ABSTRACT
Background
Genes in metabolic and nutrient signaling pathways play important roles in lifespan in model organisms and human longevity.
Objective
The aim of this study was to examine the relation of a quantitative measure of healthy diet to gene expression in a community-based cohort.
Methods
We used the 2015 Dietary Guidelines for Americans Adherence Index (DGAI) score to quantify key dietary recommendations of an overall healthy diet. Our current analyses included 2220 Offspring participants (mean age 66 ± 9 y, 55.4% women) and 2941 Third-Generation participants (mean age 46 ± 9 y, 54.5% women) from the Framingham Heart Study. Gene expression was profiled in blood through the use of the Affymetrix Human Exon 1.0 ST Array. We conducted a transcriptome-wide association study of DGAI adjusting for age, sex, smoking, cell counts, and technical covariates. We also constructed a combined gene score from genes significantly associated with DGAI.
Results
The DGAI was significantly associated with the expression of 19 genes (false discovery rate <0.05). The most significant gene, ARRDC3, is a member of the arrestin family of proteins, and evidence in animal models and human data suggests that this gene is a regulator of obesity and energy expenditure. The DGAI gene score was associated with body mass index (P = 1.4 × 10−50), fasting glucose concentration (P = 2.5 × 10−11), type 2 diabetes (P = 1.1 × 10−5), and metabolic syndrome (P = 1.8 × 10−32).
Conclusions
Healthier diet was associated with genes involved in metabolic function. Further work is needed to replicate our findings and investigate the relation of a healthy diet to altered gene regulation.
Introduction
Genes in metabolic and nutrient signaling pathways play important roles in lifespan in model organisms and human exceptional longevity (1–3). The 9 hallmarks of aging biology are linked to adverse metabolic changes (4). Dietary manipulation has been considered as an important regulator to delay age-associated molecular changes (5). Preliminary data suggest that a calorie-restricted diet can play such a role in humans, and the benefits on aging-related outcomes have been demonstrated in short-term trials (6). Together these data suggest that dietary manipulation is useful to promote metabolic fitness and extend human health and longevity.
The National Cancer Institute initiated the Dietary Patterns Methods Project to examine the relation of dietary patterns to mortality in several large and diverse US cohorts. Healthy diet is associated with significant reductions in cardiovascular disease, cancer, and type 2 diabetes (7). The Dietary Guidelines for Americans (DGA) (8) were developed to help Americans make healthy food and beverage choices, with the ultimate goal of promoting health and preventing disease. We used the 2015 Dietary Guidelines for Americans Adherence Index (DGAI) (9) score as a quantified measure of key DGA recommendations to obtain an objective measure of healthy diet. The DGAI includes 1) 14 energy-specific components, with a higher score reflecting greater intake and variety of vegetables, fruit, and protein sources; appropriate intake of grains and dairy foods; and a lower intake of “empty calories”; and 2) 10 healthy-choice components with higher scores reflecting higher intakes of dietary fiber; a higher proportion of grains, fruits, dairy, and meats as whole grains, whole fruits, low-fat dairy, and lean meat, respectively; and lower intake of saturated fats and sodium. Given that gene expression is affected by both genetic and environmental factors, we hypothesize that diet could affect gene expression. The objective of our study was to examine the relation of healthy diet with gene expression in a community-based cohort. We also created a diet gene expression score to understand potential molecular mechanisms linking diet with health.
Methods
Study participants
Participants were from the Framingham Heart Study (FHS) Offspring and Third-Generation cohorts (10, 11). Offspring cohort participants who attended the eighth examination (2005–2008, n = 3021) and Third Generation cohort participants who attended the second examination (2008–2011, n = 3411) and completed a food-frequency questionnaire (FFQ) were eligible for this study. The FFQ includes 126 food items. Participants were requested to report their typical consumption of each of these food items during the past year. Participants were excluded if FFQ or other covariate data were missing, incomplete, or invalid (n = 379 for Offspring cohort; n = 259 for Third-Generation cohort), or missing gene expression data (n = 422 for Offspring cohort; n = 211 for Third-Generation cohort). The flowchart of participants is shown in Supplemental Figure 1. Participants provided written informed consent, and the study was approved by the Boston University Medical Center Institutional Review Board.
DGAI
Participants completed the Harvard semiquantitative FFQ as part of the routine examination. This FFQ has been validated against diet records in other cohorts (12, 13). Data from the FFQ were used to compute the 2015 DGAI. FFQs with ≥13 missing food items or with estimated daily energy intake <600 kcal/d, or >4000 kcal/d for women or, >4200 kcal/d for men were deemed incomplete or invalid, respectively, and were excluded. The DGAI assesses adherence to the key dietary recommendations in the 2015 DGA (14), with higher scores representing better adherence to the guidelines. Each component is assessed on a continuous scale with a value ranging from 0 for complete nonadherence to 1 for perfect adherence to the DGA recommendations. For ease of use and interpretability, the final DGAI has been standardized to a range of 0–100 points.
Fourteen energy-specific components are used to assess adherence to food intake, variety, and empty calorie recommendations based on estimated energy requirements. Participants were first assigned to a specific energy level based on their individual estimated energy requirement according to the equation in the Dietary Reference Intakes for energy (15). Intakes for fruit, dark green vegetables, red and orange vegetables, legumes, starchy vegetables, other vegetables, meat/poultry/eggs, seafood, nuts/seeds/soy, grains, dairy, a variety of fruits and vegetables, a variety of protein sources, and empty calories are assessed on a continuous scale (each component score range 0–1). The 10 healthy choices assess adherence to nutrient and food quality recommendations that are independent of energy requirements. The 10 items include recommendations for sodium, total fat, saturated fat, trans fat, dietary fiber, whole grains, whole fruit, fat-free/low-fat dairy, lean meat, and alcohol, and are scored as above on a continuous 0–1 scale. A penalty is assigned in the scoring for overconsumption of 5 energy-specific food groups considered to be energy-dense (starchy vegetables, grains, dairy, meat/poultry/eggs, and nuts/seeds/soy) if consumption is greater than the recommended intake as listed in https://health.gov/dietaryguidelines/2015/guidelines/appendix-3/. More specifically, consumption of energy-dense foods greater than recommended intakes results in a diminished score from the maximum value of 1 to a score of 0 when consumption reaches ≥2 times the recommended intake.
Gene expression profiling: Offspring and Third-Generation
The details of gene expression profiling were described previously (16). In brief, fasting whole-blood samples were collected in PAXgene blood tubes (PreAnalytiX) during Offspring exam 8 and Third-Generation exam 2. The Affymetrix Human Exon 1.0 ST Array (Affymetrix, Inc.) was used for expression profiling. The signal intensities were summarized by robust multiarray average method (17) and the expression level was log2-transformed. The gene annotations were obtained from Affymetrix NetAffx Analysis Center (version 31). A total of 17,873 unique transcripts were used for downstream analysis.
Covariates
At each FHS examination, height and weight were obtained by technicians according to standardized protocols, and BMI was calculated as weight in kilograms divided by height in meters squared (kg/m2). Two measurements of resting blood pressure were obtained by the physician, and hypertension was considered present if the average blood pressure measurement was ≥140/90 mmHg or the participant reported taking antihypertensive medications. Fasting laboratory measurements include glucose, total cholesterol, LDL cholesterol, HDL cholesterol, and triglycerides. Type 2 diabetes was defined as a fasting plasma glucose ≥126 mg/dL from a single measurement or treatment with medications. Current smokers were defined as smoking ≥1 cigarettes/d during the year prior to the examination. Pack-years was defined as the number of packs of cigarettes smoked per day times the number of years the person has smoked. Participants reported the number of hours per day spent in sleep, sedentary, slight, moderate, and heavy activities to calculate the physical activity index (18). Prevalent cardiovascular disease was defined as coronary heart disease, stroke, or intermittent claudication according to previously established criteria (19). The majority of cancer self-reports were verified with pathology reports.
Statistical analysis
The primary outcome is the expression of 17,873 genes. Linear mixed-effects models were used to test associations between DGAI and gene expression, with DGAI the exposure variable and gene expression the dependent measure. Family relatedness was treated as a random variance–covariance factor in the models. Models were adjusted for age, sex, smoking (measured by pack-years), imputed cell counts, and technical covariates. We used pack-year for the smoking covariate because the effects of smoking on gene expression persist after smoking cessation (20–22). Smoking was included as a covariate because smoking has been associated with lower diet scores (23, 24). Given that the gene expression was measured from whole blood, we adjusted the proportions of each cell type, which were imputed from the measured cell counts based on the gene expression data as described previously (16). In our secondary analysis, we additionally adjusted for other potential confounders, including BMI, current smoking, hypertension, type 2 diabetes, total cholesterol, HDL cholesterol, lipid treatment, physical activity index, prevalent cardiovascular disease, and energy intake.
All the analyses were performed with the R software package (https://www.r-project.org/). The association of gene expression and DGAI was tested by the “lmekin” function within the “kinship2” package. To account for multiple testing we calculated the false discovery rate (FDR) by the Benjamini–Hochberg procedure (25) as implemented in the R function “p.adjust”. Significant transcripts were defined as FDR <0.05.
DGAI gene score
We developed a combined gene score from genes significantly associated with DGAI. The score for sample i is defined as 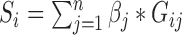 , where n is the number of genes significantly associated with DGAI,
, where n is the number of genes significantly associated with DGAI, 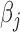 is the estimate of effect size for gene j, and
is the estimate of effect size for gene j, and 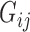 is the expression level of gene j for sample i. The DGAI gene score was scaled with mean 0 and SD 1. We then investigated the cross-sectional association of DGAI gene score with BMI and fasting glucose concentration through the use of linear mixed-effects models. Participants with type 2 diabetes were excluded from the analysis of fasting glucose concentration. We also tested the association of DGAI gene score with type 2 diabetes and metabolic syndrome through the use of generalized estimating equations. Metabolic syndrome was defined as the presence of ≥3 of the following criteria: 1) triglyceride ≥150 mg/dL; 2) blood pressure ≥130/85 mmHg or the use of blood pressure medications; 3) HDL ≤40 mg/dL in men or ≤50 mg/dL in women; 4) blood glucose ≥100 mg/dL or the use of diabetes medications; 5) waist girth ≥35 inches in women, ≥40 inches in men. All these assessments were performed at the same time when blood samples for gene expression profiling were collected (Offspring cohort exam 8 and Third-Generation cohort exam 2).
is the expression level of gene j for sample i. The DGAI gene score was scaled with mean 0 and SD 1. We then investigated the cross-sectional association of DGAI gene score with BMI and fasting glucose concentration through the use of linear mixed-effects models. Participants with type 2 diabetes were excluded from the analysis of fasting glucose concentration. We also tested the association of DGAI gene score with type 2 diabetes and metabolic syndrome through the use of generalized estimating equations. Metabolic syndrome was defined as the presence of ≥3 of the following criteria: 1) triglyceride ≥150 mg/dL; 2) blood pressure ≥130/85 mmHg or the use of blood pressure medications; 3) HDL ≤40 mg/dL in men or ≤50 mg/dL in women; 4) blood glucose ≥100 mg/dL or the use of diabetes medications; 5) waist girth ≥35 inches in women, ≥40 inches in men. All these assessments were performed at the same time when blood samples for gene expression profiling were collected (Offspring cohort exam 8 and Third-Generation cohort exam 2).
We next examined the association between DGAI gene score with metabolic traits in later life. The analysis was restricted to Offspring participants who had completed an additional examination (exam 9) after gene expression profiling (exam 8). Similar to the cross-sectional association study, the prospective study was also adjusted for age and sex.
Network analysis
A dense module-searching strategy (26) was used to build gene interaction networks related to DGAI scores. Experimentally validated gene interactions were downloaded from the PINA database (27). Each gene was assigned a z score 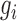 = |z(P value)|, in which the P value represents the association between the gene expression and the DGAI. Each module starts with one of the genes significantly associated with DGAI (defined as the seed genes). Neighboring genes were then added to the module sequentially if they satisfied 2 criteria: 1) the neighboring gene interacted directly with ≥1 gene in the current module; 2) the addition of the neighboring gene to the module would increase the overall module score (28), which is defined as
= |z(P value)|, in which the P value represents the association between the gene expression and the DGAI. Each module starts with one of the genes significantly associated with DGAI (defined as the seed genes). Neighboring genes were then added to the module sequentially if they satisfied 2 criteria: 1) the neighboring gene interacted directly with ≥1 gene in the current module; 2) the addition of the neighboring gene to the module would increase the overall module score (28), which is defined as 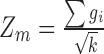 , where k is the number of genes in the module, and
, where k is the number of genes in the module, and 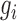 is the score of gene i. The addition of neighboring genes could increase both the numerator (
is the score of gene i. The addition of neighboring genes could increase both the numerator (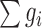 ) and denominator (
) and denominator (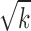 ) of the module score, so only those neighboring genes with large z scores (meaning strong association with the DGAI) could increase the module score and thus be added to the module. All the neighboring genes of the current module would be iteratively added until no more neighboring genes could be added to the current module. Each seed gene would create a module that contains multiple genes. These modules were highly overlapping, so they were merged together to build an interaction subnetwork to represent the overall association of the DGAI (26). The enrichment of these genes in biological pathways was then assessed with WebGestalt (29), a web-based pathway analysis tool. Significant pathways were defined as those with FDR <0.05.
) of the module score, so only those neighboring genes with large z scores (meaning strong association with the DGAI) could increase the module score and thus be added to the module. All the neighboring genes of the current module would be iteratively added until no more neighboring genes could be added to the current module. Each seed gene would create a module that contains multiple genes. These modules were highly overlapping, so they were merged together to build an interaction subnetwork to represent the overall association of the DGAI (26). The enrichment of these genes in biological pathways was then assessed with WebGestalt (29), a web-based pathway analysis tool. Significant pathways were defined as those with FDR <0.05.
Results
The baseline characteristics of the 2220 Offspring participants (mean age 66 ± 9 y, 55.4% women) and 2941 Third-Generation participants (mean age 46 ± 9 y, 54.5% women) in the study sample are shown in Table 1, and the baseline characteristics by cohort and sex are shown in Supplemental Table 1. The mean DGAI was very similar between Offspring and Third-Generation participants (60.5 and 60.9, respectively). As expected, women generally had a higher DGAI than men in both generations.
TABLE 1.
Clinical characteristics of Framingham Offspring participants at exam 8 and Third-Generation participants at exam 21
| Characteristics | Offspring participants (n = 2220) | Third-Generation participants (n = 2941) |
|---|---|---|
| Age, y | 66 ± 9 | 46 ± 9 |
| Women, % | 1230 (55.4) | 1603 (54.5) |
| DGAI | 60.5 ± 11.6 | 60.9 ± 11.2 |
| BMI, kg/m2 | 28 ± 5 | 28 ± 6 |
| Current smoking, % | 171 (7.7) | 103 (3.5) |
| Hypertension, % | 1405 (63.3) | 653 (22.2) |
| Type 2 diabetes, % | 362 (16.3) | 139 (4.7) |
| Total cholesterol, mg/dL | 186 ± 37 | 187 ± 36 |
| HDL cholesterol, mg/dL | 58 ± 18 | 60 ± 18 |
| Lipid treatment, % | 975 (43.9) | 476 (16.2) |
| Physical activity index | 35.3 ± 5.3 | 36.4 ± 6.6 |
| Pack-years | 15.0 ± 21.1 | 9.5 ± 17.3 |
| Cardiovascular disease, % | 356 (16.0) | 73 (2.5) |
| Energy intake, kcal/day | 1870± 640 | 1990± 634 |
Data are represented as means ± SDs for continuous values, or n (%) for categoric values. DGAI, Dietary Guidelines for Americans Adherence Index.
We examined the association of DGAI with transcriptome-wide gene expression in combination of Offspring and Third Generation participants (n = 5161). As shown in Table 2 and Figure 1, the DGAI was significantly associated with the expression of 19 genes after correction for multiple testing (FDR <0.05). Among these genes, 16 were positively associated with DGAI, whereas the remaining 3 genes were negatively associated with DGAI. The most significant gene was ARRDC3 (P = 1.6 × 10−6), a gene that encodes a member of the arrestin family of proteins involved in regulation of G protein–mediated signaling. The association was largely similar after excluding ever-smokers (Supplemental Table 2). We also performed secondary analysis by adjusting for additional potential confounders (see Methods). All 19 top genes remained nominally significant, although the associations were attenuated, suggesting that confounders have marginal effects on the gene association.
TABLE 2.
Top genes associated with DGAI in combined sample of Framingham Heart Study Offspring and Third-Generation participants (n = 5161)1
| Primary model | Secondary model2 | ||||||
|---|---|---|---|---|---|---|---|
| Gene | β | SE | P value | FDR | β | SE | P value |
| ARRDC3 | 0.0016 | 0.0003 | 1.8 × 10−6 | 0.020 | 0.0011 | 0.0003 | 7.9 × 10−4 |
| SIX4 | 0.0012 | 0.0003 | 3.1 × 10−6 | 0.020 | 0.0010 | 0.0003 | 1.3 × 10−4 |
| PKN2 | 0.0011 | 0.0002 | 4.1 × 10−6 | 0.020 | 0.0008 | 0.0002 | 4.1 × 10−4 |
| RNF141 | 0.0016 | 0.0003 | 4.5 × 10−6 | 0.020 | 0.0011 | 0.0003 | 1.7 × 10−3 |
| NOLC1 | −0.0013 | 0.0003 | 1.1 × 10−5 | 0.034 | −0.0012 | 0.0003 | 1.1 × 10−4 |
| STAG1 | 0.0008 | 0.0002 | 1.3 × 10−5 | 0.034 | 0.0006 | 0.0002 | 4.4 × 10−4 |
| DNAJA1 | −0.0012 | 0.0003 | 1.5 × 10−5 | 0.034 | −0.0011 | 0.0003 | 1.8 × 10−4 |
| MIER1 | 0.0010 | 0.0002 | 1.5 × 10−5 | 0.034 | 0.0009 | 0.0002 | 1.6 × 10−4 |
| G0S2 | 0.0027 | 0.0006 | 1.7 × 10−5 | 0.034 | 0.0024 | 0.0006 | 2.1 × 10−4 |
| PTP4A1 | 0.0012 | 0.0003 | 2.5 × 10−5 | 0.037 | 0.0009 | 0.0003 | 1.8 × 10−3 |
| EXOC8 | 0.0010 | 0.0002 | 2.5 × 10−5 | 0.037 | 0.0008 | 0.0002 | 9.5 × 10−4 |
| FAM116A | 0.0010 | 0.0002 | 2.7 × 10−5 | 0.037 | 0.0009 | 0.0002 | 4.6 × 10−4 |
| HIST1H2AE | 0.0027 | 0.0007 | 2.7 × 10−5 | 0.037 | 0.0022 | 0.0007 | 1.0 × 10−3 |
| IER3 | 0.0012 | 0.0003 | 3.1 × 10−5 | 0.039 | 0.0010 | 0.0003 | 7.5 × 10−4 |
| SNX13 | 0.0010 | 0.0002 | 3.5 × 10−5 | 0.042 | 0.0008 | 0.0002 | 1.4 × 10−3 |
| CYB5R4 | 0.0011 | 0.0003 | 4.8 × 10−5 | 0.050 | 0.0007 | 0.0003 | 4.5 × 10−3 |
| RFWD2 | 0.0010 | 0.0003 | 5.0 × 10−5 | 0.050 | 0.0009 | 0.0003 | 2.3 × 10−4 |
| CCDC126 | 0.0016 | 0.0004 | 5.3 × 10−5 | 0.050 | 0.0012 | 0.0004 | 4.0 × 10−3 |
| HSPH1 | −0.0013 | 0.0003 | 5.3 × 10−5 | 0.050 | −0.0012 | 0.0003 | 2.1 × 10−4 |
Genes with FDR <0.05 are listed. The primary model was adjusted for age, sex, pack-years, imputed cell counts, and technical covariates. In comparison with the primary model, the secondary model was additionally adjusted for BMI, current smoking, hypertension, type 2 diabetes, total cholesterol, HDL cholesterol, lipid treatment, physical activity index, prevalent cardiovascular disease, and energy intake. DGAI, Dietary Guidelines for Americans Adherence Index; FDR, false discovery rate.
FIGURE 1.
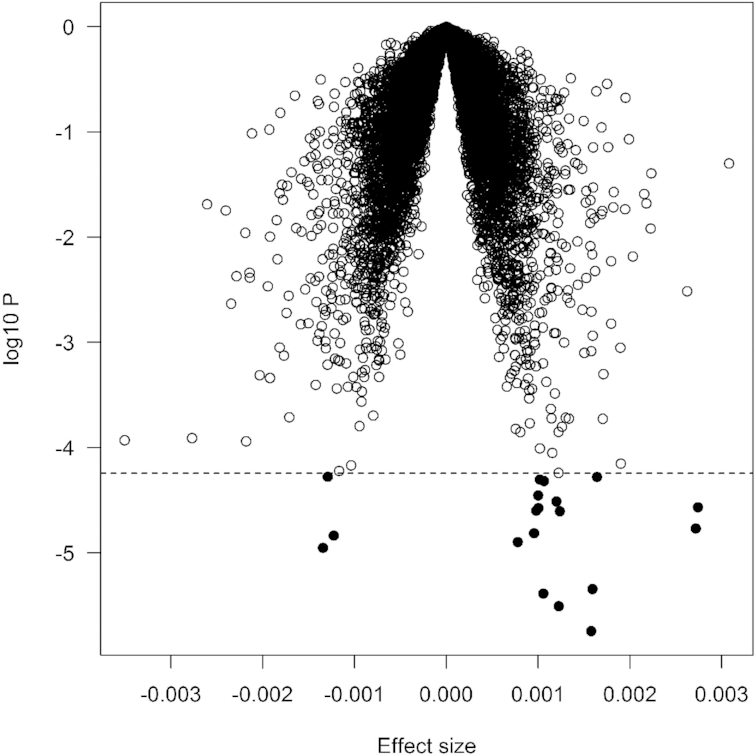
Volcano plot of association with DGAI. Each dot represents 1 gene. The x-axis represents the β estimate of each gene, whereas the y-axis represents the log10P. Positive effects indicate that the genes were positively associated with DGAI, whereas negative effects indicate that the genes were negatively associated with DGAI. The dashed line indicates false discovery rate <0.05, which is equivalent to P < 5.5 × 10−5. DGAI, Dietary Guidelines for Americans Adherence Index.
We then created a DGAI gene score by weighting the association of each gene with DGAI and tested its association with multiple traits. As shown in Table 3, the DGAI gene score was significantly associated with BMI (P = 1.4 × 10−50), fasting glucose concentration (P = 2.5 × 10−11), type 2 diabetes (P = 1.1 × 10−5), and metabolic syndrome (P = 1.8 × 10−32).
TABLE 3.
Association of DGAI gene score with different metabolic traits at the time when gene expression was profiled for Offspring and Third-Generation participants (n = 5161)
| Metabolic traits | β | SE | P value |
|---|---|---|---|
| BMI | −1.10 | 0.07 | 1.4 × 10−50 |
| Fasting glucose concentration | −0.83 | 0.12 | 2.5 × 10−11 |
| Type 2 diabetes (n = 441) | −0.25 | 0.06 | 1.1 × 10−5 |
| Metabolic syndrome (n = 1820) | −0.43 | 0.04 | 1.8 × 10−32 |
The analyses were adjusted for age and sex. DGAI, Dietary Guidelines for Americans Adherence Index.
We also performed a prospective study to examine if DGAI gene score was associated with future metabolic traits. The analysis was restricted to Offspring participants who attended examination (exam 9) on average 6 y after gene expression profiling (exam 8). Individuals with prevalent type 2 diabetes (n = 207) and prevalent metabolic syndrome (n = 837) at exam 8 were excluded from prospective analysis for type 2 diabetes and metabolic syndrome, respectively. At exam 9, there were 70 individuals who met the criteria for type 2 diabetes (1497 without type 2 diabetes) and 123 for metabolic syndrome (815 without metabolic syndrome). Interestingly, the DGAI gene score was still associated with type 2 diabetes (β −0.38, SE 0.13, P = 3.6 × 10−3) but not metabolic syndrome (β−0.02, SE 0.11, P = 0.88).
We then built a gene-interaction subnetwork to examine the interaction between the diet-related genes. As shown in Figure 2, the subnetwork is comprised of 61 nodes and 121 edges, whereas each node represents 1 gene, and each edge represents the interaction between 2 genes. One pivotal gene is YWHAZ. The gene was not associated with the DGAI (P = 0.42), but it was connected to 13 neighboring genes, including 12 genes that were nominally associated with DGAI. Supplemental Tables 3 and 4 show the top enriched biological processes. Many genes were involved in cell cycle (P = 5.6 × 10−5) and endocytosis pathway (P = 7.5 × 10−5).
FIGURE 2.
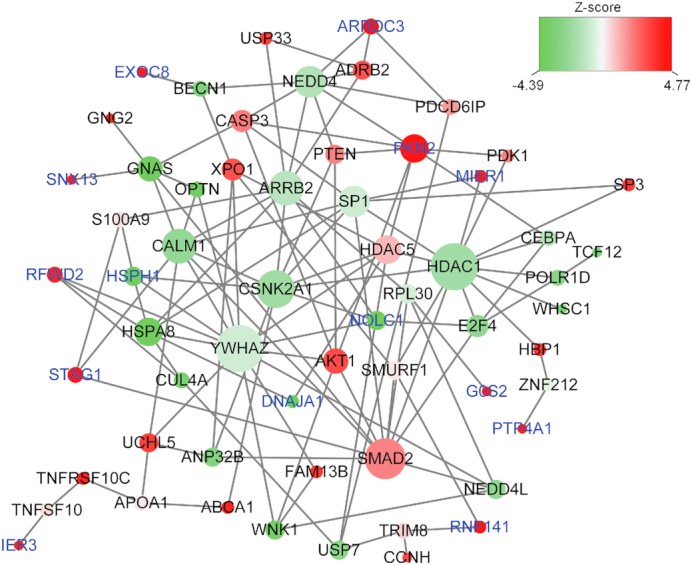
Diet-related subnetwork derived from protein–protein interaction. Each node represents 1 gene, whereas each edge represents the interaction between 2 genes. The nodes are colored to represent their association with dietary score: red represents a strong positive association, and green represents a strong negative association. The node size is proportional to the number of edges that the node connects to. Gene labels in blue represent those that were significantly associated with DGAI (false discovery rate <0.05).
Discussion
In our community-based study, we conducted a transcriptome-wide association study of dietary quality phenotype and found the DGAI was associated with the expression of 19 genes. The DGAI gene score was cross-sectionally associated with several metabolic parameters in our sample, including BMI, fasting glucose, type 2 diabetes, and metabolic syndrome. The DGAI gene score was also associated with incident type 2 diabetes in the older participants. Our findings provide a connection between diet quality, gene expression, and metabolism, and require replication in independent samples. Given the importance of metabolism to aging biology (4), the genes may provide insights into the link between healthy diet and aging or age-related disease.
The most significant gene, ARRDC3, is a member of the arrestin family of proteins and is expressed in >20 human tissues, including the brain, heart, lung, skeletal muscle, pancreas, and liver. The encoded protein plays a role in regulation of breast cancer growth and progression (30) and is a tumor suppressor. A genome-wide linkage scan for human obesity identified an association in males but not females for BMI in a locus containing a single gene, ARRDC3 (31). In mouse models, decreased ARRDC3 levels were associated with increased energy expenditure and increased thermogenesis of adipose tissue with resultant protection from obesity (31). One potential mechanism is an increase in B-adrenergic signaling in adipose tissue with decreased ARRDC3 because ARRDC3 directly interacts with the B-adrenergic receptors. Adipocyte-specific Arrdc3-null mice were observed to have improved glucose tolerance (32). Taken together, these emerging data suggest a role for this gene in metabolism; however, further work is needed to fully characterize the role of ARRDC3 in metabolic regulation and metabolic disease (33).
Additional genes among the 19 genes significantly associated with DGAI include those that regulate metabolic processes. G0S2 (G0/G1 switch 2) inhibits adipose triglyceride lipase to regulate lipolysis and fatty acid availability in adipocytes (34). This gene also plays a role in regulating triglyceride metabolism in the liver (35). G0S2 may be a useful therapeutic target for treatment of obesity-related metabolic disorders (35). A second gene, PKN2 (protein kinase N2), altered glucose and lipid metabolism via primary human skeletal muscle cells (36). In searching the genome-wide association study catalog, genetic variants in several genes linked to healthy diet were reported to have associations with metabolic traits. Genetic variants within the PKN2 region were associated with human height (37, 38), pulse pressure (39), and liver enzyme concentrations (40). Genetic variants within STAG1 were associated with human height (38), BMI (41), C-reactive protein and HDL (42), and coronary artery disease (43). Genetic variants within IER3 were associated with type 1 diabetes (44). Taken together, several of the genes associated with the DGAI score appear to have associations with metabolic health.
Our network and pathway analysis link the DGAI score genes to biological pathways with diverse cellular functions. In the diet-related gene subnetwork analysis, YWHAZ was found to connect to multiple genes in the network, including 3 of the 19 significant DGAI genes, NOLC1, RFWD2 and HSPH1, although was not itself associated with DGAI score (P = 0.42). YWHAZ encodes a highly conserved protein tyrosine 3-mono-oxygenase that is involved in the regulation of insulin sensitivity and glucose tolerance (45). The expression of YWHAZ remains stable in nonalcoholic fatty liver disease (46). It also plays an important role in the development of gastric cancer (47) and ovarian cancer (48, 49). The transforming growth factor β signaling pathway was among the top pathways identified and is known to regulate a range of cellular functions. This pathway included both SMAD2 (P = 0.05) and E2F4 (P = 0.01) nominally associated with DGAI score and involved in energy consumption and metabolism (50–53).
The FFQ allows an efficient and cost-effective assessment of usual, long-term dietary intake in large population studies. However, this method is based on self-reported recall of diet, a major limitation as FFQs are prone to error. Such error would generally be nondifferential in nature and result in weakening of observed associations. However, there is a considerable literature on the validity of the Harvard FFQ, which we used in this study, for assessing food intakes, comparing food intake assessed via FFQ and food intake based on multiple-day diet records. Salvini et al. (54) compared food intake from four 7-d diet records completed over the course of 1 y and an FFQ completed at the end of the year in women. The median correlations for the 2 methods were 0.63 for consumption of all fruits and vegetables; 0.58 for meats, fish, and eggs; 0.78 for dairy products; 0.79 for ready-to-eat cereals; 0.71 and 0.77 for white and dark breads, respectively; and 0.94 for beer, 0.90 for wine, and 0.84 for liquor consumption. A second study in men (13) compared food intakes from two 7-d diet records completed ∼6 mo apart and an FFQ completed ∼3 mo after the second diet record. Median correlations between methods were 0.77 for total fruits; 0.46 for total vegetables; 0.70 for meats, fish, and eggs; 0.71 for dairy; 0.86 for ready-to-eat cereals; 0.45 and 0.37 for white and dark breads, respectively; and 0.88 for beer, 0.81 for wine, and 0.78 for liquor consumption. These validation studies suggest that, for the most foods, the FFQ provides a valid assessment of usual intake. Moreover, to improve the validity of the dietary data, we excluded participants whose dietary data were considered to be unreliable based on extreme reported energy intakes (<600 kcal/d, or >4000 kcal/d for women or >4200 kcal/d for men) or large numbers of food items left blank (>12 food items).
The strengths of our study are that it involved a well-phenotyped community-based cohort to construct the healthy diet score, and the covariates were comprehensively assessed, permitting adjustment for known confounders in our models. We acknowledge several limitations of our study. Our study samples comprised predominantly individuals of European ancestry. It is thus unclear if similar associations are apparent in individuals of other racial/ethnic backgrounds. The current transcriptomic study is an association study, and we cannot infer causality. Our findings need to be replicated in an independent sample. Our measurements of diet and gene expression were taken at a single point over the adult life course. We did not examine other dietary recommendations known to be associated with reduced rates of mortality and chronic disease (55), nor did we study specific macronutrients (carbohydrate, fats, proteins) shown to be related to mortality in the Prospective Urban Rural Epidemiology Study (56) as well as in other large studies.
In conclusion, our study found that healthier diet was significantly associated with the expression of 19 genes. The expression profile of these genes was also associated with metabolism, suggesting potential molecular mechanisms for future investigation that may link healthy diet with outcomes in older adults.
Supplementary Material
Acknowledgments
The authors’ contributions were as follows—HL and JMM: designed the study and wrote the paper; HL, KLL, and XM: performed the statistical analysis; GTR, LMT, and PFJ: measured the healthy diet scores; DL: measured the gene expression profiles; HL: had primary responsibility for the final content; and all authors: read and approved the final manuscript. The authors declare no conflicts of interest.
Notes
Supported by NIH grants R56AG029451 (JMM) and P30AG038070 (The Jackson Laboratory Nathan Schock Center of Excellence in the Basic Biology of Aging). HL was partly supported by Boston University Digital Health Initiative, Boston University Alzheimer's Disease Pilot Grant (P30-AG013846), and the National Center for Advancing Translational Sciences, National Institutes of Health, through BU-CTSI grant 1UL1TR001430. FHS gene expression profiling was funded through the Division of Intramural Research (Levy), National Heart, Lung, and Blood Institute, NIH, Bethesda, MD. This work was also supported in part by the USDA—Agricultural Research Service (ARS), agreement 58-1950-4-003. The Framingham Heart Study is supported by the National Heart, Lung, and Blood Institute's Framingham Heart Study (contracts N01-HC-25195 and HHSN268201500001I). The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the NIH; the US Department of Health and Human Services; or the USDA.
Supplemental Tables 1–4 and Supplemental Figure 1 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: DGA, Dietary Guidelines for Americans; DGAI, Dietary Guidelines for Americans Adherence Index; FDR, false discovery rate; FFQ, food-frequency questionnaire; FHS, Framingham Heart Study.
References
- 1. Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–12. [DOI] [PubMed] [Google Scholar]
- 2. Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328(5976):321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105(37):13987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lopez-Otin C, Galluzzi L, Freije JMP, Madeo F, Kroemer G. Metabolic control of longevity. Cell. 2016;166(4):802–21. [DOI] [PubMed] [Google Scholar]
- 5. Bertozzi B, Tosti V, Fontana L. Beyond calories: an integrated approach to promote health, longevity, and well-being. Gerontology. 2017;63(1):13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ravussin E, Redman LM, Rochon J, Das SK, Fontana L, Kraus WE, Romashkan S, Williamson DA, Meydani SN, Villareal DT et al.. A 2-year randomized controlled trial of human caloric restriction: feasibility and effects on predictors of health span and longevity. J Gerontol A Biol Sci Med Sci. 2015;70(9):1097–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwingshackl L, Hoffmann G. Diet quality as assessed by the Healthy Eating Index, the Alternate Healthy Eating Index, the Dietary Approaches to Stop Hypertension score, and health outcomes: a systematic review and meta-analysis of cohort studies. J Acad Nutr Diet. 2015;115(5):780–800. e5. [DOI] [PubMed] [Google Scholar]
- 8. US Department of Health and Human Services. USDoAaUSDoHaH. Dietary Guidelines for Americans. Washington (DC): US Government Printing Office; 2010. [Google Scholar]
- 9. Sauder KA, Proctor DN, Chow M, Troy LM, Wang N, Vita JA, Vasan RS, Mitchell GF, Jacques PF, Hamburg NM et al.. Endothelial function, arterial stiffness and adherence to the 2010 Dietary Guidelines for Americans: a cross-sectional analysis. Br J Nutr. 2015;113(11):1773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4(4):518–25. [DOI] [PubMed] [Google Scholar]
- 11. Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D'Agostino RB Sr., Fox CS, Larson MG, Murabito JM et al.. The Third Generation cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165(11):1328–35. [DOI] [PubMed] [Google Scholar]
- 12. Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC. Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 1992;135(10):1114–26.; discussion 27–36. [DOI] [PubMed] [Google Scholar]
- 13. Feskanich D, Rimm EB, Giovannucci EL, Colditz GA, Stampfer MJ, Litin LB, Willett WC. Reproducibility and validity of food intake measurements from a semiquantitative food frequency questionnaire. J Am Diet Assoc. 1993;93(7):790–6. [DOI] [PubMed] [Google Scholar]
- 14. US Department of Agriculture and US Department of Health and Human Services. Dietary Guidelines for Americans, 2010. 7th edition, Washington (DC): US Government Printing Office; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panel on Macronutrients, Food and Nutrition Board, Institute of Medicine. Dietary Reference Intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein and amino acids (macronutrients). Washington (DC): National Academies Press; 2005. [Google Scholar]
- 16. Joehanes R, Zhang X, Huan T, Yao C, Ying SX, Nguyen QT, Demirkale CY, Feolo ML, Sharopova NR, Sturcke A et al.. Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol. 2017;18(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. [DOI] [PubMed] [Google Scholar]
- 18. Kiely DK, Wolf PA, Cupples LA, Beiser AS, Kannel WB. Physical activity and stroke risk: the Framingham Study. Am J Epidemiol. 1994;140(7):608–20. [DOI] [PubMed] [Google Scholar]
- 19. Tsao CW, Vasan RS. Cohort profile: the Framingham Heart Study (FHS): overview of milestones in cardiovascular epidemiology. Int J Epidemiol. 2015;44(6):1800–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW et al.. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3(2):e1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Staaf J, Jonsson G, Jonsson M, Karlsson A, Isaksson S, Salomonsson A, Pettersson HM, Soller M, Ewers SB, Johansson L et al.. Relation between smoking history and gene expression profiles in lung adenocarcinomas. BMC Med Genomics. 2012;5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang L, Lee JJ, Tang H, Fan YH, Xiao L, Ren H, Kurie J, Morice RC, Hong WK, Mao L. Impact of smoking cessation on global gene expression in the bronchial epithelium of chronic smokers. Cancer Prev Res (Phila). 2008;1(2):112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ervin RB. Healthy Eating Index scores among adults, 60 years of age and over, by sociodemographic and health characteristics: United States, 1999–2002. Adv Data. 2008(395):1–16. [PubMed] [Google Scholar]
- 24. Alkerwi A, Baydarlioglu B, Sauvageot N, Stranges S, Lemmens P, Shivappa N, Hebert JR. Smoking status is inversely associated with overall diet quality: findings from the ORISCAV-LUX study. Clin Nutr. 2017;36(5):1275–82. [DOI] [PubMed] [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 26. Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein–protein interaction networks. Bioinformatics. 2011;27(1):95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S, Wu J. PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012;40(Database issue):D862–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18 Suppl 1:S233–40. [DOI] [PubMed] [Google Scholar]
- 29. Wang J, Duncan D, Shi Z, Zhang B. Web-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arakaki AKS, Pan WA, Lin H, Trejo J. The alpha-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling. J Biol Chem. 2018;293(9):3350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, Zhang Y, Dobrin R, Cohen DE, Larsen PR et al.. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metabolism. 2011;14(5):671–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carroll SH, Zhang E, Wang BF, LeClair KB, Rahman A, Cohen DE, Plutzky J, Patwari P, Lee RT. Adipocyte arrestin domain-containing 3 protein (Arrdc3) regulates uncoupling protein 1 (Ucp1) expression in white adipose independently of canonical changes in beta-adrenergic receptor signaling. PLoS One. 2017;12(3):e0173823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patwari P, Lee RT. An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab. 2012;23(5):216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X, Heckmann BL, Campbell LE, Liu J. G0S2: a small giant controller of lipolysis and adipose-liver fatty acid flux. Biochim Biophys Acta. 2017;1862(10 Pt B):1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y, Zhang Y, Qian H, Lu J, Zhang Z, Min X, Lang M, Yang H, Wang N, Zhang P. The g0/g1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PLoS One. 2013;8(8):e72315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruby MA, Riedl I, Massart J, Ahlin M, Zierath JR. Protein kinase N2 regulates AMP kinase signaling and insulin responsiveness of glucose metabolism in skeletal muscle. Am J Physiol Endocrinol Metab. 2017;313(4):E483–E91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S et al.. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z et al.. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O'Reilly PF, Cabrera CP, Warren HR, Rose LM et al.. Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension. 2017, 70, e4–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE et al.. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43(11):1131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K et al.. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet. 2017;49(10):1458–67. [DOI] [PubMed] [Google Scholar]
- 42. Ligthart S, Vaez A, Hsu YH, , Stolk R, Uitterlinden AG, Hofman A, Alizadeh BZInflammation Working Group of the CHARGE Consortium, PMI-WG XCP, LifeLines Cohort Study; et al.; Inflammation Working Group of the CHARGE Consortium, PMI-WG XCP, LifeLines Cohort Study Bivariate genome-wide association study identifies novel pleiotropic loci for lipids and inflammation. BMC Genomics. 2016;17:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122(3):433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tomer Y, Dolan LM, Kahaly G, Divers J, D'Agostino RB Jr., Imperatore G, Dabelea D, Marcovina S, Black MH, Pihoker C et al.. Genome wide identification of new genes and pathways in patients with both autoimmune thyroiditis and type 1 diabetes. J Autoimmun. 2015;60:32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lim GE, Piske M, Lulo JE, Ramshaw HS, Lopez AF, Johnson JD. Ywhaz/14-3-3zeta deletion improves glucose tolerance through a GLP-1-dependent mechanism. Endocrinology. 2016;157(7):2649–59. [DOI] [PubMed] [Google Scholar]
- 46. Bruce KD, Sihota KK, Byrne CD, Cagampang FR. The housekeeping gene YWHAZ remains stable in a model of developmentally primed non-alcoholic fatty liver disease. Liver Int. 2012;32(8):1315–21. [DOI] [PubMed] [Google Scholar]
- 47. Liu XX, Ye H, Wang P, Zhang Y, Zhang JY. Identification of 1433zeta as a potential biomarker in gastric cancer by proteomicsbased analysis. Mol Med Rep. 2017;16(5):7759–65. [DOI] [PubMed] [Google Scholar]
- 48. Kim HJ, Sung SH, Kim CY, Bae MK, Cho MS, Kim YH, Kim SC, Ju W. 14-3-3zeta overexpression is associated with poor prognosis in ovarian cancer. Yonsei Med J. 2018;59(1):51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hong L, Chen W, Xing A, Wu D, Wang S. Inhibition of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) overcomes drug resistance and tumorigenicity in ovarian cancer. Cell Physiol Biochem. 2018;49(1):53–64. [DOI] [PubMed] [Google Scholar]
- 50. Seong HA, Manoharan R, Ha H. Smad proteins differentially regulate obesity-induced glucose and lipid abnormalities and inflammation via class-specific control of AMPK-related kinase MPK38/MELK activity. Cell Death Dis. 2018;9(5):471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Casalena G, Bottinger E, Daehn I. TGFbeta-induced actin cytoskeleton rearrangement in podocytes is associated with compensatory adaptation of mitochondrial energy metabolism. Nephron. 2015;131(4):278–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seong HA, Manoharan R, Ha H. Coordinate activation of redox-dependent ASK1/TGF-beta signaling by a multiprotein complex (MPK38, ASK1, SMADs, ZPR9, and TRX) improves glucose and lipid metabolism in mice. Antioxid Redox Signal. 2016;24(8):434–52. [DOI] [PubMed] [Google Scholar]
- 53. Lee BK, Bhinge AA, Iyer VR. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011;39(9):3558–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Salvini S, Hunter DJ, Sampson L, Stampfer MJ, Colditz GA, Rosner B, Willett WC. Food-based validation of a dietary questionnaire: the effects of week-to-week variation in food consumption. Int J Epidemiol. 1989;18(4):858–67. [DOI] [PubMed] [Google Scholar]
- 55. Greenlee H, Strizich G, Lovasi GS, Kaplan RC, Biggs ML, Li CI, Richardson J, Burke GL, Fitzpatrick AL, Fretts AM et al.. Concordance with prevention guidelines and subsequent cancer, cardiovascular disease, and mortality: A longitudinal study of older adults. Am J Epidemiol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dehghan M, Mente A, Zhang X, Swaminathan S, Li W, Mohan V, Iqbal R, Kumar R, Wentzel-Viljoen E, Rosengren A et al.. Associations of fats and carbohydrate intake with cardiovascular disease and mortality in 18 countries from five continents (PURE): a prospective cohort study. Lancet. 2017;390(10107):2050–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


