Summary
In C. elegans nematodes, components of liquid-like germ granules were shown to be required for transgenerational small RNA inheritance. Surprisingly, we show here that mutants with defective germ granules can nevertheless inherit potent small RNA-based silencing responses, but some of the mutants lose this ability after many generations of homozygosity. Animals mutated in pptr-1, which is required for stabilization of P granules in the early embryo, display extraordinarily strong heritable RNAi responses, lasting for tens of generations. Intriguingly, the RNAi capacity of descendants derived from mutants defective in the core germ granule proteins MEG-3 and MEG-4 is determined by the genotype of the ancestors and changes transgenerationally. Further, whether the meg-3/4 mutant alleles were present in the paternal or maternal lineages leads to different transgenerational consequences. Small RNA inheritance, rather than maternal contribution of the germ granules themselves, mediates the transgenerational defects in RNAi of meg-3/4 mutants and their progeny. Accordingly, germ granule defects lead to heritable genome-wide mis-expression of endogenous small RNAs. Upon disruption of germ granules, hrde-1 mutants can inherit RNAi, although HRDE-1 was previously thought to be absolutely required for RNAi inheritance. We propose that germ granules sort and shape the RNA pool, and that small RNA inheritance maintains this activity for multiple generations.
Keywords: non-Mendelian inheritance, germ granules, P granules, small RNAs, endogenous siRNAs, RNAi, meg-3/4, pptr-1, hrde-1, transgenerational inheritance
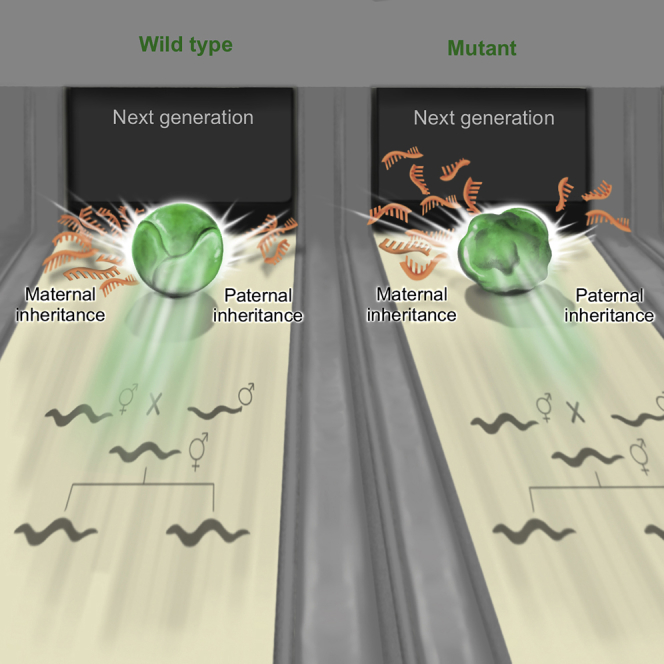
Graphical Abstract
Highlights
-
•
Disrupting germ granules leads to deviations from Mendelian inheritance
-
•
The gene silencing capacity of germ granule mutants changes transgenerationally
-
•
Paternal and maternal lineages transmit distinct germ granule-dependent small RNAs
-
•
Ancestral germ granules determine the small RNA pools of future generations
RNA-rich germ granules are highly conserved and required for germline functions. Lev et al. show that ancestral germ granules shape the gene silencing capacity and small RNA pools of future generations. Altering germ granules in paternal and maternal lineages generates distinct transgenerational dynamics that deviate from Mendelian inheritance.
Introduction
C. elegans worms synthesize small RNAs that can regulate gene expression and exert physiological changes across multiple generations [1, 2]. Environmental challenges can shape the pool of heritable germline endogenous small interfering RNAs (endo-siRNAs), leaving a mark that persists in the progeny [3, 4]. Similarly, exogeneous administration of double-strand RNA (dsRNA) triggers exogenous siRNA-mediated transgenerational silencing of cognate genes [5]. Typically, dsRNA-triggered heritable responses persist for 3–5 generations [6, 7], owing to a dedicated pathway that regulates the duration of each heritable response [7, 8]. Maintenance of RNAi over generations involves specialized argonaute proteins [9, 10, 11], amplification of the small RNAs by RNA-dependent RNA polymerases [12, 13] (RdRPs), and chromatin modifiers [8, 14, 15, 16]. Many of the factors that participate in transgenerational small RNA inheritance reside in germline-specific ribonucleoprotein complexes termed germ granules [17, 18, 19, 20, 21, 22, 23].
P granules are RNA-rich, cytoplasmic, perinuclear liquid-like condensates [24], which are maternally deposited to the oocyte [25, 26] and following fertilization are asymmetrically segregated to the P lineage designated to become the germline [27]. P granules are required for maintaining the identity of germ cells throughout development, by preventing ectopic expression of somatic genes [28, 29, 30]. P granules are localized adjacent to nuclear pores, to survey the expression of mRNA molecules exiting the nucleus [22, 31, 32]. Various P granule-associated proteins were found to be required for RNAi or for RNAi inheritance, including the P granule core-forming proteins PGL-1 [33, 34], MEG-3, and MEG-4 [20], and the VASA homolog GLH-1 [35]. Several argonaute proteins are also localized to P granules including PRG-1 [11], WAGO-1 [19], and CSR-1 [17].
Recently, P granules were found to be part of a tri-condensate assemblage that also includes the Mutator granule (or M granule) and a newly discovered Z granule [20]. The Z granules co-localize with P granules, but during the mitotic stage in the adult germline the two granules separate, while remaining adjacent to each other [20]. Z granules contain the WAGO-4 argonaute [20, 21] and the ZNFX-1 protein [20, 22] needed for proper small RNA amplification on targeted mRNAs and for balanced epigenetic inheritance [22]. Both proteins were shown to be required for RNAi inheritance [20, 21, 22].
The formation and stabilization of germ granules is a tightly regulated process [24]. PPTR-1 is a regulatory subunit of the conserved PP2A phosphatase holoenzyme [36] that stabilizes maternally deposited P granules in the early embryo, regulating the correct segregation of the granules to the germline blastomere across cell divisions [37]. In pptr-1 mutants, P granules are evenly distributed throughout the embryo, and are reduced drastically in number and size [37, 38]. PPTR-1 interacts with MEG-3 and MEG-4, which are intrinsically disordered serine-rich proteins that function redundantly to stabilize the condensed phase of P granules via binding of RNA and PGL-1 [39, 40]. In comparison to wild-type animals, meg-3/4 zygotes display an 89% loss of P granules [39].
In this manuscript, we examined the contribution of P and Z granules to RNAi inheritance. Surprisingly, we observed enhanced RNAi inheritance in pptr-1 and meg-3/4 germ granule-defective mutants. Our experiments revealed that pptr-1 mutants consistently display enhanced RNAi inheritance, while the RNAi inheritance capacity of meg-3/4 mutants is affected by history, as it is determined by the ancestral, rather than the current, meg-3/4 genotype. While P granules are known to be maternally contributed, we report that germ granule morphological changes are not inherited transgenerationally and therefore these entities do not carry the heritable memory. Instead, we propose that germ granules generate transgenerational changes via shaping of the heritable pool of small RNAs.
Results
Germ Granule Integrity Is Not Required for RNAi Inheritance
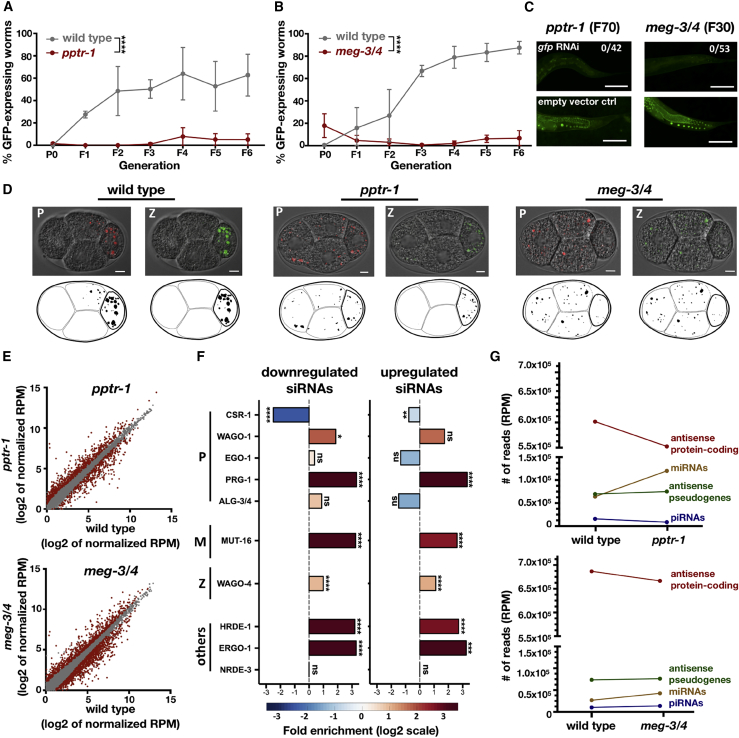
To study the role of germ granules in small RNA inheritance, we crossed different germ granule mutants to worms that contain a single-copy gfp driven by the mex-5 promoter in the germline. This gfp transgene is commonly used as a target for RNAi inheritance experiments [5, 7, 14, 35]. Surprisingly, when we tested pptr-1 and meg-3/4 mutants, we found that both strains exhibit enhanced RNAi inheritance. pptr-1 mutants displayed extremely strong heritable responses, persisting for more than 70 generations (Figures 1A–1C). Stronger RNAi inheritance in pptr-1 and meg-3/4 mutants was observed also when we used the endogenous oma-1 gene as a target (Figure S1). This was tested by crossing the mutants with the redundant and temperature-sensitive dominant lethal oma-1(zu405) allele. Heritable silencing of this allele is necessary for survival of the oma-1 mutant at the restrictive temperature [6]. After validating previous reports showing that pptr-1 and meg-3/4 mutants indeed display highly defective embryonic germ granules (Figures 1D and S1; Videos S1, S2, and S3), we conclude that germ granule integrity is not necessary for RNAi inheritance in the germline.
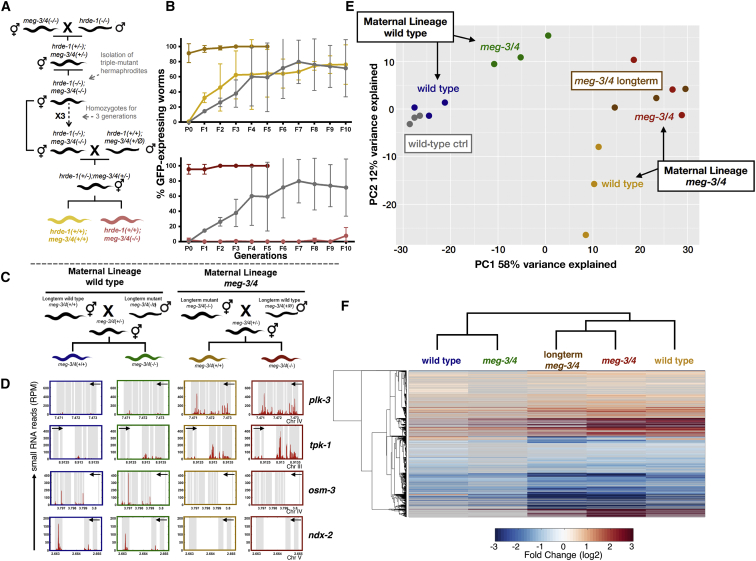
Figure 1.
Germ Granule Mutants Exhibit Enhanced RNAi Inheritance and Display Severe Mis-expression of Endogenous Small RNAs in the Germline
(A and B) Worms of the indicated genotypes containing a transgene expressing GFP in the germline (Pmex-5::gfp) were exposed to dsRNA complementary to gfp, to initiate gene silencing via RNAi. The percentage of worms expressing GFP (y axis) was assessed over generations (x axis). Shown are mean ± SD from three independent experiments. Mutant meg-3/4 worms (B) expressing Pmex-5::gfp were generated by crossing Pmex-5::gfp hermaphrodites with meg-3/4 males, and exposed to anti-gfp dsRNA two generations after homozygosity. Mutant pptr-1 worms (A) expressing Pmex-5::gfp were generated by crossing Pmex-5::gfp males with pptr-1 hermaphrodites, and exposed to anti-gfp dsRNA multiple generations after homozygosity. p values were determined via two-way ANOVA. ∗∗∗∗p < 10−4.
(C) Representative pictures of the germline in the descendants of mutated worms treated with gfp RNAi (up) or empty vector (down). The genotype and generation after RNAi initiation are indicated above the pictures. The number of gfp-expressing worms out of all examined worms is indicted in white. Scale bar, 80 μm.
(D) Representative pictures of embryos expressing pgl-1::rfp (red) and wago-4::gfp (green), and their corresponding binarized representations further used for analysis (bottom). The genotypes of the embryos are indicated above the pictures. Scale bar, 5 μm.
(E) Expression of small RNAs in the germline of pptr-1 and meg-3/4 mutants (y axis) compared to wild-type animals (x axis). Shown are the average expression values (log2 of RPM) of small RNAs targeting all genomic elements (each dot represents one annotation). Red dots, genomic elements with differential expression values of targeting small RNAs (analyzed via DESeq2, adjusted p < 0.1).
(F) X-fold enrichment and depletion values (log2, bar graphs and color-coded) for protein coding genes that displayed upregulated (left) or downregulated (right) expression levels of small RNA targeting them. Shown are results for genes with differential levels in both pptr-1 and meg-3/4 mutants compared to wild-type (see also Figure S3). We tested the enrichment for the list of genes differentially targeted by small RNAs against lists of genes known to be targeted by endogenous small RNAs of specific pathways (y axis; see STAR Methods for details). p values for enrichment were calculated using 10,000 random gene sets identical in size to the tested group (see STAR Methods for details). Enrichments were considered significant if p < 0.05. ∗∗∗∗p < 10−4, ∗∗p < 0.01, ∗p < 0.05; ns, p > 0.05.
(G) Global analysis of the various small RNA species in dissected germlines of pptr-1 and meg-3/4 mutants. Shown are the mean normalized total number of reads (RPM) aligned to the denoted small RNA species. See also Figures S1–S3, Data S1, and Videos S1, S2, and S3.
Wild type embryos expressing pgl-1::rfp (P granules) and wago-4::gfp (Z granules) were photographed via time-lapse imaging, at the 2-cell stage. Shown are maximal Z projections of the merged bright field, 488nm and 561nm channels.
Wild type embryos expressing pgl-1::rfp (P granules) and Pmex-5::gfp::h2b (chromatin) were photographed via time-lapse imaging, from the 2-cell to 4-cell stages. Shown are maximal Z projections of the merged bright field, 488nm and 561nm channels.
meg-3/4 mutant embryos expressing pgl-1::rfp (P granules) and Pmex-5::gfp::h2b (chromatin) were photographed via time-lapse imaging, from the 4-cell to 6-cell stages. Shown are maximal Z projections of the merged bright field, 488nm and 561nm channels.
To strengthen this conclusion, we examined the ability of pgl-1 mutants that we also crossed to the gfp transgene to inherit RNAi. pgl-1 mutants are also defective in P granule assembly [41]. Although these worms were reported multiple times to be resistant to germline RNAi [33, 34], we found that pgl-1 mutants are competent in RNAi inheritance (Figure S2; see more below). The Z granule argonaute WAGO-4 was also shown to be required for inheritance of dsRNA-derived exogenous siRNAs [20, 21]. Our results show that WAGO-4 is indeed required for RNAi inheritance, but only for initiation of the heritable RNAi response, in parents that are directly exposed to dsRNA. Specifically, wago-4(−/−) mutant progeny that derive from RNAi-exposed wago-4(+/−) parents can efficiently inherit RNAi for multiple generations (Figure S2). This last result shows that the consequences of the activity of WAGO-4 are preserved in further generations even in WAGO-4’s absence. In contrast, as was reported by others in the past [9, 10, 11, 14], we found that the nuclear argonaute HRDE-1 is required for maintenance of the heritable RNAi response (Figure S2).
PPTR-1 and MEG-3/4 Are Required for Maintaining Balanced Expression of Different Small RNA Pathways in the Germline
To understand how germ granules regulate small RNA levels, we sequenced small RNAs from dissected germlines of germ granule-defective meg-3/4 and pptr-1 mutants. We found that a large number of protein-coding genes (1,777 in meg-3/4 and 1,059 in pptr-1) were targeted by differential levels of cognate endo-siRNAs in the mutants compared to wild-type (DESeq2 adjusted p < 0.1; Figure 1E; Data S1). To focus our analysis on the specific effects of disruption of germ granules, we analyzed the set of genes that were targeted by differentially expressed small RNAs in both pptr-1 and meg-3/4 mutants (Figure S3A). We found a disruption in endo-siRNAs that bind the nuclear argonaute HRDE-1 (fold enrichment of 9.2× for downregulated small RNAs and 6.6× for upregulated small RNAs, p < 10−4; Figure 1F). Small RNAs that depend on or bind to proteins localized to germ granules were also substantially disturbed, such as endo-siRNAs associated with P granule proteins (the WAGO-1 [19] and the PRG-1 [42] argonautes), the M granule protein MUT-16 [43], and the Z granule argonaute WAGO-4 [21] (Figure 1F). Similar results were obtained when examining pptr-1 and meg-3/4 separately (Figure S3B). Previous studies have shown that different small RNA pathways compete over limiting biogenesis factors [7, 8, 44, 45, 46]. To understand why germ granule mutants display enhanced dsRNA-induced RNAi inheritance, we sequenced small RNAs from gonads of wild-type and pptr-1 mutants, six generations after exposure to anti-gfp dsRNA. We found more than 3-fold increase in the levels of exogenous-derived small RNAs targeting the gfp transgene in pptr-1 mutants (Figure S3C), while the global levels of endogenous small RNAs targeting protein-coding genes were decreased (Figure 1G). Altogether our data suggest that proteins that affect germ granule integrity maintain the balance between different endogenous and exogenous small RNA pathways.
The Function of meg-3/4 in the Ancestors Determines the Ability of the Progeny to Inherit RNAi in a Transgenerational Manner
We noticed that the ability of meg-3/4 double mutants to inherit RNAi was gradually lost after multiple generations, in homozygous mutants that derived from heterozygous parents (Figure S2B). Eventually, these mutants lost the ability to respond to RNAi. We observed the same gradual decline in RNAi responsiveness in pgl-1 mutants (Figure S2A), but not in pptr-1 mutants (Figure 1A). We reasoned that in previous studies researchers have found that meg-3/4 and pgl-1 mutants are refractory to RNAi [20, 33, 34] because mutants that have been homozygous for many generations have been examined.
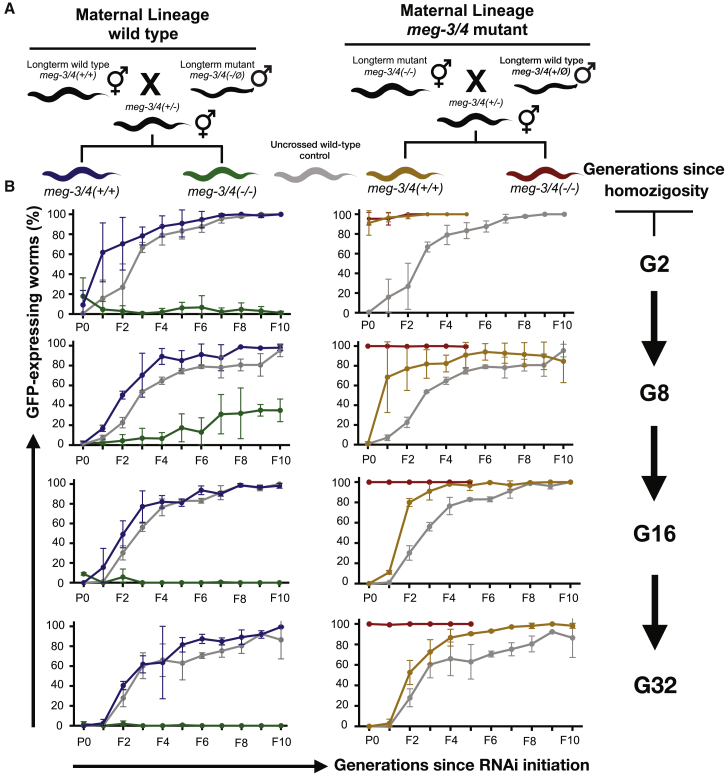
To study this intriguing transgenerational dynamic, we attempted to restore the capacity of meg-3/4 mutants to perform RNAi in two different ways: we crossed wild-type worms with RNAi-defective meg-3/4 mutants (homozygous for multiple generations), and since germ granules are maternally deposited, we introduced the mutated alleles via either the maternal or paternal germline (Figure 2A). We isolated F2 meg-3/4(−/−) homozygous mutants and F2 meg-3/4(+/+) wild-type animals and examined the capacity of their progeny to initiate RNAi inheritance responses (Figure 2B). As will be elaborated below, our results clearly demonstrate that (1) the ability of the progeny to inherit RNAi is determined by the genotype of the ancestors, (2) the maternal and paternal lineages have different contributions, and (3) the potency of the phenotypes transmitted from meg-3/4 mutants changes over generations. Unlike meg-3/4 mutants, pptr-1 mutants consistently showed enhanced inheritance of RNAi in all the experiments we performed, independently of the parent’s genotype (Figure S4).
Figure 2.
The Ancestral Germ Granule Function Determines the Ability of the Progeny to Inherit RNAi
(A) Schematic diagram depicting the crosses performed to determine the ancestral contribution to RNAi inheritance in the descendants. Long-term meg-3/4 mutants (hermaphrodites and males) were outcrossed using wild-type worms of the opposite sex. All worms contained a transgene expressing GFP in the germline (Pmex-5::gfp).
(B) Homozygote descendants of the crosses depicted in (A) were exposed to dsRNA complementary to gfp, to initiate gene silencing via RNAi. To test the transgenerational dynamics of the ability to respond to RNAi, naive descendants were exposed to dsRNA at different time points, 2, 8, 16, and 32 generations following homozygosity. The proportion of GFP-expressing worms (y axis, out of 40∼100 worms for each group and time point) was determined over ten generations (x axis) following exposure to RNAi. The colors depict the genotypes and lineages according to the scheme in (A). Shown are mean ± SD from two independent experiments (two independent ancestral crosses). In each row (initiation at different generations following homozygosity), all groups were tested side by side, and therefore the same values for the wild-type control groups are displayed on both left and right panels. Wild-type and meg-3/4(−/−) in the maternal wild-type lineage for the G2 generation (top left panel) are replicates from Figure 1B, for convenience of visualization purposes only. See also Figures S4 and S5.
We found that when the great grandmother was meg-3/4 mutant, the mutant progeny did not respond to RNAi at all (Figure 2B, right). Moreover, even the wild-type progeny that derived from such crosses lost their RNAi inheritance ability and regained their RNAi responsiveness only after multiple generations (Figure 2B, right). While the RNAi defect was stable in the mutants, we found that repetitive crossing of meg-3/4 mutant mothers to wild-type males improves the capacity of meg-3/4 mutants to respond to RNAi (Figure S5). This suggested that the male germline contributes to the heritable phenotype.
When the great grandfather was meg-3/4 mutant, we found that both mutants and wild-type great grandchildren were always responsive to RNAi, and this phenotype lasted many generations (>8) in all experiments (Figure 2B, left; Figure S2B).
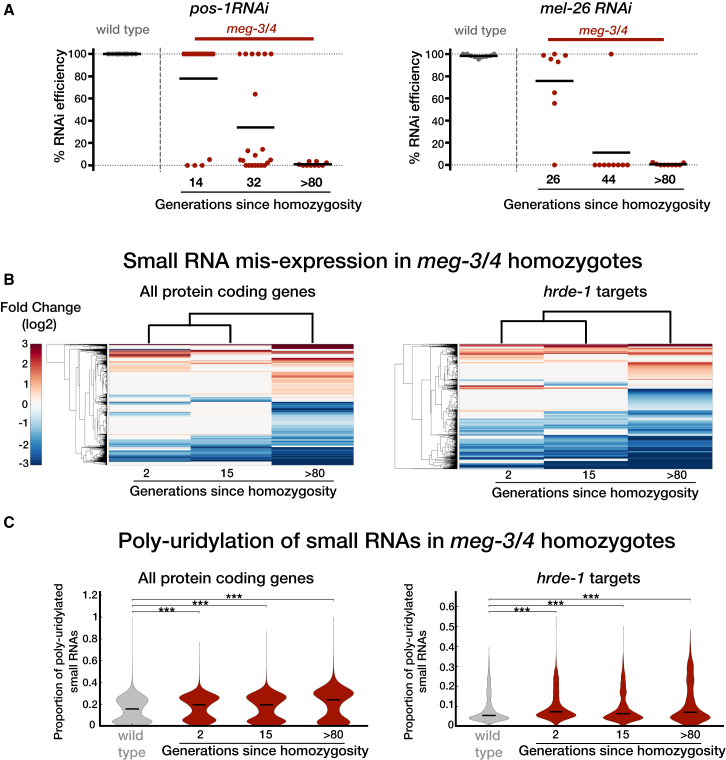
In some experiments, however, the descendants of meg-3/4 fathers remained responsive to RNAi for more generations (Figure 2B, left panel as described above). We hypothesized that even these descendants gradually lose their RNAi responsiveness, but that the assay is not sensitive enough to detect it. It is known that transgenes can be very sensitive to transgenerational RNAi [15, 47, 48] and therefore to study these dynamics further we tested RNAi responses against two endogenous germline-expressed target genes (pos-1 and mel-26). These experiments revealed that these meg-3/4 mutants are indeed RNAi competent for a few generations but gradually lose the ability to respond to RNAi (Figure 3A). Moreover, this transgenerational deterioration is reflected also in the small RNA pools of these animals: we dissected gonads of meg-3/4 mutants (that inherited the mutations from the paternal lineage) and sequenced small RNAs at three time points: 2, 15, and >80 generations after homozygosity. These data show dramatic genome-wide accumulative mis-expression of small RNAs that target protein-coding genes (Figure 3B). Specifically, we found transgenerational accumulation of mis-expressed small RNAs that are associated with the HRDE-1 (Figure 3B) and WAGO-4 pathways (Figure S6). We also found that the levels of poly-uridylation of small RNAs that target protein-coding genes increased over generations (Figures 3C and S6). Moreover, even HRDE-1-small RNAs, which are typically not poly-uridylated [49], exhibited accumulative poly-uridylation (Figure 3C). Previous studies suggested that poly-uridylation is involved in sorting of small RNAs to different argonautes [21, 49] and also in degradation [50] of small RNAs.
Figure 3.
meg-3/4 Mutants Accumulate Defects in Germline RNAi Initiation and Mis-expression of Endo-siRNAs over Generations
(A) The transgenerational dynamics of the ability of meg-3/4 mutants to respond to RNAi targeting endogenous germline genes. Wild-type (gray) and meg-3/4 (red) worm homozygotes for the indicated amount of generations (x axis) were allowed to lay eggs on plates containing dsRNA-producing bacteria targeting the germline-expressed genes pos-1 (left) and mel-26 (right). Shown are the percentages of eggs that did not hatch into larvae (y axis), indicating the penetrance of the RNAi effect. Each dot represents one tested plate (biological replicate) and bars represent the mean. Each group was tested in two independent experiments.
(B) Two-way hierarchical clustering of the different examined biological groups and genes targeted by significantly differential levels of small RNAs (compared to wild-type). Small RNA samples were obtained from dissected germlines of meg-3/4 mutants that were homozygote for 2, 15, or >80 generations (x axis). Clustering of small RNAs targeting all protein-coding genes (left panel) and small RNAs targeting genes known to be targeted by small RNAs associated with the HRDE-1 argonaute (right panel [9]) is presented. Each row represents one gene. Only genes targeted by significantly differential levels of small RNAs in at least one sample were included in the analysis (analyzed with DEseq2, adjusted p < 0.1). Each line is color-coded according to the fold change in expression. Lines depicting no significant differential expression appear in gray.
(C) An analysis of poly-uridylation of small RNAs in meg-3/4 mutants across generations. The distribution of the average fractions of poly-uridylated small RNAs against individual genes is shown for each of the examined samples. Data for small RNA samples from extracted germlines of wild-type and meg-3/4 mutants that were homozygote for 2, 15, or >80 generations (x axis) are shown. The fraction of poly-uridylated small RNAs was calculated as the number of reads with untemplated 3′Us out of the total aligning reads against a specific gene (only genes with ≥5 RPM in at least one sample were included). ∗∗∗p < 10−3, ∗∗∗p < 10−4, Wilcoxon rank-sum test.
For the >80 generations group in (B) and (C), data from two replicates were analyzed and a third replicate was left out since it displayed substantially less depth (3 × 105 reads compared to >107 reads in all other samples). See also Figure S6.
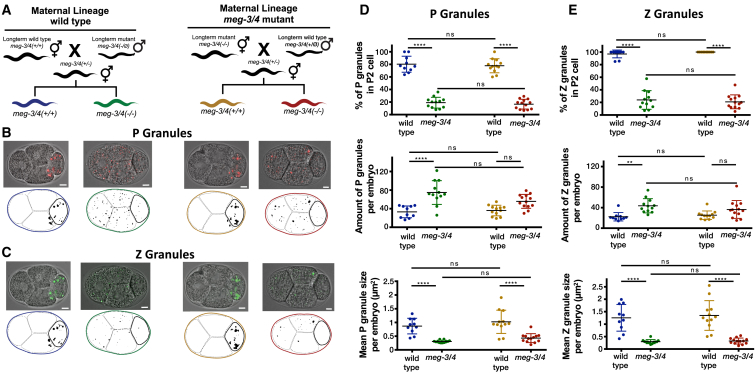
The Integrity of the Progeny’s Germ Granules Is Not Affected by the Ancestral Genotype
As described above, we found differences in the RNAi responsiveness of animals derived from paternal and maternal meg-3/4 mutant lineages. Germ granules are maternally deposited [25, 26], and therefore we tested if inheritance of the germ granules themselves could explain these differences. We performed similar crosses (see scheme in Figure 4A) and examined if the morphology, abundance, and distribution of the germ granules changed in the different isolated progeny. In contrast to the RNAi inheritance phenotype of the progeny, which depended on the ancestors’ meg-3/4 genotype, wild-type progeny had normal germ granules and meg-3/4 progeny had defective germ granules (Figures 4B–4E). Moreover, we found that the P granule characteristics are similar in both RNAi responsive and non-responsive progeny of the same lineage, and the levels of silencing did not correlate with the P granules’ morphology (Figure S7). In summary, we could not detect significant differences in the characteristics of the germ granules of G2-G5 isogenic animals that derived from wild-type or mutant ancestries.
Figure 4.
The Morphology of the Germ Granules in the Progeny Is Not Affected by the Ancestral Genotype
(A) Genotypes and maternal lineages of different tested groups are encoded in colors similarly to Figure 2.
(B) Representative pictures of embryos expressing pgl-1::rfp at the 4-cell stage with fluorescent P granules and their corresponding binarized representations further used for analysis (bottom); the P2 cell (germline lineage) is emphasized with a thicker line. Scale bar, 5 μm.
(C) Representative pictures of embryos expressing wago-4::gfp at the 4-cell stage with fluorescent Z granules and their corresponding binarized representations further used for analysis (bottom); the P2 cell (germline lineage) is emphasized with a thicker line. Scale bar, 5 μm.
(D and E) Characterization of germ granules in embryos at the 4-cell stage. Each dot represents one analyzed embryo. Data shown for P granules (D) and Z granules (E). Top: percentage of granules in the P2 cell out of total granules. Middle: amount of granules per embryo. Bottom: mean size of granules per embryo. Bars represent mean ± SD. p values were determined via two-way ANOVA with Tukey post hoc correction for multiple comparisons. ∗∗∗∗p < 10−4, ∗∗p < 0.01; ns, p > 0.05. See also Figures S1 and S7.
Endogenous Small RNAs Mediate the Transgenerational Defects of meg-3/4 Mutants
Germ granules are not deposited in the sperm; however, we observed that RNAi inheritance defects can transmit via meg-3/4(−/Ø) males to wild-type progeny. The defects that were transmitted via the paternal lineage lasted for multiple generations, but were weaker in comparison to defects transmitted via the maternal lineage (Figure 2B, left). Unlike germ granules, small RNAs are inherited from both the sperm and the oocyte, and therefore we examined whether small RNAs could mediate the transgenerational inheritance of the RNAi defects. The nuclear germline argonaute HRDE-1 is necessary for the inheritance of many endo-siRNAs and dsRNA-derived small RNAs. Small RNA sequencing data from pptr-1 and meg-3/4 mutants indicated that HRDE-1-associated small RNAs were disturbed (Figures 1F, 3B, and S3B). We reasoned that inheritance of endogenous small RNAs via the HRDE-1 pathway may transmit the defective RNAi phenotype to the descendants of meg-3/4 hermaphrodites. To test this possibility, we crossed meg-3/4 mutant hermaphrodites with hrde-1 mutant males, isolated meg-3/4;hrde-1 triple mutant progeny, and 3 generations later re-introduced the functional hrde-1 allele by crossing with wild-type males (see scheme in Figure 5A). We found that removing and restoring the functional hrde-1 allele erased the defects inherited from meg-3/4 mutant hermaphrodites, rescuing the ability of the progeny to inherit RNAi (Figure 5B). Multiple crosses with wild-type males also affect RNAi resistance, albeit to a lesser extent, independently of HRDE-1 function (Figure S5). In summary, we posit that transgenerational effects that result from defective ancestral germ granules are inherited by HRDE-1-dependent endogenous small RNAs.
Figure 5.
Endogenous Small RNAs Mediate the Transgenerational Defects of Germ Granule Mutants
(A) Schematic diagrams depicting the crosses performed to obtain the data appearing in (B). Left: hermaphrodites mutated in the meg-3/4 genes for >80 generations were crossed with hrde-1(−/−) males. Triple mutant F2 progeny of this cross were isolated and maintained for three generations prior to crossing with wild-type males. Wild-type (light yellow) and meg-3/4 (light red) F2 progeny resulting from this cross were isolated and tested for gfp RNAi on the second generation of homozygosity.
(B) The percentage of GFP-expressing worms (y axis) over generations (x axis) after exposure to gfp dsRNA in descendants of worms where hrde-1 was erased for three generations. The colors depict the genotype and lineages according to scheme in (A). Both groups as well as uncrossed wild-type controls (gray) were tested side-by-side, and therefore the same wild-type data appear in both panels. Shown are mean ± SD from two independent experiments. Data for the groups with no hrde-1 history (dark yellow and dark red) are replicates from Figure 2B, for convenience of visualization purposes only. Reset versus non-reset groups in both panels display statistically significant differences in silencing (p < 10−4) based on two-way ANOVA with Bonferroni post hoc correction for multiple analysis.
(C) Crosses were performed similarly to the scheme in Figure 2A.
(D) Distributions of normalized small RNA counts (y axis) as function of genomic location (x axis) of small RNAs targeting the genes indicated on the right. Exons are represented as gray and black arrows pointing to the direction of transcription. Small RNA libraries were produced from extracted germlines. The genotype and lineage of the tested animals are color-coded according to the scheme in (C).
(E) A principal component analysis (PCA) projection of germline small RNA samples based on normalized small RNA counts against annotated genes. Each point represents one independent replicate. Corresponding genotype and lineage are indicated and color-coded.
(F) Two-way hierarchical clustering of the different biological groups and genes targeted by significantly differential levels of small RNAs. The clustering is based on the differentially expressed small RNAs compared to wild-type control samples. Each row represents one gene. Only genes targeted by significantly differential levels of small RNAs in at least one sample were included in the analysis (analyzed with DEseq2, adjusted p < 0.1). Each line is color-coded by the fold change in expression, where cells depicting no significant differential expression appear in gray. Biological groups are color-coded according to the scheme in (C) and appear above the columns.
Next, we sequenced small RNAs from the germline of wild-type and meg-3/4 mutants derived from either paternal or maternal lineages (two generations after homozygosity; see scheme in Figure 5C) and compared their small RNAs to wild-type and to meg-3/4 mutants that had been homozygous for many generations (>80G). Principle component analysis (PCA) of the global small RNA expression levels shows that wild-type and >80G meg-3/4 mutant worms exhibit profoundly distinct small RNA pools. Strikingly, the small RNA expression patterns of the recently isolated wild-type and meg-3/4 worms grouped together according to their maternal ancestry (the meg-3/4 genotype of the great grandmother; Figures 5D–5F). The progeny’s own meg-3/4 genotype had only minor contribution to the samples’ grouping (Figure 5E). Hierarchical clustering analysis of the differentially expressed small RNAs also showed that the recently isolated wild-type and mutant worms cluster according to their maternal ancestry (Figure 5F).
These results further suggest that inheritance of mis-expressed endogenous small RNAs, rather than maternal contribution of defective granules, transmits across generations the abnormal RNAi phenotype via the meg-3/4 mutant maternal lineage.
Disrupting the Germ Granules Allows HRDE-1-Independent Inheritance of RNAi
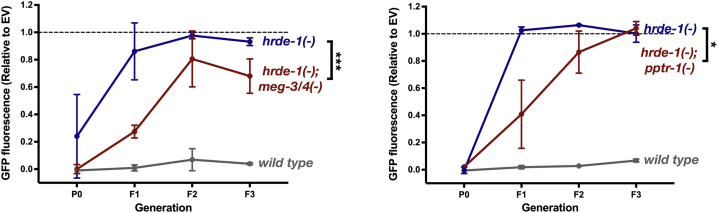
The HRDE-1 argonaute was shown in numerous studies to be absolutely essential for dsRNA-induced RNAi inheritance [9, 10, 11, 14, 51, 52]. As we found strong RNAi inheritance in germ granule mutants despite severe defects in HRDE-1-associated small RNAs, we hypothesized that when the germ granules are defective, RNAi might be inherited via alternative routes. To test this, we performed RNAi inheritance experiments in hrde-1;pptr-1 double mutants and hrde-1;meg-3/4 triple mutants. To enhance the sensitivity of the anti-gfp RNAi silencing assay, we quantified the exact levels of GFP expression (as was conducted in [8, 15, 21, 53]). We found that hrde-1;meg-3/4 triple mutants, and to a lesser extent also hrde-1;pptr-1 double mutants, can weakly inherit RNAi (Figure 6). Together, our results suggest the intriguing possibility that germ granules function in sorting of heritable small RNAs, and that changes in small RNA assortment can persist for multiple generations with extensive consequences to gene regulation in the progeny.
Figure 6.
Mutants with Defective Germ Granules Inherit RNAi in the Absence of HRDE-1
GFP fluorescence (y axis) of adult worms with the indicated genotype over generations (x axis), following exposure to gfp dsRNA. Fluorescence in each group was normalized to the mean fluorescence value of the corresponding isogenic control worms originally exposed to empty vector. Shown are mean ± SD of two independent pptr-1 experiments (left) and three independent meg-3/4 experiments (right), in which 25∼90 animals were analyzed for each group and generation. Tested strains derived from crosses of hrde-1 hermaphrodites with pptr-1 or meg-3/4 males (all with a Pmex-5::gfp transgene in the background) and the experiments were performed up to 6 generations since homozygosity. p values were determined via two-way ANOVA with Bonferroni post hoc correction for multiple comparisons to hrde-1. ∗∗∗p < 0.001, ∗p < 0.05.
Discussion
Close to a century ago it was observed that the cytoplasm contains droplets [54]. These droplets were later found to be RNA-rich granules, and characterized in the germline of more than 80 species [55]. Ingenious studies conducted over the years have elucidated the many crucial roles that germ granules play, and especially the intriguing ways by which these granules shape the transcriptome. Here we show that the influence of germ granule proteins on RNAi and small RNA inheritance changes dynamically over the course of multiple generations, shedding new light on germ cell biology.
Our study revises the current model according to which intact germ granule composition is essential for RNAi and small RNA inheritance. We demonstrate that mutants in which the germ granules are massively disrupted can exhibit potent RNAi inheritance (Figure 1). We assume that the transgenerationally dynamic nature of RNAi in germ granule mutants has not been noted before, since in the past researchers have assayed mutants that have been homozygous for many generations. We found that the ability of the progeny to inherit RNAi is determined by the genotype of the ancestors, and that the maternal and paternal lineages have different contributions (Figure 2). We discovered that the heritable effects that accumulate due to disruption of germ granule genes can last for multiple generations (Figures 2 and 3).
Both heritable small RNAs and maternally contributed germ granules could potentially mediate the inheritance of the RNAi defects to the progeny of meg-3/4 mutants. Several lines of evidence suggest that transgenerational inheritance of small RNAs mediates the heritable defects: (1) the integrity of germ granules in the progeny is unaffected by changes to the ancestors’ germ granules (Figure 4); (2) the P granule’s morphology and distribution does not correlate with RNAi capacity (Figure S7); (3) some of the effects can transmit through the paternal lineage, although germ granules are only inherited from the mother (Figure 2, left); (4) the small RNA pools of the progeny are more closely related to the small RNA pools of the great grandmothers regardless of the progeny’s genotype (Figure 5); and (5) removing and then restoring the HRDE-1 argonaute, important for small RNA inheritance, re-sets the heritable effects of germ granule mutants (Figure 5). The dynamic interactions between the germ granules and heritable small RNAs determine eventually how gene regulation would be inherited across generations [2, 56].
More generally, our results show that genetically identical animals can have strikingly different phenotypes because of their ancestry. First, we provide evidence that recently outcrossed animals differ from animals that were outcrossed many generations ago due to epigenetic effects. Second, outcrossing using wild-type males or hermaphrodites generates different long-term phenotypes.
Overall, pptr-1 and meg-3/4 mutants had similar effects on RNAi inheritance, small RNA expression, and germ granule morphology. However, the mutants differed in several aspects: while meg-3/4 mutants eventually become refractory to RNAi inheritance (after many generations), pptr-1 mutants are consistently responsive to RNAi, and exhibit enhanced RNAi inheritance. In contrast to meg-3/4 mutants, the RNAi capacity of the pptr-1 mutants is not affected by the maternal ancestry (Figure S4). In addition, pptr-1 mutants exhibited greater reduction in the levels of small RNAs that target protein coding genes (Figure 1G). These differences can stem from the fact that PPTR-1, a subunit of the PP2A phosphatase holoenzyme, affects many other targets (PP2A has ∼460 putative targets in C. elegans [57]). Additionally, we noticed that the Z granules of pptr-1 and meg-3/4 mutants differ. Whereas in meg-3/4 mutants the Z granules fail to segregate toward the P2 cell, these granules segregate normally in pptr-1 mutants (although the Z granules are smaller in both cases; Figures 1, 4, and S1B).
It is very challenging to account for non-germ-granule-related or indirect functions of the proteins examined in this work. For example, it is possible that these proteins indirectly regulate other genes that function in RNAi inheritance, even genes that encode for proteins that reside outside of granules. Interestingly, we found that genes that affect different epigenetic processes [7] are targeted by small RNAs in meg-3/4 mutants (38/285 epigenetic genes, 3.8× fold enrichment, p < 0.00001). Similarly, we cannot rule out the possibility that in the above-mentioned mutants, the typical germ granule proteins that we assayed, PGL-1 and WAGO-4 (used also as germ granule markers in [20, 37, 39]), are replaced by other unknown components that have different functions. Nevertheless, our results show that the integrity of the germ granules is not required for RNAi inheritance, and that mutating different genes required for the integrity of the germ granules leads to similar effects on endogenous small RNA expression.
In summary, we suggest a new heritable small RNA-based model to account for the function of germ granules in maintaining the balanced expression of different endogenous small RNA species. The imbalance in endogenous small RNAs in germ granule mutants can result in abnormal exogenous RNAi responses, characterized by unusual intensity, duration, and inheritance of small RNAs via alternative routes (independently of HRDE-1). In meg-3/4 mutants the imbalance exacerbates across generations and eventually hinders novel exogenous dsRNA-induced RNAi responses (see Figure 3 and model in Figure 7). Poly-uridylation of small RNAs by the germ granule localized protein CDE-1 was previously suggested to regulate the stability of small RNAs [50] or their sorting [21] to different argonautes. Interestingly, we detected an aggravating ectopic poly-uridylation of small RNAs in meg-3/4 mutants (Figure 3C) that correlates with their inability to respond to dsRNA. Previous studies have shown that the morphology of the germ granules is affected by environmental changes (for instance, temperature [58, 59]), and also that temperature shifts trigger changes in heritable small RNAs [3, 60]. Therefore, germ granules could be important mediators of transgenerationally small RNA-based heritable stress responses.
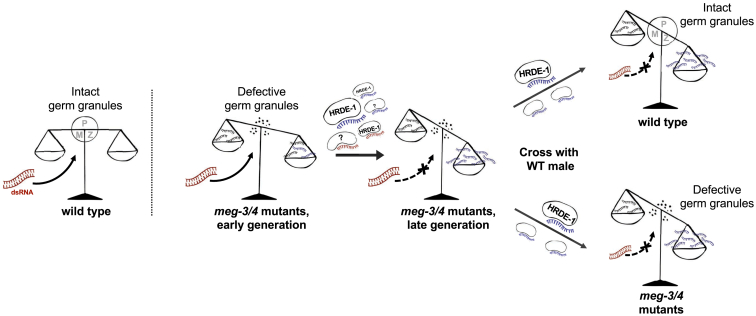
Figure 7.
A Schematic Model Describing the Roles of Germ Granules in Small RNA Inheritance
Our results suggest that germ granules shape the heritable small RNA pool. Mutants in the meg-3/4 core germ granule proteins accumulate small RNA defects over generations, resulting in abnormal processing of exogenous RNAi responses. The RNAi defects are inherited from meg-3/4 (−/−) maternal lineages to the progeny via endogenous HRDE-1-dependent small RNAs, and not by contribution of the germ granules themselves. In addition, exogenous RNAi-derived small RNAs can be inherited transgenerationally through alternative routes in meg-3/4 mutants (independent of HRDE-1). The scale represents the balance between different small RNA populations. The circle in the middle of the scales represents intact germ granules (the P, Z, and M granules) that “keep the scale balanced.” The genotype of the animal is shown below each corresponding scale. The presence of a non-interrupted black arrow originating from dsRNA (represented in red) signifies that the animal can respond to exogenous RNAi. Dotted lines that crossed out by an “X” symbol indicate that this animal lost its ability to respond to exogenous RNAi. The arrows between scales represent transitions between generations. Red and blue single-stranded RNA molecules depict exogenous RNAi-derived siRNAs and endogenous siRNAs, respectively.
Finally, prior to Mendel’s realization that traits are transmitted as discrete units of information, it was thought that parents’ heritable materials mix, as liquids do. According to the now obsolete theory known as “Blending Inheritance,” the heritable factors “dilute” in the progeny [61]. We find that disruption of maternally deposited liquid condensates [24] leads to heritable changes that dilute eventually. While the modern mechanistic explanation is obviously very different from that used before the re-discovery of Mendel, we propose that the properties of RNA-carrying liquid-like granules lead to deviations from Mendelian inheritance.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trizol Reagent | Life Technologies | 15596026 |
| Ultra Pure Glycogen | ThermoFisher | 10814010 |
| Mouth aspirator and Microcapillary tubes | Sigma | P0674 |
| Levamisole hydrochloride | Sigma | L0380000 |
| Phenol Chloroform Isoamyl | Sigma | P2069 |
| Heavy Phase Lock tube | QuantaBio | 23028330 |
| miRNeasy Mini kit | QIAGEN | 217004 |
| RNA 5′ Polyphosphatase | Epicenter | RP8092H |
| NEBNext Multiplex Small RNA Library Prep Set for Illumina | New England Biolabs | E7300 |
| TapeStation screen tapes | Agilent | 5067-5582; 5067-5588 |
| E-Gel 4% agarose | Life Technologies | G401004 |
| Deposited Data | ||
| Small RNA Seq data | This study | GEO: GSE128112 |
| Experimental Models: Organisms/Strains | ||
| mex-5::gfp::h2b::tbb-2(mjIs134 II) | Eric Miska lab | SX1263 |
| pptr-1(tm3103) | CGC | JH2787 |
| pptr-1(tm3103); mex-5::gfp::h2b::tbb-2 | This study | BFF39 |
| meg-3(tm4259);meg-4(ax2026) | CGC | JH3225 |
| meg-3(tm4259);meg-4(ax2026); mex-5::gfp::h2b::tbb-2 | This study | BFF40 |
| oma-1(zu405) | CGC | TX20 |
| oma-1(zu405); pptr-1(tm3103) | This Study | BFF41 |
| pgl-1(bn101) | CGC | SS579 |
| pgl-1(bn101); mex-5::gfp::h2b::tbb-2 | This study | BFF42 |
| wago-4(tm1019) | Scott Kennedy Lab | YY1083 |
| wago-4(tm1019); mex-5::gfp::h2b::tbb-2 | This Study | BFF43 |
| pgl-1::3xflag::tagrfp(gg547) | Scott Kennedy Lab | YY967 |
| wago-4(tor117[wago-4::gfp::flag II]) | This study | JMC223 |
| pgl-1::3xflag::tagrfp(gg547); wago-4(tor117[wago-4::gfp::flag II]) | This study | BFF44 |
| meg-3(tm4259);meg-4(ax2026); wago-4(tor117[wago-4::gfp::flag II]); pgl-1::3xflag::tagrfp(gg547) | This study | BFF45 |
| pptr-1(tm3103); pgl-1::3xflag::tagrfp(gg547); wago-4(tor117[wago-4::gfp::flag II]) | This Study | BFF46 |
| pgl-1::3xflag::tagrfp(gg547); mex-5::gfp::h2b::tbb-2 | This Study | BFF47 |
| hrde-1(tm1200) | Scott Kennedy | YY538 |
| hrde-1(tm1200);mex-5::gfp::h2b::tbb-2 | This Study | BFF48 |
| hrde-1(tm1200);meg-3(tm4259);meg-4(ax2026); mex-5::gfp::h2b::tbb-2 | This study | BFF49 |
| oma-1(zu405);meg-3(tm4259);meg-4(ax2026) | This study | BFF50 |
| hrde-1(tm1200);pptr-1(tm3103); mex-5::gfp::h2b::tbb-2 | This study | BFF51 |
| meg-3(tm4259);meg-4(ax2026);pgl-1::3xflag::tagrfp(gg547); mex-5::gfp::h2b::tbb-2 | This study | BFF52 |
| Software and Algorithms | ||
| FastQC | [62] | N/A |
| Cutadapt | [63] | N/A |
| Shortstack | [64] | N/A |
| HTSeq count | [65] | N/A |
| R Deseq2 | [66] | N/A |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Oded Rechavi (odedrechavi@gmail.com). Worm strains generated in this study are available upon request.
Experimental Model and Subject Details
Cultivation of worms
All strains were maintained according to standard methods [67]. Unless noted otherwise, we performed all experiments at 20 degrees. Strains with the oma-1(zu405) temperature-sensitive allele were maintained at 15 degrees, and transferred to 20 degrees (restrictive temperature) for the relevant RNAi experiments. All the strains we used are listed in the Key Resources Table.
Method Details
RNAi experiments
HT115 E. coli bacteria bearing the dsRNA-expression bacteria and empty-vector control bacteria were grown overnight in LB medium supplemented with 0.25 μg/mL of Carbenicillin. Bacteria were seeded onto RNAi plates: NGM plates supplemented with isopropyl β-D-1- thiogalactopyranoside (IPTG, 1 mM) and Carbenicillin (0.25 μg/mL). A day later, ∼10 adult hermaphrodites were placed on the plates, and were allowed to lay eggs for ∼4 hours. The progeny laid on this occasion was considered the P0 treated generation. For anti-gfp RNAi inheritance experiments, adult P0 young worms (40∼120 per group) were collected for imaging, and for production of the next generation via synchronized egg-laying. For all generations (other than the P0 treated generation), synchronization was performed on the same day of imaging (4 days after egg-laying), by allowing 20 adult worms of each tested group (x2 plates) to lay eggs for ∼2 hours on NGM plates seeded with standard OP50 bacteria. For anti-oma-1 RNAi inheritance experiments, in each generation twelve individual larval (L4) staged worms were placed in individual wells of a 12-well plate. Four days later the number of fertile worms was assessed (at least one progeny) and twelve individual L4 progeny worms were chosen from the most fertile well to continue to the next generation. For anti-pos-1 and anti-mel-26 RNAi experiments, larval (L4) staged worms were placed on RNAi plates. From the resulting adult progeny, individual adults were isolated to fresh RNAi plates, one adult per plate. These plates were assayed two days later.
Images of the worms were collected using a BX63 Olympus microscope with x10 magnification, and were used to evaluate GFP expression.
Granules imaging and analysis
Embryos were extracted from young (day 1) adult hermaphrodites. Embryos at the 2-cell stage were chosen and immediately mounted on freshly made 3% agarose pads for imaging. All imaging was carried at 20°C on a Nikon Ti-2 eclipse microscope equipped with a 100X CFI Plan-Apo 1.45 NA objective (Nikon, Tokyo, Japan) and CSU-W1 spinning-disk confocal head (Yokogawa Corporation, Tokyo, Japan). Embryo samples were excited using 488 and 561 nm DPSS-Laser (Gataca, France) and images acquired using Prime95B sCMOS camera (Photometrics, Tucson, AZ). Image acquisition was controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). We photographed 4 cell stage embryos, using ∼30 z stacks per embryo with three channels: Bright field, 488 and 561.
Analysis of granules was carried out using the imageJ2 open source software. First, we split the channels and then merged the z stakes using the “Maximal intensity z projection” function. Next, we binarized the fluorescent signal using automated threshold using the “Maximal entropy” method. In the next step we used the “Analyze particles” function to automatically locate the granule foci. Finally, we measured the number and size of the located granule foci. To calculate the percentage of granules in the P2 cell, we manually defined the P2 and embryo regions of interest. Next, we re-applied the “Analyze particles” function to count number of foci in the P2 cell and the whole embryo regions. Foci < 0.1 μm2 were excluded from the analysis of P granules.
Small RNA sequencing
Hermaphrodites were collected on the first day of adulthood, washed 4 times in M9 buffer, and transferred in a microscope slide with egg buffer (1M HEPES, 5M NaCl, 1M MgCl2, 1M CaCl2, 1M KCl and 20% tween-20) containing 2mM levamisole. Worms were cut right under the pharynx with a gauge needle, and the evading gonads separated from the body. Gonads were collected from the slide into an Eppendorf tube on ice prior the addition of 300 μL Trizol (Life Technologies). For RNA extraction from gonads of meg-3/4 and corresponding wild-type samples the miRNeasy Mini kit (QIAGEN) kit was used. To extract RNA from gonads of pptr-1 mutants and corresponding wild-type samples the following phenol-choloroform protocol was used: 60 μL of chloroform was added to the samples, which were then transferred to a pre-spun Heavy Phase Lock tube and centrifuged at 16,000 g for 12 min at 4C. We transferred the aqueous phase to a new Heavy Phase Lock tube, and added 1:1 Phenol:Chloroform:Isoamyl Alcohol, before a centrifugation round at 16,000 g for 15 min at RT. The aqueous phase was transferred to an eppendorf tube, to which we added 1:1 pure ethanol and 20 μg Glycogen (Thermo Fisher), and let incubate overnight in −20C. Samples were then centrifuged at 16,000 g for 30 min at 4C, then washed twice by removing the supernatant and adding ice-cold 70% ethanol. After the last wash and removal of ethanol, the samples were air-dried for 6 min, and resuspended in 10 μL RNase-free water. 150 ng of each sample was treated with RNA 5′ polyphosphatase (Epicenter) to ensure small RNA capture independently of 5′ phosphorylation status. Libraries were prepared using NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs) according to the manufacturer’s protocol. We measured the samples concentration via Qubit, and tested their quality on an Agilent 2200 TapeStation. Samples were then pooled together and run on a 4% agarose E-Gel (Life Technologies), and the 140-160 nt length bands were excised and purified using MiniElute Gel Extraction Kit.Samples were tested for their quality and concertation on an Agilent 2200 TapeStation and then pooled together. Pools were separated on a 4% agarose E-Gel (Life Technologies) and the 140–160 nt length bands were excised and purified using MiniElute Gel Extraction kit (QIAGEN). Libraries were run on an Illumina NextSeq500 sequencer.
Quantification and Statistical Analysis
Analysis of GFP silencing
For photographing of the worms ∼60 animals were mounted on a 2% agarose slides and paralyzed in a drop of M9 with 0.05% tetramisole. The worms were photographed with 10x objective using a BX63 Olympus microscope (exposure time of 200 ms and gain of 2). Analysis of the photographed worms was done using Image2 software.
For binary GFP silencing analysis, we manually scored the photographs for the numbers of worms having or lacking any observable germline GFP signal.
For quantification of GFP fluorescence levels, we manually defined in each worm the area of the oocyte and three background regions. Next we calculated a CTCF value as follows: CTCF = Integrated density values of the oocyte X – (area of measured oocyte ∗ mean fluorescence of background regions). We normalized the CTCF value to the average CTCF value obtained from photographs of control animals of the same genotype, generation and age which were fed on control plates.
Quantification of anti-oma-1 RNAi experiments
Each generation we counted the fraction of worms (out of 12) that had progeny.
Quantification of anti-pos-1 and anti-mel-26 experiments
The number of hatched larva and eggs were counted per assayed RNAi-treated worm, and the percentage of hatched larva was calculated.
Small-RNA Seq analysis
Illumina fastq output files were assessed for quality with FastQC [62]. Cutadapt [64] was then used for adaptor removal: “cutadapt -m 15–discard-untrimmed -a [adaptor sequence] input.fastq” Clipped reads were aligned to the ce11 assembly of C. elegans genome using Shortstack [63]: “–mismatches 0–readfile input.fq genome_reference.fa”.
Reads were filtered to keep reads of 20 to 23 in length. Next, we counted reads aligning in an anti-sense orientation to genes based on the corresponding Ensembl .gff file, via the HTseq [65]: “-m HTSeq.scripts.count–stranded=reverse–mode=intersection-nonempty input.sam GENES.gff.” We then used the HTseq output file as an input for DESeq2 [66]. We defined differentially expressed genes using cutoff of adjusted p value < 0.1. Full DESeq2 results comparing pptr-1 and meg-3/4 mutants to wild-type are available in Data S1.
Bioinformatic gene enrichment analysis
The enrichment values denote the ratio between the observed representation of a specific gene set within a defined differentially expressed genes group, to the expected one, i.e., the representation of the examined gene set among all protein-coding genes in C. elegans. We performed the analysis on 8 gene sets: (1) 1587 targets of HRDE-1 endo-siRNAs [9], (2) 4191 targets of CSR-1 [17], (3) MUT-16-dependent endo-siRNAs [43], (4) WAGO-4 targets [21], (5) EGO-1-dependent endo-siRNAs [68], (6) WAGO-1 targets [19], (7) ERGO-1-dependent endo-siRNAs, (8) ALG-3/4 targets [18]. The putative PRG-1 targets were defined as genes for which, in at least one transcript, the ratio of the # 22G-RNA reads at piRNA target sites between wild-type to prg-1 is at least 2 (linear scale). (10) NRDE-3 targets [69].
Enrichment of a given gene set i in pptr-1-dependent endo-siRNAs was calculated according to the formula:
We calculated p values using 10,000 random gene groups identical in size to that of genes targeted by pptr-1-dependent endo-siRNAs.
Bioinformatics analysis of different small RNA species
Briefly, reads were filtered to sizes of 20-23 in length. Next, we counted reads using HTseq in the sense, and again in the anti-sense orientations based on ce11 Ensembl .gff file. Reads were normalized to the total amounts of aligned reads, and the sum of reads aligning to each small RNA specie was calculated: for small RNAs targeting protein-coding and pseudogenes, reads aligned in the anti-sense orientation were used. For miRNAs and piRNAs, read aligning in the sense orientation were used.
Poly-U small RNA analysis
The total small RNA library were submitted to iterating cycles of alignment against the C. elegans genome (using Shortstack [57], perfect match). We first aligned the original reads after the removal of the adaptors. We then applied three consecutive iterations in which we trimmed one T nucleotide from the 3′ prime UTR of reads that fail to align in the previous round, and re-aligned these trimmed reads using Shortstack with the same parameters. Overall, we performed 3 such iterations and thus detected up to 3 occurrences of un-templated poly-uridine additions over individual reads. We next used HTseq [58] to count the perfectly-matched and poly-uridylated reads that aligned in antisense orientation to the C. elegans’ protein coding genes, while separating between the original reads and the the uridylated ones. Finally, we calculated the fraction of poly-uridylation as the number of reads with untemplated 3′Ts out of the total aligning reads against a specific gene. For the analysis of the mean fractions of poly-U values we considered only protein coding genes for which we detected at least 5 RPM (reads per million) in at least one of the examined samples.
Hierarchical clustering analysis
Hierarchical clustering analysis was performed using MATLAB’s clustergram function based on DESeq2 output data.
Principle component analysis
Principle component analysis was performed using the plotPCA function of DESeq2 [66].
Statistical analysis
For RNAi experiments and germ granule characterization, statistical tests were performed via GraphPad Prism 6. Details about the tests and corrections for multiple comparisons appear in the corresponding figure legends.
Data and Code Availability
The raw sequencing files and processed data generated during this study are available under GEO: GSE128112.
Acknowledgments
We thank all the Rechavi lab members for fruitful discussions. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Eric Miska and Scott Kennedy for kindly providing us with strains. Special thanks to Dror Cohen for the illustrations that he contributed. O.R. is thankful to the Adelis Foundation grant #0604916191. This work was funded by ERC grant #335624 and the Israel Science Foundation (grant #1339/17).
Author Contributions
I.L., I.A.T., Y.M., and O.R. conceived the project, designed the experiments, and wrote the manuscript; I.L., I.A.T., and Y.M. performed the experiments and analyzed the results with the assistance of A.N., G.W., O.A., O.B., I.B.S., and S.A.; A.N. and R.Z.-B. provided imaging expertise and resources; H.G., I.L., I.A.T., Y.M., and S.A. generated and analyzed the sequencing data; and U.S. and J.M.C. provided the wago-4::gfp strain.
Declaration of Interests
The authors declare no competing interests.
Published: August 1, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.07.054.
Contributor Information
Itamar Lev, Email: itamai.et@gmail.com.
Itai Antoine Toker, Email: itaitoker@gmail.com.
Yael Mor, Email: yael.moryosef@gmail.com.
Oded Rechavi, Email: odedrechavi@gmail.com.
Supplemental Information
DESeq2 fold-change results for the detection of differentially expressed small RNAs targeting specific genes in the germline. For both analyses, the wild-type group was used as ‘Reference’ (untreated) and the mutant group as ‘Comparison’ (treated). Data S1A: pptr-1 versus wild type. Data S1B: meg-3/4 versus wild type. The meg-3/4 mutants in Data S1B derived from crosses between mutant males and wild-type hermaphrodites, and gonads were extracted two generations after homozygosity.
References
- 1.Perez M.F., Lehner B. Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 2019;21:143–151. doi: 10.1038/s41556-018-0242-9. [DOI] [PubMed] [Google Scholar]
- 2.Rechavi O., Lev I. Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr. Biol. 2017;27:R720–R730. doi: 10.1016/j.cub.2017.05.043. [DOI] [PubMed] [Google Scholar]
- 3.Ni J.Z., Kalinava N., Chen E., Huang A., Trinh T., Gu S.G. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin. 2016;9:3. doi: 10.1186/s13072-016-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rechavi O., Houri-Ze’evi L., Anava S., Goh W.S.S., Kerk S.Y., Hannon G.J., Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vastenhouw N.L., Brunschwig K., Okihara K.L., Müller F., Tijsterman M., Plasterk R.H.A. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 6.Alcazar R.M., Lin R., Fire A.Z. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houri-Ze’evi L., Korem Y., Sheftel H., Faigenbloom L., Toker I.A., Dagan Y., Awad L., Degani L., Alon U., Rechavi O. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 8.Lev I., Seroussi U., Gingold H., Bril R., Anava S., Rechavi O. MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 2017;27:1138–1147. doi: 10.1016/j.cub.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luteijn M.J., van Bergeijk P., Kaaij L.J.T., Almeida M.V., Roovers E.F., Berezikov E., Ketting R.F. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirayama M., Seth M., Lee H.C., Gu W., Ishidate T., Conte D., Jr., Mello C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pak J., Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 13.Sijen T., Steiner F.A., Thijssen K.L., Plasterk R.H.A. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 14.Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C., Doebley A.L., Goldstein L.D., Lehrbach N.J., Le Pen J. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lev I., Gingold H., Rechavi O. H3K9me3 is required for inheritance of small RNAs that target a unique subset of newly evolved genes. eLife. 2019;8:338582. doi: 10.7554/eLife.40448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinava N., Ni J., Gajic Z., Ushakov H., Gu S. Caenorhabditis elegans heterochromatin factor SET-32 plays an essential role in transgenerational establishment of nuclear RNAi-mediated epigenetic silencing. bioRxiv. 2018 doi: 10.1016/j.celrep.2018.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claycomb J.M., Batista P.J., Pang K.M., Gu W., Vasale J.J., van Wolfswinkel J.C., Chaves D.A., Shirayama M., Mitani S., Ketting R.F. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conine C.C., Batista P.J., Gu W., Claycomb J.M., Chaves D.A., Shirayama M., Mello C.C. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu W., Shirayama M., Conte D., Jr., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan G., Fields B.D., Spracklin G., Shukla A., Phillips C.M., Kennedy S. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature. 2018;557:679–683. doi: 10.1038/s41586-018-0132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu F., Feng X., Chen X., Weng C., Yan Q., Xu T., Hong M., Guang S. A cytoplasmic Argonaute protein promotes the inheritance of RNAi. Cell Rep. 2018;23:2482–2494. doi: 10.1016/j.celrep.2018.04.072. [DOI] [PubMed] [Google Scholar]
- 22.Ishidate T., Ozturk A.R., Durning D.J., Sharma R., Shen E.Z., Chen H., Seth M., Shirayama M., Mello C.C. ZNFX-1 functions within perinuclear nuage to balance epigenetic signals. Mol. Cell. 2018;70:639–649.e6. doi: 10.1016/j.molcel.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips C.M., Montgomery T.A., Breen P.C., Ruvkun G. MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 2012;26:1433–1444. doi: 10.1101/gad.193904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 25.Hird S.N., Paulsen J.E., Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- 26.Cheeks R.J., Canman J.C., Gabriel W.N., Meyer N., Strome S., Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Strome S., Wood W.B. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 28.Updike D.L., Knutson A.K.A., Egelhofer T.A., Campbell A.C., Strome S. Germ-granule components prevent somatic development in the C. elegans germline. Curr. Biol. 2014;24:970–975. doi: 10.1016/j.cub.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strome S., Updike D. Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 2015;16:406–416. doi: 10.1038/nrm4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strome S. Specification of the germ line. WormBook. 2005:1–10. doi: 10.1895/wormbook.1.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheth U., Pitt J., Dennis S., Priess J.R. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Updike D.L., Hachey S.J., Kreher J., Strome S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 2011;192:939–948. doi: 10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spike C.A., Bader J., Reinke V., Strome S. DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development. 2008;135:983–993. doi: 10.1242/dev.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert V.J.P., Sijen T., van Wolfswinkel J., Plasterk R.H.A. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spracklin G., Fields B., Wan G., Vijayendran D., Wallig A., Shukla A., Kennedy S. Identification and characterization of C. elegans RNAi inheritance machinery. Genetics. 2017 doi: 10.1534/genetics.116.198812. Published online May 22, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padmanabhan S., Mukhopadhyay A., Narasimhan S.D., Tesz G., Czech M.P., Tissenbaum H.A. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–951. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallo C.M., Wang J.T., Motegi F., Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 2010;330:1685–1689. doi: 10.1126/science.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J.-X., Cipriani P.G., Mecenas D., Polanowska J., Piano F., Gunsalus K.C., Selbach M. In vivo interaction proteomics in Caenorhabditis elegans embryos provides new insights into P granule dynamics. Mol. Cell. Proteomics. 2016;15:1642–1657. doi: 10.1074/mcp.M115.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J.T., Smith J., Chen B.C., Schmidt H., Rasoloson D., Paix A., Lambrus B.G., Calidas D., Betzig E., Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith J., Calidas D., Schmidt H., Lu T., Rasoloson D., Seydoux G. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife. 2016;5:5. doi: 10.7554/eLife.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanazawa M., Yonetani M., Sugimoto A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J. Cell Biol. 2011;192:929–937. doi: 10.1083/jcb.201010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagijn M.P., Goldstein L.D., Sapetschnig A., Weick E.-M., Bouasker S., Lehrbach N.J., Simard M.J., Miska E.A. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips C.M., Montgomery B.E., Breen P.C., Roovers E.F., Rim Y.S., Ohsumi T.K., Newman M.A., van Wolfswinkel J.C., Ketting R.F., Ruvkun G., Montgomery T.A. MUT-14 and SMUT-1 DEAD box RNA helicases have overlapping roles in germline RNAi and endogenous siRNA formation. Curr. Biol. 2014;24:839–844. doi: 10.1016/j.cub.2014.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang J.J., Hunter C.P. Tissue specificity of Caenorhabditis elegans enhanced RNA interference mutants. Genetics. 2011;188:235–237. doi: 10.1534/genetics.111.127209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkies P., Ashe A., Le Pen J., McKie M.A., Miska E.A. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 2013;23:1258–1270. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D., Tu S., Stubna M., Wu W.-S., Huang W.-C., Weng Z., Lee H.-C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 2018;359:587–592. doi: 10.1126/science.aao2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sapetschnig A., Sarkies P., Lehrbach N.J., Miska E.A. Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 2015;11:e1005078. doi: 10.1371/journal.pgen.1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Albuquerque B.F.M., Placentino M., Ketting R.F. Maternal piRNAs are essential for germline development following de novo establishment of endo-siRNAs in Caenorhabditis elegans. Dev. Cell. 2015;34:448–456. doi: 10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 50.van Wolfswinkel J.C., Claycomb J.M., Batista P.J., Mello C.C., Berezikov E., Ketting R.F. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Kalinava N., Ni J.Z., Peterman K., Chen E., Gu S.G. Decoupling the downstream effects of germline nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis elegans. Epigenetics Chromatin. 2017;10:6. doi: 10.1186/s13072-017-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pak J., Maniar J.M., Mello C.C., Fire A. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell. 2012;151:885–899. doi: 10.1016/j.cell.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akay A., Di Domenico T., Suen K.M., Nabih A., Parada G.E., Larance M., Medhi R., Berkyurek A.C., Zhang X., Wedeles C.J. The helicase Aquarius/EMB-4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Dev. Cell. 2017;42:241–255.e6. doi: 10.1016/j.devcel.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrews E.A. On the structure of protoplasm. Science. 1895;2:893–899. [Google Scholar]
- 55.Eddy E.M. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 56.Holoch D., Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hertz E.P.T., Kruse T., Davey N.E., López-Méndez B., Sigurðsson J.O., Montoya G., Olsen J.V., Nilsson J. A conserved motif provides binding specificity to the PP2A-B56 phosphatase. Mol. Cell. 2016;63:686–695. doi: 10.1016/j.molcel.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 58.Zhang G., Wang Z., Du Z., Zhang H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018;174:1492–1506.e22. doi: 10.1016/j.cell.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Putnam A., Cassani M., Smith J., Seydoux G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 2019;26:220–226. doi: 10.1038/s41594-019-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belicard T., Jareosettasin P., Sarkies P. The piRNA pathway responds to environmental signals to establish intergenerational adaptation to stress. BMC Biol. 2018;16:103. doi: 10.1186/s12915-018-0571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weldon W.F.R. Mendel’s laws of alternative inheritance in peas. Biometrika. 1902;1:228–254. [Google Scholar]
- 62.Andrews S. Babraham Bioinformatics; 2010. FastQC-A Quality Control tool for High Throughput Sequence Data. [Google Scholar]
- 63.Axtell M.J. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 65.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maniar J.M., Fire A.Z. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr. Biol. 2011;21:449–459. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X., Xu F., Mao H., Ji J., Yin M., Feng X., Guang S. Nuclear RNAi contributes to the silencing of off-target genes and repetitive sequences in Caenorhabditis elegans. Genetics. 2014;197:121–132. doi: 10.1534/genetics.113.159780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild type embryos expressing pgl-1::rfp (P granules) and wago-4::gfp (Z granules) were photographed via time-lapse imaging, at the 2-cell stage. Shown are maximal Z projections of the merged bright field, 488nm and 561nm channels.
Wild type embryos expressing pgl-1::rfp (P granules) and Pmex-5::gfp::h2b (chromatin) were photographed via time-lapse imaging, from the 2-cell to 4-cell stages. Shown are maximal Z projections of the merged bright field, 488nm and 561nm channels.
meg-3/4 mutant embryos expressing pgl-1::rfp (P granules) and Pmex-5::gfp::h2b (chromatin) were photographed via time-lapse imaging, from the 4-cell to 6-cell stages. Shown are maximal Z projections of the merged bright field, 488nm and 561nm channels.
DESeq2 fold-change results for the detection of differentially expressed small RNAs targeting specific genes in the germline. For both analyses, the wild-type group was used as ‘Reference’ (untreated) and the mutant group as ‘Comparison’ (treated). Data S1A: pptr-1 versus wild type. Data S1B: meg-3/4 versus wild type. The meg-3/4 mutants in Data S1B derived from crosses between mutant males and wild-type hermaphrodites, and gonads were extracted two generations after homozygosity.
Data Availability Statement
The raw sequencing files and processed data generated during this study are available under GEO: GSE128112.