Abstract
Endoplasmic reticulum (ER) tubules and sheets conventionally correspond to smooth and rough ER, respectively. The ratio of ER tubules-to-sheets varies in different cell types and changes in response to cellular conditions, potentially impacting the functional output of the ER. To directly test whether ER morphology impacts vesicular trafficking, we increased the tubule-to-sheet ratio in three different ways, by overexpressing Rtn4a, Rtn4b, or REEP5. Only Rtn4a overexpression increased exocytosis, but not overall levels, of several cell surface and secreted proteins. Furthermore, Rtn4a depletion reduced cell surface trafficking without affecting ER morphology. Similar results were observed in three different mammalian cell lines, suggesting that Rtn4a generally enhances exocytosis independently of changes in ER morphology. Finally, we show that Rtn4a levels modulate cell adhesion, possibly by regulating trafficking of integrins to the cell surface. Taking the results together, we find that altering ER morphology does not necessarily affect protein trafficking, but that Rtn4a specifically enhances exocytosis.
INTRODUCTION
How organelle morphology affects function is a fundamental question in cell biology. In this study, we investigate whether the morphology of the endoplasmic reticulum (ER) affects exocytosis. The ER network is composed of flat sheets and curved tubules interconnected by three-way junctions (Shibata et al., 2006). While flat ER sheets accommodate large polyribosomes on their surface and play major roles in protein translation, folding, and modification, ER tubules exhibit low ribosome density and are involved in lipid synthesis, carbohydrate metabolism, calcium homeostasis, and interorganellar contacts (Park and Blackstone, 2010; Goyal and Blackstone, 2013). ER exit sites are specialized regions of the tubular ER where vesicle budding occurs, followed by trafficking to the Golgi and travel through the secretory pathway (Lippincott-Schwartz et al., 2000).
Cells with specialized functions are enriched in specific ER morphologies. For example, pancreatic acinar cells and plasma cells, which produce large amounts of protein, are mostly populated with polyribosome-studded ER sheets. In contrast, cells involved in carbohydrate metabolism (e.g., hepatocytes), steroid hormone synthesis (e.g., adrenal cortical cells), and Ca+2 signaling (e.g., muscle cells) are enriched in smooth tubular ER (Black, 1972; Shibata et al., 2006; Friedman and Voeltz, 2011; Goyal and Blackstone, 2013). Thus, while correlations between ER morphology and function have been described in certain specialized cell types and in response to cellular conditions, the question remains whether ER morphology directly affects the functional output of the ER.
Multiple proteins contribute to the unique structures characteristic of different ER domains. Proteins with coiled-coil domains, such as CLIMP-63, kinectin, and p-180, support ER sheets (Shibata et al., 2010; Goyal and Blackstone, 2013). ER tubules are curved by proteins of the reticulon (Rtn) and DP1/REEP/Yop1p families, which are thought to wedge into the outer leaflet of phospholipid bilayers to induce membrane curvature (Voeltz et al., 2006; Goyal and Blackstone, 2013). There are four mammalian reticulon genes (RTN1, RTN2, RTN3, and RTN4/Nogo) with conserved C-terminal hydrophobic hairpins termed reticulon-homology domains (Yang and Strittmatter, 2007). Rtn4a is the largest protein and was originally identified as an inhibitor of neurite outgrowth and axonal regeneration in the central nervous system (Chen et al., 2000; GrandPré et al., 2000). While most reticulons, including Rtn4a and its shorter splice variants Rtn4b and Rtn4c, are enriched in the nervous system, they are also ubiquitously expressed in all tissues (Ramo et al., 2016). Among the REEP family, DP1/Yop1p and REEP5/6 contain transmembrane hairpin domain architectures similar to reticulons and are expressed in different mammalian cell types (Papadopoulos et al., 2015; Grumati et al., 2017).
In our study, we manipulated the ER tubule-to-sheet ratio to test whether this broadly impacts protein exocytosis, a functional role ascribed to ER exit sites of the smooth ER. We overexpressed the tubule-shaping proteins Rtn4a/Nogo-A, Rtn4b, or REEP5 in cultured mammalian cells, a previously validated approach to alter ER morphology (Voeltz et al., 2006; Shibata et al., 2008). We examined a number of cell surface and secreted proteins, finding that while their total levels were unchanged upon overexpressing these curvature-stabilizing proteins, only Rtn4a increased their trafficking through the secretory pathway. We show that this effect on exocytosis is not due to a more tubulated ER, but is rather a specific function of Rtn4a. Thus, our data suggest that altering ER morphology does not necessarily influence vesicular protein trafficking, but that Rtn4a has a specific function in promoting exocytosis.
RESULTS AND DISCUSSION
Overexpression of Rtn4a, but not Rtn4b or REEP5, increases trafficking of cell surface proteins without changing their overall expression levels
HeLa cells were transiently transfected with plasmids expressing Rtn4a-GFP, GFP-Rtn4b, mCherry-REEP5, or GFP-NLS as a control, and overexpression of each tubule-shaping protein was confirmed by Western blot and immunofluorescence (Supplemental Figure S1, A–H). Rtn4a, Rtn4b, and REEP5 levels were increased 3.5-, 6.9-, and 1.4-fold, respectively (Supplemental Figure S1, A, B, E, F, G, and H). To quantify the conversion of ER sheets into tubules induced by these curvature-stabilizing proteins, total ER was visualized using a KDEL marker (Supplemental Figure S1I). Perinuclear ER was more abundant in control cells, while Rtn4a, Rtn4b, and REEP5 overexpression resulted in a more dispersed and tubulated ER (Supplemental Figure S1J). To directly quantify ER sheet volume, we immunostained cells for the ER sheet marker CLIMP63 (Supplemental Figure S1K). Rtn4a, Rtn4b, and REEP5 overexpression reduced ER sheet volume 2.9-, 2.3-, and 3.3-fold, respectively (Supplemental Figure S1L).
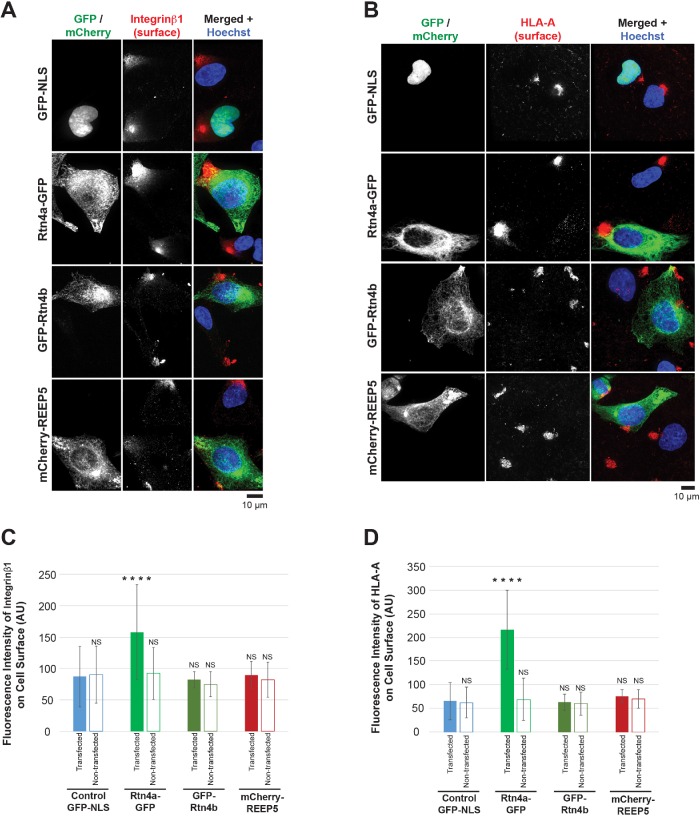
To assess how these changes in ER morphology might affect protein trafficking, we focused on two cell surface plasma membrane proteins whose transit through the secretory pathway has been well studied, integrin β1 and MHC class I/HLA-A (Jones et al., 1996; Sun et al., 2009). We performed immunostaining of nonpermeabilized cells to detect only cell surface–localized proteins (Supplemental Figure S2, A–C). While REEP5 and Rtn4b did not affect the amount of surface-localized integrin β1 and HLA-A, Rtn4a increased cell surface localization of integrin β1 and HLA-A by 1.8- and 3.2-fold, respectively (Figure 1, A–D). Clustering of integrin β1 and HLA-A on the cell surface has been previously reported (Matko et al., 1994; Clark et al., 1998), potentially corresponding to focal adhesion sites (Fourriere et al., 2019). Importantly, total levels of integrin β1 and HLA-A were unchanged upon overexpressing these tubule-shaping proteins (Supplemental Figure S2, D–I). Taken together, these data show that altering ER morphology does not generally affect protein trafficking, but that Rtn4a promotes trafficking of two membrane proteins to the cell surface without influencing their overall expression levels.
FIGURE 1:
Overexpression of Rtn4a, but not Rtn4b or REEP5, increases cell surface localization of integrin β1 and HLA-A. HeLa cells were transiently transfected with plasmids expressing GFP-NLS as a control, Rtn4a-GFP, GFP-Rtn4b, or mCherry-REEP5. (A) Nonpermeabilized cells were stained for surface-localized integrin β1. Here and in some other figures both GFP and mCherry were pseudocolored green to facilitate comparison of merged images. (B) Nonpermeabilized cells were stained for surface-localized HLA-A. (C) Integrin β1 surface fluorescence staining intensity was quantified for 30–66 transfected and nontransfected cells per condition. (D) HLA-A surface fluorescence staining intensity was quantified for 35–69 transfected and nontransfected cells per condition. All representative images are maximum intensity projections of confocal z-stacks. Error bars represent SD. ****, p ≤ 0.0001; NS, not significant.
Rtn4a promotes trafficking of cell surface proteins independently of effects on ER morphology
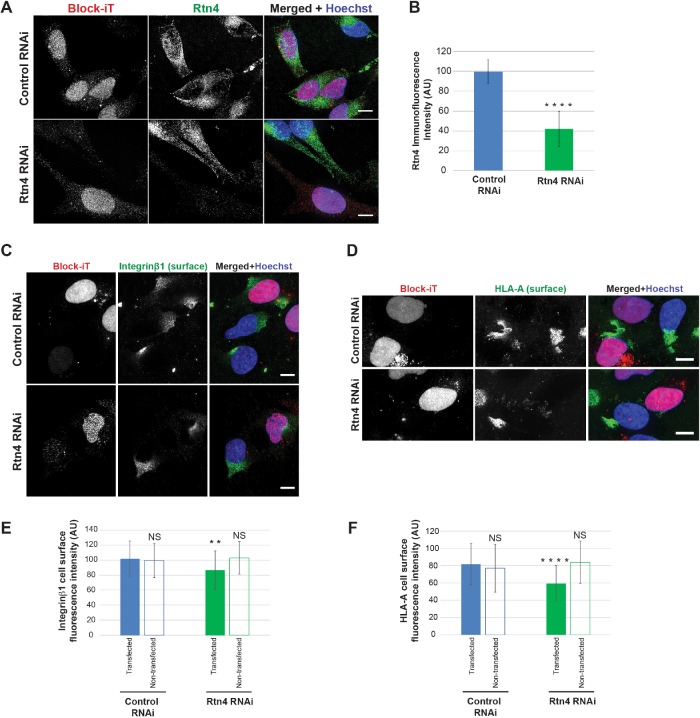
Of the three ER tubulating proteins tested in this study, only overexpression of Rtn4a increased trafficking of cell surface proteins. To validate this result, we depleted Rtn4 by small interfering RNA (siRNA) in HeLa cells (Figure 2, A and B), reducing Rtn4a levels 4.3-fold based on immunoblotting (Supplemental Figure S2, J–L). Rtn4 knockdown reduced cell surface levels of integrin β1 and HLA-A by 1.2- and 1.3-fold, respectively (Figure 2, C–F). Consistent with previous studies showing that Rtn1, 3, and 4 must be codepleted to convert ER tubules into sheets (Anderson and Hetzer, 2008), we observed no change in ER sheet volume in Rtn4 knockdown cells (Supplemental Figure S2, M and N). This result suggests that Rtn4a influences protein trafficking independently of any effect on ER morphology. Collectively, these data show that altered ER morphology and protein trafficking to the cell surface can be uncoupled, implying Rtn4a has a unique ER morphology-independent function in trafficking.
FIGURE 2:
Rtn4 knockdown decreases cell surface localization of integrin β1 and HLA-A. HeLa cells were transiently cotransfected with siRNA against Rtn4 and Block-iT fluorescent control or with Block-iT alone. (A) Cells were immunostained for Rtn4. (B) Rtn4 immunofluorescence intensity was quantified for 30–36 cells per condition. (C) Nonpermeabilized cells were stained for surface-localized integrin β1. (D) Nonpermeabilized cells were stained for surface-localized HLA-A. (E) Integrin β1 surface fluorescence staining intensity was quantified for 39–51 transfected and nontransfected cells per condition. (F) HLA-A surface fluorescence staining intensity was quantified for 34–51 transfected and nontransfected cells per condition. All representative images are maximum intensity projections of confocal z-stacks. Scale bars are 10 µm. Error bars represent SD. ****, p ≤ 0.0001; **, p ≤ 0.01; NS, not significant.
Rtn4a accelerates ER–to–cell surface trafficking
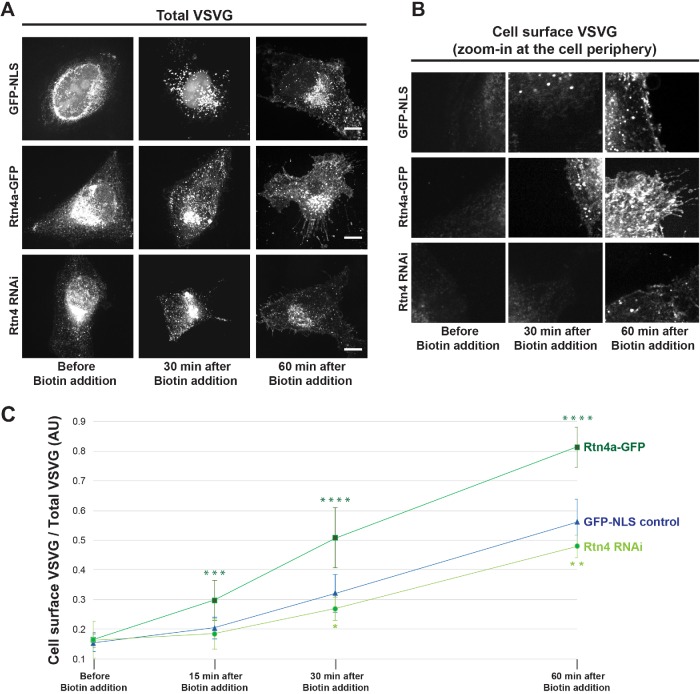
To complement our steady-state measurements of protein trafficking to the cell surface, we used the RUSH system to monitor the kinetics of arrival of a fluorescent cargo to the cell surface through the exocytic pathway. HeLa cells were transiently cotransfected with the VSVG-RUSH construct encoding VSVG-mCherry and streptavidin-Ii (Boncompain et al., 2012), along with GFP-NLS, Rtn4a-GFP, or Rtn4 siRNA. Before biotin addition, VSVG-mCherry is trapped in the ER. After biotin addition and VSVG release from the ER, we fixed cells at different time points and quantified the amount of cell surface–localized VSVG normalized to total VSVG. Consistent with Rtn4a levels influencing the rate of protein trafficking to the cell surface, we found that VSVG reached the cell surface faster upon Rtn4a overexpression and slower in Rtn4 knockdown cells (Figure 3, A–C).
FIGURE 3:
Rtn4a levels modulate the kinetics of VSVG-mCherry trafficking to the cell surface. HeLa cells were transiently cotransfected with a plasmid expressing GFP-NLS as a control, a plasmid expressing Rtn4a-GFP, or siRNA against Rtn4 along with the RUSH construct Str-Ii_VSVG-SBP-mCherry. ER-trapped VSVG was released by addition of 40 µM D-Biotin to the growth media. Cells were fixed before biotin addition and at 15, 30, or 60 min after biotin addition. (A) Representative images of maximum intensity projections of confocal z-stacks showing total VSVG-mCherry. (B) Representative images of single confocal z-planes at the cell periphery to visualize cell surface–localized VSVG-mCherry. (C) Fluorescence intensity of VSVG-mCherry was quantified from the cell surface z-plane and normalized to the total VSVG-mCherry fluorescence intensity measured from the maximum intensity projection of the same cell for 8–12 cells per condition and time point. Scale bars are 10 µm. Error bars represent SD. ****, p ≤ 0.0001; ***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05.
Rtn4a levels influence cell adhesion and exocytosis of soluble cargoes without inducing ER stress
Given that Rtn4a promotes exocytosis of integrin β1 to the cell surface, we next asked whether this was physiologically relevant in the context of cell adhesion (Chen et al., 1994). We altered Rtn4a expression levels in HeLa cells and measured cell attachment to a collagen-coated surface. While Rtn4a overexpression enhanced cell adhesion by 1.5-fold, Rtn4 knockdown reduced cell adhesion by 1.3-fold (Figure 4).
FIGURE 4:
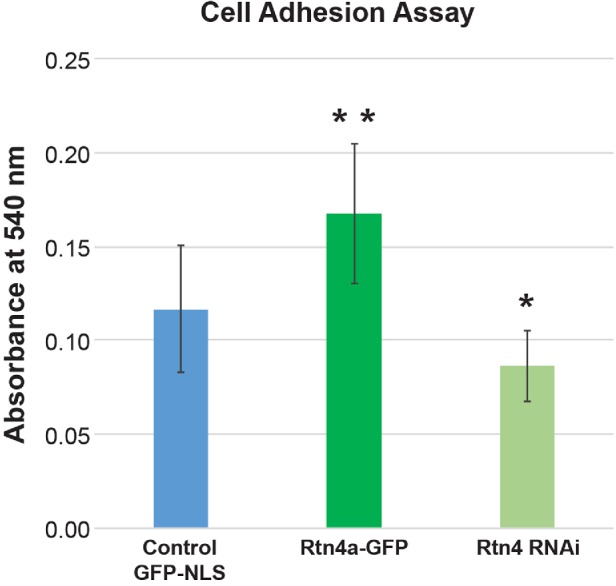
Rtn4a levels modulate cell adhesion. HeLa cells were transiently transfected with a plasmid expressing GFP-NLS as a control, a plasmid expressing Rtn4a-GFP, or siRNA against Rtn4. Cells were allowed to attach to collagen-coated dishes for 30 min. Unattached cells were removed, and attached cells were fixed, stained with crystal violet, and lysed. Cell lysate absorbance was measured at 540 nm (see Materials and Methods). Averages from nine independent experiments are shown. Error bars represent SD. **, p ≤ 0.01; *, p ≤ 0.05.
Observed effects of Rtn4a overexpression on cell surface transport were not restricted to HeLa cells, as Rtn4a overexpression increased cell surface trafficking, but not levels, of integrin β1 and HLA-A in MRC-5 cells, a noncancerous lung fibroblast cell line (Supplemental Figure S3, A–H). To test whether Rtn4a promotes exocytosis of soluble proteins in addition to cell surface membrane proteins, we examined the secreted and intracellular levels of fibulin-5 (FBLN5) and thrombospondin-1 (TSP1). FBLN5 and TSP1 are both extracellular matrix components expressed and secreted by many cell types (Crawford et al., 1998; Albig and Schiemann, 2005). Rtn4a overexpression increased the amount of FBLN5 and TSP1 secreted into the media (Supplemental Figure S3, I and L), without affecting their overall expression levels (Supplemental Figure S3, K and N). While the intracellular FBLN5 concentration was unchanged upon Rtn4a overexpression (Supplemental Figure S3J), the intracellular TSP1 concentration was reduced (Supplemental Figure S3M), suggesting TSP1 might be trafficked faster than FBLN5. These data show that Rtn4a promotes exocytosis of soluble secreted proteins in addition to membrane-bound proteins. Rtn4a overexpression did not cause ER stress as evidenced by constant levels of the ER chaperones calnexin, ERp72, and GRP78 (Supplemental Figure S3, O–R). Thus, Rtn4a overexpression increases trafficking of cell surface and secreted cargoes, without having dominant negative effects on cell function.
Rtn4a levels also influence integrin β1 trafficking in neuroblastoma cells
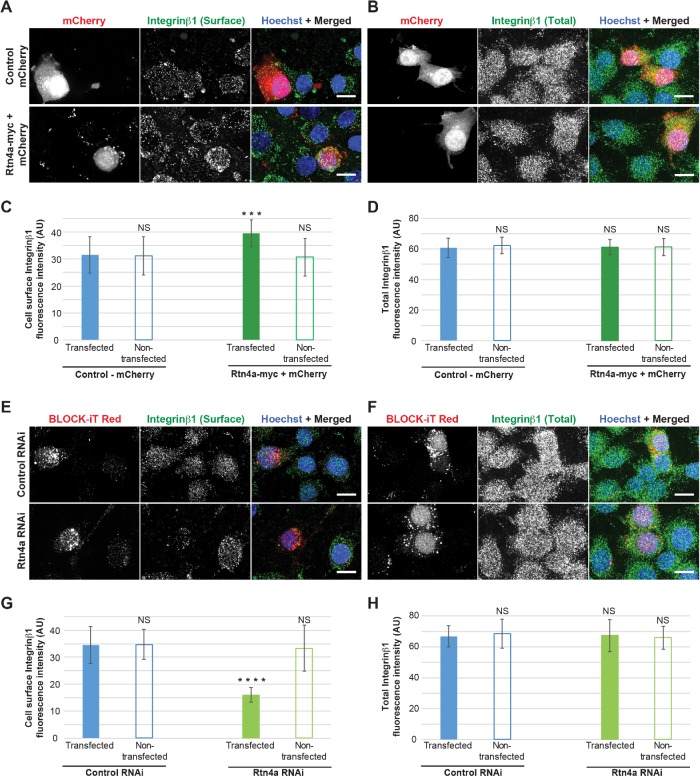
Our Rtn4a overexpression experiments tended to induce more pronounced trafficking effects than Rtn4 knockdown. Given that Rtn4a levels are relatively low in HeLa cells, we wondered whether Rtn4a knockdown might reduce exocytosis to a greater extent in a cell line with higher endogenous Rtn4a levels. To test this idea, we repeated a subset of our experiments using undifferentiated Neuro-2a cells, a neuroblastoma cell line that expresses high levels of Rtn4a (Mi et al., 2014). While Rtn4a-myc overexpression increased integrin β1 cell surface staining 1.3-fold (Figure 5, A–D, and Supplemental Figure S4, A–D), Rtn4a knockdown decreased cell surface localization of integrin β1 by 2.2-fold without changing total integrin β1 levels (Figure 5, E–H and Supplemental Figure S4, E–H). Integrin β1 clustering was less pronounced in Neuro-2a cells compared with HeLa and MRC-5 cells, perhaps because of differences in cell type and/or morphology.
FIGURE 5:
Rtn4a levels modulate cell surface localization of integrin β1 in Neuro-2a cells. (A–D) Undifferentiated Neuro-2a cells were transiently cotransfected with plasmids expressing Rtn4a-myc and mCherry or with mCherry alone as a control. To minimize concerns that the C-terminal GFP tag of Rtn4a-GFP used in other experiments might affect Rtn4a function, here we used the smaller myc tag. (A) Nonpermeabilized cells were stained for surface-localized integrin β1. (B) Permeabilized cells were stained for total integrin β1. (C) Integrin β1 surface fluorescence staining intensity was quantified for 22–30 transfected and nontransfected cells per condition. (D) Total integrin β1 fluorescence intensity was quantified for 18–26 transfected and nontransfected cells per condition. (E–H) Undifferentiated Neuro-2a cells were transiently cotransfected with siRNA specific to Rtn4a and Block-iT fluorescent control or with Block-iT alone. (E) Nonpermeabilized cells were stained for surface-localized integrin β1. (F) Permeabilized cells were stained for total integrin β1. (G) Integrin β1 surface fluorescence staining intensity was quantified for 25–28 transfected and nontransfected cells per condition. (H) Total integrin β1 fluorescence intensity was quantified for 20–24 transfected and nontransfected cells per condition. All representative images are maximum intensity projections of confocal z-stacks. Scale bars are 10 µm. Error bars represent SD. ****, p ≤ 0.0001; ***, p ≤ 0.001; NS, not significant.
Taking the results together, we report that Rtn4a levels influence protein trafficking to the cell surface in cancer epithelial, normal fibroblast, and neuroblastoma cell lines. This effect appears to be specific to Rtn4a and not related to changes in ER morphology because Rtn4a knockdown reduced trafficking of membrane proteins without affecting ER morphology and increasing the tubule-to-sheet ratio by Rtn4b or REEP5 overexpression did not affect exocytosis. It is also worth noting that altering ER morphology did not affect the expression levels of the various trafficked cargoes we tested and that converting ER tubules into sheets by CLIMP63 overexpression did not affect synthesis or trafficking of integrin β1 and HLA-A (unpublished data). Thus, while there are specialized cell types that are enriched in rough ER sheets or smooth tubular ER (Black, 1972; Shibata et al., 2006; Friedman and Voeltz, 2011; Goyal and Blackstone, 2013), altering ER morphology in cells with a more conventional ER tubule-to-sheet ratio has little effect on exocytosis.
If ER morphology per se does not influence protein trafficking, how might Rtn4a specifically promote exocytosis? Intriguingly, REEP5 and Rtn4b overexpression did not alter protein transport to the cell surface, while previous studies implicated REEP1 and Yop1p in vesicular trafficking (Calero et al., 2001; Saito et al., 2004). It may be that certain members of the reticulon and DP1/REEP/Yop1p families have unique structural elements, biophysical properties, or binding partners that facilitate exocytosis. Future studies will elucidate where Rtn4a acts in the secretory pathway and which Rtn4a domains are responsible for enhanced vesicular transport.
Our results with Rtn4a add to a growing literature on the role of reticulons in protein trafficking. Overexpression of RTN1C in rat PC12 adrenal tumor cells increased exocytosis of human growth hormone, mediated by interactions with the SNARE proteins syntaxin-1, -7, -13, and VAMP2 (Steiner et al., 2004; Di Sano et al., 2012). RTN3 has been shown to play a role in retrograde protein transport from the cis-Golgi to the ER. Notably, overexpression of RTN3 in HeLa cells delayed trafficking of VSVG from the ER to the cell surface (Wakana et al., 2005). Combining these results with our own, perhaps Rtn3 and Rtn4a have antagonizing effects on the secretory pathway. Previous studies support a role for Rtn4a in neuronal protein secretion. Rtn4a mRNA is highly expressed in the supraoptic nucleus and paraventricular nucleus of the rat hypothalamus, both regions being highly active for neuroendocrine secretion (Hasegawa et al., 2005), and RTN4A knockdown decreased dopamine release in rat PC12 cells (Xiong et al., 2008). Thus, while Rtn4a/Nogo-A has been implicated in protein secretion in the nervous system, our results show that Rtn4a plays a more general role in exocytosis and cell adhesion. Future research will elucidate the potential physiological links between Rtn4a and protein trafficking.
MATERIALS AND METHODS
Plasmids and siRNA
The Rtn4a-GFP (pAcGFP1-N1 Rtn4a; Shibata et al., 2008) and Rtn4a-myc (pcDNA3.1 Rtn4a-myc; GrandPré et al., 2000) constructs were gifts from Gia Voeltz (University of Colorado, Boulder). The pmCherry-C2 and mCherry-REEP5 (pmCherry-C2 REEP5; Schlaitz et al., 2013) constructs were gifts from Anne Schlaitz (Zentrum für Molekulare Biologie der Universität Heidelberg). The RUSH construct Str-Ii_VSVG-SBP-mCherry was a gift from Franck Perez (Institut Curie; Boncompain et al., 2012). The GFP-Rtn4b expression plasmid (pDL34) was described previously (Jevtic´ and Levy, 2015). The control GFP-NLS plasmid was from Invitrogen (V821-20). To knock down expression of RTN4 in HeLa cells, we used a DsiRNA (IDT): sense 5′-CUGGAAUCUGAAGUUGCUAUAUCUG-3′, and antisense 5′-CAGAUAUAGCAACUUCAGAUUCCAG-3′. This DsiRNA sequence is based on a previously described siRNA against RTN4 (Anderson and Hetzer, 2008). To knock down expression of RTN4A in Neuro-2a cells, we used a DsiRNA (IDT) specific for the mouse RTN4A isoform: sense 5′-AUCUGAACCAGUUGACUUAUUUAGT-3′, and antisense 5′-ACUAAAUAAGUCAACUGGUUCAGAUUC-3′.
Mammalian tissue culture and transfections
HeLa cells were obtained from the American Type Culture Collection, Neuro-2a mouse neuroblastoma cells were a gift from Baskaran Thyagarajan (University of Wyoming), and MRC-5 normal human lung fibroblast cells were a gift from Jason Gigley (University of Wyoming). Cells were verified to be mycoplasma-free (ThermoFisher Scientific; #M7006). HeLa and MRC-5 cells were cultured in Eagle’s MEM supplemented with 10% vol/vol fetal bovine serum (FBS) and 50 IU/ml penicillin/streptomycin at 37°C in 5% CO2. Undifferentiated Neuro-2a cells were cultured in DMEM-F12 medium supplemented with 10% vol/vol FBS and 50 IU/ml penicillin/streptomycin at 37°C in 5% CO2.
For transient transfection of plasmids, cells were seeded at 3 × 105 cells/well in six-well plates and grown to 70–90% confluency. Plasmid DNA (2.5 µg per well) was transfected using Lipofectamine 3000 (Invitrogen), following the manufacturer’s protocol. For Neuro-2a cells, the mCherry-C2 plasmid was used as a control and cotransfected with Rtn4a-myc to identify transfected cells. For transient siRNA transfections, cells were grown in six-well plates to 60–80% confluency. Rtn4 siRNA (25 pmol per well for HeLa cells) and Rtn4a siRNA (100 pmol per well for Neuro-2a cells) were transfected using Lipofectamine RNAiMAX (Invitrogen), following the manufacturer’s protocol. BLOCK-iT Alexa Fluor Red Fluorescent Control (Invitrogen) was used as a control and cotransfected with siRNAs to identify transfected cells. The average transfection efficiency for each plasmid and siRNA was calculated from three to five independent experiments. Transfection efficiencies ranged from 62% to 76% for HeLa cells and 40% to 51% for Neuro-2a cells. To prepare cells for immunofluorescence, 24 h after transfection cells from each well were trypsinized using 450 µl of 1X trypsin-EDTA solution (Sigma) and transferred onto two acid-washed 18-mm square coverslips in 35-mm dishes with 2 ml of fresh culture medium. Culture medium was removed 12 h later, and coverslips were processed for immunofluorescence. To prepare cell pellets for whole-cell lysates, medium was removed from the well 24 h posttransfection and replaced with 2 ml/well of fresh culture medium. Cells were trypsinized 12 h later, collected by centrifugation, and washed twice in phosphate-buffered saline (PBS). For RUSH assay Rtn4 knockdown and for enzyme-linked immunosorbent assay (ELISA) samples, HeLa cells were nucleofected using a Lonza 4D-Nucleofector device and Amaxa SE Cell Line 4D-Nucleofector X Kit (Lonza; V4XC-1024), according to the manufacturer’s protocol and using 5.8 × 105 cells per nucleofection.
Immunofluorescence and RUSH assay
After 36 h of transfection (and 12 h after nucleofection), coverslips were washed twice with PBS, fixed with 4% paraformaldehyde (PFA) for 15 min, and then subjected to three 5-min washes with PBS. Cells were permeabilized for 7–10 min with 0.25% Triton X-100 and washed three times with PBS for 5 min each. Blocking was performed for 1 h at room temperature with 10% normal goat serum (Sigma) supplemented with 0.3 M glycine, and then incubated overnight at 4°C with primary antibody diluted in 1.5% normal goat serum. Cells were washed three times in PBS for 5 min each and incubated at room temperature for 1 h with secondary antibodies diluted in 1.5% normal goat serum supplemented with 2.5 µg/ml Hoechst. Cells were then washed three times in PBS for 5 min each, followed by two brief washes with sterile water. Finally, coverslips were mounted in Vectashield (Vector Labratories) and sealed with nail polish. The following secondary antibodies were used at 1:500 dilutions: Alexa Fluor 488 and Alexa Fluor 568 conjugated goat anti-rabbit immunoglobulin G (IgG) (H+L) and anti-mouse IgG (H+L; Invitrogen), and Alexa Fluor 405 conjugated goat anti-rabbit IgG (H+L; Abcam; 175652). The Triton X-100 incubation step was omitted for cell surface staining to avoid cell permeabilization and for the RUSH experiments. For the RUSH assay, transfected cells were trypsinized 24 h after transfection and seeded onto acid-washed coverslips. ER-trapped VSVG-mCherry was released 12 h later by adding 40 µM D-Biotin to the media. Cells were fixed at different time points and processed for immunofluorescence. One set of cells was fixed without adding D-Biotin to the media.
Confocal microscopy
Imaging was performed with a spinning-disk confocal microscope based on an Olympus IX71 microscope stand equipped with a five-line LMM5 laser launch (Spectral Applied Research) and switchable two-fiber output to allow imaging through either a Yokogawa CSU-X1 spinning-disk head or TIRF illuminator. Confocal images were acquired with an ORCA-Flash4.0 V2 Digital CMOS C11440-22CU camera (ImagEM; Hamamatsu) using an Olympus PlanAPO 100×/1.40 oil objective (for fixed cells) or Olympus UPlanFLN 60×/0.90 dry objective (for live cells). z-Axis focus was regulated using a piezo Pi-Foc (Physik Instrumente), and multiposition imaging was performed using a motorized Ludl stage. Image acquisition and all system components were controlled using MetaMorph software (Molecular Devices). All images were acquired using the same exposure time for a given channel and experimental condition.
Quantification from fixed cell imaging
Unless otherwise noted, z-stacks were acquired for each cell using a 0.2 µm z-slice thickness. Z-stacks were converted into maximum intensity projections in ImageJ and thresholded using MetaMorph. Mean fluorescence intensity per cell was measured and averaged for a given condition. KDEL staining distribution was quantified from maximum intensity projections in ImageJ by drawing three straight lines per cell (25 µm in length) from the nuclear envelope to the cell periphery and measuring pixel intensities along the lines. Pixel intensities along the line scans were then averaged for all cells. To quantify ER sheet volumes from CLIMP63 immunostaining, z-stacks were reconstructed and thresholded in 3D using MetaMorph, and the voxel volume was calculated based on the thresholded isosurface. To measure the fluorescence intensity of VSVG-mCherry on the cell surface, the mean VSVG-mCherry fluorescence intensity was quantified for the outermost z-plane on the cell surface and normalized to the mean VSVG-mCherry fluorescence intensity quantified from the maximum intensity projection of the same z-stack. For publication, images were cropped and pseudocolored using ImageJ, but were otherwise unaltered.
Western blots
Whole-cell lysates were prepared from tissue culture cell pellets at 36 h posttransfection using SDS–PAGE sample buffer supplemented with benzonase nuclease (Sigma; E1014) and boiled for 5 min. Proteins were separated on 4–20% SDS–PAGE gels and transferred to polyvinylidene difluoride membrane. Membranes were blocked in Odyssey PBS Blocking Buffer (Li-Cor; 927-40000). Primary and secondary antibodies were diluted in Odyssey PBS Blocking Buffer supplemented with 0.2% Tween-20. Anti-β-actin was used as a loading control. The secondary antibodies used were anti-mouse IRDye-680RD (Li-Cor; 925-68070) and anti-rabbit IRDye-800CW (Li-Cor; 925-32211) at 1:20,000. Blots were scanned on a Li-Cor Odyssey CLx instrument and band intensity quantification was performed with ImageStudio. Rtn4a band intensities were normalized to actin band intensities.
Primary antibodies are listed in Table 1.
TABLE 1:
Primary antibodies.
| Antibody and source | Dilution |
|---|---|
| Rtn4a/Nogo-A; Abcam (ab62024) | IF, 1:50; WB, 1:500 |
| Integrin β1; Invitrogen (PA5-29606; for HeLa) | IF, 1:100 |
| Integrin β1; Bethyl Laboratories (A303-735A; for HeLa) | WB, 1:1000 |
| Integrin β1; Santa Cruz Biotechnology (sc-53711; for Neuro-2a) | IF, 1:50; WB, 1:200 |
| Climp63/CKAP4; Invitrogen (PA5-42926) | IF, 1:100 |
| HLA-A; Santa Cruz Biotechnology (sc-390473) | IF, 1:60; WB, 1:200 |
| KDEL; Santa Cruz Biotechnology (sc-58774) | WB, 1:1000 |
| β-actin (rabbit); RevMAb Biosciences (31-1013-00) | WB, 1:1000 |
| β-actin (mouse); Abcam (ab8224) | WB, 1:1000 |
| ERp72; ProSci (8211) | WB, 1:1000 |
| GRP78; Assay Biotech (C0217) | WB, 1:500 |
| REEP5; Abcam (ab76451) | IF, 1:100 |
| Rtn4/Nogo; Santa Cruz Biotechnology (sc-271878) | IF, 1:75 |
| Rtn4; Aviva Systems Biology (ARP46812_P050) | WB, 1:500 |
ELISA for secreted proteins
Postnucleofection, cells were immediately seeded into 12-well plates containing 900 µl of medium per well. The conditioned medium was removed 12 h later, cleared by centrifugation, and the supernatant was collected. The cells from each well were trypsinized, counted using a hemocytometer, and lysed in 50 µl of RIPA buffer. Both the media supernatant and whole-cell lysate were subjected to sandwich ELISA using the FBLN5 Human ELISA Kit from Fine Test (EH0772) and the TSP1 Human ELISA Kit from Invitrogen (BMS2100), following the manufacturers’ protocols. ELISA plates were read at 450 nm using a Perkin Elmer Wallac 1420 Victor2 Microplate Reader (provided by Don Jarvis at the University of Wyoming). Whole-cell lysate was diluted 1:10 in ELISA buffer. FBLN5 and TSP1 concentrations were quantified in media (secreted) and whole-cell lysate (intracellular) from standard curves. Total protein amounts present in 900 µl of media and 50 µl of cell lysate were calculated. These values were normalized to the number of live cells per well. Secreted and intracellular protein amounts were added to determine the total protein amount per cell. Data from three independent experiments were averaged. Average HeLa nucleofection efficiencies for GFP-NLS and Rtn4a-GFP were 72 and 82%, respectively (calculated from three independent experiments).
Adhesion assay
The cell adhesion assay was performed as described in Sun et al. (2009). In brief, cells were trypsinized 36 h posttransfection, washed several times with PBS, and counted. Cells (4 × 105) were pelleted, resuspended in serum-free EMEM, and plated in 24-well plates coated with collagen type-I solution (20 µg/ml). After 30 min, floating and loosely adhered cells were removed with three PBS washes, and the adhered cells were fixed with 3.75% PFA. Fixed cells were stained for 5 min with 0.25% crystal violet and washed twice with PBS. The stained cells were lysed in 70% ethanol and absorbance of the cleared cell lysate was measured at 540 nm using a Biotek Synergy H4 Microplate Reader (provided by Grant Bowman at the University of Wyoming).
Statistics
Averaging and statistical analysis were performed for independently repeated experiments. Two-tailed Student’s t tests assuming equal variances were performed using GraphPad Software to evaluate statistical significance. The p values, number of independent experiments, and error bars are denoted in the figure legends.
Supplementary Material
Acknowledgments
Research in the Levy lab is supported by funding from the National Institutes of Health/National Institute of General Medical Sciences (R01GM113028 and P20GM103432) and the American Cancer Society (RSG-15-035-01-DDC). We thank David Fay, Amy Navratil, and Karen White for critical reading of the manuscript.
Abbreviations used:
- ER
endoplasmic reticulum
- FBLN5
fibulin-5
- Rtn
reticulon
- TSP1
thrombospondin-1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-03-0159) on July 18, 2019.
REFERENCES
- Albig AR, Schiemann WP. (2005). Fibulin-5 function during tumorigenesis. Future Oncol , 23–35. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. (2008). Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol , 911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black VH. (1972). The development of smooth‐surfaced endoplasmic reticulum in adrenal cortical cells of fetal guinea pigs. Am J Anat , 381–417. [DOI] [PubMed] [Google Scholar]
- Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, Mercanti V, Jollivet F, Raposo G, Perez F. (2012). Synchronization of secretory protein traffic in populations of cells. Nat Methods , 493–498. [DOI] [PubMed] [Google Scholar]
- Calero M, Whittaker GR, Collins RN. (2001). Yop1p, the yeast homolog of the polyposis locus protein 1, interacts with Yip1p and negatively regulates cell growth. J Biol Chem , 12100–12112. [DOI] [PubMed] [Google Scholar]
- Chen MS, Huber AB, van der Haar ME, Frank M, Schnell L, Spillmann AA, Christ F, Schwab ME. (2000). Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for monoclonal antibody IN-1. Nature , 434–439. [DOI] [PubMed] [Google Scholar]
- Chen Q, Kinch MS, Lin TH, Burridge K, Juliano R. (1994). Integrin-mediated cell adhesion activates mitogen-activated protein kinases. J Biol Chem , 26602–26605. [PubMed] [Google Scholar]
- Clark MA, Hirst BH, Jepson MA. (1998). M-cell surface β1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer’s patch M cells. Infect Immun , 1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, Boivin GP, Bouck N. (1998). Thrombospondin-1 is a major activator of TGF-β1 in vivo. Cell , 1159–1170. [DOI] [PubMed] [Google Scholar]
- Di Sano F, Bernardoni P, Piacentini M. (2012). The reticulons: guardians of the structure and function of the endoplasmic reticulum. Exp Cell Res , 1201–1207. [DOI] [PubMed] [Google Scholar]
- Fourriere L, Kasri A, Gareil N, Bardin S, Bousquet H, Pereira D, Perez F, Goud B, Boncompain G, Miserey-Lenkei S. (2019). RAB6 and microtubules restrict protein secretion to focal adhesions. J Cell Biol , 2215–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Voeltz GK. (2011). The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol , 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal U, Blackstone C. (2013). Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta , 2492–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GrandPré T, Nakamura F, Vartanian T, Strittmatter SM. (2000). Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature , 439–444. [DOI] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Hölper S, Mari M, Harwardt M-LI, Yan R, Müller S, Reggiori F, Heilemann M, Dikic I. (2017). Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife , e25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T, Ohno K, Sano M, Omura T, Omura K, Nagano A, Sato K. (2005). The differential expression patterns of messenger RNAs encoding Nogo-A and Nogo-receptor in the rat central nervous system. Brain Res Mol Brain Res , 119–130. [DOI] [PubMed] [Google Scholar]
- Jevtic´ P, Levy DL. (2015). Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr Biol , 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Wiertz E, Sun L, Fish KN, Nelson JA, Ploegh HL. (1996). Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA , 11327–11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, Hirschberg K. (2000). Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol , 557–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matko J, Bushkin Y, Wei T, Edidin M. (1994). Clustering of class I HLA molecules on the surfaces of activated and transformed human cells. J Immunol , 3353–3360. [PubMed] [Google Scholar]
- Mi Y, Gao X, Ma Y, Gao J, Wang Z, Jin W. (2014). A novel centrosome and microtubules associated subcellular localization of Nogo-A: implications for neuronal development. Int J Biochem Cell Biol , 1–6. [DOI] [PubMed] [Google Scholar]
- Papadopoulos C, Orso G, Mancuso G, Herholz M, Gumeni S, Tadepalle N, Jüngst C, Tzschichholz A, Schauss A, Höning S. (2015). Spastin binds to lipid droplets and affects lipid metabolism. PLos Genet , e1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Blackstone C. (2010). Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep , 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramo O, Kumar D, Gucciardo E, Joensuu M, Saarekas M, Vihinen H, Belevich I, Smolander OP, Qian K, Auvinen P, Jokitalo E. (2016). NOGO-A/RTN4A and NOGO-B/RTN4B are simultaneously expressed in epithelial, fibroblast and neuronal cells and maintain ER morphology. Sci Rep , 35969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. (2004). RTP family members induce functional expression of mammalian odorant receptors. Cell , 679–691. [DOI] [PubMed] [Google Scholar]
- Schlaitz AL, Thompson J, Wong CC, Yates JR, Heald R. (2013). REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell , 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. (2010). Mechanisms determining the morphology of the peripheral ER. Cell , 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA. (2006). Rough sheets and smooth tubules. Cell , 435–439. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. (2008). The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem , 18892–18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner P, Kulangara K, Sarria JC, Glauser L, Regazzi R, Hirling H. (2004). Reticulon 1-C/neuroendocrine-specific protein-C interacts with SNARE proteins. J Neurochem , 569–580. [DOI] [PubMed] [Google Scholar]
- Sun W, Hu W, Xu R, Jin J, Szulc ZM, Zhang G, Galadari SH, Obeid LM, Mao C. (2009). Alkaline ceramidase 2 regulates β1 integrin maturation and cell adhesion. FASEB J , 656–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell , 573–586. [DOI] [PubMed] [Google Scholar]
- Wakana Y, Koyama S, Nakajima K, Hatsuzawa K, Nagahama M, Tani K, Hauri HP, Melancon P, Tagaya M. (2005). Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem Biophys Res Commun , 1198–1205. [DOI] [PubMed] [Google Scholar]
- Xiong N-X, Pu J-Z, Zhao H-Y, Zhang F-C. (2008). Effect of Nogo-A gene inhibition on dopamine release in PC12 cells. Neuro Endocrinol Lett , 884–888. [PubMed] [Google Scholar]
- Yang YS, Strittmatter SM. (2007). The reticulons: a family of proteins with diverse functions. Genome Biol , 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.