Abstract
Infections with highly pathogenic avian influenza viruses (HPAIV) in humans lead to systemic disease associated with cytokine storm and multiorgan failure. In this study we aimed to identify the role of monocytes for the host response to HPAIV infection. Using genome-wide microarray analysis, we surprisingly demonstrate a reduced immune response of human monocytes to HPAIV H5N1 compared to human influenza A viruses. In bioinformatic analyses we could reveal a potential role of the Rar-related orphan receptor alpha (RORα) for the gene expression pattern induced by H5N1. RORα is known as an inhibitor of NF-κB signaling. We provide evidence that in monocytes RORα is activated by H5N1, resulting in inhibited NF-κB signaling. Using murine Hoxb8-immortalized RORα−/−, monocytes rescued NF-κB signaling upon H5N1 infection, confirming the biological relevance of RORα as an H5N1-induced mediator of monocytic immunosuppression. In summary, our study reveals a novel RORα-dependent escape mechanism by which H5N1 prevents an effective inflammatory response of monocytes blocking NF-κB-dependent gene expression.
Key Words: Inflammation, Monocytes, Influenza infection, Transcription factor profiling
Introduction
H5N1 belongs to the highly pathogenic avian influenza viruses (HPAIV) which have successfully overcome the host species barrier and are able to cause systemic infections in humans. In contrast to seasonal IAV that are usually restricted to the lung, HPAIV are able to evoke a systemic inflammatory response syndrome upon infection and ultimately cause death by multiorgan failure. The highest viral loads are found in the lung and the alveolar epithelium. In the lung, strong infiltration of blood-derived macrophages is observed in infected patients and cytokine levels in blood and tissues are extremely enhanced. More than 60% of reported human cases of H5N1 infections are fatal [1, 2, 3, 4]. Besides epithelial cells, several cell types have been shown to be highly susceptible to IAV infection [3, 4, 5, 6, 7, 8, 9, 10, 11, 12]. Neuro- and endothelial cell tropism have been described for H5N1, contributing to systemic infections [11, 13, 14, 15]. Recent studies by our group have revealed specific response patterns to HPAIV infection in endothelial cells indicating that these cells may be important mediators of cytokine burst and antiviral response mechanisms [12, 16]. The pattern of gene expression in endothelial cells with strong induction of proinflammatory and antiviral genes is strongly regulated by NF-κB and in the case of H5N1 infections specifically modulated by transcriptional regulators like HMGA1 and NFATC4 [12, 16]. Very recent data published by Teijaro et al. [17] present endothelial cells as key players of cytokine amplification during influenza infection. Additionally, our group was able to reveal very distinct response patterns in influenza-infected human blood-derived macrophages [18]. Here we observed a suppression of inflammatory and antiviral responses by HPAIV via inhibition of inflammasome activation indicating HPAIV to bypass a major source of first immune responses against viral infection.
Basically, monocytes represent another well-established cellular source for cytokine production and antiviral defense. Human monocytes have been shown to be principally susceptible to IAV infections since they produce type I interferons [19] as well as various cytokines like TNF-α, IL-6 and IL-1β, and chemokines such as CCL 3, 4 and 5 upon infection with the H1N1 isolate A/PR8/34 [8, 20]. However, no investigations have demonstrated the infectibility of human peripheral blood monocytes with HPAIV. Hitherto existing publications focus on HPAIV infection of differentiated human macrophages. Results concerning cytokine production of human macrophages in response to H5N1 infection are rather inconsistent and depend very much on the type of H5N1 isolate [4, 21, 22].
Employing an unbiased genome-wide analysis of the gene expression patterns induced in monocytes after infection with different influenza strains (H1N1, H7N7, H5N1, H1N1/v and WSN/33) we now provide evidence that HPAIV efficiently replicate but circumvent an effective inflammatory response in blood monocytes. We demonstrate that activation of the transcription factor RAR-related orphan receptor alpha (RORα) results in inhibition of NF-κB-mediated gene induction during H5N1 infection.
Materials and Methods
Cell Preparation and Culture
Human monocytes were isolated from buffy coats of unrelated healthy blood donors as described earlier [23, 24]. Cells were cultivated in Teflon bags in McCoy's modified medium (Biochrom AG, Berlin, Germany) supplemented with 1% glutamine, 1% penicillin-streptomycin and 15% fetal bovine serum overnight. The next day human monocytes were transferred to uncoated culture dishes for experiments.
Virus Preparation and Exposure to Human Monocytes
The HPAIV strain A/Thailand/KAN-1/2004 (H5N1) isolated from a fatal human case was used with permission from Dr. Pilaipan Puthavathana (Bangkok, Thailand). The avian influenza virus A/FPV/Bratislava/79 (H7N7, fowl plague virus) and the human influenza virus strain A/Puerto-Rico/8/34 (Giessen variant) were taken from the strain collection of the Institute of Molecular Virology (Münster, Germany) and were initially provided by the Institute of Virology in Giessen, Germany. The low pathogenic human influenza strain A/WSN/33 was taken from the strain collection of the Institute of Molecular Virology (Münster, Germany). A/Hamburg/04/2009 (H1N1/v) was obtained from the German National Reference Centre for Influenza (Brunhilde Schweiger, Robert-Koch-Institute, Berlin, Germany). The virus was isolated from clinical specimens. Viruses were propagated on Madin Darby canine kidney (MDCKII) cells.
In order to ensure comparable infection rates, human monocytes were exposed to 0.5 MOI (multiplicity of infection) of virus and differentiated murine ER-Hoxb8 cells were infected with 5 MOI of virus for gene expression experiments. All infections were performed in serum-free McCoys modified monocyte culture medium (Biochrom AG) supplemented with 1% glutamine, 1% penicillin-streptomycin and 3% bovine albumin. Viruses were added to the infection medium for the whole infection period. Afterwards cells were washed twice with PBS and used for further analysis.
Quantitative Real-Time RT-PCR
Total cellular RNA was isolated using RNeasy kit (Qiagen, Hilden, Germany). cDNA was synthesized from 1 µg of total RNA using RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot, Germany). Specific primers for each gene (for sequences see online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000346706) were designed using the Primer Express software package (Applied Biosystems, Foster City, Calif., USA) and obtained from MWG Biotech (Ebersberg, Germany). qRT-PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) [12] and data acquired with the ABI PRISM 7900 (Applied Biosystems) provided by the Integrated Fuctional Genomics department of the University of Münster, Germany. Gene expression was normalized to the endogenous housekeeping control gene GAPDH and relative expression of respective genes was calculated using the comparative threshold cycle method [25].
DNA Microarray Hybridization and Statistical Data Analyses
Total cellular RNA was isolated from 3 independent experiments. Samples were processed for microarray hybridization using Affymetrix Human Genome 133 Plus 2.0 Gene Arrays as described earlier [12] with the kind help of the Integrated Functional Genomics department of the University of Münster, Germany. Principal component analyses, gene clustering and promoter analyses were performed as described before [12, 16]. Microarray data are MIAME compliant and deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zhmdlsiqskwsgjk&acc=GSE35283).
Western Blot
Cells were lysed in RIPA buffer containing protease and phosphatase inhibitors [26]. For nuclear and cytosolic extract preparation, incubation with a high salt buffer was followed by incubation with a buffer containing low concentration of NaCl. SDS-PAGE and Western blot staining was performed as described earlier [27] using mouse mAB M1 (GA2B, AbD Serotec, Oxford, UK), rabbit pAB NF-κB p65, rabbit pAB RORα (both Abcam, Cambridge, Mass., USA), rabbit pAB IκBα (c-21, Santa Cruz Biotechnology, Heidelberg, Germany), mouse mAB α-tubulin (ICN, Solon, Ohio, USA), goat pAB lamin B (Santa Cruz Biotechnology) and mouse mAB β-actin (Sigma, Munich, Germany). The protein bands were visualized using the ECL (200 µl 250 mM Luminol, 90 µl 90 mMp-cumar acid, 2 ml 1M Tris pH 8.5, 7.1 µl 35% H2O2) system.
Intracellular FACS Staining
5 × 105 monocytes were harvested, washed and fixed with 4% paraformaldehyde at room temperature for 20 min. Pellets were treated with permeabilization buffer (0.1% saponin, 1% FCS, PBS) for 10 min at room temperature followed by incubation with FITC-labeled anti-NP antibody (mouse mAB, MCA400, clone AA5H, Serotec, Oxford, UK) for 1 h at room temperature. The samples were washed once with permeabilization buffer and fluorescence was determined in the Fl3 channel using a FACScalibur (Becton Dickinson, Franklin Lakes, N.J., USA).
Plaque Assay
Plaque assays were performed as described earlier [12]. For quantification of viral particle release in the supernatants of infected monocytes a very low MOI of 0.001 was used in order to ensure cell survival over longer time periods. In the case of Hoxb8 cells 0.5 MOI were used. Virus-induced plaques were visualized by staining with neutral red.
Mice
Heterozygous (RORαsg/J) male mice were purchased from Jackson Laboratories (Bar Harbor, Me., USA) and paired with female C57/Bl6 mice (Jackson Laboratories). RORα+/sg pairs were bred at the Institute of Pharmacology, University of Münster, Germany, and offspring was genotyped by PCR of tail DNA according to the instructions provided by Jackson Laboratories. RORαsg/J pairs were then bred to obtain knockout fetuses. Wt littermates of the same age were bred for wt fetuses.
ER-Hoxb8 RORα−/− Monocyte Cell Line Establishment and Infection
We isolated liver stem cells from RORα knockout fetuses on day 14.5-16.5 and cultivated them in RPMI + 15% FCS, 20 ng/ml IL-6, 10 ng/ml IL-3 and 1% SCF for 4 days. Cells were transfected with ER-Hoxb8 retrovirus, selected and cultivated as described before [28, 29]. Stem cells were isolated from bone marrow of C57BL/6 wt mice and processed in the same manner. Cells were differentiated in petri dishes in the absence of estradiol for 3 days before performance of experiments. Cells were infected with 5 MOI of influenza viruses for most experiments in the same manner described for human cells.
Ethics Statement
The taking of blood samples from humans and cell isolation were conducted with approval of the local ethics committee (ethics advisory board of the Ärztekammer Westfalen-Lippe and Medical Faculty of Westfaelische Wilhelms-Universitaet Münster). Human blood samples were taken from healthy blood donors who provided written informed consent for the collection of samples and subsequent cell isolation and analysis. All animals were handled in strict accordance with good animal practice and animal keeping. Experiments were supervised by the veterinary office of Münster (Veterinary Bureau of Münster) and performed according to approved protocols of the animal welfare committee of the University of Münster (Münster, Germany). All experiments were performed with the approval of the State Review Board Nordrhein-Westfalen (Germany) according to the German law for animal welfare (Tierschutzgesetz).
Statistical Analyses
The results of all qRT-PCR experiments were assessed by rank-sum test and are shown as means ± SEM.
Results
Primary Human Blood Monocytes Are Susceptible to Influenza Infection
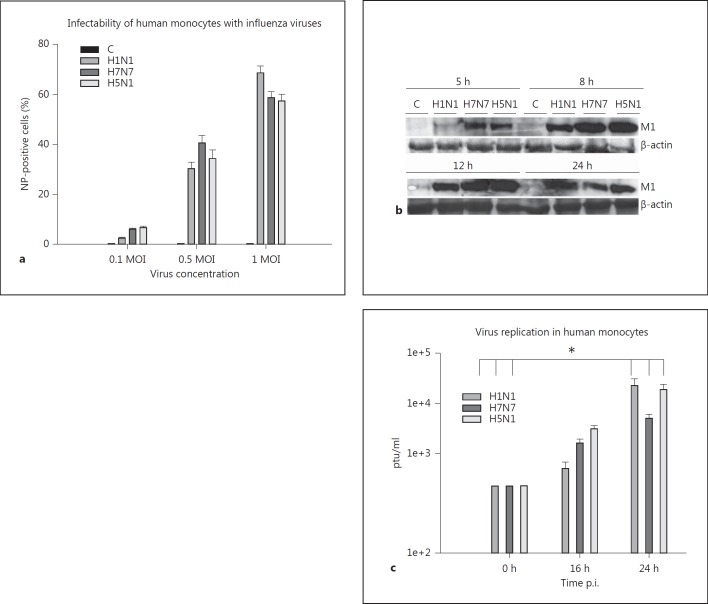
We infected primary human blood monocytes with three different influenza virus strains, a low pathogenic human strain, A/PR8/34 (H1N1) and two HPAIVs, A/FPV/Bratislava/79 (H7N7) and A/Thailand/KAN-1/2004 (H5N1). The infectibility of monocytes with these viruses was confirmed by intracellular flow cytometric analysis of viral nucleoprotein (NP) expression. Efficient viral infection after 5 h was obtained using a viral dosage of 0.5 MOI. These experimental conditions led to about 40% of infected monocytes. An MOI of 1 resulted in further increase of infection rates up to 80% (fig. 1a) but led to higher cell death rates during the course of infection (data not shown). We chose 0.5 MOI as the condition for further gene expression experiments. Additionally, efficient infection was confirmed by immunoblotting of the viral protein M1 (fig. 1b). Similar amounts of M1 protein could be detected after the first replication cycle (8 h) indicating that all three virus strains are able to replicate and to induce protein translation in human monocytes.
Fig. 1.
Infectability of human monocytes with influenza viruses and virus replication. a Human monocytes were infected with 0.1, 0.5 and 1 MOI of H1N1, H7N7 and H5N1. Viral NP was detected by flow cytometry 5 h p.i. and analyzed with Cell Quest pro software. The percentage of NP-positive cells corresponds with the amount of infected cells. Data represent means ± SEM of three independent experiments. b Lysates of uninfected (C) and H1N1-, H7N7- and H5N1-infected monocytes (0.5 MOI) were immunoblotted for viral protein M1 (25 kD) 5, 8, 12 and 24 h p.i. Immunostaining of β-actin (42 kD) served as a protein loading control. One representative blot out of 3 independent experiments is shown. c Replication of viral particles within human monocytes was detected in a plaque assay performed with supernatants of H1N1-, H7N7- and H5N1-infected monocytes 16 and 24 h p.i. (0.001 MOI). The viral load at the beginning of the experiment (input) is also shown. Experiments were performed with n = 3. Bars represent means ± SEM. * p < 0.05.
The supernatants of infected monocytes were analyzed in a plaque assay in order to detect viral replication in the cells. Cells were infected with 0.001 MOI ensuring their survival for longer infection times. The replication rate was measured 16 and 24 h postinfection (p.i.) and we observed a significant viral replication rate of all three virus strains 24 h p.i. (fig. 1c). Taken together, our results show that human blood monocytes are susceptible to influenza infection and that influenza viruses efficiently propagate in these host cells.
Immune Responses of Human Monocytes Are Suppressed during HPAIV Infection
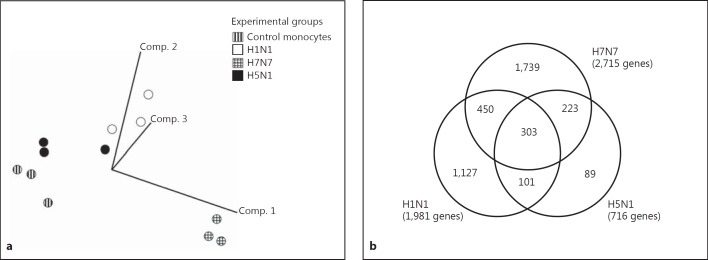
Human monocytes isolated from three independent blood donors were analyzed 5 h p.i. with H1N1, H7N7 and H5N1 in a genome-wide comparative systems biology approach. As downregulation of genes is significantly influenced by ‘cap-snatching’ mechanisms and not representative for a specific antiviral host response [30, 31], we restricted our analyses to upregulated genes. The threshold used for analysis of upregulated genes was a fold change of 2.0 and a p value of 0.05. Principal component analysis (PCA) of upregulated genes data, performed as described earlier [16, 32], allowed the unbiased display of four gene expression patterns obtained from uninfected control cells and cells infected with H1N1, H7N7 and H5N1 IAV, respectively (fig. 2a). PCA clearly showed the reproducibility of gene expression changes induced by the respective viral strain in human monocytes of different donors. Moreover, H1N1, H7N7 and H5N1 IAV infection led to specific gene expression patterns distinctively located within the three-dimensional vector space.
Fig. 2.
Gene array analysis of influenza-infected human monocytes. a PCA comparing gene profiles of uninfected control monocytes, H1N1-, H7N7- and H5N1-infected monocytes obtained from three independent blood donors (infection with 0.5 MOI for 5 h). Vector clouds representing gene profiles of individual experiments are positioned in a three-dimension vector space according to variance to each other. b Venn diagram indicating the number and overlap of genes induced 5 h p.i. by H1N1, H7N7 and H5N1. Data are based on microarray analyses of three independent experiments.
We found a total of 1,981 genes induced by H1N1, 1,127 (56.8%) of them specifically only by H1N1 and not by H7N7 and H5N1. 2,715 genes were induced by H7N7, 1,739 (64.1%) of them specifically. H5N1 led to upregulation of only 716 genes, 89 (12.4%) of them were H5N1 specifically induced (fig. 2b). Compared to H1N1 and H7N7, the number of H5N1-induced genes is strikingly low.
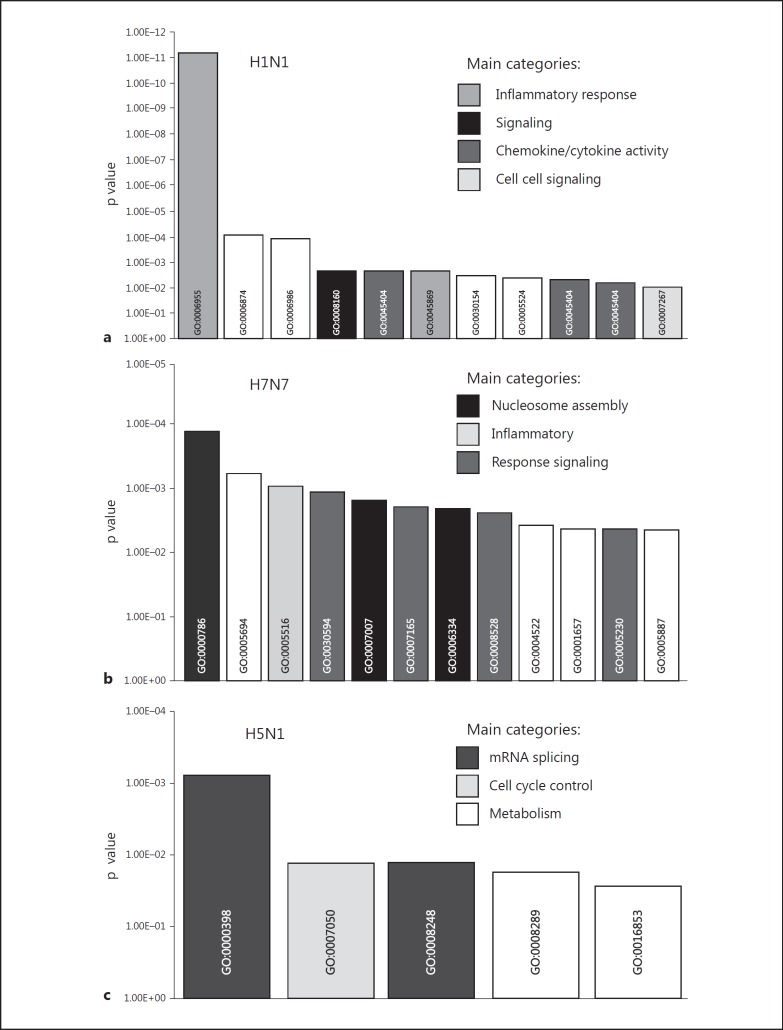
We next performed a ‘functional clustering’ analysis [12, 16] to characterize differences in gene expression patterns ‘qualitatively’ on a statistically proven basis. We focused on the group of specifically induced genes, respectively, and assigned them to gene ontology groups based on guidelines of the Gene Ontology Consortium. We found that genes belonging to interferon, immune, defense and antiviral response were clearly overrepresented in the spectrum of specifically H1N1-induced genes (fig. 3a). In contrast, infection with highly pathogenic influenza virus H5N1 led to a rather unspecific pattern of overrepresented functionally related gene clusters (mRNA splicing, cell cycle control) which could not be related to inflammatory and antiviral responses (fig. 3c). Functional clustering of the H7N7-induced gene expression revealed an intermediate pattern of overrepresented genes involved in nucleosome assembly, inflammatory response and signaling (fig. 3b).
Fig. 3.
Overrepresentation of functional gene groups. Functional gene groups are plotted according to Gene Ontology annotations significantly overrepresented in: genes only upregulated by H1N1 (a), genes only upregulated by H7N7 (b) and genes only upregulated by H5N1 (c). Significance of overrepresentation (y-axis) was determined by applying Fisher's exact test.
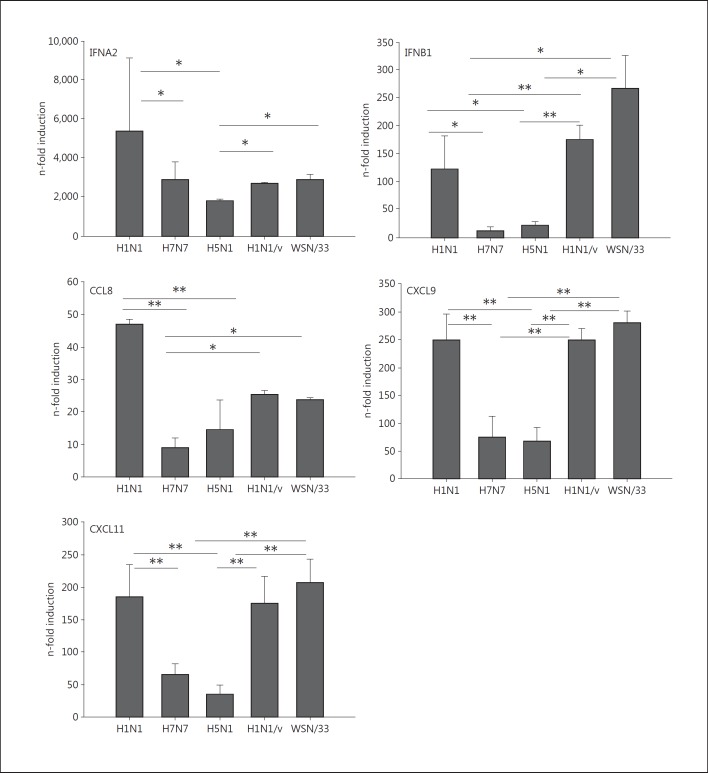
In summary, our results point to a reduced immune and antiviral response of human monocytes during infection with HPAIV, especially upon H5N1 infections. Quantitative RT-PCR experiments confirmed this response pattern and revealed stronger mRNA inductions of antiviral genes like type IFNA2 and IFNB1 as well as chemokines like CCL8, CXCL11 and CXCL9 by H1N1 5 h p.i. compared to H7N7 and H5N1 (fig. 4). With respect to these inflammatory genes, diminished induction was comparable between H7N7- and H5N1-infected monocytes. For confirming the HPAIV-specific inhibition of the immune response in human monocytes, we performed experiments with two further human influenza A viruses, A/Hamburg/04/2009 (H1N1/v) and A/WSN/33. Both strains showed significantly stronger gene induction than HPAIV, confirming the HPAIV-specific reduction of proinflammatory and antiviral responses in human blood monocytes.
Fig. 4.
Cyto- and chemokine patterns in influenza-infected monocytes. Human monocytes were infected with 0.5 MOI H1N1, H7N7, H5N1, H1N1/v and WSN/33 for 5 h. Gene induction was measured by qRT-PCR. Induction of IFNA-mRNA, IFNB1-mRNA, CCL8-mRNA, CXCL11-mRNA and CXCL9-mRNA was measured by qRT-PCR. All experiments were performed with n = 6. Bars represent means ± SEM. * p < 0.05; ** p < 0.01.
H5N1 Infection Activates RORα in Human Monocytes
To identify virus-specific transcription factors eventually mediating immunosuppression in HPAIV-infected human monocytes we performed promotor analyses (CARRIE). This in silico approach searches for transcription factor binding sites overrepresented in the promotors of H1N1-, H7N7- or H5N1-upregulated genes compared to all genes represented on the microarray (transcription factor profiles; table 1). Among all transcription factors identified, the nuclear receptor RORα was the only one known to exert inhibitory effects on immune response programs [33, 34]. Moreover, RORα-binding sites were computed to be only overrepresented in the promotors of H5N1-induced genes suggesting that particularly for this strain, which is the most pathogenic for humans out of the three strains tested, RORα might play a crucial role in inhibiting the immune response of monocytes.
Table 1.
Transcription factor profiles
| p value | |
|---|---|
| H1N1-induced gene profile | |
| Interferon-stimulated response element | 1.77e–06 |
| GATA-binding factor 1 | 1.05e–05 |
| Serum response factor | 1.27e–04 |
| HNF-3alpha | 8.57e–04 |
| H7N7-induced gene profile | |
| Transcriptional repressor CDP | 4.25e–13 |
| Octamer factor 1 | 2.12e–09 |
| Cut-like homeodomain protein | 5.44e–08 |
| POU1F1 | 5.44e–08 |
| Fork head box J 2 | 4.08e–07 |
| Myogenic MADS factor MEF-2 | 4.08e–07 |
| RSRFC4 | 8.14e–07 |
| C/EBPgamma | 1.40e–06 |
| HMG IY | 3.07e–06 |
| IRF | 3.07e–06 |
| Myocyte enhancer factor | 3.34e–06 |
| Fork head RElated ACtivator-7 | 4.93e–06 |
| Cellular and viral TATA box elements | 5.77e–06 |
| Nuclear factor of activated T-cells | 1.22e–05 |
| Pit-1 | 1.22e–05 |
| Sex-determining region Y gene product | 1.22e–05 |
| HOXA4 | 1.22e–05 |
| Cart-1 (cartilage homeoprotein 1) | 1.53e–05 |
| Interferon-stimulated response element | 2.29e–05 |
| Pbx-1 | 2.29e–05 |
| FOX | 3.02e–05 |
| HFH-4 | 5.82e–05 |
| Retroviral TATA box | 6.22e–05 |
| Crx | 6.48e–05 |
| Homeo domain factor Pbx-1 | 6.83e–05 |
| ICSBP | 7.36e–05 |
| NKX6-1 | 1.27e–04 |
| Meis-1a/HOXA9 heterodimeric binding | 2.17e–04 |
| STAT6 | 2.18e–04 |
| Meis-1b/HOXA9 heterodimeric binding | 2.42e–04 |
| TEF | 3.59e–04 |
| GATA-6 | 5.23e–04 |
| STAT5a | 5.24e–04 |
| Yin and Yang 1 | 5.42e–04 |
| CHX10 | 6.69e–04 |
| STAT4 | 7.47e–04 |
| Cell division control protein 5 | 7.77e–04 |
| POU3F2 | 8.31e–04 |
| HFH-3 (HNF3/fork head homolog 3) | 9.17e–04 |
| H5N1-induced gene profile | |
| octamer factor 1 | 6.01e–08 |
| HOXA4 | 2.56e–06 |
| RSRFC4 | 4.43e–06 |
| RAR-related orphan receptor alpha1 | 1.54e–05 |
| cAMP-responsive element binding protein 1 | 1.68e–05 |
| STAT6 | 3.93e–05 |
| Activator protein 1 | 5.49e–05 |
| Interferon regulatory factor 7 | 8.65e–05 |
| STAT5a | 2.55e–04 |
| Fork head box O1 | 2.71e–04 |
| Nuclear factor of activated T-cells | 3.08e–04 |
| Fork head box D3 | 3.15e–04 |
| Fork head box J 2 | 6.07e–04 |
| POU-factor Tst-1/Oct-6 | 7.48e–04 |
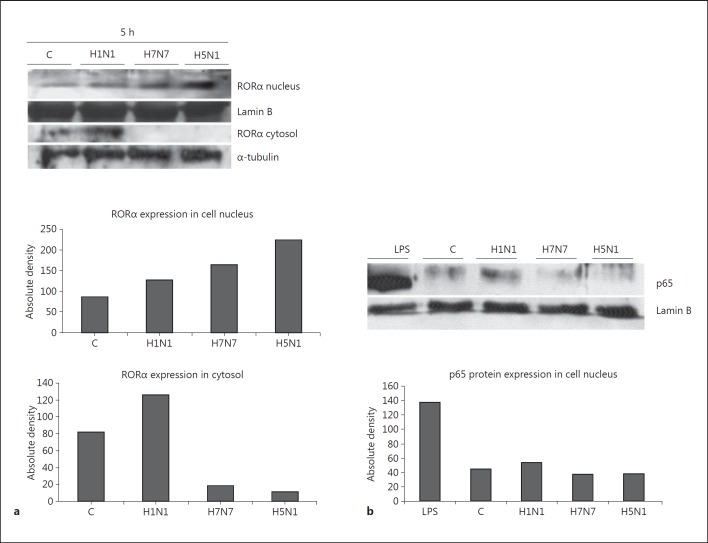
RORα protein was well expressed in uninfected as well as in IAV-infected monocytes (fig. 5a). However, nuclear translocation of RORα was strongest in H5N1-infected monocytes, poor after H7N7 infection and virtually undetectable in H1N1-infected monocytes. This finding of strong activation of the RORα signaling pathway by H5N1 virus supports our hypothesis of RORα being essentially involved in regulating the immune response program elicited by H5N1 in human monocytes.
Fig. 5.
RORα expression correlates negatively with NF-κB activation. a Whole cell lysates and nuclear extract lysates of uninfected control monocytes (C) and monocytes infected with H1N1, H7N7 and H5N1 for 5 h (MOI 0.5) were immunoblotted for RORα (68 kD). α-tubulin served as a protein loading control for cytosolic protein and lamin B as a control for nuclear extracts. One representative blot out of 3 independent experiments is shown. Absolute density of protein lanes has been measured in ImageJ software and is shown for the representative blot. b Immunoblotting of lysates of uninfected control (C) monocytes and monocytes infected with H1N1, H7N7 and H5N1 for 5 h. Nuclear extracts were immunoblotted for the NF-κB subunit p65 (65 kD). Lysates of monocytes stimulated with LPS are shown as positive control. Lamin B served as a protein loading control. One representative blot out of 3 independent experiments is shown. Absolute density of protein lanes has been measured in ImageJ software and is shown for the representative blot.
H5N1 Inhibits NF-kB Signaling in Monocytes and Activates RORα
Since RORα has been shown to exert its immunosuppressive effect by impeding the translocation of the p65-subunit of NF-κB into the nucleus [33] we analyzed p65 translocation in our settings (fig. 5b). Lipopolysaccharide (LPS)-treated monocytes served as a positive control showing strong nuclear p65 accumulation. In nuclear extracts of monocytes infected with H1N1 p65 was detectable, whereas in H7N7 and H5N1-infected monocytes virtually no p65 could be detected in the nucleus. Obviously, HPAIV inhibit the translocation of NF-κB depending on the ability to activate RORα, which is best achieved by H5N1 virus (fig. 5a).
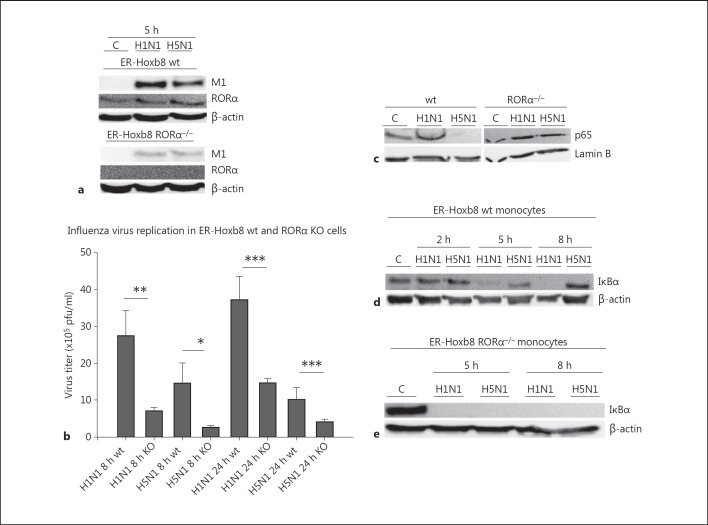
Subsequently, we used a knockout approach to demonstrate a functional dependence of the immunosuppressive properties of H5N1 in monocytes on RORα activation. RORα knockout mice (RORα−/−) show a heavy restriction in their general state of health associated with early or even embryonic death which precludes their use for virus infections. We therefore established an ER-Hoxb8 monocytic cell line derived from liver cells of RORα knockout fetuses to receive a stable knockout system. Estrogen-regulated expression of oncogene HoxB8 induces immortalization of monocyte and macrophage progenitors which execute normal differentiation after ER-HoxB8 inactivation [28, 29]. We characterized these cells by flow cytometry demonstrating expression of murine monocyte/macrophage lineage markers such as GR1, Ly6c, F4/80, CD11b and CD43, and lack of granulocyte marker Ly6g or DC marker CD11c (online suppl. fig. 1). Corresponding HoxB8-immortalized wt cells were used as controls. Cells were infected with 5 MOI of influenza virus H1N1 or H5N1. As expected we could detect RORα in wt cells with a slight increase of expression upon viral infection, whereas no RORα expression could be detected in ER-Hoxb8 RORα−/− monocytes (fig. 6a). Viral protein M1 expression was diminished in RORα−/− cells compared to wt cells (fig. 6a) and a plaque assay confirmed better replication of both viruses in wt cells compared to RORα−/− monocytes (fig. 6b). Apparently, RORα affects universally efficient propagation of different IAV strains.
Fig. 6.
NF-κB activation in RORα−/− and wt monocytes after H1N1 and H5N1 infection. a Immunoblotting of lysates of uninfected control (C) wt and RORα−/− monocytes and monocytes infected with H1N1 and H5N1 for 5 h. Lysates were immunoblotted for viral protein M1 and RORα. β-actin served as protein loading control. One representative blot out of 3 independent experiments is shown. b Plaque assay of murine wt and RORα−/− ER-Hoxb8-immortalized monocytes infected with 0.5 MOI of H1N1 and H5N1 for 8 and 24 h. Experiments were performed with n = 5. Bars represent means ± SEM. * p < 0.05; ** p < 0.01; *** p < 0.001. c Immunoblotting of nuclear extracts of uninfected (C) wt and RORα−/− monocytes and wt and RORα−/− monocytes infected with H1N1 and H5N1 for 2 h (5 MOI). Lysates were immunoblotted for the NF-κB subunit p65. Lamin B served as a loading control. The blot represents three independent experiments. Immunoblotting of lysates of uninfected control murine ER-Hoxb8 wt monocytes (C) and wt monocytes infected with H1N1 and H5N1 for 2, 5 and 8 h, respectively. Lysates were immunoblotted for IκBα (40 kD). β-actin served as a protein loading control. One representative blot out of 3 independent experiments is shown. d Immunoblotting of lysates of uninfected control (C) RORα−/− monocytes and H1N1- and H5N1-infected RORα−/− monocytes for 2, 5 and 8 h, respectively. Lysates were immunoblotted for IκBα (40 kD). β-actin served as a protein loading control. One representative blot out of 3 independent experiments is shown.
In this cell system, impaired p65 nuclear translocation in wt cells upon H5N1 infection compared to H1N1 could be confirmed. In RORα knockout cells, however, p65 was activated after both, H1N1 and H5N1 infection (fig. 6c).
IκBα is a major inhibitor of the NF-κB signaling pathway. It is readily expressed in untreated wt cells and completely and lastingly downregulated after H1N1 infection (fig. 6d), consistent with proper NF-κB activation (fig. 6c). However, wt cells infected with H5N1 keep on expressing IκBα (fig. 6d) which is in accordance with continuous inhibition of NF-κB-signaling (fig. 6c). On the other hand, in RORα knockout monocytes IκBα expression was neither detectable after H1N1 infection nor after H5N1 infection (fig. 6d). Our data broadly implicate a coherence between inhibition of NF-κB signaling and activation of RORα.
Blocking RORα Prevents H5N1-Mediated Inhibition of the Monocytic Immune Response
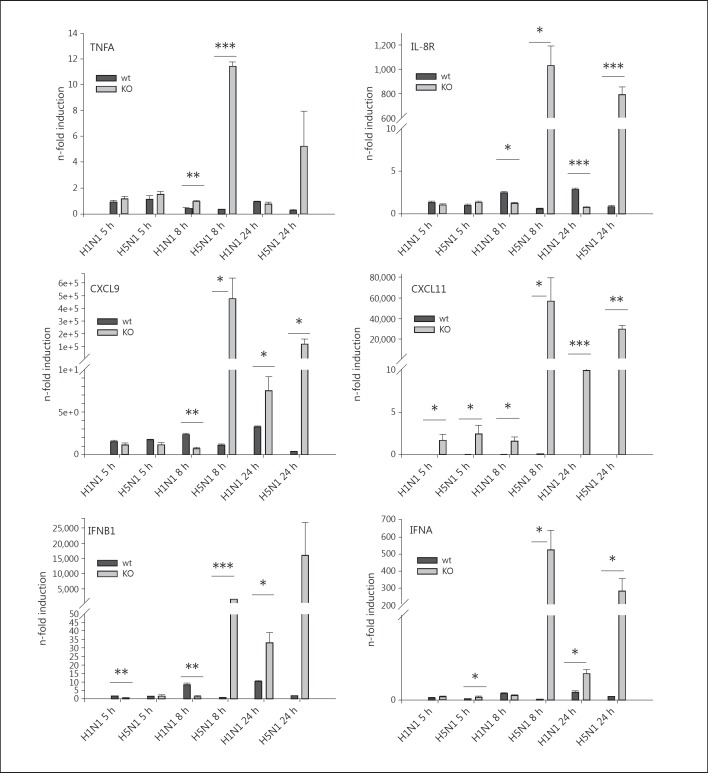
Finally, we infected ER-Hoxb8 RORα−/− and wt monocytes with 5 MOI of H1N1 and H5N1 for 5, 8 and 24 h and performed qRT-PCR for proinflammatory (TNF, IL8R) and antivirally (CXCL9, CXCL11, IFNB1, IFNA2) acting genes. In accordance to our results in primary monocytes, also in wt cells these genes were mostly less induced after H5N1 infection than after H1N1 infection, becoming most evident 24 h p.i. in this cell system (fig. 7). RORα knockout had virtually no influence on this gene expression pattern except for CXCL11, which was slightly higher expressed in H1N1-infected RORα−/− monocytes. However, with respect to H5N1 infections the knockdown of RORα resulted in strong transcriptional inductions of all of these genes (fig. 7). Data confirm the specific role of RORα in H5N1 infections to be essentially involved in the suppression of the inflammatory and antiviral response of monocytes.
Fig. 7.
Gene induction patterns in ER-Hoxb8 monocytes. ER-Hoxb8 wt (black bars) and RORα−/− (gray bars) monocytes were infected with H1N1 and H5N1 for 5, 8 and 24 h, and lysates were used for qRT-PCR. Induction of TNF-α-mRNA, IL-8R-mRNA, CXCL9-mRNA, CXCL11-mRNA, IFNB1-mRNA and IFNA-mRNA was measured by qRT-PCR. Experiments were performed with n = 4. Boxes represent means ± SEM. * p < 0.05; ** p < 0.01; *** p < 0.001.
Discussion
HPAIV which have successfully adapted to the human host show characteristic symptoms not observed during seasonal influenza infections. The most important features of HPAIV infections are cytokine storm and systemic spreading of infection [1, 2, 3, 4]. Different cell types of the innate and adaptive immune system contribute to antiviral defense and cytokine production, and some viruses have obviously developed strategies to overcome the early phase of inflammatory responses facilitating their systemic spreading.
In earlier studies our group revealed a significant role of endothelial cells for development of the observed cytokine storm [12], a finding which is in line with a recent publication by Teijaro et al. [17]. Still, the mechanisms lying behind systemic spreading of disease remained unsolved until a recent study of our group found HPAIV to bypass effective immune responses of human blood-derived macrophages [18]. Missing expression of the viral protein M2 leads to inhibition of inflammasome activation and thus an unexpected reduction of inflammatory and antiviral gene expression. Consequently, development of certain cell-type-specific mechanisms contributes to the high pathogenicity of HPAIV and their ability to cause systemic disease.
In the present study we analyzed the role of human blood monocytes for the inflammatory response to influenza infection. We compared infection of monocytes with the low pathogenic human influenza virus H1N1 (PR8) and human influenza viruses H1N1/v and H1N1 (WSN/33) on the one hand, and with HPAIVs H7N7 (FPV) and H5N1 (KAN-1) on the other hand. We demonstrated that all three virus strains are able to infect and replicate in monocytes. There is a slight advantage for H7N7 and H5N1 compared to H1N1 in viral protein expression and replication within the first hours of infection, obviously one of the pathogenic features involved in the shutdown of proinflammatory responses of monocytes to HPAIV. Through analyzing the gene expression patterns of monocytes in response to influenza virus infection, we have confirmed earlier studies showing that monocytes induce expression of certain chemokines and cytokines in response to H1N1 infection [8, 20]. However, we found a surprisingly impaired inflammatory and antiviral response of monocytes upon H5N1 infection. The strongly reduced immune response during H5N1 infection was unexpected since H5N1 patients are characterized by uncontrolled release of cytokines [1, 35]. Human monocytes would have been probable origins for strong cytokine production. Cell death could be excluded as a possible reason for reduced gene induction. Cell death experiments showed similar death rates of monocytes infected with H1N1 and H5N1 (data not shown). In addition, we could not find upregulation of proapoptotic genes in microarray analysis 5 h p.i. Therefore, induction of cell death does not influence expression data at the time point of array analysis.
Since avoidance of inflammatory activation of infiltrating monocytes could be a sophisticated escape strategy of H5N1 facilitating systemic spreading of disease we plunged into the background molecular mechanisms. Our bioinformatic workup of IAV-induced gene expression patterns pointed to RORα as a candidate transcription factor employed by H5N1 to suppress the inflammatory response of human monocytes. Recent findings demonstrated that RORα interferes with NF-κB signaling by activating the inhibitor of NF-κB, IκBα [33]. A role for RORα for the inflammatory response in vivo could be shown by the group of Stapleton et al. [34] who found an increased susceptibility of RORα knockout mice to LPS infection. However, they could not confirm the link between RORα and NF-κB inhibition in these mice. A biological relevance of RORα in the context of viral infections has not been described so far. We now demonstrate for the first time that RORα is activated in human monocytes by HPAIV, especially H5N1 virus, and that activation of RORα is associated with a diminished translocation of NF-κB in infected human monocytes. Diminished NF-κB translocation could be shown to be coupled with expression, and therefore missing phosphorylation, of IκBα, which is in line with the mechanism described for RORα in human primary smooth-muscle cells [33]. Of special interest are RORα-mediated inhibitory effects on type 1 interferons and chemokines like CXCL9 and CXCL11, which point to a completely new role of RORα for the antiviral immune response. Employing ER-Hoxb8 RORα−/− monocytes impressively discloses that blocking of RORα rescues the monocytic immune response allowing proper NF-κB activation and adequate inflammatory and antiviral gene responses upon H5N1 infection.
As a basic principle, NF-κB is an essential constituent of the IFN-β enhanceosome [36]. In airway epithelial cells as well as lungs of influenza-infected ferrets and mice [37, 38, 39] influenza A virus infection is associated with expression of NF-κB-dependent gene products such as proinflammatory cytokines. With respect to HPAIV, in particular H5N1, our group has demonstrated repeatedly that in endothelial cells activation of NF-κB is indispensable for the gene response [12, 16]. Independent of the cell type, proper NF-κB signaling seems to be inevitable for a strong H5N1-induced cytokine and antiviral immune responses. In monocytes, the activation of NF-κB in the context of HPAIV infections has not been examined yet. Accordingly, we now demonstrate that failure of NF-kB-activation results in a complete lack of inflammatory and antiviral actions.
Considering the combination of failure of human monocytes to respond properly to H5N1 infection but efficient IAV replication of all strains examined in this cell type, we conclude that H5N1 has developed a sophisticated escape mechanism. So, H5N1 virus is able to suppress inflammatory and antiviral actions of one of the earliest defense barriers. Monocytes have been shown to be the main infiltrate in lungs during high-pathogenicity IAV infection in mice [40]. Therefore, monocytes are not only important defenders beyond the lung epithelial barrier, but already come into contact with the virus in the state of local infection. The inability of monocytes to fulfill effective immune response contributes to systemic spreading of HPAIV infection beyond the epithelial barrier.
Taken together, we have deciphered a novel mechanism by which H5N1 is enabled to escape the primary innate immune response of human monocytes. In this cell type H5N1 interferes with the NF-κB signaling pathway most probably due to RORα activation. In this way H5N1 virus overcomes host responses in the early stage of infection which eventually promotes viral spreading in the organism and contributes to the severity of HPAIV infections in comparison with seasonal influenza infections.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
We thank Ursula Nordhues for technical assistance during microarray experiments and the Department of Integrated Functional Genomics (IFG) of the University of Münster for microarray hybridization as well as for supply of technical equipment. The Hoxb8 retrovirus was kindly provided by the laboratory of Georg Haecker (University Hospital of Freiburg, Germany). This work was supported by the German Federal Ministry of Education and Research (BMBF, FluResearchNet, grant No. 01KI07130) and the Interdisciplinary Center of Clinical Research of the University of Münster (Ro2/004/10).
References
- 1.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, Naghdaliyev A, Peiris JS, Shindo N, Soeroso S, Uyeki TM. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 2.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 3.Chan MC, Cheung CY, Chui WH, Tsao SW, Nicholls JM, Chan YO, Chan RW, Long HT, Poon LL, Guan Y, Peiris JS. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir Res. 2005;6:135. doi: 10.1186/1465-9921-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge KF, Gordon S, Guan Y, Peiris JS. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 5.Gong JH, Sprenger H, Hinder F, Bender A, Schmidt A, Horch S, Nain M, Gemsa D. Influenza A virus infection of macrophages: enhanced tumor necrosis factor-alpha (TNF-alpha) gene expression and lipopolysaccharide-triggered TNF-alpha release. J Immunol. 1991;147:3507–3513. [PubMed] [Google Scholar]
- 6.Herold S, von Wulffen W, Steinmueller M, Pleschka S, Kuziel WA, Mack M, Srivastava M, Seeger W, Maus UA, Lohmeyer J. Alveolar epithelial cells direct monocyte transepithelial migration upon influenza virus infection: impact of chemokines and adhesion molecules. J Immunol. 2006;177:1817–1824. doi: 10.4049/jimmunol.177.3.1817. [DOI] [PubMed] [Google Scholar]
- 7.Hinder F, Schmidt A, Gong JH, Bender A, Sprenger H, Nain M, Gemsa D. Influenza A virus infects macrophages and stimulates release of tumor necrosis factor-alpha. Pathobiology. 1991;59:227–231. doi: 10.1159/000163651. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann P, Sprenger H, Kaufmann A, Bender A, Hasse C, Nain M, Gemsa D. Susceptibility of mononuclear phagocytes to influenza A virus infection and possible role in the antiviral response. J Leukoc Biol. 1997;61:408–414. doi: 10.1002/jlb.61.4.408. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann P, Schmidtke M, Stelzner A, Gemsa D. Suppression of proinflammatory cytokines and induction of IL-10 in human monocytes after coxsackievirus B3 infection. J Med Virol. 2001;64:487–498. doi: 10.1002/jmv.1076. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 11.Klenk HD. Infection of the endothelium by influenza viruses. Thromb Haemost. 2005;94:262–265. doi: 10.1160/TH05-04-0264. [DOI] [PubMed] [Google Scholar]
- 12.Viemann D, Schmolke M, Lueken A, Boergeling Y, Friesenhagen J, Wittkowski H, Ludwig S, Roth J. H5N1 virus activates signaling pathways in human endothelial cells resulting in a specific imbalanced inflammatory response. J Immunol. 2011;186:164–173. doi: 10.4049/jimmunol.0904170. [DOI] [PubMed] [Google Scholar]
- 13.Ocana-Macchi M, Bel M, Guzylack-Piriou L, Ruggli N, Liniger M, McCullough KC, Sakoda Y, Isoda N, Matrosovich M, Summerfield A. Hemagglutinin-dependent tropism of H5N1 avian influenza virus for human endothelial cells. J Virol. 2009;83:12947–12955. doi: 10.1128/JVI.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perkins LE, Swayne DE. Pathobiology of A/chicken/Hong Kong/220/97 (H5N1) avian influenza virus in seven gallinaceous species. Vet Pathol. 2001;38:149–164. doi: 10.1354/vp.38-2-149. [DOI] [PubMed] [Google Scholar]
- 15.Szretter KJ, Gangappa S, Lu X, Smith C, Shieh WJ, Zaki SR, Sambhara S, Tumpey TM, Katz JM. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol. 2007;81:2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmolke M, Viemann D, Roth J, Ludwig S. Essential impact of NF-κB signaling on the H5N1 influenza A virus-induced transcriptome. J Immunol. 2009;183:5180–5189. doi: 10.4049/jimmunol.0804198. [DOI] [PubMed] [Google Scholar]
- 17.Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB, Rosen H. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friesenhagen J, Boergeling Y, Hrincius E, Ludwig S, Roth J, Viemann D. Highly pathogenic avian influenza viruses inhibit effective immune responses of human blood-derived macrophages. J Leukoc Biol. 2012;92:11–20. doi: 10.1189/jlb.0911479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronni T, Sareneva T, Pirhonen J, Julkunen I. Activation of IFN-alpha, IFN-gamma, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J Immunol. 1995;154:2764–2774. [PubMed] [Google Scholar]
- 20.Kaufmann A, Salentin R, Meyer RG, Bussfeld D, Pauligk C, Fesq H, Hofmann P, Nain M, Gemsa D, Sprenger H. Defense against influenza A virus infection: essential role of the chemokine system. Immunobiology. 2001;204:603–613. doi: 10.1078/0171-2985-00099. [DOI] [PubMed] [Google Scholar]
- 21.Lee SM, Gardy JL, Cheung CY, Cheung TK, Hui KP, Ip NY, Guan Y, Hancock RE, Peiris JS. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS One. 2009;4:e8072. doi: 10.1371/journal.pone.0008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakabe S, Iwatsuki-Horimoto K, Takano R, Nidom CA, Quynh Le Mt, Nagamura-Inoue T, Horimoto T, Yamashita N, Kawaoka Y. Cytokine production by primary human macrophages infected with highly pathogenic H5N1 or pandemic H1N1 2009 influenza viruses. J Gen Virol. 2011;92:1428–1434. doi: 10.1099/vir.0.030346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barczyk K, Ehrchen J, Tenbrock K, Ahlmann M, Kneidl J, Viemann D, Roth J. Glucocorticoids promote survival of anti-inflammatory macrophages via stimulation of adenosine receptor A3. Blood. 2010;116:446–455. doi: 10.1182/blood-2009-10-247106. [DOI] [PubMed] [Google Scholar]
- 24.Ehrchen J, Steinmuller L, Barczyk K, Tenbrock K, Nacken W, Eisenacher M, Nordhues U, Sorg C, Sunderkotter C, Roth J. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood. 2007;109:1265–1274. doi: 10.1182/blood-2006-02-001115. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Saint DA. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal Biochem. 2002;302:52–59. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- 26.Viemann D, Strey A, Janning A, Jurk K, Klimmek K, Vogl T, Hirono K, Ichida F, Foell D, Kehrel B, Gerke V, Sorg C, Roth J. Myeloid-related proteins 8 and 14 induce a specific inflammatory response in human microvascular endothelial cells. Blood. 2005;105:2955–2962. doi: 10.1182/blood-2004-07-2520. [DOI] [PubMed] [Google Scholar]
- 27.Viemann D, Goebeler M, Schmid S, Klimmek K, Sorg C, Ludwig S, Roth J. Transcriptional profiling of IKK2/NF-κB- and p38 MAP kinase-dependent gene expression in TNF-alpha-stimulated primary human endothelial cells. Blood. 2004;103:3365–3373. doi: 10.1182/blood-2003-09-3296. [DOI] [PubMed] [Google Scholar]
- 28.Kirschnek S, Vier J, Gautam S, Frankenberg T, Rangelova S, Eitz-Ferrer P, Grespi F, Ottina E, Villunger A, Hacker H, Hacker G. Molecular analysis of neutrophil spontaneous apoptosis reveals a strong role for the pro-apoptotic BH3-only protein Noxa. Cell Death Differ. 2011;18:1805–1814. doi: 10.1038/cdd.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- 30.Engelhardt OG, Fodor E. Functional association between viral and cellular transcription during influenza virus infection. Rev Med Virol. 2006;16:329–345. doi: 10.1002/rmv.512. [DOI] [PubMed] [Google Scholar]
- 31.Katze MG, Krug RM. Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells. Mol Cell Biol. 1984;4:2198–2206. doi: 10.1128/mcb.4.10.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delerive P, Monte D, Dubois G, Trottein F, Fruchart-Najib J, Mariani J, Fruchart JC, Staels B. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2001;2:42–48. doi: 10.1093/embo-reports/kve007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stapleton CM, Jaradat M, Dixon D, Kang HS, Kim SC, Liao G, Carey MA, Cristiano J, Moorman MP, Jetten AM. Enhanced susceptibility of staggerer (RORαsg/sg) mice to lipopolysaccharide-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2005;289:L144–L152. doi: 10.1152/ajplung.00348.2004. [DOI] [PubMed] [Google Scholar]
- 35.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim CU, Lew W, Williams MA, Liu H, Zhang L, Swaminathan S, Bischofberger N, Chen MS, Mendel DB, Tai CY, Laver WG, Stevens RC. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc. 1997;119:681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 37.Bernasconi D, Amici C, La FS, Ianaro A, Santoro MG. The IκB kinase is a key factor in triggering influenza A virus-induced inflammatory cytokine production in airway epithelial cells. J Biol Chem. 2005;280:24127–24134. doi: 10.1074/jbc.M413726200. [DOI] [PubMed] [Google Scholar]
- 38.Cameron CM, Cameron MJ, Bermejo-Martin JF, Ran L, Xu L, Turner PV, Ran R, Danesh A, Fang Y, Chan PK, Mytle N, Sullivan TJ, Collins TL, Johnson MG, Medina JC, Rowe T, Kelvin DJ. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J Virol. 2008;82:11308–11317. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kash JC, Basler CF, Garcia-Sastre A, Carter V, Billharz R, Swayne DE, Przygodzki RM, Taubenberger JK, Katze MG, Tumpey TM. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J Virol. 2004;78:9499–9511. doi: 10.1128/JVI.78.17.9499-9511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kok WL, Denney L, Benam K, Cole S, Clelland C, McMichael AJ, Ho LP. Pivotal advance: invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza A virus infection. J Leukoc Biol. 2012;91:357–368. doi: 10.1189/jlb.0411184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data