Abstract
Allosteric regulation of protein function is widespread in biology, but challenging for de novo protein design as it requires explicit design of multiple states with comparable free energies. We explore the possibility of de novo designing switchable protein systems through modulation of competing inter and intra-molecular interactions. We design a static, five-helix “Cage” with a single interface that can interact either intra-molecularly with a terminal “Latch” helix or inter-molecularly with a peptide “Key”. Encoded on the Latch are functional motifs for binding, degradation, or nuclear export that function only when the Key displaces the Latch from the Cage. We describe orthogonal Cage-Key systems that function in vitro, in yeast and in mammalian cells with up to 40-fold activation of function by Key. The design of switchable protein function controlled by induced conformational change is a milestone for de novo protein design and opens up new avenues for synthetic biology and cell engineering.
There has been considerable progress in the de novo design of stable protein structures based on the principle that proteins fold to their lowest free energy state. These efforts have focused on maximizing the free energy gap between the desired structure and all other structures, and have resulted in a wide range of stable proteins that exclusively populate the designed state1–4. Designing proteins that can switch conformations is more challenging, as multiple states must have sufficiently low free energies relative to the unfolded state, and free energy differences between the states must be small enough that switching can be toggled by an external input5,6. Recent advances in designing systems with multiple states include a transmembrane ion transporter7 and Gβ1 variants that dynamically exchange between two related conformations8; however, a method for the de novo design of modular, tunable protein systems that switch conformational states in the presence of an external input has not yet been achieved.
We set out to design de novo switchable protein systems guided by the following considerations. First, programming free energy differences between two states is more straightforward in a system governed by inter- and intra-molecular competition at the same site than by allosteric activation at distant sites9–11 because many of the residue-level interactions can be similar if not identical. Second, a stable protein framework with an extended binding surface available for the competing interactions is more programmable and less likely to engage in off-target interactions than a framework that only becomes ordered upon binding12,13. These features are described by the abstract system in Fig 1a, which undergoes thermodynamically-driven switching between binding-competent and binding-incompetent states. A Latch (blue) contains a peptide sequence (orange) that can bind a Target (yellow) unless blocked by intramolecular interactions to a Cage (cyan); a Key (green) that binds more tightly to the Cage outcompetes the Latch, allowing the peptide to bind the Target. The behavior of such a system is governed by binding equilibrium constants for the individual subreactions (Fig. 1a): Kopen, the dissociation of Latch from Cage; KLT, the binding of Latch to Target; and KCK, the binding of Cage to Key. When the Latch-Cage interaction is too weak (Fig. 1b, red and orange curves, see Supplementary Information), the system binds Target in the presence of little to no Key and the fold induction by Key is low, while when the Latch-Cage interaction is too strong (purple curve), the system only partially binds Target even at high Key concentrations. The Latch-Cage interaction affinity that gives optimal switching (Fig. 1b, blue curve left, green curve right) is a function of the Latch-Target binding affinity.
Figure 1. Design of the LOCKR system.

a, The Switch, composed of a Cage (cyan) and Latch (blue) with a functional motif (orange), has a thermodynamic transition to the open state able to bind Key (green) or Target (yellow). b, Numerical solutions of the model in (a) for different values of KLT (1 nM, left; 50 nM, right) and Kopen (0.1, red; 0.001, orange; 1e-5, green; 1e-7, blue; 1e-9, purple) with KCK fixed at 1 nM. c, Conversion of 5L6HC3 to monomeric frameworks. In LOCKR (right), the double mutant V223S/I238S allows the Key to bind. d, Guanidinium chloride denaturation of trimer (dark blue), monomer (cyan), truncated five-helix framework (red), and LOCKR (green) monitoring mean residue ellipticity (MRE) at 222 nm. Repeated 3 times with similar results. e, Small-angle X-ray scattering (SAXS) Kratky plots for the monomeric frameworks are similar to that of the input trimer, with the greatest deviation for the five-helix framework. Colors continued from (d). f, Key-GFP was added to monomeric frameworks immobilized onto a plate via a hexahistidine tag; after washing, binding was measured by GFP fluorescence (mean of n=3 technical replicates, error bars indicate s.d).
LOCKR Design
To implement the switchable system of Fig. 1a, we chose structural features amenable to tuning the affinities of the Cage-Latch and Cage-Key interactions over a wide dynamic range. Helical interfaces are dominated by sidechain-sidechain interactions, which can be more readily tuned than the backbone hydrogen-bonding interactions between β-strands14. To allow fine control over the specificity and relative affinities of the Cage-Latch and Cage-Key interactions, we chose to design interfaces containing buried hydrogen bond networks; as in DNA base-pairing, specificity can be altered by minor changes to hydrogen bond donors and acceptors15. As a starting point, a designed homo-trimer of -helical hairpins mediated by hydrogen bond networks (5L6HC3_115) was connected into two monomeric frameworks by designing short unstructured loops between the subunits (Fig. 1c). In the five-helix framework, there is an open binding site for a sixth helix added in trans; this site is filled by a sixth helix in cis in the six-helix framework.
The five-helix (Cage) and six-helix (Cage plus Latch) designs expressed in E. coli were largely monomeric by size-exclusion chromatography (Extended Data 1), and remain folded up to 5 M guanidine hydrochloride (Fig. 1d). Small-angle x-ray scattering (SAXS) spectra of the connected designs are similar to that of the starting trimer and indicative of well-folded proteins16 (Fig. 1e, Supplementary Table 1; the greater deviations for the five-helix design likely reflects the loss of a helix). The five-helix framework, but not the six-helix framework, bound GFP fused sixth-helix in a pull-down assay (Fig. 1f); the latter result is expected since if the interfaces are otherwise identical and the connecting linker unstrained, the intramolecular interaction should outcompete its intermolecular counterpart because of the reduced entropic cost of formation of intramolecular interactions. To enable the Key to outcompete the Latch, we tuned Kopen by incorporating mutations in the Latch that weaken its interaction with the Cage17–19. A Cage-Latch framework with two serine substitutions in the Latch (V223S/I238S) bound Key nearly as strongly as the five-helix Cage without the Latch (Fig. 1f, Extended Data 2); the two serines likely weaken the Cage-Latch interaction by decreasing the helical propensity of the Latch and increasing the cost of desolvating the Latch when it binds the Cage. In the absence of Key, the Latch is bound to the Cage as in the original monomer (their SAXS spectra are nearly identical and closely match those of the design models; Fig. 1e, Extended Data 1), but the guanidine hydrochloride denaturation midpoint and ΔGfolding are more similar to the truncated five-helix design indicating the mutations are destabilizing (Fig. 1d,e; Extended Data 1). We call such Cage-Latch frameworks Switches, and the Switch-Key pair LOCKR for Latching Orthogonal Cage-Key pRoteins for the remainder of this article.
LOCKR Inducible Bim-Bcl2 Binding
We reasoned that a functional peptide sequence embedded in the Latch could be rendered inactive until Key binding frees the Latch from the Cage, and that activation could be tuned by modulating the thermodynamic parameters outlined in Fig. 1a. To install function into LOCKR, we selected the Bim-Bcl2 interaction central to apoptosis as a model system, and sought to cage Bim such that binding to Bcl2 only occurred in the presence of Key. Three Bcl2 binding peptides20,21 (Extended Data 3a) were chosen to sample a range of KLT and the base LOCKR structure was manipulated to sample a range of KCK and Kopen. The three sequences were embedded in the Latch by sampling different helical registers such that residues involved in binding to Bcl2 are sequestered in the Cage-Latch interface (Extended Data 3b,c), optimizing for the burial of hydrophobic residues and surface exposure of polar residues. (Supplementary Information). To enable sampling of a broader range of Kopen and KCK values, we expanded the available interaction surface area for the Cage:Latch and Cage:Key interactions by lengthening the helices in the Switch by 5-, 9-, or 18-residues (Extended Data 3d), taking advantage of the modular nature of parametric de novo helical bundles2,22,23. Increasing Cage:Latch affinity (decreasing Kopen) makes the system more “off” in absence of Key, and extending the Key to increase affinity for the Cage could allow it to better outcompete the Latch once Kopen is appropriately tuned (increase Kopen relative to KCK). A design caging full length Bim with Cage, Latch and Key each extended by 18-residues is fully off in the absence of Key (Fig. 2a left). Strongest inducible binding was observed with a Latch truncated by 9 residues and a full-length Key (Fig. 2a right); the Key buries more surface area and hence outcompetes the Latch for Cage binding. Addition of Key activated Target binding by BimSwitch over forty-fold (Fig. 2b), comparable to or better than naturally occurring protein interaction switches24–26.
Figure 2. BimLOCKR design and activation.
a, Bio-layer interferometry (BLI) measurement of BimLOCKR (250 nM) binding to immobilized Bcl2 in the presence and absence of 5 µM Key. Bim is tightly caged in the absence of Key; introduction of the toehold (right) allows key to outcompete Latch leading to Bcl2 binding. b, BLI measurement of Key-dependent binding of 250 nM BimLOCKR to Bcl2. Purple is 3 µM Key, then a three-fold dilution of the Key through blue, cyan, green, yellow, and orange; control without Key in red. c, Bcl2 binding by BimLOCKR as a function of Key concentration. BLI data was fit for the different length Keys to obtain equilibrium sensor response. BLI experiments (b,c) repeated three times with similar results. d, Bcl2 binding of BimSwitcha (dark blue), BimSwitchb (blue), and BimSwitchc (light blue) designs in response to cognate Key, measured by BLI and normalized to Rmax. Repeated twice with similar results. e, Bcl2 binding in BLI experiments for each Switch at 250 nM, Key at 5 µM; data points are average Rmax of two replicates.
According to the model in Fig. 1a, the range of Key concentrations over which BimSwitch activates is controllable by tuning KCK and KLT. We investigated using BLI (Biolayer Interferometry) to monitor binding to Bcl2 in response to different length Keys, hence different KCK (Fig. 2c). These activate BimSwitch at different concentrations: a 40-residue Key produces no activation (pale green), a 45-residue Key activates with an EC50 of 230 +/− 58 nM (green), and the full-length 58 residue Key activates with an EC50 of 27.0 +/− 2.8 nM (dark green, Fig. 2b,c). To probe how LOCKR activation depends on KLT, we used different affinity targets: Bcl2, BclB, and Bak bind non-caged Bim with Kds of 0.17 nM21, 20 nM21, and 4.2 μM (Extended Data 4a), respectively. Consistent with the Fig. 1a model, activation of BclB binding requires higher Key concentrations than activation of Bcl2 binding, while Bak does not bind in this range of Key concentrations (Extended Data 4b). The equilibria involved in activation are indeed sensitive to small changes in the binding free energy of both Key and Target.
To enable independent caging and specific unlocking of different protein functions in the same cell, we sought to create orthogonal Switch-Key pairs by incorporating different hydrogen bond networks at the Cage-Latch/Key interface through Rosetta design27,28 (see Supplementary Information). Designs BimLOCKRb and BimLOCKRc show 22-fold and 8-fold activation with their cognate Keys (Fig. 2d). The original BimLOCKR (BimLOCKRa) and the new designs are mutually orthogonal; each Switch is activated only by its cognate Key at concentrations up to 5 μM (Fig. 2e), illustrating the power of the buried hydrogen bond network approach to achieving specificity.
LOCKR inducible protein degradation
We sought to couple Switch activation to protein degradation in living cells by caging the cODC degron29. The caging strategy employed for Bim was used to embed three variants of cODC into Switcha: the wild-type sequence, wild-type with a proline removed (since proline destabilizes -helices), and the central dipeptide CA (Extended Data 5). Designs were characterized in S. cerevisiae using a dual-inducible expression system30 to independently titrate Switch fused to yellow fluorescent protein (YFP) expression and Key fused to blue fluorescent protein (BFP) expression (Fig. 3a). Key induced degradation was dependent on the presence of the degron in the Switch, and was not observed when YFP was fused to either BimSwitcha or Switcha (Extended Data 5). We optimized the amount of inducible degradation by varying the Switch toehold length to tune Kopen (Extended Data 5), and found the Switch with proline removed and a 12-residue toehold had the largest dynamic range (hereafter referred to as degronSwitcha). Keys of different lengths (43 residues versus 55 residues) produced a similar dynamic range of Switch activation, but a higher Key concentration was required for maximal activation for the shorter Key (Fig. 3b; Extended Data 6), as in the case of BimLOCKR in vitro (Fig. 2d), suggesting the Fig 1a model holds in living cells. Key fluorescence was independent of degronSwitcha concentration (Extended Data 6), suggesting the Key is not co-degraded with degronSwitcha. We next examined the dynamics of activation, and found that the amount of YFP-degronSwitcha starts decreasing shortly after induction of Key, reaching a new steady-state in ~3 hours (Fig 3c). Taking into account the rates of synthesis (Supplementary Information), we estimate a 24 minute half-life for activated degronSwitcha, which is similar to the previously measured 11–30 minutes29 for the cODC degron.
Figure 3. Testing functionality of degronLOCKR in live cells.
a, Dual-induction system used in S. cerevisiae to test degronLOCKR function. b, Dose response of YFP-degronSwitcha and Keya-BFP at 50nM E2 as a function of Pg induction. YFP, normalized to no Pg; BFP normalized to max Pg. Lines connecting data are a guide to the eye. c, Dynamics of degronLOCKR using an automated flow cytometry platform. Cells were grown to steady-state at 50nM E2 then induced with Pg to express Keya-BFP at t0hrs. Lines represent moving average taken over three data points. d, Dose response of orthogonal degronLOCKRs as a function of Pg. YFP-degronSwitcha and RFP-degronSwitchc were expressed constitutively in the same cell with either Keya-BFP (left) or Keyc-BFP (right) expressed using Pg. YFP-degronSwitcha, RFP-degronSwitchc and either Keya-BFP or Keyc-BFP were normalized to no Pg (RFP, YFP) or max Pg (BFP). Lines connecting data are a guide to the eye. e, Asymmetric RFP-degronSwitcha was expressed in HEK293T cells with and without Key. Flow cytometry distribution of RFP fluorescence for a representative sample indicates decreased RFP expression in the presence of Key. Geometric mean of RFP expression is quantified in the bar plot. Data in all panels represent mean ± s.d. of three biological replicates.
We next designed orthogonal degronLOCKRs to enable Key-induced degradation of different proteins in the same cell by installing the proline-removed cODC degron in LOCKRb and LOCKRc. YFP fusions of these designs were expressed together with each Key variant fused to cyan fluorescent protein (CFP). degronLOCKRa and degronLOCKRc were strongly activated by their cognate Keys, but not by each other’s Key (LOCKRb did not activate; Extended Data 7). To test the orthogonality of degronLOCKRs in the same cell, we constitutively coexpressed degronLOCKRa and degronLOCKRc fused to YFP and red fluorescent protein (RFP), respectively, and titrated expression of each Key. Expression of Keya led to selective degradation of YFP but not RFP, and expression of Keyc, to selective degradation of RFP but not YFP (Fig. 3d).
To evaluate degronLOCKR function in mammalian cells, we expressed degronSwitcha fused to mCherry RFP in human HEK293T cells, and measured RFP fluorescence in the presence and absence of Key. A redesigned asymmetric degronSwitcha with an 8-residue toehold (see Supplementary Information; Extended Data 8) triggered a 11-fold reduction in mean RFP fluorescence in the presence of Key (Fig. 3e).
degronLOCKR control of gene expression in live cells
We sought to use degronLOCKR to modulate the intracellular concentration of a synthetic transcription factor and dCas9 in yeast. We independently expressed a zinc-finger based synthetic transcription factor (synTF)31 fused to RFP-degronSwitcha, and Keya-BFP-NLS. Activity of the synTF was measured using YFP produced by the synTF promoter (Fig. 4a, left). Key induction triggered a 61% decrease in RFP (the transcription factor), and an 82% decrease in YFP (the transcription factor target), respectively (Fig. 4a, right; Extended Data 9a). To investigate the generality of transcriptional control by degronLOCKR, an activating dCas9-VP64 fusion32 was fused to RFP-degronSwitcha and targeted to a tet operator site with a constitutively expressed sgRNA to induce expression of YFP (Fig. 4b, left). A 78% reduction of RFP and 41% reduction of YFP was observed upon induction of Key (Fig. 4b, right; Extended Data 9b). Together, these results demonstrate the modularity and functionality of degronLOCKR for controlling the stability of proteins in live cells.
Figure 4. Controlling gene expression using degronLOCKR in yeast.

a, (Left) Dual-induction system used to determine the effect of degronLOCKRa on a synthetic transcription factor (synTF). (Right) Dose response of YFP, SynTF-RFP-degronSwitcha and Keya-BFP-NLS at 31.25nM E2 as a function of Pg induction, normalized to no Pg (YFP, RFP) or max Pg (BFP). b, (Left) Dual-induction system used to determine the effect of degronLOCKRa on a dCas9-VP64 targeted to the pTet7x promoter. (Right) Dose response of YFP, dCas9-VP64-RFP-degronSwitcha and Keya-BFP-NLS at 31.25 nM E2 as a function of Pg induction, normalized to no Pg (YFP, RFP) or max Pg (BFP). Data in all panels represent mean ± s.d. of three biological replicates. Lines connecting data are a guide to the eye.
LOCKR inducible nuclear export
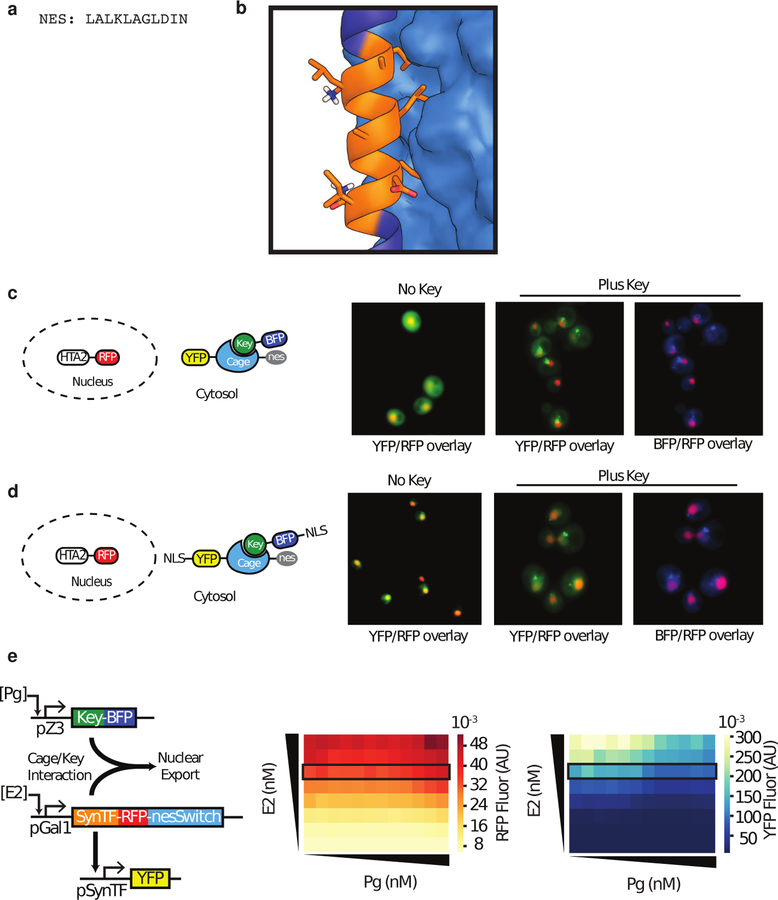
To investigate inducible control over nuclear localization, we caged a nuclear export sequence (NES33) in Switcha (Extended Data 10a,b) using the same strategy as for Bim and cODC, and fused the resulting nesSwitcha to YFP with a nuclear localization sequence34. An RFP-histone fusion (HTA2) was constitutively expressed in the same yeast cells to act as a nuclear marker (Fig. 5a). YFP co-localized with RFP in the nucleus in the absence of Keya-BFP, but upon expression of Keya-BFP the YFP fluorescence becomes more cytosolic, likely due to uncaging of the nuclear export signal (Fig. 5b; Extended Data 10c, d).
Figure 5. Controlling protein localization using nesLOCKR in yeast.
a, Key-induced nuclear export of NLS-YFP-nesSwitcha. The nucleus is marked by the histone HTA2-RFP. b, Fluorescence microscopy showing co-localization of NLS-YFP-nesSwitcha (green) with nuclear HTA2-RFP (red) fluorescence when no Keya-BFP is expressed (top), compared to a more diffuse NLS-YFP-nesSwitcha fluorescent signal observed outside of the nucleus when Keya-BFP is expressed (bottom). Images shown are representative of n=3 biological replicates. c, (Left) Dual-induction system used to determine the effect of nesLOCKRa on a synthetic transcription factor (synTF). (Right) Dose response of YFP, synTF-RFP-nesSwitcha and Keya-BFP at 31.25 nM E2 as a function of Pg induction, normalized to no Pg (RFP, YFP), maximum Pg (BFP). Data represent mean ± s.d. of three biological replicates. Lines connecting data are a guide to the eye.
Next we used nesLOCKR to control the nuclear localization of synTF to modulate its activation of the pSynTF promoter. Using the dual-induction system, we expressed synTF-RFP-nesSwitcha and Keya-BFP in the same cell as a pSynTF-YFP reporter, and observed that induction of Key caused a 33% decrease in YFP signal, indicating successful activation of nesLOCKR and exclusion of synTF from the nucleus (Fig. 5c; Extended Data 10e). Together, these results demonstrate our ability to cage different functional peptide motifs in live cells, highlighting the modularity and utility of LOCKR.
Conclusions
The design of tunable and generalizable protein switches is a considerable advance for de novo protein design. In the switchable LOCKR system described here, a designed key added in trans induces a large conformational change in a designed cage that unlocks protein function. We demonstrate the power and generality of LOCKR by caging three distinct functions: in vitro, the proapoptotic peptide Bim binding to Bcl2, and in cells, protein degradation mediated by the cODC degron and protein localization via a nuclear export sequence. The modularity and hyperstability of de novo designed proteins enables tuning of Switch activation over a broad dynamic range by modulating the strength of the competing Cage-Key and Cage-Latch interfaces. Moving forward, LOCKR provides a general approach for controlling function that should be applicable to a wide range of proteins and synthetic biology challenges.
It is instructive to compare LOCKR to regulatory systems in nature that utilize autoinhibition and efforts to co-opt those systems for engineered protein switches. Activation of apoptosis by the pro-apoptotic proteins Bak and Bax can be triggered by displacement of auto-inhibitory interactions35, analogous to Key activation in LOCKR. Actin nucleation is modulated by N-WASP, which has an autoinhibited actin nucleating Arp2/3 binding domain that is released upon binding to the activators Cdc42 and PIP236. A number of proteins, including N-WASP, have been repurposed to control non-cognate functions in a switchable, inducible manner37,38, but the LOCKR system has several advantages. First, LOCKR is a universal platform to cage and then activate functionalities at will ranging from inducible activation of high-affinity protein-protein interactions to controlled degradation or localization of an attached cargo. Second, for any functional modality, many cargoes can be regulated: here we couple key-induced LOCKR-gated degradation to fluorescent protein levels both directly through fusion and indirectly through fusion to an activating transcription factor; kinases can also be controlled in the same way (see accompanying paper). Strategies that rely on repurposing natural proteins have modularity and tunability limited by the evolved functions and ligands of these existing proteins, whereas altering the affinities of LOCKR components is tunable based on simple design principles that are generally irrespective of the functional modality or application. Our use of a toehold for tuning helical displacement is reminiscent of DNA strand displacement technology39,40, but unlike nucleic acid based approaches such as, genetic toggles41, or riboswitches42,43 (which have largely focused on controlling transcription), LOCKR systems can be readily integrated with the many diverse processes controlled by proteins. Viewed in this light, LOCKR brings to proteins the programmability of DNA switching technology, with the added advantages of tunability and flexibility over rewired natural protein systems, and ready interfacing with biological machinery over DNA nanotechnology [add reference to paper #2]. More generally, the domain of sophisticated environmentally sensitive and switchable function no longer belongs exclusively to naturally occurring proteins.
Methods
PCR mutagenesis and isothermal assembly
All primers for mutagenesis were ordered from Integrated DNA Technologies (IDT). Mutagenic primers were designed to anneal >18bp on either side of the site for mutagenesis with the desired mutation encoded in the primer. PCR was used to create fragments upstream and downstream of the mutation site with >20bp overlap with the desired pET vector. The resulting amplicons were isothermally assembled into either pET21b, pET28b, or pET29b restriction digested with XhoI and NdeI and transformed into chemically competent E. coli XL1-Blue cells. Monoclonal colonies were sequenced with Sanger sequencing. Sequence verified plasmid was purified using Qiagen miniprep kit and transformed into chemically competent E. coli BL21(DE3)Star, BL21(DE3)Star-pLysS cells (Invitrogen), or Lemo21(DE3) cells (NEB) for protein expression.
Synthetic gene construction
Synthetic genes were ordered from Genscript Inc. (Piscataway, NJ, USA) and delivered in pET 28b+, pET21b+, or pET29b+ E. coli expression vectors, inserted at the NdeI and XhoI sites of each vector. For pET28b+ constructs, synthesized DNA was cloned in frame with the N-terminal hexahistidine tag and thrombin cleavage site and a stop codon was added at the C-terminus. For pET21b+ constructs, a stop codon was added at the C-terminus such that the protein was expressed with no hexahistidine tag. For pET29b+ constructs, the synthesized DNA was cloned in frame with the C-terminal hexahistidine tag. Plasmids were transformed into chemically competent E. coli BL21(DE3)Star, BL21(DE3)Star-pLysS cells (Invitrogen), or Lemo21(DE3) cells (NEB) for protein expression.
Bacterial protein expression and purification
Starter cultures were grown in Lysogeny Broth (LB) or Terrific Broth II (TBII) overnight in the presence of 50 μg/mL carbenicillin (pET21b+) or 30 μg/mL (for LB) to 60 μg/mL (for TBII) kanamycin (pET28b+ and pET29b+). Starter cultures were used to inoculate 500 mL of Studier TBM-5052 autoinduction media containing antibiotic and grown at 37 ℃ for 24 hours. Cells were harvested by centrifugation at 4000 rcf for 20 minutes at 4 ℃ and resuspended in lysis buffer (20 mM Tris, 300 mM NaCl, 20 mM Imidazole, pH 8.0 at room temperature), then lysed by microfluidization in the presence of 1 mM PMSF. Lysates were cleared by centrifugation at 24,000 rcf for at least 30 minutes at 4 ℃. Supernatant was applied to Ni-NTA (Qiagen) columns pre-equilibrated in lysis buffer. The column was washed twice with 15 column volumes (CV) of wash buffer (20 mM Tris, 300 mM NaCl, 40 mM Imidazole, pH 8.0 at room temperature), followed by 15 CV of high-salt wash buffer (20 mM Tris, 1 M NaCl, 40 mM Imidazole, pH 8.0 at room temperature) then 15 CV of wash buffer. Protein was eluted with 20 mM Tris, 300 mM NaCl, 250 mM Imidazole, pH 8.0 at room temperature. Proteins were further purified by gel filtration using FPLC and a Superdex™ 75 Increase 10/300 GL (GE) size exclusion column, pooling fractions containing monomeric protein.
Size-exclusion Chromatography, Multi-Angle Light Scattering (SEC-MALS)
SEC-MALS experiments used a Superdex™ 75 Increase 10/300 GL (GE) size exclusion column connected to a miniDAWN TREOS multi-angle static light scattering and an Optilab T-rEX (refractometer with EXtended range) detector (Wyatt Technology Corporation, Santa Barbara CA, USA). Protein samples were injected at concentrations of 3–5 mg/mL in TBS (pH 8.0). Data was analyzed using ASTRATM (Wyatt Technologies) software to estimate the weight average molar mass (Mw) of eluted species, as well as the number average molar mass (Mn) to assess monodispersity by polydispersity index (PDI) = Mw/Mn.
Circular dichroism (CD) measurements
CD wavelength scans (260 to 195 nm) and temperature melts (25 to 95 ℃) were measured using an AVIV model 420 CD spectrometer. Temperature melts monitored absorption signal at 222 nm and were carried out at a heating rate of 4 ℃/min. Protein samples were at 0.3 mg/mL in PBS pH 7.4 in a 0.1 cm cuvette. Guanidinium chloride (GdmCl) titrations were performed on the same spectrometer with an automated titration apparatus in PBS pH 7.4 at 25 ℃, monitored at 222 nm with protein sample at 0.03 mg/mL in a 1cm cuvette with stir bar. Each titration consisted of at least 40 evenly distributed concentration points with one minute mixing time for each step. Titrant solution consisted of the same concentration of protein in PBS + GdmCl. GdmCl concentration was determined by refractive index.
Small angle X-ray scattering (SAXS)
Samples were exchanged into SAXS buffer (20 mM Tris, 150 mM NaCl, 2% glycerol, pH 8.0 at room temperature) via gel filtration. Scattering measurements were performed at the SIBYLS 12.3.1 beamline at the Advanced Light Source. The X-ray wavelength (λ) was 1.27 Å and the sample-to-detector distance of the Mar165 detector was 1.5 m, corresponding to a scattering vector q (q = 4π*sin(θ/λ) where 2θ is the scattering angle) range of 0.01 to 0.59 Å−1. Data sets were collected using 34 0.2 second exposures over a period of 7 seconds at 11 keV with protein at a concentration of 6 mg/mL. Data were also collected at a concentration of 3 mg/mL to determine concentration-dependence; all presented data was collected at the higher concentration as no concentration-dependent aggregation was observed. Data from 32 exposures was averaged separately over the Gunier, Parod, and Wide-q regions depending on signal quality over each region and frame. The averages were analyzed using the ScÅtter software package to analyze data and report statistics (Extended Data Table 1). FoXS was used to compare design models to experimental scattering profiles and calculate quality of fit (X) values. The hexahistidine tags and thrombin cleavage sites on the N-termini of LOCKR proteins were modeled using Rosetta Remodel so that the design sequence matched that of the experimentally tested protein. To capture conformational flexibility of these residues, 100 independent models were generated, clustered, and the cluster center of the largest cluster was selected as a representative model for FoXS fitting without bias.
GFP pulldown assay
His-tagged LOCKR was expressed per the above protocol from pET28b+ while the Key was expressed fused to superfolder GFP (sfGFP) without a his-tag in pET21b+. The his-tagged LOCKR was purified to completion and dialyzed into TBS (20 mM Tris, 150 mM NaCl, pH 8.0 at room temperature); the Key-GFP remained as lysate for this assay. 100 μL LOCKR at >1 μM was applied to a 96-well black Pierce® Nickel Coated Plate (ThermoFisher) and incubated at room temperature for 1 hour. Sample was discarded from the plate and washed 3x with 200 μL TBST (TBS + 0.05% Tween-20). 100 μL of lysate containing Key-GFP was added to each well and incubated at room temperature for 1 hour. Sample was discarded from the plate and washed 3x with 200 μL TBST (TBS + 0.05% Tween-20). The plate was washed 1x with TBS, and 100 μL of TBS was added to each well. sfGFP fluorescence was measured on a Molecular Devices SpectraMax M5 plate reader or BioTek Synergy Neo2 plate reader; fluorescence was measured at 485 nm excitation and 530 nm emission, with a bandwidth of 20 nm for excitation and emission.
Bio-Layer Interferometry (BLI)
BLI measurements were made on an Octet® RED96 System (ForteBio) with streptavidin (SA) coated biosensors and all analysis was performed within ForteBio Data Analysis Software version 9.0.0.10. Assays were performed with protein diluted into HBS-EP+ Buffer from GE (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20, 0.5% non-fat dry milk, pH 7.4 at room temperature). Biotinylated Bcl2 was loaded onto the SA tips to a threshold of 0.5 nm programmed into the machine’s protocol. Baseline was obtained by dipping the loaded biosensors into HBS-EP+ buffer; association kinetics were observed by dipping into wells containing defined concentrations of LOCKR and Key, then dissociation kinetics were observed by dipping into the buffer used to obtain the baseline. Kinetic constants and response at equilibrium were computed by fitting a 1:1 binding model.
Construction of DNA circuits
Hierarchical golden gate assembly was used to assemble plasmids for yeast strain construction using the method in Lee et al.44. Individual parts had their BsaI, BsmBI, and NotI cut sites removed to facilitate downstream assembly and linearization. Parts were either generated via PCR or purchased as gBlocks from IDT. These parts were assembled into transcriptional units (promoter-gene-terminator) on cassette plasmids. These cassettes were then assembled together to form multi-gene plasmids for insertion into the yeast genome.
Yeast strains and growth media
The base S. cerevisiae strain used in all experiments was BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). All yeast cultures were grown in YPD media (10 g/L Bacto Yeast Extract, 20 g/L Bacto peptone, 20 g/L dextrose) or synthetic complete medium (SDC) (6.7 g/L Bacto-yeast nitrogen base without amino acids, 2 g/L complete supplement amino acid mix, 20 g/L dextrose). Selection of auxotrophic markers (URA3, LEU2, and/or HIS3) was performed on synthetic complete medium with the appropriate dropout amino acid mix. All yeast strains used in this work are listed in Supplementary Table 4 and 5.
Estradiol and Progesterone induction
Yeast strains were grown overnight by picking a single colony from a plate into YPD media. Saturated culture was diluted 1:500 in fresh SDC and aliquoted into individual wells of a 2 mL 96 well storage block (Corning) for a three hour outgrowth at 30 ℃ and 900 RPM in a Multitron shaker (Infors HT). Estradiol (Sigma-Aldrich) and progesterone (Fisher Scientific) were prepared at a 10x concentration by making the appropriate dilutions into SDC from a 3.6 mM estradiol and 3.2 mM progesterone stock solution. After the three hour outgrowth, 50 μl of estradiol and progesterone inducer were added to the 96 well block in the appropriate combinations and the block was returned to the shaker. Flow cytometry measurement was performed after six hours of incubation for all experiments, except for those involving synTF or dCas9, which was allowed to incubate for 12 hours due to the additional transcriptional step in the system.
Mammalian Cell Culture and Lentiviral Transduction
HEK293T cells (from ATCC® CRL-3216™) were maintained in DMEM (Dulbecco’s Modified Eagle Medium, Gibco) supplemented with 10% Fetal Calf Serum (SAFC) and passaged every ~3 days. Cell line was not authenticated nor tested for mycoplasma contamination. Pantropic VSV-G pseudotyped lentivirus was produced via transfection of Lenti-X 293T cells (Clontech #11131D) with a custom pHR’SIN:CSW transgene expression vector and the viral packaging plasmids pCMVdR8.91 and pMD2.G using Fugene HD (Promega). At 48 hr, viral supernatant was harvested and the HEK293T cells were exposed to the virus for 24 hr. Transductions were performed in triplicate. All plasmids used for HEK293T experiments are listed in Supplementary Table 6.
Description of automated flow cytometry and continuous culture system
Hardware
We adapted an existing automated experimental platform45 to perform variable concentration small molecule induction and long-term culturing. Yeast cultures were grown in 50 mL optically clear conical tubes (Falcon) that were held in eight custom temperature-controlled, magnetically stirred chambers. Liquid handling was accomplished using a 14 position stream selector (VICI Cheminert) and two syringe pumps (Cavro XCalibur Pump, TECAN) of a BD High-Throughput Sampler. Commands to the HTS were controlled using LABVIEW 2013. This setup allowed for periodic sampling and dilution of individual cultures. Each sampling period consisted of three main steps: 1) send sample to flow cytometer for measurement, 2) extract culture and send to waste, and 3) replenish culture with fresh media at desired hormone concentration. Each sampling period can be designated to either induce cultures to a new higher hormone concentration or to maintain desired hormone concentration. A sampling frequency of 24 minutes and a dilution volume of 3 mL were used.
Yeast culture
Yeast strains were grown overnight by picking a single colony from a plate into YPD media. Saturated culture was diluted 1:200 into fresh SDC. Cultures were grown for 2 hours in glass tubes at 30 ˚C and 250 RPM in a Innova 44 shaker (New Brunswick). Cultures were then diluted to 0.01 OD600 in fresh SDC and aliquoted into individual 50 mL optically clear conical tubes (Falcon) at a total volume of 30 mL YPD. Another one hour outgrowth was performed in bioreactors with magnetically-controlled stir bars at 30 ˚C. All SDC media was supplemented with 5,000 U/mL Penicillin Streptomycin (Thermo-Fisher).
Estradiol and progesterone induction to test degronLOCKR dynamics
A 1X concentration was determined by the highest desired hormone concentration at which to test strains (30 nM E2 and 50 nM Pg, respectively). A solution of E2 and SDC media was created at a 10X concentration to bring pre-induced cultures to a desired concentration in one sampling period. A second solution of Pg and SDC media was created at a 10X concentration to induce Key expression after degSwitch-YFP expression reached steady-state. SDC media was prepared at three different concentrations of hormone: (1) 10X E2/no Pg, (2) 1X E2/no Pg, (3) 1X E2/10X Pg, and (4) 1X E2/1X Pg. After a one hour outgrowth in bioreactors (t=−6 hr), the first induction was performed to achieve E2 concentration by extracting 3 mL from all cultures and replenishing with (1). After E2 induction, sampling proceeded as described above (see Hardware). All sampling periods following the first induction time point included sending a sample to the cytometer for measurement, extracting 3 mL from all cultures, and replenishing cultures with (2). During the second induction time point (t=0 hr), cultures were induced with (3) to activate Key expression. This induction was followed by the same procedure as the first induction, except that hormone concentrations were maintained by (4). Controls (no activated Key expression) did not undergo a second induction and, instead, continued to be replenished by (2).
Flow cytometry
Yeast experiments
Analysis of fluorescent protein expression was performed using a BD LSRII flow cytometer (BD Biosciences) equipped with a high-throughput sampler. Yeast cultures were diluted in TE before running through the instrument to obtain an acceptable density of cells. YFP (Venus) fluorescence was measured using the FITC channel, RFP (mScarlet) was measured using the PE-Texas Red channel, and BFP (mTagBFP2) was measured using the DAPI channel. For steady-state measurements, 5,000–10,000 events were collected per sample. For dynamic measurements, 2,000–10,000 events were collected per sample. Fluorescence values were calculated as the height (H) measurement for the appropriate channel and normalized to cell size by dividing by side scatter (SSC-H). All analysis of yeast flow cytometry data was performed in Python 2.7 using the package FlowCytometryTools v0.5.0 and custom scripts.
HEK293T experiments
Analysis of fluorescent protein expression was performed using a BD Fortessa flow cytometer (BD Biosciences) equipped with a high-throughput sampler. Cells were harvested and washed twice in PBS before running through the instrument in PBS+5% FBS. RFP (mCherry) fluorescence was measured using the PE-CF594 channel and BFP (tagBFP) was measured using the BV 421 channel. 50,000 events were collected per sample. Live cells were gated according to FSC-A and SSC-A, and singlets were gated according to SSC-A and SSC-H. Analysis of HEK293T flow cytometry data was performed using FlowJo v10.
Fluorescence microscopy
Saturated culture was diluted 1:100 in fresh SC media followed by a 3 hour outgrowth at 30 ℃ with shaking at 700 RPM in a Multitron shaker (Infors HT). Estradiol (Sigma-Aldrich) and progesterone (Fisher Scientific) were prepared at a 20x concentration by making the appropriate dilutions into SC media from a 3.6 mM estradiol and 3.2 mM progesterone stock solution. Cells were induced with estradiol and/or progesterone at a final concentration of 200 μM and 250 μM, respectively. After 8 hours of growth, cells were resuspended in 1x PBS and imaged on a Zeiss Axio Observer Z1 microscope with X-Cite Series 120 fluorescent lamp and Hamamatsu Orca-Flash 4.0 Digital Camera.
Structural visualization and figures
All structural images for figures were generated using PyMOL46.
Extended Data
Extended Data Figure 1: Biophysical data from LOCKR design.
a) Size Exclusion Chromatography for the Monomer, Truncation, and LOCKR designs on Superdex 75. Peaks indicated by vertical dashed lines represent monomeric protein used in downstream characterization and functional assays. SEC repeated three times with similar results. b) Circular dichroism spectroscopy to determine protein stability upon heating and chemical denaturant, guanidinium chloride. Top row: full wavescan at 25°C (blue), 75°C (orange), 95°C (red), then cooled to 25°C (cyan). Middle row: guanidinium chloride melts also shown overlapped in Figure 1d. Bottom row: fraction folded was converted to equilibrium constant, then to ΔGunfolding value. The linear unfolding region, marked by vertical lines in middle row, was fit to determine the ΔGfolding for each design. Repeated four times with similar results. c) SAXS spectra (black) referenced in Figure 1e fit to Rosetta design models (red) using FoXS with chi-values referenced in the upper right.
Extended Data Figure 2: GFP pulldown assay finds mutations for LOCKR.
Different putative LOCKR constructs were adhered via 6x-His tag to a Ni coated 96-well plate, Key-GFP was applied, and excess washed. Resulting mean fluorescence values represent Key-GFP bound to LOCKR constructs. The truncation was used as a positive control, since the Key binds to the open interface. The monomer as a negative control since it does not bind the Key. Error bars represent the standard deviation of three technical replicates, because Key-GFP was not purified from bacterial lysate leading to minor technical variability.
Extended Data Figure 3: Caging Bim-related sequences.
a) Three Bcl2 binding sequences were grafted onto the Latch. aBcl2 is a single helix from a designed Bcl2 binder (pdb: 5JSN) where non Bcl2-interacting residues were reverted back to the standard LOCKR Latch sequence, shown as dashes. pBim is the partial Bim sequence where only Bcl2-interacting residues are grafted onto the Latch. Bim is the full consensus sequence of the BH3 domain. b) LOCKR (left) with the Latch in dark blue. The helical Bim sequence is taken from the Bim/Bcl2 interaction and grafted onto the Latch c) Left: Bcl2 (tan) binding to Bim (orange) from pdb:2MV6 with pBim residues shown as sticks. Center: a well caged graft where important binding residues are caged. Right: a poor graft where Bcl2 binding residues are exposed and polar surface residues are against the Cage interface. d) Tuning BimLOCKR. aBcl2, pBim, and Bim were caged to varying degrees of success. Early versions of the switch, with aBcl2 and pBim did not efficiently cage Bcl2 binding in the off state. They also only weakly bound the Key leading to small dynamic range. The Cage and Key was extended by 5, 9, and 18 residues in an attempt to provide a larger interface to tightly hold the Latch in the off state and provide a larger interface for Key binding to increase the dynamic range of activation. Mutations on the Latch, identified in Extended Data 2, and providing toeholds for Key binding were the two strategies employed to tune the switch. In graphs, “off” refers to 250–310 nM switch an absence of Key while “on” refers to excess Key added. The height of the bar graph shows the Req as measured by Bio-layer interferometry.
Extended Data Figure 4: Validation of model in Figure 1a.
a) Measurement of Bim:Bak affinity. Bio-layer interferometry (BLI) at three concentrations gives on and off rates for Bim:Bak binding, yielding the constants shown on right. Mean shown with standard deviation of four technical replicates to account for variablility in drift on the BLI instrument. b) BLI measurement of BimLOCKRa (400 nM) binding to Bcl2 (gold), BclB (yellow), and Bak (lighter yellow - BimLOCKR at 1 μM) as Key is added to solution. Normalized due to differences in Rmax for Bcl2 and BclB on the tip. c) BLI measurement of BimLOCKRa binding to Keya immobilized on the tip. Open circles are with no Bcl2 present, gold points are with Bcl2 present at 500 nM.
Extended Data Fig 5: Caging cODC sequences.
a) Three variations of the cODC degron to Cage. Variations meant to tune Kopen by removing the destabilizing proline (noPro) and minimizing mutations to the Latch (CA only). b) Predicted models of the full and noPro cODC sequences (orange) threaded onto the Latch (dark blue). Thread position chosen such that the cysteine residue needed for degradation is sequestered against the Cage (light blue). Proline highlighted in red in the full cODC mutated to an isoleucine in the noPro variant. c) Comparing the stability of YFP fused to cODC variants caged in Switcha to an empty Switcha and to bimSwitcha. The dual-inducible system from Fig 3a was used to express the various YFP-Switcha fusions (solid lines and dots) via pGAL1 and E2, and Keya-BFP via pZ3 and Pg. YFP (Venus) alone, YFP fused to the WT cODC (cODC) or YFP fused to the proline removed cODC (cODC noPro), were also expressed using pGal1 and E2 (dashed lines). Cells were induced with a saturating dose of E2 (50 nM) and Pg was titrated in from 0–200 nM. Fluorescence was measured at steady-state using a flow cytometer; data represent mean ± s.d. of three biological replicates. Lines connecting data are a guide to the eye. A moderate decrease in YFP fluorescence was observed as a function of Pg for the full cODC variant, whereas only a small decrease was observed for the proline removed and CA only. No decrease in fluorescence was observed as a function of Key induction for YFP alone, empty Switcha, or bimSwitcha. d) Tuning toehold lengths of degronLOCKRa. The dual-inducible system from Fig 3a was used to express the various YFP-Switcha fusions via pGal1 and E2, and Keya-BFP via pZ3 and Pg. YFP fused to the proline-removed cODC (cODC no Pro) was also expressed using pGal1 and E2 (dashed line). Cells were induced with a saturating dose of E2 (50 nM) and Pg was titrated in from 0–200 nM. Fluorescence was measured at steady-state using a flow cytometer; data represent mean ± s.d. of three biological replicates. Lines connecting data are a guide to the eye. (Left) cODC variants alone to show dynamic range of Full cODC. (Right) Extending toehold on proline-removed version from 9 to 12 and 16aa. Proline-removed with 12aa toehold shows the greatest dynamic range of all the Switches tested.
Extended Data Figure 6: YFP (a) and BFP (b) expression corresponding to Fig 3b.

0–50nM E2 and 0–200nM Pg were used to induce expression of YFP-degronSwitcha and Keya (Full-length or truncated)-BFP, respectively. Fluorescence was measured at steady-state using a flow cytometer. Heatmaps depict mean fluorescence and are a representative sample of three biological replicates. E2 dose (50nM) depicted in Fig 3b is indicated with the black rectangle on the heatmaps. YFP fluorescence was normalized to the maximum fluorescence (50nM E2, 0nM Pg). BFP expression was not dependent on expression of the Switch, suggesting the Key does not co-degrade with the Switch.
Extended Data Figure 7: degronLOCKRa-d orthogonality.

All combinations of pTDH3-YFP-degronSwitch and pTDH3-Key-CFP were tested. Fluorescence was measured at steady-state using a flow cytometer. YFP fluorescence was averaged across three biological replicates. Percentage degradation was calculated by subtracting the mean YFP-degronSwitch fluorescence with the given Key-CFP coexpressed from the YFP-degronSwitch fluorescence without any Key expressed and normalizing by the YFP-degronSwitch fluorescence without any Key expressed. degronSwitcha is activated strongly by Keya and weakly by Keyb. degronSwitchc is activated strongly by Keyc and weakly by Keyb. Because degronSwitcha and degronSwitchc are not activated by Keyc and Keya respectively, we consider these two to be an orthogonal pair.
Extended Data Figure 8: Comparison of different degronSwitch variants in HEK293T cells.
Fluorescence of RFP-degronSwitch variants in the presence and absence of Key-BFP were measured using flow cytometry. Original symmetric design was compared against a new asymmetric design. Two toehold lengths were tested for each variant. Data in bar graph represents geometric mean ± s.d. of three biological replicates. Histograms are depicted for a representative sample. Asymmetric cage with a t8 toehold demonstrates the largest dynamic range.
Extended Data Figure 9: YFP and RFP expression for synTF (a) and dCas9-VP64 (b).
assay corresponding to Fig 4 as a function of E2 (0–125 nM) and Pg (0–100 nM). YFP fluorescence represents transcriptional output of either synTF or dCas9-VP64 and RFP fluorescence represents fluorescence of either synTF or dCas9-VP64. Fluorescence was measured at steady-state using flow cytometry. Heatmaps depict mean fluorescence and are a representative sample of three biological replicates. E2 dose (31.25nM) depicted in Fig 5 is indicated with the black rectangle on the heatmaps.
Extended Data Figure 10: Design and characterization of nesLOCKR.
a) NES used in this report. b) The NES (orange) caged on the helical Latch (dark blue, cartoon) with hydrophobic residues sequestered against the Cage (light blue, surface) c) (Left) Schematic of cytosolic YFP-nesSwitcha and Key-BFP with nuclear marker HTA2-RFP. (Right) YFP fluorescence shows the expected cytosolic distribution when YFP-nesSwitcha is expressed with no NLS (left) but punctae of YFP fluorescence is observed when both YFP-nesSwitcha and Key-BFP are expressed in the cytosol, which we assume is due to aggregation of the nesSwitcha. Key-BFP fluorescence is co-localized to YFP-nesSwitcha fluorescence. d) (Left) Schematic of NLS-YFP-nesSwitcha with Key-BFP-NLS with nuclear marker HTA2-RFP. (Right) YFP-nesSwitcha is localized to the nucleus when expressed with the strong (SV40) NLS. When Key-BFP is expressed with a moderately strong NLS, the same pattern of cytosolic YFP punctae formation is observed as when Key-BFP is expressed without a NLS (Figure 5b), indicating that uncaging of the NES is independent of NLS on Key-BFP localization. Key-BFP-NLS fluorescence is co-localized to NLS-YFP-nesSwitcha fluorescence e) YFP and RFP expression for synTF assay corresponding to Fig 5c as a function of E2 (0–125 nM) and Pg (0–500 nM). Fluorescence was measured at steady-state using flow cytometry. Heatmaps depict mean fluorescence and are a representative sample of three biological replicates. E2 dose (31.25nM) depicted in Fig 5c is indicated with the black rectangle on the heatmaps.
Supplementary Material
Acknowledgements
RAL was supported by Bruce and Jeannie Nordstrom thanks to the Patty and Jimmy Barrier Gift for the Institute for Protein Design Directors Fund. SEB is supported by a Career Award at the Scientific Interface from Burroughs Wellcome Fund. AHN was supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program. MJL was supported by a Washington Research Foundation Innovation Postdoctoral Fellowship and a Cancer Research Institute Irvington Fellowship from the Cancer Research Institute. SAXS data were collected at the Advanced Light Source (ALS) at LBNL, supported by the following grants from NIH (P30 GM124169–01, ALS-ENABLE P30 GM124169, and S10OD018483), NCI SBDR (CA92584) and DOE-BER IDAT (DE-AC02–05CH11231). This work was supported by the Defense Advanced Research Projects Agency, Contract No. HR0011–16-2–0045 to HE-S The content and information does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred. HE-S. is a Chan-Zuckerberg investigator. Thank you to L. Carter and the Protein Production Facility in the Institute for Protein design for protein used in this study. Thanks to S. Bermeo, A. Quijano Rubio, B. Basanta, M. Chevalier, A. Bonny, and J. Pedro-Fonseca for help and advice.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Competing Interests: RAL, SEB, DB, WRPN, and MJL have filed a provisional patent describing the design, composition, and function of LOCKR switches, keys, and scaffolds; RAL, AHN, SEB, MJL, DB, and HE-S have filed a provisional patent application describing the design, composition, and function of degronLOCKR constructs; DB, RAL, SEB, and MJL hold equity in Lyell Immunopharma, Inc. DB holds equity in Sana Biotechnology.
Code Availability
Python scripts, bash scripts, and Rosetta Design XMLs are available for download at https://github.com/BobbyLangan/DeNovoDesignofBioactiveProteinSwitches
Data Availability
The authors declare that all data supporting the findings of this study are available within the paper (and its Supplementary Information files), but original data that supports the findings are available from the corresponding authors upon reasonable request. Plasmids encoding LOCKR scaffolds (non-functional Switches and Keys), BimLOCKR, degronLOCKR, and nesLOCKR can be found on Addgene (plasmids 127416–127424, 127200–127206 and 127246).
References
- 1.Huang P-S, Boyken SE & Baker D The coming of age of de novo protein design. Nature 537, 320–327 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Huang P-S et al. High thermodynamic stability of parametrically designed helical bundles. Science 346, 481–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunette TJ et al. Exploring the repeat protein universe through computational protein design. Nature 528, 580–584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocklin GJ et al. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambroggio XI & Kuhlman B Design of protein conformational switches. Curr. Opin. Struct. Biol 16, 525–530 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Choi JH, Laurent AH, Hilser VJ & Ostermeier M Design of protein switches based on an ensemble model of allostery. Nat. Commun 6, 6968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joh NH et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey JA, Damry AM, Goto NK & Chica RA Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol 13, 1280–1285 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Ha J-H & Loh SN Protein conformational switches: from nature to design. Chemistry 18, 7984–7999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J & Nussinov R Allostery: An Overview of Its History, Concepts, Methods, and Applications. PLoS Comput. Biol 12, e1004966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raman S, Taylor N, Genuth N, Fields S & Church GM Engineering allostery. Trends Genet 30, 521–528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins JR, Diboun I, Dessailly BH, Lees JG & Orengo C Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Singh GP, Ganapathi M & Dash D Role of intrinsic disorder in transient interactions of hub proteins. Proteins 66, 761–765 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Dou J et al. De novo design of a fluorescence-activating β-barrel. Nature (2018). doi: 10.1038/s41586-018-0509-0 [DOI] [PMC free article] [PubMed]
- 15.Boyken SE et al. De novo design of protein homo-oligomers with modular hydrogen-bond network–mediated specificity. Science 352, 680–687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyer KN et al. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol. Biol 1091, 245–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming PJ & Rose GD Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci 14, 1911–1917 (2005).15937286 [Google Scholar]
- 18.Chothia C & Janin J Principles of protein–protein recognition. Nature 256, 705 (1975). [DOI] [PubMed] [Google Scholar]
- 19.Moreira IS, Fernandes PA & Ramos MJ Hot spots—A review of the protein--protein interface determinant amino-acid residues. Proteins: Struct. Funct. Bioinf 68, 803–812 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Delgado-Soler L, Pinto M, Tanaka-Gil K & Rubio-Martinez J Molecular Determinants of Bim(BH3) Peptide Binding to Pro-Survival Proteins. J. Chem. Inf. Model 52, 2107–2118 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Berger S et al. Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigoryan G & Degrado WF Probing designability via a generalized model of helical bundle geometry. J. Mol. Biol 405, 1079–1100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crick FHC The Fourier transform of a coiled-coil. Acta Crystallogr 6, 685–689 (1953). [Google Scholar]
- 24.Ikeda K, Watanabe Y, Ohto H & Kawakami K Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell. Biol 22, 6759–6766 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giesecke AV, Fang R & Joung JK Synthetic protein-protein interaction domains created by shuffling Cys2His2 zinc-fingers. Mol. Syst. Biol 2, 2006.2011 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehtanz M, Schmidt H-M, Warthorst U & Steger G Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol. Cell. Biol 24, 2153–2168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhlman B & Baker D Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. U. S. A 97, 10383–10388 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leaver-Fay A et al. Chapter nineteen - Rosetta3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules. in Methods in Enzymology (eds. Johnson ML & Brand L) 487, 545–574 (Academic Press, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi J, Chen H, Hoyt MA & Coffino P Structural elements of the ubiquitin-independent proteasome degron of ornithine decarboxylase. Biochem. J 410, 401–407 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Aranda-Díaz A, Mace K, Zuleta I, Harrigan P & El-Samad H Robust Synthetic Circuits for Two-Dimensional Control of Gene Expression in Yeast. ACS Synth. Biol 6, 545–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khalil AS et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell 150, 647–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Pinera P et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Güttler T et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol 17, 1367–1376 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Kosugi S et al. Six Classes of Nuclear Localization Signals Specific to Different Binding Grooves of Importin α. J. Biol. Chem 284, 478–485 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Westphal D, Dewson G, Czabotar PE & Kluck RM Molecular biology of Bax and Bak activation and action. Biochim. Biophys. Acta 1813, 521–531 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Prehoda KE, Scott JA, Mullins RD & Lim WA Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science 290, 801–806 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Dueber JE, Yeh BJ, Bhattacharyya RP & Lim WA Rewiring cell signaling: the logic and plasticity of eukaryotic protein circuitry. Curr. Opin. Struct. Biol 14, 690–699 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Stein V & Alexandrov K Synthetic protein switches: design principles and applications. Trends Biotechnol 33, 101–110 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Zhang DY & Winfree E Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc 131, 17303–17314 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Linko V & Dietz H The enabled state of DNA nanotechnology. Curr. Opin. Biotechnol 24, 555–561 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Gardner TS, Cantor CR & Collins JJ Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Wittmann A & Suess B Engineered riboswitches: Expanding researchers’ toolbox with synthetic RNA regulators. FEBS Lett 586, 2076–2083 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Isaacs FJ, Dwyer DJ & Collins JJ RNA synthetic biology. Nat. Biotechnol 24, 545–554 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Lee ME, DeLoache WC, Cervantes B & Dueber JE A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol 4, 975–986 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Harrigan P, Madhani H & El-Samad H Real time genetic compensation operationally defines the dynamic demands of feedback control. bioRxiv 244020 (2018). doi: 10.1101/244020 [DOI] [PMC free article] [PubMed]
- 46.DELANO & L, W. The PyMOL Molecular Graphics System http://www.pymol.org (2002).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.