Summary
The Drosophila Taiman (Tai) protein is homologous to the human steroid-receptor coactivators SRC1–3 and activates transcription in complex with the 20-hydroxyecdysone (20E) receptor (EcR). Tai has roles in intestinal homeostasis, germline maintenance, cell motility and proliferation through interactions with EcR and the coactivator Yorkie (Yki). Tai also promotes invasion of tumor cells in adjacent organs, but this pro-invasive mechanism is undefined. Here we show that Tai expression transforms sessile pupal wing cells into an invasive mass that penetrates the adjacent thorax during a period of high 20E. Candidate analysis confirms a reliance on elements of the 20E and Hippo pathways, such as Yki and the Yki-Tai target dilp8. Screening the Tai-induced wing transcriptome detects enrichment for innate immune factors, including the Spätzle (Spz) family of secreted Toll ligands that induce apoptosis during cell competition. Tai-expressing wing cells induce immune signaling and apoptosis among adjacent thoracic cells, and genetic reduction of spz, Toll or the rpr/hid/grim pro-apoptotic factors each suppresses invasion, suggesting an intercellular Spz-Toll circuit supports killing-mediated invasion. Modeling these interactions in larval epithelia confirms that Tai kills neighboring cells via a mechanism involving Toll, Spz factors, and the Spz-inhibitor Necrotic. Tai-expressing cells evade death signals by repressing the immune deficiency (IMD) pathway, which operates in parallel to Toll to control NFkB activity, and independently regulates JNK activity. In sum, these findings suggest that Tai promotes competitive cell killing via Spz-Toll, and that this killing mechanism supports pathologic intertissue invasion in Drosophila.
Keywords: Taiman, invasion, apoptosis, Toll, IMD, innate immunity, 20E, ecdysone, Yki, Hippo
Graphical Abstract
Introduction
Certain tissues in developing organisms fuse with neighboring structures to generate elements of the mature body plan. These programmed fusion events occur in the mammalian optic cup, palate, heart, neural tube, eyelids and body wall[1]. In the invertebrate Drosophila melanogaster, fusions between epithelial primordia generate bilaterally symmetric structures such as the adult thorax[2]. Fused tissues often make shared contributions to a common structure. However, fusion can also involve intercalation of tissues that retain distinct identities, such as during local tumor invasion or syncytiotrophoblast invasion into the uterine wall following implantation[3].
The human steroid receptor coactivators SRC-1,2,3 (also Nuclear Receptor Coactivator/NCOA-1,2,3) interact with nuclear hormone receptors, and their overexpression is in some cases associated with invasion into surrounding stroma (rev. in [4]). The sole Drosophila SRC/NCOA homolog, Taiman (Tai), promotes proliferation via direct interaction with the Hippo pathway coactivator protein Yorkie (Yki)[5, 6] but also supports local invasion of RasV12-scribble transformed epithelial cells[7]. Tai and its cognate binding partner the ecdysone receptor (EcR) are required for invasive behavior of border cells (BCs) in the oocyte[8]. However, it is not known whether a common mechanism underlies these pathologic and developmental forms of Tai-dependent invasion.
Here we define the role of Tai in an intertissue invasion model. Tai overexpression causes distal wingtip cells to breach the thoracic cuticle and penetrate deeply into underlying tissue. Genetic and transcriptomic analyses of this phenomenon reveal links to known Tai- interacting pathways, but also uncover requirements for the Toll innate immune pathway and the RHG (Reaper, Grim Hid) pro-apoptotic factors. Wing-into-thorax invasion correlates with elevated Toll-reporter activity and apoptosis in surrounding tissue, and is suppressed by reducing Toll signaling or RHG expression. Modeling this phenomenon in larval wings confirms that Tai- induced killing of neighboring cells requires the Toll receptor and Spz ligands, and involves induction of hid and rpr mRNAs. Invasive Tai-expressing (Tai+) wing cells evade death, but can be killed by derepression of the IMD pathway, which operates in parallel to Toll to regulate NFκB factors and independently regulates JNK activity (rev. in [9]). In sum, these data indicate that Toll signals are necessary for Tai-induced killing of neighbors and invasion, and that the threshold for IMD activation may determine the sensitivity of Tai+ cells to pro-apoptotic signals that operate locally at boundaries with normal cells.
Result
Tai expressing wing cells penetrate the thoracic cuticle
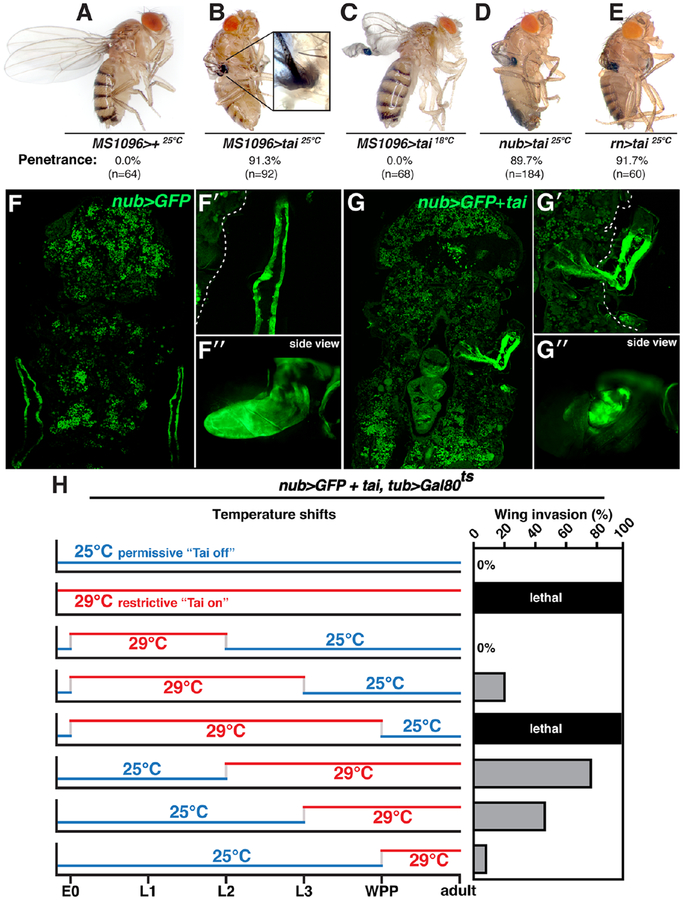
Tai overexpression in multiple tissues and stages results in pupal lethality[6]. To observe the effect of Tai on a single adult tissue, a UAS-tai transgene was expressed in committed wing cells with MS1096-Gal4 (Bx-Gal4), which is active in the larval and pupal pouch[10]. MS1096>tai is ~90% lethal at the pharate stage (n=83), and survivors eclose with wingtips embedded into thoracic cuticle anterior to the haltere (Figure 1A–B). Wingtip embedding is highly penetrant (>90% at 25°C) and accompanied by a raised ring of cuticle around the site of wing:thorax contact (inset in Figure 1B). Consistent with temperature-sensitivity of Gal4 activity, MS1096>tai wingtip embedding is suppressed at 18°C (Figure 1C).
Figure 1. tai-expressing wings penetrate thoracic cuticle.
(A-E) Adults expressing UAS-tai from MS1096, nubbin (nub), or rotund (rn) Gal4 lines at 25°C or 18°C. Penetrance is indicated (n=# wings). (F-G) 18hr APF nub>GFP and nub>tai,GFP pupal cryosections. Magnified top (F’,G’) and side (F”,G”) views of GFP+ wing tissue (dotted line denotes cuticle). (H) nub>tai,tub>Gal80ts shifts between 25°C (blue) and 29°C (red). E0=0hr embryo, L1/2/3=1st, 2nd or 3rd instar; WPP=white prepupa. Invasion frequencies (%) are indicated (‘lethal’ = no adults). See also Figure S1.
Penetration of adult wing tissue into the thorax (hereafter ‘invasion’) has not to our knowledge been reported previously. A defect in wing eversion out of the thorax, which is complete 4–6hrs after puparium formation (APF)[11], could be mistaken for invasion (e.g. [12]). However 12hr APF MS1096>tai,GFP wings are visible as enlarged discs located just under the operculum cuticle (dotted line, Figure S1A–B), indicating successful eversion. Two additional pouch Gal4 lines, nubbin-Gal4 (nub) and rotund-Gal4 (rn) also produce wingtip invasion with UAS-tai (Figure 1D–E). Wingtip invasion could not be reproduced by overexpressing EcR (EcR.A), Myc or stat92E, or by RNAi of EcR-associated repressor Smrter (Smr) or the growth suppressors hippo or Pten (Figure S1C), indicating that disc hypertrophy is not sufficient for invasion. To confirm that Tai+ wing cells penetrate the cuticle, resin-embedded adults were sectioned and visualized by toluidine blue staining (Figure S1D–E). MS1096>+ sections show an unbroken thoracic cuticle and intact flight muscle. By contrast, MS1096>tai wings penetrate the cuticle and epidermis, and contact underlying tissue. Large multivesicular cells cluster near the breach (red arrowheads in Figure S1E). Together, these data indicate that Tai+ wing discs evert normally but subsequently invade the thorax, and Tai is somewhat unique in its ability to cause this effect.
To assess invasion in pupal animals that die pre-eclosion, UAS-green fluorescent protein (GFP) was used as a live-cell marker to track invasion in 18hr APF cryosections (nub>GFP,tai). Wing tissue is visible in nub>GFP animals as two closely apposed GFP-positive (+) sheets lying along the thorax (Figure 1F–F’). Lateral sections (1F’,’side view’) confirm that nub>GFP wings develop normally, with highest GFP expression in the distal blade. Equivalent 18hr APF nub>GFP,tai sections show GFP+ tissue projecting through thoracic cuticle toward the midline (Figure 1G–G’). In side-view, GFP+ tissue projects into the plane-of-section (Figure 1G”). Tai+ cells inside the thorax express the Tai/Yki regulated transcriptional reporter diap1-lacZ [5, 6] (Figure S1F–G). These data indicate that Tai+ wing cells remain alive inside the thorax and elevate expression of a Tai-induced gene.
To define when Tai is required to drive invasion, a temperature-sensitive Gal80 transgene was used to restrict nub-Gal4 activity to specific developmental intervals (nub>tai,tub>Gal80ts) (Figure 1H). Rearing at 25°C (permissive) prevented invasion, while rearing at 29°C (restrictive) was fully lethal. Animals 25°C→29°C shifted at L2 eclose with ~80% invasion, while those 25°C→29°C shifted at L3 display ~50% invasion. Inverse 29°C→25°C shifts only produce invasion thru early L3. A 25°C→29°C shift at white prepupal stage (WPP) leads to 5–10% wing invasion. In sum, these data indicate that Tai exerts its pro-invasive effect during the early-L3 to early-WPP period, which coincides with a rise in levels of 20E (20-hydroxyecdysone), the active form of Ec [13].
Tai wing-invasion is dependent on Ec and Hippo pathways
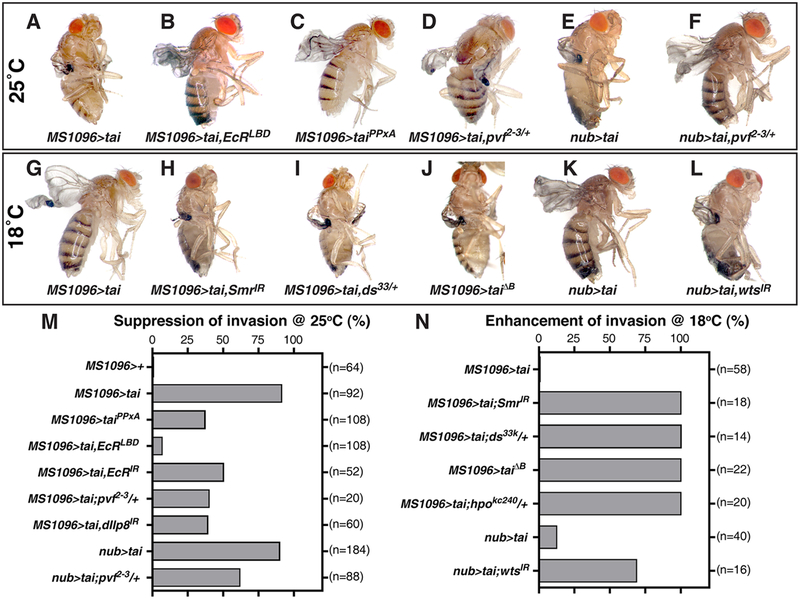
In light of links between Tai and the 20E-EcR, Hippo, and Pvf/Pvr pathways[5, 6, 8, 14], alleles corresponding to each of these pathways were tested for effects on wingtip invasion. Tai- invasion penetrance is reduced by EcR RNAi or a transgene expressing the EcR ligand-binding domain (EcRLBD)[15] (Figure 2A–B,M). Smrter RNAi is not sufficient for invasion (Figure S1C but cooperates with UAS-tai to drive invasion at 18°C (Figure 2G–H,N). A version of Tai that cannot bind Yki (UAS-taiPPxA) has reduced invasive ability (Figure 2C,M) and reciprocally, RNAi of the Yki-inhibitor warts (wts) enhances Tai invasion at 18°C (Figure 2L,N). Alleles of Hippo components dachsous and hippo (ds33 and hpoKS240), or deletions that uncover each gene (Df(2L)ED105 and Df(2R)ED3728), also act as invasion enhancers (Figure 2I,N and Table S1). The suppressive effect of a third deletion identified in a small-scale Df screen, Df(2L)BSC291 (Table S1), was mapped to an internal deletion uncovering the PDGF/VEGF-related ligands Pvf2 and Pvf3 (Figure 2D–F,M), which parallels a requirement for Pvf1 in Tai-dependent BC migration[14]. Collectively, these data show that Tai-driven invasion involves the 20E, Hippo and Pvf pathways. Intriguingly, a weak yki transgene (UAS-yki, chr2) cooperates with wts heterozygosity to produce embedded wingtips (MS1096>ykichr2,wtsx1/+) (Figure S1C), indicating that Yki hyperactivity may be sufficient to induce invasion.
Figure 2. Modification of the tai invasive phenotype by Hippo and EcR pathway alleles.
Adults of the indicated genotypes reared at 25°C (A-F) or 18°C (G-L), and paired quantification of suppression (M) or enhancement (N) of invasion frequency (n=# wings). See also Table S1.
Identification of Tai-induced transcripts in invasive wing discs
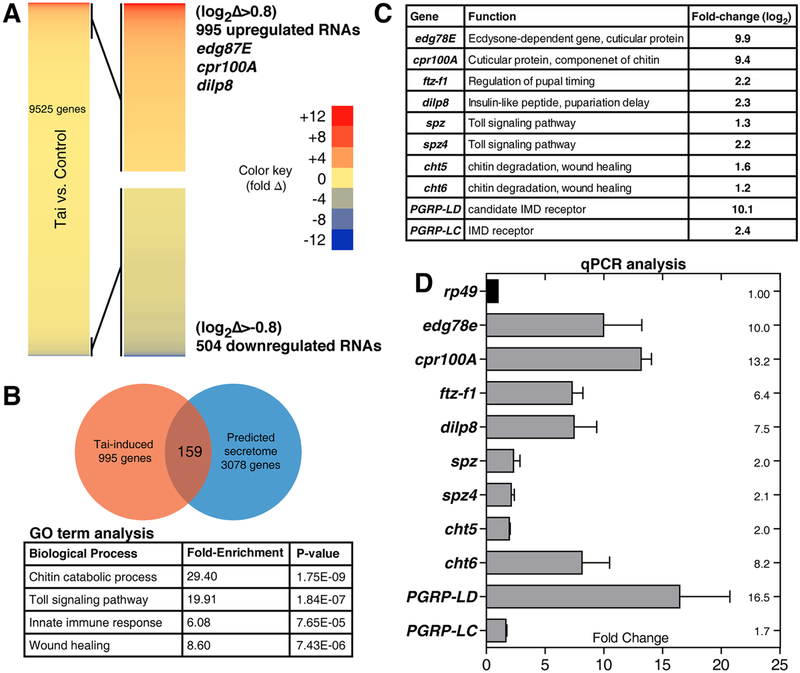
To identify Tai-induced mRNAs in wing cells, rRNA-depleted RNA from engrailed>tai late L3 wing discs was analyzed by high throughput sequencing (HTS as in ref.[6]). en>tai L3 wing discs are enlarged but non-invasive (Figure S2), and thus provide an opportunity to separate Tai- induced mRNAs from mRNAs induced indirectly by the invasive process. HTS identified 995 transcripts ≥2-fold elevated (log2Δ>0.8) in en>tai vs. en>+ samples (Figure 3A, Data S1, and Table S2), including 20E-responsive transcripts Edg78E, Eip93F, ftz-f1 and Cyp18a1[16–19], the Yki targets upd3 and insulin-like peptide-8 (dIlp8) (rev. in [20]), and the Yki-Tai target piwi, which reactivates the piRNA pathway in wts−,RasV12 wing cells[6, 21]. Induction of a small group of these mRNAs was confirmed in pre-invasion MS0196>tai discs (6hr APF) by quantitative real-time PCR (qPCR) (Figure 3C–D). dIlp8 depletion (MS1096>tai,dIlp8RNAi) reduced invasion (Figure 2M), thus validating the HTS data and indicating that invasion may require a dIlp8- mediated developmental delay similar to that following tissue damage[22, 23].
Figure 3. Identification of Tai-regulated mRNAs in larval and pupal wing discs.
(A) Heat-map of abundance changes in 9525 mRNAs in en>tai,GFP vs. en>GFP (Tai:Ctrl) L3 wing discs, with 995 upregulated >+0.8(log2)-fold and 504 downregulated <−0.8(log2)-fold. (B) Top GO-terms among 995 upregulated transcripts, with fold-enrichment and p-values. Venn diagram depicts 159 factors in overlap of Tai-induced mRNAs and predicted secretome (ref.[23]). (C) Fold-change of selected factors by RNA-HTS and (D) by qPCR of 6hr APF MS1096>tai wing disc RNAs standardized to rp49. Error bars = standard deviation (SD) of three biological replicates. See also Figure S2, Data S1 and Tables S1–4.
To focus on secreted factors that facilitate invasion, Tai-induced wing mRNAs were compared to the predicted Drosophila secretome[23]. Gene ontology (GO) analysis of the 159 overlapping mRNAs revealed enrichment in chitin catabolism, Toll signaling, innate immune response, and wound healing categories (Figure 3B and Table S3). qPCR of pre-invasion (6hr APF) MS0196>tai discs confirmed elevated expression of selected mRNAs, including the antimicrobial peptide (AMP) Diptericin-A (DptA), the Toll ligands spätzle (spz) and spz4, the immune deficiency (IMD) pathway receptor Peptidoglycan recognition protein-LC (PGRP-LC), and the chitinases cht-5 and 6 (cht5, cht6) mRNAs (Figure 3D). The IMD and Toll pathways act through the NF-κB homologs Dorsal, Dif and Relish[9, 24] to control expression of secreted AMPs in some contexts and pro-apoptotic genes in others[25]. Cht6 is produced by pupal wing cells and sculpts wing-hair cuticle[26]. Collectively, these data suggest that the microenvironment of Tai+ wing discs is characterized by altered Toll/IMD signaling and chitinase activities. Immune activation in the absence of a pathogen (termed ‘sterile inflammation’) is associated with invasive Drosophila tumors, primarily as a consequence of tissue damage and basement membrane degradation[27]. However, Tai-induced expression of Toll and IMD mRNAs precedes invasion, suggesting a causative role.
Tai-expressing wing cells elicit a systemic immune response
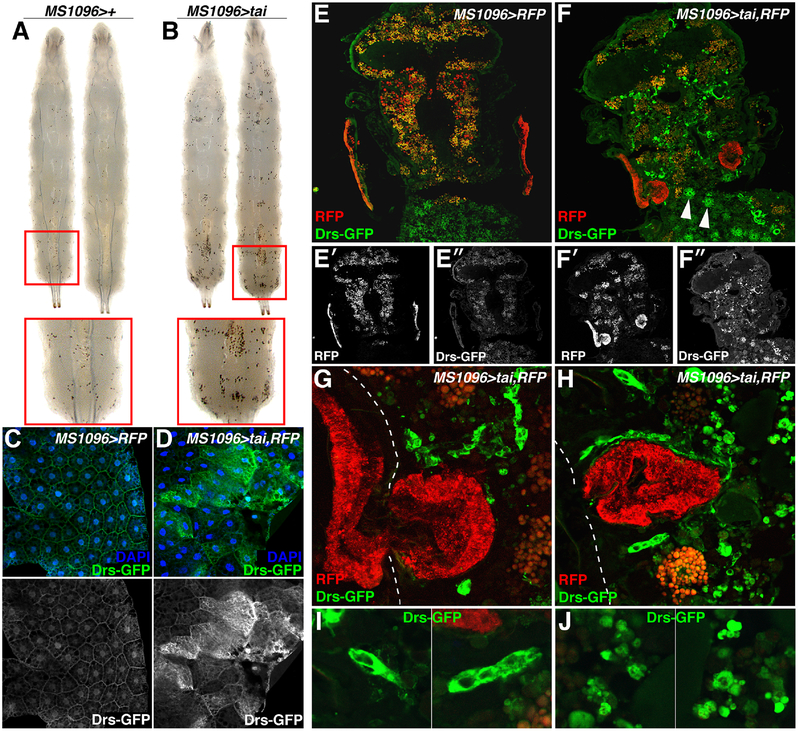
Numbers of the blood cells, lamellocytes, plasmatocytes, and crystal cells, increase in response to immune activation[28]. To test the effect of Tai on immune status, MS1096>tai larvae were briefly heated to 70°C, which activates prophenoloxidase and makes crystal cells visible as black dots[29]. Sessile crystal cells concentrate in posterior segments, and their numbers are visibly increased in MS1096>tai larvae (Figure 4A–B). Hemolymph smears also detect increased nucleated cells in MS1096>tai larvae (Figure S3). Direct analysis of immune signaling in the larval fat body detects elevated expression of the NFkB activity reporter Drosomycin-GFP (Drs- GFP)[30] in MS1096>tai animals (Figure 4C–D). Thus, Tai+ wing cells trigger immune responses in the hematopoietic system and FB before invasion occurs.
Figure 4. Wing-expression of tai activates Toll in other tissues.
MS1096>+ (A) and MS1096>tai (B) larvae processed to visualize crystal cells. Red boxes highlight dorsal posterior views. (C,D) Drosomysin-GFP (Drs-GFP; green) and DAPI (blue) in FB of MS1096>+ and MS1096>tai larvae. Lower panels show Drs-GFP greyscale. (E-F) Drs- GFP (green) in cryosections of 24hr APF (E) MS1096>RFP or (F) MS1096>tai,RFP pupae. Arrows (F) indicate FB-like cells. Lower panels (E’-E”and F’-F”) show single channels. (G-H) Magnified views of RFP (red) and Drs-GFP (green) inside 24hr APF MS1096>tai,RFP;Drs- GFP thoraxes, and (I-J) large GFP+ cells and smaller, fragmented GFP+ cells. Dotted white line indicates external surface of the thorax. See also Figure S3.
Drs-GFP was also assessed during invasion in MS1096>tai,RFP at 14–16hr APF (Figure 4E–J). Drs-GFP is low in controls (MS1096>RFP), but widely induced in MS1096>tai,RFP pupal cryosections (Figure 4E–F) in cells that resemble adult FB cells (white arrows, Figure 4F) and smaller cells inside the thorax and head capsule. Large Drs-GFP positive cells cluster around or envelop Tai+ tumors (green, Figure 4G–I). Smaller Drs-GFP positive cells located along the invasion axis appear fragmented (Figure 4J). Notably, Drs-GFP is not induced in Tai+ wing cells, indicating that immune activation is not an autonomous effect of Tai, but rather involves a paracrine effect on neighbors.
Genetic requirement for Toll components for invasion
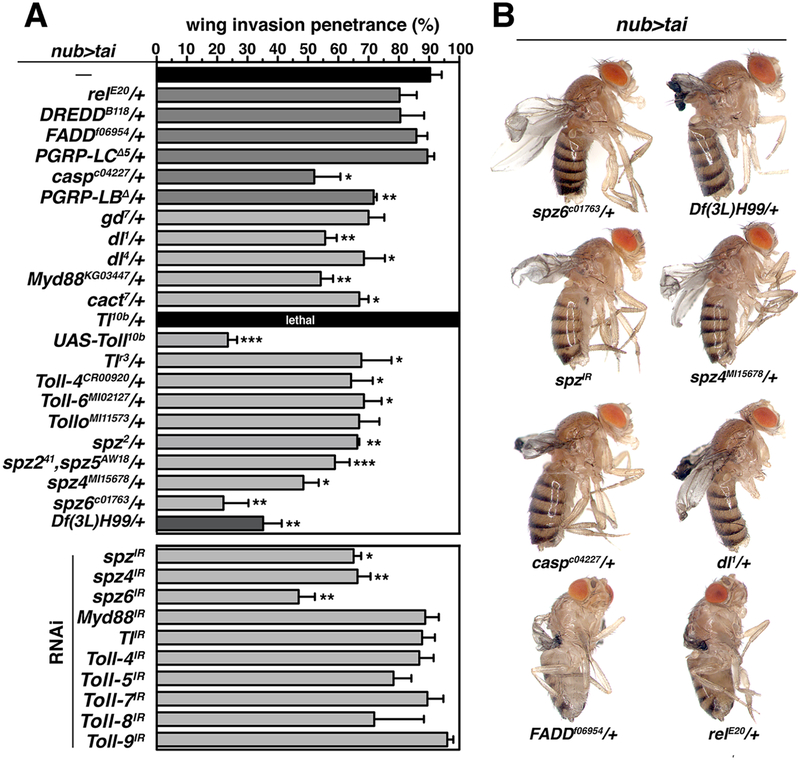
To test whether Toll/IMD pathways support Tai invasion, alleles Toll and IMD factors were tested for modification of nub>tai invasion (Figure 5A–B). Many IMD alleles (relE20, FADDf06954, DreddB118 and PGRP-LCΔ5) had no significant effect, but Toll alleles Tollr3, dl1, dl4, Myd88KG03447, spz2, spz4MI15678, spz6c01763 and Df(3L)spz5Aw18,NT141 (a compound spz2/spz5 allele) all dominantly suppressed nub>tai invasion, as did deletions uncovering Myd88 and dl (Table S1). One copy of the hyperactive Toll10b allele[31] had the inverse effect of complete lethality with nub>tai (nub>tai,Toll10b/+) similar to constitutive Tai expression at 29°C (nub>tai,tub>Gal80ts). However, UAS-Toll10b expression only in Tai+ cells (nub>tai+Toll10b) suppressed nub>tai lethality and invasion (Figure 5A), implying that lethal enhancement of nub>tai by genomic Toll10b/+ reflects effects in non-wing tissue. In support of this hypothesis, RNAi of Toll ligands spz, spz4 or spz6 in Tai+ cells suppressed invasion, while RNAi of multiple Toll genes did not (Figure 5A). RNAi of cht5/6 also suppressed invasion (Figure S4). These data indicate that nub>tai sensitizes the organism to increased Toll activity, and that nub>tai cells require spz and chitinases to promote Toll-dependent invasion.
Figure 5. Tai-driven wing invasion is sensitive to dosage of IMD/Toll pathway factors.
(A) Penetrance (%) of wing invasion in the background of indicated alleles relative to nub>tai alone (black fill). Error bars denote SD. Significance: *p<0.05; **p<0.01; ***p<0.001. (B) Examples of adults from select genotypes in (A). See also Figure S4 and Table S1.
Invasion of Tai-transformed wing cells requires apoptosis
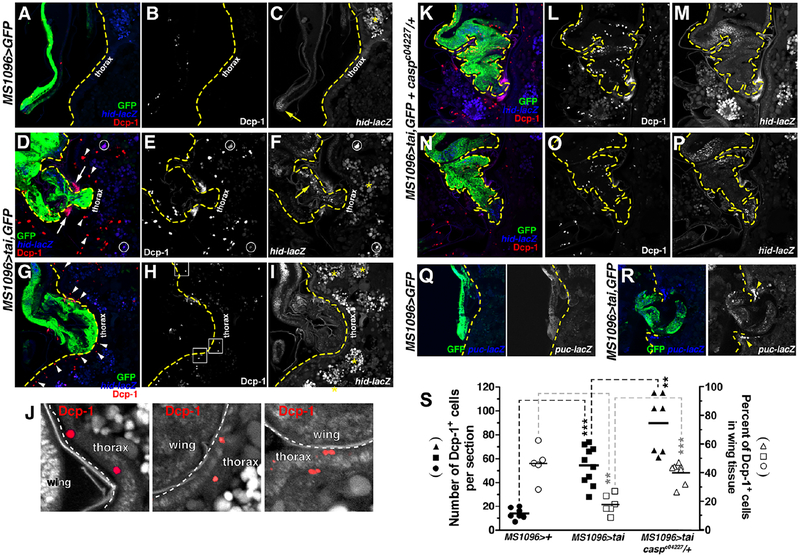
Myc-induced cell competition in the larval wing requires Spz production by ‘winner’ cells and killing of ‘losers’ by Toll-responsive NF-κB factors[25]. Epidermal cells underneath the cuticle are sensitive to apoptosis induced by loss of necrotic (nec), a secreted inhibitor of Spz processing[32], suggesting that Spz-mediated killing may occur during Tai invasion. Consistent with this hypothesis, the H99 deletion removing the RHG pro-apoptotic genes (reaper, hid, grim) dominantly suppressed nub>tai invasion (Figure 5A). Probing cryosections with anti-cleaved Dcp-1 caspase (Dcp-1+) (Figure 6A–J) shows apoptotic cells are rare in controls (12.3±4.7 per section) but abundant in 15–18hr APF MS1096>tai,GFP pupae (54.2±14.7), with a majority of these additional apoptotic events located in the thorax (Figure 6S). At a pre-invasion stage (Figure 6G–I), Dcp-1+ cells are observed in the epidermal cells under the cuticle (arrowheads in Figure 6G; boxed regions in Figure 6J). During invasion (15–18hr APF), some Dcp-1+ appears within Tai+ wings, but a significant amount is located in non-GFP+ thoracic tissue (Figure 6B vs. E,H). Dcp-1+ appears on either side of Tai+ tissue as it penetrates cuticle (arrows, Figure 6D) and in deeper regions of the thorax (arrowheads). A puc-lacZ reporter of the IMD-regulated JNK pathway[33] is also elevated flanking the invasion site (arrowheads, Figure 6Q–R). Some Dcp- 1+ cells also co-express a hid-lacZ reporter (circles, Figure 6D–F), which responds to both JNK and Toll[9]. Collectively these data indicate that thoracic cells undergo caspase-regulated death during Tai-driven invasion, and that RHG apoptotic factors support invasion. Given the rapid clearance of apoptotic cells in vivo, quantification of apoptosis in single cryosections is likely an underestimate of total of apoptotic events during invasion.
Figure 6. Non-autonomous death of neighbors during invasion.
Cleaved Dcp-1 (red), hid-lacZ (blue), and GFP (green) in APF MS1096>GFP (A-C), MS1096>tai,GFP (D-J) or (K-P) MS1096>tai,GFP+casparc04227/+ 18hr cryosections. Yellow dashes (A-I) indicate thoracic cuticle. White arrows mark Dcp-1+ cells at invasion site; arrowheads mark Dcp-1+ deeper in the thorax. Circles mark Dcp-1+/hid-lacZ double-positive cells. Yellow arrow (C) indicates wingtip hid-lacZ. Boxed regions in (H) are magnified in (J). White dashes indicate wing:thorax boundary. Dcp-1+ epidermal cells are red. Yellow asterisks (C,F,I) denote background lipid fluorescence. Yellow dashes (K-P) outline pupal wing. (Q-R) GFP (green) and puc-lacZ (blue) in MS1096>GFP (Q) or MS1096>tai,GFP (R) 18hr APF cryosections. Yellow dashes mark thoracic cuticle. White arrowheads mark puc-lacZ+ cells at invasion site. (S) Dcp-1+ quantification (black fill) and distribution in wing vs. thorax (open symbols) in indicated genotypes. Black bars mark averages. Significance values: **p<0.01; ***p<0.001. See also Figure S5.
hid-lacZ is normally expressed in non-apoptotic cells at the larval dorsoventral (D/V) boundary and hinge (Figure S5A), and pupal wingtip(Figure 6C, yellow arrow) of control discs. In MS1096>tai 15–18hr APF, hid-lacZ+ wingtip cells are among the first to breach the cuticle (Figure S5B–D), and appear to be followed by lacZ-negative cells that progressively displace the hid-lacZ cells to the side of the invading mass (Figure S5E–M). This lineage-tracing data provide evidence that Tai-driven invasion is an ordered process, with D/V boundary or hinge cells apparently serving as leaders in early stages.
The IMD-inhibitor Caspar promotes survival of Tai-transformed wing tissue
The asymmetric death of thoracic cells (Figure 6S) implies that Tai+ wing cells resist invasion- associated killing through an undefined mechanism. While screening candidate modifiers, three suppressor mutations, caspc04227, PGRP-LBΔ and cact7, were identified in IMD/Toll inhibitors (Figure 5A). Based on the suppressive effect of removing Toll activators (e.g. dl, MyD88, Tl, spz), alleles of these inhibitors would be expected to enhance, rather than suppress, invasion. In light of this paradox, we tested the alternative hypothesis that the strongest of these suppressors caspc04227, which affects an inhibitor of the IMD pathway[34], prevents invasion by enhanced killing of Tai+ cells. Anti-Dcp-1 staining confirmed this hypothesis: caspc0422 heterozygosity increases Dcp-1+ in 15–18hr APF MS0196>tai,GFP cryosections from 54.2±14.7 to 89.1±22, with much of this increase occurring within Tai-positive wing tissue (Figure 6K–P,S). Thus, lowering the threshold for IMD activation sensitizes Tai+ cells to immune killing.
Tai-induced killing within the wing epithelial sheet
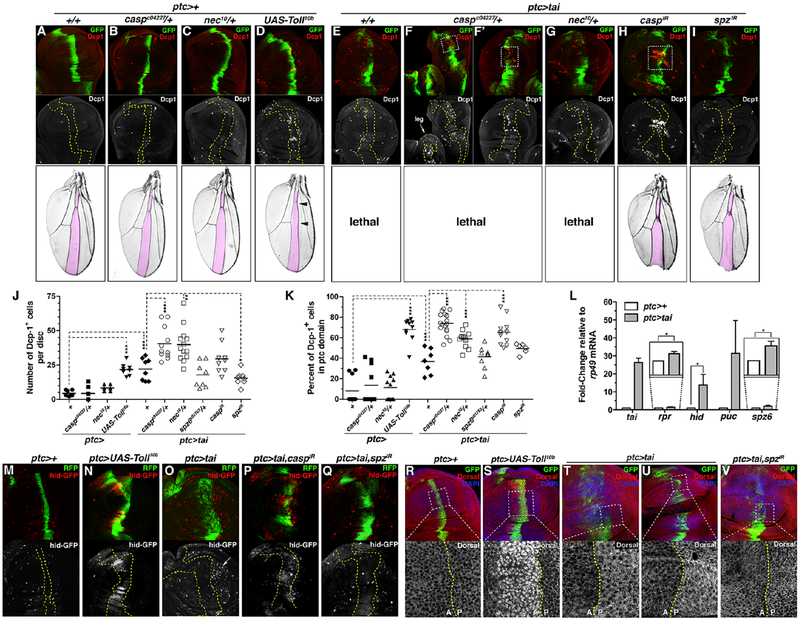
To test whether Tai-induced killing also occurs within a continuous epithelium, the patched- Gal4 (ptc-Gal4) driver was used to express tai in a graded stripe along the AP boundary of the larval wing disc. As in the wing:thorax system, tai expression (ptc>tai,GFP) increases apoptosis across the tissue (mean Dcp-1+ 22/disc, n=8 vs. ptc>GFP mean 4 Dcp-1+/disc, n=7), with most death outside the ptc domain (Figure 7E,J–K). Within the pouch, numbers of hid-GFP+ cells are elevated in the region immediately posterior to ptc>tai cells (Figure 7O), and qPCR of these discs detects elevated expression of hid, puc, rpr and spz mRNAs by qPCR (Figure 7L). Analysis of puc-lacZ and the second AP-1 reporter TRE-lacZ confirms moderate, autonomous JNK activation by Tai (Figure S6E,G–I).
Figure 7. Tai induced apoptosis within the wing epithelium.
Paired adult wings and larval expression of cleaved Dcp-1 (red) and GFP (green) in (A) ptc>GFP, (B) ptc>GFP,caspc04227/+, (C) ptc>GFP,nec10/+, (D) ptc>Toll10b,GFP, (E) ptc>tai,GFP, (F-F’) ptc>tai,GFP,caspc04227/+, (G) ptc>tai,GFP,nec10/+, (H) ptc>tai,GFP,caspIR, and (G) ptc>tai,GFP,spzIR. Arrow in (F) indicates leg disc. Yellow dashes outline ptc-Gal4 domain. (J) Total Dcp-1+ cells/disc and (K) fraction within ptc>GFP domain. Black bars mark averages. Significance values: *p<0.05; **p<0.01; ***p<0.001.(L) qPCR of tai, rpr, hid, puc, and spz6 in ptc>+ (white) and ptc>tai (grey) wing discs relative to rp49. Errors bars: SD of three biological replicates. (M-Q)hid-GFP (red) and RFP (green) in (M) ptc>RFP, (N) ptc>UAS-Toll10b,RFP, (O) ptc>tai,RFP, (P) ptc>tai,caspIR,RFP, and (Q) ptc>tai,spzIR,RFP wing discs. (R-V) Anti-Dorsal (red), GFP (green), and DAPI (blue) in (R) ptc>GFP control, (S) ptc>UAS-Toll10b,GFP, (T-U) ptc>tai,GFP, (V) and ptc>tai,spzIR discs. Bottom panels (R-V) are magnified views of dashed boxes. Dotted lines indicate anterior:posterior (AP) boundary. See also Figures S6–7.
Pouch cells adjacent ptc>tai cells also show a consistent increase in Dorsal protein levels, mainly in the cytoplasm (Figure 7T–U). This non-autonomous ptc>tai effect on Dorsal is moderate relative to ptc>Toll10b, which elevates Dorsal in nuclei (Figure 7S), induces hid-GFP (Figure 7N) and puc-lacZ (Figure 6SF; but not Dpt-lacZ and rpr-lacZ, see Figure 6SA–D), elevates Dcp-1+ (mean 22 cells/disc, n=8), and shrinks the L3-L4 intervein area of adult wings (Figure 7D, pink overlay and arrowheads). However, cytoplasmic accumulation of Dorsal in ptc>tai discs is not inconsistent with this Dorsal being ‘active’. In fact, endogenous Dorsal in larval discs is mainly cytoplasmic at steady-state (Figure 7R and see ref.[35]) but nonetheless promotes apoptosis of wing cells in response to basal microbial infection common to standard culture conditions[36] like those used here. Under these conditions, simply elevating Dorsal levels is sufficient to increase wing cell apoptosis[36], suggesting that increased Dorsal in ptc>tai discs (Figure 7T–U) is also likely to enhance killing. In sum, these data indicate that Tai+ cells increase levels of a Toll-responsive transcription factor in neighboring wing cells that has an established, pro-apoptotic role in this cell type.
Consistent with Toll-mediated apoptosis, the pattern and extent of Tai-induced apoptosis in wing discs is suppressed by reducing spz6 expression (ptc>tai;spz601763/+) across the epithelium (Figure 7J, mean=17 Dcp-1+ cells/disc, n=8) or depleting spz (ptc>tai,spzIR) within Tai+ cells (Figure 7I–K, mean=15 Dcp-1+ cells/disc, n=8). spz RNAi also reduced hid-GFP induction among adjacent pouch cells (Figure 7O vs. Q), rescued ptc>tai lethality (Figure 7I), and reduced the effect of ptc>tai on Dorsal levels in adjacent pouch cells (Figure 7V). Collectively, these genetic and molecular data indicate that spz genes are required for both local and systemic phenotypes induced by Tai.
Requirement for casp and nec in survival of cells in ptc>tai discs
As in the invasion system, Tai+ wing disc cells require casp for survival. Systemic casp reduction (ptc>tai;caspc04227/+) elevates apoptosis in wing discs (mean=40 Dcp-1+ cells/disc, n=11) with 75% of these deaths in the ptc domain. Depleting casp (ptc>tai,caspIR) within Tai+ cells has a similar effect (mean=30 Dcp-1+ cells/disc, n=14), with 65% of these deaths in the ptc domain (Figure 7F–F’,H,J–K). These additional apoptotic events concentrate in the pouch (boxed in Figure 7F,F’,H) and correlate with elevated hid-GFP expression in ptc>tai,caspIR cells. ptc>tai is lethal but ptc>tai,caspIR animals are viable and produce adult wings with small L3-L4 compartments (Figure 7A,H and S7). Thus, killing ptc>tai cells by reducing casp or lowering their ability to produce spz are each sufficient to block ptc>tai systemic effects. The importance of the Spz-Toll axis is further supported by interactions between tai and nec. One copy of the nec10 allele dominantly enhances the overall level of ptc>tai apoptosis to a very similar degree as caspc04227 (Figure 7G, J, mean=40 Dcp-1+ cells/disc, n=13). However, these additional apoptotic cells are distributed more evenly between Tai+ and non-expressing cells: only 58% of total apoptotic events in ptc>tai,GFP;nec10/+ discs occur among Tai+/GFP+ cells (Figure 7K). Thus, elevated pro-Spz processing across the ptc>tai,GFP epithelial sheet promotes killing of Tai+ cells and normal neighbors, while casp heterozygosity preferentially kills Tai+ cells. These effects of spz, casp, and nec alleles on Tai-induced apoptosis are consistent with a model in which Toll plays a more significant role in Tai-induced killing of neighboring cells during disc development, while low IMD pathway activity protects Tai+ cells from immune-associated apoptosis during Tai-driven cell competition.
Discussion
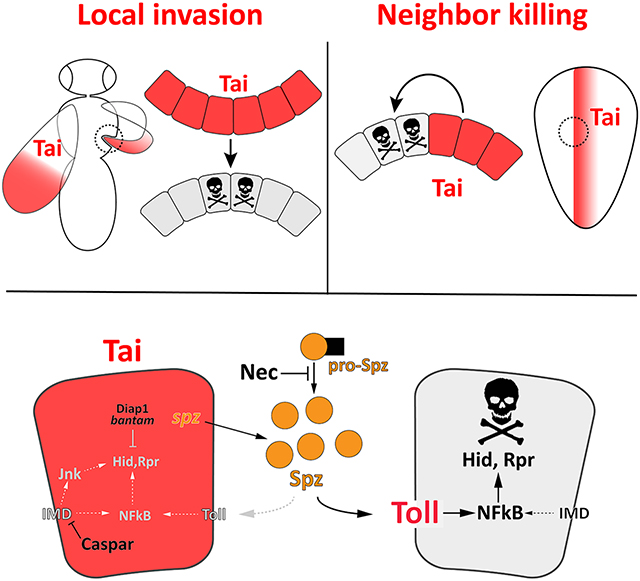
Here we show that Tai engages innate immune signals to transform sessile wing cells into an invasive mass that uses an apoptotic mechanism to penetrate the thorax during a period of high 20E[13]. Genetic tests confirm dependencies on 20E and Hippo pathway factors, including yki and the shared Yki-Tai target gene dIlp8 [6, 37], but unbiased RNA sequencing of invasive cells detects enrichment for innate immune factors, including members of the Spz family of Toll ligands. Tai expression in wing cells elevates Spz/Toll-responsive NFkB reporters in FB cells prior to invasion and in thoracic epidermal cells during invasion. Multiple Toll pathway alleles suppress invasion, and RNAi of Spz ligands within Tai+ cells block invasion, while RNAi of cytoplasmic Toll components (i.e. Toll, Myd88, Dorsal) does not, suggesting a Spz-Toll circuit between wing:thorax cells. Toll induces apoptosis in the context of Myc-induced competition[25], and the suppressive effect of RHG loss argues that apoptosis is required for Tai-driven invasion. Indeed, rates of death are increased in thoracic and epidermal cells adjacent to Tai+ invading tissue. Modeling these intercellular interactions in the larval wing epithelium confirms that tai expression elevates hid/rpr and kills neighbor cells, and that this is enhanced by elevating Toll (Toll10b or nec10) and suppressed by reducing Toll (Tollr3) or autonomously depleting spz. Neighbor killing is associated with elevated levels of the Spz/Toll-responsive transcription factor Dorsal, which has an established pro-apoptotic role in wing epithelial cells in response to basal microbial infection[36]. This Dorsal is mainly cytoplasmic but is nonetheless active, likely due to Crm1-dependent nucleocytoplasmic shuttling[38]. Significantly, an allele of the IMD-inhibitor casp selectively kills Tai+ cells in the wing epithelium and invasive models, suggesting that evasion of local pro-apoptotic signals by Tai+ cells requires repression of IMD, which induces both JNK and NFkB. The suppressive effect of the PGRP-LBΔ allele, which encodes an enzyme that degrades bacterial PGNs and protects flies from innocuous infection[39, 40], suggests basal infection may enhance IMD killing of Tai+ cells, although this hypothesis is untested here. Overall, these data argue that Tai+ wing cells use Spz-Toll to kill neighboring cells, and that differential sensitivity to Toll/IMD contributes to these asymmetric fates.
The pro-invasive effect of Tai in wing cells is notably different from its role border cell (BC) invasion through the nurse cell cluster[8]. BC migration does not involve nurse cell apoptosis, nor has it been linked to IMD or Toll. Rather, the ability of Tai to induce apoptosis of neighbors resembles Toll-mediated killing by Myc ‘super competitors’[25]. Both phenomena are induced by an oncoprotein, occur within an epithelial sheet, and involve non-cell autonomous apoptosis that requires Spz-Toll signals. However, a Tai transgene drives wing intertissue invasion, while a dMyc transgene does not (Figure S2). This difference may be due to the pupal 20E pulse, which coincides with developmentally programmed interdisc fusion[41] and to elevated chitin turnover on the apical surface of pupal wing cells[26]. A combination of these effects may enable Tai+ pupal wing cells to penetrate thoracic cuticle and signal to underlying epidermal cells in a way that Myc+ cells cannot.
How are Tai+ cells spared the apoptotic fate of neighbors? In some competition scenarios, winner vs. loser status is determined by limiting survival factors (e.g. Dpp)[42]. In others, survival is determined by a unidirectional killing signal acting on neighbors but not source cells[25]. In the case of Tai+ cells, survival is likely enhanced by elevated expression of the anti- apoptotic genes diap1 and bantam[5, 6]. However, Tai-induced sensitivity to nec and casp reveals elements of an asymmetric killing mechanism operating between cells in ptc>tai discs. Reduced nec enhances death of Tai+ and non-expressing cells alike, indicating that enhanced Spz processing can elevate Toll past a threshold necessary to kill Tai+ cells. This could be due to generalized increase in Spz levels, or altered local distribution of Spz ligands (as in [43]).
Interestingly, the Tai-interactor Yki, which also induces wingtip embedding (Figure 2A), promotes expression of the Dorsal/Dif inhibitor cactus in wing cells[44]. This is predicted to raise the threshold for NFkB activation and enhance survival during Spz-driven wing cell competition. The lethal effect of nec heterozygosity implies that Tai+ cells also rely on maintenance of a sublethal dose of Toll/NFkB activity to survive, which could be why a cact allele reduces Tai-induced invasion, although this hypothesis is untested. Tai+ cells are also sensitive to derepression of IMD, which bifurcates to induce NFkB factors in parallel to Toll, and JNK/AP-1 activity independent of Toll[9]. Elevated IMD activity could kill Tai+ cells by enhancing autocrine induction of NFkB past an apoptotic threshold, similar to nec heterozygosity. However, the lack of a pro-apoptotic effect of casp reduction on adjacent normal cells (see Fig 7F) suggests that JNK/AP-1 is also involved. Upregulation of JNK/AP-1 activity (Figure S6) may sensitize Tai+ cells to mutations in factors (e.g. casp) that further elevate JNK activity. Thus, survival of Tai+ cells appears to rely on maintenance of a sub-lethal dose of NFkB and JNK.
Although cell competition can suppress tumors by eliminating potentially cancerous cells in Drosophila and mice[45, 46], emerging evidence suggests competition-induced death can also promote invasion and metastasis. Clones of cells lacking the Drosophila Adenomatous polyposis coli (Apc) tumor suppressor require death of neighbors to expand within the gut epithelium[47], and neighbor killing is also required for invasion of cells co-overexpressing the EGFR and miR-8 oncogenes[48]. Features of Myc-induced cell competition have also been found among normal cells bordering invasive human cancers, leading to the hypothesis that competition-induced death enables these cancers to grow and colonize new sites[49].
Local inflammatory signaling mediated by Toll-like receptors (TLRs) can have either pro- or anti-tumor effects in cancers[50]. These alternative outcomes are likely a product of immune interactions between cancer cells and adjacent stroma. Work presented here indicates that the fate of Tai+ cells also depends on immune signals active at clonal boundaries with normal cells. Tai appears to engage a mechanism that resembles Toll-dependent killing by dMyc super-competitors, and evasion of these local pro-death signals is required for Tai+ cells to retain ‘winner’ status. Shifting this system to favor elevated IMD signaling transforms Tai+ cells from ‘winners’ into ‘losers’ relative to surrounding normal cells, reminiscent of the pro- or anti-tumor effects of TLRs. In future, this Drosophila model of Tai function may provide insight into immune-based interactions that contribute to the competitive advantage of cancer cells overexpressing Tai homologs.
STAR Methods for Byun et. al.
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ken Moberg (kmoberg@emory.edu).
This study generated one new Drosophila transgenic line (UAS-EcR-LBD). This stock is not available via central repositories yet to but will be deposited at the Bloomington Drosophila Stock Center in the near future. It will also be maintained as a live stock in the Moberg laboratory in perpetuity. We will share the stock with reasonable coverage by requestor for processing and shipping the line.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal model:
Drosophila melanogaster were maintained under standard culture conditions at 25°C in 12hr light:dark cycles in humidity controlled incubators, unless otherwise noted (e.g. for temperature shift experiments). Experiments used a mixture of male and female animals at the developmental stages indicated in the main text.
METHODS DETAILS
Strategy for randomization and/or stratification
Animals of both sexes were used in all experiments. Samples were not blinded at any stage of the study. For each genotype, 10 or more discs per experiment were processed and imaged for microscopy figures; representative images from two or three separate experiments are shown. RNA analysis (qPCR) was performed in biological triplicates as indicated.
Genetics & deficiency screen stocks:
All crosses were maintained at 25°C unless otherwis e noted. Alleles used are referred to in Key Resources Table (BDSC stock numbers indicated). For the small-scale modifier screen, a subset of deletion lines from the chromosome 2L and 2R deficiency (Df) kits (BDSC; as in Table S1) were crossed with MS1096-Gal4,UAS-tai/TM6B,tub>Gal80 in duplicate at 18°C and 25°C. Multiple wings (see “n” in Figures and Legends) of F1 progeny from multiple replicates (2 or more) were scored by visual inspection under light microscopy. Temperature shifts were performed as described in text.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-ß-Gal | Promega | Z3781 |
| Rabbit anti-GFP | ThermoFisher Scientific | A-6455 |
| Rabbit anti-DCP-1 | Cell Signaling | 9578S |
| Mouse-anti-Dorsal | DSHB | 7A4 |
| Goat anti-rabbit-FITC | Jackson ImmunoResearch | 111–095–003 |
| Goat anti-mouse-Cy3 | Jackson ImmunoResearch | 115–166–003 |
| Goat anti-mouse-Cy5 | Jackson ImmunoResearch | 115–175–146 |
| Goat anti-rat Cy3 | Jackson ImmunoResearch | 112–165–167 |
| Goat anti-rabbit Cy5 | Jackson ImmunoResearch | 111–175–144 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Toluidine Blue O | Sigma-Aldrich | T3260–25G |
| Triton™ X-100 | Sigma-Aldrich | 9002–93–1 |
| 20% Paraformaldehyde | Electron Microscopy Sciences | 15713 |
| Trizol™ | Invitrogen | 15596018 |
| DAPI | Invitrogen | D1306 |
| Vectashield | Vector Labs | H-1000 |
| Critical Commercial Assays | ||
| RNeasy mini kit | Qiagen | 74106 |
| Diff-Quik stain kit | Electron Microscopy Sciences | 26096–25 |
| SuperScript™ III RT | ThermoFisher Scientific | 18080093 |
| TruSeq RNA Sample Preparation Kit v2 | Illumina | 15026495 |
| TruSeq SBS kit v3-HS | Illumina | FC-401–3001 |
| TruSeq Cluster Kit v2-cBot-HS | Illumina | N/A |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: MS1096-Gal4 | BDSC | #8860 |
| D. melanogaster: nubbin-Gal4 | BDSC | #42699 |
| D. melanogaster: rotund-Gal4 | BDSC | #7405 |
| D. melanogaster: en-Gal4 | BDSC | #30564 |
| D. melanogaster: ptc-Gal4 | BDSC | #2017 |
| D. melanogaster: UAS-tai | BDSC | #6378 |
| D. melanogaster: UAS-taiΔB | BDSC | #28273 |
| D. melanogaster: UAS-taiPPXA | Moberg Lab stock, published in reference [6] | N/A |
| D. melanogaster: UAS-wts-IR | BDSC | #34064 |
| D. melanogaster: pvf2–3 | R.Read, Emory University | N/A |
| D. melanogaster: ds33k | BDSC | #288 |
| D. melanogaster: UAS-smr-IR | BDSC | #34087 |
| D. melanogaster: drs-GFP,dipt-lacZ | R.Jones, Emory University | N/A |
| D. melanogaster: relE20 | BDSC | #9457 |
| D. melanogaster: FADDf06954 | BDSC | #19026 |
| D. melanogaster: DREDDB118 | BDSC | #55712 |
| D. melanogaster: gd7 | BDSC | #3109 |
| D. melanogaster: myd88KG03447 | BDSC | #14091 |
| D. melanogaster: caspc04227 | BDSC | #11373 |
| D. melanogaster: dl1 | BDSC | #3236 |
| D. melanogaster: dl4 | BDSC | #7096 |
| D. melanogaster: UAS-spz-IR | BDSC | #28538 |
| D. melanogaster: UAS-spz4-IR | BDSC | #60044 |
| D. melanogaster: UAS-spz6-IR | BDSC | #57510 |
| D. melanogaster: NT141,spz5AW18 | BDSC | #64069 |
| D. melanogaster: spz4MI15678 | BDSC | #61127 |
| D. melanogaster: spz6c01763 | BDSC | #10719 |
| D. melanogaster: UAS-PTEN-IR | BDSC | #33643 |
| D. melanogaster: hidW05014 (hid-lacZ) | T.T.Su, Univ. of Colorado Boulder | N/A |
| D. melanogaster: UAS-dmyc | BDSC | #64759 |
| D. melanogaster: UAS-EcRLBD | This paper | N/A |
| D. melanogaster: UAS-EcR-IR | BDSC | #29374 |
| D. melanogaster: UAS-dilp8-IR | VDSC | v102604 |
| D. melanogaster: hpokc240 | BDSC | #25090 |
| D. melanogaster: Df(3L)H99 | BDSC | #1576 |
| D. melanogaster: UAS-cht5-IR | BDSC | #57512 |
| D. melanogaster: UAS-cht6-IR | BDSC | #54823 |
| D. melanogaster: UAS-Tl-IR | BDSC | #31477 |
| D. melanogaster: nec10 | BDSC | #4288 |
| D. melanogaster: UAS-stat92E | BDSC | #20181 |
| D. melanogaster: wtsx | BDSC | #44251 |
| D. melanogaster: UAS-yki | D.J. Pan, UT Southwestern | N/A |
| D. melanogaster: UAS-EcR.A | BDSC | #6470 |
| D. melanogaster: UAS-hpo-IR | BDSC | #35176 |
| D. melanogaster: th-lacZ (diap1-lacZ) | BDSC | #12093 |
| D. melanogaster: Tl10b | BDSC | #30914 |
| D. melanogaster: UAS-tl10b | BDSC | #58987 |
| D. melanogaster: Tlr3 | BDSC | #3238 |
| D. melanogaster: TlMI00181 | BDSC | #30652 |
| D. melanogaster: 18wΔ7–35 | BDSC | #4372 |
| D. melanogaster: spz2 | BDSC | #3115 |
| D. melanogaster: UAS-Toll-4-IR | BDSC | #28543 |
| D. melanogaster: UAS-Toll-5-IR | BDSC | #29533 |
| D. melanogaster: UAS-Toll-7-IR | BDSC | #30488 |
| D. melanogaster: UAS-Toll-8-IR | VDRC | v9431 |
| D. melanogaster: UAS-Toll-9-IR | BDSC | #34853 |
| D. melanogaster: Toll-4CR00920 | BDSC | #79358 |
| D. melanogaster: Toll-6MI02127 | BDSC | #34467 |
| D. melanogaster: TolloMI11573 | BDSC | #56343 |
| D. melanogaster: PGPR-LBΔ | BDSC | #55715 |
| D. melanogaster: PGRP-LCΔ5 | BDSC | #36323 |
| D. melanogaster: UAS-casp-IR | BDSC | #44027 |
| D. melanogaster: hid-GFP | BDSC | #50751 |
| D. melanogaster: rpr-LacZ | BDSC | #58793 |
| 2L & 2R Deficiency Kit | BDSC | N/A |
| Oligonucleotides for PCR: see Table S4 | ||
| Software and Algorithms | ||
| Fiji | Open source | imagej.net/Fiji |
| Primer3PLlus | NIH/NHGRI | http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi |
| Tophat2 v2.0.12 | Daehwan Kim & Steven Salzberg, Johns Hopkins University | https://ccb.jhu.edu/software/tophat/index.shtml |
| cuffdiff v2.2.1 | Trapnell Lab, Univ. of Washington | http://cole-trapnell-lab.github.io/cufflinks/ |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Galaxy | Open source web | https://usegalaxy.org/ |
RNA sequencing and analysis
Total RNA (2–4ug) extracted from L3 wing discs per genotype was subjected to high-throughput sequencing (HTS) as described previously[6]. Briefly, wandering stage L3 discs of each genotype (en>GFP and en>GFP,tai) were dissected in 1x PBS, and cleaned of other tissue. Discs transferred using tweezers to hold attached trachea into fresh 1xPBS and then quickly deposited in a tube of Trizol(™) at 4°C. Multiple discs were collected in two-hour dissection sessions and frozen in Trizol at −80°C until ~80 total discs had been collected for each genotype. Total RNA was extracted according to manufacturer’s instructions, and then utilized for library construction with the Illumina TruSeq RNA Sample Preparation Kit v2 following the manufacturer’s instructions. Libraries were sequenced by the Yerkes Genomics Core with an Illumina HiScan platform. Cluster generation was performed with Illumina TruSeq cluster kit v2-cBot-HS. Single-read 50 bp sequencing was completed with Illumina TruSeq SBS kit v3-HS. Reads were aligned using Tophat2 v2.0.12, and RPKM expression values from different conditions were extracted and compared by cuffdiff v2.2.1 using Refseq gene models. Resulting data from a single HTS replicate is summarized in Excel file Data S1. Overlap of Data S1 with the predicted secretome was performed using Galaxy opensource software.
Adult imaging and pupal cryosections
Adult flies were frozen at −20°C for >2 hours and i maged on a Leica DFC500 CCD camera. Multiple focal planes were merged and processed to generate a final image. For pupal cryosections, white pre-pupae (WPP) were washed in 1x phosphate buffered saline (PBS), aged 18hr in fresh vials, then detached and glued onto a glass slide with nail polish. After 20min drying, the pupal case was removed, and the pupa was transferred to 4% paraformaldehyde in PBS-T 0.1% (Triton-X100) and incubated at 4°C overnight. Pupae were then rinsed in PBS-T 0.1%, incubated at 4°C overnight in 15% sucrose/PBS-T 0.1%, and then overnight in 30% sucrose/PBS-T 0.1% solution. Pupae that has sunk to the bottom were embedded in Optimal Cutting Temperature (OCT) resin and thin-sectioned onto charged slides for immunofluorescence microscopy.
Immunofluorescence microscopy
Immunostaining and confocal microscopy were performed using standard procedures. Briefly, discs were dissected in 1x phosphate buffered saline (PBS), fixed 20min in 4% paraformaldehyde at room temperature (RT), rinsed 5x in 1x PBS, then permeabilized for 30 min at RT in 1xPBS+0.3% Triton X-100. Samples were then rinsed 2x in 1xPBS, resuspended in primary antibody diluted (according to manufacturer instructions) in 1xPBS+0.1% Triton X-100 plus in 10% normal goat serum (NGS; Jackson Immunologicals). Samples were incubated overnight at 4°C, rinsed 5x in 1xPBS+0.1% Triton X-100, then incubated overnight at 4°C in secondary antibody+10% NGS in 1xPBS+0.1% Triton X-100. Following 5x washes in 1xPBS+0.1% Triton X-100, discs were mounted in Vectashield. OCT sections mounted on coverslips were processed similarly but with these changes: primary and secondary antibody solutions were placed on top of tissue slices+coverslips as drops and incubated overnight in humidified chambers. Washes were done in small trays and coverslips then mounted on glass slides in Vectashield. Discs and OCT slices were imaged on a Zeiss LSM7–10 or Olympus FV1000 system. Images were processed with Fiji and Photoshop software. Refer to Key Resources Table for a complete list of antibodies and reagents used for microscopy.
Quantitative reverse-transcription PCR (qPCR)
For pupal wing RNAs: WPP were isolated, washed, sex sorted, and transferred to a new vial for aging. After 6 hours, pupal wings are dissected and dissolved in 0.5mL of TRIzol. For L3 wing imaginal disc RNAs: discs from wandering L3 larvae were dissected and dissolved in TRIzol. RNA isolation was done using standard protocol utilizing TRIZOL and Qiagen RNeasy kit. cDNA library preparation was done using SuperScript™-III RT kit from ThermoFisher. cDNAs generated with Superscript III RT and random primers (Invitrogen) were analyzed by qPCR with exon-junction spanning primers with SYBR Green I Master Mix (Roche) on a Roche LightCycler 480 in triplicate. Primers were designed using Primer3Plus and purchased from IDT Technologies. Quantitation was performed in technical triplicate on three biological replicates. Refer to Key Resources Table for a complete list of primers.
Differential Quik (Diff-Quik) hemocyte counts
Wandering L3 larvae were washed, transferred in a glass well in 15 μL of 1xPBS, and exsanguinated using tweezers. 5 μL of PBS/hemocytes mix was dropped onto a glass slide and dried completely, then stained with Diff-Quik stain kit™ (EMS #26096–25) and imaged. Blue stained nuclei were counted by light microscopy across multiple fields.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unpaired student t-test (GraphPad Prism™) was used to analyze significance between data sets. Unless noted, significance values in Text and Figure Legends are denoted by asterisks as follows: *=0.01<p<0.05, **=0.001<p<0.01, ***=p<0.001. Normal distributions were expected; no methods were used to determine whether the data met assumptions of the statistical approach.
In Figure 1A–E, ‘n’ indicates total number of adults scored.
In Figures 1H, 2M–N, 5A, ‘n’ indicates total number of wings scored.
DATA AND CODE AVAILABILITY
The RNA sequencing data (en>GFP vs. en>GFP+tai) supporting the current study can be accessed at GEO (NCBI) under accession # GSE133307.
Supplementary Material
Data S1: Fold-changes in RNA abundance in Tai+ L3 wing discs. Related to STAR Methods. RNA sequencing dataset showing fold-changes of detected transcripts in control (en>GFP) versus experimental (en>GFP,tai) samples of L3 wing discs. Provided as an Excel file.
Table S2. Candidate Tai induced mRNAs. Related to Figure 3. Alphabetical gene list of mRNAs with FPKM log[2]Δ>0.8 between control (en>GFP) and Tai-expressing (en>GFP,tai) samples.
Table S3. Overlap between the group of candidate Tai-induced mRNAs and the predicted Drosophila secretome. Related to Figure 3. Alphabetical gene list of mRNAs with log[2]Δ>0.8 between control (en>GFP) and Tai-expressing (en>GFP,tai) samples that are also included in the predicted Drosophila secretome (as in reference [S1]).
Acknowledgements
We thank Developmental Studies Hybridoma Bank (DSHB), Bloomington Drosophila Stock Center (BDSC), Harvard Transgenic RNAi Project (TRiP) and Vienna Drosophila Resource Collection (VDRC) for antibodies and stocks, and the Emory Integrated Cellular Imaging (ICI) Core for imaging expertise. Drs-GFP,dipt-lacZ gift of R. Jones. hidW05014 (hid-lacZ) gift of T-T. Su. Pvf2−3 gift of R. Read. TRE-GFP-16 gift of D. Bohmann. Cryosectioning protocol was adapted from one provided by D. Katz We thank members of K.M. and P.J. laboratories for discussions. Funding to K.M. GM123136 and GM121967.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare no competing interests.
References
- 1.Ray HJ, and Niswander L (2012). Mechanisms of tissue fusion during development. Development 139, 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Usui K, and Simpson P (2000). Cellular basis of the dynamic behavior of the imaginal thoracic discs during Drosophila metamorphosis. Dev Biol 225, 13–25. [DOI] [PubMed] [Google Scholar]
- 3.West RC, Bouma GJ, and Winger QA (2018). Shifting perspectives from “oncogenic” to oncofetal proteins; how these factors drive placental development. Reprod Biol Endocrinol 16, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dasgupta S, and O’Malley BW (2014). Transcriptional coregulators: emerging roles of SRC family of coactivators in disease pathology. J Mol Endocrinol 53, R47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Yin M-X, Wu W, Dong L, Wang S, Lu Y, Xu J, Wu W, Li S, Zhao Y, et al. (2016). Taiman acts as a coactivator of Yorkie in the Hippo pathway to promote tissue growth and intestinal regeneration. Cell Discovery 2, 16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Robinson BS, Xu W, Yang L, Yao B, Zhao H, Byun PK, Jin P, Veraksa A, and Moberg KH (2015). The ecdysone receptor coactivator Taiman links Yorkie to transcriptional control of germline stem cell factors in somatic tissue. Dev Cell 34, 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkins M, Potier D, Romanelli L, Jacobs J, Mach J, Hamaratoglu F, Aerts S, and Halder G (2016). An Ectopic Network of Transcription Factors Regulated by Hippo Signaling Drives Growth and Invasion of a Malignant Tumor Model. Curr Biol 26, 2101–2113. [DOI] [PubMed] [Google Scholar]
- 8.Bai J, Uehara Y, and Montell DJ (2000). Regulation of invasive cell behavior by aiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell 103, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 9.Kleino A, and Silverman N (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Developmental and comparative immunology 42, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann CJ, and Cohen SM (1996). Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development 122, 1781–1789. [DOI] [PubMed] [Google Scholar]
- 11.Diaz de la Loza MC, and Thompson BJ (2017). Forces shaping the Drosophila wing. Mech Dev 144, 23–32. [DOI] [PubMed] [Google Scholar]
- 12.Srivastava A, Pastor-Pareja JC, Igaki T, Pagliarini R, and Xu T (2007). Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc Natl Acad Sci U S A 104, 2721–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riddiford LM, Cherbas P, and Truman JW (2000). Ecdysone receptors and their biological actions. Vitamins and hormones 60, 1–73. [DOI] [PubMed] [Google Scholar]
- 14.McDonald JA, Pinheiro EM, and Montell DJ (2003). PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development 130, 3469–3478. [DOI] [PubMed] [Google Scholar]
- 15.Kozlova T, and Thummel CS (2002). Spatial patterns of ecdysteroid receptor activation during the onset of Drosophila metamorphosis. Development 129, 1739–1750. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H, Hirose S, and Ueda H (2002). BetaFTZ-F1 dependent and independent activation of Edg78E, a pupal cuticle gene, during the early metamorphic period in Drosophila melanogaster. Dev Growth Differ 44, 419–425. [DOI] [PubMed] [Google Scholar]
- 17.Huet F, Ruiz C, and Richards G (1995). Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development 121, 1195–1204. [DOI] [PubMed] [Google Scholar]
- 18.Woodard CT, Baehrecke EH, and Thummel CS (1994). A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell 79, 607–615. [DOI] [PubMed] [Google Scholar]
- 19.Rewitz KF, Yamanaka N, and O’Connor MB (2010). Steroid hormone inactivation is required during the juvenile-adult transition in Drosophila. Dev Cell 19, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misra JR, and Irvine KD (2018). The Hippo Signaling Network and Its Biological Functions. Annu Rev Genet 52, 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagegaltier D, Falciatori I, Czech B, Castel S, Perrimon N, Simcox A, and Hannon GJ (2016). Oncogenic transformation of Drosophila somatic cells induces a functional piRNA pathway. Genes Dev 30, 1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garelli A, Gontijo AM, Miguela V, Caparros E, and Dominguez M (2012). Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336, 579–582. [DOI] [PubMed] [Google Scholar]
- 23.Colombani J, Andersen DS, and Leopold P (2012). Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336, 582–585. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay SA, and Wasserman SA (2014). Conventional and non-conventional Drosophila Toll signaling. Developmental and comparative immunology 42, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer SN, Amoyel M, Bergantinos C, de la Cova C, Schertel C, Basler K, and Johnston LA (2014). An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science 346, 1258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler PN, Sobala LF, Thom D, and Nagaraj R (2013). dusky-like is required to maintain the integrity and planar cell polarity of hairs during the development of the Drosophila wing. Developmental biology 379, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaukat Z, Liu D, and Gregory S (2015). Sterile inflammation in Drosophila. Mediators of inflammation 2015, 369286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons B, and Foley E (2016). Cellular immune defenses of Drosophila melanogaster. Dev Comp Immunol 58, 95–101. [DOI] [PubMed] [Google Scholar]
- 29.Rizki TM, Rizki RM, and Grell EH (1980). A mutant affecting the crystal cells inDrosophila melanogaster. Wilhelm Roux’s archives of developmental biology 188, 91–99. [DOI] [PubMed] [Google Scholar]
- 30.Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, and Hoffmann JA (1998). A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J 17, 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider DS, Hudson KL, Lin TY, and Anderson KV (1991). Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev 5, 797–807. [DOI] [PubMed] [Google Scholar]
- 32.Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, and Reichhart JM (1999). Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science 285, 1917–1919. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, and Martinez-Arias A (1998). puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 12, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim M, Lee JH, Lee SY, Kim E, and Chung J (2006). Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci U S A 103, 16358–16363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mindorff EN, O’Keefe DD, Labbe A, Yang JP, Ou Y, Yoshikawa S, and van Meyel DJ (2007). A gain-of-function screen for genes that influence axon guidance identifies the NF-kappaB protein dorsal and reveals a requirement for the kinase Pelle in Drosophila photoreceptor axon targeting. Genetics 176, 2247–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Germani F, Hain D, Sternlicht D, Moreno E, and Basler K (2018). The Toll pathway inhibits tissue growth and regulates cell fitness in an infection-dependent manner. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boone E, Colombani J, Andersen DS, and Léopold P (2016). The Hippo signalling pathway coordinates organ growth and limits developmental variability by controlling dilp8 expression. Nature communications 7, 13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth P, Xylourgidis N, Sabri N, Uv A, Fornerod M, and Samakovlis C (2003). The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J Cell Biol 163, 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paredes JC, Welchman DP, Poidevin M, and Lemaitre B (2011). Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779. [DOI] [PubMed] [Google Scholar]
- 40.Charroux B, Capo F, Kurz CL, Peslier S, Chaduli D, Viallat-Lieutaud A, and Royet J (2018). Cytosolic and Secreted Peptidoglycan-Degrading Enzymes in Drosophila Respectively Control Local and Systemic Immune Responses to Microbiota. Cell Host Microbe 23, 215–228 e214. [DOI] [PubMed] [Google Scholar]
- 41.Bate M.a. A. AM (1993). The Development of Drosophila melanogaster, Volume I and I, (Cold Spring Harbor, NY: CSHL Press; ). [Google Scholar]
- 42.Moreno E, and Basler K (2004). dMyc transforms cells into super-competitors. Cell 117, 117–129. [DOI] [PubMed] [Google Scholar]
- 43.Alpar L, Bergantinos C, and Johnston LA (2018). Spatially Restricted Regulation of Spatzle/Toll Signaling during Cell Competition. Dev Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Zheng Y, Yin F, Yu J, Silverman N, and Pan D (2016). Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila. Cell 164, 406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto M, Ohsawa S, Kunimasa K, and Igaki T (2017). The ligand Sas and its receptor PTP10D drive tumour-suppressive cell competition. Nature 542, 246–250. [DOI] [PubMed] [Google Scholar]
- 46.Martins VC, Busch K, Juraeva D, Blum C, Ludwig C, Rasche V, Lasitschka F, Mastitsky SE, Brors B, Hielscher T, et al. (2014). Cell competition is a tumour suppressor mechanism in the thymus. Nature 509, 465–470. [DOI] [PubMed] [Google Scholar]
- 47.Suijkerbuijk SJ, Kolahgar G, Kucinski I, and Piddini E (2016). Cell Competition Drives the Growth of Intestinal Adenomas in Drosophila. Current biology : CB 26, 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eichenlaub T, Cohen SM, and Herranz H (2016). Cell Competition Drives the Formation of Metastatic Tumors in a Drosophila Model of Epithelial Tumor Formation. Current biology : CB 26, 419–427. [DOI] [PubMed] [Google Scholar]
- 49.Di Giacomo S, Sollazzo M, de Biase D, Ragazzi M, Bellosta P, Pession A, and Grifoni D (2017). Human Cancer Cells Signal Their Competitive Fitness Through MYC Activity. Scientific reports 7, 12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao S, Zhang Y, Zhang Q, Wang F, and Zhang D (2014). Toll-like receptors and prostate cancer. Front Immunol 5, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Fold-changes in RNA abundance in Tai+ L3 wing discs. Related to STAR Methods. RNA sequencing dataset showing fold-changes of detected transcripts in control (en>GFP) versus experimental (en>GFP,tai) samples of L3 wing discs. Provided as an Excel file.
Table S2. Candidate Tai induced mRNAs. Related to Figure 3. Alphabetical gene list of mRNAs with FPKM log[2]Δ>0.8 between control (en>GFP) and Tai-expressing (en>GFP,tai) samples.
Table S3. Overlap between the group of candidate Tai-induced mRNAs and the predicted Drosophila secretome. Related to Figure 3. Alphabetical gene list of mRNAs with log[2]Δ>0.8 between control (en>GFP) and Tai-expressing (en>GFP,tai) samples that are also included in the predicted Drosophila secretome (as in reference [S1]).
Data Availability Statement
The RNA sequencing data (en>GFP vs. en>GFP+tai) supporting the current study can be accessed at GEO (NCBI) under accession # GSE133307.