysis are already available and in use, not only for dementia imaging but also for other neurological conditions such as stroke and multiple sclerosis. There is growing clinical interest in these tools. It is however important to recognise that regulatory marking (e.g. CE in the European Economic Area, or United States Food and Drug Administration) does not necessarily mean that a solution has been fully validated. Regulatory priorities are to demonstrate that the solution reliably produces reproducible results and is therefore no direct risk of harm to patients, so the process largely prioritises documentation and framework implementation for device deployment. Clinical import or efficacy, which are key parts of the QNI framework, are not the focus of regulatory approval and therefore in isolation CE marking may provide false reassurance regarding the appropriateness of introducing a product into clinical use.32,33
Step 3: communicating QIB results
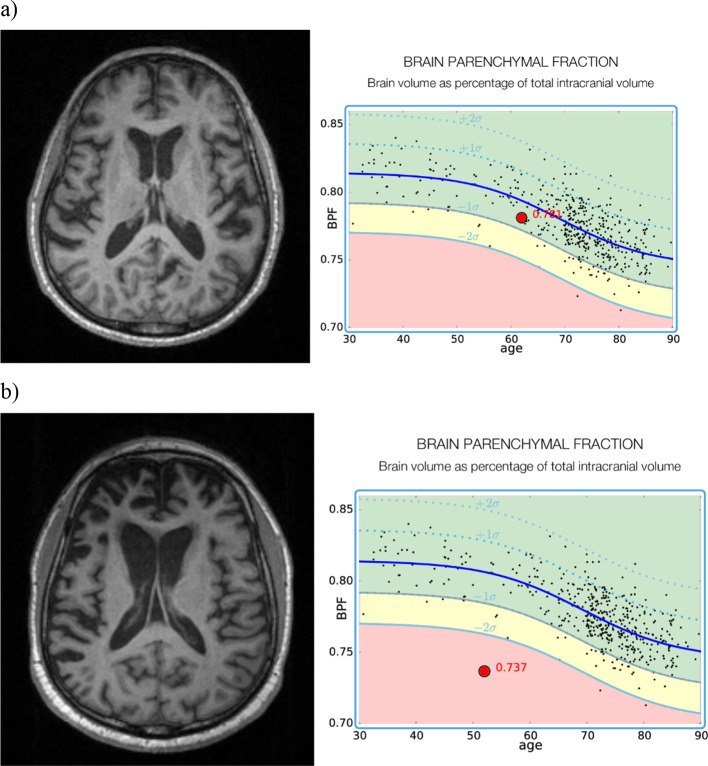
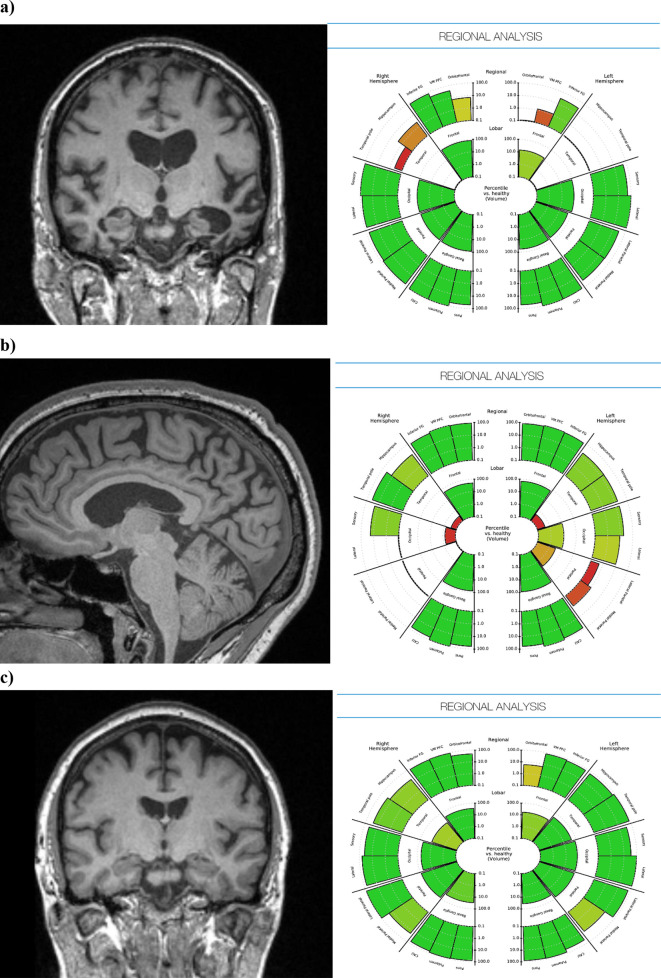
Presenting the quantitative outputs of these tools in a clinically meaningful way is key to their translational success, and to this end we have developed our own solution, one of many possibilities, for graphical representations of brain volumetry for use in dementia reporting. We provide our own example of a data presentation option in Figures 1 and 2, which is currently undergoing QNI framework validation. The graph in Figure 1 displays single-subject brain parenchymal fraction (BPF) values, visualized against an age-dependent normative dataset of 468 radiologically normal control subjects (age 30–90, median 69). BPF was derived for all using Geodesic Information Flows, GIF.34 The subject’s BPF is superimposed along the normative curve for easy comparison with the predicted normative range for individuals of the same age. In Figure 2, we provide an example of how individual subject’s volumetric data can be represented in an easily accessible way by lobe and pre-defined anatomic sub region, in an adaptation of a "bullseye" graphical display previously published,35 modified to display lobar and sublobar regions as a percentile of the GM volume normative value in each labelled region (Figure 2).
Figure 1. .
A normative data set of BPF has been generated from 468 normal control subjects aged 30–90 years. Mean and standard deviation BPF are shown with the solid and dotted blue lines. The subject’s BPF (large red dot) is placed along the normative curve for easy comparison with normal control subjects. Examples shown are of (a) normal control; (b) a subject with FTD. BPF, brain parenchymal fraction; FTD, frontotemporal dementia.
Figure 2. .
Patient-specific anatomical volumes and respective normative data can also be generated by brain region and presented as an easily interpreted and clinically useful graphic report. Examples from patients with (a) established bilateral medial temporal atrophy, (b) posterior cortical atrophy, and (c) healthy appearing brain.
Step 4: validation
The validation of brain segmentation methods is challenging, due to the absence in general of ground truth data: most validation studies have been based on cross-validation with the performance of alternative automated techniques, using data from the same source and with the same restrictions (e.g. single-site and vendor, precise scanning parameters). For repurposing research-developed QIBs into robust neuroradiological tools, it is important to note that most QIB research has focussed on group-level discrimination, which does not necessarily mean that the technique under consideration will offer sufficient sensitivity to support single-subject classification inference.36,37 Another challenge is that much of the available data used for the development and validation of QIBs in controlled research studies may poorly represent relevant clinical populations by neglecting comorbidities, socioeconomic status and education,38 i.e. the real-world scenario of patients with neurological comorbidities and other structural brain abnormalities.
Step 5: workflow integration
A software platform including image data identification and routing functionality is required to support integration of the QIB analysis and report generation into the hospital electronic information systems, including the PACS. A means of identifying examination images series appropriate for analysis is required, in our case implanted using customized DICOM series labels. Careful consideration should be given to the problem of clinical case stratification, so that reports are generated and interpreted in the appropriate context of conventional imaging and clinical history, and unwarranted quantitative reports are not generated. Introduction of QIBs into clinical dementia reporting may require modifications to the referral process. DICOM image tags could be queried by the quantification module to ensure that the report is generated for the correct compliant (in our case three-dimensional T 1 weighted volume) series. In our design, the quantitative report will appear as an additional series within PACS to preserve its place in the patient’s record.
Step 6: in-use evaluation
This should initially occur at a departmental level, ideally with engagement overseen by a key senior neuroradiologist. Initial training and technical systematic evaluation should be followed by a small pilot evaluation involving experienced radiologists who are familiar with dementia imaging reporting. They should report as per their normal routine, and by comparison corroborate the consistency and reliability of the quantitative report. Feedback should be gathered on any discrepancy or technical difficulty encountered. Following this pilot, all reporting radiologists in the department would be expected to adopt the report as part of their dementia imaging assessment. This prerequisites training and engagement not only of the radiologist team but also of the referring clinical teams. Audit of reporting efficiency and patient management pathway timelines would provide measures of service impact in comparison to previous practice. Ultimately, higher-level in-use evaluation of the outcome benefits of quantitative reporting will require larger-scale, multicentre studies once adoption by single centres has become well established.
Outlook
The QNI framework seeks to address the many and varied challenges that exist to translation of QIB reporting to the clinical setting. There remain some fundamental obstacles to translating these promising imaging biomarkers into clinical practice, and the QNI framework provides a structured technical and clinical validation process to address these. Lack of large-scale and rigorous technical and clinical validation, and over reliance on CE marking for quick commercial deployment, are major potential pitfalls in the field. The greatest challenge may be the circular problem of establishing clear evidence of clinical and socioeconomic benefit, without prior wide-scale adoption. This is especially challenging for quantification of conditions that do not currently have disease-modifying treatments available, as ground truth is not known, and other indicators must be relied on when assessing the added value of a diagnostic aid.
Imaging and clinical biomarkers are adapting our perception of dementia from a largely clinical and post-mortem diagnosis of exclusion to a more systematic biological paradigm. It is important to balance the potential that these imaging biomarkers hold with the realisation that, within the dementia field, they are still in the early stages of validation. We anticipate that the QNI framework will expedite this validation and facilitate incorporation of QIBs for dementia into clinical reporting workflows, to support radiological assessment and positively impact on patient management. There is growing potential for QIBs to find application across many of the spectrum of indications within neuroradiological practice, providing objective quantification of for example tumour volumes, lesion load in multiple sclerosis, and signal hyperintensity in hippocampal sclerosis. Looking ahead, potential developments include clinical introduction of multimodal imaging QIBs, radiomic modelling and big data mining, which, when combined with other types of patient data, may allow for predictive modelling of clinical progression39 and even delivery of individualized precision healthcare across many disease areas.40 The QNI framework could provide the common basis for the clinical translation of these developing methods in a similar way to current volumetric segmentation techniques.
Expediting these promising developments will require consensus upon and adoption of a robust translational pathway, exemplified in our QNI framework. Collaborative approaches to the technical challenges of protocol harmonization, data sharing, and algorithm development will underpin these developments, and there remains the need to afford substantially higher priority to thorough pre- and in-use clinical evaluation of quantitative techniques in the hands of practising neuroradiologists.
Footnotes
Acknowledgements: The QNI project is supported by funding from the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Tarek Yousry, John Thornton and Frederik Barkhof have contributed equally to this study and should be considered as senior authors.
Contributor Information
Olivia Goodkin, Email: o.goodkin@ucl.ac.uk.
Hugh Pemberton, Email: h.pemberton@ucl.ac.uk.
Sjoerd B Vos, Email: s.vos@ucl.ac.uk.
Ferran Prados, Email: f.carrasco@ucl.ac.uk.
Carole H Sudre, Email: carole.sudre@kcl.ac.uk.
James Moggridge, Email: j.moggridge@nhs.net.
M. Jorge Cardoso, Email: m.jorge.cardoso@kcl.ac.uk.
Sebastien Ourselin, Email: sebastien.ourselin@kcl.ac.uk.
Sotirios Bisdas, Email: s.bisdas@ucl.ac.uk.
Mark White, Email: mark.white4@nhs.net.
Tarek Yousry, Email: t.yousry@ucl.ac.uk.
John Thornton, Email: john.thornton@ucl.ac.uk.
Frederik Barkhof, Email: f.barkhof@ucl.ac.uk.
REFERENCES
- 1. Sullivan DC, Obuchowski NA, Kessler LG, Raunig DL, Gatsonis C, Huang EP, et al. Metrology standards for quantitative imaging biomarkers. Radiology 2015; 277: 813–25. doi: 10.1148/radiol.2015142202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bosco P, Redolfi A, Bocchetta M, Ferrari C, Mega A, Galluzzi S, et al. The impact of automated hippocampal volumetry on diagnostic confidence in patients with suspected Alzheimer's disease: a European Alzheimer's disease Consortium study. Alzheimers Dement 2017; 13: 1013–23. doi: 10.1016/j.jalz.2017.01.019 [DOI] [PubMed] [Google Scholar]
- 3. REGULATION (EU) 2017/745 OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL of 5 April 2017 on medical Devices, amending Directive 2001/83/EC, regulation (EC) NO 178/2002 and regulation (EC) NO 1223/2009 and repealing Council directives 90/385/EEC and 93/42/EEC. Off J Eur Union 2017: 60. [Google Scholar]
- 4. Pesapane F, Volonté C, Codari M, Sardanelli F. Artificial intelligence as a medical device in radiology: ethical and regulatory issues in Europe and the United States. Insights Imaging 2018; 9: 745–53. doi: 10.1007/s13244-018-0645-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clarke LP, Sriram RD, Schilling LB. Imaging as a biomarker: standards for change measurements in therapy workshop summary. Acad Radiol 2008; 15: 501–30. [DOI] [PubMed] [Google Scholar]
- 6. Vernooij MW, Pizzini FB, Schmidt R, Smits M, Yousry TA, Bargallo N, et al. Dementia imaging in clinical practice: a European-wide survey of 193 centres and conclusions by the ESNR Working group. Neuroradiology 2019; 61: 633–42. doi: 10.1007/s00234-019-02188-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vernooij MW, Jasperse B, Steketee R, Koek M, Vrooman H, Ikram MA, et al. Automatic normative quantification of brain tissue volume to support the diagnosis of dementia: a clinical evaluation of diagnostic accuracy. Neuroimage Clin 2018; 20(July): 374–9. doi: 10.1016/j.nicl.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klöppel S, Yang S, Kellner E, Reisert M, Heimbach B, Urbach H, et al. Voxel-wise deviations from healthy aging for the detection of region-specific atrophy. Neuroimage Clin 2018; 20: 851–60. doi: 10.1016/j.nicl.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Winblad B, Amouyel P, Andrieu S, Ballard C, Brayne C, Brodaty H, et al. Defeating Alzheimer's disease and other dementias: a priority for European science and society. Lancet Neurol 2016; 15: 455–532. doi: 10.1016/S1474-4422(16)00062-4 [DOI] [PubMed] [Google Scholar]
- 10. Cunningham EL, McGuinness B, Herron B, Passmore AP, Dementia PAP. Dementia.. Ulster Med J 2015; 84: 79–87. [PMC free article] [PubMed] [Google Scholar]
- 11. Braak H, Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol Aging 1995; 16: 271–8 discussion 278-84. doi: 10.1016/0197-4580(95)00021-6 [DOI] [PubMed] [Google Scholar]
- 12. Risacher SL, Saykin AJ. Neuroimaging biomarkers of neurodegenerative diseases and dementia. Semin Neurol 2013; 33: 386–416. doi: 10.1055/s-0033-1359312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harper L, Barkhof F, Scheltens P, Schott JM, Fox NC. An algorithmic approach to structural imaging in dementia. Journal of Neurology, Neurosurgery & Psychiatry 2014; 85: 692–8. doi: 10.1136/jnnp-2013-306285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wattjes MP. Structural MRI. Int Psychogeriatr 2011; 23 Suppl 2(S2): S13–S24 Sep 15. doi: 10.1017/S1041610211000913 [DOI] [PubMed] [Google Scholar]
- 15. Staffaroni AM, Elahi FM, McDermott D, Marton K, Karageorgiou E, Sacco S, et al. Neuroimaging in dementia. Semin Neurol 2017; 37: 510–37. doi: 10.1055/s-0037-1608808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McEvoy LK, Brewer JB. Quantitative structural MRI for early detection of Alzheimer's disease. Expert Rev Neurother 2010; 10: 1675–88. doi: 10.1586/ern.10.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salvatore C, Cerasa A, Castiglioni I. Mri characterizes the progressive course of AD and predicts conversion to Alzheimer's dementia 24 months before probable diagnosis. Front Aging Neurosci 2018; 10: 135. doi: 10.3389/fnagi.2018.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caroli A, Frisoni GB. Quantitative evaluation of Alzheimer’s disease. Expert Rev Med Devices 2009; 6: 569–88. doi: 10.1586/erd.09.35 [DOI] [PubMed] [Google Scholar]
- 19. Harper L, Fumagalli GG, Barkhof F, Scheltens P, O'Brien JT, Bouwman F, et al. Mri visual rating scales in the diagnosis of dementia: evaluation in 184 post-mortem confirmed cases. Brain 2016; 139(Pt 4): 1211–25. doi: 10.1093/brain/aww005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC, et al. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res 2009; 6: 347–61. doi: 10.2174/156720509788929273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whitwell JL, Shiung MM, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, et al. Mri patterns of atrophy associated with progression to AD in amnestic mild cognitive impairment. Neurology 2008; 70: 512–20. doi: 10.1212/01.wnl.0000280575.77437.a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, et al. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging 2004; 25: 1197–203. doi: 10.1016/j.neurobiolaging.2003.12.007 [DOI] [PubMed] [Google Scholar]
- 23. Devanand DP, Liu X, Tabert MH, Pradhaban G, Cuasay K, Bell K, et al. Combining early markers strongly predicts conversion from mild cognitive impairment to Alzheimer's disease. Biol Psychiatry 2008; 64: 871–9. doi: 10.1016/j.biopsych.2008.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cedazo-Minguez A, Graff C, Johansson G, Jönsson L, Kivipelto M, Tjernberg LO, et al. The Lancet Neurology Commission Defeating Alzheimer’s disease and other dementias: a priority for European science and society.. The Lancet Neurology 2016; 15 Vol.. [DOI] [PubMed] [Google Scholar]
- 25. Scheltens P, Leys D, Barkhof F, Huglo D, Weinstein HC, Vermersch P, et al. Atrophy of medial temporal lobes on MRI in "probable" Alzheimer's disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry 1992; 55: 967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pereira JB, Cavallin L, Spulber G, Aguilar C, Mecocci P, Vellas B, et al. Influence of age, disease onset and ApoE4 on visual medial temporal lobe atrophy cut-offs. J Intern Med 2014; 275: 317–30. [DOI] [PubMed] [Google Scholar]
- 27. Frisoni GB, Boccardi M, Barkhof F, Blennow K, Cappa S, Chiotis K, et al. Strategic roadmap for an early diagnosis of Alzheimer's disease based on biomarkers. Lancet Neurol 2017; 16: 661–76. doi: 10.1016/S1474-4422(17)30159-X [DOI] [PubMed] [Google Scholar]
- 28. Ten Kate M, Barkhof F, Boccardi M, Visser PJ, Jack CR, Lovblad K-O, et al. Clinical validity of medial temporal atrophy as a biomarker for Alzheimer's disease in the context of a structured 5-phase development framework. Neurobiol Aging 2017; 52: 167–82. doi: 10.1016/j.neurobiolaging.2016.05.024 [DOI] [PubMed] [Google Scholar]
- 29. Despotović I, Goossens B, Philips W. Mri segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med 2015; 2015: 1–23. doi: 10.1155/2015/450341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuda H. Mri morphometry in Alzheimer's disease. Ageing Res Rev 2016; 30: 17–24. doi: 10.1016/j.arr.2016.01.003 [DOI] [PubMed] [Google Scholar]
- 31. Chard DT, Jackson JS, Miller DH, Wheeler-Kingshott CAM. Reducing the impact of white matter lesions on automated measures of brain gray and white matter volumes. J Magn Reson Imaging 2010; 32: 223–8. doi: 10.1002/jmri.22214 [DOI] [PubMed] [Google Scholar]
- 32. Feldman MD, Petersen AJ, Karliner LS, Tice JA. Who is responsible for evaluating the safety and effectiveness of medical devices? the role of independent technology assessment. J Gen Intern Med 2008; 23(Suppl 1): 57–63 23 Suppl. doi: 10.1007/s11606-007-0275-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mishra S. FDA, CE mark or something else?-Thinking fast and slow. Indian Heart J 2017; 69: 1–5. doi: 10.1016/j.ihj.2016.11.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cardoso MJ, Modat M, Wolz R, Melbourne A, Cash D, Rueckert D, et al. Geodesic information flows: Spatially-Variant graphs and their application to segmentation and fusion. IEEE Trans Med Imaging 2015; 34: 1976–88. doi: 10.1109/TMI.2015.2418298 [DOI] [PubMed] [Google Scholar]
- 35. Sudre CH, Gomez Anson B, Davagnanam I, Schmitt A, Mendelson AF, Prados F, et al. Bullseye's representation of cerebral white matter hyperintensities. Journal of Neuroradiology 2018; 45: 114–22. doi: 10.1016/j.neurad.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arbabshirani MR, Plis S, Sui J, Calhoun VD. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. Neuroimage 2017; 145(Pt B): 137–65. doi: 10.1016/j.neuroimage.2016.02.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brewer JB. Fully-automated volumetric MRI with normative ranges: translation to clinical practice. Behav Neurol 2009; 21: 21–8. doi: 10.1155/2009/616581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teipel S, Kilimann I, Thyrian JR, Kloppel S, Hoffmann W. Potential role of neuroimaging markers for early diagnosis of dementia in primary care. Curr Alzheimer Res 2018; 15: 18–27. doi: 10.2174/1567205014666170908093846 [DOI] [PubMed] [Google Scholar]
- 39. Rhodius-Meester HFM, Liedes H, Koikkalainen J, Wolfsgruber S, Coll-Padros N, Kornhuber J, et al. Computer-Assisted prediction of clinical progression in the earliest stages of AD. Alzheimers Dement 2018; 10: 726–36. doi: 10.1016/j.dadm.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–77. doi: 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]