Abstract
The TP53 tumor suppressor gene encodes a DNA-binding transcription factor that regulates multiple cellular processes including cell growth and cell death. The ability of p53 to bind to DNA and activate transcription is tightly regulated by post-translational modifications and is dependent on a reducing cellular environment. Some p53 transcriptional target genes are involved in regulation of the cellular redox homeostasis, e.g. TIGAR and GLS2. A large fraction of human tumors carry TP53 mutations, most commonly missense mutations that lead to single amino acid substitutions in the core domain. Mutant p53 proteins can acquire so called gain-of-function activities and influence the cellular redox balance in various ways, for instance by binding of the Nrf2 transcription factor, a major regulator of cellular redox state. The DNA-binding core domain of p53 has 10 cysteine residues, three of which participate in holding a zinc atom that is critical for p53 structure and function. Several novel compounds that refold and reactivate missense mutant p53 bind to specific p53 cysteine residues. These compounds can also react with other thiols and target components of the cellular redox system, such as glutathione. Dual targeting of mutant p53 and redox homeostasis may allow more efficient treatment of cancer.
Keywords: p53, redox regulation, mutation, oxidative stress, Nrf2, thiols, cancer therapy
Introduction
p53 has occupied a central position in cancer research during the last three decades. One reason for the fame of p53 is the frequent mutation of the TP53 gene in human tumors (Soussi and Wiman, 2015). Also, p53 has continued to fascinate and surprise investigators through its involvement in a wide range of diverse cellular processes (Vousden and Prives, 2009), and by the discovery of multiple p53 isoforms with as yet poorly understood functions (Marcel et al., 2011). The p53 protein is a transcription factor that activates transcription of genes that regulate, for instance, cycle arrest, DNA repair, metabolism, apoptosis, senescence, and autophagy (Vousden and Prives, 2009). All these pathways can presumably contribute to p53-mediated tumor suppression.
Genome sequencing of more than 3000 tumors representing 12 common tumor types revealed TP53 mutations in 42% of the cases (Kandoth et al., 2013; Soussi and Wiman, 2015). However, mutation frequencies vary greatly between different tumor types, ranging from 2.2% in renal clear cell carcinoma to 95% in high-grade serous ovarian carcinoma (Kandoth et al., 2013). Unlike most other tumor suppressor genes which usually carry inactivating mutations in tumors, e.g. truncations and deletions, most TP53 mutations (~75%) are missense mutations that result in single amino acid substitutions and expression of a full-length but functionally deficient protein (Petitjean et al., 2007). The majority of the missense mutations are localized in the DNA binding domain, resulting in loss of DNA binding and transactivation of downstream target genes. Mutant p53 may also acquire novel so called gain-of-function activities (GOFs), such as binding to other cellular proteins and promiscuous transcriptional transactivation (Brosh and Rotter, 2009; Sabapathy and Lane, 2018). Thus, TP53 mutation may result in not only loss-of-function of p53 tumor suppressor activity and inactivation of co-expressed wild-type p53 due to a dominant-negative effect during tetramerization, but also a wide spectrum of tumor-promoting GOF effects.
In this review, we shall discuss how p53 is regulated by the cellular redox milieu and in turn regulates various antioxidant and pro-oxidant cellular pathways, and how mutant p53 can affect redox homeostasis. Moreover, we will review ongoing efforts to develop novel anticancer drugs that restore normal function to missense mutant p53 by cysteine binding. These compounds can also target the cellular antioxidant system, which may contribute to their anticancer effect.
NADPH-dependent antioxidant and redox signaling systems
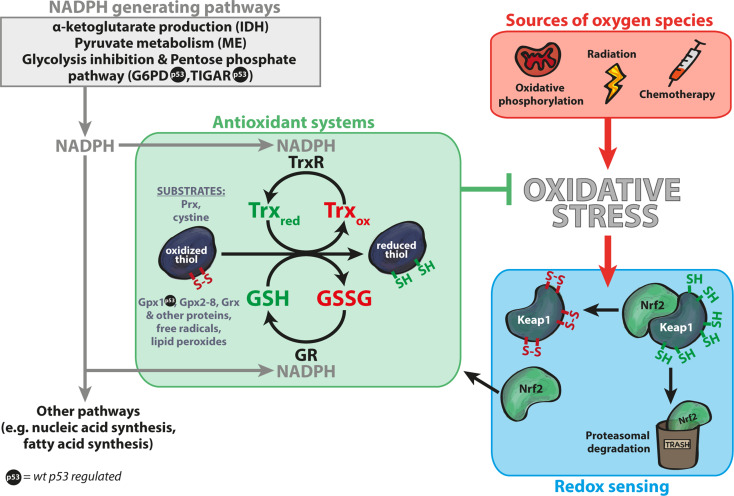
Reative oxygen species are produced in cells mainly by mitochondria and cytochrome P450 enzymes, but can also be induced by for example xenobiotics and radiation (Holmstrom and Finkel, 2014; He et al., 2017). Excessive amounts of oxygen species disrupt redox homeostasis and can lead to e.g. lipid peroxidation, DNA damage and cell death (He et al., 2017; Maiorino et al., 2018). Figure 1 shows an overview of redox regulation and sources of oxygen species, including cellular pathways that produce the major reducing agent nicotinamide adenine dinucleotide phosphate (NADPH). The cell employs several antioxidant systems to balance oxidative conditions. While catalase and superoxide dismutases (SOD) neutralize oxidant species (O2−, H2O2) without consuming significant amounts of NADPH, the two major antioxidant systems thioredoxin (Trx) and glutathione (GSH) utilize NADPH for their dithiol-disulfide exchange reactions (Figure 1) (He et al., 2017; Miller et al., 2018). Major sources of cellular NADPH are enzymes such as malic enzymes (ME), isocitrate dehydrogenases (IDH), and the pentose phosphate pathway (PPP) where glucose-6-phosphate dehydrogenase (G6PD) catalyzes the rate-limiting step.
Figure 1.
Overview of redox homeostasis. The two main antioxidant systems in the cell are the Trx and GSH systems (green box). p53 regulates several components of the antioxidant systems, marked by  . Oxidative phosphorylation in mitochondria is a major source of oxygen species, but such molecules can also be induced by radiation and chemotherapy (red box). Cells can adapt to oxidative stress by activation of the master antioxidant transcription factor Nrf2 that upregulates expression of genes with AREs, such as genes in the GSH and Trx systems. Nrf2 levels are low under unstressed conditions due to proteasomal degradation induced by the KEAP1 protein. Oxidation of KEAP1 cysteines upon oxidative stress inhibits KEAP1 ubiquitin ligase activity, leading to Nrf2 accumulation (blue box) and activation of ARE genes. The Trx system consists of TrxR and Trx and reduces substrates such as Prx and cystine. GSH is used as a cofactor by glutathione GST, Grx, and GPx. GSSG is restored by GR. Both antioxidant systems are dependent on NADPH as a supplier of electrons, since TrxR and GR utilize NADPH as a cofactor. Major NADPH-generating pathways include IDH through α-ketoglutarate production, ME through pyruvate metabolism, and the PPP (gray box). NADPH availability is regulated through the p53 target TIGAR that promotes NADPH production via PPP and through p53-mediated inhibition of G6PD that controls the first and rate-limiting step of PPP.
. Oxidative phosphorylation in mitochondria is a major source of oxygen species, but such molecules can also be induced by radiation and chemotherapy (red box). Cells can adapt to oxidative stress by activation of the master antioxidant transcription factor Nrf2 that upregulates expression of genes with AREs, such as genes in the GSH and Trx systems. Nrf2 levels are low under unstressed conditions due to proteasomal degradation induced by the KEAP1 protein. Oxidation of KEAP1 cysteines upon oxidative stress inhibits KEAP1 ubiquitin ligase activity, leading to Nrf2 accumulation (blue box) and activation of ARE genes. The Trx system consists of TrxR and Trx and reduces substrates such as Prx and cystine. GSH is used as a cofactor by glutathione GST, Grx, and GPx. GSSG is restored by GR. Both antioxidant systems are dependent on NADPH as a supplier of electrons, since TrxR and GR utilize NADPH as a cofactor. Major NADPH-generating pathways include IDH through α-ketoglutarate production, ME through pyruvate metabolism, and the PPP (gray box). NADPH availability is regulated through the p53 target TIGAR that promotes NADPH production via PPP and through p53-mediated inhibition of G6PD that controls the first and rate-limiting step of PPP.
The Trx and GSH pathways and the PPP are upregulated by Nuclear factor erythroid 2-related factor 2 (Nrf2) (Cebula et al., 2015), a transcription factor that controls numerous genes containing antioxidant response elements (AREs). Under non-stressed (reducing) cellular conditions, Kelch-like ECH-associated protein 1 (Keap1) negatively regulates Nrf2 by acting as an adapter for a CUL3 E3 ligase that targets Nrf2 for proteasomal degradation (Rojo de la Vega et al., 2018). However, Keap1 contains multiple redox-sensitive cysteine residues that can be targeted by oxidants, which affects its conformation and disrupts its ability to inhibit Nrf2 (Figure 1).
The Trx system consists of Thioredoxin reductase (TrxR), which predominantly reduces Trx or Thioredoxin-related protein of 14 kDa (TRP14) (Pader et al., 2014; Cebula et al., 2015). Trx and TRP14 in turn reduce a wide range of proteins, e.g. ribonucleotide reductase, methionine sulfoxide reductase and peroxiredoxins (Prxs), and also low-molecular-weight substrates such as cystine. Prxs neutralize H2O2 and the reduction of cystine increases intracellular cysteine which indirectly supports GSH synthesis. The tripeptide glutathione or L-γ-glutamyl-L-cysteinyl-glycine (GSH) reacts with electrophilic and oxidizing species and is utilized as a cofactor and electron donor by glutathione S-transferases (GST), glutaredoxins (Grx1–5) and glutathione peroxidases (GPx1–8) (Brigelius-Flohe and Maiorino, 2013). Glutathione reductase (GR) reduces oxidized GSH (GSSG) to GSH (Figure 1).
Redox regulation of wild-type p53
The p53 protein has 10 cysteine (Cys) residues located in the DNA-binding core domain (residues 100–300). Three cysteines, Cys176, Cys238, and Cys242, along with His179, hold a zinc atom that bridges the L2 and L3 loops and is crucial for proper folding of p53 (Cho et al., 1994; Rainwater et al., 1995; Meplan et al., 2000). Early studies demonstrated that p53 binding to DNA in vitro requires a strong reducing environment (Hainaut and Milner, 1993). Subsequent work in many laboratories has confirmed that p53 is subject to redox regulation (Bykov et al., 2009). The Apurinic/apyrimidinic endonuclease 1/reduction-oxidation factor 1 (APE1/Ref-1) was shown to stimulate p53 DNA binding in vitro and enhance p53-dependent transcription in living cells, presumably by both redox-dependent and independent mechanisms (Jayaraman et al., 1997). Moreover, p53 activity is dependent on the TrxR1–Trx system, one of the two main antioxidant systems in cells. Deletion of the TrxR1 gene in budding yeast or fission yeast inhibited p53-dependent cell growth suppression (Casso and Beach, 1996; Pearson and Merrill, 1998). Trx enhances p53 DNA binding and transactivation (Figure 2), both directly and indirectly via Ref-1 (Ueno et al., 1999). Trx and/or Ref-1 also augmented p53-mediated induction of the p53 target p21, while overexpression of a mutant Trx, lacking reducing activity, inhibited p53-dependent induction of p21 upon cisplatin treatment. Knockdown of TrxR1 in human breast cancer cells caused accumulation of oxidized Trx, which was associated with increased p53 levels and DNA binding (Seemann and Hainaut, 2005).
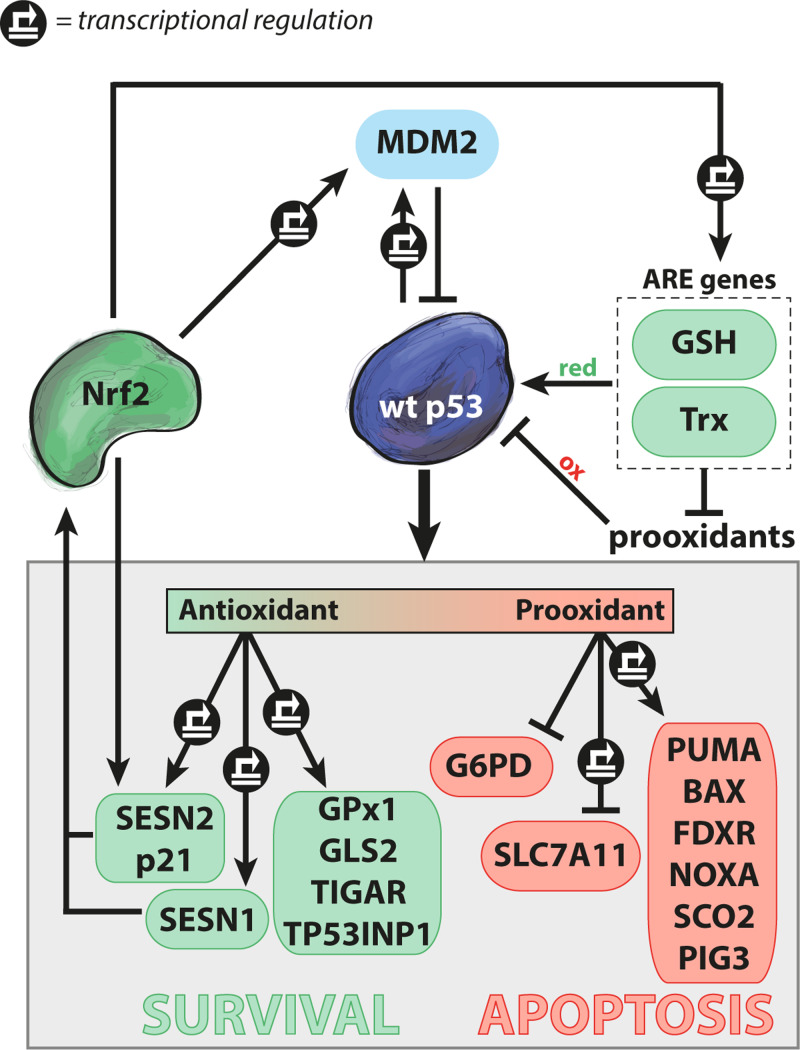
Figure 2.
Regulation of redox homeostasis by wild-type p53. Normal wild-type (wt) p53 function is dependent on a reducing environment, while oxidative conditions can inhibit p53. p53 antioxidant activity induces targets such as GPx1, GLS2, TIGAR, and TP53INP. In addition, p53 induces p21 and SESN2 that stabilize the master antioxidant regulator Nrf2, which leads to transactivation of ARE genes and a reducing milieu via GSH and Trx. p53 also induces pro-oxidant targets such as Puma, Bax, Noxa, PIG3, FDXR, and SCO2, and thereby promotes cell death by apoptosis. Furthermore, p53 can transrepress the SLC7A11 gene and directly inhibit G6PD via protein binding. Nrf2 induces MDM2 that targets p53 for degradation in the proteasome, thus antagonizing p53 activity. Green and red indicate antioxidant and pro-oxidant activities, respectively. Transcriptional regulation is marked by  .
.
In addition, the crosstalk between the Trx system and p53 involves Thioredoxin interacting protein (TXNIP), a negative regulator of Trx that also interacts with p53 (Suh et al., 2013; Yoshihara et al., 2014). TXNIP dissociates from Trx upon oxidative stress, while its binding to p53 is enhanced (Jung et al., 2013). TXNIP has also been shown to stabilize p53 by interacting with human ecdyoneless (hEcd), a protein that inhibits mouse double minute (MDM2)-dependent degradation of p53 (Suh et al., 2013).
Several studies have shown that S-glutathionylation of p53 cysteines can affect p53 function. The addition of GSH to protein thiol groups protects against irreversible oxidative modifications. S-glutathionylation of proteins can be reversible. Yeast Trx has been shown to exhibit deglutathionylase activity (Greetham et al., 2010). Mass spectrometry demonstrated that cysteines 124, 141, and 182 in human p53 can be glutathionylated, and that S-glutathionylation inhibits p53 DNA binding (Velu et al., 2007). S-glutathionylation of Cys141 increased markedly upon treatment with oxidative agents or chemotherapeutic drugs (Yusuf et al., 2010). Selective glutathionylation of monomeric and dimeric p53 has been demonstrated in the brain of Alzheimer’s disease patients (Di Domenico et al., 2009).
Thus, p53 DNA binding and function as transcription factor is clearly dependent on reducing conditions. Oxidation inhibits p53 (Figure 2). However, the exact role of oxidation-reduction of specific p53 cysteines needs further investigation. Also, as will be discussed below, p53 activates expression of several genes whose products regulate the redox balance in cells, suggesting that p53 may influence its own redox status.
Wild-type p53 regulates cellular redox homeostasis
As shown in Figure 2, p53 stimulates expression of both pro-survival genes with antioxidant properties and genes with pro-apoptotic and pro-oxidant properties. A study in three cancer cell lines of different origin showed that p53 activated by the MDM2 inhibitor Nutlin induces transcription of around one hundred target genes (Andrysik et al., 2017). p53 activates genes involved in multiple cellular functions, including classical targets such as MDM2, the cell cycle inhibitor p21 (CDKN1A) and the pro-apoptotic Bax and Puma genes, and genes involved in redox homeostasis. Interestingly, promoters of p53-regulated genes with antioxidant functions appear to be sensitive to low levels of p53, whereas pro-oxidant and pro-apoptotic p53 target genes are activated in response to higher p53 levels upon more extensive stress, and with a delay as compared to the pro-survival genes (Polyak et al., 1997; Sablina et al., 2005; Wu et al., 2017).
The p53 core transcriptional program (Andrysik et al., 2017) includes at least five early response target genes known to directly or indirectly regulate metabolism and the cellular antioxidant milieu: TIGAR (Lee et al., 2014), Sestrins 1 and 2 (SESN1/2) (Bae et al., 2013), tumor protein p53-induced nuclear protein 1 (TP53INP1) (Cano et al., 2009), and p21 (Chen et al., 2009). p53-induced TIGAR inhibits glycolysis and enhances PPP flux, resulting in more intracellular NADPH reductive power (Lee et al., 2014). Increased TIGAR expression has been shown to preserve mitochondrial function and decrease overall levels of intracellular oxidation (Li et al., 2014). Transcriptional transactivation of SESN1/2 and p21 causes stabilization and activation of Nrf2 (Chen et al., 2009; Bae et al., 2013). SESN1/2 influence cellular redox status through the interaction with the Nrf2 antagonist Keap1, promoting its degradation. In addition, the p53 target p21 competes with Keap1 for Nrf2 binding. Stabilized Nrf2 translocates to the nucleus and controls basal and inducible expression of more than 200 genes through binding to AREs (Rojo de la Vega et al., 2018). Accordingly, Nrf2 may at least in part mediate p53’s antioxidant pro-survival activity. In response to cellular stress, nuclear TP53INP1 facilitates p53 transcriptional activity by direct physical interaction with p53 and different kinases (Saadi et al., 2015). TP53INP1 also maintains mitochondrial integrity, and TP53INP loss was shown to cause pro-oxidant conditions in cells from TP53INP knockout mice (Cano et al., 2009; Saadi et al., 2015).
GPx1 and mitochondrial glutaminase (GLS2) are additional transcriptional targets of p53 that contribute to p53’s antioxidant activity (Fischer, 2017). GPx1, ubiquitously expressed in the cytosol and mitochondria, catalyzes the reduction of hydrogen peroxide using GSH as cofactor (Brigelius-Flohe and Maiorino, 2013). GPx1 protects against oxidative DNA damage and inhibits insulin signaling. p53-dependent transactivation of GLS2 promotes mitochondrial respiration by catalyzing the hydrolysis of glutamine to glutamate, subsequently increasing the levels of the citric acid cycle (TCA) substrate α-ketoglutarate (Hu et al., 2010). Fumarate is downstream of α-ketoglutarate in the TCA cycle and can inhibit Keap1, thereby resulting in elevated free Nrf2 (Linehan and Rouault, 2013). Furthermore, glutamate is also a precursor of GSH and is exported in exchange for imported cystine through solute carrier family 7 member 11 (SLC7A11).
As discussed above, three p53 targets, SESN1/2 (Bae et al., 2013) and p21 (Chen et al., 2009), have been reported to interfere with Keap1–Nrf2 complex, thus enhancing Nrf2 stability and activity. Interestingly, Nrf2 also induces transcription of SESN2 (Shin et al., 2012), p21 (Jana et al., 2018), and MDM2 (Todoric et al., 2017) (Figure 2). The induction of SESN2 and p21 suggests potential feedback regulatory mechanisms between SESN2/p21 and Nrf2. Nrf2-dependent regulation of the p53 antagonist MDM2 (You et al., 2011) and suppression of TXNIP (He and Ma, 2012) can sustain low p53 protein levels and transcriptional activity. While low levels of p53 can induce Nrf2, conditions that lead to high p53 protein levels seem to compromise Nrf2 activity (Faraonio et al., 2006; Tung et al., 2015). The suppression of Nrf2 might, at least in part, be due to p53-mediated inhibition of Sp1-dependent Nrf2 transcription (Tung et al., 2015). However, the exact mechanism requires further investigation.
Nrf2 induces an array of genes controlling cellular thiol-disulfide status, for instance SLC7A11, GSTs, Sulfiredoxin (Srx), TrxR1, GPx4, and several genes involved in NADPH and glutathione synthesis (Raghunath et al., 2018; Rojo de la Vega et al., 2018). The Nrf2 pathway is frequently mutated in cancer, either the Nrf2 gene itself or its regulator Keap1 (Kandoth et al., 2013; Jeong et al., 2017), resulting in stabilized Nrf2, increased expression of Nrf2 target genes, and elevated capacity to cope with oxidative stress. Interestingly, and as will be discussed further below, mutant p53 has been shown to bind Nrf2 and modulate its transactivation capacity (Walerych et al., 2016; Liu et al., 2017a; Lisek et al., 2018).
The p53 transcriptional program (Andrysik et al., 2017) also includes mitochondrial function-associated genes that can promote oxidative stress. Stress-induced apoptosis and pro-oxidant functions of p53 are associated with mitochondrial leakage of oxidant species (Holmstrom and Finkel, 2014). The p53 target and Bcl-2 family protein BAX induces mitochondrial outer membrane permeabilization and cytochrome C release, which leads to caspase activation and apoptotic cell death. The p53 target PUMA activates BAX and the pro-apoptitc Bcl-2 family protein BAK (Adams and Cory, 2018). Another p53 target, NOXA (PMAIP1), modulates mitochondrial function by inhibiting pro-survival Bcl-2 family proteins Mcl-1 and Bfl-1 (Shibue et al., 2003; Fischer, 2017) (Figure 2).
Other p53 targets with pro-oxidant functions include tumor protein p53 inducible protein 3 (TP53I3 or PIG3), cytochrome C oxidase assembly protein 2 (SCO2) and ferredoxin reductase (FDXR) (Figure 2). PIG3 is a NADPH-quinone oxidoreductase, and a potent generator of harmful oxidant species, functioning as a DNA damage response sensor (Flatt et al., 2000; Sablina et al., 2005). p53-dependent PIG3 induction is delayed as compared to induction of p21 and MDM2 (Szak et al., 2001). SCO2 induces production of reactive oxygen species and activates apoptosis via the ASK-1 kinase pathway (Madan et al., 2013). FDXR is a mitochondrial protein involved in iron–sulfur cluster formation and the transfer of electrons from NADPH to cytochrome P450 enzymes. Overexpression of FDXR increases the sensitivity to hydrogen peroxide-induced cell death (Zhang et al., 2017), while FDXR knockdown causes disturbed iron homeostasis (Shi et al., 2012). In addition to its ability to activate transcription of pro-oxidant genes, p53 can prevent G6PD dimer formation through direct protein binding in the cytoplasm, and thus inhibit NADPH production (Jiang et al., 2011) (Figure 2).
Ferroptosis, a form of cell death characterized by iron and lipid hydroperoxide accumulation, has been proposed to have an important role in p53-mediated tumor suppression (Jiang et al., 2015). Ferroptotic cell death is regulated by the transcription factors p53 and Nrf2 through multiple mechanisms (Stockwell et al., 2017; Maiorino et al., 2018; Tarangelo et al., 2018). Sensitivity to ferroptosis has been associated with iron homeostasis, polyunsaturated fatty acid metabolism, and cellular availability of cysteine, GSH, and NADPH (Stockwell et al., 2017). Nrf2-dependent transactivation of antioxidant genes mediates reduction of lipid peroxides and prevents ferroptosis (Maiorino et al., 2018). p53, on the other hand, has been shown to modulate ferroptosis both positively and negatively (Jiang et al., 2015; Tarangelo et al., 2018). p53-dependent regulation of dipeptidyl-peptidase-4 (DPP4) (Xie et al., 2017) and p21 (Chen et al., 2009; Tarangelo et al., 2018) has been shown to delay the onset of ferroptosis, whereas transactivation of spermidine/spermine N1-acetyltransferase 1 (SAT1) (Ou et al., 2016) and GLS2 (Gao et al., 2015), and negative regulation of SLC7A11 (Jiang et al., 2015; Wang et al., 2016b), stimulates ferroptosis. SLC7A11 dictates extracellular and intracellular cystine/cysteine redox states by importing cystine with a 1:1 counter-transport of glutamate (Banjac et al., 2008). Intracellular cysteine availability is a limiting factor in GSH synthesis.
GSH, present in cells at millimolar concentrations, has a key role in thiol redox chemistry. p53 can stimulate the production of GSH through several target genes and pathways, e.g. TIGAR (Lee et al., 2014), GLS2 (Hu et al., 2010; Suzuki et al., 2010), SESN1/2 (Bae et al., 2013), and p21-dependent activation of Nrf2. p53 downregulation increases DNA oxidation and oxygen species formation (Sablina et al., 2005). p53 null mice die at 4–6 months of age, due to the development of lymphomas and other tumors. Interestingly, diet supplemented with N-acetylcysteine, which supplies extra cysteine for GSH synthesis, lowered the tumor incidence and substantially increased the survival of p53 null mice (Sablina et al., 2005). Cysteine can be supplied through different pathways depending on tissue type. In certain cells cysteine is synthesized from homocysteine and serine by the transsulfuration pathway (Hayano et al., 2016). Interestingly, stimulating the transsulfuration activity delayed the onset of ferroptosis. An integrative multi-omics analysis, comparing metabolomics and transcriptomic data from p53 wild-type and p53-depleted cells, identified changes in sulfur and nucleotide metabolism (Huang et al., 2018). Cancer cells depleted of p53 exhibited lower levels of GSH, taurine and S-adenocylmethionine, and higher levels of methionine. Hence, p53 status seems to influence the methionine cycle and the transsulfuration pathways. Moreover, p53-depleted cancer cells failed to proliferate in a serine-deficient environment (Maddocks et al., 2013), which was explained by insufficient intracellular GSH levels.
Mutant GOF and redox regulation
The fact that most TP53 mutations in tumors are missense mutations rather than truncating mutations or deletions argues convincingly that expression of mutant p53 provides a selective advantage during tumor development. Both dominant-negative effects on wild-type p53 and various GOF activities have been associated with mutant p53 (Brosh and Rotter, 2009; Sabapathy and Lane, 2018). The GOF activities include interactions with other transcription factors, e.g. p63, and transactivation of illegitimate target genes, e.g. c-Myc, leading to activation of survival pathways as well as metabolic shifts. Some GOF activities may result in elevated oxidative stress, a characteristic trait of cancer cells (Gorrini et al., 2013). In this context, it is particularly interesting to note that mutant p53 interacts with and entraps the master antioxidant regulator Nrf2 (Walerych et al., 2016; Liu et al., 2017a; Lisek et al., 2018). However, this interaction is complex and both positive and negative regulation of Nrf2 by mutant p53 has been demonstrated, as illustrated in Figure 3. Non-small cell lung cancers (NSCLC) carrying mutant p53 but not Nrf2 or Keap1 mutations exhibited higher levels of Nrf2 mRNA than wild-type p53 tumors (Tung et al., 2015). These patients also had a worse response to cisplatin treatment as compared to patients with wild-type p53 tumors. Similarly, oncogenes such as Kras, Braf, and Myc can promote increased transcription of Nrf2 and its antioxidant downstream targets, which might lead to a more reduced cellular milieu (DeNicola et al., 2011). Furthermore, one study showed that Nrf2 expression in AML is driven by NFκB signaling and that Nrf2 expression is downregulated by NFκB inhibitors (Rushworth et al., 2012). Thus, as mutant p53 prolongs TNF-α-induced NFκB signaling (Cooks et al., 2013), it is conceivable that mutant p53 can upregulate Nrf2 via NFκB.
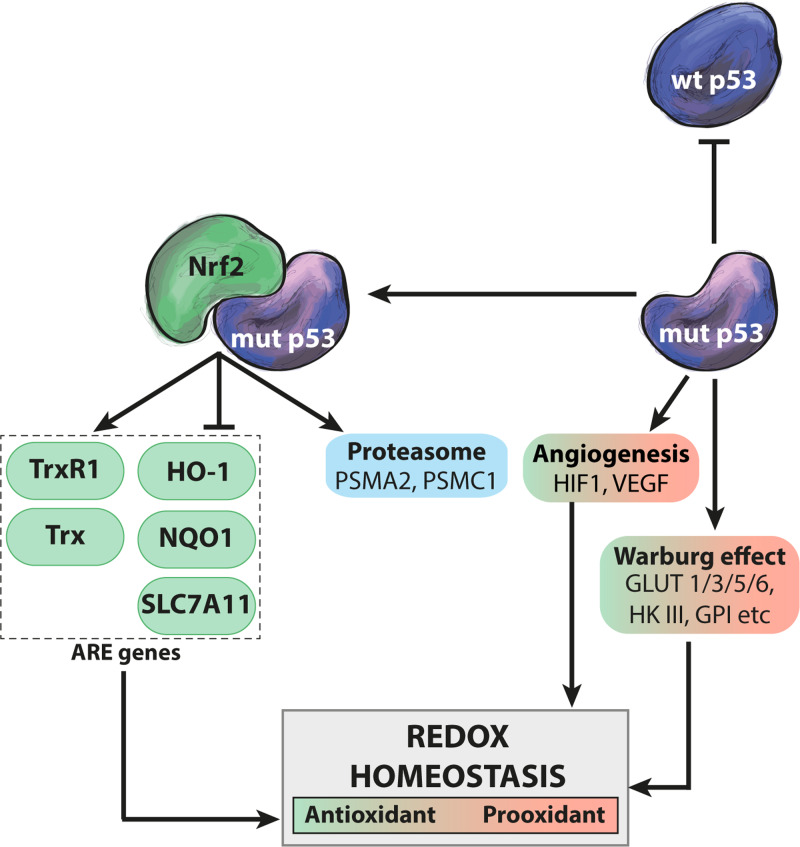
Figure 3.
Multiple effects of mutant p53 on redox homeostasis. Mutant p53 interacts with Nrf2 and disrupts the ARE transcription program governed by Nrf2, leading to activation of some genes (cytosolic TrxR1 and Trx) and repression of others (HO-1, NQO1, and SLC7A11). The mutant p53–Nrf2 interaction also causes induction of genes such as the PSMA2 and PSMC1 proteasome subunit genes. Mutant p53 promotes shifts in glycolytic metabolism (the Warburg effect) via upregulation of GLUT 1/3/5/6, HKIII, and GPI and stimulates angiogenesis via HIF1 and VEGF, affecting redox homeostasis. Mutant p53 may also exert a dominant-negative effect on co-expressed wild-type p53, thus inhibiting its regulation of the redox balance. Green and red indicate antioxidant and pro-oxidant activities, respectively.
Counteracting oxidative stress in order to maintain a reduced environment is essential as accumulation of oxygen species is harmful to the cell and can initiate cancer (Valko et al., 2006). The first responders to oxidative stress are usually called in by Nrf2 via ARE genes. Inactivation of Nrf2 is associated with decreased expression of phase 2 detoxifying enzymes such as NAD(P)H:quinone oxireductase (NQO1) which neutralize reactive electrophiles (Ramos-Gomez et al., 2001) (Figure 3). Studies have shown that NQO1 levels are elevated in cancer compared to healthy tissues (Belinsky and Jaiswal, 1993) and that NQO1 stabilizes wild-type p53, especially under oxidative stress (Asher et al., 2002). Hot spot p53 mutants show increased binding to NQO1 as compared to wild-type p53 (Asher et al., 2003). On the other hand, Kalo et al. (2012) found that while Nrf2 is induced in R273H mutant p53-carrying tumor cells upon oxidative stress, the Nrf2 antioxidant response is impaired, as shown by decreased expression of NQO1 and heme oxygenase 1 (HO-1) (Figure 3). Despite this decreased antioxidant capacity and elevated levels of oxidative stress, cells with mutant p53 are in general more resistant to chemotherapy (Sabapathy and Lane, 2018).
Del Sal and colleagues (Walerych et al., 2016) demonstrated that the interaction of mutant p53 with Nrf2 affects expression of proteasome genes. Mutant p53 induces a proteasome signature associated with poor prognosis in breast cancer, and the recruitment of mutant p53 to the promoters of the PSMA2 and PSMC1 proteasome subunit genes is dependent on the interaction with Nrf2 (Figure 3). Immunoprecipitation of wild-type p53 failed to detect an interaction with NRF2 and wild-type p53 does not affect expression of proteosome subunits, suggesting that this interaction is a mutant p53 GOF activity (Walerych et al., 2016). Moreover, mutant p53 has been shown to entrap Nrf2 on the promoter of the Nrf2 downstream target SLC7A11, resulting in decreased SLC7A11 expression (Liu et al., 2017a) (Figure 3). Cells overexpressing SLC7A11 are dependent on glucose as an energy source, due to extensive export of glutamate (Koppula et al., 2018), but are insensitive to the GSH de novo synthesis inhibitor L-buthionine-sulfoximine, as result of their increased levels of cystine/cysteine. As SLC7A11 controls both cystine/cysteine redox cycling and the availability GSH building blocks, the repression of SLC7A11 renders mutant p53 cells more sensitive to oxidative assaults. This creates an Achilles’s heel that could be exploited therapeutically (see further below).
The role of Nrf2 in the context of cancer is complex as it clearly has both tumor-suppressive and tumor-promoting effects (Rojo de la Vega et al., 2018). Mutant p53 represses the Nrf2 antioxidant response (Kalo et al., 2012; Liu et al., 2017a) (Figure 3), but it would seem more beneficial from the point of view of the tumor cell to activate Nrf2 in order to protect against oxidative stress. Antioxidants have been shown to promote tumor progression (Sayin et al., 2014). Also, the effects of the interaction between mutant p53 and Nrf2 are probably dependent on the cellular context and the specific p53 mutation. Mutant p53 increases Nrf2 localization to the nucleus and directs it to specific AREs where it induces transcription of antioxidants such as Trx and TrxR1 (cytosolic), while other antioxidant targets such as HO-1 are repressed (Lisek et al., 2018) (Figure 3).
In addition, mutant p53 upregulates FOXM1 (Tanaka et al., 2018), which plays an important role in counteracting oxidative stress by inducing SOD2, catalase, and PRDX3 (Park et al., 2009). On the other hand, decreased SOD2 expression was associated with higher mortality in hepatocellular carcinoma patients with mutant p53 (Wang et al., 2016a), further emphasizing the complex and context-dependent impact of mutant p53 on redox homeostasis.
The p53 inhibitor apoptosis stimulating protein of p53 (iASPP) positively regulates Nrf2 by promoting its accumulation and nuclear translocation (Ge et al., 2017). iASPP is suppressed by miR-124 (Liu et al., 2013), which is upregulated by wild-type p53. Mutant p53 fails to induce miR-124, resulting in upregulation of iASPP in cells lacking wild-type p53 (Liu et al., 2017b) and thus induction of Nrf2. However, miR-124 also stimulates NFκB signaling (Liu et al., 2013), and hence wild-type p53 could potentially induce Nrf2 through this pathway (Rushworth et al., 2012).
The cancer cell environment is characterized by altered metabolism. In general, cancer cells rely on aerobic glycolysis rather than oxidative phosphorylation, a phenomenon referred to as the Warburg effect (Vander Heiden et al., 2009). Mutant p53 stimulates the Warburg effect by inducing glucose transporters GLUT 5/6 and GLUT 3 through NFκB signaling, and GLUT 1 by promoting its translocation to the plasma membrane. Several glycolytic enzymes, i.e. HK III, GPI, GAPDH, PGK1, enolase 1, and PDK1, are also induced by mutant p53. This results in a higher rate of glucose uptake and glycolysis (Gomes et al., 2018). Such shifts in the metabolic environment affect the redox homeostasis, for example as the TCA cycle generates fumarate that rescues Nrf2 from Keap1 (Linehan and Rouault, 2013), but also the availability of NADPH (Rojo de la Vega et al., 2018). Moreover, mutant p53 can stimulate lipid metabolism by binding and activating the transcription factor SREBP and thereby inducing the mevalonate pathway (Freed-Pastor et al., 2012). SREBP induces the expression of the antioxidant gene HO-1 (Kallin et al., 2007).
Another important aspect of the tumor microenvironment is the insufficient blood supply resulting in hypoxic regions. The ability to stimulate vascularization or angiogenesis is therefore critical for tumor growth. Hot spot p53 mutants were shown to enhance angiogenesis by oxidative stress-induced HIF1/VEGF signaling, while wild-type p53 rather blocked angiogenesis (Khromova et al., 2009). Many of the HIF-1α targets overlap with Nrf2 transcriptional targets and VEGF can induce Nrf2, supporting the notion that angiogenesis can affect redox homeostasis (Rojo de la Vega et al., 2018). Furthermore, cancer cells rely on glutamine availability to sustain proliferation and to produce glutathione to counteract oxidative stress. Cells with mutant p53 are more resistant to glutamine deprivation compared to wild-type p53-carrying cells (Tran et al., 2017).
Pharmacological targeting of mutant p53 and redox homeostasis
Therapeutic targeting of p53 in cancer is a growing field with a potentially great impact on cancer therapy in the future. For tumors that carry missense mutant p53, the main strategy is to restore normal conformation and function to inactive mutant p53, which is often expressed at high levels in tumor cells. A number of mutant p53-reactivating compounds have been identified and characterized, and at least one, APR-246, is now being tested in the clinic (Bykov et al., 2018).
A major challenge in targeting mutant p53 for cancer therapy is the heterogeneity of the target (Sabapathy and Lane, 2018). There are two main types of missense p53 mutants, so called DNA contact mutants such as His273 that to a large extent retain wild-type conformation but in which amino acid residues that make direct contact with DNA are substituted, and so called structural mutants, such as His175, in which amino acid substitutions in p53’s core domain cause global unfolding and loss of specific DNA binding. Restoration of DNA binding by creating new DNA contacts is a plausible approach for DNA contact mutants, whereas thermodynamic stabilization should restore structural p53 mutants, as shown for the temperature-sensitive Ala143 mutant (Zhang et al., 1994). From a theoretical point of view, preferential binding of a small molecule to the folded rather than the unfolded p53 core domain should shift the equilibrium between unfolded and folded states towards the folded state according to the law of mass action (Bullock and Fersht, 2001).
Screening chemical libraries with protein assays based on recombinant p53 refolding or cellular assays with differential growth suppression in cells lacking or expressing missense mutant p53 as a readout have identified a number of compounds with ability to target mutant p53, including CP31398, PRIMA-1, APR-246, 3-benzoylacrylic acid (3BA), and PK11007. Other mutant p53-targeting compounds, e.g. PK083, ZMC1, PK7088, stictic acid, and KSS-9, have been identified by rational design, database analysis and/or molecular modeling (Bykov et al., 2018). Here we shall focus on compounds that target cysteines in mutant p53 (Figure 4). CP-31398 (Foster et al., 1999), 3BA (Kaar et al., 2010), and methylene quinuclidinone (MQ), the active product generated by non-enzymatic conversion of PRIMA-1 and APR-246 (Bykov et al., 2002; Lambert et al., 2009), have thiol-binding properties (Bykov et al., 2018) due to their ability to participate in the reaction of nucleophilic addition, or more exactly, the addition by Michael (Michael, 1887). Aromatic nucleophilic substitution is another type of thiol alkylation, as demonstrated for the sulfonylpyrimidines PK11000 and PK11007 (Bauer et al., 2016). The prime targets for these electrophilic compounds are nucleophiles, i.e. molecules or residues with ability to donate an unshared electron pair. Deprotonated cysteine or selenocysteine groups are the strongest nucleophiles in cells (Pace and Weerapana, 2013). These residues are abundant among cellular proteins and particularly enzymes that often have a thiol group in their catalytic center. Thus, compounds of this type are likely to have multiple cellular targets, and so treatment should have substantial effects at the cellular and organismal level beyond the p53 signaling network. For example, APR-246 inhibits TrxR1 (Peng et al., 2013), thioredoxin, glutaredoxin and ribonucleotide reductase (Haffo et al., 2018) and depletes cellular GSH (Tessoulin et al., 2014; Mohell et al., 2015; Bykov et al., 2016).
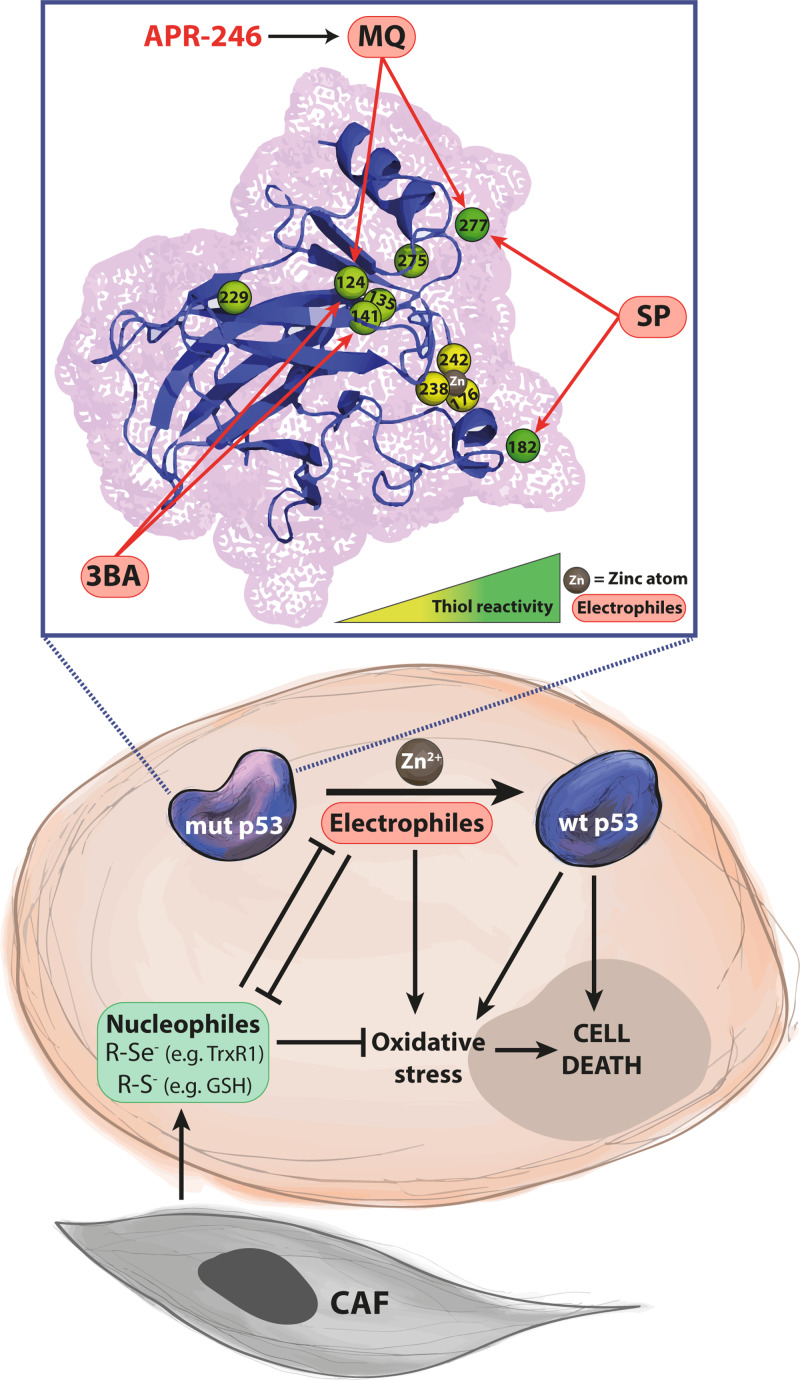
Figure 4.
Mutant p53 rescue and induction of oxidative stress and cell death by thiol-binding small molecules. The DNA binding core domain of p53 has 10 cysteine residues with variable degrees of reactivity (illustrated by the yellow–green gradient). Cys182 and Cys277, located on the surface of p53, are highly reactive, whereas Cys176, Cys238, and Cys242 are protected from oxidation by coordinating a zinc atom (marked in brown). Mutations in p53 can cause protein unfolding and exposure of cysteine residues. Several mutant p53-reactivating compounds are electrophiles that bind to cysteines in mutant p53, leading to thermostabilization and induction of p53 target genes, and ultimately tumor cell death. The compound 3-benzoylacrylic acid (3BA) has been shown to bind preferentially to Cys124 and Cys141. Sulfonylpyrimidines (SP) such as PK11007 attack Cys182 and Cys277. Methylene quinuclidinone (MQ), the conversion product of APR-246 and PRIMA-1, targets Cys277 and Cys124. This type of compounds can also react with other nucleophiles in the cell, mainly protein cysteines (R-S–) and selenocysteines (R-Se–), and GSH. MQ has been shown to deplete GSH and inhibit the selenoprotein TrxR1 as well as Trx and Grx, leading to oxidative stress that presumably contributes to tumor cell death. Zinc chelation by the compound ZMC1 reactivates certain forms of mutant p53 and also has redox effects. Stromal cells in the tumor microenvironment, e.g. cancer-associated fibroblasts (CAFs), have been shown to produce nucleophiles that may contribute to cancer cell resistance to alkylating chemotherapeutic agents. Upper panel shows a combined cartoon/mesh of the wild-type p53 core structure from 1tup.pdb crystal structure file (Cho et al., 1994) uploaded from the NCBI library (https://www.ncbi.nlm.nih.gov) and modified by open-source PyMOL molecular graphics software (DeLano Scientific).
The 10 cysteines in p53’s core domain are not equally reactive (indicated by the yellow to green gradient in Figure 4). Steric factors, accessibility to solvent and local environment will affect thiol reactivity (Kaar et al., 2010). Cys277 and Cys182 are located on the surface of wild-type p53 and are therefore open for electrophilic attack, whereas for instance Cys176 is buried in the hydrophobic core and not exposed on folded p53. Cys135, Cys141, and Cys275 also have poor solvent accessibility (Scotcher et al., 2011). As already mentioned, Cys176, Cys238, and Cys242 have a critical role for normal p53 protein folding. Coordination of a zinc atom prevents their oxidation (Cho et al., 1994; Rainwater et al., 1995; Meplan et al., 2000). Flexibility of the wild-type p53 structure allows modification at Cys124, Cys135, and Cys141, located in the L1/S3 pocket (Wassman et al., 2013). Mutations that result in local or global structural distortion can expose additional cysteine residues that are normally buried in the wild-type fold.
Molecular modeling suggested that Cys124 is a potential target for MQ, the conversion product of APR-246, as well as p53-targeting Michael acceptors MIRA-1 and STIMA-1 (Wassman et al., 2013). The Michael acceptor 3BA targets Cys124 and Cys141 in several p53 mutant proteins, while sulfonylpyrimidines such as PK11007 bind preferentially to Cys182 and Cys277 (Bauer et al., 2016). Cys277 is also a prime binding site for MQ and required for MQ-mediated thermostabilization of recombinant His175 and His273 mutant p53 (Zhang et al., 2018b) (Figure 4). However, both Cys277 and Cys124 are important for reactivation of His175 mutant p53 in living cells as assessed by induction of apoptosis and ability to upregulate p53 target genes.
Although Michael acceptor functionality has been shown to be important for mutant p53 reactivation, other chemical groups, such as aldehydes, imines and primary alcohols (upon oxidation) may also provide soft electrophile properties. Some Michael acceptors show high cellular toxicity. The optimal balance between toxicity and mutant p53 refolding capability appears to depend on the combination of functional groups where Michael acceptor functionality together with imine and primary alcohols will allow attenuated cytotoxicity and sufficient p53 refolding and reactivation. The presence of an aldehyde group is correlated with p53 thermostabilization (Zhang et al., 2018a). This suggests the possibility of designing novel compounds with high selectivity for mutant p53-expressing tumor cells based on judicious combination of the indicated chemical groups.
APR-246 was shown to inhibit the selenocysteine-containing enzyme TrxR1 via MQ (Peng et al., 2013). This reaction presumably disrupts the electron flow from oxidation of NADPH to reduction of Trx, thus rendering the enzyme a pure NADPH oxidase that promotes an oxidative environment. The ability of APR-246, via MQ, to bind and deplete GSH (Tessoulin et al., 2014; Mohell et al., 2015) and inhibit Grx and Trx (Haffo et al., 2018) will have the same effect on the redox balance, and is consistent with the identification of SLC7A11 as marker for decreased sensitivity to APR-246 (Liu et al., 2017a). However, it is difficult to assess the importance of the effect of APR-246/MQ on the redox system for the anti-tumor effect, since higher cysteine concentrations will also compete with p53 cysteine binding required for mutant p53 reactivation. Cys-binding sulfonylpyrimidines such as PK11007 have been shown to target the redox system in a similar manner (Bauer et al., 2016). These effects are presumably beneficial for cancer therapy. As compared to normal cells, tumor cells are characterized by a more oxidative intracellular milieu due to enhanced proliferation and metabolic rates in which reactive oxygen species are generated as byproducts. Thus, their redox balance is already pushed to the limit, and a further increase in oxidant species may therefore tip them over the edge and trigger cell death (Gorrini et al., 2013).
In this context, it is also interesting to note that the tumor microenvironment has been shown to produce cystine and other thiol-containing compounds that can boost GSH synthesis in tumor cells and thereby enhance drug resistance (Zhang et al., 2012; Wang et al., 2016c; Cheteh et al., 2017). This redox-dependent tumor-protective effect of the microenvironment is a possible therapeutic target in cancer.
Reactivation of mutant p53 should not only restore normal p53 function but could also synergize with other therapeutic approaches including chemotherapy and radiotherapy which to a considerable extent may induce tumor cell death via wild-type p53. In agreement with this idea, APR-246 has been shown to synergize with chemotherapeutic drugs such as adriamycin and cisplatin (Bykov et al., 2005; Mohell et al., 2015). GSH synthesis, including import of cystine by SLC7A11, and conjugation reactions are important mechanisms of drug resistance exploited by tumors (Lewerenz et al., 2013). Thus, depletion of GSH by APR-246/MQ may contribute to the observed synergy with chemotherapeutic drugs. Furthermore, APR-246 synergizes with the proteasome inhibitor Carfilzomib in triple-negative breast cancer cells by blocking mutant p53 binding to Nrf2, which leads to decreased transcription of proteasome genes and therefore increased sensitivity to Carfilzomib (Walerych et al., 2016). Of note, the Nrf2 binding region in mutant p53 has been mapped to amino acids residues 98–128 (Lisek et al., 2018), suggesting that MQ binding to Cys124 might disrupt this interaction.
As discussed above, p53 is a metalloprotein that requires a Zn atom for proper folding and activity (Hainaut and Mann, 2001). Consistent with the key role of Zn, chelating agents are detrimental for the function of wild-type p53. However, in an interesting twist, the mild chelating agent ZMC1 with ionophore capacity has been shown to raise intracellular Zn concentrations to overcome decreased Zn binding affinity due to mutation in or close to the Zn coordination site in p53 (Yu et al., 2017, 2018). This promotes refolding of His175 mutant p53 to its functional state as assessed by activation of p53 target genes and induction of tumor cell death. Several other structural p53 mutants can be restored in a similar manner. ZMC1 has also been shown to affect cellular redox homeostasis, which might add to its anti-cancer effect (Yu et al., 2018).
Conclusions and perspectives
p53 is redox-sensitive transcription factor that regulates a number of genes with antioxidant or pro-oxidant properties (Figure 2), and thereby influences the cellular redox balance. This regulation can go in both directions, depending on various factors, including p53 protein levels. Induction of p53 in response to DNA damage or oncogenic stress triggers p53-dependent apoptosis associated with release of oxygen species from mitochondria. Thus, wild-type p53-mediated redox effects are clearly highly relevant for p53-mediated tumor suppression, although their exact role needs further study. Interestingly, mutant p53 interferes with redox homeostasis in multiple ways, which may contribute to malignant progression (Figure 3). Thus, continued investigation of the redox effects of mutant p53 is crucial for a better understanding of how mutant p53 drives tumor development.
This is also highly relevant in a therapeutic context. Targeting of cysteines in mutant p53 can thermostabilize the core domain and promote correct folding and reactivation, ultimately leading to the elimination of tumor cells. Thiol-reactive compounds such as PK11007 and MQ (APR-246) not only reactivate mutant p53 but target cellular antioxidant components such as TrxR and GSH as well, leading to oxidative stress (Figure 4). Since redox homeostasis is an Achilles’s heel of tumor cells, dual targeting of mutant p53 and the redox balance may allow more efficient elimination of tumor cells. The redox effects may also explain the observed strong synergies between mutant p53-reactivating compounds and conventional chemotherapeutic drugs. This strategy is currently tested in clinical trials with APR-246.
Acknowledgements
We thank Lars Abrahmsen, Aprea Therapeutics AB, for valuable comments.
Funding
Research in the K.G.W. laboratory is supported by the Swedish Research Council (Vetenskapsrådet), the Swedish Cancer Society (Cancerfonden), the Swedish Childhood Cancer Fund (Barncancerfonden), Radiumhemmets Forskningsfonder, the Knut and Alice Wallenberg Foundation, the European Research Council, Aprea Therapeutics AB, and Karolinska Institutet.
Conflict of interest
K.G.W. and V.J.N.B. are co-founders and shareholders of Aprea Therapeutics AB, a company that develops p53-based cancer therapy including APR-246. K.G.W. is a member of its Clinical Advisory Board. K.G.W. has received a salary from Aprea Therapeutics AB.
References
- Adams J.M., and Cory S. (2018). The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 25, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrysik Z., Galbraith M.D., Guarnieri A.L., et al. (2017). Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res. 27, 1645–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Lotem J., Kama R., et al. (2002). NQO1 stabilizes p53 through a distinct pathway. Proc. Natl Acad. Sci. USA 99, 3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Lotem J., Tsvetkov P., et al. (2003). P53 hot-spot mutants are resistant to ubiquitin-independent degradation by increased binding to NAD(P)H:quinone oxidoreductase 1. Proc. Natl Acad. Sci. USA 100, 15065–15070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S.H., Sung S.H., Oh S.Y., et al. (2013). Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 17, 73–84. [DOI] [PubMed] [Google Scholar]
- Banjac A., Perisic T., Sato H., et al. (2008). The cystine/cysteine cycle: a redox cycle regulating susceptibility versus resistance to cell death. Oncogene 27, 1618–1628. [DOI] [PubMed] [Google Scholar]
- Bauer M.R., Joerger A.C., and Fersht A.R. (2016). 2-Sulfonylpyrimidines: Mild alkylating agents with anticancer activity toward p53-compromised cells. Proc. Natl Acad. Sci. USA 113, E5271–E5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky M., and Jaiswal A.K. (1993). NAD(P)H:quinone oxidoreductase1 (DT-diaphorase) expression in normal and tumor tissues. Cancer Metastasis Rev. 12, 103–117. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R., and Maiorino M. (2013). Glutathione peroxidases. Biochim. Biophys. Acta 1830, 3289–3303. [DOI] [PubMed] [Google Scholar]
- Brosh R., and Rotter V. (2009). When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer 9, 701–713. [DOI] [PubMed] [Google Scholar]
- Bullock A.N., and Fersht A.R. (2001). Rescuing the function of mutant p53. Nat. Rev. Cancer 1, 68–76. [DOI] [PubMed] [Google Scholar]
- Bykov V.J.N., Eriksson S.E., Bianchi J., et al. (2018). Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 18, 89–102. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Issaeva N., Shilov A., et al. (2002). Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8, 282–288. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Lambert J.M., Hainaut P., et al. (2009). Mutant p53 rescue and modulation of p53 redox state. Cell Cycle 8, 2509–2517. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Zache N., Stridh H., et al. (2005). PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene 24, 3484–3491. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Zhang Q., Zhang M., et al. (2016). Targeting of mutant p53 and the cellular redox balance by APR-246 as a strategy for efficient cancer therapy. Front. Oncol. 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano C.E., Gommeaux J., Pietri S., et al. (2009). Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 69, 219–226. [DOI] [PubMed] [Google Scholar]
- Casso D., and Beach D. (1996). A mutation in a thioredoxin reductase homolog suppresses p53-induced growth inhibition in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 252, 518–529. [DOI] [PubMed] [Google Scholar]
- Cebula M., Schmidt E.E., and Arner E.S. (2015). TrxR1 as a potent regulator of the Nrf2-Keap1 response system. Antioxid. Redox Signal. 23, 823–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Sun Z., Wang X.J., et al. (2009). Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell 34, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheteh E.H., Augsten M., Rundqvist H., et al. (2017). Human cancer-associated fibroblasts enhance glutathione levels and antagonize drug-induced prostate cancer cell death. Cell Death Dis. 8, e2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y., Gorina S., Jeffrey P.D., et al. (1994). Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265, 346–355. [DOI] [PubMed] [Google Scholar]
- Cooks T., Pateras I.S., Tarcic O., et al. (2013). Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 23, 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola G.M., Karreth F.A., Humpton T.J., et al. (2011). Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domenico F., Cenini G., Sultana R., et al. (2009). Glutathionylation of the pro-apoptotic protein p53 in Alzheimer’s disease brain: implications for AD pathogenesis. Neurochem. Res. 34, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraonio R., Vergara P., Di Marzo D., et al. (2006). p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J. Biol. Chem. 281, 39776–39784. [DOI] [PubMed] [Google Scholar]
- Fischer M. (2017). Census and evaluation of p53 target genes. Oncogene 36, 3943–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt P.M., Polyak K., Tang L.J., et al. (2000). p53-dependent expression of PIG3 during proliferation, genotoxic stress, and reversible growth arrest. Cancer Lett. 156, 63–72. [DOI] [PubMed] [Google Scholar]
- Foster B.A., Coffey H.A., Morin M.J., et al. (1999). Pharmacological rescue of mutant p53 conformation and function. Science 286, 2507–2510. [DOI] [PubMed] [Google Scholar]
- Freed-Pastor W.A., Mizuno H., Zhao X., et al. (2012). Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Monian P., Quadri N., et al. (2015). Glutaminolysis and Transferrin Regulate Ferroptosis. Mol. Cell 59, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W., Zhao K., Wang X., et al. (2017). iASPP is an antioxidative factor and drives cancer growth and drug resistance by competing with Nrf2 for Keap1 binding. Cancer Cell 32, 561–573.e566. [DOI] [PubMed] [Google Scholar]
- Gomes A.S., Ramos H., Soares J., et al. (2018). p53 and glucose metabolism: an orchestra to be directed in cancer therapy. Pharmacol. Res. 131, 75–86. [DOI] [PubMed] [Google Scholar]
- Gorrini C., Harris I.S., and Mak T.W. (2013). Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12, 931–947. [DOI] [PubMed] [Google Scholar]
- Greetham D., Vickerstaff J., Shenton D., et al. (2010). Thioredoxins function as deglutathionylase enzymes in the yeast Saccharomyces cerevisiae. BMC Biochem. 11, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffo L., Lu J., Bykov V.J.N., et al. (2018). Inhibition of the glutaredoxin and thioredoxin systems and ribonucleotide reductase by mutant p53-targeting compound APR-246. Sci. Rep. 8, 12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P., and Mann K. (2001). Zinc binding and redox control of p53 structure and function. Antioxid. Redox Signal. 3, 611–623. [DOI] [PubMed] [Google Scholar]
- Hainaut P., and Milner J. (1993). Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 53, 4469–4473. [PubMed] [Google Scholar]
- Hayano M., Yang W.S., Corn C.K., et al. (2016). Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 23, 270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He T., Farrar S., et al. (2017). Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell. Physiol. Biochem. 44, 532–553. [DOI] [PubMed] [Google Scholar]
- He X., and Ma Q. (2012). Redox regulation by nuclear factor erythroid 2-related factor 2: gatekeeping for the basal and diabetes-induced expression of thioredoxin-interacting protein. Mol. Pharmacol. 82, 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom K.M., and Finkel T. (2014). Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411–421. [DOI] [PubMed] [Google Scholar]
- Hu W., Zhang C., Wu R., et al. (2010). Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl Acad. Sci. USA 107, 7455–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Long Z., Lin W., et al. (2018). Integrative omics analysis of p53-dependent regulation of metabolism. FEBS Lett. 592, 380–393. [DOI] [PubMed] [Google Scholar]
- Jana S., Patra K., Jana J., et al. (2018). Nrf-2 transcriptionally activates P21(Cip/WAF1) and promotes A549cell survival against oxidative stress induced by H2O2. Chem. Biol. Interact. 285, 59–68. [DOI] [PubMed] [Google Scholar]
- Jayaraman L., Murthy K.G., Zhu C., et al. (1997). Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 11, 558–570. [DOI] [PubMed] [Google Scholar]
- Jeong Y., Hoang N.T., Lovejoy A., et al. (2017). Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov. 7, 86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Du W., Wang X., et al. (2011). p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 13, 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Kon N., Li T., et al. (2015). Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Kim M.J., Kim D.O., et al. (2013). TXNIP maintains the hematopoietic cell pool by switching the function of p53 under oxidative stress. Cell Metab. 18, 75–85. [DOI] [PubMed] [Google Scholar]
- Kaar J.L., Basse N., Joerger A.C., et al. (2010). Stabilization of mutant p53 via alkylation of cysteines and effects on DNA binding. Protein Sci. 19, 2267–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallin A., Johannessen L.E., Cani P.D., et al. (2007). SREBP-1 regulates the expression of heme oxygenase 1 and the phosphatidylinositol-3 kinase regulatory subunit p55 gamma. J. Lipid Res. 48, 1628–1636. [DOI] [PubMed] [Google Scholar]
- Kalo E., Kogan-Sakin I., Solomon H., et al. (2012). Mutant p53R273H attenuates the expression of phase 2 detoxifying enzymes and promotes the survival of cells with high levels of reactive oxygen species. J. Cell Sci. 125, 5578–5586. [DOI] [PubMed] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromova N.V., Kopnin P.B., Stepanova E.V., et al. (2009). p53 hot-spot mutants increase tumor vascularization via ROS-mediated activation of the HIF1/VEGF-A pathway. Cancer Lett. 276, 143–151. [DOI] [PubMed] [Google Scholar]
- Koppula P., Zhang Y., Zhuang L., et al. (2018). Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. 38, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.M., Gorzov P., Veprintsev D.B., et al. (2009). PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15, 376–388. [DOI] [PubMed] [Google Scholar]
- Lee P., Vousden K.H., and Cheung E.C. (2014). TIGAR, TIGAR, burning bright. Cancer Metab. 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewerenz J., Hewett S.J., Huang Y., et al. (2013). The cystine/glutamate antiporter system xc− in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 18, 522–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Sun M., Cao L., et al. (2014). A TIGAR-regulated metabolic pathway is critical for protection of brain ischemia. J. Neurosci. 34, 7458–7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan W.M., and Rouault T.A. (2013). Molecular pathways: Fumarate hydratase-deficient kidney cancer—targeting the Warburg effect in cancer. Clin. Cancer Res. 19, 3345–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisek K., Campaner E., Ciani Y., et al. (2018). Mutant p53 tunes the NRF2-dependent antioxidant response to support survival of cancer cells. Oncotarget 9, 20508–20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Chen W., Lei S., et al. (2017. b). Wild-type and mutant p53 differentially modulate miR-124/iASPP feedback following pohotodynamic therapy in human colon cancer cell line. Cell Death Dis. 8, e3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.S., Duong C.P., Haupt S., et al. (2017. a). Inhibiting the system xc−/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat. Commun. 8, 14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Zhao H., Yao H., et al. (2013). MicroRNA-124 regulates the proliferation of colorectal cancer cells by targeting iASPP. Biomed Res. Int. 2013, 867537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Madan E., Gogna R., Kuppusamy P., et al. (2013). SCO2 induces p53-mediated apoptosis by Thr845 phosphorylation of ASK-1 and dissociation of the ASK-1-Trx complex. Mol. Cell. Biol. 33, 1285–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D., Berkers C.R., Mason S.M., et al. (2013). Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorino M., Conrad M., and Ursini F. (2018). GPx4, lipid peroxidation, and cell death: discoveries, rediscoveries, and open issues. Antioxid. Redox Signal. 29, 61–74. [DOI] [PubMed] [Google Scholar]
- Marcel V., Dichtel-Danjoy M.L., Sagne C., et al. (2011). Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 18, 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meplan C., Richard M.J., and Hainaut P. (2000). Redox signalling and transition metals in the control of the p53 pathway. Biochem. Pharmacol. 59, 25–33. [DOI] [PubMed] [Google Scholar]
- Michael A. (1887). Ueber die Addition von Natriumacetessig- und Natriummalonsäureäthern zu den Aethern ungesättigter Säuren. J. Prakt. Chem. 35, 349–356. [Google Scholar]
- Miller C.G., Holmgren A., Arner E.S.J., et al. (2018). NADPH-dependent and -independent disulfide reductase systems. Free Radic. Biol. Med. 127, 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohell N., Alfredsson J., Fransson A., et al. (2015). APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis. 6, e1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y., Wang S.J., Li D., et al. (2016). Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl Acad. Sci. USA 113, E6806–E6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.J., and Weerapana E. (2013). Diverse functional roles of reactive cysteines. ACS Chem. Biol. 8, 283–296. [DOI] [PubMed] [Google Scholar]
- Pader I., Sengupta R., Cebula M., et al. (2014). Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and S-denitrosylase. Proc. Natl Acad. Sci. USA 111, 6964–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Carr J.R., Wang Z., et al. (2009). FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. 28, 2908–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G.D., and Merrill G.F. (1998). Deletion of the Saccharomyces cerevisiae TRR1 gene encoding thioredoxin reductase inhibits p53-dependent reporter gene expression. J. Biol. Chem. 273, 5431–5434. [DOI] [PubMed] [Google Scholar]
- Peng X., Zhang M.Q., Conserva F., et al. (2013). APR-246/PRIMA-1MET inhibits thioredoxin reductase 1 and converts the enzyme to a dedicated NADPH oxidase. Cell Death Dis. 4, e881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A., Mathe E., Kato S., et al. (2007). Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 28, 622–629. [DOI] [PubMed] [Google Scholar]
- Polyak K., Xia Y., Zweier J.L., et al. (1997). A model for p53-induced apoptosis. Nature 389, 300–305. [DOI] [PubMed] [Google Scholar]
- Raghunath A., Sundarraj K., Nagarajan R., et al. (2018). Antioxidant response elements: discovery, classes, regulation and potential applications. Redox Biol. 17, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainwater R., Parks D., Anderson M.E., et al. (1995). Role of cysteine residues in regulation of p53 function. Mol. Cell. Biol. 15, 3892–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M., Kwak M.K., Dolan P.M., et al. (2001). Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl Acad. Sci. USA 98, 3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo de la Vega M., Chapman E., and Zhang D.D. (2018). NRF2 and the hallmarks of cancer. Cancer Cell 34, 21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth S.A., Zaitseva L., Murray M.Y., et al. (2012). The high Nrf2 expression in human acute myeloid leukemia is driven by NF-κB and underlies its chemo-resistance. Blood 120, 5188–5198. [DOI] [PubMed] [Google Scholar]
- Saadi H., Seillier M., and Carrier A. (2015). The stress protein TP53INP1 plays a tumor suppressive role by regulating metabolic homeostasis. Biochimie 118, 44–50. [DOI] [PubMed] [Google Scholar]
- Sabapathy K., and Lane D.P. (2018). Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 15, 13–30. [DOI] [PubMed] [Google Scholar]
- Sablina A.A., Budanov A.V., Ilyinskaya G.V., et al. (2005). The antioxidant function of the p53 tumor suppressor. Nat. Med. 11, 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin V.I., Ibrahim M.X., Larsson E., et al. (2014). Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 6, 221ra215. [DOI] [PubMed] [Google Scholar]
- Scotcher J., Clarke D.J., Weidt S.K., et al. (2011). Identification of two reactive cysteine residues in the tumor suppressor protein p53 using top-down FTICR mass spectrometry. J. Am. Soc. Mass Spectrom. 22, 888–897. [DOI] [PubMed] [Google Scholar]
- Seemann S., and Hainaut P. (2005). Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene 24, 3853–3863. [DOI] [PubMed] [Google Scholar]
- Shi Y., Ghosh M., Kovtunovych G., et al. (2012). Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim. Biophys. Acta 1823, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T., Takeda K., Oda E., et al. (2003). Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 17, 2233–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin B.Y., Jin S.H., Cho I.J., et al. (2012). Nrf2-ARE pathway regulates induction of Sestrin-2 expression. Free Radic. Biol. Med. 53, 834–841. [DOI] [PubMed] [Google Scholar]
- Soussi T., and Wiman K.G. (2015). TP53: an oncogene in disguise. Cell Death Differ. 22, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell B.R., Friedmann Angeli J.P., Bayir H., et al. (2017). Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H.W., Yun S., Song H., et al. (2013). TXNIP interacts with hEcd to increase p53 stability and activity. Biochem. Biophys. Res. Commun. 438, 264–269. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Tanaka T., Poyurovsky M.V., et al. (2010). Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl Acad. Sci. USA 107, 7461–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak S.T., Mays D., and Pietenpol J.A. (2001). Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 21, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Zhao M., Tang L., et al. (2018). Gain-of-function mutant p53 promotes the oncogenic potential of head and neck squamous cell carcinoma cells by targeting the transcription factors FOXO3a and FOXM1. Oncogene 37, 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarangelo A., Magtanong L., Bieging-Rolett K.T., et al. (2018). p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep. 22, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessoulin B., Descamps G., Moreau P., et al. (2014). PRIMA-1Met induces myeloma cell death independent of p53 by impairing the GSH/ROS balance. Blood 124, 1626–1636. [DOI] [PubMed] [Google Scholar]
- Todoric J., Antonucci L., Di Caro G., et al. (2017). Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell 32, 824–839.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.Q., Lowman X.H., Reid M.A., et al. (2017). Tumor-associated mutant p53 promotes cancer cell survival upon glutamine deprivation through p21 induction. Oncogene 36, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung M.C., Lin P.L., Wang Y.C., et al. (2015). Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget 6, 41692–41705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M., Masutani H., Arai R.J., et al. (1999). Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J. Biol. Chem. 274, 35809–35815. [DOI] [PubMed] [Google Scholar]
- Valko M., Rhodes C.J., Moncol J., et al. (2006). Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 160, 1–40. [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., and Thompson C.B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velu C.S., Niture S.K., Doneanu C.E., et al. (2007). Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry 46, 7765–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden K.H., and Prives C. (2009). Blinded by the light: the growing complexity of p53. Cell 137, 413–431. [DOI] [PubMed] [Google Scholar]
- Walerych D., Lisek K., Sommaggio R., et al. (2016). Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat. Cell Biol. 18, 897–909. [DOI] [PubMed] [Google Scholar]
- Wang W., Kryczek I., Dostal L., et al. (2016. c). Effector T cells abrogate stroma-mediated chemoresistance in ovarian. Cancer Cell 165, 1092–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.J., Li D., Ou Y., et al. (2016. b). Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 17, 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Yin C., Li X.X., et al. (2016. a). Reduced SOD2 expression is associated with mortality of hepatocellular carcinoma patients in a mutant p53-dependent manner. Aging 8, 1184–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassman C.D., Baronio R., Demir O., et al. (2013). Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat. Commun. 4, 1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Ye H., Tang Z., et al. (2017). p53 dynamics orchestrates with binding affinity to target genes for cell fate decision. Cell Death Dis. 8, e3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhu S., Song X., et al. (2017). The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 20, 1692–1704. [DOI] [PubMed] [Google Scholar]
- Yoshihara E., Masaki S., Matsuo Y., et al. (2014). Thioredoxin/Txnip: redoxisome, as a redox switch for the pathogenesis of diseases. Front. Immunol. 4, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A., Nam C.W., Wakabayashi N., et al. (2011). Transcription factor Nrf2 maintains the basal expression of Mdm2: an implication of the regulation of p53 signaling by Nrf2. Arch. Biochem. Biophys. 507, 356–364. [DOI] [PubMed] [Google Scholar]
- Yu X., Blanden A., Tsang A.T., et al. (2017). Thiosemicarbazones functioning as zinc metallochaperones to reactivate mutant p53. Mol. Pharmacol. 91, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Kogan S., Chen Y., et al. (2018). Zinc metallochaperones reactivate mutant p53 using an ON/OFF switch mechanism: a new paradigm in cancer therapeutics. Clin. Cancer Res. 24, 4505–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf M.A., Chuang T., Bhat G.J., et al. (2010). Cys-141 glutathionylation of human p53: studies using specific polyclonal antibodies in cancer samples and cell lines. Free Radic. Biol. Med. 49, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bergman J., Wiman K.G., et al. (2018. a). Role of thiol reactivity for targeting mutant p53. Cell Chem. Biol. 25, 1219–1230 e1213. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bykov V.J.N., Wiman K.G., et al. (2018. b). APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis. 9, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Guo X.Y., Hu G.Y., et al. (1994). A temperature-sensitive mutant of human p53. EMBO J. 13, 2535–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qian Y., Zhang J., et al. (2017). Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev. 31, 1243–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Trachootham D., Liu J., et al. (2012). Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat. Cell Biol. 14, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]