Abstract
Salmonella enterica represents an enterobacterial species including numerous serovars that cause infections at, or initiated at, the intestinal epithelium. Many serovars also act as facultative intracellular pathogens with a tropism for phagocytic cells. These bacteria not only survive in phagocytes but also undergo de facto replication therein. Phagocytes, through the activities of phagocyte NADPH-dependent oxidase and inducible nitric oxide synthase, are very proficient in converting molecular oxygen to reactive oxygen (ROS) and nitrogen species (RNS). These compounds represent highly efficient effectors of the innate immune defense. Salmonella is by no means resistant to these effectors, which may stand in contrast to the host niches chosen. To cope with this paradox, these bacteria rely on an array of detoxification and repair systems. Combination these systems allows for a high enough tolerance to ROS and RNS to enable establishment of infection. In addition, salmonella possesses protein factors that have the potential to dampen the infection-associated inflammation, which evidently results in a reduced exposure to ROS and RNS. This review attempts to summarize the activities and strategies by which salmonella tries to cope with ROS and RNS and how the bacterium can make use of these innate defense factors.
Keywords: Reactive oxygen species, Reactive nitrogen species, Salmonella enterica, Phagocyte
Reactive Oxygen Species
Reactive oxygen species (ROS) are commonly present in various habitats occupied by living organisms. ROS are formed in the path of abiotic processes but also by living cells themselves, for example through photosynthesis and the activity of the respiratory electron transport chain in mitochondria and bacterial cytoplasmic membranes [1, 2]. As such, ROS cause damage to most, if not all, biomolecules [3, 4], including oxidation of amino acids, vitamins, lipids, nucleotides, and DNA, with damage to the later promoting mutations [5]. Indeed, deliberate production of ROS appears as a very ancient host strategy for coping with pathogens [6, 7, 8, 9, 10, 11] and for acquisition of nutrients [12]. Upon contact with microbes, excessive production of ROS might also become detrimental to the host itself. For example, ROS contribute to endotoxemic shock [13, 14], whereas Jurkat T cells undergo necroptosis upon contact with pathogenic amoebae as a result of a strong ROS response [15]. Likewise, the nematode Caenorhabditis elegans is killed during infection with S. Typhimurium through a massive ROS response filling the whole nematode, almost resembling a primordial septic shock [16].
Even pathogens themselves might produce ROS to their own disadvantage. Several classes of bacteriocidal antibiotics, including cell wall synthesis inhibitors, have been suggested to induce ROS production in bacteria via triggering of the tricarboxylic acid cycle, and ultimately hyperactivation of the electron transport chain with concomitant ROS production [17]. Concomitantly it has been proposed that wall synthesis inhibitors, for example, in part mediate their antibacterial effect through endogenous ROS production. Intestinal bacteria will inevitably be exposed to bile. Interestingly bile induces production of ROS and a genetic ROS response signature in salmonella [18]. Host antibacterial peptidoglycan recognition proteins, an additional group of effectors in our innate immunity barrier, also induce ROS stress in target bacteria, which likely contribute to the antibacterial activity of peptidoglycan recognition proteins [19].
From the host side, be it a mammal or a plant, the NADPH-dependent oxidases (in mammals NADPH-dependent phagocyte oxidase [Phox]) act as a major source of superoxide [6]. In this molecular oxygen (O2) is converted into superoxide anions (O2–) which may subsequently decompose into hydrogen peroxide (H2O2), hydroxyl radicals (HO•), and eventually water [3]. Also, phagocyte inducible nitric oxide synthase (iNOS) produce large amounts of the reactive nitrogen species (RNS) nitric oxide (NO) from L-arginine, NADPH, and molecular oxygen. NO may eventually be converted into peroxynitrite (ONOO–) in the presence of ROS [20]. Apart from causing damage to biomolecules, NO also acts as a biological transmitter causing, among other things, vasodilation, a condition characteristic of septic shock. Thus, production of ROS and RNS may be useful arms in innate antimicrobial defense, but at the same time it is weaponry to be regulated and used with care. Likewise, successful pathogens somehow have to cope with ROS and RNS in order to prevail in a host.
Salmonella enterica and Salmonellosis
Many serovars of Salmonella enterica act as facultative intracellular pathogens that cause intestinal and invasive diseases in humans and animals. The infection is acquired via the oral route, whereafter the bacteria invade the intestinal epithelium. In human typhoid fever, caused by the human-specific serovar Typhi (S. Typhi), the bacteria proceed deeper to infect the liver, spleen, gall bladder, and bone marrow [21]. This form of salmonellosis is also called the typhoidal variant of the disease. Relapses and establishment of persistent carriage, in quite a proportion of the convalescents, are also hallmarks of typhoid fever [22]. Despite modern hygiene and treatment regimens, it has been estimated that there are around 22 million cases of typhoid fever annually [23]. Also, the increasing spread and frequency of multiresistance to antibiotics in S. Typhi is becoming alarming [24].
Serovar Typhimurium (S. Typhimurium) in turn causes a more localized, nontyphoidal and usually self-healing inflammatory intestinal infection in humans. This serovar, being more promiscuous in terms of host range, is capable of causing disease in various animals, including mice. In mice, the infection is invasive and resembles typhoid fever [9, 21, 25]. To cause disease in mice, S. Typhimurium has to survive in macrophages [26], being a cell type highly proficient in generating ROS and RNS. S. Typhimurium is in addition genetically tractable and infects and replicates in professional phagocytic cells. These details have put S. Typhimurium in a key position for sorting out facets of bacterial intracellular parasitism, and factors that adapt bacteria to ROS and RNS. The picture that emerges from such studies reveals an image of salmonella being equipped with an array of enzymes and reducing compounds aimed at detoxifying ROS and repairing ROS-induced damage.
S. Typhimurium is closely related to Escherichia coli and consequently shares with E. coli a relatively large conserved core genome coding for “house-keeping” functions. In addition, S. Typhimurium, as S. Typhi, is equipped with numerous virulence genes often contained in smaller or larger horizontally acquired genetic elements named salmonella pathogenicity islands (SPI). SPI1 and SPI2, respectively, play a key role in allowing salmonella to invade the intestinal epithelium and to replicate in professional phagocytic cells [27, 28, 29]. SPI1 and SPI2 each code for a type III protein secretion system used for translocating so-called effector proteins into the host cell upon bacteria-host contact. The effector proteins as a rule have very specific functions that interfere with central host cell activities, including actin polymerization, signal transduction, and vesicular trafficking [27, 28, 29]. Many serovars, including S. Typhimurium, also possess a virulence plasmid characterized by the spv virulence genes coding for SPI2 effector proteins [29, 30]. Selected SPI1 and SPI2 effector proteins, as well as the invasion process itself, will also affect the inflammatory response and consequently ROS and RNS production.
In contrast to E. coli, S. Typhimurium lacks a capsule, while S. Typhi may express the Vi capsular antigen. However, salmonella expresses a lipopolysaccharide with an O antigen; the latter is an important virulence determinant in protection against complement opsonization and phagocytosis [31]. However, the lipid A portion of the LPS molecule is also an activator of innate immunity responses, including induction of iNOS expression [32] and enforcement of the oxidative burst [33]. In addition, salmonella is capable of expressing multiple adhesins that in concert contribute to infection of the mouse [34] and to biofilm formation that adds to the persistent carriage state [35, 36].
Salmonella and ROS
Be it the typhoidal or the nontyphoidal form of salmonellosis, already at the intestinal epithelium S. enterica is recognized by pattern recognition molecules and confronted with professional phagocytic cells, eventually resulting in exposure to ROS and RNS [8, 21]. Both ROS and RNS create a central barrier against salmonellosis in the mouse model. Mice not capable of producing an oxidative burst (phox–/– mice) quickly succumb to challenge doses of S. Typhimurium otherwise coped with by corresponding phox+/+ mice [9]. Likewise, humans suffering from chronic granulomatous disease due to a lack of functional phagocyte oxidase show increased sensitivity to invasive, sometimes unorthodox, forms of salmonellosis caused by nontyphoidal servoriants [37, 38, 39].
Detoxification of ROS
The superoxide anion generated by the phagocyte NADPH oxidase is charged at neutral pH and thus does not readily diffuse through lipid membranes [40]. However, as such, superoxide could still cause oxidative damage in the periplasmic space [41]. In response, salmonella has periplasmic superoxide dismutases capable of degrading superoxide [42]. Of these, the periplasmic SodCI is needed for virulence of S. Typhimurium in phox-proficient mice [43, 44, 45] but not in phox-deficient mice, further pointing to the role of superoxide in restricting proliferation of S. Typhimurium [43]. The importance of SodCI in this context is also highlighted by the fact that the corresponding gene (sodCI) becomes upregulated even when S. Typhimurium replicates in nonactivated murine monocytic cells [45, 46] and the fact that sodCI is comprised of the salmonella PhoP/PhoQ virulence regulon [47] that also includes the SPI1, SPI2, and the spv genes.
S. Typhimurim replicates in an endosomal compartment that is estimated to become moderately low in pH [46, 48]. At these acidities the superoxide anion may become protonated and the superoxide start to diffuse into the bacterial cytoplasm [40]. To cope with protonated superoxide diffusing through the cytoplasmic membrane, the bacterium also codes for 2 cytoplasmic superoxide dismutases, i.e., SodA and SodB. An sodA mutant shows moderately decreased survival in murine monocyte-like cells and a slight attenuation in mice [49].
During in vitro oxidative or bile stress, S. Typhimurium upregulates the sitABCD and mntH manganese transport systems [50]. This likely escalates import of Mn++ to suppor SodA (an enzyme needing Mn++), leading to an accompanying enhanced superoxide degradation. Indeed, Mn++ uptake promotes S. Typhimurim survival in the inflamed gut in a mouse model for enterocolitis [51].
Hydrogen peroxide produced by the superoxide dismutases, and any hydrogen peroxide in the close vicinity of salmonella, diffuses more readily through lipid bilayers and is possibly even transported though aquaporins into the cytosol [52]. Nevertheless, this ROS species is met by an array of cytoplasmic enzymes degrading hydrogen peroxide to water and molecular oxygen. These enzymes include 3 catalases (i.e., KatE, KatG, and KatN), and 3 peroxidases (i.e., AhpC, Tpx, and TsaA) [53, 54], yet only an S. Typhimurium mutant simultaneously lacking all 3 catalases as well as ahpC and tsaA has shown hydrogen peroxide sensitization and a replication defect in mice, bone marrow-derived murine macrophages, and murine monocytes like RAW264.7 cells [53]. However, genetic complementation with katG or tsaA alone restored hydrogen peroxide tolerance and replication in murine RAW264.7 monocytic cells. This points to a high degree of redundancy with regard to the capacity to degrade hydrogen peroxide [55].
While not essential for S. Typhimurium intracellular replication, Tpx alone does promote hydrogen peroxide tolerance and increases the intracellular replication propensity in phagocytes [54]. However, this contribution of Tpx to intracellular replication was seen only in IFN-γ activated cells. The use of a Phox inhibitor abrogated the need for Tpx for intracellular replication, indeed pointing a role of Tpx in protecting against ROS.
A twist to detoxification of hydrogen peroxide comes from the observation that the ABC-type efflux pump MacAB adds to hydrogen peroxide tolerance in S. Typhimurium and promotes intracellular replication in murine monocyte like J774 cells [56]. Also, MacAB promotes replication of S. Typhimurium in the liver of infected mice. In J774 cells not capable of mounting a respiratory burst, MacAB does not add to intracellular fitness. At first glance one would expect the efflux pump to export hydrogen peroxide from the bacterial cytoplasm. However, intriguingly, a macAB mutant revealed a markedly reduced capacity to degrade hydrogen peroxide in vitro, implicating a role of MacAB in degradation rather than in efflux of hydrogen peroxide.
Thiol Chemistry
The periplasmic space poses a special interest with regard to salmonella oxidative stress tolerance, as protein disulfide formation of gram-negative bacteria is conducted in this compartment. This is achieved with the aid of Dsb proteins using Cys-X-X-Cys motifs that undergo oxidation-reduction cycles in forming and breaking disulfide bonds [57, 58]. In this way DsbA acts as a somewhat unspecific oxidoreductase, primarily creating disulfide bonds, DsbB and DsbD act as cytoplasmic membrane electron donors, and DsbC conducts “proof- reading” of disulfide bond formation. In addition, the S. Typhimurium chromosome codes for the DsbL and DsbI proteins, and these are paralogues for, respective ly, DsbA and DsbB [59]. Furthermore, S. Typhimurium also codes for the ScsABCD proteins containing Cys-X-X-Cys motifs and for the SrgA disulfide oxidoreductase. ScsB is a homologue of DsbD and has the capacity to reduce ScsC [60], while SrgA assists in formation of the plasmid-encoded Pef-fimbriae [61]. The S. Typhimurium periplasmic disulfide oxidases DsbA and SrgA also participate in assembly of the virulence-associated SPI2 protein secretion system [62], while motility relies on DsbA [63].
In the case of oxidative stress one could expect the occurrence of nonenzymatically oxidized thiols and concomitantly an increase in wrongly matched disulfide bridges in periplasmic proteins. As the disulfide oxidases act through disulfide bond formation, the SPI2, Pef, and flagellar supramolecules could be indirect targets of an oxidative attack, e.g., through effects on DsbA and SrgA. Also, exposure to peptidoglycan recognition proteins generates thiol stress in S. Typhimurium, contributing to the afore mentioned antibacterial effect of these proteins [19].
That said, surprisingly, mutational inactivation of DsbC in S. Typhimurium does not come with major in vitro sensitization to oxidative substances or NO donors [64]. Also, a dsbC deletion mutant does not exhibit any apparent attenuation in virulence in BALB/c mice. This could be explained by the presence of the several additional proteins mentioned above that could, or do, take part in disulfide bond formation in the periplasm. Thus, there seems to be redundancy in S. Typhimurium with regard to periplasmic (oxido)reductases. That said, when the scsABCD genes were deleted in a dsbC proficient background, the mutant not only remained virulent but also showed enhanced replication in murine monocyte-like RAW264.7 cells [65]. A rational explanation for this would be that the Dsb system(s) supporting SPI2 assembly competes with the Scs system for redox equivalents, thus contributing to a more efficient SPI2 activity in the absence of the Scs system. Still, S. Typhimurium lacking ScsB becomes sensitized to copper chloride [65]. That copper chloride acts as a disulfide catalyst in vitro [66], and de facto conducts disulfide formation of periplasmic proteins in E. coli [67], points to a role of the Scs system in restoring wrong disulfide formation upon oxidative stress.
Apart from housing catalases and peroxidases, the bacterial cytoplasm includes the highly reducing enzyme thioredoxin 1 (TrxA). TrxA also operates through a Cys-X-X-Cys motif and assists the Dsb system and ribonucleotide reductase [68]. Still, a trxA mutant of S. Typhimurium mutant did not reveal any obvious in vitro sensitization to oxidative compounds or NO donors [64]. Even in a very poor medium the tolerance for NO donors was the same for the wild type and a trxA mutant. This might appear somewhat surprising as ribonucleotide reductase generates dideoxynucleotides through a tyrosine-associated radical mechanism inhibited by NO [69, 70]. However, the trxA the mutant showed a severe replication defect in cultured phagocytic cells and mice due to an inability to translocate virulence-associated SPI2 effector proteins [64].
In part the apparent redundancy of cytoplasmic catalases, peroxidases, and dismutases could be explained by the strong reductant glutathione (contained in mM concentrations in a reduced form in the cytoplasm). Glutathione acts through oxidation of its own thiol group to form an oxidized dimer. Genetic depletion of glutathione synthesis in S. Typhimurium caused marked in vitro sensitization to paraquat and hydrogen peroxide but only when the bacteria were grown in medium mimicking the intravacuolar compartment for salmonella (low pH, poor in nutrients and magnesium [64]). At first glance one would expect the cytoplasm to be a niche protected by glutathione from ROS. However, E. coli also possesses a CydDC transport system that shuffles glutathione into the periplasmic space [71], implying that glutathione may add to oxidoprotection in the periplasm as well. S. Typhimurium contains the cyd homologues, which become upregulated under oxidative stress, albeit not as strongly as many canonical oxidoprotectant genes [50].
Like ROS, the reducing gas hydrogen sulfide (H2S) is commonly present in biotic habitats, and it is also produced by many bacterial species. Hydrogen sulfide has also been proposed to act as a general protectant against various classes of antibiotics mechanistically through oxidoprotective mechanisms [72]. A classical diagnostic parameter for S. Typhimurium in the microbiological laboratory is its ability to produce large amounts of hydrogen sulfide [73]. Thus, it would not be surprising if S. Typhimurium also applies hydrogen sulfide when coping with ROS. Indeed, the genes for thiosulfate reductase, CysIJ involved in hydrogen sulfide production, become upregulated upon in vitro hypochlorite stress [74] and peroxide stress [50], as well as under in vitro conditions that mimic the environment of the salmonella-containing intracellular vacuole [50].
Damage Repair
Two additional classes of reductases have been identified as adding tolerance to ROS and virulence in S. Typhimurium through reduction of oxidized sulfur groups. In this biotin sulfoxide reductase converts biothine sulfoxide back to biothine. Biotin sulfoxide reductase adds to hydrogen peroxide tolerance in S. Typhimurium, as well as to the ability to replicate in murine monocytic cells [75]. Likewise, methionine sulfoxide reductase MsrA, that generates methionine from methionine sulfoxide, improves hydrogen peroxide tolerance and increases the fitness of S. Typhimurium in IFN-γ-activated murine monocytic cells, as well as in mice [76].
Apart from oxidizing sulfhydryl and sulfur groups, ROS also causes the conversion of aspartate to iso-aspartate, a reaction reverted by isoaspartate methyl transferase. In S. Typhimurium the gene for this enzyme, i.e., pimt, is needed for full tolerance to ROS and for growth in IFN-γ-activated peritoneal murine macrophages [77]. Inhibition of Phox by apocynin decreased the need for pimt. This would support the notion that the role of pimt indeed originates from coping with Phox-generated ROS.
The effect of ROS on nucleic acids is dual. First, nucleic acids act as a target for ROS-induced damage causing strand breaks, mutagenesis, and modification of nucleotide bases [3, 78, 79]. Indeed, components of the DNA repair machinery, such as recA, lexA, and sulA appear to be of high importance for virulence and ROS tolerance in S. Typhimurium [79, 80, 81, 82]. In S. Typhimurium, the lack of RecA, a protein needed for DNA repair and induction of the SOS response, results in substantial sensitization to ROS and a strong attenuation with regard to virulence in mice [80]. Likewise, bacterial RNA has been implicated as a main target for ROS [78]. In this, the ribonuclease polynucleotide phosphorylase has been proposed to protect E. coli against hydrogen peroxide through degradation of oxidized RNA. However, an S. Typhiumurium mutant lacking functional polynucleotide phosphorylase does not reveal increased sensitization to hydrogen peroxide (unpubl. res.). However, the S. Typhimurium polynucleotide phosphorylase participates in the regulation of SPI1 and SPI2 gene expression [83] and could thus indirectly contribute to ROS adaptation.
A second line of effects caused by ROS on DNA is at the gene regulatory level. E. coli possess redox-sensing transcriptional regulators, such as OxyR, SoxR, and SoxS, that regulate, for example, the expression of the ahpC, ahpF, katG, and sodA genes, as well as genes involved in DNA repair and methionine synthesis [84]. In this, OxyR acts through the formation of an intermolecular Cys-X-X-Cys cysteine bridge to sense oxidation. Transcriptomic profiling of S. Typhimurium shows that basically the homologues of the whole E. coli OxyR/SoxR/SoxS regulon, in terms of upregulated genes, are also induced in S. Typhimurium upon oxidative shock [50]. Nevertheless, genetic depletion of OxyR in S. Typhimurium does not affect the ability of the mutant to survive in neutrophils [85], while the Sox regulon seems to be required for S. Typhimurium tolerance to paraquat [82].
In E. coli, dps codes for a DNA-binding ferritin-like protein that becomes highly abundant in the stationary phase [86]. Dps binds DNA and confers increased resistance to oxidative stress [87, 88, 89], possibly by physically protecting DNA from ROS-induced damage. Upon oxidative and nitrosative stress S. Typhimurium strongly upregulates the expression of its dps [50]. The gene is also needed for S. Typhimurium survival in primary murine macrophages and for virulence in mice [89]. Apart from a possible role in directly protecting DNA, Dps could also add to oxidoprotection through scavenging of iron, thus preventing Fenton reactions.
Hypochlorite is an ROS produced by neutrophils in response to infection, and a compound found in many disinfectants. E. coli possess a transcriptional regulator, i.e., YjiE, that responds to hypochlorite and confers hypochlorite tolerance [90]. A homologue for yjiE is present in S. Typhimurium and yijE expression becomes activated upon oxidative stress [50], implying that S. Typhimurium may also have a hypochlorite-protection regulon.
Nitric Oxide
As implied above, when salmonella encounters professional phagocytes these cells may become activated and start producing RNS (NO) with the aid of iNOS and ROS present. Like ROS, RNS may cause a multitude of damages, such as protein nitrosylation or formation of metal complexes. Mice lacking iNOS become sensitized to S. Typhimurium, albeit not to the same extent as phox(–/–) mice [9]. Still, murine macrophages and dendritic cells infected with S. Typhimurium and simultaneously treated with iNOS inhibitors lose their ability to control intracellular replication of the bacteria [46, 91]. All of this implies that salmonella should have measures to cope with RNS. Indeed, an S. Typhimurium mutant lacking sodCI, apart from being sensitized to superoxide, becomes highly sensitized to a mixture of superoxide and an NO donor [42]. A rational explanation for this could be that SodCI prevents the formation of peroxynitrite through degradation of superoxide radicals.
Flavohemoglobins belongs to the hemoglobin superfamily and consists of 2 domains, i.e., a FAD-binding oxidoreductase domain and a heme-containing domain. E. coli and S. Typhimurium both encode for the flavohemoglobin Hmp. It protects against NO by oxidizing NO into nitrate under aerobic conditions [92, 93, 94, 95]. Hmp is also needed for the survival of S. Typhimurium in human macrophages [94] and the hmp gene becomes induced as S. Typhimurium replicates in murine macrophages [46].
E. coli furthermore possesses the NorRVW (previously YgaAKD) NO protection system [96]. This system converts NO into nitrous oxide (N2O) at lower oxygen tensions. The corresponding genes are present in S. Typhimurium, with norV and norW being strongly induced for expression upon in vitro-induced NO stress [50].
Intriguingly, De Groote et al. [97] noted that one could increase NO tolerance in S. Typhimurium by deleting the genes for the stress tolerance sigma factor RpoS or the Dpp dipeptide transport system. The mechanism(s) behind these observations remains to be sorted out, but one possible explanation is that the sigma factor and peptide transport systems would strongly distort fitness if nitrosylated by RNS. While RpoS activates katE expression, expression of rpoS itself is not induced upon either peroxide or NO stress [50].
Preventing ROS and RNS Production
While invasion of S. Typhimurium of epithelial cells results in an inflammatory response trough SPI effector proteins, i.e., LPS and flagellin, selected effector proteins also possess a potential anti-inflammatory activity. For example, the SPI1 effector protein AvrA (from avirulence protein A) prevents NF-kB nuclear translocation [98]. This would prevent the expression of several cytokines and iNOS in phagocytes. Likewise, several SPI2 effector proteins, such as SpvC, SseL, and SspH1, pose functions potentially downregulating inflammatory activation of the infected host cells [29], with, for example, SpvC dephosphorylating phosphor-threonine from phospho-ERK [99]. S. Typhi, the human-adapted serovar that causes typhoid fever, codes for a cytolethal-distending toxin (CDT) not present in S. Typhimurium. The toxin acts by being a nuclease. Surprisingly though, when the S. Typhi CDT is implanted in S. Typhimurium the intestinal inflammation score is highly reduced in orally infected mice [100]. Expression of CDT also causes downregulation of the expression of cytokines and iNOS in intestinal epithelial tissue. This implies that salmonella through its CDT dampen intestinal inflammation and likely thereby exposure of salmonella to ROS and RNS.
It is also possible to isolate phagocyte-adapted mutants of S. Typhimurium that downregulate NO production with an accompanying increased intracellular growth capacity [91]. The mechanism(s) remains to be solved, but for all such mutants isolated the effect appeared to be on iNOS activity rather than on iNOS expression. As iNOS relies on L-arginine for NO production, one tentative mechanism would be to, one way or another, deplete the phagocyte from L-arginine.
Biofilm Formation
Biofilms can be defined as microbial multicellular communities embedded in a macromolecular mass produced by a single or different microbial species therein [36, 101, 102]. The benefits of living in such communities are probably many, but from a perspective of clinical microbiology one could imagine the following: adherence to abiotic and biotic surfaces, the ability to resist host immune defense, and an increased tolerance to antimicrobial compounds [103].
Salmonella is capable of forming biofilms on gall bladder stones, which is believed to promote establishment of carriage [35], despite constant exposure to bile and hence ROS stress as stipulated above. S. Typhimurium biofilm formation is characterized by a decreased expression of motility (planktonic mode), accompanied by the expression biofilm extracellular matrix components such as amyloid curli fimbriae and cellulose fibers (sessile mode) [36]. In conjunction with the second messenger cyclic-di-GMP-associated gene regulatory network, the CsgD gene regulator plays a key role in regulation of biofilm formation. Several observations connect biofilm formation with oxidative stress. As S. Typhimurium enters biofilm formation, apart from upregulating csgD it also upregulates genes associated with ROS stress [104]. In parallel, hypochlorite stress induces in S. Typhimurium a transcriptomic signature suggestive of an adaptation towards sessile biofilm formation [74]. Growth of S. Typhimurium on the oxidative surface of a redox-active conducting polymer also enhanced biofilm formation [105]. This could all imply that, at least under some redox stress conditions, S. Typhimurium prefers to shift from the planktonic to the sessile biofilm mode to adapt to ROS. This would be consistent with the induction of biofilm formation in, for example, Campylobacter jejuni [106] and Staphylococcus aureus [107] as a response to oxidative stress.
Biofilm formation also connects to redox in the sense that deleting either dsbA or dsbB results in CsgD-dependent upregulation of biofilm formation in S. Typhimurium [108]. Furthermore, this upregulation relies on the c-di-GMP phosphodiesterase STM3615, thus linking periplasmic protein thiol chemistry to biofilm formation and c-di-GMP.
Making Use of ROS
By expressing pathogen-associated microbial patterns, such as flagellin and LPS, S. Typhimurium is clearly itself responsible for evoking inflammation [32]. Intriguingly, salmonella evidently makes use of an inflammatory response with an accompanying ROS production. For one thing, intestinal inflammation induced by S. Typhimurium infection skews the intestinal microbial flora in mice, such that it favors salmonella colonization [109]. This competitive advantage may be further potentiated in that the inflammation-associated ROS production creates tetrathionate, a sulfur oxanion, in the intestine from preexisting sulfur compounds [110]. Tetrathionate in turn can be used as a respiratory electron sink by S. Typhimurium [110, 111], thus in principle allowing S. Typhimurium to take advantage of the oxidative potential of ROS for its own respiration in an otherwise anaerobic or hypoxic environment.
S. Typhimurium can also sense oxidative/nitrosative stress with the aim of inducing virulence. The virulence-associated SPI2 response regulator SsrB becomes S-nitrosylated at Cys 203 upon NO stress [112]. While it did not affect the expression of selected SPI2 genes, a mutant with an ssrRB allele lacking the critical SsrB Cys residue showed decreased fitness in a murine infection model. This could suggest that ssrB regulates virulence factor genes outside of SPI2 but in response to RNS.
Intriguingly, S. Typhimurium seems to be able to sense the neurotransmitter adrenaline followed by induction of sodA [113]. The connection may appear farfetched, but adrenaline, like selected other neurotransmitters, participates in regulation of inflammatory responses [114]. Thus, while acting more to dampen inflammation, adrenaline could still be used by S. Typhimurium to sense an inflammatory environment, potentially being enriched in ROS.
Thus, ROS production clearly is a double-edged sword from the perspective of both salmonella and its host. ROS is obviously needed for protection against salmonellosis, as evidenced by infection experiments with phox–/– mice and by the increased prevalence of invasive salmonellosis in patients suffering from chronic granulomatous disease. S. Typhimurium in turn translocates SPI1 and SPI2 effector proteins during the infection which have the potential to dampen an inflammatory response [96, 97], possibly to prevent too early a clearance, yet host-derived ROS and RNS adds to its in vivo replication potential. Furthermore, the so-called “typhoid” CDT-like toxin dampened intestinal inflammation in a mouse infection model [100]. Thus, salmonella seems to try to balance the host response (Fig. 1) to allow a certain degree of ROS response to increase its fitness and to allow induction of virulence genes.
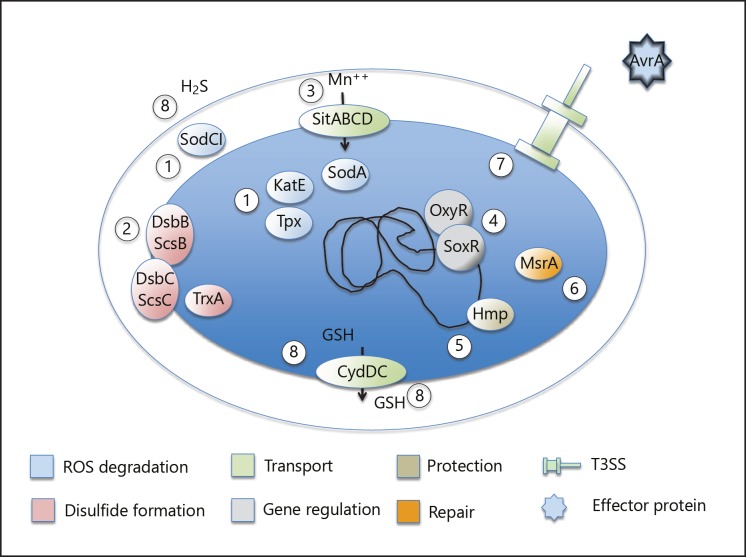
Fig. 1.
Summary of strategies by which S. Typhimurium copes with oxidative stress: (1) degradation of ROS before they act on target molecules, as exemplified by superoxide dismutases (SodA and SodCI), catalase (KatE), and thiol peroxidase (Tpx); (2) balancing periplasmic disulfide bond formation, as exemplified by the thioredoxin, Dsb, and Scs systems; (3) transport of cofactors for detoxifying enzymes; (4) redox-sensing gene regulatory systems lodging on, e.g., superoxide disumutase genes; (5) direct protective or iron-scavenging systems (Hmp) or (6) repairing enzymes (methionine sulfoxide reductase MsrA); (7) application of secreted effector proteins such as AvrA with the potential to downregulate an inflammatory response; and (8) production and transport of small-molecular reducing compounds to appropriate locations.
Disclosure Statement
The author declares no commercial interest or conflict of interest.
Acknowledgement
This work was supported by Vetenskapsrådet (the Swedish Research Council) grant Dnr 4-30 16-2013 (M.R.) and by the Umeå Centre for Microbial Research (UCMR) Linnaeus Program (grant Dnr 349-2007-8673 from Vetenskapsrådet).
References
- 1.Aluri HS, Simpson DC, Allegood JC, Hu Y, Szczepanek K, Gronert S, et al. Electron flow into cytochrome c coupled with reactive oxygen species from the electron transport chain converts cytochrome c to a cardiolipin peroxidase: role during ischemia-reperfusion. Biochim Biophys Acta. 2014 Nov;1840((11)):3199–207. doi: 10.1016/j.bbagen.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frick K, Schulte M, Friedrich T. Reactive oxygen species production by Escherichia coli respiratory complex I. Biochemistry. 2015 May;54((18)):2799–801. doi: 10.1021/acs.biochem.5b00160. [DOI] [PubMed] [Google Scholar]
- 3.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57((1)):395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999 Apr;2((2)):188–94. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 5.Sakai A, Nakanishi M, Yoshiyama K, Maki H. Impact of reactive oxygen species on spontaneous mutagenesis in Escherichia coli. Genes Cells. 2006 Jul;11((7)):767–78. doi: 10.1111/j.1365-2443.2006.00982.x. [DOI] [PubMed] [Google Scholar]
- 6.Glian'ko AK, Ishchenko AA. [Structural and functional characteristics of plant NADPH oxidase: a review] Prikl Biokhim Mikrobiol. 2010 Sep-Oct;46((5)):509–18. [PubMed] [Google Scholar]
- 7.Kerchev PI, Fenton B, Foyer CH, Hancock RD. Plant responses to insect herbivory: interactions between photosynthesis, reactive oxygen species and hormonal signalling pathways. Plant Cell Environ. 2012 Feb;35((2)):441–53. doi: 10.1111/j.1365-3040.2011.02399.x. [DOI] [PubMed] [Google Scholar]
- 8.Mastroeni P, Vazquez-Torres A, Fang FC, Xu Y, Khan S, Hormaeche CE, et al. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J Exp Med. 2000 Jul;192((2)):237–48. doi: 10.1084/jem.192.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mastroeni P, Grant AJ. Spread of Salmonella enterica in the body during systemic infection: unravelling host and pathogen determinants. Expert Rev Mol Med. 2011 Apr;13:e12. doi: 10.1017/S1462399411001840. [DOI] [PubMed] [Google Scholar]
- 10.Quie PG, White JG, Holmes B, Good RA. In vitro bactericidal capacity of human polymorphonuclear leukocytes: diminished activity in chronic granulomatous disease of childhood. J Clin Invest. 1967 Apr;46((4)):668–79. doi: 10.1172/JCI105568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales J, Kadota Y, Zipfel C, Molina A, Torres MA. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J Exp Bot. 2016 Mar;67((6)):1663–76. doi: 10.1093/jxb/erv558. [DOI] [PubMed] [Google Scholar]
- 12.Cohen R, Jensen KA, Houtman CJ, Hammel KE. Significant levels of extracellular reactive oxygen species produced by brown rot basidiomycetes on cellulose. FEBS Lett. 2002 Nov;531((3)):483–8. doi: 10.1016/s0014-5793(02)03589-5. [DOI] [PubMed] [Google Scholar]
- 13.Cauwels A, Rogge E, Janssen B, Brouckaert P. Reactive oxygen species and small-conductance calcium-dependent potassium channels are key mediators of inflammation-induced hypotension and shock. J Mol Med (Berl) 2010 Sep;88((9)):921–30. doi: 10.1007/s00109-010-0633-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattanaik U, Prasad K. Reactive oxygen species and endotoxic shock: effect of dimethylthiourea. J Cardiovasc Pharmacol Ther. 2001 Jul;6((3)):273–85. doi: 10.1177/107424840100600308. [DOI] [PubMed] [Google Scholar]
- 15.Song KJ, Jang YS, Lee YA, Kim KA, Lee SK, Shin MH. Reactive oxygen species-dependent necroptosis in Jurkat T cells induced by pathogenic free-living Naegleria fowleri. Parasite Immunol. 2011 Jul;33((7)):390–400. doi: 10.1111/j.1365-3024.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- 16.Sem X, Rhen M. Pathogenicity of Salmonella enterica in Caenorhabditis elegans relies on disseminated oxidative stress in the infected host. PLoS One. 2012;7((9)):e45417. doi: 10.1371/journal.pone.0045417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Acker H, Coenye T. Role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017 Jun;25((6)):456–66. doi: 10.1016/j.tim.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Walawalkar YD, Vaidya Y, Nayak V. Response of Salmonella Typhi to bile-generated oxidative stress: implication of quorum sensing and persister cell populations. Pathog Dis. 2016 Nov;74((8)):ftw090. doi: 10.1093/femspd/ftw090. [DOI] [PubMed] [Google Scholar]
- 19.Kashyap DR, Rompca A, Gaballa A, Helmann JD, Chan J, Chang CJ, et al. Peptidoglycan recognition proteins kill bacteria by inducing oxidative, thiol, and metal stress. PLoS Pathog. 2014 Jul;10((7)):e1004280. doi: 10.1371/journal.ppat.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Y, Zweier JL. Superoxide and peroxynitrite generation from inducible nitric oxide synthase in macrophages. Proc Natl Acad Sci USA. 1997 Jun;94((13)):6954–8. doi: 10.1073/pnas.94.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004 Sep;2((9)):747–65. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 22.Barrett FC, Knudsen JD, Johansen IS. Cases of typhoid fever in Copenhagen region: a retrospective study of presentation and relapse. BMC Res Notes. 2013 Aug;6((1)):315. doi: 10.1186/1756-0500-6-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization Initiative for vaccine research. http://who.int/vaccine_research/diseases/diarrhoeal/index7.html. [Google Scholar]
- 24.Threlfall EJ, Fisher IS, Berghold C, Gerner-Smidt P, Tschäpe H, Cormican M, et al. Trends in antimicrobial drug resistance in Salmonella enterica serotypes Typhi and Paratyphi A isolated in Europe, 1999-2001. Int J Antimicrob Agents. 2003 Nov;22((5)):487–91. doi: 10.1016/s0924-8579(03)00262-0. [DOI] [PubMed] [Google Scholar]
- 25.Broz P, Ohlson MB, Monack DM. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes. 2012 Mar-Apr;3((2)):62–70. doi: 10.4161/gmic.19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986 Jul;83((14)):5189–93. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrbar K, Mirold S, Friebel A, Stender S, Hardt WD. Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium. Int J Med Microbiol. 2002 Feb;291((6-7)):479–85. doi: 10.1078/1438-4221-00156. [DOI] [PubMed] [Google Scholar]
- 28.Kuhle V, Hensel M. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol Life Sci. 2004 Nov;61((22)):2812–26. doi: 10.1007/s00018-004-4248-z. [DOI] [PubMed] [Google Scholar]
- 29.Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 2012 May;158((Pt 5)):1147–61. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 30.Gulig PA, Danbara H, Guiney DG, Lax AJ, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993 Mar;7((6)):825–30. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 31.Mäkelä PH, Hovi M, Saxén H, Muotiala A, Rhen M. Role of LPS in the pathogenesis of salmonellosis. In: Nowotny A, Spitznagel JJ, Ziegler EJ, editors. Endotoxin research. Elsevier Science Publishers Biomedical Division; 1990. pp. pp. 537–46. [Google Scholar]
- 32.Khan SA, Everest P, Servos S, Foxwell N, Zähringer U, Brade H, et al. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998 Jul;29((2)):571–9. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen GT, Gree ER, Mecsas J. Neutrophils to the rescue: mechanisms of NADPH oxidase activation and bacterial resistance. Front. Cell. Infect. Microbiol. 2017 doi: 10.3389/fcimb.2017.00373. http://frontiersin.org/articles/10.3389/fcimb.2017.00373/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Velden AW, Bäumler AJ, Tsolis RM, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998 Jun;66((6)):2803–8. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simm R, Ahmad I, Rhen M, Le Guyon S, Römling U. Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 2014;9((11)):1261–82. doi: 10.2217/fmb.14.88. [DOI] [PubMed] [Google Scholar]
- 36.Crawford RW, Rosales-Reyes R, Ramírez-Aguilar ML, Chapa-Azuela O, Alpuche-Aranda C, Gunn JS. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci USA. 2010 Mar;107((9)):4353–8. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burniat W, Toppet M, De Mol P. Acute and recurrent salmonella infections in three children with chronic granulomatous disease. J Infect. 1980 Sep;2((3)):263–8. doi: 10.1016/s0163-4453(80)90746-x. [DOI] [PubMed] [Google Scholar]
- 38.Finoccio A, Claps A, Serafinelli J, Salfa S, Longo D, Di Matteo D, et al. Chronic granulomatous disease with Salmonella brain abcesses. J Pediatr Infect Dis. 2014;33((5)):525–8. doi: 10.1097/INF.0000000000000270. [DOI] [PubMed] [Google Scholar]
- 39.Mouy R, Fischer A, Vilmer E, Seger R, Griscelli C. Incidence, severity, and prevention of infections in chronic granulomatous disease. J Pediatr. 1989 Apr;114((4 Pt 1)):555–60. doi: 10.1016/s0022-3476(89)80693-6. [DOI] [PubMed] [Google Scholar]
- 40.Korshunov SS, Imlay JA. A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of Gram-negative bacteria. Mol Microbiol. 2002 Jan;43((1)):95–106. doi: 10.1046/j.1365-2958.2002.02719.x. [DOI] [PubMed] [Google Scholar]
- 41.Craig M, Slauch JM. Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One. 2009;4((3)):e4975. doi: 10.1371/journal.pone.0004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Groote MA, Ochsner UA, Shiloh MU, Nathan C, McCord JM, Dinauer MC, et al. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997 Dec;94((25)):13997–4001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999 Jul;33((1)):167–76. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 44.Uzzau S, Bossi L, Figueroa-Bossi N. Differential accumulation of Salmonella[Cu, Zn] superoxide dismutases SodCI and SodCII in intracellular bacteria: correlation with their relative contribution to pathogenicity. Mol Microbiol. 2002 Oct;46((1)):147–56. doi: 10.1046/j.1365-2958.2002.03145.x. [DOI] [PubMed] [Google Scholar]
- 45.Krishnakumar R, Craig M, Imlay JA, Slauch JM. Differences in enzymatic properties allow SodCI but not SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium strain 14028. J Bacteriol. 2004 Aug;186((16)):5230–8. doi: 10.1128/JB.186.16.5230-5238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003 Jan;47((1)):103–18. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 47.Golubeva YA, Slauch JM. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutase SodCI is a member of the PhoPQ regulon and is induced in macrophages. J Bacteriol. 2006 Nov;188((22)):7853–61. doi: 10.1128/JB.00706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Audia JP, Webb CC, Foster JW. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int J Med Microbiol. 2001 May;291((2)):97–106. doi: 10.1078/1438-4221-00106. [DOI] [PubMed] [Google Scholar]
- 49.Tsolis RM, Bäumler AJ, Heffron F. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect Immun. 1995 May;63((5)):1739–44. doi: 10.1128/iai.63.5.1739-1744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013 Dec;14((6)):683–95. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Diaz-Ochoa VE, Lam D, Lee CS, Klaus S, Behnsen J, Liu JZ, et al. Salmonella mitigates oxidative stress and thrives in the inflamed gut by evading calprotectin-mediated manganese sequestrion. Cell Host Microbe. 2016 Jun;19((6)):814–25. doi: 10.1016/j.chom.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bienert GP, Schjoerring JK, Jahn TP. Membrane transport of hydrogen peroxide. Biochim Biophys Acta. 2006 Aug;1758((8)):994–1003. doi: 10.1016/j.bbamem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 53.Hébrard M, Viala JP, Méresse S, Barras F, Aussel L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol. 2009 Jul;191((14)):4605–14. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horst SA, Jaeger T, Denkel LA, Rouf SF, Rhen M, Bange FC. Thiol peroxidase protects Salmonella enterica from hydrogen peroxide stress in vitro and facilitates intracellular growth. J Bacteriol. 2010 Jun;192((11)):2929–32. doi: 10.1128/JB.01652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aussel L, Zhao W, Hébrard M, Guilhon AA, Viala JP, Henri S, et al. Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst. Mol Microbiol. 2011 May;80((3)):628–40. doi: 10.1111/j.1365-2958.2011.07611.x. [DOI] [PubMed] [Google Scholar]
- 56.Bogomolnaya LM, Andrews KD, Talamantes M, Maple A, Ragoza Y, Vazquez-Torres A, et al. The ABC-type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress. MBio. 2013 Oct;4((6)):e00630–13. doi: 10.1128/mBio.00630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Åslund F, Beckwith J. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J Bacteriol. 1999 Mar;181((5)):1375–9. doi: 10.1128/jb.181.5.1375-1379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denoncin K, Collet J-F. Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antiox. Red. Sign. 2013 doi: 10.1089/ars.2012.4864. Doi 10.1089/ars2012.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin D, Kim B, Slauch JM. DsbL and DsbI contribute to periplasmic disulfide bond formation in Salmonella enterica serovar Typhimurium. Microbiology. 2009 Dec;155((Pt 12)):4014–24. doi: 10.1099/mic.0.032904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouwman CW, Kohli M, Killoran A, Touchie GA, Kadner RJ, Martin NL. Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J Bacteriol. 2003 Feb;185((3)):991–1000. doi: 10.1128/JB.185.3.991-1000.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho SH, Parsonage D, Thurston C, Dutton RJ, Poole LB, Collet JF, et al. A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. MBio. 2012 Apr;3((2)):e00291–11. doi: 10.1128/mBio.00291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miki T, Okada N, Danbara H. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J Biol Chem. 2004 Aug;279((33)):34631–42. doi: 10.1074/jbc.M402760200. [DOI] [PubMed] [Google Scholar]
- 63.Turcot I, Ponnampalam TV, Bouwman CW, Martin NL. Isolation and characterization of a chromosomally encoded disulphide oxidoreductase from Salmonella enterica serovar Typhimurium. Can J Microbiol. 2001 Aug;47((8)):711–21. [PubMed] [Google Scholar]
- 64.Bjur E, Eriksson-Ygberg S, Åslund F, Rhen M. Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2006 Sep;74((9)):5140–51. doi: 10.1128/IAI.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anwar N, Sem XH, Rhen M. Oxidoreductases that act as conditional virulence suppressors in Salmonella enterica serovar Typhimurium. PLoS One. 2013 Jun;8((6)):e64948. doi: 10.1371/journal.pone.0064948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williamson MB. Catalysis of formation of mixed disulfides between cystine and beta-globulins by copper ions. Biochem Biophys Res Commun. 1970 May;39((3)):379–83. doi: 10.1016/0006-291x(70)90587-5. [DOI] [PubMed] [Google Scholar]
- 67.Hiniker A, Collet JF, Bardwell JC. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem. 2005 Oct;280((40)):33785–91. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- 68.Brignole EJ, Ando N, Zimanyi CM, Drennan CL. The prototypic class Ia ribonucleotide reductase from Escherichia coli: still surprising after all these years. Biochem Soc Trans. 2012 Jun;40((3)):523–30. doi: 10.1042/BST20120081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lepoivre M, Fieschi F, Coves J, Thelander L, Fontecave M. Inactivation of ribonucleotide reductase by nitric oxide. Biochem Biophys Res Commun. 1991 Aug;179((1)):442–8. doi: 10.1016/0006-291x(91)91390-x. [DOI] [PubMed] [Google Scholar]
- 70.Lepoivre M, Flaman J-M, Bobe P, Lemaire G, Henry Y. Quenching of the tyrosyl free radical of ribonucleotide reductase by nitric oxide. 1994;269:21891–21897. [PubMed] [Google Scholar]
- 71.Pittman MS, Robinson HC, Poole RK. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J Biol Chem. 2005 Sep;280((37)):32254–61. doi: 10.1074/jbc.M503075200. [DOI] [PubMed] [Google Scholar]
- 72.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011 Nov;334((6058)):986–90. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 73.Kaufmann F. Kopenhagen: Muksgaard; 1961. Die bakteriologie der Salmonella species. [Google Scholar]
- 74.Wang S, Phillippy AM, Deng K, Rui X, Li Z, Tortorello ML, et al. Transcriptomic responses of Salmonella enterica serovars Enteritidis and Typhimurium to chlorine-based oxidative stress. Appl Environ Microbiol. 2010 Aug;76((15)):5013–24. doi: 10.1128/AEM.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denkel LA, Horst SA, Rouf SF, Kitowski V, Böhm OM, Rhen M, et al. Methionine sulfoxide reductases are essential for virulence of Salmonella typhimurium. PLoS One. 2011;6((11)):e26974. doi: 10.1371/journal.pone.0026974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Denkel LA, Rhen M, Bange FC. Biotin sulfoxide reductase contributes to oxidative stress tolerance and virulence in Salmonella enterica serovar Typhimurium. Microbiology. 2013 Jul;159((Pt 7)):1447–58. doi: 10.1099/mic.0.067256-0. [DOI] [PubMed] [Google Scholar]
- 77.Kumawat M, Pesingi PK, Agarwal RK, Goswami TK, Mahawar M. Contribution of protein isoaspartate methyl transferase (PIMT) in the survival of Salmonella Typhimurium under oxidative stress and virulence. Int J Med Microbiol. 2016 Jun;306((4)):222–30. doi: 10.1016/j.ijmm.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 78.Wu J, Jiang Z, Liu M, Gong X, Wu S, Burns CM, et al. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry. 2009 Mar;48((9)):2012–20. doi: 10.1021/bi801752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu M, Gong X, Alluri RK, Wu J, Sablo T, Li Z. Characterization of RNA damage under oxidative stress in Escherichia coli. Biol Chem. 2012 Mar;393((3)):123–32. doi: 10.1515/hsz-2011-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buchmeier NA, Libby SJ, Xu Y, Loewen PC, Switala J, Guiney DG, et al. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Invest. 1995 Mar;95((3)):1047–53. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Humphrey S. NacVicar T, Stevenson A, Roberts M, Humphrey TJ, Jepson MA. SulA-induced filamentation in Salmonella enterica serovar Typhimurium: effects on SPI-1 expression and epithelial invasion. J Appl Microbiol. 2011;111:185–96. doi: 10.1111/j.1365-2672.2011.05022.x. [DOI] [PubMed] [Google Scholar]
- 82.Pomposiello PJ, Demple B. Identification of SoxS-regulated genes in Salmonella enterica serovar typhimurium. J Bacteriol. 2000 Jan;182((1)):23–9. doi: 10.1128/jb.182.1.23-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clements MO, Eriksson S, Thompson A, Lucchini S, Hinton JC, Normark S, et al. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc Natl Acad Sci USA. 2002 Jun;99((13)):8784–9. doi: 10.1073/pnas.132047099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seo SW, Kim D, Szubin R, Palsson BO. Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Reports. 2015 Aug;12((8)):1289–99. doi: 10.1016/j.celrep.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 85.Papp-Szabò E, Firtel M, Josephy PD. Comparison of the sensitivities of Salmonella typhimurium oxyR and katG mutants to killing by human neutrophils. Infect Immun. 1994 Jul;62((7)):2662–8. doi: 10.1128/iai.62.7.2662-2668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Almirón M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992 Dec;6((12b 12B)):2646–54. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 87.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999 Oct;181((20)):6361–70. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol. 2002 Feb;43((3)):771–82. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- 89.Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun. 2004 Feb;72((2)):1155–8. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gebendorfer KM, Drazic A, Le Y, Gundlach J, Bepperling A, Kastenmüller A, et al. Identification of a hypochlorite-specific transcription factor from Escherichia coli. J Biol Chem. 2012 Feb;287((9)):6892–903. doi: 10.1074/jbc.M111.287219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eriksson S, Björkman J, Borg S, Syk A, Pettersson S, Andersson DI, et al. Salmonella typhimurium mutants that downregulate phagocyte nitric oxide production. Cell Microbiol. 2000 Jun;2((3)):239–50. doi: 10.1046/j.1462-5822.2000.00051.x. [DOI] [PubMed] [Google Scholar]
- 92.Hausladen A, Gow A, Stamler JS. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc Natl Acad Sci USA. 2001 Aug;98((18)):10108–12. doi: 10.1073/pnas.181199698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bang IS, Liu L, Vazquez-Torres A, Crouch ML, Stamler JS, Fang FC. Maintenance of nitric oxide and redox homeostasis by the salmonella flavohemoglobin hmp. J Biol Chem. 2006 Sep;281((38)):28039–47. doi: 10.1074/jbc.M605174200. [DOI] [PubMed] [Google Scholar]
- 94.Stevanin TM, Poole RK, Demoncheaux EA, Read RC. Flavohemoglobin Hmp protects Salmonella enterica serovar typhimurium from nitric oxide-related killing by human macrophages. Infect Immun. 2002 Aug;70((8)):4399–405. doi: 10.1128/IAI.70.8.4399-4405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crawford MJ, Goldberg DE. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem. 1998 May;273((20)):12543–7. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- 96.Gardner AM, Gardner PR. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J Biol Chem. 2002 Mar;277((10)):8166–71. doi: 10.1074/jbc.M110470200. [DOI] [PubMed] [Google Scholar]
- 97.De Groote MA, Granger D, Xu Y, Campbell G, Prince R, Fang FC. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc Natl Acad Sci USA. 1995 Jul;92((14)):6399–403. doi: 10.1073/pnas.92.14.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Collier-Hyams LS, Zeng H, Sun J, Tomlinson AD, Qin Bao Z, Chen H, et al. Salmonella AvrA effector protein inhibits key inflammatory, anti-apoptotic NF-kB pathway. J Immunol. 2002;169:2846–50. doi: 10.4049/jimmunol.169.6.2846. [DOI] [PubMed] [Google Scholar]
- 99.Mazurkiewicz P, Thomas J, Thompson JA, Liu M, Arbibe L, Sansonetti P, et al. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol Microbiol. 2008 Mar;67((6)):1371–83. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Del Bel Belluz L, Guidi R, Pateras IS, Levi L, Mihaljevic B, Rouf SF, et al. The typhoid toxin promotes host survival and the establishment of a persistent asymptomatic infection. PLoS Pathog. 2016 Apr;12((4)):e1005528. doi: 10.1371/journal.ppat.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shaw RK, Lasa I, García BM, Pallen MJ, Hinton JC, Berger CN, et al. Cellulose mediates attachment of Salmonella enterica Serovar Typhimurium to tomatoes. Environ Microbiol Rep. 2011 Oct;3((5)):569–73. doi: 10.1111/j.1758-2229.2011.00263.x. [DOI] [PubMed] [Google Scholar]
- 102.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016 Aug;14((9)):563–75. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 103.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010 Apr;35((4)):322–32. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 104.White AP, Weljie AM, Apel D, Zhang P, Shaykhutdinov R, Vogel HJ, et al. A global metabolic shift is linked to Salmonella multicellular development. PLoS One. 2010 Jul;5((7)):e11814. doi: 10.1371/journal.pone.0011814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gomez-Carretero S, Libberton B, Rhen M, Richter-Dahlfors A. Redox-active conducting polymers modulate Salmonella biofilm formation by controlling availability of electron acceptors. NPJ Biofilms Microbiomes. 2017 Sep;3((1)):19. doi: 10.1038/s41522-017-0027-0. 10.1038/s41522.017-0027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oh E, Kim JC, Jeon B. Stimulation of biofilm formation by oxidative stress in Campylobacter jejuni under aerobic conditions. Virulence. 2016 Oct;7((7)):846–51. doi: 10.1080/21505594.2016.1197471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kulkarni R, Antala S, Wang A, Amaral FE, Rampersaud R, Larussa SJ, et al. Cigarette smoke increases Staphylococcus aureus biofilm formation via oxidative stress. Infect Immun. 2012 Nov;80((11)):3804–11. doi: 10.1128/IAI.00689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anwar N, Rouf SF, Römling U, Rhen M. Modulation of biofilm-formation in Salmonella enterica serovar Typhimurium by the periplasmic DsbA/DsbB oxidoreductase system requires the GGDEF-EAL domain protein STM3615. PLoS One. 2014 Aug;9((8)):e106095. doi: 10.1371/journal.pone.0106095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007 Oct;5((10)):2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010 Sep;467((7314)):426–9. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Winter SE, Bäumler AJ. A breaking feat. Gut Microbes. 2011;2((1)):58–60. doi: 10.4161/gmic.2.1.14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Husain M, Jones-Carson J, Song M, McCollister BD, Bourret TJ, Vázquez-Torres A. Redox sensor SsrB Cys203 enhances Salmonella fitness against nitric oxide generated in the host immune response to oral infection. Proc Natl Acad Sci USA. 2010 Aug;107((32)):14396–401. doi: 10.1073/pnas.1005299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Karavolos MH, Spencer H, Bulmer DM, Thompson A, Winzer K, Williams P, et al. Adrenaline modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC Genomics. 2008 Oct;9((1)):458. doi: 10.1186/1471-2164-9-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tracey KJ. The inflammatory reflex. Nature. 2002 Dec;420((6917)):853–9. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]