Abstract
Myelin basic protein (MBP) is required for normal myelin compaction and is implicated in both experimental and human demyelinating diseases. In this study, as an initial step in defining the regulatory network controlling MBP transcription, we located and characterized the function of evolutionarily conserved regulatory sequences. Long-range human-mouse sequence comparison revealed over 1 kb of conserved noncoding MBP 5′ flanking sequence distributed into four widely spaced modules ranging from 0.1 to 0.4 kb. We demonstrate first that a controlled strategy of transgenesis provides an effective means to assign and compare qualitative and quantitative in vivo regulatory programs. Using this strategy, single-copy reporter constructs, designed to evaluate the regulatory significance of modular and intermodular sequences, were introduced by homologous recombination into the mouse hprt (hypoxanthine-guanine phosphoribosyltransferase) locus. The proximal modules M1 and M2 confer comparatively low-level oligodendrocyte expression primarily limited to early postnatal development, whereas the upstream M3 confers high-level oligodendrocyte expression extending throughout maturity. Furthermore, constructs devoid of M3 fail to target expression to newly myelinating oligodendrocytes in the mature CNS. Mutation of putative Nkx6.2/Gtx sites within M3, although not eliminating oligodendrocyte targeting, significantly decreases transgene expression levels. High-level and continuous expression is conferred to myelinating or remyelinating Schwann cells by M4. In addition, when isolated from surrounding MBP sequences, M3 confers transient expression to Schwann cells elaborating myelin. These observations define the in vivo regulatory roles played by conserved noncoding MBP sequences and lead to a combinatorial model in which different regulatory modules are engaged during primary myelination, myelin maintenance, and remyelination.
Keywords: myelin basic protein, transcription, HPRT transgenesis, oligodendrocyte, Schwann cell, remyelination
Introduction
The myelin sheath is a specialized glial membranous organelle essential for rapid and energy efficient action potential conduction. Perturbations of the sheath, as in multiple sclerosis (MS), may also lead to axonal degeneration (Bjartmar et al., 2003). The process of remyelination is invariably prolonged compared with primary myelin deposition and often results in thin sheaths with only partial recovery of conductive properties. Thus, an understanding of the mechanisms that control myelination and remyelination are key to the development of therapeutic strategies to treat demyelinating diseases.
The search for potential regulators of glial cell development and axon-glia signaling has led to the identification of numerous candidate transcription factors, including the basic helix-loop-helix proteins Olig1 and Olig2, the homeodomain proteins Tst-1/SCIP/Oct-6 and Nkx6.2/Gtx, the zinc finger protein Krox-20/Egr2, and the HMG-domain protein Sox-10 (Topilko et al., 1994; Bermingham et al., 1996; Awatramani et al., 1997; Britsch et al., 2001; Zhou and Anderson, 2002). However, the upstream glial signaling pathways and the full complement of interacting transcription factors remain to be elucidated.
Mouse transgenesis provides an ideal system to examine regulatory phenotypes under normal and experimental conditions. Using this approach, various lengths of 5′ flanking sequence from the proteolipid protein, myelin basic protein (MBP), protein zero, peripheral myelin protein 22, and 2′,3′ cyclic-nucleotide 3′-phosphodiesterase genes have been demonstrated to target reporter expression to oligodendrocytes or Schwann cells (Foran and Peterson, 1992; Gow et al., 1992; Messing et al., 1992; Wight et al., 1993; Gravel et al., 1998; Wrabetz et al., 1998; Maier et al., 2002). However, identifying cis-regulatory sequences by this method has been limited by variability associated with copy number and position effects at random transgene insertion loci. The recent description of a controlled transgenesis strategy permitting single-copy construct insertion by homologous recombination at the hprt (hypoxanthine-guanine phosphoribosyltransferase) locus resolves these issues, allowing for higher-resolution interconstruct comparisons (Bronson et al., 1996; Vivian et al., 1999; Cvetkovic et al., 2000).
Regulatory sequences are thought to constitute a small fraction of the noncoding portion of the mammalian genome (Waterston et al., 2002). Computational sequence analysis and interspecies comparisons have been used to identify conserved noncoding sequences with potential gene regulatory properties. To date, however, evolutionarily conserved sequences associated with only a few loci have been assigned regulatory functions (Oeltjen et al., 1997; Gottgens et al., 2000; Loots et al., 2000; Pennacchio et al., 2001).
In this study, we combined human-mouse noncoding sequence comparison with targeted transgenesis at the hprt locus to identify the in vivo cis-regulatory network controlling MBP, an essential constituent of central and peripheral myelin (Peterson and Bray, 1984; Readhead et al., 1987). We find that the regulatory programming of MBP is accounted for by four widely spaced conserved sequence modules. Each module confers a distinct quantitative and spatiotemporal expression pattern defining specific regulatory programs for primary myelination, myelin maintenance, and remyelination after injury. Together, our observations highlight this combined approach as a valuable tool for assigning qualitative and quantitative in vivo regulatory phenotypes and provide a basis for modeling the cis-regulatory system controlling myelin protein gene expression.
Materials and Methods
Isolation of human and mouse MBP clones. Human MBP PAC clone 248_D_12 was isolated by screening the RPCI.1 human PAC library with chromosome 18 STS marker WI-9286 and was validated with Golli/MBP exon-specific primers. PAC DNA was prepared from 2L cell cultures in LB-Broth containing 30 μg/ml kanamycin. DNA was isolated using a midi-prep kit following the instructions of the manufacturer (Qiagen, Hilden, Germany). PAC DNA was sheared using a sonicator to an average of 2 kb fragments. The ends of the fragments were repaired with Mung Bean Nuclease (New England Biolabs, Beverly, MA). The fragments were gel purified twice to select for 2 kb fragments and ligated into an M13mp18 vector digested with SmaI. The reactions were then transformed into XL-2 competent cells (Stratagene, La Jolla, CA). Individual plaques of M13 subclones were grown for 16 hr at 37°C in 0.5 ml of 2X YT with 10 μl of log-phase TG-1 bacterial cells. Single-strand M13 DNA for sequencing was obtained from 100 μl of the culture supernatant using magnetic beads (PerSeptive Biosystems, Foster City, CA) following the instructions of the manufacturer. Isolation of a mouse MBP genomic lambda DashII clone (∼15 kb) was described previously (Foran and Peterson, 1992).
Sequencing. Sequencing of the M13 clones was done with the Dye Primer Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA). Sequencing reactions included ∼400 ng of M13 DNA in 1.5 and 3 μl of reaction mixture for each of the four labeled primers. The thermal cycling parameters were as folows: 15 cycles at 96°C for 10 sec, 55°C for 5 sec, and 70°C for 1 min, followed by 15 cycles at 96°C for 10 sec and 70°C for 1 min. The four reactions were mixed and purified with an ethanol precipitation. Samples were loaded and run on an ABI 377 Sequencer according to the instructions of the manufacturer (Applied Bio-systems). A portion of the mouse sequence (-12 to -3 kb) was obtained by walking using specific oligonucleotide primers and analyzed on a LICOR sequencer. Ambiguous mouse and human sequences were verified using the DyeTerminator system (Applied Biosystems) with specific primers. The sequences were supplemented with available sequences present in GenBank, followed by assembly into contigs and analysis with the STADEN (version 1997.1) and MacVector 7.0 (Accelrys, San Diego, CA) software packages.
Generation of reporter constructs. A -3.1 kb mouse MBP/lacZ construct (Foran and Peterson, 1992) was used to derive constructs containing -165 bp (EheI), -300 bp (StuI), -377 bp (ApaI), or -794 bp (BseRI) of proximal promoter sequence driving lacZ expression. A -9.4 kb construct was derived by the addition of a 0.44 kb PvuII-SacII fragment to a previously described -8.9 kb MBP/lacZ construct (clone 8) (Forghani et al., 2001). Clone 8 was also used to isolate a -5.8 to -3.1 kb KpnI-XbaI fragment encompassing M3. Deletion of a 0.6 kb fragment containing M3 was performed by digestion with AvrII-BglII (spanning -5.0 to -4.4 kb). Both -5.8 to -3.1 kb fragments (either containing M3 or not) were in turn ligated upstream of the -300 bp proximal promoter sequence coupled to the coding region of lacZ. All of the above MBP-promoted constructs also included either the complete previously described Schwann cell enhancer 1 (SCE1) sequence (SacII-SacI fragment from -8.9 to -8.3 kb (Forghani et al., 2001) or truncated versions of 0.2 kb SCE1 (BstEII-BglII from -8.8 to -8.6 kb), 0.5 kb SCE1 (SacII-NaeI from -8.9 to -8.4 kb), or 0.44 kb of upstream SCE2 sequence (PvuII-SacII from -9.4 to -8.9 kb (Forghani et al., 2001), as shown in Fig. 1C.
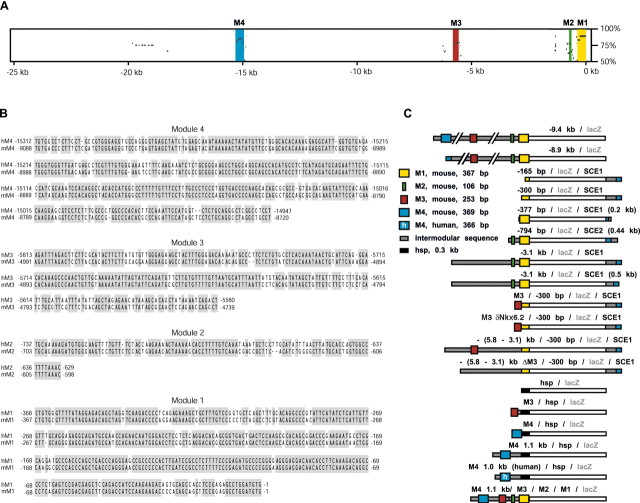
Figure 1.
Human-mouse sequence comparison identifies putative regulatory modules evaluated in reporter constructs. A, Alignment of human (25 kb) and mouse (12 kb) MBP 5′ flanking sequences using the PIPMaker software. The human genomic sequence is represented on the x-axis, and its percentage identity to the mouse sequence (from 50 to 100%) is represented on the y-axis. This analysis reveals similarly ordered and oriented conserved sequence modules. Modular sequences, represented in distinct colors, are designated as module 1 through 4 (M1 to M4) proceeding 5′ from the proximal promoter. In addition, an interspersed repeat lies at -19 kb of the human sequence. B, ClustalW alignment (MacVector 7.0) reveals that the individual modules vary in length from 106 to 369 bp in the mouse (lower limit of 75% sequence identity over at least 100 bp). Numbering is from the initiator ATG. C, Schematic representation, drawn to scale, of reporter constructs inserted in single copy at the mouse hprt locus. Enlarged boxes, colored as in A, correspond to the conserved modules in their entirety. Intermodular MBP sequences and HSP68 promoter sequences are represented in gray and black, respectively. SCE1 and SCE2 sequences (Forghani et al., 2001) are both represented in blue and gray. Human M4 is denoted by the symbol h. All constructs are ligated to a lacZ reporter gene.
Constructs containing M3 alone were generated by ligation of a 0.38 kb BtgI-AvaII fragment upstream of either -300 bp of proximal promoter sequence (as above) or a 0.3 kb HSP68 (heat shock protein) minimal promoter ligated to lacZ (clone p610ZA; a gift from R. Kothary, University of Ottawa, Ottawa, Ontario, Canada). To test whether the two putative Nkx6.2/Gtx sites on the (+) strand of M3 are involved in targeting function, complementary M3 sequence encoding oligonucleotides bearing mutations that abolish Nkx6.2/Gtx binding (GTTAATGC → GTTGGCGC and TTTAATTC → TTTGGCCC; custom made by Sheldon Biotechnology Centre, McGill University, Montreal, Quebec, Canada) (Awatramani et al., 1997) were annealed and cloned into M3. The mutated sequence (M3 δnkx6.2) was subsequently ligated to the 5′ end of -300 bp/SCE1. The resultant mutant (M3 δnkx6.2/-300 bp/SCE1) was confirmed by sequencing of both strands.
M4-containing sequences were similarly ligated upstream of the heterologous HSP68 minimal promoter: namely, a mouse 1.1 kb PvuII-PvuII fragment (spanning -9.4 to -8.3 kb), a mouse 0.4 kb BstXI-AvrII fragment (spanning -9.1 to -8.7 kb), and a human 1.0 kb BssSI-EcoRI fragment (spanning -15.5 to -14.5 kb). A construct containing modular sequences with minimal surrounding sequence was generated by sequential ligation of M4 1.1 kb and M3 (0.38 kb fragment) upstream of -794 bp of proximal promoter sequence ligated to lacZ.
All of the above constructs were directionally inserted into a slightly modified version of the hprt targeting vector pMP8SKB (a gift from Sarah Bronson, Pennsylvania State University, Hershey, PA).
Cell culture and electroporation. BK4-embryonic stem (ES) cells bearing the hprt docking site (also a generous gift from Sarah Bronson) or a derivative on a mixed genetic background (C57BL/6 and 129) generated in our laboratory were grown on gamma-irradiated murine embryonic fibroblasts in high-glucose DMEM (Invitrogen, Gaithersburg, MD) supplemented with 15% heat-inactivated FBS, 1% l-glutamine, 1% MEM amino acids, 1% sodium pyruvate, 1% penicillin-streptomycin, 0.1 mm 2-mercaptoethanol, and 50 μl of leukemia inhibitory factor (Invitrogen). A quantity of 5-7 × 106 BK4-ES cells were electroporated with 40 μg of linearized DNA (250 V and 500 μF, Gene Pulser II; Bio-Rad, Hercules, CA). Homologous recombinants were selected on HAT-supplemented medium, containing 0.1 mm hypoxanthine, 0.0004 mm aminopterin, and 0.016 mm thymidine (Sigma, St. Louis, MO). HAT-resistant colonies were picked 14-21 d later for propagation.
Generation of transgenic mice. All experiments involving animals were conducted in accordance with McGill University animal care guidelines. Targeted ES cells were injected into C57BL/6-derived blastocysts that were then transplanted into the uteri of recipient females. Resulting chimeric males were bred with C57BL/6 females, and the F1 agouti female offspring were backcrossed with C57BL/6 males. Genotyping was performed by PCR analysis of genomic DNA with lacZ coding sequence-specific primers.
Demyelination-nerve injury models. To induce spinal cord demyelination, adult female mice (2-3 months of age) were injected intrathecally at the L4-L5 level with 0.3 μg of cholera toxin subunit B (CTB)-saporin (Advanced Targeting Systems, San Diego, CA) dissolved in sterile 0.9% saline. Tissues were analyzed after a 6 week recovery period. To induce peripheral nerve regeneration, adult male or female mice (2-3 months of age) were anesthetized, and unilateral sciatic nerve crushes were performed (two times for 10 sec each) at the midthigh level with a number 5 forceps. Sciatic nerves were analyzed at 14 or 21 d post crush (dpc) injury.
Histochemical detection of β-galactosidase activity. Histochemical staining was performed as described previously (Forghani et al., 2001). Briefly, mice were anesthetized and perfused transcardially with 0.5% paraformaldehyde and 2.5% glutaraldehyde in 0.1 m phosphate buffer, pH 7.4. After postfixation for 1 additional hour, whole mounts or tissue sections (brain, spinal cord, or dorsal root ganglia) were incubated at 37°C in staining solution containing 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm magnesium chloride, 0.02% Nonidet P-40, and 0.4 mg/ml Bluo-Gal (Sigma).
Microscopy. Stained whole-mount specimens and spinal cord vibratome sections were viewed on a Wild M5A stereomicroscope (Leica, Wetzlar, Germany) and photographed with a Zeiss (Oberkochen, Germany) AxioCam HRc. Adjacent vibratome sections were embedded in Epon. After trimming, semithin (1 μm) and thin (10 nm) sections were prepared using an ultramicrotome (Reichert Jung) and processed for light microscopy and electron microscopy (Phillips CM-10), respectively.
Quantitation of β-galactosidase expression levels. Cervical spinal cords were dissected from C57BL/6 backcrossed postnatal day 18 (P18) mice and homogenized with two 10 sec bursts of a Polytron homogenizer in 10 vol (0.7 ml) of lysis solution (100 mm potassium phosphate and 0.2% Triton X-100). Total protein concentrations were then measured for all extracts in triplicate by the Bradford microassay procedure (Bio-Rad). Two separate methodologies were used for quantitation of activity levels. Absolute activity levels differed in the two techniques, but relative levels were conserved. The Galacto-Star chemiluminescent assay system (Applied Biosystems) with readings performed on a Revelation MLX luminometer (Dynex Technologies, Chantilly, VA) was used following the instructions of the manufacturer for comparison of expression levels from independently derived transgenic lines bearing identical constructs (see Fig. 4 A, inset). The fluorescent substrate DDAO galactoside [9H-(1,3-dichloro-9,9-dimethylacridin-2-one-7-yl)galactopyranoside] (Molecular Probes, Eugene, OR) with quantitation using the Typhoon phosphorimager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA) was used for comparison of activity levels between different constructs (see Fig. 4 A). Standard curves were generated by assaying serial dilutions of β-galactosidase (Roche Diagnostics, Hertfordshire, UK) in triplicate.
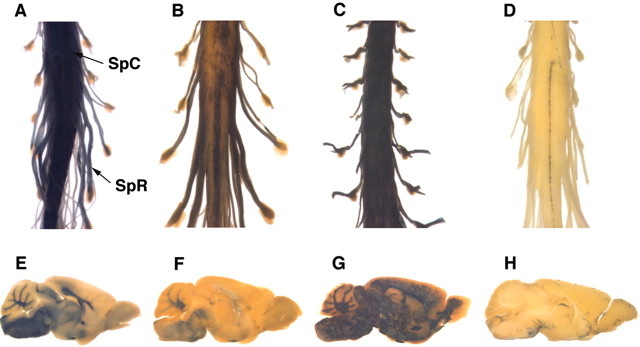
Figure 4.
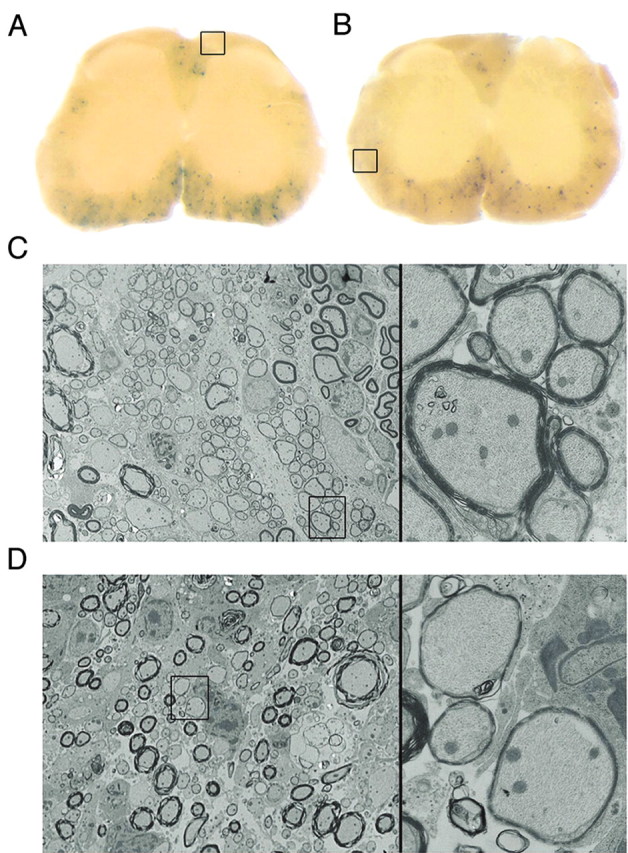
M1 and M2 do not drive expression in newly myelinating oligodendrocytes in adults. A, B, lacZ reporter constructs bearing -3.1 kb/SCE1 (0.5 kb) or -794 bp/SCE2 (0.44 kb), respectively, fail to express in newly myelinating adult oligodendrocytes 6 weeks after spinal cord demyelination induced by intrathecal injection of CTB-saporin (Jasmin et al., 2000). C, D, Sections adjacent to histochemical preparations were processed for electron microscopy with boxed areas in A and B sampled in C and D, respectively. Variable β-galactosidase labeling is observed across the section, but areas devoid of labeling contain profiles typical of newly myelinating oligodendrocytes (boxed areas at 900× shown at 6300× on right).
Results
Human-mouse sequence comparison reveals four conserved modules
To search for evolutionarily conserved MBP regulatory elements, human (25 kb) and mouse (12 kb) 5′ flanking sequences were sequenced and compared. Figure 1A shows a percentage identity plot (PIP) generated by the PIPMaker program (Schwartz et al., 2000), with the human sequence represented on the x-axis. This comparison revealed a relatively high level of interspecies polymorphism equivalent to that observed at the β-globin locus (Hardison et al., 1997). Using the threshold criteria of 75% identity over 100 bp, four conserved modules were identified. These range in length from 106 to 369 bp (Fig. 1B) and demonstrate percentage identities of 87, 78, 79, and 82% (M1 to M4, respectively). All intermodular sequences are longer in human than in mouse, with additional human-mouse conservation present only in short sequences (<30 bp), typically adjacent to the modules (as defined by the above criteria).
We further evaluated the modular sequences for the presence of known regulatory elements linked to MBP regulation. We searched for such elements using both MacVector 7.0 software and the Transcription Factor Database (TRANSFAC; http://transfac.gbf.de/TRANSFAC/) (Wingender et al., 2001). Over 250 experimentally characterized murine cis-regulatory sites were recognized in M1 to M4, including putative high-affinity recognition sites for the glial-associated factors Krox-20/Egr2, Sox-10, and Nkx6.2/Gtx.
The hprt locus does not deregulate MBP-promoted reporter gene expression in myelinating glia
To determine whether regulatory function is conferred by the conserved noncoding MBP sequences, we applied a controlled transgenesis strategy permitting direct interconstruct comparisons of both qualitative and quantitative regulatory phenotypes. Reporter constructs containing different configurations of modular and intermodular sequence were inserted, in single-copy and in a known orientation, at a predetermined site at the hprt locus (Bronson et al., 1996; Vivian et al., 1999; Cvetkovic et al., 2000) (Fig. 1C). We show first that the chosen hprt docking site provides a transcriptionally favorable environment with representative constructs displaying spatiotemporal expression patterns paralleling that observed with multiple independently derived random insertion transgenic lines. A relatively weak MBP promoter (-3.1 kb) (Foran and Peterson, 1992) drives readily detectable reporter gene expression (Fig. 2A,B,E,F), whereas stronger promoters (-9.4 kb, Fig. 2C,G; and -8.9 kb, see below) drive markedly higher expression levels. These hprt-docked constructs also contain the previously described SCE1 sequence, which, as again expected from numerous random insertion lines (Forghani et al., 2001), targets continuous high-level Schwann cell expression.
Figure 2.
The hprt-docking site is permissive and does not deregulate MBP targeting in myelinating glia. A, E, As predicted from pronuclear lines (Foran and Peterson, 1992; Forghani et al., 2001), a lacZ reporter construct driven by -3.1 kb of proximal promoter sequence and SCE1 is expressed in spinal cord (SpC) and brain oligodendrocytes and spinal root (SpR) Schwann cells of P18 mice (arrows). B, F, As observed with randomly integrated constructs, -3.1 kb-mediated oligodendrocyte expression gradually shuts off in adults, whereas high-level SCE1-mediated Schwann cell expression is maintained (shown at 6 weeks). C, G, A reporter construct driven by -9.4 kb, and thus containing the full complement of recognized evolutionarily conserved sequence, is expressed at high levels in myelinating cells throughout development and maturity (shown at 12 weeks). D, H, LacZ driven by the 0.3 kb HSP68 minimal promoter alone is expressed at high levels only in CNS endothelial cells, with no detectable expression in either oligodendrocytes or Schwann cells (shown at 6 weeks).
Despite ubiquitous expression of the endogenous hprt gene, its 5′ flanking sequence provides a relatively neutral environment. A sensitive hprt-docked enhancer trap construct, regulated by 0.3 kb of the HSP68 promoter, is expressed only in cardiomyocytes (data not shown) and blood vessels (including in the CNS) (Fig. 2D,H) and, at trace levels, in spinal cord dorsal gray matter (data not shown). Thus, hprt-associated enhancers do not drive expression in myelinated glia or neurons. Although additional features of the hprt-docking site chromatin and regulatory environment remain to be elucidated, this circumstance supports the use of direct interconstruct regulatory program comparisons made throughout this investigation.
Multiple pathways mediate oligodendrocyte targeting during development
The design of reporter constructs analyzed here was based on the modular structure of conserved MBP 5′ flanking sequence. Subsequent analysis of reporter gene expression was performed in either tissue sections or whole-mount preparations of spinal cords and brain (oligodendrocytes), or spinal roots and sciatic nerves (Schwann cells). Expression was analyzed mainly at P18, corresponding to peak MBP mRNA accumulation in the CNS (Mathisen et al., 1993), at multiple mature ages when stable and lower MBP mRNA levels are observed, and in the remyelinating CNS and PNS of mature mice.
To begin, we evaluated the putative regulatory function of M1 and M2 sequences using a series of proximal promoter sequences extending to -794 bp. No oligodendrocyte expression was detected from constructs regulated by either -165 bp (data not shown) or -300 bp (Fig. 3F, see below) of proximal promoter sequence. In contrast, extending the promoter to -377 bp, and thus including the complete M1 sequence, leads to expression in oligodendrocytes during primary myelination (Fig. 3A), which shuts off by approximately P30 (data not shown). The sequence extending to -794 bp, and thus including M2, results in higher levels of reporter gene expression at P18 (Fig. 3B, see below) and extends the period of expression (Fig. 3C) (construct also differs in content of truncated Schwann cell enhancer sequences ligated 3′ of lacZ). Like the M1-regulated construct, expression in oligodendrocytes is transient, and, at 3 months of age, only a small oligodendrocyte subpopulation expresses the -794 bp regulated reporter (data not shown).
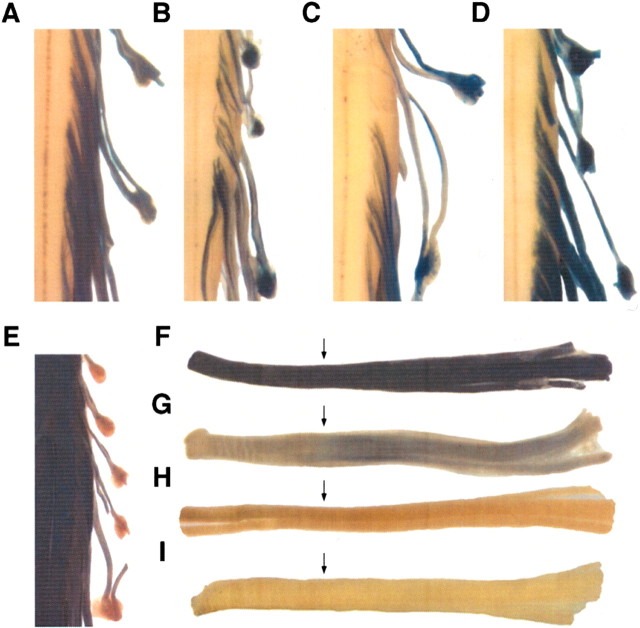
Figure 3.
Multiple pathways mediate oligodendrocyte targeting during primary myelination. A, B, lacZ regulated by -377 bp/SCE1 (0.2 kb), which includes M1, or -794 bp/SCE2 (0.44 kb), which includes both M1 and M2, is expressed in spinal cord oligodendrocytes at P18. C, -794 bp/SCE2 (0.44 kb) mediated oligodendrocyte expression gradually shuts off in the adult (shown at 6 weeks), in a pattern similar to that seen for the -3.1 kb/SCE1 construct. D, E, M3 has autonomous oligodendrocyte targeting ability. A 0.38 kb M3 fragment ligated to the HSP68 minimal promoter (0.3 kb) drives high-level expression in spinal cord oligodendrocytes (shown at P18 and 24 weeks, respectively). F, A truncated version of M1 (-300 bp) does not drive oligodendrocyte expression (shown at P18). G, H, Addition of either 0.38 or 2.7 kb (spanning from -5.8 to -3.1 kb) module 3-containing sequences to -300 bp MBP results in high-level and continuous oligodendrocyte expression throughout development and adulthood (shown at P18). I, An otherwise identical construct, deleted of a 0.6 kb sequence spanning M3 (from -5.3 to -4.7 kb), fails to express in oligodendrocytes throughout development and adulthood (shown at P18). Constructs represented in F-I contain the SCE1 sequence ligated 3′ of the lacZ gene, accounting for expression in Schwann cells (Forghani et al., 2001).
To determine whether M3 has regulatory function, a series of five constructs was evaluated. In contrast to M1- or M2-containing constructs, a construct bearing the 0.38 kb M3-containing sequence, ligated to the heterologous HSP68 promoter, was expressed at high levels not only in juvenile mice but throughout maturity and senescence (Fig. 3D,E). Furthermore, an identical CNS expression pattern is observed when either the 0.38 or 2.7 kb (from -5.8 to -3.1 kb) M3-containing sequence is ligated to the nontargeting (minimal) -300 bp MBP promoter (Fig. 3F-H). Finally, a construct otherwise identical to that bearing the -5.8 to -3.1 kb sequence but deleted of a 0.6 kb M3-containing fragment fails to express in oligodendrocytes at any age (Fig. 3I).
Together, these results demonstrate that major oligodendrocyte targeting functions reside within the first three MBP modules, corresponding to a total of 0.7 kb of conserved sequence. Depending on the stage of CNS maturation, M1, M2, and M3 all contribute to oligodendrocyte expression. Whereas M1- and M2-containing constructs are expressed robustly in oligodendrocytes of juvenile mice during primary myelination, M3 alone drives continuous expression in the mature CNS.
M1 and M2 fail to target expression during adult CNS remyelination
A demyelinating insult in the mature CNS results in local maturation of oligodendrocyte progenitors that subsequently initiate expression of myelin genes (including MBP) and partially remyelinate denuded axon segments. Among MBP-regulated constructs that express in oligodendrocytes during primary development, the -3.1 kb regulated construct fails to be reactivated in such newly myelinating adult brain oligodendrocytes, whereas a -8.9 kb regulated construct is expressed robustly (B. Finsen and A. C. Peterson, unpublished observations). Using a novel spinal cord demyelination strategy (Jasmin et al., 2000), we show here that neither the -3.1 kb (Fig. 4A,C) nor the -794 bp sequence (Fig. 4B,D) drive expression in newly myelinating oligodendrocytes analyzed at 6 weeks after the demyelinating insult. β-Galactosidase histochemical labeling and electron microscopic analysis of adjacent spinal cord cross-sections reveals no detectable activity in oligodendrocytes elaborating thin compact myelin sheaths typical of adult remyelination. Together, these results suggest that M3, essential for expression throughout myelin maintenance, may also be required for MBP expression during myelin repair in the adult CNS.
M3 and M4 targeting to myelinating Schwann cells depends on combinatorial interactions
MBP also is expressed by Schwann cells myelinating PNS axons and the previously designated 0.6 kb SCE1 sequence (Forghani et al., 2001) located at -8.9 to -8.3 kb partially overlaps M4 (-9.1 to -8.7 kb). All hprt-docked constructs containing M4, including minimally promoted constructs, express in myelinating Schwann cells during both primary development and regeneration after sciatic nerve crush (Fig. 5A,B,F). Furthermore, constructs regulated by the human M4 sequence also express robustly, demonstrating multiple levels of human-mouse conservation in the mechanism regulating Schwann cell MBP expression (Fig. 5C,D).
Figure 5.
M3 and M4 individually target expression to myelinating Schwann cells. A-C, Reporter constructs M4 1.1 kb/hsp, M4/hsp, and M4 1.0 kb (human)/hsp all demonstrate high-level expression in spinal root Schwann cells throughout primary development and in the adult (all shown at 12 weeks). D, A randomly inserted M4 1.0 kb (human)/hsp reporter construct similarly results in high-level Schwann cell expression (shown at 12 weeks). E, The M3/hsp reporter construct drives high-level expression in spinal root Schwann cells but only during active myelin formation in primary development (shown at P10). F, The M4 1.1 kb/hsp construct is expressed at high levels in both mature (left of arrow) and remyelinating (right of arrow) Schwann cells (shown at 21 dpc). G, H, The M3/hsp construct also drives expression in Schwann cells during the initial phase of remyelination after sciatic nerve crush injury but gradually shuts off (portion of nerve to right of arrow), shown at 14 and 21 dpc, respectively. I, The control HSP68-promoted construct fails to express in either mature or remyelinating Schwann cells (shown at 14 dpc).
Our combined data provide strong evidence that major MBP regulatory subprograms are conferred through each of the modules identified by sequence conservation. In addition, in the course of these functional studies, evidence of combinatorial regulatory control extending beyond individual modules was encountered. Previous investigations showed that MBP 5′ flanking sequences initiating at the proximal promoter must extend into M4 to confer Schwann cell expression (Foran and Peterson, 1992; Gow et al., 1992; Forghani et al., 2001). Thus, it was unexpected that a construct bearing only M3 sequences would drive expression in Schwann cells. Nonetheless, when isolated from adjacent intermodular sequences, M3 drives robust, albeit transient, Schwann cell expression during both development and sciatic nerve remyelination (Fig. 5E,G,H). Because the control HSP68 construct is not expressed in either developing or remyelinating sciatic nerve Schwann cells (Fig. 5I), this activity is attributable to M3 alone.
Additional evidence for combinatorial regulatory control was revealed for M4-containing constructs. When divorced from additional MBP sequence, M4-containing constructs express ectopically in satellite glia of dorsal root ganglia (Fig. 5A-D and data not shown), whereas the -9.4 kb construct shows no such expression (Fig. 2C). The M4 1.1 kb/M3/M2/M1 construct (which contains all the modules with only minimal surrounding sequence) reveals a regulation program similar to that conferred by contiguous MBP sequences extending to -9.4 kb and, as observed in the chimeric preparation shown in Figure 6, appears not to express in satellite glia. Thus, elements serving to constrain the spectrum of glial cell types expressing MBP also likely reside within modular sequences.
Figure 6.

Combined M1 to M4 targeting profile parallels that of -9.4 kb construct. The M4 1.1kb/M3/M2/M1 construct is expressed at high levels throughout development and maturity in both oligodendrocytes and Schwann cells but not in dorsal root ganglia (shown here in a chimera at 6 weeks).
The unmasked M3 Schwann cell enhancer activity and the M4-mediated satellite cell targeting suggest that either intermodular or modular sequences can serve to restrict the cell-specific enhancer function contained in modular sequences. Because combinatorial regulatory activity underlies the above two targeting phenotypes, it raised the possibility that such interactions might also regulate quantitative phenotypes.
Quantitative comparison of hprt-docked reporter gene expression levels
For the well characterized endo-16 locus of sea urchin (Yuh et al., 1998, 2001; Bolouri and Davidson, 2002), quantitative functions have been shown to play a major role in the overall cis-regulatory program. To determine whether MBP regulatory sequences similarly control a variety of complex quantitative phenotypes, we first set out to validate quantitative comparisons between hprt-docked constructs by determining whether independently derived transgenic lines bearing identical constructs accumulate similar levels of β-galactosidase. Figure 7 (inset) shows that pairs of transgenic lines derived from independent embryonic stem cell clones bearing -3.1 kb/SCE1, -8.9 kb, or -9.4 kb constructs display similar β-galactosidase activity levels (across the broad range of quantitative phenotypes predicted from the histochemical preparations). Thus, we conclude that differences in interconstruct β-galactosidase activity levels can be reliably ascribed to the complement of regulatory elements contained within each construct.
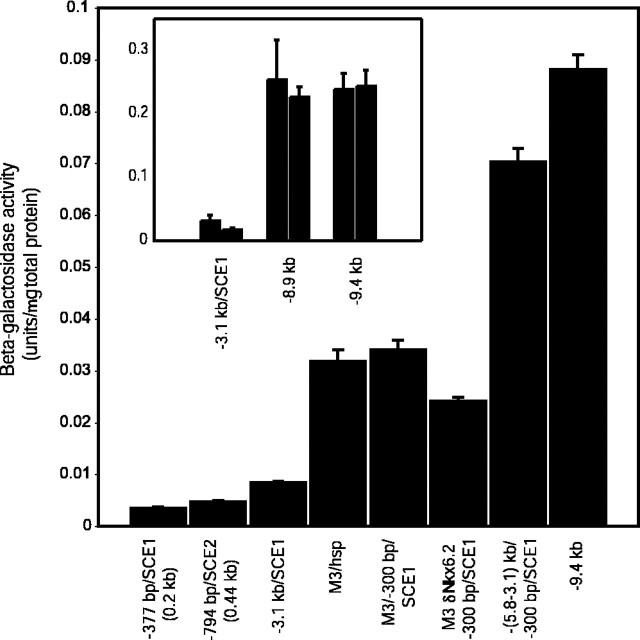
Figure 7.
Multiple comparisons of β-galactosidase activity levels between transgenic lines validates quantitative analysis of hprt-docked reporter genes. Inset demonstrates that independently derived transgenic lines bearing the same construct at the hprt locus accumulate similar levels of β-galactosidase (units per micrograms of total protein) in oligodendrocytes (comparisons between pairs of transgenic lines derived from independent ES clones bearing -3.1 kb/SCE1, -8.9 kb, and -9.4 kb constructs are shown). Comparisons of the expression levels achieved by the -9.4 kb reporter construct and constructs regulated by variously truncated MBP sequences demonstrate that significant enhancer activity is associated with all three oligodendrocyte targeting modules.
Our investigations reveal extensive quantitative modulation because mice bearing the various constructs demonstrated a wide range of oligodendrocyte β-galactosidase activity levels (Fig. 7). There is an ∼25-fold difference between the expression levels of the -377 bp/SCE1 (0.2 kb) and -9.4 kb transgenic lines. In particular, significant enhancement appears to be conferred through modular activity mediated by M3. For example, the M3/hsp and M3/-300 bp/SCE1 lines have β-galactosidase activity levels approximately eightfold that of -794 bp/SCE2 (0.44 kb) and 36% that of the -9.4 kb line. In addition, certain intermodular sequences also appear to be associated with significant quantitative activity; the (-5.8 to -3.1 kb)/-300 bp/SCE1 transgenic line displays ∼2.1-fold β-galactosidase activity compared with the M3/-300 bp/SCE1 line, possibly related to short conserved sequences lying adjacent to M3.
To identify functional regulatory elements in vivo, we extended our quantitative analysis to examine the potential contribution of putative binding sites for transcription factors already implicated in oligodendrocyte maturation. We initially focused on putative sites within M3, because we show that it is the dominant regulator of transgene expression in mature oligodendrocytes. Nkx6.2/Gtx is an oligodendrocyte-specific homeodomain protein known to avidly bind TAAT-containing consensus sites (Awatramani et al., 1997, 2000). Two evolutionarily conserved putative Nkx6.2/Gtx binding sites (both containing the essential TAAT core) on the (+) strand of M3 were mutated in a manner to abolish binding (Awatramani et al., 1997, 2000). The resultant M3 δNkx6.2/-300 bp/SCE1 construct (bearing the M3 sequence mutations GTTAATGC → GTTGGCGC and TTTAATTC → TTTGGCCC) retains continuous oligodendrocyte targeting activity (data not shown). However, quantitative analysis at P18 reveals a statistically significant decrease (∼35%) in oligodendrocyte β-galactosidase activity levels (Fig. 7). Thus, Nkx6.2/Gtx (or related Nkx homeodomain protein) mediated regulation through these elements is not essential for M3 oligodendrocyte targeting function, but transcriptional efficiency in vivo is finely modulated through transcription factor binding at one or both of these two homeodomain sites.
Discussion
Multiple modules differentially regulate MBP expression in oligodendrocytes
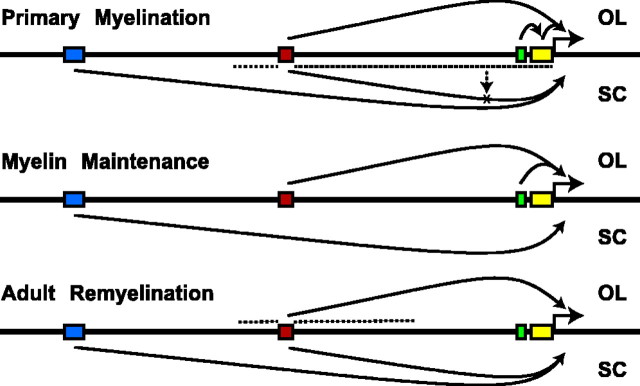
In this investigation, we validated and applied an efficient strategy for the in vivo identification and functional characterization of MBP regulatory sequences. Initially, comparison of large regions of human and mouse MBP noncoding sequences identified four widely spaced conserved noncoding modules, totaling ∼1 kb in length. Using a novel targeted transgenesis paradigm that allows for single-copy insertion at the hprt locus, each module was shown to confer a unique cell-targeting and quantitative gene programming function, seemingly irrespective of absolute spacing from the transcriptional start site. A model summarizing the basic combinatorial targeting and quantitative programs of the MBP regulatory system is presented in Figure 8. Together, our results demonstrate that three MBP modules differentially control major cell-state-dependent targeting activities and expression levels during CNS primary myelination and myelin maintenance, whereas modular and intermodular interactions contribute to maintain glial specificity.
Figure 8.
Modular organization of the MBP cis-regulatory system. Schematic representation of recognized regulatory programs conferred by MBP modular sequences. Targeting activity is represented by solid arrows in oligodendrocytes (OL) and Schwann cells (SC). Potential intermodular regulatory activity is represented by dotted lines.
Several in vivo studies have focused on the MBP proximal promoter region and together have demonstrated that short segments are sufficient to activate oligodendrocyte-specific and developmentally regulated reporter gene expression (Turnley et al., 1991; Gow et al., 1992; Miskimins et al., 1992; Goujet-Zalc et al., 1993; Wrabetz et al., 1998). Other studies, using similar promoters, have revealed ectopic expression in a number of non-neural tissues (Yoshioka et al., 1991; Asipu et al., 2001). In this study of hprt-docked constructs, the shortest segment mediating oligodendrocyte-specific targeting was the M1-containing -377 bp sequence, and it conferred activity only during the postnatal period encompassing primary CNS myelin deposition. Truncation of M1 (-300 bp, Fig. 3F; -165 bp, data not shown) led to complete loss of oligodendrocyte targeting. It is notable that the first 138 bp of the MBP promoter also is a widely expressed exon included in an overlapping transcriptional unit (Campagnoni et al., 1993). Thus, conservation in this portion of M1 likely includes constraints imposed by this coding function. In contrast to the present findings, a previous study (Goujet-Zalc et al., 1993) using random insertion transgenesis reported oligodendrocyte targeting from a construct regulated by the same -300 bp promoter sequence. However, construct multimerization and enhancer trapping are commonly observed at random insertion transgene loci, potentially leading to novel regulatory element interactions (Heard et al., 1999).
Extension of proximal promoter sequence to include M2 as well as M1 (with the -794 bp construct) increases oligodendrocyte reporter gene expression (∼37%) at P18 and modestly extends the period of expression, at least in a subpopulation of oligodendrocytes. Only M3-containing constructs were uniformly expressed at high levels in oligodendrocytes throughout primary development, adulthood, and senescence. At P18, the M3/hsp construct (which contains no other MBP sequence except the 0.38 kb fragment encompassing M3) was expressed at approximately eightfold the level of the -794 bp construct and at ∼36% that of the -9.4 kb construct (which contains the entire complement of modules). Thus, M3 appears to be the primary regulator of MBP expression both in terms of enhancing expression levels during primary myelin deposition and independently regulating expression during myelin maintenance.
These findings are consistent with previous reports demonstrating that proximal promoter sequences, up to -1.3 and -3 kb in length, targeted low-level expression to myelinating oligodendrocytes that was attenuated with maturation (Foran and Peterson, 1992; Miskimins et al., 1992; Wrabetz et al., 1998). In contrast, a randomly integrated -1.9 kb proximal promoter sequence did confer high-level oligodendrocyte reporter gene expression in adult mice (Gow et al., 1992). Unpredicted effects of copy number and integration site or the presence of introns could account for this difference. Alternatively, the -1.9 kb promoter contains a unique combination of enhancer and repressor elements not revealed by the current analysis. We consider the latter scenario less likely because the intervening region between -1.9 kb and M2 contains no modular sequence, and there is minimal difference between the quantitative phenotypes of the -3.1 kb and -794 bp-promoted constructs.
Throughout this investigation, hprt targeting made it possible to use interconstruct comparisons to assign precise quantitative phenotypes. Analysis of these quantitative results suggests that most of the differences may derive from a simple additive relationship among the regulatory sequences. Notably, addition of the activities derived from constructs regulated by individual modular sequences results in a value approximating one-half that observed for the contiguous -9.4 kb sequence. This difference may indicate either the influence of combinatorial relationships or the exclusion of relevant sequence from the defined modules. The relatively large quantitative difference observed for M3 alone versus M3 in the context of an additional 2.3 kb of surrounding sequence is consistent with the latter circumstance and suggests that not all functional regulatory sequences are grouped into distinct modules.
Among the few factors directly implicated in the coordinated activation of myelin gene expression, the homeodomain protein Nkx6.2/Gtx in particular has been shown to be expressed exclusively in differentiated oligodendrocytes (Awatramani et al., 1997). Notably, its temporal expression profile parallels that of myelin-specific mRNAs in multiple cell states (Awatramani et al., 1997; Sim et al., 2000). In addition, in vitro assays have demonstrated that Nkx6.2/Gtx binds to a number of recognition sites within both MBP and proteolipid protein proximal promoters, although none of the MBP putative sites previously examined encode a high-affinity TAAT core (Awatramani et al., 1997, 2000). In contrast, M3 contains three such TAAT core domains, and a construct bearing mutations in the two sites contained within the (+) strand shows decreased expression levels at P18 (Fig. 7). Thus, Nkx6.2/Gtx (or related homeodomain proteins) appears to function within a complex network of factors impinging on M3 in oligodendrocytes. Investigations are ongoing to unmask the contributions to transcriptional efficiency by the remaining TAAT as well as other putative binding sites within M3.
Newly myelinating adult oligodendrocytes do not recapitulate the developmental recruitment of MBP modules
CNS lesions in experimental demyelination and in diseases such as MS show delayed and frequently incomplete remyelination (Franklin, 2002). Although no susceptibility locus has so far been unequivocally identified for MS, strong evidence points to the involvement of genetic factors (Pihlaja et al., 2003). Abortive adult remyelination could result from deficient recruitment of oligodendrocyte precursors (which involves both migration and proliferation), a relative decrease in the rate of differentiation of recruited precursors into myelination competent adult oligodendrocytes, altered myelin protein gene regulation, or a combination thereof (Franklin et al., 1997; Gensert and Goldman, 1997).
Our results suggest that the M3 elements that independently mediate expression throughout myelin maintenance in the uninjured adult may also be recruited for expression in newly myelinating oligodendrocytes in the adult after a demyelinating insult. Although M1 and M2 drive expression during development in the CNS, they neither maintain expression in the adult nor reinitiate expression after a demyelinating injury (Fig. 4). The differential MBP modular recruitment exhibited by newly myelinating adult oligodendrocytes suggests that the limited remyelination capacity seen throughout the CNS, across species and in numerous disease states, may thus involve critical differences in the programming of essential myelin genes compared with primary development, possibly reflecting different transcription factor repertoires.
Schwann cells demonstrate a distinct pattern of modular recruitment
We showed previously that the M4-overlapping SCE1 sequence was both necessary and sufficient for developmentally regulated targeting to myelinating Schwann cells (Forghani et al., 2001). The experimental paradigm used here revealed additional Schwann cell enhancer activity within M3. This activity transiently targets expression to primary myelinating and adult remyelinating Schwann cells, coincident in both cases with the rapid phase of myelin deposition. In contrast to M4, which can target robustly during development and remyelination independent of any other MBP sequences, M3 activity is revealed only when it is isolated from other MBP sequences. Although its Schwann cell enhancer role in the context of the endogenous locus remains to be established, M3 interacts specifically with Schwann cell transcription factors during myelin elaboration and therefore can be used effectively as a probe or marker for factors expressed at this maturation stage.
Conclusion
Our observations identify a complex of sequence motifs through which MBP expression is differentially controlled in the CNS and PNS during primary myelination, myelin maintenance, and de novo myelin formation in the adult. In addition to providing a tool to target precisely predetermined levels of exogenous gene products to myelinating glia, we believe it likely that such motifs will serve as effective probes leading to the identification of the relevant transcription factors engaged in such different glial states as seen during development, maturity, or after myelin perturbations secondary to genetic alterations or injury. Through such associations, linking myelin gene regulation to cellular signaling may then be possible (Stolt et al., 2002).
Finally, although it is likely that many strategies will be used in the functional analysis of regulatory sequences, this investigation demonstrates the effectiveness of the controlled hprt transgenesis technique in assigning both qualitative and quantitative in vivo regulatory phenotypes. With the emergence of the complete genome sequences for human, mouse, and other organisms (Lander et al., 2001; Venter et al., 2001; Waterston et al., 2002), comparative analysis can be applied on a genome-wide scale (Deloukas et al., 2001; Mural et al., 2002). As similar levels of functional understanding are achieved with additional model loci, new insights into genome organization and the structure of regulatory networks appear likely.
Footnotes
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and Multiple Sclerosis (MS) Society of Canada and program grants from the MS Foundation of Canada and Genome Quebec. H.F.F. and R.F. were supported through CIHR Doctoral Fellowships. P.L. was supported by a CIHR Postdoctoral Fellowship. T.J.H. is a recipient of an Investigator Award from the CIHR and a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund. This work was performed in collaboration with the Royal Victoria Hospital Transgenic Facility. We dedicate this paper to its Director, Dr. N. Bachnou, deceased, 2002. We thank P. Valera, I. Tretjakoff, H. Furtenbacher, T. Fernandez, C. Dy, and J. Fortin for their excellent technical support.
Correspondence should be addressed to Alan Peterson, Royal Victoria Hospital, Molecular Oncology Group H5.35, Montreal, Quebec, Canada H3A 1A1. E-mail: alan@devbiol2.molonc.mcgill.ca.
Copyright © 2003 Society for Neuroscience 0270-6474/03/2310214-10$15.00/0
References
- Asipu A, Mellor AL, Blair GE ( 2001) The specificity of the myelin basic protein gene promoter studied in transgenic mice. Biochem Biophys Res Commun 288: 809-818. [DOI] [PubMed] [Google Scholar]
- Awatramani R, Scherer S, Grinspan J, Collarini E, Skoff R, O'Hagan D, Garbern J, Kamholz J ( 1997) Evidence that the homeodomain protein Gtx is involved in the regulation of oligodendrocyte myelination. J Neurosci 17: 6657-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awatramani R, Beesley J, Yang H, Jiang H, Cambi F, Grinspan J, Garbern J, Kamholz J ( 2000) Gtx, an oligodendrocyte-specific homeodomain protein, has repressor activity. J Neurosci Res 61: 376-387. [DOI] [PubMed] [Google Scholar]
- Bermingham Jr JR, Scherer SS, O'Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG ( 1996) Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev 10: 1751-1762. [DOI] [PubMed] [Google Scholar]
- Bjartmar C, Wujek JR, Trapp BD ( 2003) Axonal loss in the pathology of MS: consequences for understanding the progressive phase of the disease. J Neurol Sci 206: 165-171. [DOI] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH ( 2002) Modeling DNA sequence-based cisregulatory gene networks. Dev Biol 246: 2-13. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M ( 2001) The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev 15: 66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O ( 1996) Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci USA 93: 9067-9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnoni AT, Pribyl TM, Campagnoni CW, Kampf K, Amur-Umarjee S, Landry CF, Handley VW, Newman SL, Garbay B, Kitamura K ( 1993) Structure and developmental regulation of Golli-mbp, a 105-kilobase gene that encompasses the myelin basic protein gene and is expressed in cells in the oligodendrocyte lineage in the brain. J Biol Chem 268: 4930-4938. [PubMed] [Google Scholar]
- Cvetkovic B, Yang B, Williamson RA, Sigmund CD ( 2000) Appropriate tissue- and cell-specific expression of a single copy human angiotensinogen transgene specifically targeted upstream of the HPRT locus by homologous recombination. J Biol Chem 275: 1073-1078. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Matthews LH, Ashurst J, Burton J, Gilbert JG, Jones M, Stavrides G, Almeida JP, Babbage AK, Bagguley CL, Bailey J, Barlow KF, Bates KN, Beard LM, Beare DM, Beasley OP, Bird CP, Blakey SE, Bridgeman AM, Brown AJ, et al. ( 2001) The DNA sequence and comparative analysis of human chromosome 20. Nature 414: 865-871. [DOI] [PubMed] [Google Scholar]
- Foran DR, Peterson AC ( 1992) Myelin acquisition in the central nervous system of the mouse revealed by an MBP-Lac Z transgene. J Neurosci 12: 4890-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forghani R, Garofalo L, Foran DR, Farhadi HF, Lepage P, Hudson TJ, Tretjakoff I, Valera P, Peterson A ( 2001) A distal upstream enhancer from the myelin basic protein gene regulates expression in myelin-forming Schwann cells. J Neurosci 21: 3780-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJ ( 2002) Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci 3: 705-714. [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Gilson JM, Blakemore WF ( 1997) Local recruitment of remyelinating cells in the repair of demyelination in the central nervous system. J Neurosci Res 50: 337-344. [DOI] [PubMed] [Google Scholar]
- Gensert JM, Goldman JE ( 1997) Endogenous progenitors remyelinate demyelinated axons in the adult CNS. Neuron 19: 197-203. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Barton LM, Gilbert JG, Bench AJ, Sanchez MJ, Bahn S, Mistry S, Grafham D, McMurray A, Vaudin M, Amaya E, Bentley DR, Green AR, Sinclair AM ( 2000) Analysis of vertebrate SCL loci identifies conserved enhancers. Nat Biotechnol 18: 181-186. [DOI] [PubMed] [Google Scholar]
- Goujet-Zalc C, Babinet C, Monge M, Timsit S, Cabon F, Gansmuller A, Miura M, Sanchez M, Pournin S, Mikoshiba K, Zalc B ( 1993) The proximal region of the MBP gene promoter is sufficient to induce oligodendroglial-specific expression in transgenic mice. Eur J Neurosci 5: 624-632. [DOI] [PubMed] [Google Scholar]
- Gow A, Friedrich Jr VL, Lazzarini RA ( 1992) Myelin basic protein gene contains separate enhancers for oligodendrocyte and Schwann cell expression. J Cell Biol 119: 605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel M, Di Polo A, Valera PB, Braun PE ( 1998) Four-kilobase sequence of the mouse CNP gene directs spatial and temporal expression of lacZ in transgenic mice. J Neurosci Res 53: 393-404. [DOI] [PubMed] [Google Scholar]
- Hardison RC, Oeltjen J, Miller W ( 1997) Long human-mouse sequence alignments reveal novel regulatory elements: a reason to sequence the mouse genome. Genome Res 7: 959-966. [DOI] [PubMed] [Google Scholar]
- Heard E, Mongelard F, Arnaud D, Avner P ( 1999) Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol 19: 3156-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin L, Janni G, Moallem TM, Lappi DA, Ohara PT ( 2000) Schwann cells are removed from the spinal cord after effecting recovery from paraplegia. J Neurosci 20: 9215-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, et al. ( 2001) Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA ( 2000) Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136-140. [DOI] [PubMed] [Google Scholar]
- Maier M, Berger P, Nave KA, Suter U ( 2002) Identification of the regulatory region of the peripheral myelin protein 22 (PMP22) gene that directs temporal and spatial expression in development and regeneration of peripheral nerves. Mol Cell Neurosci 20: 93-109. [DOI] [PubMed] [Google Scholar]
- Mathisen PM, Pease S, Garvey J, Hood L, Readhead C ( 1993) Identification of an embryonic isoform of myelin basic protein that is expressed widely in the mouse embryo. Proc Natl Acad Sci USA 90: 10125-10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing A, Behringer RR, Hammang JP, Palmiter RD, Brinster RL, Lemke G ( 1992) P0 promoter directs expression of reporter and toxin genes to Schwann cells of transgenic mice. Neuron 8: 507-520. [DOI] [PubMed] [Google Scholar]
- Miskimins R, Knapp L, Dewey MJ, Zhang X ( 1992) Cell and tissue-specific expression of a heterologous gene under control of the myelin basic protein gene promoter in transgenic mice. Brain Res Dev Brain Res 65: 217-221. [DOI] [PubMed] [Google Scholar]
- Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GL, Wides R, Halpern A, Li PW, Sutton GG, Nadeau J, Salzberg SL, Holt RA, Kodira CD, Lu F, Chen L, Deng Z, Evangelista CC, Gan W, Heiman TJ, Li J, et al. ( 2002) A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661-1671. [DOI] [PubMed] [Google Scholar]
- Oeltjen JC, Malley TM, Muzny DM, Miller W, Gibbs RA, Belmont JW ( 1997) Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res 7: 315-329. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, Krauss RM, Rubin EM ( 2001) An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294: 169-173. [DOI] [PubMed] [Google Scholar]
- Peterson AC, Bray GM ( 1984) Hypomyelination in the peripheral nervous system of shiverer mice and in shiverer in equilibrium normal chimaera. J Comp Neurol 227: 348-356. [DOI] [PubMed] [Google Scholar]
- Pihlaja H, Rantamaki T, Wikstrom J, Sumelahti ML, Laaksonen M, Ilonen J, Ruutiainen J, Pirttila T, Elovaara I, Reunanen M, Kuokkanen S, Peltonen L, Koivisto K, Tienari PJ ( 2003) Linkage disequilibrium between the MBP tetranucleotide repeat and multiple sclerosis is restricted to a geographically defined subpopulation in Finland. Genes Immun 4: 138-146. [DOI] [PubMed] [Google Scholar]
- Readhead C, Popko B, Takahashi N, Shine HD, Saavedra RA, Sidman RL, Hood L ( 1987) Expression of a myelin basic protein gene in transgenic shiverer mice: correction of the dysmyelinating phenotype. Cell 48: 703-712. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W ( 2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Hinks GL, Franklin RJ ( 2000) The re-expression of the homeodomain transcription factor Gtx during remyelination of experimentally induced demyelinating lesions in young and old rat brain. Neuroscience 100: 131-139. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M ( 2002) Terminal differentiation of myelinforming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16: 165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P ( 1994) Krox-20 controls myelination in the peripheral nervous system. Nature 371: 796-799. [DOI] [PubMed] [Google Scholar]
- Turnley AM, Morahan G, Okano H, Bernard O, Mikoshiba K, Allison J, Bartlett PF, Miller JF ( 1991) Dysmyelination in transgenic mice resulting from expression of class I histocompatibility molecules in oligodendrocytes. Nature 353: 566-569. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, et al. ( 2001) The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Vivian JL, Klein WH, Hasty P ( 1999) Temporal, spatial and tissue-specific expression of a myogenin-lacZ transgene targeted to the Hprt locus in mice. Biotechniques 27: 154-162. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, et al. ( 2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wight PA, Duchala CS, Readhead C, Macklin WB ( 1993) A myelin proteolipid protein-LacZ fusion protein is developmentally regulated and targeted to the myelin membrane in transgenic mice. J Cell Biol 123: 443-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, Pruss M, Schacherer F, Thiele S, Urbach S ( 2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res 29: 281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrabetz L, Taveggia C, Feltri ML, Quattrini A, Awatramani R, Scherer SS, Messing A, Kamholz J ( 1998) A minimal human MBP promoter-lacZ transgene is appropriately regulated in developing brain and after optic enucleation, but not in shiverer mutant mice. J Neurobiol 34: 10-26. [DOI] [PubMed] [Google Scholar]
- Yoshioka T, Feigenbaum L, Jay G ( 1991) Transgenic mouse model for central nervous system demyelination. Mol Cell Biol 11: 5479-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH ( 1998) Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science 279: 1896-1902. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH ( 2001) Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control. Development 128: 617-629. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ ( 2002) The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109: 61-73. [DOI] [PubMed] [Google Scholar]