Abstract
Chitinase-like proteins (CLPs) of the 18 glycosyl hydrolase family retain structural similarity to chitinases but lack enzymatic activity. Although CLPs are upregulated in several human disorders that affect regenerative and inflammatory processes, very little is known about their normal physiological function. We show that an insect CLP (Drosophila imaginal disc growth factor 3, IDGF3) plays an immune-protective role during entomopathogenic nematode (EPN) infections. During these infections, nematodes force their entry into the host via border tissues, thus creating wounds. Whole-genome transcriptional analysis of nematode-infected wild-type and Idgf3 mutant larvae have shown that, in addition to the regulation of genes related to immunity and wound closure, IDGF3 represses Jak/STAT and Wingless signaling. Further experiments have confirmed that IDGF3 has multiple roles in innate immunity. It serves as an essential component required for the formation of hemolymph clots that seal wounds, and Idgf3 mutants display an extended developmental delay during wound healing. Altogether, our findings indicate that vertebrate and invertebrate CLP proteins function in analogous settings and have a broad impact on inflammatory reactions and infections. This opens the way to further genetic analysis of Drosophila IDGF3 and will help to elucidate the exact molecular context of CLP function.
Key Words: Chitinase-like proteins, Imaginal disc growth factor, Hemolymph clot, Wound healing, Nematode infection, Insect immunity
Introduction
Tissue repair and regeneration are fundamental processes required for the replacement of dead or damaged cells after injury and during inflammatory processes. Normal healing in mammals, which leaves no permanent traces, is tightly regulated and requires a strict balance between the de novo production of and degradation of extracellular matrix. In case this balance is disturbed or the stimulus persists, this leads to a fibroblastic stage which involves the sustained formation of extracellular matrix and tissue remodeling, and may leave a permanent scar (fibrosis). In addition to their involvement during regenerative processes, fibroses may also be part of immune reactions that involve the formation of larger inflammatory cell aggregates (granulomas) against abiotic objects and parasites, such as nematodes [1, 2]. When left unchecked, fibrosis formation potentially leads to permanent damage, resulting in morbidity and mortality [1]. Examples of the latter situation include cardiovascular diseases and organ-wide lesions affecting the skin, liver, kidney and lung [1] as well as aberrant tissue repair in cancers [3]. Tissue regeneration and immune responses against certain nematodes are dominated by a T-helper 2 cell (Th2)-based response, which includes the Th2 cytokines IL-4, IL-5 and IL-13 [2]. During both fibrosis formation and the response against nematodes, members of the 18 glycosyl hydrolase family are strongly induced.
Insect innate immune systems use effector mechanisms that resemble mammalian granulomas. They include capsules which are made against larger intruders, such as parasitoid eggs, nodules against large quantities of bacteria and hemolymph clots that form at the entry site of insect (entomo)pathogenic nematodes (EPNs) [4]. During all 3 reactions, blood cells (hemocytes) aggregate and release cytokines and extracellular matrix components. This may attract additional hemocytes or, in some cases, lead to the differentiation of additional hemocyte types. Subsequently, a cellular aggregate forms, which ultimately melanizes.
The 18 glycosyl hydrolase family is well conserved across animal orders. It comprises enzymatically active members that degrade chitin as well as chitinase-like proteins (CLPs), which may still bind to chitin or related polysaccharides but lack enzymatic activity (e.g. human Chi3l1/YKL40 and mouse Ym1) [2]. CLPs are often dysregulated in patients with various disorders such as asthma, chronic obstructive pulmonary disease (COPD), rheumatoid arthritis, cancer, diabetes and atherosclerosis; they effect inflammatory responses and tissue remodeling, and can serve as useful diagnostic markers [5]. Despite their association with disease, molecular insight into their physiological function and their contribution to disease etiology emerged only recently. Human Chi3l1 contributes to the augmentation of bacterial killing, the regulation of cell death, inflammation and remodeling [6, 7]. This involves the formation of a complex of IL-13, the IL-13 receptor alpha 2 and Chi3l1 [6]. Dysregulation of CLPs may influence the Th1/Th2 balance, shifting macrophages towards the M2 phenotype and activating MAP kinase and Akt signaling as well as the Wnt pathway [6]. A complex regulatory role for Chi3l1 in the progression of idiopathic pulmonary fibrosis was recently revealed [7]. Chi3l1 levels were increased in patients with idiopathic pulmonary fibrosis, which is associated with distortion of the lung architecture and compromised lung function. Importantly, the Chi3l1 levels correlated with the disease progression. The specific role of Chi3l1 was further studied in a mouse fibrosis model involving bleomycin-treatment and inducible expression of human Chi3l1. YKL-40 induction after bleomycin treatment had a protective role early on after injury, but at later stages, elevated Chi3l1 levels had profibrotic effects [7]. The 3 mouse CLP members were recently studied in an infection model that involved migration of a nematode through the lung epithelia. CLPs were found to limit parasite survival, although this came at the expense of the host and led to an increase in lung damage [8]. Both innate immune cells and γ/δ T cells were involved [8] (review [9]).
Drosophila imaginal disc growth factors (IDGFs) comprise a small family of 6 secreted glycoproteins with sequence similarity (approx. 56%) to mammalian CLPs [10]. Like mammalian CLPs, they lack the amino acids required for enzymatic activity, resulting in the loss of chitinase activity [11]. The founding member of this family, named DS47, was identified by Kirkpatrick et al. [10] in 1995 as a secreted product of S2 cells, a cell line exhibiting macrophage-like properties. The in vivo function of this family is not known, but IDGF1 and IDGF2 were implicated in the stimulation of growth and motility of Drosophila cells in vitro [12].
Drosophila IDGFs are produced in the fat body and hemocytes, the two major immune tissues in flies [10, 12, 13]. Their concentration in the hemolymph increases after infection or parasitization [4, 14]. Strong transcriptional upregulation of IDGF family members was detected in several published microarray studies of the response to infection and injury [13, 15]. Micrococcus luteus and Escherichia coli infection (but not fungal infection) induces Idgf3 expression in adults [13, 16]. These findings indicate that IDGFs contribute to immune responses.
In this study, we analyzed the function of Drosophila IDGF3, with a special focus on immunity. We found that Idgf3 mutants are homozygous-viable, and have defects in hemolymph clotting, which is the earliest response of Drosophila larvae after injury and upon nematode entry. Transcription profiling suggested that IDGF3 is involved in the regulation of innate defense mechanisms and signal pathways connected to wound healing as well as regenerative processes including Wingless (Wg) and Jak/STAT signaling, both implicated in the formation of fibrotic lesions in mammals. In addition, IDGF3 has further effects on the fly immune and regenerative response: Idgf3 mutants show an increased mortality after nematode infections and increased time requirements during wound healing. This is consistent with the proposed immune and regenerative function of mammalian CLPs. Altogether, this suggests that, similar to human Chi3l1, Drosophila IDGF3 is a key regulator of the epithelial response to injury and infection.
Materials and Methods
Drosophila Strains and Production of Transgenic Lines
For standard procedures, flies were raised on a cornmeal-yeast-agar-sugar diet with 0.3% Nipagin at 25°C. Transgenic flies carrying Idgf3::GFP and UAS-Idgf3-myc constructs were generated (see below).
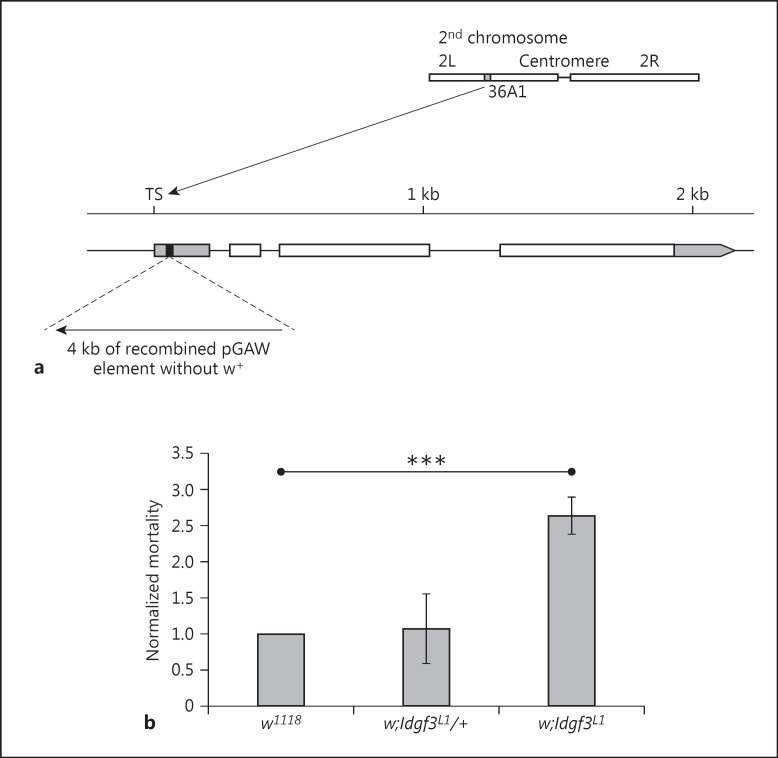
The Idgf3L1 mutant was generated by mobilization of the P-element, NP2446, which is located at +45 bp after transcription start of the Idgf3 gene. The Idgf3L1 mutant contains a partially excised and recombined P-element which has lost the w+ marker and contains a small duplication of 8 bp in transcription start of the Idgf3 gene (fig. 1a), leading to a complete loss of transcription (online suppl. fig. S1; www.karger.com/doi/10.1159/000442351 for all online suppl. material). The Idgf3L1 mutants are viable under homozygous conditions. The recessive lethal dac7 mutant [17], which covers the Idgf1–3 cluster and the 3′ end of the Dachshund gene, was kindly provided by Graeme Mardon (Baylor College of Medicine, Houston). The following fly lines were obtained from the Bloomington Stock Center: Idgf3NP2446, Act-Gal4::PR/TM6BTb[18], hmlΔ-Gal4 (III) and the wild-type lines Canton S and w1118, which served as our controls.
Fig. 1.
IDGF3 mutants are more sensitive to EPN infection. a Schematic diagram of the Idgf3 gene in the mutant Idgf3L1, which was generated by imprecise excision of NP2446, a P-element located 45 bp after transcription start of Idgf3. TS = Transcription start of the Idgf3 gene. Black box represents the 8 bp duplication at the P-element insertion site, the gray boxes represent the nontranslated region and the white boxes represent the coding regions. b Idgf3 mutant larvae (w;Idgf3L1) are sensitive to EPN infections 48 h after infection. Heterozygotes in the Idgf3 mutation (w;Idgf3L1/+) have similar mortality to the control (w1118). Differences compared to control genotype were analyzed using the Student t test. *** p < 0.001, only significant results are shown; error bars represent SEM.
For overexpression and rescue experiments, either hml-Gal4 driver was used (it mimics the natural expression of Idgf3 in hemocytes) or progesterone-inducible Act-Gal4::PR driver (it mimics inducible expression of Idgf3 as a response to infections). For activation of Act-Gal4::PR driver larvae of the experimental genotype w;Act-Gal4::PR/UAS-Idgf3-myc and control genotype w;Act-Gal4::PR/+ (driver only) were transferred for 12 h to fly food containing 5 µg/ml of mifepristone (Sigma-Aldrich) according to Rogulja and Irvine [18].
Transgenic Constructs
To produce a UAS-Idgf3-myc construct wild-type cDNA for the Idgf3 gene from the EST-clone, GH07453 from the pOT2 vector was amplified by PCR using Pfu DNA polymerase (Fermentas) and the following primers, Idgf3EcoF: TGAATTCATCATGACTGGCTCTCTTTGGCTC and Idgf3R1: CTTCTGAGATGAGTTTTTGTTCGAGAAGTCGATACTTGATGGCG in the first step. The PCR product was used as a template in a subsequent PCR reaction with the same forward primers Idgf3EcoF and MycXba: GTTCTAGATCACAGATCCTCTTCTGAGATGAGTTTTTGTTC as reverse primers, extending the sequence for the myc tag and XbaI cutting site. The final PCR product was cut using EcoR/XbaI and cloned into the pUAST vector. Stable stocks with UAS-Idgf3-myc were produced using standard P-element transformation of the w1118 fly strain. A homozygous-viable insertion on the 3rd chromosome was used for further experiments.
Recombineering of GFP into Idgf3 in Flyfos Construct
The Idgf3::GFP fusion construct is based on a fosmid library clone pFlyFos (ID = 026931) obtained from Pavel Tomancak [19] which contains the Idgf1–3 gene cluster including regulatory elements. The genomic region of the Idgf3 gene was modified by recombineering in E. coli in vivo by use of the Red/ET recombination technology according to the protocol in Ejsmont et al. [19], with the following modifications: the GFP protein was connected to the C-terminus of Idgf3 via a 2 × TY1 tag. The PCR cassette for tagging was amplified with Phusion polymerase (NEB). The plasmid containing the tagging cassette (2 × TY1 tag, C-terminus GFP and KanR) was used as a template. For the generation of the PCR cassette, oligonucleotides containing 50-bp homology arms for recombineering followed by the linker region of amplification were used; forward: GCACAAACGATCGCTTCCCCATGCTGCGCGCCATCAAGTATCGACTTCTCGAAGTGCATACCAATCAGGACCCGC and reverse: TGGACTGGAGAAGTTGGCTTAGAGAAGTTGGCTTAGAGAAGTCGGCTTACTTGTCGTCGTCATCCTTGTAGTCA. The recombined fosmid was purified with a Plasmid Midiprep kit (Macherey-Nagel). Since pFlyfos contains an attB site, a stable stock with an extra copy of the Idgf3::GFP fusion gene using the native promoter was produced using PhiC31 integrase-mediated transgenesis into the attP40 insertion site on the left arm of the 2nd chromosome.
Real Time RT-PCR
Total RNA from 15 flies per sample was isolated using the RNA Blue reagent (Top-Bio). The RNA was further purified with the NucleoSpin RNA II kit (Macherey-Nagel) including an on-column digestion step with rDNase I. One microgram of total RNA was reverse-transcribed at 42°C using oligo(dT)17 and PrimeScript reverse transcriptase (Takara). The PCR reaction volume was 20 µl, containing 5 µl of diluted cDNA and reaction mix [ExTaq Hot Start polymerase (0.75 units; Takara), ExTaq buffer and dNTPs (200 µM each), Cyber green 1:25,000 and the primers (400 nM each)]. The amplification was carried out on a Rotor-Gene 3000 (Corbet Research) for 40 cycles (94°C for 20 s; 60°C for 30 s; 72°C for 30 s) following an initial denaturation/Taq activation step (95°C for 2 min). Each sample was analyzed in triplicate. Primers (sequences shown in online suppl. table S5) were designed with Lasergene PrimerSelect software (DNASTAR) to assure that each amplicon encompassed an exon/intron boundary. The product size was confirmed by melting analysis. Data were analyzed and quantified with Rotor-Gene 6 analysis software. Relative values were normalized to the rp49 cDNA and standardized to the w1118 sample. All results are presented with means and SEM from 4 independent biological samples.
Nematode Infections
Nematode infections were performed in duplicate in 96-well plates as described previously, and analyzed using the Student t test [4].
Experimental Design and Preparation of Microarray Samples
We analyzed 2 different genotypes, i.e. a control and a null mutation in the Idgf3 gene. All genotypes were generated as progeny from the shared parental genotype w;Idgf3L1/+;UAS-Idgf3-myc/UAS-Idgf3-myc, which should minimize genetic background variation in our samples. All samples (Idgf3 mutant and control) and replicates were processed in 1 experiment. Age-matched third-instar larvae (96 h after egg-laying) were analyzed. For the control and mutant genotypes, we used samples infected by EPNs for 2 h and analyzed these 6 h afterwards as described in the study by Arefin et al. [4]. The summary of the microarray analysis is shown in online supplementary table S6. Samples were frozen and stored at −80°C till RNA extraction.
Total RNA from whole larvae was extracted using RiboZol RNA extraction reagent (Amresco) according to the manufacturer's protocol, and subsequently cleaned with the NucleoSpin RNA II kit (Macherey-Nagel). The quality and concentration of the RNA were measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was analyzed in an Agilent 2100 bioanalyzer. We included only samples with an intact RNA profile.
Expression Profiling
The Affymetrix GeneChip® Drosophila genome 2.0 array system was used for microarray analysis following the standard protocol: 100 ng RNA was amplified with GeneChip 3′ IVT express kit (Affymetrix), and 10 μg of labeled cRNA was hybridized to the chip according to the manufacturer's instructions.
Statistical Analysis of Array Data
Analysis was performed in triplicate and analyzed as previously described [4]. Although all data were processed in parallel, the transcriptome of the control larvae was already available in a previous study [4] and that of the Idgf3 mutants is made available as part of this study. The transcription data are MIAME-compliant and deposited in the ArrayExpress database (www.ebi.ac.uk/arrayexpress; accession Nos. E-MTAB-1542 [4] and E-MTAB-3478). To identify significantly perturbed pathways, we performed signal pathway impact analysis (SPIA) [20] on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [21]: genes with |logFC| > 2 and a p value <0.05 were considered differentially transcribed. Twelve genes had significantly changed after nematode infection in the controls had been validated before by quantitative RT-PCR [4].
Bead Aggregation Assay
The bead aggregation assay was performed according to Lesch et al. [22] with slight modifications. Briefly, 2.5 µl of hemolymph was collected from 6 late third-instar larvae (120 h after egg collection) and mixed for 30 s with 10 µl of Drosophila Ringer and 5 µl of bead suspension (tosylactivated Dynabeads M-280, Dynal Biotech ASA) at a concentration of approximately 2.0 × 109 beads/ml. Prior to usage, the beads were washed in 10× PBS and blocked in 0.1% BSA in 1× PBS overnight according to the manufacturer's instructions. Pictures were taken with a Leica MZ FLIII fluorescence stereomicroscope associated with a Panasonic DMC-G2 camera, and analyzed and quantified with the ImageJ graphics program with the module Analyze Particles.
Clotting and Wounding Assays
Clots were prepared using the ‘hanging drop’ method [22] and stained either natively with FITC-conjugated peanut agglutinin (PNA, Sigma-Aldrich) or fixed with 4% p-formaldehyde and stained with antibodies and other fluorescent dyes. The following antibodies and dyes were used: rabbit anti-IDGF3 (1:50 dilution, for a further description see below), rabbit polyclonal anti-GFP (1:2,000 dilution, Life Technologies), AlexaFluor 568-conjugated anti-rabbit (1:1,000 dilution, Invitrogen), FITC-conjugated anti-rabbit (1:200 dilution, Life Technologies) and DAPI (1:1,000 dilution, Sigma-Aldrich). Confocal images were taken with a Zeiss LSM 780 microscope.
Wounding assays were performed according to Burra et al. [23]. Feeding third-instar larvae (96 h after egg-laying) were injured with a tungsten needle (125 µm in diameter). For the survival experiment, wounded larvae were transferred to vials with fly food and the number of dead and pupated larvae was scored approximately every 12 h. Results were analyzed in R statistical software with the log-rank test. For localization of IDGF3:GFP, the wound site was observed under a Leica MZ FLIII fluorescence stereomicroscope and scored every hour for a period of 8 h. Images were taken with a Hamamatsu ORCA-ER camera (C4742-95) attached to a Zeiss Axioplan 2 microscope.
The anti-IDGF3 antibody was raised in rabbits against 14 amino acid long peptide C-EQRHLAQITSMKER (cysteine linked to the carrier protein), which was specific for IDGF3, by the GenScript company.
Results
IDFG3 Mutants Are More Sensitive to EPN Infection
Idgf1-3 were identified as immune response genes by several microarray studies [4, 13, 15]. In order to elucidate the role of IDGFs in immunity, we generated an Idgf3 mutant for which a P-element, integrated close to transcription start of Idgf3 gene, was available (fig. 1a; see Materials and Methods). RT-PCR analysis of a mutant obtained by imprecise excision of the P-element showed that the mutant did not express the Idgf3 gene (online suppl. fig. S1). Mutant larvae showed a normal phenotype. Idgf3 adult flies were viable but showed wing defects, in line with the proposed function as a growth factor (online suppl. fig. S2). Adult males were also sterile and females exhibited lower fecundity (online suppl. fig. S3). To test whether Idgf3 has an immune function, we followed the survival of Idgf3 mutants after infection with EPNs (Heterorhabditis bacteriophora and their symbiotic bacteria Photorhabdus luminescens) [4]. Mutant larvae infected with EPNs had a 2.5-fold higher mortality rate at day 2 after infection when compared to the control larvae (fig. 1b). These results showed that IDGF3 has a protective function against nematodes.
Transcriptome Analysis Identifies Pathways That Are Regulated by IDGF3 during EPN Infection
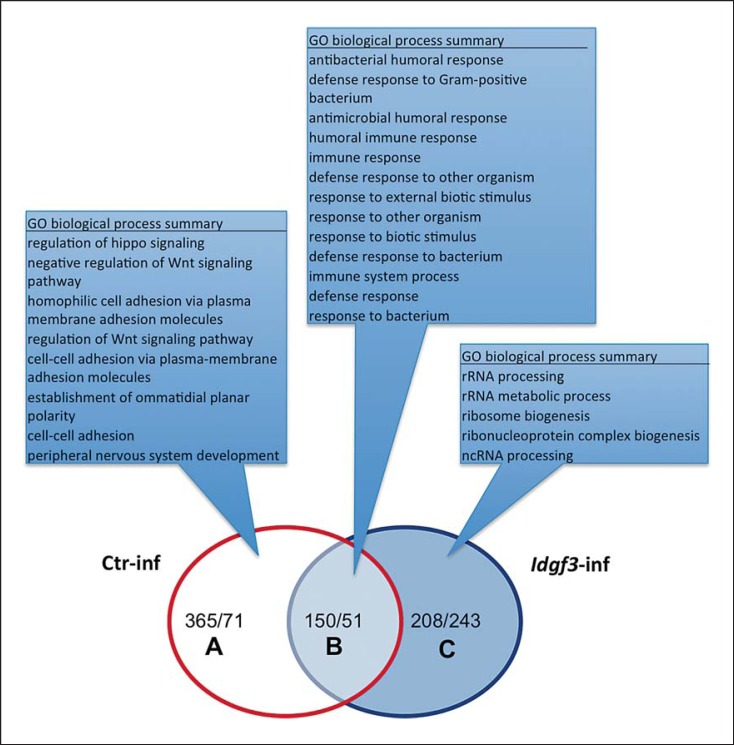
In order to map IDGF3-regulated genes, we compared the transcriptome of nematode-infected and naïve Idgf3 mutants to controls with an identical genetic background (online suppl. fig. S4) [4]. The underlying rationale was to identify genes and pathways that contribute to the difference in susceptibility towards nematodes between normal and mutant larvae. Analysis of the biological process gene ontology terms (AmiGO 2) for genes that were differentially regulated when comparing naïve wild-type and mutant larvae did not detect any enriched terms, in line with the lack of any dramatic phenotype of the mutants at this stage. In contrast, the infection of both mutant and normal larvae led to a highly significant induction of immune-related genes (fig. 2: overlapping group in the Venn diagram; online suppl. table S1: complete list of 150 induced genes and 51 downregulated ones). Enriched GO categories in this group almost exclusively included those with a function in immunity (fig. 2; online suppl. tables S1, S2). Most nematode-regulated genes shared directionality of their regulation and even showed similar levels of induction when compared in EPN-infected controls and mutant larvae (online suppl. table S1: genes shared after infection). The only exceptions were 2/201 genes (CG32379 and CG10621), a carboxypeptidase and a homocysteine S-methyltransferase, respectively, which were regulated in opposite directions. Altogether, this confirmed our previous findings that EPN infection induces a strong immune response [4], and showed at the same time that most of the immune response does not depend on the presence of IDGF3. However, there are some notable exceptions, namely 3 antimicrobial peptides which are amongst the 5 most strongly induced genes in infected control larvae but are not induced in Idgf3 mutants (fig. 2: category A; for a complete list, see online suppl. table S1: ctr infection-specific). They include 2 small peptides (immune-induced peptides, recently named Bomanins [24]) and, most significantly, Drosomycin, the signature gene for the Toll pathway. The dataset from infected Idgf3 mutants allowed us to comprehensively identify those genes that are induced in an IDGF3-dependent manner upon EPN infections (online suppl. tables S3, S4). GSEA analysis of this group showed enrichment for pathways that are involved in developmental processes in Drosophila. In flies, many of the processes identified regulate cellular activities that are required during the development of the nervous system and/or the imaginal discs. Several of these have been implicated in regenerative processes in mammals including Wg signaling (Wnt or Wg in flies), Hedgehog (Hh) and Jak/STAT signaling. SPIA (table 1) showed that Wg and Jak/STAT signaling are significantly downregulated upon EPN infection in control larvae but not in infected mutants. Completely in line with this, individual inspection of the genes that were induced after infection of wild-type larvae identified several negative regulators of both Wg and Jak/STAT signaling (online suppl. table S4). The Wg pathway includes naked cuticle[25], sulfated[26], shifted[27], nemo[28] and Rho1[29], whereas the Jak/STAT signaling involves 2 members of the SOCS family [30] and a PI3-kinase (CG4141), which acts in a regulatory loop upon axonal injury [31].
Fig. 2.
Microarray expression analysis after nematode infections of Idgf3 mutants and control larvae. Idgf3 mutants and control larvae were infected with H.bacteriophora containing GFP-labeled P.luminescens (Materials and Methods) and differentially expressed genes classified using GO term enrichment analysis in AmiGO 2. RNA samples were analyzed with Affymetrix expression arrays (genome 2.0). The Venn diagram shows the number of significantly regulated genes (|log2FC| > 1, q < 0.05) compared to noninfected control (Ctr-inf) samples of the corresponding genotype.
Table 1.
SPIA of KEGG pathways
| Group | KEGG ID | Title | gDET | gALL | gSIG_P | pG_P | Status_P |
|---|---|---|---|---|---|---|---|
| EPNi in controls | dme04630 | Jak-STAT signaling pathway | 18 | 21 | 6 | 0.00206 | inhibited |
| dme04310 | Wnt signaling pathway | 78 | 92 | 9 | 0.0198 | inhibited | |
| EPNi in Idgf3 mutants | n.s. |
gALL = Known number of genes defined in Drosophila genome; gDET = number of genes belonging to a given pathway; gSIG = number of significantly regulated genes (|logFC| >2 and p <0.05); n.s. = no significant pathway detected.
Additional induced genes were also notable, including myoblast city (Mbc), which functions during syncytium formation in other contexts and may support the formation of the epithelial syncytia found at the wound edges [32]. Altogether, these results establish IDGF3 as a novel regulator of EPN-induced immune and regenerative processes.
IDGF3 Mutants Have Clotting Defects
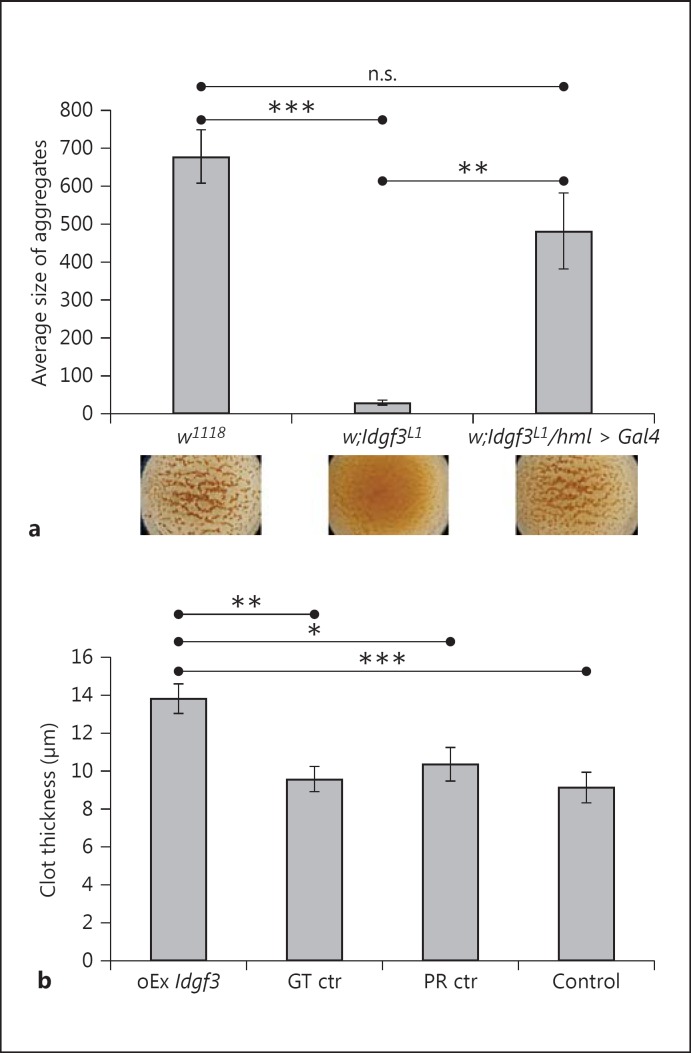
Many of the genes previously identified to protect Drosophila from nematode infection are involved in clotting, wound sealing or encoding extracellular matrix components [4, 33, 34]. To determine whether Idgf3 mutants exhibit clotting defects, we used a previously established bead aggregation assay [22]. Compared to the controls, the mutant hemolymph showed very poor bead aggregation (compare the lower part of fig. 3a and the quantification above). When we verified the ultrastructure of clots from the mutant hemolymph, we found that it completely lacked the fibrous clot matrix observed in normal larvae (online suppl. fig. S5). The ectopic expression of IDGF3 in hemocytes in the mutant background rescued the ability to aggregate beads (fig. 4a) and restored clot fiber formation (online suppl. fig. S5C), showing that although IDGF3 is expressed in other organs especially in the fat body, its expression in hemocytes is sufficient to drive clot formation. To further confirm a role for IDGF3 in clotting, we used an inducible system to overexpress the protein (details in Materials and Methods). Clot samples from overexpressing larvae showed increased lectin staining and extended fiber formation (online suppl. fig. S6). This also led to larger aggregates of clot matrix, which were not detected in normal clots (online suppl. fig. S6B). In addition, the average thickness of the clots prepared from larvae overexpressing IDGF3 was increased compared to all the controls (fig. 3b). Altogether, this indicates that the extent of clot formation correlates with the level of IDGF3 expression.
Fig. 3.
IDGF3 mutants show defects in clot formation. a Hemolymph preparations were used to detect clot formation using a previously described bead aggregation assay [22]. Idgf3L1 mutants lacked clot formation visible through a lack of bead aggregation (middle part of the figure underneath the diagram and quantification in the diagram) compared to controls (w1118). The clotting defect was rescued by ectopic expression of Idgf3 in hemocytes (Idgf3L1/hml > Idgf3). Data were analyzed using one-way ANOVA and the Tukey test. n.s. = Not significant. ** p < 0.01, *** p < 0.001; error bars represent SEM from 4 independent preparations. b Ubiquitous inducible overexpression of Idgf3 leads to more extensive clot formation measured as thickness of the clot. Control larvae (GT ctr = nontreated genotype control, PR ctr = mifepristone-treated driver control and Control = nontreated driver control) and overexpression larvae (oEx Idgf3) preparations of clots were stained with FITC-conjugated PNA and the thickness of the clots was measured using a confocal microscope. Experimental genotype: w;UAS-Idgf3/Act-Gal4:PR; driver control genotype: w;Act-Gal4:PR/+. The flies were treated 12 h with mifepristone (5 μg/ml) for 12 h. Data were analyzed using one-way ANOVA and the Tukey test. * p < 0.05, ** p < 0.01, *** p < 0.001; error bars represent SEM from 5 independent preparations measured repeatedly at multiple spots of the grid.
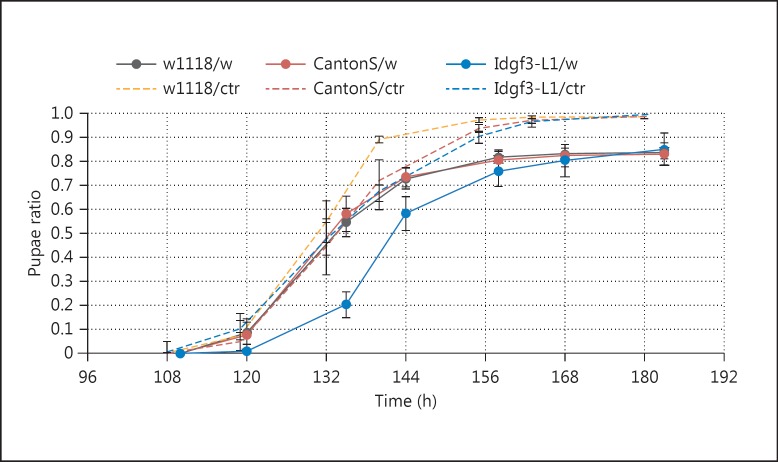
Fig. 4.
IDGF3 mutant larvae exhibit delayed development and pupate later after wounding in comparison with control larvae. Wounded Idgf3L1 mutant larvae (Idgf3-L1/w, solid blue line) delayed pupation for approximately 10 h and differed significantly (log-rank test; p < 0.01) compared to other wounded control larvae w1118 (w1118/w, solid grey line) and Canton S (CantonS/w, solid red line). Survival of all genotypes after wounding was equal at around 85%. The fraction of pupated nonwounded controls (/ctr) is shown as dashed lines.
IDGF3 Promotes Wound Healing
To test the effects of IDGF3 on wound healing, we injured mutant and wild-type larvae with a tungsten needle and followed their recovery (fig. 4). Both groups survived wounding equally well in the long term, in line with previous observations that the absence of a hemolymph clot has limited effects on survival after wounding, even in mutants where immunity is clearly affected [34]. They also both showed a developmental delay due to wounding, similar to that observed after the induction of wounds upon irradiation or in genetically induced wounds as well as in a Drosophila tumor model [35, 36]. Strikingly, though, the developmental delay was more pronounced in the mutant, showing that IDGF3 supports a swift recovery after wounding (fig. 4). Most likely as a consequence of reduced clotting, the scab in Idgf3 mutants was also less confined (online suppl. fig. S7). Altogether, this provides functional proof for the contribution of IDGF3 to wound healing, in line with our GO analysis of the transcriptome data.
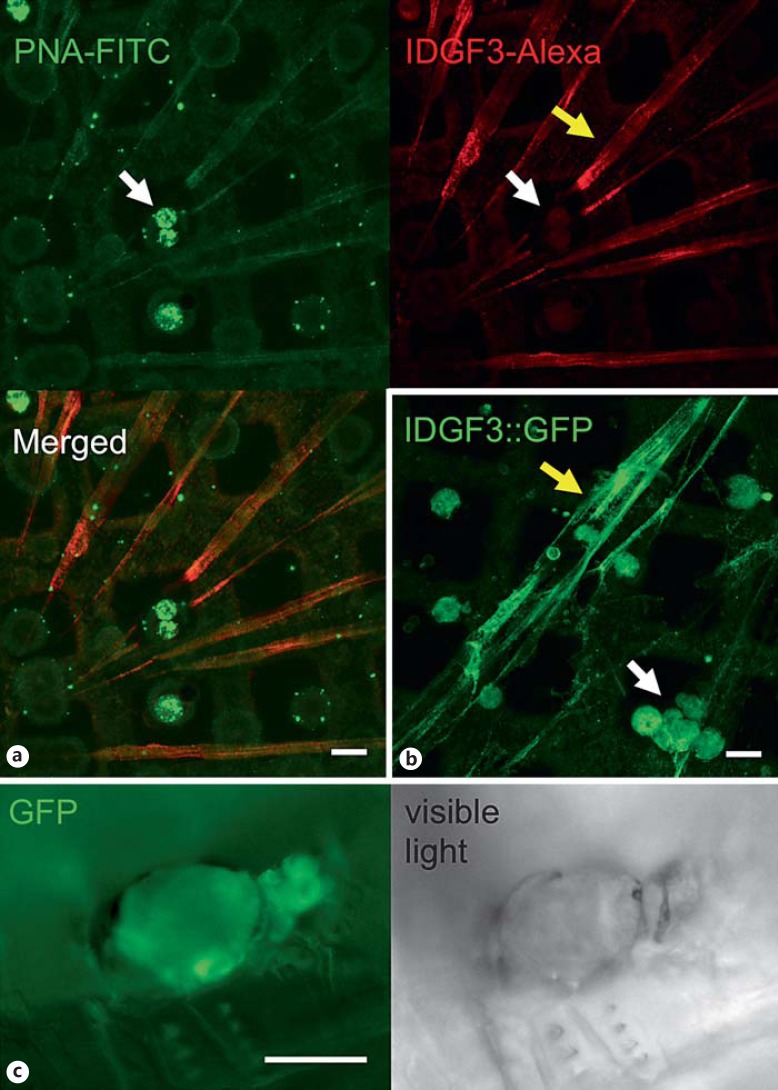
IDGF3 Is Part of Hemolymph Clots and Is Present at Wound Sites
Since the extent of clot formation seemed to depend on the concentration of IDGF3 in the hemolymph, we wondered whether IDGF3 itself was present in the clot. Clots from wild-type larvae, prepared using previously established methods [37], were labeled with an antibody against IDGF3 (fig. 5a). Double staining of the clot using PNA (which binds several clot components via their carbohydrate moiety [22]) and the IDGF3-specific antibody showed partial overlap including both clot fibers and hemocytes. Similarly, a GFP-tagged version of IDGF3 both localized to clot fibers and accumulated in wounds afflicted by mechanical injury (fig. 5b, c). The pattern that we observed is compatible with the idea that IDGF3 is an integral part of hemolymph clots, which form after wounding. Altogether, our results suggest that IDGF3 has an important role in Drosophila wound closure and contributes to wound healing and regenerative reactions that protect Drosophila larvae against nematode infections.
Fig. 5.
IDGF3 is part of the hemolymph clot and is present in wounds. a Clot fibers were prepared as described before [52]. They were caught on grids, fixed, and without any prior detergent treatment, stained with PNA-FITC (upper left), and an IDGF3-specific antibody (IDGF3-Alexa; upper right). Pictures were recorded using confocal microscopy. Scale bar: 15 µm. Both PNA and IDGF3 were detected on the surface of hemocytes (white arrows) and in clot fibers (yellow arrows). In a double-staining, the PNA signal on the surface of hemocytes is stronger, but IDGF3 staining is stronger in the fibers (Merged). b Clot fibers and hemocytes visualized using an IDGF3-GFP fusion protein show a stronger signal in hemocytes because of the presence of intracellular IDGF3. Scale bar: 15 μm. c IDGF3 accumulates in clots at the wound site. GFP-tagged IDGF3 was followed using fluorescence microscopy of larvae 6 h after wounding with tungsten needles. Scale bar: 50 µm.
Discussion
We describe a novel and unanticipated function for IDFG3, one of the Drosophila members of the CLPs. We show that IDGF3 is an integral part of the hemolymph clot and contributes to the protection against EPNs. The latter finding provides further support for the clot's function in immunity [38, 39]. Notably, the mouse CLP member Ym1 is amongst the most strongly induced proteins upon nematode infections in mice [40], and its function in the protection against nematodes was strongly suggested in recent studies of the 3 mouse CLP members [8].
To study the effects of IDGF3 in larvae infected with EPNs in an unbiased way, we performed transcriptional profiling of infected wild-type and mutant Drosophila larvae. We found a large set of genes that are induced during nematode infection in both cases. Many of these are immune genes including antimicrobial peptides; this confirms previous results [4] and suggests that at least part of the protection against EPNs is caused by antibacterial activity, which acts upon the nematodes' symbiotic bacteria that are released during the infection. A subfraction of immune genes is induced in an IDGF3-dependent manner, in line with the proposed immune modulatory role of chitinase family members [8].
Enrichment analysis using SPIA identified additional pathways that are regulated via IDGF3. Amongst these, Wg and Jak/STAT, which are downregulated, are notable. Both pathways have been linked to fibrotic lesions in mammals and we show clearly that they are modulated by CLP signaling. Using AmiGO analysis of the array data, manual analysis of individual regulated genes and SPIA, we found a number of negative regulators of Wg signaling that were induced and additional genes that have antiproliferative effects. This is in line with results from previous studies of wound healing in Drosophila and other insects where proliferation was not observed [41]. Of note, among the Wg regulators induced, we identified shifted, the Drosophila ortholog of mammalian Wnt inhibitory factor, which, in the fruit fly, has been shown to control the activity of Hh rather than Wg [27]. Thus, IDGF signaling may lead to both Wg repression and Hh activation. In vertebrates, Wg and Hh have dual functions during development and skin repair [42]. Our findings suggest that there is a different function for Wg and Jak/STAT signaling during EPN infections when compared to other Drosophila models of injury such as during gut regeneration. Both upon feeding bacteria and chemical irritants (including bleomycin), canonical Wg and Jak/STAT signaling in gut epithelia are required to induce stem cell proliferation [43, 44], indicating that, although the dual use of the pathways may be conserved between epithelia, the outcome appears to depend on the tissue and/or regulation via IDGF3 and the clot. An example for a potential key regulator is provided by neijre, the Drosophila homolog of histone acetyltransferase CREB-binding protein, which we found to be induced, and which acts as a bimodal regulator of Wg signaling [45]. Taken together, the pattern of induction that we observed after nematode infection is compatible with previous findings, i.e. that cuticular wounding in flies activates cellular activities that help to close the wound rather than activate cell proliferation [41]. Future work will enable us to localize the differential regulation of genes/pathways to individual cells/tissues. The site of damage, neighboring tissues [46] and more distant sites [47, 48] are potential targets.
Our functional analysis shows that IDGF3 supports a swift recovery after wounding. This means that, upon wounding and in the context of nematode infections, IDGF3 plays a positive role, similar to mammalian CLPs at an early stage of regeneration after wounding or upon infection. It will be of great interest to study how Drosophila IDGF3 affects the development of fibrotic lesions and the reaction against chronic states such as tumors; Drosophila models are available for both of these [49, 50, 51]. For the mechanism of action of IDGF3, we envisage the following 3, not necessarily exclusive, scenarios. (1) Based on its carbohydrate (chitin)-binding activity, it may mediate the interaction between the clot and the wound edges where chitin has become exposed due to damage to the underlying basement membrane and the epithelia; this idea is supported by the accumulation of IDGF3 that we observed at the wound edges (fig. 5c). (2) Since Drosophila IDGFs are known to act as cytokines, IDFG3 may bind to carbohydrate moieties present on hemocytes, and, similar to vertebrate Chi3l1, regulate cellular activities. (3) Similar to what has been proposed for mouse CLPs, Drosophila IDGFs may act as pattern recognition molecules that bind chitin or related carbohydrates on the surface of nematodes and other parasites, activating immune effector mechanisms [8]. Combinations of these mechanisms can be envisaged, e.g. while present as a soluble protein in the hemolymph, IDGF3 may act as a cytokine, and upon incorporation into the clot, it may act as a structural component.
Our results have the potential to aid our understanding of the dual role mammalian CLPs play during chronic states such as the development of fibrotic lesions [7]. Our list of IDGF3-dependent genes that are induced upon infection provides a genome-wide source of further targets for the functional analysis of wound healing and subsequent regenerative processes.
Taken together, the work presented here provides experimental evidence for a protective function of CLPs during wounding and immune reactions. This opens the way for using the genetically tractable Drosophila model to study these processes in normal and pathological contexts and also their contribution to resistance against parasites.
Disclosure Statement
The authors declare no competing interests.
Supplementary Material
Supplementary data
Acknowledgements
We thank Dr. Michael Williams from Uppsala University for critical reading of the manuscript. We are grateful to the service laboratory at IMG and especially to Martina Chmelikova for technical assistance. The authors' work is supported by the Swedish Research Council (VR-2010-5988 to U.T.), the Swedish Foundation for International Cooperation in Research and Higher Education (STINT, IG2011-2042 to U.T.), the Knut and Alice Wallenberg Foundation (KAW2012.0058), the Swedish Cancer Foundation (CAN 2010/553 to U.T.) and the Czech Science Foundation (grant No. GA14-27816S to M.Z.).
References
- 1.Meneghin A, Hogaboam CM. Infectious disease, the innate immune response, and fibrosis. J Clin Invest. 2007;117:530–538. doi: 10.1172/JCI30595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony RM, Rutitzky LI, Urban JF, Jr, Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nat Rev Immunol. 2007;7:975–987. doi: 10.1038/nri2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco C, Hess S. Recent proteomic advances in developmental, regeneration, and cancer governing signaling pathways. Proteomics. 2015;15:1014–1025. doi: 10.1002/pmic.201400368. [DOI] [PubMed] [Google Scholar]
- 4.Arefin B, Kucerova L, Dobes P, Markus R, Strnad H, Wang Z, Hyrsl P, Zurovec M, Theopold U. Genome-wide transcriptional analysis of Drosophila larvae infected by entomopathogenic nematodes shows involvement of complement, recognition and extracellular matrix proteins. J Innate Immun. 2014;6:192–204. doi: 10.1159/000353734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, Elias JA. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 2011;73:479–501. doi: 10.1146/annurev-physiol-012110-142250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He CH, Lee CG, Dela Cruz CS, Lee CM, Zhou Y, Ahangari F, Ma B, Herzog EL, Rosenberg SA, Li Y, Nour AM, Parikh CR, Schmidt I, Modis Y, Cantley L, Elias JA. Chitinase 3-like 1 regulates cellular and tissue responses via IL-13 receptor alpha2. Cell Rep. 2013;4:830–841. doi: 10.1016/j.celrep.2013.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, Peng H, Sun H, Peng X, Tang C, Gan Y, Chen X, Mathur A, Hu B, Slade MD, Montgomery RR, Shaw AC, Homer RJ, White ES, Lee CM, Moore MW, Gulati M, Geun Lee C, Elias JA, Herzog EL. Chitinase 3-like 1 suppresses injury and promotes fibroproliferative responses in mammalian lung fibrosis. Sci Transl Med. 2014;6:240ra276. doi: 10.1126/scitranslmed.3007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland TE, Logan N, Ruckerl D, Humbles AA, Allan SM, Papayannopoulos V, Stockinger B, Maizels RM, Allen JE. Chitinase-like proteins promote IL-17-mediated neutrophilia in a tradeoff between nematode killing and host damage. Nat Immunol. 2014;15:1116–1125. doi: 10.1038/ni.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muallem G, Hunter CA. ParadYm shift: Ym1 and Ym2 as innate immunological regulators of IL-17. Nat Immunol. 2014;15:1099–1100. doi: 10.1038/ni.3032. [DOI] [PubMed] [Google Scholar]
- 10.Kirkpatrick RB, Matico RE, McNulty DE, Strickler JE, Rosenberg M. An abundantly secreted glycoprotein from Drosophila melanogaster is related to mammalian secretory proteins produced in rheumatoid tissues and by activated macrophages. Gene. 1995;153:147–154. doi: 10.1016/0378-1119(94)00756-i. [DOI] [PubMed] [Google Scholar]
- 11.Varela PF, Llera AS, Mariuzza RA, Tormo J. Crystal structure of imaginal disc growth factor-2. A member of a new family of growth-promoting glycoproteins from Drosophila melanogaster. J Biol Chem. 2002;277:13229–13236. doi: 10.1074/jbc.M110502200. [DOI] [PubMed] [Google Scholar]
- 12.Kawamura K, Shibata T, Saget O, Peel D, Bryant PJ. A new family of growth factors produced by the fat body and active on Drosophila imaginal disc cells. Development. 1999;126:211–219. doi: 10.1242/dev.126.2.211. [DOI] [PubMed] [Google Scholar]
- 13.Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffmann JA, Hetru C. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci USA. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vierstraete E, Verleyen P, Sas F, Van den Bergh G, De Loof A, Arckens L, Schoofs L. The instantly released Drosophila immune proteome is infection-specific. Biochem Biophys Res Commun. 2004;317:1052–1060. doi: 10.1016/j.bbrc.2004.03.150. [DOI] [PubMed] [Google Scholar]
- 15.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and IMD pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mardon G, Solomon NM, Rubin GM. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- 18.Rogulja D, Irvine KD. Regulation of cell proliferation by a morphogen gradient. Cell. 2005;123:449–461. doi: 10.1016/j.cell.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Ejsmont RK, Sarov M, Winkler S, Lipinski KA, Tomancak P. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat Methods. 2009;6:435–437. doi: 10.1038/nmeth.1334. [DOI] [PubMed] [Google Scholar]
- 20.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, Kim CJ, Kusanovic JP, Romero R. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesch C, Goto A, Lindgren M, Bidla G, Dushay MS, Theopold U. A role for hemolectin in coagulation and immunity in Drosophila melanogaster. Dev Comp Immunol. 2007;31:1255–1263. doi: 10.1016/j.dci.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Burra S, Wang Y, Brock AR, Galko MJ. Using Drosophila larvae to study epidermal wound closure and inflammation. Methods Mol Biol. 2013;1037:449–461. doi: 10.1007/978-1-62703-505-7_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemmons AW, Lindsay SA, Wasserman SA. An effector peptide family required for Drosophila Toll-mediated immunity. PLoS Path. 2015;11:e1004876. doi: 10.1371/journal.ppat.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, Meng F, Ma D, Xie W, Fang M. Bridging Decapentaplegic and Wingless signaling in Drosophila wings through repression of naked cuticle by Brinker. Development. 2013;140:413–422. doi: 10.1242/dev.082578. [DOI] [PubMed] [Google Scholar]
- 26.You J, Belenkaya T, Lin X. Sulfated is a negative feedback regulator of Wingless in Drosophila. Dev Dyn. 2011;240:640–648. doi: 10.1002/dvdy.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glise B, Miller CA, Crozatier M, Halbisen MA, Wise S, Olson DJ, Vincent A, Blair SS. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev Cell. 2005;8:255–266. doi: 10.1016/j.devcel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Zeng YA, Verheyen EM. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development. Development. 2004;131:2911–2920. doi: 10.1242/dev.01177. [DOI] [PubMed] [Google Scholar]
- 29.Greer ER, Chao AT, Bejsovec A. Pebble/ECT2 RhoGEF negatively regulates the Wingless/Wnt signaling pathway. Development. 2013;140:4937–4946. doi: 10.1242/dev.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stec WJ, Zeidler MP. Drosophila SOCS proteins. J Signal Transduct. 2011;2011:894510. doi: 10.1155/2011/894510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doherty J, Sheehan AE, Bradshaw R, Fox AN, Lu TY, Freeman MR. PI3K signaling and Stat92E converge to modulate glial responsiveness to axonal injury. PLoS Biol. 2014;12:e1001985. doi: 10.1371/journal.pbio.1001985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Losick VP, Fox DT, Spradling AC. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol. 2013;23:2224–2232. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyrsl P, Dobes P, Wang Z, Hauling T, Wilhelmsson C, Theopold U. Clotting factors and eicosanoids protect against nematode infections. J Innate Immun. 2011;3:65–70. doi: 10.1159/000320634. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Wilhelmsson C, Hyrsl P, Loof TG, Dobes P, Klupp M, Loseva O, Morgelin M, Ikle J, Cripps RM, Herwald H, Theopold U. Pathogen entrapment by transglutaminase-A conserved early innate immune mechanism. PLoS Path. 2010;6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halme A, Cheng M, Hariharan IK. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol. 2010;20:458–463. doi: 10.1016/j.cub.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauling T, Krautz R, Markus R, Volkenhoff A, Kucerova L, Theopold U. A Drosophila immune response against Ras-induced overgrowth. Biol Open. 2014;3:250–260. doi: 10.1242/bio.20146494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherfer C, Karlsson C, Loseva O, Bidla G, Goto A, Havemann J, Dushay MS, Theopold U. Isolation and characterization of hemolymph clotting factors in Drosophila melanogaster by a pullout method. Curr Biol. 2004;14:625–629. doi: 10.1016/j.cub.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 38.Loof TG, Schmidt O, Herwald H, Theopold U. Coagulation systems of invertebrates and vertebrates and their roles in innate immunity: the same side of two coins? J Innate Immun. 2011;3:34–40. doi: 10.1159/000321641. [DOI] [PubMed] [Google Scholar]
- 39.Theopold U, Krautz R, Dushay MS. The Drosophila clotting system and its messages for mammals. Dev Comp Immunol. 2014;42:42–46. doi: 10.1016/j.dci.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Nair MG, Gallagher IJ, Taylor MD, Loke P, Coulson PS, Wilson RA, Maizels RM, Allen JE. Chitinase and Fizz family members are a generalized feature of nematode infection with selective upregulation of Ym1 and Fizz1 by antigen-presenting cells. Infect Immun. 2005;73:385–394. doi: 10.1128/IAI.73.1.385-394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Razzell W, Wood W, Martin P. Swatting flies: Modelling wound healing and inflammation in Drosophila. Dis Model Mech. 2011;4:569–574. doi: 10.1242/dmm.006825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bielefeld KA, Amini-Nik S, Alman BA. Cutaneous wound healing: recruiting developmental pathways for regeneration. Cell Mol Life Sci. 2013;70:2059–2081. doi: 10.1007/s00018-012-1152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takashima S, Mkrtchyan M, Younossi-Hartenstein A, Merriam JR, Hartenstein V. The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature. 2008;454:651–655. doi: 10.1038/nature07156. [DOI] [PubMed] [Google Scholar]
- 44.Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J. 2007;26:2284–2294. doi: 10.1038/sj.emboj.7601667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kux K, Pitsouli C. Tissue communication in regenerative inflammatory signaling: lessons from the fly gut. Front Cell Infect Microbiol. 2014;4:49. doi: 10.3389/fcimb.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nam HJ, Jang IH, You H, Lee KA, Lee WJ. Genetic evidence of a redox-dependent systemic wound response via Hayan protease-phenoloxidase system in Drosophila. EMBO J. 2012;31:1253–1265. doi: 10.1038/emboj.2011.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeishi A, Kuranaga E, Tonoki A, Misaki K, Yonemura S, Kanuka H, Miura M. Homeostatic epithelial renewal in the gut is required for dampening a fatal systemic wound response in Drosophila. Cell Rep. 2013;3:919–930. doi: 10.1016/j.celrep.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Zang Y, Wan M, Liu M, Ke H, Ma S, Liu LP, Ni JQ, Carlos Pastor-Pareja., J Plasma membrane overgrowth causes fibrotic collagen accumulation and immune activation in Drosophila adipocytes. Elife. 2015;4:e07187. doi: 10.7554/eLife.07187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pastor-Pareja JC, Xu T. Dissecting social cell biology and tumors using Drosophila genetics. Ann Rev Gen. 2013;47:51–74. doi: 10.1146/annurev-genet-110711-155414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Kounatidis I, Ligoxygakis P. Drosophila as a model to study the role of blood cells in inflammation, innate immunity and cancer. Front Cell Infect Microbiol. 2013;3:113. doi: 10.3389/fcimb.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindgren M, Riazi R, Lesch C, Wilhelmsson C, Theopold U, Dushay MS. Fondue and transglutaminase in the Drosophila larval clot. J Insect Physiol. 2008;54:586–592. doi: 10.1016/j.jinsphys.2007.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data