Abstract
Toll-like receptor 9 (TLR9) trafficking from the endoplasmic reticulum (ER) into endolysosomes is critical for eliciting cytidine-phosphate-guanosine (CpG) DNA-mediated immune responses. ADP-ribosylation factor 3 (ARF3) is a member of the Ras superfamily, which is crucial for a wide variety of cellular events including protein trafficking. In this study, we found that the inhibition of ARF3 by dominant mutants and siRNA impaired CpG oligodeoxynucleotide (ODN)-mediated responses whereas cells expressing the constitutively active ARF3 mutant enhanced CpG ODN-induced NF-κB activation and cytokine production. Further experiments with MyD88-overexpressing fibroblast cells transfected with a dominant-negative mutant and a constitutively active mutant of ARF3 demonstrated that ARF3 regulated CpG ODN-mediated signaling upstream of MyD88. Additional studies have shown that ARF3 inhibition impairs TLR9 trafficking from the ER into endolysosomes, thereby inhibiting the functional cleavage of TLR9, although it has no significant effect on CpG ODN uptake. Furthermore, activated ARF3 is associated with Unc93B1 and TLR9, suggesting that ARF3 conducts TLR9 trafficking by forming the TLR9-Unc93B1-ARF3 complex. Overall, our findings demonstrate that a novel ARF3 axis pathway mediates CpG ODN-induced responses by regulating TLR9 trafficking.
Key Words: Toll-like receptor 9 trafficking, CpG oligodeoxynucleotide-mediated responses, ADP-ribosylation factor 3
Introduction
Toll-like receptor 9 (TLR9) is responsible chiefly for innate immune responses from the recognition of unmethylated 2′-deoxyribo(cytidine-phosphate-guanosine, CpG) DNA motifs, frequently present in bacteria and viruses but rare in mammalian cells, for host defense. An analysis of TLR9-deficient mice revealed that TLR9 is essential for proinflammatory cytokine production and other inflammatory responses, and it also plays a role in the induction of Th1 adaptive immune responses and the proliferation of B cells. Two crucial steps, CpG ODN uptake into endosomes and TLR9 trafficking from the endoplasmic reticulum (ER) to endosomes, are required for the activation of microbial or viral CpG DNA-driven immunostimulatory activity. Although TLR9 is required for immunity against foreign infections, the inappropriate activation of TLR9 leads to acute and chronic inflammation, in addition to autoimmune diseases such as systemic lupus erythematosus [1, 2]. The intracellular localization of TLR9 is a key factor for preventing the recognition of self-components. TLR9 is carried from the ER to the endolysosomes to interact with microbial CpG DNA from infectious viruses or bacteria [3, 4, 5]. The proper localization of TLR9 and the regulation of the trafficking route are crucial for the encounter of respective ligands as well as the subsequent inflammatory responses. Therefore, the regulatory mechanisms of the trafficking and identifying of TLR9 at appropriate destinations warrant investigation. Unc93B1, an ER-resident transmembrane glycoprotein, has been reported to be a TLR9 carrier protein, and facilitates TLR9 loading into coat protein complex (e.g. COPII) vesicles that directly transport from the ER to the Golgi apparatus via the conventional secretory pathway [6, 7, 8, 9]. However, the precise mechanisms of Unc93B1-mediated TLR9 trafficking require further investigation.
ADP-ribosylation factors (ARFs) are members of the Ras superfamily of 20-kDa guanine nucleotide-binding proteins. ARFs are ubiquitously expressed, and havemultiple roles in the regulation of cellular functions including endocytosis and protein trafficking. Six related gene products exist, ARF1-ARF6, which are categorized into 3 classes based on their sequence homology, class I consisting of ARF1, ARF2 and ARF3, class II consisting of ARF4 and ARF5 and class III consisting of ARF6 [10]. The function of ARF proteins depends on the binding and hydrolyzing of GTP through a cycle between the active GTP-bound form (ARF-GTP) and the inactive GDP-bound form (ARF-GDP). A common feature of GTPase ARFs cycles between a GTP and GDP-bound state is regulated by guanine exchange factors (GEFs) and GTPase-activating proteins. Replacement of the glutamine (71) residue with leucine constitutively locks ARF3 in the GTP-bound form by sequestering GTPase-activating protein binding, which is a dominant active form of ARF3. In contrast, a substitution of threonine (31) with asparagine, ARF3T31N, leads to interference with GTP binding and maintains ARF3 in an inactive state [11]. Although ARFs have critical functions in cellular processes, studies demonstrating the precise role of each ARF in cellular biological responses have been limited, because of a lack of specific inhibitors to individual ARFs. The fungal metabolite brefeldin A has been reported to block ARFs, except for ARF6, which is transported to the Golgi body by preventing their association with the Golgi membrane and coat protein complex formation. The dependence of the ARF inhibitor brefeldin A on CpG ODN/TLR9 signaling indicates that brefeldin A-sensitive ARFs impaired CpG ODN-induced NF-κB activation by blocking TLR9 trafficking through the Golgi body [5, 12], suggesting that brefeldin A-sensitive ARFs play a critical role in CpG ODN-mediated responses. Although the roles of individual brefeldin A-sensitive ARFs in TLR9-mediated signaling remain obscure, brefeldin A-resistant ARF6 has been found to affect CpG ODN/TLR9-mediated responses by regulating cellular CpG ODN uptake [13].
Three types of stimulatory CpG ODNs, types A, B and C, have been identified and characterized, each with distinct effects on the TLR9-mediated immune response. TLR9 plays a critical role in unmethylated CpG DNA/ODN-induced innate immunity, and is linked with a role in adaptive immunity because it triggers the activation of various immune cell types. Thus, understanding the TLR9 trafficking pathway will shed light on how the immune response is activated, and is important for developing specific therapies that can efficiently fight against autoimmune diseases. In our study, we used dominant mutants and small interfering RNA (siRNA) to determine the physiological roles of ARF3 in CpG ODN/TLR9-mediated responses, in addition to the molecular mechanisms via which ARF3 regulates CpG ODN/TLR9-mediated responses. Our findings indicate that ARF3 is involved in CpG ODN/TLR9-mediated responses by regulating the trafficking and proteolytic processing of TLR9.
Materials and Methods
Reagents
Phosphorothioate-modified CpG1585 ODN, GpC1585 ODN, CpG1668 ODN, CpG1668 ODN-FITC, GpC1668 ODN, CpG1826 ODN, GpC1826 ODN, CpG2395 ODN and GpC2395 ODN were purchased from InvivoGen. Anti-Akt, anti-phospho-Akt (Ser473), anti-p38, anti-phospho-p38, anti-NF-κB and anti-phospho-NF-κB antibodies were from Cell Signaling Technology. Anti-TLR9 and anti-TLR9 (C-term) were obtained from InvivoGen or Zymed Laboratories Inc., respectively. Anti-actin and anti-HA antibodies were purchased from Millipore. Anti-Unc93B was from Imgenex. The following antibodies were used for immunofluorescence staining: anti-TLR9-FITC (Imgenex), anti-LAMP-1 (BD Pharmingen), DyLight 488-conjugated goat anti-mouse IgG, and DyLight 549-conjugated donkey anti-rat IgG (Jackson ImmunoResearch Laboratories).
Cell Culture and Treatment
Mouse RAW264.7 macrophages and human embryonic kidney 293T (HEK293T) cells were obtained from the American Type Culture Collection. The RAW264.7 and HEK293T cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, 200 mmol/l L-glutamine and 50 μM β-mercaptoethanol in a humidified atmosphere of 5% CO2 at 37°C. The medium was changed every 2 days for all of the experiments.
The cells were typically preincubated with or without inhibitors for 30 min before CpG ODN treatment, unless specified otherwise. The duration of CpG ODN treatment varied with the experiment. CpG ODNs with CpG dinucleotides mimic the immunostimulatory activity of bacterial DNA and induce TLR9 signaling whereas GpC ODNs contain GpC dinucleotides instead, and do not induce TLR9 activity. GpC ODNs were designed as a negative control for the TLR9 agonist CpG ODNs in this study.
Plasmacytoid Dendritic Cells and Monocyte/Macrophage Preparation
Normal 8-week-old male BALB/c mice from the National Laboratory Animal Center (Taipei, Taiwan, ROC) were sacrificed for the experiments through instant neck dislocation. Monocytes/macrophages and plasmacytoid dendritic cells (pDCs) were then prepared from splenocytes, as previously described [14, 15]. In brief, the spleen cells were depleted of erythrocytes in 0.25× HBSS for 15 s followed by the addition of 2× HBSS and centrifugation. The cell pellet was suspended in RPMI1640 medium, and then the pDCs and monocytes/macrophages were isolated with the use of a pDC isolation kit and CD11b microbeads, respectively (Miltenyi Biotec), according to the manufacturer's instructions.
Plasmid Constructions
The expression plasmids for human MyD88 and its dominant-negative mutant (MyD88-DN) were constructed by inserting PCR-amplified cDNA fragments into the pcDNA3.1(-) expression vector (Invitrogen), as previously described [15]. Briefly, the MyD88 cDNA was generated by RT-PCR using total RNA of the THP-1 cell line as a template and the following oligonucleotides as primers: 5′-GGA TCC ATG GCT GCA GGA GGT CCC GGC-3′ and 5′-AAG CTT CTC AGG GCA GGG ACA AGG CCT-3′. As the primers incorporate BamHI and HindIII sites, the PCR product was then cloned into BamHI and HindIII digested pcDNA3.1(-) to generate pcDNA3.1(-)-MyD88. To create its MyD88-DN construct, i.e. pcDNA3.1(-)-MyD88-DN, a truncated version of MyD88 expressing the N-terminal death domain, pcDNA3.1(-)-MyD88 was used as a template for PCR amplification using forward primer containing the BamHI restriction site (5′-GGA TCC ATG GCT GCA GGA GGT CCC GGC-3′) and a reverse primer containing the HindIII restriction site (5′-AAG CTT AAT GCT GGG TCC CAG CTC CAG). The PCR product was cloned into BamHI- and HindIII-digested pcDNA3.1(-).
Stable Transfectants
The HEK293T and mouse RAW264.7 cells were transfected with the plasmids pcDNA3-mTLR9, pcDNA3-ARF1T31N- HA, pcDNA3-ARF3-HA, pcDNA3-ARF3T31N-HA or pcDNA3- ARF3Q71L-HA with the use of FuGENE 6 or FuGENE HD (Roche Molecular Biochemicals), according to the manufacturer's instructions. Two days after transfection, a G418 antibiotic (1.0 mg/ml) was used for clonal selection. The protein expression of ARFs in their respective overexpression stable RAW264.7 cell lines including RAW-pcDNA, RAW-ARF3, RAW-ARF3TN, RAW-ARF3QL and RAW-ARF1T31N were measured by Western blot analysis (online suppl. fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000430785).
RNA Interference
Double-stranded RNAs were synthesized (Invitrogen). The following siRNA sequences were used for mouse ARF1 and mouse ARF3: 5′-CAG AAC ACC CAA GGC TTG ATC TTC G-3′ and 5′-CCG UGC UGU AUU GUC ACA AGC ACA U-3′, respectively. The negative control sequence was 5′-CCG UGU CGU UAC ACU CGA AAG UCA U-3′. The following siRNA sequences were used for human ARF1: 5′-CCT GAT CTT CGT GGT GGA CAG CAA T-3′, human ARF3: 5′-CCA TTG GGT TCA ATG TGG AGA CAG T-3′ and human ARF6: 5′-CGG CAU UAC ACU GGG AUU-3′. Cells were transfected with siRNA duplexes with the use of Lipofectamine 2000 (Invitrogen). Lipofectamine 2000 was diluted in DMEM without a serum and antibiotics, and incubated for 5 min at room temperature. The siRNA was diluted in DMEM without a serum and antibiotics. The siRNA was then added to the diluted Lipofectamine 2000, and incubated for 30 min at room temperature. After incubation, the transfection complex was added to the cells in a dropwise manner. The transfection medium was removed 6 h later. The cells were then washed twice with phosphate-buffered saline (PBS), and maintained in a culture medium for 48–72 h.
Luciferase Reporter Assay
Cells in 24-well plates were cotransfected with 0.05 μg p5 × NF-κB-luc (Stratagene) and 0.01 μg pcDNA3.1-β-galactosidase in addition to pcDNA3.1, pcDNA3-ARF1T31N-HA, pcDNA3-ARF3T31N-HA, pcDNA3-ARF3Q71L-HA, pcDNA3.1(-)-MyD88 or pcDNA3.1(-)-MyD88-DN with FuGENE 6 (Roche Molecular Biochemicals) overnight, as previously described [15]. The cells were incubated with or without 1.0 μM CpG ODN for 8 h, and then lysed. The NF-κB luciferase activity assays were performed according to the procedures recommended by the manufacturer (Promega). The β-galactosidase activity was used to normalize the data.
Cytokine-Specific ELISA
Cytokine levels in the culture supernatants and serum were determined in 96-well microtiter plates by using a specific sandwich ELISA (Biosource), as previously described [13].
Western Blot Analysis
Cellular proteins were resolved using 5–20% SDS-PAGE, and were transferred to nitrocellulose membranes before blotting with specific antibodies, as previously described [16].
Confocal Microscopy
The cells were fixed by incubation in PBS containing 3.5% paraformaldehyde at room temperature for 10 min. They were washed extensively, and then blocked with an Image-iT FX signal enhancer (Invitrogen) for 30 min at room temperature. They were subsequently permeabilized with 0.1% Triton X-100 for 1.5 min at room temperature, washed with PBS and then stained with anti-TLR9-FITC and anti-LAMP-1 at room temperature. After 1 h of staining, they were washed and counterstained with DyLight 488-conjugated goat anti-mouse IgG and DyLight 549-conjugated donkey anti-rat IgG for 1 h. All cells were visualized under a Leica TCS SP5 laser scanning confocal microscope.
CpG ODN Uptake
In brief, ODN1668-FITC-stimulated cells were washed with PBS, and surface-bound FITC cells were quenched using 0.04% Trypan blue. The fluorescence intensity of ODN1668-FITC cellular uptake was measured at 530 nm with flow cytometry (BD Biosciences) at an excitation wavelength of 488 nm, and was expressed as the mean value of the fluorescence intensity of 5,000 cells.
Statistical Analysis
All values were considered as the mean ± standard deviation (SD). The statistical significance of differences between the treatment groups was calculated using the Student t test. p < 0.05 was considered statistically significant.
Results
The Role of ARF3 in CpG ODN/TLR9-Mediated Responses
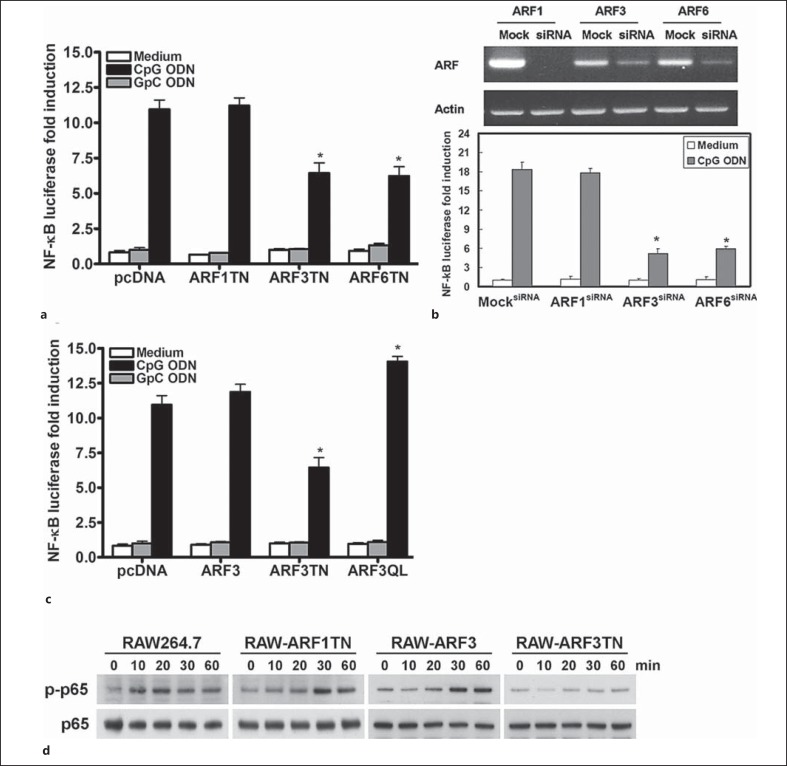
Brefeldin A-sensitive class I ARF1 and ARF3 have been implicated in numerous types of intracellular membrane vesicle trafficking events [10, 17, 18], so we evaluated the influence of ARF1 and ARF3 on CpG ODN/TLR9-mediated responses. First, we examined the role of ARF1 and ARF3 in CpG ODN-stimulated NF-κB activation. The TLR9-deficient HEK293T cell line treated with mouse TLR9-specific B-class CpG1668 ODN caused little, if any, increase in the NF-κB luciferase activity (data not shown). However, after the mouse TLR9 gene was stably integrated into HEK293T cells to form mTLR9-293T cells, the NF-κB luciferase activity was greatly increased by CpG1668 ODN but not control GpC1668 ODN (fig. 1a). This is consistent with our previous report that inhibition of brefeldin A-resistant ARF6 suppressed CpG1668 ODN-mediated NF-κB activation (fig. 1a, b). Additionally, the CpG1668 ODN-induced NF-κB luciferase activity decreased with the transient expression of the dominant-negative form of ARF3 (ARF3T31N), but not with the dominant-negative form of ARF1 (ARF1T31N) in mTLR9-293T cells (fig. 1a). To further examine the role of ARF3 in TLR9-mediated NF-κB activation, siRNAs against human ARF1 and ARF3 were used to knock down ARF1 and ARF3 expression in the mTLR9-293T cells. The CpG1668 ODN-induced NF-κB activation was impaired in cells transfected with specific ARF3 siRNA, but not in those transfected with ARF1 siRNA (fig. 1b). In contrast to the inhibitory effect of ARF3T31N on CpG1668 ODN-mediated NF-κB activation, the overexpression of constitutively active human ARF3 in RAW264.7 cells enhanced CpG1668 ODN-mediated NF-κB luciferase activity (fig. 1c). In addition to the NF-κB luciferase results, the phosphorylation level of NF-κB p65 (Ser536) induced by CpG1668 ODNs decreased in RAW264.7 cells stably expressing ARF3T31N, but not in RAW-ARF3 and RAW-ARF1T31N cells (fig. 1d).
Fig. 1.
ARF3 is involved in CpG ODN-induced NF-κB activation and phosphorylation. a mTLR9-293T cells were transiently transfected with an empty vector, pcDNA3.1, and ARF1T31N or ARF3T31N plasmids plus p5 × NF-κB luciferase overnight, and then stimulated with 1.0 μM GpC1668 ODN (negative control) or CpG1668 ODN for 8 h. The NF-κB luciferase activities were then measured. Data represent the mean ± SD from 3 experiments. * p < 0.05 for ARF6T27N vs. pcDNA3.1. b The mTLR9-293T cells were transfected with scramble siRNA (MocksiRNA), mouse ARF1 siRNA (ARF1siRNA), ARF3 siRNA (ARF3siRNA) or ARF6 siRNA (ARF6siRNA) for 48 h, and then incubated in medium or a medium supplemented with CpG1668 ODN for 8 h, as indicated. * p < 0.01 for ARF3siRNA or ARF6siRNA vs. scramble. c RAW264.7 cells were transiently transfected with pcDNA3.1, ARF3 and ARF3T31N or ARF3Q71L plasmids plus p5 × NF-κB luciferase overnight, and then stimulated with 1.0 μM GpC ODN (negative control) or CpG1668 ODN for 8 h. The NF-κB luciferase activities were then measured. Data represent the mean ± SD from 3 experiments. * p < 0.05 for ARF3T27N or ARF3Q71L vs. pcDNA3.1. d RAW264.7, RAW-ARF1T31N, ARF3 and ARF3T31N cells were treated with 1 μM CpG1668 ODN for the indicated times. Cell lysates were immunoblotted with phospho-NF-κB p65- (Ser536) or NF-κB p65-specific antibodies, as indicated. The experiment was repeated 3 times, and yielded similar results.
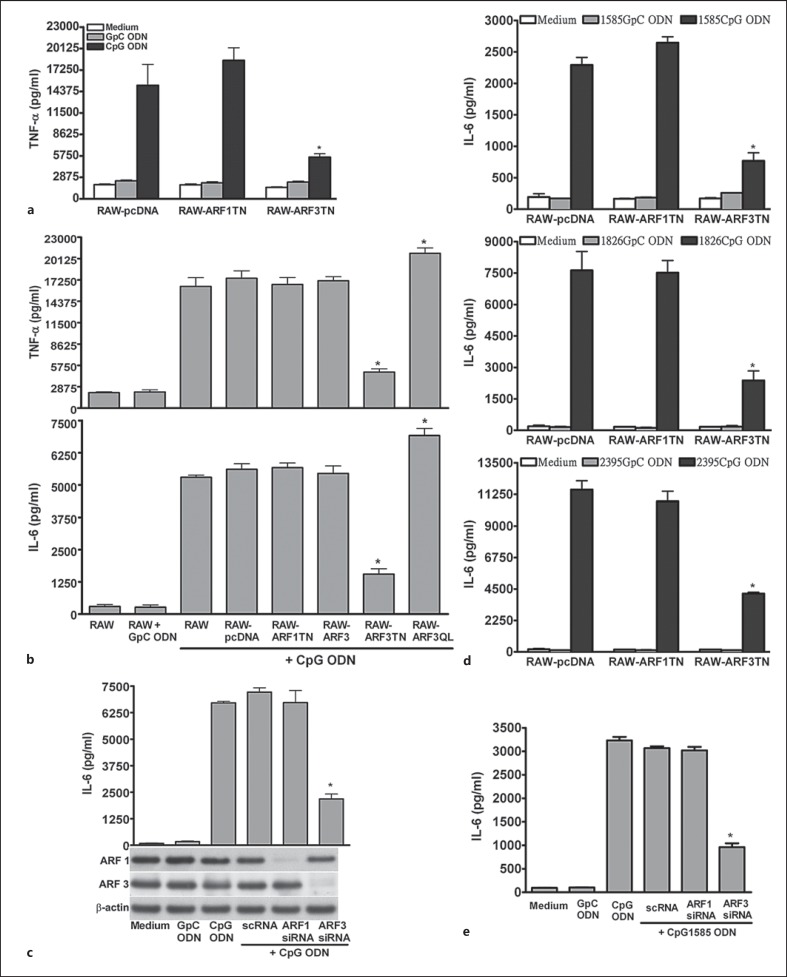
NF-κB is a key factor in CpG1668 ODN-mediated cytokine production, so we examined the role of ARF3 in CpG1668 ODN-induced cytokine production. Cytokine production (e.g. TNF-α and IL-6) in cells after 24 h of CpG1668 ODN treatment was increased compared with that in cells treated with GpC1668 ODN. The CpG1668 ODN-induced production of TNF-α and IL-6 was impaired in RAW-ARF3T31N compared with the control RAW264.7, RAW-pcDNA, RAW-ARF3 and RAW-ARF1T31N cells (fig. 2a, b). In addition, RAW264.7 cells expressing constitutively active human ARF3 enhanced the production of TNF-α and IL-6 induced by CpG1668 ODN (fig. 2b). These results support the proposal that the inhibitory effect of ARF3T31N on CpG1668 ODN-mediated cytokine production is not due to the nonspecific expression of ARF3, but to the specific blocking of ARF3 activation. The crucial role of ARF3 in TLR9-mediated responses was evident in primary cells such as mouse spleen pDCs and monocytes/macrophages. The production of IL-6 induced by CpG1668 ODN was impaired in mouse pDCs and spleen monocytes/macrophages transfected with ARF3 siRNA directed to the mouse ARF3 gene compared with those of cells transfected with scramble siRNA and ARF1 siRNA (fig. 2c; online suppl. fig. 2). It has been shown that 3 classes of synthetic CpG-ODNs, classes A, B and C, activate cells through TLR9 but elicit different responses. To determine the role of ARF3 in the 3 classes of CpG ODN-mediated responses, we stimulated RAW-pcDNA, RAW-ARF1TN and RAW-ARF3TN cells with A-type CpG1585 ODN, B-type CpG1826 ODN and C-type CpG2395 ODN. The induction of IL-6 and TNF-α in RAW264.7 cells stimulated by CpG1585 ODN, CpG1826 ODN or CpG2395 ODN was strongly reduced in RAW-ARF3TN cells compared with RAW-pcDNA and RAW-ARF1TN cells (fig. 2d; online suppl. fig. 3). In addition, the production of IL-6 and IFN-β induced by CpG1585 ODN and CpG1826 ODN was inhibited in ARF3 knockdown pDCs compared with pDCs transfected with control siRNA and ARF1-specific siRNA (fig. 2e; online suppl. fig. 4). Overall, these results suggest that ARF3, but not ARF1, is required for TLR9-mediated immune activation including NF-κB activation and cytokine production.
Fig. 2.
ARF3 is involved in CpG ODN-induced TNF-α and IL-6 production. RAW264.7, RAW-pcDNA, RAW-ARF1T31N, ARF3, ARF3T31N and ARF3Q71 cells were stimulated with medium only, GpC1668 ODN or CpG1668 ODN for 20 h, as indicated. a, b The TNF-α and IL-6 levels in the culture supernatants were measured using ELISA. * p < 0.001 for ARF3T31N or ARF3Q71L vs. RAW-pcDNA. c The pDCs were transfected with scramble siRNA, mouse ARF1 siRNA or ARF3 siRNA for 48 h, and then underwent stimulation with only medium, GpC ODN or CpG ODN for 20 h. d RAW-pcDNA, RAW-ARF1T31N and ARF3T31N cells were stimulated with medium only, GpC1585 ODN, CpG1585 ODN, GpC1826 ODN, CpG1826 ODN, GpC2395 ODN or CpG2395 ODN for 20 h. e The pDCs were transfected with scramble siRNA, mouse ARF1 siRNA or ARF3 siRNA for 48 h, and then stimulated with medium, GpC1585 ODN or CpG1585 ODN for 20 h. The IL-6 levels in the culture supernatants were measured using ELISA. Data represent the mean ± SD of 3 experiments. * p < 0.001 for ARF3 siRNA vs. scRNA or for ARF3T31N vs. RAW-pcDNA.
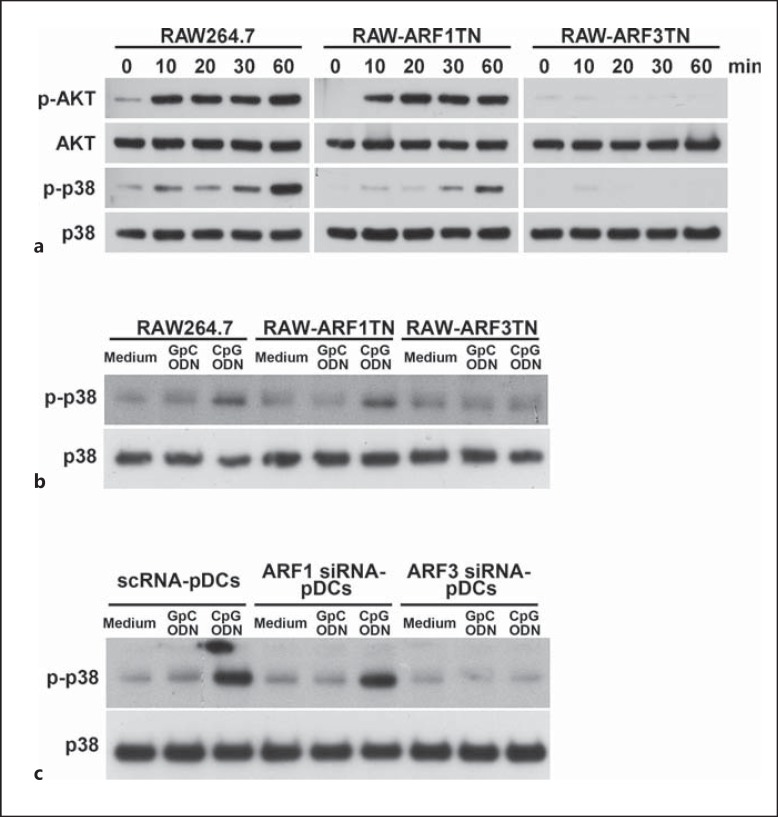
The activation of immune cells by bacterial CpG DNA or CpG ODN occurs through the TLR9 signaling cascade that regulates immune responses through the activation of numerous kinases, most notably p38 MAPK and PI3K. To evaluate the influence of ARF3 on the CpG ODN-mediated activation of kinases, we measured the phosphorylation level of kinases (e.g. p38 MAPK and AKT) in RAW264.7, RAW-ARF1T31N and ARF3T31N cells. CpG1668 ODN strongly triggered the phosphorylation of p38 MAPK and AKT in RAW264.7 and RAW-ARF1T31N cells whereas the inducing of kinase activation by CpG1668 ODN was impaired in RAW-ARF3T31N (fig. 3). To examine whether ARF3 involved in A-type CpG1585 ODN-induced p38 activation in RAW264.7 cells, we stimulated RAW264.7 cells with CpG1585 ODN for 30 min. The induction of p38 phosphorylation in RAW264.7 cells stimulated by CpG1585 ODN was strongly reduced in RAW-ARF3TN cells but not in RAW-ARF1TN cells (fig. 3b). A similar reduction in p38 phosphorylation was observed in pDCs transfected with ARF3 siRNA compared with those transfected with scramble siRNA and ARF1 siRNA (fig. 3c). These results suggest that ARF3 regulation in CpG ODN/TLR9-mediated signaling occurs upstream from these kinases.
Fig. 3.
The influence of ARF3 in the CpG ODN-mediated activation of kinases. a RAW264.7, RAW-ARF1T31N and RAW-ARF3T31N cells were treated with 1.0 μM CpG1668 ODN for the indicated times. b RAW264.7, RAW-ARF1T31N and RAW-ARF3T31N cells were stimulated with medium, 1.0 μM GpC1585 ODN or CpG1585 ODN for 30 min. c The pDCs were transfected with scramble siRNA, mouse ARF1 siRNA or ARF3 siRNA for 48 h, and then underwent stimulation with medium only, GpC ODN or CpG ODN for 30 min. Cell lysates were immunoblotted with Akt-, phospho-Akt-, p38 MAPK-, phospho-p38 MAPK- or β-actin-specific antibodies. The experiment was repeated twice, and yielded similar results.
CpG ODN Stimulation Promotes the Activation of ARF3
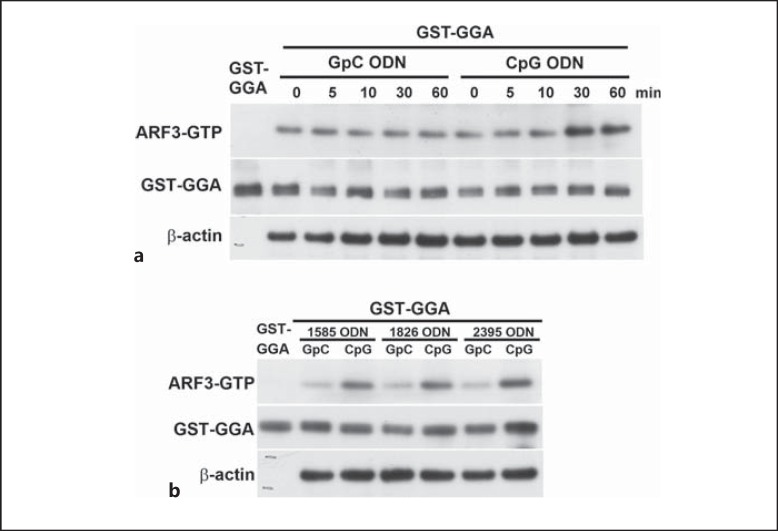
As we demonstrated, ARF3 plays a critical role in CpG ODN-mediated responses. We next examined the activation of ARF3 with CpG ODN treatment. To evaluate the activation profile of ARF3, we monitored the levels of GTP-bound ARF3 upon CpG1668 ODN stimulation by using the ARF-binding domain of GGA1 protein fused to GST, which binds the active form of ARF3 (ARF3-GTP). At different times during CpG1668 ODN stimulation, the RAW264.7 cells expressing C-terminal HA-tagged wild-type (WT) ARF3 were lysed on ice, and the activated ARF3 was pulled down by GST-GGA1 (fig. 4a). A variable degree of basal level of active ARF3 (ARF3-HA-GTP) was observed in untreated or GpC1668 ODN-treated cells and, furthermore, the ARF3-HA-GTP level was steadily increased in RAW-ARF3 cells treated with CpG1668 ODN compared to cells treated with GpC1668 ODN (fig. 4a). No precipitation of ARF3 was detected when GST alone was used in the pull-down assay (data not shown). In addition, ARF3 activation was also induced by other types of CpG ODN including CpG1585 ODN, CpG1826 ODN and CpG2935 ODN (fig. 4b). Taken together, these results suggest that CpG ODNs enhance ARF3 activation, thereby advancing TLR9 signaling.
Fig. 4.
ARF3 activation upon CpG ODN stimulation. a RAW-ARF3 cells were treated with either GpC1668 ODN or CpG1668 ODN for the indicated times. b RAW-ARF3 cells were treated with either GpC1585 ODN, CpG1585 ODN, GpC1826 ODN, CpG1826 ODN, GpC2395 ODN or CpG2395 ODN for 30 min. The experiment was repeated twice with similar results.
ARF3 Participates in the CpG ODN-Mediated TLR9 Signaling Pathway
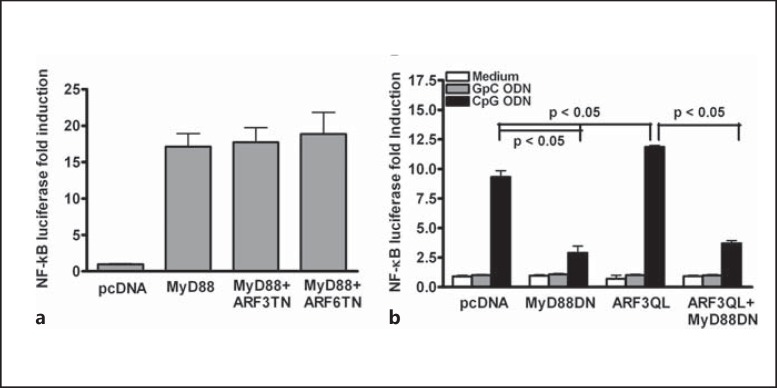
As demonstrated, ARF3 is involved in CpG ODN-mediated responses. We next attempted to delineate the ARF3 sequence of action in the TLR9 signaling pathway. MyD88 is a crucial downstream adaptor in the TLR9 signaling pathway [19, 20]. To determine whether ARF3 exerts its function upstream or downstream of MyD88, HEK293T cells were transiently cotransfected with a WT MyD88 expression plasmid and ARF3T31N-HA or ARF6T27N-HA plasmids. The overexpression of MyD88 induced NF-κB luciferase activity. The overexpression of ARF3T31N or ARF6T27N had no substantial effect on MyD88-induced NF-κB activation (fig. 5a). In contrast, CpG1668 ODN-induced NF-κB activation was impaired in cells expressing MyD88-DN (fig. 5b). CpG1668 ODN-mediated NF-κB activation was increased in ARF3Q71L-overexpressed cells compared with cells transfected with the control vector pcDNA3.1; however, the dominant-negative mutant in MyD88 (MyD88-DN) expression inhibited the enhancement of CpG ODN-mediated NF- κB activation induced by ARF3Q71L overexpression (fig. 5b). These results suggest that ARF3 regulates CpG ODN-mediated signaling upstream of MyD88.
Fig. 5.
ARF3 regulates CpG ODN-induced NF-κB activation upstream of MyD88. a WT MyD88-overexpressing 293T cells were transiently transfected with various concentrations of ARF3T31N or ARF6T27N plus p5× NF-κB luciferase. After 24 h of transfection, NF-κB luciferase activities were measured. b The mTLR9-293T cells were transiently transfected with pcDNA3.1 or ARF3Q71L plasmids plus p5 × NF-κB luciferase with/without the MyD88-DN plasmid overnight, and then stimulated with 1.0 μM GpC ODN (negative control) or CpG ODN for 8 h. The NF-κB luciferase activities were then measured. Data represent the mean ± SD from 3 experiments.
ARF3 Is Involved in TLR9 Trafficking and Proteolysis
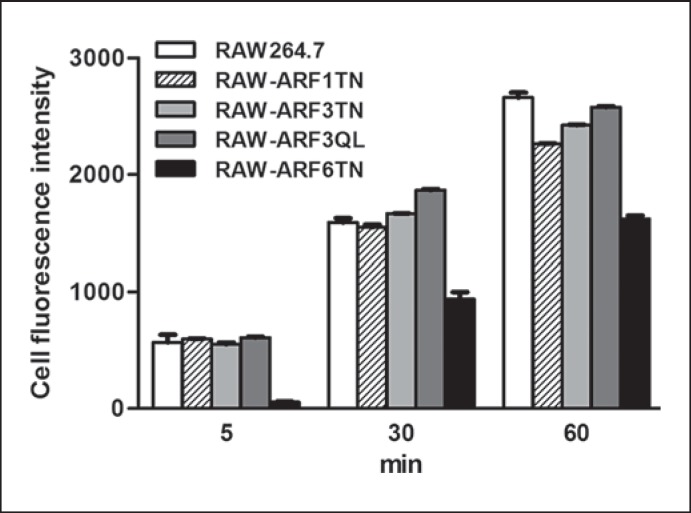
The CpG ODN uptake and the trafficking of TLR9 from the ER to endolysosomes are two critical steps that occur upstream of MyD88 in the TLR9 signaling pathway to elicit innate immune responses. In our previous studies, we demonstrated that ARF6 specifically modulates TLR9-mediated responses by regulating the uptake of CpG ODN [13, 21]. To investigate whether ARF3 participates in cellular CpG ODN uptake, we used flow cytometry to compare the levels of CpG ODN-FITC uptake in RAW264.7, RAW-ARF1T31N, RAW-ARF3T31N, RAW-ARF3Q71L and RAW-ARF6T27N cells. The level of CpG ODN-FITC decreased significantly in RAW-ARF6T27N cells, but not in RAW-ARF1T31N and RAW-ARF3T31N cells (fig. 6). Dominant-negative ARF3 mutants had no significant effect on the level of CpG ODN uptake, suggesting that the activation of TLR9 signaling impeded by the dominant-negative form of ARF3 may function through TLR9 trafficking, rather than through CpG ODN uptake.
Fig. 6.
Role of ARF3 in cellular CpG ODN uptake. RAW264.7, RAW-ARF1T31N, RAW-ARF3T31N, RAW-ARF3Q71L or RAW-ARF6T27N cells were incubated with 0.5 μM CpG ODN-FITC for the indicated times. The cell fluorescence intensity of CpG ODN-FITC uptake was measured using flow cytometry. Data represent the mean ± SD of 3 experiments.
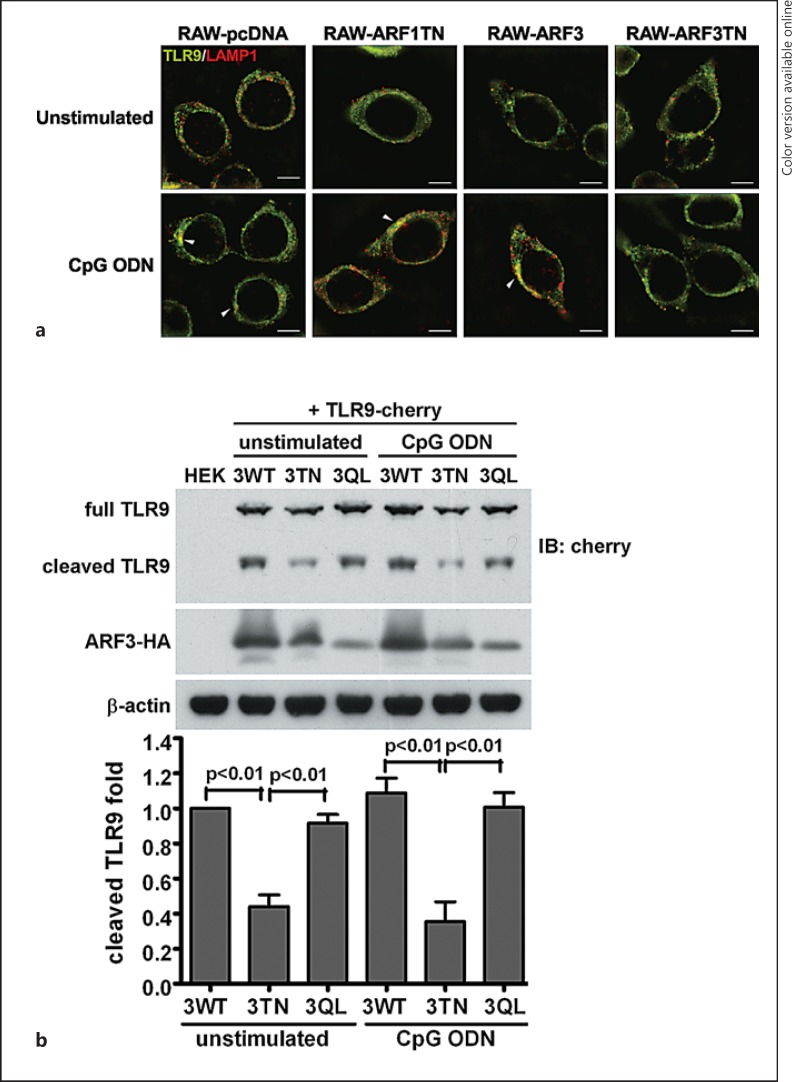
In resting cells, TLR9 chiefly resides at the ER, and subsequently translocates to endolysosome compartments after CpG ODN stimulation [3, 4]. To ascertain the subcellular distribution of TLR9 in RAW-ARF3T31N cells, we observed TLR9 proteins through confocal microscopy in differential types of stably ARFs-expressing RAW264.7 cells. After 1-μM CpG ODN stimulation, TLR9 was colocalized with LAMP1, an endolysosome-resident protein, in RAW-pcDNA, RAW-ARF1T31N and RAW-ARF3 cells (fig. 7a, white arrows). The TLR9 was not translocated to endolysosome compartments in stimulated RAW-ARF3T31N cells, indicating that the ARF3T31N mutant interfered with the trafficking of TLR9 from the ER to the endolysosomes (fig. 7a). These results suggest that ARF3 is involved in CpG ODN-mediated responses via the regulation of TLR9 trafficking.
Fig. 7.
Stable expression of ARF3T31N interferes with both the trafficking and proteolytic processing of TLR9. a RAW-pcDNA, RAW-ARF1T31N, RAW-ARF3 or RAW-ARF3T31N cells were treated with or without CpG ODN (1.0 μM) for 2 h. The cells were then fixed, permeabilized and labeled with anti-TLR9 (FITC: green) and anti-LAMP1 (DyLight 549: red) antibodies. These images are representative of 3 independent experiments. Scale bar: 5 μm. b The cell lysates of 293T cells, TLR9- cherry-expressing 293T cells transiently transfected with pcDNA3.1, ARF3-HA, ARF3T31N-HA or ARF3Q71L-HA plasmids incubated in the culture medium or a medium supplemented with 1.0 μM CpG ODN for 2 h were subjected to Western blot analysis with TLR9- (anti-cherry), ARF3- (anti-HA) or β-actin-specific antibodies. The bottom panel shows quantitative analysis by densitometry. Error bars denote mean ± SD of 3 experiments.
Recent studies have indicated that full-length TLR9 traffic migrates from the ER to endolysosomes, and matures immediately through proteolytic cleavage processing [22, 23]. Functional cleaved TLR9 recruits MyD88 to activate signal transduction, so we attempted to evaluate the involvement of ARF3 in functional receptor regulation. Full-length and cleaved TLR9 were detected in ARF3 and ARF3Q71L-overexpressing HEK293T cells, and the level of cleaved TLR9 was significantly lower in ARF3T31N-overexpressing cells (fig. 7b). These results suggest that ARF3T31N suppresses TLR9 trafficking, thereby impairing TLR9 proteolysis.
As shown in figure 7, TLR9 failed to translocate to the endolysosome in RAW-ARF3T31N cells after CpG ODN stimulation, and the level of cleaved TLR9 decreased significantly in ARF3T31N-overexpressing HEK293T cells, regardless of whether they underwent CpG ODN stimulation. In addition, the level of cleaved TLR9 in WT ARF3-overexpressing HEK293T cells treated with CpG ODN increased slightly compared with unstimulated cells, and the level of cleaved TLR9 was found to be greater in the ARF3Q71L-expressing cells, but lower in the ARF3T31N-expressing cells (fig. 7b). Densitometric analysis on the immunoblots for assessing the cleaved TLR9 level revealed that this level was reduced by 68% in ARF3TN-expressing cells compared to in ARF3-overexpressing cells (fig. 7b). Overall, these results suggest that ARF3 activation is required for CpG-ODN/TLR9-mediated signaling by regulating TLR9 trafficking and functional receptor formation.
ARF3 Is Associated with the Unc93B1-TLR9 Complex
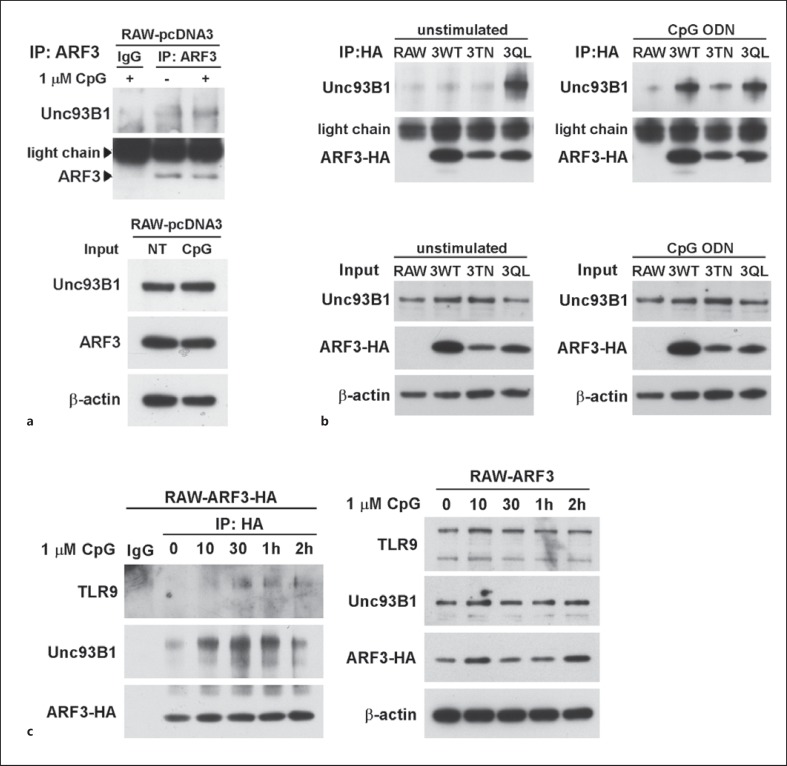
The Unc93B1 protein is an ER-resident protein responsible for TLR9 sorting, and acts as a carrier protein to transport TLR9 from the ER to endolysosomes [7, 8]. To ascertain whether ARF3 is involved in Unc93B1 expression and the formation of the Unc93B1-TLR9 complex, we applied an immunoprecipitation assay to explore the interactions of ARF3 with Unc93B1 and TLR9. The Unc93B1 protein was associated with endogenous ARF3 and WT ARF3-HA after CpG ODN stimulation in RAW264.7 and RAW-ARF3 cells, respectively (fig. 8a, b). The association profile emerged in a time-dependent manner (fig. 8c). A high association of Unc93B1 and ARF3 was detected in RAW-ARF3Q71L cells with or without CpG ODN stimulation (fig. 8b). By contrast, Unc93B1 was absent in immunoprecipitates from RAW-ARF3T31N cell lysates (fig. 7b). Furthermore, TLR9 was immunoprecipitated with ARF3-HA after CpG ODN stimulation in RAW-ARF3 immunoprecipitates (fig. 8c). Overall, these results demonstrate that ARF3 activity is responsible for the formation of the Unc93B1-TLR9 complex, implicating that ARF3 is involved in TLR9 trafficking.
Fig. 8.
Activation of ARF3 is associated with TLR9 and Unc93B1 in CpG-ODN-stimulated RAW264.7 cells. a Upper panel: RAW264.7 cells were treated with or without CpG1668 ODN (1.0 μM) for 30 min. The cells were then lysed on ice, and were subjected to an immunoprecipitation assay with anti-ARF3. The precipitated level of endogenous ARF3 and Unc93B1 was determined using immunoblotting with anti-ARF3 or anti-Unc93B1. Lower panel: total input was determined using immunoblotting. The experiment was repeated twice, and yielded similar results. b Upper panel: RAW264.7, RAW-ARF3, RAW-ARF3T31N and RAW-ARF3Q71L cells were treated with or without CpG1668 ODN (1.0 μM) for 30 min. Cell lysates were subjected to an immunoprecipitation assay with anti-HA. The precipitated level of ARF3-HA and endogenous Unc93B1 was determined using immunoblotting with anti-HA or Unc93B1. Lower panel: total input was determined using immunoblotting. The experiment was repeated twice, and yielded similar results. c Left: RAW-ARF3 cells were treated with CpG1668 ODN for the indicated times. The cell lysates were lysed on ice and then subjected to an immunoprecipitation assay with anti-HA. The immunoprecipitates were determined using immunoblotting with anti-TLR9 and anti-Unc93B1. Right: total input was determined with immunoblotting. The experiment was repeated twice, yielding similar results.
Discussion
Small GTPase ARFs play multiple roles in the regulation of cellular functions, including endocytosis and protein trafficking, by triggering the recruitment of coat protein complexes for forming vesicles. The fungal metabolite brefeldin A is a well-known inhibitor of ARF proteins (except for class III ARF6), and it blocks the Golgi transport by preventing ARF association with the Golgi membrane and coat protein complex formation [24, 25, 26]. Brefeldin A has been reported to block TLR9 trafficking through the Golgi apparatus, but had no substantial effect on CpG DNA endocytosis [5]. Our previous report indicated that brefeldin A-resistant ARF6 is involved in CpG ODN/TLR9-mediated responses by the regulation of CpG ODN uptake [13]. Accordingly, this evidence leads us to hypothesize that brefeldin A-sensitive ARFs and brefeldin A-resistant ARF6 may exhibit different regulatory mechanisms in TLR9-mediated signaling.
The brefeldin A-sensitive class I ARFs, ARF1 and ARF3, are thought to play a critical role in cellular trafficking. In this study, we used siRNA knockdown and dominant-negative approaches to demonstrate that ARF3, not ARF1, is involved in CpG ODN/TLR9-mediated responses including NF-κB activation and cytokine production. Our results indicate that CpG ODN-induced inflammatory responses in RAW264.7 cells and primary cells are reduced by the blockade of ARF3, but not that of ARF1. This means that only brefeldin A-sensitive class I ARF3 is involved in the TLR9 signaling pathway, rather than the entire class I ARF subfamily. These findings reveal that class I ARF1 does not have functions that are parallel with ARF3 in TLR9-mediated responses, despite both of them localizing chiefly to the Golgi complex and being redundantly required for the integrity of recycling endosomes [27]. We suppose that ARF3, but not ARF1, involvement in TLR9 signaling may be due to specific regulatory protein ARF-GEFs, which play a key role in inducing ARF activation. Different ARF-GEF subfamilies localize to distinct cellular compartments, and activate their target ARFs [28, 29]. These various physiological actions of ARF-GEFs lead ARF1 and ARF3 to play special roles in regulating different membrane traffic events. Activated ARF1 is involved in forming COPI vesicles in retrograde transport, whereas activated ARF3 is involved in forming COPII vesicles in anterograde transport [30]. The TLR9 trafficking is routed from the ER to endolysosomes in anterograde transport direction. ARF3, but not ARF1, may participate in TLR9 signaling, and our findings imply that its participation may occur during the receptor transport step.
CpG ODNs can be classified into 2 major classes: type A (CpG-A ODNs) and type B (CpG-B ODNs) [31]; a previous report suggested that there is a marked difference of endosome retention time between them [32]. In general, CpG-A ODNs are effective in activating NK cells and stimulating pDCs and macrophages to produce high levels of type I IFN and inflammatory cytokines, respectively [33, 34], whereas CpG-B ODNs primarily stimulate the proliferation of B cells and induce the activation of pDCs and monocyte/macrophages [33, 35]. The data from our studies reveal that inhibition of ARF3 suppressed the CpG ODN-mediated (both classes) production of inflammatory cytokines and IFN-β by macrophage and BMDCs, suggesting that ARF3 may act as important regulator of TLR9 signaling. To understand the regulation of ARF3 in TLR9 signaling, we delineated the sequence action of ARF3 in the TLR9 signaling pathway stepwise from the downstream immune responses to the upstream internalization of CpG ODNs. In the series of cellular and biochemical studies, our findings revealed that ARF3 functions upstream of MyD88 recruitment, and does not participate in CpG ODN uptake to regulate TLR9-driven immune responses, thus suggesting that the dominant-negative form of ARF3 can potentially impede the activation of TLR9 signaling by interfering with TLR9 trafficking from the ER to endosomes but not with CpG ODNs uptake and delivery. The cellular localization and the physiological role of ARF3 also support this implication. The trafficking of TLR9 and other intracellular TLRs from the ER to endolysosomes is facilitated by Unc93B1, a multipass transmembrane protein that resides in the ER [7, 8]. More recent evidence has demonstrated that Unc93B1 controls multiple steps involved in TLR9 trafficking, facilitating TLR9 loading into COPII vesicles to directly transport from the ER to the Golgi apparatus via the conventional secretory pathway [9, 36]. The localization and the membrane traffic route of ARF3 are similar to those of Unc93B1, which reasonably suggests that ARF3 may be involved in Unc93B1-mediated TLR9 trafficking from the ER to the Golgi. Here, we are the first to provide evidence that ARF3 is associated with Unc93B1 and TLR9, implicating ARF3 in conducting TLR9 trafficking by forming the TLR9-Unc93B1-ARF3 complex.
During the TLR9 translocation process, full-length TLR9 traffics through the Golgi complex, and is routed to endolysosome compartments, where it is cleaved into functional receptors by proteases [22, 23]. This process infers a critical regulation of the receptor function. The cleaved form of TLR9 obtained through the proteolytic cleavage process in endolysosomes is required for activating signal cascades. Therefore, the localization and trafficking of TLR9 are related to a crucial regulation that determines the onset of a substantial amount of signal transduction. We elucidated that ARF3 activation is required for CpG ODN/TLR9-mediated signaling by regulating TLR9 trafficking and thus affects the formation of functional cleaved TLR9 (fig. 7, 8).
We demonstrated that TLR9 translocation and CpG ODN-induced responses increased upon activation of ARF3 and decreased upon inhibition of ARF3 (fig. 1, 2, 3, 7, 8). These results indicate that the TLR9 translocation is positively correlated with the cellular activity of ARF3. Furthermore, the treatment of cells with CpG ODN enhanced TLR9 translocation and CpG ODN-induced responses, suggesting that CpG ODN induces ARF3 activation thereby controlling TLR9 translocation. We reasoned that there is a basal level of active ARF3 that can initiate TLR9 trafficking upon CpG ODN stimulation. The presence of this basal level is supported by the fact that we were able to detect a low level of ARF3-GTP in the ARF3 activation assay (fig. 4). Furthermore, treatment with CpG ODN but not GpC ODN steadily increased the ARF3-GTP level in RAW-ARF3 cells (fig. 4). Taken together, these finding strongly suggest that the basal level of active ARF3 is responsible for initiation of TLR9 trafficking, thereby inducing CpG ODN-mediated responses and ARF3 activation. The propagating ARF3 activation by CpG ODN may thus lead to a further increase of TLR9 trafficking and its responses.
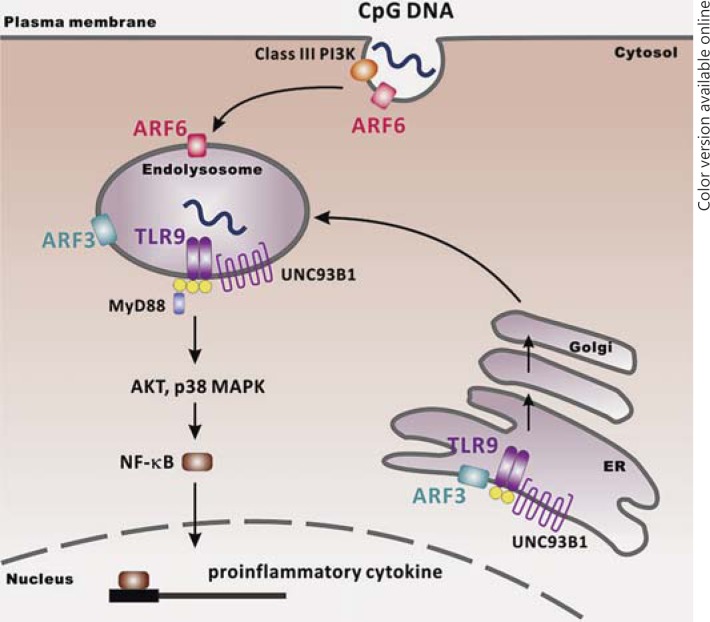
In this study, we demonstrated that brefeldin A-sensitive ARF3 is involved in TLR9-mediated signaling by regulating TLR9 trafficking. We delineated the regulatory mechanism, showing that ARF3 plays a key role in the regulation of the TLR9-Unc93B1 complex, which has further implications for conducting TLR9 to pass through the Golgi complex to localize at endolysosomes via the general secretory pathway (fig. 9). Understanding the regulation of TLR9 localization and trafficking will shed light on how autoimmune responses originate, and will be of importance for developing specific therapies. Discovering a new drug for ARF3 inhibition will potentiate an effective remedy in autoimmune diseases.
Fig. 9.
Schematic illustration of the signaling mechanisms by which both ARF3 and ARF6 regulate CpG ODN/TLR9-mediated signaling.
Disclosure Statement
The authors declare no competing financial interests.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported in part by National Health Research Institutes (Taiwan) intramural grants and National Science Council (Taiwan) grants 98-2320-B-400-007-MY3 and 102-2320-B-400 -012 -MY3 (C.-C.K.).
References
- 1.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen SR, Shlomchik MJ. Regulation of lupus-related autoantibody production and clinical disease by Toll-like receptors. Semin Immunol. 2007;19:11–23. doi: 10.1016/j.smim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 4.Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173:1179–1183. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chockalingam A, Brooks JC, Cameron JL, Blum LK, Leifer CA. TLR9 trafficks through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol Cell Biol. 2009;87:209–217. doi: 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. The Unc93B1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 7.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. Unc93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 9.Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, Barton GM. Unc93B1 mediates differential trafficking of endosomal TLRs. Elife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 11.Dascher C, Balch WE. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem. 1994;269:1437–1448. [PubMed] [Google Scholar]
- 12.Kuo CC, Kuo CW, Liang CM, Liang SM. A transcriptomic and proteomic analysis of the effect of CpG-ODN on human THP-1 monocytic leukemia cells. Proteomics. 2005;5:894–906. doi: 10.1002/pmic.200401144. [DOI] [PubMed] [Google Scholar]
- 13.Wu JY, Kuo CC. Pivotal role of ADP-ribosylation factor 6 in Toll-like receptor 9-mediated immune signaling. J Biol Chem. 2012;287:4323–4334. doi: 10.1074/jbc.M111.295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo CC, Liang SM, Liang CM. CpG-B oligodeoxynucleotide promotes cell survival via up-regulation of Hsp70 to increase Bcl-xL and to decrease apoptosis-inducing factor translocation. J Biol Chem. 2006;281:38200–38207. doi: 10.1074/jbc.M605439200. [DOI] [PubMed] [Google Scholar]
- 15.Kuo CC, Liang CM, Lai CY, Liang SM. Involvement of heat shock protein (Hsp)90 beta but not Hsp90 alpha in antiapoptotic effect of CpG-B oligodeoxynucleotide. J Immunol. 2007;178:6100–6108. doi: 10.4049/jimmunol.178.10.6100. [DOI] [PubMed] [Google Scholar]
- 16.Lee GL, Chang YW, Wu JY, Wu ML, Wu KK, Yet SF, Kuo CC. TLR 2 induces vascular smooth muscle cell migration through cAMP response element-binding protein-mediated interleukin-6 production. Arterioscler Thromb Vasc Biol. 2012;32:2751–2760. doi: 10.1161/ATVBAHA.112.300302. [DOI] [PubMed] [Google Scholar]
- 17.Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 18.Randazzo PA, Inoue H, Bharti S. Arf GAPs as regulators of the actin cytoskeleton. Biol Cell. 2007;99:583–600. doi: 10.1042/bc20070034. [DOI] [PubMed] [Google Scholar]
- 19.Burns K, Martinon F, Esslinger C, Pahl H, Schneider P, Bodmer JL, Di Marco F, French L, Tschopp J. MyD88, an adapter protein involved in interleukin-1 signaling. J Biol Chem. 1998;273:12203–12209. doi: 10.1074/jbc.273.20.12203. [DOI] [PubMed] [Google Scholar]
- 20.Kuo CC, Lin WT, Liang CM, Liang SM. Class I and III phosphatidylinositol 3′-kinase play distinct roles in TLR signaling pathway. J Immunol. 2006;176:5943–5949. doi: 10.4049/jimmunol.176.10.5943. [DOI] [PubMed] [Google Scholar]
- 21.Wu JY, Kuo CC. TLR9-mediated ARF6 activation is involved in advancing CpG ODN cellular uptake. Commun Integr Biol. 2012;5:316–318. doi: 10.4161/cib.20182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 25.Presley JF, Ward TH, Pfeifer AC, Siggia ED, Phair RD, Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 26.Sciaky N, Presley J, Smith C, Zaal KJ, Cole N, Moreira JE, Terasaki M, Siggia E, Lippincott-Schwartz J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J Cell Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo Y, Hanai A, Nakai W, Katoh Y, Nakayama K, Shin HW. ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct Funct. 2012;37:141–154. doi: 10.1247/csf.12015. [DOI] [PubMed] [Google Scholar]
- 28.Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell. 2004;15:1487–1505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouratou B, Biou V, Joubert A, Cohen J, Shields DJ, Geldner N, Jurgens G, Melancon P, Cherfils J. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics. 2005;6:20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manolea F, Chun J, Chen DW, Clarke I, Summerfeldt N, Dacks JB, Melancon P. Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors. Mol Biol Cell. 2010;21:1836–1849. doi: 10.1091/mbc.E10-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 32.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 33.Verthelyi D, Zeuner RA. Differential signaling by CpG DNA in DCs and B cells: not just TLR9. Trends Immunol. 2003;24:519–522. doi: 10.1016/s1471-4906(03)00243-6. [DOI] [PubMed] [Google Scholar]
- 34.Lenert P, Rasmussen W, Ashman RF, Ballas ZK. Structural characterization of the inhibitory DNA motif for the type A (D)-CpG-induced cytokine secretion and NK-cell lytic activity in mouse spleen cells. DNA Cell Biol. 2003;22:621–631. doi: 10.1089/104454903770238094. [DOI] [PubMed] [Google Scholar]
- 35.Hartmann G, Battiany J, Poeck H, Wagner M, Kerkmann M, Lubenow N, Rothenfusser S, Endres S. Rational design of new CpG oligonucleotides that combine B cell activation with high IFN-alpha induction in plasmacytoid dendritic cells. Eur J Immunol. 2003;33:1633–1641. doi: 10.1002/eji.200323813. [DOI] [PubMed] [Google Scholar]
- 36.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2012;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data