Abstract
BACKGROUND:
The human placenta has traditionally been viewed as sterile and microbial invasion of this organ has been associated with adverse pregnancy outcomes. Yet, recent reports employing sequencing techniques have reported that the human placenta at term contains a unique microbiota. These conclusions have been based on the results derived from sequencing placental samples. However, such an approach carries the risk of capturing background contaminating DNA (from DNA extraction kits, PCR reagents, and laboratory environments) when studying low microbial biomass samples.
OBJECTIVE:
To determine whether the human placenta delivered at term in patients without labor undergoing Cesarean delivery harbors a resident microbiota (“the assemblage of microorganisms present in a defined niche or environment”).
STUDY DESIGN:
This cross-sectional study included placentas from 29 women who had a Cesarean delivery without labor at term. The study also included technical controls to account for potential background contaminating DNA, including in DNA extraction kits, PCR reagents, and laboratory environments. Bacterial profiles of placental tissues and background technical controls were characterized and compared using bacterial culture, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomic surveys.
RESULTS:
1) Twenty-eight of 29 placental tissues had a negative culture for microorganisms. The microorganisms retrieved by culture from the remaining sample were likely contaminants because corresponding 16S rRNA genes were not detected in the same sample; 2) quantitative real-time PCR did not indicate greater abundances of bacterial 16S rRNA genes in placental tissues than in technical controls. Therefore, there was no evidence of the presence of microorganisms above background contamination of reagents in placentas; 3) 16S rRNA gene sequencing did not reveal consistent differences in the composition or structure of bacterial profiles between placental samples and background technical controls; and 4) most of the bacterial sequences obtained from metagenomic surveys of placental tissues were from cyanobacteria, aquatic bacteria, or plant pathogens, and were thus not likely to be present in the human placenta. Coprobacillus, which constituted 30.5% of the bacterial sequences obtained through metagenomic sequencing of placental samples, was not identified in any of the 16S rRNA gene surveys of these samples. These observations cast doubt as to whether this organism is really present in the placenta of patients at term not in labor.
CONCLUSIONS:
A resident microbiota could not be identified in human placentas delivered at term from women without labor using multiple modes of microbiologic inquiry. A consistently significant difference in the abundance and/or presence of a microbiota between placental tissue and background technical controls could not be found. All cultures of placental tissue except one did not yield bacteria. Incorporating technical controls for potential sources of background contaminating DNA for studies of low microbial biomass samples, such as the placenta, is necessary for deriving reliable conclusions.
Keywords: bacterial culture, in utero colonization, low microbial biomass samples, microbiome, microorganism, bacteria, PCR, next-generation sequencing, placenta, pregnancy, sterile tissues, sterile womb, reagent contamination
Condensation sentence:
A microbiota could not be demonstrated using multiple modes of microbiologic inquiry in placentas of women who delivered at term without labor
INTRODUCTION
Culture-independent sequencing technologies provide insight into the diversity of microbial communities inhabiting the human body1-3, as well as other ecosystems such as soil4, 5 and oceans6-8. Studies derived from the Human Microbiome Project indicate that different human body sites are populated by site-specific microbiota (“the assemblage of microorganisms present in a defined niche or environment”9)1, 2. For example, the microbiota of the vagina10-14 is different from that of the gut15, 16 and oral cavity17, 18. The microbial burden of each of these body sites is large19-21, and samples derived from these niches are considered to have a high microbial biomass21, 22. Results obtained with sequencing technologies of these samples are largely qualitatively consistent with those derived from cultivation techniques (i.e., while molecular surveys of these sites typically capture far more microbial diversity than culture-based surveys do, many of the prominent microbes in the molecular surveys have also been recovered through culture from these same sites)23-27. In contrast, samples derived from sites with a low microbial biomass can give results which are difficult to distinguish from DNA present in reagents used for extraction, amplification, and sequence library preparation for molecular microbiology studies22, 28-31.
Several reports have demonstrated that commercially-available kits used to characterize the microbiota contain microbial DNA similar to that found in soil or water samples28, 29, and that this can affect the results of studies of low microbial biomass samples using 16S rRNA gene amplicon or metagenomic sequencing22, 28, 29, 31-33. DNA contamination of reagents is unavoidable, given the ubiquity of microorganisms and the fact that many reagents are products of microbial processes and engineering30. Therefore, the claim that body sites with a low microbial biomass have bacteria based on the analysis of 16S rRNA gene surveys and metagenomic studies requires rigorous exclusion of reagent contamination to avoid experimental artifacts and incorrect conclusions22, 30, 31.
The challenge of studying low microbial biomass samples is particularly important in the female reproductive tract, as several investigators have viewed the endometrial cavity34, 35, amniotic cavity36-57, and placenta32, 58, 59 of healthy women as “sterile”60-63. With the application of molecular microbiologic techniques, the sterility of these sites outside cases of infection has been questioned64-82, and functional hypotheses for potential mutualistic relationships between microbiota and their human hosts are being considered78, 83-87.
With respect to the placenta, microorganisms can invade the amnion and chorion88-96, as well as the villous tree97-116. This is often associated with complications of pregnancy, such as preterm labor117-142, preterm PROM143-146, cervical insufficiency147-154, clinical chorioamnionitis155-172, and congenital infections97-102, 111, 113, 115, 129, 173-187. The concept that most placentas have a microbial community (“The Placenta Harbors a Unique Microbiome”) emerged after a pioneering report which used sequencing techniques to analyze a large number of placentas64. Shortly after that report, questions were raised about this claim188; yet, other investigators using high-throughput sequencing strategies have also reported the presence of a microbiota in the placenta65-75. The interpretation of these data has become a subject of controversy22, 32, 63, 189, 190, given the recognition that reagents used in molecular microbiologic techniques have their own microbiome (termed the “kitome”)22, 28-30, 32, 191. Recently, investigators have called for the application of rigorous and systematic methods to address DNA contamination in low microbial biomass samples22, 30, 31.
The objective of this study was to determine whether a microbiota exists in term placentas without labor delivered by Cesarean section, using multiple complementary modes of microbiologic inquiry such as cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomic approaches.
MATERIALS AND METHODS
Study design
This was a cross-sectional study in which the placenta was sampled from women not in labor at term (February - June 2016). The inclusion criteria were: 1) Cesarean delivery without labor at term (≥ 38 weeks); 2) singleton gestation; and 3) no antibiotic administration in the month prior to delivery, as determined by history and review of medical records. Each subject did, however, receive intraoperative prophylaxis prior to Cesarean delivery [cefazolin or, if allergic, gentamicin and clindamycin], given the evidence that antimicrobial administration reduces perioperative complications192-194. Exclusion criteria consisted of multiple gestations, preterm delivery, fetal anomalies, and evidence of clinical infection.
The presence of bacteria in the placenta was determined using: 1) cultivation; 2) 16S rRNA gene quantitative real-time PCR; 3) 16S rRNA gene sequencing; and 4) metagenomic sequencing. Placental histopathological examinations were conducted according to protocols established by the Perinatology Research Branch59, 93. The collection of samples and their utilization for research was approved by the Human Investigations Committee of Wayne State University and the Institutional Review Board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and all subjects provided written informed consent for participation.
Sample collection
Following Cesarean delivery, the placenta was placed in a sterile collection container with a sealed cover (Medline Standard C-Section Pack-LF; Mundelein, IL) within the sterile operating field. The placenta was taken directly to a biological safety cabinet within one of two nearby rooms in Hutzel Women’s Hospital, wherein study personnel (ADW, KRT), donning sterile surgical gowns, full hoods, and powder-free exam gloves (Kimberly-Clark; Roswell, GA), and using individually packaged, sterile, disposable scalpels (Surgical Design; Lorton, VA), forceps (TWD Scientific; Pleasant Prairie, WI), and surgical scissors (Sklar Instruments; West Chester, PA) collected a 1.5 cm2 core sample through the placenta (i.e., amnion and chorionic plate through to basal plate). The tissue sample was taken halfway between the umbilical cord insertion point and the edge of the placental disk, along the line representing the longest distance from the cord insertion point to the edge of the disk. The tissue sample was transferred to a sterile polystyrene Petri dish (Fisher Scientific, FB0875712; Waltham, MA) and divided into three approximately equal aliquots, with each aliquot traversing the amnion, chorionic plate, villous tree, and basal plate. One aliquot was placed in a sterile 5.0 ml conical tube (Denville Scientific; Holliston, MA) on ice and stored at −80° C within one hour of initial placental collection. The two remaining aliquots were placed into Anaerobic Transport Medium Surgery Packs (Anaerobe Systems; Morgan Hill, CA) and 0.85% sterile saline solution tubes (Thermo Scientific; Waltham, MA) for anaerobic and aerobic cultures, respectively.
Bacterial culture of placental tissues
Placental tissue aliquots within anaerobic and aerobic transport containers were delivered to the Detroit Medical Center University Laboratories Microbiology Core, wherein they were processed the same day. To assess viability of a placental microbiota, placental tissues were homogenized and inoculated on growth media (trypticase soy agar with 5% sheep blood, chocolate agar, MacConkey’s agar) under aerobic and anaerobic conditions, and used in an assay for genital mycoplasmas. Detailed information on the cultivation protocols and taxonomic characterization of resultant bacterial cultivars is available in Supplemental Methods (Section 1).
DNA extraction from placental tissues
DNA extraction was performed to identify bacteria with molecular microbiologic techniques. During the process, study personnel wore sterile surgical gowns and gloves, surgical masks (Kimberly-Clark Soft Touch II; Roswell, GA), and used individually packaged, sterile, and disposable scalpels and forceps (TWD Scientific, DF 8988P-SPT; Pleasant Prairie, WI). For each placental tissue specimen, the chorionic plate (including a minimal amount of villous tissue) was separated from the placental villous tree, which remained attached to the basal plate. Genomic DNA was extracted from blocks of tissue containing: 1) the amnion and the chorionic plate, and 2) the villous tree and basal plate. The extraplacental chorioamniotic membranes were not sampled. DNA was extracted from the placental tissues (0.1 to 0.2 g) and background technical controls using the MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories, 12888), according to the manufacturer’s protocol. The DNA extraction kit used, and the mass of placental tissue from which DNA was extracted, were similar to those used in prior studies addressing the issue of a placental microbiota32, 64. Background technical controls included extractions performed on: 1) DNA extraction kits without placental tissue, processed exactly as the placental samples (N = 6); 2) extraction kits whose bead tubes had been exposed to a biological safety cabinet for 20 minutes during placental biopsy collection or processing (N = 16 samples from three biosafety cabinets); and 3) extraction kits whose bead tubes had been exposed for 20 minutes to an operating room or microbiology laboratory utilized in this study (N = 21 samples from three operating rooms and three laboratories). These control samples therefore represented either five or six technical controls reflecting each potential source of background DNA contamination (i.e. extraction kits, three biosafety cabinets, three laboratories, and operating rooms), with the three contiguous operating room environments being treated as a single potential contamination source. DNA concentrations of placental tissue and background technical control samples were 42.0 ± 18.5 SD ng/μl and ≤ 0.03 ng/μl, respectively. Purified DNA was stored at −20° C.
16S rRNA gene sequencing of DNA extracted from placental tissue and background technical control samples
The 16S rRNA gene is widely used as a phylogenetic marker to identify bacterial types present in clinical samples. A table of polymerase chain reaction (PCR) 16S rRNA gene primers used in this study is available in the Supplemental Methods (Section 2). We initially used the standard PCR and Illumina MiSeq (San Diego, CA) protocols described below; however, this approach did not produce sufficient quantities of amplified DNA to generate sequence libraries from placental tissue or technical controls, and thus for 16S rRNA gene profile comparisons (see Supplemental Methods Section 3; Supplemental Figure 1). Therefore, due to the very low microbial biomass in these human tissue samples, purified bacterial DNA was amplified using a nested PCR approach195, 196.
Nested PCR has been recently used to characterize low biomass microbiota in the lungs of mice197, sheep198, and chickens199, and in the middle ear fluid of children200, 201. The first round in the nested PCR process included 20 cycles with each reaction containing 0.6 μM each of the 16S rRNA gene broad-range primers 27f-CM (5’-AGA GTT TGA TCM TGG CTC AG-3’) and 1492R (5’-ACG GCT ACC TTG TTA CGA CTT -3’)202, 203, 12.5 μl of 2X GoTaq Green Master Mix (Promega, Madison, WI), and 3.0 μl purified DNA. Thermocycling was initiated by a five-minute incubation at 95° C. Cycling parameters were 94° C for 30 seconds, 50° C for 30 seconds, and 72° C for 120 seconds. Products were then diluted 1:15 in nuclease-free water (Promega, Madison, WI).
Amplification and sequencing of the V4 region of the 16S rRNA gene was performed at the University of Michigan’s Center for Microbial Systems (Ann Arbor, MI) using the dual indexing sequencing strategy developed by Kozich et al204. Sequencing was performed on the Illumina MiSeq platform, using a MiSeq Reagent Kit V2 500 cycles (Illumina MS102-2003), according to the manufacturer’s instructions with modifications found in Kozich et al204, 205. AccuPrime High Fidelity Taq (Life Technologies 12346094) was used instead of AccuPrime Pfx SuperMix. Each PCR reaction (20 μl) contained 1.0 μM of each primer, 2.5 μl template DNA, 0.15 μl AccuPrime HiFi Polymerase, and DNase-free water to produce a final volume of 20 μl.
PCR was performed using the following conditions: 95 °C for two minutes, followed by 30 cycles of 95 °C for 20 seconds, 55 °C for 30 seconds, and 72 °C for five minutes, with an additional elongation at 72 °C for 10 minutes. Sequencing libraries were prepared according to Illumina’s protocol for Preparing Libraries for Sequencing on the MiSeq (15039740 Rev. D) for 2nM or 4nM libraries. FASTQ files were generated for paired end reads. Sample-specific MiSeq run files have been deposited on the NCBI Sequence Read Archive (BioProject ID PRJNA397876).
Processing of 16S rRNA gene sequence data
Mothur software (v1.39.5) was used to assemble paired-read contiguous sequences, trim, filter, and align sequences, identify and remove chimeras, assign sequences to bacterial taxonomies, and cluster sequences into operational taxonomic units (OTUs) based on percent nucleotide similarity (97% and 99%)206. Detailed information on sequence processing is available in Supplemental Methods (Section 4).
Sequencing of DNA extracts of all samples and controls yielded 5,316,687 sequences. They clustered into 480 (209 singletons) and 35,503 (23,892 singletons) OTUs using 97% and 99% sequence similarity cutoffs, respectively. The mean number of sequences for the placental tissue and technical control samples was 50,783 (range 509 – 92,052) and 55,145 (2,572 – 111,361), respectively. All raw count data for this study are available as supplemental material (Supplemental Data 1).
Using an OTU nucleotide similarity cutoff of 97%, the Good’s coverage values of all but one placental sample exceeded 99.7%; the exception was 98.8% (sample 25AC). Good’s coverage values of all technical control samples exceeded 99.8%. For analyses of alpha diversity (microbial diversity within a sample), individual sample libraries were subsampled to the depth of the second least represented sample (1997 sequences), and the least represented sample (509 sequences for 25AC) was excluded. After subsampling for alpha diversity analyses, Good’s coverage values of placental and technical control samples exceeded 99.4%.
Quantitative real-time PCR (qPCR) of the 16S rRNA genes in DNA extracts of placental tissues and background technical controls
Bacterial DNA abundance within the samples was determined via quantitative real-time PCR (qPCR) amplification of the V1 – V2 region of the 16S rRNA gene as described by Dickson et al207, with minor modifications. These included the use of a degenerative forward primer (27f-CM: 5’-AGA GTT TGA TCM TGG CTC AG-3’) and a degenerate probe containing locked nucleic acids (+) (BSR65/17: 5’-56FAM-TAA +YA+C ATG +CA+A GT+C GA-BHQ1-3’). Amplifications were performed using an annealing temperature of 50°C to minimize amplification bias and to allow for a greater number of potential bacterial types, such as Lactobacillus and Gardnerella species203. Detailed information on the qPCR protocols are provided in the Supplemental Methods (Section 5).
Metagenomic sequencing of extracted DNA from placental samples and background technical controls
In contrast to sequencing surveys targeting a specific bacterial gene (e.g. 16S rRNA gene), a metagenomic survey entails sequencing all of the genes in a clinical sample and assigning the protein-coding genes of bacterial origin to particular bacterial taxa. Nine placental and 11 technical control samples underwent metagenomic sequencing using the Illumina HiSeq 4000, 150-base paired-end read protocol at the University of Michigan’s DNA Sequencing Core (Ann Arbor, MI). The placental samples included amnion & chorionic plate and villous tree & basal plate samples from each of four subjects (Subjects #14, 15, 22, and 30), and a villous tree & basal plate sample from one subject (Subject #19). The technical control samples included eight biological safety cabinet and three blank extraction kit samples. Metagenomic sequence data were processed using MG-RAST208. Bacterial taxonomic assignments were made using the GenBank database and the default MG-RAST parameters. Detailed information on the metagenomic sequencing and sequence data processing protocols are available in Supplemental Methods (Sections 6 & 7). All raw genus-level count data are available as supplemental material (Supplemental Data 2).
Secondary DNA extractions and molecular analyses of placental tissues
After the primary 16S rRNA gene sequencing analyses did not yield evidence of a placental microbiota (see Results), secondary analyses were conducted to ensure that the primary sequencing results were not due to cross-contamination between DNA extracted from placental tissues and background technical controls during processing, or exclusively due to the use of a nested PCR approach for bacterial DNA amplification.
Secondary DNA extractions were performed on the collective villous tree & basal plate portion of each of the 29 placental samples. The extraction protocol was the same as that described above except that at least four blank extraction kit controls were included in each of four rounds of extractions of the placental samples. Specifically, in the first three rounds of extractions, we processed eight placental and four technical control samples. In the fourth round, we processed five placental and five technical control samples. We additionally completed a fifth round of extractions composed entirely of 12 blank extraction kit controls. The blank extraction controls were not exposed to the atmospheres of the biological safety cabinets or the laboratories; they were processed exactly as the placental samples. DNA concentrations of placental tissue and blank extraction control samples were 56.0 ± 24.3 SD ng/μl and ≤ 0.03 ng/μl, respectively. Purified DNA was stored at −20° C.
The secondary DNA extractions were used for 16S rRNA gene sequencing using three amplification approaches: standard PCR, nested PCR, and touchdown PCR. For standard PCR, we aimed to generate the 16S rRNA gene profiles of DNA extracted from placental samples and background technical controls using 30, 35, and 40 amplification cycles. For nested PCR, we used a different primer pair for the first round of amplifications from that used in the primary analysis in this study, and aimed to generate 16S profiles for these samples using 5, 10, and 20 cycles in the first round of amplification. The different primer set, 341F/1061R (Supplemental Methods, Section 2) was used for the first round of nested PCR in an attempt to eliminate potential under-representation209 or selection against single bacterial species or groups of species210 in placental samples. Specifically, in silico studies querying these selected primers against taxonomically diverse sequences in three popular 16S rRNA gene databases (i.e. Greengenes211, RDP212, and SILVA213) have shown these selected primers to be highly conserved209, 214. Lastly, we aimed to generate 16S rRNA gene profiles for these samples using touchdown PCR215-217. Touchdown PCR can increase the sensitivity of PCR reactions in cases of very low microbial biomass and high background concentrations of host DNA215-217. Touchdown PCR was recently used to characterize the microbiota of the lung216-220, brain221, and blood219 of mice and humans. The PCR started with two minutes at 95° C, followed by (i) a touchdown PCR for 20 seconds at 95° C, 15 seconds at the annealing temperature (which was 60° C in the first cycle and dropped 0.3° C with each additional cycle), and five minutes at 72° C, and then (ii) 20 cycles of a standard PCR with 20 seconds at 95° C, 15 seconds at 55° C, and five minutes at 72° C, with a final elongation step at 72° C for 10 minutes.
All template DNA was diluted three-fold and transferred to the University of Michigan’s Center for Microbial Systems (Ann Arbor, MI) for sequence library processing. Sequence library construction was done using the dual indexing sequencing strategy developed by Kozich et al204. All reactions included 4 μl of template DNA. Based on visual inspection of amplified products using gel electrophoresis, sequence library generation was unsuccessful using 30 and 35 cycles of standard PCR. Sequence library generation was also unsuccessful using five and 10 cycles in the initial amplification round for nested PCR. Therefore, for the secondary 16S rRNA gene analyses, we generated sequence libraries for placental samples and background technical controls using 40 rounds of standard PCR, nested PCR with 20 initial rounds of amplification, and touchdown PCR. Sample-specific MiSeq run files have been deposited on the NCBI Sequence Read Archive (BioProject ID PRJNA397876), and all raw count data for the secondary analyses are provided as supplemental material (Supplemental Data 3). Sequence data processing for the secondary analyses proceeded as described above and in the Supplemental Methods (Section 4). The analyses presented here are of sequence data clustered into operational taxonomic units (OTUs) based on a percent nucleotide similarity of 97%. Results did not substantively differ using a 97% or 99% nucleotide similarity; therefore, only the results using 97% are presented for the secondary analyses. Raw data from sequence clustering based on a percent nucleotide similarity of 99% are provided in Supplemental Data 3.
The abundances of 16S rRNA gene copies in each placental sample and blank extraction control in this secondary analysis were determined using quantitative real-time PCR, as described above with minor alterations. Specifically, all samples were diluted three-fold prior to analysis, each sample reaction was performed in triplicate, and, if a sample did not pass the threshold of quantification by 40 cycles, its cycle of quantification (Cq) value was assigned as 40.
Statistical analysis
16S rRNA gene profile alpha and beta diversity:
Alpha diversity (i.e., diversity within a single sample) was assessed using Chao1 richness and Simpson heterogeneity indices222, 223. Alpha diversity indices were calculated using Mothur software (v1.39.5)206 and statistically evaluated using Kruskal-Wallis tests and Mann-Whitney pairwise comparisons, if applicable, in PAST (v2.17c)224-226.
Beta diversity (i.e., diversity between two samples) was assessed using Jaccard and Bray-Curtis similarity indices to reflect 16S rRNA gene profile composition and structure, respectively. Bray-Curtis values were calculated using percent relative abundance data for OTUs within samples. Beta diversity was visualized through Principal Coordinates Analyses (PCoA) and heat maps, and statistically evaluated using non-parametric MANOVA (NPMANOVA)225-227, with 9999 permutations. PCoA plots and NPMANOVA tests were conducted using PAST software (v2.17c and 3.14)224, and heat maps were generated via Matrix2png228.
Linear discriminant analysis effect size, or LEfSe229, was used to identify any OTUs that differ in relative abundance between the placental tissue and background technical control samples. Sourcetracker (v1.0)230 was used to estimate the percentage of OTUs in placental samples whose origin could be explained by their distribution in the background technical controls. For this analysis, we removed doubleton and singleton OTUs from the dataset.
16S rRNA gene qPCR:
To assess differences in 16S rDNA abundance between amnion & chorionic plate and villous tree & basal plate samples among the 29 subjects, differences in the cycle of quantification (Cq) were evaluated with paired t-tests. To assess variation in bacterial burden among individual sample types (i.e., amnion & chorionic plate, villous tree & basal plate, operating rooms and laboratories, biosafety cabinets, and blank DNA extraction kits), ANOVA tests, or Welch F tests in the case of unequal variances, were used for global assessment of variation in Cq, followed by Tukey’s pairwise comparisons225, 226. When data were not normally distributed, we used Kruskal-Wallis tests and Mann-Whitney pairwise comparisons. Statistical analyses were performed using PAST software (v2.17c)224.
RESULTS
Patient Characteristics
Table 1 describes the demographic and clinical characteristics of the patients in this study. None of the placentas included in this study presented fetal or maternal inflammatory lesions, defined as stage 3 and/or grade 2 maternal and/or fetal inflammatory responses59, 231.
Table 1.
Descriptive and clinical characteristics of the 29 subjects included in this study.
| Median | IQRa | |
|---|---|---|
| Age (yrs) | 29.0 | 25.5 – 33.0 |
| BMIb (kg/m2) | 32.8 | 24.7 – 36.1 |
| Parity | 2 | 1 – 2 |
| GAc at Delivery (wks) | 39.1 | 39.0 – 39.3 |
| Birthweight (g) | 3450 | 3063 – 3905 |
| Raced | ||
| African American | 21 (80.8 %) | |
| White | 5 (19.2 %) | |
| Clinical indications | ||
| Repeat elective Cesarean | 23 (79.3 %) | |
| Large for gestational age fetus | 3 (10.3 %) | |
| Breech presentation | 2 (6.9 %) | |
| Myoclonus dystonia | 1 (3.4%) |
Interquartile range
Body Mass Index; unreported for 7 subjects
Gestational Age
Race was self-reported by subjects; 3 subjects chose not to report
Bacterial culture of placental tissues
Twenty-eight of the 29 placental tissue samples did not yield any bacterial cultivars. One tissue sample (subject #25) yielded three colonies in the primary zone of the 5% sheep blood agar plate incubated aerobically: Bacillus circulans, Bacillus pumilus, and Brevibacterium casei. It did not yield colonies on other media under aerobic or anaerobic conditions or yield growth of genital mycoplasmas. Exact matches (i.e., 100% nucleotide similarity) to the V4 region of the 16S rRNA genes of the three isolates recovered on the sheep blood agar plate were not found among any of the sequences from the primary (13,766 sequences; Good’s coverage > 99.9 %) or the secondary (98,392 sequences; Good’s coverage > 99.9 %) MiSeq 16S rRNA gene surveys of subject 25’s placental tissues.
16S rRNA gene surveys of placental tissue and background technical control samples
Alpha diversity:
There was no variation in OTU richness among the amnion & chorionic plate samples and the room, hood, and blank extraction kit controls (Chao1 index; Kruskal-Wallis test; H = 4.114, p = 0.248), nor was there variation among the villous tree & basal plate samples and the various controls (H = 3.871, p = 0.274). There was also no variation in OTU heterogeneity between the placental and technical control samples (Simpson index; amnion & chorionic plate: H = 3.384, p = 0.336; villous tree & basal plate: H = 2.531, p = 0.470).
Beta diversity:
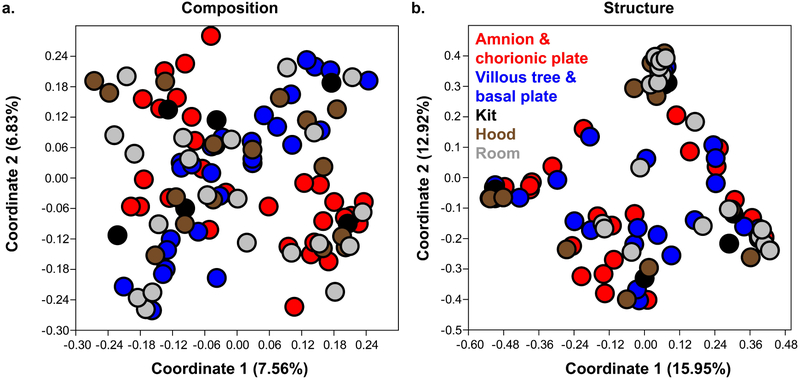
There was no variation in the composition or structure of 16S rRNA gene profiles among the three biological safety cabinets (NPMANOVA; Jaccard: F = 0.846, p = 0.781; Bray-Curtis: F = 0.880, p = 0.572), or among the different rooms used for sample processing (Jaccard: F = 0.882, p = 0.833; Bray-Curtis: F = 0.916, p = 0.602). Profile similarities among the amnion & chorionic plate and the villous tree & basal plate samples and the three different types of technical controls (i.e. blank extraction kits, biosafety cabinets, rooms) are illustrated in Figure 1. 16S rRNA gene profiles did not consistently vary among the amnion & chorionic plate samples, blank extraction kits, biological safety cabinets, and processing rooms (Figure 1; Table 2). Similarly, 16S rRNA gene profiles did not vary among the villous tree & basal plate samples, blank extraction kits, biological safety cabinets, and room controls (Figure 1; Table 2). Neither the 16S rRNA gene profiles of the amnion & chorionic plate samples nor those of the villous tree & basal plate samples differed from those of the blank extraction kits specifically (Table 2). These same patterns were found when using an OTU nucleotide similarity cutoff of 99% (Supplemental Figure 2; Supplemental Table 1).
Figure 1. Principal Coordinates Analyses (PCoA) illustrating similarity in 16S rRNA gene profiles among amnion & chorionic plate, villous tree & basal plate, and technical control samples:
a. Plot of similarity in profile composition among placental and control samples based on the Jaccard index; b. Plot of similarity in profile structure among placental and control samples based on the Bray-Curtis index. Operational taxonomic units (OTUs) were generated using a 97% sequence similarity cutoff and the primary 16S rRNA gene nested PCR data set.
Table 2. Non-parametric MANOVA (NPMANOVA) analyses showing lack of variation in 16S rRNA gene profiles among amnion & chorionic plate, villous tree & basal plate, and room, hood, and blank extraction kit technical control samples.
Operational taxonomic units (OTUs) were generated using a 97% sequence similarity cutoff. 16S profile composition and structure were characterized using Jaccard and Bray-Curtis indices, respectively. Results of overall global effect analyses are presented along with the results of pairwise comparisons involving placental samples. P-values for these permutation tests were not adjusted for multiple pairwise comparisons, as this can be overly conservative. However, for pairwise tests that were statistically significant, we do present the Bonferroni corrected p-value in parentheses.
| Composition | Structure | ||||
|---|---|---|---|---|---|
| F | p-value | F | p-value | ||
| Amnion & chorionic plate | Global | 1.080 | 0.261 | 1.128 | 0.270 |
| Rooms | 1.367 | 0.060 | 2.211 | 0.028 (0.077) | |
| Hoods | 1.310 | 0.108 | 1.190 | 0.275 | |
| Kits | 1.018 | 0.412 | 0.545 | 0.873 | |
| Villous tree & basal plate | Global | 1.051 | 0.335 | 1.222 | 0.189 |
| Rooms | 1.450 | 0.037 (0.223) | 2.513 | 0.007 (0.043) | |
| Hoods | 1.149 | 0.231 | 1.072 | 0.351 | |
| Kits | 0.944 | 0.552 | 0.875 | 0.529 | |
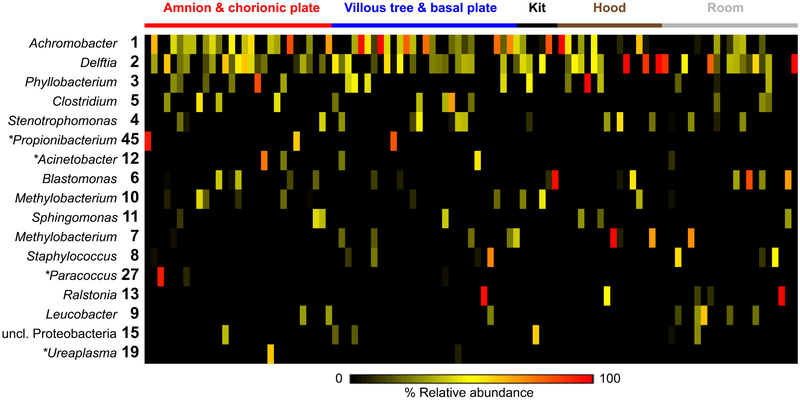
Sixteen of the 18 prominent OTUs (i.e., those having an average relative abundance ≥ 1%) among the placental samples were confidently classified at the genus level (Figure 2). These OTUs were Achromobacter, Delftia, Phyllobacterium, Clostridium, Propionibacterium, Stenotrophomonas, Acinetobacter, Blastomonas, Methylobacterium, Sphingomonas, Paracoccus, Ralstonia, Staphylococcus, Leucobacter, and Ureaplasma. These 18 prominent OTUs accounted for 90.0 and 86.4% of total sequences obtained from the placental tissue samples and background technical controls, respectively. Fourteen of these 18 prominent placental OTUs were also prominent among the control samples (Figure 2). The four exceptions (OTUs classified as Acinetobacter, Paracoccus, Propionibacterium, and Ureaplasma) were OTUs that were either widely present among the technical control samples but at low relative abundances or that were abundant in only one to a few placental tissue samples. A full description of the distribution and relative abundances of these OTUs among placental samples and technical controls is provided in Supplemental Results (Section 1).
Figure 2. Heat map illustrating similarity in percent relative abundances of prominent operational taxonomic units (OTUs) among placental samples and technical controls.
Prominent OTUs were defined as those having an average relative abundance ≥ 1% among the placental samples. OTUs were generated using a 97% sequence similarity cutoff and the primary 16S rRNA gene nested PCR data set. Asterisks indicate OTUs that were prominent in placental samples but not in controls.
Linear discriminant analysis effect size (LEfSe) indicated that four OTUs (classified as Achromobacter, Blastococcus, Methylobacterium, and Caldalkalibacillus) were more relatively abundant among the amnion & chorionic plate samples than the technical controls, and that three OTUs (classified as Achromobacter, Burkholderiales, and Herbaspirillum) were more relatively abundant among the villous tree & basal plate samples than the controls (Supplemental Figure 3). The distribution and relative abundances of these OTUs among placental samples and technical controls is discussed in detail in Supplemental Results (Section 2).
SourceTracker analyses indicated that a median of 99.7 (50.7 IQR) and 99.9% (8.2 IQR) of OTUs in the amnion & chorionic plate and villous tree & basal plate samples, respectively, could be confidently attributed to contaminating DNA in blank extraction kits, PCR reagents, and/or the rooms used for sample processing. Furthermore, when defining the core microbiota as those OTUs present in at least one-half of the samples of a particular sampling group69, 72, every core OTU in the amnion & chorionic plate and villous tree & basal plate samples was also a core OTU in the hood and blank extraction kit control samples (see Supplemental Results, Section 3).
Real-time qPCR assays of 16S rRNA gene copy abundances in the placental tissues and background technical controls
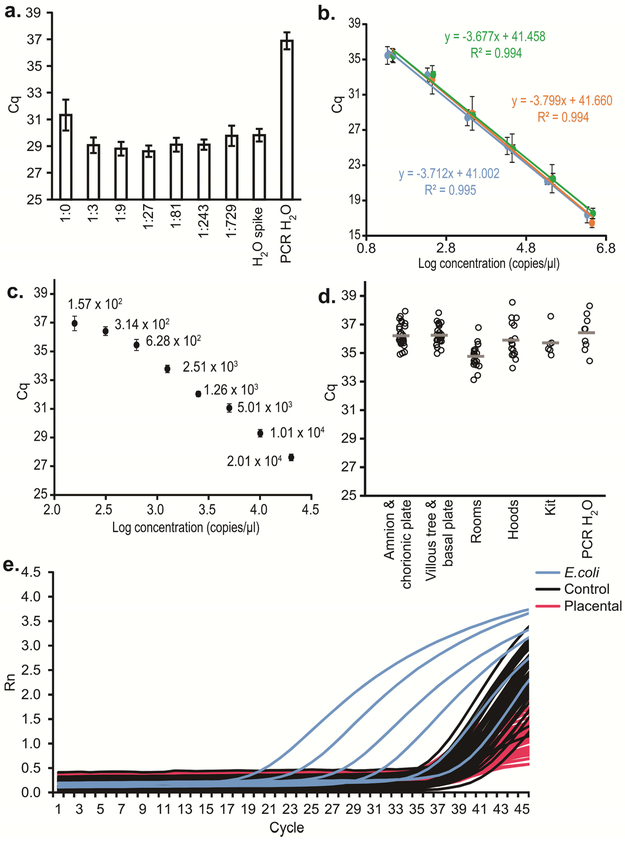
Analysis of cycle of quantification (Cq) values generated for broad-range standard curves included across all qPCR runs indicated that the average amplification efficiency of the assay was 85.44 ± 1.91% SD. The regression curves were linear over a range of 101 to 106 gene copies, with slopes ranging from −3.88 to −3.62 and R2 values ≥ 0.980 (Figure 3b). Analysis of Cq values generated for the narrow-range standard curve ranging from 2.01 × 104 to 1.57 × 102 revealed that standard deviation values reached 0.506 cycles for the most dilute replicate reactions (Figure 3c), indicating that the limits of detection and quantification for the assay were between 1.57 × 102 and 3.14 × 102 copies (Figure 3c).
Figure 3. Quantitative PCR (qPCR) analyses illustrating similarity in 16S rRNA gene abundance among amnion & chorionic plate, villous tree & basal plate, and technical control samples:
a. Comparison of quantification cycle (Cq) values (mean ± SD) of serially diluted placental genomic DNA samples spiked with equal concentrations (5.7 × 103 copies per reaction) of genomic DNA from Echerichia coli ATCC 25922, illustrating that amplification inhibition is eliminated by diluting samples with nuclease-free water by a factor of 1:3 or more; b. Standard curves for three 10-fold dilution series (2.82 × 106 – 2.82 × 101 copies, 2.12 × 106 – 2.12 × 101 copies, and 2.97 × 106 – 2.97 × 101 copies) of E. coli ATCC 25922 16S rDNA (mean Cq values across all qPCR runs); c. Standard curve for a 2-fold dilution series (mean Cq values) of E. coli ATCC 25922 DNA illustrating a limit of detection for the qPCR assay between 1.57 × 102 and 3.14 × 102 16S rDNA copies per reaction (20 μl), as indicated by a standard deviation of replicate dilution samples above 0.5 cycles; d. Comparison of mean 16S rDNA qPCR Cq values for placental and control samples; e. Amplification curves from placental samples, technical controls, and the serial dilution series of E. coli DNA described in Figure panel b.
Quantitative real-time PCR revealed that 16S rDNA abundances within the majority of the placental and background technical control samples were beyond the detection and quantification limits of the qPCR assay (Figure 3d,e). There were no differences in Cq between the amnion & chorionic plate and villous tree & basal plate samples (paired t-test; N=29, t = −0.4851, p = 0.631). For the background technical control samples, there was no variation in Cq values among the location-specific control samples from the rooms (ANOVA; N=21, F = 0.0084, p = 0.999) or from the individual biological safety cabinets (N=16, F = 0.0630, p = 0.939). Therefore, these samples were combined within their respective groups for comparison with the amnion & chorionic plate and villous tree & basal plate samples. Variation in Cq values was observed among the amnion & chorionic plate samples and room, hood, kit, and water samples (Welch F test; N=81, F = 7.683, p = 0.0005), and among the villous tree & basal plate samples and controls (F = 9.572, p = 0.0001). In both cases, the variation was due to the room control samples having lower Cq values (i.e., higher rDNA abundances) than the placental and water samples (Tukey’s pairwise comparisons; amnion & chorionic plate vs. rooms: Q = 4.544, p = 0.016; villous tree & basal plate vs. rooms: Q = 5.108, p = 0.005; water vs. rooms: Q = 5.773, p = 0.001). Cq values did not differ between amnion & chorionic plate samples and blank extraction kits (t-test; t = −1.093, p = 0.282). They also did not differ between villous tree & basal plate samples and blank extraction kits (t = −1.535, p = 0.134). When a subset (N=32/43) of total control samples was diluted 1:9, there were no differences between the amnion & chorionic plate and villous tree & basal plate samples and technical controls (t-tests; amnion & chorionic plate: t = −0.296, p = 0.768; villous tree & basal plate: t = 0.048, p = 0.962). Differences were then also absent between the placental tissue samples and a subset (N=13/21) of room control samples (t-tests; amnion & chorionic plate: t = 0.018, p = 0.985; villous tree & basal plate: t = 0.354, p = 0.725).
Metagenomic surveys of placental tissues
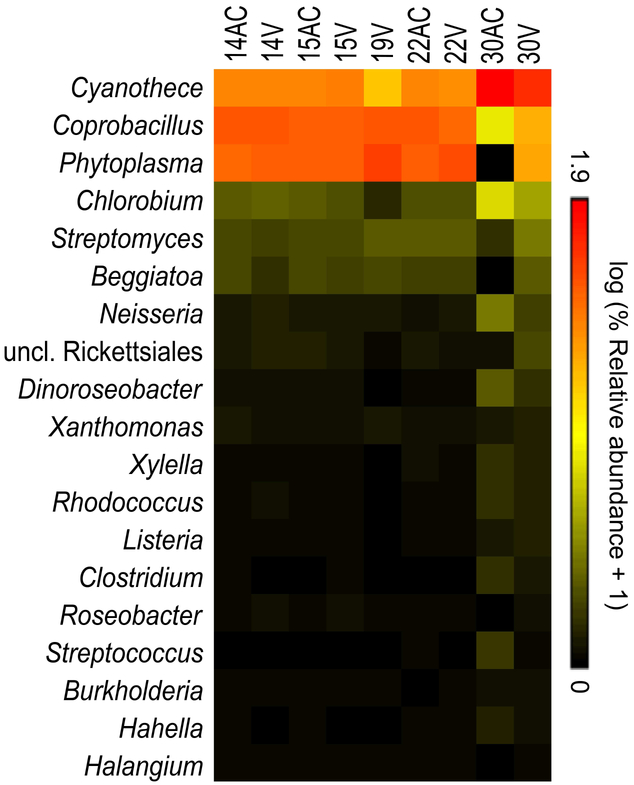
At least 43,000,000 sequence reads were obtained from each of nine placental tissue samples (61,027,678 ± 9,572,214 SD). On average, 0.05% of these sequences were classified as bacterial in origin. Good’s coverage values (99.6% ± 0.004 SD) indicated that the bacterial profiles of these samples were thoroughly characterized from a taxonomic standpoint. The survey identified 267 bacterial genera, with 19 having an average relative abundance of ≥ 0.1% (Figure 4). Only five genera had an average relative abundance ≥ 1.0%: Cyanothece, Coprobacillus, “Candidatus Phytoplasma,” Chlorobium, and Streptomyces. Escherichia was present in each placental sample, with an average relative abundance of 0.05%. The functions of bacterial genes were broadly characterized as metabolism (amino acid, carbohydrate, vitamin, and energy metabolism), and genetic (DNA translation, replication, repair, and degradation) and environmental (membrane transport and signal transduction) processing.
Figure 4. Heat map illustrating relative abundances of prominent bacterial genera among placental samples as determined by metagenomic sequencing.
Prominent genera were here defined as those having an average relative abundance ≥ 0.1% among the placental samples. AC and V indicate amnion & chorionic plate and villous tree & basal plate samples, respectively.
Given the necessary differences in metagenomic library preparation for the placental tissue and technical control samples, their broad bacterial profiles cannot be compared in a quantitative manner. However, it is reasonable to inquire if there are genera consistently identified in placental tissue samples that were not also widely present in the sequenced background technical controls. There were 36 genera present in all nine sequenced placental tissue samples, and 89 genera present in at least half. Each of these genera was present in all 11 sequenced background technical controls. Of the 267 total genera, or approximate genus-level taxa, identified in placental tissue samples, only one was not found in every control sample: an unclassified Myxococcales, present in one placental sample with an abundance < 0.01%.
Of the prominent genera identified in the primary 16S rRNA gene sequencing analysis (Figure 2), only Clostridium was present in placental metagenomic bacterial profiles at an average relative abundance ≥ 0.1% (Figure 4). Achromobacter, Clostridium, Propionibacterium, Staphylococcus and Stenotrophomonas were present in the metagenomic profiles of at least half of the placental samples. However, each of these genera was also present in the metagenomic profiles of all 11 sequenced background technical controls.
Secondary 16S rRNA gene sequencing and quantitative real-time PCR analyses
16S rRNA gene surveys using standard PCR:
The median number of sequences obtained from the 29 placental samples was 89 [IQR: 15 – 3210], and no blank extraction kit controls yielded more than 100 quality sequences. Of the 29 placental samples, only 31% (9/29) yielded at least 1,000 quality sequences. They had Good’s coverage values exceeding 99%. Their microbial profiles included eight prominent OTUs (i.e. average relative abundance ≥ 1%) (Supplemental Figure 4). Pelomonas and Sphingomonas were most consistently abundant. These genera represented two of the three OTUs present in at least half of the nine placental samples. The remaining core OTU (OTU001) was Escherichia. It was present in each of the nine placental samples with a median relative abundance of 0.07% [IQR: 0.02 – 0.13%]. Although the blank extraction kits had poor sequence yield, their bacterial profiles were dominated by Escherichia (median: 67%, IQR: 41 – 100%). Indeed, OTU001 was detected in 27/28 blank extraction kit controls yielding sequence data.
Neither Pelomonas nor Sphingomonas were detected in the bacterial profiles of the nine placental tissues characterized through metagenomic sequencing in the primary analyses described above.
16S rRNA gene surveys using nested PCR with a different primer pair for the first round of amplifications than was used in the primary analysis:
Fifty-seven of 58 placental and blank extraction kit control samples yielded ≥ 1,000 sequences with a Good’s coverage value exceeding 99%. One blank extraction kit sample yielded 423 sequences and was excluded from analyses. The remaining placental samples and technical controls yielded 80,492 ± 27,721 SD and 77, 670 ± 79,160 SD quality sequences, respectively. These sequences clustered into 207 OTUs. For alpha diversity analyses, each sample was subsampled to a depth of 4,020 sequences. Alpha diversity did not differ between blank extraction controls processed alongside (N = 16) or independent of (N = 12) placental samples (Mann-Whitney; Chao1: U = 67.5, p = 0.192; Simpson: U = 90.0, p = 0.799). The richness (U = 10.5, p < 0.0001) and heterogeneity (U = 67.0, p = 0.0001) of blank extraction kit control samples, although very low, were greater than those of placental tissue samples (Supplemental Figure 5).
Extraction controls processed alongside placental samples did not have a different bacterial profile than those processed alone (NPMANOVA; Jaccard: F = 0.863, p = 0.810; Bray Curtis: F = 0.577, p = 0.940), indicating that bacterial signals obtained from blank extraction kit samples were not simply due to DNA cross-contamination from placental tissue samples during processing. The bacterial profiles of placental tissue samples and blank extraction kit controls did differ in both composition and structure (Supplemental Table 2; Supplemental Figure 6). However, OTU001, classified as Escherichia, accounted for 99.0% and 97.6% of the sequences obtained from placental samples and extraction controls, respectively. OTU009, an Enterococcus, was also found in all samples, with an average relative abundance of 0.11% and 0.33% among placental samples and blank extraction kit controls, respectively. OTU102, a Clostridium, was the only other OTU with an average relative abundance ≥ 0.1% among the placental tissue samples, and it was detected in only 3/29 of these samples. In addition to the two Escherichia and Enterococcus OTUs, OTU185, a Shewanella, was a third core OTU (i.e. present in at least half of samples) among placental tissue samples. LEfSe analyses indicated that OTU001, Escherichia, was the only OTU that was more relatively abundant among placental samples than technical controls. SourceTracker analysis indicated that a median of 100% [IQR: 99 – 100] of the OTUs present in the 16S rRNA gene profiles of placental samples could be explained by their distribution among the profiles of technical controls.
16S rRNA gene surveys using touchdown PCR:
Twenty-four of 29 placental tissue samples and 28/29 blank extraction kit controls yielded ≥ 1,000 sequences with Good’s coverage values exceeding 99%. The other samples were excluded from analyses. The remaining placental and extraction control samples yielded 14,602 ± 12,641 SD and 38,817 ± 35,710 SD quality sequences, respectively. These sequences clustered into 350 OTUs. For alpha diversity analyses, each sample was subsampled to a depth of 1060 sequences. Alpha diversity did not differ between controls processed alongside (N = 17) or independent of (N = 11) placental samples (Mann-Whitney; Chao1: U = 81.5, p = 0.587; Simpson: U = 78.0, p = 0.480). Alpha diversity also did not differ between placental samples and extraction controls (Chao1: U = 168.5, p = 0.354; Simpson: U = 190.0, p = 0.728).
The bacterial profiles of background extraction controls did not differ between controls processed alongside placental samples or alone (NPMANOVA; Jaccard: F = 1.216, p = 0.083; Bray Curtis: F = 0.867, p = 0.672). There was variation in the composition of bacterial profiles based on sample type and round of extraction (Supplemental Table 3; Supplemental Figure 7). Specifically, there was a modest observed difference in bacterial profile composition between placental sample and blank extraction controls in the first round of extractions (F = 1.506, p = 0.040), but not in the second (F = 1.032, p = 0.394), third (F = 1.211, p = 0.122), or fourth (F = 0.900, p = 0.734) round of extractions. In the first round, 5/6 and 4/6 of the placental samples contained OTU015 (Ralstonia) and OTU034 (an uncl. Enterobacteriaceae), respectively. These OTUs were not present in any of the four blank extraction controls processed in round one. There was no difference in the structure of bacterial profiles between placental tissue samples and blank extraction controls (Supplemental Table 3; Supplemental Figure 7).
There were 21 prominent OTUs (i.e. average relative abundance ≥ 1%) among placental samples (Supplemental Figure 8). Eight of these OTUs were also prominent among blank extraction control samples. None of the 13 OTUs prominent among placental samples but not prominent among technical control samples were present in more than 21% (5/24) of the placental samples. Linear discriminant analysis effect size (LEfSe) indicated that three OTUs were more relatively abundant among placental samples than blank extraction controls (Supplemental Figure 9). These OTUs were 15 (Ralstonia), 17 (Chthoniobacter), and 41 (Anaerococcus). OTUs 15 and 17 were among the prominent OTUs for blank extraction control samples. OTU041 was not prominent among either placental or technical control samples. It was present in 5/24 placental samples, with an average relative abundance of 1.79%. OTU041 was not present in any of the 17 blank extraction control samples processed alongside placental samples. However, it did account for 6.4% of the sequences from one blank extraction control processed independently of placental samples.
There were five core OTUs (i.e. present in at least half of samples) among placental samples. Three of the five were also core OTUs among blank extraction controls (OTUs 1, 2, and 3). The exceptions were OTU015 (Ralstonia) and OTU017 (Chthoniobacter), which were nonetheless prominent among technical controls. Neither Ralstonia nor Chthoniobacter were detected in the bacterial profiles of the nine placental tissues characterized through metagenomic sequencing in the primary analyses described above.
SourceTracker analyses indicated that a median of 24% (IQR: 0 – 76%) of OTUs in the placental samples could be attributed to background DNA contamination in the extraction kits and/or PCR reagents. The large degree of observed variation was due to whether or not the bacterial profiles of placental samples were dominated by one of the four most prominent OTUs among the placental samples (OTUs 3, 8, 15, and 2; Supplemental Figure 8). Among the 12/24 placental samples that derived at least 25% of their sequences from one of these four OTUs, 75% (IQR: 55 – 95%) of their OTUs could be attributed to background DNA contamination. The profiles of the ten remaining placental samples were each dominated by a different OTU (Supplemental Figure 8). These OTUs were only sporadically present among the technical controls, so their distribution among the placental samples could not be attributed to background DNA contamination based on SourceTracker analyses (Median = 0, IQR = 0).
Quantitative real-time PCR:
The secondary qPCR analysis did not indicate the presence of bacteria in placental samples. While an increase in overall reaction efficiency was observed (96.7%) for the secondary qPCR analysis compared to the primary analysis, the sensitivity of the assay remained ~150 copies. As in the primary qPCR analysis, the vast majority of the placental and background technical control samples were beyond the detection limits of the assay. Mean cycle of quantification (Cq) values for both placental sample and background technical controls were greater than 37 cycles (Supplemental Figure 10). There was no difference in cycle of quantification (Cq) values between blank extraction kit controls processed alongside (N = 17) or independent of (N = 12) placental samples (t-test; t = 1.579, p = 0.126). Therefore, bacterial signals in blank extraction kit samples were not simply due to DNA cross-contamination from placental tissue samples during processing.
COMMENT
Principal findings of the study:
1) Cultivation of the placental tissues did not yield viable bacteria in 28/29 cases; in the case in which it did, the microorganisms were not detected by 16S rRNA gene sequencing; 2) quantitative real-time PCR did not indicate greater abundance of bacterial 16S rRNA genes in placental tissues than in technical controls (laboratory environments and reagents); 3) 16S rRNA gene sequencing did not reveal consistent differences in the composition or structure of bacterial profiles between placental samples and technical controls; and 4) metagenomic surveys of placental tissues largely yielded bacterial sequences from cyanobacteria, aquatic bacteria, and plant pathogens – microbes ecologically unlikely to populate the human placenta. The identification of Coprobacillus, Streptomyces, and other potentially clinically relevant genera in the metagenomic data, while intriguing, was not consistent with their absence or extreme rarity in the multiple 16S rRNA gene surveys of these samples. Overall, we did not find consistent evidence that the human placenta harbors a unique microbiota because microbial signals derived from placental tissues were similar to those observed in technical controls.
The claim that “the placenta harbors a unique microbiome”
In 2014, a key publication reported the results of a study of 320 placentas using 16S rRNA gene sequencing and of a subset of these (n=48) that also underwent metagenomic sequencing64. The authors characterized “a unique placental microbiome niche composed of nonpathogenic commensal microbiota from the Firmicutes, Tenericutes, Proteobacteria, Bacterioidetes, and Fusobacteria phyla”64. Placental microbiota profiles were more similar to those of the human oral cavity than those of the vagina, gut, and skin (Figure 1 in Aagaard et al64). Escherichia coli was most abundant in the placenta, followed by Bacteroides spp., Propionibacterium acnes, Neisseria lactamica, and Staphylococcus epidermidis (Figure 2 in Aagaard et al64). However, cultures were not used in this study; therefore, there is no information about the viability of the microbes from which sequences were detected. Quantitative real-time PCR was also not part of the study; nonetheless, the authors emphasized that the placenta was a low microbial biomass site64.
This publication stimulated research into the existence of a placental microbiota. Twelve additional studies, listed in Table 3, have interrogated placental samples at term using sequence-based techniques to determine, at least in part, whether or not there is a placental microbiota32, 65-75. Eleven of these studies have concluded that there is evidence of a placental microbiota at term based on 16S rRNA gene sequencing and/or metagenomics65-75. Thus, the existence of a placental microbiota has become a majority view in perinatal microbiology at this time.
Table 3.
Description of prior 16S rRNA gene or metagenomic studies of the human placental microbiota at term
| Study | Author | Year | Central research question(s) |
Mode of delivery (sample size) |
Type of sample |
Molecular microbiology methods |
Was culture used? |
Were DNA contamination controls included? |
|---|---|---|---|---|---|---|---|---|
| 1 | Aagaard et al64 | 2014 | Is there a placental microbiota, and does it vary with antenatal infection and preterm birth? |
Term Cesarean (N = 53) Term Vaginal (N= 178) Preterm Cesarean (N = 20) Preterm Vaginal (N = 69) |
Villous tree Collected <1 hour after delivery |
16S rRNA gene sequencing Metagenomic sequencing (subset of 48 subjects |
No | One blank extraction kit was processed per 11 placental samples. These blanks did not generate noticeable bands of amplified DNA and thus were not routinely sequenced. Reagents from a limited number of blanks were sequenced and their bacterial profiles reflected airway or non-human sources (data not provided). |
| Conclusions: | There is a placental microbiota at term and preterm, irrespective of mode of delivery. The placental microbiota shares similarity with the microbiota of the oral cavity. The placental microbiota differs between women delivering preterm and at term. The placental microbiota differs between women with and without a remote history of antenatal infection. |
|||||||
| 2 | Doyle et al65 | 2014 | Does the placental microbiota differ between preterm and term deliveries? |
Term Cesarean without labor (N = 4) Term Vaginal (N = 6) Preterm Vaginal (N = 14) |
Amnion and chorion The time between delivery and processing was not provided |
16S rRNA gene sequencing | No | No |
| Conclusions: | There is a placental microbiota at term and preterm, irrespective of mode of delivery. Nevertheless, the microbial profiles of placental tissues differ between Cesarean and vaginal deliveries. The placental microbiota differs between term and preterm deliveries. |
|||||||
| 3 | Antony et al66 | 2015 | Does the placental microbiota vary with maternal obesity or excess gestational weight gain and, if so, does its profile differ between preterm and term intervals? | Cesarean (N = 54) Vaginal (N = 183) 62/237 (26.2%) subjects delivered preterm |
Villous tree Processed immediately upon delivery |
16S rRNA gene sequencing Metagenomic sequencing (subset of 37 subjects) |
No | This study was a subgroup analysis of Study 1. |
| Conclusions: | There is a placental microbiota at term and preterm. Among women delivering preterm, differences in the placental microbiota exist between women with and without excess gestational weight gain. |
|||||||
| 4 | Zheng et al67 | 2015 | Is the placental microbiota at term associated with neonatal birth weight? |
Term Vaginal normal birth weight (N = 12) Term Vaginal low birth weight (N = 12) |
Villous tree Processed immediately upon delivery |
16S rRNA gene sequencing | No | No |
| Conclusions: | There is a placental microbiota at term. The placental microbiota differs between low birth weight and normal birth weight neonates. |
|||||||
| 5 | Bassols et al68 | 2016 | Is the placental microbiota at term associated with gestational diabetes? |
Term Vaginal without gestational diabetes (N = 11) Term Vaginal with gestational diabetes (N = 11) |
Villous tree Processed immediately upon delivery |
16S rRNA gene sequencing | No | No |
| Conclusions: | There is a placental microbiota at term. The placental microbiota differs between women with and without gestational diabetes. |
|||||||
| 6 | Collado et al69 | 2016 | Is the fetal gut colonized in utero by microbes from the amniotic cavity, placenta and/or maternal gut? |
Term Cesarean without labor (N = 15) |
Placental tissue (unspecified) Processed immediately upon delivery |
Denaturing gradient gel electrophoresis (DGGE) 16S rRNA gene sequencing |
Anaerobic culture was used Propionibacterium & Staphylococcus were cultured from the placenta |
No |
| Conclusions: | There is a placental microbiota at term. The microbiota of the meconium shares similarities with the microbiota of the placenta and amniotic fluid. |
|||||||
| 7 | Lauder et al32 | 2016 | Is there a placental microbiota at term? |
Term Cesarean (N = 1) Term Vaginal (N = 5) |
Fetal side biopsy of the placenta Maternal side biopsy of the placenta (basal plate) Processed immediately upon delivery |
Quantitative real-time PCR (qPCR) 16S rRNA gene sequencing |
No | Laboratory air swabs (N = 11) Sterile swabs (N = 8) Blank extraction kits (N = 8) The controls were incorporated into statistical analyses. |
| Conclusions: | Microbial signatures in placental tissues could not be distinguished from those of technical controls. | |||||||
| 8 | Prince et al70 | 2016 | Does the microbiota of the placental membranes vary in association with preterm birth and chorioamnionitis? |
Term Cesarean without chorioamnionitis (N = 4) Term Vaginal without chorioamnionitis (N = 11) Term Cesarean with chorioamnionitis (N = 3) Term Vaginal with chorioamnionitis (N = 9) Preterm Cesarean without chorioamnionitis (N = 2) Preterm Vaginal without chorioamnionitis (N = 11) Preterm Cesarean with chorioamnionitis (N = 5) Preterm Vaginal with chorioamnionitis (N = 26) |
Swabs of the chorion and/or villous membranes adjacent to the fetal side of the placenta Processed upon delivery |
Metagenomic sequencing Targeted PCR of Ureaplasma and Mycoplasma serovars |
Culture ofUreaplasma and Mycoplasma was used Ureaplasma was cultured from the chorion of 8 subjects with chorioamnionitis, and from 2 subjects delivering preterm without chorioamnionitis. Ureaplasma was not cultured from the chorion of subjects delivering at term without chorioamnionitis. |
Only samples with high sequence yield and without concern for contamination were included in analysis (specific methodology not provided). |
| Conclusions: | There is a placental microbiota at term and preterm, irrespective of mode of delivery. Variation in the placental microbiota is associated with preterm birth and the presence and severity of chorioamnionitis. |
|||||||
| 9 | Doyle et al71 | 2017 | Is there variation in the placental microbiota associated with gestational age, neonatal size, or chorioamnionitis? What is the origin of the placental microbiota (i.e., oral cavity or vagina)? |
1097 Subjects Unreported percentages of subjects delivered via Cesarean, preterm, or with chorioamnionitis |
Amnion and chorion A sample of placental tissue at full thickness Some samples were processed immediately upon delivery; others were processed 1 to 24 hours later, after being kept at room temperature |
16S rRNA gene sequencing Quantitative real-time PCR (qPCR) |
No | Reagents from one blank extraction kit were processed and sequenced for every 10 extractions. OTUs1 detected in these negative controls were removed from the data set. Only placental samples that had a positive qPCR value (equivalent to 40 CFU/μl) were sequenced2; 68.1% of amnion-chorion and 46.8% of placental samples had a positive qPCR value. A delay in sample processing increased the likelihood of a positive qPCR value. |
| Conclusions: | There is a placental microbiota at term. The placental microbiota has more overlap with the microbiota of the vagina than the oral cavity. Variation in the placental microbiota is associated with severe chorioamnionitis and delivery of a smaller neonate. |
|||||||
| 10 | Gomez-Arango et al72 | 2017 | What is the origin of the placental microbiota in overweight and obese pregnant women (i.e., oral cavity or gut)? |
Term Cesarean (N = 17) Term Vaginal (N = 20) 13 women were overweight, and 24 were obese |
Fetal side biopsy of the placenta Processed within 1 hour of delivery |
16S rRNA gene sequencing | No | The reagents from one blank extraction kit and one PCR amplification control were pooled and sequenced for each kit type (each sample type – placenta, oral swab, stool – was processed using a different kit type). OTUs detected in these negative controls were removed from the data set. |
| Conclusions: | There is a placental microbiota at term, irrespective of mode of delivery. The placental microbiota was more similar to that of the maternal oral than the intestinal environment, yet it was distinct from each. |
|||||||
| 11 | Parnell et al73 | 2017 | Is there a placental microbiota at term, and does it vary between the fetal membranes, placental villi and basal plate? |
Term Cesarean (N = 34) Term Vaginal (N = 23) |
Amnion and chorion Villous tree Basal plate Processed within 12 hours of delivery |
16S rRNA gene sequencing Quantitative real-time PCR (qPCR) |
No | Blank extraction kits (N = 8) Water controls (N =5) OTUs detected in these negative controls were removed from the data set. Only placental samples that had a positive qPCR value (>34 16S rRNA gene copies / μl) were included in sequence data analyses. |
| Conclusions: | There is a placental microbiota at term, irrespective of mode of delivery. The placental microbiota differs between the amnion-chorion and the basal plate. There may not be a resident microbiota in the villous tree. |
|||||||
| 12 | Zheng et al74 | 2017 | Does the placental microbiota differ between cases of fetal macrosomia and controls? |
Term Cesarean without fetal macrosomia (N = 10) Term Cesarean with fetal macrosomia (N = 10) |
Villous tree Processed immediately upon delivery |
16S rRNA gene sequencing | No | The amplification of 16S rDNA from blank extraction kits did not generate noticeable bands of amplified DNA. Reagents from these kit controls were not sequenced. |
| Conclusions: | There is a placental microbiota at term. The placental microbiota differs between cases of fetal macrosomia and normal birth weight controls. |
|||||||
| 13 | Leon et al75 | 2018 | Does the placental microbiota differ between preterm and term deliveries? |
Term Cesarean (N = 81) Term Vaginal (N = 84) Preterm Cesarean (N = 55) Preterm Vaginal (N = 36) |
Placental parenchyma (N = 356 samples) Villous tissue (N = 44 samples) Processed upon delivery |
16S rRNA gene sequencing | No | A blank extraction kit was processed for each round of extractions (Reagents from 19 kits that yielded ≥500 sequences were analyzed). OTUs with ≥2 reads in ≥2 kit controls were removed (excluding Lactobacillus, Veillonella & Mycoplasma). |
| Conclusions: | There was large overlap between the bacterial profiles of placental samples and technical controls. However, there is a placental microbiota at term and preterm. Mode of delivery impacted the microbial profiles of placental tissues. Although there was not a unique preterm placental microbiota, some bacteria (i.e., Ureaplasma and Mycoplasma) were enriched in preterm placentas. |
|||||||
OTU: Operational Taxonomic Unit
CFU: Colony Forming Unit
The limitations of molecular microbiologic techniques in low microbial biomass sites
Questions have emerged about the interpretation of microbiology studies based solely on sequencing techniques22, 28-30. The detection of a nucleotide sequence from a bacterium or virus is not the same as the identification of a microorganism. These sequences can represent microbial breakdown products in the body (e.g. DNA from dead microbes)188 or background DNA contamination (e.g. present in DNA extraction kits, PCR reagents, and laboratory environments) 22, 30, 31. Therefore, the demonstration of a microbiota requires: 1) microbial signals beyond contamination, 2) reproducibility across methods (sequencing, qPCR, culture, and microscopic detection of the microorganisms in tissues, for example, through fluorescence in situ hybridization), and 3) ecological plausibility22.
The microbial signals derived from the placenta are not distinguishable from those of technical controls
Lauder et al32 sampled the placental tissues, vagina, and oral cavity of six women delivering at term. For each woman, control samples included swabs waved in the air within the laboratory, sterile swabs, and blank extraction kits32. Using both qPCR and 16S rRNA gene sequencing, the bacterial profiles of placental tissues were not distinguishable from those of controls32. In contrast, the profiles of vaginal and oral samples differed from controls32. More recently, using 16S rRNA gene sequencing, de Goffau et al22 showed that the microbial signals derived from placental tissues were largely due to the DNA extraction kits used. Additionally, in a recent sequence-based survey of targeted eukaryotic microbes in placental tissues, Lager et al232 determined that sequenced DNA was due to technical artifacts and background DNA contamination rather than a true signal of a placental microbiota. These studies highlight the need for addressing DNA contamination in sequence-based surveys and for using complementary techniques, such as cultivation22.
The findings of the study in the context of other reports
In this study, placental samples from 29 women who had a Cesarean delivery at term without labor were examined for the presence of a placental microbiota. We included 72 background technical controls and employed multiple complementary modes of inquiry: bacterial culture, 16S rRNA gene qPCR, 16S rRNA gene sequencing, and metagenomic surveys. Our results are consistent with those of Lauder et al32, de Goffau et al22, and Lager et al232 in that we did not find evidence of a placental microbiota. The results are discussed below in detail.
Bacterial culture:
The results of culture were negative. Only one (3.4%) of the placental cultures was positive and the detected bacteria were Bacillus circulans, Bacillus pumilus, and Brevibacterium casei. Bacillus and Brevibacterium species are widespread bacteria that can be human commensals and opportunistic pathogens233-240. However, the 16S rRNA genes of the three cultured bacteria were not detected in the placental sample using molecular techniques, suggesting that, in this study, these bacteria were laboratory contaminants. The congruence between the primers utilized in the primary nested PCR analysis (27F, 1492R) and the sequences of the bacterial cultivars is unknown due to the methods used to amplify the V4 region of their 16S rRNA genes. Nonetheless, the 16S rRNA genes of these bacteria had exact matches to the primers used in the secondary nested PCR analysis (341F/1061R; 515F/806R) and the primers used in the secondary standard and touchdown PCR analyses (515F/806R). Therefore, if Bacillus circulans, Bacillus pumilus, and Brevibacterium casei were present in the placental tissue sample, we should have detected their 16S rRNA gene sequences. In addition, this placenta, like others in this study, did not present severe/moderate acute inflammatory responses in the histopathologic examination.
Quantitative real-time PCR:
Consistent with Lauder et al32, qPCR analyses in this study indicated that placental tissue samples did not have a greater abundance of 16S rRNA gene copies than technical controls. Indeed, the abundances of 16S rRNA gene copies in both placental samples and controls were below the limit of detection in the qPCR assay.
16S rRNA gene sequencing:
16S rRNA gene sequencing revealed similarity in the microbial profiles among placental tissues, blank extraction kits, biological safety cabinets, and laboratory controls. In the primary 16S rRNA gene nested PCR analysis and the secondary 16S touchdown PCR analysis, the structures of the microbial profiles of placental tissues and technical controls did not differ. In the secondary 16S rRNA gene nested PCR analysis, the microbial profiles of placental tissues and controls were significantly different. However, 99% and 97.6% of the sequences obtained from placental tissues and controls, respectively, belonged to Escherichia. Escherichia was also widely present, although not highly abundant, in the primary 16S nested PCR analysis, the secondary 16S touchdown PCR analysis, and the secondary 16S standard PCR analysis. Escherichia, especially E. coli, has been previously identified as a principal member of the placental microbiota using molecular surveys64, 66, 70. In a recent study, microbes were cultured from the fetal side of 20.7% (379/1832) of placentas obtained from Cesarean deliveries at term without clinical chorioamnionitis; 13.5% (247/1832) of the placental samples yielded E. coli cultures241. A valuable addition to that study would have been species-specific qPCR and/or 16S rRNA gene or metagenomic sequencing of the cultured placental samples to demonstrate that the absolute and relative abundances of E. coli were indeed greater in samples yielding E. coli cultures than in those that did not241. This would provide verification of the culture results. In the current study, molecular signals of Escherichia were as widely distributed and relatively abundant among technical controls as among placental tissues, and Escherichia was not cultured from any of the placental tissues.
In addition to the community level analyses, linear discriminant analysis effect size (LEfSe) was used to identify operational taxonomic units (OTUs) that were more relatively abundant in placental tissues than in technical controls (Table 4). Other than Escherichia, the identified genera have been detected in low relative abundance in only a few prior sequence-based studies of placental microbiota (Table 4). Most of these bacteria are also considered common DNA contaminants in sequence-based studies (Table 4).
Table 4.
Genera indicated by Linear discriminant analysis Effect Size (LEfSe) as being more relatively abundant in placental tissues than technical controls
| Data set | Genus | Ecological and clinical description and the reported occurrence of the genus in prior sequence-based studies of the human placenta at term |
Has the genus been documented as a DNA contaminant in prior sequence-based studies? |
|---|---|---|---|
| Primary 16S rRNA gene nested PCR: amnion & chorionic plate | |||
| Achromobacter | Generally aquatic and soil bacteria but they can be infectious agents in immunocompromised hosts267, 268, especially cystic fibrosis patients269. Achromobacter has been identified in one placental microbiota study at low abundance (0.05%)74, and in three others at low, yet unreported, abundances66, 68, 72. | Yes28 | |
| Blastococcus | Typically associated with rocks or marine environments270, 271. An isolate was obtained from human stool272. Blastococcus was identified in one placental microbiota study at low abundance (<0.01%)69. | Yes29 | |
| Methylobacterium | Generally aquatic and soil bacteria but they can be infectious agents in immunocompromised hosts273. Methylobacterium was identified in a prior placental microbiota study at low abundance (<0.01%)69. | Yes28, 32, 72 | |
| Caldalkalibacillus | Thermoalkaliphilic environmental bacteria274, 275. Caldalkalibacillus has not been identified in prior studies of a placental microbiota. | No | |
| Primary 16S rRNA gene nested PCR: villous tree & basal plate | |||
| Achromobacter | Generally aquatic and soil bacteria but they can be infectious agents in immunocompromised hosts267, 268, especially cystic fibrosis patients269. Achromobacter has been identified in one placental microbiota study at low abundance (0.05%)74, and in three others at low, yet unreported, abundances66, 68, 72. | Yes28 | |
| Herbaspirillum | Typically found in soils but can be opportunistic pathogens of immunocompromised hosts276, 277. Herbaspirillum was identified in a prior placental microbiota study at a low, yet unreported, abundance68. | Yes28 | |
| Secondary 16S rRNA gene nested PCR: villous tree & basal plate | |||
| Escherichia | Common human commensals and opportunistic pathogens, including of the urogenital and reproductive tracts278-280. Escherichia has been reported as a principal member of the placental microbiota in three studies64, 66, 70, at low abundances in two others (<0.05%)67, 69, and at a low, yet unreported, abundance in another68. | Yes28, 75 | |
| Secondary 16S rRNA gene touchdown PCR: villous tree & basal plate | |||
| Ralstonia | Common environmental and aquatic bacteria that can be agents of nosocomial infections281, 282. Ralstonia was widespread among placental samples in two prior studies70, 73, at low abundance in a third (<0.01%)69, and at a low, yet unreported, abundance in a fourth72. | Yes22, 28, 57,75 | |
| Chthoniobacter | This genus has a single species, a soil bacterium283, 284. Chthoniobacter was identified in a low, yet unreported, abundance in one prior study of a placental microbiota70. | No | |
| Anaerococcus | Human commensals that can be opportunistic pathogens, including in the urogenital tract285-287. Anaerococcus was identified in low abundance (0.02%) in one prior study69, and present at low, yet unreported, abundances in two others68, 75. | Yes29 | |
Metagenomic sequencing:
The metagenomics data obtained in the current study were also consistent with DNA contamination having a marked influence on the microbial profiles of placental tissues. Specifically, 63.4% of the bacterial sequences recovered from placental tissues came from Cyanothece, “Candidatus Phytoplasma,” and Chlorobium. In a recent commentary emphasizing the effect of DNA contamination in microbiome studies, it was recommended that data from sequence-based investigations of low microbial biomass environments be interpreted through the lens of microbial ecology22. One example the authors provide is to consider that sequence data indicating that photosynthetic bacteria inhabit internal organs in the human body ought to be questioned because residency in these organs precludes photosynthesis. Cyanothece is a photosynthetic cyanobacterium242, and Chlorobium is a photosynthetic green sulfur bacterium243. Furthermore, members of “Candidatus phytoplasma” are obligate plant pathogens restricted to the phloem of plants and phloem-feeding insect vectors. Among the remaining 16 prominent (≥0.1%) bacterial genera identified through metagenomic sequencing of placental tissues, there were aquatic bacteria (Beggiatoa, Roseobacter, Hahella, Halangium), additional plant pathogens (Xanthomonas, Xylella), and an algal symbiont (Dinoroseobacter). Therefore, it is unlikely that the human placenta is a suitable niche for these microorganisms.
Some metagenomic data warrant discussion. Coprobacillus represented 30.5% of the bacterial sequences identified in placental samples. Although Coprobacillus has been detected in two sequence-based studies of term and preterm placentas at low abundance66, 75, this microorganism was not detected in any of our 16S rRNA surveys. Although the primers used to target the 16S rRNA gene in the first round of amplification in the primary and secondary nested PCRs (27F/1492R; 341F/1061R) were not an exact match for Coprobacillus cateniformis (JCM 10604), the only member of the genus Coprobacillus244, the primers used for the secondary standard and touchdown PCRs (515F/806R) were a perfect match for this bacterium. Therefore, if Coprobacillus was present in placental tissues, and if its 16S rRNA gene sequence was similar to that of the lone characterized representative of this genus (i.e., Coprobacillus cateniformis), we should have identified it in the standard 16S rRNA gene PCR and touchdown PCR analyses.
Streptomyces represented, on average, 1% of bacterial sequences obtained from placental tissues through metagenomic sequencing. Although Streptomyces was previously identified in placentas at term using sequencing techniques64, 66, 69, in the current study, only two 16S rRNA gene sequences in all the 16S surveys of placental tissues were assigned to Streptomyces. However, given that our 16S rRNA gene V4 primers (515F/806R) were perfect matches for 98.6% (580/588) of the type strains of Streptomyces included in the Ribosomal Database Project212, we should have detected these microorganisms more frequently in the standard 16S rRNA gene PCR and touchdown PCR analyses. Other bacterial genera (>0.1% average relative abundance) identified through metagenomics in this study that were previously detected in sequence-based studies of placental tissues at term were Neisseria64, 66, Rhodococcus64, 67, Clostridium71, Streptococcus70, 72, and Burkholderia66. Nevertheless, in the current study, sequences for these microorganisms were detected in all placental samples and in all background technical control samples. It is noteworthy that these sequences have been previously reported as DNA contaminants in sequence-based studies28, 29, 31. There is thus not sufficient evidence to conclude that bacterial signals identified through metagenomic sequencing represent evidence of a placental microbiota or bacterial ecosystem in this organ.
Similar to the placenta, there is a lack of evidence for an amniotic fluid microbiota in normal pregnancy at term without labor
Some studies claim that amniotic fluid is not sterile and has a microbiota similar to that of the placenta69, 245. However, recent studies have shown that there is not an amniotic fluid microbiota in normal term pregnancies63. For example, in a prospective investigation of 344 asymptomatic women between 15 and 22 weeks of gestation, amniotic fluid samples were negative for the presence of genital mycoplasmas, bacteria, or fungi, using species-specific and broad-range PCR techniques54. Furthermore, in a recent study of 10 women who underwent elective Cesarean deliveries without labor, the bacterial loads (assessed through digital droplet PCR of the 16S rRNA gene) of amniotic fluid samples were comparable to those of background technical controls57. Also, these amniotic fluid samples did not yield bacterial isolates. Conversely, amniotic fluid samples from 14 women with prior rupture of membranes had bacterial loads ten times higher than those of technical controls, and these samples yielded bacterial isolates 50% of the time57. In addition to these recent clinical investigations, a logical argument against the existence of either a placental or an amniotic fluid microbiota is the generation of germ-free mammals through sterile Cesarean delivery and germ-free technology (incubators, water, food, etc.)63, 246. This has also been extended to a human infant affected by severe combined immunodeficiency syndrome246-249.
The lack of a microbiota in the placenta or amniotic fluid does not exclude fetal exposure to microbial products
The absence of a resident microbiota in the placenta or amniotic fluid does not rule out exposure of the fetus to microbial metabolites. Using germ-free pregnant mice that were transiently gestational-colonized with Escherichia coli HA107 (a genetically engineered bacterium250), Gomez de Agüero et al showed that microbial metabolites are transferred from the mother to the fetus through the placenta251. Yet, no live microorganisms were found in the placenta or the offspring251. Fetal exposure to microbial metabolites from the mother was, in part, mediated by antibodies, given that antibody-deficient dams (JH−/− mice) had a reduced concentration of such microbial products251. These microbial metabolites shaped the innate immune system of the offspring as evidenced by an increased number of intestinal group 3 innate lymphoid cells (ILC3s) and macrophages in neonates born to transiently gestational-colonized dams251. Of interest, fetal ILC3s are present in the amniotic cavity where they seem to participate in the host defense mechanisms against microbial invasion252, 253. In addition, Gomez de Agüero showed that neonates born to transiently gestational-colonized mothers had an enhanced ability to clear bacteria251, suggesting that microbial metabolites of the mother influence the innate immune fitness of the offspring.
Strengths of the current study
First, in our attempt to determine whether or not there is a placental microbiota during normal pregnancy, we limited our investigation to women who delivered at term without labor. Thereby, we avoided the introduction of bacteria into the amniotic cavity during labor at term157, 254. Second, we used samples collected after Cesarean delivery to prevent microbial colonization or 16S rRNA gene contamination of placental tissues during vaginal delivery75. Third, we excluded placentas from preterm gestation given that molecular surveys have identified a potential placental microbiota linked to preterm delivery64, 65, 70, 75. Fourth, we used multiple modes of inquiry: bacterial culture, 16S rRNA gene quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomic surveys. We further bolstered our initial 16S rRNA gene sequencing analyses with secondary analyses using alternative amplicon-library generation techniques. Each approach used in this study has its own strengths and weaknesses; however, these approaches are complementary, and their ultimate agreement here provided a more robust conclusion than any of them could have provided in isolation. Fifth, we included thorough controls for potential background DNA contamination, including conducting numerous extractions without any biological template, extractions after exposure to circulating air within our biological safety cabinets, and extractions after exposure to our broader operating rooms and laboratory environments. Importantly, we incorporated the sequence data from these background technical controls into graphical and statistical analyses.
Limitations of the current study
First, all subjects necessarily received intraoperative antibiotic prophylaxis, typically cefazolin, at Cesarean delivery, so we cannot rule out a subsequent inhibitory influence on cultivation results192, 194, 255-262. Second, for part of the study we used nested PCR, an approach that can facilitate amplifying the very low concentrations of bacterial DNA present in relation to the high background concentrations of host DNA196, 197. Given that nested PCR entails two separate rounds of amplification, it can increase the likelihood of amplification bias and can thereby promote similarity among characterized bacterial profiles196, 263. However, this would require that characterized samples contain the same preferentially amplified gene variants. In our primary 16S rRNA gene analysis, our sample coverage was thorough, and we amplified and characterized 16S rRNA gene variants from numerous genera previously identified in prior molecular surveys of placental tissues (i.e., Acinetobacter, Actinomyces, Bacillus, Bacteroides, Burkholderia, Clostridium, Corynebacterium, Enterococcus, Escherichia, Fusobacterium, Lactobacillus, Lactococcus, Mycobacterium, Neisseria, Prevotella, Propionibacterium, Pseudomonas, Rhodococcus, Staphylococcus, Streptococcus, and Ureaplasma)64, 66, 67, 69-73. However, no gene variants from these genera were more widely distributed among placental tissues than among background technical controls. Also, we followed up this primary analysis with secondary analyses, in which we utilized a second, highly conserved primer pair for the first round of amplifications in nested PCR, and additionally employed standard PCR and touchdown PCR approaches. We further complemented these approaches with metagenomic surveys of placental tissues, which minimize amplification bias264. Third, we focused exclusively on bacteria; eukaryotic pathogens and viruses were not targeted. Fourth, our study did not use morphological techniques, such as fluorescence in situ hybridization (FISH)265, to visualize bacterial cells in placental tissues. However, using FISH, we did not detect bacteria in the placental tissues of a different set of women who had elective Cesarean deliveries at term without labor (Alexander Swidsinski, Universitätsmedizin Berlin, Personal Communication). Fifth, a valuable negative control here would have been extraction of alternative presumed sterile human tissues, thereby controlling for any potential influence of competition between host and microbial DNA during extraction and amplification processes. Nevertheless, the specific kit we used to perform extractions, and the masses of placental tissues we conducted extractions on, were consistent with prior studies investigating the existence of a placental microbiota32, 64. A positive control would have been extraction of alternative human tissues with a confirmed very low microbial biomass, such as the lung216, 217. Such negative and positive control tissues would require the use of animal models. An alternative approach would be to include placental tissue samples spiked with known numbers of bacterial cells, to ascertain the specific limits of microbial detection in the study22, 266.
Criteria to establish the presence of a resident microbiota in low biomass sites such as the placenta
A fundamental question that emerges from the debate about the existence of a unique placental microbiota is: what are the requirements to demonstrate the presence of such a microbial ecosystem? The existence of a microbiota would be supported by the following evidence:
1. Identification of microbial DNA sequences in tissues or fluids through multiple modes of inquiry, such as 16S rRNA gene sequencing and metagenomics, and the profiles of these microbial DNA sequences are distinct from those detected in technical controls (e.g. DNA extraction kits, PCR reagents, laboratory environments).
2. Confirmation of microbial burden through quantitative real-time PCR.
3. Demonstration of the viability of the microorganisms, either through culture or the transcription of specific microbial genes.
4. Visualization of the microorganisms in tissues or fluids using microscopic techniques, such as fluorescence in situ hybridization with eubacterial or, ideally, species-specific probes.
5. Residency of the microorganisms in the tissues or fluids is ecologically plausible in that they are likely to survive in the niche in which they have been found.
Conclusions
Through multiple modes of microbiology inquiry, we did not find consistent and reproducible evidence of the existence of a placental microbiota at term. We have not definitively shown that microbes do not inhabit the placentas of term pregnancies; it is difficult to prove the null hypothesis and there are limits of detection inherent in the contemporary survey techniques we employed. However, using multiple investigative approaches, incorporating technical controls, and focusing on placental tissues obtained through Cesarean delivery at term without labor, we detected no consistent evidence for bacterial communities in the placental tissues beyond the signals also present in the technical controls. This study bolsters the arguments for the necessity of substantively incorporating technical control samples into studies of very low microbial biomass22, 30, such as those targeting a placental microbiota22, 32, 75, and for starting with the null hypothesis that microbial signals in these biological samples are background DNA contamination22, 30, 32, 63. Optimizing cultivation techniques, in concert with molecular survey approaches, will be important in evaluating the existence of a microbiota in low microbial biomass body sites22, 63, 189.
Supplementary Material
AJOG at a Glance:
A. Why was this study conducted?
To examine whether there was evidence to support the existence of a microbiota in placentas delivered at term without labor via Cesarean section.
B. What are the key findings?
Placentas did not have a microbial DNA abundance exceeding that of background technical controls.
16S rRNA gene sequencing did not reveal consistent differences in the composition or structure of bacterial profiles between samples of the placenta and technical controls.
Cultures were negative in 28/29 placentas.
Metagenomic analysis of placental tissues identified microbial DNA sequences not found with other methods.
C. What does this study add to what is already known?
The findings of this study do not support the existence of a placental microbiota in patients who delivered at term without labor.
Acknowledgements
We acknowledge the contributions of Dr. Paul Lephart, Mr. Timothy Burger, and Ms. Maureen Taylor of the Detroit Medical Center’s Microbiology Laboratory, who made possible the conduct of cultivation assays. Ms. Judy Opp of the University of Michigan was helpful with the sequencing of 16S rRNA gene amplicons and metagenomic surveys.
This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U. S. Department of Health and Human Services (NICHD/NIH/DHHS), and, in part, with federal funds from the NICHD/NIH/DHHS under Contract No. HHSN275201300006C. This research was also supported by the Wayne State University Perinatal Research Initiative in Maternal, Perinatal and Child Health.
GLOSSARY
Human microbiota
- Microbiota
The microbes present in a defined environment.
- Microbiome
Microbiome has two common definitions. It can refer to the genomic content of a microbiota, or more comprehensively to a habitat, its microbiota, and the genomic content of this microbiota.
Microbial alpha diversity
- Alpha diversity
Diversity within a single microbial community. In this study, we characterized alpha diversity through two indices, Chao1 and Simpson. The Chao1 index estimates the richness of microbial communities, and the Simpson Index estimates their heterogeneity.
- Richness
Number of different microbial types (e.g. species) in a mixed microbial community.
- Chao1 index
An estimate of microbial community richness. It is calculated as Sobs + (F12 / 2F2), wherein Sobs is the number of species in the sample, F1 is the number of species that are singletons, and F2 is the number of species that are doubletons. A singleton is any species occurring only once in a sample, while a doubleton is any species occurring twice in a sample
- Heterogeneity
A measure of microbial community alpha diversity that takes into account both the richness and evenness (i.e., relative abundances) of community members.
- Simpson index
An estimate of microbial community heterogeneity. It is calculated as Σ (ni (ni-1) / N (N – 1)), wherein ni is the number of individuals in the ith species, and N is the total number individuals sampled.
Microbial beta diversity
- Beta diversity
Diversity between two, or among multiple, microbial communities. In this study, we characterized beta diversity through two indices, Jaccard and Bray-Curtis. The Jaccard index describes the composition of microbial communities. Specifically, it describes the extent to which two communities share the same species. The Bray-Curtis index describes the structure of microbial communities. It describes not only the extent to which two communities share the same species, but also the extent to which the species they do share are present in the same relative abundances in the two communities.
- Jaccard index
A measure of similarity in composition (i.e., shared species membership) between two microbial communities. It is calculated as a / (a + b + c), wherein a is the number of shared species between the two communities, b is the number of species unique to the first community, and c is the number of species unique to the second community.
- Bray-Curtis index
A measure of similarity in structure (i.e., shared species membership and relative abundances of shared species) between two microbial communities. It is calculated as 2W / (N1 + N2), wherein W is the sum of the lower values of the two abundances for species shared between the two communities, N1 is the number of individuals sampled in the first community, and N2 is the number of individuals sampled in the second community.
Characterizing microbial diversity through 16S ribosomal RNA gene sequencing
- 16S rRNA gene
A housekeeping and phylogenetic marker gene present in almost all bacteria. It is critical in protein manufacturing and is therefore highly conserved. Nevertheless, it has regions of hypervariability. The conserved regions of the gene evolve slowly and can therefore serve as targets for PCR primers, while the hypervariable regions afford researchers information on the evolutionary relationships among bacterial types. 16S rRNA gene surveys are very commonly used to characterize the bacterial types (e.g. genera) within mixed bacterial communities in clinical and environmental samples.
- 16S rRNA gene survey
Characterization of mixed bacterial communities in samples based upon patterns in the presence and/or relative abundance of variants of the 16S rRNA gene, a phylogenetic marker gene present in almost all bacteria.
- Mothur
A software program providing quality filtering, alignment, clustering, and taxonomic classification of DNA sequence reads, such as variants of the 16S rRNA gene. Clustering entails grouping sequence reads into operational taxonomic units (OTUs) based on their percent nucleotide similarity.
- Operational taxonomic unit (OTU)
A group of DNA sequence reads, for example of the 16S rRNA gene, that share a certain percent nucleotide similarity (e.g. 97%). OTUs are generally viewed as bacterial types or variants. OTUs are commonly used in microbiome studies because many 16S rRNA gene variants amplified and sequenced from mixed microbial communities cannot be confidently assigned to taxa with a high degree of resolution (e.g. genus or species identity).
- Good’s coverage
An estimate of the extent to which a microbial community has been sufficiently sampled. With respect to next-generation sequencing surveys, for each community, Good’s coverage is calculated as (1 – (# singleton OTUs / # total sequences for sample)) × 100%. It reveals the percentage of sequence reads in a sample that were not in singleton OTUs, with a higher percentage indicating higher sample coverage.
- Singleton
An operational taxonomic unit (OTU) represented by only one sequence read in the entire dataset. A read is a single sequenced amplicon of the targeted gene (e.g. 16S rRNA gene).
- Doubleton
An operational taxonomic unit (OTU) represented by only two sequence reads in the entire dataset.
- Nested PCR
A modified polymerase chain reaction (PCR) approach aimed at reducing nonspecific amplification and increasing recovery of target amplicons, for example amplicons of the 16S rRNA gene. In the first round of PCR, a large gene fragment is targeted for amplification. In the second round of PCR, a smaller gene fragment within this larger fragment is targeted for amplification. The second round of PCR selects against non-specific amplicons and promotes targeted gene products.
- Touchdown PCR
A modified PCR approach aimed at reducing the initial amplification of nonspecific sequences during early steps of amplification by using a relatively high primer annealing temperature in relation to the melting point of the primers. As cycling proceeds, the annealing temperature is incrementally decreased allowing for increased amplification efficiency. The increased initial specificity of the reaction at higher annealing temperatures permits amplification of sequences with the greatest primer specificity to outcompete amplification of nonspecific sequences as cycling proceeds at lower annealing temperatures.
Characterizing microbial diversity through metagenomic sequencing
- Metagenomics survey
Characterization of bacterial communities in samples based upon patterns in the presence and/or relative abundance of all genes of bacterial origin. In contrast to surveys based on phylogenetic marker genes, like the 16S rRNA gene, all genomic DNA in samples is sequenced. Those sequences determined to be of bacterial origin are taxonomically classified, often times even at the species level, and the metabolic and functional potential of sampled mixed bacterial communities can be characterized.
- MG-RAST (Meta Genome Rapid Annotation using Subsystem Technology)
An analysis server for quality filtering, taxonomically classifying, and functionally annotating and comparing metagenomic datasets.
Quantifying microbial abundance through quantitative real-time PCR
- Quantitative real-time PCR
A molecular technique that monitors the amplification of a targeted gene (e.g. 16S rRNA gene) in real-time, across multiple cycles of PCR. In this study, it was used to compare the relative abundances of 16S rRNA gene copies in placental and background technical control samples.
- Cycle of quantification (Cq)
In quantitative real-time PCR, the cycle number at which a sample’s amplification curve exceeds a predefined minimum threshold based on background fluorescence levels. It is the point at which the signal from the sample has exceeded a baseline level for the assay. The more abundant the targeted gene is within a sample, the lower the sample’s Cq value will be.
- Degenerate primer
A PCR primer sequence in which one or more nucleotide positions has several possible bases. It enables capturing variation in nucleotide combinations for a target gene (e.g. 16S rRNA gene) within mixed microbial communities.
Footnotes
Disclosure statement: The authors report no conflicts of interest.
Disclaimer for authors employed by the U.S. Federal Government: Dr. Roberto Romero has contributed to this work as part of his official duties as an employee of the U.S. Federal Government.
Paper Presentation Information: Not applicable.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009;326:1694–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knight R, Callewaert C, Marotz C, et al. The Microbiome and Human Biology. Annu Rev Genomics Hum Genet 2017;18:65–86. [DOI] [PubMed] [Google Scholar]
- 4.Fierer N Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 2017;15:579–90. [DOI] [PubMed] [Google Scholar]
- 5.Delgado-Baquerizo M, Oliverio AM, Brewer TE, et al. A global atlas of the dominant bacteria found in soil. Science 2018;359:320–25. [DOI] [PubMed] [Google Scholar]
- 6.Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004;304:66–74. [DOI] [PubMed] [Google Scholar]
- 7.Sunagawa S, Coelho LP, Chaffron S, et al. Ocean plankton. Structure and function of the global ocean microbiome. Science 2015;348:1261359. [DOI] [PubMed] [Google Scholar]
- 8.Mende DR, Bryant JA, Aylward FO, et al. Environmental drivers of a microbial genomic transition zone in the ocean's interior. Nat Microbiol 2017;2:1367–73. [DOI] [PubMed] [Google Scholar]
- 9.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome 2015;3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamont RF, Sobel JD, Akins RA, et al. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG 2011;118:533–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108 Suppl 1:4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero R, Hassan SS, Gajer P, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome 2014;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero R, Hassan SS, Gajer P, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014;2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stout MJ, Zhou Y, Wylie KM, Tarr PI, Macones GA, Tuuli MG. Early pregnancy vaginal microbiome trends and preterm birth. Am J Obstet Gynecol 2017;217:356 e1–56 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science 2016;352:560–4. [DOI] [PubMed] [Google Scholar]
- 17.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol 2010;192:5002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, He J, Xue J, et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol 2015;17:699–710. [DOI] [PubMed] [Google Scholar]
- 19.Evaldson G, Heimdahl A, Kager L, Nord CE. The normal human anaerobic microflora. Scand J Infect Dis Suppl 1982;35:9–15. [PubMed] [Google Scholar]
- 20.Jespers V, Menten J, Smet H, et al. Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol 2012;12:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Goffau MC, Lager S, Salter SJ, et al. Recognizing the reagent microbiome. Nat Microbiol 2018;3:851–53. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasan S, Munch MM, Sizova MV, et al. More Easily Cultivated Than Identified: Classical Isolation With Molecular Identification of Vaginal Bacteria. J Infect Dis 2016;214 Suppl 1:S21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Browne HP, Forster SC, Anonye BO, et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 2016;533:543–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau JT, Whelan FJ, Herath I, et al. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med 2016;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sizova MV, Hohmann T, Hazen A, et al. New approaches for isolation of previously uncultivated oral bacteria. Appl Environ Microbiol 2012;78:194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson H, Rybalka A, Moazzez R, Dewhirst FE, Wade WG. In vitro culture of previously uncultured oral bacterial phylotypes. Appl Environ Microbiol 2015;81:8307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog 2016;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, Hofstaedter CE, Zhao C, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017;5:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh RL, Nelson MT, Pope CE, et al. How low can we go? The implications of low bacterial load in respiratory microbiota studies. Pneumonia (Nathan) 2018;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauder AP, Roche AM, Sherrill-Mix S, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016;4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bender JM, Li F, Adisetiyo H, et al. Quantification of variation and the impact of biomass in targeted 16S rRNA gene sequencing studies. Microbiome 2018;6:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler B Value of endometrial cultures in sterility investigation. Fertil Steril 1958;9:269–73. [DOI] [PubMed] [Google Scholar]
- 35.Teisala K Endometrial microbial flora of hysterectomy specimens. Eur J Obstet Gynecol Reprod Biol 1987;26:151–5. [DOI] [PubMed] [Google Scholar]
- 36.Stroup PE. Amniotic fluid infection and the intact fetal membrane. Obstet Gynecol 1962;19:736–9. [PubMed] [Google Scholar]
- 37.Harwick HJ, Iuppa JB, Fekety FR Jr. Microorganisms and amniotic fluid. Obstet Gynecol 1969;33:256–9. [PubMed] [Google Scholar]
- 38.Prevedourakis C, Papadimitriou G, Ioannidou A. Isolation of pathogenic bacteria in the amniotic fluid during pregnancy and labor. Am J Obstet Gynecol 1970;106:400–2. [DOI] [PubMed] [Google Scholar]
- 39.Prevedourakis CN, Strigou-Charalabis E, Kaskarelis DB. Bacterial invasion of amniotic cavity during pregnancy and labor. Obstet Gynecol 1971;37:459–61. [PubMed] [Google Scholar]
- 40.Lewis JF, Johnson P, Miller P. Evaluation of amniotic fluid for aerobic and anaerobic bacteria. Am J Clin Pathol 1976;65:58–63. [DOI] [PubMed] [Google Scholar]
- 41.DiGiulio DB, Romero R, Amogan HP, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One 2008;3:e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiGiulio DB, Gervasi M, Romero R, et al. Microbial invasion of the amniotic cavity in preeclampsia as assessed by cultivation and sequence-based methods. J Perinat Med 2010;38:503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiGiulio DB, Gervasi MT, Romero R, et al. Microbial invasion of the amniotic cavity in pregnancies with small-for-gestational-age fetuses. J Perinat Med 2010;38:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiGiulio DB, Romero R, Kusanovic JP, et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol 2010;64:38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gervasi MT, Romero R, Bracalente G, et al. Viral invasion of the amniotic cavity (VIAC) in the midtrimester of pregnancy. J Matern Fetal Neonatal Med 2012;25:2002–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gervasi MT, Romero R, Bracalente G, et al. Midtrimester amniotic fluid concentrations of interleukin-6 and interferon-gamma-inducible protein-10: evidence for heterogeneity of intra-amniotic inflammation and associations with spontaneous early (<32 weeks) and late (>32 weeks) preterm delivery. J Perinat Med 2012;40:329–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romero R, Miranda J, Chaiworapongsa T, et al. A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am J Reprod Immunol 2014;71:330–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romero R, Miranda J, Chaiworapongsa T, et al. Sterile intra-amniotic inflammation in asymptomatic patients with a sonographic short cervix: prevalence and clinical significance. J Matern Fetal Neonatal Med 2014:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Combs CA, Gravett M, Garite TJ, et al. Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am J Obstet Gynecol 2014;210:125 e1–25 e15. [DOI] [PubMed] [Google Scholar]
- 50.Romero R, Miranda J, Chaiworapongsa T, et al. Prevalence and clinical significance of sterile intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Reprod immunol 2014;72:458–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romero R, Miranda J, Chaemsaithong P, et al. Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med 2015;28:1394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh KJ, Kim SM, Hong JS, et al. Twenty-four percent of patients with clinical chorioamnionitis in preterm gestations have no evidence of either culture-proven intraamniotic infection or intraamniotic inflammation. Am J Obstet Gynecol 2017;216:604 e1–04 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pacora P, Romero R, Erez O, et al. The diagnostic performance of the beta-glucan assay in the detection of intra-amniotic infection with Candida species. J Matern Fetal Neonatal Med 2017:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowlands S, Danielewski JA, Tabrizi SN, Walker SP, Garland SM. Microbial invasion of the amniotic cavity in midtrimester pregnancies using molecular microbiology. Am J Obstet Gynecol 2017;217:71 e1–71 e5. [DOI] [PubMed] [Google Scholar]
- 55.Gomez-Lopez N, Romero R, Panaitescu B, et al. Inflammasome activation during spontaneous preterm labor with intra-amniotic infection or sterile intra-amniotic inflammation. Am J Reprod immunol 2018:e13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim ES, Rodriguez C, Holtz LR. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 2018;6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rehbinder EM, Lodrup Carlsen KC, Staff AC, et al. Is amniotic fluid of women with uncomplicated term pregnancies free of bacteria? Am J Obstet Gynecol 2018;219:289 e1–89 e12. [DOI] [PubMed] [Google Scholar]
- 58.Aquino TI, Zhang J, Kraus FT, Knefel R, Taff T. Subchorionic fibrin cultures for bacteriologic study of the placenta. Am J Clin Pathol 1984;81:482–6. [DOI] [PubMed] [Google Scholar]
- 59.Romero R, Kim YM, Pacora P, et al. The frequency and type of placental histologic lesions in term pregnancies with normal outcome. J Perinat Med 2018;46:613–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010;107:11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shin H, Pei Z, Martinez KA 2nd, et al. The first microbial environment of infants born by C-section: the operating room microbes. Microbiome 2015;3:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 2016;22:250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perez-Munoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the "sterile womb" and "in utero colonization" hypotheses: implications for research on the pioneer infant microbiome. Microbiome 2017;5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med 2014;6:237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doyle RM, Alber DG, Jones HE, et al. Term and preterm labour are associated with distinct microbial community structures in placental membranes which are independent of mode of delivery. Placenta 2014;35:1099–101. [DOI] [PubMed] [Google Scholar]
- 66.Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obstet Gynecol 2015;212:653 e1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng J, Xiao X, Zhang Q, Mao L, Yu M, Xu J. The Placental Microbiome Varies in Association with Low Birth Weight in Full-Term Neonates. Nutrients 2015;7:6924–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bassols J, Serino M, Carreras-Badosa G, et al. Gestational diabetes is associated with changes in placental microbiota and microbiome. Pediatr Res 2016;80:777–84. [DOI] [PubMed] [Google Scholar]
- 69.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 2016;6:23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prince AL, Ma J, Kannan PS, et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol 2016;214:627 e1–27 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doyle RM, Harris K, Kamiza S, et al. Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLoS One 2017;12:e0180167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Nitert MD. Contributions of the maternal oral and gut microbiome to placental microbial colonization in overweight and obese pregnant women. Sci Rep 2017;7:2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parnell LA, Briggs CM, Cao B, Delannoy-Bruno O, Schrieffer AE, Mysorekar IU. Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Sci Rep 2017;7:11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng J, Xiao XH, Zhang Q, et al. Correlation of placental microbiota with fetal macrosomia and clinical characteristics in mothers and newborns. Oncotarget 2017;8:82314–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leon LJ, Doyle R, Diez-Benavente E, et al. Enrichment of clinically relevant organisms in spontaneous preterm delivered placenta and reagent contamination across all clinical groups in a large UK pregnancy cohort. Appl Environ Microbiol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitchell CM, Haick A, Nkwopara E, et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol 2015;212:611 e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franasiak JM, Werner MD, Juneau CR, et al. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet 2016;33:129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moreno I, Codoner FM, Vilella F, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol 2016;215:684–703. [DOI] [PubMed] [Google Scholar]
- 79.Verstraelen H, Vilchez-Vargas R, Desimpel F, et al. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1–2 region of the 16S rRNA gene. PeerJ 2016;4:e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen C, Song X, Wei W, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun 2017;8:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.TAO X, Franasiak J, Zhan Y, et al. Characterizing the endometrial microbiome by analyzing the ultra-low bacteria from embryo transfer catheter tips in IVF cycles: Next generation sequencing (NGS) analysis of the 16S ribosomal gene. Human Microbiome Journal 2017;3:15–21. [Google Scholar]
- 82.Kyono K, Hashimoto T, Nagai Y, Sakuraba Y. Analysis of endometrial microbiota by 16S ribosomal RNA gene sequencing among infertile patients: a single-center pilot study. Reprod Med Biol 2018;17:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prince AL, Chu DM, Seferovic MD, Antony KM, Ma J, Aagaard KM. The perinatal microbiome and pregnancy: moving beyond the vaginal microbiome. Cold Spring Harb Perspect Med 2015;5:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giudice LC. Challenging dogma: the endometrium has a microbiome with functional consequences! Am J Obstet Gynecol 2016;215:682–83. [DOI] [PubMed] [Google Scholar]
- 85.Moreno I, Franasiak JM. Endometrial microbiota-new player in town. Fertil Steril 2017;108:32–39. [DOI] [PubMed] [Google Scholar]
- 86.Pelzer E, Gomez-Arango LF, Barrett HL, Nitert MD. Review: Maternal health and the placental microbiome. Placenta 2017;54:30–37. [DOI] [PubMed] [Google Scholar]
- 87.Benner MFG, Joosten I, van der Molen RG. How uterine microbiota might be responsible for a receptive, fertile endometrium. Human Reproduction Update In Press. [DOI] [PubMed] [Google Scholar]
- 88.Pankuch GA, Appelbaum PC, Lorenz RP, Botti JJ, Schachter J, Naeye RL. Placental microbiology and histology and the pathogenesis of chorioamnionitis. Obstet Gynecol 1984;64:802–6. [PubMed] [Google Scholar]
- 89.Quinn PA, Butany J, Taylor J, Hannah W. Chorioamnionitis: its association with pregnancy outcome and microbial infection. Am J Obstet Gynecol 1987;156:379–87. [DOI] [PubMed] [Google Scholar]
- 90.Hillier SL, Krohn MA, Kiviat NB, Watts DH, Eschenbach DA. Microbiologic causes and neonatal outcomes associated with chorioamnion infection. Am J Obstet Gynecol 1991;165:955–61. [DOI] [PubMed] [Google Scholar]
- 91.Hecht JL, Onderdonk A, Delaney M, et al. Characterization of chorioamnionitis in 2nd-trimester C-section placentas and correlation with microorganism recovery from subamniotic tissues. Pediatr Dev Pathol 2008;11:15–22. [DOI] [PubMed] [Google Scholar]
- 92.Kim MJ, Romero R, Gervasi MT, et al. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab Invest 2009;89:924–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM. Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol 2015;213:S29–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garmi G, Okopnik M, Keness Y, Zafran N, Berkowitz E, Salim R. Correlation between Clinical, Placental Histology and Microbiological Findings in Spontaneous Preterm Births. Fetal Diagn Ther 2016;40:141–9. [DOI] [PubMed] [Google Scholar]
- 95.Sweeney EL, Kallapur SG, Gisslen T, et al. Placental Infection With Ureaplasma species Is Associated With Histologic Chorioamnionitis and Adverse Outcomes in Moderately Preterm and Late-Preterm Infants. J Infect Dis 2016;213:1340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lannon SMR, Adams Waldorf KM, Fiedler T, et al. Parallel detection of lactobacillus and bacterial vaginosis-associated bacterial DNA in the chorioamnion and vagina of pregnant women at term. J Matern Fetal Neonatal Med 2018:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garcia AG. Fetal Infection in Chickenpox and Alastrim, with Histopathologic Study of the Placenta. Pediatrics 1963;32:895–901. [PubMed] [Google Scholar]
- 98.Purtilo DT, Bhawan J, Liao S, Brutus A, Yang JP, Balogh K. Fatal varicella in a pregnant woman and a baby. Am J Obstet Gynecol 1977;127:208–9. [DOI] [PubMed] [Google Scholar]
- 99.Garcia AG, Marques RL, Lobato YY, Fonseca ME, Wigg MD. Placental pathology in congenital rubella. Placenta 1985;6:281–95. [DOI] [PubMed] [Google Scholar]
- 100.Garcia AG, Basso NG, Fonseca ME, Outani HN. Congenital echo virus infection--morphological and virological study of fetal and placental tissue. J Pathol 1990;160:123–7. [DOI] [PubMed] [Google Scholar]
- 101.Qureshi F, Jacques SM. Maternal varicella during pregnancy: correlation of maternal history and fetal outcome with placental histopathology. Hum Pathol 1996;27:191–5. [DOI] [PubMed] [Google Scholar]
- 102.Benirschke K, Coen R, Patterson B, Key T. Villitis of known origin: varicella and toxoplasma. Placenta 1999;20:395–9. [DOI] [PubMed] [Google Scholar]
- 103.Satosar A, Ramirez NC, Bartholomew D, Davis J, Nuovo GJ. Histologic correlates of viral and bacterial infection of the placenta associated with severe morbidity and mortality in the newborn. Hum Pathol 2004;35:536–45. [DOI] [PubMed] [Google Scholar]
- 104.Onderdonk AB, Delaney ML, DuBois AM, Allred EN, Leviton A, Extremely Low Gestational Age Newborns Study I. Detection of bacteria in placental tissues obtained from extremely low gestational age neonates. Am J Obstet Gynecol 2008;198:110 e1–7. [DOI] [PubMed] [Google Scholar]
- 105.Onderdonk AB, Hecht JL, McElrath TF, et al. Colonization of second-trimester placenta parenchyma. Am J Obstet Gynecol 2008;199:52 e1–52 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cardenas I, Mor G, Aldo P, et al. Placental viral infection sensitizes to endotoxin-induced preterm labor: a double hit hypothesis. Am J Reprod Immunol 2011;65:110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stout MJ, Conlon B, Landeau M, et al. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol 2013;208:226 e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Whidbey C, Harrell MI, Burnside K, et al. A hemolytic pigment of Group B Streptococcus allows bacterial penetration of human placenta. J Exp Med 2013;210:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mouillet JF, Ouyang Y, Bayer A, Coyne CB, Sadovsky Y. The role of trophoblastic microRNAs in placental viral infection. Int J Dev Biol 2014;58:281–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bayer A, Delorme-Axford E, Sleigher C, et al. Human trophoblasts confer resistance to viruses implicated in perinatal infection. Am J Obstet Gynecol 2015;212:71 e1–71 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jurado KA, Simoni MK, Tang Z, et al. Zika virus productively infects primary human placenta-specific macrophages. JCI Insight 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quicke KM, Bowen JR, Johnson EL, et al. Zika Virus Infects Human Placental Macrophages. Cell Host Microbe 2016;20:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rosenberg AZ, Yu W, Hill DA, Reyes CA, Schwartz DA. Placental Pathology of Zika Virus: Viral Infection of the Placenta Induces Villous Stromal Macrophage (Hofbauer Cell) Proliferation and Hyperplasia. Arch Pathol Lab Med 2017;141:43–48. [DOI] [PubMed] [Google Scholar]
- 114.Sheridan MA, Yunusov D, Balaraman V, et al. Vulnerability of primitive human placental trophoblast to Zika virus. Proc Natl Acad Sci U S A 2017;114:E1587–E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stanek J Placental infectious villitis versus villitis of unknown etiology. Pol J Pathol 2017;68:55–65. [DOI] [PubMed] [Google Scholar]
- 116.Arora N, Sadovsky Y, Dermody TS, Coyne CB. Microbial Vertical Transmission during Human Pregnancy. Cell Host Microbe 2017;21:561–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Romero R, Mazor M, Wu YK, et al. Infection in the pathogenesis of preterm labor. Semin Perinatol 1988;12:262–79. [PubMed] [Google Scholar]
- 118.Romero R, Emamian M, Quintero R, et al. The value and limitations of the Gram stain examination in the diagnosis of intraamniotic infection. Am J Obstet Gynecol 1988;159:114–9. [DOI] [PubMed] [Google Scholar]
- 119.Romero R, Roslansky P, Oyarzun E, et al. Labor and infection. II. Bacterial endotoxin in amniotic fluid and its relationship to the onset of preterm labor. Am J Obstet Gynecol 1988;158:1044–9. [DOI] [PubMed] [Google Scholar]
- 120.Romero R, Avila C, Santhanam U, Sehgal PB. Amniotic fluid interleukin 6 in preterm labor. Association with infection. J Clin Invest 1990;85:1392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Romero R, Jimenez C, Lohda AK, et al. Amniotic fluid glucose concentration: a rapid and simple method for the detection of intraamniotic infection in preterm labor. Am J Obstet Gynecol 1990;163:968–74. [DOI] [PubMed] [Google Scholar]
- 122.Romero R, Shamma F, Avila C, et al. Infection and labor. VI. Prevalence, microbiology, and clinical significance of intraamniotic infection in twin gestations with preterm labor. Am J Obstet Gynecol 1990;163:757–61. [DOI] [PubMed] [Google Scholar]
- 123.Romero R, Yoon BH, Mazor M, et al. The diagnostic and prognostic value of amniotic fluid white blood cell count, glucose, interleukin-6, and gram stain in patients with preterm labor and intact membranes. Am J Obstet Gynecol 1993;169:805–16. [DOI] [PubMed] [Google Scholar]
- 124.Gravett MG, Witkin SS, Haluska GJ, Edwards JL, Cook MJ, Novy MJ. An experimental model for intraamniotic infection and preterm labor in rhesus monkeys. Am J Obstet Gynecol 1994;171:1660–7. [DOI] [PubMed] [Google Scholar]
- 125.Kundsin RB, Leviton A, Allred EN, Poulin SA. Ureaplasma urealyticum infection of the placenta in pregnancies that ended prematurely. Obstet Gynecol 1996;87:122–7. [DOI] [PubMed] [Google Scholar]
- 126.Espinoza J, Goncalves LF, Romero R, et al. The prevalence and clinical significance of amniotic fluid 'sludge' in patients with preterm labor and intact membranes. Ultrasound Obstet Gynecol 2005;25:346–52. [DOI] [PubMed] [Google Scholar]
- 127.Kusanovic JP, Espinoza J, Romero R, et al. Clinical significance of the presence of amniotic fluid 'sludge' in asymptomatic patients at high risk for spontaneous preterm delivery. Ultrasound Obstet Gynecol 2007;30:706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Romero R, Schaudinn C, Kusanovic JP, et al. Detection of a microbial biofilm in intraamniotic infection. Am J Obstet Gynecol 2008;198:135 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cardenas I, Means RE, Aldo P, et al. Viral infection of the placenta leads to fetal inflammation and sensitization to bacterial products predisposing to preterm labor. J Immunol 2010;185:1248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cao B, Stout MJ, Lee I, Mysorekar IU. Placental Microbiome and Its Role in Preterm Birth. Neoreviews 2014;15:e537–e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Allen-Daniels MJ, Serrano MG, Pflugner LP, et al. Identification of a gene in Mycoplasma hominis associated with preterm birth and microbial burden in intraamniotic infection. Am J Obstet Gynecol 2015;212:779 e1–79 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chaemsaithong P, Romero R, Korzeniewski SJ, et al. A point of care test for the determination of amniotic fluid interleukin-6 and the chemokine CXCL-10/IP-10. J Matern Fetal Neonatal Med 2015;28:1510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chaemsaithong P, Romero R, Korzeniewski SJ, et al. A rapid interleukin-6 bedside test for the identification of intra-amniotic inflammation in preterm labor with intact membranes. J Matern Fetal Neonatal Med 2016;29:349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park JY, Romero R, Lee J, Chaemsaithong P, Chaiyasit N, Yoon BH. An elevated amniotic fluid prostaglandin F2alpha concentration is associated with intra-amniotic inflammation/infection, and clinical and histologic chorioamnionitis, as well as impending preterm delivery in patients with preterm labor and intact membranes. J Matern Fetal Neonatal Med 2016;29:2563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Boldenow E, Gendrin C, Ngo L, et al. Group B Streptococcus circumvents neutrophils and neutrophil extracellular traps during amniotic cavity invasion and preterm labor. Sci Immunol 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Racicot K, Kwon JY, Aldo P, et al. Type I Interferon Regulates the Placental Inflammatory Response to Bacteria and is Targeted by Virus: Mechanism of Polymicrobial Infection-Induced Preterm Birth. Am J Reprod Immunol 2016;75:451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gomez-Lopez N, Romero R, Garcia-Flores V, et al. Amniotic fluid neutrophils can phagocytize bacteria: A mechanism for microbial killing in the amniotic cavity. Am J Reprod Immunol 2017;78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gomez-Lopez N, Romero R, Xu Y, et al. Are amniotic fluid neutrophils in women with intraamniotic infection and/or inflammation of fetal or maternal origin? Am J Obstet Gynecol 2017;217:693 e1–93 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oh KJ, Hong JS, Romero R, Yoon BH. The frequency and clinical significance of intra-amniotic inflammation in twin pregnancies with preterm labor and intact membranes. J Matern Fetal Neonatal Med 2017:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chaemsaithong P, Romero R, Docheva N, et al. Comparison of rapid MMP-8 and interleukin-6 point-of-care tests to identify intra-amniotic inflammation/infection and impending preterm delivery in patients with preterm labor and intact membranes(). J Matern Fetal Neonatal Med 2018;31:228–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kusanovic JP, Romero R, Martinovic C, et al. Transabdominal collection of amniotic fluid "sludge" and identification of Candida albicans intra-amniotic infection. J Matern Fetal Neonatal Med 2018;31:1279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Varrey A, Romero R, Panaitescu B, et al. Human beta-defensin-1: A natural antimicrobial peptide present in amniotic fluid that is increased in spontaneous preterm labor with intra-amniotic infection. Am J Reprod Immunol 2018;80:e13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Romero R, Quintero R, Oyarzun E, et al. Intraamniotic infection and the onset of labor in preterm premature rupture of the membranes. Am J Obstet Gynecol 1988;159:661–6. [DOI] [PubMed] [Google Scholar]
- 144.Romero R, Ghidini A, Mazor M, Behnke E. Microbial invasion of the amniotic cavity in premature rupture of membranes. Clin Obstet Gynecol 1991;34:769–78. [DOI] [PubMed] [Google Scholar]
- 145.Gomez R, Romero R, Edwin SS, David C. Pathogenesis of preterm labor and preterm premature rupture of membranes associated with intraamniotic infection. Infect Dis Clin North Am 1997;11:135–76. [DOI] [PubMed] [Google Scholar]
- 146.Chaemsaithong P, Romero R, Korzeniewski SJ, et al. A point of care test for interleukin-6 in amniotic fluid in preterm prelabor rupture of membranes: a step toward the early treatment of acute intra-amniotic inflammation/infection. J Matern Fetal Neonatal Med 2016;29:360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Romero R, Gonzalez R, Sepulveda W, et al. Infection and labor. VIII. Microbial invasion of the amniotic cavity in patients with suspected cervical incompetence: prevalence and clinical significance. Am J Obstet Gynecol 1992;167:1086–91. [DOI] [PubMed] [Google Scholar]
- 148.Hassan S, Romero R, Hendler I, et al. A sonographic short cervix as the only clinical manifestation of intra-amniotic infection. J Perinat Med 2006;34:13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Romero R, Kusanovic JP, Espinoza J, et al. What is amniotic fluid 'sludge'? Ultrasound Obstet Gynecol 2007;30:793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee SE, Romero R, Park CW, Jun JK, Yoon BH. The frequency and significance of intraamniotic inflammation in patients with cervical insufficiency. Am J Obstet Gynecol 2008;198:633 e1–8. [DOI] [PubMed] [Google Scholar]
- 151.Chaiworapongsa T, Hong JS, Hull WM, Romero R, Whitsett JA. Amniotic fluid concentration of surfactant proteins in intra-amniotic infection. J Matern Fetal Neonatal Med 2008;21:663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Oh KJ, Lee SE, Jung H, Kim G, Romero R, Yoon BH. Detection of ureaplasmas by the polymerase chain reaction in the amniotic fluid of patients with cervical insufficiency. J Perinat Med 2010;38:261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Paules C, Moreno E, Gonzales A, Fabre E, Gonzalez de Aguero R, Oros D. Amniotic fluid sludge as a marker of intra-amniotic infection and histological chorioamnionitis in cervical insufficiency: a report of four cases and literature review. J Matern Fetal Neonatal Med 2016;29:2681–4. [DOI] [PubMed] [Google Scholar]
- 154.Kim YM, Park KH, Park H, Yoo HN, Kook SY, Jeon SJ. Complement C3a, But Not C5a, Levels in Amniotic Fluid Are Associated with Intra-amniotic Infection and/or Inflammation and Preterm Delivery in Women with Cervical Insufficiency or an Asymptomatic Short Cervix (</= 25 mm). J Korean Med Sci 2018;33:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gibbs RS, Blanco JD, St Clair PJ, Castaneda YS. Quantitative bacteriology of amniotic fluid from women with clinical intraamniotic infection at term. J Infect Dis 1982;145:1–8. [DOI] [PubMed] [Google Scholar]
- 156.Hauth JC, Gilstrap LC 3rd, Hankins GD, Connor KD Term maternal and neonatal complications of acute chorioamnionitis. Obstet Gynecol 1985;66:59–62. [PubMed] [Google Scholar]
- 157.Romero R, Nores J, Mazor M, et al. Microbial invasion of the amniotic cavity during term labor. Prevalence and clinical significance. J Reprod Med 1993;38:543–8. [PubMed] [Google Scholar]
- 158.Newton ER. Chorioamnionitis and intraamniotic infection. Clin Obstet Gynecol 1993;36:795–808. [DOI] [PubMed] [Google Scholar]
- 159.Gibbs RS. Management of clinical chorioamnionitis at term. Am J Obstet Gynecol 2004;191:1–2. [DOI] [PubMed] [Google Scholar]
- 160.Tita AT, Andrews WW. Diagnosis and management of clinical chorioamnionitis. Clin Perinatol 2010;37:339–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Romero R, Chaiworapongsa T, Savasan ZA, et al. Clinical chorioamnionitis is characterized by changes in the expression of the alarmin HMGB1 and one of its receptors, sRAGE. J Matern Fetal Neonatal Med 2012;25:558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Romero R, Miranda J, Kusanovic JP, et al. Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques. J Perinat Med 2015;43:19–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Romero R, Chaemsaithong P, Korzeniewski SJ, et al. Clinical chorioamnionitis at term II: the intra-amniotic inflammatory response. J Perinat Med 2016;44:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Romero R, Chaemsaithong P, Korzeniewski SJ, et al. Clinical chorioamnionitis at term III: how well do clinical criteria perform in the identification of proven intra-amniotic infection? J Perinat Med 2016;44:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mazaki-Tovi S, Vaisbuch E. Clinical chorioamnionitis--an ongoing obstetrical conundrum. J Perinat Med 2016;44:1–4. [DOI] [PubMed] [Google Scholar]
- 166.Romero R, Chaemsaithong P, Docheva N, et al. Clinical chorioamnionitis at term IV: the maternal plasma cytokine profile. J Perinat Med 2016;44:77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Romero R, Chaemsaithong P, Docheva N, et al. Clinical chorioamnionitis at term V: umbilical cord plasma cytokine profile in the context of a systemic maternal inflammatory response. J Perinat Med 2016;44:53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Romero R, Chaemsaithong P, Docheva N, et al. Clinical chorioamnionitis at term VI: acute chorioamnionitis and funisitis according to the presence or absence of microorganisms and inflammation in the amniotic cavity. J Perinat Med 2016;44:33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Maddipati KR, Romero R, Chaiworapongsa T, et al. Clinical chorioamnionitis at term: the amniotic fluid fatty acyl lipidome. J Lipid Res 2016;57:1906–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Martinez-Varea A, Romero R, Xu Y, et al. Clinical chorioamnionitis at term VII: the amniotic fluid cellular immune response. J Perinat Med 2017;45:523–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fouks Y, Many A, Orbach R, et al. Is There a Role for Placental Cultures in Cases of Clinical Chorioamnionitis Complicating Preterm Premature Rupture of Membranes? Am J Perinatol 2017;34:867–73. [DOI] [PubMed] [Google Scholar]
- 172.Chaiyasit N, Romero R, Chaemsaithong P, et al. Clinical chorioamnionitis at term VIII: a rapid MMP-8 test for the identification of intra-amniotic inflammation. J Perinat Med 2017;45:539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mostoufi-zadeh M, Driscoll SG, Biano SA, Kundsin RB. Placental evidence of cytomegalovirus infection of the fetus and neonate. Arch Pathol Lab Med 1984;108:403–6. [PubMed] [Google Scholar]
- 174.Yoon BH, Jun JK, Romero R, et al. Amniotic fluid inflammatory cytokines (interleukin-6, interleukin-1beta, and tumor necrosis factor-alpha), neonatal brain white matter lesions, and cerebral palsy. Am J Obstet Gynecol 1997;177:19–26. [DOI] [PubMed] [Google Scholar]
- 175.Yoon BH, Kim CJ, Romero R, et al. Experimentally induced intrauterine infection causes fetal brain white matter lesions in rabbits. Am J Obstet Gynecol 1997;177:797–802. [DOI] [PubMed] [Google Scholar]
- 176.Yoon BH, Romero R, Kim CJ, et al. High expression of tumor necrosis factor-alpha and interleukin-6 in periventricular leukomalacia. Am J Obstet Gynecol 1997;177:406–11. [DOI] [PubMed] [Google Scholar]
- 177.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am J Obstet Gynecol 1998;179:194–202. [DOI] [PubMed] [Google Scholar]
- 178.Yoon BH, Romero R, Park JS, et al. The relationship among inflammatory lesions of the umbilical cord (funisitis), umbilical cord plasma interleukin 6 concentration, amniotic fluid infection, and neonatal sepsis. Am J Obstet Gynecol 2000;183:1124–9. [DOI] [PubMed] [Google Scholar]
- 179.Euscher E, Davis J, Holzman I, Nuovo GJ. Coxsackie virus infection of the placenta associated with neurodevelopmental delays in the newborn. Obstet Gynecol 2001;98:1019–26. [DOI] [PubMed] [Google Scholar]
- 180.Kim CJ, Yoon BH, Romero R, et al. Umbilical arteritis and phlebitis mark different stages of the fetal inflammatory response. Am J Obstet Gynecol 2001;185:496–500. [DOI] [PubMed] [Google Scholar]
- 181.Yoon BH, Romero R, Shim JY, Shim SS, Kim CJ, Jun JK. C-reactive protein in umbilical cord blood: a simple and widely available clinical method to assess the risk of amniotic fluid infection and funisitis. J Matern Fetal Neonatal Med 2003;14:85–90. [DOI] [PubMed] [Google Scholar]
- 182.Gotsch F, Romero R, Kusanovic JP, et al. The fetal inflammatory response syndrome. Clin Obstet Gynecol 2007;50:652–83. [DOI] [PubMed] [Google Scholar]
- 183.Korzeniewski SJ, Romero R, Cortez J, et al. A "multi-hit" model of neonatal white matter injury: cumulative contributions of chronic placental inflammation, acute fetal inflammation and postnatal inflammatory events. J Perinat Med 2014;42:731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kim SM, Romero R, Lee J, Chaemsaithong P, Docheva N, Yoon BH. Gastric fluid versus amniotic fluid analysis for the identification of intra-amniotic infection due to Ureaplasma species. J Matern Fetal Neonatal Med 2016;29:2579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee J, Romero R, Lee KA, et al. Meconium aspiration syndrome: a role for fetal systemic inflammation. Am J Obstet Gynecol 2016;214:366 e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mor G Placental Inflammatory Response to Zika Virus may Affect Fetal Brain Development. Am J Reprod Immunol 2016;75:421–2. [DOI] [PubMed] [Google Scholar]
- 187.Kapisi J, Kakuru A, Jagannathan P, et al. Relationships between infection with Plasmodium falciparum during pregnancy, measures of placental malaria, and adverse birth outcomes. Malar J 2017;16:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kliman HJ. Comment on "the placenta harbors a unique microbiome". Sci Transl Med 2014;6:254le4. [DOI] [PubMed] [Google Scholar]
- 189.Hornef M, Penders J. Does a prenatal bacterial microbiota exist? Mucosal Immunol 2017;10:598–601. [DOI] [PubMed] [Google Scholar]
- 190.Willyard C Baby’s first bacteria: The womb was thought to be sterile; some scientists argue it’s where the microbiome begins. Nature 2018;553:264–66.29345664 [Google Scholar]
- 191.Weiss S, Amir A, Hyde ER, Metcalf JL, Song SJ, Knight R. Tracking down the sources of experimental contamination in microbiome studies. Genome Biol 2014;15:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Elkomy MH, Sultan P, Drover DR, Epshtein E, Galinkin JL, Carvalho B. Pharmacokinetics of prophylactic cefazolin in parturients undergoing cesarean delivery. Antimicrob Agents Chemother 2014;58:3504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Smaill FM, Grivell RM. Antibiotic prophylaxis versus no prophylaxis for preventing infection after cesarean section. Cochrane Database Syst Rev 2014:CD007482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Maggio L, Nicolau DP, DaCosta M, Rouse DJ, Hughes BL. Cefazolin prophylaxis in obese women undergoing cesarean delivery: a randomized controlled trial. Obstet Gynecol 2015;125:1205–10. [DOI] [PubMed] [Google Scholar]
- 195.Fan Z-Y, Li X-R, Mao D-P, Zhu G-F, Wang S-Y, Quan Z-X. Could nested PCR be applicable for the study of microbial diversity? World Journal of Microbiology and Biotechnology 2009;25:1447–52. [Google Scholar]
- 196.Yu G, Fadrosh D, Goedert JJ, Ravel J, Goldstein AM. Nested PCR Biases in Interpreting Microbial Community Structure in 16S rRNA Gene Sequence Datasets. PLoS One 2015;10:e0132253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kostric M, Milger K, Krauss-Etschmann S, et al. Development of a Stable Lung Microbiome in Healthy Neonatal Mice. Microb Ecol 2018;75:529–42. [DOI] [PubMed] [Google Scholar]
- 198.Glendinning L, Wright S, Pollock J, Tennant P, Collie D, McLachlan G. Variability of the Sheep Lung Microbiota. Appl Environ Microbiol 2016;82:3225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Glendinning L, McLachlan G, Vervelde L. Age-related differences in the respiratory microbiota of chickens. PLoS One 2017;12:e0188455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Neeff M, Biswas K, Hoggard M, Taylor MW, Douglas R. Molecular Microbiological Profile of Chronic Suppurative Otitis Media. J Clin Microbiol 2016;54:2538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sillanpaa S, Kramna L, Oikarinen S, et al. Next-Generation Sequencing Combined with Specific PCR Assays To Determine the Bacterial 16S rRNA Gene Profiles of Middle Ear Fluid Collected from Children with Acute Otitis Media. mSphere 2017;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991;173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 2008;74:2461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dickson RP, Erb-Downward JR, Freeman CM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS One 2014;9:e97214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 2008;9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang Y, Qian PY. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One 2009;4:e7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009;37:D141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007;35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ong SH, Kukkillaya VU, Wilm A, et al. Species identification and profiling of complex microbial communities using shotgun Illumina sequencing of 16S rRNA amplicon sequences. PLoS One 2013;8:e60811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 1991;19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 2015;6:e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann Am Thorac Soc 2015;12:821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Venkataraman A, Bassis CM, Beck JM, et al. Application of a neutral community model to assess structuring of the human lung microbiome. MBio 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol 2016;1:16113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dickson RP, Erb-Downward JR, Freeman CM, et al. Bacterial Topography of the Healthy Human Lower Respiratory Tract. MBio 2017;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Singer BH, Dickson RP, Denstaedt SJ, et al. Bacterial Dissemination to the Brain in Sepsis. Am J Respir Crit Care Med 2018;197:747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Magurran AE. Measuring Biological Diversity. Malden, MA: Blackwell Publishing. [Google Scholar]
- 223.Magurran AE. Biological Diversity: Frontiers in Measurement and Assessment. New York, NY: Oxford University Press. [Google Scholar]
- 224.HAMMER O, HARPER DAT, RYAN PD. PAST: PAleontological STatistics software package for education and data analysis. Palaeontologia Electronica 2001;4:1–9. [Google Scholar]
- 225.Quinn GP, Keough MJ. Experimental Design and Data Analysis for Biologists. Cambridge: Cambridge University Press. [Google Scholar]
- 226.Gotelli NJ, Ellison AM. A Primer of Ecological Statistics. Sunderland, MA: Sinauer Associates, Inc.. [Google Scholar]
- 227.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology 2001;26:32–46. [Google Scholar]
- 228.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics 2003;19:295–6. [DOI] [PubMed] [Google Scholar]
- 229.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Knights D, Kuczynski J, Charlson ES, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 2011;8:761–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Khong TY, Mooney EE, Ariel I, et al. Sampling and Definitions of Placental Lesions: Amsterdam Placental Workshop Group Consensus Statement. Arch Pathol Lab Med 2016;140:698–713. [DOI] [PubMed] [Google Scholar]
- 232.Lager S, de Goffau MC, Sovio U, et al. Detecting eukaryotic microbiota with single-cell sensitivity in human tissue. Microbiome 2018;6:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Logan NA. Bacillus species of medical and veterinary importance. J Med Microbiol 1988;25:157–65. [DOI] [PubMed] [Google Scholar]
- 234.Workowski KA, Flaherty JP. Systemic Bacillus species infection mimicking listeriosis of pregnancy. Clin Infect Dis 1992;14:694–6. [DOI] [PubMed] [Google Scholar]
- 235.Farrar WE, Reboli AC. The genus Bacillus - Medical. Prokaryotes 2006;4:609–30. [Google Scholar]
- 236.Mallozzi M, Viswanathan VK, Vedantam G. Spore-forming Bacilli and Clostridia in human disease. Future Microbiol 2010;5:1109–23. [DOI] [PubMed] [Google Scholar]
- 237.Kumar VA, Augustine D, Panikar D, et al. Brevibacterium casei as a cause of brain abscess in an immunocompetent patient. J Clin Microbiol 2011;49:4374–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ting JY, Wong SS, Chee YY, Wong KY, Tsoi NN, Lau AS. Bacillus bloodstream infections in a tertiary perinatal centre: an 8-year study. Am J Perinatol 2013;30:309–15. [DOI] [PubMed] [Google Scholar]
- 239.Bal ZS, Sen S, Karapinar DY, Aydemir S, Vardar F. The first reported catheter-related Brevibacterium casei bloodstream infection in a child with acute leukemia and review of the literature. Braz J Infect Dis 2015;19:213–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mushtaq A, Chen DJ, Strand GJ, et al. Clinical significance of coryneform Gram-positive rods from blood identified by MALDI-TOF mass spectrometry and their susceptibility profiles - a retrospective chart review. Diagn Microbiol Infect Dis 2016;85:372–76. [DOI] [PubMed] [Google Scholar]
- 241.Zhu L, Luo F, Hu W, et al. Bacterial Communities in the Womb During Healthy Pregnancy. Front Microbiol 2018;9:2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bandyopadhyay A, Elvitigala T, Welsh E, et al. Novel metabolic attributes of the genus cyanothece, comprising a group of unicellular nitrogen-fixing Cyanothece. MBio 2011;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Eisen JA, Nelson KE, Paulsen IT, et al. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc Natl Acad Sci U S A 2002;99:9509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kageyama A, Benno Y. Coprobacillus catenaformis gen. nov., sp. nov., a new genus and species isolated from human feces. Microbiol Immunol 2000;44:23–8. [DOI] [PubMed] [Google Scholar]
- 245.Chu D, Stewart C, Seferovic M, et al. 26: Profiling of microbiota in second trimester amniotic fluid reveals a distinctive community present in the mid trimester and predictive of the placental microbiome at parturition. Am J Obstet Gynecol 2017;216:S18–S19. [Google Scholar]
- 246.Kirk RG. "Life in a germ-free world": isolating life from the laboratory animal to the bubble boy. Bull Hist Med 2012;86:237–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Barnes RD, Tuffrey M, Cook R. A "germfree" human isolator. Lancet 1968;1:622–3. [DOI] [PubMed] [Google Scholar]
- 248.Barnes RD, Fairweather DV, Reynolds EO, Tuffrey M, Holliday J. A technique for the delivery of a germfree child. J Obstet Gynaecol Br Commonw 1968;75:689–97. [DOI] [PubMed] [Google Scholar]
- 249.Barnes RD, Fairweather DV, Holliday J, et al. A germfree infant. Lancet 1969;1:168–71. [DOI] [PubMed] [Google Scholar]
- 250.Hapfelmeier S, Lawson MA, Slack E, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010;328:1705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, et al. The maternal microbiota drives early postnatal innate immune development. Science 2016;351:1296–302. [DOI] [PubMed] [Google Scholar]
- 252.Marquardt N, Ivarsson MA, Sundstrom E, et al. Fetal CD103+ IL-17-Producing Group 3 Innate Lymphoid Cells Represent the Dominant Lymphocyte Subset in Human Amniotic Fluid. J Immunol 2016;197:3069–75. [DOI] [PubMed] [Google Scholar]
- 253.Gomez-Lopez N, Romero R, Xu Y, et al. The immunophenotype of amniotic fluid leukocytes in normal and complicated pregnancies. Am J Reprod Immunol 2018;79:e12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Seong HS, Lee SE, Kang JH, Romero R, Yoon BH. The frequency of microbial invasion of the amniotic cavity and histologic chorioamnionitis in women at term with intact membranes in the presence or absence of labor. Am J Obstet Gynecol 2008;199:375 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Moellering RC Jr., Swartz MN Drug therapy: The newer cephalosporins. N Engl J Med 1976;294:24–8. [DOI] [PubMed] [Google Scholar]
- 256.DiPiro JT, Vallner JJ, Bowden TA Jr., Clark BA, Sisley JF Intraoperative serum and tissue activity of cefazolin and cefoxitin. Arch Surg 1985;120:829–32. [DOI] [PubMed] [Google Scholar]
- 257.Craig WA. Interrelationship between pharmacokinetics and pharmacodynamics in determining dosage regimens for broad-spectrum cephalosporins. Diagnostic Microbiology and Infectious Disease 1995;22:89–96. [DOI] [PubMed] [Google Scholar]
- 258.Pevzner L, Swank M, Krepel C, Wing DA, Chan K, Edmiston CE Jr. Effects of maternal obesity on tissue concentrations of prophylactic cefazolin during cesarean delivery. Obstet Gynecol 2011;117:877–82. [DOI] [PubMed] [Google Scholar]
- 259.Stitely M, Sweet M, Slain D, et al. Plasma and tissue cefazolin concentrations in obese patients undergoing cesarean delivery and receiving differing pre-operative doses of drug. Surg Infect (Larchmt) 2013;14:455–9. [DOI] [PubMed] [Google Scholar]
- 260.Ahmadzia HK, Patel EM, Joshi D, et al. Obstetric Surgical Site Infections: 2 Grams Compared With 3 Grams of Cefazolin in Morbidly Obese Women. Obstet Gynecol 2015;126:708–15. [DOI] [PubMed] [Google Scholar]
- 261.Swank ML, Wing DA, Nicolau DP, McNulty JA. Increased 3-gram cefazolin dosing for cesarean delivery prophylaxis in obese women. Am J Obstet Gynecol 2015;213:415 e1–8. [DOI] [PubMed] [Google Scholar]
- 262.Groff SM, Fallatah W, Yang S, et al. Effect of Maternal Obesity on Maternal-Fetal Transfer of Preoperative Cefazolin at Cesarean Section. J Pediatr Pharmacol Ther 2017;22:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 1996;62:625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Brooks JP, Edwards DJ, Harwich MD Jr., et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol 2015;15:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Steel JH, Malatos S, Kennea N, et al. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr Res 2005;57:404–11. [DOI] [PubMed] [Google Scholar]
- 266.Minich JJ, Zhu Q, Janssen S, et al. KatharoSeq Enables High-Throughput Microbiome Analysis from Low-Biomass Samples. mSystems 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hearn YR, Gander RM. Achromobacter xylosoxidans. An unusual neonatal pathogen. Am J Clin Pathol 1991;96:211–4. [DOI] [PubMed] [Google Scholar]
- 268.Swenson CE, Sadikot RT. Achromobacter respiratory infections. Ann Am Thorac Soc 2015;12:252–8. [DOI] [PubMed] [Google Scholar]
- 269.Edwards BD, Greysson-Wong J, Somayaji R, et al. Prevalence and Outcomes of Achromobacter Species Infections in Adults with Cystic Fibrosis: a North American Cohort Study. J Clin Microbiol 2017;55:2074–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Urzi C, Salamone P, Schumann P, Rohde M, Stackebrandt E. Blastococcus saxobsidens sp. nov., and emended descriptions of the genus Blastococcus Ahrens and Moll 1970 and Blastococcus aggregatus Ahrens and Moll 1970. Int J Syst Evol Microbiol 2004;54:253–9. [DOI] [PubMed] [Google Scholar]
- 271.Hezbri K, Louati M, Nouioui I, et al. Blastococcus capsensis sp. nov., isolated from an archaeological Roman pool and emended description of the genus Blastococcus, B. aggregatus, B. saxobsidens, B. jejuensis and B. endophyticus. Int J Syst Evol Microbiol 2016;66:4864–72. [DOI] [PubMed] [Google Scholar]
- 272.Pfleiderer A, Lagier JC, Armougom F, Robert C, Vialettes B, Raoult D. Culturomics identified 11 new bacterial species from a single anorexia nervosa stool sample. Eur J Clin Microbiol Infect Dis 2013;32:1471–81. [DOI] [PubMed] [Google Scholar]
- 273.Kovaleva J, Degener JE, van der Mei HC. Methylobacterium and its role in health care-associated infection. J Clin Microbiol 2014;52:1317–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Xue Y, Zhang X, Zhou C, et al. Caldalkalibacillus thermarum gen. nov., sp. nov., a novel alkalithermophilic bacterium from a hot spring in China. Int J Syst Evol Microbiol 2006;56:1217–21. [DOI] [PubMed] [Google Scholar]
- 275.Zhao W, Zhang CL, Romanek CS, Wiegel J. Description of Caldalkalibacillus uzonensis sp. nov. and emended description of the genus Caldalkalibacillus. Int J Syst Evol Microbiol 2008;58:1106–8. [DOI] [PubMed] [Google Scholar]
- 276.Regunath H, Kimball J, Smith LP, Salzer W. Severe Community-Acquired Pneumonia with Bacteremia Caused by Herbaspirillum aquaticum or Herbaspirillum huttiense in an Immune-Competent Adult. J Clin Microbiol 2015;53:3086–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Suwantarat N, Adams LL, Romagnoli M, Carroll KC. Fatal case of Herbaspirillum seropedicae bacteremia secondary to pneumonia in an end-stage renal disease patient with multiple myeloma. Diagn Microbiol Infect Dis 2015;82:331–3. [DOI] [PubMed] [Google Scholar]
- 278.Karch H, Tarr PI, Bielaszewska M. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol 2005;295:405–18. [DOI] [PubMed] [Google Scholar]
- 279.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol 2004;2:123–40. [DOI] [PubMed] [Google Scholar]
- 280.Foxman B Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 2014;28:1–13. [DOI] [PubMed] [Google Scholar]
- 281.Waugh JB, Granger WM, Gaggar A. Incidence, relevance and response for Ralsfonia respiratory infections. Clin Lab Sci 2010;23:99–106. [PMC free article] [PubMed] [Google Scholar]
- 282.Ryan MP, Adley CC. Ralstonia spp.: emerging global opportunistic pathogens. Eur J Clin Microbiol Infect Dis 2014;33:291–304. [DOI] [PubMed] [Google Scholar]
- 283.Sangwan P, Chen X, Hugenholtz P, Janssen PH. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl Environ Microbiol 2004;70:5875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Kant R, van Passel MW, Palva A, et al. Genome sequence of Chthoniobacter flavus Ellin428, an aerobic heterotrophic soil bacterium. J Bacteriol 2011;193:2902–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol 2001;51:1521–8. [DOI] [PubMed] [Google Scholar]
- 286.Murphy EC, Frick IM. Gram-positive anaerobic cocci--commensals and opportunistic pathogens. FEMS Microbiol Rev 2013;37:520–53. [DOI] [PubMed] [Google Scholar]
- 287.Africa CW, Nel J, Stemmet M. Anaerobes and bacterial vaginosis in pregnancy: virulence factors contributing to vaginal colonisation. Int J Environ Res Public Health 2014;11:6979–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.