Abstract
Gene fusions are one of the most common genomic alterations in pediatric cancer. Many fusions encode oncogenic drivers and play important roles in cancer diagnosis, risk stratification, and treatment selection. We report the development and clinical validation of a large custom-designed RNA sequencing panel, CHOP Fusion panel, using anchored multiplex PCR technology. The panel interrogates 106 cancer genes known to be involved in nearly 600 different fusions reported in hematological malignancies and solid tumors. The panel works well with different types of samples, including formalin-fixed, paraffin-embedded samples. The panel demonstrated excellent analytic accuracy, with 100% sensitivity and specificity on 60 pediatric tumor validation samples. In addition to identifying all known fusions in the validation samples, three unrecognized, yet clinically significant, fusions were also detected. A total of 276 clinical cases were analyzed after the validation, and 51 different fusions were identified in 104 cases. Of these fusions, 16 were not previously reported at the time of discovery. These fusions provided genomic information useful for clinical management. Our experience demonstrates that CHOP Fusion panel can detect the vast majority of known and certain novel clinically relevant fusion genes in pediatric cancers accurately, efficiently, and cost-effectively; and the panel provides an excellent tool for new fusion gene discovery.
Fusion genes are formed by combining parts of two different genes into a novel one that typically encodes for an in-frame chimeric protein, including key domains specified by the parental genes. Gene fusions are derived from chromosomal rearrangements, including translocation, duplication, deletions, inversion, or altered transcription. Although the exact biological functions of recurrent fusion proteins may not be fully understood in all cases, the clinical impact of many has been established definitively.1, 2, 3
Gene fusions occur in all types of human malignancies, and they are present in tumors that accounted for approximately 20% of human cancer deaths in 2017.1, 4, 5 Many recurrent fusions are established to be strong drivers of tumorigenesis and progression. Novel therapies targeting these driver fusions in cancer have achieved outstanding outcomes. For example, all-trans-retinoic acid therapy against RARA-associated fusions has made acute promyelocytic leukemia one of the most curable types of leukemia.6, 7 Imatinib and related tyrosine kinase inhibitors dramatically extend the lifespan of patients with chronic myeloid leukemia, a disease defined by BCR-ABL1 fusion.2, 8 Targeting NTRK gene fusions with tropomyosin receptor kinase inhibitors has shown a high percentage of durable responses in NTRK fusion-positive pediatric solid tumors.9, 10 In addition to their therapeutic significance, many gene fusions are pathognomonic diagnostic markers. PML-RARA fusion is diagnostic for acute promyelocytic leukemia;7 fusions between TMPRSS2 and ETS transcription factor family members are found in 70% of prostate cancers11; KIAA1549-BRAF is a unique recurrent fusion in pediatric low-grade gliomas,12 and the presence of a EWSR1-FLI1 fusion is diagnostic for Ewing sarcoma.5 Chimeric transcripts encoded by specific fusion genes can also be used to monitor treatment response and residual disease. BCR-ABL1 transcript levels are routinely monitored in chronic myeloid leukemia patients receiving tyrosine kinase inhibitor treatment, and specific response landmarks have been defined and are used in clinical management.13 Furthermore, many fusions are important prognostic biomarkers. RUNX1-RUNX1T1 fusion characterizes a subtype of acute myeloid leukemia with favorable outcome and prolonged median survival.14 In contrast, most KMT2A fusion genes are poor prognostic markers in different leukemias.15, 16 Pediatric cancers harbor a lower mutation rate compared with adult cancers, and fusion genes are common disease- or subtype-defining oncogenic drivers in pediatric leukemias, solid tumors, and central nervous system (CNS) tumors.17
Traditional methods for fusion transcript or fusion protein detection include chromosomal karyotyping, fluorescence in situ hybridization, RT-PCR, and immunohistochemistry stain.18, 19, 20 Although each of these technologies has specific technique limits, a common limitation is poor compatibility with multiplexing, preventing these methods from interrogating multiple fusion genes simultaneously, which is especially important when the diagnosis is unclear.1, 19, 21, 22 The emergence of next-generation sequencing (NGS) technology and modern computational tools allows sequencing numerous genes in parallel and facilitates the identification of fusion. We have developed an NGS-based test that is capable of interrogating many fusion partner genes for known and novel fusions in a variety of tumors simultaneously. Herein, we present the development and clinical validation of a large custom-designed fusion panel, Children's Hospital of Philadelphia (CHOP) Fusion panel, using RNA sequencing. Detailed analytical performance and clinical utility of the panel are also discussed.
Materials and Methods
Validation and Clinical Samples
Previously characterized samples from our laboratory and the Children's Oncology Group with known fusion transcripts were used for validating CHOP Fusion panel. Different tissue types commonly used for genomic diagnosis were used, including bone marrow (BM)/peripheral blood (PB), fresh or fresh frozen tissue, and formalin-fixed, paraffin-embedded (FFPE) samples from different leukemias, CNS tumors, and non-CNS solid tumors. The validation study was performed in a double-blinded manner (F.C., F.L.), so that both the testing personnel and analytic personnel had no knowledge of the fusions present in the samples analyzed. Clinical tumor samples from patients referred for genomic testing were also tested with the panel to evaluate the clinical validity and clinical utility.
Panel Design
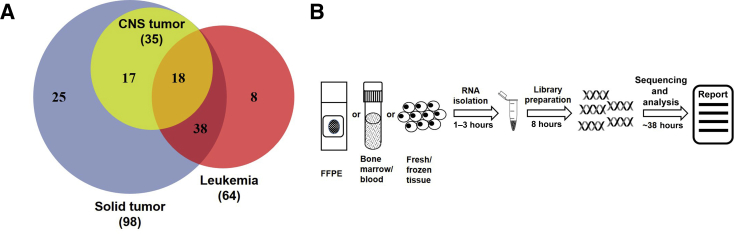
A total of 106 genes known to be involved in cancer-associated fusion genes were examined using Anchored Multiplex PCR technology, powered by unidirectional gene-specific primers, sample indexes, and molecular barcodes for multiplex targeted RNA sequencing (ArcherDX, Inc., Boulder, CO). Target-specific primers covering 586 exons were custom designed to identify known fusion transcripts and potential novel fusion transcripts associated with 106 cancer genes. The genes targeted in this panel are listed in Supplemental Table S1 and Figure 1A. To identify potential fusions involving Ig genes, multiple probes were designed to cover the entire partner genes, such as MYC, CRLF2, and EPOR1, enabling assessments of gene expressions.
Figure 1.
Schematic diagram of Children's Hospital of Philadelphia (CHOP) Fusion panel design and workflow. A: Venn diagram of genes of interest in leukemia and non–central nervous system (CNS) solid tumor and CNS tumors. B: CHOP Fusion panel workflow, from sample to report. FFPE, formalin fixed, paraffin embedded.
Nucleotide Extraction
Total RNA from BM/PB or fresh/frozen tissues was extracted using the RiboPure RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA) or the RNeasy Mini Kit and TissueLyser-LT (Qiagen, Hilden, Germany), respectively, according to the manufacturer's instructions. Total nucleic acid was extracted from FFPE samples using the Agencourt FormaPure Kit (Beckman Coulter, Indianapolis, IN), following the user manual with a few modifications, including eliminating DNase treatment, using 70°C heat block instead of 72°C water bath, and adding a reverse cross-linking step by incubating at 80°C for 60 minutes. The minimum concentration of 7.5 ng/μL of single-stranded RNA measured by Qubit (Thermo Fisher Scientific) is required for library preparation.
Library Preparation and Sequencing
Libraries were constructed using Archer Universal RNA Reagent Kit version 2 (ArcherDX, Inc.) with 150 ng of RNA or 250 ng of total nucleic acid. The workflow is illustrated in Figure 1B. RNA quality control was performed after first-strand cDNA synthesis using real-time quantitative PCR (qPCR). Libraries were quantified using KAPA Biosystems qPCR kit for Illumina, according to the user guide (KAPA Biosystems, Wilmington, MA). After quantification, the barcoded libraries were pooled at equimolar concentrations and sequenced on Illumina MiSeq platform with MiSeq version 2 300-cycle reagent kit (Illumina, San Diego, CA).
Sequence Data Analysis
Paired-end sequence data were uploaded to Archer Analysis software version 4 (ArcherDX, Inc.), installed on the on-site virtual machine server for analysis using CHOP Fusion panel region of interest. PCR duplicates were removed by consolidating reads with the same molecular barcode into a single consensus read. The minimum of 150,000 unique, mapped reads is required to pass quality control for downstream analysis. Each unique read, considered as a fusion-supporting read, has to satisfy two conditions: it spans two separate genes; and at least a 23-bp sequence is mapped on either side of an apparent break point. The criteria of strong evidence for a fusion include five or more unique fusion-supporting reads (approximately 10% of total reads) and three unique start sites, reflecting random ligation of the universal adapter. The unique reads mapped to the consensus sequence were visualized directly in Archer Analysis software using the JBrowse genome browser23 (Evolutionary Software Foundation, Berkeley, CA).
Fusion Confirmation
Novel fusions were confirmed by nested PCR, followed by Sanger sequencing. The primers were designed using Primer3.24 The first strand of cDNA was synthesized using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA). PCR was assayed using Qiagen Fast Cycling PCR kit, according to the manufacturer's manual. The PCR product was sequenced on an ABI 3730 DNA analyzer (Thermo Fisher Scientific). Confirmation primers used in the study are summarized in Table 1. Previously unrecognized fusion genes in validation samples and all fusions identified in the clinical samples were orthogonally confirmed.
Table 1.
Primer Sequences for Fusion Confirmation and CRLF2 and EPOR Expression
| Gene or gene fusion | PCR reaction | Forward primer | Reverse primer |
|---|---|---|---|
| ZNF274-JAK2 | PCR1 | 5′-GGTTTTACCCCGGAAGAGTG-3′ | 5′-GCAGGAAGCTGATGCCTATC-3′ |
| PCR2 | 5′-TGTAAAACGACGGCCAGTAGGGAAGTGATGCTGGAGAA-3′ | 5′-CAGGAAACAGCTATGACCTGCAGATTTCCCACAAAGTG-3′ | |
| PAX5-ZCCHC7 | PCR1 | 5′-ACACCAACAAGCGCAAGAGA-3′ | 5′-GGCTGGACAGGAATACAGGA-3′ |
| PCR2 | 5′-TGTAAAACGACGGCCAGTACTTCCTCCGGAAGCAGATG-3′ | 5′-CAGGAAACAGCTATGACCCAGATAATGTTTTTGTTGGCTGAA-3′ | |
| ATF7IP-PDGFRB | PCR1 | 5′-CCTTGCCAAATCCCACTAAA-3′ | 5′-TTTCATCGTGGCCTGAGAAT-3′ |
| PCR2 | 5′-TGTAAAACGACGGCCAGTGCCACAAGGACTTCTTTACCC-3′ | 5′-CAGGAAACAGCTATGACCCCTTCCATCGGATCTCGTAA-3′ | |
| CENPC-ABL1 | PCR1 | 5′-ACAGAGCAAGGCCAGAATGT-3′ | 5′-ACACCATTCCCCATTGTGAT-3′ |
| PCR2 | 5′-TGTAAAACGACGGCCAGTTGTATTCAGTCACCAAGCAAAGA-3′ | 5′-CAGGAAACAGCTATGACCACGAAAAGGTTGGGGTCATT-3′ | |
| NUP153-ABL1 | PCR1 | 5′-CATCTCTGACTCCTTCTGGTGA-3′ | 5′-CCACCGTCAGGCTGTATTTC-3′ |
| PCR2 | 5′-TGTAAAACGACGGCCAGTTGACACCCGGACAAAATAGAG-3′ | 5′-CAGGAAACAGCTATGACCGGGGGACACACCATAGACAG-3′ | |
| SPTAN1-ABL1 | PCR1 | 5′-ACCGCTTCAAGGAACTCTCA-3′ | 5′-TGGTGTCCTCCTTCAAGGTC-3′ |
| PCR2 | 5′-GTAAAACGACGGCCAGTCAACCCTTAGGCGTCAGAAG-3′ | 5′-CAGGAAACAGCTATGACGGATAATGGAGCGTGGTGAT-3′ | |
| CRLF2 | RT-PCR | 5′-ATGCCAGCAAATACTCCAGGAC-3′ | 5′-AAGGTAGTTGGTGCACTGGTCAT-3′ |
| EPOR | RT-PCR | 5′-ATCCTCGTGGTCATCCTGGTG-3′ | 5′-GGATGCCAGGCCAGATCTTC-3′ |
| ACTB | RT-PCR | 5′-CCTGAACCCCAAGGCCAAC-3′ | 5′-ACAGCCTGGATAGCAACGTACA-3′ |
Real-Time Quantitative PCR for Gene Expression
The expression level of CRLF2 and EPOR in samples with CRLF2- or EPOR-associated fusions and control samples was evaluated by qPCR. The qPCR was performed using iTaq Universal SYBR Green Supermix 2X (Bio-Rad) and ABI QuantStudio Real-Time PCR thermocycler (Thermo Fisher Scientific), with the PCR parameters of 95°C for 2 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 30 seconds. Melting experiments were performed after the last PCR cycle by increasing by 0.2°C every 5 seconds from 60°C to 95°C. Melting curves were analyzed by QuantStudio Real-Time PCR Software version 1.3 (Thermo Fisher Scientific). The PCR primers for CRLF2 and ACTB (internal control) can be found in Table 1.
Statistical Analysis
The coverage of each gene on the panel was calculated using the median of unique reads of all exons in each gene. The heat map for visualization of gene expression was generated by Archer Analysis software using four built-in housekeeping gene (VCP, GPI, RAB7A, and CHMP2A). Gene expression was calculated on the basis of the ratio of unique reads between target gene and housekeeping gene VCP using raw data from CHOP Fusion panel. Samples in the validation study were grouped on the basis of tissue types: BM/PB, fresh/frozen tissue, and FFPE samples. The distributions of on-target rate (%) of reads and deduplication ratio of three tissue types were evaluated using pairwise Wilcoxon rank sum tests against the hypothesis of equal medians of each group in the tests. P ≤ 0.001 was considered significant.
Results
Analytical Performance
Coverage and Deduplication Ratio
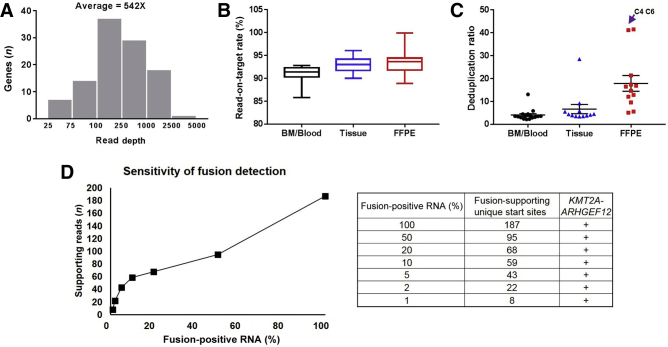
To evaluate the analytical performance of CHOP Fusion panel, libraries were generated from 60 validation samples, including 34 known fusion-positive samples (27 from the Children's Oncology Group and 7 from our laboratory), 3 negative controls, and 23 clinical samples with no prior knowledge about the presence or absence of a fusion. These samples represented three major specimen types in clinical practice (namely, BM/PB, fresh/frozen tissues, and FFPE samples). The coverage (unique reads per gene) of all genes ranged from 25 to >4000 unique reads, with an average of 542 unique reads per gene (Figure 2A). Over 92% of reads were successfully aligned to the desired region of interest, indicating a strong enrichment for the region of interest. The on-target rates were 90.9% for BM/PB, 93.0% for fresh/frozen tissues, and 93.5% for FFPE samples (Figure 2B). A pairwise Wilcoxon test showed no significant difference between any of the two groups. Deduplication ratios were used to assess the complexity of cDNA libraries. The average deduplication ratios for BM/PB, fresh/frozen tissues, and FFPE samples were 4.1, 6.7, and 17.9, respectively (Figure 2C). The higher deduplication ratio of FFPE samples was largely contributed by two samples with extremely poor RNA quality (C4 and C6 in Figure 2C), which were used in the validation study purposely to evaluate the performance of the panel on poor-quality samples. Overall, CHOP Fusion panel yielded high on-target rates and high library complexity in validation samples.
Figure 2.
Analytical performance of Children's Hospital of Philadelphia (CHOP) Fusion panel. A: Coverage of all targeted genes. B: Comparison of read-on-target rate among bone marrow (BM)/blood, fresh/frozen tissue, and formalin-fixed, paraffin-embedded (FFPE) samples. C: Comparison of deduplication ratio among bone marrow/blood, fresh/frozen tissue, and FFPE samples. D: Detection limit of CHOP Fusion panel using a serial dilution assay.
Analytic Sensitivity and Specificity
Of the 34 known fusion-positive samples and 3 negative controls, no fusions were found in the three negative controls. For the 34 positive samples, all fusion genes previously identified by other technologies (RNA sequencing, fluorescence in situ hybridization, or RT-PCR) were detected, demonstrating 100% sensitivity. In addition, three new fusions (STIL-TAL1 in V14, PAX5-ZCCHC7 in V17, and PAX5-JAK2 in V26), which were missed by previous tests, were detected (Table 2). These fusions were considered clinically significant and confirmed by RT-PCR. Twenty-three additional clinical samples were tested using CHOP Fusion panel, and seven fusion genes were detected and confirmed by RT-PCR or fluorescence in situ hybridization. In total, 44 fusion genes were detected in 60 samples of various tissue types from a spectrum of tumors (Table 2). Remarkably, 9 of the 44 fusions had, to the best of our knowledge at the time of detection, never been reported before, all of which were confirmed by nested PCR, followed by Sanger sequencing (Supplemental Figure S1). Furthermore, a KIAA1549-BRAF and a CIC-DUX4 were detected from two FFPE samples with extremely poor RNA quality, indicated by high deduplication ratios (Figure 2C), reflecting high analytic sensitivity of the assay.
Table 2.
All Fusions Identified by CHOP Fusion Panel in Validation Samples
| Sample ID | Fusion detected | Junction exons | Clinical indication | Sample type |
|---|---|---|---|---|
| V1 | NUP214-ABL1 | NUP214 (NM_005085.3):exon 32/ABL1 (NM_005157.5):exon2 | Acute lymphoblastic leukemia | Bone marrow |
| V2 | ETV6-ABL1 | ETV6 (NM_001987.4):exon 5/ABL1 (NM_005157.5):exon 2 | Acute lymphoblastic leukemia | Bone marrow |
| V3 | ZMIZ1-ABL1 | ZMIZ1 (NM_020338.3):exon 18/ABL1 (NM_005157.5):exon 2 | Acute lymphoblastic leukemia | Bone marrow |
| V4 | RANBP2-ABL1 | RANBP2 (NM_006267.4):exon 18/ABL1 (NM_005157.5):exon 2 | Acute lymphoblastic leukemia | Bone marrow |
| V5 | RCSD1-ABL2 | RCSD1 (NM_052862.3):exon 3/ABL2 (NM_007314.3):exon 5 | Acute lymphoblastic leukemia | Bone marrow |
| V6 | ZC3HAV1-ABL2 | ZC3HAV1 (NM_020119.3):exon 12/ABL2 (NM_007314.3):exon 3 | Acute lymphoblastic leukemia | Bone marrow |
| V7 | SSBP2-CSF1R | SSBP2 (NM_012446.3): exon 15/CSF1R (NM_005211.3):exon 12 | Acute lymphoblastic leukemia | Bone marrow |
| V8 | EBF1-PDGFRB | EBF1 (NM_024007.3): exon 15/PDGFRB (NM_002609.3):exon 11 | Acute lymphoblastic leukemia | Bone marrow |
| V9 | BCR-JAK2 | BCR (NM_004327.3):exon 1/JAK2 (NM_004972.3):exon 19 | Acute lymphoblastic leukemia | Bone marrow |
| V10 | PAX5-JAK2 | PAX5 (NM_016734.2):exon 5/JAK2 (NM_004972.3):exon 19 | Acute lymphoblastic leukemia | Bone marrow |
| V11 | RCSD1-ABL1 | RCSD1 (NM_052862.3):exon 2/ABL1 (NM_005157.5):exon 4 | Acute lymphoblastic leukemia | Bone marrow |
| V12 | FOXP1-ABL1 | FOXP1 (NM_032682.5):exon 19/ABL1 (NM_005157.5):exon 4 | Acute lymphoblastic leukemia | Bone marrow |
| V13 | NUP153-ABL1∗ | NUP153 (NM_005124.3):exon 11/ABL1 (NM_005157.5):exon 4 | Acute lymphoblastic leukemia | Bone marrow |
| V14 | SPTAN1-ABL1∗ | SPTAN1 (NM_003127.3): exon 2/ABL1 (NM_005157.5):exon 4 | Acute lymphoblastic leukemia | Bone marrow |
| STIL-TAL1 | STIL (NM_003035.2): exon 1/TAL1 (NM_003189.5):exon 5 | Acute lymphoblastic leukemia | Bone marrow | |
| V15 | P2RY8-CRLF2 | P2RY8 (NM_178129.4): exon 1/CRLF2 (NM_022148.3): exon 1 | Acute lymphoblastic leukemia | Bone marrow |
| V16 | EPOR-IGH | EPOR (NM_000121.3): exon 8/IGH (NC_000014):exon 1 | Acute lymphoblastic leukemia | Bone marrow |
| V17 | IGH-CRLF2 | ?/CRLF2 (NM_022148.3):exon 1 | Acute lymphoblastic leukemia | Bone marrow |
| PAX5-ZCCHC7∗ | PAX5 (NM_016734.2):exon 6/ZCCHC7 (NM_032226.2):exon 3 | |||
| V18 | ETV6-JAK2 | ETV6 (NM_001987.4):exon 4/JAK2 (NM_004972.3):exon 17 | Acute lymphoblastic leukemia | Bone marrow |
| V19 | TERF2-JAK2 | TERF2 (NM_005652.4):exon 8/JAK2 (NM_004972.3):exon 19 | Acute lymphoblastic leukemia | Bone marrow |
| V20 | ATF7IP-PDGFRB∗ | ATF7IP (NM_018179.4):exon 9/PDGFRB (NM_002609.3):exon 11 | Acute lymphoblastic leukemia | Bone marrow |
| V21 | ETV6-NTRK3 | ETV6 (NM_001987.4):exon 5/NTRK3 (NM_002530.3):exon 15 | Acute lymphoblastic leukemia | Bone marrow |
| V22 | CENPC-ABL1∗ | CENPC (NM_001812.2):exon 5/ABL1 (NM_005157.5):exon 2 | Acute lymphoblastic leukemia | Bone marrow |
| V23 | SSBP2-JAK2 | SSBP2 (NM_012446.3):exon 15/JAK2 (NM_004972.3):exon 13 | Acute lymphoblastic leukemia | Bone marrow |
| V24 | ZNF274-JAK2∗ | ZNF274 (NM_133502.2):exon 4/JAK2 (NM_004972.3):exon 17 | Acute lymphoblastic leukemia | Bone marrow |
| V25 | ATF7IP-JAK2 | ATF7IP (NM_018179.4):exon 13/JAK2 (NM_004972.3):exon 17 | Acute lymphoblastic leukemia | Bone marrow |
| V26 | PAG1-ABL2 | PAG1 (NM_018440.3):exon 8/ABL2 (NM_007314.3):exon 5 | Acute lymphoblastic leukemia | Bone marrow |
| PAX5-JAK2 | PAX5 (NM_016734.2):exon 5/JAK2 (NM_004972.3):exon 19 | |||
| V27 | ZEB2-PDGFRB | ZEB2 (NM_014795.3):exon 9/PDGFRB (NM_002609.3):exon 9 | Acute lymphoblastic leukemia | Bone marrow |
| V28 | KIAA1549-BRAF | KIAA1549 (NM_020910.2):exon 15/BRAF (NM_004333.4):exon 9 | Brain tumor | FFPE |
| V29 | KIAA1549-BRAF | KIAA1549 (NM_020910.2):exon 16/BRAF (NM_004333.4):exon 9 | Brain tumor | FFPE |
| V30 | KIAA1549-BRAF | KIAA1549 (NM_020910.2): exon 15/BRAF (NM_004333.4):exon 9 | Brain tumor | FFPE |
| V31 | EPOR-IGH | EPOR (NM_000121.3):exon 8/IGH (NC_000014):exon 1 | Acute lymphoblastic leukemia | Bone marrow |
| V32 | P2RY8-CRLF2 | P2RY8 (NM_178129.4):exon 1/CRLF2 (NM_022148.3):exon 1 | Acute lymphoblastic leukemia | Bone marrow |
| V33 | P2RY8-CRLF2 | P2RY8 (NM_178129.4):exon 1/CRLF2 (NM_022148.3):exon 1 | Acute lymphoblastic leukemia | Bone marrow |
| V34 | IGH-CRLF2 | ?/CRLF2 (NM_022148.3):exon 1 | Acute lymphoblastic leukemia | Bone marrow |
| C1 | EWSR1-CREB1 | EWSR1 (NM_005243.3):exon 7/CREB1 (NM_004379.4):exon 6 | Angiomatoid fibrous histiocytoma | FFPE |
| C2 | NOTCH1-ROS1∗ | NOTCH1 (NM_017617.3):exon 30/ROS1 (NM_002944.2):exon 34 | Angiosarcoma | FFPE |
| C3 | KMT2A-ARHGEF12 | KMT2A (NM_005933.3):exon 10/ARHGEF12 (NM_015313.2):exon 12 | Burkitt lymphoma | Frozen tissue |
| C4 | CIC-DUX4 | CIC (NM_015125.4):exon 20/DUX4 (NM_033178.2):exon 1 | Small round cell malignancy | FFPE |
| C5 | WFS1-PLAG1∗ | WFS1 (NM_006005.3):exon 1/PLAG1 (NM_002655.2):exon 3 | Anaplastic astrocytoma | Frozen tissue |
| C6 | KIAA1549-BRAF | KIAA1549 (NM_020910.2):exon 16/BRAF (NM_004333.4):exon 9 | Brain tumor | FFPE |
| C7 | GOLGA5-JAK2∗ | GOLGA5 (NM_005113.3):exon 10/JAK2 (NM_004972.3):exon 19 | Acute lymphoblastic leukemia | Bone marrow |
All NM_numbers are searchable in Nuccore (https://www.ncbi.nlm.nih.gov/nuccore, last accessed May 30, 2019). Fusions not detected by previous tests are shown in bold.
CHOP, Children's Hospital of Philadelphia; FFPE, formalin fixed, paraffin embedded; ID, identification.
Novel fusions.
Detection Limit
To evaluate the detection limit of CHOP Fusion panel, serial dilution experiments were performed. An RNA sample with a KMT2A-ARHGEF12 fusion was serially diluted with RNA from a normal control to generate various levels of mosaicism, from 100% to 1% (Figure 2D). The panel successfully detected the fusion with 1% fusion-positive RNA in this sample. In addition, a linear positive correlation was demonstrated between tumor RNA percentage and numbers of fusion-supporting reads with unique start sites (Figure 2D).
Gene Expression Analysis for CRLF2 and EPOR
CRLF2 and EPOR often fuse with the promoter or enhancer of Ig genes, and these fusions could be missed by RNA sequencing. Because these fusions lead to increased CRLF2 or EPOR expression, the presence of a gene fusion was inferred using gene expression levels. The expression levels of these genes were measured by comparing the numbers of unique reads in the genes with that of the housekeeping gene VCP. The heat map generated by CHOP Fusion panel from three CRLF2 fusion-negative samples and four positive samples harboring CRLF2 rearrangements (P2RY8-CRLF2 or IGH-CRLF2) provided quantitative evidence of potential CRLF2 fusions at the expression level: barely detectable CRLF2 expression in CRLF2 fusion-negative samples and clearly increased expression in CRLF2 fusion-positive samples (Figure 3A). The levels of gene expression are in complete concordance between the NGS data and the qPCR results. Similar expression patterns of EPOR were also observed on two samples harboring EPOR-IGH fusions (Figure 3B).
Figure 3.
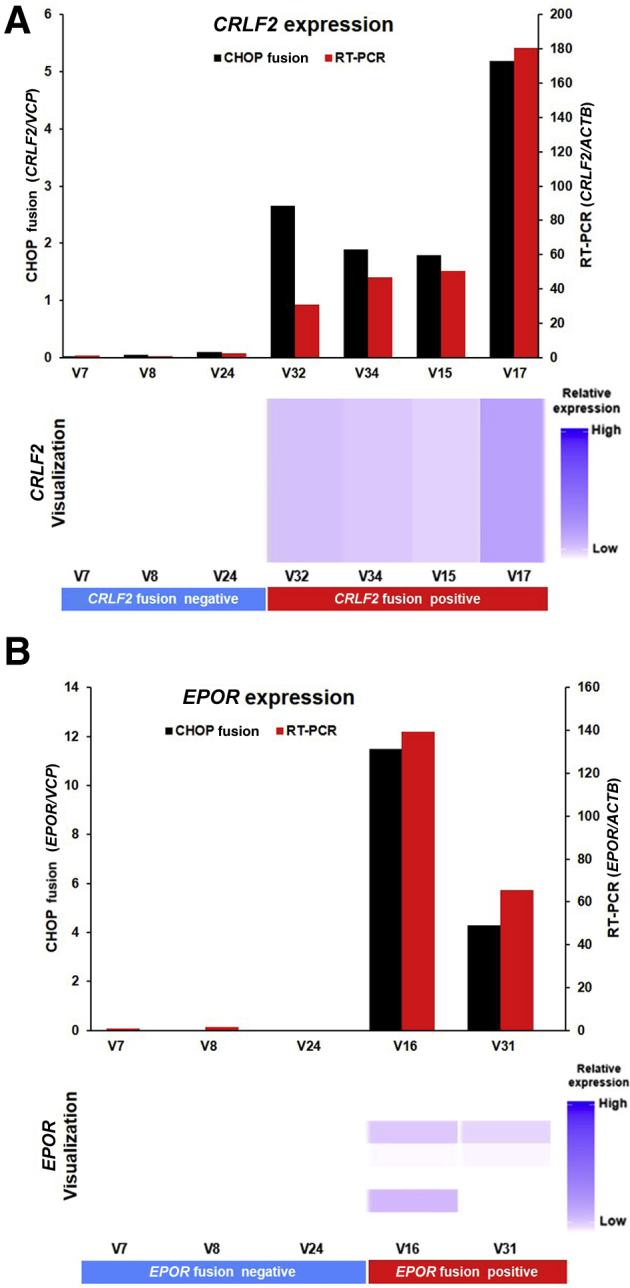
CRLF2/EPOR overexpression as a result of promoter fusion. Children's Hospital of Philadelphia (CHOP) Fusion panel detects the overexpression of CRLF2 (A) and EPOR (B) caused by promoter fusion consistent with RT-PCR. Top panels: Gene expression level by CHOP Fusion panel (normalized against housekeeping gene VCP) or by RT-PCR (normalized with housekeeping gene ACTB). Bottom panels: Visualization of gene expression level using Archer Analysis software. version 4
High Yield of Clinically Significant Fusions
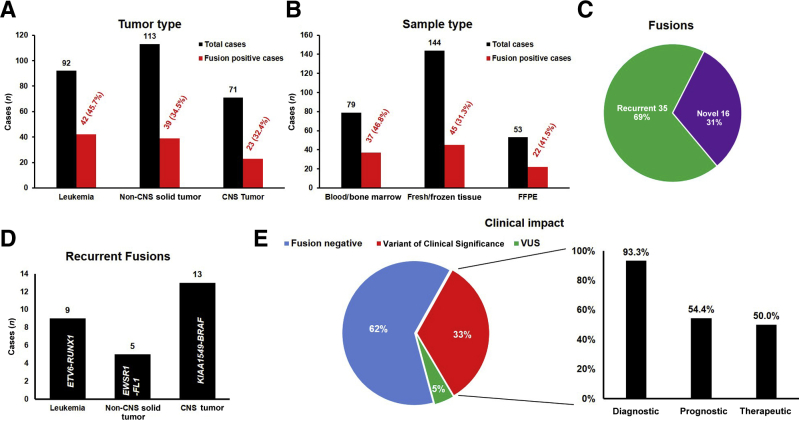
After completing the validation study with 60 samples, CHOP Fusion panel was evaluated for clinical utility using 276 consecutive clinical cancer samples, including 92 leukemias, 113 non-CNS solid tumors, and 71 CNS tumors. Of the 276 samples, 104 (37.7%) were found to harbor at least one fusion gene. Although a higher percentage (45.7%) of leukemia patients carrying positive findings compared with that of non-CNS solid tumor (34.5%) and CNS tumor (32.4%) patients was observed, there was no statistically significant difference among the three groups (Figure 4A). Of the 53 FFPE samples, 22 were positive for a fusion (41.5%), which was a similar detection rate when compared with BM/PB (37/79; 46.8%; P > 0.05) and fresh/frozen tissues (45/144; 31.2%; P > 0.05) (Figure 4B). Among the 51 unique fusions, 35 were known recurrent fusions and the remaining 16 had not been reported in the literature at the time of detection (Figure 4C). The fusion with highest frequency was ETV6-RUNX1 in leukemia (9 examples), EWSR1-FL1 in non-CNS solid tumors (5 examples), and KIAA1549-BRAF in CNS tumors (13 examples) (Figure 4D), consistent with the high frequencies of associated diseases [ie, acute lymphoblastic leukemia (ALL), Ewing sarcoma, and low-grade glioma in pediatric patients]. Of patients, 33% (90/276) were found to carry clinically significant fusions based on the Association for Molecular Pathology/American Society of Clinical Oncology/College of American Pathologists guidelines for somatic variant interpretation and reporting.25 Of patients, 5% (14/276) had fusions of uncertain clinical significance. Among the patients harboring fusions of clinical significance, 50.0% (45/90) had fusions impacting patient treatment, 93.3% (84/90) had fusions providing genomic evidence for diagnosis, and 54.4% (49/90) had fusions of prognostic significance (Figure 4E and Supplemental Table S2).
Figure 4.
Clinical utility of Children's Hospital of Philadelphia (CHOP) Fusion panel in 276 clinical cases. A: Fusion-positive cases in different cancer groups: leukemia, non–central nervous system (CNS) solid tumors, and CNS tumors. B: Fusion-positive cases in different sample types: bone marrow/blood, fresh/frozen tissue, and formalin-fixed, paraffin-embedded (FFPE) samples. C: Distribution of recurrent and novel fusions detected by CHOP Fusion panel. D: Most abundant recurrent fusions detected in leukemia, non-CNS tumors, and CNS tumors. E: Clinical significance of all fusions detected. VUS, variant of uncertain significance.
Discussion
As an increasing number of clinically significant fusions are being identified, and the recognition that a cancer gene may have a variety of fusion partners with varying break points, an efficient method allowing the detection of many recurrent and novel fusions in different tumors simultaneously is critically important. We developed and clinically validated a large custom-designed fusion assay, CHOP Fusion panel, which covers 106 major fusion-associated cancer genes and 586 known fusion transcripts and is capable of identifying potential novel fusions using NGS-based RNA sequencing. To the best of our knowledge, this is the largest pan-cancer fusion panel published so far. Although the panel is clinically validated for pediatric cancers, it has broad applicability for most adult cancer–related fusions as well.
CHOP Fusion Panel Demonstrates Excellent Analytic Performance on Different Tissues
CHOP Fusion panel demonstrated outstanding analytical performance characteristics. Of the 60 validation samples, no fusions were detected in three negative controls and all previously identified fusions were correctly detected. Ten additional fusions, including three from Children's Oncology Group samples and seven from 23 clinical samples, were orthogonally confirmed by RT-PCR. These results demonstrate 100% specificity and 100% sensitivity of the assay for the validation samples. To assess the detection limit of the assay, serial dilutions of a sample with KMT2A-ARHGEF12 fusion were performed; the assay could detect this fusion with 1% of fusion-positive RNA (Figure 2D). It is important to point out that this fusion with average unique supporting reads was purposely chosen for the detection limit study. Although this detection limit represents most of the fusions, the detection limit could be <1% for fusions with higher levels of expression, such as ETV6-RUNX1, or may not be as low as 1% for fusions with a lower-level expression, such as BCL6-associated fusions. Although CHOP Fusion panel demonstrates an excellent detection limit, it is not recommended for quantitative monitoring of minimal residual disease for clinical decision making in its current design. However, the assay provides detailed sequence information about the fusions detected, enabling the design of personalized RT-qPCR testing to trace treatment response, monitor molecular minimal residual disease, and identify early disease relapse.26
FFPE tissue specimens are one of the most common types of tissues for genomic profiling. FFPE samples with different levels of degradation were selectively tested to determine how well CHOP Fusion panel performs with FFPE samples and the minimum RNA quality required. The results showed a comparable read-on-target ratio among the libraries generated from BM/PB, fresh/frozen tissue, and FFPE samples (Figure 2B), indicating excellent enrichment of targeted transcripts regardless of RNA sources. The library complexity of FFPE samples was generally poorer than that of other specimen types, which was reflected by the significantly higher deduplication ratio (Figure 2C). Nonetheless, the fusion panel was able to detect fusions in 41.5% of FFPE clinical samples, comparable to the detection rates of other sample types (Figure 4B). It is worth mentioning that samples C4 and C6 had poor RNA quality, evidenced by a high presequencing qPCR value (Ct values, 32.0 and 31.6, respectively; laboratory cutoff Ct, ≤28.5) and a high deduplication ratio (>40; laboratory cutoff, ≤20); nevertheless, CIC-DUX4 and KIAA1549-BRAF were identified with 128 and 14 unique fusion-supporting reads, respectively, as well as 4 and 10 unique fusion-supporting starting sites in the respective samples.
Open-Ended Anchored Multiplex PCR Technology Leads to High Yield of Clinically Significant Fusions
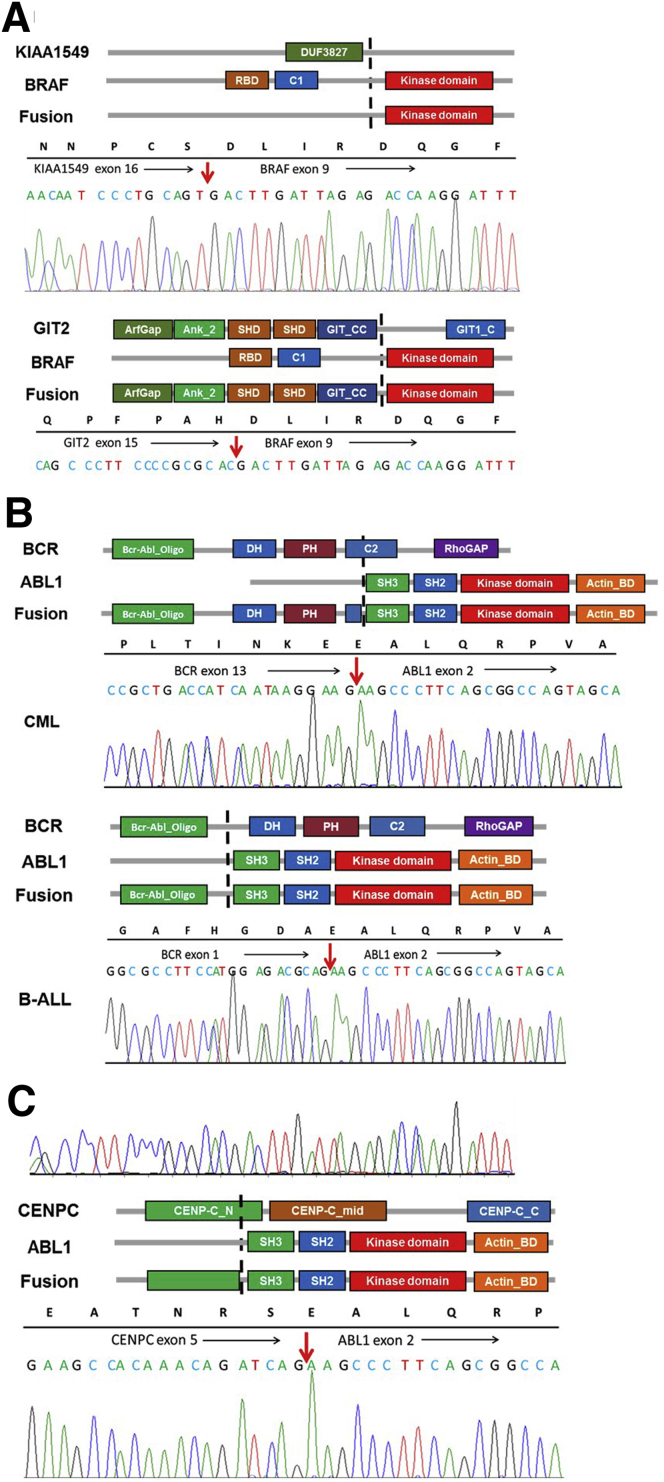
Unlike RT-PCR or NGS technologies that require opposing primers, Anchored Multiplex PCR technology uses unidirectional gene-specific primers on the major translocation partner genes, allowing for the detection of multiple different fusions partners. For example, KMT2A has >100 different known translocation partners with numerous translocation break points. Targeting all known translocation break points on the KMT2A gene using the Anchored Multiplex PCR technology can theoretically detect all known KMT2A-associated fusions. In addition, the open-ended nature of the technology enables the detection of not only previously described fusions but also previously unrecognized fusions, leading to significantly increased detection rates. Of 51 unique fusions identified in this study, 16 (31.4%) had not been reported at the time of detection (Figure 4C). In addition, CHOP Fusion panel provides detailed fusion information (ie, where the two genes fuse together), as well as their sequence information, which can be important for determining the clinical significance of novel fusions. If novel fusions identified contain protein domains with well-studied functions in the recurrent fusions, the function of the novel fusions can be reasonably assumed. KIAA1549-BRAF is the most common fusion gene in pediatric low-grade gliomas, predominantly associated with pilocytic astrocytoma (70%). This fusion gene keeps the C-terminal serine/threonine-protein kinase B-Raf (BRAF) protein kinase domain with a substituted N-terminal region missing the BRAF autoregulatory domain, resulting in constitutive activation of the BRAF kinase.12 Although most BRAF fusions in pilocytic astrocytoma were KIAA1549-BRAF, with the one combining KIAA1549 exon 16 to BRAF exon 9 (KIAA1549exon16-BRAFexon9) the most common, CHOP Fusion panel also detected a novel fusion, GIT2exon15-BRAFexon9, in a patient suspected of having pilocytic astrocytoma (Figure 5A). The GIT2-BRAF fusion retained the BRAF kinase domain and lost the N-terminal BRAF autoregulatory domain, presumably leading to constitutive activation of the BRAF kinase. A similar fusion, GIT2exon14-BRAFexon9, was later reported by Helgager et al27 in a patient with pilocytic astrocytoma. We have since detected additional novel BRAF fusions in pediatric low-grade gliomas (F.L., K.C., L.F.S., M.M.L., unpublished data). Another example was a CENPC-ABL1 fusion in a patient with ALL (Figure 5, B and C), which was later reported in a Children's Oncology Group study on high-risk B-cell ALL.28 Because all genes in this panel are involved in recurrent fusions associated with cancer development/progression, the significance of the novel fusions (fusions of these genes with new partners) detected by our panel can be reasonably presumed in most cases.
Figure 5.
Schematic diagram of protein domains of selected fusions, detailed fusion sequences, and Sanger confirmation. A: KIAA1549-BRAF and GIT2-BRAF in pilocytic astrocytoma patients. B: Two different BCR-ABL1 fusions in a chronic myeloid leukemia (CML) and a B-cell acute lymphoblastic leukemia (B-ALL) patient. C:CENPC-ABL1 in a B-ALL patient. Black arrows indicate the direction of reading frame; red arrows, the break points on mRNA level.
CHOP Fusion Panel Provides Genomic Evidence That Impacts Clinical Care
After completion of the assay validation, 276 pediatric tumors were tested with CHOP Fusion panel, and 106 fusions in 104 patients were detected, 84.9% (90/106) of which were classified as clinically significant. Of the clinically significant fusions, 50% (45/90) impacted patient treatment, including potential targeted therapy in 40 cases, such as targeting the Janus kinase (JAK)-STAT pathway in Philadelphia chromosome–like ALL (Ph-like ALL) with EPOR-associated fusions (Figure 4E). Of the clinically significant fusions, 93% (84/90) provided genomic evidence for disease diagnosis (Figure 4E), including nine cases (10%) in which the results altered the original diagnosis. For example, a novel EWSR1-CREB3L3 fusion was found in a patient diagnosed with atypical Ewing sarcoma, which led to the change of diagnosis to sclerosing epithelioid fibrosarcoma and an altered treatment plan for the patient. Of the clinically significant fusions, 54% (49/90) were of prognostic significance (Figure 4E), such as TCF3-HLF fusions associated with extremely poor outcomes and ETV6-RUNX1 fusions related to good prognosis in ALL. The information not only aided in stratifying patient risk, but also helps with treatment decision making.
Because of the nature of RNA sequencing, Ig gene (IGH, IGK, and IGL) associated fusions may or may not be detected by CHOP Fusion panel, depending on the break points on these gene. These fusions take advantage of the active promoter or enhancer of the Ig genes to increase the expression of the partner genes.29, 30, 31 To detect these fusions, the partner genes, such as MYC, CRLF2, and EPOR genes, with gene-specific primers to evaluate gene expression were heavily targeted (Figure 3). This assay not only successfully revealed the overexpression of CRLF2 and EPOR in fusion-positive cases, but also provided consistent quantitative expression comparable to the result of qPCR (Figure 3). Suspected fusions should be confirmed using clinically validated orthogonal methods. In addition, CHOP Fusion panel can provide the gene expression signature for research purposes.
In summary, CHOP Fusion panel is an accurate, efficient, and cost-effective NGS-based RNA sequencing assay for identifying fusion transcripts in pediatric cancers. It can replace most of currently used fluorescence in situ hybridization and RT-PCR assays for fusion detection and molecular diagnosis of childhood cancers.
Acknowledgments
We thank the Children's Oncology Group, especially Dr. Julie Gastier-Foster, for providing valuable validation samples; and Dr. Robert Domes for thoughtful review and feedback on the manuscript.
Footnotes
Supported by the Department of Pathology and Laboratory Medicine and Center for Childhood Cancer Research, Children's Hospital of Philadelphia; National Cancer Institute grant 1K08CA184418 (S.K.T.); and NIH grant U2CHL138346 (A.C.R., P.B.S., and M.M.L.).
F.C. and F.L. contributed equally to this work.
Disclosures: S.P.H. has received honoraria from Amgen, Jazz Pharmaceuticals, and Erytech, and consulting fees from Novartis; S.K.T. has received research funding from Incyte Corp. and Gilead Sciences and is on the Scientific Advisory Board for Aleta Biotherapeutics.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.05.006.
Supplemental Data
Additional novel fusions identified in validation samples. Black arrows indicate the direction of reading frame; red arrows, the break points on the mRNA level. UTR, untranslated region.
References
- 1.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Mertens F., Johansson B., Fioretos T., Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 3.Teixeira M.R. Recurrent fusion oncogenes in carcinomas. Crit Rev Oncog. 2006;12:257–271. doi: 10.1615/critrevoncog.v12.i3-4.40. [DOI] [PubMed] [Google Scholar]
- 4.Annala M.J., Parker B.C., Zhang W., Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340:192–200. doi: 10.1016/j.canlet.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M.M., Ewton A.A., Smith J.L. Using cytogenetic rearrangements for cancer prognosis and treatment (pharmacogenetics) Curr Genet Med Rep. 2013;1:99–112. [Google Scholar]
- 6.Melnick A., Licht J.D. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 7.McCulloch D., Brown C., Iland H. Retinoic acid and arsenic trioxide in the treatment of acute promyelocytic leukemia: current perspectives. Onco Targets Ther. 2017;10:1585–1601. doi: 10.2147/OTT.S100513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Druker B.J. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 9.Cocco E., Scaltriti M., Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15:731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laetsch T.W., DuBois S.G., Mascarenhas L., Turpin B., Federman N., Albert C.M., Nagasubramanian R., Davis J.L., Rudzinski E., Feraco A.M., Tuch B.B., Ebata K.T., Reynolds M., Smith S., Cruickshank S., Cox M.C., Pappo A.S., Hawkins D.S. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018;19:705–714. doi: 10.1016/S1470-2045(18)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J.E., Shah R.B., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 12.Jones D.T., Kocialkowski S., Liu L., Pearson D.M., Backlund L.M., Ichimura K., Collins V.P. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes T., Deininger M., Hochhaus A., Branford S., Radich J., Kaeda J., Baccarani M., Cortes J., Cross N.C., Druker B.J., Gabert J., Grimwade D., Hehlmann R., Kamel-Reid S., Lipton J.H., Longtine J., Martinelli G., Saglio G., Soverini S., Stock W., Goldman J.M. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson P., Gao J., Chang K.S., Look T., Whisenant E., Raimondi S., Lasher R., Trujillo J., Rowley J., Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 15.Manola K.N. Cytogenetics of pediatric acute myeloid leukemia. Eur J Haematol. 2009;83:391–405. doi: 10.1111/j.1600-0609.2009.01308.x. [DOI] [PubMed] [Google Scholar]
- 16.Rubnitz J.E., Crist W.M. Molecular genetics of childhood cancer: implications for pathogenesis, diagnosis, and treatment. Pediatrics. 1997;100:101–108. doi: 10.1542/peds.100.1.101. [DOI] [PubMed] [Google Scholar]
- 17.Ma X., Liu Y., Liu Y., Alexandrov L.B., Edmonson M.N., Gawad C., Zhou X., Li Y., Rusch M.C., Easton J., Huether R., Gonzalez-Pena V., Wilkinson M.R., Hermida L.C., Davis S., Sioson E., Pounds S., Cao X., Ries R.E., Wang Z., Chen X., Dong L., Diskin S.J., Smith M.A., Guidry Auvil J.M., Meltzer P.S., Lau C.C., Perlman E.J., Maris J.M., Meshinchi S., Hunger S.P., Gerhard D.S., Zhang J. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555:371–376. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gainor J.F., Shaw A.T. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist. 2013;18:865–875. doi: 10.1634/theoncologist.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertens F., Tayebwa J. Evolving techniques for gene fusion detection in soft tissue tumours. Histopathology. 2014;64:151–162. doi: 10.1111/his.12272. [DOI] [PubMed] [Google Scholar]
- 20.Mino-Kenudson M., Chirieac L.R., Law K., Hornick J.L., Lindeman N., Mark E.J., Cohen D.W., Johnson B.E., Janne P.A., Iafrate A.J., Rodig S.J. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qadir M.A., Zhan S.H., Kwok B., Bruestle J., Drees B., Popescu O.E., Sorensen P.H. ChildSeq-RNA: a next-generation sequencing-based diagnostic assay to identify known fusion transcripts in childhood sarcomas. J Mol Diagn. 2014;16:361–370. doi: 10.1016/j.jmoldx.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Xu T., Wang H., Huang X., Li W., Huang Q., Yan Y., Chen J. Gene fusion in malignant glioma: an emerging target for next-generation personalized treatment. Transl Oncol. 2018;11:609–618. doi: 10.1016/j.tranon.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3: new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., Nikiforova M.N. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Y.Y., Stern J.W., Jubelirer T.F., Wertheim G.B., Lin F., Chang F., Gu Z., Mullighan C.G., Li Y., Harvey R.C., Chen I.M., Willman C.L., Hunger S.P., Li M.M., Tasian S.K. Clinical efficacy of ruxolitinib and chemotherapy in a child with Philadelphia chromosome-like acute lymphoblastic leukemia with GOLGA5-JAK2 fusion and induction failure. Haematologica. 2018;103:e427–e431. doi: 10.3324/haematol.2018.192088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helgager J., Lidov H.G., Mahadevan N.R., Kieran M.W., Ligon K.L., Alexandrescu S. A novel GIT2-BRAF fusion in pilocytic astrocytoma. Diagn Pathol. 2017;12:82. doi: 10.1186/s13000-017-0669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reshmi S.C., Harvey R.C., Roberts K.G., Stonerock E., Smith A., Jenkins H., Chen I.M., Valentine M., Liu Y., Li Y., Shao Y., Easton J., Payne-Turner D., Gu Z., Tran T.H., Nguyen J.V., Devidas M., Dai Y., Heerema N.A., Carroll A.J., 3rd, Raetz E.A., Borowitz M.J., Wood B.L., Angiolillo A.L., Burke M.J., Salzer W.L., Zweidler-McKay P.A., Rabin K.R., Carroll W.L., Zhang J., Loh M.L., Mullighan C.G., Willman C.L., Gastier-Foster J.M., Hunger S.P. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children's Oncology Group. Blood. 2017;129:3352–3361. doi: 10.1182/blood-2016-12-758979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Look A.T. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 30.Dyer M.J., Akasaka T., Capasso M., Dusanjh P., Lee Y.F., Karran E.L., Nagel I., Vater I., Cario G., Siebert R. Immunoglobulin heavy chain locus chromosomal translocations in B-cell precursor acute lymphoblastic leukemia: rare clinical curios or potent genetic drivers? Blood. 2010;115:1490–1499. doi: 10.1182/blood-2009-09-235986. [DOI] [PubMed] [Google Scholar]
- 31.Mussolin L., Basso K., Pillon M., D'Amore E.S., Lombardi A., Luzzatto L., Zanesco L., Rosolen A. Prospective analysis of minimal bone marrow infiltration in pediatric Burkitt's lymphomas by long-distance polymerase chain reaction for t(8;14)(q24;q32) Leukemia. 2003;17:585–589. doi: 10.1038/sj.leu.2402828. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional novel fusions identified in validation samples. Black arrows indicate the direction of reading frame; red arrows, the break points on the mRNA level. UTR, untranslated region.