ABSTRACT
The endoplasmic reticulum (ER) is the main site of cellular protein and calcium homeostasis, as well as lipid synthesis in eukaryotic cells. Reticulophagy is the selective clearance and degradation of ER components and membranes by the cellular autophagy machinery. Recently, 2 groups (the laboratories of Noboru Mizushima and Wade Harper) independently identified the previously uncharacterized protein TEX264 (testis expressed gene 264) as a major receptor for selective reticulophagy in mammalian cells. Here we highlight and integrate the major findings of their recent work.
Abbreviations: AIM: Atg8-interacting motif; AP-MS: affinity purification-mass spectrometry; ATL3: atlastin GTPase 3; Baf A1: bafilomycin A1; CCPG1: cell cycle progression 1; CRISPR: clustered regularly interspaced short palindromic repeats; GABARAP: gamma-aminobutyric acid receptor associated protein; GFP: green fluorescent protein; GyrI: gyrase inhibitor; IDR: intrinsically disordered region; IP: immunoprecipitation; KO: knockout; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule-associated protein 1 light chain 3; MEF: mouse embryonic fibroblast; MS: mass spectrometry; MTOR: mechanistic target of rapamycin kinase; RB1CC1/FIP200: RB1-inducible coiled-coil 1; RFP: red fluorescent protein; RNAi: RNA interference; RTN3: reticulon 3; RTN3L: long isoform of RTN3; siRNA: small interfering RNA; SARS: selective autophagy receptors; ss: signal sequence; TEM: transmission electron microscopy, TEX264: testis expressed gene 264; TMT: tandem mass tagging
KEYWORDS: Autophagy, lysosome, macroautophagy, selective autophagy, stress
Reticulophagy is the selective clearance and degradation of the endoplasmic reticulum (ER) by the cellular macroautophagy/autophagy machinery [1]. In mammalian cells, phosphati-dylinositol-3-phosphate-enriched ER subdomains are the initial membrane donor sites for autophagosome biogenesis [2,3]. During selective autophagy, unique cargo is targeted for autophagic degradation through receptor-mediated interactions between selective autophagy receptors (SARS) and Atg8-family proteins (MAP1LC3A/B/B2/C and GABARAP/L1/L2) via LC3-interacting regions (LIRs) of the receptors. The canonical LIRs consist of a consensus motif [W/F/Y]-X-X-[L/I/V], where X is any amino acid [4,5].
Thus far, 2 reticulophagy receptors – Atg39 and Atg40 – have been identified in the yeast Saccharomyces cerevisiae [6]. Atg39 is required for reticulophagy of perinuclear ER, whereas Atg40 is required for reticulophagy of cortical and cytoplasmic ER, and shows the most functional similarity to the mammalian receptor RETREG1/FAM134B (further described below) [6]. Both Atg39 and Atg40 contain verified Atg8-interacting motifs (AIMs) and interact with the selective autophagy scaffold protein Atg11 in yeast. Atg39 and Atg40 function as reticulophagy receptors in response to nitrogen starvation and rapamycin treatment. An additional factor, Lnp1, facilitates both the localization of Atg40 and ER membrane rearrangements [7]. More recently, the COPII-cargo adaptor complex Sfb3/Lst1-Sec23 was shown to function with Atg40 to selectively target ER subdomains for autophagy [8].
At least 5 different reticulophagy receptors have been identified in mammals, including RETREG1/FAM134B [9], SEC62 [10], the long (L) isoform of RTN3 (reticulon 3) [11], CCPG1 (cell cycle progression 1) [12], and more recently, ATL3 (atlastin GTPase 3) [13]. There is collective evidence that these receptors may exert spatiotemporal control over selective regions of the ER during reticulophagy. RETREG1 and RTN3L are reticulon-type proteins involved in autophagy-mediated membrane turnover of ER sheets and tubules, respectively, during nutrient deprivation [9,11]; SEC62 is an ER-resident protein, which functions in response to ER stress [10]; ATL3 mediates tubular ER fusion and degradation [13]; and CCPG1 is a resident ER membrane protein involved in the clearance and degradation of peripheral ER [12]. However, outstanding questions have remained unanswered; especially regarding reticulophagy regulation, and how certain subdomains of the ER are selectively targeted for degradation, while others are largely excluded.
Here, we highlight 2 back-to-back articles [14,15] that identified the previously uncharacterized protein TEX264 (testis expressed gene 264) as a major receptor for reticulophagy in mammalian cells. TEX264 is a single-pass transmembrane ER-resident protein, consisting of an N-terminal hydrophobic region, cytosolic gyrase inhibitor (GyrI)-like domain, and a C-terminal unstructured intrinsically disordered region (IDR) [14,15]. TEX264 is tethered to the ER by its N-terminal transmembrane segment [15]. The central GyrI-like domain and C-terminal region are in the cytosol [15].
To identify novel factors (either receptors or substrates) involved in selective autophagy, the Mizushima lab performed a differential LC3B-interactome screen with wild-type LC3B and a LIR recognition-deficient mutant (LC3BK51A) [14]. Using co-immunopreciption (co-IP) coupled with a mass spectrometry (MS) approach with either LC3B or LC3BK51A, Chino et al. identified 87 novel factors that associate with LC3B but not the LIR-deficient mutant [14]. The authors focused on further characterizing TEX264 because of its high binding score with LC3B [14]. Furthermore, the role of TEX264 in autophagy had not been previously explored.
The Harper lab identified TEX264 by global quantitative proteome analysis using tandem mass tagging (TMT) and synchronous precursor selection-tandem mass spectrometry/MS/MS (MS3) [16] during MTOR (mechanistic target of rapamycin kinase) inhibition with Torin1 or amino acid deprivation in HEK293T cells with and without ATG7 or RB1CC1 (RB1-inducible coiled-coil 1) [15]. An et al. chose to focus on TEX264, as it was previously uncharacterized and expression is reduced in response to amino acid deprivation or MTOR inhibition, in an ATG7- or RB1CC1-dependent manner, to levels comparable to the selective autophagy receptor SQSTM1 [15].
Both groups identified TEX264 as an ER-resident protein [14,15]. Using a TEX264-GFP plasmid, Chino and colleagues found that TEX264 forms punctate structures that colocalize with LC3B and fails to do so with an LC3B LIR mutant during nutrient starvation (induced by amino acid and serum deprivation) [14]. TEX264 also colocalizes with phagophore markers WIPI2 and RB1CC1, suggesting that TEX264 may function at early stages of autophagosome formation [14]. An et al. verified the localization of TEX264 to the ER through CRISPR-Cas9 genome tagging with EGFP in multiple cell types [15]. Further analyses also verified that TEX264 colocalizes with LC3B, and is therefore incorporated within autophagosomes [15].
Both groups also identified the C-terminal LIR motif (F273EEL) of TEX264 [14,15]. Chino et al. verified that C-terminal amino acids 273–276 form a bona fide LIR motif by mutating specific residues (TEX264F273A, TEX264L276A) and performing co-IPs with LC3B; the interaction was abolished with the mutants [14]. Similarly, An and colleagues examined the LIR mutant TEX264F273A, and found that the mutant fails to mobilize to punctate structures (in contrast to TEX264) when cells are starved [15].
TEX264 is trafficked from the ER to the lysosomes as part of autophagic flux [14,15]. When mouse embryonic fibroblast (MEF) cells expressing TEX264 are treated with the auto-phagy flux inhibitor bafilomycin A1 (BafA1), the frequency of TEX264-positive puncta increases, and TEX264 colocalizes with the lysosomal marker LAMP1 when cells are starved for amino acids and serum [14]. Together, Chino and co-workers suggest that TEX264 associates with forming autophagosomes through the C-terminal LIR, and undergoes lysosomal degradation during autophagy flux [14]. The authors verified that TEX264 accumulates in the presence of BafA1 and in the absence of RB1CC1 (in knockout MEF cells), supporting the conclusion that TEX264 is targeted to the lysosomes in an autophagy-dependent manner [14].
An et al. generated a TEX264 fusion with Keima (a pH-sensitive reporter that is resistant to degradation by lysosomal hydrolases [17]) to reveal that TEX264 undergoes trafficking from the ER to the lysosome during basal autophagy. This process is upregulated during conditions of nutrient deprivation or MTOR inhibition [15]. Flux of TEX264 through the autophagy pathway is dependent on components involved in the canonical pathway, including the Atg8-family protein conjugation system, the ULK1-RB1CC1 kinase complex, and the class III phosphatidylinositol 3-kinase complex [15]. The autophagy-dependent lysosomal turnover of TEX264 was confirmed to be dependent on the LIR motif [15]. The authors observed the appearance of LC3A nearby 3-way junctions of ER tubules, followed by the emergence of TEX264, and the colocalization of LC3A and TEX264 [15]. This result is intriguing because it is in agreement with a previous model for autophagosome biogenesis in mammalian cells, where the phagophore may emerge from structures resembling 3-way junctions in the ER [18].
As TEX264 is a transmembrane protein that localizes to the ER and is targeted for autophagy-dependent lysosomal degradation, Chino et al. examined whether TEX264 could be a candidate receptor for reticulophagy [14]. To test this hypothesis, the authors generated a doxycycline-inducible reticulophagy reporter comprised of an N-terminal ER signal sequence (ss), followed by tandem monomeric RFP and GFP sequences and the ER retention sequence KDEL [14]. In principle, when autophagy is activated by conditions such as amino acid deprivation, the RFP fragment is released by lysosomal hydrolases, and the GFP signal is quenched within the acidic lysosomal environment. Thus, the release of RFP can be used as a readout for reticulophagy flux. Using this method, the authors induced the reporter by doxycycline treatment and depleted TEX264 from HeLa cells using CRISPR-Cas9 [14]. They observed that in the absence of TEX264, reticulophagy (as measured by the release of free RFP with the doxycycline-inducible reporter ssRFP-GFP-KDEL) is impaired under both nutrient-replete and starvation conditions [14]. Conversely, reticulophagy flux is restored when TEX264 is reintroduced into the cells, but fails to be rescued with a LIR-deficient mutant. This result along with fluorescence microscopy data validated TEX264 as a reticulophagy receptor that interacts with LC3 [14]. Chino and colleagues note that TEX264 is more ubiquitously expressed than previously identified mammalian reticulophagy receptors [14], and a closer examination of the TEX264 distribution shows that this receptor is expressed in all primary mouse tissues [14].
To determine the relative contribution of known reticulophagy receptors, Chino et al. used small interfering RNA (siRNA)-mediated knockdown to simultaneously deplete HeLa cells of RETREG1, CCPG1, RTN3L, SEC62 and TEX264 [14]. When multiple receptors (RETREG1, CCPG1, RTN3L and SEC62, but not TEX264) are depleted, reticulophagy still occurs to significant levels (>50%) [14]. Depletion of all identified receptors concurrently significantly reduces reticulophagy to levels comparable to when a key component of the initiation complex, RB1CC1, is depleted [14]. Consistent with this finding, An et al. examine how the global proteome during amino acid starvation is affected by the loss of TEX264 [15]. The authors estimate that ~50% of reticulophagy flux during starvation in 293T cells may be attributed to TEX264 alone [15], and increased levels of TEX264 can enhance basal and starvation-induced reticulophagy in a LIR-dependent manner [15]. Collectively, these data suggest that TEX264 is a major receptor for reticulophagy in mammals [14,15].
Additionally, Chino and coworkers, determined the relative binding efficiency between TEX264 and Atg8-family proteins by co-IP, and found a preference for binding to LC3A, GABARAP, and GABARAPL1 in HEK293T cells [14]. Using unbiased affinity purification-mass spectrometry (AP-MS) followed by label-free quantification precursor ions, An et al. verified that TEX264 interacts most with LC3B and GABARAP under both untreated and amino-acid starved conditions in a LIR-dependent manner [15]. However, when Atg8-family proteins are expressed at comparable levels, TEX264 associates with LC3A, LC3B, and GABARAPL1 [15] Together, these data indicate that the in vivo association between TEX264 and Atg8-family proteins may be dependent on their differential expression in various cell types [14,15].
Transmission electron microscopy (TEM) analysis by the Mizushima lab showed the presence of ribosomes spaced between the inner phagophore membrane and ER membrane [14]. Given the fact that reticulophagy receptors link both membranes, and the size of a ribosome is approximately 20 nm, the authors considered how a small molecule such as TEX264 can span the distance of more than 20 nm between the ER and phagophore membrane [14]. Chino et al. assumed that the answer to this question can be found in the presence of an unfolded structure [14]. Indeed, the PSIPRED (http://bioinf.cs.ucl.ac.uk/index.php?id=780 [19]) algorithm used by the Mizushima lab predicts a long IDR at the C terminus of TEX264 [14], in agreement with An et al. who also predicted the C-terminal unstructured region (residues 185–313) [15]. IDRs are dynamic and flexible protein regions lacking stable secondary and/or tertiary structures, and thereby serving as regulatory domains for critical cellular processes [20,21]. These regions also harbor short linear motifs that are important protein interaction modules through which multiple proteins with very diverse amino acid sequences can fit a binding pocket on one globular surface [22]. The LIR/AIM is one example of a short linear motif [23]. Therefore, it is not surprising that the LIR motifs of the other reticulophagy receptors, CCPG1, RETREG1, RNT3L, and SEC62, are also located in the IDRs predicted by PSIPRED [14].
To probe the idea that the length of the TEX264 IDR is essential for overcoming the spatial gap resulting from the size of ribosomes, in order to link the ER and phagophore membrane, Chino et al. generated 2 truncation mutants both containing the LIR motif but shortened to various degrees, by either 32 (∆230–261) or 56 (∆206–261) amino acids. Only the mutant carrying the smaller truncation is able to restore reticulophagy and to colocalize with LC3 [14]. To probe the specificity of the amino acid sequence in the TEX264 IDR, the authors inserted a portion of the ATG13 IDR (amino acid residues 191–248 [24]) into TEX264, by replacing the TEX264 segment 206–261. This approach structurally retained an IDR, but changed its amino acid composition. The reticulophagy function and LC3 colocalization of the fused mutant was rescued [14]. This intriguing result supports the conclusion that the length of the IDR, not the specific amino acid sequence, is essential for bridging the spatial gap that exists in between 2 globular surfaces, the TEX264 GyrI-like domain in the proximity of the ER membrane on one side, and the LIR-binding hydrophobic pocket of LC3 conjugated to the phagophore membrane on the other side (Figure 1) [14]. Without this molecular mechanism involving an IDR linker, TEX264 cannot function as a reticulophagy receptor [14].
Figure 1.
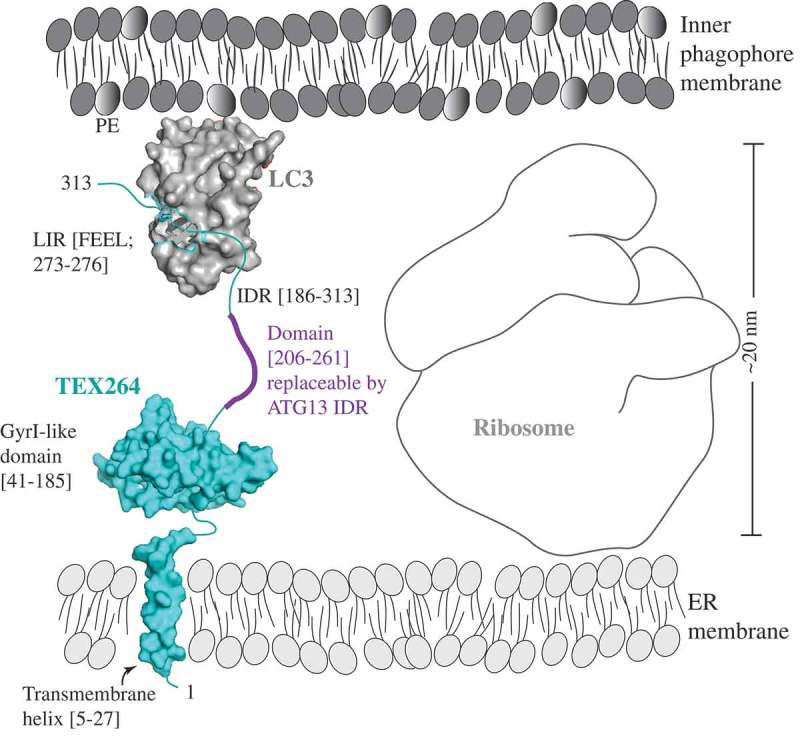
A model of the reticulophagy receptor TEX264 (teal) attached to the ER membrane via the transmembrane helix (residues 5–27). The gyrase inhibitor (GyrI)-like domain (residues 41–185) of TEX264 is followed by the IDR (residues 186–313) that binds via the LIR motif (F273EEL) to LC3 (gray), which is conjugated to PE at the inner phagophore membrane. The ribosome size of approximately 20 nm creates the spatial gap between the ER and inner phagophore membrane. The IDR of the small molecule TEX264 is essential for bridging this gap and for the function of TEX264 as a reticulophagy receptor. The length of the IDR linker (residues 206–261), rather than the specific amino acid sequence, is a critical factor, as shown by the finding that this segment of the TEX264 IDR is functionally replaceable by the ATG13 IDR (residues 191–248). To visualize the LIR-LC3 complex, the crystal structure of LC3 bound to the LIR of SQSTM1 was used (PDB ID: 2ZJD). The transmembrane helix and GyrI-like domain of TEX264 have been modeled using the Phyre2 server with 72% and 99.7% confidence by the single highest-scoring template c3a0hJ and d1jyha, respectively.
An et al. propose a model for the role of TEX264 during reticulophagy [15]. In this model, the expansion of the phagophore during the early stages of autophagy enables Atg8-family proteins to interact with TEX264 on ER tubules through trans membrane interactions [15]. However, as the authors note, the mechanisms contributing to the separation of the forming autophagosome from the ER and subsequent phagophore closure have yet to be defined [15]. Chino et al. propose that the transmembrane receptors (TEX264, CCPG1 and SEC62) may function to link ER and phagophore membranes, whereas reticulon-type receptors (such as RETREG1 and RTN3L) may serve to remodel or fragment the ER prior to its engulfment by phagophores [14]. Therefore, the authors suggest that these receptors may function in a hierarchical order [14]. In contrast, An and coworkers suggest that TEX264 functions independently, and that there may be functional redundancy between known reticulophagy receptors [15], consistent with what others have noted [9–12]. Together, these findings provide insight into the regulation of reticulophagy and how selectivity of unique ER subdomains for degradation may be achieved.
Funding Statement
This work was supported by the National Institute of General Medical Sciences [GM131919].
Acknowledgments
The authors apologize to those whose work was not included here due to space limitations.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. PubMed PMID: 26799652; PubMed Central PMCID: PMCPMC4835977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nascimbeni AC, Giordano F, Dupont N, et al. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. Embo J. 2017. July 14;36(14):2018–2033. PubMed PMID: 28550152; PubMed Central PMCID: PMCPMC5509996. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008. August 25;182(4):685–701. PubMed PMID: 18725538; PubMed Central PMCID: PMCPMC2518708. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008. December;13(12):1211–1218. PubMed PMID: 19021777; eng. [DOI] [PubMed] [Google Scholar]
- [5].Birgisdottir AB, Lamark T, Johansen T.. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013. August 1;126(Pt 15):3237–3247. PubMed PMID: 23908376; eng. [DOI] [PubMed] [Google Scholar]
- [6].Mochida K, Oikawa Y, Kimura Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015. June 18;522(7556):359–362. PubMed PMID: 26040717. eng. [DOI] [PubMed] [Google Scholar]
- [7].Chen S, Cui Y, Parashar S, et al. ER-phagy requires Lnp1, a protein that stabilizes rearrangements of the ER network. Proc Natl Acad Sci U S A. 2018. July 3;115(27):E6237–e6244. PubMed PMID: 29915089; PubMed Central PMCID: PMCPMC6142256. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cui Y, Parashar S, Zahoor M, et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science (New York, NY). 2019. July 5;365(6448):53–60. PubMed PMID: 31273116; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Khaminets A, Heinrich T, Mari M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015. June 18;522(7556):354–358. PubMed PMID: 26040720. eng. [DOI] [PubMed] [Google Scholar]
- [10].Fumagalli F, Noack J, Bergmann TJ, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016. November;18(11):1173–1184. PubMed PMID: 27749824. eng. [DOI] [PubMed] [Google Scholar]
- [11].Grumati P, Morozzi G, Holper S, et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 2017. June 15;6 PubMed PMID: 28617241; PubMed Central PMCID: PMCPMC5517149. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smith MD, Harley ME, Kemp AJ, et al. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell. 2018. January 22;44(2):217–232.e11. PubMed PMID: 29290589; PubMed Central PMCID: PMCPMC5791736. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen Q, Xiao Y, Chai P, et al. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr Biol. 2019. March 4;29(5):846–855.e6. PubMed PMID: 30773365. eng. [DOI] [PubMed] [Google Scholar]
- [14].Chino H, Hatta T, Natsume T, et al. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol Cell. 2019. June 6;74(5):909–921.e6. PubMed PMID: 31006538. eng. [DOI] [PubMed] [Google Scholar]
- [15].An H, Ordureau A, Paulo JA, et al. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol Cell. 2019. June 6;74(5):891–908.e10. PubMed PMID: 31006537. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McAlister GC, Nusinow DP, Jedrychowski MP, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014. July 15;86(14):7150–7158. PubMed PMID: 24927332; PubMed Central PMCID: PMCPMC4215866. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Katayama H, Kogure T, Mizushima N, et al. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011. August 26;18(8):1042–1052. PubMed PMID: 21867919; eng. [DOI] [PubMed] [Google Scholar]
- [18].Hayashi-Nishino M, Fujita N, Noda T, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009. December;11(12):1433–1437. PubMed PMID: 19898463; eng. [DOI] [PubMed] [Google Scholar]
- [19].McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000. April;16(4):404–405. PubMed PMID: 10869041; eng. [DOI] [PubMed] [Google Scholar]
- [20].Darling AL, Uversky VN. Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front Genet. 2018;9:158 PubMed PMID: 29780404; PubMed Central PMCID: PMCPMC5945825. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Uversky VN. Functional roles of transiently and intrinsically disordered regions within proteins. Febs J. 2015. April;282(7):1182–1189. PubMed PMID: 25631540; eng. [DOI] [PubMed] [Google Scholar]
- [22].Van Roey K, Uyar B, Weatheritt RJ, et al. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem Rev. 2014. July 9;114(13):6733–6778. PubMed PMID: 24926813; eng. [DOI] [PubMed] [Google Scholar]
- [23].Popelka H, Klionsky DJ. Analysis of the native conformation of the LIR/AIM motif in the Atg8/LC3/GABARAP-binding proteins. PubMed PMID: 26565669; PubMed Central PMCID: PMCPMC4835208. eng Autophagy. 2015;1112:2153–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yamamoto H, Fujioka Y, Suzuki SW, et al. The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev Cell. 2016. July 11;38(1):86–99. PubMed PMID: 27404361. [DOI] [PubMed] [Google Scholar]


