Abstract
The principal α subunit of voltage-gated sodium channels is associated with auxiliary β subunits that modify channel function and mediate protein-protein interactions. We have identified a new β subunit termed β4. Like the β1-β3 subunits, β4 contains a cleaved signal sequence, an extracellular Ig-like fold, a transmembrane segment, and a short intracellular C-terminal tail. Using TaqMan reverse transcription-PCR analysis, in situ hybridization, and immunocytochemistry, we show that β4 is widely distributed in neurons in the brain, spinal cord, and some sensory neurons.β4 is most similar to theβ2 subunit (35% identity), and, like theβ2 subunit, the Ig-like fold of β4 contains an unpaired cysteine that may interact with the α subunit. Under nonreducing conditions, β4 has a molecular mass exceeding 250 kDa because of its covalent linkage to Nav1.2a, whereas on reduction, it migrates with a molecular mass of 38 kDa, similar to the mature glycosylated forms of the otherβ subunits. Coexpression ofβ4 with brain Nav1.2a and skeletal muscle Nav1.4 α subunits in tsA-201 cells resulted in a negative shift in the voltage dependence of channel activation, which overrode the opposite effects ofβ1 andβ3 subunits when they were present. This novel, disulfide-linked β subunit is likely to affect both protein-protein interactions and physiological function of multiple sodium channel α subunits.
Keywords: sodium channel, auxiliary subunit, β4, cDNA cloning, tissue distribution, disulfide link, voltage dependence of activation
Introduction
Voltage-gated sodium channels purified from brain are composed of a pore-forming α subunit and associated β subunits (Catterall, 2000). To date, nine functional α subunits have been identified (Nav1.1-1.9; Goldin et al., 2000). They are large proteins (∼2000 amino acids), with 24 transmembrane helices arranged in four homologous domains. Each domain is composed of six transmembrane helices surrounding a reentrant hydrophobic loop that is believed to form the outer pore of the channel (Catterall, 2000). Two β subunits were originally identified as proteins copurifying with the brain α subunits (Catterall, 2000). β1 and β2 are related proteins with a large extracellular Ig-like domain, a single transmembrane domain, and a short cytoplasmic tail (Isom et al., 1992, 1995). A notable difference is that β1is noncovalently associated with α subunits and has four cysteines in its extracellular domain that contribute to the Ig-like fold, whereas β2 has five extracellular cysteines and forms a disulfide bond to the α subunit.
The β subunits display several important functional properties. First, in Xenopus oocytes, expression of α subunits alone results in functional sodium channels, but these do not exhibit the “fast-gating” kinetic parameters of sodium channels in neurons. However, coexpression of β1 and β2 subunits results in a small acceleration of the rate of activation and a substantial acceleration of the rate of inactivation during test pulses, which can be modeled as a shift from a slow-gating mode to a fast-gating mode in the presence of β subunits (Isom et al., 1992, 1995). Second, interactions of β1 and β2 with ankyrin, tenascin-R, and neurofascin are likely to be responsible for targeting sodium channel complexes to nodes of Ranvier, which is essential for the saltatory conduction of action potentials in myelinated nerves (Srinivasan et al., 1998; Malhotra et al., 2000; Ratcliffe et al., 2001). Finally, the importance of the β subunits is underscored by the association of generalized epilepsy with febrile seizures plus (GEFS+) with a mutation in the β1 gene (SCN1B) that disrupts one of the cysteines responsible for the structure of the Ig-like fold, resulting in slower inactivation and slower recovery from inactivation of sodium channels (Wallace et al., 1998). Other febrile epilepsy syndromes are also associated with mutations in the Nav1.1 channel (SCN1A; Escayg et al., 2000; Claes et al., 2001; Wallace et al., 2001).
Recently, we and others have identified a third β subunit based on structural and sequence homology to β1 and β2 (Morgan et al., 2000; Qu et al., 2001). The β3 subunit modulates the sodium channel gating when expressed in Xenopus oocytes or in mammalian cell systems (Morgan et al., 2000; Qu et al., 2001). It causes a positive shift in the voltage dependence of channel activation and increased persistent sodium current on expression in the human embryonic cell line tsA-201 (Qu et al., 2001). β3 also binds neurofascin and may be involved in sodium channel clustering at nodes of Ranvier (Ratcliffe et al., 2001). These findings raised the possibility that further β subunits might exist. Here, we report the identification of the fourth β subunit, designated β4. The sodium channel β4 subunit is most highly related to the β2 subunit, although it also shares substantial sequence similarity with β1 and β3. Its distinct localization and function indicate that it may differentially affect sodium channel protein-protein interactions and physiological function in the neurons and other cell types in which it is expressed.
Materials and Methods
Bioinformatics, cloning, and sequence analysis. To identify additional sodium channel β subunits, the protein sequences of the human sodium channel β1-β3 subunits were used in a TBLASTN (Altschul et al., 1997) search of the Millennium Pharmaceuticals database of expressed sequence tags (ESTs) and the public dbEST database. Several human clones were identified on the basis of primary homology to the sodium channel β2 subunit, and these were sequenced. The complete 4489 bp cDNA (GenBank accession number AY149967) was found to contain a single open reading frame encoding a protein of 228 amino acids plus a 3649 bp 3′ untranslated region. BLAST search identified the genomic sequence in human chromosome 11 clones AC068591 and AP002800. The human β4 sequence was aligned with the human genomic sequence, and five exons were identified and confirmed by examination of the splice donor and acceptor sites.
A partial rat clone encoding 95 amino acid residues at the 3′ end of the open reading frame and >3500 bp of the 3′ untranslated region was also sequenced and used to generate the TaqMan reagents and in situ hybridization reagents (see below). In addition, the full-length coding sequences of rat and mouse β4 subunits were deduced from genomic sequencing data. BLAST search revealed the genomic sequence in AC129680, AC129457, and AC136555 (rat) and in AC122504 and AC122305 (mouse). The human β4 sequence was aligned with the rat and mouse genomic sequences to identify the exons, which were confirmed by examination of the splice junctions. The deduced full-length rat and mouse β4 sequences were deposited in GenBank (BK001030 and BK001031, respectively).
Tissue distribution of β4 mRNA. Adult male Sprague Dawley rats (250 gm) were anesthetized with ketamine (50 mg/kg) plus xylazine (10 mg/kg) and exsanguinated. Brain, spinal cord, dorsal root ganglion, superior cervical ganglion, heart, gastrocnemius muscle, and selected peripheral organs were removed. Expression in the nervous system and peripheral tissues was assessed by TaqMan real-time quantitative reverse transcription (RT)-PCR (Medhurst et al., 2000). mRNA was extracted, and cDNA was synthesized by reverse transcription (SuperScript preamplification system; Invitrogen, Carlsbad, CA) as described (Qu et al., 2001). PCR was performed using TaqMan universal PCR master mix on the ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA), with the following probe and primers designed with Primer Express software (Applied Biosystems): sense, 5′-ACTCTGCTACGATCTTCCTCCAA-3′; antisense, 5′-GCAGACGAGAAGTCCGATGAC-3′; and fluorescent probe, 5′-CCGCCTACCACAGCCAGGATGATG-3′. β4 mRNA levels were expressed as arbitrary values relative to the 18 S ribosomal RNA.
Twelve-micrometer fresh-frozen sections were prepared from rat tissues (brain, spinal cord, dorsal root ganglion, heart, and gastrocnemius muscle). In situ hybridization was performed with 35S-labeled rat β4 cRNA probes as described (Stahl et al., 1999; Qu et al., 2001).
Immunocytochemical localization of β4 subunits. The rabbit polyclonal anti-β4 antibody was generated against the peptide EGTVKNEKSDPKVTLKD corresponding to amino acids 51-67 of the predicted amino acid sequence of the mature β4 protein (Fig. 1 A) coupled through an additional C-terminal Cys residue to keyhole limpet hemocyanin (Affinity Bioreagents, Golden, CO). The rabbit polyclonal anti-β2 antibody was described previously (Ratcliffe et al., 2000). Tissue sections were prepared as described (Westenbroek et al., 1998). Free-floating tissue sections were rinsed in Tris buffer for 30 min, rinsed in Tris-buffered saline (TBS) for 30 min, blocked in 2% avidin in TBS for 30 min, rinsed in TBS for 30 min, blocked in 2% biotin in TBS for 30 min, and finally rinsed in TBS. The sections were then incubated in peptide affinity-purified primary anti-β2 (diluted 1:15) or anti-β4 (diluted 1:25) antibodies for 36 hr at 4°C. The sections were rinsed in TBS for 1 hr, incubated in biotinylated goat anti-rabbit IgG for 1 hr at 37°C, rinsed in TBS for 1 hr, incubated in avidin D fluorescein for 1 hr at 37°C, rinsed, mounted onto gelatin-subbed slides, coverslipped with Vectashield, and viewed with a Bio-Rad (Hercules, CA) MRC 600 microscope located in the W. M. Keck Imaging Facility at the University of Washington.
Figure 1.

Amino acid sequence and tissue distribution of the sodium channel β4 subunit. A, Alignment of human sodium channel β4 and β2 (hβ4 AY149967, hβ2 AAF21472) subunits using Clustal W. Identical residues are shown as white letters against black, and similar residues are black letters on gray. The solid box indicates the N-terminal signal sequence predicted for β4 and determined for β2 (Isom et al., 1995). Numbering of the amino acid sequence (above) is relative to the predicted first residue of the mature β4 protein after cleavage of the signal peptide. The asterisks show conserved cysteine residues that form a disulfide bond in the Ig-like domain. The solid triangle indicates the conserved cysteine in β2 and β4, which may be covalently linked to sodium channel α subunits. The dotted box indicates the predicted transmembrane domains. B, Expression of the sodium channelβ4 subunit in the CNS and peripheral tissues. TaqMan RT-PCR was used to quantify β4 subunit mRNA, expressed as arbitrary units relative to the 18 S RNA internal reference. Lane 1, Brain; lane 2, spinal cord; lane 3, dorsal root ganglion; lane 4, superior cervical ganglion; lane 5, hairy skin; lane 6, gastrocnemius muscle; lane 7, heart; lane 8, kidney; lane 9, liver; lane 10, lung; lane 11, spleen; lane 12, aorta; lane 13, adrenal gland; lane 14, salivary gland; lane 15, thyroid; lane 16, prostate; lane 17, thymus; lane 18, trachea; lane 19, testes; lane 20, colon.
Sodium channel expression and electrophysiological recording. Plasmids pCDM8-rIIA (containing the cDNA fragment of the full-length rat Nav1.2a α subunit; Linford et al., 1998) and pCDM8-rH1 (containing the cDNA of the full-length rat heart Nav1.5 sodium channel α subunit; Qu et al., 1994) have been described. The rSKM1 cDNA encoding the Nav1.4 α subunit (Featherstone et al., 1998) was a kind gift from Dr. Peter Ruben (Utah State University) and was subcloned into pCDM8 to produce pCDM8-rSKM1. The full-length human β4 cDNA was subcloned into mammalian expression vectors to create pCDNA3.1/B4 (Invitrogen, San Diego, CA) or pIRES-EGFP/B4 (Clontech, Palo Alto, CA). To facilitate efficient detection of heterologously expressed sodium channel subunits, one copy of an influenza virus hemagglutinin (HA) peptide (YPYDVPDYA) was appended to the cytoplasmic N-terminus of Nav1.2a (designated pCDM8 HA-rIIA) and to the C-terminus of β4 (designated pCDNA3.1/B4-HA) by PCR. All cDNA fragments that were subcloned after PCR amplification were verified by DNA sequencing.
tsA-201 cells, a subclonal line of human embryonic kidney 293 cells, were maintained as described (Herlitze et al., 1996). Sodium channel expression plasmids were transiently transfected using the calcium phosphate coprecipitation method. Twelve hours later, transfected cells were replated at low density for electrophysiological recordings. For biochemical studies, transfected cells were harvested 40 hr after transfection, and membrane proteins were isolated as described previously (Zhong et al., 1999). Immunoprecipitation reactions were performed by combining 500 μg of total protein with 30 μg of affinity-purified anti-SP20 antibody recognizing the sodium channel α subunit or control IgG antibody and incubated for 2 hr at 4°C in 50 mm Tris-Cl, 150 mm NaCl, 1 mm PMSF, and 1% Triton X-100, pH 8.0. Protein A-Sepharose beads were added, incubated overnight, and washed extensively, and the immunoprecipitated proteins were eluted by boiling in SDS-PAGE sample buffer containing 100 mm dithiothreitol. For elution of the bound complex under nonreducing conditions, proteins bound to the washed Sepharose beads were denatured in 8 m urea for 10 min at 20°C. Samples were divided into two equal portions and boiled for 5 min after adding SDS-PAGE sample buffer with and without 5% β-mercaptoethanol. Proteins were resolved using SDS-PAGE and transferred onto nitrocellulose, and HA tags were revealed with monoclonal antibody HA11 (Covance, Princeton, NJ)
Whole-cell voltage-clamp experiments were performed on tsA-201 cells that were transfected with Nav1.2a sodium channel α subunit plasmid pCDM8-rIIA with or without β2 or β4 subunit cDNAs in pIRESEGFP. Transfected cells were identified by fluorescence. Recording solutions and voltage protocols have been described previously (Mantegazza et al., 2001). The bath solution contained (in mm): 140 NaCl, 2 CaCl2, 2 MgCl2, and 10 HEPES, pH adjusted to 7.4 with NaOH. The pipette solution contained (in mm): 120 Cs-aspartate, 5 NaCl, 2 MgCl2, 10 EGTA, and 10 HEPES, pH adjusted to 7.3 with CsOH. Conductance-voltage (g-V) relationships were calculated from the current-voltage (I-V) relationships according to the extended Ohm's law: g =INa/(V - ENa), where INa is the peak Na+ current measured at potential V, and ENa is the calculated Nernst equilibrium potential. The normalized g-V relationships and inactivation curves were fit with a Boltzmann distribution: 1/(1 + exp[(V - V1/2)/k]), where V1/2 is the voltage at which half-activation or half-inactivation occurred, and k is the slope factor. Statistical results are reported as means ± SEM. Statistical comparisons were done using Student's t test or ANOVA followed by Tukey's post test, with p < 0.05 as the criterion for significance.
Results
Cloning and sequence analysis of the β4 subunit
The discovery of the sodium channel β3 subunit (Morgan et al., 2000; Qu et al., 2001) suggested the existence of an extended family of related sodium channel auxiliary subunits. We searched the EST databases for novel sequences with homology to the known β subunits. We identified several human clones representing a single cDNA with highest homology to the β2 subunit (Fig. 1A). The novel β4 subunit is 228 amino acid residues in length and shares 35% amino acid identity with the β2 subunit but only 20-22% identity with β1 and β3. The β4 subunit is composed of five exons covering 19.5 kb, whereas the β2 subunit has only four exons. Interestingly, human β4 maps to 11q23 ∼12.5 kb downstream of human β2 (Eubanks et al., 1997), and the genomic organization of the two subunits is very similar. The first three exons of a β4 subunit are completely conserved with those of a β2 subunit. However, the equivalent of the fourth exon is split by an intron of 4 kb into exons 4 and 5 of the β4 gene. The human β3 gene is located on chromosome 11q24 and also consists of five exons with a similar organization, suggesting multiple gene duplication events.
Rat and mouse β4 subunit sequences were deduced from genomic sequences, and the predicted proteins share 80% amino acid identity with the human protein. Although the open reading frame encodes a 228-amino acid protein, the cDNA is 4489 bp in length because of a 3649 bp 3′ untranslated region, similar to the extended 3′ untranslated regions of the β1, β2 (Isom et al., 1992, 1995), and β3 (GenBank number AF378093) subunits. The significance of these large 3′ untranslated regions is unclear at present.
Like the other β subunits, the β4 subunit is a type 1 membrane protein. Hydropathy analysis indicates that the N-terminal 30 amino acid residues form a hydrophobic domain that is likely to be a cleaved signal sequence. The predicted N terminus of the mature β4 protein (Leu-1; Fig. 1A) aligns precisely with the experimentally established N terminus of mature β2 (Isom et al., 1995). The mature protein has a large extracellular domain, which is predicted to form an Ig-like fold, a single transmembrane α helix, and a short cytoplasmic C-terminal region. There are three cysteines in the Ig-like fold. The cysteines at positions 23 and 101 of the predicted mature protein (Fig. 1A) are completely conserved in all other β subunits as well as in other V-type Ig-like folds, and these have been proposed to form an intramolecular disulfide bond that stabilizes the structure of the extracellular domain. Mutation of one of the corresponding cysteines in the β1 subunit leads to a febrile epilepsy syndrome (Wallace et al., 1998). The additional cysteine at position 28 of the mature β4 protein is conserved in the disulfide-linked β2 subunit but not in β1 or β3 and therefore may form the disulfide linkage to the α subunit.
Tissue and cellular distribution of β4 mRNA
To determine the tissue distribution of the sodium channel β4 subunit, we performed TaqMan quantitative RT-PCR using a panel containing 20 different rat tissues (Fig. 1B). Expression was highest in the dorsal root ganglia (lane 3), and lower levels were detected in brain, spinal cord, skeletal muscle, and heart (lanes 1, 2, 6, 7). Notably, there was no expression of β4 in the superior cervical ganglion. The β4 subunit was expressed primarily in excitable tissues, with no cDNA amplification detected from non-excitable tissue samples under our conditions except a low signal in skin. TaqMan analysis of a more restricted panel of human tissues gave essentially similar results (data not shown).
Regional expression and cellular distribution of the β4 subunit in the nervous system were analyzed by in situ hybridization using rat tissue sections. The β4 subunit was expressed in a restricted pattern throughout the cerebral cortex (Fig. 2A). Expression was observed in pyramidal cells, mainly in layer V, and in other cortical neurons in layers III and VI. No expression was detected in laminae I, II, or IV. β4 was present at high levels in cerebellar Purkinje cells (Fig. 2B) and deep cerebellar nuclei (data not shown). Within the thalamus, β4 was detected at high levels in the nucleus reticularis (Fig. 2C) and at lower levels in various other nuclei but was absent from the posterior and ventrolateral thalamic nuclei. In the hippocampal region, β4 was expressed at very low levels in the pyramidal cells and in a small number of hilar neurons in the dentate gyrus (Fig. 2D). However, there was no expression in dentate granule neurons. Within the striatum, β4 was expressed in essentially all neurons in the caudate putamen but in only a few cells in the globus pallidus (Fig. 2E). Other areas expressing β4 include lateral and medial septal nuclei, the piriform cortex, the diagonal band, the magnocellular preoptic nucleus, and the bed nucleus of the stria terminalis. In the mesencephalic area, β4 was present at high levels in the substantia nigra reticulata (but not in substantia nigra compacta), the red nucleus, mammillary nuclei, deep mesencephalic nuclei, and a subpopulation of neurons in the superior colliculus and medial geniculate nucleus (data not shown). Very high levels were observed throughout the pons and brainstem, including the majority of motor nuclei as well as the reticular nuclei and the spinal trigeminal nuclei. There was little or no expression in central glial cells.
Figure 2.
Distribution of the sodium channel β4 subunit mRNA in the brain. Sodium channel mRNA distribution is shown in dark field autoradiograms of rat tissue sections. A, Cerebral cortex; B, Purkinje cell layer of the cerebellum; C, nucleus reticularis of the thalamus (indicated by the dotted line); D, hippocampus (arrows indicate the width of the pyramidal cell layer); E, caudate putamen (CP) and globus pallidus (GP). Scale bars, 100 μm.
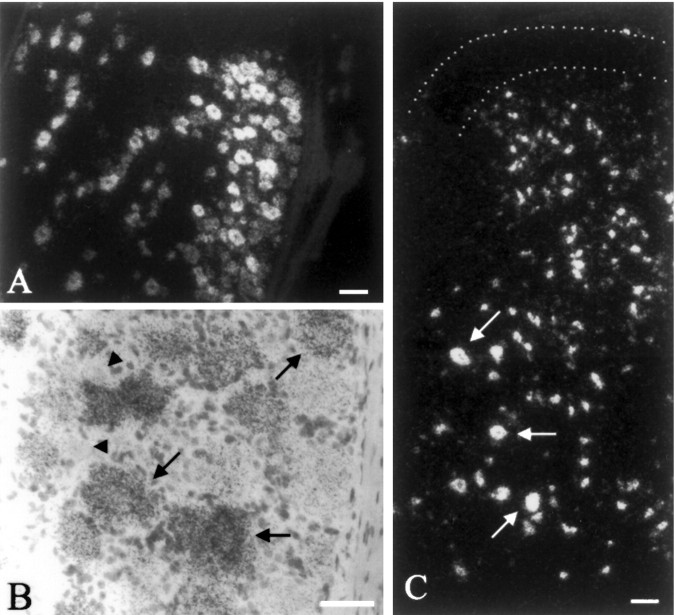
The majority of sensory neurons in the dorsal root ganglia (Fig. 3A,B) and trigeminal ganglion (data not shown) expressed β4 mRNA. High levels were expressed in the large proprioceptive neurons, whereas small and intermediate-sized cells typically displayed lower levels. Labeling was observed in a subpopulation of small (presumably nociceptive) sensory neurons, but many others did not express β4. No β4 expression was detected in sympathetic neurons in the superior cervical ganglion (data not shown). In the spinal cord, β4 mRNA was detected in ventral horn motor neurons and interneurons of the intermediate zone and deep laminae of the dorsal horn (Fig. 3C). No expression was detected in the superficial laminae of the dorsal horn (laminae I and II) or in lamina X around the central canal.
Figure 3.
Expression of the β4 subunit in the spinal cord and sensory neurons. A, Dark-field image of dorsal root ganglion neurons. B, Bright-field image of dorsal root ganglion neurons showing high expression in the majority of neurons (arrows) and absence of labeling in some small-diameter neurons (arrowheads). C, Dark-field micrograph showing labeling of neurons in most laminae of the spinal cord, including ventral horn motor neurons (arrows). Dotted line, Superficial laminae I and II of the dorsal horn. Scale bars, 100 μm.
Cellular localization of the β4 subunit
To examine the cellular localization of the β4 protein, we raised a polyclonal antipeptide antibody directed to the extracellular domain and immunostained sections of rat tissue. This anti-β4 antibody was highly specific for β4 subunit proteins as assayed by immunoblot of membrane proteins isolated from tsA-201 cells transfected with individual β1-β4 subunits (Fig. 4). In general, protein expression was in accordance with the in situ hybridization data. For comparison, we also used a previously characterized antibody to determine the distribution of the β2 subunit (Ratcliffe et al., 2000). Control sections omitting the primary anti-β4 or anti-β2 antisera were essentially blank (Fig. 5C, inset). Both β2 and β4 were expressed together in many areas of the brain, but β2 showed more widespread distribution. In the hippocampus, β4 protein was found in isolated pyramidal neurons but was absent from the dentate gyrus apart from occasional hilar neurons (Fig. 5A,C). In contrast, β2 protein was highly expressed in hippocampal pyramidal cells and in the hilus and granule cells of the dentate gyrus (Fig. 5B,D).
Figure 4.
Analysis of anti-β4 antibody specificity. TsA-201 cells were transfected individually with β1-β4 expression plasmids. Purified membrane proteins (10 μg) were fractionated by SDS-PAGE, immunoblotted, and probed using anti-β4 antibody.
Figure 5.
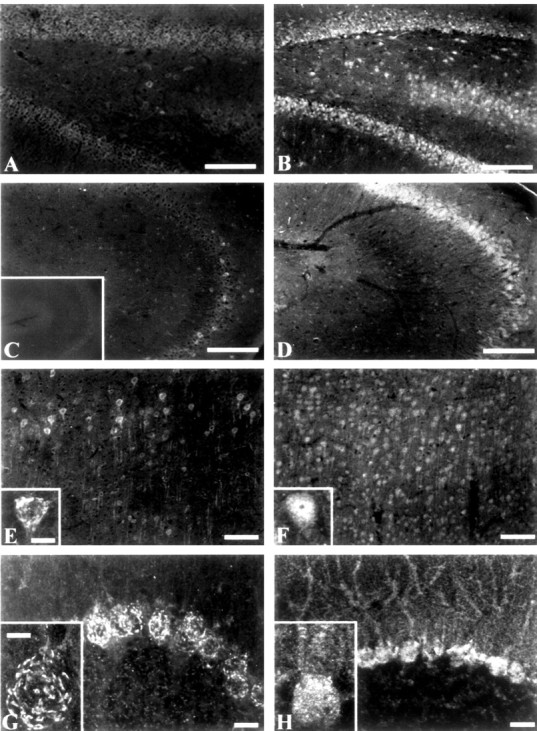
Localization of β2 and β4 subunits in the hippocampus, cerebral cortex, and cerebellum. Tissue sections were stained with either anti-β4 (A, C, E, G) or anti-β2(B, D, F, H). A, B, Dentate gyrus of the hippocampus. Scale bar, 250 μm. C, D, CA3 region of the hippocampus. Scale bar, 250 μm. C, inset, Typical control of an adjacent slice without primary antibody. E, F, Low-magnification views of dorsal cerebral cortex. Scale bar, 100 μm. Insets, High-magnification views of layer V neurons. Scale bar, 25 μm. G, H, Purkinje cell layer of the cerebellum. Scale bars, 50 μm. Insets, High-magnification views of cerebellar Purkinje cells. Scale bar, 25 μm.
In the cerebral cortex, β4 subunit protein was observed in a subset of cell bodies and processes of pyramidal neurons in layer V as well as cells in layers III and VI (Fig. 5E). A greater percentage of cortical neurons stained for β2, and staining was uniform in all layers (Fig. 5F). In particular, β2 staining of neurons of layer V was uniform throughout the cell body and apical dendrites (Fig. 5F, inset), whereas staining by anti-β4 was found in a reticular pattern on the surface of the cell body, as suggested by a z series of 1 μm steps through the neuron (Fig. 5E, inset). In the cerebellum, strong staining by β4 was observed in a reticular pattern near the surface of cerebellar Purkinje cells (Fig. 5G, inset), as indicated by a z series (data not shown), and in the deep cerebellar nuclei (data not shown). Anti-β2 also stained cerebellar Purkinje cells, but it was more uniformly distributed over the soma and dendrites (Fig. 5H). Staining of cell bodies in the deep cerebellar nuclei by anti-β2 was also observed (data not shown).
In the striatum, strong β4 protein expression was observed in neuronal cell bodies and fiber tracts in the caudate nucleus. In contrast, β4 was generally absent in the globus pallidus but was expressed at low levels in occasional cells (Fig. 6A). Staining by anti-β2 was less prominent than that of β4 in the caudateputamen. In contrast to β4, staining by anti-β2 was also prominent in cells and fiber tracts of the globus pallidus (Fig. 6B). This differential staining of the striatum is clear evidence of the specificity of this anti-β4 antibody for β4 subunits. In the thalamus, both β2 and β4 were expressed by neurons in the nucleus reticularis (Fig. 6C,D). As for β4 mRNA, prominent staining of β4 subunits was observed throughout the pons, brainstem, and trigeminal nucleus (data not shown).
Figure 6.
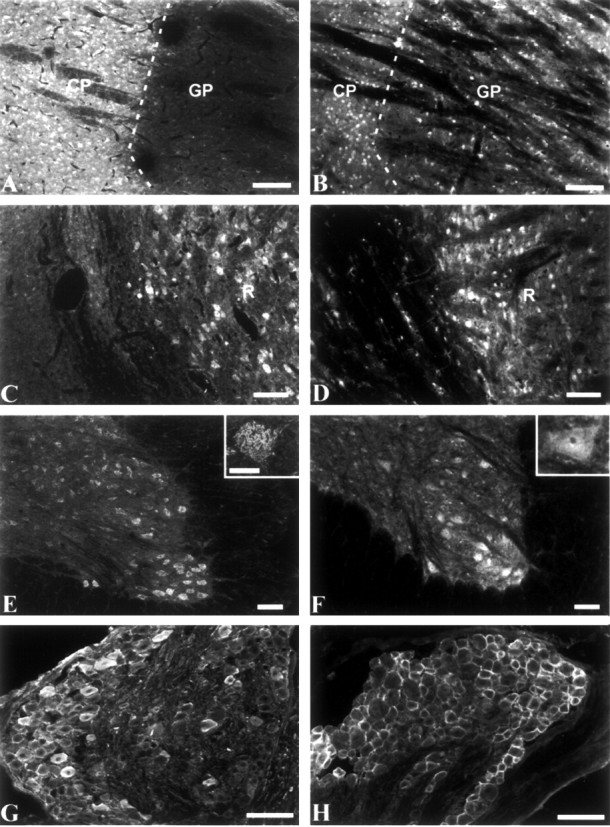
Immunocytochemical localization of β2 and β4 in the basal ganglia and thalamus. Tissue sections were stained with either anti-β4 (A, C, E, G) or antiβ2 (B, D, F, H). A, B, Caudate-putamen (CP) and globus pallidus (GP). C, D, Reticular nucleus of the thalamus. Scale bars, 100 μm. E, F, Spinal cord, low magnification. Scale bars, 250 μm. Insets, Spinal motor neurons, high magnification. Scale bar, 50 μm. G, H, Low-magnification views of dorsal root ganglion. Scale bars, 100 μm.
A strong reticular pattern of staining by anti-β4 was observed on motor neurons of the ventral horn of the spinal cord (Fig. 6E). Staining by anti-β2 was more uniform (Fig. 6F). In the dorsal root ganglia, strong staining by both anti-β4 and anti-β2 antibodies was observed on the cell surface of medium and large sensory neurons, with anti-β2 labeling a larger fraction of the cells (Fig. 6G,H). Expression of β2 by dorsal root ganglion neurons was confirmed by in situ hybridization (data not shown).
Association of the β4 subunit with the Nav1.2a α subunit in tsA-201 cells
To determine whether the β4 subunit forms a complex with sodium channel α subunits, we expressed β4 and Nav1.2a α subunits in tsA-201 cells. To facilitate detection, the β4 subunit was HA-tagged at its C terminus, and the Nav1.2a α subunit was HA-tagged at its N terminus. Membrane proteins from transfected cells were solubilized in Triton X-100 and immuno-precipitated with anti-SP20, a polyclonal antibody recognizing the sodium channel α subunit, or a control antibody. Immunoblot analysis of the immuno-precipitated proteins with the anti-HA11 monoclonal antibody detected immunoreactive products of two different sizes (Fig. 7A). As expected, anti-SP20 immunoprecipitated the HA-tagged Nav1.2a α subunit migrating at >250 kDa as well as faster-migrating bands near 38 kDa, which correspond to the HA-tagged β4 subunit. Neither the Nav1.2a α or β4 subunits were evident in parallel controls using nonimmune rabbit IgG. Three immunoreactive bands near 38 kDa are apparent in the Western blot. This size is greater than the 22 kDa predicted for the β4 subunit based on its amino acid sequence. The molecular mass of ∼38 kDa and the multiple protein bands are expected for N-linked glycosylation at multiple sites, as observed for β1 and β2 isolated from rat brain (Messner and Catterall, 1985).
Figure 7.
Association of Nav1.2a with β4 when coexpressed in tsA-201 cells. A, Coexpression of N-terminal HA-tagged Nav1.2a α subunits and C-terminal HA-tagged β4 subunits in tsA-201 cells. Solubilized proteins were immunoprecipitated with anti-SP20 recognizing the Nav1.2a α subunit, resolved by SDS-PAGE, and immunoblotted using an anti-HA11 monoclonal antibody. Immunoreactive products of ∼250 and 38 kDa were observed, corresponding to Nav1.2a α and β4 subunits, respectively. B, Coexpression of untagged Nav1.2a α subunit with HA-tagged β4. Sodium channel complexes were immunoprecipitated from solubilized proteins using anti-SP20, denatured, and separated by SDS-PAGE under nonreducing (left) or reducing (right) conditions in the presence of 5% β-mercaptoethanol and immunoblotted with anti-HA11.
In purified preparations of rat brain sodium channels, the β2 subunit is disulfide-linked to the α subunit (Catterall, 2000). Because the proposed site of disulfide linkage to the α subunit is conserved (Cys-28 in β4 and Cys-26 in β2), we performed immunoprecipitation experiments to determine whether β4 was also disulfide-linked to sodium channel α subunits. For these experiments, we used the untagged Nav1.2a α subunit so that the HA-tagged β4 subunit could be detected specifically with the anti-HA11 monoclonal antibody. Sodium channel complexes were immunoprecipitated with anti-SP20, and denatured proteins were prepared under nonreducing and reducing conditions (Fig. 7B). Under nonreducing conditions in the absence of β-mercapto ethanol, the β4 subunit was covalently associated with Nav1.2a in a complex of >250 kDa. On reduction with β-mercaptoethanol, the HA-immunoreactive β4 band shifted from >250 kDa to ∼38 kDa, indicating that the β4 subunit is covalently associated with sodium channel α subunits expressed in tsA-201 cells via a disulfide bond.
Functional effects of the β4 subunit
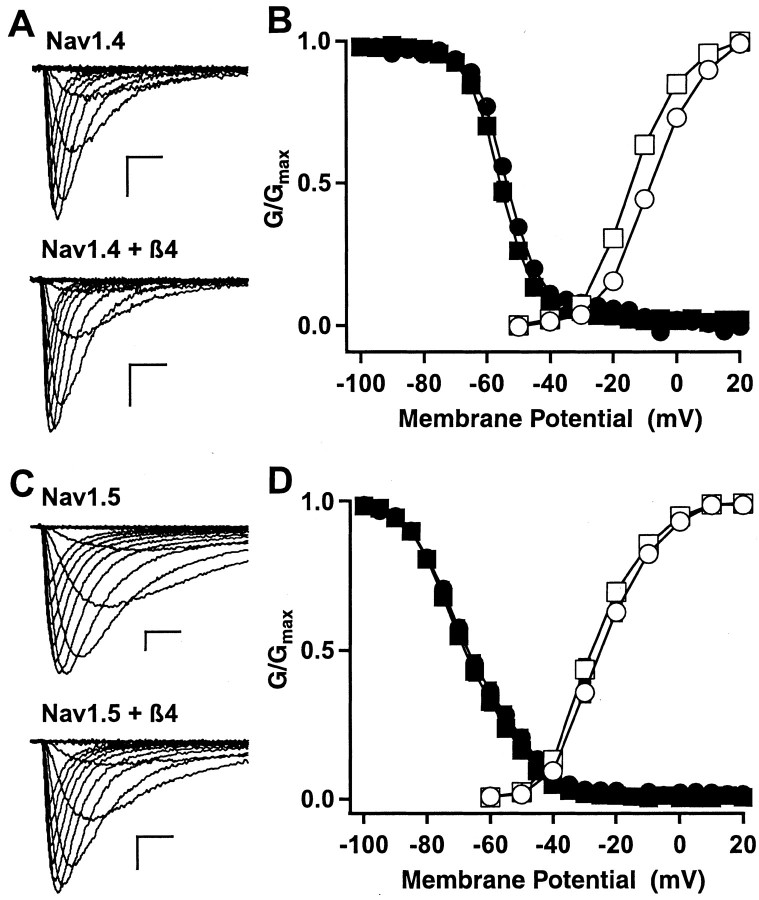
To examine the functional effect of β4 auxiliary subunits, we recorded whole-cell sodium currents generated by Nav1.2a subunits expressed without and with β4 subunits in tsA-201 cells (Fig. 8A). The voltage dependence of activation of peak sodium currents was significantly shifted in the hyperpolarizing direction without a concomitant shift in the voltage dependence of inactivation when the β4 subunit was coexpressed (Fig. 8B). The midpoints of activation were -4.2 ± 0.5 mV (n =27) for Nav1.2a, -6.9 ± 0.8 mV (n =9) for Nav1.2a with β2, and -11.2 ± 0.8 mV (n =18) for Nav1.2a with β4. This -7mV shift in the voltage dependence of activation for β4 was significantly greater than that produced by the β2 subunit as well as being significantly different from control (p < 0.01). The β2 subunit produced a -2.7 mV shift in the midpoint of voltage dependence of activation that was not statistically different from that of the α subunit alone. In contrast to the effect on activation, neither β2 or β4 caused a significant hyperpolarizing shift in the voltage dependence of inactivation [Nav1.2a, V1/2 -42.2 ± 1.2 mV (n =20); Nav1.2a with β2, V1/2 -42.9 ± 1.8 mV (n =9); and Nav1.2a with β4, V1/2 =-45.9 ± 1.6mV (n =9)]. Thus, coexpression of β4 alone with the sodium channel α subunit in tsA-201 cells results in a significant hyperpolarization in the voltage dependence of activation.
Figure 8.
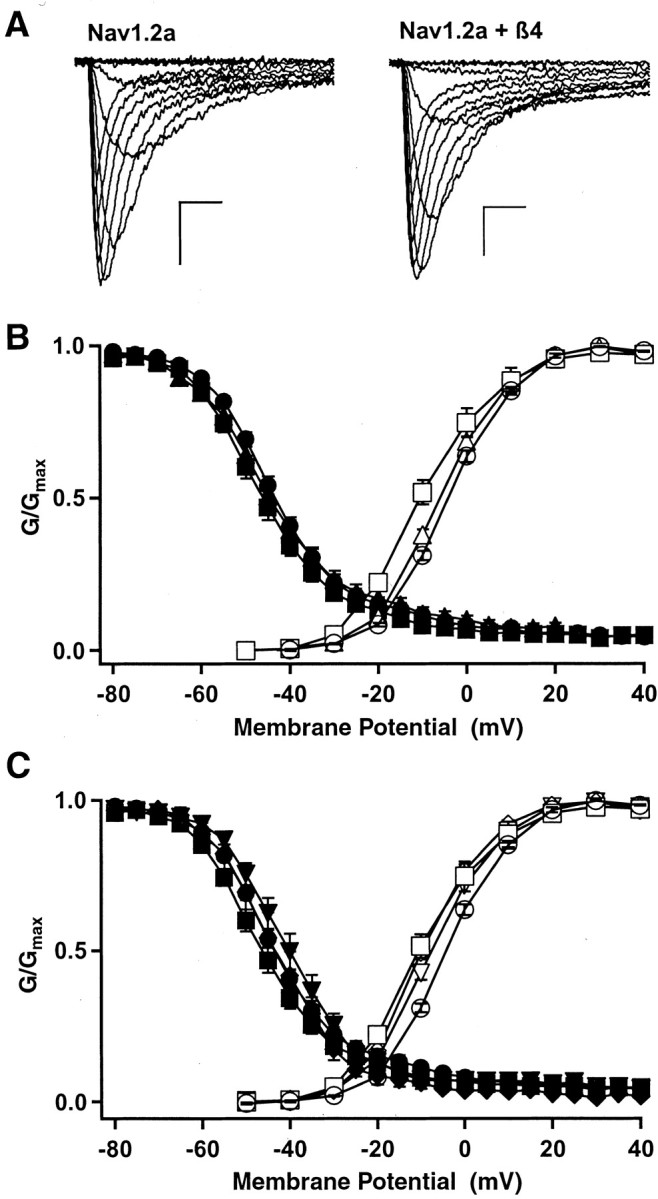
Effects of coexpression of β subunits on the functional properties of the Nav1.2a subunit. A, Families of sodium current traces recorded from a tsA-201 cell transfected with the Nav1.2a α subunit alone (left) or coexpressed with β4 (right). Calibration: 1 nA, 1 msec. B, Conductance-voltage (open symbols) and steady-state (filled symbols) inactivation curves for cells transfected with Nav1.2a α alone (circles), Nav1.2a α with β2 (triangles), and Nav1.2a α with β4 (squares). C, Conductance-voltage (open symbols) and steady-state inactivation (filled symbols) relationships for Nav1.2a α alone (circles), Nav1.2a α with β4 (squares), Nav1.2a α with β1 and β4 (inverted triangles), and Nav1.2a α with β3 and β4 (diamonds).
When expressed in neurons, the sodium channel α subunit is likely to be associated with a β1 or β3 subunit in addition to β2 or β4 (Catterall, 2000). Therefore, we also examined the effect of coexpression of the β4 subunit in combination with either β1 or β3 subunits on sodium currents conducted by the Nav1.2a α subunit. In tsA-201 cells, the β1 and β3 subunits cause a positive shift in the voltage dependence of activation (Qu et al., 2001) (data not shown). In contrast, when β4 was combined with β1or β3, the voltage dependence of activation was similar to that produced by transfection of β4 alone (Fig. 8C). Thus, β4 overrides the effect of β1 or β3 subunits to shift the voltage dependence of activation to more positive potentials in this cell system. These combinations of subunits, like β4 alone, had no significant effect on the voltage dependence of inactivation. Thus, β4 expressed in combination with β1or β3 is very much like β4 alone in producing a negative shift in the voltage dependence of activation and little effect on the voltage dependence of inactivation.
We also examined effects of the β4 subunit on the function of the skeletal muscle Nav1.4 and the cardiac Nav1.5 α subunits because there was strong expression of β4 in these tissues (Fig. 1B). Coexpression of β4 with Nav1.4 caused a modest negative shift in the voltage dependence of activation with little or no effect on the voltage dependence of inactivation (Fig. 9A,B), much as was observed for the Nav1.2a α subunit (Fig. 8B). In contrast, coexpression of the β4 α subunit with the cardiac Nav1.5 α subunit had little effect on channel kinetics or voltage dependence when expressed in tsA-201 cells (Fig. 8C,D).
Figure 9.
Effects of β4 coexpression on the functional properties of Nav1.4 and Nav1.5 α subunits. A, Families of sodium current traces recorded from cells expressing the Nav1.4 α subunit alone (top) or coexpressed with β4 (bottom). Calibration: 1 nA, 1 msec. B, Conductance-voltage (open symbols) and inactivation (filled symbols) relationships for Nav1.4 α alone (circles) and Nav1.4 α with β4 (squares). The midpoints of activation curves were -8.0 ± 0.5 mV (n = 10) for Nav1.4 and -14.1 ± 0.6 mV (n = 12) for Nav1.4 with β4. The midpoints of inactivation curves were -52.8 ± 0.5 mV (n = 7) for Nav1.4 and -55.4 ± 0.7 mV (n = 8) for Nav1.4 with β4. C, Families of sodium current traces recorded from cells expressing the Nav1.5 α subunit alone (top) or coexpressed with β4 (bottom). Calibration: 1 nA, 1 msec. D, Conductance-voltage (open symbols) and inactivation (filled symbols) relationships for Nav1.5 α alone (circles) and Nav1.5 α with β4 (squares). The midpoints of activation curves were -24.9 ± 0.9 mV (n = 13) for Nav1.5 and -27.5 ± 1.2 mV (n = 12) for Nav1.5 with β4. The midpoints of inactivation curves were -66.4 ± 1.1 mV (n = 14) for Nav1.5 and -67.8 ± 1.2 mV (n = 12) for Nav1.5 with β4.
Discussion
The sodium channel β subunit family
We have identified a new β subunit of voltage-gated sodium channels, β4, which is structurally and functionally related to the previously characterized β2 subunit. Biochemical characterization of purified brain sodium channels indicated that they constitute a heterotrimeric complex of pore-forming α subunits with both covalently and noncovalently linked β subunits (Catterall, 2000). The noncovalently linked β subunits include β1 (Isom et al., 1992) and β3 (Morgan et al., 2000; Qu et al., 2001). Our findings indicate that the disulfide-linked moieties consist of β2 (Isom et al., 1995) and the novel β4 subunit identified here.
Structurally, β4 is like the three previously cloned β subunits, with an N-terminal cleaved signal sequence, an extracellular V-type Ig-like fold, a transmembrane domain, and an intracellular region that might participate in protein-protein interactions. However, β4 has the highest amino acid sequence similarity to the β2 subunit (35%). Like the β2 subunit, β4 contains an extra-cellular unpaired cysteine that could form a disulfide bond with the α subunit, and our results show that β4 associates with the α subunit via a reducible disulfide bond when coexpressed in tsA-201 cells.
In functional studies, the β4 subunit caused negative shifts in the voltage dependence of activation of rat brain Nav1.2a and skeletal muscle Nav1.4 channels but did not affect the voltage dependence of inactivation. It had little functional effect on the cardiac Nav1.5 channel. These effects contrast with those observed for the β2 subunit, which did not shift the voltage dependence of activation, and the β1 and β3 subunits, which cause a positive shift in the voltage dependence of activation in tsA-201 cells. Moreover, coexpression of the β4 subunit with the β1 and β3 subunits overrode their effects, resulting in sodium channels with negatively shifted voltage dependence of activation as observed for β4 coexpression alone. These results indicate that the β4 subunit will have an important effect in the neurons and other excitable cells in which it is expressed, even if those cells express other β subunits.
Although the functional effects of auxiliary β subunits are relatively small in these expression studies in heterologous mammalian cells, they are likely to be critical in tuning the functional properties of sodium channels to impart appropriate electrical excitability to neurons. Because excitatory postsynaptic potentials depolarize neurons by only a few millivolts, shifts of this magnitude in the voltage dependence of activation of sodium channels will have a major effect on the response of a neuron to its synaptic inputs, determining in an all-or-none manner whether those neurons fire action potentials in response to typical EPSPs. The effects of the β4 subunits described here will contribute to tuning the electrical properties of neurons by allowing sodium channel activation at more negative voltages. The resulting action potentials generated in response to mild depolarizing stimuli would be manifested as increased neuronal sensitivity to excitatory inputs in cells expressing the β4 subunit. Additional complexity will be found in cells expressing multiple subtypes of sodium channel principal and auxiliary subunits.
Expression pattern of β4 versus β2 sodium channel auxiliary subunits
The expression pattern of the β4 subunit in rat tissues is perhaps most properly compared with that of the β2 subunits because they are most similar in amino acid sequence, and both subunits are thought to be disulfide-linked to sodium channel α subunits. Therefore, the β2 and β4 subunits might substitute for each other in particular tissues or neuronal cell types.
In the CNS, expression patterns of β2 and β4 often overlap. In the cerebellum, β2 and β4 strongly label Purkinje cells and deep cerebellar nuclei, consistent with previous reports of β2 in situ hybridization (Gastaldi et al., 1998; Levy-Mozziconacci et al., 1998). In the brainstem, both β2 and β4 exhibited strong staining in motor nuclei. In the spinal cord, β2 and β4 are both highly expressed in the motor neurons of the ventral horn.
There are also numerous areas of differential expression of β2 and β4 at the cellular level. In the cerebral cortex, β2 expression is more widespread in different layers than that of β4. In the globus pallidus, β2 is widely expressed, but β4 is not. In the hippocampus, β2 is expressed strongly in CA1 and CA3 pyramidal cells and cells of the hilus but less prominently in dentate granule cells (Gastaldi et al., 1998), whereas β4 is expressed in only a small number of pyramidal cells and hilar neurons. Thus, although expression of β2 and β4 subunits overlap in many brain regions, in others, such as the hippocampus, cerebral cortex, and particularly the globus pallidus, their distributions are primarily complementary. This complementary distribution may indicate selective association with distinct sodium channel α subunits in those regions and may differentially influence sodium channel properties or dictate differences in secondary protein-protein interactions.
Possible functions of β4 in sodium channel localization and cell adhesion
Sodium channel β1-β3 subunits have been proposed to have important functions in sodium channel localization and cell adhesion (Isom et al., 1995; Catterall, 2000; Ratcliffe et al., 2000; Isom, 2002). They interact with extracellular matrix and cell adhesion molecules such as contactin (Isom et al., 1992; Malhotra et al., 2000; Kazarinova-Noyes et al., 2001), neurofascin (Ratcliffe et al., 2001), and tenascin (Srinivasan et al., 1998; Xiao et al., 1999). β subunits also participate in homophilic cell interactions and interact with ankyrin, thus localizing channels to sites of cell-cell interaction (Malhotra et al., 2000, 2002). These roles are also likely for the β4 subunit described here because it is predicted to form an extracellular Ig-like fold that might be implicated in such intermolecular interactions. Moreover, modulatory molecules such as receptor protein-tyrosine phosphatase β are recruited to sodium channel complexes through interactions with β subunits (Ratcliffe et al., 2000). Presumably, β4 subunits will be involved in a similar but distinct array of intracellular and extracellular interactions and are likely to play key roles in specific targeting and regulation of sodium channels.
Footnotes
This work was supported by National Institutes of Health Research Grants NS34802 (T.S.) and NS25704 (W.A.C.).
Correspondence should be addressed to Dr. William A. Catterall, University of Washington, Mail Stop 357280, Seattle, WA 98195-7280. E-mail: wcatt@u.washington.edu.
P. S. DiStefano's and R. Curtis's present address: Elixir Pharmaceuticals, Inc., 1 Kendall Square, Building 1000, Cambridge, MA 02139.
Copyright © 2003 Society for Neuroscience 0270-6474/03/237577-09$15.00/0
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ ( 1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA ( 2000) From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26: 13-25. [DOI] [PubMed] [Google Scholar]
- Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P ( 2001) De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet 68: 1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A ( 2000) Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet 24: 343-345. [DOI] [PubMed] [Google Scholar]
- Eubanks J, Srinivasan J, Dinulos MB, Disteche CM, Catterall WA ( 1997) Structure and chromosomal localization of the β2 subunit of the human brain sodium channel. NeuroReport 8: 2775-2779. [DOI] [PubMed] [Google Scholar]
- Featherstone DE, Fujimoto E, Ruben PC ( 1998) A defect in skeletal muscle sodium channel deactivation exacerbates hyperexcitability in human paramyotonia congenita. J Physiol (Lond) 506: 627-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaldi M, Robaglia-Schlupp A, Massacrier A, Planells R, Cau P ( 1998) mRNA coding for voltage-gated sodium channel β2 subunit in rat central nervous system: cellular distribution and changes following kainate-induced seizures. Neurosci Lett 249: 53-56. [DOI] [PubMed] [Google Scholar]
- Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Netter YB, Noda M, Tamkun MM, Waxman SG, Wood JN, Catterall WA ( 2000) Nomenclature of voltage-gated sodium channels. Neuron 28: 365-368. [DOI] [PubMed] [Google Scholar]
- Herlitze S, Garcia DE, Mackie K, Hille B, Scheuer T, Catterall WA ( 1996) Modulation of Ca2+ channels by G-protein βγ subunits. Nature 380: 258-262. [DOI] [PubMed] [Google Scholar]
- Isom LL ( 2002) The role of sodium channels in cell adhesion. Front Biosci 7: 12-23. [DOI] [PubMed] [Google Scholar]
- Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA ( 1992) Primary structure and functional expression of the β1 subunit of the rat brain sodium channel. Science 256: 839-842. [DOI] [PubMed] [Google Scholar]
- Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BF, Scheuer T, Catterall WA ( 1995) Structure and function of the β2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell 83: 433-442. [DOI] [PubMed] [Google Scholar]
- Kazarinova-Noyes K, Malhotra JD, McEwen DP, Mattei LN, Berglund EO, Ranscht B, Levinson SR, Schachner M, Shrager P, Isom LL, Xiao ZC ( 2001) Contactin associates with Na+ channels and increases their functional expression. J Neurosci 21: 7517-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Mozziconacci A, Alcaraz G, Giraud P, Boudier JA, Caillol G, Couraud F, Autillo-Touati A ( 1998) Expression of the mRNA for the β2 subunit of the voltage-dependent sodium channel in rat CNS. Eur J Neurosci 10: 2757-2767. [DOI] [PubMed] [Google Scholar]
- Linford NJ, Cantrell AR, Qu Y, Scheuer T, Catterall WA ( 1998) Interaction of batrachatoxin with the local anesthetic receptor site in transmembrane segment IVS6 of the voltage-gated sodium channel. Proc Natl Acad Sci USA 95: 13947-13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra JD, Kazen-Gillespie K, Hortsch M, Isom LL ( 2000) Sodium channel β subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell-cell contact. J Biol Chem 275: 11383-11388. [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Koopmann MC, Kazen-Gillespie KA, Fettman N, Hortsch M, Isom LL ( 2002) Structural requirements for interaction of sodium channel β1 subunits with ankyrin. J Biol Chem 277: 26681-26688. [DOI] [PubMed] [Google Scholar]
- Mantegazza M, Yu FH, Catterall WA, Scheuer T ( 2001) Role of the C-terminal domain in inactivation of brain and cardiac sodium channels. Proc Natl Acad Sci USA 98: 15348-15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhurst AD, Harrison DC, Read SJ, Campbell CA, Robbins MJ, Pangalos MN ( 2000) The use of TaqMan RT-PCR assays for semiquantitative analysis of gene expression in CNS tissues and disease models. J Neurosci Methods 98: 9-20. [DOI] [PubMed] [Google Scholar]
- Messner DJ, Catterall WA ( 1985) The sodium channel from rat brain: separation and characterization of subunits. J Biol Chem 260: 10597-10604. [PubMed] [Google Scholar]
- Morgan K, Stevens EB, Shah B, Cox PJ, Dixon AK, Lee K, Pinnock RD, Hughes J, Richardson PJ, Mizuguchi K, Jackson AP ( 2000) β3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci USA 97: 2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Rogers J, Tanada T, Scheuer T, Catterall WA ( 1994) Modulation of cardiac Na+ channels expressed in a mammalian cell line and in ventricular myocytes by protein kinase C. Proc Natl Acad Sci USA 91: 3289-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Curtis R, Lawson D, Gilbride K, Ge P, DiStefano PS, Silos-Santiago I, Catterall WA, Scheuer T ( 2001) Differential modulation of sodium channel gating and persistent sodium currents by the β1, β2, and β3 subunits. Mol Cell Neurosci 18: 570-580. [DOI] [PubMed] [Google Scholar]
- Ratcliffe CF, Qu Y, McCormick KA, Tibbs VC, Dixon JE, Scheuer T, Catterall WA ( 2000) A sodium channel signaling complex: modulation by associated receptor protein tyrosine phosphatase β. Nat Neurosci 3: 437-444. [DOI] [PubMed] [Google Scholar]
- Ratcliffe CF, Westenbroek RE, Curtis R, Catterall WA ( 2001) Sodium channel β1 and β3 subunits associate with neurofascin through their extracellular immunoglobulin-like domain. J Cell Biol 154: 427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, Schachner M, Catterall WA ( 1998) Interaction of voltage-gated sodium channels with the extracellular matrix molecules tenascin-C and tenascin-R. Proc Natl Acad Sci USA 95: 15753-15757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl A, Hirsch DJ, Gimeno RE, Punreddy S, Ge P, Watson N, Patel S, Kotler M, Raimondi A, Tartaglia LA, Lodish HF ( 1999) Identification of the major intestinal fatty acid transport protein. Mol Cell 4: 299-308. [DOI] [PubMed] [Google Scholar]
- Wallace RH, Wang DW, Singh R, Scheffer IE, George ALJ, Phillips HA, Saar K, Reis A, Johnson EW, Sutherland GR, Berkovic SF, Mulley JC ( 1998) Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel β1 subunit gene SCN1B. Nat Genet 19: 366-370. [DOI] [PubMed] [Google Scholar]
- Wallace RH, Scheffer IE, Barnett S, Richards M, Dibbens L, Desai RR, Lerman-Sagie T, Lev D, Mazarib A, Brand N, Ben-Zeev B, Goikhman I, Singh R, Kremmidiotis G, Gardner A, Sutherland GR, George Jr AL, Mulley JC, Berkovic SF ( 2001) Neuronal sodium-channel α1-subunit mutations in generalized epilepsy with febrile seizures plus. Am J Hum Genet 68: 859-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenbroek RE, Hoskins L, Catterall WA ( 1998) Localization of Ca2+ channel subtypes on rat spinal motor neurons, interneurons, and nerve terminals. J Neurosci 18: 6319-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZC, Ragsdale DS, Malhotra JD, Mattei LN, Braun PE, Schachner M, Isom LL ( 1999) Tenascin-R is a functional modulator of sodium channel β subunits. J Biol Chem 274: 26511-26517. [DOI] [PubMed] [Google Scholar]
- Zhong H, Yokoyama CT, Scheuer T, Catterall WA ( 1999) Reciprocal regulation of P/Q-type Ca2+ channels by SNAP-25, syntaxin and synaptotagmin. Nat Neurosci 2: 939-941. [DOI] [PubMed] [Google Scholar]