Abstract
Cell-to-cell signaling via small molecules is an essential process to coordinate behavior in single species within a community, and also across kingdoms. In this review, we discuss the quorum sensing (QS) systems used by the opportunistic pathogen Pseudomonas aeruginosa to sense bacterial population density and fitness, and regulate virulence, biofilm development, metabolite acquisition, and mammalian host defense. We also focus on the role of N-acylhomoserine lactone-dependent QS signaling in the modulation of innate immune responses connected together via calcium signaling, homeostasis, mitochondrial and cytoskeletal dynamics, and governing transcriptional and proteomic responses of host cells. A future perspective emphasizes the need for multidisciplinary efforts to bring current knowledge of QS into a more detailed understanding of the communication between bacteria and host, as well as into strategies to prevent and treat P. aeruginosa infections and reduce the rate of antibiotic resistance.
Keywords: Host-pathogen interactions, Bacteria, Cell-to-cell signaling, Quorum sensing, Pseudomonas aeruginosa, N-acyl homoserine lactones, Biofilms, Innate immunity, Mucosal surfaces, Proteogenomics
Introduction
Quorum sensing (QS) is a cell-to-cell communication that allows bacteria to recognize the population density by producing and sensing small diffusible signaling molecules. This form of bacterial intercellular signaling coordinates gene regulation and controls numerous cooperative behaviors, including biofilm formation, virulence traits, metabolic demands, and host-microbe interactions [1]. Multicellular eukaryotic organisms have coexisted with bacteria for approximately 2 billion years under evolutionary pressure and both represent remarkable examples of adaptive evolution. Our body meets numerous potential pathogens daily, and during the first critical hours and days, the outcome of infections and development of disease depend on the properties of both bacterial community and our innate immune system [2]. Epithelial surface linings of the mucosa, e.g., in the gut and lung, provide physical and immune barriers between the host and pathogens and external environment. Tight junctions (TJ) and adherens junctions (AJ) are specialized transmembrane protein complexes located between neighboring epithelial cells and associated with the cytoskeleton and regulatory proteins [3]. The apical surface of the epithelial cells in the intestine and lung is equipped with two types of projections, microvilli versus cilia, and covered with a mucus layer made of glycoproteins that protects against mechanical, chemical, and microbial agents. The lumen of the gut and the lung is also largely inhospitable for microbes because of the presence of antimicrobial factors, such as lysozyme, α-defensin, and lactoferrin. Moreover, macrophages and neutrophils are activated by bacterial products (e.g., cell wall components, formylated peptides, flagellin) or immune stimuli (e.g., complement components, cytokines, antibodies) can quickly phagocyte and kill bacterial pathogens at the early onset of infection, and dendritic cells in mucosa activate the adaptive immune response, including T and B cells [3]. Using QS communication, bacteria can coordinate their social behavior, influencing host cell activities in a noninvasive manner. The early events in the communication between bacteria and host cells may happen even before the bacteria bind to and enter host cells and then spread further. Research over the past 2 decades has greatly increased our understanding of how bacterial QS communication orchestrate cooperative behaviors of bacteria during host-microbe interactive coexistence. These advances open up new frontiers of the sociomicrobiology research field aiming to find new strategies for the prevention and treatment of bacterial infections.
Pseudomonas aeruginosa
Pseudomonas aeruginosa is a social, nonfermentative opportunistic Gram-negative bacterium that inhabits diverse environments. Normally considered to be commensals on the host body, bacteria can establish themselves as opportunistic pathogens. Being highly adaptable, invasive, toxigenic, and able to colonize various surfaces and tissues, P. aeruginosa can cause severe nosocomial outbreaks and also threaten local and systemic infections in patients with compromised underlying health conditions, particularly in those with ventilator-associated pneumonia, burn wounds, cystic fibrosis, and bloodstream infections [4]. As other members in a large group of so-called ESKAPE pathogens (E nterococcus faecium,S taphylococcus aureus,K lebsiella pneumonia,A cinetobacter baumannii,P seudomonas aeruginosa, and E nterobacter species), it is capable of escaping from the action of multiple drugs and represents a new paradigm of a “superbug” in pathogenesis, transmission, and antibiotic resistance [5]. Therefore, traditional therapeutic options for P. aeruginosa have become limited and finding novel alternative prevention and treatment strategies is an urgent priority.
QS Signaling Networks in P. aeruginosa
Concept of QS
The general concept of QS is that small-signal molecules broadcast the information about cell density in the bacterial population and allow collective coordinated production of costly extracellular items. This gives beneficial cooperation with each other, behavior as a powerful multicellular community, and performance of tasks, which would be impossible for single cells [1]. Sophisticated QS networks exist in both Gram-negative and Gram-positive bacteria, and they communicate via many different circuits and various signal molecules. Despite the diversity of QS mechanisms, there are common items among these networks. Many of the QS-controlled social factors and cooperative behaviors are conserved and include biofilm formation, virulence traits, and metabolic demands [6].
P. aeruginosa QS Systems
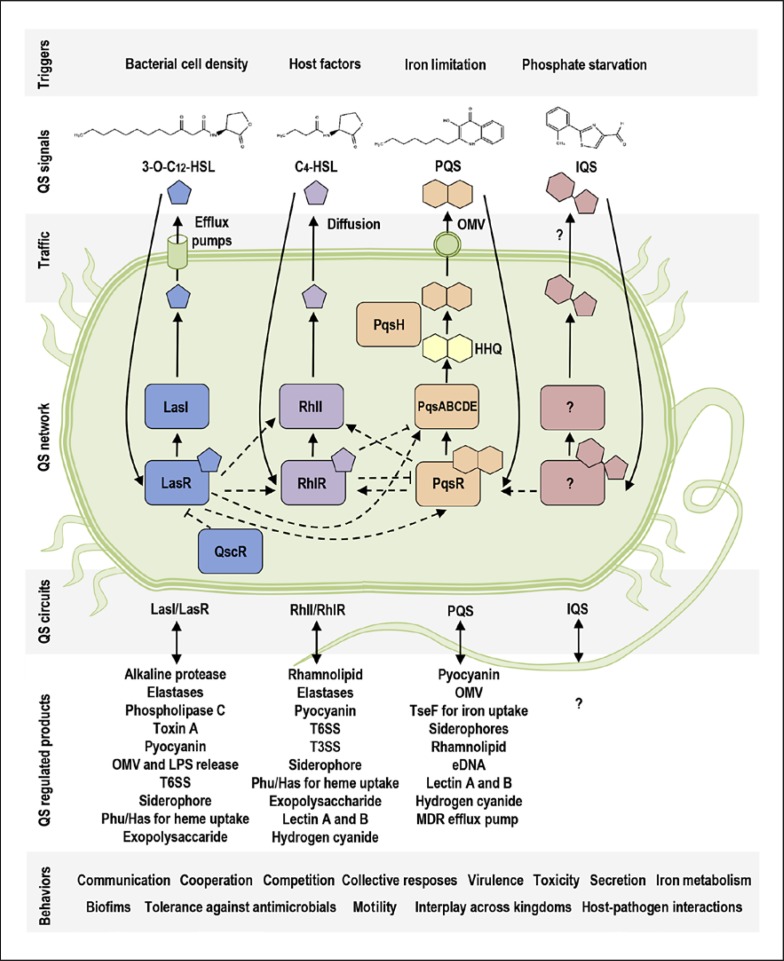
P. aeruginosa harbors one of the most complex QS systems (Fig. 1), equipped with at least four distinct but deeply intertwined and subordinated circuits [6]. There are two N-acylhomoserine lactone (AHL) circuits, LasI/LasR and RhlI/RhlR; both are homologs of LuxI/LuxR type, and both are activated by an increased cell density within a bacterial population (Fig. 1). Each AHL-dependent system is composed of a LuxI-type synthase and a LuxR-type receptor [6]. LasI produces a molecule, N-3-oxo-dodecanoyl-L-homoserine lactone (3O-C12-HSL), which is detected by the cytoplasmic receptor LasR. RhlI produces N-butyryl-L-homoserine lactone (C4-HSL), which is recognized by the cytoplasmic receptor RhlR. LasR and RhlR are transcriptional regulators and together control the activation of more than 300 genes in the P. aeruginosa genome [7]. In addition, there is a 3O-C12-HSL-binding receptor QscR, a LuxR homolog which not only controls its own set of genes but also inactivates LasR and RhlR and thereby represses many Las- and Rhl-dependent genes resulting in prevention of QS responses before the bacteria reach a quorum in a population [8].
Fig. 1.
The QS system in P. aeruginosa. The scheme illustrates (in different colors) four main circuits of the QS system: AHL-dependent circuits, Las and Rhl, which are interconnected to the PQS circuit, and IQS. They can be triggered by certain stimuli, such as bacterial cell density or iron limitation, etc. Chemical structures are shown for four main QS signal molecules, 3-O-C12-HSL, C4-HSL, PQS, and IQS. Furthermore, the scheme shows QS-regulated products and cooperative behaviors.
An additional level of complexity is added by the third QS circuit, the P. aeruginosa quinolone signal (PQS) system (Fig. 1), which is interconnected to the AHL-dependent signaling and can be triggered by iron limitation within bacterial population [9]. Here, PqsABCDE produces the precursor 2-heptyl-4-quinolone (HHQ), and PqsH catalyzes conversion of HHQ to 2-heptyl-3-hydroxy-4-quinolone (PQS), detected by the receptor PqsR [10]. Phosphate starvation can activate the production of yet another QS molecule, 2-(2-hydroxyphenyl)-thiasole-4-carbaldehyde, involved in the integrated QS (IQS) system, which in turn activates the expression of pqs genes [11]. It has been proposed that the IQS signal molecule is synthesized by AmbBCDE [11], but recent results imply that IQS is a byproduct in the synthesis of siderophore pyochelin [12]. The bacterial receptor for IQS still remains elusive. In addition, in biofilm communities, bacteria can communicate using long-range electrical signals via ion channels [13]. Host factors can crosstalk with microbiota via QS [14] and trigger AHL- and PQS-dependent QS communication (Fig. 1), for example neurotransmitter serotonin, estrogen steroid, and stress hormones [15, 16].
The profile of QS molecules formed by P. aeruginosa when growing in vitro as a planktonic culture is dominated by a high concentration of C4-HSL (30 µM), an intermediate amount of PQS-family molecules and 3O-C12-HSL (1.5–10 µM), and smaller concentrations of other AHL-family members (0.1–1 µM), which differ in the length of the fatty acid chain from C4 to C14 [17]. In biofilms growing in vitro, the concentration of QS signals can be much higher, for example, 3-O-C12-HSL accumulates at up to 300–600 µM [18]. The functional QS system has a high impact on the pathogenesis of bacteria. Thus, in experimental models of P. aeruginosa infections in mice, such as burn wounds and pneumonia, bacterial strains containing mutations or deletions in one or several of the QS genes were obviously less virulent than wild-type bacteria [19, 20, 21]. These observations have led to further studies with focus on fundamental insights into the molecular basis of QS, bacterial pathogenesis, host-pathogen interactions, and novel strategies to combat bacterial infections. The nature of QS communication was also assessed using chemically synthetized small molecules that are structurally and functionally identical to natural AHL and PQS existing in P. aeruginosa cultures. These were used both in in vitro and in vivo studies in a variety of models and systems (Fig. 2), and mainly at around 1- to 300-µM concentrations [20, 22, 23, 24], thereby relating to different scenarios of communication between microorganisms and hosts in environments inhabited by planktonic bacteria and in biofilms. Chemical tools were designed and applied to find out and visualize human targets for QS molecules (Fig. 3, 4) and study the effects and pathways of QS signaling on host cells [25]. Novel inhibitors and activators of QS receptors and synthases were developed and included different classes, such as agonists, antagonists, partial agonists, and those that mimic native QS compounds, many of which have been reviewed recently [26]. Indeed, QS blockers can target certain points of QS circuits and thereby inhibit bacterial virulence. It is proposed that resistance to QS inhibitors will develop slowly [27], making them promising antivirulence agents and potential alternatives to traditional antibiotics [28, 29].
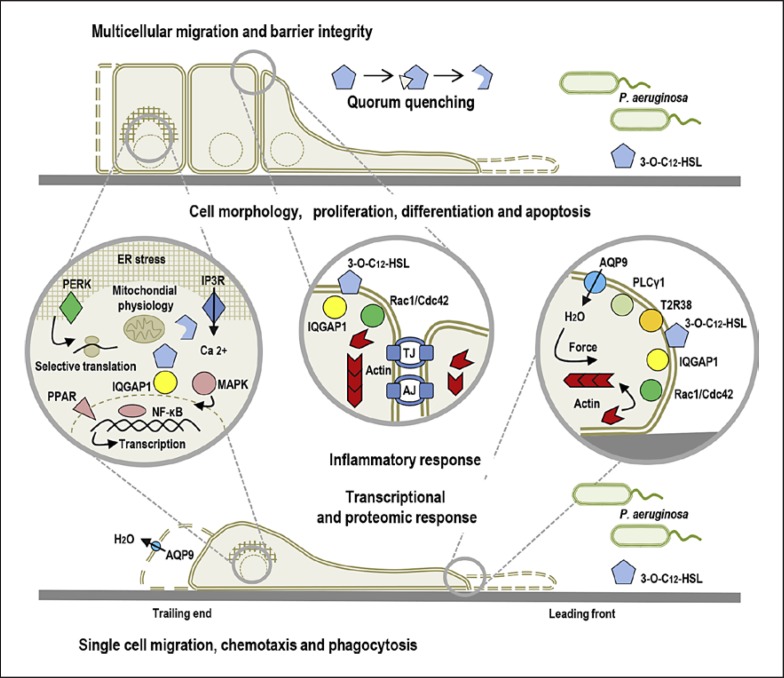
Fig. 2.
QS communication during host-microbe interaction. The scheme illustrates the impact of P. aeruginosa 3O-C12-HSL on the mammalian host at cellular and molecular levels. The model also shows that 3O-C12-HSL can associate and target several membrane-associated proteins, e.g., scaffold IQGAP1, sensory T2R38, and nuclear PPAR, which promote interaction with signaling cascades and cellular processes. These are connected together by calcium signaling, homeostasis, and mitochondrial and cytoskeletal dynamics, for example, and govern transcriptional and proteomic responses.
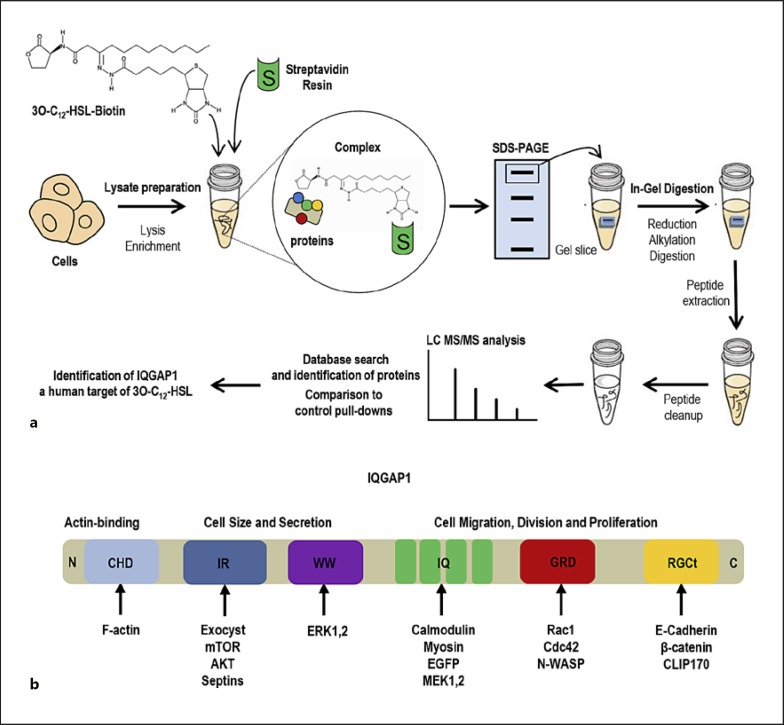
Fig. 3.
IQGAP1 as a human target of 3O-C12-HSL. a A pull-down approach with a biotin-conjugated 3O-C12-HSL probe and mass spectrometry-based proteomic analysis used to identify the target IQGAP1. For more details on methods, refer to Karlsson et al. [25]. b Schematic structure of IQGAP1 with several domains that interact with many different proteins and thereby regulate diverse cytoskeletal reorganization and cell signaling events.
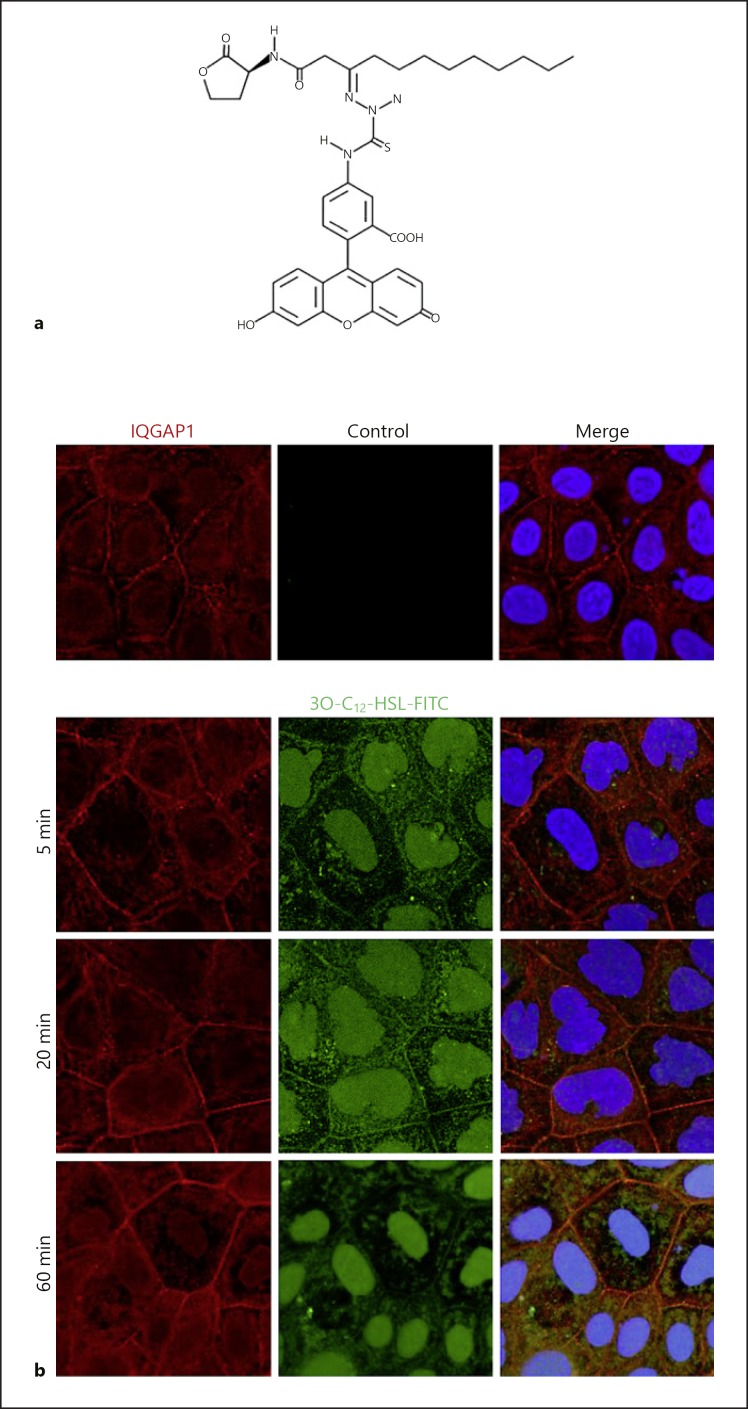
Fig. 4.
Imaging of IQGAP1 and 3O-C12-HSL in human intestinal epithelial cells. a Synthetic fluorescently tagged 3O-C12-HSL-FITC used for imaging. b Caco-2 human intestinal epithelial cells were treated with 1 µM of 3O-C12-HSL-FITC (green) or 0.018% DMSO as a diluent control for 5, 20, or 60 min. Samples were fixed, and then the IQGAP1 was immunolabelled (Atto647N; red) and the nucleus stained with DAPI (blue), as described previously [25]. The samples were visualized by confocal imaging. The image size is 67.6 × 67.6 µm and pixel size is 0.13 µm.
QS-Regulated Factors That Impact P. aeruginosa Communities and Pathogenicity
Motility and Adhesion
P. aeruginosa can exist either as free-swimming planktonic organisms or in biofilm communities associated to surfaces or tissues. Planktonic bacteria differ from those living within the early or confluent biofilm stages in terms of their transcriptome [30] and proteome profiles [31, 32]. When planktonic, P. aeruginosa moves using a single polar flagellum, an essential part of its swimming motility in aqueous environments and its direction is driven by chemotaxis towards or away from the gradient of concentration of environmental stimuli. Bacterial adhesion is mediated by both specific and nonspecific interactions. Nonspecific forces, including hydrophobicity and electrostatic charge, for example, are influenced by the cell surface and milieu composition. Specific interaction and adhesion are mediated by proteins, lipids, and sugars in the cell membrane. Bacteria possess polar type 4 pili, which are the main adhesins but also mediate a flagellum-independent surface translocation, called twitching motility. QS molecules produced by LuxS-family synthase are involved in biogenesis and function of pili and flagella in Salmonella, Vibrio, and Escherichia coli, but not in P. aeruginosa [33, 34]. However, QS system controls social cellular motility in P. aeruginosa, known as swarming, mostly via the rhamnolipid (RHL) production (Fig. 1). Both flagella and pili are potent virulence factors enabling the bacteria to move to and bind to host cells or mucus layers, sense mechanical features of their environment and facilitate the early stage of biofilm development at the surface [35]. Pseudomonas adhesins recognize different components of the host extracellular matrix, i.e., collagen, fibrinogen, elastin, and laminin. Bacterial adhesins can interact with host transmembrane proteins, called cell adhesion molecules, such as integrins, selectins, cadherins, and immunoglobulins for binding and sometimes for cell entry.
Lipopolysaccharide
Adhesion initiates a closer physical contact between tissue surfaces and bacterial traits, such as lipopolysaccharide (LPS), a dominant complex glycolipid of the outer membrane in Gram-negative bacteria, a highly potent activator of the innate immune system and a key player in pathogenesis. The classical LPS molecule has a three-domain structure: the conservative lipid A, the hydrophobic moiety that anchors LPS to the outer membrane; the more variable core oligosaccharide, which together with lipid A maintains the integrity of the outer membrane; and the hypervariable O-antigen polysaccharide, which is a polymer of repeating oligosaccharides connected to the core with direct contact to external milieu [36]. Various modifications in LPS biosynthesis and structure, such as loss of O-antigen and lipid A modifications, occur during adaptation of pathogens to chronic infections in respiratory and gastrointestinal tracts, which results in alteration of host immune responses and promotes bacterial persistence [37]. Typically, LPS is anchored to the bacterial outer membrane via lipid A, but it can also be released from the cells to external milieu during normal growth and AHL-dependent QS system is involved in this release (Fig. 1) [38]. The LPS provides an effective permeability barrier against antibiotics and antimicrobial peptides. The last-resort antibiotic colistin, also known as polymyxin E, binds to LPS with high affinity; LPS modifications lead to the development of antibiotic resistance and changes in bacterial fitness and virulence potential [39]. LPS is able to enhance bacterial cell surface hydrophobicity, and therefore increase P. aeruginosa motility and impact adhesion and biofilm formation [40]. Flagellin and LPS of P. aeruginosa can associate with host pattern recognition receptors, e.g., Toll-like receptors (TLR5 and TLR4) expressed on different host cells, and thereby initiate an inflammatory response via the NF-κB signaling pathway [4].
Enzymes and Exotoxins
P. aeruginosa is able to secrete a large variety of enzymes and exotoxins, which have established roles as virulence factors that damage tissue, provide nutrition supply for the bacteria, increase bacterial survival, actively subvert immune responses, and promote the development of an infection and further spreading of bacteria to underlying tissues and the circulatory system.
In particular, proteases possess proteolytic activity against many different types of substrate in host tissues, e.g., complement components, mucins, fibrin, cytokines, immunoglobulins (Ig), and epithelial junctions [4]. Pseudomonas produces several extracellular proteases that serve for these purposes and are often associated with in vivo and in vitro infections [41, 42]. Three of them – alkaline protease (APR), elastase LasA and LasB – are produced by P. aeruginosa under the control of the Las and Rhl circuits in the QS system (Fig. 1) [43]. APR and elastases are exported via the type 1 and type 2 secretion systems (T1SS and T2SS, respectively), the general secretory mechanism in Gram-negative bacteria [44]. These enzymes are zinc metalloproteases and require calcium for proteolytic activity against various types of substrates and to combat host immune responses. Thus, APR degrades the C2 component of the complement system, preventing its activation via the classical and lectin pathways and thereby inhibiting phagocytosis and bacterial clearance [45]. Similarly, elastases cleave complement components and also elastin, collagen, IgG, mucin, and surfactant proteins to escape from phagocytosis and clearance [46]. Pseudomonas proteases can mimic the activity of neutrophil elastase by cleaving peptides from host thrombin that inhibits inflammatory responses [47].
Three extracellular phospholipases C (PLC), PLC-H, PLC-N, and PLC-B, hydrolyze lipids in host cell membranes, e.g., sphingomyelin and phosphatidylcholine, and pulmonary surfactant. The production of one of them, PLC-B, but not two others, is under control by AHL-dependent QS-system (Fig. 1) [48, 49]. PLC are secreted via twin arginine translocase pathway and T2SS [44]. The activity of this single virulence factor in P. aeruginosa was directly linked with effects on lung function during infection [50]. PLC-B has one more specific function: it is required for the directed twitching motility of P. aeruginosa towards a gradient of certain phospholipids, such as phosphatidylcholine and phosphatidylethanolamine, which are strong chemoattractants for prokaryotes [51, 52].
Pseudomonas exotoxin A (ETA) belongs to the two-component AB family, where the B subunit has cell binding activity and the A subunit has enzymatic activity [53]. ETA is secreted from cytoplasm into the extracellular space through T2SS [54]. Once secreted, ETA specifically binds its B subunit to lipoprotein receptor-related CD91 and gets into the host cell via clathrin-coated regions of the plasma membrane [55]. Once in endosomes, ETA has two pathways open to reach the endoplasmic reticulum (ER): lipid-dependent sorting and the KDEL receptor-mediated pathway, both of which go through the Golgi apparatus [56, 57]. Then, ETA goes from the ER into the cytosol, ADP-ribosylates the eukaryotic elongation factor-2 (eEF-2) on the ribosomes, inactivate it, and thereby block the translocation of mRNA and protein synthesis in the host cell. The regulation of ETA is complex and related to glucose and iron metabolism and the Las/Rhl circuits of the QS system (Fig. 1) [58].
P. aeruginosa secretes a number of virulence factors known as phenazines, and one of them is pyocyanine (PYO). It is an aromatic compound that contributes to the green color of P. aeruginosa cultures. PYO is toxic due its ability to increase intracellular levels of reactive oxygen species, in particular superoxide, which leads to oxidative stress, NAD(P)H depletion and enzyme inhibition, DNA damage, and cell death. Numerous studies have disclosed the importance of PYO in the pathophysiology of P. aeruginosa infection in different tissues and biological systems [59]. All three QS systems, Las, Rhl, and PQS, play a role in the regulation of PYO production (Fig. 1) [48], and a recent study uncovered a higher level of its complexity [60].
Protein Secretion Systems
Secreted proteins and enzymes are large hydrophilic molecules which need to be transported via either spherical 50- to 250-nm outer membrane vesicles (OMV), or assemblies of specialized macromolecular complexes, called protein secretion systems. OMV are multifunctional structures that bleb from the outer membrane of bacteria and typically include proteins, phospholipids, nucleic acids, LPS, QS signals, ions, and metabolites. These extracellular structures are considered as a type zero secretion system (T0SS), an additional and independent traffic system [61]. Six classical types of protein secretion systems have been identified in Pseudomonas, termed T1SS to T6SS, and they are conserved in Gram-negative bacteria. [44]. Both secretion modes, T1SS-T6SS and T0SS, are essential for the interaction between bacteria and their environments, used to deliver virulence factors, and linked to QS signaling in P. aeruginosa (Fig. 1) [44, 62]. Furthermore, PQS in a strong interaction with LPS is required for OMV formation [63] and the Las circuit controls the release of OMV in extracellular milieu [38]. Moreover, PQS signal molecules in OMV can hijack iron ions from the extracellular medium via the OMV-associated metal-binding protein TseF, secreted by the type 6 secretion system (T6SS). The delivery of PQS-iron complexes into recipient bacterial cells goes through interaction between the TseF (T6SS effector for iron uptake), ferric pyochelin receptor FtpA, and porin OprF [62, 64]. In parallel, QS enables differential regulation of the expression of genes at the loci of T6SS [65]. The expression of T6SS in P. aeruginosa is positively regulated by the AHL-dependent Las and Rhl circuits of the QS system during transition from exponential to stationary phase growth, and modulates bacteria entry into epithelial cells [66]. Furthermore, the expression of exoensyme S regulon exoS, which comprises genes for T3SS and four effector proteins, ExoS, T, U, and Y, is coordinated by the Rhl circuit of the QS system [67]. They are GTPase-activating proteins and ADP-ribosyltransferase, and able to induce cell death, inhibit phagocytosis during pneumonia, and lead to disruption of the pulmonary-vascular barrier, allowing bacterial dissemination into the bloodstream [68].
Iron Uptake
Efficient uptake of iron by bacteria is another important factor, along with virulence, which allows colonization of the host and establishment of infection [69]. Like the majority of organisms, P. aeruginosa needs iron to grow. However, it is confronted with a problem of iron availability in a mammalian host because iron is sequestered in host proteins. Here, the largest part of iron is present in the heme molecules found in intracellular hemoproteins, such as hemoglobin, myoglobin, and cytochromes. Also, iron can be reversible when bound to extracellular glycoproteins, such as lactoferrin, ferritin, and transferrin. P. aeruginosa uses multiple iron uptake strategies [70], and at least three of them – the siderophore system (pyoverdine and pyochelin), heme uptake system, and Feo system – are controlled via different QS circuits (Fig. 1) and triggered by bacterial cell density and iron limitation [71, 72, 73]. Pyoverdine (PVD) binds iron and displaces it from transferrin, and it can also act as a signaling molecule: when iron-bound, it causes the upregulation of exotoxin A, proteases, and PVD itself [4]. PVD is essential to P. aeruginosa to cause infections [74] and involved in the establishment of biofilms [69]. The heme uptake system, represented by Phu- and Has-components and positively regulated by Las and Rhl circuits of QS, allows P. aeruginosa to take up heme from hemoglobin and utilize it as a source of iron [71]. Uptake of Fe2+ through Feo system demands the involvement of QS-controlled phenazines, such as PYO, which are able to reduce Fe3+ bound to host proteins to Fe2+ [73]. The uptake of Fe2+ is important when bacteria grow not as planktonic but rather as biofilms in microaerobic or aerobic conditions and lower pH, which is a common situation during lung infections.
Biofilms
Higher organisms, including humans, are colonized by microorganisms, either as free-living bacterial cells or biofilms, which can be associated with normal microflora, persistent infections, and with the contamination of medical devices [75]. Biofilms are robust, organized social communities of microorganisms connected to each other and to a surface and encased by extracellular polymeric substances (EPS), as a matrix. The biofilm lifestyle is very distinct from that of planktonic bacterial cells as this ecosystem includes a completely new, higher level of physical and social interactions between community members and the matrix, and can be viewed as a biogenic habitat on the microscale [76]. Typically, biofilms have a high cell density, ranging between 108 and 1011 cells/g wet weight [77], and a heterogenic community, comprising many species with different gene expressions and metabolic and physiological activities that fluctuate over time [76]. EPS comprises most of the biomass of the biofilm rather than cells, and is mainly composed of polysaccharides, lipids, proteins, and extracellular DNA (eDNA) [78]. Matrix components provide structural stability, facilitate liquid and nutrient transport, and support robust integrity to a biofilm, including tolerance against antimicrobials [79], stress factors, and immune cells [75, 80]. The environment of biofilms provide a perfect milieu for intercellular communication. Indeed, the concentration of QS signals can be up to 1,000-fold higher in biofilms existing in the lung or gut than in environments inhabited by planktonic bacteria. Thus, 3-O-C12-HSL accumulates at 300–600 µM in biofilms growing in vitro [18]. Moreover, the QS system, together with other signaling systems, is involved and required for biofilm development in P. aeruginosa [76, 81].
The process of bacterial biofilm development consists of three key stages: attachment and transition from planktonic to sessile lifestyle; growth of microcolonies, biofilm maturation and differentiation; and detachment and dispersal of cells to inhabit new sites [82]. For microcolony and early biofilm formation, bacterial twitching motility and the ability to produce RHL and iron chelating siderophores are required, which are elements controlled by AHL and PQS circuits of the QS system (Fig. 1) [83, 84]. RHL is a glycolipid surfactant, which is essential for optimal interactions between the substratum and bacterial cells, and therefore also for the swarming motility [83] and early biofilm formation [84]. This biosurfactant maintains the pores and channels between microcolonies to facilitate liquid and nutrient transport within mature biofilm [85]. It has furthermore been reported to have antimicrobial properties against other bacteria, viruses, and fungi, and thereby give an advantage for P. aeruginosa in the competition and niche colonization in the tissues. RHL is a potent virulence factor since it is able to disrupt epithelial junctions, kill neutrophils, and inhibit phagocytosis [4].
The PQS system is also responsible for increased synthesis of eDNA, which interacts with EPS and is needed for its stability and resistance. P. aeruginosa requires nucleic acids mostly during early biofilm formation, while taking advantage of capsular and aggregative polysaccharides at several development stages [86]. One of them, aggregative glucose-rich PEL exopolysaccharide, facilitates stability, adherence, and compactness of the biofilm and cell-to-cell associations, and is controlled by the AHL-dependent QS system [87]. In the matrix, PEL can also serve a protective role, which relies on its ability to bound with and inhibit antibiotics [88]. Moreover, two QS-dependent carbohydrate-binding proteins, lectin A and B, are located in the outer membrane and important for cell recognition, adhesion, and proper biofilm formation [89, 90]. There are several accessory matrix components that support biofilm life: iron siderophores, cyclic glucans, LPS, and OMV [69]. Thus, many QS-regulated factors have an impact on biofilm formation and development at all stages of an infectious process (Fig. 1).
Phenotypic and genotypic alterations can occur in P. aeruginosa over time during acute and chronic infections and environmental changes. Thus, bacteria isolated from patients with an established chronic infection are usually nonmotile, nonadhesive, less cytotoxic, and less inflammatory than the strains isolated from the patients with acute infection. Other common changes include some mutations in the circuits of the QS system, downregulated T3SS and structural changes in LPS which results in alteration of host immune responses and promote bacterial persistence. However, strains from chronic infections more readily form colonies and biofilms, as they are usually more mucoid and overexpress exopolysaccharides [4].
Traffic and Human Targets for P. aeruginosa 3O-C12-HSL
Diffusion and Active Transport in Bacterial and Host Cells
QS molecules differ in their ability to diffuse across the plasma membrane or the membrane of subcellular organelles. For instance, P. aeruginosa AHL with a short fatty acid chain containing 4–6 carbons are freely diffusible across the membrane, in contrast to those with a long acyl chain, 3O-C12-HSL, and PQS, due to the larger size and hydrophobic nature of the latter [91]. The PQS transport across the bacterial membrane is facilitated by membrane transporters and bacterial OMV that also contain other bacterial products [64]. The active traffic of 3O-C12-HSL across cell membranes is driven by members of a large family of proteins, called the ATP binding cassette (ABC) transporter. For example, multidrug-resistant (MDR) efflux pumps, such as MexAB-OprM and MexGHI-OpmD, are involved in the active traffic of 3O-C12-HSL and PQS, respectively [91, 92]. MDR efflux pumps can furthermore extrude a wide range of substrates, not just antibiotics and toxic compounds, but also bacterial endogenous QS signal molecules, indicating that these membrane transporters are essential in bacterial pathogenesis and host-pathogen interactions. In human cells, the ABC transporter, ABCA1, was proposed to be involved in the active efflux of 3O-C12-HSL. Thus, microarray analysis of the transcriptional response of human lung epithelial cells revealed that mRNA levels for several genes involved in xenobiotic metabolism and drug transport were increased after exposure to 3O-C12-HSL [93].
IQGAP1 Is a Human Target for 3O-C12-HSL
In mammalian cells, long acyl chain AHL with an intact lactone ring are able to interact with phospholipids and diffuse across membrane systems, where membrane microdomains, such as caveolae and lipid rafts, may facilitate this process [94] via interactions with a sub-membrane organization of filamentous actin, actin-binding proteins, and anchoring membrane proteins. The overall plasma membrane fitness and the dynamic of these interactions, including those of the actin cytoskeleton, can influence the shape of microdomain compartments and the capacity of transport and receptor signaling [95]. The entry of 3O-C12-HSL into the host cell across the plasma membrane is followed by interaction and colocalization with the IQ-motif-containing GTPase-activation protein (IQGAP1) and paralleled by phosphorylation of Rac1 and Cdc42 and essential changes in the actin cytoskeleton network (Fig. 2); these events also trigger alterations in human epithelial cell migration and wound healing [25]. IQGAP1 was identified as a human target for bacterial 3O-C12-HSL using a pull-down approach with the biotin-conjugated 3O-C12-HSL probe and mass spectrometry-based proteomic analysis [25] (Fig. 3a). IQGAP1 is a 189-kDa multidomain signaling protein involved in many important cellular processes and functions (Fig. 3b). It orchestrates diverse cytoskeletal rearrangements and cell signaling events for cell cycle, polarization, migration, and adhesion, for example [96]. IQGAP1 localizes at the plasma membrane in the leading edge of migrating cells, in the cytoplasm and also at the cytoplasmic face of the nuclear envelope [97]. IQGAP1 interacts with a variety of proteins, including actin, calmodulin, Rac1, Cdc42, β-catenin, E-cadherin, myosin, exocyst, ERK, MAPK, and mTOR [96, 97]. Using a fluorescent dye-conjugated 3O-C12-HSL probe (Fig. 4a) and nanoscale super-resolution microscopy (Fig. 4b), it was demonstrated that 3O-C12-HSL colocalizes with and targets IQGAP1 to modulate the migration of human epithelial cells [25].
Recognition of 3O-C12-HSL by Sensory and Nuclear Receptors
The P. aeruginosa 3O-C12-HSL activates and associates with the chemosensory G-protein-coupled receptor T2R38 (Fig. 2) on leukocytes of different origins [98, 99]. Still, the role of sensory receptors in innate immunity processes and others beyond the taste is not fully resolved. P. aeruginosa 3O-C12-HSL may also bind to nuclear peroxisome proliferator-activated receptors (PPAR) to modulate DNA binding activity and transcription of NF-κB-dependent genes (Fig. 2) [100].
Thus, by passing through the plasma membrane in host cells, 3O-C12-HSL can associate and target several membrane-associated proteins, which promote interactions with intracellular molecules and signaling cascades, and further with RNA and DNA processes in eukaryotic cells.
P. aeruginosa 3O-C12-HSL Regulates an Ensemble of Host Functions
During the last 2 decades, many investigators have indeed shown that P. aeruginosa 3O-C12-HSL has multiple effects on mammalian cells and acts via different signaling pathways (Fig. 2), and the recent advances have been reviewed [101, 102].
Inflammatory Response
Earlier studies were focused on the role of P. aeruginosa 3O-C12-HSL in modulation of innate and adaptive immune responses and inflammatory signaling. It affects both pro- and anti-inflammatory responses, inhibiting the production of cytokine TNF-α, IL-12 [20, 103, 104, 105], and increasing the secretion of interleukins, e.g. IL-6, IL-8 [106, 107, 108, 109], IL-10 [103, 104, 109], and IL-1β [109]. In addition, 3O-C12-HSL interferes with T cell proliferation [105] and blocks the differentiation of the Th1 and Th2 cells [110], which play important roles in the regulation of immune responses (Fig. 2). Transcriptome, systems biology, and network analyses revealed activation of multiple innate immune pathways in lung epithelial cells by 3O-C12-HSL [93, 107].
Host Transcription
Moreover, 3O-C12-HSL has been shown to be a strong activator of inflammatory response via the phosphorylation of MAPK (Fig. 2) and induction of transcriptional factor NF-κB signaling [106]. NF-κB is a major transcription factor that controls many genes, regulating both the innate and adaptive immune response during infections, and incorrect regulation of NF-κB may lead to the development of cancer, inflammatory, and autoimmune diseases. In the classical pathway, bacterial compounds (LPS, flagellin, lipoteichoic acid, RNA, or DNA) are recognized by TLR as specific membrane-bound pattern recognition receptors, which leads to NF-κB activation and targeted gene expression. However, 3O-C12-HSL does not interact with TLR expressed on immune cells and thus likely activates host cells via the mechanism distinct from the TLR pathway [111]. It was later shown that 3O-C12-HSL activates the ER stress transducer protein kinase RNA-like ER kinase (PERK), increases phosphorylation of the eukaryotic translation elongation factor eI-F2α leading to the selective translation and thereby affecting protein synthesis, particularly IL-8 [108]. 3O-C12-HSL triggers the unfolded protein response and increases the expression of its certain target genes, being negatively correlated with LPS-induced NF-κB activation [112].
Mitochondrial Physiology, Pro-Apoptosis, and Quorum Quenching
The P. aeruginosa 3O-C12-HSL also influences mitochondrial physiology [113, 114] by depolarizing the membrane potential and releasing mitochondrial cytochrome C into the cytosol; it was concluded that 3O-C12-HSL has a pro-apoptotic effect via activating caspases 3/7 and 8 without the involvement of Bak/Bax [114]. Indeed, it appears to trigger apoptosis in certain types of cells, such as mouse and human fibroblasts, monocyte-, and neutrophil-like tumor cell lines [115]. Still, polarized human epithelial cells [25, 116] and human primary macrophages and neutrophils [101, 117] did not reveal any apoptosis-like changes after 3O-C12-HSL-treatment. Maturity and polarization of epithelial sheets seem to protect them against the pro-apoptotic effect of 3O-C12-HSL [116], but do not prevent loss of barrier integrity and repair capacity [25, 101]. Polarized epithelial monolayers, in contrast to nonpolarized cells, are also able to degrade 3O-C12-HSL using membrane-associated paraoxonase 2 [116] that catalyzes the opening of the lactone ring but also has antioxidant activity and benefits mitochondrial function in the response to oxidative stress and facilitates the responses of the innate immune system. Inactivation of AHL, or so-called quorum quenching by enzymes (Fig. 2), has also been described in cells of other origins, for example in bacteria, fungi, plants, and various mammalian cells. Different quorum-quenching strategies have been applied as promising tools in the areas of medicine, agriculture, and anti-biofouling [29].
Calcium Signaling
P. aeruginosa 3O-C12-HSL initiates strong and rapid calcium signaling in neutrophils, fibroblasts, and epithelial cells [22, 24, 107, 118] (Fig. 2). Ca2+ is a second messenger regulating many processes, being as diverse as gene transcription, muscle contraction, epithelial integrity, immune defense, cell motility, and phagocytosis. The calcium signaling proteome is tissue- and cell-specific and suited to a particular demand and function. In neutrophils and epithelial cells, 3O-C12-HSL can induce calcium influx from intracellular stores such as ER and actin cytoskeleton relying on the inositol 1,4,5-triphosphate receptors (IP3R) [22, 24] (Fig. 2).
Directional Cell Migration: Chemotaxis
As a response to external cues, such as a soluble gradient of cytokines, growth factors, and bacterial traits, the host cell can initiate directional migration, also defined as chemotaxis, where a cellular steering system will guide the cell towards the higher concentration. Host cells can utilize different migratory patterns depending on cell morphology, cytoskeletal organization, cell-extracellular matrix interaction, and extracellular stimuli [119]. During migration, the cell is polarized in the direction of motion, possessing leading and trailing edges, blebbing, wide and thin membrane protrusions, the lamellipodium and small finger-like projections, the filopodia. Within the human body, we can define single cell and multicellular, collective migration (Fig. 2); both can be modulated by P. aeruginosa 3O-C12-HSL [25, 101], which we will discuss more below.
Single Cell Migration
Rapidly moving cells, such as neutrophils, macrophages, and fibroblasts, utilize the pattern of single cell migration. They can either crawl in the amoeba manner (neutrophils) or move in the mesenchymal mode, utilizing a multistep cycle of protrusion, adhesion, and retraction (fibroblasts, macrophages) [119]. Human neutrophils and macrophages are fast-migrating phagocytes controlling inflammation and infection at the very early contact with bacteria. Thus, P. aeruginosa 3O-C12-HSL and 3O-C10-HSL, but not C4-HSL, strongly promote their chemotaxis [24, 101, 120] and increase phagocytic capacity in a dose-dependent manner [101, 121]. Moreover, P. aeruginosa with a complete QS system elicits a more intensive phagocytosis in macrophages than the lasI/rhlI mutant lacking both 3O-C12-HSL and C4-HSL [23]. Both chemotaxis and phagocytosis require rapid changes in cell size and morphology driven by homeostasis and the transport of ions and water across the membrane via water channels called aquaporins (AQP). Thus, 3O-C12-HSL enhances the cell area, volume, protrusive activity, and AQP9 expression and structural dynamics in moving macrophages [117]. Furthermore, AQP, together with ion channels and other transporters, mediate the influx of ions and water across the cell membrane (Fig. 2); water creates an increased local intracellular hydrostatic pressure [122], which forces the membrane to extend, allowing the polymerizing actin cytoskeleton to fill the protrusion and promotes cell migration [117, 123, 124]. AHL causes filamentous actin accumulation in the leading edge of migrating phagocytes, as regulated by calcium signaling, activation of phospholipase Cγ1, and the Rho family of small GTPases, such as Rac1 and Cdc42 [24, 101], which also control the formation of membrane protrusion activity. Through these mechanisms, leukocytes can quickly migrate to the site of AHL and mount a more effective phagocytosis of bacteria. However, in contrast to the stimulatory effect on macrophages and neutrophils, 3O-C12-HSL has been shown to suppress chemotaxis, degranulation, and cytokinesis in mast cells [125], key effectors of allergic processes. Thus, the interactions between QS signals and innate immune cells are rather complex, and while 3O-C12-HSL is recognized as a stimuli for macrophages and neutrophils, mast cells are likely targets for its suppressive action and may contribute to the pathogenicity of P. aeruginosa and its ability to avoid host defense.
Multicellular Migration
Multicellular migration, like epithelial sheet migration, occurs in the processes such as wound healing and renewal of epithelial cell monolayers in the skin and the gut (Fig. 2). This type of directional locomotion is highly influenced by the fitness of the whole epithelial sheet: differences in morphology, dynamics of cellular junctions, and abilities to respond to the external cues surrounding cells [119]. P. aeruginosa 3O-C12-HSL, but not C4-HSL, induces loss of barrier integrity and disruption of TJ and AJ complexes associated with actin cytoskeleton (Fig. 2) in polarized intestinal [101, 126] and airway epithelial cells [116]. These processes are being affected by calcium-dependent signaling [22], the MAPK cascade [127], and activation of metalloproteases via protease-activated receptor (PAR) and lipid rafts [126]. TJ and AJ are multiprotein complexes composed of many transmembrane and cytoplasmic proteins associated with cytoskeleton, transport proteins, and a variety of regulatory and signaling proteins. TJ and AJ complexes include, for example, occludin-zonula occludens (ZO), cadherin-catenin, claudin-ZO, and occludin-tricellulin. 3O-C12-HSL-induced perturbation in barrier integrity was associated with changes in the phosphorylation status of TJ and AJ proteins [22, 128]. Being highly dynamic structures, cell junctions can alter barrier integrity and permeability upon a broad variety of stimuli, including immune cells, bacteria, cytokines, toxins, and oxidative stress. Normally and after an injury, the epithelial sheets undergo a process of renewal and wound healing, which are dependent on the balance of collective cell migration, proliferation, and differentiation [3]. P. aeruginosa 3O-C12-HSL, encoded by the lasI gene, modulates wound healing in a dose-dependent manner in an in vitro model of polarized human epithelial cells [25] and rat in vivo model of skin sheets [129]. Low doses of short fatty acid molecule C4-HSL, encoded by the rhlI gene, also promote wound healing, which was paralleled with cyclooxygenase-driven neutrophils infiltration [130]. As in cases of phagocyte activity, 3O-C12-HSL induced an increase in epithelial sheet migration that was also associated with activation of the Rho family of small GTPases and actin cytoskeleton reorganization [25].
In summary, research over recent decades has suggested that P. aeruginosa 3O-C12-HSL signals, through different mechanisms, can interfere with RNA and DNA processes, protein synthesis, calcium signaling, homeostasis, and mitochondrial and cytoskeletal dynamics in host cells (Fig. 2). In these ways, P. aeruginosa communication via 3O-C12-HSL can perturb an ensemble of important cellular processes in the host, such as cell morphology, proliferation, differentiation, and apoptosis, and thereby many biological activities and functions.
Conclusion and Outlook
Our knowledge about the cellular and molecular mechanisms involved in bacteria-host crosstalk is constantly expanding. In this review, we have discussed how P. aeruginosa use QS systems to regulate its virulence, biofilms, and metabolic demands, and thereby promote infection in a host (Fig. 1). We have also addressed the role of P. aeruginosa AHL-dependent QS signaling in the modulation of a diverse array of host functions with a focus on innate immune responses via different targets and mechanisms (Fig. 2). Interference with QS, termed quorum quenching, is a new and promising avenue to combat bacterial pathogenicity and biofilm formation, and thereby enlarge the therapeutic arsenal against bacterial infections and reduce the rate of antibiotic resistance. This naturally occurring strategy has been extensively studied and reviewed by many researches [28, 29]. Further breakthroughs will require multidisciplinary tools to investigate the consequences of host-microbe interactions and insights from both pathogen and host perspectives.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgements
We apologize to the numerous researchers whose work could not be cited because of space restrictions. We thank our co-authors for the excellent collaboration and colleagues at Linköping University for enthusiastic commitment, and especially Prof. Karl- Eric Magnusson for helpful discussions. We thank Dr. Hans Blom and Prof. Hjalmar Brismar (Advanced Light Microscopy, SciLifeLab, Stockholm, Sweden). We also thank Prof. Peter Konradsson (Linköping University, Sweden) and Cayman Chemical (Ann Arbor, MI, USA) for help in the design and synthesis of AHL according to our propositions. Our work was supported by the Swedish Research Council, the European Science Foundation (TraPPs Euromembrane project), Euro-BioImaging Proof-of Concept Studies, Magnus Bergvalls Foundation, and the Faculty of Health Sciences, Linköping University.
References
- 1.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017 Nov;551((7680)):313–20. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol. 2010 Feb;8((2)):117–28. doi: 10.1038/nrmicro2295. [DOI] [PubMed] [Google Scholar]
- 3.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014 Mar;14((3)):141–53. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 4.Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013 Apr;67((3)):159–73. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 5.Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013 Mar;11((3)):297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 6.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016 Aug;14((9)):576–88. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006 Apr;296((2-3)):73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006 May;188((9)):3365–70. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000 May;182((10)):2702–8. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Cámara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011 Mar;35((2)):247–74. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, et al. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol. 2013 May;9((5)):339–43. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 12.Ye L, Cornelis P, Guillemyn K, Ballet S, Hammerich O. Structure revision of N-mercapto-4-formylcarbostyril produced by Pseudomonas fluorescens G308 to 2-(2-hydroxyphenyl)thiazole-4-carbaldehyde [aeruginaldehyde] [aeruginaldehyde] Nat Prod Commun. 2014 Jun;9((6)):789–94. [PubMed] [Google Scholar]
- 13.Prindle A, Liu J, Asally M, Ly S, Garcia-Ojalvo J, Süel GM. Ion channels enable electrical communication in bacterial communities. Nature. 2015 Nov;527((7576)):59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev. 2015 Jul;39((4)):509–21. doi: 10.1093/femsre/fuu010. [DOI] [PubMed] [Google Scholar]
- 15.Knecht LD, O'Connor G, Mittal R, Liu XZ, Daftarian P, Deo SK, et al. Serotonin Activates Bacterial Quorum Sensing and Enhances the Virulence of Pseudomonas aeruginosa in the Host. EBioMedicine. 2016 Jul;9:161–9. doi: 10.1016/j.ebiom.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007 Mar;3((3)):e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortori CA, Dubern JF, Chhabra SR, Cámara M, Hardie K, Williams P, et al. Simultaneous quantitative profiling of N-acyl-L-homoserine lactone and 2-alkyl-4(1H)-quinolone families of quorum-sensing signaling molecules using LC-MS/MS. Anal Bioanal Chem. 2011 Jan;399((2)):839–50. doi: 10.1007/s00216-010-4341-0. [DOI] [PubMed] [Google Scholar]
- 18.Charlton TS, de Nys R, Netting A, Kumar N, Hentzer M, Givskov M, et al. A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: application to a model bacterial biofilm. Environ Microbiol. 2000 Oct;2((5)):530–41. doi: 10.1046/j.1462-2920.2000.00136.x. [DOI] [PubMed] [Google Scholar]
- 19.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999 Nov;67((11)):5854–62. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith RS, Harris SG, Phipps R, Iglewski B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol. 2002 Feb;184((4)):1132–9. doi: 10.1128/jb.184.4.1132-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang HB, DiMango E, Bryan R, Gambello M, Iglewski BH, Goldberg JB, et al. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996 Jan;64((1)):37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vikström E, Bui L, Konradsson P, Magnusson KE. Role of calcium signalling and phosphorylations in disruption of the epithelial junctions by Pseudomonas aeruginosa quorum sensing molecule. Eur J Cell Biol. 2010 Aug;89((8)):584–97. doi: 10.1016/j.ejcb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Holm A, Karlsson T, Vikström E. Pseudomonas aeruginosa lasI/rhlI quorum sensing genes promote phagocytosis and aquaporin 9 redistribution to the leading and trailing regions in macrophages. Front Microbiol. 2015 Sep;6:915. doi: 10.3389/fmicb.2015.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson T, Musse F, Magnusson KE, Vikström E. N-Acylhomoserine lactones are potent neutrophil chemoattractants that act via calcium mobilization and actin remodeling. J Leukoc Biol. 2012 Jan;91((1)):15–26. doi: 10.1189/jlb.0111034. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson T, Turkina MV, Yakymenko O, Magnusson KE, Vikström E. The Pseudomonas aeruginosa N-acylhomoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 2012;8((10)):e1002953. doi: 10.1371/journal.ppat.1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welsh MA, Blackwell HE. Chemical probes of quorum sensing: from compound development to biological discovery. FEMS Microbiol Rev. 2016 Sep;40((5)):774–94. doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdt JP, Blackwell HE. Competition studies confirm two major barriers that can preclude the spread of resistance to quorum-sensing inhibitors in bacteria. ACS Chem Biol. 2014 Oct;9((10)):2291–9. doi: 10.1021/cb5004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsen TH, Tolker-Nielsen T, Givskov M. Bacterial Biofilm Control by Perturbation of Bacterial Signaling Processes. Int J Mol Sci. 2017 Sep;18((9)):18. doi: 10.3390/ijms18091970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev. 2016 Jan;40((1)):86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 30.Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, Littler E, et al. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics. 2006 Jun;7((1)):162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park AJ, Murphy K, Krieger JR, Brewer D, Taylor P, Habash M, et al. A temporal examination of the planktonic and biofilm proteome of whole cell Pseudomonas aeruginosa PAO1 using quantitative mass spectrometry. Mol Cell Proteomics. 2014 Apr;13((4)):1095–105. doi: 10.1074/mcp.M113.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nigaud Y, Cosette P, Collet A, Song PC, Vaudry D, Vaudry H, et al. Biofilm-induced modifications in the proteome of Pseudomonas aeruginosa planktonic cells. Biochim Biophys Acta. 2010 Apr;1804((4)):957–66. doi: 10.1016/j.bbapap.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000 Dec;64((4)):694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JH, Rhee JE, Park U, Ju HM, Lee BC, Kim TS, et al. Identification and functional analysis of vibrio vulnificus SmcR, a novel global regulator. J Microbiol Biotechnol. 2007 Feb;17((2)):325–34. [PubMed] [Google Scholar]
- 35.Haiko J, Westerlund-Wikström B. The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2013 Oct;2((4)):1242–67. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83((1)):99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 37.Moskowitz SM, Ernst RK. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell Biochem. 2010;53:241–53. doi: 10.1007/978-90-481-9078-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura S, Higashiyama Y, Izumikawa K, Seki M, Kakeya H, Yamamoto Y, et al. The roles of the quorum-sensing system in the release of extracellular DNA, lipopolysaccharide, and membrane vesicles from Pseudomonas aeruginosa. Jpn J Infect Dis. 2008 Sep;61((5)):375–8. [PubMed] [Google Scholar]
- 39.Da Silva GJ, Domingues S. Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii. Antibiotics (Basel) 2017 Nov;6((4)):6. doi: 10.3390/antibiotics6040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruhal R, Antti H, Rzhepishevska O, Boulanger N, Barbero DR, Wai SN, et al. A multivariate approach to correlate bacterial surface properties to biofilm formation by lipopolysaccharide mutants of Pseudomonas aeruginosa. Colloids Surf B Biointerfaces. 2015 Mar;127:182–91. doi: 10.1016/j.colsurfb.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 41.Tingpej P, Smith L, Rose B, Zhu H, Conibear T, Al Nassafi K, et al. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol. 2007 Jun;45((6)):1697–704. doi: 10.1128/JCM.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nouwens AS, Beatson SA, Whitchurch CB, Walsh BJ, Schweizer HP, Mattick JS, et al. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology. 2003 May;149((Pt 5)):1311–22. doi: 10.1099/mic.0.25967-0. [DOI] [PubMed] [Google Scholar]
- 43.Toder DS, Gambello MJ, Iglewski BH. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol Microbiol. 1991 Aug;5((8)):2003–10. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 44.Filloux A. Protein Secretion Systems in Pseudomonas aeruginosa: An Essay on Diversity, Evolution, and Function. Front Microbiol. 2011 Jul;2:155. doi: 10.3389/fmicb.2011.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, van Strijp JA, et al. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J Immunol. 2012 Jan;188((1)):386–93. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- 46.Alcorn JF, Wright JR. Degradation of pulmonary surfactant protein D by Pseudomonas aeruginosa elastase abrogates innate immune function. J Biol Chem. 2004 Jul;279((29)):30871–9. doi: 10.1074/jbc.M400796200. [DOI] [PubMed] [Google Scholar]
- 47.van der Plas MJ, Bhongir RK, Kjellström S, Siller H, Kasetty G, Mörgelin M, et al. Pseudomonas aeruginosa elastase cleaves a C-terminal peptide from human thrombin that inhibits host inflammatory responses. Nat Commun. 2016 May;7:11567. doi: 10.1038/ncomms11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003 Apr;185((7)):2080–95. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003 Apr;185((7)):2066–79. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wargo MJ, Gross MJ, Rajamani S, Allard JL, Lundblad LK, Allen GB, et al. Hemolytic phospholipase C inhibition protects lung function during Pseudomonas aeruginosa infection. Am J Respir Crit Care Med. 2011 Aug;184((3)):345–54. doi: 10.1164/rccm.201103-0374OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barker AP, Vasil AI, Filloux A, Ball G, Wilderman PJ, Vasil ML. A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol Microbiol. 2004 Aug;53((4)):1089–98. doi: 10.1111/j.1365-2958.2004.04189.x. [DOI] [PubMed] [Google Scholar]
- 52.Kearns DB, Robinson J, Shimkets LJ. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol. 2001 Jan;183((2)):763–7. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Odumosu O, Nicholas D, Yano H, Langridge W. AB toxins: a paradigm switch from deadly to desirable. Toxins (Basel) 2010 Jul;2((7)):1612–45. doi: 10.3390/toxins2071612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voulhoux R, Taupiac MP, Czjzek M, Beaumelle B, Filloux A. Influence of deletions within domain II of exotoxin A on its extracellular secretion from Pseudomonas aeruginosa. J Bacteriol. 2000 Jul;182((14)):4051–8. doi: 10.1128/jb.182.14.4051-4058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kounnas MZ, Morris RE, Thompson MR, FitzGerald DJ, Strickland DK, Saelinger CB. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A. J Biol Chem. 1992 Jun;267((18)):12420–3. [PubMed] [Google Scholar]
- 56.Hessler JL, Kreitman RJ. An early step in Pseudomonas exotoxin action is removal of the terminal lysine residue, which allows binding to the KDEL receptor. Biochemistry. 1997 Nov;36((47)):14577–82. doi: 10.1021/bi971447w. [DOI] [PubMed] [Google Scholar]
- 57.Smith DC, Spooner RA, Watson PD, Murray JL, Hodge TW, Amessou M, et al. Internalized Pseudomonas exotoxin A can exploit multiple pathways to reach the endoplasmic reticulum. Traffic. 2006 Apr;7((4)):379–93. doi: 10.1111/j.1600-0854.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- 58.Michalska M, Wolf P. Pseudomonas Exotoxin A: optimized by evolution for effective killing. Front Microbiol. 2015 Sep;6:963. doi: 10.3389/fmicb.2015.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall S, McDermott C, Anoopkumar-Dukie S, McFarland AJ, Forbes A, Perkins AV, et al. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins (Basel) 2016 Aug;8((8)):8. doi: 10.3390/toxins8080236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higgins S, Heeb S, Rampioni G, Fletcher MP, Williams P, Cámara M. Differential Regulation of the Phenazine Biosynthetic Operons by Quorum Sensing in Pseudomonas aeruginosa PAO1-N. Front Cell Infect Microbiol. 2018 Jul;8:252. doi: 10.3389/fcimb.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guerrero-Mandujano A, Hernández-Cortez C, Ibarra JA, Castro-Escarpulli G. The outer membrane vesicles: secretion system type zero. Traffic. 2017 Jul;18((7)):425–32. doi: 10.1111/tra.12488. [DOI] [PubMed] [Google Scholar]
- 62.Gallique M, Bouteiller M, Merieau A. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front Microbiol. 2017 Jul;8:1454. doi: 10.3389/fmicb.2017.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol. 2008 Jul;69((2)):491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, et al. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 2017 Mar;8:14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lesic B, Starkey M, He J, Hazan R, Rahme LG. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology. 2009 Sep;155((Pt 9)):2845–55. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, et al. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem. 2012 Aug;287((32)):27095–105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hogardt M, Roeder M, Schreff AM, Eberl L, Heesemann J. Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbiology. 2004 Apr;150((Pt 4)):843–51. doi: 10.1099/mic.0.26703-0. [DOI] [PubMed] [Google Scholar]
- 68.Rangel SM, Diaz MH, Knoten CA, Zhang A, Hauser AR. The Role of ExoS in Dissemination of Pseudomonas aeruginosa during Pneumonia. PLoS Pathog. 2015 Jun;11((6)):e1004945. doi: 10.1371/journal.ppat.1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA. 2005 Aug;102((31)):11076–81. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol. 2013 Nov;3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arevalo-Ferro C, Hentzer M, Reil G, Görg A, Kjelleberg S, Givskov M, et al. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ Microbiol. 2003 Dec;5((12)):1350–69. doi: 10.1046/j.1462-2920.2003.00532.x. [DOI] [PubMed] [Google Scholar]
- 72.Stintzi A, Evans K, Meyer JM, Poole K. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett. 1998 Sep;166((2)):341–5. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 73.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006 Sep;61((5)):1308–21. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 74.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996 Feb;64((2)):518–23. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mulcahy LR, Isabella VM, Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb Ecol. 2014 Jul;68((1)):1–12. doi: 10.1007/s00248-013-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016 Aug;14((9)):563–75. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 77.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016 Aug;14((8)):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012 Jul;36((4)):893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh S, Singh SK, Chowdhury I, Singh R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol J. 2017 Apr;11((1)):53–62. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bjarnsholt T, Jensen PO, Burmølle M, Hentzer M, Haagensen JA, Hougen HP, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005 Feb;151((Pt 2)):373–83. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 81.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998 Apr;280((5361)):295–8. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 82.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56((1)):187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 83.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006 Dec;62((5)):1264–77. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 84.Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2007 Mar;189((6)):2531–9. doi: 10.1128/JB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davey ME, Caiazza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003 Feb;185((3)):1027–36. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006 Feb;59((4)):1114–28. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 87.Sakuragi Y, Kolter R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol. 2007 Jul;189((14)):5383–6. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, et al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011 Jan;7((1)):e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winzer K, Falconer C, Garber NC, Diggle SP, Camara M, Williams P. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J Bacteriol. 2000 Nov;182((22)):6401–11. doi: 10.1128/jb.182.22.6401-6411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rampioni G, Falcone M, Heeb S, Frangipani E, Fletcher MP, Dubern JF, et al. Unravelling the Genome-Wide Contributions of Specific 2-Alkyl-4-Quinolones and PqsE to Quorum Sensing in Pseudomonas aeruginosa. PLoS Pathog. 2016 Nov;12((11)):e1006029. doi: 10.1371/journal.ppat.1006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pearson JP, Van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999 Feb;181((4)):1203–10. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aendekerk S, Diggle SP, Song Z, Høiby N, Cornelis P, Williams P, et al. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology. 2005 Apr;151((Pt 4)):1113–25. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- 93.Bryan A, Watters C, Koenig L, Youn E, Olmos A, Li G, et al. Human transcriptome analysis reveals a potential role for active transport in the metabolism of Pseudomonas aeruginosa autoinducers. Microbes Infect. 2010 Nov;12((12-13)):1042–50. doi: 10.1016/j.micinf.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davis BM, Jensen R, Williams P, O'Shea P. The interaction of N-acylhomoserine lactone quorum sensing signaling molecules with biological membranes: implications for inter-kingdom signaling. PLoS One. 2010 Oct;5((10)):e13522. doi: 10.1371/journal.pone.0013522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laude AJ, Prior IA. Plasma membrane microdomains: organization, function and trafficking. Mol Membr Biol. 2004 May-Jun;21((3)):193–205. doi: 10.1080/09687680410001700517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 2015 Apr;16((4)):427–46. doi: 10.15252/embr.201439834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith JM, Hedman AC, Sacks DB. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015 Mar;25((3)):171–84. doi: 10.1016/j.tcb.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maurer S, Wabnitz GH, Kahle NA, Stegmaier S, Prior B, Giese T, et al. Tasting Pseudomonas aeruginosa Biofilms: Human Neutrophils Express the Bitter Receptor T2R38 as Sensor for the Quorum Sensing Molecule N-(3-Oxododecanoyl)-l-Homoserine Lactone. Front Immunol. 2015 Jul;6:369. doi: 10.3389/fimmu.2015.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gaida MM, Dapunt U, Hänsch GM. Sensing developing biofilms: the bitter receptor T2R38 on myeloid cells. Pathog Dis. 2016 Apr;74((3)):74. doi: 10.1093/femspd/ftw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, et al. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol. 2008 Jul;190((13)):4408–15. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holm A, Vikström E. Quorum sensing communication between bacteria and human cells: signals, targets, and functions. Front Plant Sci. 2014 Jun;5:309. doi: 10.3389/fpls.2014.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu YC, Chan KG, Chang CY. Modulation of Host Biology by Pseudomonas aeruginosa Quorum Sensing Signal Molecules: messengers or Traitors. Front Microbiol. 2015 Nov;6:1226. doi: 10.3389/fmicb.2015.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hooi DS, Bycroft BW, Chhabra SR, Williams P, Pritchard DI. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect Immun. 2004 Nov;72((11)):6463–70. doi: 10.1128/IAI.72.11.6463-6470.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Glucksam-Galnoy Y, Sananes R, Silberstein N, Krief P, Kravchenko VV, Meijler MM, et al. The bacterial quorum-sensing signal molecule N-3-oxo-dodecanoyl-L-homoserine lactone reciprocally modulates pro- and anti-inflammatory cytokines in activated macrophages. J Immunol. 2013 Jul;191((1)):337–44. doi: 10.4049/jimmunol.1300368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Skindersoe ME, Zeuthen LH, Brix S, Fink LN, Lazenby J, Whittall C, et al. Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol Med Microbiol. 2009 Apr;55((3)):335–45. doi: 10.1111/j.1574-695X.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- 106.Smith RS, Fedyk ER, Springer TA, Mukaida N, Iglewski BH, Phipps RP. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J Immunol. 2001 Jul;167((1)):366–74. doi: 10.4049/jimmunol.167.1.366. [DOI] [PubMed] [Google Scholar]
- 107.Mayer ML, Sheridan JA, Blohmke CJ, Turvey SE, Hancock RE. The Pseudomonas aeruginosa autoinducer 3O-C12 homoserine lactone provokes hyperinflammatory responses from cystic fibrosis airway epithelial cells. PLoS One. 2011 Jan;6((1)):e16246. doi: 10.1371/journal.pone.0016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grabiner MA, Fu Z, Wu T, Barry KC, Schwarzer C, Machen TE. Pseudomonas aeruginosa quorum-sensing molecule homoserine lactone modulates inflammatory signaling through PERK and eI-F2α. J Immunol. 2014 Aug;193((3)):1459–67. doi: 10.4049/jimmunol.1303437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Holban AM, Bleotu C, Chifiriuc MC, Bezirtzoglou E, Lazar V. Role of Pseudomonas aeruginosa quorum sensing (QS) molecules on the viability and cytokine profile of human mesenchymal stem cells. Virulence. 2014 Feb;5((2)):303–10. doi: 10.4161/viru.27571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ritchie AJ, Whittall C, Lazenby JJ, Chhabra SR, Pritchard DI, Cooley MA. The immunomodulatory Pseudomonas aeruginosa signalling molecule N-(3-oxododecanoyl)-L-homoserine lactone enters mammalian cells in an unregulated fashion. Immunol Cell Biol. 2007 Nov-Dec;85((8)):596–602. doi: 10.1038/sj.icb.7100090. [DOI] [PubMed] [Google Scholar]
- 111.Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Wood MR, et al. N-(3-oxo-acyl)homoserine lactones signal cell activation through a mechanism distinct from the canonical pathogen-associated molecular pattern recognition receptor pathways. J Biol Chem. 2006 Sep;281((39)):28822–30. doi: 10.1074/jbc.M606613200. [DOI] [PubMed] [Google Scholar]
- 112.Zhang J, Gong F, Li L, Zhao M, Song J. Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl) homoserine lactone attenuates lipopolysaccharide-induced inflammation by activating the unfolded protein response. Biomed Rep. 2014 Mar;2((2)):233–8. doi: 10.3892/br.2014.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tao S, Luo Y, Bin He, Liu J, Qian X, Ni Y, et al. Paraoxonase 2 modulates a proapoptotic function in LS174T cells in response to quorum sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone. Sci Rep. 2016 Jul;6((1)):28778. doi: 10.1038/srep28778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schwarzer C, Fu Z, Shuai S, Babbar S, Zhao G, Li C, et al. Pseudomonas aeruginosa homoserine lactone triggers apoptosis and Bak/Bax-independent release of mitochondrial cytochrome C in fibroblasts. Cell Microbiol. 2014 Jul;16((7)):1094–104. doi: 10.1111/cmi.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tateda K, Ishii Y, Horikawa M, Matsumoto T, Miyairi S, Pechere JC, et al. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect Immun. 2003 Oct;71((10)):5785–93. doi: 10.1128/IAI.71.10.5785-5793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Losa D, Köhler T, Bacchetta M, Saab JB, Frieden M, van Delden C, et al. Airway Epithelial Cell Integrity Protects from Cytotoxicity of Pseudomonas aeruginosa Quorum-Sensing Signals. Am J Respir Cell Mol Biol. 2015 Aug;53((2)):265–75. doi: 10.1165/rcmb.2014-0405OC. [DOI] [PubMed] [Google Scholar]
- 117.Holm A, Magnusson KE, Vikström E. Pseudomonas aeruginosa N-3-oxo-dodecanoyl-homoserine Lactone Elicits Changes in Cell Volume, Morphology, and AQP9 Characteristics in Macrophages. Front Cell Infect Microbiol. 2016 Mar;6:32. doi: 10.3389/fcimb.2016.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shiner EK, Terentyev D, Bryan A, Sennoune S, Martinez-Zaguilan R, Li G, et al. Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell Microbiol. 2006 Oct;8((10)):1601–10. doi: 10.1111/j.1462-5822.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 119.De Pascalis C, Etienne-Manneville S. Single and collective cell migration: the mechanics of adhesions. Mol Biol Cell. 2017 Jul;28((14)):1833–46. doi: 10.1091/mbc.E17-03-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wagner C, Zimmermann S, Brenner-Weiss G, Hug F, Prior B, Obst U, et al. The quorum-sensing molecule N-3-oxododecanoyl homoserine lactone (3OC12-HSL) enhances the host defence by activating human polymorphonuclear neutrophils (PMN) Anal Bioanal Chem. 2007 Jan;387((2)):481–7. doi: 10.1007/s00216-006-0698-5. [DOI] [PubMed] [Google Scholar]
- 121.Vikström E, Magnusson KE, Pivoriūnas A. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)-L-homoserine lactone stimulates phagocytic activity in human macrophages through the p38 MAPK pathway. Microbes Infect. 2005 Dec;7((15)):1512–8. doi: 10.1016/j.micinf.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 122.Chengappa P, Sao K, Jones TM, Petrie RJ. Intracellular Pressure: A Driver of Cell Morphology and Movement. Int Rev Cell Mol Biol. 2018;337:185–211. doi: 10.1016/bs.ircmb.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 123.Loitto VM, Karlsson T, Magnusson KE. Water flux in cell motility: expanding the mechanisms of membrane protrusion. Cell Motil Cytoskeleton. 2009 May;66((5)):237–47. doi: 10.1002/cm.20357. [DOI] [PubMed] [Google Scholar]
- 124.Schwab A, Fabian A, Hanley PJ, Stock C. Role of ion channels and transporters in cell migration. Physiol Rev. 2012 Oct;92((4)):1865–913. doi: 10.1152/physrev.00018.2011. [DOI] [PubMed] [Google Scholar]
- 125.Khambati I, Han S, Pijnenburg D, Jang H, Forsythe P. The bacterial quorum-sensing molecule, N-3-oxo-dodecanoyl-L-homoserine lactone, inhibits mediator release and chemotaxis of murine mast cells. Inflamm Res. 2017 Mar;66((3)):259–68. doi: 10.1007/s00011-016-1013-3. [DOI] [PubMed] [Google Scholar]
- 126.Eum SY, Jaraki D, Bertrand L andrás IE, Toborek M. Disruption of epithelial barrier by quorum-sensing N-3-(oxododecanoyl)-homoserine lactone is mediated by matrix metalloproteinases. Am J Physiol Gastrointest Liver Physiol. 2014 Jun;306((11)):G992–1001. doi: 10.1152/ajpgi.00016.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vikström E, Tafazoli F, Magnusson KE. Pseudomonas aeruginosa quorum sensing molecule N-(3 oxododecanoyl)-l-homoserine lactone disrupts epithelial barrier integrity of Caco-2 cells. FEBS Lett. 2006 Dec;580((30)):6921–8. doi: 10.1016/j.febslet.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 128.Vikström E, Bui L, Konradsson P, Magnusson KE. The junctional integrity of epithelial cells is modulated by Pseudomonas aeruginosa quorum sensing molecule through phosphorylation-dependent mechanisms. Exp Cell Res. 2009 Jan;315((2)):313–26. doi: 10.1016/j.yexcr.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 129.Nakagami G, Morohoshi T, Ikeda T, Ohta Y, Sagara H, Huang L, et al. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in pressure ulcer infection in rats. Wound Repair Regen. 2011 Mar-Apr;19((2)):214–22. doi: 10.1111/j.1524-475X.2010.00653.x. [DOI] [PubMed] [Google Scholar]
- 130.Kanno E, Kawakami K, Miyairi S, Tanno H, Suzuki A, Kamimatsuno R, et al. Promotion of acute-phase skin wound healing by Pseudomonas aeruginosa C4 -HSL. Int Wound J. 2016 Dec;13((6)):1325–35. doi: 10.1111/iwj.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]