To the Editor:
RNA sequencing (RNA-seq) is used to measure gene expression levels across the transcriptome for a huge variety of samples. For example, RNA-seq has been applied to study gene expression in individuals with rare diseases1, in hard-to-obtain tissues2 or for rare forms of cancer3. Recently, enormous RNA-seq datasets have been produced in the GTEx (Genotype-Tissue Expression) study4, which comprises 9,662 samples from 551 individuals and 54 body sites, and in the Cancer Genome Atlas (TCGA) study, which comprises 11,350 samples from 10,340 individuals and 33 cancer types. Public data repositories, such as the Sequence Read Archive (SRA), host >50,000 human RNA-seq samples. It is estimated that these repositories are likely to double in size every 18 months5. Deposited data are provided as raw sequencing reads, which are costly for standard academic labs researchers to analyze. Efforts have been made to standardize and publish ready-to-analyze summaries of both DNA sequencing6 and exome-sequencing7 data. Adopting a similar approach for archived RNA-seq data, we have developed recount2, which comprises >4.4 trillion uniformly processed and quantified RNA-seq reads.
Many researchers rely on processed forms of publicly available data, such as gene counts, for statistical methods development and re-analysis of candidate genes. Although these quantified data are sometimes available through the Gene Expression Omnibus8, there are no requirements to deposit these data, nor are data always processed with standard or complete pipelines9–13. Five years ago, we began to address this problem by summarizing RNA-seq data into concise gene count tables and making these processed data and metadata available as Bioconductor14 ExpressionSet objects with one documented processing pipeline. Together this formed an RNA-seq resource named ReCount15 that contained 8 billion reads from 18 studies. ReCount was used in the development of the DESeq2 (ref. 16), voom17 and metagenomeSeq18 methods for differential expression and normalization, compilation of co-expression networks19 and to study the effect of ribosomal DNA dosage on gene expression20. The amount of archived RNA-seq data has massively increased over the past five years. To meet the needs of researchers, we have produced recount2, which contains >4.4 trillion uniformly processed and quantified RNA-seq reads that are derived from in excess of 70,603 human RNA-seq samples deposited in the SRA, GTEx and TCGA projects aligned with Rail-RNA21,22.
The recount2 resource summarizes expression data for genes, exons, exon–exon splice junctions and base-level coverage (Supplementary Methods), which enables multiple downstream analyses, including testing for differential expression of potentially unannotated transcribed sequence23. A searchable interface is available at this site (https://jhubiostatistics.shinyapps.io/recount/) and via the accompanying Bioconductor package (http://bioconductor.org/packages/recount).
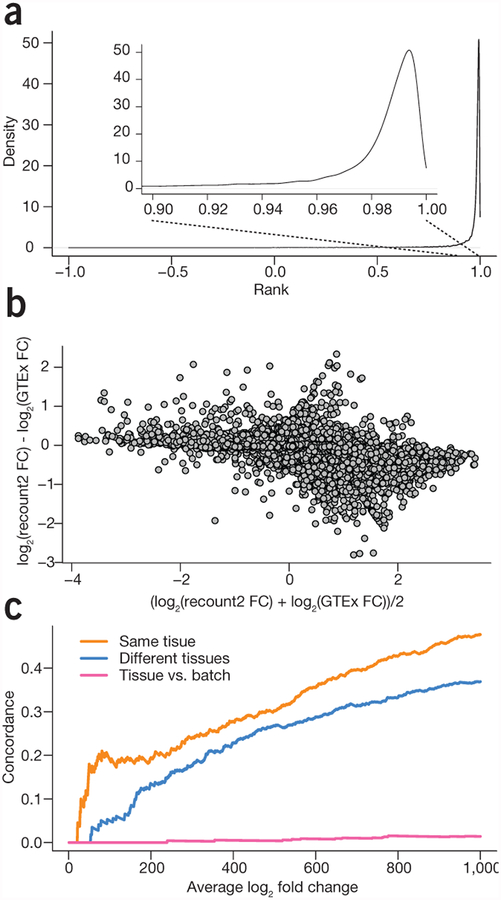
We first compared recount2-processed data with the publicly available data from the GTEx project, which comprises 9,662 samples from >250 individuals24 to demonstrate that our processing pipeline produced gene counts similar to the published counts (Supplementary Methods). We downloaded the official release of the gene counts from the GTEx portal and compared them with the recount2 gene counts (Supplementary Note 1, Section 4). For protein coding genes, the gene expression levels that we estimated using the recount2 pipeline had a median (IQR) correlation of 0.987 (0.971, 0.993) with the v6 release from GTEx (Fig. 1a and Supplementary Note 1, Section 4). A differential expression analysis comparing colon and whole blood samples using the gene expression measurements from recount2 matched the results obtained using the v6 release from the GTEx portal (r2 = 0.92 between fold changes for recount2 and GTEx v6 counts for protein coding genes; Fig. 1b and Supplementary Note 1, Section 5). These results suggest that recount2 produces directly comparable gene counts to one of the largest published studies.
Figure 1.
Meta-analysis and study comparison facilitated by recount2. (a) The distribution of correlations between gene expression estimates for GTEx v6 from the GTEx portal and the counts calculated in recount2 for protein coding genes. The gene expression counts are highly correlated between both quantifications for almost all genes. (b) A comparison of the fold changes for differential expression between colon and whole blood using the quantifications from GTEx and from recount2 for protein coding genes. The majority of genes have a similar fold-change between the two analyses. (c) A concordance versus rank (i.e., ‘concordance at the top’, CAT) plot showing comparisons between a meta-analysis tissue comparison of whole blood and colorectal tissue in data from the sequence read archive and the GTEx project. When comparing the same tissues, there is a strong concordance between differential expression results on public data and GTEx (orange), less concordance when different tissues are compared (blue) and almost none when comparing different analyses (pink).
The advantage of using the recount2 version of GTEx data is that all data are identically processed, therefore enabling integrated analyses of multiple datasets. To illustrate how recount2 can be used to investigate or validate cross-tissue differences using publicly available data, we computed expression differences comparing samples from healthy colon tissue and whole blood from healthy individuals (Supplementary Methods, Supplementary Note 2, Section 1.8 and Supplementary Code 1). Control samples were used to limit differences observed to those due to tissue type and not disease status. Colon control samples were used from studies SRP029880 (a study of colorectal cancer25, n = 19) and SRP042228 (a study of Crohn’s disease26, n = 41). Whole blood control samples were used from SRP059039 (virus-induced diarrhea, unpublished, n = 24), SRP059172 (a study of blood biomarkers for brucellosis, unpublished, n = 47) and SRP062966 (a study of lupus, unpublished, n = 18). After filtering genes to include only those with an average normalized count of at least 5 across samples (to restrict to genes that were expressed well above the limit of detection), we carried out gene-level differential expression analysis using limma27 and voom17 (Supplementary Note 2, Section 1.8).
To validate the meta-analysis results, we evaluated whether we had found similar patterns of differential expression between the same tissues collected as part of a single project. We selected all of the colon and whole blood samples from the GTEx project (n = 376 and 456, respectively) and performed the same analysis, adjusting for batch effects by including the reported batch from GTEx as a covariate in the linear model. We then computed rank-based concordance, examining the fraction of the top differentially expressed genes that were included in both analyses. Approximately 20% of the top 100 genes from the two analyses were concordant (Fig. 1c and Supplementary Note 2, Section 1.9).
As a comparison and to provide context for this result, we performed two additional comparisons. First, we used GTEx lung data (n = 374) in place of the colon data and computed differentially expressed genes compared with whole blood. In this case, only ~5% of the top 100 differentially expressed genes were shared in the top 100 genes from our multi-study analysis (Supplementary Note 2, Section 1.9). Second, to represent concordance results expected for a comparison of unrelated things, we used ranked coefficients for batch instead of for tissue and saw very little concordance. These comparisons show that we can use the resources found in recount2 to perform a valid tissue-specific meta-analysis without generating the necessary data in-house, which would add considerable time and expense, provided that samples were even available to analyze.
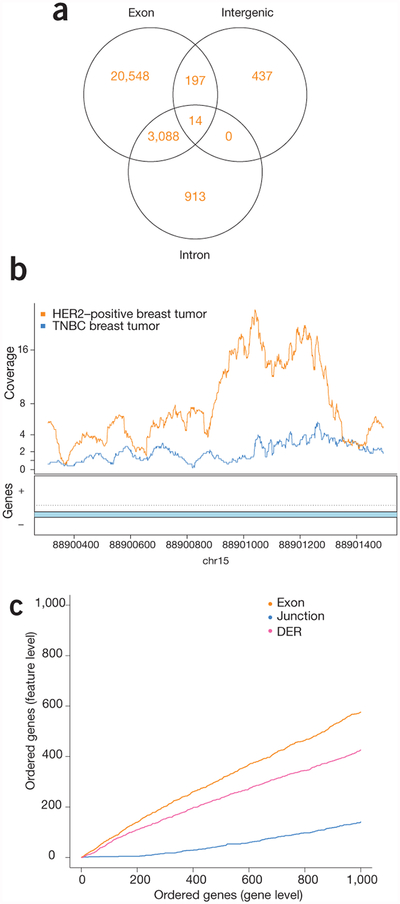
The recount2 pipeline enables an in-depth characterization of transcriptional differences across biological conditions. To illustrate this using data from breast cancer subtypes, we first chose HER2-positive and triple-negative breast cancer (TNBC) samples from study SRP032789 (TNBC, n = 6; HER2-positive, n = 5)28, and extracted feature-level expression across genes, exons, junctions and expressed regions, finding widespread expression differences by subtype (Table 1, Supplementary Note 3 and Supplementary Methods). Of these significant differentially expressed regions (DERs) found with derfinder23, 1,350 did not overlap any annotated exons (Fig. 2a and Supplementary Note 4, Section 4), demonstrating that 5% of DERs detected would not be reported using annotation-dependent methods of expression estimation. These DERs would only be identified when a quantification method not reliant on annotation was used. Such quantifications are made readily available within recount2 (Fig. 2b).
Table 1.
Differentially expressed features comparing TNBC versus HER2-positive breast cancer samples at a false-discovery rate of 5%
| Feature level | Higher expression in TNBC | Higher expression in HER2-positive | Total differentially expressed features |
|---|---|---|---|
| Genes | 1,362 | 1,612 | 2,974 |
| Exons | 14,546 | 13,159 | 27,705 |
| Exon-exon junctions | 1,732 | 18,073 | 19,805 |
| Expressed regions | 11,414 | 13,783 | 25,197 |
Figure 2.
Multi-feature-level differential expression analysis is facilitated by recount2. (a) Venn diagram showing the number of expressed regions detected that overlap exons, intergenic regions and intronic regions, including expressed regions that overlap multiple annotation types. Differential expression occurs outside of previously annotated protein-coding regions. (b) An example of a region on chromosome 15 showing differential expression between breast cancer subtypes in an annotated intron. The lines show the average coverage in each group across samples. (c) CAT plot of concordance between gene-level analysis and then exon-, junction- and region-level (DER) analyses shows high concordance across the different feature levels.
We further summarized junctions and exons at the gene level using the resulting differential expression P-values, and 73% of the top 100 genes were shared at the gene- and exon-level analyses. In comparison, expressed regions and exon–exon junction analyses shared only 58% and 4% of the top 100 features, respectively (Fig. 2c and Supplementary Note 3, Section 5). Furthermore, to validate the differential expression findings, we compared the gene-level results from study SRP032789 with an independent study (SRP019936 (ref. 29); TNBC, n = 8; HER2-positive, n = 7; Supplementary Note 5). Expression analysis was carried out as described above, identifying 3,434 genes as differentially expressed (q < 0.05, Supplementary Note 5, Section 4). Given the low concordance (8% among the top 1,000 genes, Supplementary Note 5, Section 5.1) between these results and those from study SRP032789, we then applied independent hypothesis weighting (IHW)30 across the two studies, which slightly improved replication rates, although sample size is limited in these two studies and thus likely thwarts our ability to see a huge increase in power using IHW (Supplementary Note 5, Section 5.2). As the data within recount2 have all been processed with the same analytical pipeline (Supplementary Code 2 and 3), the analytical burden on the user is minimized when comparing across datasets.
The recount2 pipeline can be used for querying, downloading and analyzing large-scale human RNA-seq datasets across more than 70,000 samples, including all of GTEx, TCGA and the SRA. We also allow users to process and upload their own experimental data to recount2 (Supplementary Methods and Supplementary Code 4). Although all recount2 samples have been processed and summarized with a single pipeline, so-called ‘batch’ effects could occur and should be considered in downstream analyses, particularly when comparing among studies. As an example, the type of library preparation is not accounted for in our processing, but we will continue to annotate these variables so they can be included in downstream analyses. By removing a large number of data processing and quantification choices potentially made by researchers, recount2 reduces the number of ‘researcher degrees of freedom’31, which can improve replication and reduce the potential for false positives created by processing pipeline differences.
Other tools have been developed to summarize publically deposited gene expression data. For example, the Expression Atlas32 provides final results that can be queried only at the gene level, Toil focuses only on curated datasets33 and other efforts focus primarily on cancer34,35. Unlike these resources, recount2 uses analysis pipelines that are annotation agnostic to process and summarize samples. For example, in junction and expressed region analyses, gene annotations are only used to label summarized data post-analysis and not to align reads or discover splice junctions—downstream analyses are therefore fully aware of unannotated splicing events36.
By providing an updateable resource of uniformly processed RNA-seq samples, together with R-based software for analysis, recount2 will enable studies that individual laboratories would otherwise not have the resources to undertake.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Kingsford and D. Filippova for their assistance in adding SHARQ metadata to recount2. recount2 data are hosted on SciServer, a collaborative research environment for large-scale data-driven science. It is being developed at, and administered by, the Institute for Data Intensive Engineering and Science at Johns Hopkins University. SciServer is funded by the National Science Foundation Award ACI-1261715. For more information about SciServer, visit http://www.sciserver.org/. We thank E. Lehnert and P. Radovic at Seven Bridges for their help accessing RNA-seq data from TCGA using the Cancer Genomics Cloud API and depositing results in a shared bucket on Amazon S3. We also thank other Seven Bridges team members for facilitating our TCGA reanalysis, including B. Dusenbery, N. Tijanic, M. Kovacevic, M. Sadoff and G. Kaushik. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health. Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI/SAIC-Frederick, Inc. (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by a supplement to University of Miami grants DA006227 and DA033684 and to contract N01MH000028. Statistical Methods development grants were made to the University of Geneva (MH090941 & MH101814), the University of Chicago (MH090951, MH090937, MH101820, MH101825), the University of North Carolina–Chapel Hill (MH090936 & MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University St. Louis (MH101810) and the University of Pennsylvania (MH101822). The data used for the analyses described in this manuscript were obtained from: the GTEx Portal on 11/21/15 and/or dbGaP accession number phs000424.v6.p1 on 11/30/15—12/04/15. Funding: B.L., J.T.L., L.C.T., S.E., A.N., M.T., K.H. and K.K. were supported by NIH R01 GM105705. L.C.T. was supported by Consejo Nacional de Ciencia y Tecnología México 351535. L.C.T. and A.E.J. were supported by 1R21MH109956. Amazon Web Services experiments were supported by AWS in Education research grants. Storage costs on S3 for TCGA runs were partially covered by a grant from Seven Bridges Genomics for use of the Cancer Genomics Cloud.
Footnotes
Editor’s note: This article has been peer-reviewed.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Albers CA et al. Nat. Genet 44, 435–439, S431–432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohen R, Dobra A, Tracy JH & Haugen E Transl. Psychiatry 4, e366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goh G et al. Nat. Genet 46, 613–617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melé M et al. Science 348, 660–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kodama Y, Shumway M & Leinonen R Nucleic Acids Res 40, D54–D56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.1000 Genomes Project Consortium et al. Nature 467, 1061–1073 (2010).20981092 [Google Scholar]
- 7.Lek M et al. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett T et al. Nucleic Acids Res 39, D1005–D1010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nookaew I et al. Nucleic Acids Res 40, 10084–10097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobin A et al. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim D et al. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engström PG et al. Nat. Methods 10, 1185–1191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar PK, Hoang TV, Robinson ML, Tsonis PA & Liang C Sci. Rep 5, 13443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman RC et al. Genome Biol 5, R80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazee AC, Langmead B & Leek JT BMC Bioinformatics 12, 449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love MI, Huber W & Anders S Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law CW, Chen Y, Shi W & Smyth GK Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulson JN, Stine OC, Bravo HC & Pop M Nat. Methods 10, 1200–1202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iancu OD et al. Bioinformatics 28, 1592–1597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbons JG, Branco AT, Yu S & Lemos B Nat. Commun 5, 4850 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Nellore A et al. Bioinformatics 10.1093/bioinformatics/btw575 (2016). [DOI]
- 22.Nellore A, Wilks C, Hansen KD, Leek JT & Langmead B Bioinformatics 32, 2551–2553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collado-Torres L et al. Nucleic Acids Res 45, e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GTEx Consortium G et al. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SK et al. Mol. Oncol 8, 1653–1666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haberman Y et al. J. Clin. Invest 124, 3617–3633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth GK in Bioinformatics and Computational Biology Solutions using R and Bioconductor 397–420 (Springer, 2005). [Google Scholar]
- 28.Eswaran J et al. Sci. Rep 3, 1689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalari KR et al. PLoS One 8, e79298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ignatiadis N, Klaus B, Zaugg JB & Huber W Nat. Methods 13, 577–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons JP, Nelson LD & Simonsohn U Psychol. Sci 22, 1359–1366 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Petryszak R et al. Nucleic Acids Res 44, D746–D752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vivian J et al. Nat. Biotechnol 35, 314–316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatlow PJ & Piccolo SR Sci. Rep 6, 39259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman M et al. Bioinformatics 31, 3666–3672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nellore A et al. Genome Biol 17, 266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.