Nitric oxide exerts an anti-ripening effect, regulating the transcriptome and the fatty acid metabolism of sweet pepper (Capsicum annuum) fruits.
Keywords: Ascorbate peroxidase, fatty acids, fruit ripening, lipid peroxidation, nitric oxide, pepper, proline, RNA-Seq, transcriptome
Abstract
Ripening is a complex physiological process that involves changes in reactive nitrogen and oxygen species that govern the shelf-life and quality of fruits. Nitric oxide (NO)-dependent changes in the sweet pepper fruit transcriptome were determined by treating fruits at the initial breaking point stage with NO gas. Fruits were also harvested at the immature (green) and ripe (red) stages. Fruit ripening in the absence of NO resulted in changes in the abundance of 8805 transcripts whose function could be identified. Among these, functional clusters associated with reactive oxygen/nitrogen species and lipid metabolism were significantly modified. NO treatment resulted in the differential expression of 498 genes framed within these functional categories. Biochemical analysis revealed that NO treatment resulted in changes in fatty acid profiling, glutathione and proline contents, and the extent of lipid peroxidation, as well as increases in the activity of ascorbate peroxidase and lipoxygenase. These data provide supporting evidence for the crucial role of NO in the ripening of pepper fruit.
Introduction
Nitric oxide (NO) is generated endogenously by plant cells and regulates the functions of many biomolecules, including proteins, nucleic acids, and lipids, either directly or through a family of derived molecules called reactive nitrogen species (RNS) (Corpas, 2016; Mata-Pérez et al., 2016). Although most studies have focused on the analysis of NO-induced protein post-translational modifications, such as S-nitrosation, tyrosine nitration, and metal nitrosylation (Corpas et al., 2008, 2019; Astier and Lindermayr, 2012; Holzmeister et al., 2015; Begara-Morales et al., 2016), accumulating data indicate that NO is also involved in gene regulation (Grün et al., 2006; Tossi et al., 2011). The development of next-generation sequencing technologies has enabled large-scale analyses of NO-responsive genes to be carried out in different plant systems. Among these technologies, RNA sequencing (RNA-Seq) has become a very useful tool for performing comparative plant transcriptome analyses under physiological and stress conditions (Begara-Morales et al., 2014; Ocaña et al., 2015; Mata-Pérez et al., 2016; Mizzotti et al., 2018).
Pepper (Capsicum annuum L.) plants belong to the Solanaceae family, which includes species such as tomatoes, whose fruits are of notable agro-economic importance worldwide. Among the five domesticated species of Capsicum, the most agronomically important is C. annuum, which has many varieties differentiated by external features such as color (green, red, orange, yellow, white, or purple), size, or shape. At the biochemical level, the capacity for biosynthesis of capsaicinoids also allows distinction between ‘hot’ and ‘sweet’ peppers. Both types of peppers have nutritional value, owing to their content of ascorbic acid (vitamin C) or carotenoids (provitamin A). The majority of studies have focused on hot peppers because capsaicinoids are bioactive molecules with biomedical and industrial applications (Egan et al., 2019; Naves et al., 2019); the number of scientific studies in sweet peppers is significantly lower. However, sweet peppers have a high agro-economical relevance in Europe, especially in south-east Spain because of its optimal climate conditions (temperature and sunlight hours per day) for growth. Over the past 10 years, biochemical data have indicated that, owing to significant modulations in key reactive oxygen species (ROS) and RNS components during ripening from the green to red stage, sweet pepper fruits develop a dynamic nitro-oxidative metabolism (Mateos et al., 2009, 2013; Chaki et al., 2015; Palma et al., 2015; Rodríguez-Ruiz et al., 2017a, b; Muñoz-Vargas et al., 2018; Chu-Puga et al., 2019). However, the molecular information in sweet pepper, especially during the transition from green to red fruits during ripening, is very scarce.
Based on pioneering work by Leshem’s group on NO and fruits (Leshem et al., 1998; Leshem and Pinchasov, 2000), exogenous NO application started to be used in research, with the main goal of extending the post-harvest stage of both climacteric and non-climacteric fruit ripening. Initially, Leshem and colleagues found that in climacteric fruits, NO and ethylene displayed opposite profiles, thus indicating their antagonism. An increasing number of studies showed that the fumigation of fruits with NO could extend their post-harvest life and also preserve their qualities (i.e. turgor, aroma, or flavor) to a different degree depending of the type of fruits (Zhu et al., 2010; Corpas and Palma, 2018). Accordingly, it has been reported that the application of NO during the post-harvest stage can delay softening of papaya (Guo et al., 2014) or provide disease resistance to some types of fungus in orange (Zhou et al., 2016) and peach (Li et al., 2017). Recently, other potential modulators such as hydrogen sulfide (H2S), melatonin, and abscisic acid (ABA) have begun to be used. Many studies have focused on the interaction of NO, H2S, ABA, and ethylene in climacteric fruits (Wu et al., 2018; Cao et al., 2018; Mukherjee, 2019). By contrast, in non-climacteric fruits, research into the role and interactions of specific phytohormones as driving forces in the ripening process is still at an early stage.
The present study has a different aim because it was designed to understand how NO gas could modulate the physiological transition of ripening (from the green to the red stage) of sweet pepper fruit, with the goal of identifying potential signal transduction networks that govern this transition. Accordingly, we provide the first de novo assembly of the sweet pepper fruit transcriptome, which allowed a comparative time-course analysis of its modulation during the transition from green to red fruits in the absence of NO and after exposure to NO gas. Data indicate that NO governs fruit ripening by affecting the transcriptome and fatty acid (FA) metabolism; this suggests that FA-derived molecules could participate in this process, since FAs are key membrane components that play diverse roles during plant growth, development, and pathogen defense, and as aroma volatile compound precursors in fruits (Ties and Barringer, 2012; Li et al., 2016; Lim et al., 2017). The analysis of other specific biochemical parameters, such as the content of proline and glutathione (both reduced and oxidized), lipid peroxidation (an oxidative stress marker), and the activity of ascorbate peroxidase (an antioxidant enzyme), also suggests that NO exerts an anti-maturation effect.
Materials and methods
Plant material
California-type sweet pepper (C. annuum L.) fruits were collected from plants grown in plastic-covered greenhouse (Syngenta Seeds, Ltd, El Ejido, Almería, Spain). Fruits from five different plants were harvested at three different developmental stages: green immature, breaking point (BP), and ripe red. The selected fruits had the same visual characteristics (size, shape, and color) for each stage.
NO treatment at the BP stage was carried out according to Palma et al. (2018). Basically, BP pepper fruits were placed in an NO-enriched atmosphere (5 ppm) in a hermetic box for 1 h, and then stored at room temperature for 3 days. Then, samples were frozen in liquid nitrogen and kept at –80 °C until processing.
RNA extraction, library preparation, and sequencing
Total RNA was isolated from pepper fruits using a two-step method based on Trizol® Reagent (Gibco BRL) and the RNAeasy Plant Mini Kit (Qiagen), following the manufacturer’s instructions. The RNA was quantified spectrophotometrically (NanoDrop, mySPEC, VWR®). Absorbance ratios at 260/280 nm and 260/230 nm were used in order to determine sample quality. RNA integrity was checked by agarose gel electrophoresis and visualized using the ChemiDoc-It®TS3 Imaging Systems (DBA Analytik Jena, USA). Total RNA samples were sequenced by the High-Throughput Sequencing Unit of the University of Malaga, Spain, in the NextSeq550 platform using 2×75 bp reads, and 40–50 million reads were generated for each replicate.
Sequence pre-processing, assembling and annotation
Raw reads were pre-processed, assembled, and evaluated using TransFlow (Seoane et al., 2018), which produces up to 190 different assemblies (including primary and reconciled transcripts) that are then evaluated to find which assembly is closer to the Arabidopsis thaliana and Populus trichocarpa complete transcriptomes. Briefly, reads were pre-processed to remove low-quality, vector, adaptor, low-complexity and contaminant sequences, and organelle DNA, and the undesired segments were trimmed using SeqTrimNext (Falgueras et al., 2010). The resulting useful reads were assembled using different assemblers based on Bruijn algorithms: Oases (Schulz et al., 2012), SOAPdenovo-Trans (Luo et al., 2012), and Ray (Boisvert et al., 2010), executing k-mers 25 and 35. The resulting contigs were clustered with CD-HIT (Fu et al., 2012) and then reconciled with CAP3 or Minimus (Sommer et al., 2007). Evaluation of each set of tentative transcripts (TTs) was performed by Principal Component Analysis (PCA) using FactoMineR (Lê et al., 2008). Finally, the set of TTs from the best transcriptome assembly (the one closest to A. thaliana and P. trichocarpa transcriptomes in the PCA) was annotated against the plant division of UniProtKB using Full-LengtherNext (Seoane et al., 2018) with default parameters (minimal identity of ≥45% and minimal TT coverage of ≥25%). This software also allowed annotation with orthologs from the proteomes of A. thaliana (TAIR 10 database, 27 416 proteins), Solanum lycopersicum (ITAG 2.4, 34 725 proteins), Solanum tuberosum (PGSC_DM_v3.4, 52 925 proteins), and C. annuum (bell pepper, UP000222542, 35 548 proteins).
Mapping and expression analysis
Useful reads were mapped on to the reconstructed transcriptome using Bowtie2 (Langmead and Salzberg, 2012). Transcript counts were obtained using Sam2count.py. Data normalization and differential expression analysis were carried out using DEgenes-Hunter (Gayte et al., 2017) with default parameters. The differential expression analysis examined the relative change in expression between the different samples using different algorithms (EdgeR, DESeq2, Limma, and NOISeq), with the requirement that any differentially expressed gene (DEG) must appear in at least three of the four algorithms. A time-course analysis of gene expression was performed using the RNA-Seq ready maSigPro package (Nueda et al., 2014), where different ripening stages were considered as their corresponding time points. Gene datasets were used for gene ontology (GO) and KEGG pathway enrichment analyses using different web tools [AgriGO v2.0 (Tian et al., 2017), PlantRegMap (Jin et al., 2017), and KOBAS 3.0 (Xie et al., 2011)]. Significantly enriched GO terms were submitted to REViGO (Supek et al., 2011) to remove redundant categories. Supplementary Fig. S1 at JXB online schematically illustrates the process of the experimental procedure used in the sequence pre-processing, assembly, and annotation of the pepper fruit transcriptome.
Preparation of samples for biochemical analyses
Samples were ground in liquid N2 using a mortar and pestle, and the resulting powder was suspended in 50 mM Tris–HCl buffer, pH 7.5, containing 0.1 mM EDTA, 2 mM DTT, 0.1% (v/v) Triton X-100, 1 mM MgCl2, and 10% (v/v) glycerol, to a final plant material/buffer ratio of 1:1 (w/v). Homogenates were filtered through two layers of Miracloth and centrifuged at 27 000 × g at 4 °C for 30 min. The supernatants were used for the enzymatic assays.
Ascorbate peroxidase (APX) activity was determined by monitoring the initial ascorbate oxidation by H2O2 at 290 nm (Hossain et al., 1984). In this case, 2 mM ascorbate was added to the homogenizing medium to preserve this activity (Miyake and Asada, 1996).
Lipoxygenase (LOX) activity was determined by an in-gel assay according to Heinish et al. (1996) with minor modifications. Samples (6 μg proteins were loaded per lane) were separated using discontinuous gel electrophoresis under non-denaturing conditions. After electrophoresis, the gel was briefly rinsed in distilled water and then incubated at 4 °C for 1 h under continuous shaking with a solution containing 0.2 M glycine-NaOH buffer (pH 9.0) and 50 μl linolenic acid (Sigma) prepared in 50 μl ethanol. The gel was then rinsed with distilled water and incubated with 20 ml staining solution containing 0.2 g N,N-dimethyl-p-phenylenediamine, 1.8 ml methanol, and 0.2 ml acetic acid. Gels were shaken at room temperature until the appearance of pink bands representing enzyme activity. Bands were quantified by using ImageJ software.
Determination of lipid peroxidation and proline and glutathione content
Lipid peroxidation was assessed by measuring the concentration of malondialdehyde (MDA) through the formation of thiobarbituric acid reactive substances (Buege and Aust, 1978). Proline was measured spectrophotometrically at 520 nm using the ninhydrin assay (Bates et al., 1973). Reduced glutathione (GSH) and oxidized glutathione (GSSG) were quantified by liquid chromatography electrospray mass spectrometry (LC-ES/MS) according to Airaki et al. (2011). Protein concentration was determined with the Bio-Rad Protein Assay (Hercules, CA, USA) using bovine serum albumin as a standard. Pairwise analysis of variance was used to detect differences between samples with the aid of the Statgraphics Centurion program.
Gas chromatography-mass spectrometry assay of fatty acids
Total lipids were extracted in triplicate based on previous methods (Bligh and Dyer, 1959; Saini and Keum, 2016) with minor modifications. For extraction, only the edible parts of the pepper fruit (pericarp) were used. Samples were ground in liquid N2 using a mortar and pestle, and 2.5 g of the resulting powder was suspended in 25 ml of a solution of chloroform and methanol (2:1, v:v) with continuous shaking for 1 h. Samples were then centrifuged at 5000 × g for 10 min at 25 °C, and the supernatants were recovered. This process was repeated at least twice until the supernatant became colorless. The supernatants obtained during the whole process were transferred to decanting ampoules, 15 ml 0.85% (w/v) NaCl was added, and the mixture was gently shaken. After decanting, the lower organic phase was collected into a pre-weighted glass tube, evaporated to dryness under a flow of N2, and the total lipid content was then determined gravimetrically. Then, the extracted material was derivatized with 1 ml boron trifluoride (BF3)-MeOH for 15 min at 60 °C to obtain the fatty acid methyl esters (FAMEs). After cooling to room temperature, 700 µl hexane and 700 µl Milli-Q water were added and mixed vigorously for 5 min. Approximately 500 µl of the organic phase was transferred to a chromatography vial for loading on to the GC-MS system. A Varian 450 GC 240 MS ion trap mass spectrometer was used with the following conditions: 1 µl injected at 300 °C and split 20:1 with 1 ml min–1 of He as carrier gas. A DB-5MS UI (30 m×0.25 mm×0.25 µm) capillary column (Agilent) was used, with the following temperature program: 50 °C for 5 min, then increasing at 10 °C min–1 to 180 °C, then 3 °C min–1 to 250 °C, and then 5 °C min–1 to 320 °C, where the temperature was held for 2 min. The mass spectrometer worked in electron ionization mode and mass spectra were acquired between 50 and 1000 arbitrary units of mass. Compounds were identified on the basis of similarity with the NIST08 mass spectral library and using standards injected in the same conditions.
Results and discussion
From the point of view of growers and consumers, fruit ripening involves a series of organoleptic changes that lead to products of good or high quality (i.e. fruits that have a desirable color, attractive flavor, palatable nature and other textural properties). On the basis of the physiological behavior of ripening, fruits are classified as either climacteric or non-climacteric. Climacteric fruits continue to ripen after harvesting; for example, harvested green tomatoes can ripen to red because green tomatoes emit ethylene and also have an increased rate of respiration. By contrast, non-climacteric fruits, such as pepper, do not ripen further once they have been harvested and do not respond to ethylene treatment. In previous studies, we demonstrated that endogenous NO was down-regulated during the transition of sweet pepper from green to red (Chaki et al., 2015; Muñoz-Vargas et al., 2018). Consequently, the main goal of the present study was to discover whether exogenously applied NO gas can modulate pepper fruit ripening as far as the whole transcriptome is concerned, and to evaluate at the biochemical and metabolic levels the functional effects of NO application. There was particular interest in the analysis of the transcriptome, which might provide routes and events that can be modulated during ripening of non-climacteric fruits.
De novo assembly of the sweet pepper fruit transcriptome
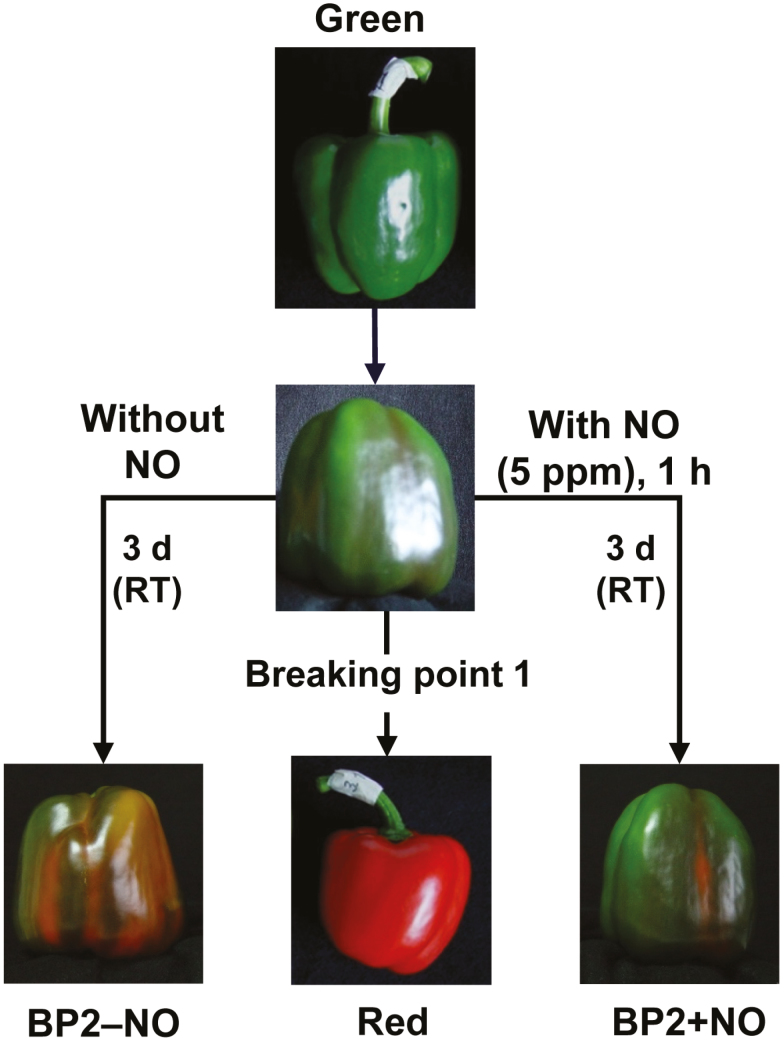
The first step in identifying the genes and functions affected by NO treatment was to construct a transcriptome that incorporates the sweet pepper fruit stages analyzed. Fruits were collected at the standard ripening stages: immature fully green (G), breaking point 1 (BP1), and ripe fully red (R). In addition, to study the effect of NO gas on fruit ripening, two further groups at a breaking point 2 (BP2) stage were included: fruits treated with 5 ppm NO for 1 hour (BP2+NO) and a parallel group of fruits not treated with NO (BP2–NO) followed by storage for 3 d at room temperature. Fig. 1 illustrates the phenotype of pepper fruits at the different ripening stages. In total, 24 cDNA libraries, comprising four independent replicates belonging to the green stage and five to each of the other stages, were generated from these samples. The libraries were sequenced and produced a total of 1 204 589 625 raw reads, which were then trimmed, resulting in 1 153 185 684 (95.73%) useful reads (Supplementary Table S1). Owing to the large number of reads, only two replicates from each ripening stage were loaded into TransFlow (Seoane et al., 2018) to obtain the best affordable de novo assembly using these reads. The best transcriptome consisted of 564 642 TTs with a mean length of 791 bp, of which 42.19% were over 500 bp in length (Supplementary Table S2).
Fig. 1.
Visual phenotype of sweet pepper (Capsicum annuum L.) fruits at different stages and treatments: immature green (G), breaking point 1 (BP1), breaking point 2 without NO-treatment (BP2–NO), breaking point 2 with NO-treatment (BP2+NO), and ripe red (R). Fruits were subjected to an NO-enriched atmosphere (5 ppm) in a hermetic box for 1 h and were then stored at room temperature (RT) for 3 d.
After annotation on the Full-LengtherNext platform, 31.13% of TTs had a clear ortholog and 34 071 were non-redundant according to their UniProtKB identifier (ID). Remarkably, 66 847 TTs encoded complete proteins, 18 397 of which had unique IDs. This suggests that, despite the clear overestimation of the total number of transcripts, full-length open reading frames (ORFs) were reconstructed for a major set of genes expressed in sweet pepper fruit. Annotation also revealed that 67.93% of TTs had no significant homology to any other plant gene, even though 49 708 TTs were predicted to code for complete, unknown proteins by TransDecoder as part of a Full-LengtherNext analysis (Supplementary Table S2). These TTs could be sweet pepper-specific transcripts, small splicing intermediates, or simply assembling artifacts (Benzekri et al., 2014; Ocaña et al., 2015). Although the libraries were not designed to detect any non-coding RNA, due to the deep sequencing analysis by the NextSeq500 platform, 4903 TTs presented significant homology to small non-coding RNA precursors (over 40 nt in length) derived from the RNAcentral database (The RNAcentral Consortium, 2017).
Suitability of the sweet pepper fruit reference transcriptome
The 564 642 TTs can be clearly inferred to include many redundancies and significant transcript fragmentation, which will hinder their further use. Because of this, we used Full-LengtherNext to select a subset of TTs that have a representative sweet pepper fruit transcriptome. This resulted in 63 359 TTs, which include the longest TT for each unique ortholog and all non-redundant, unknown TTs that code for a complete protein (Supplementary Table S2). The mean TT length of this representative transcriptome was 1560 bp, with 55 172 TTs of over 500 bp (results not shown). Quality enhancement was first evaluated by mapping the original set of reads (in Supplementary Table S1) on to the reference transcriptome. Given that the representative transcriptome accounts for only 11.22% of the full set of TTs, a mean of 56.23% reads was found to be mapped (Supplementary Table S1, last column), indicating a 5-fold mapping enrichment, which demonstrates the suitability of the representative transcriptome.
The representative transcriptome was further evaluated according to the annotation level of different protein sets (Table 1). First, annotation in the Plant division of UniProtKB revealed that the reference transcriptome contained 29 716 TTs for different proteins, 17 281 of which coded for a complete protein, and included 13 631 TTs that had no ortholog but were predicted to code for an unidentified protein. These data show that the quality of the representative transcriptome was enhanced with respect to the complete set of TTs. Another independent source of annotation was C. annuum (bell pepper), which has the closest sequenced genome to sweet pepper (Table 1); it was not included as a source of annotation in the Plant division of UniProtKB, as its proteome status has not been reviewed. A total of 46 924 TTs were homologous to bell pepper proteins, but covered only 18 674 different UniProtKB IDs (52.5% of the bell pepper proteome), 12 726 of which contain the complete ORF. The pepper fruit transcriptome was not expected to contain fewer different transcripts than a complete transcriptome. However, surprisingly, the Plant division of the UniProtKB database provided more orthologs than the bell pepper proteome, suggesting that the representative sweet pepper fruit transcriptome contained TTs not yet described in the bell pepper proteome. It would therefore appear to be more appropriate to use our de novo transcriptome than the unreviewed bell pepper proteome/transcriptome.
Table 1.
Statistical characterization of the representative transcriptome annotations using different sets of proteins: plant division of UniProtKB, Capsicum annuum (bell pepper), Arabidopsis thaliana, Solanum lycopersicum (tomato), and Solanum tuberosum (potato)
| Property | Plant division | Bell pepper | Arabidopsis | Tomato | Potato |
|---|---|---|---|---|---|
| TTs with annotation | 49 701 | 46 924 | 36 481 | 47 270 | 50 324 |
| Unique IDs | 29 716 | 18 674 | 14 060 | 18 051 | 23 030 |
| TTs including a complete ORF | 26 968 | 24 878 | 19 482 | 25 357 | 30 345 |
| Unique IDs for complete ORFs | 17 281 | 12 726 | 10 081 | 12 754 | 16 416 |
| TT without ortholog | 13 631 | 16 404 | 26 874 | 16 080 | 13 022 |
ID, Identifier; ORF, open reading frame; TT, tentative transcripts.
To facilitate subsequent functional analysis, the reference transcriptome was also annotated with curated proteomes of the model species A. thaliana and other species closely related to sweet pepper, such as tomato and potato. The A. thaliana proteome, only 45.8% of whose proteins have an ortholog in sweet pepper, was the poorest source of orthologs (Table 1). However, despite the missing information on 26 874 TTs, many bioinformatic tools for functional analysis capable of handling A. thaliana annotations can be very useful. Superior results were obtained with the S. lycopersicum proteome, 53.2% of whose proteins (a proportion similar to that for bell pepper proteins) were able to annotate 18 051 different TTs, with only 16 060 remaining unannotated (Table 1). The results for the S. tuberosum proteome, which provided the largest number of unique orthologs using one single proteome, were even more striking (Table 1). For this species, 23 030 different UniProtKB IDs were actually used, with only 13 022 sweet pepper fruit TTs remaining unannotated (the lowest figure of all; Table 1). This, together with the large number of TTs coding for a complete ORF, clearly indicates that our reference transcriptome provides a suitable representation of the sweet pepper proteome/transcriptome, and it was therefore used for the RNA-Seq studies.
Differentially expressed genes under NO gas treatment
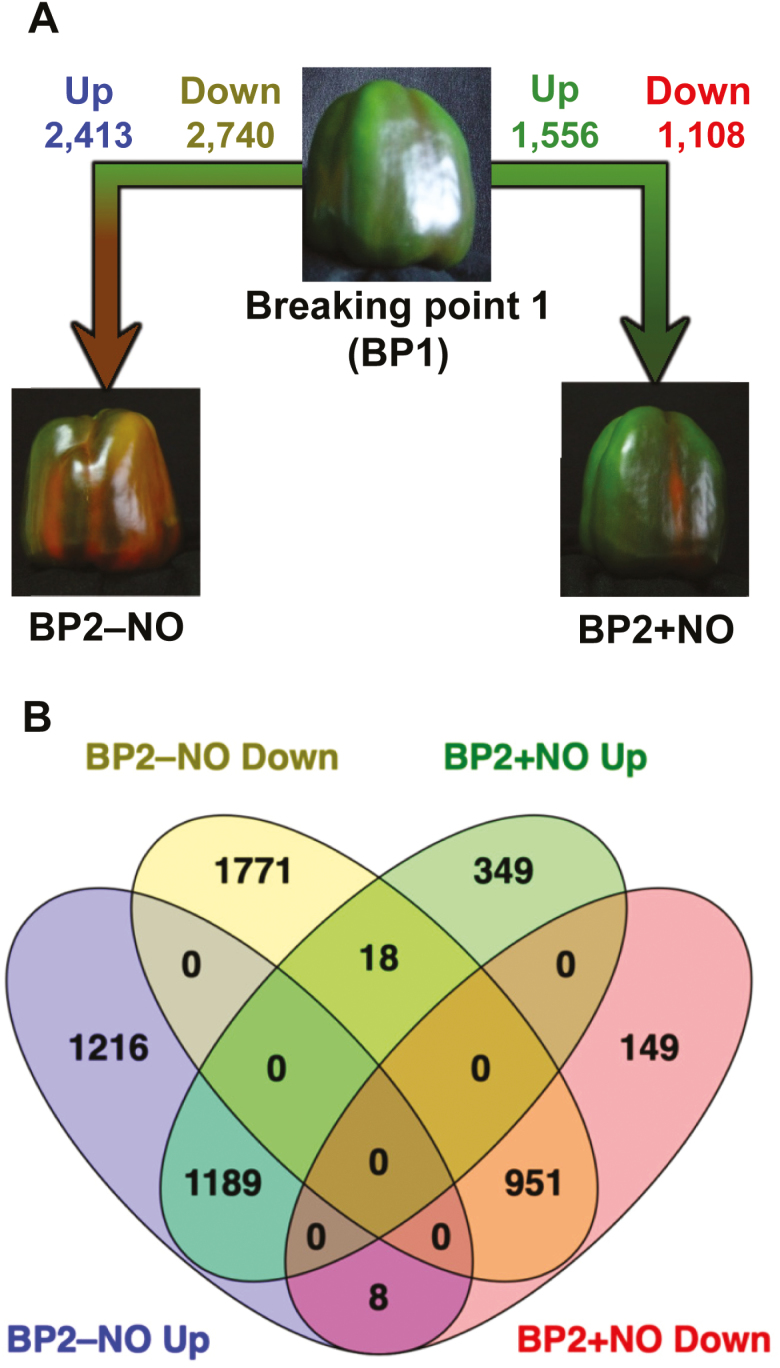
Although the effect of NO treatment could be studied by comparing the differences in gene expression between BP2–NO and BP2+NO pepper fruits, our experimental design (Fig. 1) facilitated a more exhaustive analysis by using the previous ripening stage (BP1) as a reference point. We identified a total of 5153 genes (2413 up-regulated and 2740 down-regulated) expressed differentially in BP1 and BP2–NO; the comparison between BP1 and BP2+NO involved 2664 genes (1556 up-regulated and 1108 down-regulated) (Fig. 2A). In addition, the genes could be divided into groups sharing different expression patterns. Table 2 summarizes eight such groups of genes; these groups enabled us to reduce the complexity of the analysis and to obtain useful biological information (Khatri et al., 2012) (Fig. 2B). In all the gene sets analyzed, a functional enrichment analysis based on GO was carried out, and the assigned GO categories together with their P values were then filtered using the REViGO web application to gather relevant information.
Fig. 2.
Analysis of gene expression during ripening and modulated by NO in sweet pepper fruits. (A) Analysis of differentially expressed genes (DEGs) between pepper fruits at breaking point 1 (BP1), breaking point 2 in untreated fruits (BP2–NO) ,and breaking point 2 in fruits treated with NO (BP2+NO). (B) Venn diagrams combining DEGs (up- and down-regulated) among pepper fruits at different stages of ripening and under NO treatment.
Table 2.
Groups of genes modulated by NO gas treatment observed in Fig. 2B
| Group | Description | Tendency | Gene number |
|---|---|---|---|
| 1 | Genes not affected by the NO treatment | Up-regulated | 1189 |
| 2 | Down-regulated | 951 | |
| 3 | Genes affected by the NO treatment | Up-regulated | 1216 |
| 4 | Down-regulated | 1771 | |
| 5 | Genes specifically affected by the NO treatment | Up-regulated | 349 |
| 6 | Down-regulated | 149 | |
| 7 | Genes whose pattern of expression was changed by the NO treatment | From up- to down-regulated | 8 |
| 8 | From down- to up-regulated | 18 |
Functions and processes influenced by NO gas treatment
Depending on the comparison between gene groups, the effects of NO gas treatment could be masked by genetic changes caused by the natural ripening process itself (Fig. 2A, left side). Up- or down-regulated genes that share the same tendency in both comparisons (BP1 versus BP2–NO and BP1 versus BP2+NO) are considered to be genes whose expression is not modulated by NO; this was the case for a total of 2140 genes (1189 up-regulated and 951 down-regulated) (Fig. 2B). With respect to biological processes where genes were up-regulated regardless of the NO gas treatment, we focused on responses to stimuli such as temperature, redox state, bacteria, oxidative stress, and ABA. We also detected primary metabolic processes including photosynthesis and cellular lipid metabolism, as well as other important mechanisms such as wax biosynthesis, leaf senescence, and protein stabilization. Conversely, in the group of down-regulated genes, we identified different categories: immune systems, developmental processes, ribosome biogenesis, and cell death.
The genes identified when comparing BP1 and BP2–NO (1216 up-regulated and 1771 down-regulated) were those whose overexpression and repression, respectively, were impeded by NO. Treatment with NO masked the overexpression of genes involved in processes such as osmotic stress responses, development, lipid oxidation, and proteolysis. Furthermore, NO prevented the repression of genes involved in methylation, respiratory burst, chloroplast protein targeting, and auxin influx.
Genes specifically affected by NO treatment
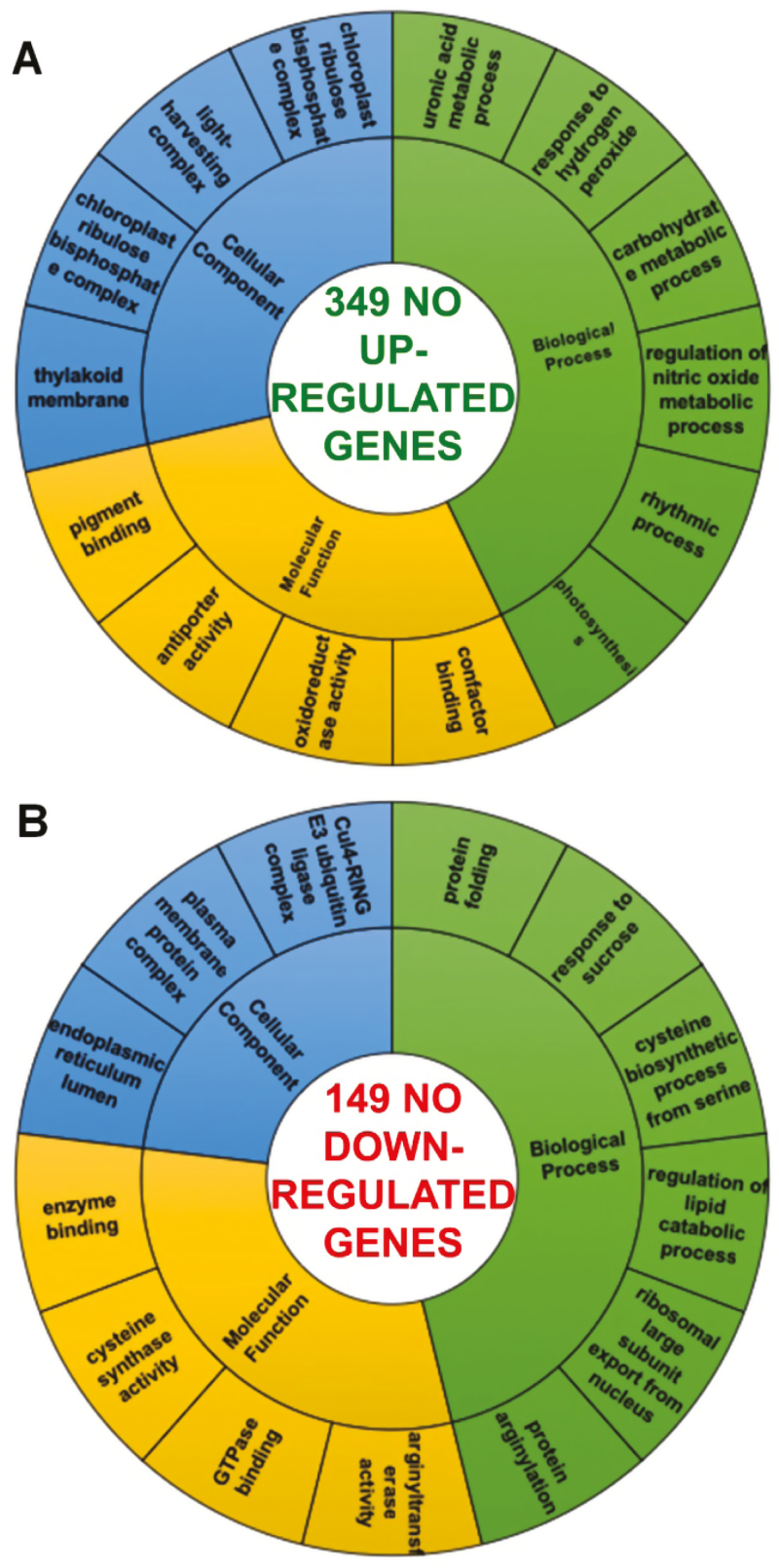
The groups of up- and down-regulated genes exclusively found in BP2+NO (349 and 149, respectively) are those whose expression is exclusively modulated by NO. Fig. 3A shows a gene set enrichment analysis of the 349 genes that were up-regulated by NO treatment. We then carried out a more precise classification to highlight genes encoding proteins, such as calmodulin, related to the regulation of NO metabolism; responses to H2O2, such as heat shock and ubiquitination proteins; and oxidoreductase activity, including thioredoxins, APX, and glutaredoxin (Wang et al., 2013; Mata-Pérez et al., 2016; Zhou et al., 2016; Niu et al., 2017; Zhang et al., 2018). We also classified the 149 down-regulated genes (Fig. 3B), among which we found genes encoding proteins with roles in biological processes involving arginylation and protein folding. Additionally, we detected molecular functions such as cysteine synthase, connected with sulfur metabolism.
Fig. 3.
Functional classification of sweet pepper fruit genes specifically affected by NO gas treatment. Gene ontology (GO) enrichment analysis was performed using PlantRegMap. Genes were classified according to biological processes, molecular functions, and cellular component categories. GO categories with their associated p-values were submitted to REViGO to remove redundant information. (A) Genes up-regulated by NO. (B) Genes down-regulated by NO.
Beyond the simple arithmetic analysis, it could be concluded that the NO gas treatment had less effect on the gene expression pattern than the post-harvest storage for 3 d. In other words, NO seems to modulate the profound genetic changes that take place throughout the ripening process of pepper fruits—a condition that perhaps also occurs in other crop fruits.
Time-course analysis of NO-treated and untreated fruits
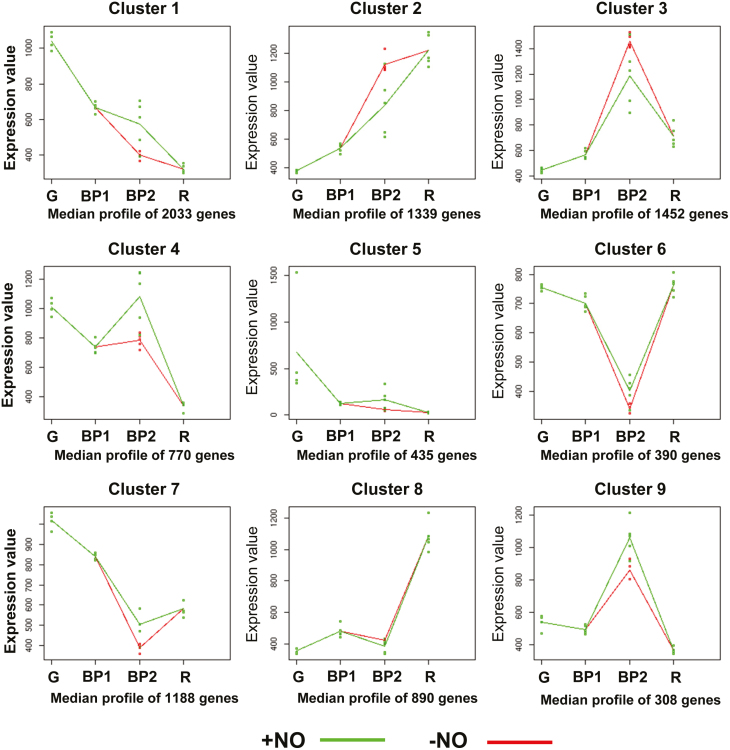
We carried out two different time-course analyses to study variations in transcriptome profiling during fruit ripening and after exposure to NO gas. We first analyzed the natural ripening process in fruits at the G, BP1, BP2–NO, and R stages, and then examined the NO-treated fruits at the stages evaluated in the first time-course analysis. In the second analysis, the BP2–NO sample was replaced by the BP2+NO sample, as both samples had similar time intervals. The maSigPro tool revealed profound temporal changes in the expression of 8805 genes grouped into nine clusters. Fig. 4 shows that NO treatment caused a significant variation in the time course of expression of many genes, especially in clusters 1, 2, 3, 4, 7, and 9. Overall, we observed that the genes in BP2+NO fruits clearly behaved differently from those in BP2–NO fruits, with the possible exception of genes in clusters 5, 6, and 8. Cluster 1 included several down-regulated genes coding for key enzymes involved in ROS and RNS metabolism, including L-galactono-1,4-lactone dehydrogenase (GalLDH), an enzyme that catalyzes the terminal step of the ascorbate biosynthesis pathway (Scherlt et al., 2012); S-nitrosoglutathione reductase, which catalyzes the NADH-dependent reduction of S-nitrosoglutathione, a mobile reservoir of NO (Leterrier et al., 2011); Fe-superoxide dismutase (Fe-SOD), which catalyzes the dismutation of the superoxide radical into H2O2; and 6-phosphogluconate dehydrogenase (6PGDH), which is part of the oxidative pentose phosphate pathway that generates the NADPH necessary for FA synthesis (Hutchings et al., 2005). Some of these enzymes have previously been found to be modulated during ripening (Mateos et al., 2013; Rodríguez-Ruiz et al., 2017a, b). Moreover, cue1/nox1 (chlorophyll a/b binding protein underexpressed 1/NO overproducer 1), which codes for a phosphoenolpyruvate/phosphate translocator of the inner plastid envelope membrane, was present in cluster 1 (Voll et al., 2003). The Arabidopsis knockout mutant (nox1) is characterized by L-arginine and citrulline accumulation and higher NO content compared with wild-type Arabidopsis (He et al., 2004). This suggests that high NO content is caused by NOS-like activity, as nox1 has high L-arginine content. Given this possibility in pepper fruits, the down-regulation of NOX1 during ripening could restore endogenous NO levels, as we previously showed that green pepper fruits have a higher NO content than red fruits (Chaki et al., 2015; Muñoz-Vargas et al 2018). This hypothesis is further corroborated by the presence of the NOA1/RIF1 (nitric oxide-associated protein 1/resistant to inhibition with fosmidomycin 1) ortholog in cluster 9, which could restore endogenous NO levels. On the other hand, although the Arabidopsis null mutant noa1 is reported to have lower NO content than wild-type Arabidopsis, this gene does not encode an NO-generating protein but instead plays an indirect role in NO accumulation (Van Ree et al., 2011). Interestingly, orthologous expression of NOA1 in pepper appears to increase in cluster 9 in the presence of exogenous NO gas, which correlates with the NO metabolism. These findings suggest that the NOA1/RIF1 protein is not essential for NO production and probably plays only an indirect role in NO accumulation.
Fig. 4.
Time-course analysis of genes showing statistically significant changes in expression throughout ripening in sweet pepper fruits exposed to NO gas. Genes were grouped into nine clusters according to their expression profiles at different ripening stages. Expression values corresponding to the ripening stages G, BP1, and R are shared by both time-courses. However, at stage BP2, the expression profile of the clustered genes is represented as either untreated fruits (red line) or fruits treated with NO (green line). The solid line indicates the mean and dots represent the SD of the expression values at each ripening stage. In all clusters the treatment with NO altered the natural post-harvest evolution of gene expression represented by the sequence G → BP1 → BP2 → R.
Moreover, genes in the first three clusters showed enriched GO terms associated with lipid metabolism, including long-chain acyl-CoA synthetase 1, peroxisomal acyl-coenzyme A oxidase 1, and biotin carboxyl carrier protein of acetyl-CoA carboxylase 1, as well as lipoxygenase 6 and phospholipase D. We therefore decided to analyze the FA profile of fruits at all stages of ripening, as well as lipid peroxidation and proline content, which are two parameters associated with different types of oxidative stress.
NO gas modulates the profiles of proline, lipid peroxidation, and glutathione, FA content, APX and LOX activity during ripening
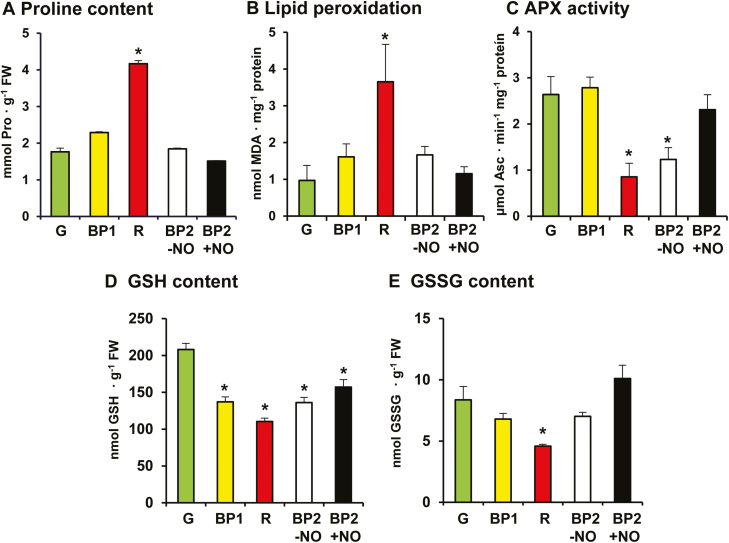
Fig. 5A shows that proline content gradually increased at the different stages of fruit ripening that were analyzed, with a maximum 2.4-fold increase in red fruit compared with green fruit. Significantly, fruit with and without NO treatment (BP2+NO and BP2–NO, respectively) both showed proline content values similar to those for green pepper; values were slightly lower in NO-treated fruits compared with untreated fruits, but the difference was not statistically significant. An increase in proline content is usually associated with a stress response mechanism (Szabados and Savouré, 2010), which is normally associated with oxidative stresses such as salinity, drought, and heavy metals (Signorelli et al., 2013; Bouthour et al., 2015; Rodríguez-Ruiz et al., 2019). However, an increase in proline content has also been linked to other physiological processes, such as senescence, as has been observed in flowers and leaves (Zhang and Becker, 2015). Previous studies of grapevine and tomato fruits also report an increase in proline content during ripening (Stines et al., 1999; Claussen et al., 2006), suggesting that it is an indicator of fruit quality. Our findings corroborate these observations of proline accumulation during ripening but also indicate that NO may negatively regulate proline biosynthesis, which opens up new questions concerning this process. These data are well correlated with the down-regulation of 6PGDH, considering that the biosynthesis of proline from glutamate requires NADPH (Signorelli and Monza, 2017).
Fig. 5.
(A) Proline content, (B) lipid peroxidation level, (C) ascorbate peroxidase (APX) activity, (D) reduced glutathione (GSH) content, and (E) oxidized glutathione (GSSG) contents of sweet pepper fruits at different stages of ripening: immature green (G), breaking point 1 (BP1), breaking point 2 with and without NO treatment (BP2+NO and BP2–NO, respectively), and ripe red (R). Lipid peroxidation was determined by the thiobarbituric acid reactive substances method, using malondialdehyde as standard. Pairwise analysis of variance was used to detect differences in comparison to green fruits. *P<0.05. (This figure is available in colour at JXB online.)
Lipid peroxidation was analyzed because this parameter is a recognized marker of oxidative stress (Yamauchi et al., 2008) characterized by an increase of MDA. Fig. 5B shows that MDA content progressively increased across the different ripening stages, with a 3.7-fold increase in red fruit compared with green fruit. Again, it is worth noting that fruit with and without NO treatment showed similar and intermediate values for MDA content, with a slightly lower value observed for NO-treated fruits compared with untreated fruits, although, again, the difference was not statistically significant. These findings confirm that the ripening process is associated with oxidative stress and that NO treatment has the capacity to reduce MDA content. An increase in lipid peroxidation has also been described during ripening of the fruit of other pepper cultivars (Martí et al., 2011) and species such as blackberry (Wang and Jiao, 2001), tomato (Jiménez et al., 2002), and peach (Huan et al., 2016), which are associated with lower antioxidant enzyme activities. These data are in good agreement with the down-regulation of Fe-SOD found in gene cluster 1, described earlier. In previous studies of pepper ripening, catalase activity was found to diminish, and this was associated with an increase in tyrosine protein nitration (Chaki et al., 2015), a recognized marker of nitro-oxidative stress (Corpas et al., 2009). Recently, we found that O2·−-generating NADPH oxidase activity is up-regulated during pepper fruit ripening (Chu-Puga et al., 2019), which correlates with the increase observed in lipid peroxidation. All these data connect well with earlier observations that NO-treated pepper fruit showed a significant increase in the activity and gene expression of GalLDH, which catalyzes the last step in the biosynthesis of ascorbic acid, whose content increased by 40% compared with untreated pepper fruit (Rodríguez-Ruiz et al., 2017a). Data pertaining to other plant species under diverse stress conditions show that exogenous NO treatment stimulates the activity of different antioxidant enzymes (Manai et al., 2014; Kharbech et al., 2017), including those in fruits; for example, during peach fruit ripening, NO treatment stimulates SOD activity (Kang et al., 2016; Corpas and Palma, 2018).
To corroborate this potential effect on antioxidant enzymes, we analyzed APX activity. Fig. 5C shows a 2.8-fold decrease in this activity in red fruits compared with green fruits. As with proline and lipid peroxidation, APX activity showed intermediate values both with and without NO treatment; in this case, APX activity was 2.7-fold higher in NO-treated fruits compared with red fruits. This positive effect of NO on APX activity closely correlates with findings of previous studies that demonstrated that APX activity is positively regulated by S-nitrosation in pea and Arabidopsis (Begara-Morales et al., 2014; Yang et al., 2015).
Fig. 5D shows that GSH content gradually decreased as ripening proceeded, with a 1.9-fold decrease in red fruit compared with green fruit. Fruits with and without NO treatment showed an intermediate GSH content, with a slightly higher value for NO-treated fruits. GSSG followed a similar pattern to that of GSH, but with values 25-fold lower (Fig. 5E). This GSH enhancement indicates that exogenous NO provides a higher antioxidant capacity, confirming what was previously reported with respect to ascorbate content (Rodríguez-Ruiz et al., 2017a). Similar increases in GSH and ascorbate levels have been described recently in sweet cherry fruit treated with NO-releasing chitosan nanoparticles (Ma et al., 2019).
Taken together, the data suggest that exogenous NO treatment has a regulatory effect on FAs by reducing the degree of lipid peroxidation, with a concomitant increase in GSH content and APX activity, which is expected to affect the mechanism involved in palliating oxidative stress associated with the ripening process.
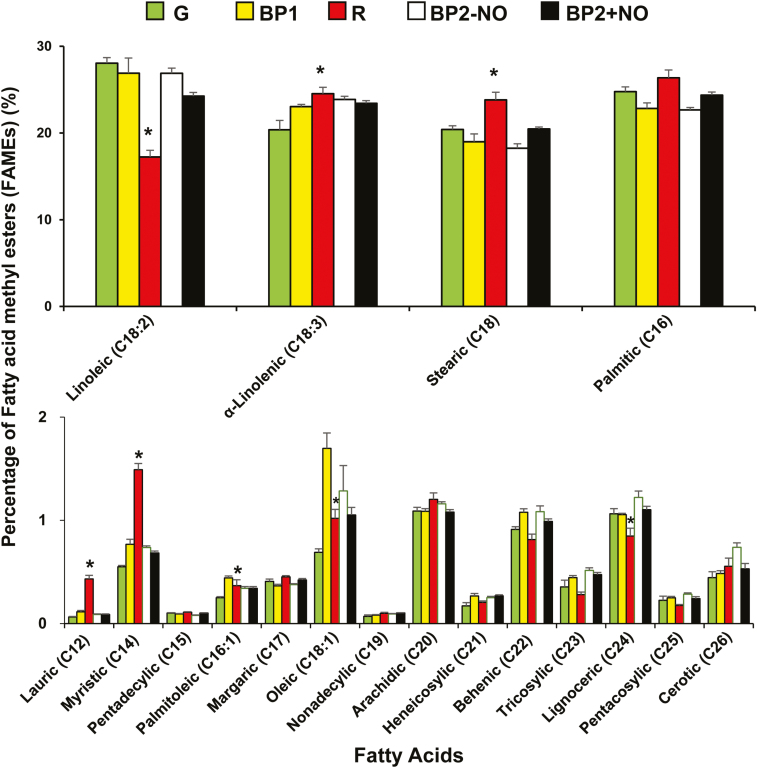
Plant FAs are involved in various functions, such as cell membrane mechanisms and acting as energy reserves, as well as providing jasmonic acid and oxylipin signaling molecules for plant defense (Kachroo and Kachroo, 2009). Given that the composition of FAs can be modulated by physiological processes and in response to biotic and abiotic stresses, FAs in pepper fruits at the different stages of ripening were identified and quantified (Fig. 6). Polyunsaturated linoleic (C18:2) and linolenic (C18:3) acids were the most abundant FAs in the pepper fruits, followed by saturated palmitic (C16) and stearic (C18) acids. We also identified other less abundant FAs ranging from C12 to C26. These data are very much in line with findings of previous studies of pepper fruits of various origins (Pérez-Gálvez et al., 1999; Sora et al., 2015; Ananthan et al., 2018) and also studies of tomato (Saini et al., 2017). During the fruit ripening process, the content of these FAs was differentially regulated, with the content of linolenic and stearic acids being observed to increase and that of linoleic acid to decrease. Moreover, as previously observed with respect to proline and MDA content (see Fig. 5), pepper fruits at BP1 generally showed intermediate values for the majority of FAs. The FA content of fruits at BP2 with and without NO treatment also showed intermediate values, with the content of specific FAs being observed to either decrease or increase in NO-treated fruits. Although the content of different FAs either increases or decreases during the ripening process, to our knowledge, no studies have found that exogenous NO treatment affects either the biosynthesis or the breakdown of these FAs in non-climacteric fruits. In contrast, climacteric peach fruits fumigated with NO and then stored at 5 °C showed an increase in the content of palmitoleic (16:1), oleic, and linolenic acids, while the linoleic acid content decreased (Zhu and Zhou, 2006). These data are in accordance with some of the FAs affected by NO in pepper fruit, supporting the assertion that FA metabolism is modulated by NO.
Fig. 6.
Fatty acid content and profile in sweet pepper fruits at different stages of ripening: immature green (G), breaking point 1 (BP1), breaking point 2 with and without NO treatment (BP2+NO and BP2–NO, respectively), and ripe red (R). Pairwise analysis of variance was used to detect differences in comparison to green fruits. *P<0.05. (This figure is available in colour at JXB online.)
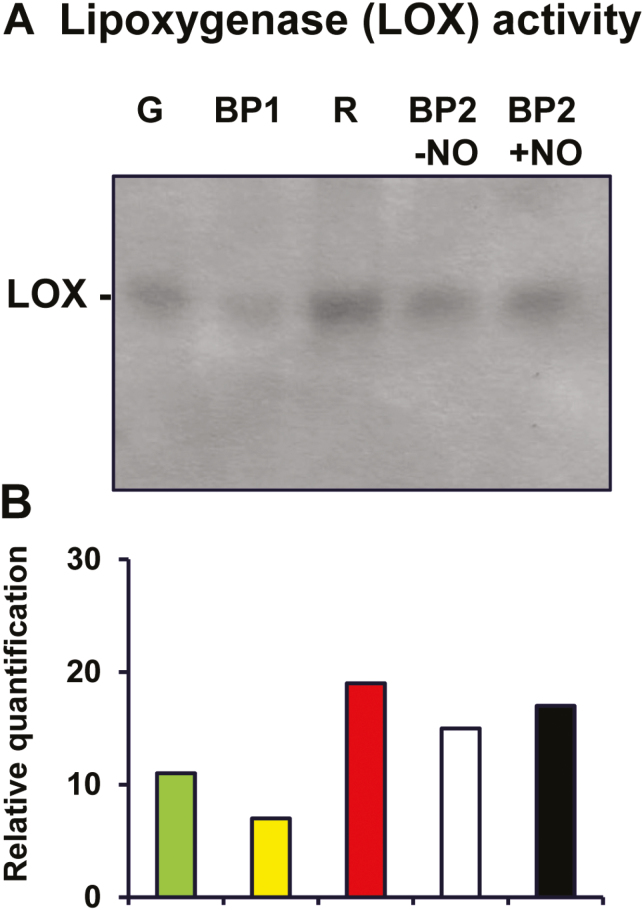
Given that linoleic and linolenic acids are the principal substrates of LOX (Siedow, 1991; Porta and Rocha-Sosa, 2002), which catalyzes the first oxygenation step of polyunsaturated FAs, we assayed LOX activity in pepper fruits at different stages of ripening by non-denaturing gel electrophoresis. LOX activity was detected as a single band (Fig. 7A). Relative quantification indicated a 1.7-fold increase in red fruit compared with green fruit (Fig. 7B). LOX activity in BP2 stage fruits with and without NO treatment showed intermediate values, with a slightly higher level of activity being observed in NO-treated fruits, although the difference was not statistically significant; this tendency was similar to that observed in the analyses of proline and MDA content in BP2–NO fruits (see Fig. 5). Several studies have indicated that LOX activity is involved in fruit ripening, which appears to be modulated by two phytohormones (ABA and ethylene) associated with aroma production and fruit quality (Zhang et al., 2009; Lv et al., 2014; Zhang et al., 2017; Del Ángel-Coronel et al., 2018).
Fig. 7.
Lipoxygenase (LOX) activity staining in non-denaturing polyacrylamide gel electrophoresis of sweet pepper fruits at different stages of ripening: immature green (G), breaking point 1 (BP1), breaking point 2 with and without NO treatment (BP2+NO and BP2–NO, respectively), and ripe red (R). (A) Representative picture of a gel stained for LOX activity. (B) Relative quantification of the band activities were determined using ImageJ software. (This figure is available in colour at JXB online.)
Conclusions
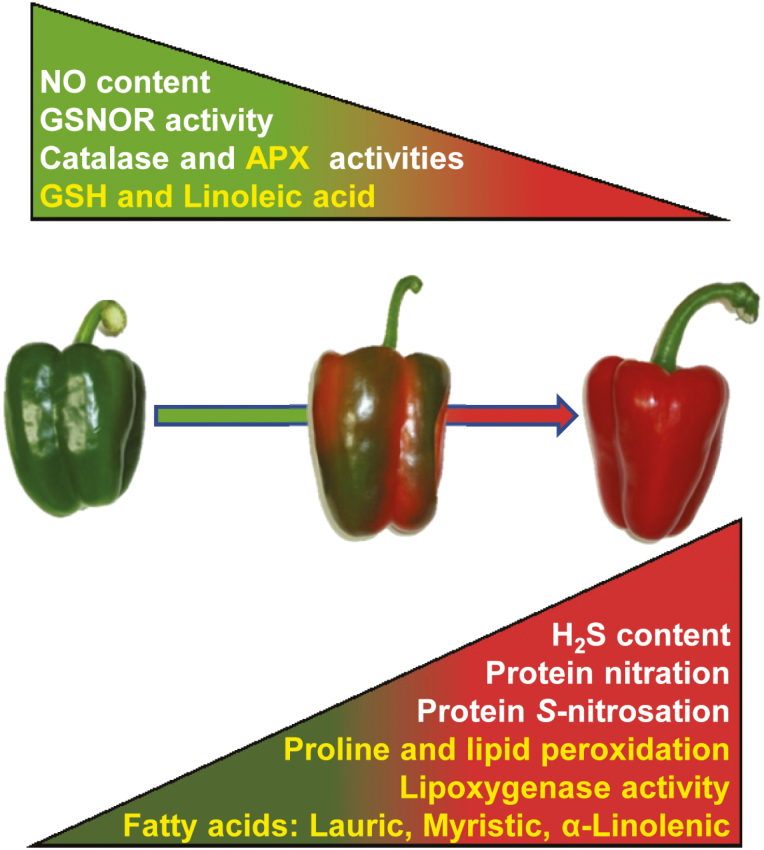
This study provides the first transcriptome of sweet pepper fruit, which could be a valuable tool for developing comparative studies of available transcriptomes of hot pepper fruits characterized to have a biosynthetic pathway for capsaicin, which is responsible for the ‘heat’ and pungency associated with hot peppers (Zhang et al., 2016; Tanaka et al., 2017). Likewise, this transcriptomic analysis will allow knowledge to be gained on the pathways that are modulated during ripening in non-climacteric fruits. On the other hand, exogenous NO application has a strong regulatory effect on a wide variety of processes during sweet pepper fruit ripening, which are regulated at the levels of gene and protein expression, enzymatic activity, and, thus, metabolite content. During ripening of sweet pepper fruits from the green to the red stage, the major effects of NO were: (i) delayed ripening, with phenotypic (color) and gene expression changes; and (ii) a regulatory effect on FAs, by reducing the degree of lipid peroxidation with a concomitant increase in antioxidant capacity (GSH, APX activity), which palliates the oxidative stress associated with the ripening process. Fig. 8 shows a working model of natural pepper fruit ripening during the transition from green to red, in which multiple parameters of nitro-oxidative metabolism are modulated. Finally, in addition to the relevance of NO as a regulator of pepper fruit ripening, which is independent of ethylene, the potential involvement of other endogenous molecules, such as H2O2, ABA, H2S, or melatonin, which could exert synergistic or antagonistic actions to NO, establishing a complex network of signals, as has been reported elsewhere (Corpas et al., 2019; Mukherjee 2019; Vithana et al., 2019), should be mentioned. The mechanisms that govern these interactions, and how they modulate the whole ripening process, should be deciphered in future research.
Fig. 8.
Working model of the metabolism of pepper fruit during ripening. Yellow text refers to the parameters reported in the present study, whereas white text refers to previously published data. APX, ascorbate peroxidase; GSH, reduced glutathione; GSNOR, S-nitrosoglutathione reductase; H2S, hydrogen sulfide.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Summary of the data obtained after sequencing of the 24 cDNA libraries.
Table S2. Statistics of the full set of tentative transcripts for the best de novo assembled transcriptome.
Fig. S1. Flow chart of the experimental procedure to analyze the transcriptome of sweet pepper fruit.
Acknowledgements
The work of FJC and JMP is supported by a European Regional Development Fund-cofinanced grant from the Ministry of Economy and Competitiveness (AGL2015-65104-P) and Junta de Andalucía (group BIO192), Spain. SGG acknowledges a ‘Formación de Personal Investigador’ contract from the Ministry of Economy and Competitiveness. The provision of pepper fruits by Syngenta Seeds Ltd (El Ejido, Almería, Spain) is acknowledged. GC-MS analyses were done at the Instrumental Technical Services of the Estación Experimental del Zaidín (CSIC) and special thanks are given to Dr Rafael Núñez-Gómez. The valuable technical assistance of Mrs María J. Campos and Mr Carmelo Ruiz-Torres is deeply acknowledged. The authors also gratefully acknowledge the computer resources and technical support provided by the Plataforma Andaluza de Bioinformática of the University of Málaga.
References
- Airaki M, Sánchez-Moreno L, Leterrier M, Barroso JB, Palma JM, Corpas FJ. 2011. Detection and quantification of S-nitrosoglutathione (GSNO) in pepper (Capsicum annuum L.) plant organs by LC-ES/MS. Plant & Cell Physiology 52, 2006–2015. [DOI] [PubMed] [Google Scholar]
- Ananthan R, Subhash K, Longvah T. 2018. Capsaicinoids, amino acid and fatty acid profiles in different fruit components of the world hottest Naga king chilli (Capsicum chinense Jacq). Food Chemistry 238, 51–57. [DOI] [PubMed] [Google Scholar]
- Astier J, Lindermayr C. 2012. Nitric oxide-dependent posttranslational modification in plants: an update. International Journal of Molecular Sciences 13, 15193–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant and Soil 39, 205–207. [Google Scholar]
- Begara-Morales JC, Sánchez-Calvo B, Chaki M, Valderrama R, Mata-Pérez C, López-Jaramillo J, Padilla MN, Carreras A, Corpas FJ, Barroso JB. 2014. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. Journal of Experimental Botany 65, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begara-Morales JC, Sánchez-Calvo B, Chaki M, Valderrama R, Mata-Pérez C, Padilla MN, Corpas FJ, Barroso JB. 2016. Antioxidant systems are regulated by nitric oxide-mediated post-translational modifications (NO-PTMs). Frontiers in Plant Science 7, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begara-Morales JC, Sánchez-Calvo B, Luque F, Leyva-Pérez MO, Leterrier M, Corpas FJ, Barroso JB. 2014. Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant & Cell Physiology 55, 1080–1095. [DOI] [PubMed] [Google Scholar]
- Benzekri H, Armesto P, Cousin X, et al. . 2014. De novo assembly, characterization and functional annotation of Senegalese sole (Solea senegalensis) and common sole (Solea solea) transcriptomes: integration in a database and design of a microarray. BMC Genomics 15, 952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. Journal of Computational Biology 17, 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthour D, Kalai T, Chaffei HC, Gouia H, Corpas FJ. 2015. Differential response of NADP-dehydrogenases and carbon metabolism in leaves and roots of two durum wheat (Triticum durum Desf.) cultivars (Karim and Azizi) with different sensitivities to salt stress. Journal of Plant Physiology 179, 56–63. [DOI] [PubMed] [Google Scholar]
- Buege JA, Aust SD. 1978. Microsomal lipid peroxidation. Methods in Enzymology 52, 302–310. [DOI] [PubMed] [Google Scholar]
- Cao S, Shao J, Shi L, Xu L, Shen Z, Chen W, Yang Z. 2018. Melatonin increases chilling tolerance in postharvest peach fruit by alleviating oxidative damage. Scientific Reports 8, 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Álvarez de Morales P, Ruiz C, Begara-Morales JC, Barroso JB, Corpas FJ, Palma JM. 2015. Ripening of pepper (Capsicum annuum) fruit is characterized by an enhancement of protein tyrosine nitration. Annals of Botany 116, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Puga A, González-Gordo S, Rodríguez-Ruiz M, Palma JM, Corpas FJ. 2019. NADPH oxidase (Rboh) activity is up regulated during sweet pepper (Capsicum annuum L.) fruit ripening. Antioxidants 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen W, Brückner B, Krumbein A, Lenz F. 2006. Long-term response of tomato plants to changing nutrient concentration in the root environment—the role of proline as an indicator of sensory fruit quality. Plant Science 171, 323–331. [DOI] [PubMed] [Google Scholar]
- Corpas FJ. 2016. Reactive nitrogen species (RNS) in plants under physiological and adverse environmental conditions: current view. In: Cánovas FM, Lüttge U, Matyssek R, eds. Progress in Botany Vol. 78 Cham: Springer International Publishing, 97–119. [Google Scholar]
- Corpas FJ, Chaki M, Fernández-Ocaña A, Valderrama R, Palma JM, Carreras A, Begara-Morales JC, Airaki M, del Río LA, Barroso JB. 2008. Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant & Cell Physiology 49, 1711–1722. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Chaki M, Leterrier M, Barroso JB. 2009. Protein tyrosine nitration: a new challenge in plants. Plant Signaling & Behavior 4, 920–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Freschi L, Rodríguez-Ruiz M, Mioto PT, González-Gordo S, Palma JM. 2018. Nitro-oxidative metabolism during fruit ripening. Journal of Experimental Botany 69, 3449–3463. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, González-Gordo S, Cañas A, Palma JM. 2019. Nitric oxide and hydrogen sulfide in plants: which comes first? Journal of Experimental Botany doi: 10.1093/jxb/erz031 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM. 2018. Nitric oxide on/off in fruit ripening. Plant Biology 20, 805–807. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Río LA, Palma JM. 2019. Impact of nitric oxide (NO) on the ROS metabolism of peroxisomes. Plants. 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Ángel-Coronel OA, León-García E, Vela-Gutiérrez G, Rojas-Reyes JO, Gómez-Lim MA, García HS. 2018. Lipoxygenase activity associated to fruit ripening and senescence in chayote (Sechium edule Jacq. Sw. cv. “virens levis”). Journal of Food Biochemistry 42, e12438. [Google Scholar]
- Egan AN, Moore S, Stellari GM, Kang BC, Jahn MM. 2019. Tandem gene duplication and recombination at the AT3 locus in the Solanaceae, a gene essential for capsaicinoid biosynthesis in Capsicum. PLOS ONE 14, e0210510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgueras J, Lara AJ, Fernández-Pozo N, Cantón FR, Pérez-Trabado G, Claros MG. 2010. SeqTrim: a high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinformatics 11, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayte IG, Moreno RB, Zonjic PS, Claros MG. 2017. DEgenes Hunter – a flexible R pipeline for automated RNA-seq studies in organisms without reference genome. Genomics and Computational Biology 3, 31. [Google Scholar]
- Grün S, Lindermayr C, Sell S, Durner J. 2006. Nitric oxide and gene regulation in plants. Journal of Experimental Botany 57, 507–516. [DOI] [PubMed] [Google Scholar]
- Guo Q, Wu B, Chen W, Zhang Y, Wang J, Li X. 2014. Effects of nitric oxide treatment on the cell wall softening related enzymes and several hormones of papaya fruit during storage. Food Science and Technology International 20, 309–317. [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, et al. . 2004. Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971. [DOI] [PubMed] [Google Scholar]
- Heinisch O, Kowalski E, Ludwig H, Tauscher B. 1996. Staining for soybean lipoxygenase activity in electrophoretic gels. European Journal of Lipid Science and Technology 98, 183–184 [Google Scholar]
- Holzmeister C, Gaupels F, Geerlof A, Sarioglu H, Sattler M, Durner J, Lindermayr C. 2015. Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. Journal of Experimental Botany 66, 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Nakano Y, Asada K. 1984. Monodehydroascorbate reductase in spinach chloroplast and its participation in regeneration of ascorbate for scavenging of hydrogen peroxide. Plant & Cell Physiology 25, 385–395. [Google Scholar]
- Huan C, Jiang L, An X, Yu M, Xu Y, Ma R, Yu Z. 2016. Potential role of reactive oxygen species and antioxidant genes in the regulation of peach fruit development and ripening. Plant Physiology and Biochemistry 104, 294–303. [DOI] [PubMed] [Google Scholar]
- Hutchings D, Rawsthorne S, Emes MJ. 2005. Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus L.). Journal of Experimental Botany 56, 577–585. [DOI] [PubMed] [Google Scholar]
- Jimenez A, Creissen G, Kular B, Firmin J, Robinson S, Verhoeyen M, Mullineaux P. 2002. Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 214, 751–758. [DOI] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. 2017. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research 45, D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Kachroo P. 2009. Fatty acid-derived signals in plant defense. Annual Review of Phytopathology 47, 153–176. [DOI] [PubMed] [Google Scholar]
- Kang R, Zhang L, Jiang L, Yu M, Ma R, Yu Z. 2016. Effect of postharvest nitric oxide treatment on the proteome of peach fruit during ripening. Postharvest Biology and Technology 112, 277–289. [Google Scholar]
- Kharbech O, Houmani H, Chaoui A, Corpas FJ. 2017. Alleviation of Cr(VI)-induced oxidative stress in maize (Zea mays L.) seedlings by NO and H2S donors through differential organ-dependent regulation of ROS and NADPH-recycling metabolisms. Journal of Plant Physiology 219, 71–80. [DOI] [PubMed] [Google Scholar]
- Khatri P, Sirota M, Butte AJ. 2012. Ten years of pathway analysis: current approaches and outstanding challenges. PLOS Computational Biology 8, e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. Journal of Statistical Software 25 doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- Leshem YY, Pinchasov Y. 2000. Non-invasive photoacoustic spectroscopic determination of relative endogenous nitric oxide and ethylene content stoichiometry during the ripening of strawberries Fragaria anannasa (Duch.) and avocados Persea americana (Mill.). Journal of Experimental Botany 51, 1471–1473. [PubMed] [Google Scholar]
- Leshem YY, Wills RBH, Veng-Va Ku V. 1998. Evidence for the function of the free radical gas-nitric oxide (NO) as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiology and Biochemistry 36, 825–833. [Google Scholar]
- Leterrier M, Chaki M, Airaki M, Valderrama R, Palma JM, Barroso JB, Corpas FJ. 2011. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signaling & Behavior 6, 789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xu C, Li-Beisson Y, Philippar K. 2016. Fatty acid and lipid transport in plant cells. Trends in Plant Science 21, 145–158. [DOI] [PubMed] [Google Scholar]
- Li G, Zhu S, Wu W, Zhang C, Peng Y, Wang Q, Shi J. 2017. Exogenous nitric oxide induces disease resistance against Monilinia fructicola through activating the phenylpropanoid pathway in peach fruit. Journal of the Science of Food and Agriculture 97, 3030–3038. [DOI] [PubMed] [Google Scholar]
- Lim GH, Singhal R, Kachroo A, Kachroo P. 2017. Fatty acid- and lipid-mediated signaling in plant defense. Annual Review of Phytopathology 55, 505–536. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, et al. . 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Rao J, Zhu Y, Chang X, Hou Y, Zhu Q. 2014. Cloning and expression of lipoxygenase genes and enzyme activity in ripening persimmon fruit in response to GA and ABA treatments. Postharvest Biology and Technology, 92, 54–61. [Google Scholar]
- Ma Y, Fu L, Hussain Z, Huang D, Zhu S. 2019. Enhancement of storability and antioxidant systems of sweet cherry fruit by nitric oxide-releasing chitosan nanoparticles (GSNO-CS NPs). Food Chemistry 285, 10–21. [DOI] [PubMed] [Google Scholar]
- Manai J, Gouia H, Corpas FJ. 2014. Redox and nitric oxide homeostasis are affected in tomato (Solanum lycopersicum) roots under salinity-induced oxidative stress. Journal of Plant Physiology 171, 1028–1035. [DOI] [PubMed] [Google Scholar]
- Martí MC, Camejo D, Vallejo F, Romojaro F, Bacarizo S, Palma JM, Sevilla F, Jiménez A. 2011. Influence of fruit ripening stage and harvest period on the antioxidant content of sweet pepper cultivars. Plant Foods for Human Nutrition 66, 416–423. [DOI] [PubMed] [Google Scholar]
- Mata-Pérez C, Begara-Morales JC, Luque F, Padilla MN, Jiménez-Ruiz J, Sánchez-Calvo B, Fierro-Risco J, Barroso JB. 2016. Transcriptomic analyses on the role of nitric oxide in plant disease resistance. Current Issues in Molecular Biology 19, 121–128. [PubMed] [Google Scholar]
- Mateos RM, Bonilla-Valverde D, del Río LA, Palma JM, Corpas FJ. 2009. NADP-dehydrogenases from pepper fruits: effect of maturation. Physiologia Plantarum 135, 130–139. [DOI] [PubMed] [Google Scholar]
- Mateos RM, Jiménez A, Román P, et al. . 2013. Antioxidant systems from pepper (Capsicum annuum L.): involvement in the response to temperature changes in ripe fruits. International Journal of Molecular Sciences 14, 9556–9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake C, Asada K. 1996. Inactivation mechanism of ascorbate peroxidase at low concentrations of ascorbate; hydrogen peroxide decomposes compound I of ascorbate peroxidase. Plant & Cell Physiology 36, 661–668. [Google Scholar]
- Mizzotti C, Rotasperti L, Moretto M, Tadini L, Resentini F, Galliani BM, Galbiati M, Engelen K, Pesaresi P, Masiero S. 2018. Time-course transcriptome analysis of Arabidopsis siliques discloses genes essential for fruit development and maturation. Plant Physiology 178, 1249–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S. 2019. Recent advancements in the mechanism of nitric oxide signaling associated with hydrogen sulfide and melatonin crosstalk during ethylene-induced fruit ripening in plants. Nitric Oxide 82, 25–34. [DOI] [PubMed] [Google Scholar]
- Muñoz-Vargas MA, González-Gordo S, Cañas A, López-Jaramillo J, Palma JM, Corpas FJ. 2018. Endogenous hydrogen sulfide (H2S) is up-regulated during sweet pepper (Capsicum annuum L.) fruit ripening. In vitro analysis shows that NADP-dependent isocitrate dehydrogenase (ICDH) activity is inhibited by H2S and NO. Nitric Oxide 81, 36–45. [DOI] [PubMed] [Google Scholar]
- Naves ER, de Ávila Silva L, Sulpice R, Araújo WL, Nunes-Nesi A, Peres LEP, Zsögön A. 2019. Capsaicinoids: pungency beyond Capsicum. Trends in Plant Science 24, 109–120. [DOI] [PubMed] [Google Scholar]
- Niu L, Yu J, Liao W, Yu J, Zhang M, Dawuda MM. 2017. Calcium and calmodulin are involved in nitric oxide-induced adventitious rooting of cucumber under simulated osmotic stress. Frontiers in Plant Science 8, 1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nueda MJ, Tarazona S, Conesa A. 2014. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 30, 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocaña S, Seoane P, Bautista R, Palomino C, Claros GM, Torres AM, Madrid E. 2015. Large-scale transcriptome analysis in faba bean (Vicia faba L.) under Ascochyta fabae infection. PLOS ONE 10, e0135143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma JM, Ruiz C, Corpas FJ. 2018. A simple and useful method to apply exogenous no gas to plant systems: bell pepper fruits as a model. Methods in Molecular Biology 1747, 3–11. [DOI] [PubMed] [Google Scholar]
- Palma JM, Sevilla F, Jiménez A, del Río LA, Corpas FJ, Álvarez de Morales P, Camejo DM. 2015. Physiology of pepper fruit and the metabolism of antioxidants: chloroplasts, mitochondria and peroxisomes. Annals of Botany 116, 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Gálvez A, Garrido-Fernández J, Mínguez-Mosquera MI, Lozano-Ruiz M, Montero-de-Espinosa V. 1999. Fatty acid composition of two new pepper varieties (Capsicum annuum L. cv. Jaranda and Jariza). Effect of drying process and nutritional aspects. Journal of the American Oil Chemists’ Society 76, 205–208. [Google Scholar]
- Porta H, Rocha-Sosa M. 2002. Plant lipoxygenases. Physiological and molecular features. Plant Physiology 130, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ruiz M, Aparicio-Chacón MV, Palma JM, Corpas FJ. 2019. Arsenate disrupts ion balance, sulfur and nitric oxide metabolisms in roots and leaves of pea (Pisum sativum L.) plants. Environmental and Experimental Botany 161, 143–156. [Google Scholar]
- Rodríguez-Ruiz M, Mateos RM, Codesido V, Corpas FJ, Palma JM. 2017a. Characterization of the galactono-1,4-lactone dehydrogenase from pepper fruits and its modulation in the ascorbate biosynthesis. Role of nitric oxide. Redox Biology 12, 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ruiz M, Mioto P, Palma JM, Corpas FJ. 2017b. S-nitrosoglutathione reductase (GSNOR) activity is down-regulated during pepper (Capsicum annuum L.) fruit ripening. Nitric Oxide 68, 51–55. [DOI] [PubMed] [Google Scholar]
- Saini RK, Keum Y-S. 2016. GC–MS and HPLC–DAD analysis of fatty acids and tocopherols in sweet peppers (Capsicum annuum L.). Journal of Food Measurement and Characterization 10, 685–689. [Google Scholar]
- Saini RK, Zamany AJ, Keum YS. 2017. Ripening improves the content of carotenoid, α-tocopherol, and polyunsaturated fatty acids in tomato (Solanum lycopersicum L.) fruits. 3 Biotech 7, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane P, Espigares M, Carmona R, et al. . 2018. TransFlow: a modular framework for assembling and assessing accurate de novo transcriptomes in non-model organisms. BMC Bioinformatics 19, 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertl P, Sunderhaus S, Klodmann J, Grozeff GE, Bartoli CG, Braun HP. 2012. L-galactono-1,4-lactone dehydrogenase (GLDH) forms part of three subcomplexes of mitochondrial complex I in Arabidopsis thaliana. Journal of Biological Chemistry 287, 14412–14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN. 1991. Plant lipoxygenase: structure and function. Annual Review of Plant Physiology and Plant Molecular Biology 42, 145–88. [Google Scholar]
- Signorelli S, Corpas FJ, Borsani O, Barroso JB, Monza J. 2013. Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicus. Plant Science 201-202, 137–146. [DOI] [PubMed] [Google Scholar]
- Signorelli S, Monza J. 2017. Identification of Δ1-pyrroline 5-carboxylate synthase (P5CS) genes involved in the synthesis of proline in Lotus japonicus. Plant Signaling & Behavior 12, e1367464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer DD, Delcher AL, Salzberg SL, Pop M. 2007. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sora GT, Haminiuk CW, da Silva MV, Zielinski AA, Gonçalves GA, Bracht A, Peralta RM. 2015. A comparative study of the capsaicinoid and phenolic contents and in vitro antioxidant activities of the peppers of the genus Capsicum: an application of chemometrics. Journal of Food Science and Technology 52, 8086–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stines AP, Naylor DJ, Høj PB, van Heeswijck R. 1999. Proline accumulation in developing grapevine fruit occurs independently of changes in the levels of delta1-pyrroline-5-carboxylate synthetase mRNA or protein. Plant Physiology 120, 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Savouré A. 2010. Proline: a multifunctional amino acid. Trends in Plant Science 15, 89–97. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nakashima F, Kirii E, Goto T, Yoshida Y, Yasuba KI. 2017. Difference in capsaicinoid biosynthesis gene expression in the pericarp reveals elevation of capsaicinoid contents in chili peppers (Capsicum chinense). Plant Cell Reports 36, 267–279. [DOI] [PubMed] [Google Scholar]
- The RNAcentral Consortium. 2017. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Research 45, D128–D134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Research 45, W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ties P, Barringer S. 2012. Influence of lipid content and lipoxygenase on flavor volatiles in the tomato peel and flesh. Journal of Food Science 77, C830–C837. [DOI] [PubMed] [Google Scholar]
- Tossi V, Amenta M, Lamattina L, Cassia R. 2011. Nitric oxide enhances plant ultraviolet-B protection up-regulating gene expression of the phenylpropanoid biosynthetic pathway. Plant, Cell & Environment 34, 909–921. [DOI] [PubMed] [Google Scholar]
- Van Ree K, Gehl B, Chehab EW, Tsai YC, Braam J. 2011. Nitric oxide accumulation in Arabidopsis is independent of NOA1 in the presence of sucrose. The Plant Journal 68, 225–233. [DOI] [PubMed] [Google Scholar]
- Vithana MD, Singh Z, Johnson SK, Gupta R. 2019. Concentrations of health-promoting phytochemicals in ripe mango fruit triggered by postharvest application of elicitors. Journal of the Science of Food and Agriculture 99, 1126–1134. [DOI] [PubMed] [Google Scholar]
- Voll L, Häusler RE, Hecker R, Weber A, Weissenböck G, Fiene G, Waffenschmidt S, Flügge UI. 2003. The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. The Plant Journal 36, 301–17. [DOI] [PubMed] [Google Scholar]
- Wang SY, Jiao H. 2001. Changes in oxygen-scavenging systems and membrane lipid peroxidation during maturation and ripening in blackberry. Journal of Agricultural and Food Chemistry 49, 1612–1619. [DOI] [PubMed] [Google Scholar]
- Wang Y, Luo Z, Du R, Liu Y, Ying T, Mao L. 2013. Effect of nitric oxide on antioxidative response and proline metabolism in banana during cold storage. Journal of Agricultural and Food Chemistry 61, 8880–8887. [DOI] [PubMed] [Google Scholar]
- Wu Q, Tao X, Ai X, Luo Z, Mao L, Ying T, Li L. 2018. Contribution of abscisic acid to aromatic volatiles in cherry tomato (Solanum lycopersicum L.) fruit during postharvest ripening. Plant Physiology and Biochemistry 130, 205–214. [DOI] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research 39, W316–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Furutera A, Seki K, Toyoda Y, Tanaka K, Sugimoto Y. 2008. Malondialdehyde generated from peroxidized linolenic acid causes protein modification in heat-stressed plants. Plant Physiology and Biochemistry 46, 786–793. [DOI] [PubMed] [Google Scholar]
- Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou JM, Zuo J. 2015. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiology 167, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Becker DF. 2015. Connecting proline metabolism and signaling pathways in plant senescence. Frontiers in Plant Science 6, 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cao S, Jin Y, Ju L, Chen Q, Xing Q, Qi H. 2017. Melon13-lipoxygenase CmLOX18 may be involved in C6 volatiles biosynthesis in fruit. Scientific Reports 7, 2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fang H, Huo J, Huang D, Wang B, Liao W. 2018. Involvement of calcium and calmodulin in nitric oxide-regulated senescence of cut lily flowers. Frontiers in Plant Science 9, 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Yin XR, Li X, Yang SL, Ferguson IB, Chen KS. 2009. Lipoxygenase gene expression in ripening kiwifruit in relation to ethylene and aroma production. Journal of Agricultural and Food Chemistry 57, 2875–2881. [DOI] [PubMed] [Google Scholar]
- Zhang ZX, Zhao SN, Liu GF, Huang ZM, Cao ZM, Cheng SH, Lin SS. 2016. Discovery of putative capsaicin biosynthetic genes by RNA-Seq and digital gene expression analysis of pepper. Scientific Reports 6, 34121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li S, Zeng K. 2016. Exogenous nitric oxide-induced postharvest disease resistance in citrus fruit to Colletotrichum gloeosporioides. Journal of the Science of Food and Agriculture 96, 505–512. [DOI] [PubMed] [Google Scholar]
- Zhu S, Sun L, Zhou J. 2010. Effects of different nitric oxide application on quality of kiwifruit during 20°C storage. International Journal of Food Science and Technology 45, 245–251. [Google Scholar]
- Zhu S, Zhou J. 2006. Effects of nitric oxide on fatty acid composition in peach fruits during storage. Journal of Agricultural and Food Chemistry 54, 9447–9452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.