Abstract
Little is known about the effects of combinatorial dietary compounds on the regulation of epigenetic mechanisms involved in breast cancer prevention. The human diet consists of a multitude of components, and there is a need to elucidate how certain compounds interact in collaboration. Withaferin A (WA), found in the Indian winter cherry and documented as a DNA methyl transferase (DNMT) inhibitor, and sulforaphane (SFN), a well-known histone deacetylase (HDAC) inhibitor found in cruciferous vegetables, are two epigenetic modifying compounds that have only recently been studied in conjunction. The use of DNMT and HDAC inhibitors to reverse the malignant expression of certain genes in breast cancer has shown considerable promise. Previously, we found that SFN + WA synergistically promote breast cancer cell death. Herein, we determined that these compounds inhibit cell cycle progression from S to G2 phase in MDA-MB-231 and MCF-7 breast cancer. Furthermore, we demonstrate that this unique combination of epigenetic modifying compounds down-regulates the levels of Cyclin D1 and CDK4, and pRB; conversely, the levels of E2F mRNA and tumor suppressor p21 are increased independently of p53. We find these events coincide with an increase in unrestricted histone methylation. We propose SFN + WA-induced breast cancer cell death is attributed, in part, to epigenetic modifications that result in the modulated expression of key genes responsible for the regulation of cancer cell senescence.
Keywords: Withaferin A, Sulforaphane, Combinatorial therapy, Adjuvant, Breast cancer, Chemotherapy
1. Introduction
Many advancements have been made with regard to breast cancer treatment and prevention and an area of prevention that has gained increasing interest is alteration of the diet. It is known that cancer can be classified as an epigenetic disease, as many cancers result from environmental factors that promote carcinogenesis as a result of aberrant expression of tumor suppressor genes [1-3]. The epigenetic impact of dietary compounds on cancer is a topic of continuous emerging interest, and there is a need to elucidate the mechanisms behind how dietary compounds are effective. Over the past several years, we have found that sulforaphane (SFN), epigallocatechin gallate, resveratrol, pterostilbene, genistein and others have chemopreventive capability, and the combination of some of these compounds is more efficient than their singular use [4-6]. More recently, we have begun to study withaferin A (WA), a steroidal lactone, in conjunction with SFN [7]. Our previous results show efficacy in the use of these compounds for breast cancer cell death, thus providing merit to study their combined effects in depth. We found there to be synergy with regard to inhibition of cell viability in MCF-7 breast cancer cells. No significant cell death was demonstrated in MCF10A control cells thus indicating the safety of these treatments. We further showed induction of BAX and reduction of BCL-2 after treatment with SFN + WA in cancer cells in addition to changes in DNMTs and HDAC1 expression. The current study has been conducted in an effort to examine regulators of cell cycle progression along with the tumor suppressor genes that are known to be aberrantly expressed in multiple cancer types.
There are several genes that have been identified as potential tumor suppressors and oncogenes; to date, p53 is one of the most studied genes correlated with the inhibition or progression of breast cancer dependent upon its wild type or mutated status, respectively [8-10]. P53 activates the tumor suppressor p21, a cyclin dependent kinase inhibitor (CKI). Studies show that DNA damage-induced p21 expression is dependent on p53 [11,12]. Though several studies report p21 to act independently of p53 in some cases [13], it is important to note that in reference to DNA damage these two genes appear to be linked. Another tumor suppressor implicated in the regulation of cell cycle progression is retinoblastoma protein (RB); RB can induce both p53 dependent-and-independent-apoptosis upon inactivation, and is a negative regulator of p21 [14,15].
Several studies indicate that p21 is responsible for the inhibition of cell cycle progression and promotion of apoptosis in some cases [16,17]. We previously reported that combined use of WA and SFN induced apoptosis in both triple negative MDA-MB-231 and ERα positive MCF-7 breast cancer cells; therefore, we hypothesized that these compounds may regulate one or more tumor suppressor genes responsible for cell cycle progression. It is important to note that these two cell lines are considerably different. MCF-7 cells were originally derived from a 69-year-old Caucasian female. These cells are slow growing and have a wild-type p53 status [18]. On the other hand, MDA-MB-231 cells are relatively aggressive in comparison to MCF-7 cells and have a mutated p53 status [19]. Our previous studies found that combinatorial SFN and WA is effective in impeding overexpressed epigenetic genes and enzymes in addition to cellular proliferation in MCF-7 and MDA-MB-231 breast cancer cell lines. Herein, we investigated whether SFN + WA-induced epigenetic changes, i.e., acetylation and methylation, result in the activation of tumor suppressor genes that in turn inhibits cell cycle progression of two breast cancer cell lines.
2. Materials and methods
2.1. Chemicals
R, S-sulforaphane (≥ 98% pure), C6H11NOS2 with a molecular weight of 177.28 g/mol, was purchased from LKT Laboratories (Minneapolis, MN). Withaferin A (≥ 95% pure), C28H38O6, has a molecular weight of 470.606 g/mol and was acquired from Sigma-Aldrich (St. Louis, MO). Compounds were diluted in dimethyl sulfoxide (DMSO), also purchased from Sigma-Aldrich, and stored in stocks of 10 mmol/L at −20 °C.
2.2. Cell Culture
Cells were cultured using DMEM 1× media supplemented with 10% total volume of fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) and 0.5% total volume of 100× penicillin-streptomycin purchased from Corning Cellgro (Corning, NY). After seeding, cells were allowed 24 h to adhere to plates and all cells used in this study were treated with over a 3-day period with either 5.0 μM SFN, 1.0 μM WA or both. Treatments were refreshed every 24 h with fresh media. A maximum of 1.2 μM of DMSO was used as a vehicle control. The breast cancer cell lines were used in this study were MCF-7 [ERα (+)] and the ERα (−) MDA-MB-231 (ATCC, Manassas, VA).
2.3. Cell cycle analysis
Flow cytometry cell cycle analysis was determined utilizing propidium iodide staining. Cells were harvested and then washed in PBS after which they were fixed with 70% ethanol which was added drop wise while vortexing. After a 30 min fixation at 4 °C, samples were washed twice in PBS and centrifuged at 850 g. Cells were then treated with approximately 50 μL of ribonuclease A at 100 μg/mL. Cells were then sent to the campus Flow Cytometry Center at the University of Alabama at Birmingham and analyzed by measuring the forward and side scatter and pulse processing excluding cell doublets.
2.4. DNA extraction
DNA extracts were prepared using the PureYield Plasmid MiniPrep System from Promega. The manufacturer's protocol was followed accordingly, then the Nano-drop 2000 was used to assess sufficient DNA yields.
2.5. Nuclear protein extraction
Nuclear extracts were prepared using the EpiQuik nuclear extraction kit from EpiGenTek (OP-0002-1) (Farmingdale, NY) and the manufacturer's procedure was followed.
2.6. Protein extraction
Protein was extracted using the TeloTAAAGG Lysis buffer purchased from Roche. Cell pellets were collected after 3-day treatments and centrifuged at approximately 8000 RPM for 5 min. Afterwards media was removed and cells were washed twice with PBS before 200 μL of the lysis buffer was added. Samples were left to incubate on ice for 30 min before centrifugation again for 20 min at 4 °C. Approximately 175 μL of lysate was then transferred to a new collection tube. Samples were stored at −80 °C and protein concentrations were later determined via Bradford assay (Bio-Rad Protein Assay, Bio-Rad; Hercules, CA).
2.7. Quantitative RT-PCR
qRT-PCR was used to determine the mRNA expression of the cell cycle genes of interest. RNA was extracted using the Qiagen RNeasy kit (Valencia, CA) and the manufacturer's instructions were followed. cDNA was made from RNA extracts using the cDNA synthesis kit from Bio-Rad (Hercules, CA). PCR reactions were completed in triplicate using 1 μL of cDNA for each sample. Both forward and reverse primers (1 μL) for the gene of interest were used along with 5 μL of SSO SYBR green from Bio-Rad and 2 μL of nuclease free water for a total volume of 10 μL. Once samples were prepared they were placed in the CFX Connect Real Time System from Bio-Rad upon which the 3-step amplification protocol was selected. Thermal cycling was initiated at 94° C for 4 min followed by 35 cycles of PCR (94 °C, 15 s; 60 °C, 30 s; 72 °C, 30 s). GAPDH was used as an endogenous control in order to calculate fold change using the ΔΔCq method as we reported previously. Primers were purchased from Integrated DNA Technologies (IDT, Coralville, IA) where forward 5′CCGTCCATGCGGAAGATC-3 and reverse 5′-GAAGACCTCCTCCTCGCACT-3′ were the sequences for Cyclin D1. The CDK4 forward primer sequence was 5′-CTT CTG CAG TCC ACA TAT GCA ACA-3′ and the reverse was 5′-CAA CTG GTC GGC TTC AGA GTT TC-3′, and finally the E2F forward and reverse primers were 5′-GTCTGGTTGCTATGGTAGCTGGC-3′; 5′-ACTCCTCGCAGATCGTCATCATCT-3′ respectively.
2.8. Western blot
Protein was loaded onto the Novex NuPage 4–12% premade Bis-Tris gel from Invitrogen and separated by electrophoresis at 200 V until the dye almost ran off the gel. Proteins were then transferred to nitrocellulose membrane using the Trans Turbo Blot from Bio-Rad. Membranes were then blocked in milk buffer [5% dry milk, Tris Buffered Saline (TBS) and 1% Tween (T)] using the Millipore SnapID (Billerica, Massachusetts). Primary antibody incubations were carried out at room temperature for no more than 30 min and membranes were washed four times with 30 mL of TBS+T before probing with secondary antibody for 15 min followed by four more washes. Immunoreactive bands were visualized using an enhanced chemiluminescence detection system (Bio-Rad). Santa Cruz Biotechnology (Dallas, TX) and Cell Signaling Technology (Danvers, MA) were the suppliers of the selected antibodies.
2.9. Global methylation activity assay
The MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit (Colorimetric) from EpiGenTek was used to assess overall DNA methylation activity from DNA extracts that were gathered using the methodology described in the above section.
2.10. Histone acetyltransferase activity/ Inhibition assay
Histone acetyltransferase enzymatic activity was determined utilizing the Histone Acetyltransferase Activity/ Inhibition Assay from EpiGenTek and the provided protocol was followed.
2.11. Histone methyltransferase activity/Inhibition assay
Histone methyltransferase (HMT) activity was assessed via the HMT Activity Assay kit from EpiGenTek. Nuclear extracts were prepared as described above and the manufacturer's protocol was used to assess HMT activity.
2.12. Statistical analysis
Error bars represent standard deviation (SD). Each assay was completed in triplicate culturing experiments with 3 or 4 technical replicates. The student's t-test was used to determine significance where a p < 0.05 is significant.
2.13. Chromatin immunoprecipitation (ChIP) analyses
ChIP assays were performed as previously described [20]. Cells were grown as described above and nuclei from cross-linked cells were resuspended in Tris/EDTA. The soluble chromatin was adjusted into RIPA buffer and precleared with salmon sperm blocked protein A beads. Immunoprecipitation was performed with 5 μg of antibodies directed against trimethylated lysine 4 of histone 3 (H3K4Me3), or IgG, as described [20]. Immune complexes were absorbed with protein A beads blocked with salmon sperm DNA. After pre-clearing and before immunoprecipitation, equal amounts of sonicated DNA (10% volume of each sample) were reserved for qPCR (input) analysis. The p21 promoter was probed with specific primers against the immunoprecipitated DNA by qPCR using primers sets based on known sequences, based on known mouse sequences. Reactions for each sample were performed in triplicate using an ABI StepOnePlus Detection System and a PCR protocol comprising an initial 10-min incubation at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60–65 °C. The raw data were analyzed using StepOnePlus software and ΔΔCt values for each gene in each sample.
3. Results
3.1. WA and SFN regulate cell cycle progression through inhibition of cell cycle genes in breast cancer cells
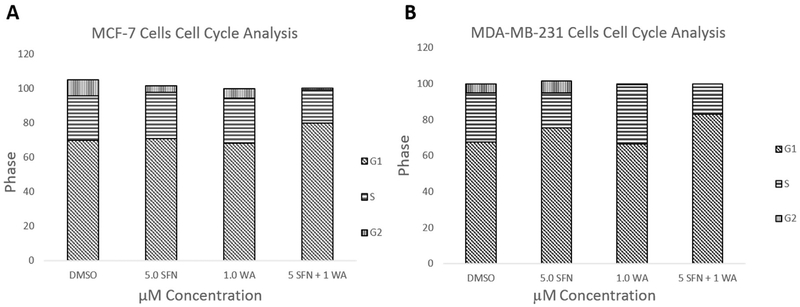
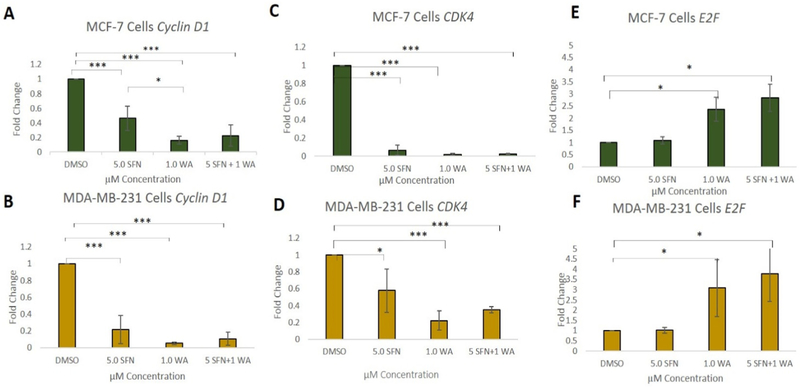
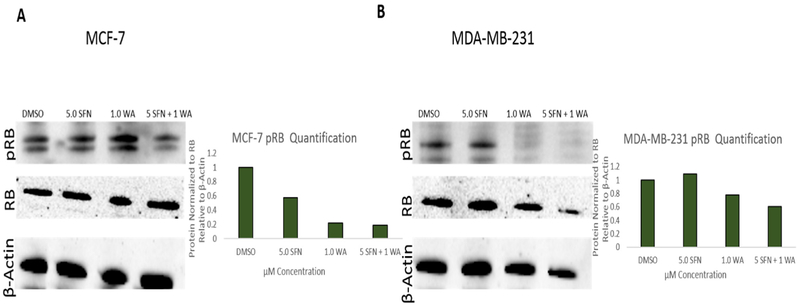
Fig. 1 demonstrates the effects of SFN and WA on the cell cycle in two vastly different breast cancer cell lines. Cells were treated as described previously [7] and as indicated in Fig. 1A. SFN and WA alone show a decrease in transition into the G2/M phase of the cell cycle, while the combination of SFN and WA arrests cells primarily at the G1 phase in MCF-7 cells. Similarly, MDA-MB-231 cells (Fig. 1B) show a decrease in transition into S phase with the incorporation of the combination of SFN and WA, with SFN appearing to be more effective than WA. Due to an increase in G1 arrest in both cell lines, we analyzed cell cyclin D1 (CCND1), cyclin D kinase 4 (CDK4), and E2F which are known to have various roles in the cell cycle. CCND1 regulates cell cycle progression through its ability to promote transition from G1 to S phase and CDK4 and E2F are also closely associated with these changes in the cell cycle. Upon analyzing changes in these genes using qRT-PCR, we found that CCND1 and CDK4 expression was significantly decreased with the incorporation of SFN and WA alone in both MCF-7 and MDA-MB-231 breast cancer cells (Fig. 2). Interestingly, the combination of these compounds was not more effective than the single dosages as we originally hypothesized. Figs. 2C and 2F show a comparable increase in E2F in both cell lines. We further observed retinoblastoma protein (RB) which is another cell cycle regulator instrumental in the regulation of the G1 check point. We found a decrease in phosphorylated RB (pRB) upon introduction of SFN and WA in both MCF-7 and MDA-MB-231 cells with the combination being most effective as seen in Fig. 3.
Fig. 1. Combinatorial SFN and WA arrest cells most abundantly at G1 in breast cancer cells:
A. Combinatorial SFN and WA arrest cells at G1 phase and prevent transition into G2 in MCF-7 cells. B. In MDA-MB-231 breast cancer cells it can be noted that the incorporation of these compounds prevent transition into G2 phase (n=3).
Fig. 2. Cell cycle genes are regulated by WA and SFN at the mRNA level in breast cancer cells:
A. Combinatorial SFN + WA down-regulate cyclin D1 in an extremely significant manner but not significantly more than the single dosages in MCF-7 breast cancer cells. B. Cyclin D1 is also down-regulated in MDA-MB-231 breast cancer cells yet the combination is not significantly different from the signal dosages. C. CDK4 in MCF-7 cells shows a downward trend with single treatments of SFN or WA although SFN + WA does not show greater significance than the single dosages. D. CDK4 in MDA-MB-231 cells is down-regulated. E. E2F gene expression in MCF-7 cells is significantly upregulated in comparison to the control with the introduction of WA and the combo. F. E2F mRNA expression in MDA-MB-231 cells is significantly increased in comparison to the control (n=3; p < 0.05 *, p < 0.01 **, p < 0.001 ***).
Fig. 3. pRB protein is inhibited by SFN and WA in MCF-7 and MDA-MB-231 cells:
A. Combinatorial SFN and WA down-regulate pRB expression in MCF-7 breast cancer cells with the combination being the most effective. Densitometry was determined using ImageJ and bar graphs represent 3 replicates relative to B-actin. B. pRB protein is down-regulated by SFN + WA in MDA-MB-231. ImageJ was used to calculate densitometry (n=3; images are representative).
3.2. WA and SFN promote changes in epigenetic regulators in MCF-7 and MDA-MB-231 cells
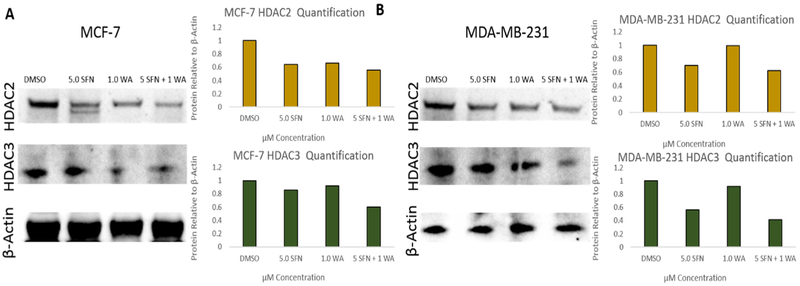
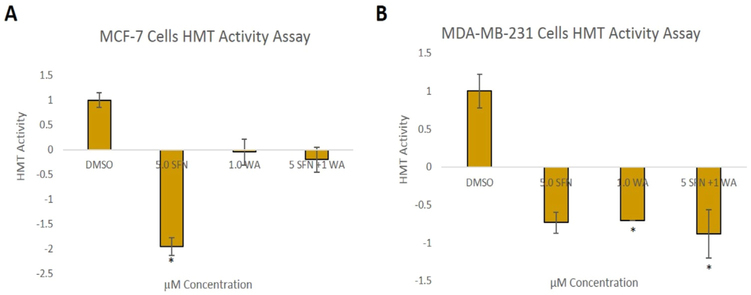
Since we previously found that combinatorial SFN and WA treatments decrease HDAC1 and overall HDAC activity as well as modulate DNMTs in breast cancer cells [7], we further studied epigenetic modulators to determine if changes in the expression of these genes are partially responsible for the breast cancer cell death reported. We show decreases in HDAC2 and HDAC3 at the protein level in both cell lines upon incorporation of SFN and WA with the combination being most effective (Fig. 4). In addition, the effects of the chosen nutritive compounds on histone methyltransferase activity (HMT) were analyzed. In Fig. 5A it can be noted that SFN alone decreased overall HMT enzymatic activity more than WA and the combination in MCF-7 cells. In contrast, HMT activity in MDA-MB-231 cells was significantly decreased by WA and the combination treatment as seen in Fig. 5B.
Fig. 4. HDAC2 and HDAC3 protein levels are down-regulated by SFN and WA in breast cancer cells:
A. Combinatorial SFN and WA downregulate HDAC2 and HDAC3 expression in MCF-7 breast cancer cells with the combination appearing to be the most effective. Densitometry was determined using ImageJ and bar graphs represent 3 replicates relative to B-actin. B. HDAC2 and HDAC3 protein is downregulated by SFN + WA in MDA-MB-231. ImageJ was used to calculate densitometry (n=3; images are representative).
Fig. 5. HMT enzymatic activity in breast cancer cells is down-regulated by natural compounds:
A. Combinatorial SFN and WA down-regulate HMT activity with SFN being most effective in MCF-7 breast cancer cells. B. Histone methyltransferase activity is down-regulated by SFN and WA in MDA-MB-231(n=3; p < 0.05*).
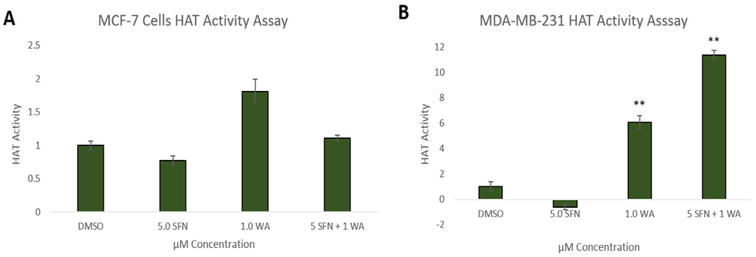
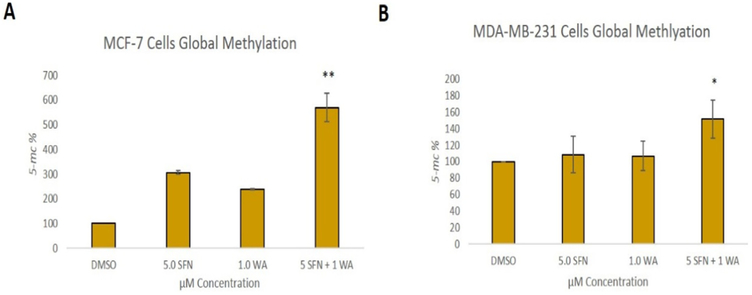
Fig. 6 shows the effects of SFN and WA on histone acetyltransferase activity (HAT) and in accordance to previous findings in our lab [21], SFN has no significant effect on HAT activity in either cell line. In contrast, WA appears to increase HAT activity in both cell lines but only significantly in MDA-MB-231 cells. The combination is not more effective than WA alone; however, there is an upward trend in HAT activity in these cells (Fig. 6B). Global methylation was determined after treatment with SFN, WA and both compounds. It is known that many different cancer types show global hypomethylation leaving the promoter region of aberrantly expressed genes and tumor suppressors to be hypermethylated [22]. We report a significant increase in global methylation with the incorporation of the combination in both MCF-7 and MDA-MB-231 cells (Fig. 7). The single dosages of SFN and WA do not appear effective in reversing global hypomethlyation in the breast cancer cells studied.
Fig. 6. Effects of SFN and WA on HAT Activity in MCF-7 and MDA-MB-231 Cells:
A. Combinatorial SFN and WA show no significant change in HAT activity in MCF-7 breast cancer cells. B. Histone acetyltransferase activity is upregulated by WA and the combination appears more effective in MDA-MB-231 but not significantly (n=3; p < 0.05*, p < 0.01**).
Fig. 7. Global methylation is increased in breast cancer cells by combinatorial WA and SFN:
A. SFN and WA alone have no significant effect on global methylation but when these two compounds are used together we report an increase in methylation in MCF-7 cells. B. Global methylation is significantly upregulated by combinatorial WA and SFN in MDA-MB-231 cells (n=3; p < 0.05 *, p < 0.01**).
3.3. WA and SFN promote changes in p53 and p21 in breast cancer cells
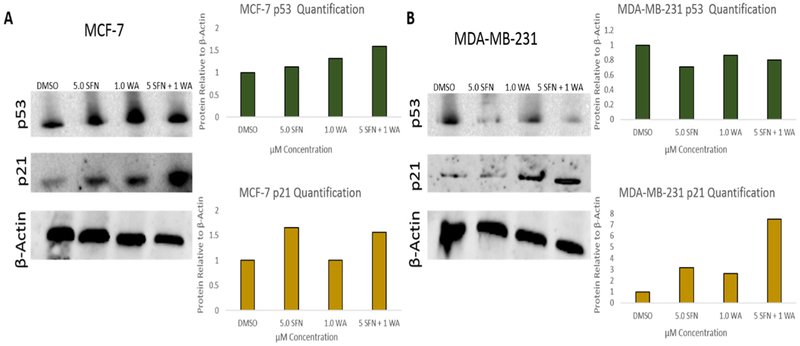
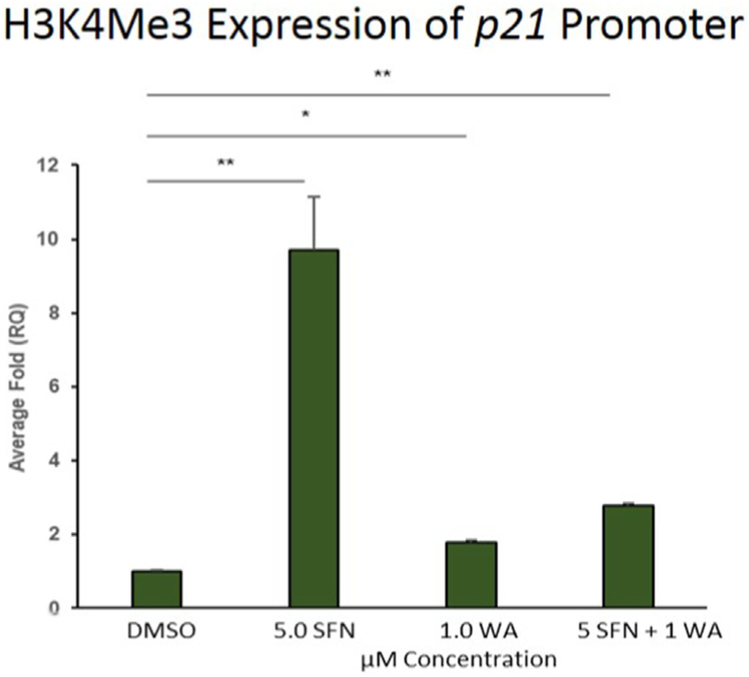
Two tumor suppressors with roles in cell cycle progression are p53 and p21 of which mutated p53 has been frequently implicated in tumor cell progression [23]. Several studies also reveal p21 to be responsible for inhibiting the cell cycle [17,24]. Upon Western blot analyses we reveal a slight increase in p53 in MCF-7 cells as well as an induction of p21 after treatment with SFN, WA and both compounds (Fig. 8A). In Fig. 8B we also show an increase in p21 expression with the combination being the most effective in MDA-MB-231 cells. Unlike the MCF-7 cells p53 is reduced in Fig. 8B. It is important to note that MDA-MB-231 cells have a high level of mutant p53 and it serves as an oncogene instead of a tumor suppressor [23]. Because p21 was re-expressed in both cell lines upon analysis we decided to determine if this regulation was attributed to changes in histone modifications at its promoter. To assess the impact of SFN, WA or both on epigenetic modifications at the p21 promoter, we performed chromatin immunoprecipitation (ChIP) assays using antibodies specific for trimethylated lysine 4 of histone (H3K4Me3), a mark which is associated with transcriptional activation. We determined that SFN or WA alone, or in combination with WA significantly increased the levels of H3K4Me3 at the p21 promoter, indicating these dietary compounds positively modify the epigenome at the promoter region (Fig. 9).
Fig. 8. Tumor suppressor proteins implicated in cell cycle progression are modulated by SFN and WA in breast cancer cells:
A. Combinatorial SFN and WA show no significant change in p53 protein expression in MCF-7 breast cancer cells; however, p21 is upregulated with the combination being most effective. B. p53 protein expression is down-regulated by SFN and the combination of SFN and WA where p21 is upregulated with the combination being the most effective (n=3; images are representative and bar graphs represent the densitometry results of 3 replicates).
Fig. 9. Activation of p21 is mediated by transcriptional activator H3K4Me3 in MDA-MB-231 cells:
ChIP assay reveals a significant increase in the expression of H3K4Me3 at the p21 promoter of triple negative breast cancer cells. Values represent average +/− SEM. One exemplar shown (n=3; p < 0.05*, p < 0.01**).
4. Discussion
Our previous study on the topic of combinatorial SFN and WA revealed a synergistic inhibition of breast cancer cell viability with limited effects on a noncancerous control cell line [7]. We found that both WA and SFN were capable of down-regulating epigenetic modifiers which led us to hypothesize that WA and SFN's ability to promote apoptosis and cell death in breast cancer cells is due to their epigenetic control of cell cycle progression. We therefore conducted cell cycle analysis in this study to determine which phase of the cell cycle breast cancer cells were impeded. As seen in Fig. 1, our results indicate that the breast cancer cells were arrested primarily at the G1 phase of the cell cycle with treatments of WA and SFN.
Since it is known that CCND1 and CDK4 are primarily responsible for the transition from G1 into S phase, we expected to observe a decrease in these two genes in the breast cancer cells that we treated. Though the combination was not more significant in the down-regulation of CCND1 and CDK4 despite an increase in arrest at G1, we do report a decrease in pRB and an increase in E2F (Fig. 2). RB is present at the promoters during quiescence, senescence and in cycling cells in which it represses G1-S genes. It could be that the repression of pRB (Fig. 3) by combinatorial WA and SFN is partially responsible for the lack of greater CCND1 and CDK4 inhibition in comparison to the single dosages. A study by Stanelle et.al describes the varying roles of E2F in the cell cycle which notes that an upregulation of the E2F-1 gene drives overall E2F expression and when overexpressed has roles in apoptosis [25]. Interestingly, a review of the literature indicates that E2F, CCND1, CDK4 and pRB form a complex with p21 that is implicit in cell cycle progression [26]. Though pRB is typically associated with negatively regulating entry into the cell cycle, there have been some studies indicating that elevated pRB loses its cell cycle inhibitory effects in cancer [27]. In addition, RB also has roles in the negative regulation of p21 [14]. If SFN and WA are working through similar mechanisms, an alternative explanation for the lack of greater inhibition of CCND1 and CDK4 in the breast cancer cells studied could be that the combination is not more effective due to an unknown mechanism having already been acted upon by the other compound. Further studies should focus on determining potential modes of action. For example, research into helicases may reveal that combinatorial WA and SFN's ability to decrease cancer cell viability and promote DNA damage is resultant from modulation of helicases thereby further inhibiting the cell cycle and promoting G1 arrest.
The tumor suppressor and cell cycle regulator p21 has been associated with impeding the cell cycle in cancer cells [28,29]. Our study shows an increase in this tumor suppressor at the protein level independent of p53 expression. The down-regulation of pRB, as shown in Fig. 3, could be at least partially responsible for the increase in expression of p21 (Fig. 8). Several studies show that the suppression of HDACs and other epigenetic enzymes are associated with the status of both p53 and p21, and it has been shown that p21-dependent G1 arrest is accompanied by RB hypophosphrylation with the incorporation of a synthetic HDAC inhibitor [30]. This adds supporting evidence that the compounds used in this study promote breast cancer cell death through their ability to impede HDACs (Fig. 4). Another study revealed HDAC inhibition to be capable of promoting p21 expression at both the gene and protein levels in addition to gene associated acetylation [31]. Further, Lagger and colleagues report a direct inhibition of p21 by HDAC1 in their study [32].
This study aimed to focus on H3K4Me3 since it is a methylation marker associated with transcriptional activation. As suspected, we report significant increases of H3K4Me3 at the p21 promoter in concert with increased p21 protein expression. The combination of SFN and WA, though effective, does not have greater significance in epigenetic modulatory abilities according to the genes we assessed in this research. Our previous study showed a synergistic inhibition of breast cancer cell proliferation, and while p21 activation serves as a partial explanation for decreased proliferation, H3K4Me3 at its promoter is not increased significantly more than SFN alone. We may find there to be a greater abundance of acetyl markers and changes in methyl markers associated with suppression of transcription by the combination. As mentioned, HDAC has roles in p21 suppression and it therefore remains feasible that the down-regulation of HDACs caused by these compounds is promoting acetylation of p21 leading to transcriptional activation.
HDACs and DNMTs are extremely important in the regulation of the cell cycle and the binding of transcription factors. Hypermethylation and hypoacetylation are typically associated with gene silencing [8] and many tumor suppressors and oncogenes are dysregulated through epigenetic modifiers that are instrumental in the regulation of a number of carcinogenic processes [33-37]. One hallmark of cancer is global DNA hypomethylation which promotes genome instability [38]. Our data indicate that combinatorial SFN and WA are capable of significantly increasing global methylation. Interestingly, our previous results showed DNMTs to be down regulated by SFN + WA [7] and other studies indicate that the inhibition of DNMTs results in decreased CpG methylation [39]. It is important to note that class I DNMTs are not the only methyltransferases. In fact, EZH2 has been reported to directly control DNA methylation and an assessment of this polycomb group protein could reveal additional information that explains the decrease in hypomethylation reported in this study [40], however the ELISA used for this assessment only covers a small percentage of CpG sites so a more comprehensive analysis is needed.
The negative regulation of HDAC1, HDAC2 and HDAC3 induced by SFN and WA are responsible for the changes seen in p53 and p21 in this study. Interestingly, combinatorial SFN and WA was more effective in the reduction of HMT activity (Fig. 5B) and induction of HAT activity in the triple-negative MDA-MB-231 cells (Fig. 6B). The combination was also more effective in p21 activation in these cells (Fig. 8B). Zupkovitz and colleagues conducted a study that confirms HDAC regulation of p21 [41] in which direct binding of HDAC1 to the p21 promoter was shown. This provides supporting evidence to our study that the down-regulation of HDACs and DNMTs reported in both this study and our previous work is linked to the epigenetic re-expression of the p21 tumor suppressor In addition to the epigenetic modifiers examined in this report further studies of the p21 promoter may reveal E2F to aide in the activation of p21 as reported by Gartel and colleagues [14]. The regulation of cell cycle progression and associated genes and epigenetic mechanisms via these compounds has been demonstrated in this study although there could be other modes of action for these compounds as well. An in vivo component testing the efficacy of WA and SFN in combination in a xenograft or transgenic mouse model will also be warranted in future studies.
5. Conclusion
The necessity of a better comprehension of the effects of multiple nutritive compounds on cancer progression is apparent. By understanding how modifiable factors such as diet and lifestyle promote an anti-cancerous epigenome, we are steps closer to identifying ways to prevent the malignancy/ occurrence of the disease. Many advancements have been made with respect to decreasing breast cancer-related mortality; although the effects of chemotherapy can be harsh and lead to numerous side effects. In addition, hormone therapies are not a viable option for individuals with triple-negative breast cancer. Our study has helped to advance the field with regard to novel approaches to cancer prevention and therapy. The results we report herein explore the effects of combinatorial SFN and WA on the cell cycle, cell cycle regulators and epigenetic modifying enzymes. We are the first to show this combination to be capable of epigenetically reactivating the p21 tumor suppressor gene. This finding serves as one explanation for the breast cancer cell death reported in our previous publication on this topic [7]. Collectively, our study has both adjuvant and chemopreventive potential since WA and SFN are effective at impeding breast cancer cell proliferation in both ERα (+) and triple-negative breast cancer cell lines.
Acknowledgements
We thank Marion Spell for his help with Cell Cycle analysis. We would also like to acknowledge the Tollefsbol lab members for their advice and support.
Funding
This work was supported by grants from the National Cancer Institute (R01 CA178441 and R01 CA204346), The Cancer Prevention and Control Training Program (R25 CA047888), the Susan G. Komen Graduate Training in Disparities Research program (GTDR15329376), the American Institute for Cancer Research (316184), and the Department of Radiation Oncology at UAB (SN, RR).
Footnotes
Conflicts of interest
The authors disclose that there are no conflicts of interest.
References
- [1].Sharma S, Kelly TK, Jones PA, Epigenetics in cancer, Carcinogenesis 31 (1) (2010) 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kelly TK, De Carvalho DD, Jones PA, Epigenetic modifications as therapeutic targets, Nat. Biotechnol. 28 (10) (2010) 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rodriguez-Paredes M, Esteller M, Cancer epigenetics reaches mainstream oncology, Nat. Med. 17 (3) (2011) 330–339. [DOI] [PubMed] [Google Scholar]
- [4].Kala R, Tollefsbol TO, A novel combinatorial epigenetic therapy using resveratrol and pterostilbene for restoring estrogen receptor-alpha (ERalpha) expression in ERalpha-negative breast cancer cells, PLoS One 11 (5) (2016) e0155057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li Y, Meeran SM, Tollefsbol TO, Combinatorial bioactive botanicals re-sensitize tamoxifen treatment in ER-negative breast cancer via epigenetic reactivation of ERalpha expression, Sci. Rep. 7 (1) (2017) 9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li Y, et al. , Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein, PLoS One 8 (1) (2013) e54369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Royston KJ, et al. , A novel combination of withaferin a and sulforaphane inhibits epigenetic machinery, cellular viability and induces apoptosis of breast cancer cells, Int. J. Mol. Sci. 18 (2017) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Deppert W, p53: oncogene, tumor suppressor, or both? in: Wagener C, Neumann S (Eds.), Molecular Diagnostics of Cancer, Springer Berlin Heidelberg, Berlin, Heidelberg, 1993, pp. 27–39. [Google Scholar]
- [9].Soussi T, Wiman KG, TP53: an oncogene in disguise, Cell Death Differ. 22 (2015) 1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Strano S, et al. , Mutant p53: an oncogenic transcription factor, Oncogene 26 (2007) 2212. [DOI] [PubMed] [Google Scholar]
- [11].Epe C, et al. , Efficacy of toltrazuril as a metaphylactic and therapeutic treatment of coccidiosis in first-year grazing calves, Parasitol. Res. 97 (Suppl 1) (2005) S127–S133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barr AR, et al. , DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression, Nat. Commun. 8 (2017) 14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Macleod KF, et al. , p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage, Genes Dev. 9 (8) (1995) 935–944. [DOI] [PubMed] [Google Scholar]
- [14].Gartel AL, et al. , Activation and repression of p21(WAF1/CIP1) transcription by RB binding proteins, Oncogene 17 (26) (1998) (p. 3463–9). [DOI] [PubMed] [Google Scholar]
- [15].Benson EK, et al. , p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes, Oncogene 33 (2013) 3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Karimian A, Ahmadi Y, Yousefi B, Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage, DNA Repair 42 (Supplement C) (2016) 63–71. [DOI] [PubMed] [Google Scholar]
- [17].Chen A, et al. , The Role of p21 in apoptosis, proliferation, cell cycle arrest, and antioxidant activity in UVB-irradiated human HaCaT keratinocytes, Med. Sci. Monit. Basic Res. 21 (2015) 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Alkhalaf M, El-Mowafy A, Overexpression of wild-type p53 gene renders MCF-7 breast cancer cells more sensitive to the antiproliferative effect of progesterone, J. Endocrinol. 179 (1) (2003) 55–62. [DOI] [PubMed] [Google Scholar]
- [19].Hui L, et al. , Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D, Oncogene 25 (2006) 7305. [DOI] [PubMed] [Google Scholar]
- [20].Nozell S, et al. , The ING4 tumor suppressor attenuates NF-κB activity at the promoters of target genes, Mol. Cell. Biol. 28 (21) (2008) 6632–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Meeran SM, Patel SN, Tollefsbol TO, Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines, PLoS One 5 (7) (2010) e11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hon GC, et al. , Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer, Genome Res. 22 (2) (2012) 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vogiatzi F, et al. , Mutant p53 promotes tumor progression and metastasis by the endoplasmic reticulum UDPase ENTPD5, Proc. Natl. Acad. Sci. 113 (52) (2016) E8433–E8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Abbas T, Dutta A, p21 in cancer: intricate networks and multiple activities, Nat. Rev. Cancer 9 (6) (2009) 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stanelle J, et al. , Gene expression changes in response to E2F1 activation, Nucleic Acids Res. 30 (8) (2002) 1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ko TC, Bresnahan WA, Thompson EA, Intestinal cell cycle regulation, in: Meijer L, Guidet S, Philippe M (Eds.), Progress in Cell Cycle Research, Springer US, Boston, MA, 1997, pp. 43–52. [DOI] [PubMed] [Google Scholar]
- [27].Giacinti C, Giordano A, RB and cell cycle progression, Oncogene 25 (2006) 5220. [DOI] [PubMed] [Google Scholar]
- [28].Gartel AL, Tyner AL, The role of the cyclin-dependent kinase inhibitor p21 in Apoptosis 1 supported in part by NIH Grant R01 DK56283 (to A. L. T.) for the p21 research and campus research board and Illinois department of public health penny severns breast and cervical cancer grants (to A. L. G.). < a id="xref-fn-1-1" class="xref-" href="#fn-1" > 1 < /a >, Mol. Cancer Ther. 1 (8) (2002) 639–649. [PubMed] [Google Scholar]
- [29].Poole AJ, et al. , Tumor suppressor functions for the Cdk inhibitor p21 in the mouse colon, Oncogene 23 (2004) 8128. [DOI] [PubMed] [Google Scholar]
- [30].Sandor V, et al. , P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228, Br. J. Cancer 83 (6) (2000) 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Richon VM, et al. , Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation, Proc. Natl. Acad. Sci. USA 97 (18) (2000) 10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lagger G, et al. , The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene, Mol. Cell Biol. 23 (8) (2003) 2669–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].da Costa Prando É, Cavalli LR, Rainho CA, Evidence of epigenetic regulation of the tumor suppressor gene cluster flanking RASSF1 in breast cancer cell lines, Epigenetics 6 (12) (2011) 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kazanets A, et al. , Epigenetic silencing of tumor suppressor genes: paradigms, puzzles, and potential, Biochim. Et. Biophys. Acta (BBA) - Rev. Cancer 1865 (2) (2016) 275–288. [DOI] [PubMed] [Google Scholar]
- [35].Smith LT, et al. , Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer, Proc. Natl. Acad. Sci. USA 103 (4) (2006) 982–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yasuda H, et al. , Distinct epigenetic regulation of tumor suppressor genes in putative cancer stem cells of solid tumors, Int. J. Oncol. 37 (6) (2010) 1537–1546. [DOI] [PubMed] [Google Scholar]
- [37].YASUDA K, et al. , ERas oncogene expression and epigenetic regulation by histone acetylation in human cancer cells, Anticancer Res. 27 (6B) (2007) 4071–4075. [PubMed] [Google Scholar]
- [38].Ferguson LR, et al. , Genomic instability in human cancer: molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition, Semin. Cancer Biol. 35 (2015) S5–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Arzenani MK, et al. , Genomic DNA hypomethylation by histone deacetylase inhibition implicates DNMT1 nuclear dynamics, Mol. Cell. Biol. 31 (19) (2011) 4119–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Viré E, et al. , The polycomb group protein EZH2 directly controls DNA methylation, Nature 439 (2005) 871. [DOI] [PubMed] [Google Scholar]
- [41].Zupkovitz G, et al. , The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation, Mol. Cell. Biol. 30 (5) (2010) 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]