Abstract
Background
Photosynthetic efficiency might be a key factor determining plant resistance to abiotic stresses. Plants can sense when growing conditions are not favorable and trigger an internal response at an early stage before showing external symptoms. When a high amount of salt enters the plant cell, the membrane system and function of thylakoids in chloroplasts could be destroyed and affect photosynthetic performance if the salt concentration is not regulated to optimal values. Oryza species have salt-tolerant and salt-sensitive genotypes; however, very few studies have investigated the genetic architecture responsible for photosynthetic efficiency under salinity stress in cultivated rice.
Results
We used an imaging-based chlorophyll fluorometer to monitor eight rice varieties that showed different salt tolerance levels for four consecutive days under control and salt conditions. An analysis of the changes in chlorophyll fluorescence parameters clearly showed the maximum quantum efficiency of PSII in sensitive varieties was significantly reduced after NaCl treatment when compared to tolerant varieties. A panel of 232 diverse rice accessions was then analyzed for chlorophyll fluorescence under salt conditions, the results showed that chlorophyll fluorescence parameters such as F0 and NPQ were higher in Japonica subspecies, ΦPSII of Indica varieties was higher than that in other subgroups, which suggested that the variation in photosynthetic efficiency was extensively regulated under salt treatment in diverse cultivated rice. Two significant regions on chromosome 5 were identified to associate with the fraction of open PSII centers (qL) and the minimum chlorophyll fluorescence (F0). These regions harbored genes related to senescence, chloroplast biogenesis and response to salt stress are of interest for future functional characterization to determine their roles in regulating photosynthesis.
Conclusions
Rice plant is very sensitive to salinity stress, especially at young seedling stage. Our work identified the distribution pattern of chlorophyll fluorescence parameters in seedlings leaf and their correlations with salt tolerance level in a diverse gene pool. We also revealed the complexity of the genetic architecture regulating rice seedling photosynthetic performance under salinity stress, the germplasm analyzed in this study and the associated genetic information could be utilized in rice breeding program.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1983-8) contains supplementary material, which is available to authorized users.
Keywords: Oryza sativa, Photosynthetic efficiency, Chlorophyll fluorescence, Salt stress, Genetic variation, Subspecies, Genome-wide association
Background
Photosynthesis is an essential process that generate energy to support plant growth. Components participating in photosynthetic machinery, such as photosynthetic pigments, photosystems, electron transport systems, gas-exchange processes and enzymes involved in carbon metabolism, are important for photosynthetic efficiency and could be potentially affected by abiotic stresses [1].
In recent years, technology to detect chlorophyll fluorescence in leaves has advanced rapidly, and its power to reflect photosynthetic efficiency in vivo needs to be examined. Light energy captured by chlorophyll molecules is used in photochemical reactions to drive photosynthesis or, if in excess, dissipated as heat (nonphotochemical quenching, NPQ) or emitted as chlorophyll fluorescence [2]. These three processes mutually compete with each other, e.g., increasing photosynthesis will lead to a decrease in extra energy dissipation; therefore, by measuring the yield of chlorophyll fluorescence, the efficiency of the photochemical reactions and the degree of heat dissipation could also be estimated.
When a high amount of salt was accumulated in the plant cell for a period of time, the membrane permeability and function of thylakoids in chloroplasts was damaged [3]; a gradual decrease in the activity of photosystems (PSI and PSII) and chlorophyll fluorescence was also observed [4]. Because the repair of photosystem II (PSII) was affected in salt-treated plants, photoinhibition was enhanced, and the photosynthetic efficiency was reduced [5]. The correlation between chlorophyll fluorescence and chloroplast ultrastructure, such as the morphology of thylakoids, under salt stress has been examined in barley and rice [4, 6, 7]. These studies showed that salt-tolerant varieties had reduced thylakoid swelling, relatively high PSII electron transport activity, slowly decreasing maximum chlorophyll fluorescence yield and delayed senescence.
Compared to its physiological and biochemical regulation, the genetic nature of photosynthetic efficiency has been less explored [8, 9]. The activity of plant photosynthesis could be expressed and quantified in many forms, such as rate of CO2 fixation (gas-exchange) per unit leaf area, production of carbohydrates or dry mass per plant, and chlorophyll fluorescence. To investigate the genetic characteristics of plant photosynthetic efficiency, researchers analyzed the properties of diverse genotypes: the response of photosynthetic light use efficiency (ΦPSII) to different light conditions in Arabidopsis [10], chlorophyll fluorescence kinetics under heat stress in wheat [11], leaf chlorophyll content in rice [12–14], photosynthetic responses of rice seedlings to salinity stress [15], cold responses of photosynthesis in Arabidopsis and maize [16, 17] and other traits (summarized in Table 1 of Flood et al. [8]). These results highlighted that photosynthetic efficiency was regulated differentially in diverse genetic backgrounds.
Table 1.
The eight accessions used in the pilot experiment
| ID | Name | Country of origin | Subspecies | Injury scoreb | Salinity tolerance levelc |
|---|---|---|---|---|---|
| 8777 | Munao PS405 | Philippines | Indica | 7.86 | S |
| IR28 | IR28 | Philippines | Indica | 5.2 | M to S |
| IR64 | IR64 | Philippines | Indica | 5.12 | M to S |
| 223 | Priano Guaira | Brazil | Japonica | 5 | M |
| Nona Bokra | Nona Bokra | India | Indica | 2.1 | M to T |
| 149 | Sinaguing | Philippines | Japonica | 2 | M to T |
| 245a | Sab Ini | Egypt | Japonica | 1.57 | M to T |
| IR66946 | FL478 | Philippines | Indica | 1 | T |
aVariety 245 was excluded from discussion due to its erratic behavior; it is likely that the tested samples were contaminated
bThe injury score was the average of three replicates
cSalinity tolerance level was assigned to five classes based on the injury score. M Moderate (Injury score = 5), S Sensitive (Injury score > = 7), T Tolerant (Injury score = 1)
Several studies have examined changes in chlorophyll fluorescence parameters under salinity stress in rice. F0 (minimum fluorescence yield in the absence of photosynthetic light) increased under the salinity stress [7, 18, 19]. Chloroplast ultrastructure, such as the thylakoid membrane, was damaged by salt stress [20]. It is possible that increased F0 values were due to the disassociation of light-harvesting complex II (LHC II) and the PSII reaction center on swollen thylakoids [7]; however, Fm (maximum fluorescence yield in the absence of photosynthetic light) was decreased in plants under salinity stress compared with control plants [7, 18]. Fv/Fm (maximum efficiency of PSII) is highly consistent (approximately 0.78–0.84) in different species under normal growth conditions; it was decreased in sensitive varieties under moderate salt stress or a long period of salt treatment [7, 15, 18, 19, 21, 22]. ΦPSII, which is also the effective PSII quantum yield, represents the fraction of absorbed energy used in photochemistry, which determines the efficiency of PSII. It is affected by the rate of electron transport or the concentrations of electron acceptors, e.g., NADP+, available at the acceptor side of PSI. The value of ΦPSII declined under moderate salt stress, and the tolerant lines exhibited higher values than the sensitive lines [15]. However, another study [23] had the opposite result; ΦPSII did not change, but the electron transport rate (ETR) declined, and they speculated that photosynthetically active radiation (PAR) was not uniform in the experiment. Another study also indicated that ΦPSII decreased severely when Fv/Fm decreased slightly under different levels of salinity stress [7]. The parameter qL is the coefficient of photochemical quenching based on a lake model in which the PSII reaction centers are connected by shared antenna [24]. qL reflects the fraction of open PSII reaction centers, which is highly correlated with ΦPSII and declines under salt stress in sensitive varieties [21]. NPQ reflects the level of excess energy dissipation as heat. A previous study showed that the NPQ value increased in tolerant varieties and decreased in sensitive varieties under salt stress [21], but in another study, NPQ in both tolerant and sensitive lines was increased substantially under salt stress [23]. It is still not clear how NPQ is related to plant salt tolerance levels.
An image-based phenotyping platform has been applied to increase the accuracy and throughput of the trait evaluation process in studying the effect of abiotic stress in various crops [25, 26]. A recent study combined digital imaging and an automated robotic system (the Plant Accelerator) to measure the relative growth rate (RGR), transpiration rate (TR) and transpiration use efficiency (TUE) in diverse rice accessions and identified several new genetic loci controlling the early responses to salinity through association studies [27]. In this work, we aimed to understand how natural genetic variation correlates with plant photosynthetic efficiency under salinity stress using a chlorophyll fluorometer. The leaf chlorophyll fluorescence of eight rice varieties that exhibited different levels of salt tolerance were monitored for four consecutive days under control and salinity conditions. The results were examined to determine how internal chlorophyll fluorescence parameters correspond to the symptoms of salt toxicity in older seedlings. Based on these findings, we further investigated a diverse panel of 232 rice cultivars for their seedling leaf chlorophyll fluorescence performance under salt conditions. The genetic architecture controlling photosynthetic efficiency was analyzed based on the association of selected chlorophyll fluorescence parameters and SNP variations.
Results
Evaluating the performance of leaf fluorescence parameters in time series under control and salinity conditions using eight rice varieties that show different salt tolerance levels
In the pilot experiment, we aimed to identify the optimal seedling stage and target leaf tissue displaying a wide range of chlorophyll fluorescence variation under salt stress. We selected eight rice varieties based on their salt tolerance level and classified them as tolerant, moderate and sensitive (Table 1).
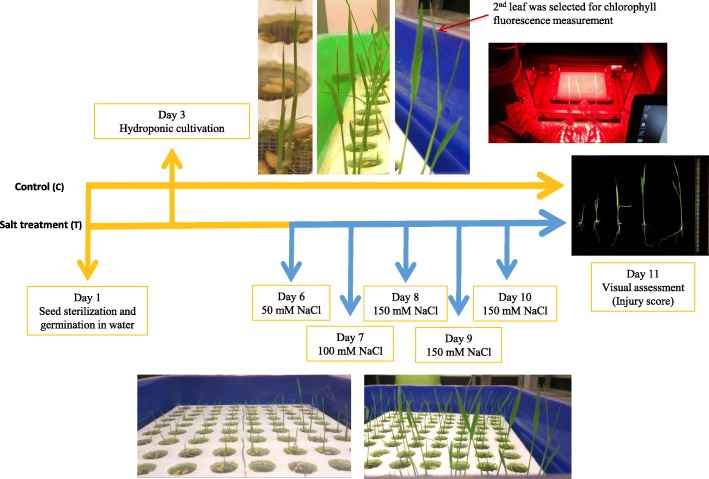
To synchronize seed germination, we soaked sterilized seeds in water at 37 °C for 2 days until the embryonic plumule protruded. After growing in the hydroponic system for 3 days, rice seedlings were treated with salt levels that increased daily (50, 100, 150 mM of NaCl) over subsequent days (days 6, 7, 8) and then maintained at 150 mM for 3 days for a visual evaluation of salt tolerance (injury score, IS) on day 11. As shown in Fig. 1, the second leaf was the major photosynthetic organ from day 6 to day 9 and was fully expanded on day 8 under both control and salt conditions. To assess the changes in the fluorescence parameters under the control and salt treatments at the young seedling stage, the second leaves of eight varieties were evaluated before starting 150 mM NaCl treatment (day 8) and 24, 48, and 72 h after 150 mM NaCl treatment (day 9, 10, 11) using a pulse-amplitude modulation (PAM) fluorometer (Fig. 1). The second leaf of the control group from the same eight varieties was evaluated at the same time point.
Fig. 1.
Experimental workflow of the rice salinity regime and phenotyping at the seedling stage. Rice seeds were sterilized and germinated at 37 °C for two days. Germinated seeds were transferred to the hydroponic system on day 3. Salinity treatment was started from 50 mM NaCl on day 6 and progressively increased to 150 mM on day 8, which allowed the seedling to acclimate to salt stress. The fully developed second leaf was selected for chlorophyll fluorescence measurement from day 8 to day 11. Salt tolerance level was evaluated and assigned an injury score on day 11
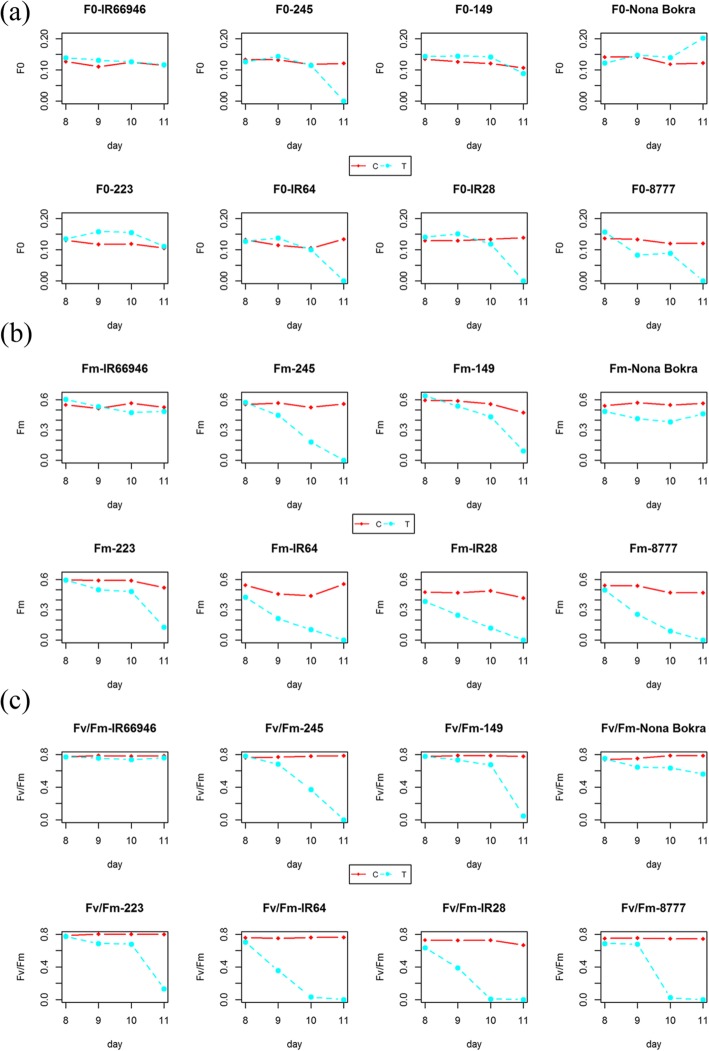
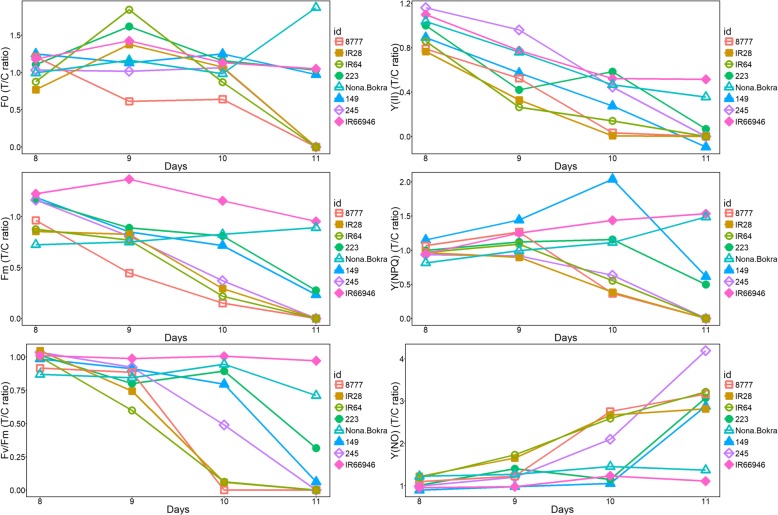
As shown in Fig. 2 and S1, the changes in six chlorophyll fluorescence parameters were relatively stable in the control group (red line) compared to the salt treatment group (blue line) from day 8 to day 11. A varietal difference was observed in the parameters of the control group such as ΦPSII, qL and NPQ, which strongly suggested that the photosynthetic potential varies in nature by genotype and could be regulated genetically. The F0, Fm and Fv/Fm values of all varieties were stable under control conditions over the observation period (red line in Fig. 2) but declined at different rates in the tested accessions except the extremely tolerant variety IR66946 under salt treatment (blue line in Fig. 2). For the parameters associated with energy partitioning, such as ΦPSII, qL and NPQ, the pattern of chlorophyll fluorescence was difficult to interpret when control and salt treatment profiles were compared side by side across varieties (Additional file 7: Figure S1). It is likely that the capacity of the photosystem was already compromised in both the control and salt treatment groups, which compounded with the natural variation in photosynthetic efficiency that already existed between genotypes. To accurately evaluate the salt tolerance level through chlorophyll fluorescence across genotypes, the ratio of treatment to control (T/C) values for each parameter was then calculated and used to infer the regulation of photosynthetic efficiency (Fig. 3; Additional file 2: Table S2). Y(II) significantly declined in the second leaves of all seedlings treated with 150 mM NaCl compared to the control seedlings from days 8 to 11. Both the quantum yield of regulated [Y(NPQ)] and nonregulated [Y(NO)] energy dissipation were elevated from day 8 to day 9 in the sensitive varieties (Fig. 3). The Y(NPQ) increased from days 8 to 11 in the tolerant varieties IR66946 and Nona Bokra but decreased from day 9 to 11 in the sensitive varieties. The NPQ was regulated in response to salt stress in moderate varieties 149 and 223 and in tolerant varieties Nona Bokra and IR66946 at day 10. The actual quantum yield of nonregulated NPQ, Y(NO), remained low in the moderate and tolerant varieties (IR66946, Nona Bokra, 149 and 223) compared to the sensitive variety 8777 under salt treatment at day 10.
Fig. 2.
Daily changes in chlorophyll fluorescence parameters in eight rice varieties under control and salt conditions. The data in (a), (b), and (c) represent F0, Fm and Fv/Fm, respectively. Each data point represents the average value of 3 replicates. Red line represents control sample; blue line represents salt treated sample
Fig. 3.
Regulation of the chlorophyll fluorescence index in eight rice varieties under salinity stress. The T/C ratio of chlorophyll fluorescence (F0, Fm, and Fv/Fm) and quantum yield (Y(II), Y(NPQ), and Y(NO)) parameters were determined in control (C) and treatment (T) groups from day 8 to day 10. Each variety is indicated by a different colored line and symbol. 149: Blue/ filled triangle, 223: green/ filled circle, 245: violet/ open diamond, 8777: orange/ open square, IR28: brown/ filled square, IR64: light green/ open circle, IR66946: pink/ close diamond, Nona Bokra: jade/ open triangle
The time point displaying the maximum difference in photosynthetic efficiency in seedling leaves of diverse varieties under salt stress
The second leaves displayed no significant visual difference between the control and salt-treated groups before the salt concentration increased to 150 mM NaCl on day 8. Twenty-four hours after 150 mM NaCl incubation began (day 9), the seedlings of the sensitive varieties 8777 and IR28 started to show weak senescence symptoms in the second leaf. Forty-eight hours after 150 mM NaCl incubation began (day 10), the moderate and sensitive varieties started withering; moderate to tolerant varieties started showing mild senescence symptoms on day 11, and the extremely tolerant varieties still looked healthy. We suspected that the photosynthetic machinery had already been affected since the first day of salt treatment (50 mM on day 6, Fig. 1); however, the phenotypic effect was not shown until the leaf tissue started senescing and dying from day 9 to day 11, which depended on the tolerance level of the variety. We closely examined the changes in the six chlorophyll fluorescence parameters across eight varieties from day 8 to day 11. The pattern of each parameter varied greatly among eight varieties in our time-series observation, suggesting that chlorophyll fluorescence is a sensitive and complex phenomenon that reflects the seedling’s salt tolerance level. Fv/Fm and ΦPSII values of salt-treated moderate and sensitive varieties were close to 0 on day 10 (Fig. 2c and Additional file 7: Figure S1), which cannot be treated as a quantitative phenotype in a genome-wide association study (GWAS). We then decided to measure six chlorophyll fluorescence parameters on day 9 in 223 diverse rice accessions under salt stress and used these measurements as quantitative traits for follow-up association analysis (Additional file 3: Table S3).
Distribution of chlorophyll fluorescence parameters under salinity stress in 232 diverse rice varieties
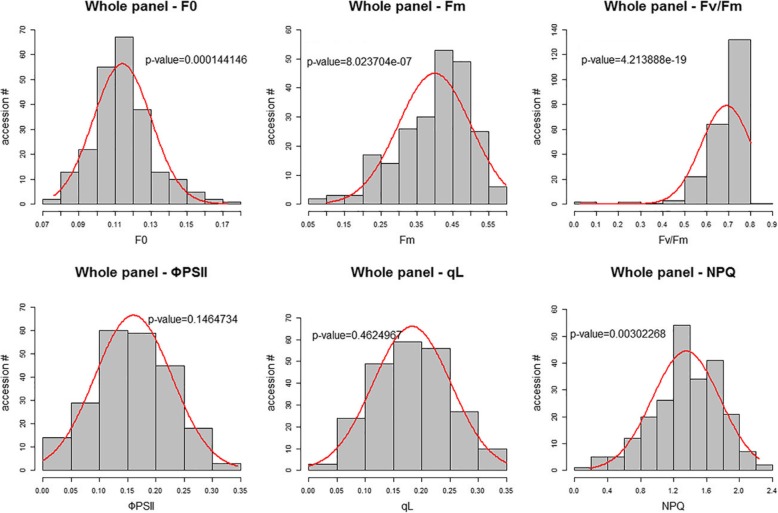
After 150 mM salt treatment for 2 days, Fv/Fm was between 0.7 and 0.8 in the majority of the accessions, as we expected; F0, Fm, ΦPSII, qL and NPQ values showed a normal distribution in 232 diverse accessions (Fig. 4). We then investigated the variation in the parameters in five subgroups and two subspecies and found that Japonica subspecies have significantly higher F0 and NPQ values than Indica subspecies (P-value < 0.0001 by Student’s t-test, Additional file 8: Figure S2); within Japonica subspecies, tropical japonica varieties have higher F0 values than other varieties, and aromatic varieties have higher NPQ values (Additional file 8: Figure S2a and f). ΦPSII of the Indica subgroup is slightly higher than that in the other four subgroups (Additional file 8: Figure S2d), and for Fm, Fv/Fm and qL, we did not detect a significant difference between the two subspecies (Additional file 8: Figure S2b, c and e). The value of chlorophyll fluorescence parameters were widely distributed in each subgroup, which could reflect the extensive genetic variation in photosynthetic efficiency in rice.
Fig. 4.
Distribution of six chlorophyll fluorescence parameters in 232 diverse varieties. Each histogram showed the distribution of chlorophyll fluorescence parameter in 232 rice accessions. The normality of each parameter was examined using the Shapiro-Wilks test. The P-value of each test is provided
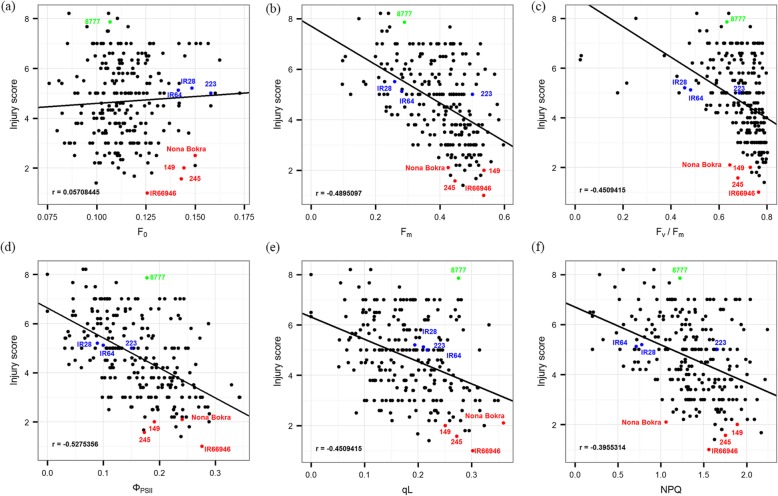
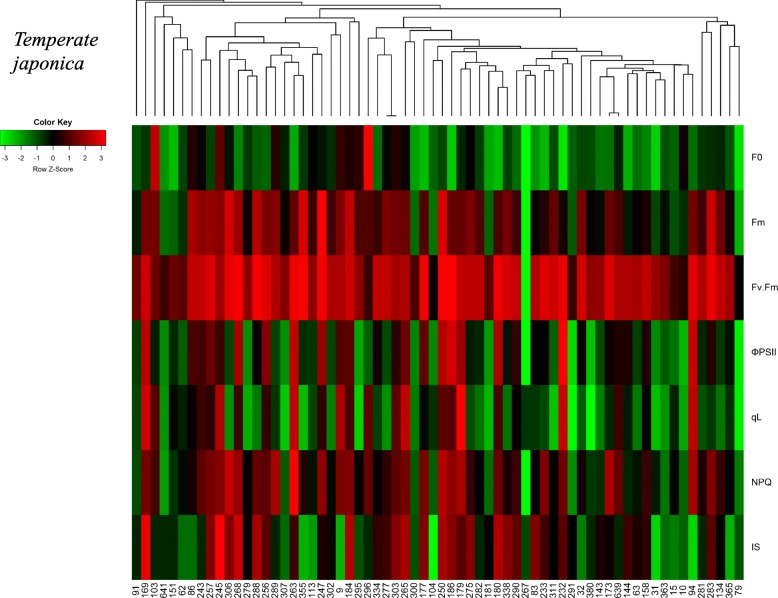
To evaluate how the chlorophyll fluorescence of these 232 rice accessions recorded on day 9 was related to their visual salt toxicity symptoms (i.e., injury score) observed on day 11, we plotted these variables in pairs and calculated their correlations (Fig. 5 and Additional file 4: Table S4). The five fluorescence parameters other than F0 were moderately correlated with injury score. Considering that chlorophyll fluorescence parameters differed among subpopulations, we investigated whether these parameters and the salt tolerance level were correlated by genetic relatedness in each subpopulation (temperate japonica in Fig. 6 and other five subpopulations in Additional file 9: Figure S3). Colors in the heatmap near red indicated that the value of each chlorophyll fluorescence parameter was higher than the average value of all accessions. For the injury score, red represented a lower injury score, which suggested that the variety was more tolerant to salinity toxicity than the other varieties. These results clearly showed that rice accessions with related genetic backgrounds do not necessarily share similar chlorophyll fluorescence patterns, which was reflected by their diverse responses to salt toxicity, strongly supporting the complexity of salt response mechanisms.
Fig. 5.
Pattern of correlation between injury score and each chlorophyll parameter. a to f showed distribution of injury score and F0, Fm, Fv/Fm, ΦPSII, qL and NPQ, respectively. Pearson correlation coefficient (r) represents the level of correlation between two traits. The variety labeled in color corresponds to the salt tolerance level described in Table 1
Fig. 6.
Genetic relationships and phenotypic distributions of varieties in temperate japonica populations. Each column represents a single variety, and the sample ID is labeled at the bottom of the heatmap. Each row represents the chlorophyll fluorescence parameter and injury score of each variety. The colors closest to red indicate that the value of each chlorophyll fluorescence parameter is higher than its average value of all accessions; for injury score, the red colors represent the lower injury scores, which suggest that the variety is more tolerant to salt toxicity. The color key of the Z-score was calculated from the distance between the raw score and the population mean in units of the standard deviation
GWAS of chlorophyll fluorescence parameters under salt stress
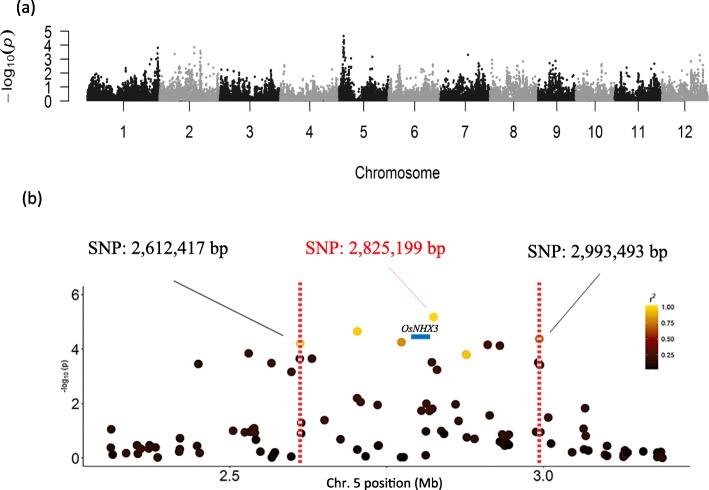
To identify the chromosomal regions that are potentially involved in the photosynthetic efficiency response to salt stress, we conducted GWAS using five fluorescence parameters in three panels – a whole panel (232 accessions, Fig. 2), a Japonica-specific panel (165 accessions, Additional file 10: Figure S4) and an Indica-specific panel (66 accessions, Additional file 11: Figure S5). Fv/Fm was excluded because of its narrow variation and skewed distribution (Fig. 2). For the whole panel, the P model, K model and P + K model were applied to reduce false-positive signals compared to the naïve model results; for Japonica-specific panels, only the K model was applied. According to the quantile-quantile (QQ) plot, the significant SNPs were selected by a P-value < 10− 4 in the P model and a P-value < 10− 3 in the K and P + K models, and the singleton SNP was not considered in further analysis. Manhattan plots and QQ plots of the SNPs associated with each chlorophyll fluorescence parameter in the whole panel and two subspecies-specific panels are provided in supplementary files (Additional file 12: Figure S6). To identify candidate genes in significant genomic regions, we calculated the linkage disequilibrium (LD) between the most significant SNP and its neighboring SNPs. The chromosomal region in high LD with a significant SNP could potentially harbor the causal gene affecting the photosynthetic efficiency under salt stress. Several photosynthesis-associated loci under salt stress were identified in this study (Table 2). Significant peaks on chromosome 1 (41.7–42.5 Mb) were detected for five different parameters (F0, Fm, ΦPSII, qL, NPQ). This region overlapped with one quantitative trait locus (QTL), and this QTL was related to seedling height under salt stress [28]. The Ygl8 gene encoding a UMP kinase (LOC_Os01g73450) was found near this significant SNP cluster. UMP kinase may control the UMP/UMP/UTP level in chloroplasts. Chlorophyll biogenesis was affected in a mutant of Ygl8, and the chloroplast ultrastructure of Ygl8 showed that the lamellae were intricately stacked and swollen [32]. Another significant peak, located on chromosome 6 from 2.9 to 3.05 Mb, was observed for four traits (Fm, ΦPSII, qL, NPQ) (Table 2). Fm and NPQ are related to the dissipation of excess energy as heat, and significant SNP peaks were detected in three chromosome regions (Table 2). Here, we used F0 and qL as examples to describe the gene mining process in detail. The most significant SNP peak associated with F0 in the whole panel and Japonica-specific panel was on chromosome 5 (Fig. 7a). We investigated the LD structure flanking the most significant SNP at 2,825,199 bp, and the region between 2,612,417 and 2,993,493 bp was identified based on the decay of the LD (Fig. 7b). According to the Plant Genes 60 database in Gramene Mart (www.gramene.org), 61 genes were annotated in this area (Additional file 5: Table S5). OsNHX3 (LOC_Os05g05590 or Os05g0148600, located at chr5: 2,777,359 - 2,783,546 bp) encoded the NHX-type Na+/H+ antiporter and was near the most significant SNP (Fig. 7b). A previous study showed that the expression of OsNHX3 was upregulated by salt treatment [35]. Another gene (LOC_Os05g05600 or Os05g0148700) next to OsNHX3 was called the “senescence-associated protein OSA15” and is located in the chloroplast, but how it regulates leaf senescence in rice needs to be examined. Two other genes (LOC_Os05g05830 or Os05g0150800 and LOC_Os05g05950 or Os05g0151400) were also predicted to target chloroplasts, but their roles in photosynthetic efficiency need to be confirmed. For qL, a total of seven significant SNP peaks (P-value < 10− 3 based on the P + K model) located among chromosomes 1, 2, 5, 6 and 12 were detected in the whole panel (Additional file 13: Figure S7a). The LD around the most significant SNP (at 26,473,167 bp) on chromosome 5 was analyzed (Additional file 13: Figure S7b). Based on the decay of the LD, the region harboring candidate genes was between 26,164,849 and 26,862,431 bp, and 113 genes were annotated in this region (Additional file 6: Table S6). Four QTLs related to root length and root dry weight under salt stress [30] and salt tolerance level [33] resided in this region (Table 2). The gene OsCBL4 (OsSOS3, LOC_Os05g45810 or Os05g0534400) encoded an EF-hand-type calcineurin B-like protein in this region (Additional file 13: Figure S7b). OsCBL4 has the same function as AtSOS3 in Arabidopsis and is involved in the regulatory pathway controlling intracellular Na+ and K+ homeostasis and salt tolerance [36].
Table 2.
The significant SNPs detected in a GWAS for five chlorophyll parameters and associated QTLs and genes
| Trait | SNP ID | Chr. | Position (IRGSP-1.0) | Statistical model | P-value | Known QTL | Known gene | |
|---|---|---|---|---|---|---|---|---|
| NPQ | id1003509 | 1 | 4,209,827 | All - P | 4.17E-07 | QKr1.1 [28]; qRNC-1, qRRNC4, qRKC1, qRRKC1, qRNa/K1–1, qRRNa/K1–1 [29] | ||
| F0 | id1004960 | 1 | 6,508,269 | IND - K | 2.17E-04 | qRL-1, qNAUP-1b [30] | ||
| qL | id1016791 | 1 | 28,594,265 | All - P + K | 7.95E-05 | qNAUP-1a, qFWSH-1 [30]; QKr1.2, Qnar/Kr1 [28] | ||
| id1016825 | 1 | 28,599,615 | All - K | 8.10E-04 | ||||
| NPQ | id1024223 | 1 | 38,162,791 | All - P | 4.95E-05 | |||
| ΦPSII | id1026852 | 1 | 41,787,616 | All - P | 3.99E-05 | qSH1–3 [31] | Ygl8 (LOC_Os01g73450, Zhu et al. [32]) | |
| id1027309 | 1 | 42,134,803 | All - K | 2.20E-04 | ||||
| NPQ | id1026852 | 1 | 41,787,616 | All - P | 4.67E-06 | qSH1–3 [31] | ||
| F0 | id1027269 | 1 | 42,082,183 | All - K | 1.01E-04 | qSH1–3 [31] | ||
| JPN - K | 7.39E-04 | |||||||
| id1027563 | 1 | 42,339,408 | All - P | 7.63E-06 | qSH1–3 [31] | |||
| All - P + K | 1.53E-04 | |||||||
| qL | id1027309 | 1 | 42,134,803 | All - P | 4.26E-05 | qSH1–3 [31] | ||
| All - K | 5.44E-04 | |||||||
| Fm | id1027821 | 1 | 42,581,986 | All - P | 1.55E-05 | qSH1–3 [31] | ||
| ΦPSII | id2001842 | 2 | 3,253,759 | All - P | 7.78E-05 | |||
| All - K | 3.05E-04 | |||||||
| All - P + K | 2.35E-04 | |||||||
| qL | id2001842 | 2 | 3,253,759 | All - K | 7.90E-04 | qSST2 [33] | ||
| NPQ | id2006687 | 2 | 16,648,368 | All - P | 4.10E-05 | qPL-2 [30]; QSsst2b, QDss2b [34] | ||
| NPQ | id2007192 | 2 | 17,927,911 | All - P | 5.81E-05 | qPL-2 [30]; QSsst2b, QDss2b [34] | ||
| F0 | id2008158 | 2 | 20,817,846 | All - P | 7.91E-05 | QDss2a, QSkc2, QKna2 [34] | ||
| All - K | 2.06E-04 | |||||||
| All - P + K | 1.41E-04 | |||||||
| NPQ | id2008679 | 2 | 21,812,170 | All - P | 5.36E-05 | QDss2a, QSkc2, QKna2 [34] | ||
| F0 | id2010410 | 2 | 24,565,561 | All - P + K | 2.62E-04 | QDss2a, QSkc2, QKna2 [34]; qRRNa/K1–2 [29] | ||
| id2010572 | 2 | 24,696,197 | All - P | 1.25E-05 | ||||
| id2010581 | 2 | 24,713,963 | All - K | 5.03E-04 | ||||
| NPQ | id2010932 | 2 | 25,236,351 | All - P | 5.50E-05 | QDss2a, QSkc2, QKna2 [34]; qRRNa/K1–2 [29] | ||
| Fm | id4000429 | 4 | 686,361 | All - P | 1.90E-05 | |||
| All - K | 7.32E-04 | |||||||
| qL | id4002349 | 4 | 5,526,924 | IND - K | 3.61E-04 | qRL4 [30] | ||
| Fm | id4002319 | 4 | 5,487,935 | IND - K | 5.13E-04 | |||
| F0 | id5001540 | 5 | 2,773,921 | All - P | 1.70E-06 | QKs5 [28] | OsNHX3 (LOC_Os05g05610, Fukuda et al. [35]) | |
| id5001584 | 5 | 2,825,199 | All - K | 9.66E-05 | ||||
| All - P + K | 6.82E-06 | |||||||
| JPN - K | 3.06E-04 | |||||||
| Fm | id5001738 | 5 | 3,064,389 | IND - K | 2.25E-05 | QSst5, QDss5a, QDss5b [34] | ||
| NPQ | IND - K | 7.46E-04 | ||||||
| qL | id5012588 | 5 | 26,473,167 | All - P | 3.40E-06 | qRL-5, qDWRO-5a, qDWRO-5b [30]; qSST5 [33] | OsCBL4 (LOC_Os05g45810, Martinez-Atienza et al. [36]) | |
| All - K | 5.61E-04 | |||||||
| All - P + K | 4.95E-04 | |||||||
| ΦPSII | id6002261 | 6 | 2,930,916 | All - P | 8.97E-07 | |||
| All - P + K | 4.12E-05 | |||||||
| id6002494 | 6 | 3,113,730 | All - K | 6.48E-05 | ||||
| IND - K | 8.00E-04 | |||||||
| Fm | id6002261 | 6 | 2,930,916 | All - P | 2.77E-05 | |||
| All - K | 5.39E-04 | |||||||
| All - P + K | 4.79E-04 | |||||||
| qL | id6002261 | 6 | 2,930,916 | All - P | 3.90E-05 | |||
| All - P + K | 5.85E-04 | |||||||
| id6002286 | 6 | 2,974,286 | All - K | 3.95E-04 | ||||
| NPQ | id6002389 | 6 | 3,042,568 | All - P | 2.10E-06 | |||
| All - P + K | 4.59E-04 | |||||||
| NPQ | id6005326 | 6 | 8,187,911 | All - P | 1.31E-08 | qDRW6 [31] | ||
| All - K | 3.98E-04 | |||||||
| All - P + K | 2.35E-04 | |||||||
| NPQ | id6006336 | 6 | 10,184,310 | All - P | 3.32E-05 | qDRW6 [31] | ||
| All - K | 1.70E-04 | |||||||
| All - P + K | 1.83E-04 | |||||||
| JPN - K | 2.29E-04 | |||||||
| NPQ | id7000336 | 7 | 1,865,306 | All - P | 3.43E-05 | |||
| F0 | id7003938 | 7 | 22,819,640 | All - P | 2.00E-05 | QSkc7; [34] qSST7 [33] | ||
| qL | id10000081 | 10 | 407,964 | JPN - K | 6.21E-04 | |||
| ΦPSII | id10003594 | 10 | 13,662,830 | All - K | 5.63E-04 | |||
| All - P + K | 4.98E-04 | |||||||
| JPN - K | 3.21E-04 | |||||||
| Fm | id10003623 | 10 | 13,782,039 | All - P | 3.03E-05 | |||
| Fm | id10006610 | 10 | 20,943,116 | All - P | 1.43E-06 | |||
| NPQ | id11008520 | 11 | 22,516,795 | All - P | 6.39E-06 | qSNC11 [37] | ||
| Fm | id11008950 | 11 | 23,178,265 | All - P | 4.04E-06 | qSNC11 [37] | ||
| Fm | id12001261 | 12 | 3,059,845 | All - K | 4.65E-05 | |||
| All - P | 1.30E-06 | |||||||
| JPN - K | 5.35E-05 | |||||||
| id12001332 | 12 | 3,223,671 | All - P + K | 6.34E-05 | ||||
| NPQ | id12001256 | 12 | 3,027,442 | All - P | 1.23E-05 | |||
| qL | id12001769 | 12 | 4,044,980 | All - P + K | 6.57E-04 | qRRNC12 [29] | ||
| ΦPSII | id12004053 | 12 | 10,470,139 | All - P | 5.14E-06 | qRRNC12 [29]; qSUR12, qSES12, qCNL12 [38] | ||
| All - K | 3.54E-04 | |||||||
| ΦPSII | id12007270 | 12 | 21,906,089 | All - P | 1.48E-04 | QSst12, QDss12 [34]; qSUR12, qSES12, qCNL12 [38] | ||
| ΦPSII | id12008292 | 12 | 23,734,278 | All - P | 2.45E-05 | qSH12.1, qSH12.2, qDSW12.1 [31]; qSUR12, qSES12, qCNL12 [38] | ||
| All - K | 4.67E-04 | |||||||
| qL | id12008234 | 12 | 23,663,599 | All - P | 5.21E-05 | qSH12.1, qSH12.2, qDSW12.1 [31]; qSUR12, qSES12, qCNL12 [38] | ||
| All - K | 8.73E-04 | |||||||
| ΦPSII | id12010084 | 12 | 27,378,310 | All - P | 1.65E-05 | qSUR12, qSES12, qCNL12 [38] | ||
| All - K | 1.76E-04 |
Fig. 7.
GWAS of F0 in 232 diverse varieties and LD pattern of the most significant SNP. a Manhattan plot showing the significance of each SNP tested by a mixed linear model. b The most significant SNP is at 2,825,199 bp. The dot color of each SNP represents its LD with the most significant SNP, and the decay of the LD is bordered by two vertical dashed lines. The location of the OsNHX3 gene is marked with a blue bar
Discussion
To explore the potential genetic resources for salinity-tolerant breeding in rice, chlorophyll fluorescence parameters could serve as reliable selection indicators, the pixelated chlorophyll fluorescence data can be used to evaluate the level of stress tolerance days before the injury symptom were expressed externally. The chlorophyll fluorescence parameters F0 and Fm were directly determined from the leaves, however, the alteration in F0 can be attributed to PSI fluorescence, which can be positively or negatively affected under stress [39]. In our study, F0 had the lowest correlation with the injury score in both the pilot experiment and the 232 varieties experiment. Among the chlorophyll fluorescence parameters, Fm had a higher correlation with the injury score in our pilot experiment than the others. However, ΦPSII had the highest correlation of the parameters with the injury score in 232 diverse varieties. Considering that the whole panel includes Indica- and Japonica-specific panels, the correlation of injury score and the six chlorophyll fluorescence parameters was better in each subgroup than in the whole. In the Indica-specific panel, the correlation of the injury score with Fm was improved from − 0.490 to − 0.617, which was also similar to the value in the pilot experiment that included five Indica rice varieties. In addition, the correlations of NPQ and ΦPSII to the injury score were also improved in the subgroups. The correlation of injury score with ΦPSII was slightly better when in the Japonica varieties alone. The pattern of chlorophyll fluorescence parameters and how they correlated with injury score may indicate the different strategies operating in Indica and Japonica rice for survival of salinity stress.
The decrease in Fm was shown to be an indicator of a PSII reaction center made inactive by the stress [40]. In addition, the increase in F0 can also be a negative trait associated with stress, which could result in the separation of LHCII and PSII or the inactivation of the PSII reaction center [41, 42]. In our pilot experiment, F0 increased for all varieties except the most sensitive variety “8777” from day 8 to day 9 under salt stress and then gradually declined until the end of the 150 mM salt treatment in most varieties (not the two extremely tolerant varieties Nona Bokra and IR66946). It is possible that the changes of F0 and Fm were due to the damages of the chloroplast, which could be seen in the ultrastructure of the membrane system [7, 18, 20]. In addition, the malfunction of proteins involves in photosystem II or I could also relate to the reduction of Fm [7]. Unlike the tolerant rice varieties, which have stable Fv/Fm and Fm values, sensitive varieties exhibited significantly decreased F0 and Fm values after day 9, which may indicate severe damage to the antenna structure or inactivation of the PSII reaction center. We suspected that the chloroplast ultrastructure of extremely sensitive variety 8777 was already severely damaged, since the seedling was already grown in 50–100 mM saline solution for 2 days (from day 6 to day 7). We anticipate that the membrane system of variety 8777 was relatively fragile compared to that of other sensitive varieties tested in our study, which was also reflected by the low Fv/Fm ratio of variety 8777.
PSII efficiency can be determined by chlorophyll fluorescence indexes, which include Y(II), Y(NPQ), and Y(NO). These indexes represent the total energy distribution in the photosynthetic reaction center in PSII. Reduction in Y(II) levels and increased Y(NPQ) and Y(NO) levels have been shown to be responses to salt stress in cucumber [43]. In our study, the dynamics of Y(II), Y(NPQ) and Y(NO) from day 9 to day 11 in the tolerant lines (IR66946, Nona Bokra) and sensitive lines (8777, IR28) strongly suggest that enhancing PSII efficiency could improve the salt tolerance; therefore, the quantum yield of energy dissipation in PSII (including Y(NPQ) and Y(NO) components) may provide a hint of the different salinity tolerance levels among diverse rice varieties. The increase in Y(NPQ) depends on the formation of a transthylakoid pH gradient, and proton pumping appears to be inefficient in the sensitive lines [44]. Low Y(NPQ) values may also represent low xanthophyll cycle activity [45, 46], while Y(NPQ) is an indicator of ΔpH-dependent dissipation of excess light energy in the PSII antennae that causes photoinhibition or the xanthophyll cycle [47–49]. Y(NO) corresponds to nonregulated dissipation of excess energy and may be used as a stress indicator [50]. Combining responses to salinity and submergence stresses, ETR, qN, qP and Y(NO) can be used as indicators to distinguish the stress tolerances of different rice genotypes [51].
Associating photosynthetic efficiency with the level of salinity tolerance is challenging, especially when photosynthetic performance and plant growth rate vary among diverse genotypes regardless of salt treatment. We know that the accumulation of excess salt in the plant cell triggered internal physiological changes before external symptoms appeared [52]. In our study, visible senescence was not detected in the salt-treated varieties during day 6 to day 9, and the fluorescence parameters of the second leaf on day 9 were partially correlated with the injury score assessed on day 11 in 232 diverse varieties. Therefore, we hypothesized that these results could be due to the existence of multiple salt tolerance mechanisms in the diverse germplasms. Three major salt tolerance mechanisms were proposed: tissue tolerance, osmotic tolerance and ion exclusion [53]. Considering that all accessions received the same amount of salt treatment from day 6, the varieties with better tissue tolerance would mobilize the sodium to vacuole more efficiently and leave the chloroplast with only minor damage to conduct photosynthesis. If the varieties mainly generate salt tolerance through ion exclusion from roots, the chlorophyll fluorescence detected in the leaf may mostly reflect the natural aging (senescence) process in plants and not the net toxic effect from salt accumulation. As described in Negrão et al. [54], it is difficult to differentiate cause-effect relationships between photosynthesis and growth reduction when quantifying the effects of salinity on photosynthesis; therefore, the phenotypes associated with chlorophyll biosynthesis could confound the interpretation of association results. Through our experimental process, we identified several chromosomal regions associated with chlorophyll fluorescence parameter variation, some significant SNPs closely linked to the genes which were previously shown to be involved in salt tolerance (e.g. Ygl8, OsNHX3, OsCBL4), however, the effects of most novel SNPs were small (R2 less than 10%) and could be partially attributed to developmental aging variation in the tested varieties. A careful experimental design that considers the natural senescence process is required when the objective is to identify the chromosomal regions that are responsive solely to salinity stress.
Conclusions
Is photosynthetic efficiency a quantitative trait and how does it vary under salinity stress? In this study, we tried to answer these two questions by phenotyping a diverse panel of rice seedling chlorophyll fluorescence under salt stress. Our results revealed the quantitative trends in photosynthetic efficiency, and a significant genetic heterogeneity in photosynthetic efficiency that implied that photosynthetic efficiency is highly genetic background-dependent was detected. Considering that the population size is relatively small in our study, we recommend that future association studies should increase the number of varieties to enrich favorable alleles; alternatively, using a biparental mapping population created by crossing tolerant and susceptible varieties that show contrasting chlorophyll fluorescence imaging patterns would allow us to dissect and identify the genetic components and genes that control photosynthetic efficiency.
Methods
Plant material and growth conditions
Eight rice accessions, including four salt-tolerant, three moderate and one salt-sensitive accessions, were evaluated in the pilot experiment (Table 1). For the GWAS experiment, 232 accessions from 71 countries were used for the study (Additional file 3: Table S3). This global panel included 5 subpopulations: 59 tropical japonica, 65 temperate japonica, 38 indica, 21 aus, 9 aromatic and 42 admixed accessions. Rice accessions were obtained from the USDA-ARS, Dale Bumpers National Rice Research Center, Stuttgart, Arkansas, Genetic Stocks Oryza Collection (www.ars.usda.gov/GSOR). Each accession was genotyped on a 44,000 SNP array as described in Zhao et al. [55]. After removing the SNPs with missing rates larger than 10% and minor allele frequencies less than 5%, a total of 29,195, 14,750, and 21,514 SNP markers were identified in the 232, 165 and 66 accessions that belong to the whole panel, Japonica-specific panel, and Indica-specific panel, respectively.
Sterilized seeds were soaked at 37 °C in water for 2 days to synchronize the germination, and germinated seeds were placed in a hydroponic culture system in a phytotron on day 3. Three replicates for each rice accession were included in each experiment. The photoperiod was set to a 12-h-day/12-h-night cycle, and the temperature was set at 28 °C / 25 °C (day/night). The light intensity was controlled at 350 μmole m− 2 s− 1. The hydroponic solution was a half-strength Kimura B solution [56]. After growing in the hydroponic system for 3 days, rice seedlings were treated with salt levels that increased daily (50, 100, 150 mM) over subsequent days (days 6, 7, 8) and then maintained at 150 mM for 3 days for a visual evaluation of salt tolerance (injury score, IS) on day 11 (Fig. 1). In comparison, the control group was grown under the same conditions without any added salt. The hydroponic solution was refreshed every 3 days, and the pH of the culture solution was adjusted to 4.9.
In the pilot experiment, chlorophyll fluorescence imaging was conducted on days 8, 9, 10, and 11 in the control and treatment groups; for the GWAS experiment, chlorophyll fluorescence imaging was conducted on day 9 in the salt treatment group only.
Chlorophyll fluorescence imaging of the seedling leaf
A PAM fluorometer (the MAXI version of IMAGING-PAM; Heinz Walz GmbH, Effeltrich, Germany) was used to capture the images reflecting several chlorophyll fluorescence parameters from a leaf. The imaging was conducted in the dark room (temperature was set at 25 °C). The rice seedlings were dark-adapted at least 30 min before taking measurements on the second leaf of the seedling. To minimize the effect of circadian rhythms on the photosynthesis efficiency, all measurements were taken at the same time during the day. Minimum fluorescence (F0) was measured under weak modulating radiation (0.5 μmol m− 2 s− 1), and maximum fluorescence (Fm) was recorded by applying a saturating pulse of radiation (2700 μmol m− 2 s− 1). To measure the light responses, the actinic light (350 μmol m− 2 s− 1) was switched on to drive photosynthesis for 5 min. Steady-state fluorescence under light illumination (Ft) was continuously monitored under weak modulating radiation (0.5 μmol m− 2 s− 1), the maximum fluorescence in the light (Fm′) was assessed by applying a saturating pulse of radiation (2700 μmol m− 2 s− 1), and the minimum fluorescence was calculated as F0’ = F0 / (Fv/Fm + F0/Fm′). These basic measurements were used to derive the relevant fluorescence parameters: Fv/Fm, ΦPSII, qL, and NPQ [57].
The actual photosynthetic efficiency [Y(II)] was calculated as described by Genty et al. [58]. The quantum yield of regulated energy dissipation in PSII [Y(NPQ)] and the quantum yield of nonregulated energy dissipation in PSII [Y(NO)] were calculated according to Kramer et al.’s [24] method.
The definition and calculation of six major chlorophyll fluorescence parameters are described below:
F0 (minimum fluorescence)
Fm (maximal fluorescence)
Fv/Fm (maximal PSII quantum yield), Fv/Fm = (Fm-F0)/Fm
ΦPSII (effective PSII quantum yield), ΦPSII = (Fm′-Ft)/Fm′
qL (coefficient of photochemical quenching), qL = (Fm′-Ft)/(Fm′-F0’) * (F0’/Ft)
NPQ (nonphotochemical quenching), NPQ = (Fm-Fm′)/Fm′
Y(II) = (Fm′-Fs)/Fm′
Y(NO) = 1/(NPQ + 1 + qL(Fm/Fo-1))
Y(NPQ) = 1-Y(II)-1/(NPQ + 1 + qL(Fm/Fo-1))
Y(II) + Y(NO) + Y(NPQ) = 1
Establish a visual standard system to evaluate the salt stress level
To quantify and compare the symptoms of salt toxicity among rice varieties, we modified the International Rice Research Institute (IRRI) standard evaluation system (SES) [59] according to our observations and assigned an injury score on a scale of 1 to 9, where a low score (near 1) represents tolerant individuals and a higher score represents sensitive individuals (Additional file 1: Table S1).
Genome-wide association analysis
Four statistical models were applied to identify the association between SNPs and trait variations: naïve, P, K, and P + K models. The naïve model was represented by Y = Xβ + ε, where Y was phenotype data, X was genotype data, β was the SNP effect and ε was random effects. The P model was represented by Y = Xβ + Pγ + ε, where P was the population structure and γ was the effect of the population structure. The K model was represented by Y = Xβ + Zu + ε, where u was the random effect of kinship and Z was a coincidence matrix. The P + K model was represented by Y = Xβ + Pγ + Zu + ε. The population structure was analyzed by principal component analysis [60]. Association analyses were conducted in TASSEL 4.0 [61] and R/GAPIT [62].
The LD between the most significant SNP and its neighboring SNPs was estimated using Tagger in Haploview 4.2 [63]. The candidate region was determined based on the LD (r2 > 0.7) between the most significant SNP and its neighboring SNPs, and the candidate genes within the region were identified using Gramene BioMart (http://ensembl.gramene.org/biomart). The SNP position was based on IRGSP 1.0 annotation.
Statistical analysis
Pearson correlation was used to estimate the level of correlation between traits. The normality of each trait was tested using the Shapiro-Wilk method. The genetic distance between the diverse accessions was calculated using the “euclidean” method, and agglomerative hierarchical clustering using the “complete” method was applied to evaluate the relationship between traits and rice accession genetic backgrounds and was plotted as a heatmap. All analyses were conducted using publicly available R packages.
Additional files
Table S1. Phenotypic evaluation to score the visual symptoms of salt toxicity at the seedling stage (DOCX 14 kb)
Table S2. The quantum yield calculated from the ratio of treatment to control (T/C) values of each parameter in eight varieties (XLSX 14 kb)
Table S3. Chlorophyll fluorescence and injury score of 232 accessions and their subpopulation assignment (XLSX 33 kb)
Table S4. Correlation between chlorophyll florescence parameters and injury score (IS) in the global panel (232 varieties) (DOCX 16 kb)
Table S5. The genes inside the LD region of the most significant SNP detected in the F0 GWAS (XLSX 75 kb)
Table S6. The genes inside the LD region of the most significant SNP detected in the qL GWAS (XLSX 112 kb)
Figure S1. Daily changes in chlorophyll fluorescence parameters (ΦPSII, qL, NPQ) in eight rice varieties under control (red line) and salt (blue line) conditions. Each data point represents the average value of 3 replicates. (PPTX 10924 kb)
Figure S2. Box plot of six chlorophyll fluorescence measurements and injury score in six subpopulations and two subspecies. Student’s t-test was used to determine whether any two subpopulations or two subspecies were different. *, **, and **** represent the significance level at P < 0.05, 0.01 and 0.0001, respectively. (PDF 979 kb)
Figure S3. Genetic relationship and phenotypic distributions of varieties in five subpopulations. Each column represents a single variety, and the sample ID is labeled at the bottom of the heatmap. Each row represents the chlorophyll fluorescence parameter and injury score of each variety. The colors close to red indicate that the value of each chlorophyll fluorescence parameter is higher than its average value over all accessions; for the injury score, the red represents the lower injury scores, which suggest that the variety is more tolerant to salt toxicity. The color key of the Z-score was calculated from the distance between the raw score and the population mean in units of the standard deviation. (PDF 1686 kb)
Figure S4. Distribution of six chlorophyll fluorescence parameters in Japonica varieties. The normality of each parameter was examined using the Shapiro-Wilks test. The P-value of each test is provided. (PPTX 7609 kb)
Figure S5. Distribution of six chlorophyll fluorescence parameters in Indica varieties. The normality of each parameter was examined using the Shapiro-Wilks test. The P-value of each test is provided. (PPTX 7606 kb)
Figure S6. Genome-wide association analysis of six chlorophyll fluorescence parameters in 232 diverse varieties or in two subspecies (Japonica and Indica). Manhattan plot showing the significance of each SNP tested by the appropriate statistical models described in “Methods”. (PDF 4610 kb)
Figure S7. Genome-wide association analysis of qL in 232 diverse varieties and LD pattern of the most significant SNP. (a) Manhattan plot showing the significance of each SNP tested by a mixed linear model. (b) The most significant SNP is at 26,473,167 bp. The dot color of each SNP represents its LD with the most significant SNP, and the decay of LD is bordered by two vertical dashed lines. The location of the OsCBL4 gene is marked with a blue bar. (PPTX 14234 kb)
Acknowledgements
We thank Dr. Susan McCouch and USDA-ARS, Dale Bumpers National Rice Research Center-GSOR for providing the rice seeds, and the specialists from the Oldinburgh Co., Ltd., Taiwan for advising regarding the operation of the IMAGING-PAM instrument.
Abbreviations
- ETR
Electron transfer rate
- GWAS
Genome-wide association study
- IRRI
International Rice Research Institute
- LD
Linkage disequilibrium
- LHC
Light-harvesting complex
- NPQ
Nonphotochemical quenching
- PAM
Pulse-amplitude modulation
- PS
Photosystem
- Q-Q
Quantile-quantile
- QTL
Quantitative trait loci
- RGR
Relative growth rate
- SES
Standard evaluation system
- TEJ
Temperate japonica
- TR
Transpiration rate
- TRJ
Tropical japonica
- TUE
Transpiration use efficiency
Authors’ contributions
YCT, KCC, TSC and CWT conceived and designed the experiments; KCC, CL, SHL optimized and performed the experiments; YCT, KCC, TSC and CWT analyzed the data and wrote the manuscript. All authors have read and approved the manuscript.
Funding
This research work was supported by the Ministry of Science and Technology Taiwan (grant numbers MOST 102–2313-B-002-001-MY3 and MOST 105–2311-B-002-024-MY3). The funding body had no role in the design of this study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yu-Chang Tsai, Email: yuchangt@ntu.edu.tw.
Kuan-Chuan Chen, Email: r02621121@ntu.edu.tw.
Tung-Shan Cheng, Email: d07621101@ntu.edu.tw.
Chuan Lee, Email: r01621106@ntu.edu.tw.
Shih-Hung Lin, Email: r01621120@ntu.edu.tw.
Chih-Wei Tung, Email: chihweitung@ntu.edu.tw.
References
- 1.Ashraf M, Harris PJC. Photosynthesis under stressful environments: an overview. Photosynthetica. 2013;51:163–190. doi: 10.1007/s11099-013-0021-6. [DOI] [Google Scholar]
- 2.Maxwell K, Johnson GN. Chlorophyll fluorescence--a practical guide. J Exp Bot. 2000;51:659–668. doi: 10.1093/jexbot/51.345.659. [DOI] [PubMed] [Google Scholar]
- 3.Wang RL, Hua C, Zhou F, Zhou QC. Effects of NaCl stress on photochemical activity and thylakoid membrane polypeptide composition of a salt-tolerant and a salt-sensitive rice cultivar. Photosynthetica. 2009;47:125–127. doi: 10.1007/s11099-009-0019-2. [DOI] [Google Scholar]
- 4.Tiwari BS, Bose A, Ghosh B. Photosynthesis in rice under a salt stress. Photosynthetica. 1998;34:303–306. doi: 10.1023/A:1006857027398. [DOI] [Google Scholar]
- 5.Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI. Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta. 2007;1767:414–421. doi: 10.1016/j.bbabio.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Belkhodja R, Morales F, Abadia A, Gomez-Aparisi J, Abadia J. Chlorophyll fluorescence as a possible tool for salinity tolerance screening in barley (Hordeum vulgare L.) Plant Physiol. 1994;104:667–673. doi: 10.1104/pp.104.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamane K, Kawasaki M, Taniguchi M, Miyake H. Correlation between chloroplast ultrastructure and chlorophyll fluorescence characteristics in the leaves of rice (Oryza sativa L.) grown under salinity. Plant Prod Sci. 2008;11:139–145. doi: 10.1626/pps.11.139. [DOI] [Google Scholar]
- 8.Flood PJ, Harbinson J, Aarts MG. Natural genetic variation in plant photosynthesis. Trends Plant Sci. 2011;16:327–335. doi: 10.1016/j.tplants.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Van Bezouw R, Keurentjes JJB, Harbinson J, Aarts MGM. Converging phenomics and genomics to study natural variation in plant photosynthetic efficiency. Plant J. 2019;97:112–133. doi: 10.1111/tpj.14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Rooijen R, Aarts MGM, Harbinson J. Natural genetic variation for acclimation of photosynthetic light use efficiency to growth irradiance in Arabidopsis. Plant Physiol. 2015;167:1412–1429. doi: 10.1104/pp.114.252239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azam FI, Chang X, Jing R. Mapping QTL for chlorophyll fluorescence kinetics parameters at seedling stage as indicators of heat tolerance in wheat. Euphytica. 2015;202:245–258. doi: 10.1007/s10681-014-1283-1. [DOI] [Google Scholar]
- 12.Takai T, Kondo M, Yano M, Yamamoto T. A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice. Rice. 2010;3:172–180. doi: 10.1007/s12284-010-9047-6. [DOI] [Google Scholar]
- 13.Wang Q, Xie W, Xing H, Yan J, Meng X, Li X, et al. Genetic architecture of natural variation in rice chlorophyll content revealed by a genome-wide association study. Mol Plant. 2015;8:946–957. doi: 10.1016/j.molp.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Lin P-C, Tsai Y-C, Hsu S-K, Ou J-H, Liao C-T, Tung C-W. Identification of natural variants affecting chlorophyll content dynamics during rice seedling development. Plant Breed. 2018;137:355–363. doi: 10.1111/pbr.12584. [DOI] [Google Scholar]
- 15.Dionisio-Sese ML, Tobita S. Effects of salinity on sodium content and photosynthetic responses of rice seedlings differing in salt tolerance. J Plant Physiol. 2000;157:54–58. doi: 10.1016/S0176-1617(00)80135-2. [DOI] [Google Scholar]
- 16.Fracheboud Y, Jompuk C, Ribaut JM, Stamp P, Leipner J. Genetic analysis of cold-tolerance of photosynthesis in maize. Plant Mol Biol. 2004;56:241–253. doi: 10.1007/s11103-004-3353-6. [DOI] [PubMed] [Google Scholar]
- 17.Ehlert B, Hincha DK. Chlorophyll fluorescence imaging accurately quantifies freezing damage and cold acclimation responses in Arabidopsis leaves. Plant Methods. 2008;4:12. doi: 10.1186/1746-4811-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutts S, Kinet JM, Bouharmont J. NaCl-induced senescence in leaves of rice (Oryza sativa L) cultivars differing in salinity resistance. Ann Bot. 1996;78:389–398. doi: 10.1006/anbo.1996.0134. [DOI] [Google Scholar]
- 19.Sarkar RK, Mahata KR, Singh DP. Differential responses of antioxidant system and photosynthetic characteristics in four rice cultivars differing in sensitivity to sodium chloride stress. Acta Physiol Plant. 2013;35:2915–2926. doi: 10.1007/s11738-013-1322-x. [DOI] [Google Scholar]
- 20.Yamane K, Rahman S, Kawasaki M, Taniguchi M, Miyake H. Pretreatment with antioxidants decreases the effects of salt stress on chloroplast ultrastructure in rice leaf segments (Oryza sativa L.) Plant Prod Sci. 2004;7:292–300. doi: 10.1626/pps.7.292. [DOI] [Google Scholar]
- 21.Lee MH, Cho EJ, Wi SG, Bae H, Kim JE, Cho JY, et al. Divergences in morphological changes and antioxidant responses in salt-tolerant and salt-sensitive rice seedlings after salt stress. Plant Physiol Biochem. 2013;70:325–335. doi: 10.1016/j.plaphy.2013.05.047. [DOI] [PubMed] [Google Scholar]
- 22.Singh DP, Sarkar RK. Distinction and characterisation of salinity tolerant and sensitive rice cultivars as probed by the chlorophyll fluorescence characteristics and growth parameters. Funct Plant Biol. 2014;41:727–736. doi: 10.1071/FP13229. [DOI] [PubMed] [Google Scholar]
- 23.Moradi F, Ismail AM. Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot. 2007;99:1161–1173. doi: 10.1093/aob/mcm052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer DM, Johnson G, Kiirats O, Edwards GE. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res. 2004;79:209–218. doi: 10.1023/B:PRES.0000015391.99477.0d. [DOI] [PubMed] [Google Scholar]
- 25.Rajendran K, Tester M, Roy SJ. Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 2009;32:237–249. doi: 10.1111/j.1365-3040.2008.01916.x. [DOI] [PubMed] [Google Scholar]
- 26.Hairmansis A, Berger B, Tester M, Roy SJ. Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice. 2014;7:16. doi: 10.1186/s12284-014-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Tamimi N, Brien C, Oakey H, Berger B, Saade S, Ho YS, et al. Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat Commun. 2016;7:13342. doi: 10.1038/ncomms13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmadi J, Fotokian MH. Identification and mapping of quantitative trait loci associated with salinity tolerance in rice (Oryza Sativa) using SSR markers. Iran J Biotechnol. 2011;9:21–30. [Google Scholar]
- 29.Sun J, Zou DT, Luan FS, Zhao HW, Wang JG, Liu HL, et al. Dynamic QTL analysis of the Na+ content, K+ content, and Na+/K+ ratio in rice roots during the field growth under salt stress. Biol Plant. 2014;58:689–696. doi: 10.1007/s10535-014-0445-2. [DOI] [Google Scholar]
- 30.Sabouri H, Sabouri A. New evidence of QTLs attributed to salinity tolerance in rice. Afr J Biotechnol. 2008;7:4376–4383. [Google Scholar]
- 31.Wang Z, Cheng J, Chen Z, Huang J, Bao Y, Wang J, et al. Identification of QTLs with main, epistatic and QTL x environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor Appl Genet. 2012;125:807–15. doi: 10.1007/s00122-012-1873-z. [DOI] [PubMed] [Google Scholar]
- 32.Zhu X, Guo S, Wang Z, Du Q, Xing Y, Zhang T, et al. Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa) BMC Plant Biol. 2016;16:134. doi: 10.1186/s12870-016-0821-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X, Yuan Z, Liu H, Xiang X, Yang L, He W, et al. Identification of salt tolerance-improving quantitative trait loci alleles from a salt-susceptible rice breeding line by introgression breeding. Plant Breed. 2015;134:653–660. doi: 10.1111/pbr.12321. [DOI] [Google Scholar]
- 34.Cheng L, Wang Y, Meng L, Hu X, Cui Y, Sun Y, et al. Identification of salt-tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice. Genome. 2012;55:45–55. doi: 10.1139/g11-075. [DOI] [PubMed] [Google Scholar]
- 35.Fukuda A, Nakamura A, Hara N, Toki S, Tanaka Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta. 2011;233:175–188. doi: 10.1007/s00425-010-1289-4. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007;143:1001–1012. doi: 10.1104/pp.106.092635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng H, Zhao H, Liu H, Wang J, Zou D. QTL analysis of Na+ and K+ concentrations in shoots and roots under NaCl stress based on linkage and association analysis in japonica rice. Euphytica. 2015;201:109–21. doi: 10.1007/s10681-014-1192-3. [DOI] [Google Scholar]
- 38.Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, et al. Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3:148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- 39.Berry JA, Downton WJS. Photosynthesis. 1982. Environmental regulation of photosynthesis; pp. 263–343. [Google Scholar]
- 40.Tikkanen M, Mekala NR, Aro EM. Photosystem II photoinhibition-repair cycle protects photosystem I from irreversible damage. Biochim Biophys Acta. 2014;1837:210–215. doi: 10.1016/j.bbabio.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Yamane Y. In: Effects of high temperatures on photosynthetic systems in higher plants. 1. Causes of the increase in the fluorescence Fo level. Mathis P, editor. Dordrecht: Springer; 1995. pp. 849–852. [Google Scholar]
- 42.Yamane Y, Kashino Y, Koike H, Satoh K. Increases in the fluorescence Fo level and reversible inhibition of photosystem II reaction center by high-temperature treatments in higher plants. Photosynth Res. 1997;52:57–64. doi: 10.1023/A:1005884717655. [DOI] [Google Scholar]
- 43.Wu Y, Jin X, Liao WBA, Hu LL, Dawuda MM, Zhao XJ, et al. 5-Aminolevulinic acid (ALA) alleviated salinity stress in cucumber seedlings by enhancing chlorophyll synthesis pathway. Front Plant Sci. 2018;9:635. doi: 10.3389/fpls.2018.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cardona T, Shao SX, Nixon PJ. Enhancing photosynthesis in plants: the light reactions. Essays Biochem. 2018;62:85–94. doi: 10.1042/EBC20170015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton P, Ruban AV, Young AJ. Regulation of the structure and function of the light harvesting complexes of photosystem II by the xanthophyll cycle. In: Frank HA, Young AJ, Britton G, Cogdell RJ, editors. The photochemistry of carotenoids. Dordrecht: Springer; 1999. pp. 271–291. [Google Scholar]
- 46.Yamamoto HY, Bugos RC, Hieber AD. Biochemistry and molecular biology of the xanthophyll cycle. In: Frank HA, Young AJ, Britton G, Cogdell RJ, editors. The photochemistry of carotenoids. Dordrecht: Springer; 1999. pp. 293–303. [Google Scholar]
- 47.Bilger W, Björkman O. Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth Res. 1990;25:173–185. doi: 10.1007/BF00033159. [DOI] [PubMed] [Google Scholar]
- 48.Ishida S, Morita KI, Kishine M, Takabayashi A, Murakami R, Takeda S, et al. Allocation of absorbed light energy in PSII to thermal dissipations in the presence or absence of PsbS subunits of rice. Plant Cell Physiol. 2011;52:1822–1831. doi: 10.1093/pcp/pcr119. [DOI] [PubMed] [Google Scholar]
- 49.Roháček K, Soukupová J, Barták M. In: Chlorophyll fluorescence: a wonderful tool to study plant physiology and plant stress. Schoefs B, editor. Kerala: Research Signpost; 2008. pp. 41–104. [Google Scholar]
- 50.Qu CX, Liu C, Gong XL, Li CX, Hong MM, Wang L, et al. Impairment of maize seedling photosynthesis caused by a combination of potassium deficiency and salt stress. Environ Exp Bot. 2012;75:134–141. doi: 10.1016/j.envexpbot.2011.08.019. [DOI] [Google Scholar]
- 51.Pradhan B, Chakraborty K, Prusty N, Deepa MAK, Chattopadhyay K, et al. Distinction and characterisation of rice genotypes tolerant to combined stresses of salinity and partial submergence, proved by a high-resolution chlorophyll fluorescence imaging system. Functional Plant Biol. 2019;46:248–261. doi: 10.1071/FP18157. [DOI] [PubMed] [Google Scholar]
- 52.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 53.Roy SJ, Negrao S, Tester M. Salt resistant crop plants. Curr Opin Biotechnol. 2014;26:115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Negrao S, Schmockel SM, Tester M. Evaluating physiological responses of plants to salinity stress. Ann Bot. 2017;119:1–11. doi: 10.1093/aob/mcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma JF, Goto S, Tamai K, Ichii M. Role of root hairs and lateral roots in silicon uptake by rice. Plant Physiol. 2001;127:1773–1780. doi: 10.1104/pp.010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker NR. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol. 2008;59:89–113. doi: 10.1146/annurev.arplant.59.032607.092759. [DOI] [PubMed] [Google Scholar]
- 58.Genty B, Briantais J-M, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta Gen Subj. 1989;990:87–92. doi: 10.1016/S0304-4165(89)80016-9. [DOI] [Google Scholar]
- 59.IRRI . Standard evaluation system for rice. Philippine: International Rice Research Institute; 2002. [Google Scholar]
- 60.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 61.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 62.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 63.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Phenotypic evaluation to score the visual symptoms of salt toxicity at the seedling stage (DOCX 14 kb)
Table S2. The quantum yield calculated from the ratio of treatment to control (T/C) values of each parameter in eight varieties (XLSX 14 kb)
Table S3. Chlorophyll fluorescence and injury score of 232 accessions and their subpopulation assignment (XLSX 33 kb)
Table S4. Correlation between chlorophyll florescence parameters and injury score (IS) in the global panel (232 varieties) (DOCX 16 kb)
Table S5. The genes inside the LD region of the most significant SNP detected in the F0 GWAS (XLSX 75 kb)
Table S6. The genes inside the LD region of the most significant SNP detected in the qL GWAS (XLSX 112 kb)
Figure S1. Daily changes in chlorophyll fluorescence parameters (ΦPSII, qL, NPQ) in eight rice varieties under control (red line) and salt (blue line) conditions. Each data point represents the average value of 3 replicates. (PPTX 10924 kb)
Figure S2. Box plot of six chlorophyll fluorescence measurements and injury score in six subpopulations and two subspecies. Student’s t-test was used to determine whether any two subpopulations or two subspecies were different. *, **, and **** represent the significance level at P < 0.05, 0.01 and 0.0001, respectively. (PDF 979 kb)
Figure S3. Genetic relationship and phenotypic distributions of varieties in five subpopulations. Each column represents a single variety, and the sample ID is labeled at the bottom of the heatmap. Each row represents the chlorophyll fluorescence parameter and injury score of each variety. The colors close to red indicate that the value of each chlorophyll fluorescence parameter is higher than its average value over all accessions; for the injury score, the red represents the lower injury scores, which suggest that the variety is more tolerant to salt toxicity. The color key of the Z-score was calculated from the distance between the raw score and the population mean in units of the standard deviation. (PDF 1686 kb)
Figure S4. Distribution of six chlorophyll fluorescence parameters in Japonica varieties. The normality of each parameter was examined using the Shapiro-Wilks test. The P-value of each test is provided. (PPTX 7609 kb)
Figure S5. Distribution of six chlorophyll fluorescence parameters in Indica varieties. The normality of each parameter was examined using the Shapiro-Wilks test. The P-value of each test is provided. (PPTX 7606 kb)
Figure S6. Genome-wide association analysis of six chlorophyll fluorescence parameters in 232 diverse varieties or in two subspecies (Japonica and Indica). Manhattan plot showing the significance of each SNP tested by the appropriate statistical models described in “Methods”. (PDF 4610 kb)
Figure S7. Genome-wide association analysis of qL in 232 diverse varieties and LD pattern of the most significant SNP. (a) Manhattan plot showing the significance of each SNP tested by a mixed linear model. (b) The most significant SNP is at 26,473,167 bp. The dot color of each SNP represents its LD with the most significant SNP, and the decay of LD is bordered by two vertical dashed lines. The location of the OsCBL4 gene is marked with a blue bar. (PPTX 14234 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.