Abstract
Southern African indigenous groups, traditionally hunter-gatherers (San) and herders (Khoekhoe), are commonly referred to as “Khoe-San” populations and have a long history in southern Africa. Their ancestors were largely isolated up until ∼2,000 years ago before the arrival of pastoralists and farmers in southern Africa. Assessing relationships among regional Khoe-San groups has been challenging due to admixture with immigrant populations that obscure past population affinities and gene flow among these autochthonous communities. We re-evaluate a combined genome-wide data set of previously published southern Africa Khoe-San populations in conjunction with novel data from Khoe-San individuals collected in Xade (Central Kalahari Game Reserve, Botswana) prior to their resettlement outside the reserve. After excluding regions in the genome that trace their ancestry to recent migrant groups, the genetic diversity of 20 Khoe-San groups fitted an isolation-by-distance model. Even though isolation-by-distance explained most genetic affinities between the different autochthonous groups, additional signals of contact between Khoe-San groups could be detected. For instance, we found stronger genetic affinities, than what would be explained by isolation-by-distance gene flow, between the two geographically separated Khoe-San groups, who speak branches of the Kx’a-language family (ǂHoan and Ju). We also scanned the genome-wide data for signals of adaptive gene flow from farmers/herders into Khoe-San groups and identified a number of genomic regions potentially introduced by the arrival of the new groups. This study provides a comprehensive picture of affinities among Khoe-San groups, prior to the arrival of recent migrants, and found that these affinities are primarily determined by the geographic landscape.
Keywords: Khoe-San, southern Africa, population structure, isolation-by-distance, adaptive gene-flow
Introduction
Southern African Khoe-San populations collectively refer to hunter-gatherer (San) and herder (Khoekhoe) communities who all speak Khoisan languages. The Khoe-San populations of southern Africa have a complex and enigmatic prehistory, but recent advances in archeological, anthropological, linguistic, and genetic research started to clarify their history (Schuster et al. 2010; Henn et al. 2011; Pickrell et al. 2012; Schlebusch et al. 2012, 2017; Mitchell and Lane 2013; Güldemann and Fehn 2014; Kim et al. 2014). The Khoisan linguistic sprachbund (where language features are shared due to horizontal transfer), includes various “click” languages, however, the major families within the sprachbund are linguistically unrelated to each other. Aside from the Hadza and Sandawe click languages from eastern Africa, all Khoisan languages are found in the southern parts of Africa and encompass three distinct, unrelated language families: Kx’a, (formerly called Northern Khoisan), Tuu (formerly Southern Khoisan), and Khoe-Kwadi (formerly Central Khoisan) (Güldemann and Fehn 2014) (supplementary fig. S1 and table S1, Supplementary Material online).
Kx’a can be subdivided into two main branches: Ju, present across the northwest Kalahari Basin (in northern Namibia, southern Angola and northern Botswana) and ǂHoan, spoken only in a small area in the central Kalahari (central Botswana), geographically isolated from the Ju languages (supplementary fig. S1, Supplementary Material online). Despite having a wider geographic distribution in the past, the Tuu languages now are restricted to Taa speakers living in the southern Kalahari (Botswana) and the ǂKhomani in the north of Northern Cape Province (South Africa) (supplementary fig. S1 and table S1, Supplementary Material online). Current-day southern Botswana, southern Namibia, and South Africa were once areas where the Tuu language family were widely spoken (Güldemann 2005; Güldemann and Fehn 2014, supplementary fig. S1 and table S1, Supplementary Material online). In South Africa, it is known that descendants of Tuu-speaker groups have adopted non-Khoisan languages and/or have been culturally assimilated by the extant surrounding populations and the mixed-ancestry Coloured population (Barnard 1992; Schlebusch et al. 2010, 2011, 2016).
Khoe-Kwadi is a Khoisan language family that is linguistically completely unrelated to the Kx’a and Tuu families (Güldemann 2008) and can be sub-structured in three language branches: Kalahari-Khoe, Khoekhoe, and Kwadi (extinct) (supplementary table S1, Supplementary Material online). Khoekhoe populations were pastoralists in historical times and the Kwadi people were proposed to have been pastoralists (Barnard 1992; Güldemann 2008; Sadr 2015), whereas the Kalahari-Khoe were historically hunter-gatherers. There are still Kalahari-Khoe groups today who hunt and gather. The Kwadi language disappeared over the last few decades and the Khoekhoe speakers that retain their language today are limited to the Nama and Hai‖om from Namibia.
On a genetic level, southern African Khoe-San groups carry high frequencies of the highly divergent L0d mitochondrial DNA lineage (Behar et al. 2008; Barbieri et al. 2013), and they have the greatest level of genetic (autosomal) diversity among worldwide populations (Jakobsson et al. 2008; Tishkoff et al. 2009; Schuster et al. 2010; Henn et al. 2011; Pickrell et al. 2012; Schlebusch et al. 2012). Their genetic variation, however, appears to be highly structured among communities (Henn et al. 2011; Pickrell et al. 2012; Schlebusch et al. 2012; Uren et al. 2016; Montinaro et al. 2017). Previous genetic research indicates that Khoe-San genetic variation is influenced by geography (Pickrell et al. 2012; Schlebusch et al. 2012; Montinaro et al. 2017) and ecology (Uren et al. 2016). Other factors, such as language and subsistence practices, were also reported to have contributed to the genetic variation among Khoe-San groups (Schlebusch et al. 2012; Montinaro et al. 2017). Many of these studies, however, did not account for admixture from neighboring groups into Khoe-San groups and therefore inferences might have been skewed by uneven amounts of admixture from the various immigrant groups.
It is quite likely that San hunter-gatherers were the only inhabitants of southern Africa during most of the prehistory of the region (Schlebusch et al. 2017; Skoglund et al. 2017), with evidence of the arrival of external groups only in the last ∼2,000 years (Mitchell and Lane 2013; Sadr 2015). Possibly, the first immigration wave into the area was from a mixed East African-Eurasian herder group (Pickrell et al. 2012; Breton et al. 2014; Schlebusch et al. 2017; Skoglund et al. 2017). According to genetic evidence, most Khoe-San show some degree of east African admixture and Khoe-speaking groups have higher East African admixture (Breton et al. 2014; Schlebusch et al. 2017). Furthermore, Schlebusch et al. (2017) established that the Amhara from Ethiopia was the best current-day representative group of the mixed East African-Eurasian group who admixed with local San groups and introduced pastoralism to southern Africa. Of particular note is the relative high frequency of the “east African” Lactase Persistence (LP) polymorphism (C-14010) within Khoekhoe speakers (Breton et al. 2014; Macholdt et al. 2014). This LP mutation confers the ability to digest milk in adulthood and is not common in the San hunter-gatherer gene-pools. Archeological findings of pastoralism start to appear in southern Africa around 2,000 years ago (Robbins et al. 2005; Smith 2008; Pleurdeau et al. 2012), and a linguistic link has been suggested between the eastern African Khoisan-speaking Sandawe group and the proto Khoe-Kwadi language (Güldemann 2008).
Shortly after the arrival of pastoralists from east Africa, the agro-pastoral Bantu expansion reached the area at around 1,500 years ago causing a significant change in the genomic composition of southern Africans (Pickrell et al. 2012; Schlebusch et al. 2012; Petersen et al. 2013; González-Santos et al. 2015). The farming societies out-competed especially San groups who, to a large extent, were replaced and/or assimilated by farming groups (Barbieri et al. 2013; Schlebusch et al. 2016). Most recently, the European colonization of southern Africa introduced infectious diseases into the area and many Khoekhoe and San communities were obliterated by, for example, smallpox and flu epidemics (Nurse et al. 1986; Owers et al. 2017). Additional factors, including unfair trading practices and war caused further loss of people, livestock, land, and culture among the Khoekhoe herders. Most South African Khoekhoe ended up as indentured laborers on farms adopting the Afrikaans Indo-European language (Elphick 1977; Barnard 1992; de Jongh 2016). Slave trade introduced additional intercontinental gene flow giving rise to complex genomic admixture patterns in current-day southern African populations (de Wit et al. 2010; Schlebusch et al. 2012; Petersen et al. 2013; Chimusa et al. 2015). A new mixed-ancestry cultural identity emerged in South Africa, namely the “Coloured” groups and they are genetically composed of Khoekhoe and San descendants with inputs from Europeans, Asians, and Bantu-speakers (de Wit et al. 2010; Schlebusch et al. 2012; Daya et al. 2013; Choudhury et al. 2017).
Assessing the regional genetic prehistory of autochthone communities from southern Africa has been challenging due to genetic contribution of immigrant populations into their original gene-pool. The mentioned migration waves had a strong impact on patterns of genome variation among Khoe-San groups, obscuring much of the past regional genetic and cultural exchange among these autochthone communities. Despite several genetic studies over the last few years, most investigations did not exclude the exogenous genetic components when analyzing Khoe-San population structure dynamics. We evaluate a combined genome-wide polymorphism data set of previously published data from southern Africa populations in conjunction with novel data, consisting of 50 individuals form a hunter-gatherer group collected at Xade, in the Central Kalahari Game Reserve (CKGR, Botswana). The Xade individuals were sampled prior the forced relocation of their settlement during the late 1980s and the ensuing admixture and cultural loss that occurred after their relocation (supplementary note 1, Supplementary Material online). The genetic diversity of a total of 20 Khoe-San groups was analyzed and great care was taken to exclude genome segments originating from immigrant farmers, herders, and colonists. With this “admixture-removed” data set of Khoe-San populations, we infer genetic affinities among Khoe-San groups and the influence of geography, linguistics, and ecology on their genomic variation in order to assess the genetic affinities among the Khoe-San prior to the impact of immigration and admixture. We also evaluate potential adaptive gene flow that Khoe-San populations might have acquired from immigrant farmer and herder gene-pools.
Results and Discussion
Overall Genetic Structure in the Khoe-San
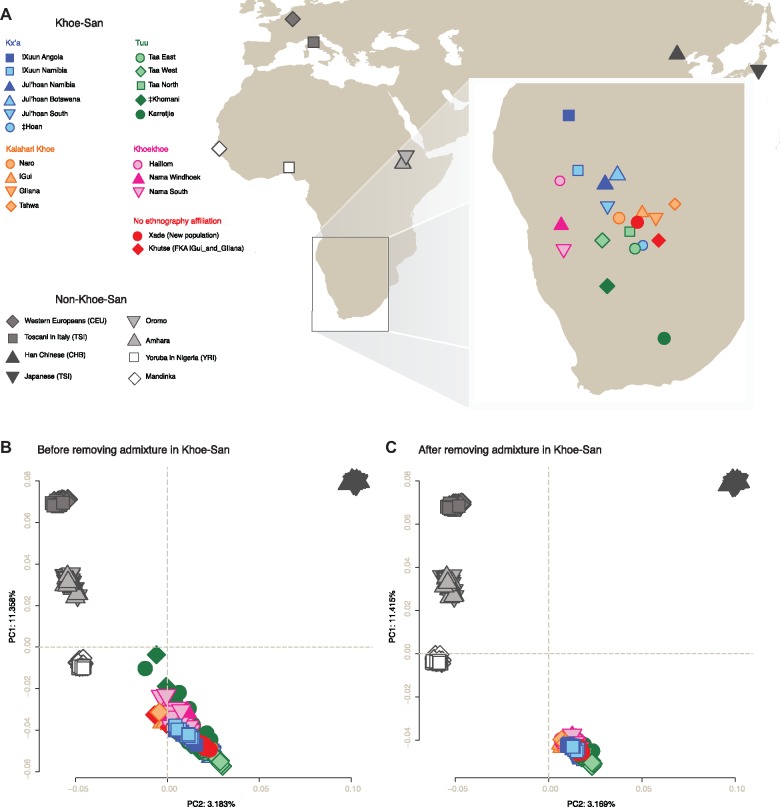
The genetic structure of the southern African Khoe-San is key to understand the pre-farming history of the region. We started by analyzing the admixture patterns among southern African populations (fig. 1A) by estimating population structure (Alexander et al. 2009), at a range of assumed numbers of clusters, from K = 2 to K = 10 (supplementary fig. S2, Supplementary Material online). Apart from the southern African Khoe-San groups (fig. 1A) and neighboring Bantu-speakers, we included Yoruba and Mandinka (representing West Africa), Amhara and Oromo (East Africa), Central Europeans and Tuscans (Europe), and the Han Chinese and Japanese (Asia) to account for admixture from external groups into the Khoe-San gene-pool (1000 Genomes Project Consortium et al. 2015; Gurdasani et al. 2015). At K = 5, the ancestry components recapitulate the five major genetic ancestries: Khoe-San (green), West African (white), East African (light-gray), European (gray), and Asian (dark-gray) (supplementary fig. S2, Supplementary Material online).
Fig. 1.
Geographic location of the samples analyzed in this study (A). PCA of the Khoe-San individuals, Eurasians, West and East Africans before (unmasked, B) and after (masked, C) applying the local ancestry pipeline (146,696 independent SNPs).
The genetic contribution from immigrant groups and the autochthonous San contribution vary among Khoe-San groups (supplementary figs. S2 and S3, Supplementary Material online). The Ju|’hoan populations from the north of Namibia and Botswana have the highest autochthonous component among the populations studied with 99.2% and 98.7% at K = 5, respectively (but see also Schlebusch et al. [2017]; this “autochthonous” component encompasses an East African ancestry component that is not visible since all modern-day Khoe-San groups carry this ancestry). K = 6 and K = 8 adds structure to the initial Khoe-San component (appearing at K = 3), separating three ancestral clusters, which previous studies reports as North, Central, and Southern San genetic components (e.g., Schlebusch et al. 2012; Uren et al. 2016; Montinaro et al. 2017). This tripartite genetic structure correlates with geography reflected in the naming scheme. Additional substructure can still be seen in further K values, mostly subgrouping neighboring Khoe-San groups.
From all the external source genetic contributions, the West African component is the most prominent in Khoe-San individuals. Originating from a region within current-day southeastern Nigeria and western Cameroon (Greenberg 1972; Nurse and Philippson 2003; Pakendorf et al. 2011; Bostoen 2018), the expansion of Bantu-speaking farmers had a great impact culturally and genetically on most of sub-Sahara Africa including the Kalahari Basin and surroundings (Tishkoff et al. 2009; Petersen et al. 2013; Li et al. 2014; González-Santos et al. 2015). At K = 5, the Western Africa component varies between 0% (for both Ju|’hoan North populations) to 74.8% in Duma San decedents (Schlebusch et al. 2016) and 87.1% in Khoekhoe-speaking Damara (supplementary fig. S2, Supplementary Material online). Damara genetic ancestry has been suggested to indicate a signature of language and cultural transition. They have primarily a Bantu-speaking genetic background but they speak a Khoisan language and practice pastoralism (Pickrell et al. 2012, 2014). Duma San and Damara populations, together with ‖Xegwi, Khwe, and Shua, were excluded from further downstream analysis due to their high level of recent admixture (mostly with Bantu-speakers).
Most Khoe-San individuals (and groups) have a small fraction of their genomes attributed to an East African ancestry component, with exception of populations from the central/south part of Kalahari Desert, such as Taa West, Taa East, ‡Hoan, and the Khutse San who do not show the component in the analyses used here. An East African admixture event in Khoe-San groups dated to ∼900–1,800 years ago was reported previously (Schlebusch et al. 2012, 2017; Pickrell et al. 2014). Khoekhoe groups show greater levels of East African ancestry compared to other groups, the two Nama populations have a fraction of around 15% and the Hai‖om ∼12% of their genomes. It has also been suggested, however, that Hai‖om-speakers might have been !Xuun hunter-gatherers who have shifted language and subsistence practice as result of contact with the Nama (Barnard 1992). Although we could not detect whether Hai‖om-speakers have acquired their moderately high proportion of East African component from direct contact with East Africans or through the Nama as intermediate, the rest of their genetic ancestry clusters with neighboring !Xuun and Ju|’hoan speaking groups.
The population from Xade Pan, which was genetically typed for the first time in this study, is a group of Khoe-San individuals from whom samples were collected in 1987 at Xade Pan, CKGR, Botswana. Although we do not have information on any specific ethnic affiliations of these individuals, Xade Pan became a permanent settlement for the |Gui and G‖ana groups in the late 1970s. The Khoe-San from Xade Pan clusters with the |Gui speakers, which indicates that they might belong to the |Gui population in accordance with the historical records (supplementary note 1, Supplementary Material online, Schlebusch 2010).
Complementary to ADMIXTURE, the genome proportions calculated by RFMix (Maples et al. 2013) vary per population, confirming the different dynamics among Khoe-San groups and groups that migrated recently into the region (supplementary fig. S3, Supplementary Material online). As a precautionary measure since there is no “unadmixed” Khoe-San group today (Schlebusch et al. 2017), we allowed RFMix to assign ancestries on the source populations as well. It is worth mentioning that Amhara and Oromo-speaking populations denoted high levels of East African ancestry in this analysis (50.7% and 51.3%, respectively). However, their relatively high proportion of West African (17.2% Amhara, 19.4% Oromo) and Eurasian (30.3% Amhara, 26.7% Oromo) ancestries were reported before (Pagani et al. 2012; Pickrell et al. 2014; Schlebusch et al. 2017).
Khoe-San-Specific Genetic Ancestry
We explored the genetic affinity of the Khoe-San groups to the external admixing sources by using principal component analysis (PCA) and ADMIXTURE before and after removing the admixture-tracts (fig. 1B and C and supplementary figs. S2 and S4, Supplementary Material online) and assessed the effect of the masking on diversity within each population (supplementary figs. S5 and S6, Supplementary Material online). In figure 1, we see the same Khoe-San individuals clustering much closer together in the PCA space after masking the genetic signatures from recent admixture (we compared their genetic affinity—after admixture removal—to different individuals from the same non-Khoe-San proxy populations to indicate the success of the admixture removal process).
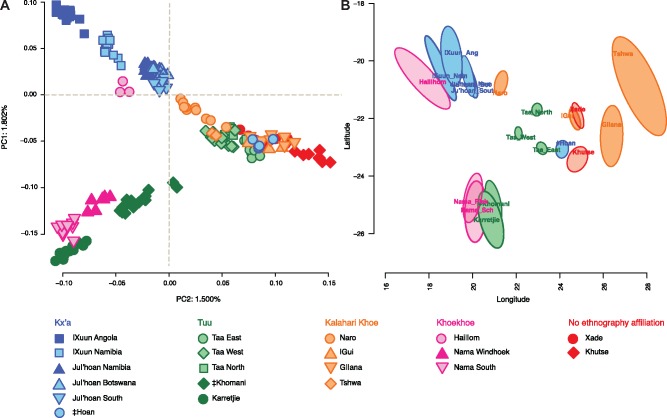
To visualize Khoe-San group affinities among each other, prior to recent admixture from pastoralists, farmers, and colonists, we reanalyzed PCA of the masked Khoe-San data set (fig. 2A). The first principal component axis (PC1) clearly reflects the North-South geographic vertices’ of the southern African landscape, whereas the PC2 outline the Central Kalahari Desert populations. Previous studies have also reported the impact of geography on the genetic variation of the southern African hunter-gatherers (e.g., Pickrell et al. 2012; Schlebusch et al. 2012; Montinaro et al. 2017). By masking non-Khoe-San genomic regions, the PCs become less affected by external admixture and the Khoe-San groups’ distribution on the PCA space correlates more strongly with their geographic area. The PC1 space is no longer driven by the least-admixed Ju|’hoan individuals as seen in nonmasked data sets (e.g., Pickrell et al. 2012; Schlebusch et al. 2012) and instead !Xuun from Angola (the most northern Khoe-San population of the study) defines the northern edge of PC1 space.
Fig. 2.
PCA of Khoe-San individuals without recent admixture (A). Geogenetic map under an isolation-by-distance model (B).
The Kx’a-speaking Ju|’hoan and !Xuun populations have their closest genetic affinity with the neighboring Khoekhoe-speaking Hai‖om, whereas the Kx’a-speaking ‡Hoan is genetically closer to the other central Kalahari populations (fig. 2A). Interestingly, the genetic variation continuum breaks with the Southern San individuals, where Nama, ‡Khomani, and Karretjie groups form their own cluster somewhat separated from other groups. However, the genetic continuum gap coincides geographically with the place where speakers of the extinct N‖ng languages once lived (Güldemann 2008).
The association between pairwise genetic distances and geographical distances was tested with a Mantel test (Mantel 1967, supplementary fig. S7, Supplementary Material online). A highly correlated r = 0.755 (P value <0.0001) was observed between genetics and geography to a greater level than previously observed in smaller data sets (r = 0.715, P value <0.0001, Schlebusch et al. [2012], and 0.65, P < 0.001, Montinaro et al. [2017]). When conditioned with language a slightly lower correlation is observed (r = 0.673, P value <0.0001) possibly explained by the language shifts that have occurred within the Khoe-San groups. The Nama groups likely moved to their current locations in relative recent times (Barnard 1992), and as such when Nama was excluded from the analysis we get an even higher Mantel correlation between genetics and geography of r = 0.885 (P value <0.0001).
Our analysis took advantage of the fact that we were able to use diploid-state, admixed-removed data sets, which allowed us to study the Khoe-San genetic affinities without the influence of non-local admixture or missing data. Typically individuals with high levels of Khoe-San ancestry (e.g., Ju|’hoan or Taa West) had a tendency to define PCAs in previous studies, due to their low levels of admixture (Schlebusch et al. 2012; Montinaro et al. 2017). Our findings confirm the major role of geography over linguistics and subsistence strategy in shaping the genetic diversity among Khoe-San groups.
The correlation between genetic variation and geography among Khoe-San groups was further explored using the Bayesian framework of the SpaceMix software (Bradburd et al. 2016). Four different models were analyzed whereby isolation-by-distance patterns are used to explain characteristics of the geographic coordinates and the genetic distances (geogenetic coordinates) of the individuals (fig. 2B and supplementary fig. S8, Supplementary Material online). In addition to the strict isolation-by-distance model, added factors such as potential admixture between Khoe-San groups and/or migration of Khoe-San groups were also evaluated. In all scenarios, the predicted and observed identity by descent covariance overlap well, reflecting support for isolation-by-distance being the major factor shaping the local patterns of genetic variation among the Khoe-San groups. In the models where the populations freely choose their location, (i.e., migration allowed), the geogenetic distribution of the populations reflects the geography well, independent of whether admixture is allowed or not (fig. 3 and supplementary fig. S8F and H, Supplementary Material online). In the Southern San populations, however, the geogenetic coordinates do not correlate well with the current geographic distribution in either of the models that allows migration. Instead, the populations cluster together and if the geogenetic map would be converted into a geographical landscape, all Southern San would be situated in the current South Africa.
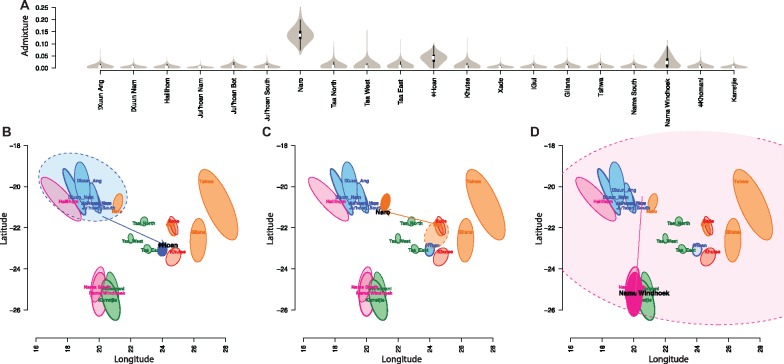
Fig. 3.
Proportions of admixture among masked Khoe-San when analyzed under the isolation-by-distance model with migration and mixture allowed (A). Geogenetic map of the Khoe-San where the source of admixture is coloured (95% CI) and admixture into the ǂHoan (B), Naro (C), and Nama Windhoek (D) populations are indicated.
From a linguistic perspective, it was reported that the Tuu language family, in particular the !Ui major group (which ‡Khomani and Karretjie ancestors once spoke), is a more unified language group when compared with other Khoisan families (Güldemann 2005). This language similarity could possibly be explained by a recent and rapid radiation of the language. A previously published mitochondrial DNA study also reported evidence of a recent and fast spread of the mtDNA L0d2a haplogroup that occurs at high frequencies in Southern Khoe-San groups (Schlebusch et al. 2013) signifying a recent expansion in Southern Khoe-San groups. We also note that the Taa speakers do not share such close genetic affinities to the other Tuu speakers of the study (fig. 3). This parallels with the geographical distribution of the Tuu languages, with Taa being in isolation from the other Tuu languages (supplementary fig. S1, Supplementary Material online, Güldemann [2005]).
The Khoekhoe-speaking Nama populations have high genetic affinities to the other Southern Khoe-San ‡Khomani and Karretjie in the geogenetic map, in line with previous findings (Schlebusch et al. 2012). This could be explained in part by the possible recent migration of Nama groups to their current-day locations (Barnard 1992).
Admixture among Khoe-San Groups and Correlation with Language
When the data are analyzed under the isolation-by-distance model where migration and admixture among Khoe-San groups are allowed, we observe limited amounts of admixture between a few Khoe-San groups (fig. 3). ǂHoan, Naro, and Nama Windhoek populations revealed levels of admixture higher than 5% (fig. 3A).
An admixture fraction of 5% (95% CI: 0.8–7.2%) is observed from the Ju-speakers into the ǂHoan. This result suggests a genetic link between the two geographically remote groups that speak Kx’a-language branches (see map in supplementary fig. S1, Supplementary Material online). There has been a debate whether proto-Kx’a-language speakers inhabited a more continuous geographical distribution over the Kalahari Basin or alternatively, whether the ǂHoan language was acquired by some sort of contact diffusion (Heine and Honken 2010). Although linguistic studies mostly support the first hypothesis, genetic studies have been unable to find evidence to support either of the theories (Pickrell et al. 2012; Barbieri et al. 2014). A possible scenario was proposed that the proto-Kx’a continuum was disrupted by spread of the Khoe languages into the Kalahari (Güldemann 2008). From figure 3B, the most likely source of the admixture found in the ǂHoan was from a population closest related to the Ju|’hoan (South) (marked by the beginning of the arrow), however the 95% confidence interval hints any Ju-speaker group as possible sources of the admixture event (in addition to the Hai‖om and Naro).
The San from Khutse were previously thought to be a mixed group drawing ancestry from |Gui and G‖ana (Schlebusch et al. 2012; Breton et al. 2014) but due to unclear ethnic identity recorded during sampling, in this study they are referred to as Khutse San (since they were collected in the Khutse Game Reserve). The Khutse Game reserve is considered to be the homeland of the ǂHoan language (Gerlach 2016). We indeed find a significant genetic link between the ǂHoan and the Khutse San to the exclusion of the Taa East (D[ǂHoan, TaaEast, Khutse, Chimp] with a Z-score of 3.27). In SpaceMix with no admixture allowed, the two populations are almost overlaying each other in the geogenetic map (supplementary fig. S8F, Supplementary Material online). We could not, however, detect any connection between Kx’a-speakers and the Khutse San. Since ethnographic information on the Khutse San is not clear, we cannot make further inferences about this group and their language connection.
Interestingly, the Naro shows high levels (∼13%, 95% CI: 9.8–21.3%) of admixture from a population close to the |Gui and Xade San. In the past, the Naro would temporarily leave their permanent camp during the wet season for food resource reasons, and their range extended to the CKGR, the homeland of the |Gui and G‖ana (Barnard 1992). This nomadic life-style practiced by the Naro could be the explanation of the observed pronounced admixture from the |Gui. Such a signal of interaction would (fig. 3C), however, have been expected to occur in both ways but, interestingly, no significant signal of admixture was observed from the Naro into the |Gui gene-pool.
The Nama Windhoek (first published in Schlebusch et al. 2012) show a relatively high level of admixture with another Khoe-San group. Even though the 95% confidence interval could not pinpoint the source of the admixture event in the Bayesian framework of SpaceMix, the iteration with the highest posterior possibly hints at a population genetically closer to the Ju|’hoan and/or Naro (tip of the arrow in fig. 3D). The possible Ju|’hoan/Naro source for the admixture seems likely due to the current location of the Nama people. Interestingly, SpaceMix could not pick up any similar signal of admixture for the Nama South (first published in Pickrell et al. [2012]). The Nama South population, however, was collected at different places that all were situated south of Nama Windhoek (who were sampled in Windhoek) and therefore likely had less contact with Ju|’hoan/Naro-speakers. The Nama Windhoek population was previously reported to carry mtDNA L0k1a lineages (Schlebusch et al. 2010), typical in hunter-gatherers from the central and northern Kalahari in Botswana but present also in Hai‖om from Namibia ( Barbieri et al. 2013, 2014). In contrast, the Nama South did not have any L0k1a mtDNA lineages in their gene-pool (Barbieri et al. 2013, 2014).
We formally tested evidence of admixture between each of the ǂHoan, Nama, and Naro, respectively, with other Khoe-San groups by using D-statistics and admixture f3 (supplementary table S2 and figs. S9–S14, Supplementary Material online). None of the f3 statistics showed negative results indicating admixture. Although D-tests seemed to be more sensitive to these events and showed a few Z-scores above 3 for each of the three populations (supplementary table S2, Supplementary Material online), generally the results seemed to be influenced by geographic proximity. It is our interpretation that f3 statistics and D-tests are affected by isolation-by-distance relationships between groups and that SpaceMix is more sensitive to detect admixture between groups while accounting for isolation-by-distance effects.
We also analyzed the effective migration surfaces obtained through Estimated Effective Migration Surfaces (EEMS) analyses (supplementary fig. S15, Supplementary Material online) (Petkova et al. 2016). The overall genetic affinities among Khoe-San groups are visualized over the spatial structure. Generally, low migration rates are illustrated between different Khoe-San groups but an indication of higher rates is visible between the Naro and |Gui in central Botswana. The increased migration rate along the west coast could be a reflection of the genetic similarity of the Nama groups to Tuu speakers and might be indicative of the recent migration of the Nama from the south (Barnard 1992). The migration surfaces also denote a reduction in migration movements around the Kalahari Desert that might indicate the presence of ecological barriers as previously reported (Uren et al. 2016). To further inspect this signal, we did a Mantel correlation of genetics and ecology (we used rainfall patterns as a proxy for ecology) and observe a moderate correlation of 0.464 (P value <0.0001). However, when we performed a partial Mantel test on genetics and geographical distances conditioned on ecology, the correlation observed (r = 0.67, P value <0.0001) is lower than when we correlate only genetic and geographical distances (r = 0.755, P value <0.0001). These results suggest that geography is the main factor explaining genetic variation within Khoe-San groups. Although findings of correlation between geography and genetic distances have been reported before (Pickrell et al. 2012; Schlebusch et al. 2012; Montinaro et al. 2017), our analyses confirm that an isolation-by-distance model explains most of the variation between Khoe-San groups.
Genomic Impact from the Contact with Immigrant Populations
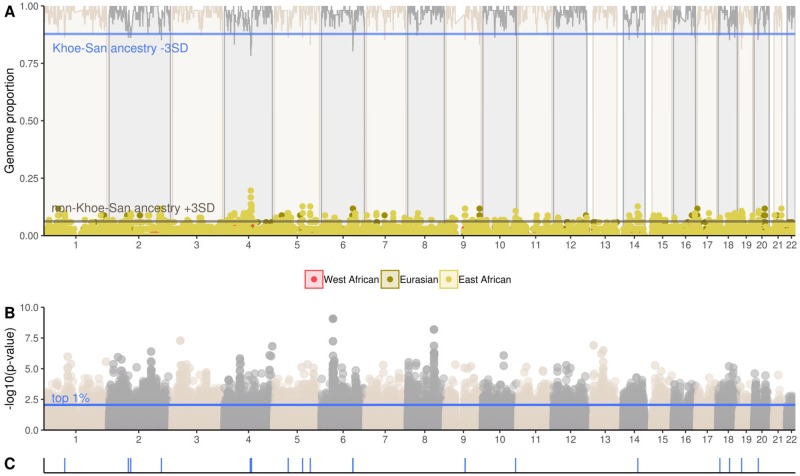
The arrival of pastoralists and farmers in southern Africa led to subsequent admixture with the San. The impact of admixture in extant Khoe-San genomes varies between groups. To evaluate whether admixture with immigrant groups brought new beneficial traits that were retained in the Khoe-San gene-pool (i.e., adaptive gene flow), we selected regions that were 3 standard deviations (SD) lower than the average of their Khoe-San ancestry (fig. 4A and supplementary figs. S16A–S21A, Supplementary Material online), and compared those regions with the top 1% of the integrated haplotype score (iHS) in the different Khoe-San groups (fig. 4B and supplementary figs. S16B–21B, Supplementary Material online).
Fig. 4.
Ancestral genomic proportions across the genomes of Ju|’hoan individuals (A). Lines at the top of the graph represent the San ancestry across the genome. Non-Khoe-San ancestry are indicated at the bottom of the graph as dots: in red (West African), yellow (East African), and olive green (Eurasian). P values of the iHSs is shown in (B) and top 1% iHS SNPs that also show a reduction (−3 SD) of Khoe-San ancestry (C).
Ju|’hoan individuals have the highest amount of San genetic ancestry among all the extant Khoe-San groups in Southern Africa (fig. 4A). With support of ancient DNA studies, it has been demonstrated that all Khoe-San groups, including Ju|’hoan-speakers had admixture (9–14%) with the mixed East African-Eurasian pastoralist group that immigrated to southern Africa in the last 2,000 years (Schlebusch et al. 2017; Skoglund et al. 2017). In total, 19 possible adaptive admixture regions have been identified (fig. 4C and supplementary table S3, Supplementary Material online). Specific regions on chromosomes 2, 4, 6, 19, and 20 show particularly low signals of autochthonous ancestry in Ju|’hoan-speakers. On chromosome 4, the San ancestry decreases dramatically between positions 103552897 to 107703545 to the lowest percentages across the genome (78.4%, −6,09 SD). After inspection of the region we found that it extends over 4 Mb and contains 17 genes (4 genes contained single nucleotide polymorphisms (SNPs) in the top 1% of the selection scan, or had these SNPs nearby, supplementary table S3, Supplementary Material online). The region with the highest non-Khoe-San ancestry overlaps with the CXXC4 gene, which is assigned to have a high East African ancestry (19.6%, 8.00 SD, fig. 4A and supplementary fig. S22, Supplementary Material online). The CXXC4 gene encodes a zinc finger domain-containing protein, mostly expressed in the hypothalamus and pituitary gland. Previous GWAS has associated this gene with low-density lipoprotein cholesterol levels (Kathiresan et al. 2007). However, the CXXC4 gene did not contain the highest iHS signals of the region and we further explored SNPs in the vicinity of the gene that also falls within the East African ancestry peak. The rs17209891 SNP, located 5.7-Mb downstream of the CXXC4 gene, have a derived allele at highest frequency in East African and Eurasians (averaging at 26.7% and 21.7%, respectively). This SNP have a frequency of 26.7% in the Khoe-San and lower frequencies in West African ancestry populations (11.2%).
Across all Khoe-San groups, only the Nama had a region in the top five iHS regions (supplementary table S4, Supplementary Material online, iHS P value 1.57 × 10−5) that also occur among the top post-admixture selection candidates (supplementary table S3, Supplementary Material online). This specific region on chromosome 18 contains the DCC gene that encodes a netrin 1 receptor, which has been associated previously with body weight changes (Fox et al. 2007) and alcoholism (Heath et al. 2011). The Nama had a very high East African ancestry assignment (58%) in this region (supplementary table S4, Supplementary Material online). The second highest iHS peak in the Nama is also particularly interesting, it departs 3.82 SD from the Eurasian ancestry mean across the genome and contains four genes, among others, an aldehyde dehydrogenase family gene. The ALDH1A2 gene has been associated with hypertension and blood pressure in African Americans (Adeyemo et al. 2009).
Previous reports identified genes related to food consumption (taste receptor TAS2R and KRTAP5 gene clusters) to be under selection in the last 2,000 years in the hunter-gatherers of southern Africa (Skoglund et al. 2017). These dates coincide with the arrival of immigrant herders into the area and, even though we did not detect the same signals under our top five regions (possibly due to the different nature of the data and populations tested) there are tentative indications of dietary adaptation in southern African autochthonous groups (e.g., the CXXC4 and DCC genes) after their contact with herders and farmers, apart from the LP mutations reported before (Breton et al. 2014; Macholdt et al. 2014).
Concluding Remarks
In this study, we investigated the pre-farming genetic structure and affinities among southern African Khoe-San populations. All the Khoe-San groups in the present study had exogenous DNA from contact with immigrant groups, however, their genetic make-up after removing genetic material from recent admixture fits an isolation-by-distance model, which mimics the southern African landscape. Although the isolation-by-distance model explains most of the affinities between Khoe-San groups, signals of admixture between different Khoe-San groups could be detected. We find a genetic link between the two geographically separated groups that speak Kx’a-languages. A fraction of 5% of the ǂHoan speaker genomes have been associated to Ju-speakers. Such a signal might be reminiscent of a connection between the proto-Kx’a-speakers that possibly were disrupted by the arrival of ancestors of Khoe-speakers into the region or, alternatively, admixture of Ju-speakers into ǂHoan speakers. With the current data, we cannot identify which of the two hypotheses explain the genetic connection that link these two groups who both speak Kx’a-languages.
Although all Khoe-San groups have non-local admixture to some extent, the level and sources of the non-Khoe-San fraction varies in each population. Previous studies have reported admixture from an East African-Eurasian group into Khoe-San groups, including Ju|’hoan populations (Pickrell et al. 2014; Schlebusch et al. 2017; Skoglund et al. 2017). It is possible that this genetic exchange introduced adaptive variants into the Khoe-San and we identified possible regions that underwent adaptive gene flow in the Nama and Ju|’hoan-speakers.
Indigenous southern African hunter-gatherers are the descendants of one of the two branches of the deepest split in the human lineage (all remaining current-day humans being descendants of the other branch). Although some Khoe-San populations have changed their way of subsistence, for example, the Nama and Hai‖om, some still retain their traditional hunter-gathering life-style. In this study, we could see that the past genetic landscape of Khoe-San groups was largely determined by geographic distances. This isolation-by-distance model of Khoe-San population structure might have extended way beyond the Kalahari Basin area in the past, stretching into the northeastern parts of the continent as suggested by results from a recent ancient DNA study (Skoglund et al. 2017). Future studies on ancient human remains across Africa that predates the invention of farming and herding practices could provide further clarification regarding the importance of the isolation-by-distance model in African prehistory.
Materials and Methods
Sampling and Genotyping
We generated novel genotype data from 50 Khoe-San individuals from Xade and 1 individual from Ghanzi in Botswana (which was assumed based on genetic affinities to be Naro), all collected in 1987 by P. Ebbesen. The samples were collected with informed consent and the study was explained to participants by means of a translator before blood samples were collected. The government of Botswana approved the collection of the blood samples for medical and population history studies. The samples were previously used in a publication of Y-chromosome polymorphisms (Batini et al. 2011).
DNA was extracted with QIAamp DNA Mini kit from Qiagen using blood serum as primary material. The extraction was done following the kit protocol guidelines with small alterations for optimization purposes. The samples were genotyped by the SNP&SEQ Technology Platform in Uppsala, Sweden, on the Illumina Omni2.5-Octo BeadChip. The data were aligned to the Human Genome built version 37 and will be made available for academic research use, through the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress, last accessed April, 2019) accession number E-MTAB-7813.
Data management and quality filtering was carried out with PLINK v.1.90 (Chang et al. 2015). Only autosomal SNPs were included in the data set and duplicate SNPs were filtered out. For merging with existing data sets, AT and CG SNPs were excluded to prevent strand flipping errors. Genotype missingness was set to 0.1 and Hardy–Weinberg Equilibrium (HWE) to 0.001. HWE was separately calculated for the Xade population and another unrelated population (Bantu-speakers—not included in this study) typed during the same genotyping run. Only the SNPs that depart from HWE in both populations were removed from the analyses (8 SNPs in total). These SNPs are strong candidates for genotyping errors. All individuals had lower than 15% missing data but due to high levels of relatedness, 17 individuals (of which 6 were first-degree related) were excluded with a pairwise Identity by State (IBS) threshold of 0.32. The final data set was composed of 33 Khoe-San from Xade and 1 Naro individual with a total of 2,190,923 SNPs.
Merging with Existent Data Sets
We merged the 33 Xade and 1 Naro individuals with previous published genotype data, creating two distinct data sets.
Low-Density SNP Data Set
The first data set focus on the Khoe-San genetic variation. Selected Khoe-San- and neighboring Bantu-speaking populations from Schlebusch et al. (2012, 2016), and Pickrell et al. (2014) were merged together with the newly generated data. Since the Pickrell data was mapped to hg36, we converted positions to hg37 with LiftOver. The Non-Southern African comparative groups YRI, CEU, TSI, CHB, and JPT were included from the 1000 Genome Project Consortium et al. (2015). We also included the East African Amhara and Oromo, the West African Mandinka and the Southern African Bantu-speakers (Sotho and Zulu) from the African Genome Variation Project (Gurdasani et al. 2015). To avoid sample-size bias in further analyzes, we randomly downsized each population to a maximum of 30 individuals. Quality filtering and data management were handled similar to the newly generated data, described above. Due to different genotyping platforms used in the different studies, the number of overlapping SNPs was reduced to 150,240 SNPs for 685 individuals.
High-Density SNP Data Set
A second data set used to assess the genomic impact from the contact with immigrant populations included all the samples mentioned above except data genotyped on the Affymetrix Human Origins array (Pickrell et al. 2014) and the three ‖Xegwi and five Duma descendants from Schlebusch et al. (2016) (because of the sample size/highly admixed nature of these populations). In total, this dense-SNP data set was composed of 472 individuals from 18 populations and 1,507,271 SNPs.
Both data sets were imputed and phased with fastPHASE v.1.4.0 (Scheet and Stephens 2006). The number of haplotype clusters was set to 25 and we use 25 runs of the Expectation–Maximization (EM) algorithm to generate the “best” haplotype guess. fastPHASE analyses were run with the related individuals included, which were discarded from the data set afterward.
Initial population structure analyses were performed for both data sets. We inferred admixture fractions with ADMIXTURE (Alexander et al. 2009) to investigate relationships among individuals. The number of clusters, K, was set from 2 to 10, replicated 100 times. The cluster-inference and visual inspection was made with Pong v.1.4.5 (Behr et al. 2016 ).
In order to investigate the genetic ancestry of the Khoe-San and possible signals of adaptive gene flow, we ran RFMix (Maples et al. 2013) on the low- and high-density SNP data sets, respectively. Based on the ADMIXTURE analyses at K = 4, we selected individuals who contained the “Khoe-San” component higher than 95%, independent of their ethnographic label. Sixty and thirty Khoe-San were selected to represent the “Khoe-San” parental source for the lower and higher SNP-dense data sets, respectively. We randomly selected similar sample sizes from Yoruba and Mandinka (to represent West Africa), Amhara and Oromo (East Africa), Central Europeans and Tuscans (Europe), and the Han Chinese and Japanese (Asia) for the RFMix analyses. We used the HapMap II genetic map as recombination map. We ran RFMix analyses with two extra iterations to account for admixture in the source populations and minimize assignment errors, we set three minimum reference haplotypes per tree node and a window size of 0.02 cM, on the low-density SNP data set, and 0.2 cM in the high-density SNP data set. We did initial tests with different admixture times in our low-density data set: default settings and 50 generations. We did not see significant differences and since admixture occurred at different time periods with different external groups, we decided to keep this parameter at its default setting. Similarly, the default settings and 50 generations were tested in the high-density SNP data set. We opted to perform the RFMix analysis with 50 generations (∼1,500 years ago) since it provided results that are more coherent with Admixture results and low-density genome proportions. Since we are not able to provide multiple generation times for the admixture event, we chose 50 generations because it coincides with the arrival of the Bantu-speakers in the area (the major admixture component) and it is time-wise the middle-most event. We did not discard the sources in the final output and the Khoe-San source individuals were analyzed along with their respective populations.
Khoe-San-Specific Ancestry Analyses
We extracted only Khoe-San-specific segments assigned by RFMix in the Khoe-San. The individuals with more than 60% of their genome assigned to Khoe-San were identified (see below). We reimputed, on a population-specific basis, the removed non-Khoe-San regions of individuals, using fastPHASE (run with similar parameters as before). We inspected the changes in heterozygosity levels before and after filtering out the non-autochthonous components and following imputation (supplementary figs. S5 and S6, Supplementary Material online) (this was only done to inspect the effect of the methods on the data—since heterozygosity values for SNP chip data are subject to ascertainment bias—they were not used to make inferences about genetic diversity). As expected, we found that heterozygosity levels decreased in all populations.
Tests regarding population-specific re-imputation were performed. We randomly selected 15 individuals from Yoruba and Mandinka, due to their close genetic affinity. We randomly set to missing 10%, 20%, 30%, 40%, and 50% of the data and imputed the missing data with fastPHASE. Comparative PCAs were done to inspect how much the missingness/imputation affected the location of the samples in the PCA space (supplementary fig. S23, Supplementary Material online). Furthermore, we tested how the set to missing and population-specific imputation would affect admixture by randomly setting to missing 10%, 20%, 30%, 40%, and 50% of Herero, Sotho, and Zulu Bantu-speakers. We chose these populations because of their known admixture into the local hunter-gatherers. Subsequently we calculated D-statistics under the D(Bantu-speaker, YRI, Ju|’hoan, Chimp) with AdmixTools (Patterson et al. 2012). Results are shown in supplementary figure S24, Supplementary Material online. Based on both these tests we decided on a cut-off for our analyses of Khoe-San individuals, with their Khoe-San ancestry not lower than 60% (supplementary fig. S25, Supplementary Material online).
The selected individuals were extracted and merged together in a data set after the imputation process. Due to possible imputation errors, a HWE filter with a threshold of 0.05 and an IBS filter of 0.32 were applied. To limit population size biases, a maximum of 15 individuals per population was included, resulting in a final data set of 196 Southern Africa Khoe-San individuals from 20 groups with 146,696 SNPs.
Population Structure
As quality check, we generated a PCA that compares Khoe-San individuals to a different set of randomly picked individuals from the same nonlocal sources, before and after the local ancestry pipeline was applied. A Khoe-San-ancestry-only PCA was also created to evaluate the genetic relationships among the Southern African hunter-gatherers and herders without the effects of recent admixture. All PCA analyses were performed with EIGENSOFT (Price et al. 2006; Patterson et al. 2006) under default settings. Since the artificial reduction of the intra-population diversity induced by the imputation might affect PCA space we re-analyzed the data without imputation using the flags “shrink mode” and “lsq mode” in smartPCA to account for data with high amounts of missing data (supplementary fig. S26, Supplementary Material online). When we compare the two PCAs (masked without imputation and masked imputed) we observe that the main genetic structure is maintained in the PCA space and we observe a high correlation between the two PCAs (Mantel correlation of 0.694 [P value <0.0001] and a procrustes scale factor of 0.824). However, individuals with low missingness (Ju|'hoan speakers and three Taa West individuals) tend to define the main PCs more than in the imputed data, indicating that the large blocks of missing data still influence the method even with adding the flags.
The relationship between genetic and geographic distances was inspected with a Mantel test. We generated a great circle geographic distance matrix for all Khoe-San populations with the R package fields v.9.0 (Nychka et al. 2017). A genetic distance matrix was also computed based on the average Euclidian distance of the individuals from each specific Khoe-San group based on the PC1, PC2, and PC3 coordinates. Each PC coordinate was weighted by their respective PC eigenvalue in order to compensate for the importance of the PC in explaining the data. Population Fst distances were calculated with smartPCA. We used the R package vegan (Oksanen et al. 2018) to calculate the Mantel correlation of the two generated distance matrices under 100,000 iterations. We also performed a partial Mantel test to account for language and ecology. For the language distance matrix, we used a simple hierarchical language family distance where all three major language families are equidistant to each other in the following fashion:
[Kx’a (Ju)(Amkoe)] [Khoe (Kalahari-Khoe)(Khoe-khoe)] [Tuu (Taa)(!Ui)]
If the populations speak the same language a value of zero was assigned; populations who speak languages within the same subfamily was assigned a value of 1; for populations that speak languages within the same major family we assigned a value of 3; and if the populations speak languages from distinct language families we differentiated them with 6 units. The populations without language affiliation (Khutse and Xade) were excluded from the analyses and ǂKhomani and Karretjie were assigned as !Ui speakers.
To assess the impact of ecology on the Khoe-San genetic variation, we created a distance matrix based on yearly rainfall average of each location. The data were extracted based on the Climate Change Knowledge Portal (http://sdwebx.worldbank.org/climateportal, January 16, 2019) for each of the coordinates of our populations. The matrix was computed based on the average yearly rainfall between 1901 and 2015.
We further investigated the genetic relatedness to geography with SpaceMix (Bradburd et al. 2016). SpaceMix analyses were tested under four different models: 1) samples do not choose their own location, nor can admix with each other (i.e., pure isolation-by-distance), 2) samples are not allowed to choose their own location but they can admix (i.e., isolation-by-distance and admixture), 3) samples choose their own location on a −180,180 × −90,90 “GeoGenetic map” but no admixture is allowed (i.e., isolation-by-distance and migration), and 4) samples choose their own locations and they draw admixture (i.e., isolation-by-distance plus migration and admixture). For the first two models, the population geographic coordinates are used as the priors. In each model, we performed an initial short run of 105 Markov Chain Monte Carlo iterations under default settings and based on the highest posterior values from the short run, a longer run of 108 iterations was initialized with draws of the posterior in every 105 chains. Similar identity by descent covariance was observed in the SpaceMix analysis of Khoe-San ancestry without imputation for all four isolation-by-distance models (supplementary fig. S27, Supplementary Material online). The covariance estimates for the observed versus expected models overlap each other completely in all four isolation-by-distance scenarios (supplementary fig. S27, Supplementary Material online), therefore we believe that the drift induced by the population-specific imputation does not influence whether the isolation-by-distance models fit the data or not.
To complement SpaceMix isolation-by-distance analyses, we used the stepping stone model from EEMS to estimate effective migration surfaces (Petkova et al. 2016). Genetic dissimilarities were calculated with bed2diffs program from EEMS software. One thousand demes were assigned in the prior settings and we run three independent runs of 107 iterations with a burnin of 5 × 106. To inspect the relationships of ǂHoan, Naro, and Nama, D-statistics and f3 were calculated with AdmixTools (Patterson et al. 2012). Neighboring populations of the target population were used, while testing for signals across the Khoe-San data set. For the D-test, the chimpanzee was used as outgroup (Chimpanzee Sequencing and Analysis Consortium 2005).
Adaptive Gene Flow and Selection Scans
iHS was analyzed using the R package REHH (Gautier et al. 2017 ). iHS was calculated with only 200,000 bp of maximum distance between two SNPs allowed. To identify the ancestral allele, we used only positions that were found basal to human, chimpanzee, bonobo, and gorilla. Based on this requirement, we performed the selection scan analyses on 1,188,247 SNPs. Peaks were identified by averaging the −log10(P value) every ten SNPs. The iHS scores were compared with RFMix results to identify overlaps in selection and admixture signals. For each chromosome, regions within the 2-Mb borders were excluded due to high recombination around the telomeres. For iHS and RFMix, the top peaks were identified and target regions were inspected on Genome Browser to identify possible genes in the target region.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to all subjects who participated in this research. We thank the Working Group of Indigenous Minorities (WIMSA) and the South African San Council for their support and for assisting and facilitating during fieldwork. We thank the reviewers for helpful comments. We also would like to thank Torsten Günther and Paul Verdu for their helpful comments and fruitful discussions. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). The facility is part of the National Genomics Infrastructure supported by the Swedish Research Council for Infrastructures and Science for Life Laboratory, Sweden. The SNP&SEQ Technology Platform is also supported by the Knut and Alice Wallenberg Foundation. The computations were performed at the Swedish National Infrastructure for Computing (SNIC-UPPMAX). C.M.S. was supported by the Swedish Research Council (#621-2014-5211) and the European Research council (ERC #759933). M.J. was supported by the Knut and Alice Wallenberg Foundation, the Swedish Research Council (#642-2013-8019), and the Göran Gustafsson Foundation.
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 5267571:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, et al. 2009. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 57:e1000564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batini C, Ferri G, Destro-Bisol G, Brisighelli F, Luiselli D, Sánchez-Diz P, Rocha J, Simonson T, Brehm A, Montano V, et al. 2011. Signatures of the preagricultural peopling processes in sub-Saharan Africa as revealed by the phylogeography of early Y chromosome lineages. Mol Biol Evol. 289:2603–2613. [DOI] [PubMed] [Google Scholar]
- Barbieri C, Güldemann T, Naumann C, Gerlach L, Berthold F, Nakagawa H, Mpoloka SW, Stoneking M, Pakendorf B.. 2014. Unraveling the complex maternal history of Southern African Khoisan populations. Am J Phys Anthropol. 1533:435–448. [DOI] [PubMed] [Google Scholar]
- Barbieri C, Vicente M, Rocha J, Mpoloka SW, Stoneking M, Pakendorf B.. 2013. Ancient substructure in early mtDNA lineages of southern Africa. Am J Hum Genet. 922:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard A. 1992. Hunters and herders of southern Africa: a comparative ethnography of the Khoisan peoples. Cambridge: Cambridge University Press. [Google Scholar]
- Behar DM, Villems R, Soodyall H, Blue-Smith J, Pereira L, Metspalu E, Scozzari R, Makkan H, Tzur S, Comas D, et al. 2008. The dawn of human matrilineal diversity. Am J Hum Genet. 825:1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S.. 2016. pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 3218:2817–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostoen K. 2018. The Bantu expansion In: Oxford research encyclopedia of African history. Oxford: Oxford University Press. [Google Scholar]
- Bradburd GS, Ralph PL, Coop GM.. 2016. A spatial framework for understanding population structure and admixture. PLoS Genet. 121:e1005703.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G, Schlebusch CM, Lombard M, Sjödin P, Soodyall H, Jakobsson M.. 2014. Lactase persistence alleles reveal partial East African ancestry of southern African Khoe pastoralists. Curr Biol. 248:852–858. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 4377055:69–87. [DOI] [PubMed] [Google Scholar]
- Chimusa ER, Meintjies A, Tchanga M, Mulder N, Seoighe C, Soodyall H, Ramesar R.. 2015. A genomic portrait of haplotype diversity and signatures of selection in indigenous southern African populations. PLoS Genet. 113:e1005052.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A, Ramsay M, Hazelhurst S, Aron S, Bardien S, Botha G, Chimusa ER, Christoffels A, Gamieldien J, Sefid-Dashti MJ, et al. 2017. Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nat Commun. 81:2062.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya M, van der Merwe L, Galal U, Möller M, Salie M, Chimusa ER, Galanter JM, van Helden PD, Henn BM, Gignoux CR, et al. 2013. A panel of ancestry informative markers for the complex five-way admixed South African Coloured Population. PLoS One 812:e82224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jongh M. 2016. A forgotten first people: the Southern Cape Hessequa. Durban (South Africa: ): The Watermark Press. [Google Scholar]
- de Wit E, Delport W, Rugamika CE, Meintjes A, Möller M, van Helden PD, Seoighe C, Hoal EG.. 2010. Genome-wide analysis of the structure of the South African Coloured Population in the Western Cape. Hum Genet. 1282:145–153. [DOI] [PubMed] [Google Scholar]
- Elphick R. 1977. Krall and Castle: Khoikhoi and the founding of White South Africa. London: Yale University Press. [Google Scholar]
- Fox CS, Heard-Costa N, Cupples LA, Dupuis J, Vasan RS, Atwood LD.. 2007. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet. 8(Suppl 1): S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Klassmann A, Vitalis R.. 2017. rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol Ecol Resour. 171:78–90. [DOI] [PubMed] [Google Scholar]
- Gerlach L. 2016. Phonetic and phonological description of the N!aqriaxe variety of ǂ'Amkoe and the impact of language contact. Berlin (Germany: ): Universitat zu Berlin. [Google Scholar]
- González-Santos M, Montinaro F, Oosthuizen O, Oosthuizen E, Busby GB, Anagnostou P, Destro-Bisol G, Pascali V, Capelli C.. 2015. Genome-wide SNP analysis of southern African populations provides new insights into the dispersal of Bantu-speaking groups. Genome Biol Evol. 79:2560–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J. 1972. Linguistic evidence regarding Bantu origins. J Afr Hist. 13:189–216. [Google Scholar]
- Güldemann T, Fehn AM.. 2014. Beyond ‘Khoisan’—historical relations in the Kalahari Basin. Current issues in linguistic theory 330. Amsterdam: John Benjamins. [Google Scholar]
- Güldemann T. 2008. A linguist’s view: khoe-Kwadi speakers as the earliest food-producers of southern Africa. S Afr Hum. 20:93–132. [Google Scholar]
- Güldemann T. 2005. Studies in Tuu (Southern Khoisan) In: Papers on Africa, languages and literatures 23. Leipzig (Germany: ): Universitat Leipzig. [Google Scholar]
- Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, et al. 2015. The African Genome Variation Project shapes medical genetics in Africa. Nature 5177534:327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, et al. 2011. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry 706:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine B, Honken H.. 2010. The Kx’a family: a new Khoisan genealogy. J Asian Afr Stud. 79:5–36. [Google Scholar]
- Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodríguez-Botigué L, Ramachandran S, Hon L, Brisbin A, et al. 2011. hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 10813:5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. 2008. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 4517181:998–1003. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, et al. 2007. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 8(Suppl 1):S17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HL, Ratan A, Perry GH, Montenegro A, Miller W, Schuster SC.. 2014. Khoisan hunter-gatherers have been the largest population throughout most of modern-human demographic history. Nat Commun. 5:5692.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Schlebusch C, Jakobsson M.. 2014. Genetic variation reveals large-scale population expansion and migration during the expansion of Bantu-speaking peoples. Proc Biol Sci. 2811793:pii: 20141448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholdt E, Lede V, Barbieri C, Mpoloka SW, Chen H, Slatkin M, Pakendorf B, Stoneking M.. 2014. Tracing pastoralist migrations to southern Africa with lactase persistence alleles. Curr Biol. 248:875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209–220. [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, Bustamante CD.. 2013. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 932:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Lane P.. 2013. The Oxford handbook of African archeology. Oxford: Oxford Handbooks. [Google Scholar]
- Montinaro F, Busby GB, Gonzalez-Santos M, Oosthuitzen O, Oosthuitzen E, Anagnostou P, Destro-Bisol G, Pascali VL, Capelli C.. 2017. Complex ancient genetic structure and cultural transitions in southern African populations. Genetics 2051:303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nychka D, Furrer R, Paige J, Sain S.. 2017. fields: Tools for spatial data. R package version 9.6. Available from: www.image.ucar.edu/∼nychka/Fields. Boulder, USA. University Corporation for Atmospheric Research. Accessed April 2019.
- Nurse D, Philippson G, editors. 2003. Towards a historical classification of the Bantu languages In: The Bantu languages. London: Routledge and Sons; p. 164–181. [Google Scholar]
- Nurse GT, Weiner JS, Jenkins T.. 1986. The peoples of southern Africa and their affinities. New York: Oxford University Press. [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, et al. 2018. vegan: community ecology package. R package version 2.5-2. Available from: https://CRAN.R-project.org/package1/4vegan. Accessed April 2019.
- Owers KA, Sjödin P, Schlebusch CM, Skoglund P, Soodyall H, Jakobsson M.. 2017. Adaptation to infectious disease exposure in indigenous Southern African populations. Proc Biol Sci. 2841852: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Kivisild T, Tarekegn A, Ekong R, Plaster C, Gallego Romero I, Ayub Q, Mehdi SQ, Thomas MG, Luiselli D et al. 2012. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet. 2012 Jul 13;91(1):83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakendorf B, Bostoen K, de Filippo C.. 2011. Molecular perspectives on the Bantu expansion: a synthesis. Lang Dyn Change 11:50–88. [Google Scholar]
- Patterson Nick, Alkes L. Price, and David Reich. 2006. Population structure and eigenanalysis. PLoS genetics 2.12:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 1923:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen DC, Libiger O, Tindall EA, Hardie R-A, Hannick LI, Glashoff RH, Mukerji M, Fernandez P, Haacke W, Schork NJ, et al. 2013. Complex patterns of genomic admixture within southern Africa. PLoS Genet. 93:e1003309.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova D, Novembre J, Stephens M.. 2016. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 481:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Patterson N, Barbieri C, Berthold F, Gerlach L, Güldemann T, Kure B, Mpoloka SW, Nakagawa H, Naumann C, et al. 2012. The genetic prehistory of southern Africa. Nat Commun. 3:1143.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Patterson N, Loh PR, Lipson M, Berger B, Stoneking M, Pakendorf B, Reich D.. 2014. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci U S A. 1117:2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleurdeau D, Imalwa E, Détroit F, Lesur J, Veldman A, Bahain JJ, Marais E.. 2012. “Of sheep and men”: earliest direct evidence of caprine domestication in Southern Africa at Leopard Cave (Erongo, Namibia). PLoS One 77:e40340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38(8):904–9. [DOI] [PubMed] [Google Scholar]
- Robbins LH, Campbell AC, Murphy ML, Brook GA, Srivastava P, Badenhorst S.. 2005. The advent of herding in southern Africa: early AMS dates on domestic livestock from the Kalahari Desert. Curr Anthropol. 464:671–677. [Google Scholar]
- Sadr K. 2015. Livestock first reached southern Africa in two separate events. PLoS One 108:e0134215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M.. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 784:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch CM, de Jongh M, Soodyall H.. 2011. Different contributions of ancient mitochondrial and Y-chromosomal lineages in ‘Karretjie people’ of the Great Karoo in South Africa. J Hum Genet. 569:623–630. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Lombard M, Soodyall H.. 2013. MtDNA control region variation affirms diversity and deep sub-structure in populations from southern Africa. BMC Evol Biol. 13:56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch CM, Malmström H, Günther T, Sjödin P, Coutinho A, Edlund H, Munters AR, Vicente M, Steyn M, Soodyall H, et al. 2017. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science 3586363:652–655. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Prins F, Lombard M, Jakobsson M, Soodyall H.. 2016. The disappearing San of southeastern Africa and their genetic affinities. Hum Genet. 13512:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MG, et al. 2012. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 215:550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch CM. 2010. Genetic variation in Khoisan-speaking populations from southern Africa. Johannesburg (South Africa: ): University of the Witwatersrand. [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. 2010. Complete Khoisan and Bantu genomes from southern Africa. Nature 4637283:943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Thompson JC, Prendergast ME, Mittnik A, Sirak K, Hajdinjak M, Salie T, Rohland N, Mallick S, Peltzer A, et al. 2017. Reconstructing prehistoric African population structure. Cell 1711:59–71.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB. 2008. Pastoral origins at the Cape, South Africa: influences and arguments. S Afr Hum. 20:49–60. [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, et al. 2009. The genetic structure and history of Africans and African Americans. Science 3245930:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren C, Kim M, Martin AR, Bobo D, Gignoux CR, van Helden PD, Möller M, Hoal EG, Henn BM.. 2016. Fine-scale human population structure in southern Africa reflects ecogeographic boundaries. Genetics 2041:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.