The evolution of photosynthesis was a significant event during the diversification of biological life. Aerobic anoxygenic photoheterotrophic bacteria (AAPB) share physiological characteristics with chemoheterotrophs and represent an important group associated with bacteriochlorophyll-dependent phototrophy in the environment. Here, characterization and evolutionary analyses were conducted for 13 bacterial strains that contained photosynthetic gene clusters (PGCs) carried by extrachromosomal replicons (ECRs) to shed light on the evolution of chlorophototrophy in bacteria. This report advances our understanding of the importance of ECRs in the transfer of PGCs within marine photoheterotrophic bacteria.
KEYWORDS: aerobic anoxygenic photoheterotrophic bacteria, AAPB, photosynthetic gene cluster, PGC, extrachromosomal replicons, ECRs, Roseobacter clade
ABSTRACT
Aerobic anoxygenic photoheterotrophic bacteria (AAPB) represent a bacteriochlorophyll a-containing functional group. Substantial evidence indicates that highly conserved photosynthetic gene clusters (PGCs) of AAPB can be transferred between species, genera, and even phyla. Furthermore, analysis of recently discovered PGCs carried by extrachromosomal replicons (exPGCs) suggests that extrachromosomal replicons (ECRs) play an important role in the transfer of PGCs. In this study, 13 Roseobacter clade genomes from seven genera that harbored exPGCs were used to analyze the characteristics and evolution of PGCs. The identification of plasmid-like and chromid-like ECRs among PGC-containing ECRs revealed two different functions: the spread of PGCs among strains and the maintenance of PGCs within genomes. Phylogenetic analyses indicated two independent origins of exPGCs, corresponding to PufC-containing and PufX-containing puf operons. Furthermore, the two different types of operons were observed within different strains of the same Tateyamaria and Jannaschia genera. The PufC-containing and PufX-containing operons were also differentially carried by chromosomes and ECRs in the strains, respectively, which provided clear evidence for ECR-mediated PGC transfer. Multiple recombination events of exPGCs were also observed, wherein the majority of exPGCs were inserted by replication modules at the same genomic positions. However, the exPGCs of the Jannaschia strains comprised superoperons without evidence of insertion and therefore likely represent an initial evolutionary stage where the PGC was translocated from chromosomes to ECRs without further combinations. Finally, a scenario of PGC gain and loss is proposed that specifically focuses on ECR-mediated exPGC transfer to explain the evolution and patchy distribution of AAPB within the Roseobacter clade.
IMPORTANCE The evolution of photosynthesis was a significant event during the diversification of biological life. Aerobic anoxygenic photoheterotrophic bacteria (AAPB) share physiological characteristics with chemoheterotrophs and represent an important group associated with bacteriochlorophyll-dependent phototrophy in the environment. Here, characterization and evolutionary analyses were conducted for 13 bacterial strains that contained photosynthetic gene clusters (PGCs) carried by extrachromosomal replicons (ECRs) to shed light on the evolution of chlorophototrophy in bacteria. This report advances our understanding of the importance of ECRs in the transfer of PGCs within marine photoheterotrophic bacteria.
INTRODUCTION
Aerobic anoxygenic photoheterotrophic bacteria (AAPB) are chlorophototrophic members of the Proteobacteria phylum that contain bacteriochlorophyll a and are widely distributed in euphotic ocean environments (1–7). AAPB account for up to 10% of the total bacterial communities in the upper ocean layers and play crucial roles in carbon and energy cycling (8–10). AAPB are facultatively phototrophic, using light as an alternative energy source, which thereby reduces the respiration of organic carbon from marine primary production by ∼2.4% to ∼5.4% (4). In addition to their ecological significance, physiological and genomic analysis of AAPB has indicated their important potential to inform on the evolution of photosynthesis (11–13). AAPB are hypothesized to have evolved after widespread oxygenation of Earth’s atmosphere ∼2.4 to ∼2.3 billion years (Ga) ago (14, 15). Purple photosynthetic bacteria are one such group of anaerobic phototrophs that are thought to represent the ancestral lineage to AAPB due to the high similarity of their photosynthetic apparatuses (13, 16, 17). However, unlike the purple photosynthetic bacteria that grow in the absence of oxygen and are mainly autotrophic, AAPB are aerobic heterotrophs that perform phototrophy as an auxiliary energy conservation strategy, which can contribute up to 20% of their total cellular metabolic energy demands (6, 18). Phylogenetic analysis of the 16S rRNA gene indicates that the distribution of AAPB is scattered throughout the Proteobacteria and that they are closely related to nonphototrophic bacteria and purple nonsulfur bacteria within the Proteobacteria (12, 13, 18).
Similarly to purple photosynthetic bacteria, photosynthetic gene clusters (PGCs) are found in AAPB. The large PGC superoperon is approximately 35 to 50 kb in length and contains the approximately 40 genes required for the biosynthesis of bacteriochlorophyll, carotenoids, photosynthetic reaction complexes, and light harvesting complexes, in addition to other regulatory functions (19–22). PGC gene sequences are highly conserved, and some essential genes within PGCs (e.g., pufL, pufM, and bchY) are typically used as molecular markers in phylogenetic analyses to classify phototrophic Proteobacteria and study their ecological diversity and evolutionary relationships (5, 22–25).
Previous comparisons of closely related strains have demonstrated that PGCs can be lost from bacterial genomes. For example, Citromicrobium sp. JL1363 lost PGCs from its genome during its evolutionary history and is now reliant solely on heterotrophy (26). Furthermore, PGCs can be transferred between proteobacterial strains and among distantly related phyla. For instance, although Erythrobacter sp. AP23 shares 99.5% 16S rRNA gene sequence identity with Erythrobacter sp. LAMA915, only the former strain contains a PGC within its genome (23). Intriguingly, the PGC of Erythrobacter sp. AP23 is closely related to that of Citromicrobium (23). Gemmatimonadetes is a novel phototrophic phylum and has been suggested to acquire its PGC from phototrophic bacteria of the Proteobacteria via a horizontal gene transfer (HGT) event (25).
Members of the Roseobacter clade are important ecological generalists within marine ecosystems, and many are AAPB (27–29). Inconsistencies within phylogenetic topologies between the 16S rRNA gene and PGCs indicate that HGT events of PGCs have occurred among members of this clade (30–33). Of note, PGC-containing extrachromosomal replicons (ECRs) have been identified within six strains of the Roseobacter clade, further indicating the possibility of HGT of PGCs among these species (33). ECRs are mobile genetic elements that carry many essential genes involved in metabolism that are essential for rapid adaptation to changing environments (34–36). Therefore, it is likely that PGCs can also be transferred via ECRs. However, a limited number of studies have focused on the role of ECRs in the HGT of PGCs (31, 33).
In this study, we analyzed 13 Roseobacter clade genomes that contain PGCs carried by extrachromosomal replicons (exPGCs). Phylogenetic and structural analyses of the exPGCs, in addition to comparisions against chromosomal PGCs (cPGCs), were used to elucidate the potential role of ECRs in the transfer of PGCs among Roseobacter clade species.
RESULTS AND DISCUSSION
General features of the 13 Roseobacter clade strains.
Thirteen Roseobacter clade strains carried PGC-containing ECRs and were affiliated with seven different genera: Tateyamaria, Jannaschia, Sulfitobacter, Roseobacter, Oceanicola, Shimia, and Nereida (Table 1). The genome sizes of the 13 strains ranged from 2.89 to 4.75 Mb, while the average level of genomic GC content ranged from 54.0% to 65.5%. Most strains contained more than two ECRs, and the largest ECR among the 13 strains was a 185-kb PGC-containing ECR in Sulfitobacter sp. AM1-D1. The 13 identified exPGCs ranged in size from 41.3 to 54.0 kb, and their average GC contents were consistent with their respective chromosomes. All PGC-containing ECRs harbored DnaA-like I replication systems except for that of Sulfitobacter guttiformis, which exhibited a RepB-III type system.
TABLE 1.
Genomic information for 13 Roseobacter strains carrying exPGCs
| Genus | Strain | Genome size (Mb) |
No. of contigs |
Genome GC content (%) |
Phototrophic plasmid size (kb) |
PS size (kb)a |
Phototrophic plasmid GC content (%) |
PGC GC content (%) |
Isolation source |
|---|---|---|---|---|---|---|---|---|---|
| Tateyamaria |
Tateyamaria sp. ANG-S1 |
4.43 | 33 | 60.6 | 139.0 | 53 | 61.37 | 62.59 | Accessory nidamental gland |
| Tateyamaria sp. syn59 | 4.42 | 32 | 61.3 | 79.9 | 52 | 63.02 | 63.23 | Seawater from the South China Sea |
|
|
Tateyamaria omphalii DOK1-4 |
4.31 | 8 | 61.98 | 77.6 | 54 | 63.18 | 63.2 | Seawater from the East Sea in South Korea |
|
| Jannaschia |
Jannaschia faecimaris DSM1004020 |
3.81 | 48 | 62.0 | 87.2 | 42 | 63.95 | 64.01 | Unknown |
|
Jannaschia pohangensis DSM19073 |
3.73 | 18 | 65.5 | 49.5 | 41.3 | 66.88 | 66.79 | Seashore sand in South Korea |
|
|
Jannaschia donghaensis CECT7802 |
3.49 | 67 | 64.6 | 67.2 | 43 | 66.01 | 66.14 | Surface sea water, coast of Dokdo, South Korea |
|
| Sulfitobacter |
Sulfitobacter sp. AM1-D1 |
4.69 | 6 | 64.9 | 185.2 | 45 | 65.56 | 65.87 | Cell culture of Alexandrium minutum |
|
Sulfitobacter noctilucicola KCTC32123 |
4.09 | 8 | 57.1 | 107.4 | 50 | 57.38 | 58.15 | Sea sparkle bloom region of Geoje Island in South Korea |
|
|
Sulfitobacter guttiformis KTCT32187 |
3.98 | 4 | 56.1 | 118.5 | 51 | 55.46 | 56.63 | Costal seawater | |
| Roseobacter |
Roseobacter litoralis Och 149 |
4.75 | 1 | 57.2 | 93 | 50 | 58.40 | 59.2 | Seaweed |
| Oceanicola | Oceanicola sp. HL-35 | 4.31 | 8 | 63.9 | 44 | 44 | 66.45 | 66.45 | Unknown |
| Shimia | Shimia sp. wx04 | 3.78 | 12 | 58.8 | 54 | 54 | 60.00 | 60.13 | Seawater from the South China Sea |
| Nereida |
Nereida ignava DSM16309 |
2.89 | 36 | 54.0 | 79 | 47 | 53.52 | 54.38 | Seawater, Mediterranean Sea |
PS, PGC superoperon.
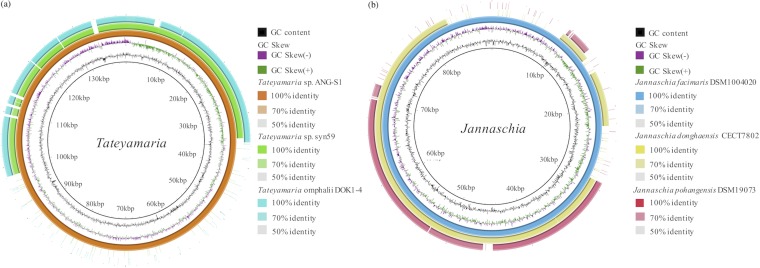
Tateyamaria spp. have been isolated from different environments, including seawater, tidal-flat sediments, and marine animals (37–41). Tateyamaria spp. have also been frequently detected in algal culture bacterial communities and can occasionally dominate such communities (42–44). The pufM gene is present in all genomes reported for this genus (as of 28 February 2019). Three strains carrying exPGCs with similar genome sizes (average, 4.4 ± 0.8 Mb) and GC contents (average, 61.30% ± 0.70%) were chosen for analysis in this study. The sizes of the ECRs carrying exPGCs ranged from 77.6 to 139.8 kb, and the exPGCs had similar sizes (average, 53.0 ± 1.0 kb). A considerable number of highly homologous genes were present in the three PGC-containing ECRs of this genus (Fig. 1). In addition, the genes present on smaller PGC-containing ECRs were mostly also present on larger PGC-containing ECRs. Phylogenetic analysis of the replication partitioning gene (parA), supported by high bootstrap values among homologs, indicated that the replication modules of the three PGC-containing ECRs were highly conserved (see Fig. S1 in the supplemental material; see also Table S1 in the supplemental material).
FIG 1.
Circular maps of the PGC-containing extrachromosomal replicons for Tateyamaria (a) and Jannaschia (b) strains. (a) Tateyamaria sp. ANG-S1 was used as the reference. Tracks (innermost to outermost) show G+C content, GC skew (G-C/G+C), Tateyamaria sp. ANG-S1 synteny, Tateyamaria sp. syn59 synteny, and Tateyamaria omphalii DOK1-4 synteny. (b) Jannaschia faecimaris DSM 1004020 was used as the reference. Tracks (innermost to outermost) show G+C content, GC skew (G-C/G+C), Jannaschia faecimaris DSM 1004020 synteny, Jannaschia donghaensis CECT 7802 synteny, and Jannaschia pohangensis DSM19073 synteny.
Phylogeny of the ParA plasmid partitioning proteins. The tree was constructed using maximum likelihood methods with 100 bootstrap replicates to evaluate node support. Only bootstrap values that are >50% are shown. Phototrophic strains carrying exPGCs are indicated in red. Accession numbers of all sequences are summarized in Table S1. Download FIG S1, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers of partitioning replication gene parA and 16S rRNA genes and whole-genome sequences of 38 AAPB strains used for phylogenetic analyses. Download Table S1, DOCX file, 0.02 MB (17.2KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Jannaschia is an ecologically important genus of AAPB (45). Indeed, 25% to 30% of all AAPB 16S rRNA gene clone sequences from samples collected in the central Baltic Sea belonged to Jannaschia-related bacteria (46). Among known Jannaschia isolates, CCS1 is the only strain observed to conduct photoheterotrophy (47, 48). Twelve Jannaschia strains were available for analysis with whole-genome sequences, with six containing PGCs; three of the six were exPGC types, and the other three were cPGC types. The PGC-containing ECRs ranged in size from 49.5 to 87.2 kb, while their exPGCs were ∼45 kb in length. The sizes of the genomes of the three Jannaschia strains carrying exPGCs ranged from 3.49 to 3.81 Mb, with GC contents ranging from 62.0% to 65.5%. The three PGC-containing ECRs of Jannaschia shared a large syntenic region comprising ∼50 kb, and the larger PGC-containing ECRs contained genes that were carried by the PGC-containing smaller ECRs, as observed for the ECRs in the Tateyamaria genomes (Fig. 1). In addition, their replicon replication modules were closely related based on a phylogenetic analysis of parA genes (Fig. S1; see also Table S1).
Sulfitobacter spp. are widely distributed in different marine environments and may play important roles in organic sulfur cycling (49–54). Culture-independent surveys of AAPB have indicated that Sulfitobacter spp. account for a significant fraction of AAPB communities in natural environments (10, 55, 56). However, only a few Sulfitobacter AAPB strains have been isolated and described (10, 51, 57). Genomes have been sequenced from only three Sulfitobacter AAPB strains, and the data revealed that their PGCs were located on ECRs. The three genomes ranged in size from 3.98 to 4.69 Mb, with GC contents ranging from 56.1% to 64.9%. The three PGC-containing ECRs exhibited sizes greater than 100 kb, while the three exPGCs were 45, 50, and 51 kb. Other than genes involved in photosynthesis, only a small number of genes were shared by PGC-containing ECRs in Sulfitobacter, which contrasted with the high degree of conservation observed for ECRs of Tateyamaria and Jannaschia. Furthermore, the parA genes within the three ECRs of Sulfitobacter were phylogenetically very distinct (Fig. S1; see also Table S1).
Roseobacter litoralis Och 149 was the first AAPB bacterium described (in 1991) and contained a linear ECR harboring a PGC (58, 59). The genus comprises only two species, R. litoralis Och 149 and R. denitrificans OCh114, with the latter containing a cPGC (58). In contrast, little is known about phototrophic species within Oceanicola, Shimia, and Nereida. Oceanicola sp. HL-35 exhibits 99% 16S rRNA gene sequence identity with Lacimonas salitoletrans TS-T30, which lacks a PGC (60). Prior to the isolation of Shimia sp. wx04, Shimia spp. were thought to be strict chemoorganotrophs based on culture-dependent investigations (61–65). Finally, Nereida ignava DSM 16093 was isolated from waters of the Mediterranean and is the sole species currently described for the Nereida genus (66, 67).
Identification of PGC-containing chromid-like ECRs and PGC-containing plasmid-like ECRs.
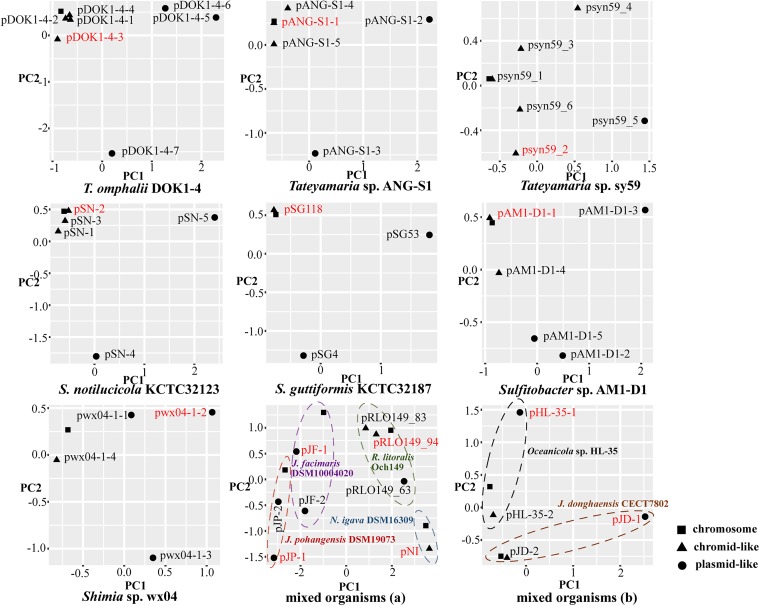
Chromids represent a novel type of ECRs that were recently described as genetic elements that are distinct from both chromosomes and plasmids (68). Since chromids typically carry essential genes, they remain more stably present than plasmids in bacteria and are considered indispensable for bacterial hosts (68). Comparison of the relative synonymous codon usage (RSCU) levels of bacterial chromosomes and ECRs can help identify if ECRs are chromids or not, as RSCU levels of chromid are similar to those of the corresponding chromosomes (34, 68, 69). Principal-component analysis (PCA) of the RSCU levels of all replicons from the 13 strains were analyzed for use in classifying elements as chromids and plasmids. The analyses indicated that eight PGC-containing ECRs (Tateyamaria sp. ANG-S1, Tateyamaria sp. syn59, T. omphalii DOK1-4, Sulfitobacter sp. AM1-D1, Sulfitobacter noctilucicola KCTC 32123, S. guttiformis KTCT 32187, R. litoralis Och 149, and N. ignava DSM16309) could be clearly assigned as chromid-like ECRs, while the other ECRs containing PGCs were provisionally classified as plasmid-like ECRs (Fig. 2; see also Table S2).
FIG 2.
Principal-component analysis of the relative synonymous codon usage (RSCU) of replicons from the 13 strains carrying exPGCs. Chromsomes, chromids, and plasmids are indicated by squares, triangles, and circles, respectively. The strains containing more than three replicons were analyzed on a strain-by-strain basis, while the rest were mixed and then analyzed. The replicons containing PGCs are indicated in red.
The extrachromosomal replicon types for 13 strains. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis.
A phylogenetic analysis based on 16S rRNA gene nucleotide sequences from 13 exPGC-containing bacterial strains and 43 reference strains was conducted with photoheterotrophs and heterotrophs to show the phylogenetic distribution of the 13 strains carrying exPGCs (Fig. S2). As observed for the cPGC-containing AAPB, the 13 exPGC-containing bacterial strains did not comprise a monophyletic phylogenetic group but were instead distributed throughout the 16S rRNA phylogenetic tree.
16S rRNA gene sequence phylogenetic analysis indicating the patchy distribution of AAPB within the Roseobacter clade. The tree was constructed using maximum likelihood methods with 100 bootstrap replicates to evaluate node support. Only bootstrap values that are >50% are shown. Phototrophic strains are indicated by bolding, and those carrying exPGCs are indicated in red. Accession numbers of all sequences are summarized in Table S1. Download FIG S2, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
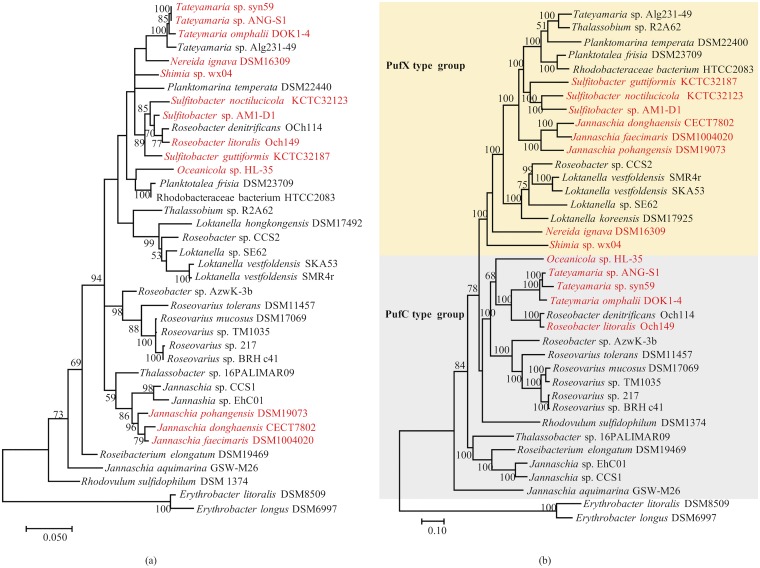
To further investigate the phylogenetic relationships of the Roseobacter clade strains, 38 photoheterotrophic bacterial genomes were subjected to phylogenetic analysis using 29 conserved PGC genes. Comparison of phylogenies based on 16S rRNA nucleotide sequences and amino acid sequences of 29 conserved genes of the PGC revealed considerable topological differences (Fig. 3). For example, the four Tateyamaria strains clustered and were shown to be closely related to the N. ignava DSM16309 strain in the 16S rRNA gene phylogenetic analysis. However, the Tateyamaria strains were using the 29 highly conserved genes of the PGC, with Tateyamaria sp. Alg231-49 closely related to Thalassonium sp. R2A62 and the other three strains associated with the Roseobacter species. Similarly, the five Jannaschia strains formed a group with (86%) bootstrap support based on the 16S rRNA phylogenetic analysis; however, they were clearly differentiated into two distant subtrees in the PGC-based phylogenetic analysis.
FIG 3.
Phylogenetic trees of 16S rRNA genes (a) and concatenation of 29 conserved PGC genes (b). Trees were constructed using maximum likelihood methods with 100 bootstrap replicates to evaluate node support. Only bootstrap values that were >50% are shown. Phototrophic strains carrying exPGCs are indicated in red. Accession numbers of all sequences are summarized in Table S1.
In addition, the PGC-based phylogenetic analysis indicated the presence of two phylogenetic groups corresponding to differences in their puf operon structures. Specifically, the groups corresponded to PufC-containing and PufX-containing groups (Fig. 3b). In particular, Tateyamaria sp. syn59, T. omphalli pDOK1-4, Tateyamaria sp. ANG-S1, R. litoralis Och149, and Oceanicola sp. HL-35 contained pufC genes, while the others contained pufX genes. Five of the exPGCs in the PufC group clustered together, suggesting common ancestry for these exPGCs. Moreover, the cPGC in Roseobacter denitrificans Och114 represented a basal clade to a subtree also comprising the five aforementioned exPGCs, thereby providing evidence for chromosomal reintegration from an ECR (33). Furthermore, the externally nested position of the exPGC from Oceanicola sp. HL-35 within this subtree indicated that the exPGC was possibly transferred from other AAPB strains and could be further transferred to other distant bacterial strains via an ECR. Nine of the 13 exPGCs belonged to three genera among the seven that were analyzed. Among these, the exPGCs from strains of the same genus were closely related phylogenetically, suggesting that the transfer of exPGCs was more likely to occur among strains within the same genus. Two types of puf operon structures were observed within the genomes of different strains within the Tateyamaria and Jannaschia genera. Among the six PGC-containing Jannaschia strains, three exPGCs were PufX types whereas the other three were PufC types. Similarly, among the four phototrophic Tateyamaria strains analyzed, one cPGC was a PufX type whereas the other three exPGCs were PufC types.
exPGC structures and arrangements.
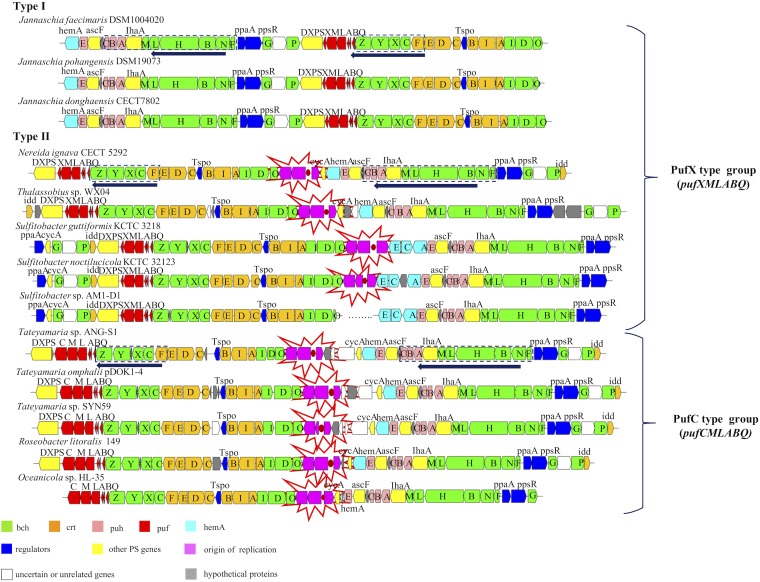
Three different structures were observed among the 13 exPGCs (Fig. 4). The 41 photosynthetic genes on the ECR of the three Jannaschia strains were organized into one superoperon with the same structure as that of the cPGC (20). In addition, the exPGC of Sulfitobacter sp. AM1-D1 was separated by more than 100 genes between bchIDO and hemECA. The other exPGCs all appeared to have been inserted by their replication modules, as previously identified (31, 33).
FIG 4.
Photosynthetic gene cluster structures and arrangements for 13 exPGCs. ECR module positions (plasmid origin replication) within the exPGC are shown in red and highlighted by stars. Arrows indicate the order of the conserved PGC genes as follows: type I, forward puh-LhaA-bchMLHNF plus forward puf-bchZYXC-crtF; type II, forward puf-bchZYXC-crtF plus forward puh-LhaA-bchMLHNF. The classifications of type PufX and type PufC groups are based on their different puf operon compositions, corresponding to pufXMLABQ and pufCMLABQ.
The arrangement of photosynthetic genes is also a key characteristic of PGCs in AAPB. Three forms of PGC arrangement have been observed in Roseobacter clade organisms based on a combination of two conserved regions (puh-LhaA-bchMLHNF and puf-bchZYXC-crtF) (20). The 13 exPGCs contained the same conserved gene order as the cPGCs and comprised two different types. The type I arrangement was exhibited by exPGCs of the three Jannaschia strains (J. pohangensis DSM19073, J. donghaensis CECT7802, and J. faecimaris DSM10004020), wherein genes were arranged as forward puh LhaA bchMLHNF plus forward puf-bchZYXC-crtF. Type II arrangements were observed for the other 10 strains, wherein arrangements followed the pattern of forward puf-bchZYXC-crtF plus forward puh-LhaA-bchMLHNF (Fig. 4).
The arrangements of exPGCs of the PufC-containing group were of type II, which exhibited the high conservation in the direction and order of all photosynthetic genes on the exPGCs. In contrast, the exPGCs in the PufX-containing group exhibited two different types of arrangements, with unique traits present in different genera. For example, hemC and hemE genes encoding tetrapyrrole biosynthesis proteins were present only in the exPGCs of Sulfitobacter. In addition, the genomic region ranging from bchG to idd was located upstream of the puf operon in Sulfitobacter, although it is typically downstream of the ppaA and ppsR regulator genes within PGCs (20). Furthermore, the three exPGCs of the Jannaschia strains lacked cytochrome c2 (cyc2) and diphosphate delta-isomerase (idi) genes. cyc2 is involved in electron transfer, while idi is involved in isoprenoid biosynthesis. The loss of these genes is not lethal for phototrophic bacteria (70, 71), but the genes are nevertheless expected to be present in the PGCs of phototrophic bacteria within the Roseobacter clade (33, 72, 73).
Evidence of ECR-mediated PGC transfer within the Roseobacter clade.
Recent studies have suggested that ECRs could be vehicles for HGT of PGCs (31, 33), albeit with limited evidence. The idea of transfer of PGCs by ECRs was supported in our analyses by the coexistence of two different types of puf operon structures (PufC and PufX types) in different strains of two genera, Tateyamaria and Jannaschia. In particular, these two types of puf operons were located on cPGCs and exPGCs, respectively. The Global Ocean Sampling expedition metageomes were the first to reveal that pufC could be replaced by pufX in AAPB and that pufC and pufX were present in different AAPB phylogroups (5, 74). Thus, phylogenetic divergence of the two types of puf operons in strains from the same genus suggested that one or both of them were introduced by other phototrophic phylogroups. Moreover, phylogenetic congruence between whole PGCs and conserved photosynthetic operons within the PGC (i.e., bchFNBHLM-IhaA-puhABC and pufMLABQ-bchZYXC-crtF) indicate that PGCs act as entire functional units rather than being subject to partial transfer between strains (Fig. S3); this is consistent with a previous study (33). ECRs are mobile genetic elements; thus, PGCs carried by ECRs are more likely to be horizontally transferred.
Phylogenetic tree of the two conserved PGC subclusters based on concatenated bchFNBHLM-IhaA-puhABC (a) and pufMLAB-bchZYXC-crtF (b) protein sequences. The tree was constructed using maximum likelihood methods with 100 bootstrap replicates to evaluate node support. Only bootstrap values that are >50% are shown. Phototrophic strains carrying exPGCs are indicated in red. Accession numbers of all sequences are summarized in Table S1. Download FIG S3, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The potential for transfer of PGC-containing ECRs.
As described above, the 13 PGC-containing ECRs were divided into two types based on their sizes and functions. Small PGC-containing ECRs within Oceanicola sp. HL-35, Shimia sp. wx04, and J. pohangensis DSM19073 carried more than 80% of the genes coding for PGCs. These ECRs are usually present as plasmids and are likely to play an important role in the transfer of phototrophic capacities among species. This is especially probable because the transfer of small plasmids achieves higher efficiencies and the three streamlined PGC-containing ECRs still appear to confer the capability of chlorophototrophy (75, 76). The acquisition of streamlined PGC-containing ECRs might enable strains to obtain new lifestyles at low costs, thereby providing advantages under certain environmental conditions (34). The other large PGC-containing ECRs also encoded proteins with various nonphotosynthetic functions. Moreover, a sox gene cluster (soxRSVYAZBCD), usually located on the bacterial chromosome, was observed on the PGC-containing chromid-like elements of N. ignava DSM1630, suggesting that the sox gene cluster might be also transferred by the ECR. Most of these large ECRs were classified as chromid-like ECRs. Consequently, these PGC-containing ECRs might preferentially be maintained in bacterial hosts rather than be transferred among hosts. Notably, PGCs carried by both plasmid-like and chromid-like ECRs have been suggested to be genomically stable because most exPGCs have been inserted by their corresponding ECR replication modules (31).
Comparison of the GC contents of the bacterial genomes, exPGCs, and PGC-containing ECRs did not reveal significant differences for any of the 13 Roseobacter clade strains (Fig. S4). Thus, the transfer of these PGC-containing ECRs into bacteria likely occurred during very distant evolutionary events or, otherwise, only between closely related species (77).
GC content correlations among 13 strains of Roseobacter species for genomic GC content and that of PGC-containing ECRs (a), genomes and exPGCs (b), and PGC-containing ECRs and exPGCs (c). Download FIG S4, TIF file, 0.3 MB (267.4KB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A scenario to explain the evolution of AAPB exPGCs in the Roseobacter clade.
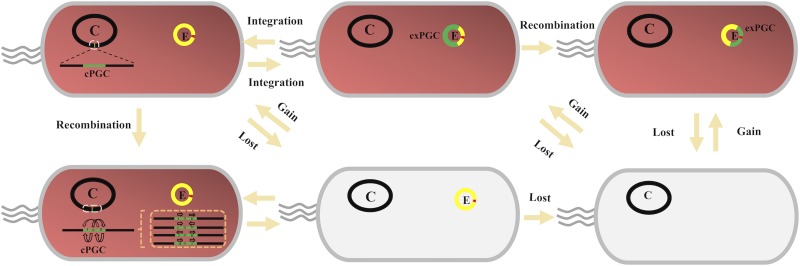
A previous scenario was suggested to explain the evolution of exPGCs in Roseobacter clade organisms, wherein a chromosomal PGC superoperon was transferred into an ECR, followed by integration of replication origin genes into the ECR (31, 34). Our analyses validate this explanation, and we further present a more detailed scenario (Fig. 5) to explain the transfer of PGCs within the Roseobacter clade after analyzing genomic and evolutionary characteristics of 13 exPGCs in this group (31). In this revised scenario, PGCs were first initially translocated from chromosomes to ECRs, as represented by the superoperon structure of exPGCs from J. faecimaris DSM1004020, J. pohangensis DSM19073, and J. donghaensis CECT 7802. Three subsequent transfer possibilities are present for exPGCs: (i) exPGCs reintegrated into chromosomes and became cPGCs, as observed for most phototrophic Roseobacter clade strains; (ii) PGC-containing ECRs were lost from strains, such that the bacteria became heterotrophs; or (iii) exPGCs carried by ECRs were subjected to further recombination and became stable within ECRs. A remarkable characteristic of the majority of exPGCs is the insertion of an ECR replication module within the PGC as a result of a series of recombination events. Such an event could have helped ensure the stability of exPGCs (31).
FIG 5.
Scenarios to explain the transfer and evolution of exPGCs within Roseobacter clade species. PGCs are indicated in green, and PGC-containing ECR origins of replication are indicated in red. Photosynthetic bacteria are indicated in light red, while non-photosynthetic bacteria are indicated in light gray. C, chromosome; E, PGC-containing ECR.
The patchy distribution of AAPB within the Roseobacter clade has been explained by two evolutionary models that invoke either loss or gain of PGCs (11–13). Given that ECRs have played a critical role in the loss or gain of PGCs during the evolutionary history of photosynthesis, the patchy distribution of AAPB within the Roseobacter clade can be plausibly explained by ECR-mediated mechanisms. To date, exPGCs have accounted for ∼20% of all PGCs in the currently available genomes of Roseobacter clade strains (Table S3), highlighting their prevalence in these organisms. It is likely that additional exPGCs carried by other strains will be identified with further generation of new bacterial genome sequences. Moreover, we suggest that the gain and loss of PGCs, as mediated by chromosomes and especially ECRs, resulted in the patchy distribution of AAPB within the Roseobacter clade.
Whole-genome sequences for 67 AAPB strains within the Roseobacter clade that are publicly available in NCBI. AAPB strains carrying exPGCs are indicated in red. Download Table S3, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the present study, the genomic characteristics and evolution of 13 PGCs carried by ECRs were analyzed. The coexistence of two types of puf operon structures within strains of the same genera provided clear evidence of the horizontal transfer of PGCs mediated by ECR. Analysis of PGC-containing plasmid-like and chromid-like ECRs indicated that exPGCs could stably exist in bacteria after transfer, highlighting the importance of phototrophic metabolism carried by ECRs for some bacteria. Furthermore, these analyses indicated that the process of gain or loss of PGCs, as mediated by ECRs, contributes to the patchy distribution of phototrophic capacities within the Roseobacter clade.
MATERIALS AND METHODS
Strain isolation.
Tateyamaria sp. syn59 and Shimia sp. wx04 were isolated from the South China Sea in April 2016 using oligotrophic medium F/2 plates (78), followed by transfer onto rich organic liquid medium (Marine Broth 2216; Difco, USA) for further isolation and cultivation. All cultures were incubated at 28°C with shaking at 200 rpm in the dark. Genomic DNA from the two strains was extracted using a TaKaRa MiniBEST universal genomic DNA extraction kit (Japan).
Genome sequencing, assembly, and annotation.
The genomes of Tateyamaria sp. syn59 and Shimia sp. wx04 were sequenced on an Illumina MiSeq platform (Illumina, USA). Specifically, 2 × 250-bp paired-end read sequencing was conducted, followed by read assembly using the Velvet program (version 2.8) (79). Prediction and annotation of open reading frames (ORFs) were conducted using a Rapid Annotation using Subsystems Technology (RAST) platform. Further, annotations of plasmids and exPGCs were validated by BLASTP searches against the National Center for Biotechnology Information (NCBI) nonredundant (nr) protein database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The whole-genome sequences of Tateyamaria sp. syn59 and Shimia sp. wx04 are available under GenBank accession numbers VCBA00000000.1 (https://www.ncbi.nlm.nih.gov/nuccore/VCDK00000000) and VCDK00000000.1 (https://www.ncbi.nlm.nih.gov/nuccore/VCBA00000000), respectively.
Retrieval of AAPB genomes from GenBank.
Genome sequence data for the other eleven Roseobacter clade strains were obtained from NCBI, including data for Tateyamaria omphalii DOK1-4 (CP019312.1); Tateyamaria sp. ANG-S1 (JWLL00000000.1); Roseobacter litoralis Och 149 (CP002623.1); Oceanicola sp. HL-35 (JAFT00000000.1); Nereida ignava DSM 16309 (CVPC00000000.1); Sulfitobacter noctilucicola KCTC 32123 (JASD00000000.1); Sulfitobacter guttiformis KCTC 32187 (JASG00000000.1); Sulfitobacter sp. AM1-D1 (CP018076.1); Jannaschia donghaensis CECT 7802 (NZ_CXSU00000000.1); Jannaschia faecimaris DSM 10004020 (FNPX00000000.1); and Jannaschia pohangensis DSM 19073 (FORA00000000.1).
Phylogenetic analysis.
Complete 16S rRNA gene sequences were extracted from the whole-genome assemblies (WGA) using the Cluster program (80). To construct a phylogeny for the PGCs, the amino acid sequences of 29 conserved photosynthetic genes within PGCs (bchI, bchD, bchO, tspO, crtC, crtD, crtF, bchC, bchX, bchY, bchZ, pufL, pufM, bchP, puCC, bchG, ppsR, bchF, bchN, bchB, bchH, bchL, bchM, IhaA, puhA, puhB, puhC, ascF, and puhE) (see Table S4 in the supplemental material) were retrieved from the query genomes and then individually aligned using ClustalW, as implemented in the BioEdit program (81). Phylogenetic analyses were conducted using RAxML 8.02 (82) and maximum likelihood (ML) methods. The robustness of the tree topologies was evaluated using bootstrap analysis with 100 replicates. Final trees were then visualized using the Interactive Tree of Life viewer and MEGA version 7.0 (83, 84).
The list of 29 conserved genes in the PGC. Download Table S4, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
All data used in this study are publicly available in GenBank. Accession numbers can be found in Table S1 and Table S3.
ACKNOWLEDGMENTS
We are grateful to J. Thomas Beatty for helpful discussion and constructive suggestions.
This work was supported by the National Key Research Programs (2018YFA0605800), the Senior User Project of RV KEXUE (KEXUE2019GZ03), the National Natural Science Foundation of China (NSFC) project (41776145, 41876150, and 91751207), and the Fundamental Research Funds for the Central Universities (20720190095).
We declare that we have no conflict of interest.
REFERENCES
- 1.Jiao N, Zhang Y, Zeng Y, Hong N, Liu R, Chen F, Wang P. 2007. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ Microbiol 9:3091–3099. doi: 10.1111/j.1462-2920.2007.01419.x. [DOI] [PubMed] [Google Scholar]
- 2.Ritchie AE, Johnson ZI. 2012. Abundance and genetic diversity of aerobic anoxygenic phototrophic bacteria of coastal regions of the Pacific Ocean. Appl Environ Microbiol 78:2858–2866. doi: 10.1128/AEM.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koblížek M. 2015. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev 39:854–870. doi: 10.1093/femsre/fuv032. [DOI] [PubMed] [Google Scholar]
- 4.Jiao N, Zhang F, Hong N. 2010. Significant roles of bacteriochlorophylla supplemental to chlorophylla in the ocean. ISME J 4:595–597. doi: 10.1038/ismej.2009.135. [DOI] [PubMed] [Google Scholar]
- 5.Yutin N, Suzuki MT, Teeling H, Weber M, Venter JC, Rusch DB, Béjà O. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ Microbiol 9:1464–1475. doi: 10.1111/j.1462-2920.2007.01265.x. [DOI] [PubMed] [Google Scholar]
- 6.Kolber ZS, Plumley FG, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblizek M, Rathgeber C, Falkowski PG. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 7.Koblížek M, Masín M, Ras J, Poulton AJ, Prášil O. 2007. Rapid growth rates of aerobic anoxygenic phototrophs in the ocean. Environ Microbiol 9:2401–2406. doi: 10.1111/j.1462-2920.2007.01354.x. [DOI] [PubMed] [Google Scholar]
- 8.Lami R, Cottrell MT, Ras J, Ulloa O, Obernosterer I, Claustre H, Kirchman DL, Lebaron P. 2007. High abundances of aerobic anoxygenic photosynthetic bacteria in the South Pacific Ocean. Appl Environ Microbiol 73:4198–4205. doi: 10.1128/AEM.02652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell MT, Mannino A, Kirchman DL. 2006. Aerobic anoxygenic phototrophic bacteria in the Mid-Atlantic Bight and the North Pacific Gyre. Appl Environ Microbiol 72:557–564. doi: 10.1128/AEM.72.1.557-564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeuf D, Cottrell MT, Kirchman DL, Lebaron P, Jeanthon C. 2013. Summer community structure of aerobic anoxygenic phototrophic bacteria in the western Arctic Ocean. FEMS Microbiol Ecol 85:417–432. doi: 10.1111/1574-6941.12130. [DOI] [PubMed] [Google Scholar]
- 11.Yurkov V, Hughes E. 2017. Aerobic anoxygenic phototrophs: four decades of mystery, p 193–214. In Modern Topics in the Phototrophic Prokaryotes. Springer, Cham. [Google Scholar]
- 12.Beatty JT. 2002. On the natural selection and evolution of the aerobic phototrophic bacteria. Photosynth Res 73:109–114. doi: 10.1023/A:1020493518379. [DOI] [PubMed] [Google Scholar]
- 13.Yurkov V, Csotonyi JT. 2009. New light on aerobic anoxygenic phototrophs, p 31–55. In The purple phototrophic bacteria, 3rd ed Springer, Dordrecht, Netherlands. [Google Scholar]
- 14.Bekker A, Holland HD, Wang PL, Rumble D III, Stein HJ, Hannah JL, Coetzee LL, Beukes NJ. 2004. Dating the rise of atmospheric oxygen. Nature 427:117. doi: 10.1038/nature02260. [DOI] [PubMed] [Google Scholar]
- 15.Holland HD. 2006. The oxygenation of the atmosphere and oceans. Philos Trans R Soc B 361:903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada K. 1995. Aerobic anoxygenic phototrophs, p 105–122. In Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 17.Yurkov VV, Beatty JT. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev 62:695–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura K, Shimada H, Ohta H, Masuda T, Shioi Y, Takamiya K-I. 1996. Expression of the puf operon in an aerobic photosynthetic bacterium, Roseobacter denitrificans. Plant Cell Physiol 37:153–159. doi: 10.1093/oxfordjournals.pcp.a028926. [DOI] [PubMed] [Google Scholar]
- 19.Beatty JT. 1995. Organization of photosynthesis gene transcripts, p 1209–1219. In Anoxygenic photosynthetic bacteria. Springer, Dordrecht, Netherlands. [Google Scholar]
- 20.Zheng Q, Zhang R, Koblížek M, Boldareva EN, Yurkov V, Yan S, Jiao N. 2011. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One 6:e25050. doi: 10.1371/journal.pone.0025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liotenberg S, Steunou AS, Picaud M, Husson FR, Astier C, Ouchane S. 2008. Organization and expression of photosynthesis genes and operons in anoxygenic photosynthetic proteobacteria. Environ Microbiol 10:2267–2276. doi: 10.1111/j.1462-2920.2008.01649.x. [DOI] [PubMed] [Google Scholar]
- 22.Swingley WD, Blankenship RE, Raymond J. 2009. Evolutionary relationships among purple photosynthetic bacteria and the origin of proteobacterial photosynthetic systems, p 17–29. In The purple phototrophic bacteria. Springer, Dordrecht, Netherlands. [Google Scholar]
- 23.Zheng Q, Lin W, Liu Y, Chen C, Jiao N. 2016. A comparison of 14 Erythrobacter genomes provides insights into the genomic divergence and scattered distribution of phototrophs. Front Microbiol 7:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y, Feng F, Medová H, Dean J, Koblížek M. 2014. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc Natl Acad Sci U S A 111:7795–7800. doi: 10.1073/pnas.1400295111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Q, Zhang R, Fogg PCM, Beatty JT, Wang Y, Jiao N. 2012. Gain and loss of phototrophic genes revealed by comparison of two Citromicrobium bacterial genomes. PLoS One 7:e35790. doi: 10.1371/journal.pone.0035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, Rinta-Kanto JM, Sharma S, Sun S, Varaljay V, Vila-Costa M, Westrich JR, Moran MA. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J 4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 28.Luo H, Moran MA. 2014. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78:573–587. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinkhoff T, Giebel H-A, Simon M. 2008. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 30.Nagashima KVP, Hiraishi A, Shimada K, Matsuura K. 1997. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J Mol Evol 45:131–136. doi: 10.1007/PL00006212. [DOI] [PubMed] [Google Scholar]
- 31.Petersen J, Brinkmann H, Bunk B, Michael V, Päuker O, Pradella S. 2012. Think pink: photosynthesis, plasmids and the Roseobacter clade. Environ Microbiol 14:2661–2672. doi: 10.1111/j.1462-2920.2012.02806.x. [DOI] [PubMed] [Google Scholar]
- 32.Raymond J, Zhaxybayeva O, Gogarten JP, Gerdes SY, Blankenship RE. 2002. Whole-genome analysis of photosynthetic prokaryotes. Science 298:1616–1620. doi: 10.1126/science.1075558. [DOI] [PubMed] [Google Scholar]
- 33.Brinkmann H, Göker M, Koblížek M, Wagner-Döbler I, Petersen J. 2018. Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae. ISME J 12:1994–2010. doi: 10.1038/s41396-018-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen J, Frank O, Göker M, Pradella S. 2013. Extrachromosomal, extraordinary and essential–the plasmids of the Roseobacter clade. Appl Microbiol Biotechnol 97:2805–2815. doi: 10.1007/s00253-013-4746-8. [DOI] [PubMed] [Google Scholar]
- 35.Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol 3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 36.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 37.Kurahashi M, Yokota A. 2007. Tateyamaria omphalii gen. nov., sp. nov., an α-Proteobacterium isolated from a top shell Omphalius pfeifferi pfeifferi. Syst Appl Microbiol 30:371–375. doi: 10.1016/j.syapm.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Sass H, Köpke B, Rutters H, Feuerlein T, Droge S, Cypionka H, Engelen B. 2010. Tateyamaria pelophila sp. nov., a facultatively anaerobic alphaproteobacterium isolated from tidal-flat sediment, and emended descriptions of the genus Tateyamaria and of Tateyamaria omphalii. Int J Syst Evol Microbiol 60:1770–1777. doi: 10.1099/ijs.0.013524-0. [DOI] [PubMed] [Google Scholar]
- 39.Jeanthon C, Boeuf D, Dahan O, Gall FL, Garczarek L, Bendif EM, Lehours AC. 2011. Diversity of cultivated and metabolically active aerobic anoxygenic phototrophic bacteria along an oligotrophic gradient in the Mediterranean Sea. Biogeosciences 8:1955–1970. doi: 10.5194/bg-8-1955-2011. [DOI] [Google Scholar]
- 40.Collins AJ, Fullmer MS, Gogarten JP, Nyholm SV. 2015. Comparative genomics of Roseobacter clade bacteria isolated from the accessory nidamental gland of Euprymna scolopes. Front Microbiol 6:11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karimi E. 2018. Metagenomics and functional genomics of bacterial symbionts of Spongia (Porifera, Dictyoceratida) specimens from the Algarvian shore (South Portugal). PhD thesis University of Algarve, Faro, Portugal. [Google Scholar]
- 42.Huggett MJ, McMahon K, Bernasconi R. 2018. Future warming and acidification result in multiple ecological impacts to a temperate coralline alga. Environ Microbiol 20:2769–2782. doi: 10.1111/1462-2920.14113. [DOI] [PubMed] [Google Scholar]
- 43.Bengtsson MM, Sjøtun K, Lanzén A, Øvreås L. 2012. Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J 6:2188–2198. doi: 10.1038/ismej.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalitnik AA, Nedashkovskaya OI, Stenkova AM, Yermak IM, Kukhlevskiy AD. 2018. Carrageenanolytic enzymes from marine bacteria associated with the red alga Tichocarpus crinitus. J Appl Phycol 30:2071–2081. doi: 10.1007/s10811-017-1355-4. [DOI] [Google Scholar]
- 45.Zeng YH, Chen XH, Jiao NZ. 2007. Genetic diversity assessment of anoxygenic photosynthetic bacteria by distance-based grouping analysis of pufM sequences. Lett Appl Microbiol 45:639–645. doi: 10.1111/j.1472-765X.2007.02247.x. [DOI] [PubMed] [Google Scholar]
- 46.Salka I, Moulisová V, Koblížek M, Jost G, Jürgens K, Labrenz M. 2008. Abundance, depth distribution, and composition of aerobic bacteriochlorophyll a-producing bacteria in four basins of the central Baltic Sea. Appl Environ Microbiol 74:4398–4404. doi: 10.1128/AEM.02447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A. 2007. Ecological genomics of marine Roseobacters. Appl Environ Microbiol 73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pujalte MJ, Lucena T, Ruvira MA, Arahal DR, Macián MC. 2014. The family rhodobacteraceae, p 439–512. In The prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 49.Prabagaran SR, Manorama R, Delille D, Shivaji S. 2007. Predominance of Roseobacter, Sulfitobacter, Glaciecola and Psychrobacter in seawater collected off Ushuaia, Argentina, sub-Antarctica. FEMS Microbiol Ecol 59:342–355. doi: 10.1111/j.1574-6941.2006.00213.x. [DOI] [PubMed] [Google Scholar]
- 50.Rooney-Varga JN, Giewat MW, Savin MC, Sood S, LeGresley M, Martin JL. 2005. Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb Ecol 49:163–175. doi: 10.1007/s00248-003-1057-0. [DOI] [PubMed] [Google Scholar]
- 51.Labrenz M, Tindall BJ, Lawson PA, Collins MD, Schumann P, Hirsch P. 2000. Staleya guttiformis gen. nov., sp. nov. and Sulfitobacter brevis sp. nov., alpha-3-Proteobacteria from hypersaline, heliothermal and meromictic Antarctic Ekho Lake. Int J Syst Evol Microbiol 50:303–313. doi: 10.1099/00207713-50-1-303. [DOI] [PubMed] [Google Scholar]
- 52.Moran MA, González JM, Kiene RP. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiology J 20:375–388. doi: 10.1080/01490450303901. [DOI] [Google Scholar]
- 53.Zachariah S, Kumari P, Das SK. 2017. Sulfitobacter pontiacus subsp. fungiae subsp. nov., isolated from coral Fungia seychellensis from Andaman Sea, and description of Sulfitobacter pontiacus subsp. pontiacus subsp. nov. Curr Microbiol 74:404–412. doi: 10.1007/s00284-017-1200-7. [DOI] [PubMed] [Google Scholar]
- 54.Pukall R, Buntefuss D, Fruhling A, Rohde M, Kroppenstedt RM, Burghardt J, Lebaron P, Bernard L, Stackebrandt E. 1999. Sulfitobacter mediterraneus sp. nov., a new sulfite-oxidizing member of the α-Proteobacteria. Int J Syst Bacteriol 49:513–519. doi: 10.1099/00207713-49-2-513. [DOI] [PubMed] [Google Scholar]
- 55.Buchan A, González JM, Moran MA. 2005. Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng Y, Dong P, Qiao Z, Zheng T. 2016. Diversity of the aerobic anoxygenic phototrophy gene pufM in Arctic and Antarctic coastal seawaters. Acta Oceanol Sin 35:68–77. doi: 10.1007/s13131-016-0877-y. [DOI] [Google Scholar]
- 57.Kwak M-J, Lee J-S, Lee KC, Kim KK, Eom MK, Kim BK, Kim JF. 2014. Sulfitobacter geojensis sp. nov., Sulfitobacter noctilucae sp. nov., and Sulfitobacter noctilucicola sp. nov., isolated from coastal seawater. Int J Syst Evol Microbiol 64:3760–3767. doi: 10.1099/ijs.0.065961-0. [DOI] [PubMed] [Google Scholar]
- 58.Shiba T. 1991. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst Appl Microbiol 14:140–145. doi: 10.1016/S0723-2020(11)80292-4. [DOI] [Google Scholar]
- 59.Kalhoefer D, Thole S, Voget S, Lehmann R, Liesegang H, Wollher A, Daniel R, Simon M, Brinkhoff T. 2011. Comparative genome analysis and genome-guided physiological analysis of Roseobacter litoralis. BMC Genomics 12:324. doi: 10.1186/1471-2164-12-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong Z-P, Liu Y, Wang F, Zhou Y-G, Liu H-C, Liu Z-P. 2015. Lacimonas salitolerans gen. nov., sp. nov., isolated from surface water of a saline lake. Int J Syst Evol Microbiol 65:4550–4556. doi: 10.1099/ijsem.0.000611. [DOI] [PubMed] [Google Scholar]
- 61.Hameed A, Shahina M, Lin S-Y, Lai W-A, Hsu Y-H, Liu Y-C, Huang Y-M, Young C-C. 2013. Shimia biformata sp. nov., isolated from surface seawater, and emended description of the genus Shimia Choi and Cho 2006. Int J Syst Evol Microbiol 63:4533–4539. doi: 10.1099/ijs.0.053553-0. [DOI] [PubMed] [Google Scholar]
- 62.Hyun D-W, Kim M-S, Shin N-R, Kim JY, Kim PS, Whon TW, Yun J-H, Bae J-W. 2013. Shimia haliotis sp. nov., a bacterium isolated from the gut of an abalone, Haliotis discus hannai. Int J Syst Evol Microbiol 63:4248–4253. doi: 10.1099/ijs.0.053140-0. [DOI] [PubMed] [Google Scholar]
- 63.Chen M-H, Sheu S-Y, Chen CA, Wang J-T, Chen W-M. 2011. Shimia isoporae sp. nov., isolated from the reef-building coral Isopora palifera. Int J Syst Evol Microbiol 61:823–827. doi: 10.1099/ijs.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 64.Choi DH, Cho BC. 2006. Shimia marina gen. nov., sp. nov., a novel bacterium of the Roseobacter clade isolated from biofilm in a coastal fish farm. Int J Syst Evol Microbiol 56:1869–1873. doi: 10.1099/ijs.0.64235-0. [DOI] [PubMed] [Google Scholar]
- 65.Nogi Y, Mori K, Uchida H, Hatada Y. 2015. Shimia sagamensis sp. nov., a marine bacterium isolated from cold-seep sediment. Int J Syst Evol Microbiol 65:2786–2790. doi: 10.1099/ijs.0.000333. [DOI] [PubMed] [Google Scholar]
- 66.Pujalte MJ, Macián MC, Arahal DR, Ludwig W, Schleifer KH, Garay E. 2005. Nereida ignava gen. nov., sp. nov., a novel aerobic marine α-proteobacterium that is closely related to uncultured Prionitis (alga) gall symbionts. Int J Syst Evol Microbiol 55:631–636. doi: 10.1099/ijs.0.63442-0. [DOI] [PubMed] [Google Scholar]
- 67.Arahal DR, Pujalte MJ, Rodrigo-Torres L. 2016. Draft genomic sequence of Nereida ignava CECT 5292 T, a marine bacterium of the family Rhodobacteraceae. Stand Genomic Sci 11:1–21. doi: 10.1186/s40793-016-0141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harrison PW, Lower RPJ, Kim NKD, Young J. 2010. Introducing the bacterial “chromid”: not a chromosome, not a plasmid. Trends Microbiol 18:141–148. doi: 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 69.Petersen J, Wagner-Döbler I. 2017. Plasmid transfer in the ocean—a case study from the Roseobacter group. Front Microbiol 8:1350. doi: 10.3389/fmicb.2017.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonnett TC, Cobine P, Sockett RE, McEwan AG. 1995. Phenotypic characterisation and genetic complementation of dimethylsulfoxide respiratory mutants of Rhodobacter sphaeroides and Rhodobacter capsulatus. FEMS Microbiol Lett 133:163–168. doi: 10.1111/j.1574-6968.1995.tb07878.x. [DOI] [PubMed] [Google Scholar]
- 71.Addlesee HA, Fiedor L, Hunter CN. 2000. Physical mapping of bchG, orf427, and orf177 in the photosynthesis gene cluster of Rhodobacter sphaeroides: functional assignment of the Bacteriochlorophyll synthetase gene. J Bacteriol 182:3175–3182. doi: 10.1128/jb.182.11.3175-3182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones MR, McEwan AG, Jackson JB. 1990. The role of c-type cytochromes in the photosynthetic electron transport pathway of Rhodobacter capsulatus. Biochim Biophys Acta 1019:59–66. doi: 10.1016/0005-2728(90)90124-m. [DOI] [PubMed] [Google Scholar]
- 73.Hahn FM, Baker JA, Poulter CD. 1996. Open reading frame 176 in the photosynthesis gene cluster of Rhodobacter capsulatus encodes idi, a gene for isopentenyl diphosphate isomerase. J Bacteriol 178:619–624. doi: 10.1128/jb.178.3.619-624.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yutin N, Béjà O. 2005. Putative novel photosynthetic reaction centre organizations in marine aerobic anoxygenic photosynthetic bacteria: insights from metagenomics and environmental genomics. Environ Microbiol 7:2027–2033. doi: 10.1111/j.1462-2920.2005.00843.x. [DOI] [PubMed] [Google Scholar]
- 75.Kreiss P, Mailhe P, Scherman D, Pitard B, Cameron B, Rangara R, Aguerre-Charriol O, Airiau M, Crouzet J. 1999. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res 27:3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan V, Dreolini LF, Flintoff KA, Lloyd SJ, Mattenley AA. 2002. The effect of increasing plasmid size on transformation efficiency in Escherichia coli. J Exp Microbiol Immunol 2:207–223. [Google Scholar]
- 77.Lassalle F, Périan S, Bataillon T, Nesme X, Duret L, Daubin V. 2015. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet 11:e1004941. doi: 10.1371/journal.pgen.1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guillard RRL, Ryther JH. 1962. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can J Microbiol 8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 79.Zerbino D, Birney E. 2008. Velvet: algorithms for de novo short read assembly using De Bruijn graphs. Genome Res 18:107–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (41):95–98. [Google Scholar]
- 81.Hall T. 2011. BioEdit: an important software for molecular biology. GERF Bull Biosci 2:60–61. [Google Scholar]
- 82.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of the ParA plasmid partitioning proteins. The tree was constructed using maximum likelihood methods with 100 bootstrap replicates to evaluate node support. Only bootstrap values that are >50% are shown. Phototrophic strains carrying exPGCs are indicated in red. Accession numbers of all sequences are summarized in Table S1. Download FIG S1, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers of partitioning replication gene parA and 16S rRNA genes and whole-genome sequences of 38 AAPB strains used for phylogenetic analyses. Download Table S1, DOCX file, 0.02 MB (17.2KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The extrachromosomal replicon types for 13 strains. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene sequence phylogenetic analysis indicating the patchy distribution of AAPB within the Roseobacter clade. The tree was constructed using maximum likelihood methods with 100 bootstrap replicates to evaluate node support. Only bootstrap values that are >50% are shown. Phototrophic strains are indicated by bolding, and those carrying exPGCs are indicated in red. Accession numbers of all sequences are summarized in Table S1. Download FIG S2, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic tree of the two conserved PGC subclusters based on concatenated bchFNBHLM-IhaA-puhABC (a) and pufMLAB-bchZYXC-crtF (b) protein sequences. The tree was constructed using maximum likelihood methods with 100 bootstrap replicates to evaluate node support. Only bootstrap values that are >50% are shown. Phototrophic strains carrying exPGCs are indicated in red. Accession numbers of all sequences are summarized in Table S1. Download FIG S3, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GC content correlations among 13 strains of Roseobacter species for genomic GC content and that of PGC-containing ECRs (a), genomes and exPGCs (b), and PGC-containing ECRs and exPGCs (c). Download FIG S4, TIF file, 0.3 MB (267.4KB, tif) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Whole-genome sequences for 67 AAPB strains within the Roseobacter clade that are publicly available in NCBI. AAPB strains carrying exPGCs are indicated in red. Download Table S3, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The list of 29 conserved genes in the PGC. Download Table S4, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2019 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All data used in this study are publicly available in GenBank. Accession numbers can be found in Table S1 and Table S3.