Abstract
Background
Dextransucrases are extracellular enzymes, which catalyze the formation of α-1→6-linked glucose polymers from sucrose. These enzymes are exclusively expressed by lactic acid bacteria, which commonly acidify the extracellular environment due to their physiology. Dextransucrases are thus confronted with steadily changing reaction conditions in regards to the environmental pH, which can further affect the amount of released dextransucrases. In this work, we studied the effect of the environmental pH on the release, the productivity and the product specificity of the dextransucrase expressed by Lactobacillus (L.) hordei TMW 1.1822. Dextransucrases were recovered as crude extracts at pH 3.5–pH 6.5 and then again used to produce dextrans at these pH values. The respectively produced dextran amounts and sizes were determined and the obtained results finally systematically correlated.
Results
Maximum dextran amounts were produced at pH 4.0 and pH 4.5, while the productivity of the dextransucrases significantly decreased at pH 3.5 and pH 6.5. The distribution of dextran amounts produced at different pH most likely reflects the pH dependent activity of the dextransucrases released by L. hordei, since different transglycosylation rates were determined at different pH using the same dextransucrase amounts. Moreover, similar hydrolysis activities were detected at all tested conditions despite significant losses of transglycosylation activities indicating initial hydrolysis prior to transglycosylation reactions. The molar masses and rms radii of dextrans increased up to pH 5.5 independently of the stability of the enzyme. The gelling properties of dextrans produced at pH 4.0 and pH 5.5 were different.
Conclusions
The presented methodological approach allows the controlled production of dextrans with varying properties and could be transferred and adapted to other microbes for systematic studies on the release and functionality of native sucrases or other extracellular enzymes.
Keywords: Lactic acid bacteria, Dextransucrase, Systematic approach, Extracellular enzyme
Introduction
Dextrans contain α–1→6-linked glucose monomers in their backbone, which can be branched at positions O2, O3 or O4. They are synthesized by extracellular dextransucrases (EC 2.4.1.5) belonging to the family 70 of glycoside hydrolases (GH 70) [1–5]. These enzymes use the energy of the glycosidic bond of their substrate sucrose for the polymerization of glucose, while fructose is continuously released [2, 6–8]. Moreover, they can hydrolyze sucrose if water is used as acceptor instead of the growing polymer chain. Dextrans are exclusively expressed by lactic acid bacteria (LAB) living in various ecological niches. The properties as well as the biosynthesis of dextran have been extensively studied in the past due to its wide use in biotechnological applications [4, 5, 9–14]. The biological functions of dextran are not completely revealed, but there are many studies concerning the role of dextran in biofilm formation [15–19]. As LAB are naturally subjected to pH fluctuations, changes in pH during growth are crucial factors influencing the release of dextransucrases as well as their activity [20–23] implying the complexity of natural dextran formation. The aim of this study was to establish a systematic approach, which gives new insights into the pH-dependent release, productivity and product specificity of dextransucrases and, beyond that, enables an improved control over the formation of specific dextran fractions. For this purpose, we used the native dextransucrase of Lactobacillus hordei TMW 1.1822, which is efficiently released in the presence of sucrose and serves as sucrose converting enzyme in the extracellular environment [24–26].
Materials and methods
Strain, media and cultivation
The strain L. hordei TMW 1.1822 isolated from water kefir was cultivated in modified MRS as described before [27], containing 10 g/L of glucose and 10 g/L fructose, 10 g/L peptone from casein, 5 g/L yeast extract, 5 g/L meat extract, 4 g/L K2HPO4, 2.6 g/L KH2HPO4, 3 g/L NH4Cl, 1 g/L Tween 80, 0.5 g/L cysteine-HCl, 0.2 g/L MgSO4∙7H2O, 0.036 g/L MnSO4 H2O and 15 g/L agar for solid media. The pH of MRS was adjusted to 6.2 and cultivations were generally performed at 30 °C. To obtain cells in the mid-exponential growth phase main cultures were inoculated with 4% (v/v) of 30 h grown pre-cultures diluted to an OD590nm = 2.0. All experiments were carried out in biological triplicates. To exclude any possible contamination during storage at − 80 °C, prior to the experimental start the strain identity was verified with matrix-assisted laser-desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS) by comparing the generated mass spectra with our in house database.
Dextran production and isolation
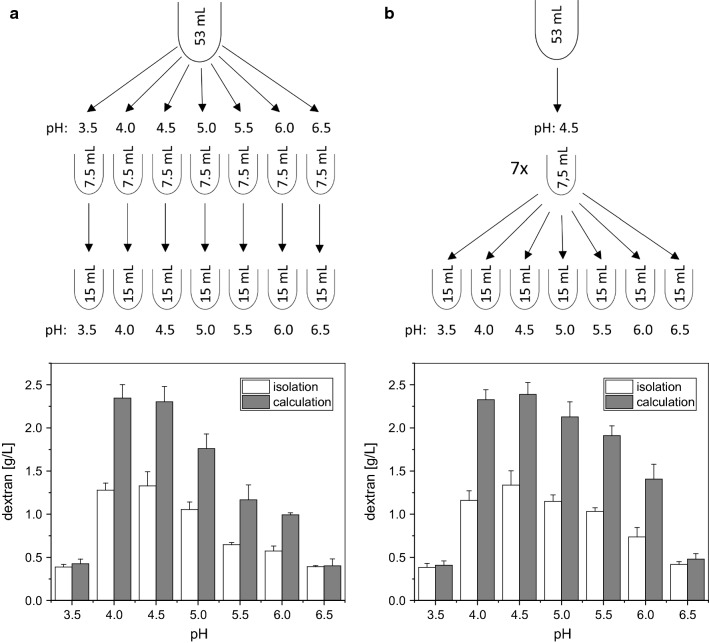
Figure 1 depicts the procedure for recovery of dextransucrase containing solutions. At first, the grown main culture (53 mL) was split in 7.5 mL tubes. Cells were then removed by centrifugation at 10,000×g for 10 and re-dissolved in equal volumes (7.5 mL) of 0.1 M citrate–phosphate buffers [12] containing 0.1 M sucrose. After 3.0 h of incubation at 30 °C the cells were removed by centrifugation (10,000×g, 10 min) and sterile filtration (0.2 µm nylon filters, Phenomenex, Germany). The pH of the acidified buffer was re-adjusted to the desired value by adding 7.5 mL of buffer of calculated pH values to reach the final dextran production pH according to McIlvaine [28]. The release of the enzyme for 3.0 h in presence of 0.1 M sucrose was either performed at pH values from 3.5 to 6.5 (steps of 0.5 units) or at pH 4.5 for all samples. Subsequently, the EPS were synthesized at 30 °C for 24 h at pH values of 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5. Enzymatic reactions were stopped by adding two volumes (15 mL) of chilled ethanol for dextran isolation or by shock freezing (− 20 °C) in case of subsequent HPLC analysis (2.4), respectively. The ethanol treated buffer solutions were stored over night at 4 °C. Precipitated dextrans were then recovered by centrifugation at 10,000×g for 10 min and subsequently redissolved in an appropriate amount of water. The water polysaccharide mixture was dialyzed against dH2O using dialysis tubing (SERVAPOR, SERVA Electrophoresis GmbH, Germany) with a MWCO (molecular weight cut off) of 3.5 kDa at 4 °C and subsequently lyophilized for at least 24 h. The isolated dextran was quantified by weighing.
Fig. 1.
Overview of the experimental setups used to characterize dextran production at different pH. Cells were grown in 53 mL mMRS medium (without sucrose) for 18 h, split up to 7.5 mL samples, centrifuged and re-dissolved in 7.5 mL buffer (0.1 M Na2HPO4/0.1 M citric acid + 0.1 M sucrose) of the desired pH values, respectively. In setup (a), dextransucrase containing supernatants were collected at pH 3.5 to 6.5, while in setup (b) the release pH for all samples was kept constant at pH 4.5. After 3.0 h of incubation in these buffers, the cells were removed by centrifugation and the supernatants were sterile-filtered. 7.5 mL buffer of the required pH to reach the final production pH were added, followed by 24 h of incubation and subsequent dextran quantification. Bars indicate the produced dextran amounts in g/L, which were either determined gravimetrically (isolated dextran) or via calculation of the totally produced amount using the calculated transglycosylation activity (24 h). Data are expressed with mean ± SD of three biological replicates
Verification of the released dextransucrase
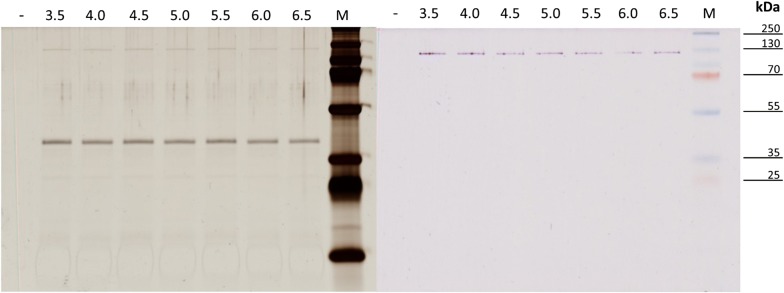
The protein composition of the samples was analyzed by vertical SDS-PAGE, carried out in a Mini-PROTEAN® Tetra Cell Electrophoresis System (Bio-Rad laboratories, Hercules, USA). A separation gel (12% (w/v)) total monomer concentration with a stacking gel (4% (w/v)) were used. Prior to application on the gel, protein samples were diluted in 2× Laemmli buffer (Sigma-Aldrich, St. Louis, USA) and denatured at 90 °C for 10 min. Separation was initially started at 80 V, 10 min and continued at 100 V for 120 min using a Power Pack 3000 unit (Bio-Rad laboratories, Hercules, USA). Visualization of proteins was conducted by silver staining as described by Blum, Beier [29]. For determination of the size of the dextransucrases and identification of the corresponding SDS-PAGE lane, an activity staining was performed. Therefore, following electrophoresis, SDS gels were washed three times for 10 min in sodium acetate buffer (20 mM, 0.3 mM CaCl2, 0.1% Tween 80, pH 5.4) at 4 °C and incubated in the same buffer, containing 5% (w/v) sucrose, at 30 °C overnight. Afterwards, the gels were washed in an aqueous solution containing 50% (v/v) methanol and 10% (v/v) acetic acid for 30 min, followed by washing in deionized water (dH2O) for 30 min. For oxidization of formed dextrans, the gels were incubated in periodic acid solution [1% (w/v) periodic acid (≥ 99.0%, Sigma-Aldrich, St. Louis, USA), 3% (v/v) acetic acid] for 45 min and subsequently washed in dH2O for 1 h. Finally, dextrans were stained by Schiff’s reagent (Sigma-Aldrich, St. Louis, USA) until discrete pink bands appeared. The gels were washed in dH2O for 5 min. If not otherwise stated, all steps were carried out at room temperature. Total protein concentrations of the buffer supernatants were determined by the Coomassie/Bradford Protein Assay Kit (Thermo Fisher Scientific, USA). The assay was performed in standard 96 microtiter plates according to the instructions of the manufacturer.
Quantification of sugars
For quantification of free sugars, high pressure liquid chromatography (HPLC) (Dionex Ultimate 3000, Thermo Fisher Scientific, USA) coupled to refractive index detection (ERC Refractomax 521, Thermo Fisher Scientific, USA) was used. Sugar separations were performed on a Rezex RPM-Monosaccharide Pb2+ column (Phenomenex Ltd., Germany) using water as mobile phase and a flow rate of 0.6 mL/min at 85 °C. The sugars were identified and quantified using sugar standards and subsequently generated calibration curves and the Chromeleon™ software (Version 6.8, Dionex, Germany).
Analysis of dextran sizes
The characterization of molecular weights and rms radii of the isolated dextrans was accomplished by asymmetric flow field-flow fractionation (AF4) coupled with multi-angle laser light scattering (MALS) (Dawn Heleos II, Wyatt Technology, Germany) analysis and UV detection (Dionex Ultimate 3000, Thermo Fisher Scientific, USA). The freeze dried dextran was at first redissolved in 0.05 M NaNO3 (which also served as the eluent) at a final concentration of 0.1 mg/mL. 100 µL of the respective sample were then injected into the separation channel, equipped with a 10 kDa cellulose membrane. Separations were performed using a detector-flow rate of 1 mL/min and a cross-flow gradient of 3 to 0.1 mL/min over 15 min, followed by 15 min of a steady cross flow of 0.1 mL/min. All chromatograms were analyzed with the software ASTRA 6.1 (Wyatt Technologies, Germany) using a dn/dc value of 0.1423 mL/g [30] and the Berry model integrated in the ASTRA software. The distributions of rms radii were calculated in the particle mode, whereas the molar mass distributions were determined by using the respective UV concentration signals of the dextrans at 400 nm [20, 23, 25]. The extinction coefficients of dextrans were determined in standard 96 microtiter plates at a wavelength of 400 nm (SPECTROstar, BMG LABTECH) according to the Beer–Lambert law.
Statistical analysis
Data were evaluated with SigmaPlot Version 12.5 (Systat Software GmbH, Germany) by one-way ANOVA. Differences in the means were considered as significant for p < 0.05 and are depicted in Additional file 1.
Results
Quantification of dextrans produced at different pH
For recovery of native dextransucrases and subsequent production of dextran at different pH, two different experimental setups were conducted (Fig. 1). As L. hordei TMW 1.1822 releases its dextransucrase only in the presence of sucrose [24], pre-cultured cells (mMRS medium) were at first re-suspended and incubated in sucrose-supplemented buffers. After 3 h of incubation in these buffers, the respective cell densities, pH values as well as the released and consumed sugars were determined. While the viable cells remained constant, a reduction in pH and the release of fructose and glucose were observed (Table 1). Afterwards, cells were separated and the dextransucrase containing supernatants were used to produce dextran at different pH for 24 h. The respective pHs were adjusted to the final production pH according to McIlvaine [28]. The produced dextrans were quantified both by weighing of isolated dextrans as well as by calculation of the total dextran amount (per L) using the equation c (transglycosylated glucose) × 162.16 g/mol (molar mass of glucose in dextran) (2.4). To visualize the crude protein extracts and to verify the presence of the dextransucrase, SDS-PAGE and subsequent silver and activity staining were performed using the supernatants recovered in setup A (Fig. 2). The dextransucrase band was similarly intense at all conditions and similar total protein concentrations were present in the tested supernatants (Table 1). In both setups, maximum dextran amounts were produced at pH 4.0 and 4.5. The lowest amounts were determined for pH 3.5 and pH 6.5. Outside of the depicted pH range, no dextran production took place, respectively (data not shown). Dextran amounts were significantly higher at pH 5.5 and 6.0 in setup B (Fig. 1b; Additional file 1), while at other pH similar amounts were produced in both approaches. The loss of dextran during the isolation process accounts for ~ 40% in between pH 4.0–pH 6.0 in both setups. At pH 3.5 and 6.5 no differences in the calculated and the isolated amounts were detectable (Fig. 1a, b). The concentration of liberated glucose, which reflects the hydrolysis activity of the dextransucrase, was similar at all test conditions (~ 12 mM glucose/27 h) (Table 1; Additional file 1).
Table 1.
Log10 CFU/mL, pH and glucose, fructose concentrations after 3 h of dextransucrase release
| Smp./pH | Log10 CFU/mL 3 h | pH 3 h | Glucose 3 h [mM] | Fructose 3 h [mM] | Glucose 27 h [mM] | Fructose 27 h [mM] | Protein [µg/mL] | |
|---|---|---|---|---|---|---|---|---|
| Setup A | 3.5 | 9.69 ± 0.17 | 3.37 ± 0.01 | 4.84 ± 0.02 | 4.90 ± 0.07 | 12.00 ± 0.29 | 14.69 ± 0.04 | 16.38 ± 2.11 |
| 4 | 9.61 ± 0.03 | 3.88 ± 0.01 | 4.91 ± 0.08 | 8.41 ± 0.19 | 12.07 ± 0.43 | 30.04 ± 1.00 | 16.39 ± 1.87 | |
| 4.5 | 9.63 ± 0.13 | 4.38 ± 0.00 | 5.03 ± 0.05 | 8.50 ± 0.32 | 12.18 ± 1.02 | 29.86 ± 0.07 | 16.40 ± 1.90 | |
| 5 | 9.64 ± 0.09 | 4.77 ± 0.01 | 5.04 ± 0.03 | 7.77 ± 0.36 | 12.16 ± 1.29 | 25.74 ± 0.52 | 16.27 ± 1.98 | |
| 5.5 | 9.71 ± 0.10 | 5.25 ± 0.01 | 4.72 ± 0.13 | 7.07 ± 0.30 | 11.34 ± 0.72 | 20.88 ± 1.73 | 15.83 ± 2.02 | |
| 6 | 9.62 ± 0.10 | 5.73 ± 0.01 | 5.22 ± 0.13 | 7.27 ± 0.11 | 11.77 ± 0.62 | 19.81 ± 0.75 | 17.12 ± 2.00 | |
| 6.5 | 9.67 ± 0.05 | 6.21 ± 0.00 | 4.89 ± 0.03 | 5.83 ± 0.20 | 11.78 ± 0.02 | 15.21 ± 0.51 | 16.60 ± 1.88 | |
| Setup B | 3.5 | 4.37 ± 0.01 | 4.84 ± 0.07 | 8.40 ± 0.14 | 11.86 ± 0.32 | 17.94 ± 0.35 | ||
| 4 | 4.37 ± 0.01 | 4.93 ± 0.02 | 8.58 ± 0.22 | 12.04 ± 0.37 | 30.04 ± 0.51 | |||
| 4.5 | 4.37 ± 0.01 | 5.03 ± 0.10 | 8.44 ± 0.54 | 12.35 ± 0.26 | 30.49 ± 0.85 | |||
| 5 | 4.37 ± 0.01 | 5.16 ± 0.06 | 8.28 ± 0.46 | 12.40 ± 0.97 | 28.66 ± 0.73 | |||
| 5.5 | 4.37 ± 0.01 | 4.99 ± 0.13 | 8.56 ± 0.57 | 11.64 ± 0.82 | 26.99 ± 0.37 | |||
| 6 | 4.37 ± 0.01 | 4.93 ± 0.09 | 7.90 ± 0.41 | 12.06 ± 0.69 | 23.71 ± 1.09 | |||
| 6.5 | 4.37 ± 0.01 | 4.71 ± 0.08 | 8.27 ± 0.87 | 11.79 ± 0.20 | 18.24 ± 0.22 |
For calculation of the totally produced dextran amounts, glucose and fructose concentrations were additionally determined at the end of dextran production (27 h). Sugar concentrations were determined according to 2.4. The protein concentrations of the buffer supernatants were determined by the Bradford assay. Data are expressed with mean ± SD of three biological replicates, respectively
Smp. sample
Fig. 2.
Silver (left) and activity (right) stained SDS-gels of the buffer supernatants obtained after 3 h of incubation of Lactobacillus hordei TMW 1.1822 at different pH (setup A); negative control (−), marker (M)
Determination of the molecular size and weight of dextran produced at different pH
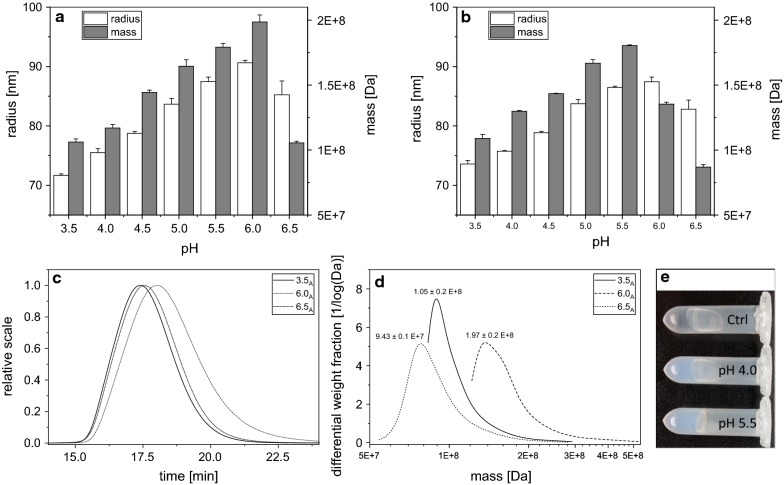
The isolated dextrans from both setups were further analyzed using AF4-MALS-UV regarding their molecular weight and rms radii. In both setups, similar tendencies were observed. The rms radii increased from pH 3.5 to 6.0 and decreased at pH 6.5 (Fig. 3a, b). The molecular weight increased to the same extent except for pH 6.0 (Fig. 3a, b). At pH 6.5, the averaged rms radii and molecular weights decreased, respectively. Furthermore, differences in the retention times were observed, as the dextrans exhibiting smaller averaged molar masses and radii eluted earlier (Fig. 3c, d). According to the principles of AF4 separation, it can thus be concluded that the hydrodynamic volumes of these dextrans increase with their molecular size and weight [31, 32] (Fig. 3c, d). Figure 3e shows the gel forming capability of dextrans of predominantly small fractions (pH 4.0) compared to the ones with bigger fractions (pH 5.5). Both dextrans were re-dissolved in a concentration of 50 g/L resulting in increased gel stability for the dextran derived from pH 5.5.
Fig. 3.
Averaged molar masses [Da] and rms radii [nm] of dextrans produced in experimental setups (a, b) (Fig. 1). c AF4 chromatograms (light scattering signal at 90°) of representative dextrans isolated from setup A. d Differential weight fractions of the dextrans depicted in Fig. 2c. Data are expressed as average of three biological replicates with ± SD, respectively. e Photographs of dextrans produced at pH 5.5 and pH 4.0 redissolved in a concentration of 5% (w/v) in water; control (ctrl): pure water
Discussion
Diverse studies reported about the sucrose-inducible or pH-dependent release of dextransucrases in LAB [22, 33–36]. We recently observed that the dextransucrase of L. hordei is released into buffer supernatants efficiently in the presence of sucrose [24]. Within the present study we wanted to inter alia verify, if the environmental pH additionally influences the dextransucrase release by L. hordei.
At first we observed that maximum dextran amounts were obtained at pH 4.0 and 4.5 in setup A (Fig. 1a), which are within the range of pH values for optimal activities of dextransucrases expressed by some other LAB [37, 38]. However, the respectively used enzyme amounts could have significantly differed in setup A, as the dextransucrases were collected at different pH. Setup B was thus used to see if the tendencies in regard to produced dextran amounts could indeed be driven by differing enzyme concentrations, at which similar overall trends were observed (Fig. 1). At pH 3.5 and pH 6.5 the decrease in produced dextran amounts was similar in both setups (Fig. 1a, b) and the amount of released glucose was similar at all tested pH conditions (Table 1: ~ 12 mM/27 h). Moreover, the total protein concentrations exhibited no significant differences (~ 16 µg/mL; Table 1/Additional file 1) and the dextransucrase recovered at different pH appeared similarly intense (Fig. 2). These findings strongly suggest that similar dextransucrase amounts were present in all assays, also considering that distinctly different numbers of identical dextransucrases could not lose transglycosylation, or rather, keep up hydrolysis activities at similar levels at non optimum pH, respectively. However, comparatively higher amounts of dextrans were produced at pH 5.0, 5.5, 6.0, if the dextransucrases had been recovered at pH 4.5 (setup B). This indicates a less reducing effect concerning the productivity and stability of the dextransucrase due to the inevitable denaturation and the concomitant loss of transglycosylation activity at non optimum pH, if it had been preliminarily recovered working at its approximate optimum pH. Furthermore, the comparable hydrolysis rates observed at all tested pH conditions imply a directed hydrolysis mechanism, which takes place independently of the loss of transglycosylation activity and, thus, most likely before the polymerization reaction.
Our data further reveal that considerable amounts of dextrans could not be isolated at more optimum pH using a standardized EPS isolation approach [39–41], which could be explained by the more frequent occurrence of short-chain dextrans, which get lost during the ethanol precipitation or dialysis of the samples. Therefore, the observed increases in averaged molar masses and rms radii (Fig. 3) with rising pH refer to the respective high molecular weight fractions of the dextrans produced by L. hordei. A similar relationship between sizes of sucrase-synthesized EPS and the fermentation pH was reported for levans synthesized by acetic acid bacteria (AAB) [23]. The reasons for the pH dependent mass increases of glucans and fructans still remain unclear. First observations, however, reveal that high molecular weight dextrans produced by L. hordei at different pH exhibit different functional properties (Fig. 3e), which coincides with the findings about levans from AAB [23]. A proper control of the pH during the production of diverse sucrase-synthesized EPS hence appears to be a decisive factor for recovery of functionally versatile glucans and fructans. Overall, similar pH dependent size distributions were obtained in both setups (Fig. 3) enabling the reproducible recovery of defined glucans independently of the initial dextransucrase activity, which however can affect the produced dextran amounts as described above. The observed decrease in averaged molar masses at pH 6.0 in setup B despite the increase in rms radii (Fig. 3) is actually not clear and subject of further investigations.
Conclusions
Our results give new insights into the pH-dependent release, functionality and product specificity of the dextransucrase expressed by L. hordei. The presented systematic approach allows the controlled production of dextran with varying properties and could be transferred and adapted to other glucan- or fructansucrase expressing microbes.
Supplementary information
Additional file 1. p-values of all relevant and discussed samples. Empty fields refer to p > 0.05.
Acknowledgements
Not applicable.
Abbreviations
- AAB
acetic acid bacteria
- AF4-MALS-UV
asymmetric flow field-flow fractionation coupled with multi-angle laser light scattering and UV detection
- EPS
exopolysaccharide
- GH 70
glycoside hydrolase 70 family
- HPLC
high-performance liquid chromatography
- LAB
lactic acid bacteria
- L.
Lactobacillus
- MWCO
molecular weight cut-off
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Smp
sample
- TMW
Technische Mikrobiologie Weihenstephan
Authors’ contributions
JS, FJ and RFV were involved in planning the experimental setup and in writing the manuscript. JS performed the experimental work, evaluated data and wrote the main text of the manuscript. JB was involved in the verification of the dextransucrase. All authors read and approved the final manuscript.
Funding
Part of this study was supported by the German Federal Ministry for Economics and Energy via the German Federation of Industrial Research (AiF) and the Industrial Association of Food Technology and Packaging (IVLV) and the Wissenschaftsförderung der Deutschen Brauwirtschaft e.V (Wifoe) project numbers AiF 18479 N/1 and AiF 19180 N. Open access publishing was supported by the German Research Foundation (DFG) and the Technical University of Munich (TUM) in the framework of the Open Access Publishing Program.
Availability of data
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable. The manuscript does not contain data collected from humans or animals.
Consent for publication
Not applicable. The manuscript does not contain any individual person’s data.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12934-019-1208-8.
References
- 1.Robyt JF, Kimble BK, Walseth TF. The mechanism of dextransucrase action: direction of dextran biosynthesis. Arch Biochem Biophys. 1974;165(2):634–640. doi: 10.1016/0003-9861(74)90291-4. [DOI] [PubMed] [Google Scholar]
- 2.Sidebotham RL. Dextrans. Adv Carbohydr Chem Biochem. 1974;30:371–444. doi: 10.1016/S0065-2318(08)60268-1. [DOI] [PubMed] [Google Scholar]
- 3.Robyt JF, Taniguchi H. The mechanism of dextransucrase action: biosynthesis of branch linkages by acceptor reactions with dextran. Arch Biochem Biophys. 1976;174(1):129–135. doi: 10.1016/0003-9861(76)90331-3. [DOI] [PubMed] [Google Scholar]
- 4.De Vuyst L, Degeest B. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol Rev. 1999;23(2):153–177. doi: 10.1111/j.1574-6976.1999.tb00395.x. [DOI] [PubMed] [Google Scholar]
- 5.Monsan P, Bozonnet S, Albenne C, Joucla G, Willemot R-M, Remaud-Siméon M. Homopolysaccharides from lactic acid bacteria. Int Dairy J. 2001;11(9):675–685. doi: 10.1016/S0958-6946(01)00113-3. [DOI] [Google Scholar]
- 6.Luzio GA, Mayer RM. The hydrolysis of sucrose by dextransucrase. Carbohydr Res. 1983;111(2):311–318. doi: 10.1016/0008-6215(83)88315-3. [DOI] [Google Scholar]
- 7.Schmid J, Sieber V, Rehm B. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol. 2015;6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naessens M, Cerdobbel A, Soetaert W, Vandamme EJ. Leuconostoc dextransucrase and dextran: production, properties and applications. J Chem Technol Biotechnol. 2005;80(8):845–860. doi: 10.1002/jctb.1322. [DOI] [Google Scholar]
- 9.Cerning J. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol Rev. 1990;7(1–2):113–130. doi: 10.1111/j.1574-6968.1990.tb04883.x. [DOI] [PubMed] [Google Scholar]
- 10.Doco T, Wieruszeski J-M, Fournet B, Carcano D, Ramos P, Loones A. Structure of an exocellular polysaccharide produced by Streptococcus thermophilus. Carbohydr Res. 1990;198(2):313–321. doi: 10.1016/0008-6215(90)84301-A. [DOI] [PubMed] [Google Scholar]
- 11.Reeves PR, Hobbs M, Valvano MA, Skurnik M, Whitfield C, Coplin D, et al. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4(12):495–503. doi: 10.1016/S0966-842X(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 12.Han YW. Microbial levan. Adv Appl Microbiol. 1990;35:171–194. doi: 10.1016/S0065-2164(08)70244-2. [DOI] [PubMed] [Google Scholar]
- 13.van Geel-Schutten GH, Flesch F, ten Brink B, Smith MR, Dijkhuizen LJAM. Biotechnology. Screening and characterization of Lactobacillus strains producing large amounts of exopolysaccharides. Appl Microbiol Biotechnol. 1998;50(6):697–703. doi: 10.1007/s002530051353. [DOI] [Google Scholar]
- 14.Sutherland IW. Microbial exopolysaccharides. Trends Biochem Sci. 1979;4(3):55–59. doi: 10.1016/0968-0004(79)90262-7. [DOI] [Google Scholar]
- 15.Baker P, Hill PJ, Snarr BD, Alnabelseya N, Pestrak MJ, Lee MJ, et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv. 2016;2(5):e1501632. doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cérantola S, Marty N, Montrozier H. Structural studies of the acidic exopolysaccharide produced by a mucoid strain of Burkholderia cepacia, isolated from cystic fibrosis. Carbohydr Res. 1996;285:59–67. doi: 10.1016/0008-6215(96)00014-6. [DOI] [PubMed] [Google Scholar]
- 17.Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol. 2008;10(6):1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 18.Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192(12):3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fels L, Jakob F, Vogel RF, Wefers D. Structural characterization of the exopolysaccharides from water kefir. Carbohydr Polym. 2018;189:296–303. doi: 10.1016/j.carbpol.2018.02.037. [DOI] [PubMed] [Google Scholar]
- 20.Prechtl RM, Wefers D, Jakob F, Vogel RF. Cold and salt stress modulate amount, molecular and macromolecular structure of a Lactobacillus sakei dextran. Food Hydrocolloids. 2018;82:73–81. doi: 10.1016/j.foodhyd.2018.04.003. [DOI] [Google Scholar]
- 21.Kim D, Robyt JF, Lee S-Y, Lee J-H, Kim Y-M. Dextran molecular size and degree of branching as a function of sucrose concentration, pH, and temperature of reaction of Leuconostoc mesenteroides B-512FMCM dextransucrase. Carbohydr Res. 2003;338(11):1183–1189. doi: 10.1016/S0008-6215(03)00148-4. [DOI] [PubMed] [Google Scholar]
- 22.Otts DR, Day DF. Dextransucrase secretion in Leuconostoc mesenteroides depends on the presence of a transmembrane proton gradient. J Bacteriol. 1988;170(11):5006–5011. doi: 10.1128/jb.170.11.5006-5011.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ua-Arak T, Jakob F, Vogel RF. Fermentation pH modulates the size distributions and functional properties of Gluconobacter albidus TMW 2.1191 levan. Front Microbiol. 2017;8:807. doi: 10.3389/fmicb.2017.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bechtner J, Wefers D, Schmid J, Vogel RF, Jakob F. Identification and comparison of two closely related dextransucrases released by water kefir borne Lactobacillus hordei TMW 1.1822 and Lactobacillus nagelii TMW 1.1827. Microbiology. 2019;165(9):956–966. doi: 10.1099/mic.0.000825. [DOI] [PubMed] [Google Scholar]
- 25.Xu D, Fels L, Wefers D, Behr J, Jakob F, Vogel RF. Lactobacillus hordei dextrans induce Saccharomyces cerevisiae aggregation and network formation on hydrophilic surfaces. Int J Biol Macromol. 2018;115:236–242. doi: 10.1016/j.ijbiomac.2018.04.068. [DOI] [PubMed] [Google Scholar]
- 26.Xu D, Bechtner J, Behr J, Eisenbach L, Geißler AJ, Vogel RF. Lifestyle of Lactobacillus hordei isolated from water kefir based on genomic, proteomic and physiological characterization. Int J Food Microbiol. 2019;290:141–149. doi: 10.1016/j.ijfoodmicro.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 27.De Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of Lactobacilli. J Appl Bacteriol. 1960;23(1):130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 28.McIlvaine TC. A buffer solution for colorimetric comparison. J Biol Chem. 1921;49(1):183–186. [Google Scholar]
- 29.Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8(2):93–99. doi: 10.1002/elps.1150080203. [DOI] [Google Scholar]
- 30.Yuryev V, Tomasik P, Bertoft E. Starch: achievements in understanding of structure and functionality. Hauppauge: Nova Science Publishers; 2007. pp. 1–315. [Google Scholar]
- 31.Gunderson JJ, Giddings JC. 14—field-flow fractionation. In: Allen G, Bevington JC, editors. Comprehensive polymer science and supplements. Amsterdam: Pergamon; 1989. pp. 279–291. [Google Scholar]
- 32.Wyatt PJ. Light scattering and the absolute characterization of macromolecules. Anal Chim Acta. 1993;272(1):1–40. doi: 10.1016/0003-2670(93)80373-S. [DOI] [Google Scholar]
- 33.Robyt JF. Mechanisms in the glucansucrase synthesis of polysaccharides and oligosaccharides from sucrose. Adv Carbohydr Chem Biochem. 1995;51:133–168. doi: 10.1016/S0065-2318(08)60193-6. [DOI] [PubMed] [Google Scholar]
- 34.Dols M, Remaud-Simeon M, Willemot RM, Vignon M, Monsan P. Characterization of the different dextransucrase activities excreted in glucose, fructose, or sucrose medium by Leuconostoc mesenteroides NRRL B-1299. Appl Environ Microbiol. 1998;64(4):1298–1302. doi: 10.1128/aem.64.4.1298-1302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Côté GL, Robyt JF. Isolation and partial characterization of an extracellular glucansucrase from Leuconostoc mesenteroides NRRL B-1355 that synthesizes an alternating (1 → 6), (1 → 3)-α-d-glucan. Carbohydr Res. 1982;101(1):57–74. doi: 10.1016/S0008-6215(00)80795-8. [DOI] [PubMed] [Google Scholar]
- 36.Prechtl RM, Janßen D, Behr J, Ludwig C, Küster B, Vogel RF, et al. Sucrose-induced proteomic response and carbohydrate utilization of Lactobacillus sakei TMW 1.411 during dextran formation. Front Microbiol. 2018;9:2796. doi: 10.3389/fmicb.2018.02796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiqui NN, Aman A, Qader SAU. Mutational analysis and characterization of dextran synthesizing enzyme from wild and mutant strain of Leuconostoc mesenteroides. Carbohydr Polym. 2013;91(1):209–216. doi: 10.1016/j.carbpol.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Rühmkorf C, Bork C, Mischnick P, Rübsam H, Becker T, Vogel RF. Identification of Lactobacillus curvatus TMW 1.624 dextransucrase and comparative characterization with Lactobacillus reuteri TMW 1.106 and Lactobacillus animalis TMW 1.971 dextransucrases. Food Microbiol. 2013;34(1):52–61. doi: 10.1016/j.fm.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Korakli M, Rossmann A, Gänzle MG, Vogel RF. Sucrose metabolism and exopolysaccharide production in wheat and rye sourdoughs by Lactobacillus sanfranciscensis. J Agric Food Chem. 2001;49(11):5194–5200. doi: 10.1021/jf0102517. [DOI] [PubMed] [Google Scholar]
- 40.Torino MI, Font de Valdez G, Mozzi F. Biopolymers from lactic acid bacteria. Novel applications in foods and beverages. Front Microbiol. 2015;6:834. doi: 10.3389/fmicb.2015.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notararigo S, Nácher-Vázquez M, Ibarburu I, Werning ML, de Palencia PF, Dueñas MT, et al. Comparative analysis of production and purification of homo- and hetero-polysaccharides produced by lactic acid bacteria. Carbohydr Polym. 2013;93(1):57–64. doi: 10.1016/j.carbpol.2012.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. p-values of all relevant and discussed samples. Empty fields refer to p > 0.05.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.