Abstract
The infiltration and subsequent in-situ subtype specification of monocytes to effector/inflammatory and repair macrophages is indispensable for tissue repair upon acute sterile injury. However, the chromatin-level mediators and regulatory events controlling this highly dynamic macrophage phenotype switch are not known. Here, we used a murine acute muscle injury model to assess global chromatin accessibility and gene expression dynamics in infiltrating macrophages during sterile physiological inflammation and tissue regeneration. We identified a heme-binding transcriptional repressor, BACH1, as a novel regulator of this process. Bach1 knockout mice displayed impaired muscle regeneration, altered dynamics of the macrophage phenotype transition and transcriptional deregulation of key inflammatory and repair-related genes. We also found that BACH1 directly binds to and regulates distal regulatory elements of these genes, suggesting a novel role for BACH1 in controlling a broad spectrum of the repair-response genes in macrophages upon injury. Inactivation of heme oxygenase-1 (Hmox1), one of the most stringently deregulated genes in the Bach1 KO, in macrophages, impairs muscle regeneration by changing the dynamics of the macrophage phenotype switch. Collectively, our data suggest the existence of a heme-BACH1-HMOX1 regulatory axis, that controls the phenotype and function of the infiltrating myeloid cells upon tissue damage, shaping the overall tissue repair kinetics.
Introduction
Cells of the innate immune systems undergo dynamic changes in population size, differentiation state and function in response to diverse insults to organismal integrity (1). Due to their role in controlling the initiation, maintenance, and resolution of wound-healing responses in different organ systems, macrophages (MFs) are considered major therapeutic targets (2, 3). However, our knowledge is fragmented on how external stimuli influences the MF phenotype switch, and how these cells employ sensory and effector functions to serve such reparatory roles. This is particularly important because the proper signaling between the participating cell types can ensure precisely timed progression of repair, while avoiding asynchrony, which can lead to regeneration delay, fibrosis and/or chronic inflammation (4, 5).
Monocyte-derived MFs are consistently detectable in injured muscles and, as demonstrated by depletion experiments using pharmacological and genetic tool, are required for muscle regeneration (6, 7). Several studies have shown that MFs are involved in all phases of regeneration, such as (a) confining the damage, (b) clearing the necrotic debris via phagocytosis and (c) contributing to repair (8). Acute damage causes the release of a myriad of chemoattractant molecules which initiate the invasion of neutrophils and monocytes from the blood stream into the muscle (9). This highly dynamic process is characterized by an in-situ transition of infiltrating monocytes to an inflammatory (Ly6Chigh) and later to a repair (Ly6Clow) MF phenotype (10), which appears to be indispensable for proper regeneration (6, 7). It is likely that the microenvironment and inter-cellular interactions are driving the inflammatory to repair phenotypic switch (11, 12). However, these dynamic changes in cellular phenotype must be also driven by transcription factors (TFs), which bind to regulatory elements and support cell type-specific gene expression.
We and others sought to identify such integrated sensory, regulatory and effector mechanisms (13), equipping a macrophage with the capacity to contribute to properly timed progression of repair. After damage, Ly6Chigh MFs produce high levels of inflammatory cytokines, such as TNF-α and IL-1β, and remove the remnants of myofibers and apoptotic cells while sustaining the activation and the proliferation of myogenic precursors (8, 14). At later time points during regeneration, Ly6Clow MFs predominate (11) and secrete cytokines and growth factors such as IL-10, TGF-β, GDF3, and IGF-1 (13, 15–18) that promote myoblast fusion and neofiber formation and assembly. In addition, previous work from our lab, and others, identified TFs and enzymes that are required for the phenotypic shift towards repair MFs and for efficient muscle regeneration. These include the nuclear receptor peroxisome proliferator-activated receptor (PPAR)-γ, CCAAT/enhancer-binding protein beta (C/EBPβ), AMP-activated protein kinase (AMPK)-α1, and MAP kinase phosphatase-1 (also known as DUSP1), (13, 15, 19–21). In contrast, other key pathways, such as those coordinated by myeloid hypoxia-inducible factors, are dispensable for the MF phenotypic shift and muscle healing after sterile injury (22). These studies show that the conversion of infiltrating muscle MFs from inflammatory to repair ones is stringently regulated with multiple pathways being involved. Moreover, gene expression studies revealed the highly dynamic nature of the muscle MF response and document that a distinct signature, primarily driven by the cellular milieu is characterizing inflammatory and repair MFs at each step of tissue injury and repair (11). How all these pathways are regulated and coordinated is still unclear.
Here, we applied an established and highly reproducible in vivo (cardiotoxin-induced) model of acute muscle injury and sterile physiological inflammation (23), to carry out an unbiased chromatin accessibility (ATAC-seq) and TF-binding motif enrichment analysis of the dynamically changing MF subpopulations involved in the regeneration process. This time course-based profiling revealed that the chromatin structure is dynamically changing during the macrophage phenotype switch, with Maf recognition elements (MAREs) being overrepresented in the opened chromatin regions of muscle-invading MFs. Based on this system-level chromatin openness analysis and complemented with transcriptomic data from the same cell populations, we identified BTB Domain And CNC Homolog 1 (BACH1), a MARE-binding and heme-regulated transcription factor, as a regulator of MF cell type specification having more pervasive roles than previously thought. Bach1 deletion (a) leads to transcriptional deregulation of critical inflammatory and repair genes, (b) impacts the MF phenotype switch, (c) impairs muscle regeneration in vivo, and (d) deregulates myoblast proliferation in-vitro. These data support a role for BACH1 in driving transient inflammatory and repair transcriptional programs in MFs during tissue injury. In addition, we show that the presence of the heme-regulated transcriptional repressor BACH1, and expression of one of its most stringently regulated targets, heme oxygenase 1 (Hmox1), by macrophages, at the site of injury, represent a regulatory axis responsible for coordinated in-situ MF phenotypic shift, and subsequently for proper and complete tissue regeneration.
Materials and Methods
Ethical approval
All animal experiments were carried out in accordance with ethical regulations and approved by the Institutional Animal Care and Use Committees at Johns Hopkins University (license no: MO18C251), BSRC “Alexander Fleming” (approved by Veterinary Service Management of the Hellenic Republic Prefecture of Attika, license no: 4400–05/07/2016), and University of Debrecen, School of Medicine (license no.: 21/2011/DEMÁB) following national, and European legislation.
Mice
Wild type 8-week old BoyJ (B6.SJL-Ptprca Pepcb/BoyJ, stock number 002014) and C57BL/6J male control mice were obtained from the Jackson Laboratories and bred under specific-pathogen free (SPF) conditions. Male Bach1-KO (obtained by Dr. Kazuhiko Igarashi at Tohoku University), and Hmox1fl/fl LysM-Cre mice (obtained by Dr. George Kollias group at BSRC-Athens), were used in the experiments (24). All irradiation experiments were performed under anesthesia in cohorts of 12 animals per experiment. Briefly, mice were anaesthetized with a single intraperitoneal dose of ketamine/xylazine (ketamine 80–100 mg/kg, xylazine 10–12.5 mg/kg). Irradiated and bone marrow-transplanted mice were maintained in an SPF status (autoclaved top filter cages) for the entire course of experimentation and antibiotics (amoxicillin antibiotic, clavulanic acid [500mg/ 125 mg/ liter of drinking water]) were administered in the drinking water for 4 weeks after transplantation to minimize bacterial contamination within the water source and potentially decrease the burden of gastrointestinal bacteria. Irradiated mice were also fed autoclaved rodent chow ad libitum. Animals that undergo irradiation for BMT typically lose a considerable amount of weight, only to gain it back relatively quickly after successful transplantation. At our institutions, weight loss of 20%, or greater was used as a rationale for euthanasia before the intended experimental endpoint according to the IACUC guidelines. When necessary and for tissue collection mice were euthanized by either isoflurane overdose (adjusted flow rate or concentration to 5% or greater) or CO2 exposure (adjusted flow rate 3 L/min) in accordance to the University of Debrecen and Johns Hopkins University’s IACUC guidelines.
Histological analysis of muscle regeneration
Muscles were removed and snap frozen in nitrogen-chilled isopentane (−160°C). 8 μm thick cryosections were cut and stained with hematoxylin-eosin (H&E) or Masson’s trichrome (Polysciences, Inc.). For each histological analysis, at least 5 slides (per condition) were selected where the total regenerative region within the CTX injured TA muscle was at least 70%. For each TA, myofibers in the entire injured area were counted and measured. H&E and Masson’s Trichrome stained muscle sections were scanned with the Mirax digital slide scanner. The cross-sectional area (CSA) and fibrosis were quantified with HALO software (Indica Labs). CSAs for these samples are reported in μm2. Fibrosis is reported as the percentage of the area (in μm2) of fibrotic stain (blue) over the total regeneration area. Quantitative analysis of necrotic/phagocytic myofibers was performed using the Panoramic Viewer software and was expressed as a percentage of the total number of myofibers. Areas of necrosis were identified based on the following histological criteria: blurring of cell borders, cytoplasmic fragmentation, caliber variation, cell distances, loss of nuclei, and increased immune cell infiltration (25). Necrotic/phagocyted myofibers were further defined as pink pale patchy fibers that are invaded by basophil single cells (macrophages). The necrotic fiber content data presented here were quantified using both immunohistochemistry (desmin staining) and histology.
Immunohistochemistry
Tissue sections were fixed and permeabilized in ice-cold acetone for 5 min and blocked for 30 minutes at 20 °C (room temperature) in PBS containing 2 % bovine serum albumin (BSA). Tissues were stained for 1 hour at room temperature using a primary antibody diluted in 2 % BSA. The primary antibodies used for immunofluorescence were rabbit anti-laminin (L9393 SIGMA) at a dilution 1/200, chicken anti-PAX7 (DSHB) at a dilution 1/20, rabbit anti-Desmin (Abcam 32362) at a dilution 1/200 and rat anti-F4/80 (Abcam 6640) at a dilution 1/200. In all cases, the primary antibody was detected using secondary antibodies (dilution 1/200) conjugated to FITC (JIR 703-095-155) or Cy3 JIR (711-165-152). The nuclei were counter stained with 0.1–1 μg/ml Hoechst. Fluorescent microscopy was performed using Carl Zeiss Axio Imager Z2 microscope equipped with lasers at 488, 568 and 633 nm. Images were analyzed and assembled using Fiji and Illustrator CS5 (Adobe).
Bone Marrow Transplantation (BMT)
Recipient congenic BoyJ mice (7 weeks old) are irradiated with 11 Gy using a Theratron 780C cobalt unit for the ablation of the recipient bone marrow. During irradiation one of the hindlimbs was shielded as described previously (12). Following the irradiation, isolated bone marrow cells (in sterile RPMI-1640 medium) flushed out the femur; tibia and humerus from donor C57Bl/6J mice are transplanted into the recipient mice by retro-orbital injection (20×106 BM cells per mice). This experimental BMT CD45 congenic model allows us to detect donor, competitor and host contribution in hematopoiesis and repopulation efficiency of donor cells (congenic mice with CD45.1 versus CD45.2). The CD45.1 and CD45.2 contribution is then detected by flow cytometry usually 8–12 weeks following the BMT. In short, a cut at the tail tip of the mice provided a drop of blood that was placed into 0.5 ml PBS + 1% FBS + 10 U/ml Heparin buffer (samples kept on ice). The cells were directly stained by 2 μl mouse anti-mouse CD45.2-FITC (clone 104) and 2 μl rat anti-mouse GR1-PE (clone RB6–8C5) antibodies (BD Pharmingen) and incubated on ice for 30 min. After 2 washes with ice-cold PBS/FBS/Heparin buffer we resuspended the cells in 0.5–1 ml FACS Lysing solution (BD Cat #349202). We incubated for 5 min at RT and then centrifuged the cells (400g, 5 min, 4°C). We run the double stained samples on FACS (BD FACS Calibur) and determined the ratio of donor cells. The repopulation is usually over 90% gated on the granulocyte fraction.
Acute sterile muscle injury
Mice were anaesthetized with isoflurane (adjusted flow rate or concentration to 1,5%) and 50 μl of cardiotoxin (12×10−6 M in PBS) (from Latoxan) was injected in the tibialis anterior (TA) muscle. Muscles were recovered for flow cytometry analysis at day 1 to day 8 post-injury or for muscle histology at day 8 to day 70 post-injury.
Macrophage cell culture for conditioned medium generation
Macrophages were obtained from bone marrow (BM) precursor cells. Briefly, total BM was obtained from mice by flushing femurs and tibiae bone marrow with DMEM. Cells were cultured in DMEM medium containing 20% FBS and 30% conditioned medium of L929 cell line (enriched in CSF-1) for 7 days. MFs were seeded (at 50000 cell/cm2 for all experiments) and were activated with IFN-γ (50 ng/ml), IL4 (10 ng/ml) and IL10 (10ng/ml) to obtain pro-inflammatory (stimulation with IFNγ), alternatively activated (IL-4), and anti-inflammatory (IL10) MFs, respectively, in DMEM containing 10% FBS medium for 3 days, as described previously (26). After washing steps, DMEM serum-free medium was added for 24 h, recovered and centrifugated to obtain MF-conditioned medium.
Myoblast C2C12 culture and proliferation assay
Murine myoblast C2C12 cells were obtained from ATCC (CRL-1772) and were maintained according to the company’s instructions. In brief, cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (growth medium) at 37°C in 5% CO2 and 95% air at 100% humidity. For proliferation assays, cells were seeded at 10000 cell/cm2 on Matrigel (1/10) and incubated for 1 day with MF-conditioned medium +2.5% FBS. Cells were then fixed with 4% PFA, incubated with anti-Ki67 antibodies for 1 hour at room temperature (15580 Abcam, dilution 1/400), and were subsequently visualized using Cy3-conjugated secondary antibodies (Jackson Immunoresearch Inc, dilution 1/200). Cells were also cultured in DMEM supplemented with 2,5% FBS and the antibiotics mentioned above; this medium was termed the proliferation medium and allowed the myoblasts to proliferate but not differentiate. The nuclei were counter stained with 0.1–1 μg/ml Hoechst. Fluorescent microscopy was performed using Carl Zeiss Axio Imager Z2 microscope equipped with lasers at 488, 568 and 633 nm. Images were analyzed for proliferation index, using Fiji.
Differentiation of bone marrow-derived macrophages
Isolation and differentiation were completed as described earlier (27). Isolated bone marrow-derived cells were differentiated for 6 days in the presence of L929 supernatant.
In vivo isolation of macrophages from muscle
Fascia of the TA was removed. Muscles were dissociated in RPMI containing 0.2% collagenase B (Roche Diagnostics GmbH) at 37°C for 1 hour and filtered through a 100 μm and a 40 μm filter. CD45+ cells were isolated using magnetic sorting (Miltenyi Biotec). For FACS, macrophages were treated with Fcγ receptor blocking antibodies and with 10% normal rat serum: normal mouse serum 1:1 mix, then stained with a combination of PE-conjugated anti-Ly6C antibody (HK1.4, eBioscience), APC-conjugated F4/80 antibody (BM8, eBioscience) and FITC-conjugated Ly6G antibody (1A8, Biolegend). Ly6Chigh F4/80low MFs, Ly6Clow F4/80high MFs and Ly6Ghigh Ly6Cmed F4/80− neutrophils were quantified. In each experiment, compared samples were processed in parallel to minimize experimental variation. Cells were analyzed on a BD FACSAria III sorter and data analysis was performed using BD FACSDIVA and FlowJo V10 software.
RNA isolation
Total RNA was isolated with TRIZOL reagent according to the manufacturer’s recommendation. 20 μg glycogen (Ambion) was added as carrier for RNA precipitation.
Real-Time Quantitative PCR (RT-qPCR)
Transcript quantification was performed by quantitative real-time reverse transcriptase (RT) polymerase chain reaction (PCR) using SYBR Green assays. RT-qPCR results were analyzed with the standard delta Ct method and results were normalized to the expression of Rps26 or Ppia. mRNA primer sequences used in transcript quantification are provided in Table 1.
Table 1.
mRNA primer sequences used in qPCR transcript quantification.
| Gene | Primer sequences | |
| Il10: | F: 5’ CAGAGCCACATGCTCCTAGA 3’ | R: 5’ TGTCCAGCTGGTCCTTTGTT 3’ |
| Igf1: | F: 5’ AGCAGCCTTCCAACTCAATTAT 3’ | R: 5’ TGAAGACGACATGATGTGTATCTTTAT 3’ |
| Dusp1: | F: 5’ TGGTTCAACGAGGCTATTGAC 3’ | R: 5’ GGCAATGAACAAACACTCTCC 3’ |
| Slc40a1: | F: 5’ ACCCATCCCCATAGTCTCTGT 3’ | R: 5’ CCGATTCTAGCAGCAATGACT 3’ |
| Ftl1: | F: 5’ AGTTTCAGAACGATCGCGGG 3’ | R: 5’ GGAAGTCACAGAGATGAGGGTC 3’ |
| Tnfsf12: | F: 5’GAGCCCCCTGAACTGAATC 3’ | R: 5’ AGGCCGGACTAGTTGTTCC 3’ |
| Cebpb: | R: 5’ TGATGCAATCCGGATCAA 3’ | R: 5’ CACGTGTGTTGCGTCAGTC 3’ |
| Spp1: | F: 5’ TCCCTCGATGTCATCCCTGT 3’ | R: 5’ TTGACTCATGGCTGCCCTTT 3’ |
| Hmox1: | F: 5’ CTGCTAGCCTGGTGCAAGATACT 3’ | R: 5’ GTCTGGGATGAGCTAGTGCTGAT 3’ |
| Il6: | F: 5’ CAAAGCCAGAGTCCTTCA 3’ | R: 5’ GGTCCTTAGCCACTCCTT 3’ |
| Arg1: | F: 5’TTTTAGGGTTACGGCCGGTG 3’ | R: 5’ CCTCGAGGCTGTCCTTTTGA 3’ |
| Socs3: | F: 5’ ATTTCGCTTCGGGACTAGC 3’ | R: 5’ AACTTGCTGTGGGTGACCAT 3’ |
| Pparg: | F: 5’ TCCATTCACAAGAGCTGACCC 3’ | R: 5’ GGTGGAGATGCAGGTTCTACT 3’ |
| Gdf3: | F: 5’ GGGTGTTCGTGGGAACCT 3’ | R: 5’ CCATCTTGGAAAGGTTTCTGTG 3’ |
Expression data processing and analysis
GSE71155 data sets were loaded into the Genespring GX software, and multiarray average summarization was carried out. Next, the lowest 5% of the entities with detectable signals were filtered out as not expressed. Duplicate entities, not/poorly annotated transcripts and transcripts reporting inconsistent expression values were also discarded. Further analysis was carried out on the filtered data set based on the RAW expression values. Heatmap was generated based on log10-transformed raw values with R software package pheatmap. Hierarchical clustering analysis was then applied by Euclidean distance measure and Ward’s clustering algorithm, to find correlated genes.
RNA sequencing (RNA-Seq) library preparation
cDNA library for RNA-Seq was generated from 100 – 400 ng total RNA using TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Briefly poly-A tailed RNA molecules were pulled down with poly-T oligo attached magnetic beads. Following purification, mRNA was fragmented with divalent cations at 85C, and then cDNA was generated by random primers and SuperScript II enzyme (Life Technologies). Second strand synthesis was performed followed by end repair, single À` base addition and ligation of barcode indexed adaptors to the DNA fragments. Adapter specific PCRs were performed to generate sequencing libraries. Libraries were size selected with E-Gel EX 2% agarose gels (Life Technologies) and purified by QIAquick Gel Extraction Kit (Qiagen). Libraries were sequenced on HiSeq 2500 instrument. Three biological replicates were sequenced for each population.
RNA-seq analysis
RNA-seq samples were analyzed using an in-house pipeline. Briefly, the 50-bp raw single reads were aligned using TopHat (28) to the mm10 genome assembly (GRCm38) and only the uniquely mapped reads were kept using ‘-max- multihits 1’ option, otherwise the default parameters were used. SAMtools (29) was used for indexing the alignment files. Coverage density tracks (wig files) for RNA-seq data were generated by igvtools with ‘count’ command and then converted into tdf files using ‘toTDF’ option. Genes with CPM>=10 (at least in one sample) were considered to be expressed. Statistically significant difference was considered as p < 0.05 from GLM test using R package edgeR(30). Heatmaps were drawn using R package pheatmap.
Gene ontology analysis
Lists of genes were analyzed using Panther tool (http://www.geneontology.org/) and the GO Enrichment Analysis to create a gene ontology (GO). GOs with p values <0.05 were selected and results were presented according to their -log10 p value.
Real-Time Quantitative PCR for enhancer RNA
RNA was isolated with Trizol reagent (Ambion). RNA was DNase-treated and reverse transcribed with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. Transcript quantification was performed by qPCR reactions using SYBR green master mix (BioRad). Transcript levels were normalized to Ppia or Actb. eRNA primer sequences and loci coordinates are provided in Table 2.
Table 2.
Enhancer RNA primer sequences and loci coordinates used in qPCR quantification.
| Gene locus | Primer sequences | Coordinates | |
| Hmox1 Eα: | F: 5’ TTGTCCTACGTGTGTGGCAG 3’ | R: 5’ GAAGGCAGGAGACTCCAGTG 3’ | chr8:75,075,818–75,096,261 |
| Hmox1 Eβ: | F: 5’ AAGGGACAGAAGGAAGCTGAT 3’ | R: 5’ GTGGGGCAGTCACTAGTATCC 3’ | chr8:75,075,818–75,096,261 |
| Hmox1 Eγ (used also for ChIP): | F: 5’ CTGTGAGTTCTGGTCCGTGG 3’ | R: 5’ ACAGGAACATCTTGGAGCCAG 3’ | chr8:75,075,818–75,096,261 |
| Hmox1 Eδ: | F: 5’ GCTAGCATGCGAAGTGAGCA 3’ | R: 5’ GCACAGCTCCGGATTCCTAAT 3’ | chr8:75,075,818–75,096,261 |
| Hmox1 Eε (used also for ChIP): | F: 5’ TGCTCAGTCTCCGTGTATGT 3’ | R: 5’ CCTGGCTTTGAGTCCATTCAT 3’ | chr8:75,075,818–75,096,261 |
| Cebpb +76kb: | F: 5’ CCAACTCCAACAAACTGCCC 3’ | R: 5’ GTCCAGGCACGACAGATGAG 3’ | chr2:167,759,270–167,766,462 |
| Slc40a1 +43kb: | F: 5’ TCACCTATGAAGCCTCCCTCA 3’ | R: 5’ AGGCATTGGCAGAAATAGGC 3’ | chr1:45,861,910–45,902,792 |
| Gdf3 −47kb: | F: 5’ TTGGGTAGAGGTGGTGTATGC 3’ | R: 5’ AGCCTCATGACCTGACTGAGA 3’ | chr6:122,643,983–122,667,639 |
| Socs3 +22kb: | F: 5’ ATGAAACCAGCCTGTGGAGAT 3’ | R: 5’ AACCTGAGAAGCTGATGGGTC 3’ | chr11:117,938,606–117,948,249 |
| Ftl1 −1kb: | F: 5’ GCTGTACGGCTCTGGAGTG 3’ | R: 5’ CCCAAATCAACAGACCACCG 3’ | chr7:45,457,542–45,463,285 |
| Arg1 −4kb: | F: 5’ ATGAGCTGGTCTCTCGTCGG 3’ | R: 5’ GGCCATGGTATGTGTTTCCC 3’ | chr10:24,929,431–24,933,681 |
| Dusp1 −25kb: | F: 5’ AGCCAGAGCAGTGAAAAGGA 3’ | R: 5’ TCCCTTGAGGCCATTTTGCT 3’ | chr17:26,531,508–26,539,010 |
| Il10 (used also for ChIP) +13kb: | F: 5’ TCCCTGAGCCACCAGATAGAT 3’ | R: 5’ ATTTAGTAGGGCTTCCCCAGC 3’ | chr1:131,036,412–131,044,131 |
| Il6 −64kb | F: 5’ GTATCGTTCCTCCCCCTTGC 3’ | R: 5’ ACTAGATATGGCCCAGGGGT 3’ | chr5:29,943,171–29,955,164 |
| Igf1 −105kb | F: 5’ GGAAACTCTGCCTGGGTCAT 3’ | R: 5’ CACCACGGGATAAGAGTCTGG 3’ | chr10:87,744,142–87,764,790 |
| Gdf3 −25kb | F: 5’ CACTCCTTTGGCTTCTGATAGTG 3’ | R: 5’ AGTTGCACACTGTGGCTTGA 3’ | chr6:122,629,871–122,641,698 |
| Spp1 −54kb | F: 5’ AGCTATAGATTGCCAGGGTTGG 3’ | R: 5’ CAGACCACCGTGATTTACCCA 3’ | chr5:104,376,093–104,388,086 |
Chromatin immunoprecipitation (ChIP)
ChIP-qPCR was performed essentially as previously described (27). anti-BACH1 antibody was a gift from Dr. Spilianakis (IMBB-FORTH, Greece). ChIP-qPCR primer sequences and loci coordinates are provided in Table 3.
Table 3.
ChIP-qPCR primer sequences and loci coordinates.
| Gene locus | Primer sequences | Coordinates | |
| Il10 +13kb: | F: 5’ TCCCTGAGCCACCAGATAGAT 3’ | R: 5’ ATTTAGTAGGGCTTCCCCAGC 3’ | chr1:131,036,412–131,044,131 |
| Dusp1 −25kb: | F: 5’ AGATGACCCAAAGGGAAGCTG 3’ | R: 5’ GCCTCCCCCACCTGACTAAT 3’ | chr17:26,531,508–26,539,010 |
| Spp1 −9kb: | F: 5’ ACACGAACAAAGGCGAAACTC 3’ | R: 5’ AGCTTCTGTGTGACTCGGC 3’ | chr5:104,421,637–104,431,579 |
| Spp1 −54kb | F: 5’ AGCCAACTTGCCCTCCATTTC 3’ | R: 5’ CAGTGGCATTGGTGGTGAGA 3’ | chr5:104,376,093–104,388,086 |
| Ftl1 −1kb: | F: 5’ GGCCCTTAGTGGAAGGGGTA 3’ | R: 5’ GGAAAACAGACCACAAGCCC 3’ | chr7:45,457,542–45,463,285 |
| Cebpb +76kb: | F: 5’ CCCAAGCTTCCCAGAACTCG 3’ | R: 5’ TGCCTTGCACCCAAAAATGC 3’ | chr2:167,759,270–167,766,462 |
| Slc40a1 +43kb: | F: 5’ GGCAGGGTCCAGGGAAACTA 3’ | R: 5’ GTGACAGAGGGACACATCGG 3’ | chr1:45,861,910–45,902,792 |
| Gdf3 −47kb: | F: 5’ ATGCTCACGCAGACTTGACT 3’ | R: 5’ ACGAGAAAATGTTGGCACAGC 3’ | chr6:122,643,983–122,667,639 |
| Gdf3 −25kb | F: 5’ AAGGGTGAGGGACTCTAGCC 3’ | R: 5’ AGGCAGCTTTGCGGTATCAT 3’ | chr6:122,629,871–122,641,698 |
| Socs3 +22kb: | F: 5’ TTTGGTTCCCCAGTGGTGTTC 3’ | R: 5’ CGGATCATAGCTTTCCCCCA 3’ | chr11:117,938,606–117,948,249 |
| Pparg −17kb: | F: 5’ CTCCTCTTTCGTCTGAGGTTTGA 3’ | R: 5’ AGCTGACAGAGAATCTGGGGA 3’ | chr6:115,336,928–115,353,899 |
| Tnfsf12 +4kb: | F: 5’ CTGTCCATGTCATGTGGCCT 3’ | R: 5’ ACTCCCCGTGAATGAAGCTG 3’ | chr11:69,681,048–69,684,986 |
| Igf1 −15kb: | F: 5’ CTTCCTTAGTAGCTGCACCAGT 3’ | R: 5’ GCAAGCCATAGGGAAGAGGAA 3’ | chr10:87,834,650–87,850,298 |
| Igf1 −105kb | F: TGTTGCATGATGTGAGCCAT | R: TGCCTGATGTGGCATTTTCAC | chr10:87,744,142–87,764,790 |
| Arg1 −4kb: | F: 5’ CCAAAGTGGCACAACTCACG 3’ | R: 5’ CATAAGGTCACGGAGGGTGG 3’ | chr10:24,929,431–24,933,681 |
| Hmox1 Eα: | F: 5’ TGGGAGGGGTGATTAGCAGA 3’ | R: 5’ TAGCTGAGGCTGAGGGAACA 3’ | chr8:75,075,818–75,096,261 |
| Hmox1 Eβ: | F: 5’ CCGGATACTAGTGACTGCCC 3’ | R: 5’ CCACTTAAGGGCATGTGGGG 3’ | chr8:75,075,818–75,096,261 |
| Il6 −64kb | F: 5’ TAGAGCACCCATTGGCTTCC 3’ | R: 5’ CCGTGCAATAGACAGGATT 3’ | chr5:29,943,171–29,955,164 |
| Il6 −0,2kb | F: 5’ ACATGCTCAAGTGCTGAGTC 3’ | R: 5’ ACTAGGAAGGGGAAAGTGTGC 3’ | chr5:30,011,114–30,021,975 |
| Prmt8 −1,7kb (negative control region) | F: 5’ CGTGAGCAGAGGTGAGGAGT 3’ | R: 5’ GGTTAACCCAAGCTTCTTGCT 3’ | chr6:127,654,175–127,738,325 |
Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq)
ATAC-seq was carried out as described earlier with minor modification (31). 20 000 cells were sorted in ice-cold PBS. Nuclei were isolated with ATAC-Lysis Buffer (10mM Tris-HCl pH7.4, 10mM NaCl, 3mM MgCl2, 0.1% IGEPAL) and were used for tagmentation using Nextera DNA Library Preparation Kit (Illumina) from 2–3 biological replicates. After tagmentation DNA was purified with MinElute PCR Purification Kit (Qiagen). Tagmented DNA was amplified with Kapa Hifi Hot Start Kit (Kapa Biosystems) using 9 PCR cycles. Amplified libraries were purified again with MinElute PCR Purification Kit. Fragment distribution of libraries was assessed with Agilent Bioanalyzer and libraries were sequenced on a HiSeq 2500 platform.
Mapping and normalization of ATAC-seq
Three replicates of muscle-derived Ly6Chigh MFs of day 1 and Ly6Chigh and Ly6Clow MFs of days 2 and 4 upon muscle injury were used for the ATAC-seq experiments (20000 sorted cells per sample). The primary analysis of ATAC-seq-derived raw sequence reads has been carried out using the newest version of ChIP-seq analysis command line pipeline (86) including the following steps: Alignment to the mm10 mouse genome assembly was done by the BWA tool (32), and BAM files were created by SAMTools (29). Signals (peaks) were predicted by MACS2 (33), artifacts were removed according to the blacklist of ENCODE (34), and filtered for further analysis by removing low mapping quality reads (MAPQ score < 10), duplicated reads and reads located in blacklisted regions. All regions derived from at least any two samples were united within 0.5kb and those summits having the highest MACS2 peak score in any sample were assigned to each region. Promoter-distal regions were selected excluding the TSS+/−0.5kb regions according to the mouse GRCm38.p1 (mm10) annotation version. In total, we identified 57,409 peaks from muscle-derived MF samples. Tag directories used by HOMER in the following steps were generated with a 120-nucleotide fragment length with makeTagDirectory (35). Genome coverage (bedgraph and tdf) files were generated by makeUCSCfile.pl (HOMER) and igvtools, respectively, and used for visualization with IGV2 (36). Coverage values were further normalized by the upper decile value detected in the consensus regions for each sample to minimize the inter-sample variance.
Differential chromatin accessibility analysis
To identify the open chromatin regions involved in muscle-derived MF differentiation, we compared the two end-point cell population of this process: Day 1 Ly6Chigh versus Day 4 Ly6Clow. DiffBind v2.6.6 (84) was used to identify differentially opened regions, with DESeq2 (method = DBA_DESEQ2, bFullLibrarySize = FALSE) (37). An ATAC-seq region was defined as differentially changed if the peak showed | log2 fold change | > 1.5 and FDR corrected p-value < 0.05. For hierarchical K-means clustering, we used the previously defined differentially opened chromatin regions. Briefly, we counted the normalized read counts of all samples across the differential sites with HOMER’s annotatePeaks.pl program (35). Sample values for each peak were scaled and used as an input for hierarchical K-means clustering in R. Clusters were visualized in R.
Motif analysis
De novo motif analysis of differentially opened chromatin regions was performed using HOMER’s findMotifsGenome.pl (-len 12 -size 200 -dumpFasta -bits -fdr) (35). Motif matrices of HOMER’s collection selected by the resulting top de novo motifs were used to calculate motif enrichments using HOMER’s annotatePeaks.pl program and plotted in R (35). Because of their palindromic nature TREs were united, and because of the partial identity TREs overlapping with MAREs were excluded. Motif logos were created with seqLogo in R (85).
Statistics
ANOVA with Bonferroni correction for multiple testing was used to determine statistical significance. Adjusted p values are stated within Fig. legends. All experiments were performed using at least three independent experiments from distinct samples. No repeated measures were performed. For RT-qPCR analyses, three biological samples were used for each condition. For FACS marker analysis, four independent samples were analyzed. At least 105 cells were counted for FACS cell populations. For the histology experiments, at least 10 biological samples were used (each animal provides 2 biological samples). For the CSA distribution, two-way ANOVA was used to mark significance for each size class. In scatter dot plots, mean and SEM are shown in addition to individual data points. In bar graphs, bars show the mean of the indicated number of samples, error bars represent SEM. Student’s t-tests and ANOVA analyses were performed in GraphPad Prism 6 (GraphPad Software) with 95% confidence intervals, and P<0.05 was considered statistically significant (P<0.05=*, P<0.01=**, P<0.001=***, P<0.0001=****).
Results
MARE binding TFs remodel the muscle-infiltrating macrophage epigenome
We chose the cardiotoxin (CTX)-induced skeletal muscle injury model to understand and dissect the role and contribution of myeloid cells, primarily MFs and their epigenomic changes in skeletal muscle regeneration. This model proved to be instrumental for the identification of exogenous mechanisms of muscle regeneration (3, 11, 13, 15, 18, 19, 21, 38–40).
First, we aimed to assess the epigenomic changes that are taking place during MF subtype specification (transition from Ly6Chigh to Ly6Clow) following acute sterile injury. We hypothesized that by using an unbiased genomic approach termed “Assay for Transposase Accessible Chromatin” coupled with sequencing (ATAC-seq)(31) we could study the chromatin structure and accessibility during the transition of inflammatory MFs to repair type ones (sorting strategy is shown on Supp. Fig. 1A), and thus identify novel regulators of this process. More specifically, we focused on comparing the chromatin state of the two distinct states of MF function (inflammatory Day 1 Ly6Chigh MFs versus repair Day 4 Ly6Clow MFs) during MF phenotype transition and muscle regeneration (Fig. 1A). We identified 5120 genomic regions that are exclusively accessible in Day 1 Ly6Chigh inflammatory MFs and 3504 sites exclusively characterizing the Day 4 Ly6Clow MFs. Next, we grouped these changes into 4 clusters based on their dynamically changing (opening/closing) patterns (Fig. 1B). Cluster 1 characterizes the MFs of the early inflammatory phase (Day 1 post injury) while Cluster 4 characterizes the MFs involved in the regeneration phase (Day 4 post injury) following the injury (Fig. 1B). Importantly, clusters 2 and 3 reveal transient chromatin accessibility and appear to be associated more with the Day 2 cell populations in which the MF phenotypic transition takes place, and thus both functionally diverse MF phenotypes exist at the same time. This global genomic analysis of ATAC-seq data helped us to assess the dynamic changes at each time point and for each subpopulation, allowing the identification of open/accessible and closed genomic loci. Focusing on well-established inflammatory (i.e. Arg1, Il1b, Cd68, Il1r2, Cd80, Nfkb1, Mcl1, Ptges, Il6, Spp1, Socs3, Cebpb) and repair (i.e. Il10, Klf4, Tgfbr1, Slc40a1, Igf1, Il4ra) related marker gene loci (gene expression is shown on Supp. Fig. 1B), our ATAC-seq experiments revealed a dynamic and extensively reorganized chromatin structure, which could be associated with most of these gene loci (representative examples for each cluster are shown on Fig. 1C). Therefore, it appears that the transcriptional changes are accompanied and probably underpinned by major transient changes in the chromatin structure (Figs. 1B–C). Altogether, these results show that MFs exhibit high level of chromatin plasticity upon CTX injury and following resolution of inflammation, characterized by precisely timed chromatin opening and closing at distinct stages of the regeneration process.
Fig. 1. MARE-motifs are overrepresented in the open chromatin of muscle-infiltrating inflammatory macrophages following acute sterile injury.
A. Differential chromatin openness (assessed by ATAC-seq) between inflammatory Day 1 Ly6Chigh versus reparatory Day 4 Ly6Clow sorted MF populations (n= at least 2 samples per group). Gating strategy for the macrophage subsets isolation is shown in Supplementary Fig. 1A. An MA-plot (log2 fold change vs. average read count) was used to visualize statistically significant changes (fold > 1.5 and FDR < 0.05) of chromatin accessibility for all counted peaks. Numbers in the upper and lower corner indicates number of upregulated and downregulated sites, respectively.
B. Heatmap representation of four defined clusters with differential ATAC-seq chromatin openness dynamics in muscle-infiltrating MF populations. Hierarchical K-means clustering was performed on variance-stabilized read counts to build a heatmap for the 8624 differentially accessible sites.
C. IGV genome browser view of distal regions of known inflammatory and regeneration-related genes according to ATAC-seq based data clustering from Ly6Chigh and Ly6Clow muscle-derived MFs recovered and sorted from regenerating muscle upon CTX injury. Examples for each cluster presented in panel B are shown.
D. Sequence motif enrichment in the four ATAC-seq clusters from muscle-infiltrating MFs. Top 3 motifs predicted/enriched are shown. Scatter plots show the mean motif density of each peak in 20 nucleotide bins for each cluster. The motif matrices used in the analysis are indicated on the top of each panel. Lines represent the loess regression model for each cluster.
E. Heatmap showing the mRNA expression pattern of the MARE-binding TFs in Ly6Chigh and Ly6Clow muscle-infiltrating MFs at the indicated days post CTX. RNA-seq expression values are visualized as log10 of the normalized expression calculated using the DEseq method.
Next, we sought to identify the transcription factors driving the uncovered dynamic reorganization of the MF chromatin. Applying our bioinformatics pipeline (27, 41), we could detect and identify sequence motifs associated with the uniquely changing chromatin regions, representing stages of the phenotypic transition. Using known motif matrices according to our de novo motif enrichments, we assigned TF motifs to the four ATAC-seq clusters (Supp. Fig. 1C). Significantly, we found that the 12-O-tetradecanoylphorbol-13-acetate (TPA) response element (TRE), specific for activator protein 1 (AP-1), and MARE motifs were enriched at the sites included in Clusters 1, 2 and 3 (Fig. 1D upper panels), while the motif of PU.1, a common macrophage-specific core TF motif, showed high enrichment in Cluster 4 (Fig. 1D lower panel). The identification of MARE, an extended variant of TRE, focused our attention to this TF family. There are twelve MARE-binding TFs identified so far (42, 43). Evaluation of RNA-seq and microarray data from in vivo isolated MFs following CTX injury revealed that only four members of the MARE-binding TF family are expressed, including Bach1, Nuclear Factor Erythroid 2 Like 2 (Nfe2l2), MAF BZIP Transcription Factor (Maf), and MAF BZIP Transcription Factor B (Mafb) (Fig. 1E). Further analyses revealed that Bach1 and Nfe2l2 are the only two that are differentially expressed, in inflammatory Ly6Chigh versus repair Ly6Clow MFs of Day 2, suggesting that these two TFs are activated in MFs as part of the injury response and during the MF phenotype switch (Fig. 1E). NFE2L2 is known to promote gene expression and have signal-dependent activity. In contrast, BACH1, inhibits gene expression driven by transcriptional activators (such as NFE2L2) and therefore functions to oppose or shape their functional/transcriptional outcome (mode of action is summarized in Supp. Fig. 2A). Thus, to understand the function of transcriptional activators (such as NFE2L2), it is essential to consider the transcriptional repressors (BACH1) that antagonize them.
In summary, our unbiased approach using ATAC-seq revealed that the process of MF subtype specification is dynamic and requires a transient early activation and late repression of gene expression programs. In addition, we identified the putative TF binding sites, regulatory elements and TFs, which are relevant in the MF phenotypic transition. We chose BACH1 for further investigation for the following reasons: (a) BACH1 is highly expressed and overrepresented in Ly6Chigh inflammatory MFs (Fig. 1E); (b) its binding motif is unexpectedly enriched specifically in the open chromatin regions of clusters 1, 2 and 3 (Fig. 1D) during the early inflammatory phase (cluster 1) and during MF phenotype switch (clusters 2 and 3); (c) BACH1 has been previously reported to be released from the chromatin in response to cell injury and inflammation, and control the expression of antioxidant proteins, such as heme oxygenase 1 (HMOX1), that protect against oxidative damage (44, 45); (d) re-analyzing publicly available datasets from various models of acute muscle injury, such as glycerol-induced, contraction-induced and freeze-induced injuries (46, 47), we found both Bach1 and Hmox1, as an indicator of BACH1 activity; to be present or induced in all these models (Supp. Figs. 2C–F), suggesting the involvement of BACH1 and its targets in acute type of injuries; (e) BACH1 is regulated by an acute damage response ligand (heme) which directly binds to BACH1 and removes its suppressive activity (44) (Supp. Fig. 2A). This latter is relevant in our model because the homeostatic response to acute injury comprises of the modulated expression of genes involved in heme and iron handling, since heme is released by damaged myocytes (38). Gene enrichment analysis on our RNA-seq data from muscle-infiltrating MFs following CTX injury, comparing the expression profile of the two distinct states of MF function (inflammatory Day 1 Ly6Chigh MFs versus repair Day 4 Ly6Clow MFs); similar to the ATAC-seq comparison, revealed iron and heme-related pathways being highly enriched (Supp. Fig. 2B). For all these reasons we hypothesized that BACH1 and its targets could have an important role in the inflammatory response in general, and the MF phenotypic transition in particular.
BACH1 is required for efficient muscle regeneration
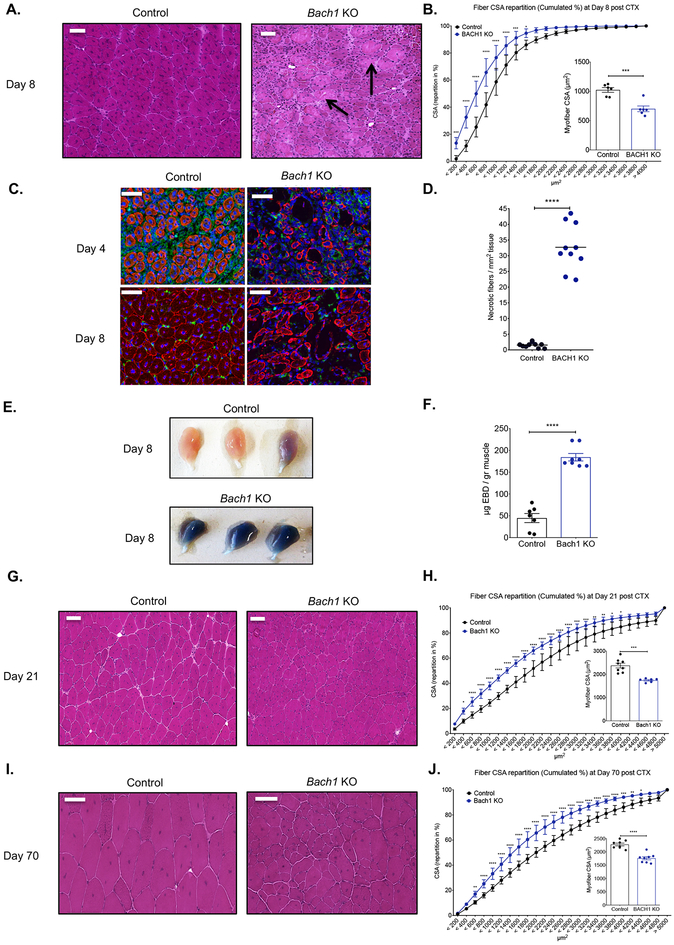
In order to assess the role of BACH1 during muscle regeneration we used the CTX injury model, and utilized an established genetic BACH1 ablation model (deletion of exon 2) (45). In this model, muscle regeneration was severely impaired at day 8 post CTX, in comparison to control muscles as shown by histological analysis (Fig. 2A). This impairment can also be illustrated by a shift to the left (towards small fiber sizes) of the distribution of the myofiber cross sectional area (CSA) (Fig. 2B), a 35% decrease in the mean CSA of regenerating myofibers (Fig. 2B inset panel), a severe impairment in regenerating myocyte organization, shown by desmin (a major intermediate filament protein) staining at day 4 and 8 post CTX (Fig. 2C), and an increase in necrotic fiber content (average increase of 30 necrotic fibers per mm2 of regenerating area compared to controls; equivalent to an increase of 1600%) (Fig. 2D). The extent of necrosis in the Bach1-KO was so extensive that we decided to validate this observation by an independent in vivo myofiber damage marker assay, utilizing Evans Blue dye (EBD) injection (48, 49). Indeed, EBD uptake was increased and could be observed both macroscopically (Fig. 2E), and by quantifying the dye uptake inside the Bach1-KO muscles at day 8 post CTX (Fig. 2F).
Fig. 2. Impaired regeneration of skeletal muscle in Bach1-deficient animals.
A. Representative images of H&E stained skeletal muscle (tibialis anterior) from WT-control, and Bach1-KO animals at day 8 post CTX-induced injury. Scale bars in the upper left corner represent 100μm. Arrows indicate persistent necrotic fibers.
B. Fiber size repartition of regenerating muscle in WT-control, and Bach1-KO animals at day 8 post CTX injury. Inset show the average fiber CSA of regenerating muscle at day 8 post CTX injury (n=6 mice per group).
C. IHC detection of desmin (red), F4/80 (green), and nuclei (blue) in WT-control and Bach1-KO at day 4 and 8 post CTX injury is shown. Scale bars in the upper left corner represent 100μm.
D. Number of necrotic fibers relative to the regeneration area (in mm2) at day 8 of regeneration in WT-control and Bach1-KO muscles is shown (n=10 muscles per group).
E. Representative macroscopical images of WT-control and Bach1-KO TA muscles at day 8 following CTX injury. Mice were administrated systemically with Evans Blue Dye (EBD) 24h before muscle harvest.
F. In vivo EBD uptake assay (shown as μg of EB dye per gr muscle) from WT-control and Bach1-KO TA muscles at day 8 post CTX (n=7–8 muscles per group).
G. and I. Representative images of H&E stained skeletal muscle (tibialis anterior) from WT-control, and Bach1-KO animals at days (G) 21 and (I) 70 post CTX-induced injury. Scale bars in the upper left corner represent 100 μm.
H. and J. Fiber size repartition of regenerating muscle in WT-control, and Bach1-KO mice at days 21 and 70 post CTX injury. Insets show the average fiber CSA of regenerating muscle at indicated timepoints post CTX injury (n=6 mice per group).
In all graphs, bars and lines represent mean ± SEM.
Next, we wanted to determine whether regeneration was still impaired at later stages of the regeneration process in the Bach1-KO. Intriguingly, both at day 21 and day 70 post CTX the Bach1-KO failed to recover to physiological levels compared to controls, as illustrated by histological analysis (Figs. 2G and 2I), a shift to the left of the distribution of the myofiber CSA (Figs. 2H and 2J), and a 33% decrease in the mean CSA of regenerating myofibers (Figs. 2H and 2J inset panels), albeit with no significant increase in collagen deposition in either timepoints (Supp. Figs. 3A–B). It is important to note that no developmental impairment was observed in Bach1-KO uninjured muscles (Supp. Figs. 3C–D), suggesting that the muscle regeneration/growth impairment is only evident after an acute injury.
Myeloid BACH1 controls the phenotypic transition of myeloid cells following CTX injury
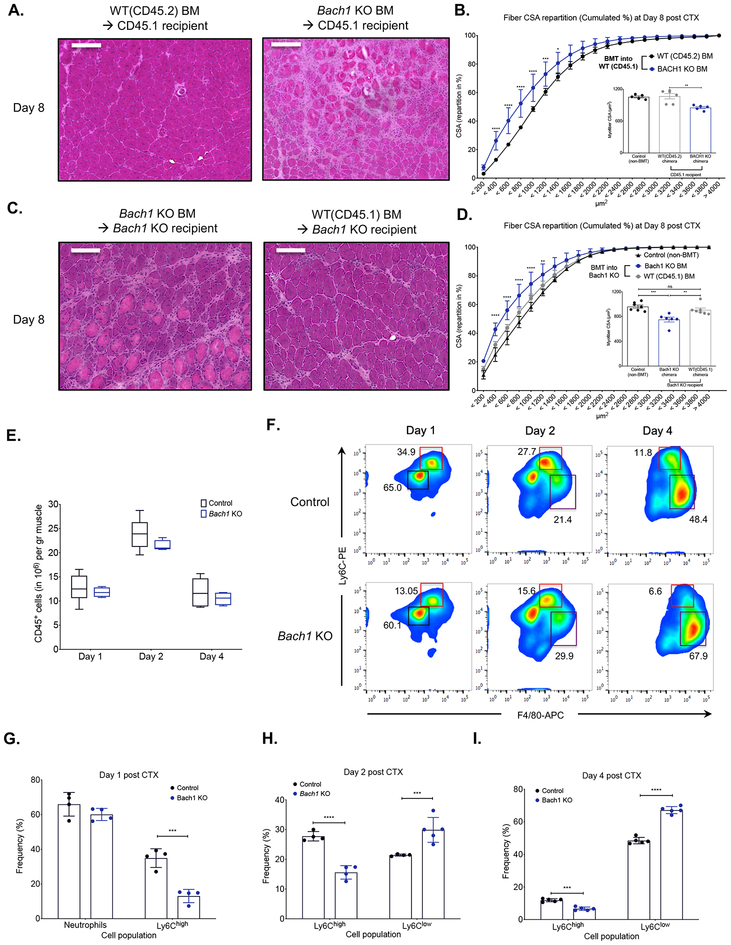
To limit the involvement of compensatory mechanisms in other tissue compartments and to ascertain the hematopoietic/myeloid cell involvement during muscle regeneration, we generated chimeric animals reconstituted with Bach1-KO bone marrows. When the Bach1-KO chimeric animals were challenged with CTX-induced injury, they exhibited a similar impairment in regeneration at day 8 (Fig. 3A), as observed previously in the full body Bach1-KO (Fig. 2A). When compared to WT bone marrow transplanted (BMT) animals, Bach1-KO chimeras contained more regenerating myofibers with smaller CSA (Fig. 3B) and the regenerating muscle was characterized by necrotic fibers; average increase of 20 necrotic fibers per mm2 of regenerating area compared to controls (Supp. Fig. 3E), which are hallmarks of defective muscle regeneration. To further validate our results, we also performed gain of function bone marrow transplantation (BMT) in which WT bone marrow was transplanted into Bach1-KO mice. According to the histological assessment (Fig. 3C), the CSA distribution (Fig. 3D) and the necrotic content (Supp. Fig. 3E) the Bach1-KO mice that received the WT BM were able to recover to control levels at day 8 following CTX with no signs of any persistent necrosis or muscle regeneration impairment. Considering the high expression of BACH1 in the myeloid compartment of the hematopoietic niche, these data further illustrate the involvement of myeloid BACH1 in the muscle regeneration process.
Fig. 3. Altered phenotypic transition of infiltrating myeloid cells in Bach1-deficient models following CTX injury.
A. Representative images of H&E stained TA skeletal muscle 8 days after CTX injury from chimeric WT BoyJ bone marrow-transplanted (BMT) animals (CD45.1 recipients) that received either WT (CD45.2) or Bach1-KO bone marrow. Scale bars in the upper left corner represent 100 μm.
B. Cumulated myofiber cross sectional area repartition and mean CSA (inset panel) at day 8 post CTX injury from WT chimeric animals transplanted with either WT (CD45.2) or Bach1-KO bone marrow (n=5 mice per group).
C. Representative images of H&E stained TA skeletal muscle 8 days after CTX injury from chimeric Bach1-KO BMT animals that received either WT (CD45.1) or Bach1-KO bone marrow. Scale bars in the upper left corner represent 100 μm.
D. Cumulated myofiber cross sectional area repartition and mean CSA (inset panel) at day 8 post CTX injury from Bach1-KO BMT animals transplanted with either WT (CD45.1) or Bach1-KO bone marrow (n=6 mice per group).
E. Number of infiltrating myeloid (CD45+) cells in regenerating muscle from WT-control, and Bach1-KO muscles at indicated timepoints post CTX injury (n=8 muscles per group).
F. Representative flow cytometry images of inflammatory and repair MFs from WT-control and Bach1-KO at indicated timepoints post CTX injury. Squares indicate the gating used for cell frequency quantification (black=PMNs, red=Ly6Chigh inflammatory MFs, purple=Ly6Clow repair MFs). Representative frequencies for each cell population are shown adjacent to each gate.
G. H., and I. Frequency (in %) of CD45+ inflammatory (Ly6Chigh F4/80low) and repair (Ly6Clow F4/80high) MFs from WT-control and Bach1-KO mice at indicated timepoints following CTX injury (n = 4 mice per group).
In all bar graphs, bars represent mean ± SEM.
Next, we asked whether the impaired muscle regeneration was caused by a defect in the cellular dynamics of the myeloid cell infiltrate during muscle regeneration. Hence, we injected both tibialis anterior (TA) muscles of Bach1-KO with CTX and isolated the myeloid cells at day 1, 2 and 4 after the injury using CD45+ magnetic bead selection and cell sorting. Interestingly, we did not find any difference in the numbers of invading myeloid cells (CD45+) at any of the timepoints we examined (Fig. 3E), thus excluding the possibility of a massively diminished myeloid cell invasion contributing to the muscle regeneration impairment we observed previously (Fig. 2). However, this finding did not exclude the possibility of a change in the cellular composition and subtype specification of the infiltrating myeloid cells. Therefore, we examined the dynamics of the infiltrating myeloid cell populations (inflammatory Ly6Chigh F4/80low and repair Ly6Clow F4/80high MFs) during the course of the regeneration period under the same conditions (Figs. 3F–I). Normally, in an uncompromised tissue, Ly6Chigh inflammatory MFs are progressively differentiating into Ly6Clow repair MFs by day 4 after CTX injury (Fig. 3F control panel). In the case of Bach1-KO the frequency of Ly6Clow F4/80high repair MFs in injured muscle at day 2 and 4 show an increase of ~40% compared to controls (Figs. 3F, 3H and 3I), revealing an enhanced conversion of inflammatory to repair MFs. It is worth noting that the frequency of neutrophils at day 1 doesn’t seem to be impacted in the absence of BACH1 (Figs. 3F and 3G). These results suggest that the accelerated MF phenotype switch alone could be an important factor contributing to the impaired clearance of necrotic debris observed histologically (Figs. 2A and 2D). In summary, we observe major differences in the frequencies and ratios of inflammatory and repair MFs, during their phenotypic transition. These data reveal a critical role for myeloid BACH1 as a damage response sensor that controls the phenotype switch in inflammatory (Ly6Chigh) and repair MFs (Ly6Clow).
Key muscle regeneration-related genes are novel BACH1 targets
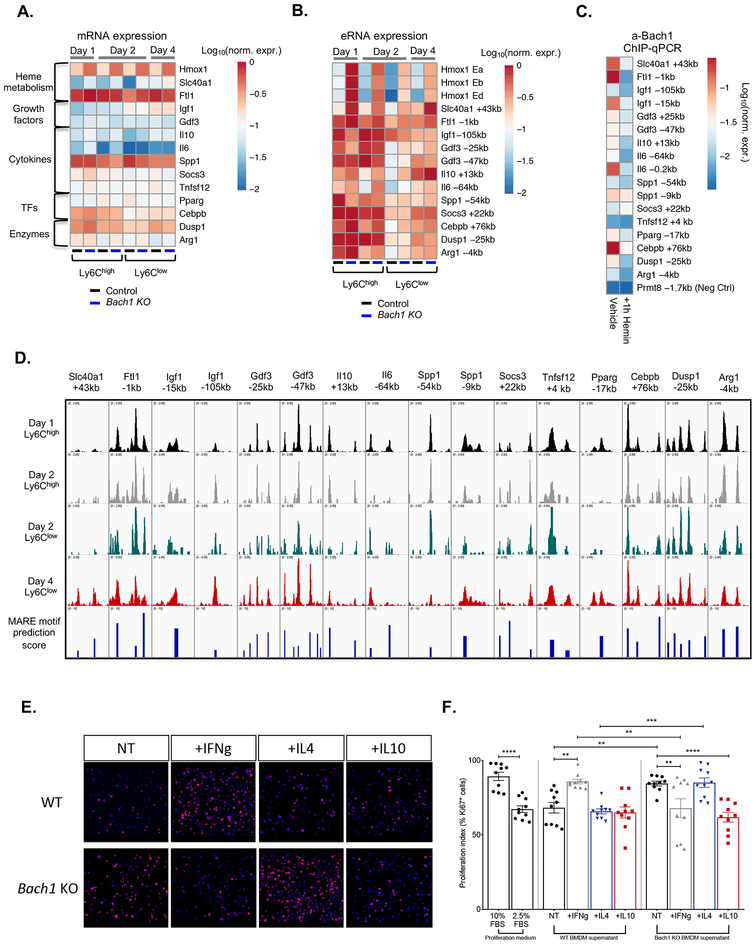
Evaluation of gene expression data from Bach1-KO muscle-infiltrating MFs revealed dysregulation of numerous genes (Il6, Il10, Dusp1, Slc40a1, Cebpb, Gdf3, Igf1, Pparg, Spp1), known to be involved in muscle regeneration and MF phenotype switching pathways (13, 15, 18, 19, 21, 38, 50) (Fig. 4A). These critical genes fall into several functional categories ranging from growth factors, transcriptions factors, enzymes, cytokines and genes related to heme metabolism (Fig. 4A). These findings suggest that BACH1 controls the expression of these key regeneration genes, potentially by directly regulating their enhancers. To test our hypothesis, we initially performed cistromic analysis to identify whether distal differentially accessible chromatin regions around these gene loci, detected by ATAC-seq, are differentially changing during the course of regeneration in the muscle infiltrating MFs, and whether we can predict in silico MARE binding motifs at these sites. Indeed, several putative enhancer regions show both differential chromatin accessibility (Fig. 4D), in line with the gene expression data (Fig. 4A), and high BACH1 binding motif scores (Fig. 4D lower panel). To validate whether these regions are accessible and active, we compared the enhancer RNAs (eRNAs) expression of these loci in WT versus Bach1-KO MFs isolated from the injured muscles (Fig. 4B). As expected, many of the enhancer RNAs around these genes are activated and differentially expressed at the various stages of the inflammatory and repair phases in WT MFs. Importantly, ablation of BACH1 heavily impacts the eRNA expression/activity as well (Fig. 4B).
Fig. 4. Dysregulation of inflammatory and muscle repair-related genes in Bach1-KO muscle-derived macrophages.
A. mRNA expression analysis of pro-inflammatory and repair markers in WT and Bach1-KO muscle-derived MFs upon CTX injury. Heatmap represents log10 of normalized mRNA expression. Each gene was normalized over Ppia (n= 3 independent experiments).
B. Enhancer RNA expression of pro-inflammatory and repair markers in WT and Bach1-KO muscle-derived MFs upon CTX injury. Heatmap represents log10 of normalized expression over Ppia (n= 3 independent experiments).
C. a-BACH1 chromatin immunoprecipitation on the putative enhancer regions in cultured BMDMs reveal BACH1 binding in all marked enhancers around the pro-inflammatory and repair markers shown in panels A and B. Heme treatment for 1 h was used to decipher the direct heme-regulated BACH1 targets. Heatmap represents log10 of normalized abundance over input (n=3 independent experiments).
D. IGV genome browser view of ATAC-seq signals from muscle-derived MFs at the indicated genomic regions upstream or downstream the corresponding gene’s TSS, showing differential peak intensities and predicted MARE motif scores.
E. and F. Effect of WT-control and Bach1-KO BMDM derived conditioned media (non-treated, IFN-g-treated, IL-4-treated or IL-10-treated) on the proliferation (assessed by Ki67 positivity) of C2C12 myoblasts (n=4 independent experiments per group). Representative immunofluorescence images per condition are shown on panel E (red marks Ki67 and blue indicates nuclei). Quantification is shown as a percentage of Ki67+ cells over total cells in the respective field of view (n=10 representative fields of view per group).
In all bar graphs, bars represent mean ± SEM.
To further validate the direct binding of these genes by BACH1, we performed anti-BACH1 chromatin immunoprecipitation (ChIP) experiments in bone marrow-derived MFs (BMDMs) before and after heme treatment. These experiments demonstrated that in untreated MFs, BACH1 is bound at multiple enhancers, at various genomic locations and distances from the transcription start site (TSS) of most of these genes (Fig. 4C). Heme treatment abolishes BACH1 binding at these sites (Fig. 4C). Together, these observations suggest that BACH1 directly regulates many key inflammatory and repair-related genes in the context of muscle regeneration (such as Igf1, Slc40a1, Il6, Il10, Gdf3, Pparg, Dusp1, Cebpb), which could further explain the severity of the Bach1-KO mice’s delayed muscle regeneration phenotype.
These results led us to test whether MF BACH1 activity confers a yet unidentified muscle progenitor proliferation-promoting or inhibiting phenotype to MFs. To test this hypothesis, we used an in vitro muscle precursor cell proliferation assay using C2C12 myoblasts. In this assay, we cultured C2C12 myoblasts with conditioned medium derived from non-treated, interferon-g (IFN-g), interleukin-4 (IL-4) or interleukin-10 (IL-10)-treated WT and Bach1-KO BMDMs, in conditions favoring cell proliferation, and measured the proliferation index by detecting Ki67+ cells by immunofluorescence. As expected, conditioned medium derived from IFN-g treated WT BMDMs increased myoblast proliferation (13, 20) (Fig. 4E WT panel). Surprisingly, conditioned medium from non-treated and IL-4 treated Bach1-KO BMDMs phenocopied the proliferation enhancing effect of inflammatory IFN-g treated WT BMDMs on myoblasts (Figs. 4E–F). At the same time conditioned media from IFN-g and IL-10-treated Bach1-KO BMDMs revealed an inhibition in myoblast proliferation. These results indicated that BACH1-ablation in MFs activates a signaling system that affects myoblast proliferation in a paracrine manner. In summary, we show that BACH1 is an acute damage signal-dependent TF having a much larger gene regulation repertoire that expands beyond iron and heme-related pathways, and with direct effects on myoblast proliferation.
Myeloid Hmox1, a direct BACH1 target, is required for the phenotypic transition of macrophages and for efficient muscle regeneration following CTX injury
To start to dissect the role of BACH1 in the regulation of this important gene set we decided to focus on BACH1-regulated genes whose activity could regulate the MF phenotype switch and ultimately muscle regeneration, the most. Therefore, we selected Hmox1 as the most highly upregulated and most stringently BACH1-dependent gene for further analysis. Gene expression measurements in heme-treated BMDMs verified the BACH1 signal-dependent activity, and that Hmox1, a gene encoding for an enzyme essential for toxic heme clearance and downstream metabolism, is a direct BACH1 target (Supp. Fig. 4A). Further analysis of the Hmox1 locus using ATAC-seq data visualization (Supp. Fig. 4B), MARE binding motif prediction scores (Supp. Fig. 4B lower panel), and BACH1 ChIP, identified the regulatory elements (Supp. Fig. 4B–C), that show extensively transcribed enhancer regions (Supp. Fig. 4D). These regions are forming an enhancer cluster spanning 12kb upstream of Hmox1 TSS (Supp. Fig. 4B). Among the 5 enhancers (Eα-Eε) identified in our experiments, only 2 (Eα and Eδ) where previously described in the literature (45). Based on motif mapping, those two enhancers (Eα and Eδ) are indeed the ones with the strongest predicted MARE binding motifs (Supp. Fig. 4B lower panel), validated experimentally by BACH1 ChIP (Supp. Fig. 4C). However, their enhancer RNA transcriptional activity is not as robust, and especially in the case of Eδ (Supp. Fig. 4D). In conclusion, BACH1 occupies a larger and more complex set of active enhancers around the Hmox1 locus than previously thought (51), and suggests that Hmox1 expression is driven by a damage response-specific enhancer cluster regulated by BACH1.
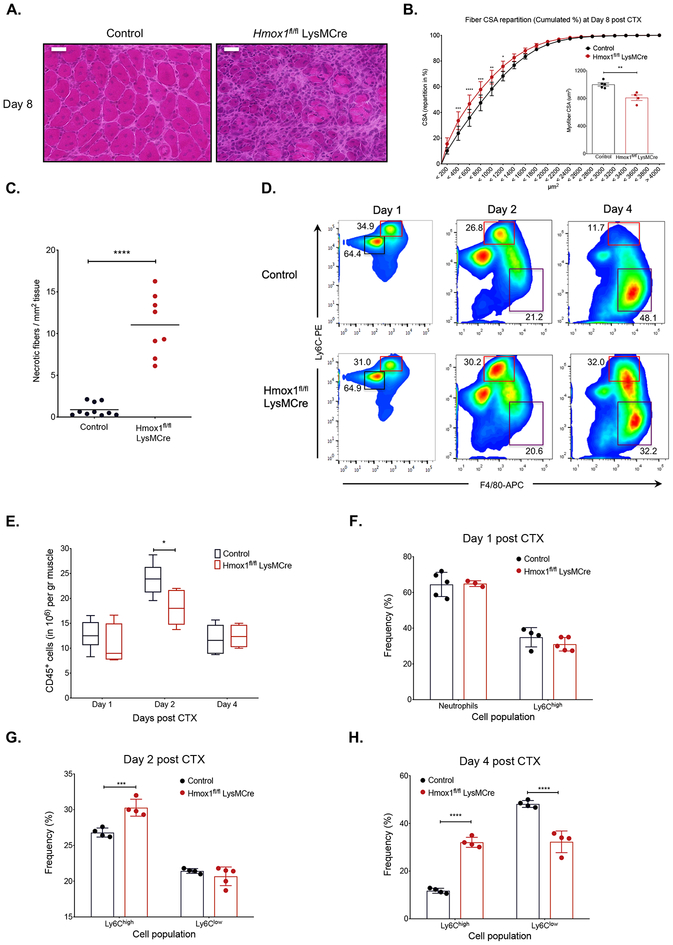
In order to delineate the functional role of macrophage Hmox1 during muscle regeneration we used our CTX injury model in myeloid-specific Hmox1 KO mice (Hmox1fl/fl LysM-Cre) (24). In this model muscle regeneration was severely impaired at day 8 post CTX, in comparison to control muscles as shown by histological analysis (Fig. 5A). These impairments can also be illustrated by a statistically significant shift to the left of the distribution of the myofiber CSA (Fig. 5B), a 20% decrease in the mean CSA of regenerating myofibers (Fig. 5B inset panel), and an increase in persistent necrotic fibers (average increase of 10 necrotic fibers per mm2 of regenerating area compared to controls) (Fig. 5C). Next, we asked whether the impaired muscle regeneration was caused by a defect in the numbers of the myeloid cell infiltrate during muscle regeneration. Isolated myeloid cells from the macrophage-specific Hmox1-KO at day 1, 2 and 4 after the injury revealed only a small reduction in the numbers of infiltrating MFs at day 2 post CTX compared to controls (Fig. 5E). This finding was unlikely to cause the observed regeneration impairment and therefore, we examined the composition dynamics of the infiltrating myeloid cell populations. We observed that in Hmox1fl/fl LysM-Cre muscles, the MF phenotypic shift is severely delayed (Fig. 5D and 5F–H). More specifically, the frequency of Ly6Chigh inflammatory MFs was higher compared to controls at days 2 (~15%) (Fig. 5G) and 4 (~160%) (Fig. 5H) post injury, revealing a potential delay in the conversion of inflammatory to repair MFs. In summary, we detect differences in the ratios of Hmox1-deficient inflammatory and repair MFs, during their phenotypic transition. These data reveal a critical role for myeloid Hmox1 in response to acute sterile injury, and collectively suggest that the BACH1-HMOX1 axis is involved in controlling the phenotype switch from inflammatory (Ly6Chigh) to repair MFs (Ly6Clow).
Fig. 5. Impaired regeneration and delayed phenotypic transition of infiltrating myeloid cells in macrophage specific Hmox1-deficient animals.
A. Representative images of H&E stained skeletal muscle (tibialis anterior) from WT-control, and Hmox1fl/fl LysM-Cre animals at day 8 post CTX-induced injury are shown. Scale bars in the upper left corner represent 100 μm.
B. Fiber size repartition of regenerating muscle in WT-control, and Hmox1fl/fl LysM-Cre animals at day 8 post CTX injury. Insets show the average fiber CSA of regenerating muscle at day 8 post CTX injury (n=4–5 mice per group).
C. Number of necrotic fibers relative to the regeneration area (in mm2) at day 8 post CTX in WT-control and Hmox1fl/fl LysM-Cre, muscles (n=8–10 muscles per group).
D. Representative flow cytometry images of inflammatory and repair MFs from WT-control, and Hmox1fl/fl LysM-Cre muscles at indicated timepoints post CTX injury. Squares indicate the gating used for cell frequency quantification (black=PMNs, red=Ly6Chigh inflammatory MFs, purple=Ly6Clow repair MFs). Representative frequencies for each cell population are shown adjacent to each gate.
E. Number of infiltrating myeloid (CD45+) cells in regenerating muscle from WT- control, and Hmox1fl/fl LysM-Cre animals at indicated timepoints post CTX injury (n= 8–10 muscles per group).
F. G., and H. Percentage of inflammatory (Ly6Chigh F4/80low) and repair (Ly6Clow F4/80high) MFs from WT control, and Hmox1fl/fl LysM-Cre muscles at indicated timepoints following CTX injury (n=4–5 mice per group).
In all graphs, bars and lines represent mean ± SEM.
Discussion
Macrophage function is supported by multi-level transcriptional control. Differentiation is achieved by linage-specific transcription factors such as PU.1, and polarization is driven by cytokine signal-dependent factors such as NFkB (nuclear factor kappa-light-chain-enhancer of activated B cells) and STAT6 (Signal Transducer And Activator Of Transcription 6) (35, 52, 53). However, the key switch(es) between tissue injury-associated inflammatory to reparatory MF phenotypes have not been uncovered. Here, we identify the heme-BACH1-HMOX1 axis as one of the regulatory modules of this process.
The phenotypic transition from inflammatory Ly6Chigh to repair Ly6Clow cells is highly correlated with the tissue regeneration kinetics and is accompanied by a dynamic crosstalk between MFs and other muscle tissue components that lead to a hierarchical chromatin and transcriptional reprogramming process likely containing tissue-specific elements. This switch in MF phenotype has been documented extensively by multiple transcriptomic and lipidomic approaches by several labs including ours (7, 10, 11, 18–20) (54). Recently muscle-infiltrating MFs were also gated based on their MHCII expression (an indicator of antigen presentation) (55). Although in that study they identified two subsets of MFs with a direct functional readout (i.e., antigen presentation) and exhibited different transcriptional programs, these subsets showed non-uniformity in expression of CD64, Tim4, CX3CR1, and CD11c. These results suggest that additional distinctions could be extrapolated based on the use of alternative markers such as MHCII, CD11b and CCR2 and thus we acknowledge that profiling at the single-cell level would be needed to fully resolve this heterogeneity and understand the different function of each macrophage subset. Nonetheless, our results shown here and previously by our lab (11–13, 54) support that Ly6Chigh and Ly6Clow MFs in regenerating muscle display distinct inflammatory profiles and thus, we believe that our CD45, Ly6C, Ly6G and F4/80 gating strategy is sufficient to discriminate between inflammatory and repair type MFs. However, the epigenomic requirements and the main transcriptional drivers of the process are not known. By using ATAC-seq on muscle-infiltrating MFs from day 1, 2 and 4 post injury, we detected differentially opening sites as a function of time containing AP-1 motifs (including the extended Maf recognition element - MARE). This intriguing finding prompted us to ask the following questions: 1) what are the MARE binding transcription factors that potentially regulate these accessible sites, and 2) how these MARE binding TFs contribute overall to the MF phenotype switch during muscle regeneration. It has been shown that during muscle damage, profound changes in the muscle expression of heme and iron metabolism-related genes can be observed (38). Muscle-infiltrating MFs carry out this function thus preventing the deleterious effects caused by its oxidizing action (56). In addition, the ability of infiltrating MFs to export iron is crucial to avoid fat accumulation during the regeneration process (38). Myoglobin, which is released due to muscle necrosis very likely impacts the expression of iron homeostatic genes which in turn have an impact on MF phenotype transition. Therefore, we narrowed down our studies to the heme-sensing transcription factor BACH1 that could act as a direct sensor of cellular environment changes during injury (such as metabolite concentration changes) and would initiate a sequence of events as part of a metabolite (heme) - receptor (BACH1) - effector (target genes including Hmox1) axis.
The transcription factor BACH1 forms heterodimers with small Maf proteins and bind to MARE, a subcategory of AP-1 motif, to act as transcriptional repressor (43). BACH1 is expressed in most cell types, but it is more highly upregulated in hematopoietic cells (MFs, dendritic cells and thymic T cells) (57, 58). Notably, BACH1 possesses a heme-binding region and thus can be directly bound by free heme that is released by many hemoproteins such as hemoglobin and myoglobin. Based on these findings, we propose a model in which free heme released from myoglobin in damaged myocytes is taken-up from MFs, binds and destabilizes BACH1, thus inhibiting its DNA binding capacity, and subsequently BACH1 is exported from nucleus and degraded in the proteasome (44) (findings are summarized in Visual abstract). Subsequently, activators (such as members of the Nrf2 TF family), competing for the same MARE sites, heterodimerizes with small Mafs and activates BACH1 target genes, such as the Hmox1 enzyme, to further degrade heme to less harmful metabolites (biliverdin, ferrous iron, and carbon monoxide) (59). The absence of BACH1 leads to constitutive expression of Hmox1 (45). Thus, BACH1 functions as a metabolite-driven regulator of heme and iron metabolism, metal detoxification and cellular signaling programs. BACH1 is ubiquitously expressed in most MFs, and has been predicted to be among the core regulators of MF identity, along with TCEF3, C/EBPα, and CREG-1 (57, 58, 60). However, contrary to expectations based on its importance and ubiquitous presence, the existing mouse model has minimal MF developmental defect and no overall physiological defects in mouse development and lifespan (61, 62). In addition, in experimental injury and inflammatory models, these animals show protective phenotypes and dampened overall inflammatory responses (61, 63, 64). However, in our model, dampening physiological inflammation will also inhibit the tightly immune-regulated events that are necessary for muscle debris clearance and regeneration.
Regarding the molecular function of BACH1 we found that it binds to a large number of accessible enhancers in the genome, based on the ATAC-seq mapping, and close to a number of critical gene loci (Il6, Il10, Cebpb, Dusp1, PPARg, Igf1, Slc40a1, Gdf3) previously shown to participate and heavily impact the regeneration process (13, 15, 18, 19, 21, 38, 50). It will be interesting to validate BACH1 binding to these sites in in vivo isolated muscle-infiltrating MFs, when technology limitations will allow for robust low cell number ChIP-seq experiments. Nonetheless, these observations substantially expand the current view of BACH1 as mere rheostat of heme-oxygenase 1 expression, and potentially suggest BACH1 involvement in an active repressing mode of genomic regions. These sites can be activated in MFs when appropriate signals become present. Here, one needs to acknowledge that in the Bach1-KO, tightly regulated BACH1 targets, like Hmox1 and other heme metabolism-related genes (Ftl1, Slc40a1), are constitutively highly expressed and their function can mask other BACH1-dependent secondary repression actions that are also dependent on other TFs. Taken together, these results suggest that proper muscle regeneration depends on tightly and timely regulated inflammatory responses following acute injury and that BACH1 likely acts to coordinate activation, phenotypic transition and resolution of inflammatory programs. BACH1’s protein structure and capacity to bind metabolites and sense the tissue micro-environment makes it an attractive model to test how such signal integrators sense different tissue stimuli and shape different transcriptional programs and diverse MF subtypes. It will also be of interest for future studies to determine the chromatin state of naïve circulating monocytes before they enter the injured muscle. It is possible that a hierarchical chromatin and transcriptional reprogramming process mediated by BACH1 takes place with genomic regulatory elements becoming de novo accessible during monocyte infiltration in the muscle. Lastly, our study urges for a more comprehensive analysis of BACH1’s molecular and transcriptional contribution to MF gene regulation.
To test whether the BACH1-HMOX1 axis contributes to MF function during muscle injury and regeneration, we found delayed regeneration in Bach1-KO and Hmox1 macrophage-specific KO mice. Notably, and in support of our results, it has been shown recently that full body Hmox1 null mice have a similar regeneration impairment with increased injury and necrosis, albeit with a hypertrophic effect at later timepoints (Hmox1-deficient satellite cells seem to be prone to activation and have higher proliferation rates) (65). BMT experiments showed that this defect is intrinsic to the immune infiltrate and not attributed to BACH1 function in muscle cells. FACS analysis showed comparable cell numbers of infiltrating myeloid cells but a reproducible higher (in the case of Bach1-KO) or lower (in the case of the MF conditional Hmox1 KO) ratio of repair (Ly6Clow) to inflammatory (Ly6Chigh) MFs at day 4 post CTX. To our knowledge only three other experimental systems; DUSP1, AMPKa1 or IGF1 deficiency in muscle-infiltrative MFs, were reported to lead to altered MF phenotype switch (18–20). Furthermore, it has been recently suggested that one of the MARE family TFs, Maf (also active in our experimental system), upon exposure to pathogens can promote an acute inflammatory response, while suppressing both NFE2L2 activity and conversion to a cytoprotective phenotype in colon CD169+ and BM-derived MFs (66).
The signal component of the proposed axis is heme. Heme is an essential molecule in myoglobin and its movement, presence, and synthesis needs to be strictly regulated due to its documented toxic effects (67). It was recently established that “free” heme has the ability to act as a signaling molecule for monocyte differentiation via activation of the transcription factor SpiC (68). Circulating Ly6Chigh monocytes enter the tissue during inflammation or injury, and differentiate into repair and anti-inflammatory MFs or pro-inflammatory and immune-stimulatory dendritic cells (DCs) (69). Thus, the ability of heme to impact the differentiation of a monocyte into MF or DC has important ramifications on immunity and tissue homeostasis. In this context, it might be important to uncover factors and pathways that control the movement of heme within distinct cellular compartments after its uptake. A minor limitation that should be taken into consideration is that injection of heme into the peritoneal cavity in vivo can cause neutrophil recruitment in a mechanism dependent on inflammasome components (70). The increase of extracellular heme is a hallmark of hemolysis or extensive cell damage (71), and while heme treatment deactivates BACH1 it could also activate the inflammasome on MFs, which in turn promotes the secretion of IL-1β that could exacerbate tissue damage and affect MF phenotype following CTX. Recent work has shown that heme-mediated activation of MFs is relevant in certain non-infectious diseases such as sickle cell disease. Upon hemolysis, released heme leads to the induction of a pro-inflammatory “M1” polarization in MFs, in a TLR4 and ROS-dependent manner (72). Heme is also a natural ligand for nuclear receptors RevErbα and RevErbβ that generally act to suppress enhancer-directed transcription in MFs, similar to BACH1. The heme-RevErbα action promotes a pro-inflammatory phenotype in human monocytes and MFs by repressing IL10 (73–77). In our chromatin openness analyses we didn’t identify a particular enrichment of RevErb binding motif, indicating that this is likely not a major mechanism in this context. However, the relevance of heme regulated factors, including RevErb, remains to be fully explored in the context of muscle injury and MF subtype specification.
Concurrently, the biological significance of Hmox1 up-regulation in MF phenotype and function remains to be fully elucidated. Regarding MF polarization, high induction of Hmox1 mediated by multiple pathways including endogenous factors and chemical agents drives the phenotypic shift to unconventional M2-like MF phenotypes; called (Mhem)/hemorrhage-associated MFs (78), and MFs generated with oxidized phospholipids (Mox) (79–81). However, the molecular mechanism of the MF phenotypic switch mediated by HMOX1 remains to be established. In our model, HMOX1 expression might become a marker protein of the phenotype switch from inflammatory to repair MFs, which might reflect a difference in intracellular redox status between the two MF subtypes (82). One also needs to acknowledge, that the Hmox1fl/fl LysM-Cre model will affect both neutrophils and monocytes/macrophages. We did not observe any differences in the number or frequency of neutrophils, but that doesn’t exclude paracrine signaling and functional intercellular interaction alterations between MFs and neutrophils that could contribute to the delay of the MF phenotype switch (83). Taken together, BACH1-mediated HMOX1 induction in MFs should be considered as a potential therapeutic approach for immunomodulation in many human diseases including skeletal muscle dystrophies.
Supplementary Material
Key points.
BACH1 and HMOX1 are required for complete muscle regeneration upon acute injury.
BACH1-HMOX1 axis is required for coordinated in-situ MF phenotype switch.
BACH1 regulates inflammatory and repair gene modules in MFs during tissue injury.
Acknowledgements
The authors acknowledge the technical assistance of Ms. Monika Porcelánné and discussions and comments on the manuscript by Dr. Istvan Szatmari (University of Debrecen), and members of the Nagy laboratory. The Bach1-KO mouse line was kindly provided by Dr. Kazuhiko Igarashi (Tohoku University) as a gift.
Funding Support: A.P., P.T. and L.N. are supported by “NR-NET” ITN PITN-GA-2013-606806 from the EU-FP7 PEOPLE-2013 program. G.N. is supported by a grant from the Hungarian Scientific Research Fund (OTKA PD124843). N.G. and L.N. are supported by “Chromatin3D” ITN funded by the European Union under the Horizon-2020 Framework Programme (Grant Agreement 622934) and a grant from Higher Education Institutional Excellence Programme (20428-3/2018/FEKUTSTRAT) of the Ministry of Human Capacities. L.N. is also supported by grants from the Hungarian Scientific Research Fund (Grants K124298, K126885, KKP129909) and by the National Institutes of Health - National Institute of Diabetes and Digestive and Kidney Diseases (R01DK115924).
Footnotes
Competing interests: None of the authors has any conflicts of interest to disclose.
Data and materials availability: The accession number for the muscle-infiltrating MFs RNA-seq and ATAC-seq data reported in this study are GSE114291 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114291) and GSE129393 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129393), respectively. Microarray analysis data of muscle-infiltrating MFs (GSE71155-https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71155), and whole muscle data following glycerol-induced, CTX-induced (GSE45577-https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45577), freeze-induced and eccentric contraction-induced injury models (GDS2701-https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5413) are publicly available.
References
- 1.Okabe Y, and Medzhitov R. 2014. Tissue-Specific Signals Control Reversible Program of Localization and Functional Polarization of Macrophages. Cell 157: 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume DA, Perlman H, Malissen B, Zelzer E, and Jung S. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chazaud B 2014. Macrophages: Supportive cells for tissue repair and regeneration. Immunobiology 219: 172–178. [DOI] [PubMed] [Google Scholar]
- 4.Tidball JG, and Villalta SA. 2010. Regulatory interactions between muscle and the immune system during muscle regeneration. AJP: Regulatory, Integrative and Comparative Physiology 298: R1173–R1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dadgar S, Wang Z, Johnston H, Kesari A, Nagaraju K, Chen YW, Hill DA, Partridge TA, Giri M, Freishtat RJ, Nazarian J, Xuan J, Wang Y, and Hoffman EP. 2014. Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. J Cell Biol 207: 139–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, and Chazaud B. 2007. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. The Journal of Experimental Medicine 204: 1057–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Melton DW, Porter L, Sarwar ZU, McManus LM, and Shireman PK. 2014. Altered Macrophage Phenotype Transition Impairs Skeletal Muscle Regeneration. The American Journal of Pathology 184: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tidball JG 2017. Regulation of muscle growth and regeneration by the immune system. Nat Rev Immunol 17: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigamonti E, Touvier T, Clementi E, Manfredi AA, Brunelli S, and Rovere-Querini P. 2013. Requirement of inducible nitric oxide synthase for skeletal muscle regeneration after acute damage. J Immunol 190: 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varga T, Mounier R, Gogolak P, Poliska S, Chazaud B, and Nagy L. 2013. Tissue LyC6- Macrophages Are Generated in the Absence of Circulating LyC6- Monocytes and Nur77 in a Model of Muscle Regeneration. The Journal of Immunology 191: 5695–5701. [DOI] [PubMed] [Google Scholar]
- 11.Varga T, Mounier R, Horvath A, Cuvellier S, Dumont F, Poliska S, Ardjoune H, Juban G, Nagy L, and Chazaud B. 2016. Highly Dynamic Transcriptional Signature of Distinct Macrophage Subsets during Sterile Inflammation, Resolution, and Tissue Repair. The Journal of Immunology 196: 4771–4782. [DOI] [PubMed] [Google Scholar]
- 12.Patsalos A, Pap A, Varga T, Trencsenyi G, Contreras GA, Garai I, Papp Z, Dezso B, Pintye E, and Nagy L. 2017. In situ macrophage phenotypic transition is affected by altered cellular composition prior to acute sterile muscle injury. J Physiol 595: 5815–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varga T, Mounier R, Patsalos A, Gogolák P, Peloquin M, Horvath A, Pap A, Daniel B, Nagy G, Pintye E, Poliska S, Cuvellier S, Larbi S. Ben, Sansbury Brian E., Spite M, Brown Chester W., Chazaud B, and Nagy L. 2016. Macrophage PPARγ, a Lipid Activated Transcription Factor Controls the Growth Factor GDF3 and Skeletal Muscle Regeneration. Immunity 45: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chazaud B, Sonnet C, Lafuste P, Bassez G, Rimaniol AC, Poron F, Authier FJ, Dreyfus PA, and Gherardi RK. 2003. Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J Cell Biol 163: 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng B, Wehling-Henricks M, Villalta SA, Wang Y, and Tidball JG. 2012. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J Immunol 189: 3669–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, and Henson PM. 1998. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest 101: 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Huang D, Saederup N, Charo IF, Ransohoff RM, and Zhou L. 2011. Macrophages recruited via CCR2 produce insulin-like growth factor-1 to repair acute skeletal muscle injury. FASEB J 25: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tonkin J, Temmerman L, Sampson RD, Gallego-Colon E, Barberi L, Bilbao D, Schneider MD, Musarò A, and Rosenthal N. 2015. Monocyte/Macrophage-derived IGF-1 Orchestrates Murine Skeletal Muscle Regeneration and Modulates Autocrine Polarization. Molecular Therapy 23: 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perdiguero E, Sousa-Victor P, Ruiz-Bonilla V, Jardi M, Caelles C, Serrano AL, and Munoz-Canoves P. 2011. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J Cell Biol 195: 307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mounier R, Theret M, Arnold L, Cuvellier S, Bultot L, Goransson O, Sanz N, Ferry A, Sakamoto K, Foretz M, Viollet B, and Chazaud B. 2013. AMPKalpha1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab 18: 251–264. [DOI] [PubMed] [Google Scholar]
- 21.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, and Nerlov C. 2009. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A 106: 17475–17480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gondin J, Théret M, Duhamel G, Pegan K, Mathieu JRR, Peyssonnaux C, Cuvellier S, Latroche C, Chazaud B, Bendahan D, and Mounier R. 2015. Myeloid HIFs Are Dispensable for Resolution of Inflammation during Skeletal Muscle Regeneration. The Journal of Immunology 194: 3389–3399. [DOI] [PubMed] [Google Scholar]
- 23.Hardy D, Besnard A, Latil M, Jouvion G, Briand D, Thépenier C, Pascal Q, Guguin A, Gayraud-Morel B, Cavaillon J-M, Tajbakhsh S, Rocheteau P, and Chrétien F. 2016. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLOS ONE 11: e0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzima S, Victoratos P, Kranidioti K, Alexiou M, and Kollias G. 2009. Myeloid heme oxygenase-1 regulates innate immunity and autoimmunity by modulating IFN-beta production. J Exp Med 206: 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Sawaf O, Fragoulis A, Rosen C, Keimes N, Liehn EA, Holzle F, Kan YW, Pufe T, Sonmez TT, and Wruck CJ. 2014. Nrf2 augments skeletal muscle regeneration after ischaemia-reperfusion injury. J Pathol 234: 538–547. [DOI] [PubMed] [Google Scholar]
- 26.Saclier M, Theret M, Mounier R, and Chazaud B. 2017. Effects of Macrophage Conditioned-Medium on Murine and Human Muscle Cells: Analysis of Proliferation, Differentiation, and Fusion. Methods Mol Biol 1556: 317–327. [DOI] [PubMed] [Google Scholar]
- 27.Daniel B, Balint BL, Nagy ZS, and Nagy L. 2014. Mapping the genomic binding sites of the activated retinoid X receptor in murine bone marrow-derived macrophages using chromatin immunoprecipitation sequencing. Methods Mol Biol 1204: 15–24. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome S Project Data Processing. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, and Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, and Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsdottir H, Robinson JT, and Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corna G, Caserta I, Monno A, Apostoli P, Manfredi AA, Camaschella C, and Rovere-Querini P. 2016. The Repair of Skeletal Muscle Requires Iron Recycling through Macrophage Ferroportin. J Immunol 197: 1914–1925. [DOI] [PubMed] [Google Scholar]
- 39.Castiglioni A, Corna G, Rigamonti E, Basso V, Vezzoli M, Monno A, Almada AE, Mondino A, Wagers AJ, Manfredi AA, and Rovere-Querini P. 2015. FOXP3+ T Cells Recruited to Sites of Sterile Skeletal Muscle Injury Regulate the Fate of Satellite Cells and Guide Effective Tissue Regeneration. PLOS ONE 10: e0128094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joe AWB, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, and Rossi FMV. 2010. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature Cell Biology 12: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagy G, Daniel B, Jonas D, Nagy L, and Barta E. 2013. A novel method to predict regulatory regions based on histone mark landscapes in macrophages. Immunobiology 218: 1416–1427. [DOI] [PubMed] [Google Scholar]
- 42.Igarashi K, Kurosaki T, and Roychoudhuri R. 2017. BACH transcription factors in innate and adaptive immunity. Nat Rev Immunol 17: 437–450. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, Wu H, Zhao M, Chang C, and Lu Q. 2016. The Bach Family of Transcription Factors: A Comprehensive Review. Clin Rev Allergy Immunol 50: 345–356. [DOI] [PubMed] [Google Scholar]
- 44.Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibahara S, Fujita H, and Igarashi K. 2001. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J 20: 2835–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, and Igarashi K. 2002. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J 21: 5216–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukjanenko L, Brachat S, Pierrel E, Lach-Trifilieff E, and Feige JN. 2013. Genomic profiling reveals that transient adipogenic activation is a hallmark of mouse models of skeletal muscle regeneration. PLoS One 8: e71084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warren GL, Summan M, Gao X, Chapman R, Hulderman T, and Simeonova PP. 2007. Mechanisms of skeletal muscle injury and repair revealed by gene expression studies in mouse models. J Physiol 582: 825–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamer PW, McGeachie JM, Davies MJ, and Grounds MD. 2002. Evans Blue Dye as an in vivo marker of myofibre damage: optimising parameters for detecting initial myofibre membrane permeability. J Anat 200: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wooddell CI, Radley-Crabb HG, Griffin JB, and Zhang G. 2011. Myofiber Damage Evaluation by Evans Blue Dye Injection. Curr Protoc Mouse Biol 1: 463–488. [DOI] [PubMed] [Google Scholar]
- 50.Serrano AL, Baeza-Raja B, Perdiguero E, Jardi M, and Munoz-Canoves P. 2008. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab 7: 33–44. [DOI] [PubMed] [Google Scholar]
- 51.Reichard JF, Motz GT, and Puga A. 2007. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res 35: 7074–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, and Glass CK. 2013. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51: 310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, and Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171. [DOI] [PubMed] [Google Scholar]
- 54.Giannakis N, Sansbury BE, Patsalos A, Hays TT, Riley CO, Han X, Spite M, and Nagy L. 2019. Dynamic changes to lipid mediators support transitions among macrophage subtypes during muscle regeneration. Nat Immunol 20: 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panduro M, Benoist C, and Mathis D. 2018. Treg cells limit IFN-gamma production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc Natl Acad Sci U S A 115: E2585–E2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hentze MW, Muckenthaler MU, Galy B, and Camaschella C. 2010. Two to tango: regulation of Mammalian iron metabolism. Cell 142: 24–38. [DOI] [PubMed] [Google Scholar]
- 57.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ, and Immunological Genome C. 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13: 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, Regev A, Koller D, Immunological Genome Project C, Best AJ, Knell J, Goldrath A, Joic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemarte-Pelletier A, Malhotra D, and Turley S. 2013. Identification of transcriptional regulators in the mouse immune system. Nat Immunol 14: 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alam MZ, Devalaraja S, and Haldar M. 2017. The Heme Connection: Linking Erythrocytes and Macrophage Biology. Front Immunol 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, Desland F, Chudnovskiy A, Mortha A, Dominguez C, Tellier J, Kim E, Dwyer D, Shinton S, Nabekura T, Qi Y, Yu B, Robinette M, Kim KW, Wagers A, Rhoads A, Nutt SL, Brown BD, Mostafavi S, Buenrostro JD, Benoist C, and Immunological Genome P. 2019. The cis-Regulatory Atlas of the Mouse Immune System. Cell 176: 897–912 e820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.So AY, Garcia-Flores Y, Minisandram A, Martin A, Taganov K, Boldin M, and Baltimore D. 2012. Regulation of APC development, immune response, and autoimmunity by Bach1/HO-1 pathway in mice. Blood 120: 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ota K, Brydun A, Itoh-Nakadai A, Sun J, and Igarashi K. 2014. Bach1 deficiency and accompanying overexpression of heme oxygenase-1 do not influence aging or tumorigenesis in mice. Oxid Med Cell Longev 2014: 757901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harusato A, Naito Y, Takagi T, Yamada S, Mizushima K, Hirai Y, Horie R, Inoue K, Fukumoto K, Hirata I, Omatsu T, Kishimoto E, Uchiyama K, Handa O, Ishikawa T, Kokura S, Ichikawa H, Muto A, Igarashi K, and Yoshikawa T. 2009. Inhibition of Bach1 ameliorates indomethacin-induced intestinal injury in mice. J Physiol Pharmacol 60 Suppl 7: 149–154. [PubMed] [Google Scholar]
- 64.Takada T, Miyaki S, Ishitobi H, Hirai Y, Nakasa T, Igarashi K, Lotz MK, and Ochi M. 2015. Bach1 deficiency reduces severity of osteoarthritis through upregulation of heme oxygenase-1. Arthritis Res Ther 17: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kozakowska M, Pietraszek-Gremplewicz K, Ciesla M, Seczynska M, Bronisz-Budzynska I, Podkalicka P, Bukowska-Strakova K, Loboda A, Jozkowicz A, and Dulak J. 2018. Lack of Heme Oxygenase-1 Induces Inflammatory Reaction and Proliferation of Muscle Satellite Cells after Cardiotoxin-Induced Skeletal Muscle Injury. Am J Pathol 188: 491–506. [DOI] [PubMed] [Google Scholar]
- 66.Kikuchi K, Iida M, Ikeda N, Moriyama S, Hamada M, Takahashi S, Kitamura H, Watanabe T, Hasegawa Y, Hase K, Fukuhara T, Sato H, Kobayashi EH, Suzuki T, Yamamoto M, Tanaka M, and Asano K. 2018. Macrophages Switch Their Phenotype by Regulating Maf Expression during Different Phases of Inflammation. J Immunol 201: 635–651. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S, and Bandyopadhyay U. 2005. Free heme toxicity and its detoxification systems in human. Toxicol Lett 157: 175–188. [DOI] [PubMed] [Google Scholar]
- 68.Haldar M, Kohyama M, So AY, Kc W, Wu X, Briseno CG, Satpathy AT, Kretzer NM, Arase H, Rajasekaran NS, Wang L, Egawa T, Igarashi K, Baltimore D, Murphy TL, and Murphy KM. 2014. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 156: 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, and Ley K. 2010. Development of monocytes, macrophages, and dendritic cells. Science 327: 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dutra FF, Alves LS, Rodrigues D, Fernandez PL, de Oliveira RB, Golenbock DT, Zamboni DS, and Bozza MT. 2014. Hemolysis-induced lethality involves inflammasome activation by heme. Proc Natl Acad Sci U S A 111: E4110–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dutra FF, and Bozza MT. 2014. Heme on innate immunity and inflammation. Front Pharmacol 5: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vinchi F, Costa da Silva M, Ingoglia G, Petrillo S, Brinkman N, Zuercher A, Cerwenka A, Tolosano E, and Muckenthaler MU. 2016. Hemopexin therapy reverts heme-induced proinflammatory phenotypic switching of macrophages in a mouse model of sickle cell disease. Blood 127: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carter EL, Gupta N, and Ragsdale SW. 2016. High Affinity Heme Binding to a Heme Regulatory Motif on the Nuclear Receptor Rev-erbbeta Leads to Its Degradation and Indirectly Regulates Its Interaction with Nuclear Receptor Corepressor. J Biol Chem 291: 2196–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, and Rastinejad F. 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 14: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, and Lazar MA. 2007. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789. [DOI] [PubMed] [Google Scholar]
- 76.Chandra V, Mahajan S, Saini A, Dkhar HK, Nanduri R, Raj EB, Kumar A, and Gupta P. 2013. Human IL10 gene repression by Rev-erbalpha ameliorates Mycobacterium tuberculosis clearance. J Biol Chem 288: 10692–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, and Glass CK. 2013. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyle JJ, Harrington HA, Piper E, Elderfield K, Stark J, Landis RC, and Haskard DO. 2009. Coronary intraplaque hemorrhage evokes a novel atheroprotective macrophage phenotype. Am J Pathol 174: 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadl A, Meher AK, Sharma PR, Lee MY, Doran AC, Johnstone SR, Elliott MR, Gruber F, Han J, Chen W, Kensler T, Ravichandran KS, Isakson BE, Wamhoff BR, and Leitinger N. 2010. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res 107: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Araujo JA, Zhang M, and Yin F. 2012. Heme oxygenase-1, oxidation, inflammation, and atherosclerosis. Front Pharmacol 3: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naito Y, Takagi T, and Higashimura Y. 2014. Heme oxygenase-1 and anti-inflammatory M2 macrophages. Arch Biochem Biophys 564: 83–88. [DOI] [PubMed] [Google Scholar]
- 82.Dobashi K, Aihara M, Araki T, Shimizu Y, Utsugi M, Iizuka K, Murata Y, Hamuro J, Nakazawa T, and Mori M. 2001. Regulation of LPS induced IL-12 production by IFN-gamma and IL-4 through intracellular glutathione status in human alveolar macrophages. Clin Exp Immunol 124: 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Uderhardt S, Martins AJ, Tsang JS, Lammermann T, and Germain RN. 2019. Resident Macrophages Cloak Tissue Microlesions to Prevent Neutrophil-Driven Inflammatory Damage. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stark R, Brown G (2011). DiffBind: differential binding analysis of ChIP-Seq peak data. Bioconductor. [Google Scholar]
- 85.Bembom O (2018). seqLogo: Sequence logos for DNA sequence alignments. R package version 1.48.0. Bioconductor. [Google Scholar]
- 86.Barta E Command line analysis of ChIP-seq results. EMBnet.journal, [S.l.], v. 17, n. 1, p. pp. 13–17, May 2011. ISSN 2226–6089. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.