Abstract
Background
Small RNAs regulate a wide variety of processes in plants, from organ development to both biotic and abiotic stress response. Being master regulators in genetic networks, their biogenesis and action is a fundamental aspect to characterize in order to understand plant growth and development. Three main gene families are critical components of RNA silencing: DICER-LIKE (DCL), ARGONAUTE (AGO) and RNA-DEPENDENT RNA POLYMERASE (RDR). Even though they have been characterized in other plant species, there is no information about these gene families in Citrus sinensis, one of the most important fruit species from both economical and nutritional reasons. While small RNAs have been implicated in the regulation of multiple aspects of plant growth and development, their role in the abscission process has not been characterized yet.
Results
Using genome-wide analysis and a phylogenetic approach, we identified a total of 13 AGO, 5 DCL and 7 RDR genes. We characterized their expression patterns in root, leaf, flesh, peel and embryo samples using RNA-seq data. Moreover, we studied their role in fruit abscission through gene expression analysis in fruit rind compared to abscission zone from samples obtained by laser capture microdissection. Interestingly, we determined that the expression of several RNA silencing factors are down-regulated in fruit abscission zone, being particularly represented gene components of the RNA-dependent DNA Methylation pathway, indicating that repression of this process is necessary for fruit abscission to take place in Citrus sinensis.
Conclusions
The members of these 3 families present characteristic conserved domains and distinct expression patterns. We provide a detailed analysis of the members of these families and improved the annotation of some of these genes based on RNA-seq data. Our data suggests that the RNA-dependent DNA Methylation pathway is involved in the important fruit abscission process in C. sinensis.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1998-1) contains supplementary material, which is available to authorized users.
Keywords: Small RNAs, DCL, AGO, RDR, Orange, Abscission, Citrus sinensis
Background
A wide variety of biological processes in plants are regulated by small RNAs, which are regulatory molecules of RNA, typically between 21 and 24 nucleotides long. In general, there are two main distinct small RNAs classes, which are known as microRNAs (miRNAs) and short-interfering RNAs (siRNAs). However, the vast number of siRNA sources described so far has rendered multiple subgroups in the siRNA class, including trans acting (ta-siRNAs), phased (pha-siRNAs), heterocromatic (hc-siRNAs) and natural antisense siRNAs (nat-siRNAs) among others [1, 2]. In plants, the biogenesis and action of these regulatory molecules depend mainly on members of the AGO, RDR and DCL families. RDRs are capable of synthesizing dsRNA using RNA as a template and DCLs are responsible for the cleavage of dsRNA into 21–24 nt long small RNAs thanks to their RNAseIII-type activity. In turn, the small RNA provide the specificity of action of the RISC complex containing the AGO factor, targeting at post-transcriptional level RNA molecules with partial or total base complementarity, which can be cleaved via the RNAseH-type activity of AGO proteins or through translation inhibition [3, 4]. In plants, RDR proteins contain one unique conserved domain named RNA-dependent RNA polymerase (RdRP) [5, 6], whereas six domains are present in DCL proteins: DEXDc, Helicase-C, RNA-binding, PAZ, RNaseIIIa, RNaseIIIb (RIBOc) and Double Stranded RNA-binding (dsRB), but one or more may be missing [7]. Within AGO proteins, there are four main domains, which are known as N-Terminal, PAZ, Mid and PIWI [8]. Different studies have reported that plant DCL, RDR and AGO gene families are normally constituted by multiple members. A total of 20 genes coding for these protein families have been identified in Arabidopsis, 28 genes in tomato and maize [5, 9], 32 genes in rice [3] and 22 in grapevine and pepper [10, 11].
Sweet orange (Citrus sinensis) is one of the most important species for fruit consumption cultivated worldwide. Its nutritional attributes for human health are well known since it is an excellent source of easy access vitamin C [12], besides the immense economic importance of this species, for which a global production of US$9 billion was estimated in 2012 [13]. From an agricultural point of view, abscission has a tremendous impact on yield, leading to high yield losses in key crops, including citrus species [14]. Small RNAs have been shown to participate in several aspects of plants growth and development, such as stress response, leaf polarity, flowering time [15–18] and resistance against diseases [19]. In the present work, we performed a genome-wide analysis in order to characterize these important protein families in Citrus sinensis. Using a phylogenetic approach, we identified and characterized the AGO, DCL and RDR gene families in orange and analyzed their expression patterns across five plant tissues, improving the gene structural annotation of five of them using RNA-seq data. Finally, we established that selected members of these families as well as additional single copy factors of the RNA-dependent DNA methylation (RdDM) pathway show differential expression in the fruit abscission zone of sweet orange samples analyzed using laser capture microdissection (LCM). A detailed analysis of the AGO, RDR and DCL gene families in Citrus sinensis is presented in this work and we provide initial evidence of an epigenetic component in the regulation of fruit abscission in this species.
Results
Identification and in silico analysis of AGO, RDR and DCL genes in orange
In order to identify the AGO, RDR and DCL gene families in orange, we gathered data from previously characterized genes in Arabidopsis, rice, poplar and tomato. (Additional file 1). Using Arabidopsis sequences as queries in BLAST analyses, we identified five genes encoding DCL proteins (CsDCLs), seven encoding RDR (CsRDRs) and thirteen encoding AGO proteins (CsAGOs) in Pythozome (www.phytozome.org) and in the “Citrus sinensis annotation project” (CSAP) (http://citrus.hzau.edu.cn/orange/) databases (Table 1). Even though most of these genes were present in both databases, several inconsistencies were detected both in their functional and structural annotation across these two genome versions, as well as poor gene structural annotation for some of them. In general, a better structural annotation consistent with RNA-seq data was observed in CSAP database, except for genes orange1.1g001771m (CsRDR3) and orange1.1g002204m (CsAGO5a), which are better annotated in Phytozome (Table 1). In order to work with the most accurate gene models, we improved the annotation of five genes using information from both databases, as well as RNA-seq data from different plant tissues (Table 1; Additional file 2). For example, the CSAP annotated gene model for CsAGO5c is missing the Mid and PIWI domains; but when we used RNAseq data to update the structural annotation of this gene, all the typical AGO domains are detected in the newly annotated gene model (Fig. 1, Additional file 2). The characteristics of all the genes identified in this study are detailed in Table 1, including gene IDs in both databases, ORF length, protein length, isoelectric point (IP) and molecular weight (Mw), while the gene structure analysis and the updated protein sequences for the corrected gene structural annotations are detailed in Additional file 2. Also, the Pfam and SMART ID for all the domains identified in the CsAGO, CsRDR and CsDCL proteins are detailed in Additional file 3 and their associated Gene Ontology terms are detailed in Additional file 4, which describe the associated biological functions of these genes.
Table 1.
Characterization of AGO, DCL and RDR genes in Citrus sinensis
| Gene Name | Gene ID Phytozome | Gene ID CSAP | Location | Protein | ||||
|---|---|---|---|---|---|---|---|---|
| ORF length (bp) | Length (AA) | PI | Mw (Da) | |||||
| DCL | CsDCL1 | orange1.1g000174m | orange1.1 t00584 | ChrUn: 6828329–6,838,566 (− strand) | 5892 | 1963 | 5.96 | 219,934 |
| CsDCL2a | orange1.1g000607m | Cs6g03520 | Chr6: 3969936–3,982,992 (− strand) | 4191 | 1396 | 7.65 | 158,488 | |
| CsDCL2b | orange1.1g003062m | Cs6g03500 | Chr6: 3910063–3,922,849 (− strand) | 4206 | 1401 | 6.48 | 158,770 | |
| CsDCL3 | orange1.1g000379m | Cs4g06370 | Chr4: 3914566–3,925,649 (+ strand) | 4959 | 1652 | 7.95 | 184,339 | |
| CsDCL4 | orange1.1g000380m | Cs4g01340 | Chr4: 255555–267,608 (+ strand) | 4902 | 1633 | 6.47 | 183,256 | |
| RDR | CsRDR1a | orange1.1g002586m | Cs2g17570 | Chr2: 14327852–14,333,995 (+ strand) | 3399 | 1132 | 7.13 | 129,642 |
| CsRDR1b | orange1.1g003789m | Cs5g14110 | Chr5: 11659292–11,663,430 (− strand) | 3249 | 1082 | 6.04 | 122,608 | |
| CsRDR1c | orange1.1g035741m | – | Scaffold00168–67,689 – 71,577 (− strand) | 2910 | 969 | 8.58 | 110,376 | |
| CsRDR2 | orange1.1g001183m | Cs5g05170 | Chr5: 3069393–3,074,020 (+ strand) | 3396 | 1131 | 6.59 | 128,958 | |
| CsRDR31 | orange1.1g001771m | Cs4g15260 | Chr4: 14278137–14,289,339 (+ strand) | 3048 | 1015 | 6.8 | 115,671 | |
| CsRDR6a | orange1.1g041430m | Cs7g05350 | Chr7: 2849984–2,855,672 (+ strand) | 3594 | 1197 | 6.13 | 136,437 | |
| CsRDR6b2 | orange1.1g048783m | Cs1g14730 | Chr1: 18022840–18,027,238 (+ strand) | 3501 | 1166 | 6.01 | 132,490 | |
| AGO | CsAGO1 | orange1.1g001466m | Cs5g16710 | Chr5: 15851375–15,859,311 (− strand) | 3222 | 1073 | 9.38 | 118,338 |
| CsAGO2a | orange1.1g012649m | Cs2g10760 | Chr2: 8058870–8,063,724 (+ strand) | 2946 | 981 | 9.29 | 110,071 | |
| CsAGO2b | – | Cs2g10770 | Chr2: 8075960–8,080,634 (+ strand) | 2946 | 981 | 9.26 | 110,304 | |
| CsAGO4a | orange1.1g002449m | Cs2g29070 | Chr2: 28612071–28,619,504 (− strand) | 2763 | 920 | 8.98 | 102,998 | |
| CsAGO4b | orange1.1g002636m | Cs3g06860 | Chr3: 9639528–9,647,482 (+ strand) | 2697 | 898 | 9.43 | 100,950 | |
| CsAGO5a1 | orange1.1g002204m | Cs7g17940 | Chr7: 13746350–13,751,613 (− strand) | 2865 | 954 | 9.27 | 106,773 | |
| CsAGO5b2 | orange1.1g003630m | Cs7g17970 | Chr7: 13774473–13,781,129 (− strand) | 2847 | 948 | 9.50 | 105,299 | |
| CsAGO5c2 | – | Cs7g17930 | Chr7: 13732122–13,738,824 (− strand) | 3021 | 1006 | 9.28 | 112,004 | |
| CsAGO5d | – | Cs6g14430 | Chr6: 15643144–15,650,122 (− strand) | 2688 | 895 | 9.23 | 99,978 | |
| CsAGO6 | orange1.1g002661m | Cs2g20520 | Chr2: 17331361–17,339,897 (− strand) | 2688 | 895 | 9.41 | 100,500 | |
| CsAGO6-like2 | orange1.1g048669m | Cs6g16080 | Chr6: 17010270–17,016,001 (− strand) | 2814 | 937 | 9.71 | 105,659 | |
| CsAGO7 | orange1.1g001684m | Cs7g03360 | Chr7: 1453333–1,457,453 (− strand) | 3093 | 1030 | 9.24 | 117,036 | |
| CsAGO10 | orange1.1g001954m | Cs9g07740 | Chr9: 4853982–4,862,787 (− strand) | 2979 | 992 | 9.34 | 111,516 | |
1 Phytozome gene structural annotation used
2 Updated gene structural annotation
Fig. 1.
Domains present in AGO, DCL and RDR protein families from C. sinensis. 1: DEXDc; 2: Helicase C; 3: RNA-binding; 4: PAZ; 5: RIBOc; 6: dsRB; 7: RdRP; 8: Gly-Rich; 9: N-terminal; 10: L1; 11: L2; 12: Mid; 13: PIWI. Representation is in amino acid (AA) scale
All seven RDR genes found in the orange genome encode proteins with the RdRP domain. The number of amino acids of the members of this family varies between 969 and 1197 (Table 1). We improved the annotation of the gene orange1.1g048783m/Cs1g14730 (CsRDR6b) using RNA-seq data from leaf, root, embryo, flesh and peel libraries (Table 1 and Additional files 2 and 5). We observed expression in all the libraries in the region annotated as intron 3 for this gene, therefore the gene structure was updated (Additional file 2). The region of the updated sequence codes for the RdDP domain of this protein, which is predicted correctly using SMART after this re-annotation (Fig. 1), while the previous version of the gene showed a truncated RdDP domain.
The CsDCL family consists of five members for which the coding sequences range between 4191 and 5892 bp and code for proteins between 1396 and 1963 amino acids (Table 1). Pfam and SMART analyses revealed that all CsDCLs showed six conserved domains: DEXDc, Helicase C, RNA-binding, PAZ and two consecutive RIBOc domains (RNaseIIIa and RNaseIIIb). Besides these, CsDCL1 and CsDCL4 presented two consecutive dsRB domains after the RIBOc domains (Fig. 1).
The thirteen identified AGO proteins have the N-terminal, L1, PAZ and PIWI domains, whereas the L2 domain is present in twelve of these proteins, but is missing in orange1.1g003630m/Cs7g17970 (CsAGO5b). Also, the Mid domain is present in all proteins, except in orange1.1g001684m/Cs7g03360 (CsAGO7) and orange1.1g001466m/Cs5g16710 (CsAGO1) is the only one starting with a Gly-Rich domain (Fig. 1).
In addition to the conserved domains present in these proteins, we also examined the presence of conserved motifs in these gene families. In Arabidopsis, AGO proteins have DDH/H or DDD/H motifs in the PIWI domain, which are responsible for their endonuclease activity [20–22]. We found the DDH/H motif in CsAGO1, CsAGO7 and CsAGO10, and the DDD/H motif was present in CsAGO2a (Additional file 6). We also identified DDH/P motifs for CsAGO4a and CsAGO4b; a DDH/S motif in CsAGO6 and DDY/H and DDY/P motifs for CsAGO5a and CsAGO5b, respectively. Finally, a D-H/T motif was observed in CsAGO6-like protein (Additional file 6), which is therefore likely to lack endonuclease activity.
We detected the conserved DECH motif in all the CsDCL proteins, which is characteristic of plant DCL proteins [23, 24] and the DLDGD motif, which is typically present in the catalytic domain of RDR proteins [6, 25], was detected in 5 out of 7 of the CsRDR proteins (Additional file 6).
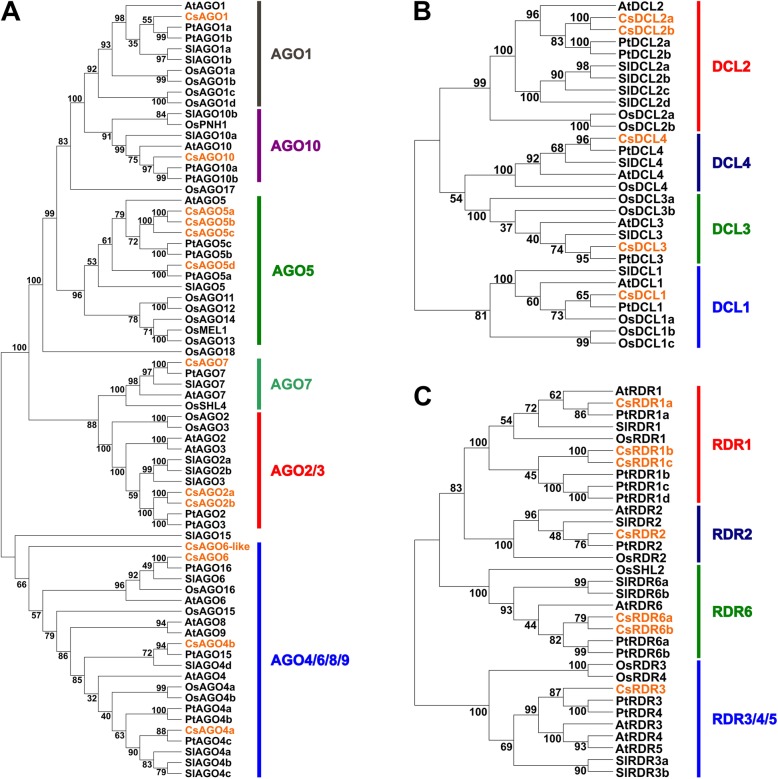
Phylogenetic analysis allows identification of DCL, RDR and AGO putative orthologues in orange
We used a phylogenetic approach to identify the putative orthologous genes from the AGO, RDR and DCL families in orange. We built an unrooted neighbor-joining tree for the three analyzed families using full length protein sequences from Arabidopsis, tomato, poplar and rice (Fig. 2). The AGO gene family was separated into six clades: AGO1, AGO10, AGO5, AGO7, AGO2/3 and AGO4/6/8/9 (Fig. 2a). We identified only one gene from the C. sinensis genome in each one of the clades containing AGO1, AGO10 and AGO7 members of the AGO family in the rest of the analyzed species, which we named CsAGO1, CsAGO10 and CsAGO7, respectively. Two AGO members were identified in the AGO2/3 clade and were named CsAGO2a and CsAGO2b, being the latter only present in the CSAP genome, sharing 93% amino acid identity with each other. Four C. sinensis proteins grouped in the AGO5 clade, sharing between 52 and 87% amino acid identity, and were named CsAGO5a, CsAGO5b, CsAGO5c and CsAGO5d, being the last two also only present in CSAP genome. Also four members of the orange AGO family grouped within the AGO4/6/8/9 clade; of these, we named two as CsAGO4a and CsAGO4b, while a third one was named CsAGO6, due to its proximity to AtAGO6 (Fig. 2a). Finally, one AGO protein in the AGO4/6/8/9 clade failed to group with other characterized proteins in this clade and was named CsAGO6-like due to its proximity to the AGO6 group (Table 1, Fig. 2a, for the corresponding gene IDs, please refer to Additional file 1 or Table 1).
Fig. 2.
Phylogenetic analysis of DCL, AGO and RDR genes from orange, Arabidopsis, poplar, tomato and rice. Unrooted Neighbor-Joining trees were obtained from multiple alignments of (a) AGO, (b) DCL and (c) RDR protein sequences using Muscle algorithm in MEGA 6.0. Bootstrap values from 1000 replicates are indicated. Gene families are divided by clades, highlighted with different colors. C. sinensis genes are colored in orange
The five CsDCL proteins were distributed in four clades: DCL1, DCL2, DCL3 and DCL4 (Fig. 2b). Two highly similar proteins with 90% of amino acid identity grouped in the DCL2 clade, and therefore were named CsDCL2a and CsDCL2b, respectively. The remaining three clades contain one CsDCL protein each, which we called CsDCL1, CsDCL3 and CsDCL4 accordingly (Table 1, Fig. 2b and Additional file 1).
Finally, the tree derived from CsRDRs sequences also consists of four clades: RDR1, RDR2, RDR6 and RDR3/4/5. The RDR1 clade contains three proteins: CsRDR1a, CsRDR1b and, only present in Phytozome genome, CsRDR1c (Table 1, Fig. 2c). Two proteins were placed in the RDR6 clade, CsRDR6a and CsRDR6b, both similar in length, with 1197 and 1145 amino acids, respectively and sharing 80% of their amino acid sequence. Finally, only one protein was located in each of the RDR2 and RDR3/4/5, which were named CsRDR2 and CsRDR3, respectively (Table 1, Fig. 2c, Additional file 1).
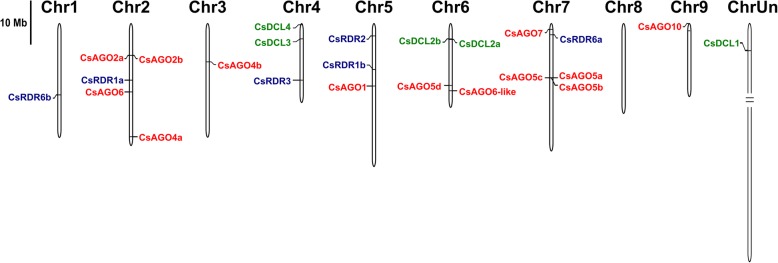
Chromosomal location of orange DCL, RDR and AGO genes
Next, we analyzed the chromosomal distribution of all the DCL, RDR and AGO genes in sweet orange using CSAP database, which provides chromosomal location (Fig. 3). The genes corresponding to four CsDCL, five CsRDR and thirteen CsAGO proteins were unevenly located in all chromosomes, except for chromosome 8 which do not harbor any members of the analyzed families. Only one gene from the AGO family is encoded in each chromosome 3 and 9, namely CsAGO4b and CsAGO10, respectively. The paralogues CsDCL2a and CsDCL2b appear very close on chromosome 6, as well as CsAGO2a and CsAGO2b on chromosome 2 and CsAGO5a, CsAGO5b and CsAGO5c on chromosome 7 (Fig. 3, Table 1). Conversely, CsRDR1a and CsRDR1b are located in chromosomes 2 and 5 respectively. Chromosomes 2 and 7 contain five gene members of the analyzed families each, while chromosomes 4 and 5 host three of them. Only CsRDR1c was not annotated in this database, thus it is not represented in Fig. 3 (Table 1), while CsDCL1 is placed in the “Unknown chromosome” (ChrUn, Fig. 3 (Table 1), which contains sequence from the C. sinensis genome that could not be assigned to any of the chromosomes yet.
Fig. 3.
Genomic distribution of CsDCL, CsRDR and CsAGO genes. Chromosomes are represented on the scale indicated on the left of the figure. Different gene families are represented with different colors. 30 Mb were removed from “ChrUn” in order to fit the figure
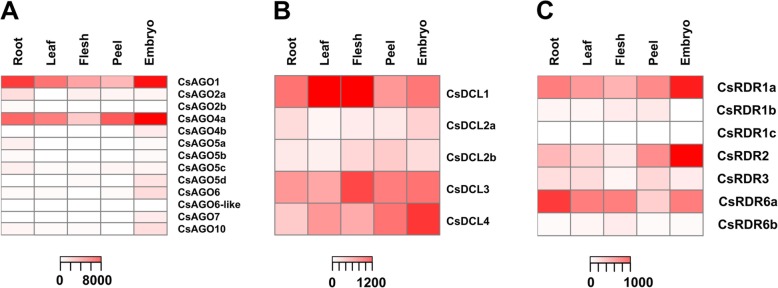
Common and specific AGO, DCL and RDR gene expression patterns in different tissues
In order to characterize the expression pattern of the different members of these families, we analyzed RNA-seq data from five different tissues: leaf, root, flesh, peel and embryo (accession numbers for the raw data are detailed in Additional file 5). We performed differential expression analyses for all tissues (Additional file 7) and represented the expression levels using one heatmap plot per family (Fig. 4). CsAGO1 and CsAGO4a showed the highest expression levels within the AGO family and both accumulate to a much higher levels than the rest of the AGO genes analyzed. In order to better appreciate differences in expression, Additional file 8 shows the same heatmap plot from Fig. 4a without these two genes. Embryo tissue evidenced the highest expression for most of the AGO genes in comparison with the rest of the analyzed tissues, except for CsAGO2a, CsAGO2b, CsAGO5a and CsAGO5c, for which expression levels in roots are slightly higher (Fig. 4a; Additional file 8). Conversely, a weak expression of most genes is detected in flesh, peel and leaf, while CsAGO2b, CsAGO5b and CsAGO6-like presented very low expression levels in all tissues (Fig. 4a, Additional file 8).
Fig. 4.
Expression levels of (a) CsAGO, (b) CsDCL and (c) CsRDR genes. Analyzed tissues were root, leaf, flesh, peel and embryo. Normalized expression for each gene family is represented with different color intensity, ranging from white to red
All CsDCL proteins are expressed in the five analyzed tissues: CsDCL2a and CsDCL2b exhibit the lowest expression among this family, while CsDCL1 has the highest expression levels, especially in leaf and flesh. On the other hand, CsDCL3 and CsDCL4 show intermediate expression, presenting the highest accumulation in flesh and embryo, respectively (Fig. 4b).
Regarding the RDR family, CsRDR1c is not detected in any of the analyzed tissues (Fig. 4c, Additional file 7), CsRDR1b accumulates weakly in flesh, peel, leaf and root and CsRDR1a is the most abundant gene of this family across these samples. CsRDR2 is highly expressed in embryo and CsRDR6a accumulates in roots preferentially, whereas CsRDR6b is weakly expressed in all tissues and shows preferential accumulation in flesh (Fig. 4c, Additional file 7).
Expression of genes involved in RdDM and ta-siRNA pathways show downregulation during orange fruit abscission
In order to gain insight into the role of RNA silencing pathways during the abscission process, we identified all the probes corresponding to small RNA-related factors present in the cDNA microarray used by [14], including the newly characterized AGO, RDR and DCL genes together with other genes coding for single-copy factors involved in the biogenesis and action of small RNAs (Fig. 5, Additional file 9). We analyzed expression data from abscission zone (AZ) in comparison to fruit rind (FR), obtained using LCM, after 12 or 24 h of ethylene treatment to induce abscission [14]. Interestingly, we detected downregulation in the expression of three members of the analyzed families: CsDCL1, CsRDR1a and CsAGO4a, as well as in three single-copy genes: the orthologue of SUPRESSOR OF GENE SILENCING 3 (CsSGS3), a component of the ta-siRNA biogenesis pathway; the orange orthologue of the subunit 2 of PolIV and Pol V (CsNRPD2) and the orthologue of SAWADEE HOMEODOMAIN HOMOLOGUE 1 (CsSHH1), which are involved in the RdDM pathway. Downregulation of these genes was also detected by qRT-PCR, except for CsRDR1 which was not tested using this method (Additional files 10 and 11). Of these, CsRDR1a, CsSHH1 and CsNRPD2 are specifically downregulated in the AZ and not in FR, both at 12 and 24 h after ethylene treatment (Fig. 5). CsSGS3 and CsAGO4a are downregulated specifically in AZ at 12 h, but at 24 h they are also downregulated in FR. Finally, CsDCL1 is downregulated both in AZ and FR after 24 h of ethylene treatment (Fig. 5). Some of these expression changes were also confirmed by qRT-PCR, and the overall tendency of downregulation of these genes was confirmed using this method (Additional file 10). These analyses show that a downregulation of several factors involved in small RNAs biogenesis and action is necessary during fruit abscission, which implies a still unexplored role for small RNAs in this important process.
Fig. 5.
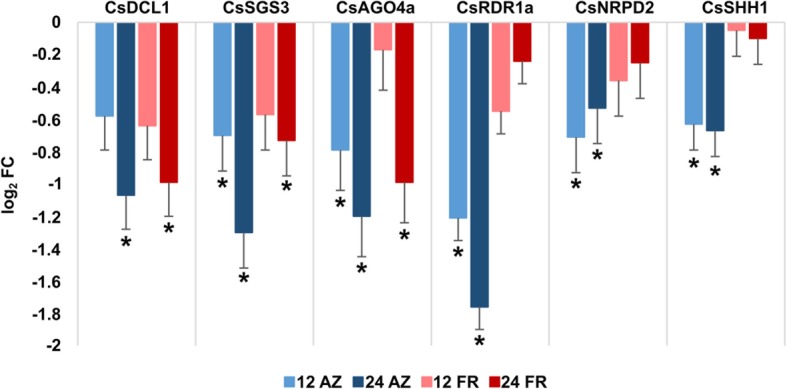
Relative expression of genes involved in RNA silencing. Comparisons between abscission zone (AZ) and fruit rind (FR) at 12 and 24 h after ethylene treatment. Values are represented relative to the expression in AZ or FR without ethylene treatment (0 h). Significant differential expression is indicated with an asterisk (* = q ≤ 0.05 and a log2 FC contrast cutoff value of ±0.5)
Discussion
RNA silencing is an ancient molecular mechanism which involves the participation of small RNAs acting at different levels in the regulation of gene expression. Plants have evolved a wide variety of pathways over different RDR-DCL-AGO combinations which have been shown to participate in several aspects of plant development [1, 2]. For example, DCL1 and AGO1 are mainly involved in the miRNA pathway, but no RDR protein is necessary for miRNA biogenesis [1, 26]. Some exceptional miRNA have evolved specialized modes of action, such as miR166 binding AGO10 exclusively to regulate shoot apical development and the also exclusive miR390/AGO7 combination in Arabidopsis to regulate AUXIN RESPONSE FACTORS 3 and 4 (ARF3 and ARF4) [27, 28]. RDR6, SGS3, AGO1 and DCL4 are the main components of the ta-siRNA pathway, responsible for the downregulation of target mRNAs at the post-transcriptional level in multiple plant species studied so far [29–31]; while RDR2, AGO4 and DCL3 participate in the RdDM pathway, mainly involved in the silencing of repetitive regions in the genome [32–34].
RDR family in Citrus sinensis present distinctive features and member-specific expression patterns
RDRs are characterized by the presence of a conserved domain required to copy single-stranded RNA into double-stranded RNA, which was detected in all the members of this family identified in this work (Fig. 1). Also present in these proteins is a conserved DLDGD motif in the catalytic site, which was detected in 5 of the CsRDRs. Within this motif, lysine is often variable, and this was the case for CsRDR3, presenting a DFDGD motif, also detected in other species such as Arabidopsis, rice, pepper and coffee orthologues [6, 35, 36], whereas CsRDR1c presented a DQDGE motif, with two amino acid substitutions, which remains to be tested for functional activity (Additional file 6). The biological function of RDR proteins is usually linked to the subsequent action of specific DCL proteins and have been found in eukaryotic genomes including plants, fungi and invertebrate animals, but not in vertebrates and insects [37]. The different RDR family members exhibit functional diversification which is usually conserved across species; for example, RDR1 proteins have been shown to play a role in antiviral defense in Arabidopsis, pepper and potato [36, 38, 39]; RDR2 plays a critical role in RNA-directed DNA methylation and repressive chromatin modification which is conserved between Arabidopsis and maize, among other species [2, 32]. In addition to their participation in virus defense, RDR6 orthologues have been shown to participate in the ta-siRNA biogenesis pathway also in several species, having a key role in plant development through the regulation of the TAS3 biogenesis pathway [29, 40]. In this study we identified seven members of the CsRDR family, including one RDR2 orthologue and one member grouping with the RDR3/4/5 clade (Fig. 3). Interestingly, we identified two RDR6 orthologues, which was also the case for tomato and other solanaceae like potato and S. commersonii [41], and three RDR1 orthologues, only seen in the citrus genome. This expansion in gene number was not observed in Arabidopsis, rice and tomato, which have only one member in the RDR1 clade (Fig. 2), whereas poplar (P. trichocarpa), a species that is evolutionary closer to orange, presents four RDR1 orthologues (Fig. 2) [42]. The members of this family showed distinct expression patterns in the tissues analyzed (Fig. 4), being CsRDR1a, CSRDR2 and CsRDR6a the most abundant. CsRDR1c was not detected in the analyzed samples, suggesting that this particular gene could be important in a different plant tissue, at different developmental stages not analyzed here, or that its expression is induced when the plant is growing in a particular environmental condition [11, 43, 44]. Is important to mention that CsRDR1c is only present in the Citrus sinensis genome published in Phytozome but not in CSAP database (Table 1), this could be due to the fact that these genomes are in early versions and discrepancies and incomplete gene structural annotations are expected. CsRDR1b, CsRDR3 and CsRDR6b exhibited low expression levels (Fig. 4), but they show tissue preferences, suggesting they could play roles under specific circumstances in plant growth and development or that their expression could be induced in response to specific environmental cues.
DCL family members in Citrus sinensis present different expression patterns
DCL endonucleases process dsRNA into small RNA duplexes with 2-nt 3′ overhangs. DCL1 is capable of recognizing imperfect stem-loop substrates present in pre-miRNA transcripts, while the rest of the DCLs found in plants are responsible for the 21-, 22 and 24-nt siRNA production [45]. Plant DCL genes form a monophyletic group spawned after the plant-animal split but before the monocot-dicot divergence 150 million years ago [46]. All the genes identified in this study harbor the DEXDc, Helicase C, RNA binding and two tandem RIBOc domains, whereas CsDCL1 and CsDCL4 showed 2 additional dsRB domains (Fig. 1). In addition, all of them presented the characteristic DECH motif (Additional file 6). The C. sinensis genome presents one member of each DCL1, DCL3 and DCL4 clades, similar to Arabidopsis and tomato, but noticeably two members of the DCL2 clade, which is also the case for the OsDCL2 proteins. CsDCL2 proteins, which are presumed responsible for the generation of 22-nt siRNAs based on their Arabidopsis counterpart, appear to have suffered tandem duplication considering the close location of CsDCL2a and CsDCL2b on chromosome 6 and the 90% identity in their amino acid sequence (Figs. 2 and 3). The different expression patterns detected for these two genes suggest they acquired distinct expression regulation and could have preferential roles in specific tissues. CsDCL1 showed expression in all tissues analyzed, which is consistent with its putative role in miRNA biogenesis [1, 26]. Similarly, CsDCL3 showed highest expression in flesh and CsDCL4 in embryo, but they are abundantly expressed in all samples analyzed, also consistent with their ubiquitous roles in RdDM and ta-siRNA biogenesis, respectively [2].
AGO family in Citrus sinensis underwent tandem duplication events during evolution
AGO proteins are the main RNA silencing effectors across kingdoms, since they possess the slicing activity required by the small RNA-mediated regulatory pathways. The AGO family expanded during plant evolution, from ancient unicellular or multicellular green algae (e.g Micromonas pusilla and Volvox carteri), where three or less AGO genes are present, to ten or more members in flowering plants. The expansion of the AGO family in plants suggests a functional diversification of AGO proteins presumably due to expanding small RNA-directed regulatory pathways [47]. Eukaryotic AGOs contain four main domains: a variable N-terminal domain and the highly conserved PAZ, MID, and PIWI domains [1]. In this study, we identified these domains in all the characterized AGO proteins, with the exception of CsAGO7 missing the Mid domain (Fig. 1), which has been implicated in the sorting of small RNAs into different AGOs [48]. AGO7 has been shown to bind miR390 exclusively in Arabidopsis [27], but the basis for this unique AGO7-miR390 association is not completely understood and remains to be studied in C. sinensis. We also analyzed the presence of the characteristic DDH/H and DDD/H motifs in these proteins, which are present in Arabidopsis AGO proteins and their function has been extensively studied, establishing they are required for the slicing activity of AGO proteins [21, 22, 49]. Some amino acid substitutions have been detected in other species such as tomato and Brassica species, including DDH/P, DDH/S, DDY/H and DDY/P motifs detected in the CsAGO proteins identified in this study [5, 25]. However, CsAGO6-like did not present this motif, suggesting that this member of the CsAGO family may lack slicing activity (Additional file 6).
We identified single CsAGO1, CsAGO6, CsAGO7 and CsAGO10, while two highly identical CsAGO2 alleles (93% amino acid identity) were detected, possibly originated by tandem duplication based on their close location on chromosome 2 (Fig. 3, Table 1). Similarly, three CsAGO5 paralogues were detected in the orange genome in close locations on chromosome 7, also suggesting duplication events to give rise to these alleles sharing more than 90% amino acid identity (Fig. 3, Table 1). CsAGO5a and CsAGO5c have slightly higher expression in roots, whereas CsAGO5b has low expression in all the samples analyzed, suggesting new allele functionalization after the duplication events (Fig. 4). CsAGO1 exhibits high accumulation in most tissues as expected for its putative role as effector of miRNA-mediated regulation. CsAGO2 has preferential expression in roots, while CsAGO4a, CsAGO5d, CsAGO6 and CsAGO10 accumulate mostly in embryo. This could be related to a demand for CsAGO4 and CsAGO6, involved in the RdDM pathway during embryo development, since this pathway is of special importance during this process and others, like gametogenesis and meiosis, where hc-siRNAs are essential to silence the repetitive regions of the genome [2]; as well as a putative role of CsAGO10 analogous to the demonstrated role of AtAGO10 in embryo development [50, 51]. Similar to CsAGO5b, very low expression was observed for CsAGO7 and CsAGO4b (Additional file 8; Fig, 4); which might indicate a preferential role of these gene products in other tissues and/or developmental stages and/or during exposure to specific environmental conditions.
Is interesting to note that co-expression of factors involved in the same regulatory pathways has been detected in our expression analysis. For example, for miRNA biogenesis and action both CsDCL1 and CsAGO1 are needed, and co-expression of these genes was detected (Fig. 4). Similarly, RdDM requires the coordinated action of CsRDR2, CsDCL3 and CsAGO4 and the ta-siRNA pathway requires CsRDR6, CsDCL4 and CsAGO1, all of which have been shown to be co-expressed in our expression analysis (Fig. 4). However, is also important to take into consideration that the biological function of the small RNAs generated by these pathways are typically necessary in most plant tissues during plant growth and development. For example, miRNAs are key regulators of several transcription factors involved in leaf and fruit development [26, 52–54], as well as TAS3-derived ta-siRNAs, which regulate auxin response through cleavage of ARF transcripts [29–31], and gene silencing at the transcriptional level which is also regulated by RNA silencing components [55–57]. Therefore, different combinations of these factors are usually ubiquitously active. Elucidation of how the different combinations of RNA silencing factors are needed during plant growth and development is key to understand how the expansion of gene members of the AGO, RDR and DCL families have contributed to their unique functionalization in different species. Is also necessary to gain insight into the subcellular localization of the different members of these families, which have been well characterized in Arabidopsis and tomato, for example, but very little is known in other species [58, 59].
Small RNA pathways are downregulated during fruit abscission
In citrus, fruit abscission represents a high percentage of annual yield losses. It is well established that plant growth hormones are deeply involved in abscission and that among them, ethylene is thought to be its natural regulator [60]. This process occurs specifically in the abscission zone (AC) of fruits and other organs, through coordinated changes in gene expression and is accelerated by ethylene treatment [14, 61, 62]. Our gene expression analysis in orange fruit abscission was oriented to analyze changes in the expression of RNA silencing-related genes, in a model system used before to analyze the molecular mechanisms underlying the abscission process induced by ethylene treatment [14, 61, 62]. Therefore we compared the expression of AGO, RDR, DCL and additional single gene factors with homology to known RNAi factors in the abscission zone of fruits (Fig. 5, Additional file 10). Interestingly, we detected a general downregulation of RNAi factors. In particular, we detected lower levels of expression for CsAGO4a, CsSHH1 and CsNRPD2 in the AZ. These genes participate in the RdDM pathway, which is an important RNAi-mediated epigenetic pathway in plants. The RdDM pathway is involved in transcriptional silencing of transposons and repetitive sequences and relies on specialized transcriptional machinery that includes the plant-specific RNA polymerases Pol IV and Pol V. Pol IV transcripts are rapidly processed into dsRNAs by RDR2, and subsequently processed into 24 nucleotide siRNAs by DCL3 and exported to the cytoplasm. In the cytoplasm, they are mainly incorporated into AGO4 containing complexes and imported back to the nucleus to target nascent transcripts transcribed by Pol V at the same loci, leading to DNA methylation [2]. Gene silencing also involves chromatin remodelation, typically through methylation of histone H3. The role of SHH1 in A. thaliana is to recognize H3K9me2 (dimethylation of lysine 9 in histone H3), which recruits Pol IV and initiates siRNA biogenesis for the maintenance of gene silencing [63]. Besides the RdDM pathway directed by 24-nt siRNAs, work in Arabidopsis showed that 21-nt ta-siRNAs can also direct RdDM. Instead of being processed by DCL4 and loaded into AGO1 to target mRNA cleavage in trans, these ta-siRNAs are processed by DCL1 and loaded into AtAGO4 or AtAGO6, directing methylation of TAS loci [64]. In this regard, the observed downregulation of CsDCL1 and CsSGS3 in FR or AZ could also contribute to a general inhibition of DNA methylation during the abscission process, as a result of the downregulation of these components of the TAS-related RdDM pathway and ta-siRNA biogenesis. The roles of RDR1 and RDR6 are usually redundant when they are involved in anti-viral defense and both produce DCL4 substrates [39]. Therefore, a role of CsRDR1 can be proposed in the context of ta-siRNA biogenesis and/or the TAS loci-related RdDM during the abscission process, instead of CsRDR6 for which no differential expression was observed. These changes in the expression of RNA silencing factors could lead to modification in the expression of TAS genes and other genomic regions regulated by the RdDM and the ta-siRNAs biogenesis pathways, presumably involved in the cellular processes participating in fruit abscission, such as cell wall disassembly [14].
The present work gives rise to interesting questions about potential epigenetic regulation of the abscission process. Further studies comprising genome-wide analysis of DNA methylation in the abscission zone and changes in small RNA population during the abscission process, together with their regulated genes are very interesting experiments that will help elucidate the molecular mechanisms involving RNA silencing during this interesting and economically important aspect of citrus biology.
Conclusions
In the present work we identified and characterized 13 AGO, 5 DCL and 7 RDR genes present in the orange genome, through a careful search and analysis of the 2 available C. sinensis genomes. Their expression patterns across five plant tissues indicate that most of these genes are ubiquitously expressed but they show distinct levels of expression in different plant tissues. We further improved the annotation of five of these genes using RNA-seq data. Finally, we established that selected members of these families as well as additional single copy factors of the RdDM pathway (CsSHH1, CsNRPD2 and CsAGO4a) show differential expression in the fruit abscission zone of sweet orange samples analyzed using LCM, providing initial evidence of an epigenetic component in the regulation of fruit abscission in this species.
Methods
Identification of candidate AGO, DCL and RDR genes in orange
Citrus sinensis genome assembly (v1.1) and protein sequences were downloaded from Phytozome v12.1 (https://phytozome.jgi.doe.gov) and CSAP V2 (http://citrus.hzau.edu.cn/orange/). Amino acids sequences of all the DCL, AGO and RDR genes of Arabidopsis thaliana were retrieved from TAIR (https://www.arabidopsis.org/) and were used to search for orange orthologous genes with Phytozome’s online BLASTP tool. Conserved domains in orange’s proteins were analyzed and annotated using the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/) and the Pfam database (31.0, https://pfam.xfam.org/). Protein’s isoelectric point and molecular weight were calculated using Compute pI/Mw (http://web.expasy.org/compute_pi/). Gene structures for Additional file 2 were created using GSDS 2.0 [65].
Phylogenetic analysis and chromosomal localization
Candidate proteins from Citrus sinensis were aligned with AGO, RDR and DCL proteins from Arabidopsis thaliana, Solanum lycopersicum and Oryza sativa using MEGA 6.0 [66]. For the estimation of a phylogenetic tree, we used the MUSCLE algorithm (gap open, − 2.9; gap extend, 0; hydrophobicity multiplier, 1.2; clustering method, UPGMB) ([67]. A phylogenetic tree was built for each protein family using the Neighbor-Joining method with the bootstrap test replicated 1000 times. A Chromosomal location image was made according to genome annotations from the “Citrus sinensis annotation project” (CSAP) (http://citrus.hzau.edu.cn/orange/), using the online tool MapGene2Chromosome V2 (http://mg2c.iask.in/mg2c_v2.0/).
Expression analysis of DCL, AGO, and RDR genes in orange tissues
We used publicly available libraries of RNA-seq to analyze the expression levels of these genes in different plant tissues. Accession numbers are detailed in Additional file 3. Raw reads were processed for removal of non-coding RNA sequences present in the Rfam 13.0 database (http://rfam.xfam.org/), using Bowtie version 1.1.1, allowing 2 mismatches [68]. Clean reads were mapped to the Citrus sinensis reference genome from CSAP or from Phytozome in the case of CsRDR1c and CsRDR6b, which are not annotated in the genome version from CSAP. We used Hisat2 software, version 2.1.0 [69], and hisat2-build to create the corresponding indexed genomes. Next, we used featureCounts 1.5.3 [70] to obtain read counts for each gene, using gene models from CSAP (csi.gene.models.gff3) or from Phytozome for CsRDR1c and CsRDR6b (Csinensis_154_v1.1.gene.gff3). Differentially expressed genes were selected as those showing a 2-fold change in expression using DESeq2 version 1.20.0 [71] with an adjusted p-value < 0.05.
Expression analysis of DCL, AGO and RDR orange genes during fruit abscission
For gene expression in the abscission zone, data obtained from [14] was used. In this work gene expression was analyzed in Washington Navel orange fruits after 0, 12 and 24 h of ethylene treatment, used to promote abscission. RNA samples from abscission zone and fruit rind cells were obtained by LCM. For gene expression analysis a cDNA microarray including 21.081 putative genes of citrus was utilized [14]. Genes with an adjusted p-value ≤0.05 and a fold change of ±2 were considered differentially expressed.
Quantitative RT-PCR analysis
Abscission zone and fruit rind tissue was dissected from Salustiana orange fruits after 0, 12 and 24 h of ethylene treatment, as described in [14]. Total RNA was prepared using Quick-zol reagent (Kalium Technologies) following manufacturer’s instructions and treated with DNase I (Promega). cDNA from 1000 ng of RNA per sample was synthesized using EasyScript (Trans) according to manufacturer’s protocol. Gene-specific primers were designed (Additional file 11) for use with TransStart Tip Green qPCR SuperMix (Trans). The specificity of all amplification products was determined using dissociation curve analyses. Relative quantification values were calculated based on three biological and three technical replicates using the 2-ΔCt method, using Ubiquitin (CsUBI) expression as normalization control. Efficiency correction was necessary just for CsSGS3 primers (which was calculated 88%), as described in [72].
Additional files
Gene ID for AGO, RDR and DCL gene families used in the phylogenetic analysis (XLSX 13 kb)
Gene structural annotation and updated protein sequences (PDF 461 kb)
Pfam and SMART domain IDs (XLSX 12 kb)
GO term annotation of the analyzed genes (XLSX 13 kb)
Accession numbers of RNA-seq libraries used in this study (XLSX 11 kb)
Motifs detected in CsAGO, CsRDR and CsDCL proteins (XLSX 13 kb)
Differential expression analysis of CsAGO, CsDCL and CsRDR genes (XLSX 10 kb)
Heatmap plots for the AGO family without CsAGO1 and CsAGO4a (TIF 3434 kb)
Microarray data of RNA silencing factors in AZ and FR from C. sinensis fruits (XLSX 13 kb)
Relative expression levels of RNA silencing genes. Transcript levels determined by qRT-PCR (mean ± SE; n = 3) compared to the expression of each gene in AZ or FR without ethylene treatment (0 h) and using CsUBI as normalization control (* p < 0.05; ** p-value< 0.001) (TIF 5450 kb)
Primers used for qRT-PCR (XLSX 11 kb)
Acknowledgements
We thank our colleagues who read the paper and shared their valuable comments and discussion. MD, AV and LD are members of the Researcher Career of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and AS is a CONICET doctoral fellow.
Abbreviations
- AGO
Argonaute
- DCL
Dicer-like
- RdDM
RNA-directed DNA Methylation
- RDR
RNA-dependent RNA Polymerase
Authors’ contributions
Conceived and designed the experiments: MD, LD, FT, AV, MT. Performed the experiments: AS, MD. Analyzed the data: AS, MD, LD, FT. Wrote the paper: AS, MD. Revised the manuscript: LD, FT, MT. All authors read and approved the final manuscript.
Funding
This research was partially supported by FONCyT grant PICT 201–0426 to Dr. Lucas Daurelio.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Agustín Sabbione, Email: agustin.sabbione@hotmail.com.ar.
Lucas Daurelio, Email: lucasdaurelio@gmail.com.
Abelardo Vegetti, Email: avegetti@fca.unl.edu.ar.
Manuel Talón, Email: talon_man@gva.es.
Francisco Tadeo, Email: tadeo_fra@gva.es.
Marcela Dotto, Phone: +54 3496 426400, Email: mdotto@fca.unl.edu.ar.
References
- 1.Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 2.Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16(12):727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapoor M, Arora R, Lama T, Nijhawan A, Khurana JP, Tyagi AK, et al. Genome-wide identification, organization and phylogenetic analysis of DICER-LIKE, ARGONAUTE and RNA-DEPENDENT RNA POLYMERASE gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics. 2008;9:1–17. doi: 10.1186/1471-2164-9-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai M, Yang GS, Chen WT, Mao ZC, Kang HX, Chen GH, et al. Genome-wide identification of DICER-LIKE, ARGONAUTE and RNA-DEPENDENT RNA POLYMERASE gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene. 2012;501:52–62. doi: 10.1016/j.gene.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, et al. The evolution and diversification of dicers in plants. FEBS Lett. 2006;580:2442–2450. doi: 10.1016/j.febslet.2006.03.072. [DOI] [PubMed] [Google Scholar]
- 8.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 9.Qian Y, Cheng Y, Cheng X, Jiang H, Zhu S, Cheng B. Identification and characterization of DICER-LIKE, ARGONAUTE and RNA-DEPENDENT RNA POLYMERASE gene families in maize. Plant Cell Rep. 2011;30:1347–1363. doi: 10.1007/s00299-011-1046-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H, Zhao K, Wang J, Chen X, Chen Z, Cai R, et al. Comprehensive analysis of DICER-LIKE, ARGONAUTE, and RNA-DEPENDENT RNA POLYMERASE dene families in grapevine (Vitis Vinifera) J Plant Growth Regul. 2015;34:108–121. doi: 10.1007/s00344-014-9448-7. [DOI] [Google Scholar]
- 11.Qin L, Mo N, Muhammad T, Liang Y. Genome-wide analysis of DCL, AGO, and RDR gene families in pepper (Capsicum Annuum L.). Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed]
- 12.Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, et al. The draft genome of sweet orange (Citrus sinensis) Nat Genet. 2013;45:59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 13.Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Bitechnol. 2015;32:656–662. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merelo P, Agustí J, Arbona V, Costa ML, Estornell LH, Gómez-Cadenas A, et al. Cell wall remodeling in abscission zone cells during ethylene-promoted fruit abscission in citrus. Front Plant Sci. 2017;8:126. doi: 10.3389/fpls.2017.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nature Genetics. 2006;38:31–37. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 16.Nag A, Jack T. Sculpting the flower; the role of microRNAs in flower development. Curr Top Dev Biol. 2010;91(C):349–378. doi: 10.1016/S0070-2153(10)91012-0. [DOI] [PubMed] [Google Scholar]
- 17.Jin D, Wang Y, Zhao Y, Chen M. MicroRNAs and their cross-talks in plant development. J Genet Genomics. 2013;40:161–170. doi: 10.1016/j.jgg.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Noman A, Fahad S, Aqeel M, Ali U, Amanullah AS, et al. miRNAs: major modulators for crop growth and development under abiotic stresses. Biotechnol Lett. 2017;39:685–700. doi: 10.1007/s10529-017-2302-9. [DOI] [PubMed] [Google Scholar]
- 19.Zhai J, Jeong DH, de Paoli E, Park S, Rosen BD, Li Y, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 21.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci. 2005;102(33):11928. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, et al. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutant. Plant Cell. 2012;24:3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Axtell MJ, Fedoroff NV. The helicase and RNaseIIIa domains of Arabidopsis DICER-LIKE1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol. 2012;159:748–758. doi: 10.1104/pp.112.193508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 25.Cao J-Y, Xu Y-P, Li W, Li S-S, Rahman H, Cai X-Z. Genome-wide identification of DICER-LIKE, ARGONAUTE, and RNA-DEPENDENT RNA POLYMERASE gene families in Brassica species and functional analyses of their Arabidopsis homologs in resistance to Sclerotinia sclerotiorum. Front Plant Sci. 2016;7:1–17. doi: 10.3389/fpls.2016.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Tianran J, Xuemei C. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017;216:1002–1017. doi: 10.1111/nph.14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, Hu F, Wang R, Zhou X, Sze S-H, Liou LW, et al. Arabidopsis ARGONAUTE 10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 30.Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, et al. Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell. 2012;24:3575–3589. doi: 10.1105/tpc.112.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dotto MC, Petsch KA, Aukerman MJ, Beatty M, Hammell M, Timmermans MCP. Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genet. 2014;10(12):e1004826. doi: 10.1371/journal.pgen.1004826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J a L, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Axtell MJ. AGO4 is specifically required for heterochromatic siRNA accumulation at pol V-dependent loci in Arabidopsis thaliana. Plant J. 2017;90:37–47. doi: 10.1111/tpj.13463. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Duttke SH, Hetzel J, Groth M, Feng S, Gallego-Bartolome J, et al. RNA-directed DNA methylation involves co-transcriptional small-RNA-guided slicing of polymerase V transcripts in Arabidopsis. Nat Plants. 2018;4:181–188. doi: 10.1038/s41477-017-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandes-Brum CN, Rezende PM, Ribeiro THC, De Oliveira RR, De Sousa Cardoso TC, Do Amaral LR, et al. A genome-wide analysis of the RNA-guided silencing pathway in coffee reveals insights into its regulatory mechanisms. PLoS One. 2017;12:1–29. doi: 10.1371/journal.pone.0176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin L, Mo N, Zhang Y, Muhammad T, Zhao G, Zhang Y, et al. CaRDR1, an RNA-dependent RNA polymerase plays a positive role in pepper resistance against TMV. Front Plant Sci. 2017;8:1–13. doi: 10.3389/fpls.2017.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zong J, Yao X, Yin J, Zhang D, Ma H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene. 2009;447:29–39. doi: 10.1016/j.gene.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Hunter LJR, Brockington SF, Murphy AM, Pate AE, Gruden K, Macfarlane SA, et al. RNA-dependent RNA polymerase 1 in potato (Solanum tuberosum) and its relationship to other plant RNA-dependent RNA polymerases. Sci Rep. 2016;6:1–11. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 40.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of. Genes Dev. 2005:2164–75. [DOI] [PMC free article] [PubMed]
- 41.Esposito S, Aversano R, D’Amelia V, Villano C, Alioto D, Mirouze M, et al. DICER-LIKE and RNA-DEPENDENT RNA POLYMERASE gene family identification and annotation in the cultivated Solanum tuberosum and its wild relative S. commersonii. Planta. 2018;248:729–743. doi: 10.1007/s00425-018-2937-3. [DOI] [PubMed] [Google Scholar]
- 42.Zhao K, Zhao H, Chen Z, Feng L, Ren J, Cai R, et al. The DICER-LIKE, ARGONAUTE and RNA-DEPENDENT RNA POLYMERASE gene families in Populus trichocarpa: gene structure, gene expression, phylogenetic analysis and evolution. J Genet. 2015;94:317–321. doi: 10.1007/s12041-015-0508-y. [DOI] [PubMed] [Google Scholar]
- 43.Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtin SJ, Kantar MB, Yoon HW, Whaley AM, Schlueter JA, Stupar RM. Co-expression of soybean Dicer-like genes in response to stress and development. Funct Integr Genomics. 2012;12:671–682. doi: 10.1007/s10142-012-0278-z. [DOI] [PubMed] [Google Scholar]
- 45.Fukudome A, Fukuhara T. Plant DICER-LIKE proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis. J Plant Res. 2017;130:33–44. doi: 10.1007/s10265-016-0877-1. [DOI] [PubMed] [Google Scholar]
- 46.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Xia R, Meyers BC, Walbot V. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr Opin Plant Biol. 2015;27:84–90. doi: 10.1016/j.pbi.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Frank F, Hauver J, Sonenberg N, Nagar B. Arabidopsis ARGONAUTE MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012;31:3588–3595. doi: 10.1038/emboj.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arribas-Hernández L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, et al. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell. 2016;28:1563–1580. doi: 10.1105/tpc.16.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tucker MR, Hinze A, Tucker EJ, Takada S, Jurgens G, Laux T. Vascular signaling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- 51.Moussian B, Schoof H, Haecker A, Jürgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X. microRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karlova R, Van Haarst JC, Maliepaard C, Van De Geest H, Bovy AG, Lammers M, et al. Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot. 2013;64:1863–1878. doi: 10.1093/jxb/ert049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.José Ripoll J, Bailey LJ, Mai Q-A, Wu SL, Hon CT, Chapman EJ, et al. microRNA regulation of fruit growth. Nat Plants. 2015;1:15036. doi: 10.1038/nplants.2015.36. [DOI] [PubMed] [Google Scholar]
- 55.Djupedal I, Ekwall K. Epigenetics: heterochromatin meets RNAi. Cell Res. 2009;19:282–295. doi: 10.1038/cr.2009.13. [DOI] [PubMed] [Google Scholar]
- 56.Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon SA, Meyers BC. Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol. 2011;14:148–155. doi: 10.1016/j.pbi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Poulsen C, Vaucheret H, Brodersen P. Lessons on RNA silencing mechanisms in plants from eukaryotic Argonaute structures. Plant Cell. 2013;25:22–37. doi: 10.1105/tpc.112.105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:642–652. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson M, Osborne D. Ethylene, the natural regulator of leaf abscission. Nature. 1970;225:1019–1022. doi: 10.1038/2251019a0. [DOI] [PubMed] [Google Scholar]
- 61.Agustí J, Merelo P, Cercós M, Tadeo FR, Talón M. Ethylene-induced differential gene expression during abscission of citrus leaves. J Exp Bot. 2008;59:2717–2733. doi: 10.1093/jxb/ern138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agustí J, Merelo P, Cercás M, Tadeo FR, Talán M. Comparative transcriptional survey between laser-microdissected cells from laminar abscission zone and petiolar cortical tissue during ethylene-promoted abscission in citrus leaves. BMC Plant Biol. 2009;9:1–20. doi: 10.1186/1471-2229-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AMS, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu L, Mao L, Qi Y. Roles of DICER-LIKE and ARGONAUTE proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 2012;160:990–999. doi: 10.1104/pp.112.200279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B. Aligning short sequencing reads with bowtie. Curr Protoc Bioinformatics. 2010. p. 1–24. [DOI] [PMC free article] [PubMed]
- 69.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9):45e–45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene ID for AGO, RDR and DCL gene families used in the phylogenetic analysis (XLSX 13 kb)
Gene structural annotation and updated protein sequences (PDF 461 kb)
Pfam and SMART domain IDs (XLSX 12 kb)
GO term annotation of the analyzed genes (XLSX 13 kb)
Accession numbers of RNA-seq libraries used in this study (XLSX 11 kb)
Motifs detected in CsAGO, CsRDR and CsDCL proteins (XLSX 13 kb)
Differential expression analysis of CsAGO, CsDCL and CsRDR genes (XLSX 10 kb)
Heatmap plots for the AGO family without CsAGO1 and CsAGO4a (TIF 3434 kb)
Microarray data of RNA silencing factors in AZ and FR from C. sinensis fruits (XLSX 13 kb)
Relative expression levels of RNA silencing genes. Transcript levels determined by qRT-PCR (mean ± SE; n = 3) compared to the expression of each gene in AZ or FR without ethylene treatment (0 h) and using CsUBI as normalization control (* p < 0.05; ** p-value< 0.001) (TIF 5450 kb)
Primers used for qRT-PCR (XLSX 11 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article and its additional files.