Abstract
Complement resistance is an important virulence trait of Yersinia enterocolitica (Ye). The predominant virulence factor expressed by Ye is Yersinia adhesin A (YadA), which enables bacterial attachment to host cells and extracellular matrix and additionally allows the acquisition of soluble serum factors. The serum glycoprotein vitronectin (Vn) acts as an inhibitory regulator of the terminal complement complex by inhibiting the lytic pore formation. Here, we show YadA-mediated direct interaction of Ye with Vn and investigated the role of this Vn binding during mouse infection in vivo. Using different Yersinia strains, we identified a short stretch in the YadA head domain of Ye O:9 E40, similar to the ‘uptake region’ of Y. pseudotuberculosis YPIII YadA, as crucial for efficient Vn binding. Using recombinant fragments of Vn, we found the C-terminal part of Vn, including heparin-binding domain 3, to be responsible for binding to YadA. Moreover, we found that Vn bound to the bacterial surface is still functionally active and thus inhibits C5b-9 formation. In a mouse infection model, we demonstrate that Vn reduces complement-mediated killing of Ye O:9 E40 and, thus, improved bacterial survival. Taken together, these findings show that YadA-mediated Vn binding influences Ye pathogenesis.
Keywords: Bacterial infection, Cell surface molecules, Complement, Yersinia adhesin A, Vitronectin
Introduction
Yersinia enterocolitica (Ye) and Yersinia pseudotuberculosis (Yps) are enteropathogens causing enteric and systemic diseases [1, 2]. Besides the chromosomally encoded adhesins invasin (Inv) and Ail [3, 4, 5], the trimeric autotransporter adhesin (TAA) Yersinia adhesin A (YadA) is the decisive factor that determines the pathogenicity of Ye[6]. YadA forms rigid fibrous structures, which protrude approximately 23 nm from the cell surface [7, 8], and mediates adhesion to extracellular matrix (ECM) proteins such as collagen, fibronectin and laminin and also to complement factors [9]. Being the prototype of the TAA family of proteins, YadA is characterized by a modular composition of several domains; the extracellularly located N-terminal head domain is followed by a connector element (also called the neck region) leading into a coiled-coil stalk. The stalk is connected to the C-terminal translocator or membrane anchor domain, consisting of 4 β-strands per monomer [9]. To form a functional adhesin on the bacterial surface, 3 YadA monomers trimerize and form the pore of the translocator domain, which is inserted into the outer membrane [10]. The translocator enables the transport of the passenger domains onto the bacterial surface, where they also form obligate trimers [9].
YadA knockout strains of Ye are avirulent and do not cause infection in a mouse model [11, 12, 13]. This striking effect has been attributed mainly to the reduced efficiency of effector protein (Yop) delivery by a dedicated type 3 secretion system (T3SS) which requires proper adhesion to host cells; loss of adherence results in the inability to resist phagocytosis [14, 15]. However, in Yps, which is more closely related to Yersinia pestis, YadA is dispensable for virulence and Yop injection [16]. YadA of Yps and Ye not only differ in their role during infection, but also in the sequence and binding repertoire of host ECM proteins and cellular receptors. YadA of Yps carries an additional stretch within its head region that enables entry into host cells [17]. This important stretch is absent in YadA of several Ye serotypes and strains. Moreover, the binding capacities of YadA differ between Ye, which binds collagen and laminin, and Yps, which binds fibronectin [18].
By interacting with several complement factors, serum resistance is an important virulence trait of Ye. It has been shown that factor H, C4b-binding protein (C4BP) and C3 bind to the YadA stalk domain and thus inhibit complement killing [19, 20]. Recently, we demonstrated a novel mechanism that contributes to serum resistance in Ye O:8 WA-314, and amended the current model of direct factor H binding to YadA0:3 and YadA0:9. We have shown that Ye binds C3b or iC3b and thereby attracts high amounts of factor H to the bacterial surface [21]. This is different from the direct binding of factor H, which was shown earlier [19, 20, 22]. Importantly, by binding these complement regulatory factors, Ye is able to interfere with complement activity by inhibiting complement-mediated killing at an early stage of the cascade.
The human glycoprotein Vn is synthesized in the liver and secreted into plasma [23], where it is present as a monomer (65 and 75 kDa) at high concentrations (200–400 µg/ml) [24]. Vn also exists as an extravascular cell-bound multimeric form in several tissues, and Vn mRNA can be detected in high concentrations in the liver, brain, heart and adipose tissue but is rare or absent in the kidney and spleen [25]. It comprises an N-terminal somatomedin-binding domain, consisting of 43 amino acid (aa) residues, followed by the host cell integrin receptor-binding motif RGD (Arg-Gly-Asp). In addition to 4 hemopexin-like domains with unknown function, Vn also contains 3 heparin-binding domains (HBDs) which span aa 82–137 (HBD-1), aa 175–219 (HBD-2) and aa 348–361 (HBD-3) [26, 27]. Vn is an important regulator of complement activity at the level of terminal complement complex (TCC) formation and a component of the ECM, and it also fulfills functions in cell migration and tissue repair [27].
At the level of TCC formation, Vn regulates complement activity by directly binding to the protein complex C5b-7 or to C9 [28]. The exact mode of regulation is not fully understood. It has been postulated, however, that Vn binds the nascent precursor complex C5b-7, resulting in a Vn-C5b-7 complex that is unable to insert into the cell membrane [27, 28]. Vn can also directly bind C9 and thereby inhibit C9 polymerization. This binding takes places through HBD-3 whereas the binding site for the nascent C5b-7 is still unknown [27, 28, 29].
A wide variety of bacteria bind Vn via various surface proteins. The respiratory pathogens Moraxella catarrhalis (Mc) and Haemophilus influenzae (Hi) as well as the urogenital pathogen Haemophilus ducreyi express proteins belonging to the TAA family. These proteins are the ubiquitous surface protein A2 (UspA2) of Mc, the Haemophilus surface fibrils (Hsf) and the Haemophilus adhesin (Hia) of Hi or the H. ducreyi serum resistance protein A (DsrA) [9, 30, 31, 32, 33, 34, 35, 36]. In the invasive bacterial pathogen Neisseriameningitidis the 3 proteins Opc, Opa and Msf interact with Vn [37, 38, 39, 40]. However, to date, no enteropathogenic bacteria have been reported to use Vn to escape complement-mediated attack and thus mediate serum resistance.
Ye has evolved a multitude of mechanisms to evade the host immune system. Amongst these, serum resistance is of uttermost importance. The significance of the complement regulator Vn in complement evasion and modulation of host cell interaction with bacterial and fungal pathogens has recently been recognized [27, 30, 31, 32, 37, 39, 40, 41, 42, 43, 44]. Ye is able to bind several regulators of complement activity. The role of Vn in Ye host cell interaction and in pathogenicity has not yet been addressed in detail, but it was shown in previous studies that YadA from Ye O:8 does not bind Vn under stringent assay conditions [45]. In this study, we systematically investigated (1) Vn binding of different Ye strains, (2) which components of Ye might enable this binding and (3) how this interaction modulates Ye serum resistance, host cell interaction and overall pathogenicity. Importantly, we were able to demonstrate a novel mechanism that facilitates Ye serum resistance mediated by the surface adhesin YadA binding to Vn. We found that subtle differences within the YadA head domain of different Yersinia strains determine the efficacy of the Vn binding. An additional stretch in Ye YadAO:9, which is similar to the ‘uptake region’ of Yps YadAYPIII[18], was identified as a crucial region for the high-affinity binding of Vn. Moreover, we located HBD-3 within Vn as the YadA binding site. Notably, bound Vn is active on the bacterial surface and protects bacteria from complement-mediated lysis by the inhibition of C9 polymerization. This mechanism allows the enhanced survival of Ye O:9 E40 during the early phase of a mouse infection in vivo.
Materials and Methods
Mice
C57BL/6 wild-type (WT) mice were purchased from Harlan Winkelmann (Horst, The Netherlands). B6.129S2(D2)-Vtntm1Dgi/J mice (http://jaxmice.jax.org/strain/004371.html) with a C57BL/6 background were purchased from Jackson Laboratories (Bar Harbor, Maine, USA). All mice were bred under specific pathogen-free conditions in individually ventilated cages with access to water and food ad libitum. Experiments were performed with female mice (aged 6–8 weeks) according to German law with the permission of the Regierungspräsidium Tübingen (permission No. H4/15).
Plasmids
Plasmids used in this study are listed in table 1.
Table 1.
Plasmids used in this study
| Plasmid name | Description | Resistance | Reference |
|---|---|---|---|
| pBla | expression of YopE aa 1–53 β-lactamase hybrid protein under control of the YopE promoter | kanamycin | 46 |
| pACYC184 EGFP | EGFP expressed under control of a constitutive tac/lac promoter | chloramphenicol | 47 |
| pASK-IBA4C_yadAO:8 | yadA from Ye O:8 WA-314 cloned into pASK-IBA4C; expression under control of an anhydrotetracycline-inducible promoter | chloramphenicol | this study |
| pASK-IBA4C_yadAO:9 | yadA from Ye O:9 E40 cloned into pASK-IBA4C; expression under control of an anhydrotetracycline-inducible promoter | chloramphenicol | this study |
| pASK-IBA4C_yadAO:9/ O:8 hybrid | plasmid for inducible expression of a hybrid protein consisting of the N-terminal aa 1–89 of yadA from Ye O:9 E40 fused to aa 55–422 of yadA from Ye O:8 WA-314; expression under control of an anhydrotetracycline-inducible promoter | chloramphenicol | this study |
| pASK-IBA4C_yadAO:9 Δuptake region | plasmid for inducible expression of yadA from Ye O:9 E40 lacking aa 60–86 comprising the uptake region; expression under control of an anhydrotetracycline-inducible promoter | chloramphenicol | this study |
Bacterial Strains and Culture Conditions
All Yersinia strains were cultivated in lysogeny broth medium with supplements (antibiotics as listed in table 2) overnight at 27°C. To promote YadA expression, a 1:20 dilution of the overnight culture was made with fresh medium and incubated for 3 h at 37°C. Moraxella strains were grown overnight at 37°C in brain-heart infusion medium. All bacteria were washed twice with PBS, and the optical density at 600 nm was determined. The number of bacteria used for the individual experimental setups are indicated in the respective sections. All bacterial strains used in this study are listed in table 2.
Table 2.
Bacterial strains used in this study
| Bacterial strain | Description | Resistance | Reference |
|---|---|---|---|
| Ye O:3 6471/76 | serotype O:3, fecal isolate, WT | – | 48 (GI:48607) |
| Ye O:8 8081 | serotype O:8, fecal isolate, WT | – | 49 (GI:122815846) |
| Ye O:8 WA-314 YadAwt | coding sequence of YadA WA-314 O:8 was reinserted into a YadA0 strain | Nal, Kan, Spec | 12 (GI:310923211) |
| Ye O:9 E40 pBla | Ye O:9 E40 Δasd transformed with pMK-Bla | Nal, Kan, Ars | 46 (GI:972903261) |
| Ye O:9 E40 ΔpYV pBla | Ye O:9 E40 Δasd without virulence plasmid transformed with pMK-Bla | Nal, Kan | 46 |
| Ye O:9 E40 ΔInv pBla | Inv mutant strain obtained by recombinational integration of suicide plasmid pMS154 into E40 Δasd, transformed with pMK-Bla | Nal, Kan, Ars, Tet | 47 |
| Ye O:9 E40 ΔYadA pBla | pYV-Δasd strain was transformed with pLJM4029 (YadA-) and with pMK-Bla | Nal, Kan, Ars, Strep | 47 |
| Ye O:9 E40 ΔInv ΔYadA pBla | pYV-Δasd ΔInv strain was transformed with pLJM4029 (YadA-) and with pMK-Bla | Nal, Kan, Ars, Tet, Strep | 47 |
| Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:8 | Ye O:9 E40 Δasd lacking expression of both YadA and Inv transformed with pASK-IBA4C yadAO:8 | Nal, Kan, Ars, Strep, Cm | this study |
| Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:9 | Ye O:9 E40 Δasd lacking expression of both YadA and Inv transformed with pASK-IBA4C yadAO:9 | Nal, Kan, Ars, Strep, Cm | this study |
| Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:9/0:8 hybrid | Ye O:9 E40 Δasd lacking expression of both YadA and Inv transformed with pASK-IBA4C_yadAO:9/O:8 hybrid | Nal, Kan, Ars, Strep, Cm | this study |
| Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:9 Δuptake region | Ye O:9 E40 Δasd lacking expression of both YadA and Inv transformed with pASK-IBA4C_yadAO:9 Δuptake region | Nal, Kan, Ars, Strep, Cm | this study |
| Ye O:9 E40 pBla eGFP | Ye O:9 E40 pBla transformed with pACYC184 EGFP | Nal, Kan, Ars, Cm | 47 |
| Ye O:3 01 | clinical isolate derived from fecal sample | – | this study |
| Ye O:3 02 | clinical isolate derived from fecal sample | – | this study |
| Ye O:3 03 | clinical isolate derived from swine (tongue) | – | this study |
| Ye O:8 04 | clinical isolate derived from fecal sample | – | this study |
| Ye O:5,27 06 | clinical isolate derived from fecal sample | – | this study |
| Ye O:5,27 07 | clinical isolate derived from fecal sample | – | this study |
| Ye O:9 08 | clinical isolate derived from fecal sample | – | this study |
| Ye O:9 09 | clinical isolate derived from fecal sample | – | this study |
| Ye O:9 10 | clinical isolate derived from fecal sample | – | this study |
| Ye O:9 11 | clinical isolate derived from fecal sample | – | this study |
| Ye O:9 12 | clinical isolate derived from fecal sample | – | this study |
| Ye O:9 13 | clinical isolate derived from blood sample | – | 50 |
| Ye O:9 14 | clinical isolate derived from fecal sample | – | this study |
| Yps YPIII | Yps WT strain, pIB1 | – | 51 |
| Yps YP46 pIB1 | yadAΔ53–83 | Kan, Amp | 18 |
| Yps YP47 pIB1 | yadA– | Kan | 17 |
| Ec omp2 + pASK-IBA4C | Ec BL21 lacking expression of ompF transformed with pASK-IBA4C | Cm | this study |
| Ec omp2 + pASK-IBA4C_yadAO:8 | Ec BL21 lacking expression of ompF transformed with pASK-IBA4C_yadAO:8 | Cm | this study |
| Ec omp2 + pASK-IBA4C_yadAO:9 | Ec BL21 lacking expression of ompF transformed with pASK-IBA4C_yadAO:9 | Cm | this study |
| Ec omp2 + pASK-IBA4C_yadAO:9/O:8 hybrid | Ec BL21 lacking expression of ompF transformed with pASK-IBA4C_yadAO:9/O:8 hybrid | Cm | this study |
| Ec omp2 + pASK-IBA4C_ yadAO:9 Δuptake region | Ec BL21 lacking expression of ompF transformed with pASK-IBA4C_yadAO:9 Δuptake region | Cm | this study |
| Mc RH4 WT | Mc WT strain | – | 52 |
| Mc RH4 ΔUspA2H | Mc lacking expression of UspA2H | Zeo | 53 |
Amp = Ampicillin? Ars = arsenite? Cm = chloramphenicol; Kan = kanamycin; Nal = nalidixic acid; Spec = spectinomycin; Strep = streptomycin; Tet = tetracycline? Zeo = zeocine.
Serum
Normal human serum (NHS) was collected from at least 4 healthy volunteers and pooled. Aliquots were stored at −80°C and thawed only once. Heat-inactivated serum (HIS) was generated by incubation at 56°C for 30 min immediately before use.
Antibodies
Antibodies used in this study are listed in table 3.
Table 3.
Antibodies used in this study
| Conjugate | Clone | Manufacturer | Working dilutions | |
|---|---|---|---|---|
| Primary antibodies | ||||
| Goat anti-factor H | polyclonal | Complement Technology | 1:100 | |
| Rabbit anti-Vn | – | polyclonal | Complement Technology | FACS 1:100; WB 1:1,000 |
| Rabbit anti-Ye YadA | – | polyclonal | Lab antibody; I. Autenrieth | 1:200 |
| Rabbit anti-Yps YadA | – | polyclonal | Lab antibody; P. Dersch | 1:200 |
| Sheep anti-Vn | – | polyclonal | AbD Serotech | 1:100 |
| Mouse anti-human C5b-9 | – | aE11 | Dako | 1:1,000 |
| Mouse anti-β subunit of E.coli RNA-polymerase | – | 8RB13 | NeoClone Biotechnology | 1:2,000 |
| Secondary antibodies | ||||
| Donkey anti-rabbit | APC | Jackson ImmunoResearch | 1:200 | |
| Goat anti-rabbit | DyLight 800 | Thermo Scientific | 1:10,000 | |
| Goat anti-rabbit | DyLight 680 | Thermo Scientific | 1:10,000 | |
| Goat anti-mouse | DyLight 680 | Thermo Scientific | 1:10,000 | |
| Rabbit anti-sheep | DyLight 800 | Thermo Scientific | 1:10,000 | |
| Rabbit anti-goat | Alexa Fluor 488 | Jackson ImmunoResearch | 1:200 | |
| Goat anti-mouse | Alexa Fluor 647 | polyclonal | Jackson ImmunoResearch | 1:2,500 |
Purified Proteins Used in This Study
Purified monomeric and multimeric Vn was purchased from BD Bioscience (Heidelberg, Germany) and Millipore (Schwalbach, Germany), respectively. Vn fragments were expressed and purified as described previously [35, 54].
Binding Assay with Serum or Purified Proteins Analyzed by Flow Cytometry
To analyze the binding of purified Vn or Vn and factor H from HIS, a total of 1 × 107 bacteria per assay were incubated with 5–50% HIS or purified Vn (1–10 µg/ml) diluted with PBS (Life Technologies, Darmstadt, Germany) in a total volume of 100 µl for 30 min at 37°C. As an internal control, each strain was also treated with PBS only. Recombinant Vn fragments were used at 4 µg/ml. After washing with 1% BSA in PBS (washing buffer), bacteria were spun down and the pellet was resuspended in 200 µl 4% paraformaldehyde in PBS for 1 h at room temperature. Bacteria were washed once again and finally incubated with primary polyclonal antibodies (pAb) directed against Vn or factor H overnight at 4°C. The next day, bacteria were washed once and incubated with suitable secondary antibodies for 1 h at room temperature. After a final washing step, bacteria were transferred to FACS tubes and analyzed with a Fortessa LSR II instrument. Data analysis was carried out using WinMDI v2.8. The PBS-only control was used to determine background staining using the same primary and secondary antibodies as for all other samples. Values obtained for the control samples were subtracted from the values obtained for the corresponding samples that were incubated in serum or purified Vn. All flow cytometry figures show background subtracted values.
Detection of Vn Binding or YadA Expression by Western Blot
To analyze Vn binding by immunoblotting, 5 × 108 bacteria (bacterial numbers were determined photometrically by measuring the optical density at 600 nm; a volume corresponding to the desired number of bacteria was harvested by centrifugation, and the bacterial pellets were then used to carry out the assay) were incubated in 100 µl of 50% HIS diluted in PBS as described above. Thereafter, bacteria were washed twice with washing buffer, once with PBS, and finally resuspended in 50 µl deionized water. For the detection of YadA, bacteria were simply washed after harvest. After the addition of 25 µl 4× Laemmli buffer (Bio-Rad Laboratories, Munich, Germany), samples were boiled for 5 min at 95°C and separated in a 10% acrylamide SDS gel (Bio-Rad Laboratories). Each lane was loaded with an equal number of bacteria. After blotting, the membranes were blocked with 3% BSA and 5% milk powder in TBS for 1 h at room temperature. They were then incubated with the desired antibodies (a complete list of antibodies and working dilutions is given in table 3) for 1 h at room temperature or at 4°C overnight, washed with 0.1% TBS-T and then incubated with the suitable secondary antibody. Fluorescence signals were recorded using a LICOR Odyssey imaging system.
Detection of Vn Binding by Blot Overlay Assay
Bacterial lysates were prepared as described above, separated by SDS-PAGE and blotted. After blocking with 5% milk, 3% BSA in PBS for 3 h at room temperature, the membrane was incubated with 7 µg/ml purified monomeric Vn in 3% BSA in PBS-T overnight at 4°C. After washing with 0.1% TBST, Vn was detected with rabbit anti-Vn pAb and a secondary DyLight 680-conjugated goat anti-rabbit pAb. Fluorescence signals were recorded using a LICOR Odyssey imaging system.
Purification of DNA from Yersinia Colonies
Yersinia strains were streaked on the LB agar plates without antibiotics. The next day, a single colony was used for DNA extraction using the Qiagen QIAmp DNA mini kit according to the manufacturer's protocol. DNA was finally eluted in 100 µl of ultrapure water.
PCR Amplification of the YadA Head Region
To test Yersinia YadA for the presence of the additional stretch (enabling the recruitment of Vn) within its head region, we used the primers YadA_Seroseq_435F (5′-gatcagtgtctctgcggcat-3′) and YadA_Seroseq_435R (5′-gccccataagtaactgccga-3′) that bind to highly conserved regions upstream and downstream of the uptake region (online suppl. fig. S1; for all online suppl. material, see www.karger.com/doi/10.1159/000449200). According to the sequence alignment, the PCR reaction should yield a fragment of 442 bp with Ye O:9 E40 or 451 bp with Yps YPIII (both harboring the uptake region of approx. 90 bp) or 337 bp with Ye O:8 WA-314 and 346 bp with Ye O:3 6471/76 and Ye O:5.27 (all 3 lacking the uptake region) and thus allow us to discriminate between YadA with and without the uptake region. We used the following PCR program: 2 min 95°C (initial denaturation), 30 s 95°C → 1 min 55°C → 30 s 68°C (repeated 29 times), 5 min 72°C (final extension) and cooling at 4°C until further processing.
Separation of PCR Products by Capillary Gel Electrophoresis
To determine the size of the PCR products, they were analyzed using a QIAxcel capillary gel electrophoresis system according to the manufacturer's protocol.
DNA Sequencing
PCR products were purified using the Promega Wizard® SV gel and PCR Clean-Up System according to the manufacturer's protocol. Subsequently, Sanger sequencing was performed by GATC using the same primers as for the PCR reaction.
Heparin Inhibition Assay
Sterile glass coverslips were coated with purified Vn (10 µg/ml) at 4°C overnight and air-dried. The coverslips were then placed in a 24-well plate and either incubated with PBS or 100 µM heparin in PBS; 5 × 107 bacteria (Ye O:9 E40 pBla EGFP) were added to each well, spun down for 5 min at 300 g and incubated for 1 h at 37°C in a humidified atmosphere. Afterwards, the supernatant was removed, and the samples were washed 2 times and finally fixed by the addition of 4% paraformaldehyde in PBS. After washing, coverslips were mounted in Mowiol, and micrograph pictures were acquired using a Zeiss LSM 510. To quantify adhesion, the number of bacteria for a given field of view (representative for the entire coverslip) was counted.
Analysis of C5b-9 Deposition by Flow Cytometry
To analyze whether bound Vn was functionally active, bacteria were incubated with Vn (10–50 μg/ml) or C4BP (10–50 µg/ml) for 30 min at 37°C. After washing, bacteria were incubated with C5b-6 (1 μg/ml) and C7 (1 μg/ml) for 10 min, and then C8 (0.4 μg/ml) and C9 (1 μg/ml) were added for 30 min at 37°C. All complement components except for Vn were from Complement Technology (Tyler, Tex., USA). Deposited C5b-9 was detected by mouse anti-human C5b-9 mAb followed by Alexa Fluor 647-conjugated goat anti-mouse pAb. After 2 additional washes, bacteria were analyzed by flow cytometry (EPICS XL-MCL; Coulter, Hialeah, Fla., USA). All incubations were kept in a final volume of 100 μl 1% BSA in PBS, and washes were performed with the same buffer. Primary and secondary pAb were added separately as negative controls for each strain analyzed.
In vitro Serum Killing Assay
To analyze the susceptibility of Ye and Yps to complement-mediated killing in human serum, 5 × 106 bacteria were incubated in 100 µl 20% NHS or HIS for 30 min at 37°C. Complement activity was stopped by adding 100 µl BHI medium and placing the samples for 5 min on ice. Afterwards, serial dilutions of the samples were prepared, plated on selective agar plates and incubated at 27°C for 48 h. The colony-forming units (CFU) were determined. The serum bactericidal effect was calculated as the survival percentage, taking the bacterial counts obtained with bacteria incubated in HIS as 100%.
In vivo Serum Killing Assay
To analyze the lytic activity of serum complement against Ye in C57BL/6 and B6.129S2(D2)-Vtntm1Dgi/J mice, the animals were infected intravenously with 1 × 107 bacteria. After 30 min, they were sacrificed by CO2 asphyxiation and blood was withdrawn from the heart. Heparin (100 µl at 100 µg/ml) (Sigma-Aldrich, Steinheim, Germany) was mixed with the blood to avoid coagulation. Serial dilutions of the samples were plated on selective agar and incubated at 27°C for 48 h. The CFU were determined by counting the colonies.
Bioinformatics and Statistical Analysis
The GI numbers or the references of the sequences used in this work are listed in table 2. Alignments were produced with Kalign or Muscle and further edited manually [55, 56]. Data are expressed as means ± SD and were analyzed with the Student t test or with one-way ANOVA for multiple comparisons as described in the figure legends. GraphPad Prism v6.0 was used to analyze the data (GraphPad Software, La Jolla, Calif., USA). Differences were considered significant if p ≤ 0.05.
Results
Ye O:9 E40 Efficiently Binds Vn
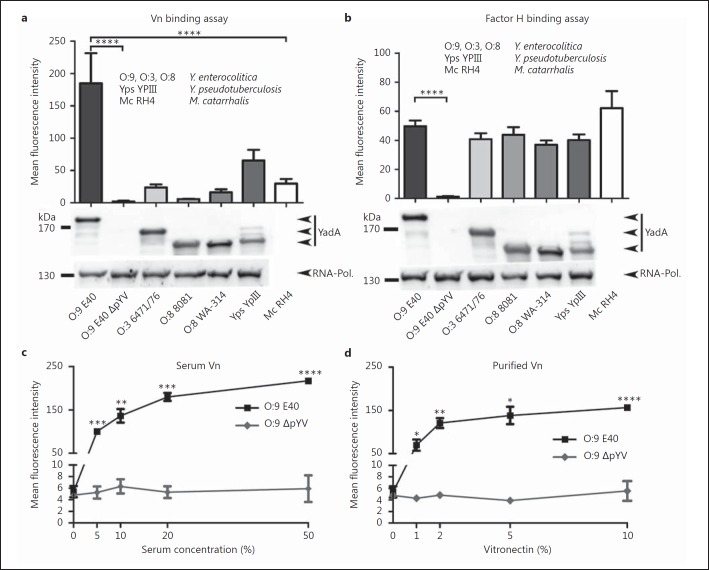
Vn plays an important role in the complement resistance of, for example, Mc, Hi and Streptococcus pneumoniae [32, 34, 44, 54, 57]. In order to test if Ye is able to bind Vn and if there are differences in the binding capacity of various Ye strains and serotypes, we incubated a set of strains in 50% HIS, washed the cells and detected Vn bound to bacteria by immunostaining with antibodies directed against Vn and subsequent flow cytometry analysis (fig. 1a). Upon incubation with HIS, we found very diverse binding properties of Ye strains compared to Mc RH4 and Yps YPIII. Mc RH4 served as a positive control [30, 34], whereas Yps YPIII was used as an additional comparator. It has been recognized that Yps YPIII YadA differs from the YadA sequences of other strains and that this difference coincides with a change in preferred ECM binding partners; this might possibly also affect the interaction with Vn (Yps YPIII YadA preferentially binds to fibronectin instead of collagen and laminin as observed with Ye) [18]. Ye O:9 E40 was able to bind exceptionally high amounts of Vn, which led to a mean fluorescence intensity of approximately 2.8 times higher than that measured with Mc RH4 (133.9 ± 33.9 vs. 47.4 ± 19.6). Yps also bound Vn, but at concentrations comparable to that of the Mc RH4 positive control (56.7 ± 11.0 vs. 47.4 ± 19.6). Ye O:8 WA-314 and Ye O:3 6471/176 also bound Vn, though to a lesser extent than Mc RH4 (approx. 54.1 or 70.7% of Mc RH4 signal). Ye O:8 8081 bound only residual amounts of Vn (6.7 ± 0.6). Interestingly, the binding of Vn to Ye O:9 E40 depended on the presence of the plasmid of Yersinia virulence (pYV) and was dose dependent (fig. 1c, d). In a plasmid-deficient strain (Ye O:9 E40 ΔpYV), Vn binding was almost abolished (6.2 ± 2.8). To test if the strain-specific binding pattern of Vn (O:9 E40 > YPIII > RH4 > O:3 > O:8 WA-314 > O:9 E40 ΔpYV = O:8 8081) is exclusive in comparison to other serum factors, we also tested the binding of factor H (fig. 1b). Factor H has been shown to interact with several discontinuous stretches within the stalk domain of YadA [20, 21, 22, 58]. Our data corroborate previous findings that the binding of factor H by Yersinia strains relies on the presence of YadA, but in contrast to Vn, there is no significant difference in binding efficiency in the various serotypes tested. This indicates different mechanisms of binding of Vn and factor H. Taken together, we found that Ye O:9 E40 is able to bind high amounts of serum-derived as well as purified Vn, although only in the presence of the pYV plasmid, in a dose-dependent manner. In contrast, Ye O:8 WA-314, 8081 and Ye O:3 6471/76 are weak Vn binders, although they also carry the pYV plasmid. This partially substantiates earlier findings that YadA-dependent Vn binding is at least weak if not nonexistent for Ye O:8 WA-314 in whole-cell adhesion assays under specific flow conditions [45].
Fig. 1.
Vn is efficiently bound by Ye O:9 E40 and Yps. a Several strains of Ye, serotype O:9 with and without virulence plasmid (O:9 E40 and O:9 E40 ΔpYV), serotype O:3 (O:3 6471/76) and serotype O:8 (O:8 8081; O:8 WA-314), and 1 Yps (Yps YPIII) WT strain were incubated with HIS, washed and subsequently analyzed for the presence of Vn on the bacterial surface by flow cytometry. Mc (Mc RH4), which is known to bind Vn and Yps, which we supposed also binds Vn, were included as a positive control for Vn binding. Ye O:9 E40, cured from the virulence plasmid (plasmid of Yersinia virulence; pYV) that encodes for the Ye T3SS, effector proteins and YadA, was included as a negative control because we surmised that Vn binding is pYV dependent. YadA protein levels were analyzed by Western blot analysis in whole-cell lysates and are shown below the bar chart (1 representative Western blot is shown). RNA polymerase protein (RNA-Pol.) was used as a loading control. YadAO:3 6471/76 has a calculated molecular weight of approximately 141 kDa (455 aa), YadAO:8 8081 of 132 kDa (422 aa), YadAO:8 WA-314 of 132 kDa (422 aa), YadAO:9 E40 of 153 kDa (487 aa), YadAYPIII of 135 kDa (434 aa) and UspA2H of approximately 272 kDa (876 aa). b To test if strain-specific differences in the binding of Vn are exclusive, we compared Vn binding levels to that of factor H. In contrast to Vn, factor H is bound in comparable amounts by all Yersinia strains tested, except for the negative control strain (O:9 E40 ΔpYV). The protein levels of YadA and the RNA polymerase as a loading control were analyzed by Western blots of whole-cell lysates and are shown below the bar chart (1 representative Western blot is shown). c Binding of serum-derived Vn to Ye O:9 E40 is dose dependent. Ye O:9 E40 and the pYV-cured version thereof were incubated with increasing serum concentrations. Afterwards, cell surface-associated Vn was quantified by flow cytometry. dYe O:9 E40 and the pYV-cured version thereof were incubated with increasing amounts of purified Vn. Afterwards, cell surface-associated Vn was quantified by flow cytometry. Binding of purified Vn to Ye O:9 E40 is dose-dependent. a–d Data are means ± SD of at least 4 individual experiments. a, b The main p values were determined by one-way ANOVA. p < 0.0001. Multiple comparisons were performed by one-way ANOVA with Dunnett's multiple-comparisons test. c, d The p values were determined by Student's t test. The error bars denote the SD. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Binding of Vn Is YadA Dependent
In order to assess whether YadA is the determinant for the binding of Vn to Yersinia, we used flow cytometry to compare Vn binding in a Ye O:9 E40 WT strain, a mutant deficient for YadA (ΔYadA), a mutant deficient for the chromosome-encoded adhesin Inv (ΔInv), the corresponding double mutant (ΔInvΔYadA; ΔΔ) and, again, the cured strain lacking the pYV plasmid (ΔpYV) (fig. 2a, left panel). We used Mc RH4 as positive control and an Mc ΔUspA2H [32] knockout strain as a negative control (fig. 2a, right panel). Our data indicate that the presence of YadA, but not of Inv, is decisive for the binding of Vn to Ye O:9 E40. Thus, in contrast to Ye O:9 E40 WT or ΔInv, Vn did not bind to ΔYadA, the ΔInvΔYadA double mutant or the pYV-cured strain. We could corroborate these findings by blot overlay assays and Western blot (fig. 2b, c). Analysis of the influence of YadA and, more specifically, a distinct region within YadA of Yps for Vn binding revealed that in Yps, YadA is also the Vn binding determinant (fig. 2a, middle panel). Moreover, the deletion of 30 aa (Δ53–83) corresponding to the uptake region in the head domain of Yps YadAYPIII abolishes Vn binding (fig. 2a, middle panel). Thus, our data demonstrate that YadA is essential for mediating Vn binding in Ye and that a stretch of 31 aa within the head region of YadAYPIII is decisive for the binding of Vn in Yps.
Fig. 2.
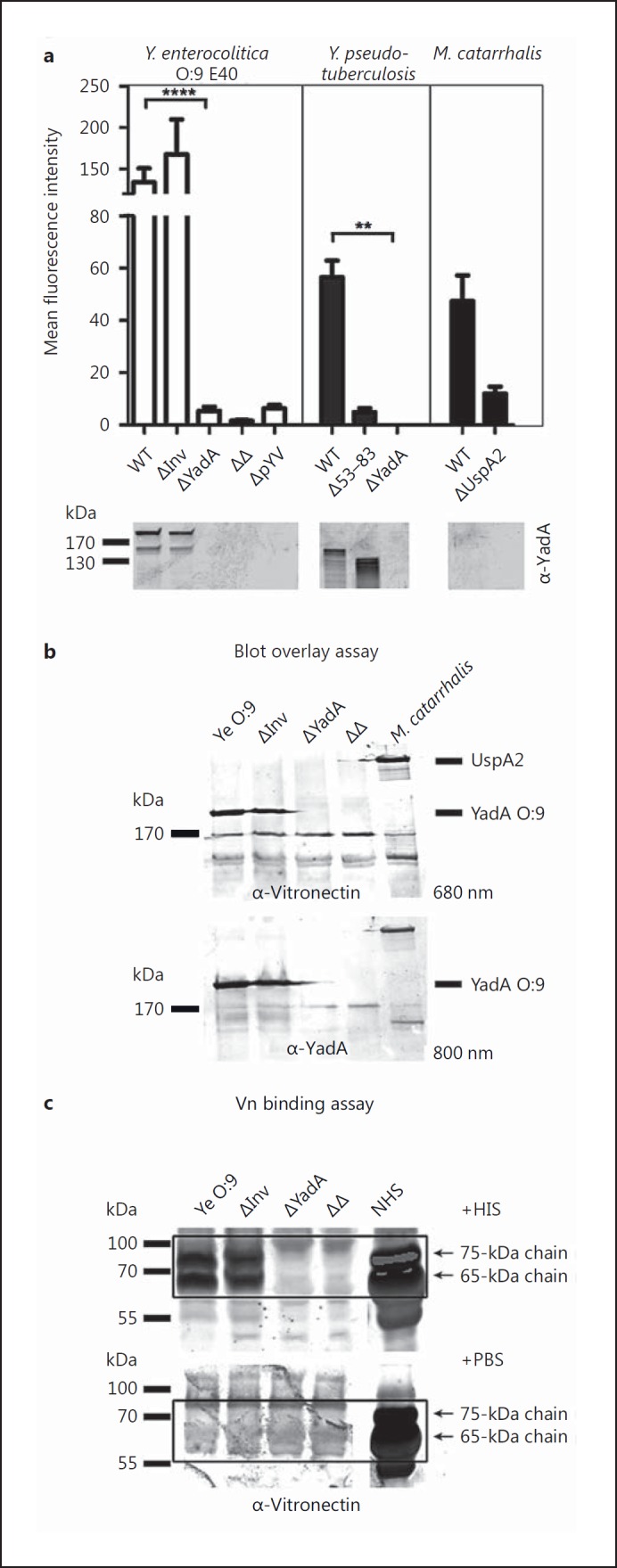
Vn binding to Ye is YadA dependent. a Left panel: a Ye O:9 E40 WT strain or strains carrying individual deletions for the adhesins Invasin (ΔInv) or YadA (ΔYadA) and a respective double knockout strain (ΔΔ) as well as a virulence plasmid-cured strain (ΔpYV) were incubated with serum and washed, and then Vn binding was quantified by flow cytometry. Middle panel: Yps YPIII WT and corresponding strains lacking expression of ΔYadA or expressing a YadA version lacking part of the head domain (Δ53–83) were included as controls. Right panel: an Mc WT strain known to bind Vn via the surface adhesin UspA2 and a corresponding strain lacking expression of UspA2 (ΔUspA2) were included as positive and negative controls. YadA protein levels were analyzed by Western blot analysis of whole-cell lysates and are shown below the bar chart (1 representative blot is shown). b A selection of the strains used in (a) was tested for Vn binding in a blot overlay assay. Vn and YadA were detected on the identical blot with specific antibodies and differently labeled secondary antibodies (emission maximum at 680 and 800 nm, respectively) simultaneously. Vn is bound only in the presence of YadA (Ye) or UspA2, respectively. c In a direct binding assay, essentially performed as in a, Vn can be detected at the expected molecular weight (65 and 75 kDa) by Western blot only in those Ye strains expressing YadA. Data are means ± SD of at least 4 individual experiments (a) or 1/3 representative experiments is shown (b, c). The main p value was determined by one-way ANOVA (a: p < 0.0001). Multiple comparisons were performed by one-way ANOVA with Dunnett's multiple-comparisons test. The error bars denote the SD. ** p < 0.01, **** p > 0.0001.
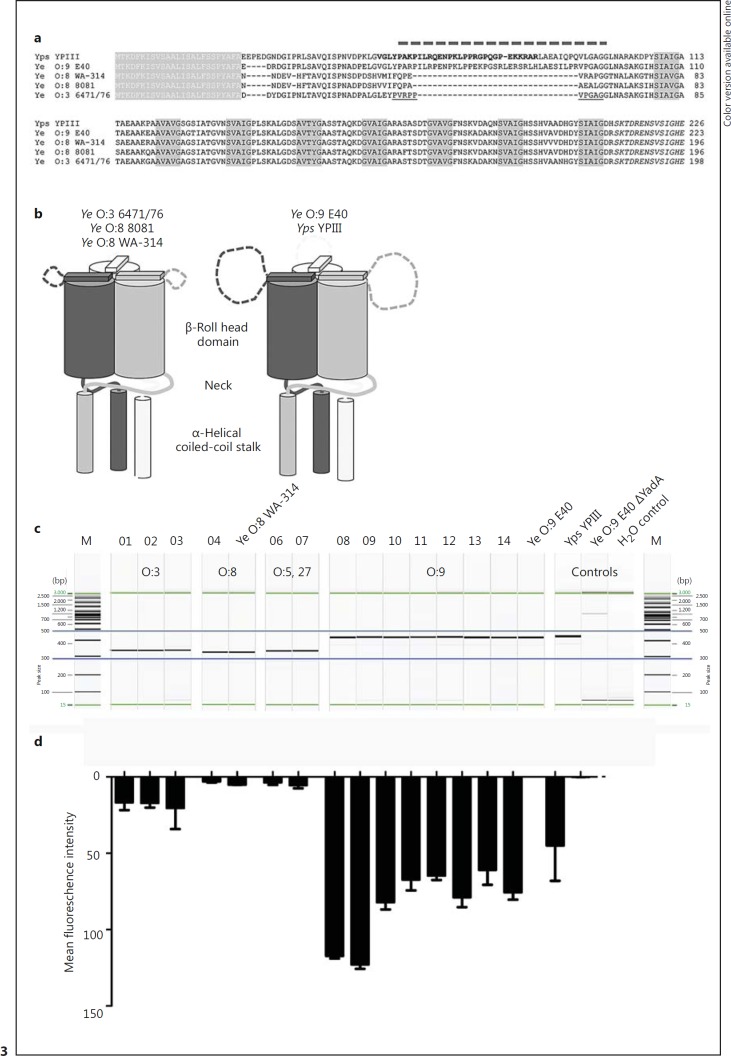
A Specific Stretch within YadA Discerns Low-Affinity Binding from High-Affinity Binding of Vn
We found that Ye expressing YadA derived from the O:8 WA-314 strain is a relatively weak binder compared to Ye O:9 E40 (fig. 1a). Therefore, we aimed to determine if other strains also carry the uptake region and also what actually discerns YadAO:9 E40 from YadAO:8 and if this difference might be causative for the discriminative Vn binding behavior. The head domain of YadAYPIII contains a stretch of sequence (uptake region) which is crucial for cell adhesion and efficient internalization of Yersinia via YadA [18]. This motif is absent in YadA of Ye O:8 but present in the Ye O:9 E40 strain (aa 56–88) (fig. 3a). It is rich in prolines and charged residues, suggesting an undefined loop structure (fig. 3b), inserted in a shorter loop that is not resolved in the crystal structure of the Ye O:3 YadA head (PDB: 1P9H) [59].
Fig. 3.
A specific region in the YadA head domain is decisive for efficient binding of Vn. a Alignment of the head of various YadA variants. White letters on gray background: signal peptide. Black letters on gray background: the canonical ‘SVAIG’ head repeats of YadA. Italics: the neck region that links the head to the coiled-coil stalk of YadA. The insertion of Yps originally proposed by Heise and Dersch [18] is displayed in bold, and is slightly shifted towards the N-terminus of YadA. The dashed line on top shows the corrected position of the insertion, based on improved alignments and the structure of the YadA head from Ye O:3, where the short insertion is not resolved (underlined region). This and the unusually high number of prolines in this region suggest that it is not structured. The long version of the insertion carries a strongly positive net charge (+5 for Yps YPIII, +4 for the Ye O:9 E40), which probably plays a role in binding to fibronectin and Vn. b Schematic view of the differences in the YadA heads. The Yps YPIII and Ye O:9 E40 variants have long insertions in an unstructured loop region close to the N-terminus of the head. c PCR products comprising the YadA head region of Ye O:8 WA-314, Ye O:9 E40 with and without YadA, Yps YPIII and clinical isolates derived from fecal samples (Ye O:3, No. 01–03; Ye O:8, No. 04; Ye O:5,27, No. 06–07 and Ye O:9, No. 08–12) or blood (No. 13) were separated by capillary gel electrophoresis. The predicted length of PCR products was as follows: Ye O:3 346 bp; Ye O:8 337 bp; Ye O:9 451 bp; Ye O:5,27 346 bp, and Yps YPIII 442 bp. Water control and a YadA-deficient strain were included as negative controls. d The strains shown in c were tested for Vn binding. Cell surface-associated Vn after incubation in HIS was quantified by flow cytometry. One of 3 representative experiments is shown.
To investigate whether this motif is present exclusively in Ye O:9 E40 or can be found also in other Yersinia strains and especially in strains isolated from clinical specimens, we carried out PCRs. We designed primers binding to rather conserved regions within the YadA sequence flanking that part of the head domain which comprises the uptake region (online suppl. fig. S1). The size of the PCR products allowed us to easily detect the presence of the uptake region. The predicted lengths of the YadA head fragments were 346 bp (Ye O:3), 337 bp (Ye O:8), 346 bp (Ye O:5,27), 451 bp (Ye O:9) and 442 bp (Yps YPIII). Strikingly, the additional stretch present in YadA of Ye O:9 E40 and Yps YPIII was present in all tested clinical isolates of serotype O:9 but absent in all other strains (belonging to the indicted serotypes; fig. 3c) that we tested. Ye O:9 E40 ΔYadA and water control were included as negative controls (fig. 3c, all strains depicted were also tested for Vn binding). Cell surface-associated Vn after incubation in HIS was quantified by flow cytometry (fig. 3d). Whereas all strains belonging to serotype O:9 (No. 08–14) and possessing the uptake region within YadA bound Vn in comparably high amounts as Ye E40 O:9, Ye strains of serotype O:3 (No. 01, 02, 03), O:8 (No. 04) and O:5,27 (No. 06, 07) turned out to be rather weak binders. Thus, we assume that the presence of the uptake region is the major determinant that allows binding of Vn and (at least in the strains we have tested) is present exclusively in the YadA of Ye strains of serotype O:9.
To test this hypothesis, we generated a YadA hybrid where we replaced the N-terminus of the head domain of YadAO:8 by that of YadAO:9 E40 (including the uptake region) and a YadAO:9 E40 deletion mutant lacking the uptake region (aa 56–88) (fig. 4a). We then compared Vn binding by flow cytometry. The strains Ye O:9 ΔYadA expressing YadAO:8 WA-314 or YadAO:9 E40 were also included in this analysis. In addition, we used Ye O:8 WA-314, Ye O:9 E40 and Ye O:9 E40 ΔYadA as controls (fig. 4b). Ectopic expression of YadAO:9 E40 was able to rescue Vn binding of Ye O:9 E40 ΔYadA. This was also true for the O:9/O:8 hybrid YadA. Additionally, deletion of the uptake region from YadAO:9 led to significantly reduced Vn binding (fig. 4b). Our data show that the uptake region of YadAO:9 E40 significantly enhances recruitment of Vn. Of note, a sequence alignment of YadA from different Yersinia strains also revealed insertions in the stalk regions of Ye YadAO:9 E40 and YadAO:3 6471/76 that are not found in YadAYPIII and YadA0:8WA-314 (online suppl. fig. S2). However, these regions show no clear association with Vn or factor H binding (fig. 1a). Finally, we wanted to assess whether cofactors expressed by Yersiniae are necessary or if YadA containing the uptake region alone is sufficient to mediate efficient binding of Vn. We tested Vn binding of E. coli omp2 [60] which ectopically expressed the YadA version described above (online suppl. fig. S3). We found that expression of YadAO:9 or the hybrid YadAO:9/O:8 is sufficient to mediate the binding of Vn. Thus, we conclude that the decisive factor for Vn binding is YadA comprising the uptake region.
Fig. 4.
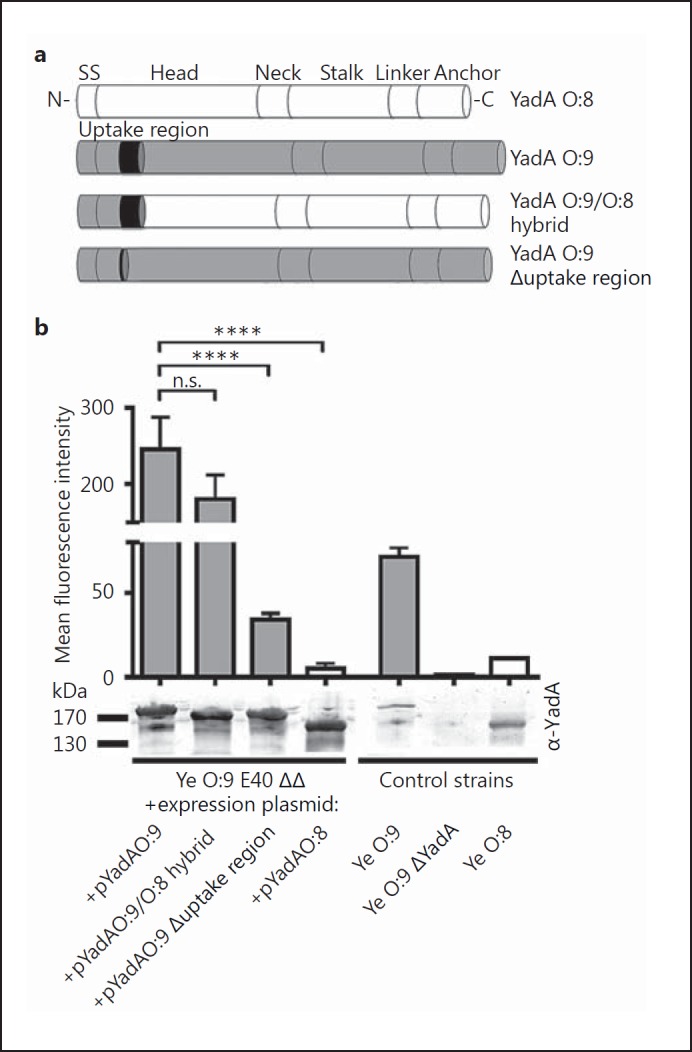
The uptake region is decisive for YadA-mediated Vn binding. Schematic representation of different YadA versions that were expressed from a plasmid in Ye O:9 E40 and analyzed for Vn binding capacity. The YadA versions tested comprise YadAO:8, YadAO:9, a YadAO:9/O:8 hybrid consisting of the O:9 head domain fused to the corresponding Ye O:8 head/stalk and membrane anchor domain (details: Material and Methods) and Ye YadAO:9 with the uptake region deleted (Δuptake region). b Flow cytometry analysis of Vn binding to different Ye strains carrying plasmids for inducible expression of the YadA versions depicted in a. As control strains, we used Ye strains expressing WT YadA from the endogenous pYV plasmid (Ye O:9 = positive control, Ye O:8) and a Ye O:9 E40 YadA-deficient strain (Ye O:9 ΔYadA = negative control). YadA protein levels were analyzed by Western blot analysis in whole-cell lysates and are shown below the bar chart. Data are means ± SD of at least 3 individual experiments (flow cytometry), or 1/3 representative experiments is shown (Western blot). The main p value was determined by one-way ANOVA (b, flow cytometry: p < 0.0001). Multiple comparisons were performed by one-way ANOVA with Dunnett's multiple-comparisons test. The error bars denote the SD. **** p > 0.0001.
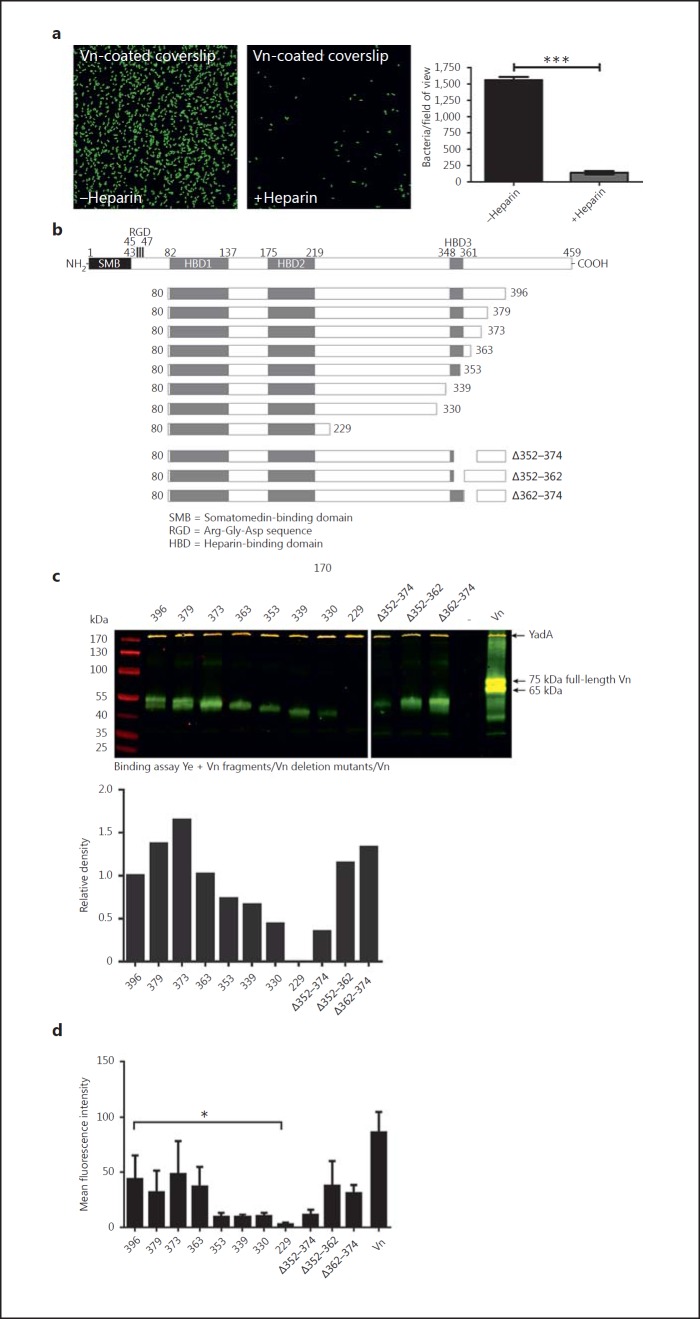
Vn Interacts with YadA via Its C-Terminal HBD-3
Previous work with Mc and Hi revealed HBD-3 as the decisive part of Vn for interaction with UspA2 or Hsf [34, 35]. Therefore, we wanted to know if this domain might also mediate the interaction of Vn with YadA. In order to test this, we first analyzed whether heparin might block the binding of Ye to Vn by occupying the HBDs. This would be a clear indicator of the involvement of one of the HBDs in the interaction with YadA. Coverslips were coated with Vn and then incubated with Ye O:9 E40, expressing enhanced green fluorescent protein for easier detection of binding, either in the presence or absence of heparin. Thereafter, coverslips were washed, fixed, mounted and analyzed by fluorescence microscopy (fig. 5a). Our results demonstrate that, in the presence of heparin, the binding of bacteria to Vn-coated coverslips is significantly reduced. Therefore, we conclude that at least one of the HBDs is involved in mediating the binding of Vn to YadA0:9 E40.
Fig. 5.
Vn interacts with YadA via its C-terminal HBD-3. a Adhesion of Ye to Vn-coated coverslips can be blocked by heparin. b Schematic representation of Vn, the C-terminal-truncated Vn molecules [54] and the Vn molecules carrying deletions within and adjacent to the HBD-3 [35] that were used for a direct binding assay. c Western blot of a binding assay of Ye O:9 E40 with full-length Vn and all fragments depicted in b. Vn fragments appear in green; YadA, which was detected simultaneously, appears in yellow bands (trimer runs at approx. 200 kDa). d Flow cytometry analysis of Vn binding to Ye O:9 E40 with full-length Vn and all fragments depicted in b. Data are means ± SD of at least 3 individual experiments (a, d), or 1/3 representative experiments is shown. The p value for the comparison with and without heparin was determined by Student's t test. The main p value was determined by one-way ANOVA (d: p < 0.0001). Multiple comparisons were performed by one-way ANOVA with Dunnett's multiple-comparisons test. The error bars denote the SD. * p < 0.05, *** p < 0.001.
To locate the sites within Vn that actually determine YadA binding, we used a set of recombinant Vn fragments (fig. 5b). These fragments essentially comprise C-terminal-truncated Vn molecules as well as deletion mutants lacking parts of HBD-3 (comprising aa 348–361) or adjacent regions. All fragments were tested for appropriate quality (online suppl. fig. S4). Our binding assay (fig. 5c) demonstrates that the fragments Vn 80–396, 80–379, 80–373 and 80–363 are efficiently bound by Ye O:9 E40. However, further C-terminal truncation, comprising either parts of or the entire HBD-3 (80–353, 80–339), led to a reduction of binding. Fragments lacking the entire HBD-3 plus the adjacent N-terminal region (80–330, 80–229) bound only weakly to Ye O:9 E40 (fig. 5c). Thus, we assume that not only HBD-3 but also the adjacent N-and especially the C-terminal approximately 10–20 aa are important for a stable interaction of Vn with Ye O:9 E40. These findings are in agreement with the fact that a Vn molecule lacking the C-terminal part of HBD-3 plus the adjacent C-terminal region (Δ352–374) is also impaired when binding to Ye O:9 E40 whereas deletion of either only part of HBD-3 (Δ352–362) or only the adjacent C-terminal region (Δ362–374) does not significantly influence binding. In conclusion, aa 331–363 are decisive for the stable interaction of Vn with Ye O:9 E40.
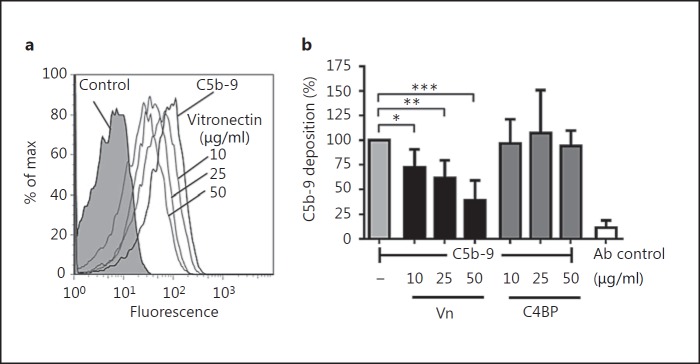
Vn Is Functionally Active and Inhibits the Terminal Pathway when Bound to the Surface of Ye
Besides modulating the adhesive properties of pathogens, Vn regulates the terminal complement pathway and blocks TCC formation. In order to test if Vn bound to Ye is functionally active and inhibits the terminal complement pathway, we assayed C5b-9 deposition in the presence of Vn bound to intact bacteria. To this end, Ye O:9 E40 was preincubated with Vn or C4BP followed by the addition of C5b-6, C7, C8 and C9. C5b-9 deposition was determined by using an anti-C5b-9 mAb and flow cytometry.
We clearly demonstrate that Vn bound to the surface of Ye O:9 E40 was functionally active and inhibited C5b-9 deposition in a dose-dependent manner (fig. 6a, b). Vn (50 µg/ml) inhibited C5b-9 deposition by 61%. C4BP, the C3 convertase inhibitor of the classical/lectin pathways, did not influence the C5b-9 deposition and thus the terminal pathway. From this, we conclude that Vn when bound to intact Ye is functionally active and inhibits the terminal complement pathway and C5b-9 deposition.
Fig. 6.
Vn is functionally active and inhibits the terminal pathway when bound to the surface of Ye. a Histogram overlay of flow cytometry analyses of TCC formation (detected by formation of the neoepitope C5b-9) on the surface of Ye O:9 E40 WT after preincubation of bacteria with PBS or different concentrations of Vn in PBS (10, 25 and 50 µg/ml). Preincubation with Vn reduces the amount of TCC that is formed. b Bar chart depicting C5b-9 deposition as percent of the amount of C5b-9 that was formed on the surface of bacteria preincubated with PBS only compared to bacteria preincubated with either Vn or C4BP at different concentrations (10, 25 and 50 µg/ml). Vn, but not C4BP, is able to reduce the formation of C5b-9. Antibody (Ab) control indicates background signal that was obtained using secondary antibody only for detection. Data are means ± SD of at least 3 individual experiments. The main p value was determined by one-way ANOVA (b: p < 0.001). Multiple comparisons were performed by one-way ANOVA with Dunnett's multiple-comparisons test. The error bars denote the SD. * p < 0.05; ** p < 0.01, *** p < 0.001.
Binding of Vn Decreases the Susceptibility to Complement-Mediated Killing by Human Serum
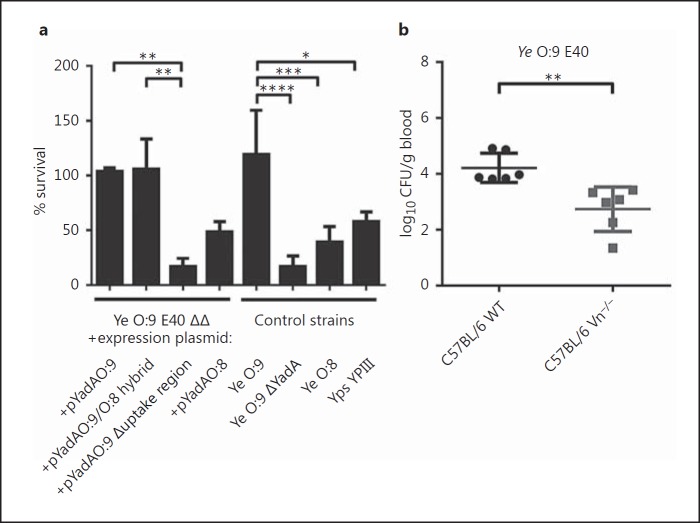
YadA-mediated serum resistance is an important virulence trait of Ye [21, 61, 62]. To analyze the importance of Vn binding for preventing complement-mediated killing, we performed serum killing assays. We incubated Ye O:9 E40, the corresponding YadA-deficient mutant (ΔYadA) and Ye O:8 WA-314 in NHS (fig. 7a, ‘control strains’). Their survival was calculated as the survival percentage, taking the bacterial counts obtained with samples incubated in HIS as 100%. Our data show that Ye O:9 E40 - a strong Vn binder - is resistant to complement-mediated killing (% survival in NHS compared to HIS 119.1 ± 40.39) whereas the Ye O:9 E40 ΔYadA mutant strain was highly susceptible for killing by the complement system (16.74 ± 9.83). Compared to Ye O:9 E40, the weak Vn binder Ye O:8 WA-314 was significantly more susceptible to complement-mediated killing (39.21 ± 7.11) than Ye O:9 E40 (fig. 7a). Furthermore, we also tested Ye O:9 E40 ΔΔ expressing either YadAO:9, YadAO:9/O:8, YadAO:9 Δuptake region or YadAO:9 for serum resistance. We found that the expression of YadAO:9 (103.6 ± 3.42) and also of the O:9/O:8 hybrid YadA (105.7 ± 27,56) conferred serum resistance comparable to that of the Ye O:9 E40 WT strain. In contrast, the serum survival was significantly reduced upon the expression of YadAO:8 (48.55 ± 9.36). Compared to all these strains, a strain expressing the YadA O:9 lacking the uptake region showed the greatest sensitivity towards serum treatment (16.8 ± 7.57). These data clearly indicate that the YadA-dependent binding of Vn plays an important role in preventing the lysis of Ye by the complement system.
Fig. 7.
Ye O:9 E40 is resistant to complement-mediated killing in vitro, and in an in vivo serum killing assay, Ye is more efficiently eliminated in the absence of Vn. a An in vitro serum killing assay using Ye O:9 E40, Ye O:9 E40 ΔYadA, Ye O:8 WA-314, Yps YPII, Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:8, Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:9, Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:9/O:8 hybrid, Ye O:9 E40 ΔΔ + pASK-IBA4C_yadAO:9 Δuptake region. The serum bactericidal effect was calculated as the survival percentage. b WT and Vn−/− mice were infected intravenously with 1 × 107Ye O:9 E40 for 30 min. After that, the mice were killed, and blood was withdrawn and plated on selective agar plates. CFU were determined by counting colonies the next day, shown as log10 CFU per gram of blood. a Data are means ± SD of at least 3 individual experiments. The main p value was determined by one-way ANOVA (p < 0.0001). Multiple comparisons were performed by one-way ANOVA with Dunnett's multiple-comparisons test. b The p value for the comparison of C57BL/6 and Vn−/− mice was determined by Student's t test. The horizontal lines denote the mean and the error bars denote the SD. * p < 0.05; ** p < 0.01, *** p < 0.001, **** p > 0.0001 (n = 6).
Mice Deficient for Vn Expression Eliminate Ye More Rapidly in Short-Term Systemic Infection
It is known that YadA is decisive for the survival of Ye upon contact with serum [6, 12]. This is one reason why YadA-deficient strains of Ye are avirulent in the mouse model [12]. However, the contribution of the YadA-dependent recruitment of Vn to the survival of Ye in a mouse model has not been addressed so far. In order to test if the presence of Vn has an influence on the survival of Ye in vivo, we infected Vn−/− and WT mice with Ye O:9 E40, sacrificed the mice 30 min after infection and determined the bacterial burden in the blood. We found that the bacterial load in the blood was significantly reduced (log10 CFU per gram of blood = 2.7 ± 0.8) for the Vn−/− mice compared to WT mice (log10 CFU per gram of blood = 4.2 ± 1.0) (fig. 7b). In line with the reduction of C5b-9 deposition on Ye by Vn, these data would suggest that Vn protects Ye from early killing in the blood stream.
Compared to YadA of Ye O:9 E40, the YadA of Ye O:8 WA-314 shows a low Vn-binding capacity. Therefore, we hypothesized that due to this low Vn-binding capacity and in contrast to our findings with Ye O:9 E40, the availability of Vn should only marginally impact the outcome of an early bloodstream infection with the Ye O:8 WA-314 strain. However, since the Ye O:9 and O:8 strains exhibit additional differences with regard to sequence and also virulence mechanisms [63, 64, 65, 66], this experiment may not solve the question of whether the uptake region actually contributes to better clearance of infection by mediating more efficient binding of Vn specifically. Therefore, we used a slightly different approach. To clearly assess the role of the uptake region and to exclude other differences between the Ye O:8 and the Ye O:9 strain tampering with the result of our experiments, we infected mice with Ye harboring pYadAO:9/8 hybrid or pYadAO:8 in the same strain background (Ye O:9 E40 ΔΔ). The basic sequence of the YadA of these strains is identical, with the exception of the part encoding the uptake region. Surprisingly, the infection of C57BL/6 WT or Vn−/− mice with Ye O:9 E40 ΔΔ + pASK-IBA4c_yadAO:8 led to a small but significant difference in bacterial counts (online suppl. fig. S5A; 5.9 ± 0.3 log10 CFU per gram of blood in WT mice vs. 6.4 ± 0.3 in Vn−/− mice). As observed previously with Ye O:9 E40, infection with Ye O:9 E40 ΔΔ harboring pASK-IBA4c_yadAO:9/O:8 hybrid revealed a significantly reduced bacterial load in the blood for the Vn−/− mice (log10 CFU per gram of blood = 4.9 ± 0.2) compared to WT mice (log10 CFU per gram of blood = 5.5 ± 0.2) (online suppl. fig. S5B). This leads to the assumption that the binding of Vn to different regions of YadA may have various implications for YadA function. While binding of Vn to the uptake region seems to increase virulence, binding of Vn to other regions of YadA might also reduce virulence.
Discussion
Complement inhibitor recruitment by bacterial cell surface proteins and adhesins is an important virulence mechanism used by many pathogens. Accordingly, several complement regulators (factor H, factor H-like protein-1 and C4BP) and complement proteins (C3b and iC3b) have been identified that interact with the Gram-negative enteropathogen Ye [19, 20, 21, 22, 61, 62, 67]. Here, we describe a novel mechanism that contributes to Ye complement resistance and the overall virulence of Ye. We show that the TAA YadA of different Yersinia species binds Vn and demonstrate that a part of the YadA head domain of YadAO:9 E40 comprising aa 56–88 binds Vn with high efficiency. Recruitment of Vn to YadA led to the reduced surface formation and deposition of C5b-9 (TCC) and thus enhanced complement resistance. Moreover, Ye O:9 E40 was completely resistant to complement-mediated killing in human serum, in contrast to the YadA-deficient strain. In addition, it turned out that, in comparison to Ye O:8 WA-314, Ye O:9 E40 is significantly more serum-resistant. Using Vn-deficient mice, we were also able to demonstrate the reduced survival of Ye O:9 E40 in the absence of Vn in an in vivo serum killing assay. Thus, the binding of Vn to the surface of Ye has a great impact on the interaction of Ye with the host.
In our experiments, we found that different strains of Ye and Yps bind Vn in a YadA-dependent manner but that different Yersinia strains exhibited divergent Vn-binding capacities. Previous studies with different Mc WT strains show that Mc also binds Vn with different affinities via UspA2 [30]. The N-terminus of the UspA2 head domain sequence displays 2 different conserved regions that may explain these Vn binding differences [68]. Furthermore, we show for the first time that Ye strains of serotype O:9 - unlike all other Ye strains we tested - exhibit an additional stretch in their YadA head domain. These strains, and to a lesser extent Yps YPIII, showed high-affinity binding to Vn while other tested Ye strains showed only low-affinity binding. Unfortunately, we were not able to correlate the ability to bind Vn and the pathogenic potential of clinical isolates due to the low frequency of Ye infection (and thus available isolates) and the fact that systemic infection with Ye happens only on rare occasions. The stretch in YadAO:9 is highly similar to the uptake region described for Yps YPIII [18], which is important for the ability of YadA to promote the invasion of Ye into host cells. Yps binds preferentially to fibronectin, but has low affinity for laminin or collagen type I, which is in contrast to the ECM protein-binding capacity of Ye which preferentially associates with collagen type I and laminin. This indicates that the uptake region may modulate the overall affinity to different ECM proteins. Sequence comparison of YadAO:9 E40 also revealed additional aa stretches in the YadA stalk domain, lacking in some other Ye strains. However, comparison of the Vn-binding capacity of different Ye and Yps strains shows no clear indication that this region may also contribute to the differences in Vn binding, since YadAO:3 6471/76 has the same insertion in the stalk region. In contrast to Vn binding, the interaction with factor H, which was shown to bind to the stalk region of YadA in Ye and Yps strains, revealed no differences [20]. This indicates that the presence or absence of the uptake region modulates affinity to Vn.
The site of interaction between Mc and Vn was mapped to the N-terminal residues 30–177 within UspA2 [34]. This region is located in the head domain of UspA2, which is similar to YadAO:9 E40. Our data show that subtle differences within the YadA protein sequence can significantly influence the protein interaction repertoire of Ye. The recruitment of such proteins to the surface of Ye may exert a significant influence on serum resistance and host cell interaction.
Localization of the Vn-binding domain within the YadA protein is a crucial step when analyzing the function of YadA in complement evasion. In contrast to complement regulator factor H or the complement component C3, which bind to the stalk domain of YadA [20], we found that Vn is bound via the YadA head domain. In Ye, the neutrophil-binding domain is located at the N-terminal part of YadA whereas the collagen-binding domain is located at the central and C-terminal part of the YadA head domain [59, 69, 70, 71, 72]. The inhibition of Vn binding with heparin was already shown for Mc and Hi. In both species, the interaction of Vn with UspA2 or Hsf was assigned to HBD-3 [34, 35]. In contrast, for Ye O:9 E40, not only HBD-3 but also the adjacent N- and C-terminal portions of Vn are decisive for the efficient interaction with YadA. We conclude that complement evasion of Ye is not limited to interactions mediated by the stalk domain but can involve the head domain of YadA, depending on the strain in question. Furthermore, the uptake region in Ye O:9 seems to provide a binding domain for Vn which strongly amplifies the binding of Vn.
Previous studies showed that recruitment of Vn by Mc or Hi inhibits C5b-9 formation to block pore formation [27]. However, analyzing the TCC formation in Ye with purified complement proteins (C5b-6, C7, C8 and C9), we showed that bound Vn inhibits the deposition of C5b-9 on the bacterial surface. Consequently, these data show that Vn bound to the bacterial surface via YadA is functionally active and inhibits the terminal pathway and thus contributes to complement resistance. Indeed, in in vitro serum killing assays, we showed that Ye O:9 E40 is the strain that sustains treatment with serum most efficiently compared to Ye O:8 and Yps YPIII. In contrast, a YadA-deficient strain of Ye O:9 E40 was susceptible to serum killing. Thus YadA-mediated binding of Vn in Ye O:9 E40 is decisive for the success of serum treatment in vitro. The situation is different in Ye O:8 WA-314. This strain is much more sensitive to serum treatment compared to Ye O:9. We know that in Ye O:8, serum resistance is mediated by the YadA-dependent recruitment of C3b/iC3b, factor H and C4BP [21, 33]. As all these factors bind to YadA and, at least for C4BP, the binding site(s) within YadA is unknown, there might be competition for binding sites, and this might lead to the binding of low levels of Vn. Still, binding of all the other negative regulators of complement can mediate serum resistance to a certain extent. A decisive role of YadA for serum resistance of Yps YPIII is rather unlikely as it has been shown that Yps serum resistance occurs independently of the presence of a virulence plasmid (that encodes YadA [73]). The known mechanisms involved in the serum resistance of Yps are the binding of C4BP and factor H via Ail [74, 75]. Nevertheless, we have shown that Yps also binds Vn via YadA. We think that in this case the recruitment of Vn has a function other than mediating serum resistance and speculate that it might be involved in, for example, the modulation of host cell targeting [66] and interaction [24].
Consequently, this should also improve the survival of Ye in vivo. Indeed, the short-term infection of Vn-deficient mice with Ye O:9 E40 revealed that Vn protects Ye from being killed by the immune system. A short-term infection of mice was used to avoid (as far as possible) the action of other virulence mechanisms such as those provided by the T3SS. According to ex vivo measurements, the injection of Yops should efficiently show its action at later time points. Therefore, the short-term mouse experiments should predominantly reflect the impact of Vn on complement killing, as the complement system is activated within seconds after infection. Thus, the mouse infection experiments provide evidence that the inhibition of TCC formation by Vn via binding to YadA indeed has biological relevance. These findings clearly demonstrate the importance of Vn binding to the uptake region for the pathogenicity of Ye. However, binding of Vn may also counteract YadA-mediated virulence, which is indicated by the slightly increased bacterial load after infection of Vn-deficient mice with Ye O:9 E40 ΔΔ expressing YadAO:8. We assume that the weak binding of Vn outside of the uptake region might interfere with the binding of other factors to YadA which are critical for YadA as a virulence factor. From an evolutionary point of view, the acquirement of the uptake region converts Vn from a factor protecting against infection to a factor mediating immune evasion.
Although individuals lacking terminal complement components are known to be more susceptible to N.meningitidis[76] but not especially to Ye infections, Vn binding is an important mechanism contributing to the overall serum resistance of Ye. Ye YadA interacts with a multitude of complement regulatory factors (C4bp, C3b, iC3b and factor H) that all contribute to serum resistance of Ye in a true infection situation. These interactions in sum finally determine the success of Ye within the host.
Taken together, our data present a novel mechanism of how YadA mediates immune evasion. By binding the HBD-3 domain of Vn, YadA containing the uptake region mediates the efficient inhibition of TCC formation and thus contributes to complement resistance and better survival of Ye. YadA is a multifunctional protein mediating complement resistance and also adhesion which, in turn, are critical for the subsequent injection of Yops into the host cells via the T3SS. Beyond bacteriolysis mediated by the assembly of the TCC, the even more important effect of Vn may be to modulate the interaction of Ye with immune cells [66]. Further studies will now address how Vn may influence adhesion, invasion and Yop injection during mouse infection.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We thank T. Späth for excellent technical assistance, A. Fruth (Wernigerode), E. Carniel (Paris), M. Skurnik (Helsinki), J. Heesemann (Munich) and D. Villinger (Frankfurt) for providing the Y.enterocolitica clinical isolates, and P. Dersch (Braunschweig) for the gift of the Y. pseudotuberculosis strains as well as the antibody against Yps YadA. This work was funded by grants from the German Research Council (DFG) within the SFB 766 to M.S.S. and I.B.A., the Swedish Medical Research Council (grant No. K2015-57X-03163-43-4, http://www.vr.se) and the Anna and Edwin Berger Foundation to K.R., and it was supported by the Gender Equality Program E.05.00390 of the University Clinics Tübingen to M.S.S.
References
- 1.Bottone EJ. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1999;1:323–333. doi: 10.1016/s1286-4579(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 2.Cover TL, Aber RC. Yersinia enterocolitica. N Engl J Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 3.Isberg RR. Mammalian cell adhesion functions and cellular penetration of enteropathogenic Yersinia species. Mol Microbiol. 1989;3:1449–1453. doi: 10.1111/j.1365-2958.1989.tb00128.x. [DOI] [PubMed] [Google Scholar]
- 4.Isberg RR. Determinants for thermoinducible cell binding and plasmid-encoded cellular penetration detected in the absence of the Yersinia pseudotuberculosis invasin protein. Infect Immun. 1989;57:1998–2005. doi: 10.1128/iai.57.7.1998-2005.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller VL, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Tahir Y, Skurnik M. YadA, the multifaceted Yersinia adhesin. Int J Med Microbiol. 2001;291:209–218. doi: 10.1078/1438-4221-00119. [DOI] [PubMed] [Google Scholar]
- 7.Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 2006;14:264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 2000;19:5989–5999. doi: 10.1093/emboj/19.22.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mühlenkamp M, Oberhettinger P, Leo JC, Linke D, Schütz MS. Yersinia adhesin A (YadA) - Beauty and beast. Int J Med Microbiol. 2015;305:252–258. doi: 10.1016/j.ijmm.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Szczesny P, Lupas A. Domain annotation of trimeric autotransporter adhesins - daTAA. Bioinformatics. 2008;24:1251–1256. doi: 10.1093/bioinformatics/btn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pepe JC, Wachtel MR, Wagar E, Miller VL. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect Immun. 1995;63:4837–4848. doi: 10.1128/iai.63.12.4837-4848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schütz M, Weiss EM, Schindler M, Hallström T, Zipfel PF, Linke D, Autenrieth IB. Trimer stability of YadA is critical for virulence of Yersinia enterocolitica. Infect Immun. 2010;78:2677–2690. doi: 10.1128/IAI.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Genaro MS, Waidmann M, Kramer U, Hitziger N, Bohn E, Autenrieth IB. Attenuated Yersinia enterocolitica mutant strains exhibit differential virulence in cytokine-deficient mice: implications for the development of novel live carrier vaccines. Infect Immun. 2003;71:1804–1812. doi: 10.1128/IAI.71.4.1804-1812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelis GR. Yersinia type III secretion: send in the effectors. J Cell Biol. 2002;158:401–408. doi: 10.1083/jcb.200205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viboud GI, Bliska JB. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Ann Rev Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 16.Hoyer-Hansen G, Behrendt N, Ploug M, Dano K, Preissner KT. The intact urokinase receptor is required for efficient vitronectin binding: receptor cleavage prevents ligand interaction. FEBS Lett. 1997;420:79–85. doi: 10.1016/s0014-5793(97)01491-9. [DOI] [PubMed] [Google Scholar]
- 17.Eitel J, Dersch P. The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed. Infect Immun. 2002;70:4880–4891. doi: 10.1128/IAI.70.9.4880-4891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heise T, Dersch P. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc Natl Acad Sci USA. 2006;103:3375–3380. doi: 10.1073/pnas.0507749103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biedzka-Sarek M, Jarva H, Hyytiäinen H, Meri S, Skurnik M. Characterization of complement factor H binding to Yersinia enterocolitica serotype O:3. Infect Immun. 2008;76:4100–4109. doi: 10.1128/IAI.00313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biedzka-Sarek M, Salmenlinna S, Gruber M, Lupas AN, Meri S, Skurnik M. Functional mapping of YadA- and Ail-mediated binding of human factor H to Yersinia enterocolitica serotype O:3. Infect Immun. 2008;76:5016–5027. doi: 10.1128/IAI.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindler MK, Schütz MS, Mühlenkamp MC, Rooijakkers SH, Hallström T, Zipfel PF, Autenrieth IB. Yersinia enterocolitica YadA mediates complement evasion by recruitment and inactivation of C3 products. J Immunol. 2012;189:4900–4908. doi: 10.4049/jimmunol.1201383. [DOI] [PubMed] [Google Scholar]
- 22.China B, Sory MP, N'Guyen BT, De Bruyere M, Cornelis GR. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect Immun. 1993;61:3129–3136. doi: 10.1128/iai.61.8.3129-3136.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preissner KT. Structure and biological role of vitronectin. Annu Rev Cell Biol. 1991;7:275–310. doi: 10.1146/annurev.cb.07.110191.001423. [DOI] [PubMed] [Google Scholar]
- 24.Boyd NA, Bradwell AR, Thompson RA. Quantitation of vitronectin in serum: evaluation of its usefulness in routine clinical practice. J Clin Pathol. 1993;46:1042–1045. doi: 10.1136/jcp.46.11.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seiffert D, Crain K, Wagner NV, Loskutoff DJ. Vitronectin gene expression in vivo. Evidence for extrahepatic synthesis and acute phase regulation. J Biol Chem. 1994;269:19836–19842. [PubMed] [Google Scholar]
- 26.Liang OD, Rosenblatt S, Chhatwal GS, Preissner KT. Identification of novel heparin-binding domains of vitronectin. FEBS Lett. 1997;407:169–172. doi: 10.1016/s0014-5793(97)00330-x. [DOI] [PubMed] [Google Scholar]
- 27.Singh B, Su YC, Riesbeck K. Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol Microbiol. 2010;78:545–560. doi: 10.1111/j.1365-2958.2010.07373.x. [DOI] [PubMed] [Google Scholar]
- 28.Milis L, Morris CA, Sheehan MC, Charlesworth JA, Pussell BA. Vitronectin-mediated inhibition of complement: evidence for different binding sites for C5b-7 and C9. Clin Exp Immunol. 1993;92:114–119. doi: 10.1111/j.1365-2249.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan M, Morris CA, Pussell BA, Charlesworth JA. Complement inhibition by human vitronectin involves non-heparin binding domains. Clin Exp Immunol. 1995;101:136–141. doi: 10.1111/j.1365-2249.1995.tb02289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attia AS, Ram S, Rice PA, Hansen EJ. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect Immun. 2006;74:1597–1611. doi: 10.1128/IAI.74.3.1597-1611.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole LE, Kawula TH, Toffer KL, Elkins C. The Haemophilus ducreyi serum resistance antigen DsrA confers attachment to human keratinocytes. Infect Immun. 2002;70:6158–6165. doi: 10.1128/IAI.70.11.6158-6165.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallström T, Trajkovska E, Forsgren A, Riesbeck K. Haemophilus influenzae surface fibrils contribute to serum resistance by interacting with vitronectin. J Immunol. 2006;177:430–436. doi: 10.4049/jimmunol.177.1.430. [DOI] [PubMed] [Google Scholar]
- 33.Leduc I, Olsen B, Elkins C. Localization of the domains of the Haemophilus ducreyi trimeric autotransporter DsrA involved in serum resistance and binding to the extracellular matrix proteins fibronectin and vitronectin. Infect Immun. 2009;77:657–666. doi: 10.1128/IAI.00819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh B, Blom AM, Unal C, Nilson B, Mörgelin M, Riesbeck K. Vitronectin binds to the head region of Moraxella catarrhalis ubiquitous surface protein A2 and confers complement-inhibitory activity. Mol Microbiol. 2010;75:1426–1444. doi: 10.1111/j.1365-2958.2010.07066.x. [DOI] [PubMed] [Google Scholar]
- 35.Singh B, Su YC, Al-Jubair T, Mukherjee O, Hallström T, Mörgelin M, Blom AM, Riesbeck K. A fine-tuned interaction between trimeric autotransporter Haemophilus surface fibrils and vitronectin leads to serum resistance and adherence to respiratory epithelial cells. Infect Immun. 2014;82:2378–2389. doi: 10.1128/IAI.01636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su YC, Jalalvand F, Mörgelin M, Blom AM, Singh B, Riesbeck K. Haemophilus influenzae acquires vitronectin via the ubiquitous protein F to subvert host innate immunity. Mol Microbiol. 2013;87:1245–1266. doi: 10.1111/mmi.12164. [DOI] [PubMed] [Google Scholar]
- 37.Sa ECC, Griffiths NJ, Virji M. Neisseria meningitidis Opc invasin binds to the sulphated tyrosines of activated vitronectin to attach to and invade human brain endothelial cells. PLoS Pathog. 2010;6:e1000911. doi: 10.1371/journal.ppat.1000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill DJ, Griffiths NJ, Borodina E, Andreae CA, Sessions RB, Virji M. Identification and therapeutic potential of a vitronectin binding region of meningococcal MSF. PLoS One. 2015;10:e0124133. doi: 10.1371/journal.pone.0124133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duensing TD, Putten JP. Vitronectin binds to the gonococcal adhesin OpaA through a glycosaminoglycan molecular bridge. Biochem J. 1998;334((Pt 1)):133–139. doi: 10.1042/bj3340133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths NJ, Hill DJ, Borodina E, Sessions RB, Devos NI, Feron CM, Poolman JT, Virji M. Meningococcal surface fibril (MSF) binds to activated vitronectin and inhibits the terminal complement pathway to increase serum resistance. Mol Microbiol. 2011;82:1129–1149. doi: 10.1111/j.1365-2958.2011.07876.x. [DOI] [PubMed] [Google Scholar]
- 41.Bergmann S, Lang A, Rohde M, Agarwal V, Rennemeier C, Grashoff C, Preissner KT, Hammerschmidt S. Integrin-linked kinase is required for vitronectin-mediated internalization of Streptococcus pneumoniae by host cells. J Cell Sci. 2009;122:256–267. doi: 10.1242/jcs.035600. [DOI] [PubMed] [Google Scholar]
- 42.Kohler TP, Gisch N, Binsker U, Schlag M, Darm K, Völker U, Zähringer U, Hammerschmidt S. Repeating structures of the major staphylococcal autolysin are essential for the interaction with human thrombospondin 1 and vitronectin. J Biol Chem. 2014;289:4070–4082. doi: 10.1074/jbc.M113.521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voss S, Hallström T, Saleh M, Burchhardt G, Pribyl T, Singh B, Riesbeck K, Zipfel PF, Hammerschmidt S. The choline-binding protein PspC of Streptococcus pneumoniae interacts with the C-terminal heparin-binding domain of vitronectin. J Biol Chem. 2013;288:15614–15627. doi: 10.1074/jbc.M112.443507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohler S, Hallström T, Singh B, Riesbeck K, Spartà G, Zipfel PF, Hammerschmidt S. Binding of vitronectin and factor H to Hic contributes to immune evasion of Streptococcus pneumoniae serotype 3. Thromb Haemost. 2015;113:125–142. doi: 10.1160/TH14-06-0561. [DOI] [PubMed] [Google Scholar]
- 45.Müller NF, Kaiser PO, Linke D, Schwarz H, Riess T, Schäfer A, Eble JA, Kempf VA. Trimeric autotransporter adhesin-dependent adherence of Bartonella henselaeBartonella quintana, and Yersinia enterocolitica to matrix components and endothelial cells under static and dynamic flow conditions. Infect Immun. 2011;79:2544–2553. doi: 10.1128/IAI.01309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Köberle M, Klein-Günther A, Schütz M, Fritz M, Berchtold S, Tolosa E, Autenrieth IB, Bohn E. Yersinia enterocolitica targets cells of the innate and adaptive immune system by injection of Yops in a mouse infection model. PLoS Pathog. 2009;5:e1000551. doi: 10.1371/journal.ppat.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller B, Mühlenkamp M, Deuschle E, Siegfried A, Mössner S, Schade J, Griesinger T, Katava N, Braunsdorf C, Fehrenbacher B, Jimenez-Soto LF, Schaller M, Haas R, Genth H, Retta SF, Meyer H, Bottcher RT, Zent R, Schütz M, Autenrieth IB, Bohn E. Yersinia enterocolitica exploits different pathways to accomplish adhesion and toxin injection into host cells. Cell Microbiol. 2015;17:1179–1204. doi: 10.1111/cmi.12429. [DOI] [PubMed] [Google Scholar]
- 48.Skurnik M. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J Appl Bacteriol. 1984;56:355–363. doi: 10.1111/j.1365-2672.1984.tb01362.x. [DOI] [PubMed] [Google Scholar]
- 49.Portnoy DA, Moseley SL, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strobel E, Heesemann J, Mayer G, Peters J, Müller-Weihrich S, Emmerling P. Bacteriological and serological findings in a further case of transfusion-mediated Yersinia enterocolitica sepsis. J Clin Microbiol. 2000;38:2788–2790. doi: 10.1128/jcm.38.7.2788-2790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolin I, Norlander L, Wolf-Watz H. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect Immun. 1982;37:506–512. doi: 10.1128/iai.37.2.506-512.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mollenkvist A, Nordström T, Hallden C, Christensen JJ, Forsgren A, Riesbeck K. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J Bacteriol. 2003;185:2285–2295. doi: 10.1128/JB.185.7.2285-2295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nordström T, Blom AM, Tan TT, Forsgren A, Riesbeck K. Ionic binding of C3 to the human pathogen Moraxella catarrhalis is a unique mechanism for combatting innate immunity. J Immunol. 2005;175:3628–3636. doi: 10.4049/jimmunol.175.6.3628. [DOI] [PubMed] [Google Scholar]
- 54.Singh B, Jalalvand F, Mörgelin M, Zipfel P, Blom AM, Riesbeck K. Haemophilus influenzae protein E recognizes the C-terminal domain of vitronectin and modulates the membrane attack complex. Mol Microbiol. 2011;81:80–98. doi: 10.1111/j.1365-2958.2011.07678.x. [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lassmann T, Sonnhammer EL. Kalign - an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hallström T, Singh B, Resman F, Blom AM, Morgelin M, Riesbeck K. Haemophilus influenzae protein E binds to the extracellular matrix by concurrently interacting with laminin and vitronectin. J Infect Dis. 2011;204:1065–1074. doi: 10.1093/infdis/jir459. [DOI] [PubMed] [Google Scholar]
- 58.Flügel A, Schulze-Koops H, Heesemann J, Kuhn K, Sorokin L, Burkhardt H, von der Mark K, Emmrich F. Interaction of enteropathogenic Yersinia enterocolitica with complex basement membranes and the extracellular matrix proteins collagen type IV, laminin-1 and -2, and nidogen/entactin. J Biol Chem. 1994;269:29732–29738. [PubMed] [Google Scholar]
- 59.Nummelin H, Merckel MC, Leo JC, Lankinen H, Skurnik M, Goldman A. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 2004;23:701–711. doi: 10.1038/sj.emboj.7600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prilipov A, Phale PS, Van Gelder P, Rosenbusch JP, Koebnik R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol Lett. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 61.Pilz D, Vocke T, Heesemann J, Brade V. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect Immun. 1992;60:189–195. doi: 10.1128/iai.60.1.189-195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biedzka-Sarek M, Venho R, Skurnik M. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect Immun. 2005;73:2232–2244. doi: 10.1128/IAI.73.4.2232-2244.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dersch P, Isberg RR. An immunoglobulin superfamily-like domain unique to the Yersinia pseudotuberculosis invasin protein is required for stimulation of bacterial uptake via integrin receptors. Infect Immun. 2000;68:2930–2938. doi: 10.1128/iai.68.5.2930-2938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deuschle E, Keller B, Siegfried A, Manncke B, Späth T, Köberle M, Drechsler-Hake D, Reber J, Böttcher RT, Autenrieth SE, Autenrieth IB, Bohn E, Schütz M. Role of beta1 integrins and bacterial adhesins for Yop injection into leukocytes in Yersinia enterocolitica systemic mouse infection. Int J Med Microbiol. 2016;306:77–88. doi: 10.1016/j.ijmm.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Duan R, Liang J, Shi G, Cui Z, Hai R, Wang P, Xiao Y, Li K, Qiu H, Gu W, Du X, Jing H, Wang X. Homology analysis of pathogenic Yersinia species Yersinia enterocolitica Yersinia pseudotuberculosis, and Yersinia pestis based on multilocus sequence typing. J Clin Microbiol. 2014;52:20–29. doi: 10.1128/JCM.02185-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maldonado-Arocho FJ, Green C, Fisher ML, Paczosa MK, Mecsas J. Adhesins and host serum factors drive Yop translocation by Yersinia into professional phagocytes during animal infection. PLoS Pathog. 2013;9:e1003415. doi: 10.1371/journal.ppat.1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirjavainen V, Jarva H, Biedzka-Sarek M, Blom AM, Skurnik M, Meri S. Yersinia enterocolitica serum resistance proteins YadA and Ail bind the complement regulator C4b-binding protein. PLoS Pathog. 2008;4:e1000140. doi: 10.1371/journal.ppat.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brooks MJ, Sedillo JL, Wagner N, Laurence CA, Wang W, Attia AS, Hansen EJ, Gray-Owen SD. Modular arrangement of allelic variants explains the divergence in Moraxella catarrhalis UspA protein function. Infect Immun. 2008;76:5330–5340. doi: 10.1128/IAI.00573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.El Tahir Y, Kuusela P, Skurnik M. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVAIG-S motifs in the amino-terminal half of the protein involved in collagen binding. Mol Microbiol. 2000;37:192–206. doi: 10.1046/j.1365-2958.2000.01992.x. [DOI] [PubMed] [Google Scholar]
- 70.Roggenkamp A, Neuberger HR, Flügel A, Schmoll T, Heesemann J. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol Microbiol. 1995;16:1207–1219. doi: 10.1111/j.1365-2958.1995.tb02343.x. [DOI] [PubMed] [Google Scholar]
- 71.Tamm A, Tarkkanen AM, Korhonen TK, Kuusela P, Toivanen P, Skurnik M. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol Microbiol. 1993;10:995–1011. doi: 10.1111/j.1365-2958.1993.tb00971.x. [DOI] [PubMed] [Google Scholar]
- 72.Roggenkamp A, Ruckdeschel K, Leitritz L, Schmitt R, Heesemann J. Deletion of amino acids 29 to 81 in adhesion protein YadA of Yersinia enterocolitica serotype O:8 results in selective abrogation of adherence to neutrophils. Infect Immun. 1996;64:2506–2514. doi: 10.1128/iai.64.7.2506-2514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perry RD, Brubaker RR. Vwa+ phenotype of Yersinia enterocolitica. Infect Immun. 1983;40:166–171. doi: 10.1128/iai.40.1.166-171.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ho DK, Riva R, Kirjavainen V, Jarva H, Ginstrom E, Blom AM, Skurnik M, Meri S. Functional recruitment of the human complement inhibitor C4BP to Yersinia pseudotuberculosis outer membrane protein Ail. J Immunol. 2012;188:4450–4459. doi: 10.4049/jimmunol.1103149. [DOI] [PubMed] [Google Scholar]
- 75.Ho DK, Riva R, Skurnik M, Meri S. The Yersinia pseudotuberculosis outer membrane protein Ail recruits the human complement regulatory protein factor H. J Immunol. 2012;189:3593–3599. doi: 10.4049/jimmunol.1201145. [DOI] [PubMed] [Google Scholar]
- 76.Ram S, Lewis LA, Rice PA. Infections of people with complement deficiencies and patients who have undergone splenectomy. Clin Microbiol Rev. 2010;23:740–780. doi: 10.1128/CMR.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data