Abstract
The cobalt(II) complex salts [Co(bpy)(az)2](PF6)2 and [Co(az)4](PF6), each bearing the unusual cis-N,N′-diphenylazodioxide ligand, were both screened as possible anticancer agents against SK-HEP-1 liver cancer cells. Both compounds were found to induce substantial apoptosis as an increasing function of concentration and time. Measurement of apoptosis-related proteins indicated that both the extrinsic and intrinsic pathways of apoptosis were activated. The apoptotic activity induced by these salts is not displayed either by simple cobalt(II) salts or complexes or by the free nitrosobenzene ligand. Additionally, these compounds did not induce apoptosis, as assessed by poly(adenosine diphosphate-ribose) polymerase cleavage, in several other cell lines.
Introduction
Since the discovery of the antitumor activity of cisplatin by Rosenberg and co-workers,1 it and other platinum complexes have seen widespread use in the treatment of a variety of cancers, and their mechanisms of action have been extensively explored.2 Other transition metals have been explored in cancer therapy, such as ruthenium2a,2b,3 osmium,2c rhodium,4,5a and iridium,5 but nonplatinum anticancer drugs in clinical use remain predominantly organic. In particular, despite the high natural abundance of cobalt6 and the extensive synthetic chemistry of its complexes, there are, to the best of our knowledge, so far no cobalt compounds in widespread clinical use other than vitamin B12 (cobalamin),7 though the cobalt(III) imine complex CTC-96 (Doxovir) has been explored as a treatment for herpes simplex virus,8 reaching phase II clinical trials.8b
Although a variety of cobalt compounds have been shown to be cytotoxic against certain cancer cell lines, those that have been demonstrated to induce apoptosis remain fairly few in number. All of these complexes feature cobalt in either the Co(II) or Co(III) oxidation state. Klegeris and co-workers prepared a series of square-planar Co(II) complexes of β-ketoaminato ligands, which increase the activation of caspase-3 and induce apoptosis in prostate cancer cells.9 Pombeiro and co-workers showed that a series of Co(II) tris(pyrazolyl)methane (“scorpionate”) complexes induced apoptosis in colorectal and liver cancer cells, with the generation of oxygen-centered radicals proposed as the mechanism of action based on their observed ability to cleave plasmid DNA.10 Chiniforoshan and co-workers showed that Co(II), Ni(II), and Cu(II) complexes of phenanthroline and the conjugate base of the natural product juglone induced early-phase apoptosis in liver, cervical, and colorectal cancer cells, likely involving intercalation with DNA.11
Gao and co-workers demonstrated that ellipsoid-shaped, bimetallic Co(II) and Ni(II) complexes with bis(pyrazolyl)phthalate bridging ligands induced apoptosis in oral epithelial carcinoma cells, with a proposed mechanism of DNA intercalation and cleavage.12 Similar behavior was observed by the same group for both mono- and bimetallic Co(II) complexes of tris(imidazolyl)benzene, with the monometallic complex being more potent.13 Ghosh and co-workers showed that a bimetallic Co(II) complex with 4,4′-azopyridine as the bridging ligand was able to induce apoptosis in osteosarcoma cells by a caspase- and p53-independent mechanism proposed to consist of DNA binding followed by the generation of reactive oxygen species (ROS).14 Simpson and co-workers demonstrated that a series of Co(II) and Ni(II) complexes of benzyl carbazate-derived imines induced apoptosis in leukemia cells, though the mechanism of apoptosis induction was not identified.15 Similarly, Katsaros and co-workers showed that a series of bimetallic Co(II) complexes with macrocyclic ligands were highly toxic to chronic myelogenous leukemia cells with little to no necrosis observed, but the mechanism of cell death (and potential apoptosis) was not identified.16
Co(III) complexes capable of inducing apoptosis include a bimetallic Co(III) dihydrazide complex with a chiral helical structure, prepared (as a racemic mixture) by Sladić and co-workers. This complex was shown to increase the fraction of early apoptotic cells in samples of breast cancer cells, but experiments with plasmid DNA indicated that it did not induce apoptosis by DNA cleavage.17 Chen and co-workers prepared a pair of Co oxoisoaporphine complexes, one Co(II) and one Co(III), and found that both induced apoptosis in cisplatin-resistant ovarian cancer cells by increasing mitochondrial ROS levels and activating caspase-3 and caspase-9. Additionally, the Co(III) complex was found to induce cell senescence by acting as a telomerase inhibitor, binding strongly to G-quadruplex DNA.18
Satyanarayana and co-workers demonstrated that a series of Co(III) complexes of an imidazole-fused phenanthroline ligand induced apoptosis in ovarian cancer cells. Experiments with plasmid DNA showed that the complexes were potentially phototoxic, able to cleave plasmid DNA when irradiated with UV light (365 nm) but not in the dark. It was not, however, determined whether their induction of apoptosis in ovarian cancer cells was photodependent.19 Filipović and co-workers prepared a series of cobalt complexes of (chalcogen)semicarbazones, with a Co(II) complex of the O-donor ligand and Co(III) complexes of the S- and Se-donor ligands. Each of these complexes was shown to induce differentiation in all-trans retinoic acid-resistant acute myeloid leukemia cells, and each was cytotoxic toward cervical cancer cells, with the O-donor and S-donor complexes inducing apoptosis by DNA damage from ROS generated by Fenton-type reactions and by G2/M cell cycle arrest, respectively.20 Qin and co-workers showed that a Co(III) complex of a substituted terpyridine induced G1-phase apoptosis in bladder cancer cells, with a mechanism involving mitochondrial membrane depolarization, caspase-3 and caspase-9 activation, and an increase in intracellular Ca2+ levels.21
An important subset of apoptosis-inducing cobalt complexes consists of the Co(III) complexes of long-chain primary amine surfactants, studied by Arunachalam and co-workers.22 These authors showed that undecylamine complexes formed micelles at low concentrations and induced apoptosis in breast cancer cells with DNA damage and mitochondrial membrane depolarization.22a Further work by the same group showed that one of these complexes caused downregulation of Bcl-2 and upregulation of p53 in breast cancer cells, though long treatment times with this complex resulted in some necrosis by oxidative stress, in addition to apoptosis.22b These groups later demonstrated that a related tetradecylamine complex induced apoptosis in cervical cancer cells with DNA fragmentation, whereas a decylamine complex induced apoptosis in liver cancer cells.22c Nagaraj and co-workers demonstrated that a related Co(III) decylamine complex with different ancillary ligands was both an effective surfactant and able to induce both apoptosis and necrosis in liver cancer cells.23
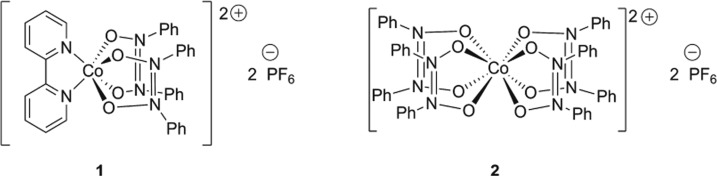
Recently, one of our groups has prepared two novel Co(II) complexes of the unusual ligand cis-N,N′-diphenylazodioxide (az): the six-coordinate, trigonal prismatic complex salt [Co(bpy)(az)2](PF6)2, where bpy = 2,2′-bipyridyl (compound 1), and the eight-coordinate, tetragonal complex salt [Co(az)4](PF6)2 (compound 2), both high-spin with three unpaired electrons per complex cation.24 The structures of 1 and 2 are shown in Scheme 1.
Scheme 1. Structures of Cobalt(II) Azodioxide Complexes 1 and 2.
The unusual coordination geometries for cobalt displayed by these complexes, combined with the rarity of crystallographically characterized azodioxide complexes of any metal,25 suggested that they may display novel reactivity, including potentially novel biological effects. We were, thus, inspired to investigate their biological activity and medicinal potential. We have now found that each of compounds 1 and 2 induces apoptosis in human liver adenocarcinoma (SK-HEP-1) cells and carried out a series of experiments to elucidate the possible mechanisms of cytotoxicity.
Results and Discussion
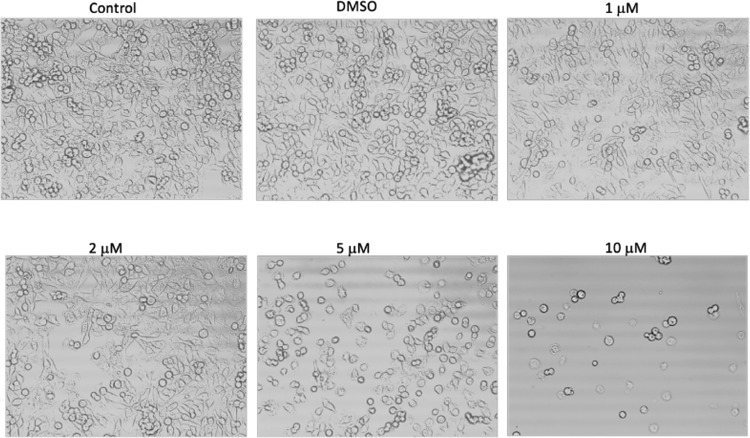
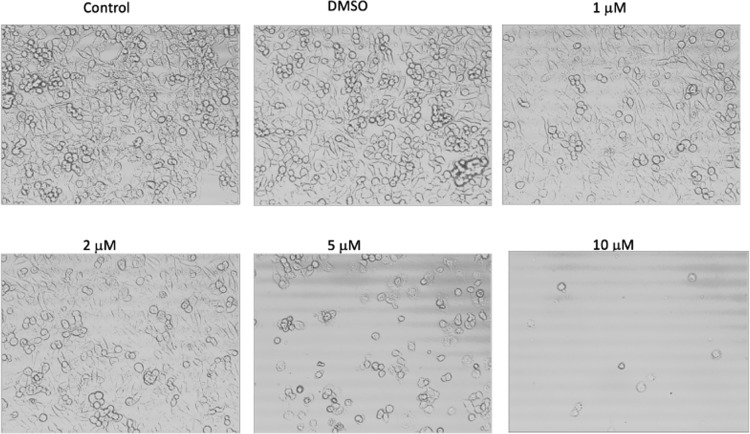
We investigated the concentration-dependent changes in the SK-HEP-1 cell morphology induced by cobalt(II) azodioxide complexes. Cells were treated with or without dimethyl sulfoxide (DMSO) (vehicle control) or compound 1 (Figure 1) or 2 (Figure 2) at concentrations of 1, 2, 5, or 10 μM for 12 h. Cell morphology was examined by phase-contrast microscopy under 20× magnification. Both compounds induced rounding and detachment of cells suggestive of cell death in the culture. An increased response by the cells to higher doses of the compounds was observed, as more dead cells detached from the culture dishes after the concentration of compound 1 or 2 was increased to 10 μM.
Figure 1.
Cell morphology after treatment with compound 1 for 12 h.
Figure 2.
Cell morphology after treatment with compound 2 for 12 h.
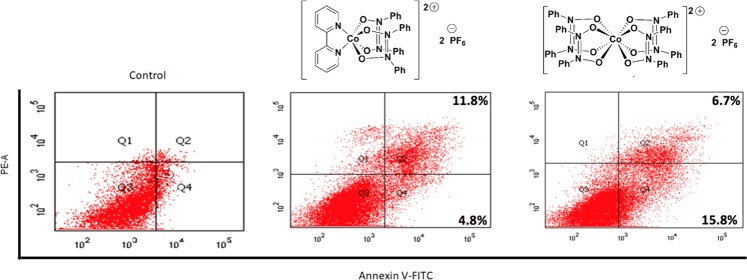
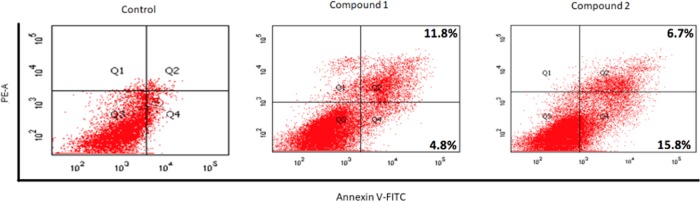
We conducted flow cytometry measurements of SK-HEP-1 cells after treatment with each of compounds 1 and 2 at 10 μM for 12 h. The percentages of apoptotic cells were significantly increased after treatment with compound 1 or 2 compared with control cells (Figure 3). After treatment with compound 1, the apoptotic population increased from 4.8 to 16.6%. With compound 2, the apoptotic population increased from 4.8 to 22.5%. These results suggest that compounds 1 and 2 are able to efficiently induce apoptosis in SK-HEP-1 cells. Fluorescence spectroscopy of compounds 1 and 2 showed that neither one displayed an emission spectrum, indicating that fluorescence from neither compound served as an artifact in the flow cytometry measurements.
Figure 3.
Flow cytometry results for treatment with compounds 1 and 2 (10 μM, 12 h).
We next investigated the molecular mechanisms underlying apoptosis induced by compounds 1 and 2. We first examined the proteolytic processing of caspases by immunoblotting analysis, as it has been demonstrated that the activation of caspases plays a vital role in apoptosis.26 Apoptosis induced in SK-HEP-1 cells was involved with the cleavage of poly(adenosine diphosphate-ribose) polymerase (PARP) and the activation of procaspase-8 and procaspase-9, as assessed by the appearance of the respective cleaved, active caspases compared to the control. The presence of each of these apoptosis-related proteins was measured after treatment with compounds 1 and 2 in both concentration-dependent and time-dependent experiments.
To determine the involvement of the caspases and PARP, we carried out concentration-dependent treatments with compounds 1 or 2 for 12 h. Interestingly, the cleavage of both procaspase-8 and procaspase-9 was observed at 5 and 10 μM concentrations of each compound. The cleavage of PARP was observed slightly at 2 μM concentrations and increased significantly at 5 and 10 μM (Figure 4).
Figure 4.
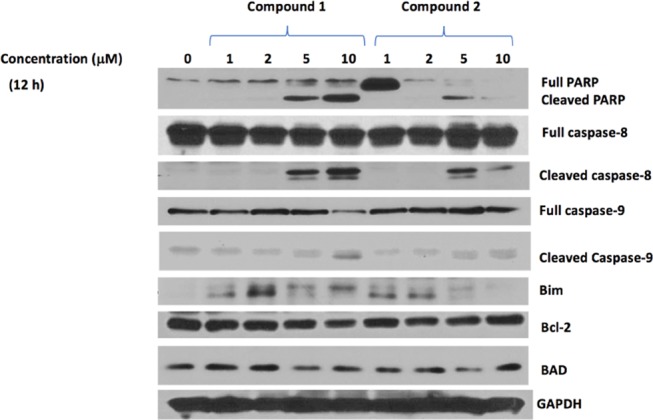
Concentration-dependent protein levels after 12 h of treatment with compounds 1 or 2.
We also investigated the effect of compounds 1 and 2 on other pro- and antiapoptotic proteins. The levels of these proteins were significantly regulated after treatment with compound 1 or 2 compared to the control, in a concentration-dependent manner (Figure 4).
In addition, we found that the apoptosis-related proteins were affected in a time-dependent manner compared to the control upon treatment with compound 1 or 2 at 2 μM concentration. Substantial amounts of cleaved PARP, caspase-8, and caspase-9 were observed as early as 6 h after treatment with 1 or 2. There are a variety of gene products involved in the process of cell apoptosis. A well-known group of these gene products is the Bcl-2 family, which includes both antiapoptotic and proapoptotic members.27 Upon the treatments, the level of Bcl-2, an antiapoptotic member, decreased over time, whereas BAD and Bim, two proapoptotic members, were upregulated (Figure 5). These data not only confirmed that compounds 1 and 2 induced the apoptosis of SK-HEP-1 cells in a concentration- and time-dependent manner but also indicated that they induced apoptosis via both the extrinsic and intrinsic pathways, since both procaspase-8 and procaspase-9 were cleaved after treatment.28 The cleavage of procaspase-8, involved in the extrinsic pathway, occurred to a greater extent than the cleavage of procaspase-9, involved in the intrinsic pathway,28 suggesting that more apoptosis proceeded by the extrinsic pathway than the intrinsic one.
Figure 5.
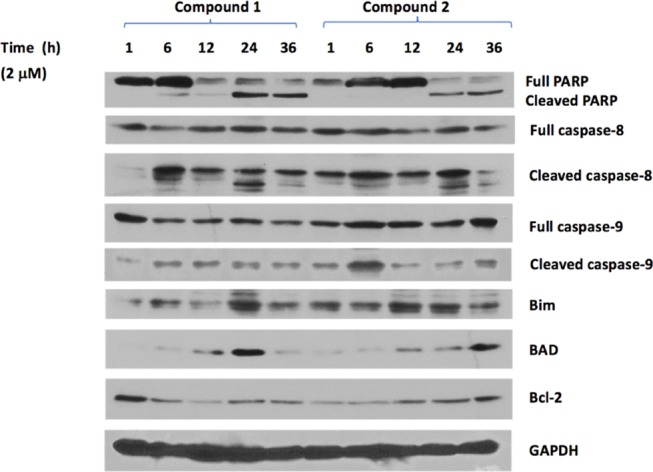
Time-dependent protein levels after treatment with compounds 1 or 2 at 2 μM concentration.
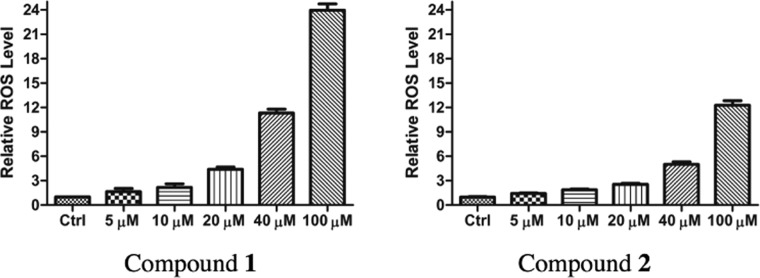
Cobalt(II) salts are known to induce oxidative DNA damage via the generation of reactive oxygen species such as hydroxyl radical and superoxide, akin to the iron-catalyzed Fenton reactions.29 Additionally, the mechanism of apoptosis induced by some cobalt complexes has been proposed to involve the generation of ROS and subsequent oxidative DNA damage.10,14,20 Thus, we decided to examine the possibility of ROS involvement in the proapoptotic activities of 1 and 2. We treated SK-HEP-1 cells with several different concentrations of 1 and 2 for an incubation time of 14 h and then ran a fluorescence-based ROS assay. This assay showed significant increases in ROS levels at high concentrations of 1 and 2, but only modest increases in ROS levels at concentrations used in the flow cytometry and western blot assays (Figure 6). These results suggest that 1 and 2 do not induce apoptosis primarily via ROS-mediated DNA damage.
Figure 6.
Effect of compounds 1 and 2 on ROS levels in SK-HEP-1 cells.
We performed the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay to assess the cytotoxicity of both 1 and 2 to SK-HEP-1. We incubated cells with 1 or 2 for 48 h at a series of concentrations (5.12 nM, 25.6 nM, 128 nM, 640 nM, 3.2 μM, 16 μM, 80 μM, 400 μM, 2 mM) and then performed the MTT assay. This assay showed a significant loss of cell viability only at 400 μM and 2 mM concentrations of 1 or 2. These are higher doses than those shown to induce substantial cell death by phase-contrast microscopy (Figures 1 and2), apoptosis and necrosis by flow cytometry (Figure 3), and the upregulation of proapoptotic proteins and downregulation of antiapoptotic proteins by western blot (Figures 4 and 5). The MTT assay has been shown to overestimate cell viability (and underestimate cytotoxicity) in certain cases,30 which may explain the apparent discrepancy between it and the other assays.
The ability of compounds 1 and 2 to induce PARP cleavage was shown to be cell line-specific. We treated several other cell lines with 2 and 10 μM concentrations of 1 and 2 for 12 h: HEK 293 (human embryonic kidney cells), HT-29 (colon cancer), MCF-7 (breast cancer), and PC-3 (prostate cancer). In none of these cases, extensive PARP cleavage was observed compared to the response of SK-HEP-1 to 1 and 2. These results suggest that 1 and 2 may allow for the selective apoptosis of liver cancer cells, with minimal harm to the surrounding tissue. The western blots from these experiments are shown in Figures S1–S4 (see the Supporting Information).
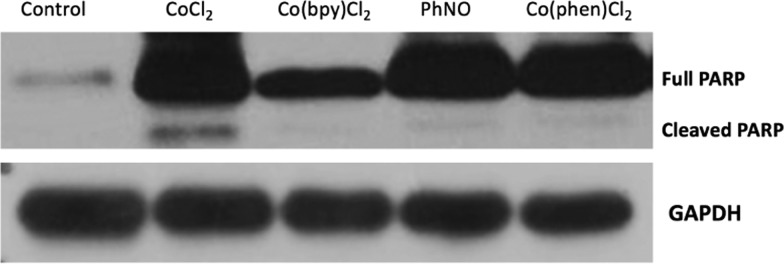
We also examined whether the starting materials used in the synthesis of compounds 1 and 2 were capable of causing apoptosis by themselves. These compounds included CoCl2, used in the synthesis of compound 2, Co(bpy)Cl2,31 used in the synthesis of compound 1, nitrosobenzene (PhNO), used in the synthesis of both compounds, and the additional compound Co(phen)Cl2,32 where phen = 1,10-phenanthroline, a compound related to Co(bpy)Cl2. Interestingly, treatment of SK-HEP-1 cells with each of these compounds for 12 h at 10 μM did not lead in any case to significant PARP cleavage compared with that observed after treatment with 1 or 2 (Figure 7). We observed minor PARP cleavage after treatment with CoCl2 but much less than that observed after treatment with 1 or 2 at this concentration and time.
Figure 7.
Effect of starting materials and Co(phen)Cl2 (12 h, 10 μM) on PARP cleavage.
Due to the small number of azodioxide complexes known, it is difficult to identify a structure–activity relationship with confidence regarding the biological activity of such complexes. As 1 and 2 are the only cobalt azodioxide complexes known, the differences in their coordination numbers and geometries cannot be compared in depth. The data do suggest, however, that the azodioxide ligand is essential to the activity. The inactivity of CoCl2, Co(bpy)Cl2, and Co(phen)Cl2 toward the induction of PARP cleavage demonstrates the neither Co(II) alone nor Co(II) bound to a π-delocalized, redox-active ligand33 such as bpy or phen is necessary to induce apoptosis. The azodioxide ligand, although stable in the solid state, dissociates to monomeric PhNO in the solution, which is also inactive toward the induction of PARP cleavage. We propose that the stabilization of dimeric azodioxide by coordination is responsible for the proapoptotic activity. Nicholas and co-workers have shown that the iron azodioxide compound [Fe(az)3](FeCl4)2 is capable of catalyzing allylic amination/C–C double-bond migration reactions of alkenes, with a mechanism proposed to involve the transfer of a PhNO moiety from a coordinated azodioxide to an alkene in a nitroso-ene-type reaction.25b PhNO transfer from 1 and 2 to unsaturated biomolecules may be involved in their proapoptotic activity. The low levels of ROS generated at the concentrations of 1 and 2 shown to induce apoptosis suggest that ROS generation through Fenton-type electron transfer reactions is at best a minor contribution to the proapoptotic activity of these compounds.
Conclusions
In this study, we have demonstrated that each of compounds 1 and 2 induces apoptosis in SK-HEP-1 cells and examined their mechanisms of action. Our results showed that the compounds induce apoptosis via both the extrinsic and intrinsic pathways, though with a preference for the extrinsic pathway. Both compounds induce the cleavage of PARP and the activation of procaspase-8 and procaspase-9. It is clear that the activation of procaspase-8 proceeded to a greater extent than that of procaspase-9. Additionally, we found that both compounds led to a significant decrease in the levels of the antiapoptotic protein Bcl-2 as well as an increase in the expression of Bim and BAD, members of the Bcl-2 family that promote apoptosis. Future work will consist of examining the pharmacological potential of 1 and 2 in mouse models of liver adenocarcinoma. Mechanistically, the effect of 1 and 2 on additional apoptosis-related proteins, such as the NEDD8-activating enzyme,34 Wee1,35 Cdc2,35b and Pin1,36 will be examined. Additionally, transition-metal azodioxide complexes synthesized in the future will be screened for potential anticancer activity against SK-HEP-1 liver cancer cells and other types of cancer cells.
Methods
Reagents and Antibodies
Compounds 1 and 2, Co(bpy)Cl2 were prepared as previously described by the Boyd group.24 Co(phen)Cl2 was prepared as described by Nami and Siddiqi.32 Antibodies to PARP, procaspase-8, cleaved caspase-8, procaspase-9, cleaved caspase-9, Bim, Bcl-2, and BAD were obtained from Cell Signaling Technology, Inc. (Danvers, MA). The antibody to GAPDH was obtained from Santa Cruz Biotechnology (Dallas, TX).
Cell Culture and Treatment
SK-HEP-1 and other cells were grown in RPMI-1640 (purchased from the Cleveland Clinic Foundation Core Facility) supplemented with 10% cosmic calf serum (HyClone) and 1% antibiotics. The cells were kept in a humidified atmosphere with 5% CO2 at 37 °C. When the cell confluence reached 70%, the cells were treated with or without compound 1 or 2 at different concentrations (0, 1, 2, 5, or 10 μM) for a given amount of time (1, 6, 12, 24, or 36 h). Compounds 1 and 2 were dissolved in DMSO before being diluted in the aqueous solution.
Morphology of Apoptotic Cells
An inverted phase-contrast fluorescence microscope (Carl Zeiss, Heidenheimer, Germany) was used to directly observe morphological changes in the cell line after treatment with 1 or 2. Cells were seeded into 6-well (12 × 104 cells/well) plates, cultured to confluence, and treated with a solution of compound 1 or 2 at the relevant concentration at 37 °C for 12 h. After the 12 h treatment, cells were washed with phosphate-buffered saline (PBS), and images were taken from five random fields for each well.
Western Blot Analysis
After treatments, cells were washed twice with ice-cold PBS and collected with a scraper. Total protein extracts were prepared by the suspension of cell pellets in Triton-X 100 lysis buffer (50 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid, 150 mM NaCl, 1% Tris-X 100, and 5 mM ethylenediaminetetraacetic acid). After centrifugation at 20 000g in a microcentrifuge at 4 °C for 10 min, the total protein in the supernatant was measured using a UV-1280 UV–visible spectrometer (Shimadzu, Kyoto, Japan). Equal amounts of protein from each sample were introduced to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After electrophoresis, proteins were transferred to a poly(vinylidene difluoride) membrane using a Bio-Rad Mini-PROTEAN 3 wet transfer unit at 100 V for 80 min. The membrane was then incubated with 5% nonfat milk in TBST buffer (1 mM Tris with pH 7.4, 150 mM NaCl, 0.2% Tween-20) for 1 h at room temperature. The membrane was then incubated with a primary antibody in TBST overnight at 4 °C. After several washes with TBST, the membrane was incubated with the relevant horseradish peroxidase-linked secondary antibody diluted 1:3000, followed by three washes. The membrane was then processed with the Pierce ECL Plus Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA) and the enhanced chemoluminescence signal acquired.
Flow Cytometry Analysis of Apoptosis
An Annexin V fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences, San Jose, CA) was used. Briefly, following treatment with compound 1 or 2 (10 μM for 12 h), SK-HEP-1 cells (1 × 105 cells/sample) were collected in cold PBS, centrifuged at 1500g at 4 °C for 5 min, washed twice with PBS, and resuspended in 1× binding buffer (100 μL/sample). Each sample was stained with Annexin V-FITC (5 μL) and PI (5 μL) in the dark at 4 °C for 15 min, and 400 μL 1× binding buffer was added. The cells were immediately analyzed with a flow cytometer equipped with the BD FACSDiva software (BD Biosciences). Annexin V-FITC–/PI– cells were identified as viable, Annexin V-FITC+/PI– cells were identified as early apoptotic, Annexin V-FITC+/PI+ cells were identified as late apoptotic, and Annexin V-FITC–/PI+ cells were identified as necrotic.37 Fluorescence spectroscopy of compounds 1 and 2, as a control, was performed on a Hitachi F-7000 fluorescence spectrometer. No emission peaks were observed for acetonitrile solutions of either compound.
Reactive Oxygen Species (ROS) Assay
A fluorometric intracellular ROS kit (Sigma-Aldrich, product number MAK143) was used to test for the production of ROS during the treatment of SK-HEP-1 cells with 1 or 2. Briefly, cells were grown overnight in the medium in a 96-well plate at 4000 cells/well. The master reaction mix was prepared according to the protocol provided by Sigma-Aldrich, and 100 μL of this mixture was added to each well. The cells were incubated for 1 h and then treated with the relevant concentrations of 1 or 2 with PBS buffer. After further incubation for 14 h, the fluorescence intensity was measured on a Hitachi F-7000 fluorescence spectrometer, with an excitation wavelength of 490 nm and an emission wavelength of 525 nm. Each treatment was performed with 16 replicates.
MTT Assay
Cells were grown overnight in the medium in a 96-well plate at 3000 cells/well. The growth medium was replaced by the relevant concentrations of 1 or 2, and the cells were incubated for 48 h. An MTT solution was prepared as a 1:3 ratio of 5 mg/mL MTT and growth medium, and 100 μL of this MTT solution was added to each well. The cells were then incubated for a further 2–4 h, after which 200 μL of DMSO was added to each well. Once all precipitates had dissolved, absorbance was measured at 570 nm. Each treatment was performed with 8 replicates.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01684.
Western blots for the determination of PARP cleavage in additional cell lines and HPLC chromatograms of compounds 1 and 2 (PDF)
Author Present Address
§ Clinical Chemistry Fellowship Program, Department of Pathology & Laboratory Medicine, University of Louisville Hospital, Louisville, Kentucky 40206, United States (N.J.A.).
Author Contributions
L.B., K.A.E., and A.M.H.S. contributed equally to this work and should be jointly considered as second authors.
K.A.E. was supported by a Graduate Student Research Award from Cleveland State University. A.Z. was supported by a Gene Regulation in Health and Disease (GRHD) Award from Cleveland State University.
The authors declare no competing financial interest.
Supplementary Material
References
- Rosenberg B.; VanCamp L.; Trosko J. E.; Mansour V. H. Platinum Compounds: a New Class of Potent Antitumour Agents. Nature 1969, 222, 385–386. 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- a Bruijnincx P. C. A.; Sadler P. J. New trends for metal complexes with anticancer activity. Curr. Opin. Chem. Biol. 2008, 12, 197–206. 10.1016/j.cbpa.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Brujinincx P. C. A.; Sadler P. J. Controlling Platinum, Ruthenium, and Osmium Reactivity for Anticancer Drug Design. Adv. Inorg. Chem. 2009, 61, 1–61. 10.1016/S0898-8838(09)00201-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Arnesano F.; Natile G. Mechanistic insight into the cellular uptake and processing of cisplatin 30 years after its approval by FDA. Coord. Chem. Rev. 2009, 253, 2070–2081. 10.1016/j.ccr.2009.01.028. [DOI] [Google Scholar]; d Jungwirth U.; Kowol C. R.; Keppler B. K.; Hartinger C. G.; Berger W.; Heffeter P. Anticancer Activity of Metal Complexes: Involvement of Redox Processes. Antioxid. Redox Signaling 2011, 15, 1085–1127. 10.1089/ars.2010.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Chakrabortty S.; Agrawalla B. K.; Stumper A.; Vegi N. M.; Fischer S.; Reichardt C.; Kögler M.; Dietzek B.; Feuring-Buske M.; Buske C.; Rau S.; Weil T. Mitochondria Targeted Protein-Ruthenium Photosensitizer for Efficient Photodynamic Applications. J. Am. Chem. Soc. 2017, 139, 2512–2519. 10.1021/jacs.6b13399. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jarman P. J.; Noakes F.; Fairbanks S.; Smitten K.; Griffiths I. K.; Saeed H. K.; Thomas J. A.; Smythe C. Exploring the Cytotoxicity, Uptake, Cellular Response, and Proteomics of Mono- and Dinuclear DNA Light-Switch Complexes. J. Am. Chem. Soc. 2019, 141, 2925–2937. 10.1021/jacs.8b09999. [DOI] [PubMed] [Google Scholar]
- a Yang G.-J.; Zhong H.-J.; Ko C.-N.; Wong S.-Y.; Vellaisamy K.; Ye M.; Ma D.-L.; Leung C.-H. Identification of a rhodium(III) complex as a Wee1 inhibitor against TP53-mutated triple-negative breast cancer cells. Chem. Commun. 2018, 54, 2463–2466. 10.1039/C7CC09384E. [DOI] [PubMed] [Google Scholar]; b Yang G.-J.; Wang W.; Mok S. W. F.; Wu C.; Law B. Y. K.; Miao X.-M.; Wu K.-J.; Zhong H.-J.; Wong C.-Y.; Wong V. K. W.; Ma D.-L.; Leung C.-H. Selective Inhibition of Lysine-Specific Demethylase 5A (KDM5A) Using a Rhodium(III) Complex for Triple-Negative Breast Cancer Therapy. Angew. Chem., Int. Ed. 2018, 57, 13091–13095. 10.1002/anie.201807305. [DOI] [PubMed] [Google Scholar]
- a Kang T.-S.; Wang W.; Zhong H.-J.; Dong Z.-Z.; Huang Q.; Mok S. W. F.; Leung C.-H.; Wong V. K. W.; Ma D.-L. An anti-prostate cancer benzofuran-conjugated iridium(III) complex as a dual inhibitor of STAT3 and NF-κB. Cancer Lett. 2017, 396, 76–84. 10.1016/j.canlet.2017.03.016. [DOI] [PubMed] [Google Scholar]; b Wang W.; Vellaisamy K.; Li G.; Wu C.; Ko C.-N.; Leung C.-H.; Ma D.-L. Development of a Long-Lived Luminescence Probe for Visualizing β-Galactosidase in Ovarian Carcinoma Cells. Anal. Chem. 2017, 89, 11679–11684. 10.1021/acs.analchem.7b03114. [DOI] [PubMed] [Google Scholar]; c Wu C.; Wu K.-J.; Liu J.-B.; Zhou X.-M.; Leung C.-H.; Ma D.-L. A dual-functional molecular strategy for in situ suppressing and visualizing of neuraminidase in aqueous solution using iridium(III) complexes. Chem. Commun. 2019, 55, 6353–6356. 10.1039/C9CC02189B. [DOI] [PubMed] [Google Scholar]
- Emsley J.Nature’s Building Blocks: An A–Z Guide to the Elements, 2nd ed.; Oxford University Press, Oxford, 2011; p 141. [Google Scholar]
- a Hall A. H.; Rumack B. H. Hydroxycobalamin/sodium thiosulfate as a cyanide antidote. J. Emerg. Med. 1987, 5, 115–121. 10.1016/0736-4679(87)90074-6. [DOI] [PubMed] [Google Scholar]; b Spence J. D.; Lees K.; Spence J. D. Nutrition and Stroke Prevention. Stroke 2006, 37, 2430–2435. 10.1161/01.STR.0000236633.40160.ee. [DOI] [PubMed] [Google Scholar]; c Wheatley C. The return of the Scarlet Pimpernel: cobalamin in inflammation II – cobalamins can both selectively promote all three nitric oxide synthases (NOS), particularly iNOS and eNOS, and, as needed, selectively inhibit iNOS and nNOS. J. Nutr. Environ. Med. 2007, 16, 181–211. 10.1080/10520290701791839. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Kalita J.; Agarwal R.; Chandra S.; Misra U. K. A study of neurobehavioral, clinical psychometric, and P3 changes in vitamin B12 deficiency neurological syndrome. Nutr. Neurosci. 2013, 16, 39–46. 10.1179/1476830512Y.0000000028. [DOI] [PubMed] [Google Scholar]; e Elliott T. R.; Guildford A. L. An In Vitro Model of Gastric Inflammation and Treatment with Cobalamin. Int. J. Inflammation 2017, 2017, 5968618 10.1155/2017/5968618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Schwartz J. A.; Lium E. K.; Silverstein S. J. Herpes Simplex Virus Type 1 Entry Is Inhibited by the Cobalt Chelate Complex CTC-96. J. Virol. 2001, 75, 4117–4128. 10.1128/JVI.75.9.4117-4128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mjos K. D.; Orvig C. Metallodrugs in Medicinal Inorganic Chemistry. Chem. Rev. 2014, 114, 4540–4563. 10.1021/cr400460s. [DOI] [PubMed] [Google Scholar]
- Gurley L.; Beloukhina N.; Boudreau K.; Klegeris A.; McNeil W. S. The synthesis and characterization of a series of cobalt(II) β-ketoaminato complexes and their cytotoxic activity towards human tumor cell lines. J. Inorg. Biochem. 2011, 105, 858–866. 10.1016/j.jinorgbio.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Silva T. F. S.; Martins L. M. D. R. S.; Guedes da Silva M. F. C.; Fernandes A. R.; Silva A.; Borralho P. M.; Santos S.; Rodrigues C. M. P.; Pombeiro A. J. L. Cobalt complexes bearing scorpionate ligands: synthesis, characterization, cytotoxicity and DNA cleavage. Dalton Trans. 2012, 41, 12888–12897. 10.1039/c2dt11577h. [DOI] [PubMed] [Google Scholar]
- Tabrizi L.; Fooladivanda M.; Chiniforoshan H. Copper(II), cobalt(II) and nickel(II) complexes of juglone: synthesis, structure, DNA interaction and enhanced cytotoxicity. Biometals 2016, 29, 981–993. 10.1007/s10534-016-9970-0. [DOI] [PubMed] [Google Scholar]
- Gao E.; Qi Z.; Qu Y.; Ding Y.; Zhan Y.; Sun N.; Zhang S.; Qiu X.; Zhu M. Two novel dinuclear ellipsoid Ni(II) and Co(II) complexes bridge by 4,5-bis(pyrazol-1-yl)phthalic acid: Synthesis, structural characterization and biological evaluation. Eur. J. Med. Chem. 2017, 136, 235–245. 10.1016/j.ejmech.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Zhu M.; Zhao H.; Peng T.; Su J.; Meng B.; Qi Z.; Jia B.; Feng Y.; Gao E. Structure and cytotoxicity of zinc (II) and cobalt (II) complexes based on 1,3,5-tris(1-imidazolyl)benzene. Appl. Organomet. Chem. 2019, 33, e4734 10.1002/aoc.4734. [DOI] [Google Scholar]
- Eskandari A.; Kundu A.; Lu C.; Ghosh S.; Suntharalingam K. Synthesis, characterization, and cytotoxic properties of mono- and di-nuclear cobalt(II) polypyridyl complexes. Dalton Trans. 2018, 47, 5755–5763. 10.1039/C8DT00577J. [DOI] [PubMed] [Google Scholar]
- Nithya P.; Rajamanikandan R.; Simpson J.; Ilanchelian M.; Govindajaran S. Solvent assisted synthesis, structural characterization and biological evaluation of cobalt(II) and nickel(II) complexes of Schiff bases generated from benzyl carbazate and cyclic ketones. Polyhedron 2018, 145, 200–217. 10.1016/j.poly.2018.02.008. [DOI] [Google Scholar]
- Katsaros N.; Katsarou M.; Sovilj S. P.; Babić-Samardžija K.; Mitić D. M. Biological Activity of Some Cobalt(II) and Molybdenum(VI) Complexes: in vitro Cytotoxicity. Bioinorg. Chem. Appl. 2004, 2, 193–207. 10.1155/S1565363304000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshkourfu R.; Čobeljić B.; Vujčić M.; Turel I.; Pevec A.; Sepčić K.; Zec M.; Radulović S.; Srdić-Radić T.; Mitić D.; Andjelković K.; Sladić D. Synthesis, characterization, cytotoxic activity, and DNA binding properties of the novel dinuclear cobalt(III) complex with the condensation product of 2-acetylpyridine and malonic acid dihydrazide. J. Inorg. Biochem. 2011, 105, 1196–1203. 10.1016/j.jinorgbio.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Qin Q.-P.; Qin J.-L.; Meng T.; Lin W.-H.; Zhang C.-H.; Wei Z.-Z.; Chen J.-N.; Liu Y.-C.; Liang H.; Chen Z.-F. High in vivo antitumor activity of cobalt oxoisoaporphine complexes by targeting G-quadruplex DNA, telomerase and disrupting mitochondrial function. Eur. J. Med. Chem. 2016, 124, 380–392. 10.1016/j.ejmech.2016.08.063. [DOI] [PubMed] [Google Scholar]
- Ravi Ch.; Dandu K.; Kumar V. R.; Kumar Y. P.; Thakur S. S.; Mohan Rao Ch.; Satyanarayana S. Synthesis, Characterization, DNA-binding, Photocleavage, Cytotoxicity of Co(III) Mixed Polypyridyl Complexes. Int. J. Pharm. Sci. Rev. Res. 2016, 41, 372–379. [Google Scholar]
- Todorović T. R.; Vukašinović J.; Portalone G.; Suleiman S.; Gligorijević N.; Bjelogrlić S.; Jovanović K.; Radulović S.; Anđelković K.; Cassar A.; Filipović N. R.; Schembri-Wismayer P. (Chalcogen)semicarbazones and their cobalt complexes differentiate HL-60 myeloid leukaemia cells and are cytotoxic towards tumor cell lines. Med. Chem. Commun. 2017, 8, 103–111. 10.1039/C6MD00501B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B.-Q.; Wang S.-L.; Qin Q.-P.; Bai Y.-X.; Tan M.-X. Synthesis, Characterization, and Cytotoxicity of the Cobalt (III) Complex with N,N-Diethyl-4-(2,2′:6′,2″-terpyridin-4′-yl)aniline. Chem. Biodiversity 2018, 15, e1800215 10.1002/cbdv.201800215. [DOI] [PubMed] [Google Scholar]
- a Kumar R. S.; Arunachalam S.; Periasamy V. S.; Preethy C. P.; Riyasdeen A.; Akbarsha M. A.. Surfactant–cobalt(III) complexes: Synthesis, critical micelle concentration (CMC) determination, DNA binding, antimicrobial and cytotoxicity studies. J. Inorg. Biochem. 2009, 103, 117–127. 10.1016/j.jinorgbio.2008.09.010 [DOI] [PubMed] [Google Scholar]; b Kumar R. S.; Riyasdeen A.; Dinesh M.; Paul C. P.; Srinag S.; Krishnamurthy H.; Arunachalam S.; Akbarsha M. A. Cytotoxic Property of Surfactant–Cobalt(III) Complexes on a Human Breast Cancer Cell Line. Arch. Pharm. 2011, 344, 422–430. 10.1002/ardp.201000144. [DOI] [PubMed] [Google Scholar]; c Kumar R. S.; Paul P.; Riyasdeen A.; Wagniéres G.; van den Bergh H.; Akbarsha M. A.; Arunachalam S. Human serum albumin binding and cytotoxicity studies of surfactant–cobalt(III) complex containing 1,10-phenanthroline ligand. Colloids Surf., B 2011, 86, 35–44. 10.1016/j.colsurfb.2011.03.012. [DOI] [PubMed] [Google Scholar]; d Nagaraj K.; Arunachalam S. Synthesis, CMC determination, nucleic acid binding and cytotoxicity of a surfactant–cobalt(III) complex: effect of ionic liquid additive. New J. Chem. 2014, 38, 366–375. 10.1039/C3NJ00832K. [DOI] [Google Scholar]
- Nagaraj K.; Murugan K. S.; Thangamuniyandi P.; Sakthinathan S. Synthesis, Micellization Behaviour, DNA/RNA Binding and Biological Studies of a Suractant Cobalt(III) Complex With Dipyrido[3,2-a:2′,4′-c](6,7,8,9-tetrahydro)phenazine. J. Fluoresc. 2014, 24, 1701–1714. 10.1007/s10895-014-1457-1. [DOI] [PubMed] [Google Scholar]
- Emhoff K. A.; Balaraman L.; Simpson S. R.; Stromyer M. L.; Kalil H. F.; Beemiller J. R.; Sikatzki P.; Eshelman T. S.; Salem A. M. H.; DeBord M. A.; Panzner M. J.; Youngs W. J.; Boyd W. C. Synthesis and Characterization of Cobalt(II) N,N′-Diphenylazodioxide Complexes. ACS Omega 2018, 3, 16021–16027. 10.1021/acsomega.8b01200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Srivastava R. S.; Khan M. A.; Nicholas K. M. A Novel Intermediate in Allylic Amination Catalyzed by Iron Salts. J. Am. Chem. Soc. 1996, 118, 3311–3312. 10.1021/ja960088+. [DOI] [Google Scholar]; b Srivastava R. S.; Nicholas K. M. On the Mechanism of Allylic Amination Catalyzed by Iron Salts. J. Am. Chem. Soc. 1997, 119, 3302–3310. 10.1021/ja964006t. [DOI] [Google Scholar]; c Lightfoot A. P.; Pritchard R. G.; Wan H.; Warren J. N.; Whiting A. A novel scandium ortho-methoxynitrosobenzene-dimer complex: mechanistic implications for the nitroso-Diels-Alder reaction. Chem. Commun. 2002, 6, 2072–2073. 10.1039/B206366B. [DOI] [PubMed] [Google Scholar]; d Fitts L. S.; Bierschenk E. J.; Hanusa T. P.; Rheingold A. L.; Pink M.; Young V. G. Jr. Selective modification of the metal coordination environment in heavy alkaline-earth iodide complexes. New J. Chem. 2016, 40, 8229–8238. 10.1039/C6NJ01713D. [DOI] [Google Scholar]; e Emhoff K. A.; Balaraman L.; Salem A. M. H.; Mudarmah K. I.; Boyd W. C. Coordination chemistry of organic nitric oxide derivatives. Coord. Chem. Rev. 2019, 396, 124–140. 10.1016/j.ccr.2019.06.011. [DOI] [Google Scholar]
- Widmann C.; Gibson S.; Johnson G. L. Caspase-dependent Cleavage of Signaling Proteins during Apoptosis: A Turn-off Mechanism for Anti-apoptotic Signals. J. Biol. Chem. 1998, 273, 7141–7147. 10.1074/jbc.273.12.7141. [DOI] [PubMed] [Google Scholar]
- Knight T.; Luedtke D.; Edwards H.; Taub J. W.; Ge Y. A delicate balance – The BCL-2 family and its role in apoptosis, oncogenesis, and cancer therapeutics. Biochem. Pharmacol. 2019, 162, 250–261. 10.1016/j.bcp.2019.01.015. [DOI] [PubMed] [Google Scholar]
- Feldstein A. E.; Gores G. J. Apoptosis in alcoholic and nonalcoholic steatohepatitis. Front. Biosci. 2005, 10, 3093–3099. 10.2741/1765. [DOI] [PubMed] [Google Scholar]
- a Beyersmann D.; Hartwig A. The Genetic Toxicology of Cobalt. Toxicol. Appl. Pharmacol. 1992, 115, 137–145. 10.1016/0041-008X(92)90377-5. [DOI] [PubMed] [Google Scholar]; b Kasprzak K. S.; Zastawny T. H.; North S. L.; Riggs C. W.; Diwan B. A.; Rice J. M.; Dizdaroglu M. Oxidative DNA Base Damage in Renal, Hepatic, and Pulmonary Chromatin of Rats after Intraperitoneal Injection of Cobalt(II) Acetate. Chem. Res. Toxicol. 1994, 7, 329–335. 10.1021/tx00039a009. [DOI] [PubMed] [Google Scholar]; c Beyersmann D.; Hartwig A. Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch. Toxicol. 2008, 82, 493–512. 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- a Jo H. Y.; Kim Y.; Park H. W.; Moon H. E.; Bae S.; Kim J.; Kim D. G.; Paek S. H. The Unreliability of MTT Assay in the Cytotoxic Test of Primary Cultured Glioblastoma Cells. Exp. Neurobiol. 2015, 24, 235–245. 10.5607/en.2015.24.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Śliwka L.; Wiktorska K.; Suchocki P.; Milczarek M.; Mielczarek S.; Lubelska K.; Cierpał T.; Łyżwa P.; Kiełbasiński P.; Jaromin A.; Flis A.; Chilmonczyk Z. The Comparison of MTT and CVS Assays for the Assessment of Anticancer Agent Interactions. PLoS One 2016, 11, e0155772 10.1371/journal.pone.0155772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M.; Reddy M. M.; Maikap G. C.; Iqbal J. Cobalt(II)-Catalyzed Conversion of Allylic Alcohols/Acetates to Allylic Amides in the Presence of Nitriles. J. Org. Chem. 1995, 60, 2670–2676. 10.1021/jo00114a013. [DOI] [Google Scholar]
- Nami S. A. A.; Siddiqi K. S. Unique keto-enol tautomerism in transition metal complexes of cyanoimidodithiocarbonate. J. Chem. Res. 2006, 2006, 563–565. 10.3184/030823406778521392. [DOI] [Google Scholar]
- a Kalyanasundaram K. Photophysics, Photochemistry and Solar Energy Conversion with Tris(bipyridyl)ruthenium(II) and Its Analogues. Coord. Chem. Rev. 1982, 46, 159–244. 10.1016/0010-8545(82)85003-0. [DOI] [Google Scholar]; b Juris A.; Balzani V.; Barigelletti F.; Campagna S.; Belser P.; von Zelewsky A. Ru(II) Polypyridine Complexes: Photophysics, Photochemistry, Electrochemistry, and Chemiluminescence. Coord. Chem. Rev. 1988, 84, 85–277. 10.1016/0010-8545(88)80032-8. [DOI] [Google Scholar]
- Wu K.-J.; Zhong H.-J.; Li G.; Liu C.; Wang H.-M. D.; Ma D.-L.; Leung C.-H. Structure-based identification of a NEDD8-activating enzyme inhibitor via drug repurposing. Eur. J. Med. Chem. 2018, 143, 1021–1027. 10.1016/j.ejmech.2017.11.101. [DOI] [PubMed] [Google Scholar]
- a Yang G.-J.; Zhong H.-J.; Ko C.-N.; Wong S.-Y.; Vellaisamy K.; Ye M.; Ma D.-L.; Leung C.-H. Identification of a rhodium(III) complex as a Wee1 inhibitor against TP53-mutated triple-negative breast cancer cells. Chem. Commun. 2018, 54, 2463–2466. 10.1039/C7CC09384E. [DOI] [PubMed] [Google Scholar]; b Yuan H.; Xie Y.-M.; Chen I. S. Y. Depletion of Wee-1 Kinase is Necessary for both Human Immunodeficiency Virus Type 1 Vpr- and Gamma Irradiation-Induced Apoptosis. J. Virol. 2003, 77, 2063–2070. 10.1128/JVI.77.3.2063-2070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.-J.; Zhong H.-J.; Yang G.; Wu C.; Huang J.-M.; Li G.; Ma D.-L.; Leung C.-H. Small Molecule Pin1 Inhibitor Blocking NF-κB Signaling in Prostate Cancer Cells. Chem. - Asian J. 2018, 13, 275–279. 10.1002/asia.201701216. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Qi Y.; Diao Q.; Wu L.; Du X.; Li Y.; Sun L. Cytotoxicity of melittin and apamin in human hepatic L02 and HepG2 cells in vitro. Toxin Rev. 2013, 32, 60–67. 10.3109/15569543.2013.852108. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.