Abstract
Lentiviral vectors (LVs) are excellent tools for gene transfer into mammalian cells. It is noteworthy that the first gene therapy treatment using LVs was approved for commercialization in 2017. The G glycoprotein from rhabdovirus vesicular stomatitis virus (VSV-G) is the glycoprotein most used to pseudotype LVs, due to its high efficiency in transducing several cell types and its resistance to viral vector purification and storage conditions. However, VSV-G expression induces cytotoxicity, which limits LV production to short periods. As alternative to VSV-G, γ-retrovirus glycoproteins (4070A derived, GaLV derived, and RD114 derived) have been used to pseudotype both γ-retroviral vectors (RVs) and LVs. These glycoproteins do not induce cytotoxicity, allowing the development of stable LV producer cells. Additionally, these LV pseudotypes present higher transduction efficiencies of hematopoietic stem cells when compared to VSV-G. Here, new 4070A-, RD114-TR-, and GaLV-TR-derived glycoproteins were developed with the aim of improving its cytoplasmic tail R-peptide cleavage and thus increase LV infectious titers. The new glycoproteins were tested in transient LV production using the wild-type or the less active T26S HIV-1 protease. The GaLV-TR-derived glycoproteins were able to overcome titer differences observed between LV production using wild-type and T26S protease. Additionally, these glycoproteins were even able to increase LV titers, evidencing its potential as an alternative glycoprotein to pseudotype LVs.
Keywords: lentiviral vectors, gene therapy, pseudotyping, envelope glycoproteins, GalV, 4070A, RD114, lentiviral, protease
Introduction
Retroviral vectors can deliver and integrate a gene of interest into a target cell genome upon cell transduction, allowing a long-term constitutive expression. In the particular case of lentiviral vectors (LVs), these are able to promote gene transfer into both dividing and non-dividing cells,1 potentially presenting lower genotoxicity than γ-retroviral vectors (γ-RVs) due to their integration pattern.2, 3 These reasons justify the growing number of gene therapy clinical trials using LVs in the past 2 decades with the aim of treating several disorders.4
LVs may be pseudotyped with heterologous viral glycoproteins acquiring alternative tropism and specific cell-entry properties.5, 6 Generally, the G glycoprotein from rhabdovirus vesicular stomatitis virus (VSV-G) is used to pseudotype LVs since, the wide distribution and high molecule number of VSV-G receptors at the cell surface allow for efficient transduction of several cell types.7, 8, 9 Additionally, LVs pseudotyped with VSV-G can be concentrated by ultracentrifugation without a substantial loss of infectivity and are resistant to several freeze-thaw cycles, which also contributes for their wide usage.6 However, the syncytium formation and consequent cytotoxicity, both resultant from VSV-G fusogenicity activation in viral producer cells, do not allow LV production for more than a few days.10, 11, 12 The VSV-G-pseudotyped LVs are usually produced transiently by co-transfection of 293T cells with LV expression cassettes or constitutively produced by LV producer stable cell lines making use of inducible promoters to drive VSV-G expression.10, 11, 12
As alternatives to VSV-G, the amphotropic murine leukemia virus (4070A), gibbon ape leukemia virus (GaLV), and feline endogenous retrovirus (RD114) glycoproteins have been used to pseudotype both γ-RVs and LVs.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 The production of γ-RVs pseudotyped with these three γ-retrovirus glycoproteins do not present major constraints. However, while the production of LVs pseudotyped with 4070A has no impact on infectious titers,23, 24, 25, 26, 27 the co-expression of HIV-1 core proteins with GaLV or RD114 glycoproteins barely results in the production of infectious LVs.28, 29, 30, 31, 32 These titer asymmetries are due to differences between cytoplasmatic tail amino-acid sequences of the glycoproteins (Figure 1A), since the fully fusogenic activation of 4070A, RD114, and GaLV is dependent on the cytoplasmatic tail R-peptide recognition and cleavage by the viral protease.33, 34, 35, 36, 37, 38, 39, 40, 41, 42 The replacement of the RD114 and GaLV cytoplasmic tail region by the one for 4070A (originating the RD114-TR and GaLV-TR, respectively) allows a high increase of LV particle infectivity.19, 23, 28, 29, 31, 32 Alternatively, just by replacing the protease cleavage sequence of RD114 with a sequence naturally cleaved by the HIV-1 protease—namely, the matrix-capsid cleavage sequence of HIV-1 Gag-Pro-Pol polyprotein (originating the RDpro glycoprotein)—it is also possible to increase the production of infectious LV particles.23, 30, 43
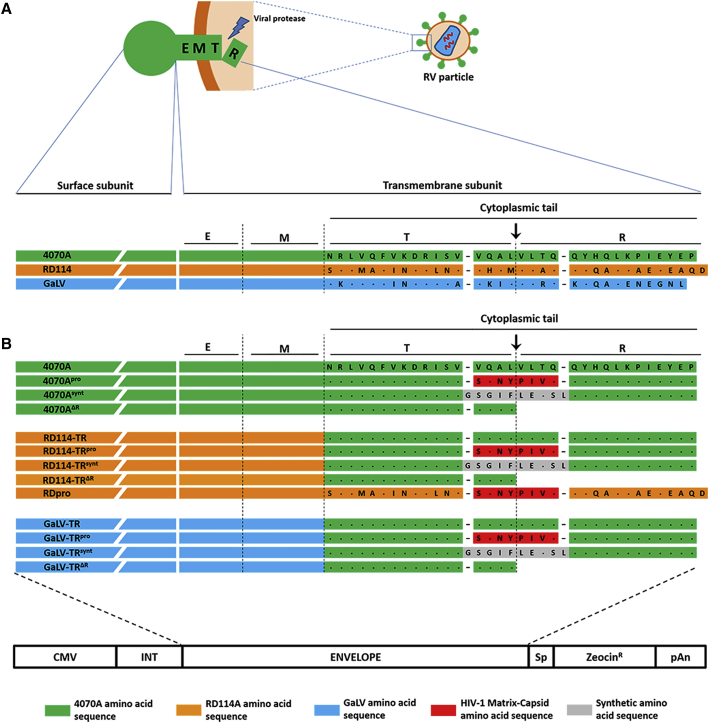
Figure 1.
Schematic Representation of Wild-Type and Engineered Viral Envelope Glycoproteins
(A) Amino-acid sequence alignment of glycoprotein cytoplasmic tail: murine leukemia virus (4070A), endogenous feline retrovirus (RD114), and gibbon ape leukemia virus SEATO strain (GaLV). (B) Amino-acid sequence alignment of the cytoplasmic tail region of the glycoproteins used in LV production and schematic representation of the envelope expression cassette. The black arrows indicate the protease cleavage site. E, ectodomain; M, transmembrane domain; T, cytoplasmic tail after R-peptide cleavage; R, R-peptide; CMV, cytomegalovirus promoter; INT, intron; Sp, spacer; pAn, polyadenylation site.
The LVs pseudotyped with the γ-retrovirus glycoproteins are able to transduce several cell types, being particularly efficient in the transduction of progenitor and differentiated hematopoietic stem cells (HSCs), when compared to the VSV-G pseudotype.19, 20, 21, 22, 28 Moreover, these LV pseudotypes are not inactivated by human serum23, 28 and may be concentrated by ultrafiltration, ultracentrifugation, or tangential flow filtration.20, 44, 45, 46 Additionally, the fusogenic activation of these glycoproteins should mainly occur after being incorporated into the viral particles (during viral maturation, when the viral particle buds out of the cell), preventing syncytium formation, which allows the development of stable cell lines able to constitutively produce LVs for several weeks.43, 47, 48, 49 Recently, we have established a stable LV producer cell line using a Gag-Pro-Pol polyprotein with a less active HIV-1 protease and the 4070A glycoprotein, able to constitutively produce 106 transducing units (TUs) per milliliter (TU ⋅ mL−1) for at least 2 months.50
The main drawback of pseudotyping LVs with γ-retrovirus glycoproteins is related to its lower infectious titers when compared to VSV-G pseudotype.20, 23, 30, 31 In this work, new versions of 4070A, GaLV-TR, and RD114-TR glycoproteins with modified sequences on the cytoplasmic tail were developed with the aim of improving glycoprotein fusogenicity activation and, consequently, increasing infectious LV titers. The impact of the modified glycoproteins in viral titers was assessed by performing transient LV production using either the wild-type or the mutated less active T26S HIV-1 protease.
Results
Modifications on γ-Retrovirus Glycoprotein Cytoplasmic Tail
The amino-acid protease cleavage sequence VQALVLTQ on the cytoplasmic tail of 4070A, RD114-TR, and GaLV-TR glycoproteins was replaced by the HIV-1 natural sequence SQNYPIVQ51 or the synthetic sequence GSGIFLETSL,52 originating the glycoproteinpro and glycoproteinsynt modified versions, respectively (Figure 1B). A truncated version of each glycoprotein with the R-peptide sequence deleted was also developed, the glycoproteinΔR, to be used as cleaved glycoprotein positive control. All the envelope glycoprotein genes were introduced in the same expression cassette to allow direct comparison between the different LV pseudotype productions (Figure 1B).
Transient Productions of LV Pseudotypes Using the Wild-Type Viral Protease
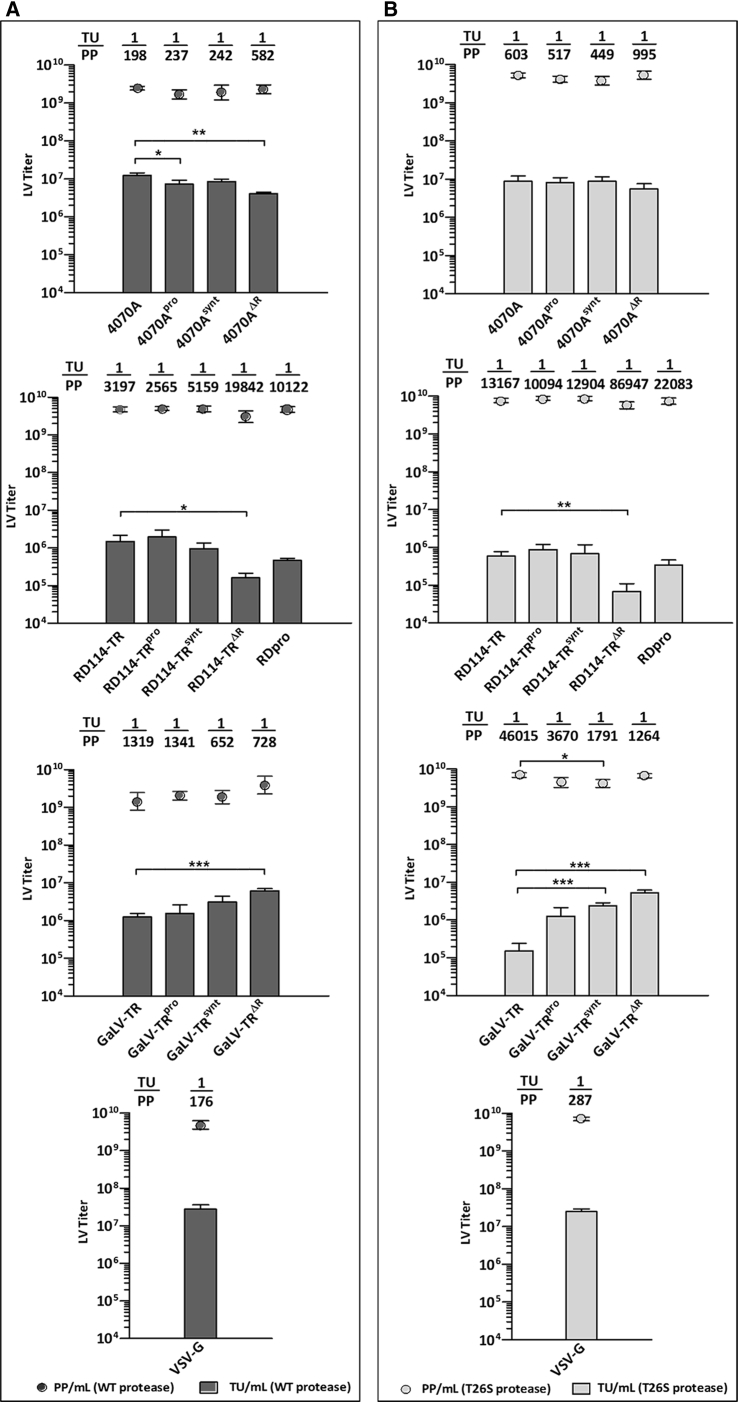
Transient production of third-generation SIN-LVs (self-inactivating lentiviral vectors) was performed for each pseudotype using the gag-pro-pol coding the wild-type HIV-1 protease. The supernatant titers (Transduction Units [TUs] and physical particles [PPs]) were assessed and compared (Figure 2A). The LV production of 4070A and RD114-TR pseudotypes generated average titers of about 1.2 × 107 TU ⋅ mL−1 and 1.5 × 106 TU ⋅ mL−1, respectively. The new glycoproteinpro and glycoproteinsynt versions of 4070A and RD114-TR did not markedly alter LV titers. However, a decrease in infectious titers was detected for production using 4070AΔR and RD114-TRΔR, leading to a 3-fold and 6-fold decrease in the TU/PP ratio when compared to the production of LVs pseudotyped with the parental 4070A and RD114-TR, respectively. In addition to RD114-derived production, the RDpro glycoprotein was also used, leading to a 3-fold decrease in the TU/PP ratio, when compared to RD114-TR production. The LV production of GaLV-TR pseudotype generated a titer of about 1.3 × 106 TU ⋅ mL−1. In the case of GaLV-TR-derived glycoproteins, while the GaLV-TRpro did not change LV titers, a 2- and 5-fold increment on functional LV titers were observed for the LV production using GaLV-TRsynt and GaLV-TRΔR glycoproteins, respectively, generating a maximum average titer of about 6.1 × 106 TU ⋅ mL−1. The LV production of VSV-G pseudotype generated an average titer of about 2.8 × 107 TU ⋅ mL−1.
Figure 2.
Transient LV Production Titers with the Engineered Envelope Glycoproteins
(A and B) Transient production titers of LVs pseudotyped with 4070A, 4070A-derived, RD114-TR, RD114-TR-derived, RDpro, GaLV-TR, GaLV-TR-derived, or VSV-G glycoproteins using the (A) HIV-1 wild-type protease or (B) HIV-1 T26S protease. The bars represent the transducing units (TUs), and the circles represent the physical particles (PPs). The titer values presented are the means ± SD of 3 independent experiments (n = 3). Statistical analysis for the comparison of LV titers was performed by using an unpaired Student t test (two-tailed). *p < 0.05; **p < 0.01; ***p < 0.001. The ratios of TUs to PPs are indicated above each set of LV titers.
Transient Productions of LV Pseudotypes Using the Less Active T26S Viral Protease
LV production, identical to that described earlier, was also performed using the gag-pro-pol with the T26S mutated protease (Figure 2B). For the viral production of 4070A-derived, RD114-derived, and VSV-G LV pseudotypes, average titer values similar to the ones previously observed were detected. However, for the LV production of GaLV-TR pseudotype, an average infectious titer of just 1.5 × 105 TU.mL−1 was observed. Nevertheless, infectious titer increases of 9-, 16-, and 35-fold were observed for GaLV-TRpro, GaLV-TRsynt, and GaLV-TRΔR LV-pseudotype production, respectively, generating a maximum average titer of about 5.3 × 106 TU ⋅ mL−1.
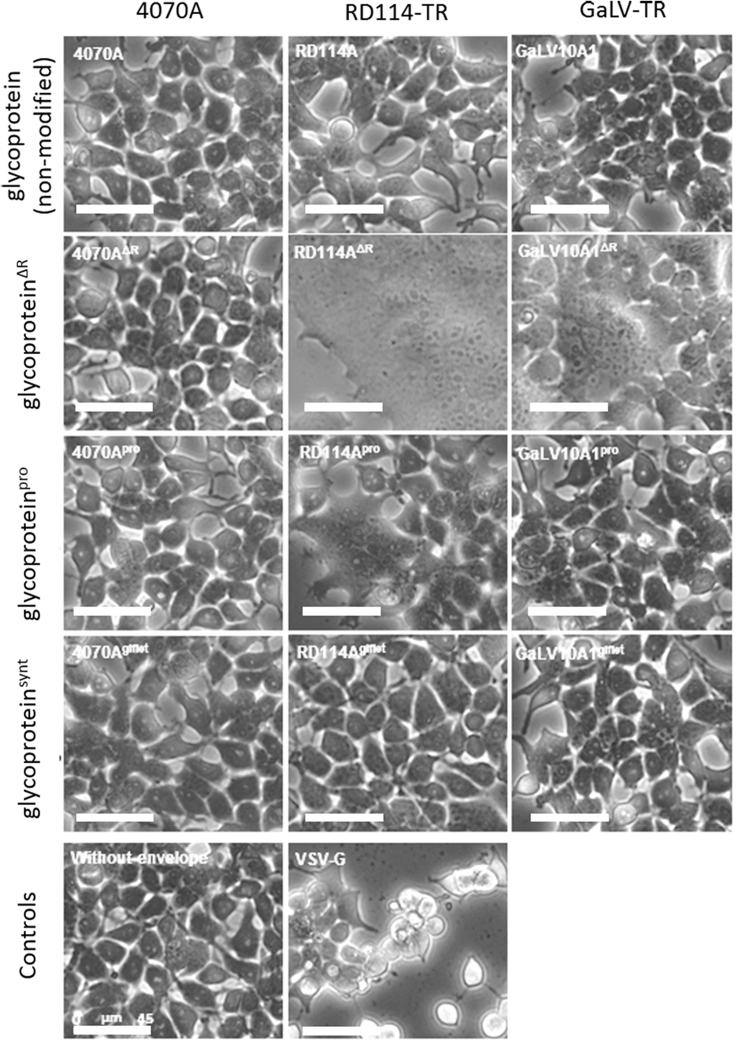
Syncytium Formation on 293T Cells Transiently Expressing the Viral Envelope Glycoproteins
Syncytium formation induced by glycoprotein expression was evaluated in 293T cells transiently transfected with the plasmids coding for envelope glycoproteins. 24 h post-transfection, cells were observed by phase-contrast microscopy (Figure 3). Syncytium and non-adherent round cells were observed in cells transfected with RD114-TRΔR, GaLV-TRΔR, and VSV-G expression cassettes. Additionally, RD114-TRpro expression also led to the formation of few syncytia. In all the other cases, no major morphological cell differences, relative to the no expression control, were observed.
Figure 3.
Representative Pictures of HEK293T Cells Transiently Expressing Envelope Glycoproteins
80× bright-field microscopy. Scale bars, 45 μm.
Discussion
The 4070A-, RD114-, and GaLV-derived envelope glycoproteins allow an efficient LV transduction of hematopoietic stem cells, conferring a more restricted tropism to LVs than VSV-G.19, 20, 21, 22, 28 Additionally, these glycoproteins can be used as an alternative to VSV-G in the development of stable constitutive LV producer cell lines since they are non-cytotoxic.43, 47, 48, 49 Despite the advantages of γ-retrovirus glycoproteins, the production of LVs pseudotyped with these usually present lower infectious titer particles when compared to the production of LVs pseudotyped with VSV-G.
To evaluate whether improved cleavage of the R-peptide in 4070A, RD114-TR, and GaLV-TR glycoproteins would translate into increased infectious titers of those LVs pseudotypes, the protease recognition sequence on the cytoplasmic tail of these glycoproteins was replaced by two others described to be efficiently cleaved by the HIV-1 protease: (1) the matrix-capsid natural cleavage sequence SQNYPIVQ, which is present in the HIV-1 Gag-Pro-Pol polyprotein,51 and (2) the GSGIFLETSL synthetic peptide.52 A truncated version of each glycoprotein without R-peptide was also developed. These truncated glycoproteins should be produced in their fully fusogenic states; thus, it would be expected that the resultant particles should exhibit the maximum infectious titer for each specific pseudotype (if no major interference on glycoprotein cellular traffic and assembling occurs).
The titer analysis of LV production using the wild-type protease evidences that the engineered 4070A-derived and RD114-TR-derived glycoproteins did not markedly alter LV titers (Figure 2A). Furthermore, the respective truncated glycoprotein versions (4070AΔR and RD114-TRΔR) impaired the production of infectious LVs. Nevertheless, the results of RD114-TR and RDpro LV production clearly indicate that RD114-TR is preferable to RDpro for producing higher titers of infectious LVs. In contrast to 4070A-derived and RD114-TR-derived glycoproteins, a 3- to 5-fold improvement in titers of infectious LVs in relation to the parental glycoprotein (GaLV-TR) was observed when using the GaLV-TRsynt and GaLV-TRΔR glycoproteins, leading to a 2-fold increase in the TU/PP ratio. These results suggest that, unlike for 4070A and RD114-TR pseudotypes, the infectivity of GaLV-TR vectors is limited by an inefficient HIV-1 protease cleavage of the glycoprotein R-peptide.
The R-peptide cleavage by the viral protease and consequent glycoprotein fusogenic activation should mainly occur during viral particle maturation, at the viral budding step (outside of the cell).53, 54 This should allow cellular glycoprotein production, intracellular trafficking, and cell membrane incoorporation in its non-fusogenic conformation, preventing counterproductive cell interactions.34, 35, 36, 37 In the specific case of the R-peptide-truncated glycoproteins developed herein, these are produced in a fusogenic active state. Thus, once at the cell-membrane surface, the glycoproteins will interact with the respective cell receptors of neighboring cells, promoting cell membrane fusion and consequent syncytia formation.29, 31, 41 This may explain the large syncytia observed in cells transfected with RD114-TRΔR and GaLV-TRΔR (Figure 3) and in the respective LV production. Nonetheless, unlike for RD114-TRΔR, the syncytia on GaLV-TRΔR LV production did not negatively affect LV titers (Figure 2A).
The impact of protease activity in the production of several LV pseudotypes was also evaluated by assessing LV titers obtained from transient LV production using the HIV-1 gag-pro-pol gene with the less active T26S mutated protease.55 Comparing the 4070A, RD114-TR, and GaLV-TR LV production titers of both proteases, it is possible to observe that the lower activity of T26S protease affects the production of infectious LVs. Whereas the less active protease led to merely a 3- to 4-fold decrease in TU/PP ratio for 4070A and RD114-TR pseudotypes, a 35-fold ratio decrease was observed with the GaLV-TR pseudotype. This decrease was mainly due to the 10-fold reduction of infectious GaLV-TR LVs when using the T26S protease. Nevertheless, the modified glycoproteins GaLV-TRpro, GaLV-TRsynt, and GaLV-TRΔR allowed the rescue of LV titers to levels identical to those obtained in production using the wild-type protease. As expected, the production titers of LVs pseudotyped with VSV-G were not affected by the T26S protease, since this glycoprotein is not dependent on viral protease cleavage to become fusogenically active.56 Together, these results demonstrate that the lower activity of T26S protease, despite not affecting Gag-Pro-Pol production and its respective viral processing, impairs the R-peptide cleavage of 4070A, RD114-TR, and GaLV-TR glycoproteins, which directly affects the infectivity of LVs. Additionally, since titers of these three LV pseudotypes were not similarly affected, despite their common cytoplasmic tail (Figure 1B), it is suggested that the glycoprotein sequence upstream regions (ectodomain + transmembrane domain of transmembrane subunit and surface subunit) may condition R-peptide recognition and cleavage by the viral protease. We hypothesize that the amino-acid differences on those glycoprotein upstream regions may promote different conformational changes in the cytoplasmic tail of 4070A, RD114-TR, and GaLV-TR glycoproteins, hampering the accessibility of viral protease to the respective cleavage sequences.
In this work, new GaLV-TR-derived glycoproteins were developed to pseudotype LVs. Those glycoproteins allowed us to transiently produce a maximum LV titer of 6.0 × 106 TU ⋅ mL−1, surpassing the titers of LV pseudotypes with GaLV-TR, RD114-TR, or RDpro. Additionally, this work evidences that glycoprotein R-peptide cleavage efficiency by viral protease limits titers of infectious LVs. The new GaLV-TRsynt and GaLV-TRpro are potential alternatives to both VSV-G or RD114-derived glycoproteins for the establishment of constitutively stable LV producer cell lines. Although further studies are required, these novel envelopes show potential for the development of alternative high-titer viral vector production platforms.
Materials and Methods
Cell Culture
HEK293T cells (CRL-11268), obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA), were used for LV production and titration. Cells were cultured in DMEM (GIBCO, Life Technologies, Paisley, UK) supplemented with 10% (v/v) fetal bovine serum (FBS) (GIBCO). Cells were maintained in an incubator at 37°C with a humidified atmosphere of 7% CO2 in air. The trypan blue exclusion method was used to assess cell concentration and viability.
Plasmids
Plasmid pRRLSIN.cPPT.PGK-GFP.WPRE (Addgene #12252, kindly provided by Dr. Didier Trono) codes for a SIN-LV genome carrying a GFP as a reporter gene and was used as a transgene in all LV production.
Plasmid pMDLg/pRRE57 (Addgene #12251, kindly provided by Dr. Didier Trono) codes for HIV-1 Gag-Pro-Pol polyprotein. The introduction of the T26S point mutation on the viral protease sequence originated the pGP(T26S)P plasmid.50 Plasmid pRSV-REV57 (Addgene #12253, kindly provided by Dr. Didier Trono) codes for the second and third exons of HIV-1 rev. These plasmids coding for the LV packaging functions were used in transient LV production.
Plasmids coding for envelope glycoproteins are as follows: pMD2.G (Addgene #12259, kindly provided by Dr. Didier Trono) codes for VSV-G; pCMV-GaLV-TR codes for a modified glycoprotein of the GaLV SEATO strain and results from the removal of 19 nt prior to the start codon of the GaLV glycoprotein from phGaLV10A1 by inverse PCR; phGaLV10A1 was kindly provided by Dr. Otto Merten (Généthon, Évry, France); pCMV-RD114-TR codes for a modified RD114 glycoprotein, amplified from the pLTR-RD114A19 plasmid (Addgene #17576, kindly provided by Dr. Jakob Reiser) and cloned into the vector resultant from phGaLV10A1 restriction with EcoRI and KasI enzymes; pCMV-RDpro codes for RDpro43 glycoprotein, which was chemically synthesized (GeneScript, Piscataway, NJ, USA) and cloned into the vector resultant from phGaLV10A1 restriction with EcoRI and KasI enzymes; pCMV-4070A codes for the amphotropic MLV glycoprotein amplified from pMonoZeo-4070A50 and cloned into the vector resultant from phGaLV10A1 restriction with EcoRI and KasI enzymes; pCMV-GaLV-TRpro, pCMV-RD114-TRpro, and pCMV-4070Apro code for the respective modified viral glycoproteins in which the viral protease cleavage sequence VQALVLTQ of the 4070A glycoprotein cytoplasmic tail was replaced by that of the HIV-1 Gag matrix-capsid, SQNYPIVQ, by inverse PCR from the parental plasmids; pCMV-GaLV-TRsynt, pCMV-RD114-TRsynt, and pCMV-4070Asynt code for the respective viral modified glycoproteins in which the viral protease cleavage sequence VQALVLTQ of the 4070A glycoprotein cytoplasmic tail was replaced by synthetic peptide GSGIFLETSL52 by performing two inverse PCRs from the parental plasmids; pCMV-GaLV-TRΔR, pCMV-RD114-TRΔR, and pCMV-4070AΔR code for the respective truncated glycoproteins in which the R-peptide was deleted from the cytoplasmic tail of the glycoproteins and replaced by a STOP codon by inverse PCR.
The primers and templates used in plasmid constructions, as well as the cloning strategies, are described in Table S1.
Transient LV Production
Transient LV production was performed by transfecting 293T cells as described by Tomás et al. (2018).50 Briefly, 6 × 104 cells per square centimeter were seeded in tissue culture flasks and, 24 hours later, transfected using linear 25 kDa polyethyleneimine (PEI; Polysciences, Hirschberg an der Bergstrasse, Germany) at a mass ratio of 1:1.5 (DNA:PEI), with the respective plasmids. The amount of each viral component per million cells was the following: 2.5 μg vector genome; 1 μg Gag-Pro-Pol; 0.25 μg Rev; 0.9 μg envelope. Medium was exchanged 24 hours after transfection. 24 hours after medium exchange, the supernatant was collected, clarified at 0.45 μm, and stored at −80°C.
Transducing LV Particle Titers
The transducing LV titer determination was performed by transducing 293T cells with the produced supernatants followed by flow cytometry analysis for GFP expression, as described by Tomás et al. (2018).50 The concentration of LV TUs (TU ⋅ mL−1) was calculated using the following equation:
Physical LV Particle Titers
The concentration of p24 LV protein in supernatants was determined by a p24 ELISA using the INNOTEST HIV Antigen mAb (Fujirebio Diagnostics, Malvern, PA, USA), following the manufacturer’s instructions. It was assumed that 1 ng p24 corresponds to 1.25 × 107 physical particles.58 The titer of LV PPs (PP ⋅ mL−1) was estimated using the following equation:
Statistical Analysis
To assess the significance of differences seen among titers of LV-pseudotype production, statistical analysis was used to evaluate data from multiple experiments using GraphPad Prism v5 (GraphPad Software, San Diego, CA, USA). For the comparison of LV titers differences on p values <0.05, using unpaired Student t test (two-tailed), were considered significant.
Author Contributions
Conceptualization, H.A.T., D.A.M., A.F.R., and A.S.C.; Investigation, H.A.T.; Writing – Original Draft, H.A.T.; Writing – Review & Editing, H.A.T., D.A.M., A.F.R., M.R.G., M.J.T.C., and A.S.C.; Supervision, M.J.T.C. and A.S.C.
Conflicts of Interest
The Instituto de Biologia Experimental e Tecnológica filed a provisional patent application related to this study.
Acknowledgments
The authors acknowledge iNOVA4Health Research Unit (LISBOA-01-0145-FEDER-007344), which is cofunded by Fundação para a Ciência e Tecnologia (FCT)/Ministério da Ciência e do Ensino Superior, through national funds, and by FEDER under the PT2020 Partnership Agreement. Financial support was received directly from FCT, Portugal (PTDC/EBB-EBI/118621/2010). H.A.T and M.R.G. acknowledge FCT for the award of the individual grants SFRH/BD/79022/2011 and SFRH/BD/90685/2012, respectively. A.F.R. acknowledges FCT for the award of the postdoctoral fellowship SFRH/BPD/111678/2015.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.08.001.
Supplemental Information
References
- 1.Vigna E., Naldini L. Lentiviral vectors: excellent tools for experimental gene transfer and promising candidates for gene therapy. J. Gene Med. 2000;2:308–316. doi: 10.1002/1521-2254(200009/10)2:5<308::AID-JGM131>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.De Palma M., Montini E., Santoni de Sio F.R., Benedicenti F., Gentile A., Medico E., Naldini L. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- 3.Beard B.C., Dickerson D., Beebe K., Gooch C., Fletcher J., Okbinoglu T., Miller D.G., Jacobs M.A., Kaul R., Kiem H.P., Trobridge G.D. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol. Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 4.Edelstein M. 2017. The Journal of Gene Medicine Clinical Trial at.http://www.abedia.com/wiley/index.html [Google Scholar]
- 5.Swanstrom R., Wills J. Synthesis, assembly, and processing of viral proteins. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor Laboratory Press; New York: 1997. pp. 263–334. [PubMed] [Google Scholar]
- 6.Verhoeyen E., Cosset F.-L. Surface-engineering of lentiviral vectors. J. Gene Med. 2004;6(Suppl 1):S83–S94. doi: 10.1002/jgm.494. [DOI] [PubMed] [Google Scholar]
- 7.Amirache F., Lévy C., Costa C., Mangeot P.-E., Torbett B.E., Wang C.X., Nègre D., Cosset F.L., Verhoeyen E. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- 8.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cronin J., Zhang X.-Y., Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merten O.-W., Hebben M., Bovolenta C. Production of lentiviral vectors. Mol. Ther. Methods Clin. Dev. 2016;3:16017. doi: 10.1038/mtm.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomás H.A., Rodrigues A.F., Alves P.M., Coroadinha A.S. Lentiviral gene therapy vectors: challenges and future directions. In: Molina F.M., editor. Gene Therapy - Tools and Potential Applications. IntechOpen; 2013. [Google Scholar]
- 12.Rodrigues A.M.P., Coroadinha A. Production of retroviral and lentiviral gene therapy vectors: challenges in the manufacturing of lipid enveloped virus. In: Xu K., editor. Viral Gene Therapy. IntechOpen; 2011. [Google Scholar]
- 13.Marandin A., Dubart A., Pflumio F., Cosset F.L., Cordette V., Chapel-Fernandes S., Coulombel L., Vainchenker W., Louache F. Retrovirus-mediated gene transfer into human CD34+38low primitive cells capable of reconstituting long-term cultures in vitro and nonobese diabetic-severe combined immunodeficiency mice in vivo. Hum. Gene Ther. 1998;9:1497–1511. doi: 10.1089/hum.1998.9.10-1497. [DOI] [PubMed] [Google Scholar]
- 14.Glimm H., Kiem H.P., Darovsky B., Storb R., Wolf J., Diehl V., Mertelsmann R., Von Kalle C. Efficient gene transfer in primitive CD34+/CD38lo human bone marrow cells reselected after long-term exposure to GALV-pseudotyped retroviral vector. Hum. Gene Ther. 1997;8:2079–2086. doi: 10.1089/hum.1997.8.17-2079. [DOI] [PubMed] [Google Scholar]
- 15.Kelly P.F., Vandergriff J., Nathwani A., Nienhuis A.W., Vanin E.F. Highly efficient gene transfer into cord blood nonobese diabetic/severe combined immunodeficiency repopulating cells by oncoretroviral vector particles pseudotyped with the feline endogenous retrovirus (RD114) envelope protein. Blood. 2000;96:1206–1214. [PubMed] [Google Scholar]
- 16.Porter C.D., Collins M.K.L., Tailor C.S., Parkar M.H., Cosset F.-L., Weiss R.A., Takeuchi Y. Comparison of efficiency of infection of human gene therapy target cells via four different retroviral receptors. Hum. Gene Ther. 1996;7:913–919. doi: 10.1089/hum.1996.7.8-913. [DOI] [PubMed] [Google Scholar]
- 17.Movassagh M., Desmyter C., Baillou C., Chapel-Fernandes S., Guigon M., Klatzmann D., Lemoine F.M. High-level gene transfer to cord blood progenitors using gibbon ape leukemia virus pseudotype retroviral vectors and an improved clinically applicable protocol. Hum. Gene Ther. 1998;9:225–234. doi: 10.1089/hum.1998.9.2-225. [DOI] [PubMed] [Google Scholar]
- 18.Kiem H.P., Heyward S., Winkler A., Potter J., Allen J.M., Miller A.D., Andrews R.G. Gene transfer into marrow repopulating cells: comparison between amphotropic and gibbon ape leukemia virus pseudotyped retroviral vectors in a competitive repopulation assay in baboons. Blood. 1997;90:4638–4645. [PubMed] [Google Scholar]
- 19.Zhang X.-Y., La Russa V.F., Reiser J. Transduction of bone-marrow-derived mesenchymal stem cells by using lentivirus vectors pseudotyped with modified RD114 envelope glycoproteins. J. Virol. 2004;78:1219–1229. doi: 10.1128/JVI.78.3.1219-1229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanawa H., Kelly P.F., Nathwani A.C., Persons D.A., Vandergriff J.A., Hargrove P., Vanin E.F., Nienhuis A.W. Comparison of various envelope proteins for their ability to pseudotype lentiviral vectors and transduce primitive hematopoietic cells from human blood. Mol. Ther. 2002;5:242–251. doi: 10.1006/mthe.2002.0549. [DOI] [PubMed] [Google Scholar]
- 21.Relander T., Johansson M., Olsson K., Ikeda Y., Takeuchi Y., Collins M., Richter J. Gene transfer to repopulating human CD34+ cells using amphotropic-, GALV-, or RD114-pseudotyped HIV-1-based vectors from stable producer cells. Mol. Ther. 2005;11:452–459. doi: 10.1016/j.ymthe.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Mühlebach M.D., Schmitt I., Steidl S., Stitz J., Schweizer M., Blankenstein T., Cichutek K., Uckert W. Transduction efficiency of MLV but not of HIV-1 vectors is pseudotype dependent on human primary T lymphocytes. J. Mol. Med. (Berl.) 2003;81:801–810. doi: 10.1007/s00109-003-0491-2. [DOI] [PubMed] [Google Scholar]
- 23.Strang B.L., Ikeda Y., Cosset F.-L., Collins M.K.L., Takeuchi Y. Characterization of HIV-1 vectors with gammaretrovirus envelope glycoproteins produced from stable packaging cells. Gene Ther. 2004;11:591–598. doi: 10.1038/sj.gt.3302189. [DOI] [PubMed] [Google Scholar]
- 24.Page K.A., Landau N.R., Littman D.R. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 26.He J., Landau N.R. Use of a novel human immunodeficiency virus type 1 reporter virus expressing human placental alkaline phosphatase to detect an alternative viral receptor. J. Virol. 1995;69:4587–4592. doi: 10.1128/jvi.69.7.4587-4592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiser J., Harmison G., Kluepfel-Stahl S., Brady R.O., Karlsson S., Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc. Natl. Acad. Sci. USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandrin V., Boson B., Salmon P., Gay W., Nègre D., Le Grand R., Trono D., Cosset F.L. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 29.Sandrin V., Muriaux D., Darlix J.-L., Cosset F.-L. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J. Virol. 2004;78:7153–7164. doi: 10.1128/JVI.78.13.7153-7164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell A.J., Jr., Fegen D., Ward M., Bank A. RD114 envelope proteins provide an effective and versatile approach to pseudotype lentiviral vectors. Exp. Biol. Med. (Maywood) 2010;235:1269–1276. doi: 10.1258/ebm.2010.010053. [DOI] [PubMed] [Google Scholar]
- 31.Christodoulopoulos I., Cannon P.M. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 2001;75:4129–4138. doi: 10.1128/JVI.75.9.4129-4138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stitz J., Buchholz C.J., Engelstädter M., Uckert W., Bloemer U., Schmitt I., Cichutek K. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology. 2000;273:16–20. doi: 10.1006/viro.2000.0394. [DOI] [PubMed] [Google Scholar]
- 33.Apte S., Sanders D.A. Effects of retroviral envelope-protein cleavage upon trafficking, incorporation, and membrane fusion. Virology. 2010;405:214–224. doi: 10.1016/j.virol.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Rein A., Mirro J., Haynes J.G., Ernst S.M., Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragheb J.A., Anderson W.F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Januszeski M.M., Cannon P.M., Chen D., Rozenberg Y., Anderson W.F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubo Y., Amanuma H. Mutational analysis of the R peptide cleavage site of Moloney murine leukaemia virus envelope protein. J. Gen. Virol. 2003;84:2253–2257. doi: 10.1099/vir.0.19126-0. [DOI] [PubMed] [Google Scholar]
- 38.Aguilar H.C., Anderson W.F., Cannon P.M. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R Peptide. J. Virol. 2003;77:1281–1291. doi: 10.1128/JVI.77.2.1281-1291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubo Y., Tominaga C., Yoshii H., Kamiyama H., Mitani C., Amanuma H., Yamamoto N. Characterization of R peptide of murine leukemia virus envelope glycoproteins in syncytium formation and entry. Arch. Virol. 2007;152:2169–2182. doi: 10.1007/s00705-007-1054-6. [DOI] [PubMed] [Google Scholar]
- 40.Schneider I.C., Eckhardt M., Brynza J., Collins M.K., Cichutek K., Buchholz C.J. Escape from R-peptide deletion in a γ-retrovirus. Virology. 2011;418:85–92. doi: 10.1016/j.virol.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 41.Bobkova M., Stitz J., Engelstädter M., Cichutek K., Buchholz C.J. Identification of R-peptides in envelope proteins of C-type retroviruses. J. Gen. Virol. 2002;83:2241–2246. doi: 10.1099/0022-1317-83-9-2241. [DOI] [PubMed] [Google Scholar]
- 42.Löving R., Li K., Wallin M., Sjöberg M., Garoff H. R-Peptide cleavage potentiates fusion-controlling isomerization of the intersubunit disulfide in Moloney murine leukemia virus Env. J. Virol. 2008;82:2594–2597. doi: 10.1128/JVI.02039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikeda Y., Takeuchi Y., Martin F., Cosset F.-L., Mitrophanous K., Collins M. Continuous high-titer HIV-1 vector production. Nat. Biotechnol. 2003;21:569–572. doi: 10.1038/nbt815. [DOI] [PubMed] [Google Scholar]
- 44.Reiser J. Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 2000;7:910–913. doi: 10.1038/sj.gt.3301188. [DOI] [PubMed] [Google Scholar]
- 45.Sena-Esteves M., Tebbets J.C., Steffens S., Crombleholme T., Flake A.W. Optimized large-scale production of high titer lentivirus vector pseudotypes. J. Virol. Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Boudeffa D., Fenard D., Mormin M., Galy A., Merten O.W. 534. Development of innovative scalable protocols for the purification of lentiviral vectors pseudotyped with GaLV-TR or mutated measles virus glycoproteins. Mol. Ther. 2015;23(Supp. 1):S214–S215. [Google Scholar]
- 47.Stornaiuolo A., Piovani B.M., Bossi S., Zucchelli E., Corna S., Salvatori F., Mavilio F., Bordignon C., Rizzardi G.P., Bovolenta C. RD2-MolPack-Chim3, a packaging cell line for stable production of lentiviral vectors for anti-HIV gene therapy. Hum. Gene Ther. Methods. 2013;24:228–240. doi: 10.1089/hgtb.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanber K.S., Knight S.B., Stephen S.L., Bailey R., Escors D., Minshull J., Santilli G., Thrasher A.J., Collins M.K., Takeuchi Y. Construction of stable packaging cell lines for clinical lentiviral vector production. Sci. Rep. 2015;5:9021. doi: 10.1038/srep09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marin V., Stornaiuolo A., Piovan C., Corna S., Bossi S., Pema M., Giuliani E., Scavullo C., Zucchelli E., Bordignon C. RD-MolPack technology for the constitutive production of self-inactivating lentiviral vectors pseudotyped with the nontoxic RD114-TR envelope. Mol. Ther. Methods Clin. Dev. 2016;3:16033. doi: 10.1038/mtm.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomás H.A., Rodrigues A.F., Carrondo M.J.T., Coroadinha A.S. LentiPro26: novel stable cell lines for constitutive lentiviral vector production. Sci. Rep. 2018;8:5271. doi: 10.1038/s41598-018-23593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tözsér J. Comparative studies on retroviral proteases: substrate specificity. Viruses. 2010;2:147–165. doi: 10.3390/v2010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beck Z.Q., Hervio L., Dawson P.E., Elder J.H., Madison E.L. Identification of efficiently cleaved substrates for HIV-1 protease using a phage display library and use in inhibitor development. Virology. 2000;274:391–401. doi: 10.1006/viro.2000.0420. [DOI] [PubMed] [Google Scholar]
- 53.Henderson L.E., Sowder R., Copeland T.D., Smythers G., Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J. Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green N., Shinnick T.M., Witte O., Ponticelli A., Sutcliffe J.G., Lerner R.A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc. Natl. Acad. Sci. USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konvalinka J., Litterst M.A., Welker R., Kottler H., Rippmann F., Heuser A.M., Kräusslich H.G. An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J. Virol. 1995;69:7180–7186. doi: 10.1128/jvi.69.11.7180-7186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao Y., Ghosh K., Epand R.F., Epand R.M., Ghosh H.P. Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low pH but not by heat or denaturant. Virology. 2003;310:319–332. doi: 10.1016/s0042-6822(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 57.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valkama A.J., Leinonen H.M., Lipponen E.M., Turkki V., Malinen J., Heikura T., Ylä-Herttuala S., Lesch H.P. Optimization of lentiviral vector production for scale-up in fixed-bed bioreactor. Gene Ther. 2018;25:39–46. doi: 10.1038/gt.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.