Gonorrhea, a sexually transmitted disease, causes substantial global morbidity and economic burden. New prevention and control measures for this disease are urgently needed, as strains resistant to almost all classes of antibiotics available for treatment have emerged. Previous reports demonstrate that cross-protection from gonococcal infections may be conferred by meningococcal serogroup B (MenB) outer membrane vesicle (OMV)-based vaccines. Among 1,525 common proteins shared across the genomes of both N. gonorrhoeae and N. meningitidis, 57 proteins were predicted to be surface expressed (outer membrane proteins [OMPs]) and thus preferred targets for vaccine development. The majority of these OMPs showed high sequence identity between the 2 bacterial species. Our results provide valuable insight into the meningococcal antigens present in the current OMV-containing MenB-4C vaccine that may contribute to cross-protection against gonorrhea and may inform next steps in gonorrhea vaccine development.
KEYWORDS: Bexsero, cross-protection, genetic similarity, MenB-4C, Neisseria gonorrhoeae, Neisseria meningitidis, vaccine development, outer membrane proteins, outer membrane vesicles
ABSTRACT
The human pathogens Neisseria gonorrhoeae and Neisseria meningitidis share high genome identity. Retrospective analysis of surveillance data from New Zealand indicates the potential cross-protective effect of outer membrane vesicle (OMV) meningococcal serogroup B vaccine (MeNZB) against N. gonorrhoeae. A licensed OMV-based MenB vaccine, MenB-4C, consists of a recombinant FHbp, NhbA, NadA, and the MeNZB OMV. Previous work has identified several abundantly expressed outer membrane proteins (OMPs) as major components of the MenB-4C OMV with high sequence similarity between N. gonorrhoeae and N. meningitidis, suggesting a mechanism for cross-protection. To build off these findings, we performed comparative genomic analysis on 970 recent N. gonorrhoeae isolates collected through a U.S surveillance system against N. meningitidis serogroup B (NmB) reference sequences. We identified 1,525 proteins that were common to both Neisseria species, of which 57 proteins were predicted to be OMPs using in silico methods. Among the MenB-4C antigens, NhbA showed moderate sequence identity (73%) to the respective gonococcal homolog, was highly conserved within N. gonorrhoeae, and was predicted to be surface expressed. In contrast, the gonococcal FHbp was predicted not to be surface expressed, while NadA was absent in all N. gonorrhoeae isolates. Our work confirmed recent observations (E. A. Semchenko, A. Tan, R. Borrow, and K. L. Seib, Clin Infect Dis, 2018, https://doi.org/10.1093/cid/ciy1061) and describes homologous OMPs from a large panel of epidemiologically relevant N. gonorrhoeae strains in the United States against NmB reference strains. Based on our results, we report a set of OMPs that may contribute to the previously observed cross-protection and provide potential antigen targets to guide the next steps in gonorrhea vaccine development.
INTRODUCTION
Neisseria gonorrhoeae and Neisseria meningitidis are obligate human pathogens that are genetically closely related, sharing between 80 and 90% genome sequence identity (1–3). However, infections with these pathogens typically have very different clinical presentations (4). N. gonorrhoeae is responsible for gonorrhea, one of the most common bacterial sexually transmitted diseases (STDs), frequently causing inflammation of the urogenital tract. N. meningitidis infections can lead to life-threatening bacterial meningitis and septicemia (5) following penetration of the bacteria through the blood-brain barrier and colonization of the meninges.
Gonorrhea results in substantial morbidity and economic burden globally, with an estimated 100 million cases worldwide. In the United States, gonorrhea is the second most common notifiable disease. More than 550,000 infections were reported to the U.S. Centers for Disease Control and Prevention (CDC) in 2017, the highest number since 1991 (6, 7). Though N. gonorrhoeae infections typically trigger urethritis in men and cervicitis in women, mucosal infections of the rectum, pharynx, and eye are also common (8–10). Gonorrhea facilitates human immunodeficiency virus (HIV) transmission through the stimulation of antiapoptotic proteins essential for the HIV life cycle (11). The emergence of N. gonorrhoeae strains that are resistant to nearly all classes of antibiotics available for treatment and the lack of an effective gonococcal vaccine (9, 10, 12, 13) underscore the urgent need for new prevention and control measures.
In contrast to polysaccharide-based vaccines targeting N. meningitidis serogroups A, C, W, and Y, protein-based vaccines have been developed for N. meningitidis serogroup B (NmB) (14, 15). NmB capsular polysaccharides contain polysialic acid structures that are similar to human neuronal glycoproteins (16); this results in poor immunogenicity and poses the risk of triggering an autoimmune response. Vaccines containing NmB outer membrane vesicles (OMVs) have been utilized in a number of countries, where they are primarily designed to match outbreak strains circulating during regional epidemics (17, 18). Previous studies suggested a decline in gonorrhea cases following the introduction of meningococcal serogroup B (MenB) OMV vaccines. Retrospective analyses of surveillance data in Cuba, Norway, and New Zealand have indicated potential cross-protection from MenB OMV vaccines against N. gonorrhoeae infections (19–21). A case-control study in New Zealand estimated that vaccinated individuals had a 31% lower risk of developing gonorrhea (20); a decision-analysis model showed a 20% assumptive reduction in gonorrhea due to the cross-protection conferred by the licensed protein-based and OMV-containing MenB-4C (Bexsero) vaccine. Recently, Semchenko and colleagues analyzed the sequence identity and conservation of MenB-4C vaccine antigens against publicly available gonococcal genomes and demonstrated the potential contribution of OMV components to the observed cross-protection (22). Decreased gonorrhea incidence could lower morbidity and rates of complications, such as HIV (23), and reduce the risk of the emergence of untreatable antibiotic-resistant gonorrhea.
MenB OMV vaccines contain outer membrane proteins (OMPs), with porin A (PorA) being immunodominant (24). However, evidence suggests that the porA gene might only be present as a pseudogene in N. gonorrhoeae and is not expressed (25, 26). Therefore, the mechanism of action for protective immunity conferred by the OMV vaccines against N. gonorrhoeae has yet to be fully explained. Depending on the bacterial species and detection methods used to assess them, OMVs may comprise more than 300 different protein components, of which up to 80% are OMPs (27). In addition to PorA, meningococcal OMPs such as PorB, FetA, and OpcA have been shown to induce protective antibody responses (18, 28, 29). Nevertheless, other OMPs and/or membrane-exposed antigens also potentially generate cross-reactivity inducing broader protection. Numerous proteins have been extracted and identified from NmB OMVs (30–35).
Reverse vaccinology, which includes genome and proteome mining, has been utilized to identify potentially surface-exposed antigens that are highly immunogenic and protective for vaccine development (36–38). The multicomponent MenB-4C vaccine contains the following 3 immunogenic antigens identified by reverse vaccinology: factor H binding protein (FHbp) fused with GNA2091, Neisseria adhesion A (NadA), and Neisseria heparin binding antigen (NhbA) fused with GNA1030 supplemented with the OMV of the New Zealand epidemic strain (NZ98/254) to warrant broader immunogenicity (39). Although the formulation is not identical to that of the New Zealand MenB OMV (MeNZB) vaccine, immunogenic OMV components included in the MenB-4C vaccine may cross-protect against N. gonorrhoeae infections.
In this study, we sequenced and analyzed a diverse genome data set of 970 N. gonorrhoeae isolates collected from Gonorrhea Isolate Surveillance Project (GISP) sites across the United States between 2014 and 2016. We identified common proteins present in both N. gonorrhoeae and N. meningitidis and assessed the diversity of MenB vaccine antigens and OMV proteins to and within the U.S. N. gonorrhoeae strain collection. Bioinformatic tools were applied to predict the subcellular localization of each identified common protein. We found a significant number of predicted OMPs present in both Neisseria species, which could potentially contribute to cross-protection against gonorrhea and may serve as candidate antigens for gonococcal vaccine development.
RESULTS
Identification of common proteins and their subcellular localizations.
N. gonorrhoeae and N. meningitidis show the closest genetic relationship within the Neisseria family (1–3, 40). In total, we identified 1,525 proteins that were common among our set of 970 N. gonorrhoeae genomes and 4 NmB references, including MenB-4C vaccine antigens and major abundant proteins previously found in OMVs (see Table S1 in the supplemental material). Using various in silico prediction tools, we predicted the subcellular localization of each common protein found in both Neisseria species. The outer membrane localization was first determined for proteins in NmB and then compared them to those of the respective gonococcal homologs. A protein received its outer membrane-associated assignment only after being confirmed by at least 2 of 3 prediction tools, along with the detection of a signal peptide. A schematic workflow of the OMP identification process is shown in Fig. 1.
FIG 1.
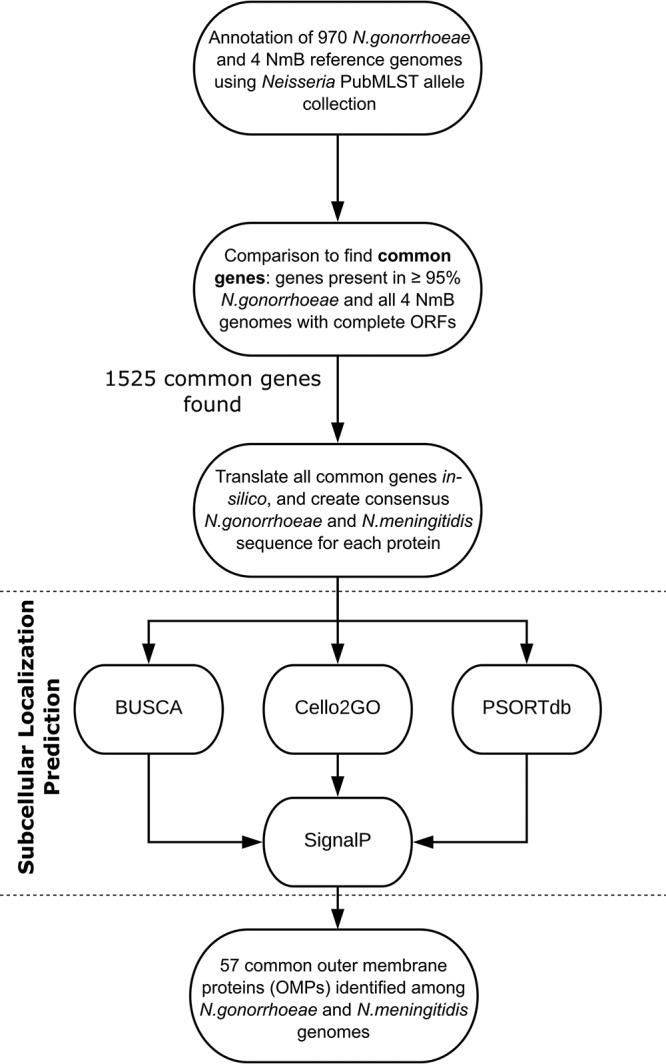
Schematic presentation of common protein identification and assessment of proteins’ subcellular localization. Ng, N. gonorrhoeae; Nm, N. meningitidis .
Common genes found in 4 NmB and 970 N. gonorrhoeae strains. Download Table S1, PDF file, 0.2 MB (228.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Among the major MenB-4C vaccine antigens, only FHbp and NhbA with their corresponding fusion proteins (GNA2091 and GNA1030, respectively) were found in all 4 NmB strains and 970 N. gonorrhoeae isolates (Table 1). A full-length NadA protein was present in the MC58 and CU385/83 NmB strains, but it was truncated in the other two NmB strains (H44/76 and NZ98/254) and absent in all N. gonorrhoeae isolates used in this study. In contrast to the NmB FHbp, the gonococcal FHbp homolog was predicted as not being expressed on the cell surface, based on its subcellular localization and the lack of a signal peptide (Table 1). The gonococcal NhbA was predicted as being surface exposed. Both fusion proteins (GNA2091 and GNA1030) were predicted as being surface exposed in N. meningitidis; GNA1030 was surface exposed in N. gonorrhoeae, while only 1 tool predicted GNA2091 to be surface exposed in N. gonorrhoeae.
TABLE 1.
Sequence analysis and subcellular localization of MenB-4C vaccine antigensa
| Vaccine antigen | NEIS no. | NmB strains (n = 4) |
N. gonorrhoeae strains (n = 970) |
||
|---|---|---|---|---|---|
| No. of isolates with full-length protein | Predicted OMP | No. of isolates with full-length protein | Predicted OMP | ||
| FHbp | NEIS0349 | 4 | + | 970 | – |
| NhbA | NEIS2109 | 4 | + | 970 | + |
| NadA | NEIS1969 | 2 | + | 0 | NA |
| OMV (PorA) | NEIS1364 | 4 | + | 0 | NA |
| GNA2091 | NEIS2071 | 4 | + | 970 | +/– |
| GNA1030 | NEIS1183 | 4 | + | 970 | + |
Cellular localization and signal peptide predictions were determined using the in silico tools BUSCA, Cello2GO, PSORTdb, and SignalP. OMP, outer membrane protein; +, predicted as OMP by at least 2 tools; –, not predicted as OMP by any of the tools; +/–, predicted as OMP by 1 tool; NA, not applicable due to the absence of an ORF.
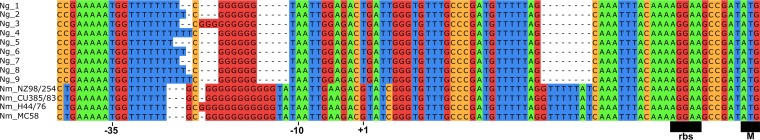
While the porA gene was identified in 966 of 970 (99.5%) N. gonorrhoeae isolates, the DNA sequences contained inactivating mutations in the promoter region (Fig. 2), along with frameshift deletions in the coding sequence resulting in premature internal stops, consistent with previous reports describing porA’s pseudogene status in N. gonorrhoeae (25, 26).
FIG 2.
The porA promoter region for N. gonorrhoeae and N. meningitidis. The putative meningococcal −10 and −35 locations as well as the ribosome binding site and the start codon are indicated. The porA promoter sequences for all 970 N. gonorrhoeae genomes were filtered down to 9 unique sequences. The bottom four rows consist of the porA promoter sequences for the N. meningitidis references. The gaps represent nucleotides that were missing from the respective sequence compared to the rest.
Common outer membrane proteins shared between NmB and N. gonorrhoeae strains.
Among the 1,525 common proteins, 57 were predicted to be OMPs (Table 2); 28 of these proteins had known biological functions, while the remaining 29 were either putative or hypothetical proteins. Of the 57 predicted OMPs, we identified 25 proteins that had been previously detected in the NZ98/254 OMV components using different laboratory approaches (24, 30, 32, 34), 12 of which were abundantly and consistently expressed across different OMV lots (32) (Table 2). These 12 proteins include some major immunogenic antigens that have been tested experimentally as potential gonorrhea vaccine candidates, such as PorB, PilQ, OpcA, and Omp85 (BamA) (9, 12, 13, 41). The remaining 34 OMPs, including the MenB-4C vaccine antigens FHbp and NhbA, had not been previously identified in NZ98/254 OMVs. Two OMV proteins (MtrE and NEIS1428) were predicted as being outer membrane exposed in N. meningitidis; however, a signal peptide was detected only by 1 of the prediction tools. A similar observation was also seen for these 2 OMV proteins in N. gonorrhoeae.
TABLE 2.
Predicted outer membrane proteins and sequence similarity in N. gonorrhoeae and NmB strains
| NEIS no. | Protein namea | Identified in OMVs | Amino acid sequence similarity (%) |
Experimentally tested as GC vaccinec | |||
|---|---|---|---|---|---|---|---|
| Between N. gonorrhoeae and NZ98/254b
|
Within N. gonorrhoeae |
||||||
| Range | Mean ± SD | Range | Mean ± SD | ||||
| NEIS2109 | NhbA | 67.5–88 | 72.8 ± 5.2 | 78.3–100 | 96.8 ± 5.0 | ||
| NEIS1183 | GNA1030 | 90.9–93.5 | 92.2 ± 0.9 | 98.4–100 | 99.1 ± 0.5 | ||
| NEIS2020 | PorB | + | 66.2–94.2 | 69.7 ± 2.9 | 65.2–100 | 91.3 ± 6.2 | + |
| NEIS1963 | FetA | + | 90.5–95 | 92.8 ± 1.4 | 89.5–100 | 96.4 ± 1.9 | |
| NEIS0408 | PilQ | + | 89.3–97.2 | 93.1 ± 2.3 | 95.7–100 | 99.2 ± 0.6 | + |
| NEIS0173 | Omp85 (BamA) | + | 95–95.8 | 95.2 ± 0.2 | 99–100 | 99.6 ± 0.2 | + |
| NEIS1783 | RmpM | + | 92.6–93.6 | 93.2 ± 0.3 | 98.7–100 | 99.4 ± 0.3 | |
| NEIS2198 | OpcA | + | 44.8–47.1 | 45.4 ± 0.6 | 97.7–100 | 99 ± 0.5 | + |
| NEIS0612 | NspA | + | 90.8–95.7 | 94 ± 0.9 | 92–100 | 98.2 ± 1.5 | + |
| NEIS1632 | MtrE | + | 93–97.5 | 94.9 ± 1.7 | 95–100 | 99.1 ± 1.3 | + |
| NEIS0586 | MafB1 | 79–80.9 | 79.5 ± 0.4 | 96–100 | 99.2 ± 1.1 | ||
| NEIS1917 | MetQ | + | 96.5–99.3 | 96.9 ± 0.7 | 97.2–100 | 99.6 ± 0.9 | + |
| NEIS0653 | ComL | + | 98.1–99.6 | 98.4 ± 0.4 | 97.8–100 | 99.4 ± 0.6 | |
| NEIS1687 | OM phospholipase A precursor | + | 98.4–98.9 | 98.6 ± 0.1 | 99.2–99.7 | 99.5 ± 0.2 | |
| NEIS1468 | LbpA | + | 86.3–96.3 | 95.5 ± 1.8 | 87.9–100 | 98.3 ± 2.4 | + |
| NEIS1487 | FkpA (macrophage infectivity protein) | + | 96.7–99.2 | 97.3 ± 0.6 | 96–100 | 98.9 ± 1.0 | |
| hmbR | HmbR | 89.5–89.6 | 89.5 ± 0.1 | 100–100 | 100 ± 0.0 | ||
| NEIS0566 | LolB | 98.1–98.5 | 98.4 ± 0.2 | 99–100 | 99.7 ± 0.3 | ||
| NEIS1418 | NlpD | 90.3–94 | 93.1 ± 0.7 | 95.4–100 | 99.2 ± 0.7 | ||
| NEIS1462 | H.8 OMP | + | 95.1–98.3 | 97.8 ± 0.8 | 88.6–100 | 97.6 ± 3.8 | |
| NEIS1933 | VacJ-related protein | 94.9–95.6 | 95.3 ± 0.2 | 95.6–100 | 98.4 ± 1.6 | ||
| NEIS0338 | Ferric siderophore receptor protein | 74.9–97.2 | 92.9 ± 7.5 | 76.6–100 | 94.3 ± 8.8 | ||
| NEIS1813 | OstA | 93.2–94.3 | 93.5 ± 0.4 | 98.9–100 | 99.2 ± 0.3 | ||
| NEIS1691 | TbpB | 60.8–86.3 | 67.3 ± 5.3 | 58.9–100 | 75.9 ± 7.7 | + | |
| NEIS1549 | AniA | 83.4–92.3 | 84.4 ± 2.4 | 95.8–100 | 99.2 ± 0.6 | + | |
| NEIS1205 | Ape1 | 94.5–97.4 | 95.3 ± 0.6 | 95.2–100 | 98.3 ± 1.3 | ||
| NEIS2124 | Lipoprotein | 98.6–99.3 | 98.9 ± 0.3 | 98.6–100 | 99.1 ± 0.4 | ||
| NEIS1063 | Putative periplasmic protein | + | 96.3–96.7 | 96.6 ± 0.2 | 99.1–100 | 99.7 ± 0.3 | |
| NEIS1066 | Putative periplasmic protein | + | 93.3–94.1 | 93.8 ± 0.3 | 98.7–100 | 99.4 ± 0.3 | |
| NEIS0275 | Putative OM solvent tolerance protein (LPS assembly protein) | + | 89.1–89.9 | 89.5 ± 0.2 | 92.6–100 | 99.4 ± 1.1 | |
| NEIS0944 | Putative OM receptor protein (TonB-dependent receptor) | + | 96.7–97.3 | 96.9 ± 0.1 | 99.4–100 | 99.7 ± 0.1 | |
| NEIS1428 | Putative OM substrate binding protein (TonB-dependent receptor) | + | 94.8–96.5 | 95.8 ± 0.4 | 97–100 | 98.6 ± 0.6 | |
| NEIS0739 | Putative amino acid permease substrate-binding protein | + | 97.1–98.5 | 97.8 ± 0.3 | 98.6–100 | 99.4 ± 0.4 | |
| NEIS1920 | Putative transglycosylase | + | 95.9–97.8 | 97.4 ± 0.3 | 96.3–100 | 99.1 ± 0.6 | |
| NEIS2112 | Putative OMP | 98.4–99.1 | 98.9 ± 0.2 | 98.1–100 | 99.5 ± 0.4 | ||
| NEIS0261 | Putative periplasmic protein | 86.2–97 | 93.5 ± 2.8 | 85.4–100 | 97.1 ± 3.7 | ||
| NEIS0504 | Putative thiamine biosynthesis protein | 96–96.3 | 96.2 ± 0.1 | 99.4–100 | 99.8 ± 0.2 | ||
| NEIS0630 | Putative periplasmic protein | 92.1–92.6 | 92.2 ± 0.2 | 99–100 | 99.4 ± 0.3 | ||
| NEIS0729 | Putative secreted protein | 91.4–96.3 | 93.9 ± 1.3 | 92.9–100 | 98 ± 1.9 | ||
| NEIS1304 | Putative membrane lipoprotein | 96.7–97.2 | 97.1 ± 0.2 | 99.5–100 | 99.8 ± 0.3 | ||
| NEIS1195 | Putative peptidyl-prolyl cis-trans isomerase A | 92.4–94.5 | 93.6 ± 0.6 | 97.3–100 | 99.1 ± 0.7 | ||
| NEIS1935 | Putative periplasmic transport protein | 91.8–92.8 | 92.2 ± 0.4 | 98.5–100 | 99.1 ± 0.4 | ||
| NEIS0196 | Putative OM lipoprotein | + | 92.8–95.2 | 93.4 ± 0.9 | 91.2–99.2 | 95.8 ± 3.6 | |
| NEIS1084 | Putative periplasmic protein | + | 93.1–95 | 93.7 ± 0.6 | 97–100 | 98.3 ± 0.8 | |
| NEIS1125 | Putative periplasmic protein | 95.3–97 | 95.8 ± 0.6 | 98.8–100 | 99.1 ± 0.3 | ||
| NEIS1172 | Putative periplasmic protein | 92.7–99.7 | 93.2 ± 1.1 | 92.7–100 | 99.1 ± 1.4 | ||
| NEIS1367 | Putative lipoprotein | 96.1–96.6 | 96.2 ± 0.3 | 98.9–100 | 99.2 ± 0.4 | ||
| NEIS1271 | Hypothetical protein | + | 95.5–97.1 | 96.4 ± 0.4 | 97.3–100 | 99 ± 0.5 | |
| NEIS1858 | Hypothetical protein | 92.4–99.1 | 94.5 ± 1.5 | 91.6–100 | 98.3 ± 1.9 | ||
| NEIS1945 | Hypothetical protein | 74.2–99.2 | 82.3 ± 8.3 | 74.0–100 | 93.5 ± 9.2 | ||
| NEIS1485 | Hypothetical protein | 97.3–100 | 97.7 ± 0.7 | 97.3–100 | 99 ± 0.9 | ||
| NEIS1546 | Hypothetical protein | 88.4–97.7 | 96.1 ± 2.2 | 87.0–100 | 97.9 ± 3.4 | ||
| NEIS1746 | Hypothetical protein | 62.7–87.7 | 67 ± 9.3 | 98.4–100 | 99 ± 0.5 | ||
| NEIS2446 | Hypothetical protein | 83.5–91.2 | 88.1 ± 2.4 | 90.1–100 | 96.5 ± 2.7 | ||
| NEIS0790 | Hypothetical protein | 74.6–80.6 | 76.7 ± 2.2 | 93–100 | 97.7 ± 2.6 | ||
| NEIS1136 | Hypothetical protein | 82.6–83.7 | 83.1 ± 0.5 | 97.8–100 | 98.9 ± 0.6 | ||
| NEIS1253 | Hypothetical protein | 91.9–97.5 | 93.4 ± 1.3 | 93.8–100 | 98.1 ± 1.5 | ||
Previously reported proteins from detergent-extracted NZ98/254 outer membrane vesicles (OMVs) (24, 30, 32, 34). Proteins in boldface are most abundantly expressed, and their expression was not significantly expressed across different OMV lots (32). OM, outer membrane; LPS, lipopolysaccharide; OMP, outer membrane protein.
Values in boldface show mean sequence similarity that is greater than 80%.
Of note, a previous report predicted the meningococcal PilE (NEIS0210) as an OMP (32); however, the prediction tools used in our study indicated extracellular/periplasmic/plasma membrane localization of PilE and absence of a signal peptide in both N. meningitidis and N. gonorrhoeae. In addition, a gene coding for OMP P1 (NEIS0073) contained an internal stop codon in all 970 N. gonorrhoeae isolates. While mafA (NEIS0596) was present in the 4 NmB and 970 N. gonorrhoeae genomes, it was fragmented across multiple contigs within the NZ98/254 genome. Furthermore, Tbp1 (NEIS1690) was only found in 89% of all N. gonorrhoeae strains, which did not meet our threshold of 95% for common protein selection. Therefore, PilE, OMP P1, MafA, and Tbp1 were not included in our list of common proteins and/or OMPs.
Sequence similarity of OMPs found within NmB and N. gonorrhoeae strain collections.
Next, we analyzed the genetic similarity of each of the 57 predicted OMPs between the two Neisseria species and within the whole N. gonorrhoeae collection to assess sequence conservation. To this end, we focused our analysis on the NZ98/254 strain, from which the OMV in the MenB-4C vaccine derives. It has been shown that vaccines against Japanese encephalitis virus may induce cross-reactivity to West Nile virus and dengue virus since their common proteins share approximately 80% and 50% amino acid sequence homology, respectively (42, 43). Based on this knowledge, we used a minimum sequence similarity of 80% as a threshold for potential cross-protection against N. gonorrhoeae (Fig. 3). The mean sequence similarity between N. meningitidis and N. gonorrhoeae for all 57 OMPs was 91.0% ± 9.8%. At the amino acid level, 50 of 57 (88%) OMPs from the NZ98/254 strain showed 83 to 99% sequence similarity to all 970 N. gonorrhoeae isolates, of which 24 proteins had previously been identified in NZ98/254 OMVs (Table 2). The remaining 7 OMPs had 45 to 79% sequence similarity to N. gonorrhoeae.
FIG 3.
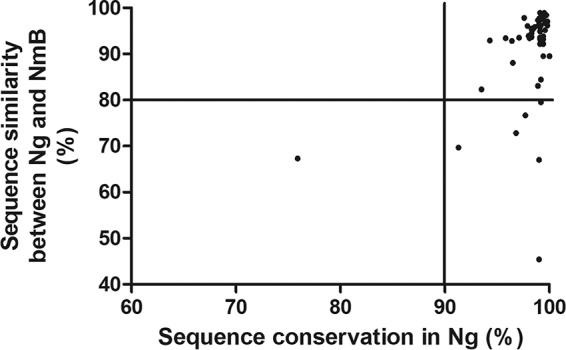
Amino acid sequence similarity of 57 common OMPs found in 970 N. gonorrhoeae and NmB (NZ98/254) strains. A value of 80% was set as a minimum level of sequence similarity that might confer cross-protection. The majority of the common OMPs had >90% sequence conservation in N. gonorrhoeae.
Among MenB-4C vaccine antigens, FHbp of the NZ98/254 strain showed a modest mean amino acid sequence similarity to N. gonorrhoeae (64%); the NhbA similarity was higher (73%) (Table 2). The 2 fusion proteins of NZ98/254 had high sequence similarity to N. gonorrhoeae, at 95% for GNA2091 and 92% for GNA1030. These 4 vaccine antigens were highly conserved (97 to 99% identity) within the N. gonorrhoeae collection. Of all the predicted OMPs, OpcA of NZ98/254 had the lowest mean similarity to N. gonorrhoeae (45%), followed by the abundant OMV protein PorB (70%), but the 2 proteins showed 99% and 91% sequence conservancy within N. gonorrhoeae, respectively (Table 2).
The majority (56 of 57) of predicted OMPs displayed 91 to 100% sequence conservation within the 970 N. gonorrhoeae isolates, with the lowest mean similarity observed for TbpB (76%) (Table 2). It is noteworthy that the sequence similarities at the amino acid and DNA levels were consistent between NZ98/254 and all N. gonorrhoeae strains, except for PorB, PilQ, and OpcA, which had ∼10% differences (Table S2). Other major OMV proteins, such as FetA, PilQ, Omp85 (BamA), RmpM, and MtrE of NZ98/298 consistently showed ≥93% mean amino acid sequence similarity to and within the N. gonorrhoeae isolates. The NZ98/298 nitrite reductase AniA, essential for N. gonorrhoeae anaerobic respiration and biofilm formation (44–46), showed 84% and 99% sequence similarity to and within N. gonorrhoeae isolates, respectively (Table 2).
Sequence similarities of common OMPs found in NmB and N. gonorrhoeae strains. Download Table S2, PDF file, 0.1 MB (112.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
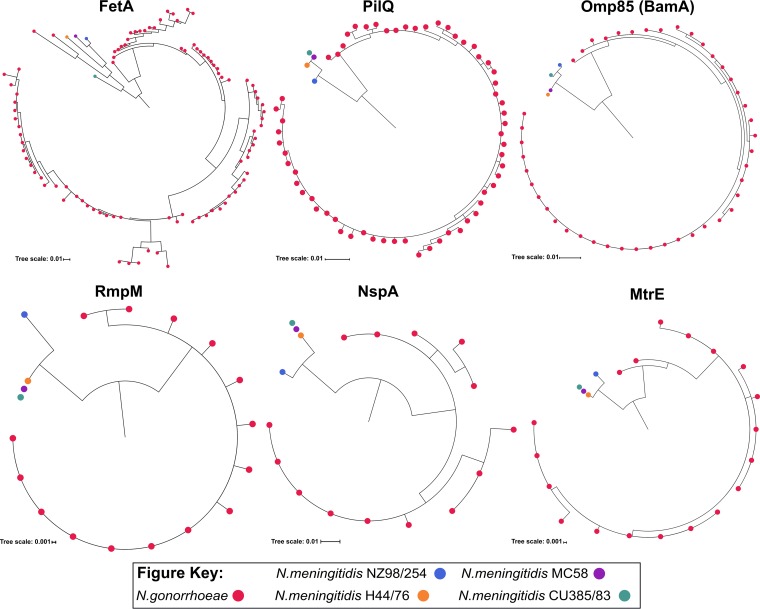
Phylogenetic analysis was also conducted to assess the sequence diversity of the OMPs found within the two Neisseria species. Six common OMPs that had previously been identified in the NZ98/254 OMV and showed amino acid sequence similarity above the 80% cutoff to and within the 970 N. gonorrhoeae strains were selected for this purpose. These OMPs were FetA, PilQ, Omp85 (BamA), RmpM, NspA, and MtrE. Our analysis indicated that PilQ, Omp85 (BamA), RmpM, NspA, and MtrE of NZ98/254 formed a distinct cluster from the other 3 NmB strains, whereas FetA of CU385/83 was distinct from the rest of the NmB group (Fig. 4). This pattern of distinction among the NmB strains was also found with other OMV proteins, such as FHbp and its fusion protein (GNA2091), NhbA, AniA, and OpcA (Fig. S1), in contrast to PorB, which formed one cluster. Consistent with the number of unique alleles, RmpM, NspA, and MtrE formed clusters that are less diverse within N. gonorrhoeae than other OMPs, particularly FetA and PorB (Fig. 4 andS1). It is noteworthy that 2 alleles of gonococcal FetA, 1 allele of NhbA, and PorB shared a cluster with the NmB ones, suggesting that these alleles are closely related.
FIG 4.
Phylogenetic clustering of FetA, PilQ, Omp85 (BamA), RmpM, NspA, and MtrE proteins found in 4 NmB and 970 N. gonorrhoeae strains. Each red dot represents sequences obtained from a unique N. gonorrhoeae allele. In total, 16 unique alleles were found within NmB strains, as follows: 4 for FetA, 3 for PilQ, 3 for Omp85 (BamA), 2 for RmpM, 2 for NspA and 2 for MtrE; there were 248 unique alleles within N. gonorrhoeae strains, as follows: 95 for FetA, 61 for PilQ, 44 for Omp85 (BamA), 15 for RmpM, 14 for NspA, and 19 for MtrE.
Phylogenetic tree of AniA, FHbp, PorB, GNA1030, GNA2091, NhbA, and OpcA. Download FIG S1, PDF file, 0.5 MB (494.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
Increasingly resistant N. gonorrhoeae strains represent a global threat, and no gonococcal vaccine is currently available (13). Two vaccine candidates tested in a full cycle of field trials in humans, a whole-cell vaccine within the Aboriginal population of Inuits in northern Canada (47) and a pilus-based vaccine for high-risk U.S. military personnel stationed in South Korea (48), were unsuccessful due to a lack of efficacy compared to placebo-treated groups. Evidence for potential cross-protection against gonorrhea conferred by MenB OMV vaccines (19–21) could reduce the disease burden. Our analysis of NmB and N. gonorrhoeae sequences revealed more than 1,500 common proteins shared across the genomes of these 2 Neisseria species. We evaluated antigens of the MenB-4C vaccine, a licensed protein-based vaccine containing OMVs of the New Zealand N. meningitidis epidemic strain (NZ98/254) (32, 34), to assess its potential cross-protective effect, particularly on the recent U.S. gonococcal strains.
Major MenB-4C vaccine antigens FHbp, NadA, and PorA are less likely to contribute to cross-protection against N. gonorrhoeae infections. The gonococcal FHbp has previously been shown to not be surface expressed and is unable to bind factor H (49). A recent study demonstrates that antibodies raised against the 3 major MenB-4C antigens did not recognize FHbp in whole-cell lysates of N. gonorrhoeae test strains (22), which confirms that the gonococcal FHbp homolog is not expressed on the cell surface. Furthermore, in accordance with previous reports (2, 25, 26), NadA was absent in all 970 N. gonorrhoeae isolates analyzed in this study, while porA’s promoter appeared to be inactive for DNA transcription. Conversely, NhbA showed moderate amino acid sequence similarity to N. gonorrhoeae strains, as shown by others (2, 22), and may therefore contribute to cross-protection to an extent. Similar levels of sequence similarity were found for NhbA of the other 3 NmB strains, including MC58, consistent with the previous finding (2). Interestingly, we found a high level of sequence similarity between NZ98/254 and N. gonorrhoeae for the 2 recombinant fusion proteins GNA2091 and GNA1030. These fusion proteins were shown to induce protective immunity in mice (50) and may therefore exert a cross-protective effect. A recent report showed that antisera raised against recombinant MenB-4C vaccine antigens recognized the gonococcal homolog of NhbA and the 2 fusion proteins (22). However, only NhbA is surface expressed in N. gonorrhoeae, while GNA2091 and GNA1030 are not (22), consistent with previous findings (51, 52). Our analysis predicted GNA1030 to be surface exposed in both N. meningitidis and N. gonorrhoeae, suggesting that laboratory experiments are required to confirm the bioinformatic data.
In pathogenic bacteria, OMPs present important virulence determinants through their direct interaction with the host immune system (especially OMPs involved in adhesion and invasion) and have therefore become major targets for vaccine development (53, 54). Our sequence and subcellular localization analyses detected 57 common OMPs that were present in both NmB and 970 U.S. N. gonorrhoeae strains used in this study. One of the factors that has hampered gonorrhea vaccine development is the high antigenic variability of surface proteins among gonococcal strains (9, 12, 41). Our results demonstrate that FHbp, NadA, and PorA in the MenB-4C vaccine would not be effective vaccine antigens against N. gonorrhoeae. However, a number of other immunogenic proteins seem to have a high level of sequence conservation and may therefore serve as potential vaccine targets. In our study, ∼98% of common OMPs present in both Neisseria species exhibit between 91% and 100% amino acid sequence similarity within 970 N. gonorrhoeae isolates. Approximately 42% of these proteins have been identified in OMV components of the NZ98/254 strain (24, 30, 32, 34). Twelve of these OMPs, including PorB, RmpM, PilQ, OpcA, FetA, Omp85 (BamA), and LbpA, were abundantly and consistently expressed across different OMV lots of the NZ98/254 strain (32), and using bioinformatic analysis, their protein homologs were also identified in publicly available N. gonorrhoeae genomes, including FA1090. Of note, while the gene encoding LbpA is expressed in the majority of N. gonorrhoeae strains, it is a pseudogene in FA1090 (22). The abundances of other OMPs in the NZ98/254 strain and their potential to serve as candidate vaccine antigens warrant further investigation.
Accurate assessment of protein expression may require optimal protein detection methods. It is worth noting that most OMV vaccines were produced by extraction of vesicles using detergents to remove endotoxin from lipopolysaccharides. This extraction method could lower the content of immunogenic proteins released in OMVs compared to detergent-free methods (55–57). Therefore, the protein composition isolated from OMVs may vary depending on the preparation process. Interestingly, the vaccine antigen FHbp was found exclusively in detergent-free OMVs (30, 56), which may explain its absence in the detergent-extracted OMVs (24, 30, 32, 34) and underscores the importance of choosing proper laboratory methods in these investigations. In regard to cross-protection, one recent finding shows that a MenB vaccine made of native OMVs elicited gonococcal bactericidal antibodies in a mouse model (57).
Quantitative proteomic analyses have been demonstrated to facilitate the identification of surface-expressed gonococcal antigens for vaccine development, such as ubiquitous proteins localized to the cell envelopes and membrane vesicles, including BamA, LptD, TamA, MetQ, and NGO2054 (58, 59). In these studies, assessment of protein subcellular localization was performed using a number of prediction tools, SoSuiGramN, PSORTb, CELLO, and SignalP, of which the last 3 tools and BUSCA were used in our study. While a combination of proteomic and bioinformatic approaches was utilized to identify surface-expressed proteins in the previous studies (58, 59), our data are strictly based on the bioinformatic analysis of bacterial genomes. Among common OMPs identified in our study, PorB, PilQ, Omp85 (BamA), OpcA, LbpA, NspA, MtrE, MetQ, TbpB, and AniA have been experimentally tested as potential gonorrhea vaccine antigens due to their immunogenic and antigonococcal properties in vitro and/or in a mouse model (9, 12, 13, 41). Previous findings describe the significance of MetQ (60) and AniA (44–46) as targets for antigonococcal vaccines and therapeutic intervention. Of those previously tested as gonorrhea vaccine candidates, PilQ, Omp85 (BamA), NspA, MtrE, MetQ, and LbpA have been identified within OMVs of the NmB strain NZ98/254 (24, 30, 32, 34) and had ≥93% amino acid sequence similarity to N. gonorrhoeae, suggesting their potential contribution to mechanisms of cross-protection. On the other hand, PorB and OpcA only showed ≤70% amino acid sequence similarity to the respective N. gonorrhoeae homologs, which may reduce their cross-protective role against gonorrhea. These 4 OMPs (PorB, PilQ, OpcA, and LbpA) were abundant in the total protein amount quantified across different NZ98/254 OMV lots (32). Interestingly, copper-containing nitrite reductase AniA, the N. gonorrhoeae pivotal anaerobic respiration factor, was shown to be expressed only in urethritis isolates from men who have sex with men (61), providing the rationale for not being detected in the NZ98/254 OMV. Among gonococcal vaccine targets that were not part of our analysis, a monoclonal antibody against the conserved lipooligosaccharide (LOS) 2C7 epitope has been shown to hold promise in preclinical testing (13, 62). Similarly, Liu and colleagues demonstrated that intravaginal immunization of N. gonorrhoeae-challenged mice with a mixture of gonococcal OMVs and microencapsulated interleukin-12 resulted in accelerated clearance of infection (63).
Taken together, our bioinformatics approach to examine the genetic similarities between NmB (particularly NZ98/254) and N. gonorrhoeae strains contributes to a better understanding of the OMPs present within the MenB-4C vaccine that may be involved in cross-protection and offers valuable insight into antigens of interest to guide next steps for gonorrhea vaccine development. Recognition of gonococcal OMV proteins by MeNZB-like OMV-induced antibodies provides an explanation for the previously observed decrease in gonococcal cases following MeNZB vaccination (22). Investigations of immunogenicity and types of immune responses are required to further investigate the contribution of host defense mechanisms induced by specific OMPs within the MenB-4C OMV against N. gonorrhoeae infections.
MATERIALS AND METHODS
Bacterial strain collection.
This study included 970 N. gonorrhoeae isolates from 27 sites participating in GISP, a national sentinel surveillance system (https://www.cdc.gov/std/gisp/default.htm), obtained between 2014 and 2016. These isolates were selected to represent the geographic, temporal, and genetic diversity among the domestic N. gonorrhoeae population from an original set of 1,518 isolates previously sequenced for surveillance and for antimicrobial susceptibility studies. The NmB reference strain (MC58) and 3 NmB epidemic strains from New Zealand (NZ98/254), Norway (H44/76), and Cuba (CU385/83) were included as meningococcal reference strains. NZ98/254 belongs to clonal complex 41/44 (CC41/44), while the other 3 NmB strains belong to CC32. The 3 NmB epidemic strains were used in the OMV-containing MenB vaccines; the licensed MenB-4C vaccine contains an OMV from NZ98/254.
DNA sequencing of N. gonorrhoeae isolates.
Genomic DNA extraction was performed using Qiagen DNeasy kit, MagNA Pure 24 system (Roche), or Promega Wizard DNA extraction kit (Promega). Sequencing libraries were generated using the Nextera XT library preparation kit (Illumina) or NEBNext Ultra DNA library prep kit for Illumina (New England BioLabs), following the manufacturers’ instructions or PulseNet protocols (https://www.cdc.gov/pulsenet/). High-quality libraries were (paired-end) sequenced on an Illumina MiSeq instrument using V2 reagents at Antibiotic Resistance Laboratory Network laboratories (in Maryland, Washington State, and Texas) and the Hawaii Department of Health or on an Illumina HiSeq instrument using HiSeq 2500 V2 reagents at the CDC Biotechnology Core Facility Branch.
Genome analysis and selection of OMPs.
Illumina sequence reads for 970 N. gonorrhoeae isolates were trimmed using Cutadapt 1.8 (64) and assembled using SPAdes 3.7.0 (65). Low-coverage contigs relative to the overall genomic coverage were discarded. The assemblies for 4 NmB strains were included in the analysis. All genomes were annotated using the Neisseria allele collection from PubMLST (66) to identify and characterize the genomic features found in the assembly. The annotation files were compared to find genes that were common to all 4 NmB strains and found at a frequency of 95% or greater (n ≥ 922) within N. gonorrhoeae genomes. The threshold of 95% represents the “soft-core genes,” which includes genes that may otherwise have not been considered due to sequencing, assembly, or annotation errors. These common gene sequences were obtained and translated using custom Python (https://www.python.org/) scripts. Any gene sequence without a complete ORF or with premature internal stops was discarded. The resulting collection of protein sequences was used to generate a consensus sequence for each locus for N. meningitidis and N. gonorrhoeae separately. The consensus protein sequences for NmB and N. gonorrhoeae were provided as input to 3 subcellular localization Web servers, Cello2Go (67), BUSCA (68), and PSORTb (69). The molecular function of the proteins was predicted using InterProScan (70) software. Two of these tools, BUSCA and PSORTb, provided signal peptide predictions, while Cello2Go did not. As a result, a third signal peptide prediction software, SignalP (71), was included. The results of these tools were combined and used to identify OMPs of interest.
Analysis of sequence similarity.
Unique protein sequences were obtained for the selected OMPs from all 970 N. gonorrhoeae and 4 NmB genomes. Protein similarity values were calculated using BLAST (72) for the following comparisons: all 4 NmB strains against all N. gonorrhoeae genomes, within all N. gonorrhoeae genomes, the NZ98/254 NmB strain against all N. gonorrhoeae genomes, and the other three NmB strains against all N. gonorrhoeae genomes.
Phylogeny analysis.
Protein-based phylogenetic trees were created for selected OMPs. Alignments were created for each OMP using Clustal Omega 1.2 (73), and these alignments were used as input for RAxML 8.2.9 (74) to generate each phylogeny. All phylogenies were visualized and annotated using the Interactive Tree of Life (75) platform.
Data availability.
All raw read data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA317462. The accession numbers of the 970 gonococcal genomes used in this study are listed in Table S3 in the supplemental material.
Accession numbers for 970 gonococcal genomes. Download Table S3, PDF file, 0.2 MB (202.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We thank the CDC Gonococcal Isolate Surveillance Project (GISP) contributors and all U.S. GISP sites for submitting gonococcal isolates to the CDC. We acknowledge Barbara Mahon, Adam Retchless, members of the CDC Meningitis and Vaccine Preventable Diseases Branch, and members of the CDC’s Division of STD Prevention for insightful discussion and technical support. We thank the CDC’s Biotechnology Core Facility Branch sequencing facility (Atlanta, GA) for their role in whole-genome sequencing.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
The Antimicrobial-Resistant Neisseria gonorrhoeae Working Group includes the following members: Sancta St. Cyr (Surveillance and Data Management Branch, Division of STD Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, GA), Matthew W. Schmerer (Laboratory Reference and Research Branch, Division of STD Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, GA), Christian Whelen and Pamela O’Brien (Hawai’i Department of Health State Laboratories Division, HI), Catherine Dominguez (Maryland Department of Health, MD); Sopheay Hun (Washington State Department of Health, WA), and Katie Kneupper (Texas Department of State Health Services, TX).
This work was supported by the Centers for Disease Control and Prevention Office of Antimicrobial Resistance and partially funded by the Combating Antibiotic-Resistant Bacteria (CARB) initiative.
Footnotes
Citation Marjuki H, Topaz N, Joseph SJ, Gernert KM, Kersh EN, Antimicrobial-Resistant Neisseria gonorrhoeae Working Group, Wang X. 2019. Genetic similarity of gonococcal homologs to meningococcal outer membrane proteins of serogroup B vaccine. mBio 10:e01668-19. https://doi.org/10.1128/mBio.01668-19.
REFERENCES
- 1.Perrin A, Bonacorsi S, Carbonnelle E, Talibi D, Dessen P, Nassif X, Tinsley C. 2002. Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species. Infect Immun 70:7063–7072. doi: 10.1128/iai.70.12.7063-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hadad R, Jacobsson S, Pizza M, Rappuoli R, Fredlund H, Olcen P, Unemo M. 2012. Novel meningococcal 4CMenB vaccine antigens–prevalence and polymorphisms of the encoding genes in Neisseria gonorrhoeae. APMIS 120:750–760. doi: 10.1111/j.1600-0463.2012.02903.x. [DOI] [PubMed] [Google Scholar]
- 3.Tinsley CR, Nassif X. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci U S A 93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todar K. 2008. Pathogenic neisseriae: gonorrhea, neonatal ophthalmia and meningococcal meningitis. Todar’s online textbook of bacteriology. http://textbookofbacteriology.net/neisseria.html.
- 5.Cohn AC, MacNeil JR, Clark TA, Ortega-Sanchez IR, Briere EZ, Meissner HC, Baker CJ, Messonnier NE, Centers for Disease Control and Prevention (CDC). 2013. Prevention and control of meningococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 62:1–28. [PubMed] [Google Scholar]
- 6.World Health Organization (WHO). 2016. Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. World Health Organization, Geneva, Switzerland: https://apps.who.int/iris/bitstream/handle/10665/44863/9789241503501_eng.pdf?sequence=1. Accessed 18 July 2018. [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC). 2016. Sexually transmitted diseases surveillance 2017. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/std/stats17/2017-STD-Surveillance-Report_CDC-clearance-9.10.18.pdf. Accessed 17 December 2018. [Google Scholar]
- 8.Dolange V, Churchward CP, Christodoulides M, Snyder L. 2018. The growing threat of gonococcal blindness. Antibiotics (Basel) 7:E59. doi: 10.3390/antibiotics7030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards JL, Jennings MP, Apicella MA, Seib KL. 2016. Is gonococcal disease preventable? The importance of understanding immunity and pathogenesis in vaccine development. Crit Rev Microbiol 42:928–941. doi: 10.3109/1040841X.2015.1105782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feily A, Namazi MR. 2010. Increasing HIV infectivity by Neisseria gonorrhea [sic]: role of host antiapoptotic proteins in enhanced transmission. AIDS 24:1237. doi: 10.1097/QAD.0b013e3283389141. [DOI] [PubMed] [Google Scholar]
- 12.Vincent LR, Jerse AE. 2018. Biological feasibility and importance of a gonorrhea vaccine for global public health. Vaccine, in press. doi: 10.1016/j.vaccine.2018.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice PA, Shafer WM, Ram S, Jerse AE. 2017. Neisseria gonorrhoeae: drug resistance, mouse models, and vaccine development. Annu Rev Microbiol 71:665–686. doi: 10.1146/annurev-micro-090816-093530. [DOI] [PubMed] [Google Scholar]
- 14.Banzhoff A. 2017. Multicomponent meningococcal B vaccination (4CMenB) of adolescents and college students in the United States. Ther Adv Vaccines 5:3–14. doi: 10.1177/2051013616681365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zlotnick GW, Jones TR, Liberator P, Hao L, Harris S, McNeil LK, Zhu D, Perez J, Eiden J, Jansen KU, Anderson AS. 2015. The discovery and development of a novel vaccine to protect against Neisseria meningitidis serogroup B disease. Hum Vaccin Immunother 11:5–13. doi: 10.4161/hv.34293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finne J, Leinonen M, Makela PH. 1983. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet ii:355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 17.Harder T, Koch J, Wichmann O, Hellenbrand W. 2017. Predicted vs observed effectiveness of outer membrane vesicle (OMV) vaccines against meningococcal serogroup B disease: systematic review. J Infect 75:81–94. doi: 10.1016/j.jinf.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang NY, Pollard AJ. 2018. The next chapter for group B meningococcal vaccines. Crit Rev Microbiol 44:95–111. doi: 10.1080/1040841X.2017.1329276. [DOI] [PubMed] [Google Scholar]
- 19.Perez O, del Campo J, Cuello M, Gonzalez E. 2009. Mucosal approaches in Neisseria vaccinology. VacciMonitor 18:53–55. [Google Scholar]
- 20.Petousis-Harris H, Paynter J, Morgan J, Saxton P, McArdle B, Goodyear-Smith F, Black S. 2017. Effectiveness of a group B outer membrane vesicle meningococcal vaccine against gonorrhoea in New Zealand: a retrospective case-control study. Lancet 390:1603–1610. doi: 10.1016/S0140-6736(17)31449-6. [DOI] [PubMed] [Google Scholar]
- 21.Whelan J, Klovstad H, Haugen IL, Holle MR, Storsaeter J. 2016. Ecologic study of meningococcal B vaccine and Neisseria gonorrhoeae infection, Norway. Emerg Infect Dis 22:1137–1139. doi: 10.3201/eid2206.151093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semchenko EA, Tan A, Borrow R, Seib KL. 2018. The serogroup B meningococcal vaccine Bexsero elicits antibodies to Neisseria gonorrhoeae. Clin Infect Dis, in press. doi: 10.1093/cid/ciy1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Régnier SA, Huels J. 2014. Potential impact of vaccination against Neisseria meningitidis on Neisseria gonorrhoeae in the United States: results from a decision-analysis model. Hum Vaccin Immunother 10:3737–3745. doi: 10.4161/hv.36221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holst J, Oster P, Arnold R, Tatley M, Næss L, Aaberge I, Galloway Y, McNicholas A, O'Hallahan J, Rosenqvist E, Black S. 2013. Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum Vaccin Immunother 9:1241–1253. doi: 10.4161/hv.24129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feavers IM, Maiden MC. 1998. A gonococcal porA pseudogene: implications for understanding the evolution and pathogenicity of Neisseria gonorrhoeae. Mol Microbiol 30:647–656. doi: 10.1046/j.1365-2958.1998.01101.x. [DOI] [PubMed] [Google Scholar]
- 26.Unemo M, Norlen O, Fredlund H. 2005. The porA pseudogene of Neisseria gonorrhoeae–low level of genetic polymorphism and a few, mainly identical, inactivating mutations. APMIS 113:410–419. doi: 10.1111/j.1600-0463.2005.apm_206.x. [DOI] [PubMed] [Google Scholar]
- 27.van der Pol L, Stork M, van der Ley P. 2015. Outer membrane vesicles as platform vaccine technology. Biotechnol J 10:1689–1706. doi: 10.1002/biot.201400395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin DR, Ruijne N, McCallum L, O’Hallahan J, Oster P. 2006. The VR2 epitope on the PorA P1.7-2,4 protein is the major target for the immune response elicited by the strain-specific group B meningococcal vaccine MeNZB. Clin Vaccine Immunol 13:486–491. doi: 10.1128/CVI.13.4.486-491.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosaheb M, Wetzler LM. 2018. Meningococcal PorB induces a robust and diverse antigen specific T cell response as a vaccine adjuvant. Vaccine 36:7689. doi: 10.1016/j.vaccine.2018.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrari G, Garaguso I, Adu-Bobie J, Doro F, Taddei AR, Biolchi A, Brunelli B, Giuliani MM, Pizza M, Norais N, Grandi G. 2006. Outer membrane vesicles from group B Neisseria meningitidis delta gna33 mutant: proteomic and immunological comparison with detergent-derived outer membrane vesicles. Proteomics 6:1856–1866. doi: 10.1002/pmic.200500164. [DOI] [PubMed] [Google Scholar]
- 31.Gil J, Betancourt LH, Sardiñas G, Yero D, Niebla O, Delgado M, García D, Pajón R, Sánchez A, González LJ, Padrón G, Campa C, Sotolongo F, Barberá R, Guillén G, Herrera L, Besada V. 2009. Proteomic study via a non-gel based approach of meningococcal outer membrane vesicle vaccine obtained from strain CU385: a road map for discovering new antigens. Hum Vaccin 5:347–356. doi: 10.4161/hv.5.5.7367. [DOI] [PubMed] [Google Scholar]
- 32.Tani C, Stella M, Donnarumma D, Biagini M, Parente P, Vadi A, Magagnoli C, Costantino P, Rigat F, Norais N. 2014. Quantification by LC-MSE of outer membrane vesicle proteins of the Bexsero vaccine. Vaccine 32:1273–1279. doi: 10.1016/j.vaccine.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Tsolakos N, Lie K, Bolstad K, Maslen S, Kristiansen PA, Hoiby EA, Wallington A, Vipond C, Skehel M, Tang CM, Feavers IM, Wedege E, Wheeler JX. 2010. Characterization of meningococcal serogroup B outer membrane vesicle vaccines from strain 44/76 after growth in different media. Vaccine 28:3211–3218. doi: 10.1016/j.vaccine.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 34.Vipond C, Suker J, Jones C, Tang C, Feavers IM, Wheeler JX. 2006. Proteomic analysis of a meningococcal outer membrane vesicle vaccine prepared from the group B strain NZ98/254. Proteomics 6:3400–3413. doi: 10.1002/pmic.200500821. [DOI] [PubMed] [Google Scholar]
- 35.Williams JN, Skipp PJ, Humphries HE, Christodoulides M, O'Connor CD, Heckels JE. 2007. Proteomic analysis of outer membranes and vesicles from wild-type serogroup B Neisseria meningitidis and a lipopolysaccharide-deficient mutant. Infect Immun 75:1364–1372. doi: 10.1128/IAI.01424-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masignani V, Rappuoli R, Pizza M. 2002. Reverse vaccinology: a genome-based approach for vaccine development. Expert Opin Biol Ther 2:895–905. doi: 10.1517/14712598.2.8.895. [DOI] [PubMed] [Google Scholar]
- 37.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 38.Rappuoli R. 2001. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine 19:2688–2691. doi: 10.1016/S0264-410X(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 39.Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. 2012. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30(Suppl 2):B87–B97. doi: 10.1016/j.vaccine.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donati C, Zolfo M, Albanese D, Tin Truong D, Asnicar F, Iebba V, Cavalieri D, Jousson O, De Filippo C, Huttenhower C, Segata N. 2016. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nat Microbiol 1:16070. doi: 10.1038/nmicrobiol.2016.70. [DOI] [PubMed] [Google Scholar]
- 41.Jerse AE, Bash MC, Russell MW. 2014. Vaccines against gonorrhea: current status and future challenges. Vaccine 32:1579–1587. doi: 10.1016/j.vaccine.2013.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz FX, Stiasny K. 2012. Flaviviruses and flavivirus vaccines. Vaccine 30:4301–4306. doi: 10.1016/j.vaccine.2011.09.114. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Gao N, Fan D, Chen H, Sheng Z, Fu S, Liang G, An J. 2016. Cross-protection induced by Japanese encephalitis vaccines against different genotypes of dengue viruses in mice. Sci Rep 6:19953. doi: 10.1038/srep19953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikora AE, Mills RH, Weber JV, Hamza A, Passow BW, Romaine A, Williamson ZA, Reed RW, Zielke RA, Korotkov KV. 2017. Peptide inhibitors targeting the Neisseria gonorrhoeae pivotal anaerobic respiration factor AniA. Antimicrob Agents Chemother 61:e00186-17. doi: 10.1128/AAC.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shewell LK, Jen FE, Jennings MP. 2017. Refinement of immunizing antigens to produce functional blocking antibodies against the AniA nitrite reductase of Neisseria gonorrhoeae. PLoS One 12:e0182555. doi: 10.1371/journal.pone.0182555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shewell LK, Ku SC, Schulz BL, Jen FE, Mubaiwa TD, Ketterer MR, Apicella MA, Jennings MP. 2013. Recombinant truncated AniA of pathogenic Neisseria elicits a non-native immune response and functional blocking antibodies. Biochem Biophys Res Commun 431:215–220. doi: 10.1016/j.bbrc.2012.12.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenberg L, Diena BB, Ashton FA, Wallace R, Kenny CP, Znamirowski R, Ferrari H, Atkinson J. 1974. Gonococcal vaccine studies in Inuvik. Can J Public Health 65:29–33. [PubMed] [Google Scholar]
- 48.Boslego JW, Tramont EC, Chung RC, McChesney DG, Ciak J, Sadoff JC, Piziak MV, Brown JD, Brinton CC Jr, Wood SW. 1991. Efficacy trial of a parenteral gonococcal pilus vaccine in men. Vaccine 9:154–162. doi: 10.1016/0264-410X(91)90147-X. [DOI] [PubMed] [Google Scholar]
- 49.Jongerius I, Lavender H, Tan L, Ruivo N, Exley RM, Caesar JJ, Lea SM, Johnson S, Tang CM. 2013. Distinct binding and immunogenic properties of the gonococcal homologue of meningococcal factor h binding protein. PLoS Pathog 9:e1003528. doi: 10.1371/journal.ppat.1003528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, Cartocci E, Ciucchi L, Di Marcello F, Ferlicca F, Galli B, Luzzi E, Masignani V, Serruto D, Veggi D, Contorni M, Morandi M, Bartalesi A, Cinotti V, Mannucci D, Titta F, Ovidi E, Welsch JA, Granoff D, Rappuoli R, Pizza M. 2006. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A 103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bos MP, Grijpstra J, Tommassen-van Boxtel R, Tommassen J. 2014. Involvement of Neisseria meningitidis lipoprotein GNA2091 in the assembly of a subset of outer membrane proteins. J Biol Chem 289:15602–15610. doi: 10.1074/jbc.M113.539510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donnarumma D, Golfieri G, Brier S, Castagnini M, Veggi D, Bottomley MJ, Delany I, Norais N. 2015. Neisseria meningitis GNA1030 is a ubiquinone-8 binding protein. FASEB J 29:2260–2267. doi: 10.1096/fj.14-263954. [DOI] [PubMed] [Google Scholar]
- 53.Rollauer SE, Sooreshjani MA, Noinaj N, Buchanan SK. 2015. Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans R Soc Lond B Biol Sci 370:20150023. doi: 10.1098/rstb.2015.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernardini G, Braconi D, Martelli P, Santucci A. 2007. Postgenomics of Neisseria meningitidis for vaccines development. Expert Rev Proteomics 4:667–677. doi: 10.1586/14789450.4.5.667. [DOI] [PubMed] [Google Scholar]
- 55.Lappann M, Otto A, Becher D, Vogel U. 2013. Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J Bacteriol 195:4425–4435. doi: 10.1128/JB.00625-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van de Waterbeemd B, Mommen GP, Pennings JL, Eppink MH, Wijffels RH, van der Pol LA, de Jong AP. 2013. Quantitative proteomics reveals distinct differences in the protein content of outer membrane vesicle vaccines. J Proteome Res 12:1898–1908. doi: 10.1021/pr301208g. [DOI] [PubMed] [Google Scholar]
- 57.Beernink PT, Ispasanie E, Lewis LA, Ram S, Moe GR, Granoff DM. 2018. A meningococcal native outer membrane vesicle vaccine with attenuated endotoxin and overexpressed factor H binding protein elicits gonococcal bactericidal antibodies. J Infect Dis 219:1130–1137. doi: 10.1093/infdis/jiy609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zielke RA, Wierzbicki IH, Baarda BI, Gafken PR, Soge OO, Holmes KK, Jerse AE, Unemo M, Sikora AE. 2016. Proteomics-driven antigen discovery for development of vaccines against gonorrhea. Mol Cell Proteomics 15:2338–2355. doi: 10.1074/mcp.M116.058800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zielke RA, Wierzbicki IH, Weber JV, Gafken PR, Sikora AE. 2014. Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol Cell Proteomics 13:1299–1317. doi: 10.1074/mcp.M113.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Semchenko EA, Day CJ, Seib KL. 2017. MetQ of Neisseria gonorrhoeae is a surface-expressed antigen that elicits bactericidal and functional blocking antibodies. Infect Immun 85:e00898-16. doi: 10.1128/IAI.00898-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taha MK, Claus H, Lappann M, Veyrier FJ, Otto A, Becher D, Deghmane AE, Frosch M, Hellenbrand W, Hong E, Parent Du Chatelet I, Prior K, Harmsen D, Vogel U. 2016. Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLoS One 11:e0154047. doi: 10.1371/journal.pone.0154047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gulati S, Zheng B, Reed GW, Su X, Cox AD, St Michael F, Stupak J, Lewis LA, Ram S, Rice PA. 2013. Immunization against a saccharide epitope accelerates clearance of experimental gonococcal infection. PLoS Pathog 9:e1003559. doi: 10.1371/journal.ppat.1003559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Hammer LA, Liu W, Hobbs MM, Zielke RA, Sikora AE, Jerse AE, Egilmez NK, Russell MW. 2017. Experimental vaccine induces Th1-driven immune responses and resistance to Neisseria gonorrhoeae infection in a murine model. Mucosal Immunol 10:1594–1608. doi: 10.1038/mi.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 65.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu C-S, Cheng C-W, Su W-C, Chang K-C, Huang S-W, Hwang J-K, Lu C-H. 2014. CELLO2GO: a Web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS One 9:e99368. doi: 10.1371/journal.pone.0099368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R. 2018. BUSCA: an integrative Web server to predict subcellular localization of proteins. Nucleic Acids Res 46:W459–W466. doi: 10.1093/nar/gky320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardy JL, Spencer C, Wang K, Ester M, Tusnády GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, Brinkman FSL. 2003. PSORTb: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res 31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zdobnov EM, Apweiler R. 2001. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 71.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 72.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Common genes found in 4 NmB and 970 N. gonorrhoeae strains. Download Table S1, PDF file, 0.2 MB (228.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequence similarities of common OMPs found in NmB and N. gonorrhoeae strains. Download Table S2, PDF file, 0.1 MB (112.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Phylogenetic tree of AniA, FHbp, PorB, GNA1030, GNA2091, NhbA, and OpcA. Download FIG S1, PDF file, 0.5 MB (494.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Accession numbers for 970 gonococcal genomes. Download Table S3, PDF file, 0.2 MB (202.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
All raw read data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA317462. The accession numbers of the 970 gonococcal genomes used in this study are listed in Table S3 in the supplemental material.
Accession numbers for 970 gonococcal genomes. Download Table S3, PDF file, 0.2 MB (202.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.