Abstract
Objective
Keratin 19 (K19) expression is a potential predictor of poor prognosis in patients with hepatocellular carcinoma (HCC). To clarify the feature of K19-proficient HCC, we traced epigenetic footprints in cultured cells and clinical materials.
Patients and Methods
In vitro, KRT19 promoter methylation was analyzed and 5-aza-2′-deoxycytidine with trichostatin A (TSA) treatment was performed. Among 564 surgically resected HCCs, the clinicopathological relevance of K19-proficent HCCs was performed in comparison with hepatocytic (HepPar-1 and arginase-1), epithelial-mesenchymal transition (E-cadherin and vimentin), biliary differentiation-associated (K7 and NOTCH-1) markers, and epigenetic markers (KRT19 promoter/long interspersed nucleotide element-1 [LINE-1] methylation status).
Results
KRT19 promoter methylation was clearly associated with K19 deficiency and 5-aza-2′-deoxycytidine with TSA treatment-stimulated K19 re-expression, implicating DNA methylation as a potential epigenetic process for K19 expression. After excluding HCCs with recurrence, TNM stage as IIIB or greater, preoperative therapy, transplantation, and combined hepatocellular cholangiocarcinoma, we assessed 125 of 564 HCC cases. In this cohort, K19 expression was found in 29 HCCs (23.2%) and corresponded with poor survival following surgery (p = 0.025) and extrahepatic recurrence-free survival (p = 0.017). Compared with K19-deficient HCCs, lower KRT19 promoter methylation level was observed in K19-proficient HCCs (p < 0.0001). Conversely, HCC with genome-wide LINE-1 hypermethylation was frequently observed in K19-proficient HCCs (p = 0.0079). Additionally, K19 proficiency was associated with K7 proficiency (p = 0.043), and reduced E-cadherin and HepPar-1 expression (p = 0.043 and p < 0.0001, respectively).
Conclusions
K19-proficient HCC exhibited poor prognosis owing to extrahepatic recurrence, with molecular signatures differing from those in conventional cancer stem cells, providing novel insights of the heterogeneity underlying tumor development.
Keywords: Keratin 19, KRT19, Hepatocellular carcinoma, Methylation, LINE-1
Introduction
Hepatocellular carcinoma (HCC) constitutes the sixth most common neoplasm and the third leading cause of cancer deaths worldwide [1, 2, 3]. Surgical resection and liver transplantation comprise the only curative treatments for patients with early-stage HCC. However, the high rate of recurrence or metastases leads to worse prognosis in HCC after curative resection [2].
The positive expression of keratin 19 (K19), a marker for biliary or hepatic progenitor cells and early hepatoblasts, has been significantly associated with poor prognosis along with stemness-related and epithelial-mesenchymal transition (EMT) features in HCC [4, 5, 6, 7, 8]. In addition, K19 proficiency in patients with HCC has frequently been associated with vascular invasion, poorly differentiated tumors, and tumor recurrence after resection, radiofrequency ablation, or transplantation [9]. However, to date the cell origin of K19-proficient HCC has remained unclear [6]. Multiple studies suggested that HCC cells with K19 proficiency were originated from hepatic progenitor cells as such cells express progenitor cell markers, have invasive potential, and exhibit chemoresistance [6, 9, 10, 11]. Conversely, others indicated that the expression of K19 in human HCCs may result from the dedifferentiation of malignant hepatocytes during continuous mutagenesis [12, 13, 14]. Furthermore, studies of liver regeneration have also found difficulties in identifying the cell origin of liver cancers including K19-proficient HCCs [15, 16].
Although the mechanisms contributing to hepatocarcinogenesis remain unclear, it is widely accepted that HCC exhibits numerous genetic abnormalities, such as chromosomal alterations, gene amplifications, and mutations, as well as epigenetic alterations [17]. For example, increased DNA methylation levels of tumor suppressor genes correlate positively with HCC development and progression [18, 19]; accordingly, a genome-wide methylation analysis has identified tumor suppressor genes in HCC [20]. However, although the gene encoding K19 (KRT19), located on chromosome 17 [21], contains a CpG island within its promoter region [22], to date there have been no reports regarding KRT19 promoter methylation in any malignancies. Thus, although K19 expression regulation mechanisms in HCC have not been fully elucidated, the presence of a promoter CpG island suggests DNA methylation as a potential epigenetic process in this malignancy.
In the present study, we attempted to reveal the features of K19-proficient HCC by tracing epigenetic footprints in cultured cells and clinical materials. Firstly, we examined epigenetic alterations and underlying molecular mechanisms of K19-positive HCC cell lines. Next, from a panel of 564 surgically resected HCCs, we clarified the clinicopathological relevance of K19-proficent HCCs by analyzing robust methylation in the KRT19 promoter region and LINE-1 elements in comparison with other cholangiocytic (K7), hepatocytic (HepPar-1 and arginase-1), EMT (E-cadherin and vimentin), and biliary differentiation-associated (NOTCH-1) markers.
Patients and Methods
Patients
We retrospectively analyzed consecutive patients with initial HCC who received surgical resection at Okayama University Hospital from January 2000 to December 2010. The histopathological diagnosis of HCC was based on the World Health Organization criteria. The clinical history, pathological reports, and hematoxylin-and-eosin-stained slides for all cases were reviewed to confirm the diagnosis. We excluded patients with a diagnosis of combined hepatocellular cholangiocarcinoma (CCA), with recurrent HCC, major vascular invasion, and with rupture or another organ invasion. We additionally excluded patients who received transplantation, noncurative resection, and preoperative therapy; e.g., transcatheter arterial chemoembolization. The tumor size was measured macroscopically after the removal of the tumor. Microvascular invasion was graded based on histopathological evaluation. The histological grade of tumor differentiation was determined according to the classification of Edmondson and Steiner (ES differentiation grade) [23]. Fibrosis of the nonneoplastic parenchyma was classified based on the Meta-analysis of Histologic Data in Viral Hepatitis (METAVIR) scoring system that assesses the degree of fibrosis ranging from F0 (no fibrosis) to F4 (cirrhosis) [24]. Tumor grade was classified according to the 7th edition of the UICC/AJCC TNM staging system [25]. Survival time was determined from the date of surgical resection. Local or remote recurrence of disease after surgery was investigated by clinical assessment and regular abdominal ultrasonography or computed tomography. The study was approved by the institutional review board of the Okayama University Hospital.
Cell Lines
K19-proficient HCC cell lines (HepG2, HuH7, and PLC/PRF/5); K19-deficient HCC cell lines (HLE and HLF); and a colon cancer cell line (HT29) as a K19-positive control were purchased from American Type Culture Collection (Manassas, VA, USA) [26]. All cell lines were cultured using Dulbecco's modified Eagle's medium (Sigma Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum, penicillin (100 IU/mL), and streptomycin (100 μg/mL) at 37°C in a humidified incubator with 5% CO2.
Immunohistochemistry Analysis
Immunohistochemistry (IHC) was performed using formalin-fixed, paraffin-embedded tissue sections of surgically resected liver specimens. After deparaffinization and blocking of the endogenous peroxidase, antigen retrieval was performed by microwaving with citrate buffer (pH 6.0) or TRIS-ethylenediaminetetraacetic acid (pH 9.0). The sections were incubated with primary antibodies against K19 (1: 200, mouse monoclonal, Cell Signaling Technology, Danvers, MA), keratin 7 (K7, 1: 100, mouse monoclonal; Daco), HepPar-1 (1: 100, mouse monoclonal; Daco, Glostrup, Denmark), arginase-1 (1: 5,000, rabbit polyclonal; Sigma-Aldrich), E-cadherin (1: 1, mouse monoclonal; Daco), and vimentin (1: 200, mouse monoclonal; Abcam, Cambridge, UK). Then, the sections were incubated with secondary antibodies conjugated to peroxidase-labeled polymer, using the EnVision system (Dako). Color development was performed using 3,3-diaminobenzidine and the sections were counterstained with hematoxylin. Negative controls were carried out by substitution of the primary antibodies with nonimmunized serum, but resulted in no signal detection. IHC results were interpreted by pathologists blinded to the corresponding clinicopathological data. K19, K7, NOTCH-1, and cytoplasmic vimentin were scored according to the percentage of tumor cells with positive staining as follows: 0 = < 5%, 1 = 5–10%, 2 = 11–20%, 3 = 21–50%, and 4 = 51–100%, and considered as positive when ≥5% of tumor cells were stained, as reported previously [4, 5, 6, 7, 8]. HepPar-1, arginase-1, and E-cadherin were considered as positive when ≥51% of tumor cells were stained [27] and they were scored using the percentage of tumor cells with positive staining as follows: 0 = 0–10%, 1 = 11–20%, 2 = 21–50%, 3 = 51–80%, and 4 = 81–100%.
Western Blotting
Protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto Immun-Blot polyvinylidene fluoride membranes (BIO-RAD, Hercules, CA, USA). After treating with 5% fat-free dried milk in 1× TBST for 1 h at room temperature, the membranes were incubated with mouse anti-human CK19 (1: 1,000; Cell Signaling Technology) for 1 h at room temperature, followed by secondary antibody for 1 h at room temperature. Actin was used as an internal positive control. Target proteins were detected using enhanced chemiluminescence (BIO-RAD).
Bisulfite Modification of DNA
Genomic DNA from the cell lines and formalin-fixed, paraffin-embedded specimens was extracted using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) and TaKaRa DEXPAT Kit (TaKaRa Bio Inc., Otsu, Japan), respectively. Approximately 1 μg DNA was subsequently modified using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA).
Bisulfite Sequencing
Polymerase chain reaction products from the KRT19 promoter were amplified by a set of primers (see online suppl. Table S1; for all online suppl. material, see www.karger.com/doi/10.1159/000490806) for bisulfite DNA cloning and sequencing. The KRT19 promoter was cloned from bisulfite-treated DNA into the pCR2.1TOPO vector using the TOPO-TA cloning system (Life Technologies, Carlsbad, CA, USA), followed by automated DNA sequencing with both the forward (F) and reverse (R) primers using an ABI 310-Avant NA sequencer (Applied Biosystems, Foster City, CA, USA).
Methylation Analysis of KRT19 and LINE-1
We performed quantitative methylation analysis for the promoter CpG islands of KRT19 and LINE-1 in tumors, their matched corresponding nontumor liver tissues, and cell lines. For KRT19 analysis, we used the high-sensitive assay for bisulfite DNA (Hi-SA), a modified combined bisulfite restriction analysis method, by which fluorescence-labeled DNA fragments are detected using a genetic analyzer [28]. The primer sequences are summarized in online supplementary Table S1, whereas the details for the remaining assays have been described previously [28, 29]. We analyzed two regions in the KRT19 promoter, termed region 1 and region 2. Region 2 contained two sites that can be restricted by HhaI. The methylation ratio was calculated by the ratio of the peak of methylated and unmethylated bands (examples are shown in online suppl. Fig. S1). We used primers for LINE-1 methylation analysis as described previously [30] with assay modification by adding a fluorescent dye to measure the methylation ratio using an ABI 310-Avant NA sequencer (see online suppl. Table S1). Additionally, we analyzed the association between methylation and expression for KRT19 of HCCs by the data obtained from cBioportal for cancer genomics (http://www.cbioportal.org/).
Demethylation Analysis
HLF, HLE, PLC/PRF/5, HepG2, and HuH7 cells were cultured with 5-aza-2′-deoxycytidine (5-aza-dC, Wako, Japan). 5-aza-dC was dissolved with phosphate-buffered saline and diluted with medium. The concentrations of 5-aza-dC were set as 1 μM according to the nadir of the promoter methylation rate in both regions 1 and 2 of KRT19 (see online suppl. Fig. S2). After 5-aza-dC treatment from day 1 to day 3, cells were treated by 100 μM trichostatin A (TSA, MERCK, Darmstadt, Germany), a potent histone deacetylase (HDAC) inhibitor, for 24 h. DNA and RNA were extracted after TSA treatment for determining methylation status and expression status by a microarray analysis (SurePrint G3 Human Gene Expression 8X60K v2, Agilent, Santa Clara, CA, USA).
Statistical Analysis
All statistical analyses were performed using JMP software (version 10.0; SAS Institute, Inc., Cary, NC, USA). We compared K19 expression status with various clinicopathological features and the results of other immunostaining using Fisher's exact test. Methylation levels in regions 1 and 2 of the KRT19 promoter were analyzed as both continuous and categorical variables (positive, methylation level in both regions 1 and 2 > 10%). Methylation levels in LINE-1 were categorized according to the mean value (≥55% methylation in LINE-1 defined as hypermethylation and < 55% defined as hypomethylation). Categorical variables were compared using Fisher's exact test. Differences between continuous variables were determined using the analysis of variance test (ANOVA). Correlation coefficient between continuous variables was nonparametrically determined (Spearman's correlation coefficient ρ). Overall survival (OS) was calculated from the date of surgical resection to the date of death owing to HCC or last follow-up for censored patients. Recurrence-free survival was calculated from the date of surgical resection to the date of the first documentation of local, regional, or distant relapse, appearance of a second primary lesion by computed tomography, and/or magnetic resonance imaging routinely performed every 6 months. Extrahepatic recurrence-free survival was calculated from the date of surgical resection to the date of the first documentation of appearance of extrahepatic recurrences by computed tomography and/or magnetic resonance imaging routinely performed every 6 months. OS, recurrence-free survival, and extrahepatic recurrence-free survival were estimated using the Kaplan-Meier method. Then, a multivariate analysis for OS was performed using a Cox proportional hazards model. All p values reported were calculated using two-sided tests and values < 0.05 were considered statistically significant.
Results
Association of KRT19 Promoter Methylation with K19 Expression in Cultured Cell Lines
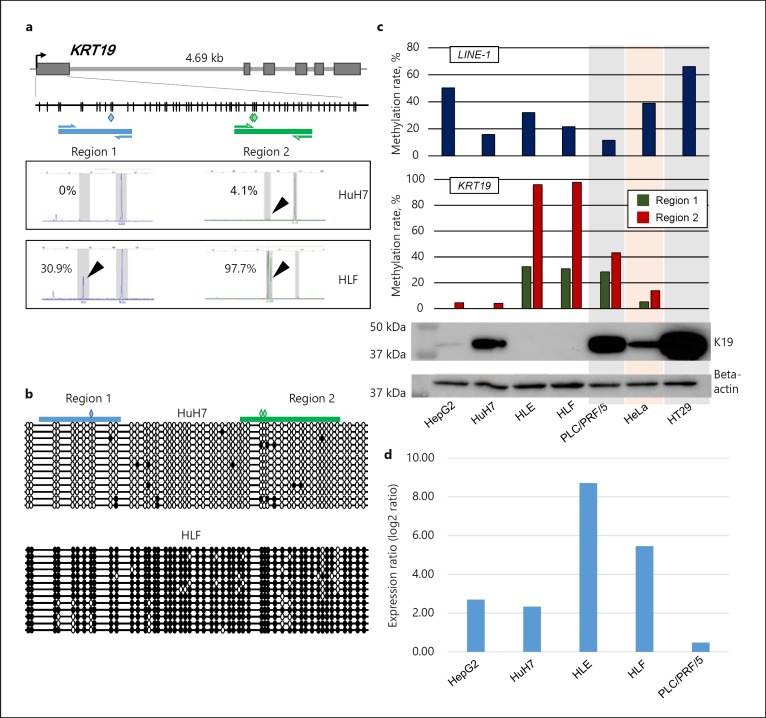
To examine methylation status in the KRT19 promoter, which contains dense CpG sites, the promoter region of KRT19 was divided into two regions (region 1 and region 2, Fig. 1a). Methylation status in the discrete regions was analyzed as a continual variable. Cloning and bisulfite sequencing were used to precisely confirm this methylation status, providing validation and further evidence that the K19-deficient HLF cells showed dense methylation throughout the KRT19 promoter CpG, whereas the K19-proficient HuH7 cells exhibited no methylation (Fig. 1b). Both regions in the KRT19 promoter were hypermethylated in K19-deficient cell lines (HLE and HLF) but were hypomethylated in K19-proficient cell clines (HepG2 and HuH7, Fig. 1c). The mean methylation ratio of K19-deficient and K19-proficient cells in region 1 was 31.7% (95% confidence interval [95% CI] 11.6–51.6) and 6.7% (−6.0 to 19.4), respectively (p = 0.0426), whereas those of K19-deficient and K19-proficient cells in region 2 were 96.8% (68.2–125.4) and 6.7% (−4.9 to 31.2), respectively (p = 0.0014). Thus, methylation levels in both regions of KRT19 were significantly inversely associated with K19 expression. We next treated 5 HCC cell lines (HepG2, HuH7, HLEE, HLF, and PLC/PRF/5) with a demethylating agent, 5′-aza-dC, and a potent HDAC inhibitor, TSA. Among them, the K19-deficient HLF and HLE cells with the higher methylation rate in the KRT19 promoter showed that, although 5′-aza-dC alone treatment caused demethylation in the KRT19 promoter but failed to recover KRT19 expression, 5′-aza-dC following TSA treatment successfully recovered expression level of KRT19 by the microarray analysis, indicating that K19 expression would be regulated by promoter methylation and histone modification (Fig. 1d).
Fig. 1.
Methylation and expression analyses of K19. a Schematic representation of the location of discrete KRT19 gene promoter regions and the results of KRT19 methylation assessment by a highly sensitive fluorescence assay for bisulfite DNA (Hi-SA). The gray squares denote the coding exon regions of the KRT19 gene. The blue and green squares represent the restriction sites for HhaI; vertical lines indicate CpG sites; thick horizontal blue and green lines represent polymerase chain reaction fragments; arrows represent primers; and black arrows point out methylated alleles cleaved by the restriction enzyme. b Cloning and sequencing of KRT19 regions 1 and 2. Polymerase chain reaction products that were amplified by primer sets for bisulfite cloning were cloned into a TOPO cloning vector and sequenced. For the two cell lines, at least 12 clones were sequenced. Empty circles indicate unmethylated CpG sites. Filled circles represent methylated CpG sites. c Expression of K19 protein in cell lines and methylation rates in the KRT19 gene and LINE-1. d Expression ratio (log2 ratio) of KRT19 messenger RNA in the 5 cell lines (HepG2, HuH7, HLE, HLF, and PLC/PRF/5). Expression ratio denotes log2 ratio obtained from the signal intensity of KRT19 messenger RNA from the cells treated with 5-aza-dC and TSA divided by the signal intensity of KRT19 messenger RNA from the cells untreated by a microarray analysis.
We also examined the genome global methylation level using LINE-1 retrotransposons, which constitute a substantial portion (approximately 17%) of the human genome and are regarded as a surrogate marker of global DNA methylation. Although the mean methylation ratio of LINE-1 of K19-proficient cells (36.5% [95% CI 2.5–60.5]) was higher than that of K19-deficient cells (26.8% [−11.2 to 64.8]), the difference was not statistically significant (p = 0.6, Fig. 1b).
Clinicopathological Features in Patients with K19-Proficient HCC
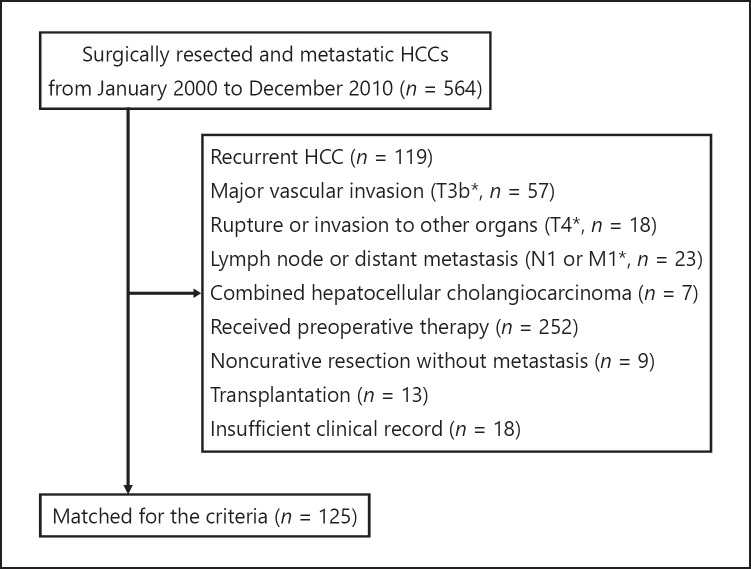
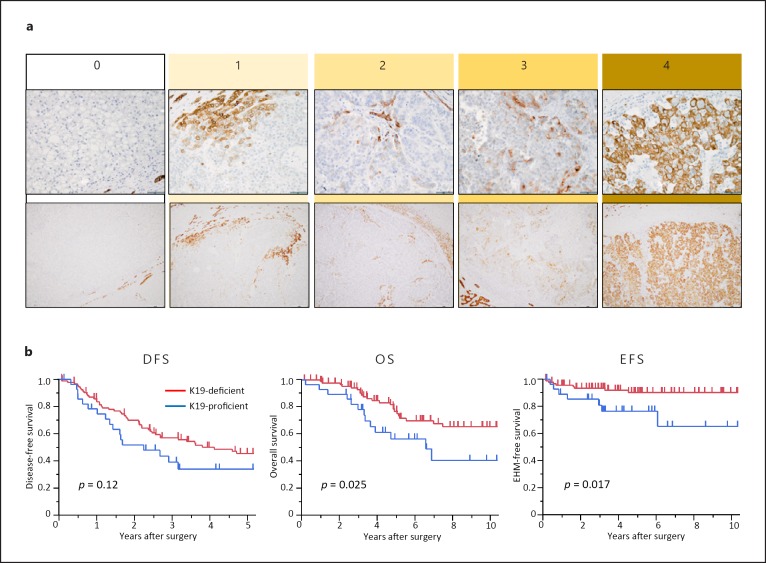
To assess the precise clinical landscapes of HCC with K19 proficiency, we retrospectively examined patients with HCC who underwent surgical resection. Among 564 patients recruited, 125 met the inclusion and exclusion criteria of this study (Fig. 2). The clinicopathological features of 125 patients are summarized in online supplementary Table S2. Of these, 113 patients (90.4%) were categorized in TNM stage I or II and no patients exhibited major vascular invasion. IHC analyses revealed that K19 expression was detected in 29 patients (23.2%) and that patients with K19-proficient HCC were significantly younger than those with K19-deficient HCC (p = 0.020; Fig. 3a; Table 1). Although a high level of serum α-fetoprotein and microvascular invasion were more frequently observed in patients with K19-proficient HCC (p = 0.021 and 0.019, respectively), K19 proficiency had no association with TNM stage, tumor size, tumor number, or differentiation. Patients with K19-proficient HCC showed poorer survival after surgery (p = 0.025) and poorer extrahepatic metastasis-free survival (p = 0.017; Fig. 3b–d). Multivariate analysis demonstrated that K19 acted as an independent prognostic factor for survival after surgery (Table 2).
Fig. 2.
STROBE diagram of the patient cohort.
Fig. 3.
Immunohistochemistry for K19 and clinical outcomes of 125 HCCs. a Examples of immunohistochemistry staining of K19 in HCCs classified into 5 groups according to the percentage of tumor cells with positive staining. b Kaplan-Meier survival curves for disease-free survival (DFS), overall survival (OS), and extrahepatic metastasis (EHM) recurrence-free survival (EFS) according to K19 expression status.
Table 1.
Comparison of clinicopathologic features between K19-proficient and deficient HCC
| Variable | All (n = 125) | K19-deficient HCC (n = 96) | K19-proficient HCC (n = 29) | p value1 |
|---|---|---|---|---|
| Age (mean ± SD), years | 65.1 (9.9) | 66.2 (8.76) | 61.3 (12.6) | 0.020 |
| Sex, n (%) | 0.027 | |||
| Female | 31 (25) | 19 (20) | 12 (41) | |
| Male | 94 (75) | 77 (80) | 17 (59) | |
| ECOG performance status, n (%) | 1.0 | |||
| 0 | 114 (91) | 87 (91) | 27 (93) | |
| 1–2 | 11 (9) | 9 (9) | 2 (7) | |
| HCV antibody, n (%) | 0.67 | |||
| Positive | 61 (50) | 48 (52) | 13 (46) | |
| Negative | 60 (50) | 45 (48) | 15 (54) | |
| Missing | 4 | 3 | 1 | |
| HBs antigen, n (%) | 0.33 | |||
| Positive | 32 (27) | 22 (24) | 10 (36) | |
| Negative | 86 (73) | 68 (76) | 18 (64) | |
| Missing | 7 | 6 | 1 | |
| AST, n (%) | 0.24 | |||
| ≥60 IU/L | 37 (31) | 26 (28) | 11 (41) | |
| <60 IU/L | 84 (69) | 68 (72) | 16 (59) | |
| Missing | 4 | 2 | 2 | |
| Total bilirubin, n (%) | 0.11 | |||
| ≥1.5 mg/dL | 9 (7) | 5 (5) | 4 (15) | |
| <1.5 mg/dL | 112 (93) | 89 (95) | 23 (85) | |
| Missing | 4 | 2 | 2 | |
| α-Fetoprotein, n (%) | 0.021 | |||
| ≥1,000 ng/mL | 14 (12) | 7 (8) | 7 (26) | |
| <1,000 ng/mL | 100 (88) | 80 (92) | 20 (74) | |
| Missing | 11 | 9 | 2 | |
| PIVKA-II, n (%) | 0.51 | |||
| ≥200 mAU/mL | 52 (46) | 38 (44) | 14 (52) | |
| <200 mAU/mL | 62 (54) | 49 (56) | 13 (48) | |
| Missing | 11 | 9 | 2 | |
| TNM stage, n (%) | 0.31 | |||
| I | 71 (57) | 58 (60) | 13 (45) | |
| II | 42 (34) | 30 (31) | 12 (41) | |
| IIIA | 12 (10) | 8 (8) | 4 (14) | |
| Tumor size, n (%) | 0.10 | |||
| ≥5 cm | 35 (28) | 23 (24) | 12 (41) | |
| <5 cm | 90 (72) | 73 (76) | 17 (59) | |
| Number of tumor, n (%) | 0.81 | |||
| Multiple | 31 (25) | 23 (24) | 8 (28) | |
| Single | 94 (75) | 73 (76) | 21 (72) | |
| Histology, n (%) | 0.11 | |||
| Poorly | 15 (12) | 9 (9) | 6 (21) | |
| Well/moderately | 110 (88) | 87 (91) | 23 (79) | |
| Microvascular invasion, n (%) | 0.019 | |||
| Present | 37 (30) | 23 (24) | 14 (48) | |
| Absent | 88 (70) | 73 (76) | 15 (52) | |
| Fibrosis stage2, n (%) | 1.0 | |||
| F0–3 | 85 (68) | 65 (68) | 20 (69) | |
| F4 | 40 (32) | 31 (32) | 9 (31) |
SD, standard deviation; HCV, hepatitis C virus; HBs, hepatitis B surface; AFP, α-fetoprotein.
ANOVA or Fisher's exact test.
New Inuyama classification which assesses the degree of fibrosis ranging from F0 (no fibrosis) to F4 (cirrhosis).
Table 2.
Univariate and multivariate analysis for overall survival
| Variables | Univariate analysisa |
Multivariate analysisa |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| K19 proficiency (vs. K19 deficiency) | 2.1 | 1.05–4.18 | 0.036 | 2.9 | 1.21–6.41 | 0.018 |
| Age ≥65 (vs. <65) | 1.6 | 0.83–3.37 | 0.16 | 1.7 | 0.75–4.00 | 0.21 |
| Female (vs. male) | 1.3 | 0.62–3.03 | 0.52 | 1.5 | 0.60–4.06 | 0.41 |
| ECOG PS ≥1 (vs. PS 0) | 3.7 | 1.07–9.65 | 0.04 | 2.70 | 0.66–8.96 | 0.15 |
| AFP ≥1,000 (vs. <1,000) | 0.7 | 0.16–1.90 | 0.50 | 0.50 | 0.10–1.84 | 0.31 |
| PIVKA-II ≥200 (vs. <200) | 1.5 | 0.74–2.93 | 0.27 | 1 | 0.45–2.35 | 0.92 |
| Number of tumor, multiple (vs. single) | 4.2 | 2.15–8.16 | <0.0001 | 3 | 1.26–6.87 | 0.013 |
| Tumor size ≥5 cm (vs. <5 cm) | 1.5 | 0.72–3.06 | 0.25 | 1.5 | 0.61–3.38 | 0.37 |
| Poor differentiation (vs. others) | 1.1 | 0.38–2.63 | 0.82 | 0.4 | 0.10–1.05 | 0.063 |
| Microvascular invasion, positive (vs. negative) | 2.70 | 1.36–5.23 | 0.0051 | 2.9 | 1.27–6.65 | 0.012 |
| Fibrosis stage F4b (vs. F0–3) | 2 | 1.03–3.88 | 0.040 | 2.1 | 0.94–4.55 | 0.070 |
HR, hazard ratio; CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; PS, performance status; AFP, α-fetoprotein; PIVKA-II, protein induced by vitamin K absence-II.
Cox proportional hazard model.
New Inuyama classification which assesses the degree of fibrosis ranging from F0 (no fibrosis) to F4 (cirrhosis).
K19 Expression and Promoter Methylation Status in the KRT19 Gene
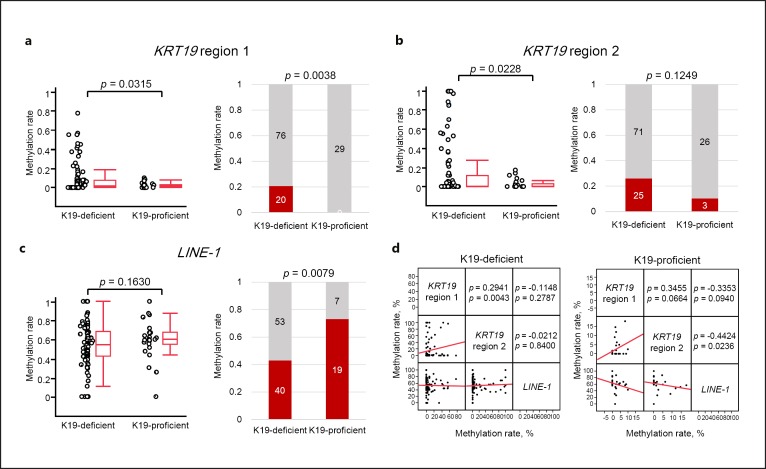
Next, we investigated the methylation levels of discrete regions in the KRT19 promoter in the 125 HCCs. Initially, methylation levels in the discrete regions obtained from fluorescent Hi-SA were analyzed as a continuous variable. In region 1, the mean methylation level was 2.3% (95% CI 1.2–3.5) among HCC tissues with K19 proficiency but 8.7% (95% CI 5.4–11.9) among HCC tissues with K19 deficiency (p = 0.0315, ANOVA; Fig. 4a). In region 2, the mean methylation levels were 2.7% (95% CI 0.9–4.5) among HCC tissues with K19 proficiency and 15.7% (95% CI 9.6–21.8) among HCC tissues with K19 deficiency (p = 0.0228, ANOVA; Fig. 4b). To define the threshold of methylation levels in region 1 and region 2 of the KRT19 promoter, we examined the methylation levels in the discrete regions of adjacent normal liver tissues. The mean methylation level in 123 adjacent normal liver tissues was 4.3% (95% CI 2.5–6.1) in region 1 and 8.3% in region 2. Therefore, we defined KRT19 methylation in both regions at 10% or more as a continuous variable (i.e., > 10% methylation as methylation-positive [methylated] and < 10% methylation as methylation-negative [unmethylated]). Using this criterion, we found that 20 (16.0%) and 28 (22.4%) of 125 HCCs could be categorized as methylated in region 1 and region 2 of the KRT19 promoter, respectively. All of the 20 HCCs (100%) categorized as methylated in region 1 showed K19 deficiency, whereas all of the 29 K19-proficient HCCs (100%) were unmethylated in region 1 (p = 0.0038, Fig. 4a). With respect to region 2, 25 of 28 HCCs (89.3%) with region 2 methylation showed K19 deficiency, whereas 26 of 29 HCCs (89.7%) with K19-proficient HCCs were unmethylated (p = 0.12, Fig. 4b). We also examined the association between methylation and expression for KRT19 of 442 HCCs by the data obtained from cBioportal for cancer genomics (see online suppl. Fig. S3). Although K19-deficient HCCs had various methylation levels in KRT19, most of K19-proficient HCCs showed lower methylation levels. Thus, HCCs with K19 proficiency demonstrated unmethylation in the discrete promoter regions of the KRT19 gene. Together with the results of our in vitro study, these findings suggest that the mechanism of K19 expression may be partially regulated by promoter methylation and histone modification in the KRT19 gene.
Fig. 4.
Methylation analyses of KRT19 and LINE-1 in 125 HCCs. a Results of methylation analysis in KRT19 region 1. Average methylation level (left panel) and methylation frequency (right panel) of KRT19 region 1 in HCCs with or without K19 proficiency. b Results of methylation analysis in KRT19 region 2. Average methylation level (left panel) and methylation frequency (right panel) of KRT19 region 2 in HCCs with or without K19 proficiency. c Results of methylation analysis in LINE-1. Average methylation level (left panel) and methylation frequency (right panel) of LINE-1 in HCCs with or without K19 proficiency. Left panel: in the box plot diagrams, the horizontal line within each box represents the median; the limits of each box are the interquartile ranges, and the whiskers are the maximum and minimum values. The p values were based on Kruskal-Wallis one-way analysis of variance on ranks and represent the statistical differences in average methylation between K19-deficient and proficient HCCs. Right panel: the numbers in each box denote the number of cases. The p values were based on Fisher's exact test. d Scatterplot matrix demonstrating the pairwise Spearman's correlation coefficient (ρ) between three analyzed loci in the cohort of 125 HCCs. The y and x axes denote methylation rates.
Association between LINE-1 Methylation and KRT19 Promoter Methylation in K19-Proficient HCCs
Evaluation of the LINE-1 methylation levels in the 119 HCC tissues demonstrated that the mean methylation level was 54.8% (95% CI 50.7–59.0; Fig. 4c). When we categorized LINE-1 methylation levels in the two groups according to the threshold of 55% (≥55% methylation defined as hypermethylation and < 55% defined as hypomethylation), hypermethylation in LINE-1 was frequently observed in K19-proficient (19 of 26 [73.1%]) compared with K19-deficient HCCs (40 of 93 [43.0%], p = 0.0079) (Fig. 4c).
We then asked whether an association exists between genome-wide methylation level and the local regions in the KRT19 promoter. For this, we performed multivariate correlation by comparing the methylation levels of LINE-1 and regions 1 and 2 in the KRT19 promoter from K19-proficient and -deficient HCCs, respectively (Fig. 4d, e). Regardless of K19 expression status, positive correlations were observed in the methylation level between regions 1 and 2 in the KRT19 promoter (Spearman's correlation coefficients ρ = 0.2941, p = 0.0043 in K19-deficient HCCs; ρ = 0.3455, p = 0.0644 in K19-proficient HCCs). In contrast, although there was no association between LINE-1 methylation and KRT19 promoter methylation levels in K19-deficient HCCs, an inverse correlation was observed in K19-proficient HCCs (ρ = −0.3353, p = 0.094 between LINE-1 and region 1; ρ = −0.4424, p = 0.0236 between LINE-1 and region 2).
Markers Associated with Expression Profiles of EMT, Hepatocytic and Biliary Differentiation
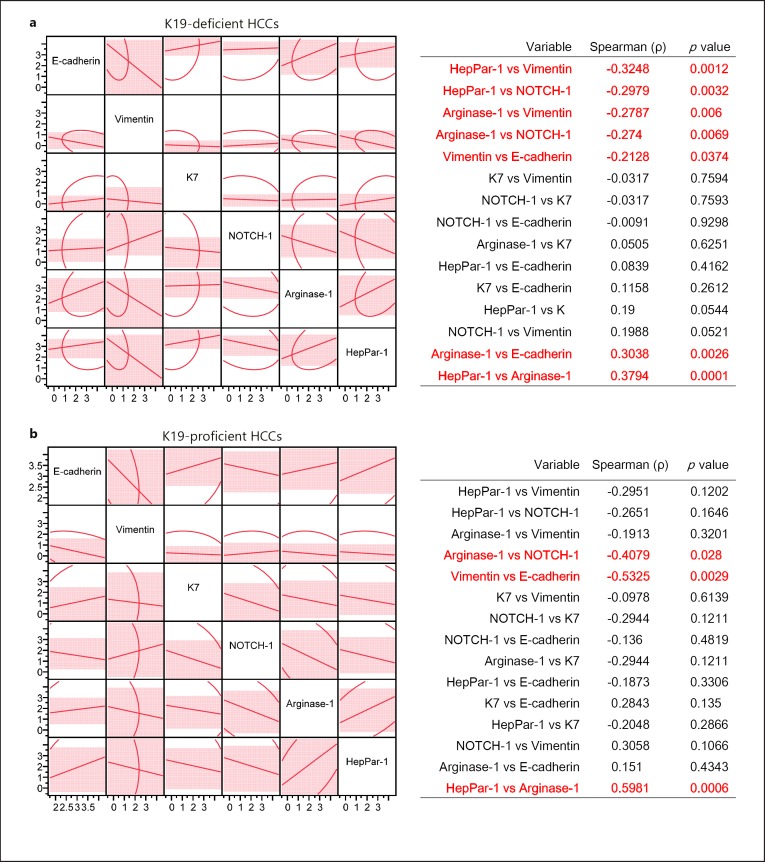
We next clarified features of HCC in relation to K19 expression status in comparison with EMT markers (E-cadherin and vimentin), hepatocytic markers (HepPar-1 and arginase-1), and markers associated with biliary differentiation (K7 and NOTCH-1). Examples of scoring of IHC staining are shown in online supplementary Figure S4. K19-proficient HCCs demonstrated increased EMT features with loss of E-cadherin (p = 0.043) and gain of vimentin expression (p = 0.084, Table 3). With respect to organ signature, findings of hepatocytic or cholangiocytic markers contrasted; although K19-proficient HCCs exhibited a decreased number of cases with positive expression of the hepatocytic markers HepPar-1 and arginase-1, they showed at least one positive result of staining for HepPar-1 or arginase-1. Thus, K19 expression correlated with an increase in EMT features accompanying K7-positive expression and a reduction of hepatocytic features. By multivariate correlations (Fig. 5), in K19-deficient HCCs, hepatocytic markers showed inverse associations with vimentin and NOTCH-1. Conversely, for HCCs with K19 proficiency, although expression of E-cadherin was decreased and that of K7 was increased, the positive association of HepPar-1 and arginase-1 expression (ρ = 0.5981, p = 0.0006) indicated that hepatocytic features were conserved.
Table 3.
Association between K19 expression status and other markers
| Variable | All (n = 125) n (%) | K19-deficient HCC (n = 96) n (%) | K19-proficient HCC (n = 29) n (%) | p value |
|---|---|---|---|---|
| E-cadherin | ||||
| Proficient | 106 (85) | 85 (89) | 21 (72) | 0.043 |
| Deficient | 19 (15) | 11(12) | 8 (28) | |
| Vimentin | ||||
| Proficient | 8 (6) | 4 (4) | 4 (14) | 0.084 |
| Deficient | 117 (94) | 92 (96) | 25 (86) | |
| K7 | ||||
| Proficient | 43 (34) | 28 (29) | 15 (52) | 0.043 |
| Deficient | 82 (66) | 68 (71) | 14 (48) | |
| NOTCH-1 | ||||
| Proficient | 72 (58) | 55 (57) | 17 (59) | 1.00 |
| Deficient | 53 (42) | 41 (43) | 12 (41) | |
| HepPar-1 | ||||
| Proficient | 90 (72) | 74 (77) | 16 (55) | 0.033 |
| Deficient | 35 (28) | 22 (23) | 13 (45) | |
| Arginase-1 | ||||
| Proficient | 87 (70) | 76 (79) | 11 (34) | <0.0001 |
| Deficient | 38 (30) | 20 (21) | 18 (66) | |
| LINE-1 methylation ratio | ||||
| Mean % (95% CI) | 53.3 (48.6–58.1) | 60.3 (52.0–68.6) | ||
| 55% or more | 59 (50) | 40 (43) | 19 (73) | 0.0079* |
| Under55% | 60 (50) | 53 (57) | 7 (27) | |
| Not analyzed | 6 | 3 | 3 | |
Fig. 5.
Correlation between 6 markers examined by immunohistochemical (IHC) staining in125 HCCs. a, b Scatterplot matrices demonstrating the pairwise Spearman's correlation coefficient (ρ) between 6 analyzed markers in the cohort of 96 K19-deficient HCCs (a) and of 29 K19-proficient HCCs (b). The y and x axes denote IHC score.
Discussion
Herein, we have shown the biological significance of HCC with K19 proficiency with relation to methylation status in the promoter region of KRT19, genome-wide methylation level of LINE-1, and EMT features. Results from clinical samples as well as our in vitro study indicated that K19-deficient HCCs demonstrated a higher methylation level in the promoter region of KRT19 compared with K19-proficient HCCs, suggesting that K19 expression might be regulated by the density of its promoter methylation. Our demethylation analysis demonstrated that re-expression of KRT19 required not only application of a demethylating agent, 5′-aza-dC, but also a HDAC inhibitor, TSA, in K19-deficient HLF and HLE cells, suggesting that epigenetic mechanisms including histone modification play an important role in K19 expression.
Consistent with previous studies [4, 5, 6, 7, 8], we confirmed the utility of K19 as a prognostic biomarker for HCC. Notably, we extracted 125 HCCs from 564 surgically resected HCCs according to exclusion criteria that excluded HCCs with major vascular invasion pathologically, which is the strongest prognostic factor for this malignancy. We also excluded recurrent HCCs for accurate survival analysis, and HCCs with preoperative transcatheter arterial chemoembolization because such treatment might influence the tumor characteristics including K19 expression on the membrane of the cancer cells. Thus, the 125 HCCs analyzed in this study were, as far as possible, homogeneous in both clinical and pathological settings. Of this cohort, K19 proficiency was observed in 23.2% (29/125) HCCs and was more frequently observed in younger, female patients. Other clinical features of K19-proficient HCCs included higher serum α-fetoprotein levels, positive for pathological microvascular invasion, and poor survival after surgery.
In this study, K19-proficient HCCs exhibited EMT features with loss of E-cadherin and gain of vimentin expression. This association between EMT features and K19 proficiency was consistent with a prior study reporting that K19-proficient HCC exhibited significantly increased EMT-related protein and mRNA expression [5]. EMT has been shown to be a pivotal mechanism contributing to cancer invasion and metastasis including HCC [31, 32]. Consistent with this hypothesis, patients with K19-proficient HCC in this and other studies [7, 33] showed poorer survival and more frequently exhibited extrahepatic metastasis.
Generally, primary liver cancers are classified into the following subtypes: HCCs, CCAs, combined HCC-CCAs, hepatoblastomas, and fibrolamellar hepatocellular carcinomas [34]. K19 is commonly expressed in two types of liver cancers, CCAs and combined HCC-CCAs; however, their cell origins and means of development are not yet sufficiently understood. Recent studies using a mouse model of hepatocyte fate tracing have revealed that CCAs may be originated from fully differentiated hepatocytes via NOTCH signaling activation [35, 36]. Combined HCC-CCAs are pathologically diagnosed based on the classical type of combined HCC-CCAs as areas of typical HCC and CCA mixed within the tumor, with the latest edition of this classification proposing a subtype with stem cell features [37].
Kawai et al. [38] reported that K19-proficient HCCs possessed cancer stem cell features, such as EMT features and the activation of the TGFb/Smad signal cascade. Zhang et al. [39] reported that HCCs expressed a stem cell marker CD133, considered to be a marker for cancer stem cells of HCC, and showed hypomethylation of the global DNA methylation marker LINE-1. Therefore, we next examined the association between K19-proficent HCCs and LINE-1 methylation levels associated with cancer stem cell features. Notably, in our cohort, K19-proficent HCCs exhibited LINE-1 hypermethylation that was significantly correlated with demethylation in the KRT19 promoter. This evidence obtained from clinical samples was also supported by our in vitro study that K19-proficient cell lines were more likely showing an increased LINE-1 methylation level (Fig. 1b). As Kim et al. [5] demonstrated that only 1.5% of HCCs showed both CD133 and K19 expression, it is reasonable to presume that the K19-proficient HCCs were not equivalent to the CD133-proficient HCCs that are associated with LINE-1 hypomethylation. With respect to IHC staining, our cohort of HCCs showed inverse correlation in expression status between NOTCH-1 and hepatocytic markers irrespective of K19 expression status. Although K19-proficient HCCs demonstrated increased EMT and cholangiocytic features and a reduced number of cases with a positive expression of hepatocytic markers, strongly conserved hepatocytic features were still observed.
This study has some limitations. For example, analyzed samples were obtained from a retrospective cohort in a single hospital. However, this study describes the novel features observed in HCCs with K19 proficiency: increased EMT features and decreased mature signatures of hepatocyte cells, increased genome-wide DNA methylation levels, and reduced promoter methylation density in the KRT19 gene. Additionally, in vitro analysis revealed that expression of K19 was regulated under promoter methylation and histone modification. Our clinical data suggest that not only may the detection of K19 proficiency in HCC be useful as a biomarker for identifying patients who have a poor prognosis with extrahepatic recurrence, but also that K19-proficient HCCs likely arise from hepatocytes or HCCs via epigenetic reprogramming, leading to EMT features. Thus, our findings provide novel insights regarding the heterogeneity underlying tumor development.
Statement of Ethics
The human tissues used in this study were approved by the institute ethics committee of Okayama University.
Disclosure Statement
The authors have no conflicts of interest that pertain to this work.
Funding Sources
This work was supported by grants from MEXT/JSPS KAKENHI (20590572, 25860409, 26462016, and 15H03034 to T.N.).
Author Contributions
N.Y. performed experiments and analyzed data supported by Y.U., F.T., K.Y., Toshi. T., Y.M., and A.N.; N.N. and M.K. performed all microarray analyses; Take. T. and Take. Y. performed all immunohistochemistry analyses. Taka. Y. and T.F. prepared all samples and summarized clinical information; Y.Y. and A.G. analyzed data and wrote the paper; T.N. obtained funding, designed the work, analyzed data, and wrote the paper.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgment
The authors would like to thank Mr. Toru Nakai and Mrs. Tae Yamanishi for the immunohistochemistry techniques.
References
- 1.Boyle P, Levin B, International Agency for Research on Cancer World Health Organization: World Cancer Report 2008. Lyon, International Agency for Research on Cancer, Distributed by WHO Press. 2008 [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 4.Wu PC, Fang JW, Lau VK, Lai CL, Lo CK, Lau JY. Classification of hepatocellular carcinoma according to hepatocellular and biliary differentiation markers. Clinical and biological implications. Am J Pathol. 1996;149:1167–1175. [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE, Cho JY, Yoo JE, Choi JS, Park YN. Human hepatocellular carcinomas with “Stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 2011;54:1707–1717. doi: 10.1002/hep.24559. [DOI] [PubMed] [Google Scholar]
- 6.Govaere O, Komuta M, Berkers J, Spee B, Janssen C, de Luca F, Katoonizadeh A, Wouters J, van Kempen LC, Durnez A, Verslype C, De Kock J, Rogiers V, van Grunsven LA, Topal B, Pirenne J, Vankelecom H, Nevens F, van den Oord J, Pinzani M, Roskams T. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut. 2014;63:674–685. doi: 10.1136/gutjnl-2012-304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uenishi T, Kubo S, Yamamoto T, Shuto T, Ogawa M, Tanaka H, Tanaka S, Kaneda K, Hirohashi K. Cytokeratin 19 expression in hepatocellular carcinoma predicts early postoperative recurrence. Cancer Sci. 2003;94:851–857. doi: 10.1111/j.1349-7006.2003.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durnez A, Verslype C, Nevens F, Fevery J, Aerts R, Pirenne J, Lesaffre E, Libbrecht L, Desmet V, Roskams T. The clinicopathological and prognostic relevance of cytokeratin 7 and 19 expression in hepatocellular carcinoma. A possible progenitor cell origin. Histopathology. 2006;49:138–151. doi: 10.1111/j.1365-2559.2006.02468.x. [DOI] [PubMed] [Google Scholar]
- 9.Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H, Thung SN, Bruix J, Newell P, April C, Fan JB, Roayaie S, Mazzaferro V, Schwartz ME, Llovet JM. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512. doi: 10.1053/j.gastro.2011.02.006. e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villanueva A, Hoshida Y, Toffanin S, Lachenmayer A, Alsinet C, Savic R, Cornella H, Llovet JM. New strategies in hepatocellular carcinoma: genomic prognostic markers. Clin Cancer Res. 2010;16:4688–4694. doi: 10.1158/1078-0432.CCR-09-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen JB, Loi R, Perra A, Factor VM, Ledda-Columbano GM, Columbano A, Thorgeirsson SS. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology. 2010;51:1401–1409. doi: 10.1002/hep.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos NP, Oliveira PA, Arantes-Rodrigues R, Faustino-Rocha AI, Colaco A, Lopes C, Gil da Costa RM. Cytokeratin 7/19 expression in N-diethylnitrosamine-induced mouse hepatocellular lesions: implications for histogenesis. Int J Exp Pathol. 2014;95:191–198. doi: 10.1111/iep.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezzoukhry Z, Louandre C, Trecherel E, Godin C, Chauffert B, Dupont S, Diouf M, Barbare JC, Maziere JC, Galmiche A. EGFR activation is a potential determinant of primary resistance of hepatocellular carcinoma cells to sorafenib. Int J Cancer. 2012;131:2961–2969. doi: 10.1002/ijc.27604. [DOI] [PubMed] [Google Scholar]
- 14.Yoneda N, Sato Y, Kitao A, Ikeda H, Sawada-Kitamura S, Miyakoshi M, Harada K, Sasaki M, Matsui O, Nakanuma Y. Epidermal growth factor induces cytokeratin 19 expression accompanied by increased growth abilities in human hepatocellular carcinoma. Lab Invest. 2011;91:262–272. doi: 10.1038/labinvest.2010.161. [DOI] [PubMed] [Google Scholar]
- 15.Itoh T, Miyajima A. Liver regeneration by stem/progenitor cells. Hepatology. 2014;59:1617–1626. doi: 10.1002/hep.26753. [DOI] [PubMed] [Google Scholar]
- 16.Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011;12:130–137. doi: 10.2174/138920211795564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida N, Kudo M, Nagasaka T, Ikai I, Goel A. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology. 2012;56:994–1003. doi: 10.1002/hep.25706. [DOI] [PubMed] [Google Scholar]
- 20.Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM, Powers S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145:1424.e1–25–1435.e1–25. doi: 10.1053/j.gastro.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schweizer J, Bowden PE, Coulombe PA, Langbein L, Lane EB, Magin TM, Maltais L, Omary MB, Parry DA, Rogers MA, Wright MW. New consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, et al. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature. 2006;440:1045–1049. doi: 10.1038/nature04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Ichida F, Tsuji T, Omata M, Ichida T, Inoue K, Kamimura T, Yamada G, Hino K, Yokosuka O, Suzuki H. New Inuyama classification; new criteria for histological assessment of chronic hepatitis. Int Hepatol Commun. 1996;6:112–119. [Google Scholar]
- 25.Edge S, Byrd D, Compton C, Fritz A, Greene F, Trotti A. ed 7. New York: Springer; 2010. AJCC Cancer Staging Manual. [Google Scholar]
- 26.Wu F, Nishioka M, Fujita J, Murota M, Ohtsuki Y, Ishida T, Kuriyama S. Expression of cytokeratin 19 in human hepatocellular carcinoma cell lines. Int J Oncol. 2002;20:31–37. [PubMed] [Google Scholar]
- 27.Yan BC, Gong C, Song J, Krausz T, Tretiakova M, Hyjek E, Al-Ahmadie H, Alves V, Xiao SY, Anders RA, Hart JA. Arginase-1: a new immunohistochemical marker of hepatocytes and hepatocellular neoplasms. Am J Surg Pathol. 2010;34:1147–1154. doi: 10.1097/PAS.0b013e3181e5dffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagasaka T, Tanaka N, Cullings HM, Sun DS, Sasamoto H, Uchida T, Koi M, Nishida N, Naomoto Y, Boland CR, Matsubara N, Goel A. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009;101:1244–1258. doi: 10.1093/jnci/djp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida K, Nagasaka T, Umeda Y, Tanaka T, Kimura K, Taniguchi F, Fuji T, Shigeyasu K, Mori Y, Yanai H, Yagi T, Goel A, Fujiwara T. Expansion of epigenetic alterations in EFEMP1 promoter predicts malignant formation in pancreatobiliary intraductal papillary mucinous neoplasms. J Cancer Res Clin Oncol. 2016;142:1557–1569. doi: 10.1007/s00432-016-2164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lertkhachonsuk R, Paiwattananupant K, Tantbirojn P, Rattanatanyong P, Mutirangura A. LINE-1 methylation patterns as a predictor of postmolar gestational trophoblastic neoplasia. Biomed Res Int. 2015;2015:421747. doi: 10.1155/2015/421747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Ma CQ, Yang Y, Wang JM, Du GS, Shen Q, Liu Y, Zhang J, Hu JL, Zhu P, Qi WP, Qian YW, Fu Y. The aPKCiota blocking agent ATM negatively regulates EMT and invasion of hepatocellular carcinoma. Cell Death Dis. 2014;5:e1129. doi: 10.1038/cddis.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang PY, Zhang JB, Zhu XD, Zhang W, Wu WZ, Tan YS, Hou J, Tang ZY, Qin LX, Sun HC. Two pathologic types of hepatocellular carcinoma with lymph node metastasis with distinct prognosis on the basis of CK19 expression in tumor. Cancer. 2008;112:2740–2748. doi: 10.1002/cncr.23488. [DOI] [PubMed] [Google Scholar]
- 34.Oikawa T. Cancer stem cells and their cellular origins in primary liver and biliary tract cancers. Hepatology. 2016;64:645–651. doi: 10.1002/hep.28485. [DOI] [PubMed] [Google Scholar]
- 35.Sekiya S, Suzuki A. Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes. J Clin Invest. 2012;122:3914–3918. doi: 10.1172/JCI63065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan B, Malato Y, Calvisi DF, Naqvi S, Razumilava N, Ribback S, Gores GJ, Dombrowski F, Evert M, Chen X, Willenbring H. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122:2911–2915. doi: 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosman FT, World Health Organization International Agency for Research on Cancer . Lyon, International Agency for Research on Cancer. ed 4 2010. WHO Classification of Tumours of the Digestive System. [Google Scholar]
- 38.Kawai T, Yasuchika K, Ishii T, Katayama H, Yoshitoshi EY, Ogiso S, Kita S, Yasuda K, Fukumitsu K, Mizumoto M, Hatano E, Uemoto S. Keratin 19: a cancer stem cell marker in human hepatocellular carcinoma. Clin Cancer Res. 2015;21:3081–3091. doi: 10.1158/1078-0432.CCR-14-1936. [DOI] [PubMed] [Google Scholar]
- 39.Zhang C, Xu Y, Zhao J, Fan L, Jiang G, Li R, Ling Y, Wu M, Wei L. Elevated expression of the stem cell marker CD133 associated with Line-1 demethylation in hepatocellular carcinoma. Ann Surg Oncol. 2011;18:2373–2380. doi: 10.1245/s10434-011-1599-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data