Replication intermediates and ends of viral DNA genomes can be recognized by the cellular DNA damage response (DDR) network as DNA damage whose repair may lead to inhibition of virus replication. Therefore, many viruses evolved mechanisms to inhibit the DDR network. We have previously shown that the adenovirus (Ad) E4orf4 protein inhibits DDR signaling, but the mechanisms were not identified. Here, we describe an association of E4orf4 with the DNA damage sensor poly(ADP-ribose) polymerase 1 (PARP-1). E4orf4 reduces phosphorylation of this enzyme and inhibits its activity. PARP-1 inhibition assists E4orf4 in reducing Ad-induced DDR signaling and improves the efficiency of virus replication. Furthermore, the ability of E4orf4, when expressed alone, to accumulate at DNA damage sites and to kill cancer cells is attenuated by chemical inhibition of PARP-1. Our results indicate that the E4orf4–PARP-1 interaction has an important role in Ad replication and in promotion of E4orf4-induced cancer-selective cell death.
KEYWORDS: DNA damage response, E4orf4 protein, PARP-1, PP2A, poly(ADP-ribose) polymerase 1, adenoviruses, cancer cell death
ABSTRACT
The adenovirus (Ad) E4orf4 protein was reported to contribute to inhibition of ATM- and ATR-regulated DNA damage signaling during Ad infection and following treatment with DNA-damaging drugs. Inhibition of these pathways improved Ad replication, and when expressed alone, E4orf4 sensitized transformed cells to drug-induced toxicity. However, the mechanisms utilized were not identified. Here, we show that E4orf4 associates with the DNA damage sensor poly(ADP-ribose) polymerase 1 (PARP-1) and that the association requires PARP activity. During Ad infection, PARP is activated, but its activity is not required for recruitment of either E4orf4 or PARP-1 to virus replication centers, suggesting that their association occurs following recruitment. Inhibition of PARP-1 assists E4orf4 in reducing DNA damage signaling during infection, and E4orf4 attenuates virus- and DNA damage-induced parylation. Furthermore, E4orf4 reduces PARP-1 phosphorylation on serine residues, which likely contributes to PARP-1 inhibition as phosphorylation of this enzyme was reported to enhance its activity. PARP-1 inhibition is important to Ad infection since treatment with a PARP inhibitor enhances replication efficiency. When E4orf4 is expressed alone, it associates with poly(ADP-ribose) (PAR) chains and is recruited to DNA damage sites in a PARP-1-dependent manner. This recruitment is required for inhibition of drug-induced ATR signaling by E4orf4 and for E4orf4-induced cancer cell death. Thus, the results presented here demonstrate a novel mechanism by which E4orf4 targets and inhibits DNA damage signaling through an association with PARP-1 for the benefit of the virus and impacting E4orf4-induced cancer cell death.
IMPORTANCE Replication intermediates and ends of viral DNA genomes can be recognized by the cellular DNA damage response (DDR) network as DNA damage whose repair may lead to inhibition of virus replication. Therefore, many viruses evolved mechanisms to inhibit the DDR network. We have previously shown that the adenovirus (Ad) E4orf4 protein inhibits DDR signaling, but the mechanisms were not identified. Here, we describe an association of E4orf4 with the DNA damage sensor poly(ADP-ribose) polymerase 1 (PARP-1). E4orf4 reduces phosphorylation of this enzyme and inhibits its activity. PARP-1 inhibition assists E4orf4 in reducing Ad-induced DDR signaling and improves the efficiency of virus replication. Furthermore, the ability of E4orf4, when expressed alone, to accumulate at DNA damage sites and to kill cancer cells is attenuated by chemical inhibition of PARP-1. Our results indicate that the E4orf4–PARP-1 interaction has an important role in Ad replication and in promotion of E4orf4-induced cancer-selective cell death.
INTRODUCTION
The DNA damage response (DDR) is a conglomerate of pathways designed to detect DNA damage and signal its presence to cell cycle checkpoints and to the repair machinery, allowing the cell to pause and mend the damage or, if the damage is too severe, to trigger apoptosis or senescence (1, 2). Proteins involved in the numerous DDR pathways fall into several major groups. These include sensors of DNA lesions, signal transducers that link DNA damage recognition to downstream effectors, and the effectors that are responsible for activation of checkpoints and for the repair process (3). Poly(ADP-ribose) polymerase 1 (PARP-1) is a sensor protein that belongs to a large family of PARP proteins and detects various types of DNA lesions, including single-strand DNA breaks, double-strand DNA breaks, and others (4–6).
PARPs use NAD+ to produce ADP-ribose groups and attach them to protein acceptors while releasing nicotinamide. PARP-1 can generate long poly(ADP-ribose) (PAR) chains up to 200 units in length and can also produce branched chains (7). PARP-1 binding to DNA breaks causes a local unfolding of an autoinhibitory domain, which allows correct binding of the substrate NAD+ and triggers PARP-1 activation, leading to rapid PAR production (8, 9). PAR is attached to PARP-1 itself and to several other proteins (10). In addition to PARP activation by DNA breaks, nucleosomes, protein interaction partners, and several types of posttranslational modifications, including phosphorylation, also enhance PARP-1 activity (5). Protein parylation regulates many cellular mechanisms, including DNA repair, transcription, mitochondrial function, and others (5). During the DNA repair process, several proteins containing conserved PAR-interacting domains are recruited to the site of DNA damage by binding parylated proteins (11, 12). These PAR-binding proteins contribute to regulation of DNA damage signal transduction. Once the DDR has been initiated, ADP-ribosylation signaling is extinguished by enzymes, such as poly(ADP-ribose) glycohydrolase (PARG), which remove the PAR chains (13, 14).
Infection by several virus families triggers the DDR because viral DNA replication products are recognized by the cell as DNA damage. Because DDR activation by viral DNA can lead to repair in the form of ligation of viral genomes, this process may result in inhibition of virus replication. To combat this antiviral defense mechanism, viruses have evolved mechanisms to inactivate DDR pathways (15, 16). Some of the most well-described mechanisms include inhibition of DDR transducers belonging to the phosphatidylinositol 3-kinase-like protein kinase (PIKK) family such as ataxia-telangiectasia mutated (ATM), ATM- and Rad3-related (ATR), and DNA-PK (15–17). Several PARPs as well as ADP ribosylation participate in the mammalian antiviral response, but they may also provide assistance at some stages of replication of certain viruses (18). Specifically, adenovirus (Ad) infection was shown to induce PARP activity, but one of the consequences of this activation, apoptosis-inducing factor-dependent nuclear fragmentation, was prevented in the presence of the Ad E1B-55K and E4orf3 proteins (19).
The Ad E4orf4 protein is a 14-kDa viral regulator which fulfills many functions that contribute to the efficiency of Ad infection. These include temporally regulated decrease of early viral gene expression, control of alternative splicing of viral RNAs, regulation of protein translation, and a contribution to inhibition of the DDR by Ad (20–30). When E4orf4 is expressed alone, in the absence of other viral proteins, it initiates a unique mode of cell death which does not require p53 or caspase activation but which can be amplified by triggering classical caspase-dependent apoptosis downstream (31–34). The mechanisms underlying E4orf4-induced cell death are conserved in several species, including Saccharomyces cerevisiae, Drosophila, and human cells (35–39). Importantly, E4orf4-induced cell death is more efficient in cancer cells in tissue culture than in normal cells (40), and E4orf4 inhibits tumorigenesis in vivo in a Drosophila cancer model (41), indicating that study of E4orf4 signaling could provide fresh insights that may contribute to the development of a new approach to cancer therapy. Many cancers are deficient in some DDR pathways, making them more sensitive to DDR inhibition. The ability of E4orf4 to inhibit DDR pathways, including those regulated by ATM and ATR (28), could therefore contribute to the cancer selectivity of E4orf4.
Protein phosphatase 2A (PP2A) is an enzymatic complex containing three subunits: a catalytic subunit, a scaffolding subunit, and one of several regulatory subunits responsible for dictating substrate specificity of the enzyme (42). Investigation of the mechanisms contributing to E4orf4 functions uncovered several E4orf4 partners, including various PP2A heterotrimers (43, 44), and all three types of PP2A subunits are present in the E4orf4-PP2A complex which possesses extensive phosphatase activity (38, 43, 44). The PP2A interaction with E4orf4 was shown to contribute to all E4orf4 functions known to date, including attenuation of the DDR (27–29, 40, 43–45).
In this work we investigated the point of interaction between E4orf4 and the DDR as well as the mechanisms by which E4orf4 inhibits DNA damage signaling. We show that E4orf4 interacts with PARP-1 and explore the consequences of this interaction during virus infection and under conditions of nonviral DNA damage.
RESULTS
E4orf4 associates with PARP-1.
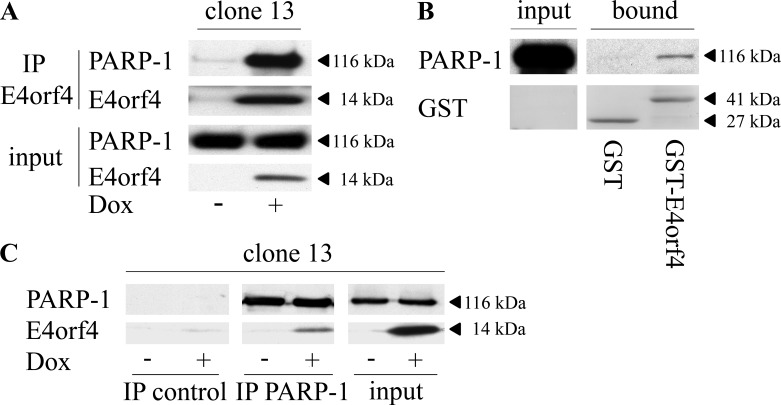
Mass spectrometry analysis of nuclear E4orf4 immune complexes revealed PARP-1 as a potential E4orf4 partner (data not shown). To validate this interaction, E4orf4 was immunoprecipitated from nuclear extracts of clone 13 cells induced by doxycycline (Dox) to stimulate E4orf4 expression or from control uninduced cells. As shown in Fig. 1A, PARP-1 coimmunoprecipitated with E4orf4 but was absent in immune complexes from the control cells. In addition, pulldown assays with glutathione S-transferase (GST)-E4orf4 or GST alone further corroborated the E4orf4–PARP-1 interaction (Fig. 1B). A reciprocal immunoprecipitation using PARP-1-specific antibodies or isotype-matched control antibodies also demonstrated that E4orf4 associated specifically with PARP-1 (Fig. 1C).
FIG 1.
E4orf4 associates with PARP-1. (A) Clone 13 cells were either induced with Dox for 4 h to stimulate E4orf4 expression or remained uninduced. The cells were harvested, and nuclear extracts were prepared and subjected to immunoprecipitation with an E4orf4-specific antibody. The presence of PARP-1 and E4orf4 in the immune complexes was determined by Western blot analysis. (B) Nuclear extracts from uninduced clone 13 cells were subjected to GST pulldown analysis with GST or GST-E4orf4 bound to glutathione beads. The presence of PARP-1 and GST proteins in the complex was determined by Western blot analysis. (C) Nuclear extracts were prepared as described for panel A and were subjected to immunoprecipitation with a monoclonal antibody against endogenous PARP-1 or with a control isotype-matched Flag-tagged antibody. The presence of E4orf4 in the immune complexes was determined by Western blot analysis. The separated parts of the blot showing the immune complexes are from the same exposed blot, but some lanes were removed from the middle. IP, immunoprecipitation.
Investigation of the interaction between E4orf4 and PARP-1.
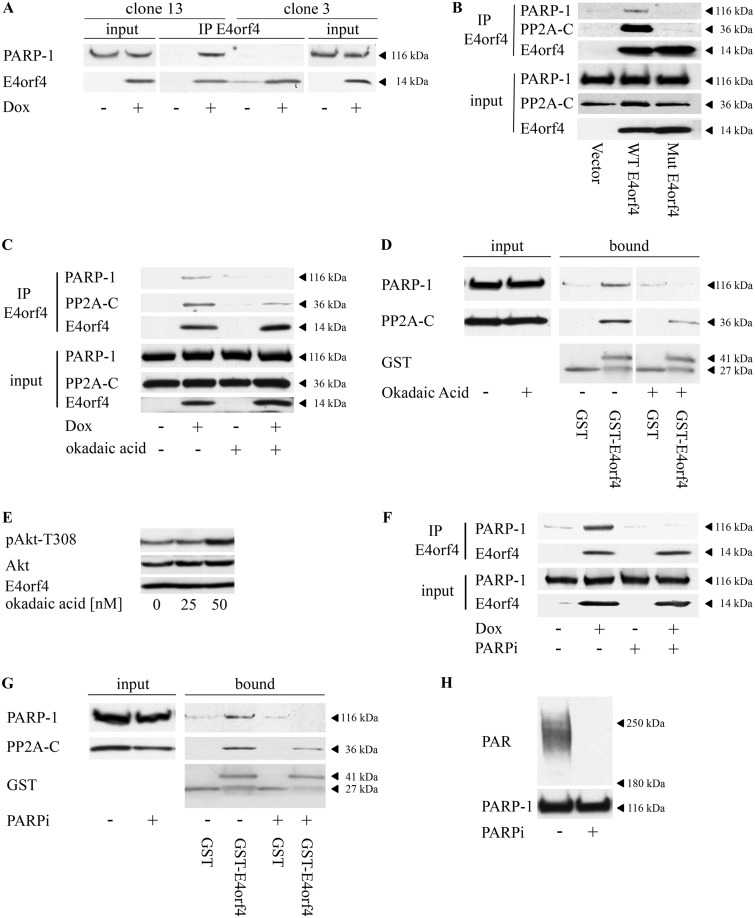
Because PP2A is an important E4orf4 partner, known to contribute to all E4orf4 functions described so far (27–29), we investigated whether PP2A also contributed to the interaction between E4orf4 and PARP-1. Cells containing Dox-inducible wild-type (WT)-E4orf4 (clone 13) or the R81A/F84A E4orf4 mutant that does not bind PP2A (clone 3) (28, 45) were induced with Dox for 4 h or remained untreated, and E4orf4 proteins were immunoprecipitated from nuclear extracts. The results shown in Fig. 2A demonstrate that PARP-1 coimmunoprecipitated with WT-E4orf4 but not with the mutant protein. Because we observed a relatively high level of leaky expression of the mutant E4orf4 protein in clone 3 cells, we also compared the ability of WT and mutant E4orf4 proteins to associate with PARP-1 in cells that were transiently transfected with the inducible E4orf4 constructs or with an empty vector. As seen in Fig. 2B, the mutant E4orf4 protein that could not bind PP2A lost its ability to bind PARP-1 under these conditions as well. To examine whether PP2A activity influenced the E4orf4–PARP-1 interaction, E4orf4 was immunoprecipitated from Dox-induced clone 13 cells or from control uninduced cells, which either were left untreated or were treated with 50 nM okadaic acid for the last hour before harvest. As seen in Fig. 2C, E4orf4 associated with PARP-1 in the absence of the PP2A inhibitor, but when the cells were treated with okadaic acid prior to harvest, the E4orf4–PARP-1 interaction was reduced to background levels. Similarly, when GST and GST-E4orf4 were incubated with cell extracts that were subjected to okadaic acid treatment prior to harvest, the GST-E4orf4–PARP-1 interaction was eliminated whereas an interaction with PP2A was maintained although at a lower level (Fig. 2D). The okadaic acid concentration used in these experiments was sufficient to inhibit PP2A, as demonstrated by its ability to increase phosphorylation of the PP2A substrate Akt (Fig. 2E). These results indicate, therefore, that an active PP2A must be present to facilitate the association of E4orf4 with PARP-1.
FIG 2.
PP2A and PARP-1 activities are required for the E4orf4-PARP-1 interaction. (A) Clone 13 cells containing Dox-inducible WT E4orf4 and clone 3 cells containing a Dox-inducible E4orf4 mutant that cannot bind PP2A (R81A/F84A) were treated with Dox for 4 h or were left untreated. Nuclear extracts were subjected to immunoprecipitation with an antibody against E4orf4. The presence of PARP-1 and E4orf4 in input lysates and in the immune complexes was analyzed by Western blotting. (B) 293T cells were transfected with plasmids that inducibly express WT E4orf4 or the R81A/F84A mutant that cannot bind PP2A (Mut E4orf4) or with the corresponding empty vector. Expression of E4orf4 proteins was induced by Dox 24 h posttransfection for 4 h. Nuclear extracts were subjected to immunoprecipitation with an E4orf4-specific antibody. The presence of proteins in the immune complexes and in input lysates was determined by Western blot analysis with the specified antibodies. (C) Clone 13 cells were induced with Dox for 3 h to stimulate E4orf4 expression or were left uninduced. The cells were then treated with 50 nM okadaic acid, a PP2A inhibitor, for another hour. Nuclear extracts were subjected to immunoprecipitation and Western blot analysis as described for panel A. (D) Nuclear extracts from uninduced clone 13 cells that were either treated with 50 nM okadaic acid for 1 h or left untreated were subjected to GST pulldown analysis with GST or GST-E4orf4 bound to glutathione beads. The presence of proteins in the complex (bound) or in input lysates was determined by Western blot analysis. The two parts of the blot containing bound proteins are from the same exposed blot, but some lanes were removed from the middle. (E) Clone 13 cells were induced by Dox and treated with the specified concentrations of okadaic acid as described for panel C. Whole-cell extracts were subjected to Western blot analysis. (F) Clone 13 cells were induced with Dox for 3 h as described for panel C and were then either treated with 4 μM the PARP inhibitor olaparib (PARPi) for 1 h or left untreated. Nuclear extracts were subjected to immunoprecipitation with an antibody against E4orf4. The presence of proteins in the immune complexes and in input lysates was analyzed by Western blotting. (G) Nuclear extracts from uninduced clone 13 cells that were either treated with 4 μM PARP inhibitor (PARPi) for 1 h or left untreated were subjected to GST pulldown analysis with GST or GST-E4orf4 bound to glutathione beads. The presence of proteins in the complex (bound) or in input lysates was determined by Western blot analysis with the specified antibodies. (H) Clone 13 cells were either treated with a PARP inhibitor (PARPi) as described for panels F and G or left untreated, and cell extracts were subjected to Western blot analysis with the indicated antibodies. Representative blots are shown in all experiments (A to H).
Next, the contribution of PARP-1 activity to the E4orf4–PARP-1 interaction was also investigated. Clone 13 cells expressing E4orf4 were subjected to a 1-h incubation with a PARP inhibitor (olaparib) prior to harvest or were left untreated. As seen in Fig. 2F, PARP-1 coimmunoprecipitated with E4orf4 in the absence of the PARP inhibitor but not in its presence. Similarly, when GST pulldown assays were performed, only PARP-1 from cells that were not treated with a PARP inhibitor associated with GST-E4orf4. In contrast, PARP inhibition only slightly affected PP2A-C binding to E4orf4 (Fig. 2G). The concentration of PARP inhibitor used in these experiments was sufficient to reduce internal parylation in clone 13 cells (Fig. 2H), which was evident even without external stimuli. Altogether, the results indicate that both PP2A and PARP activities are required to facilitate the E4orf4–PARP-1 association.
PARP-1 and E4orf4 accumulate at viral replication centers independently of each other.
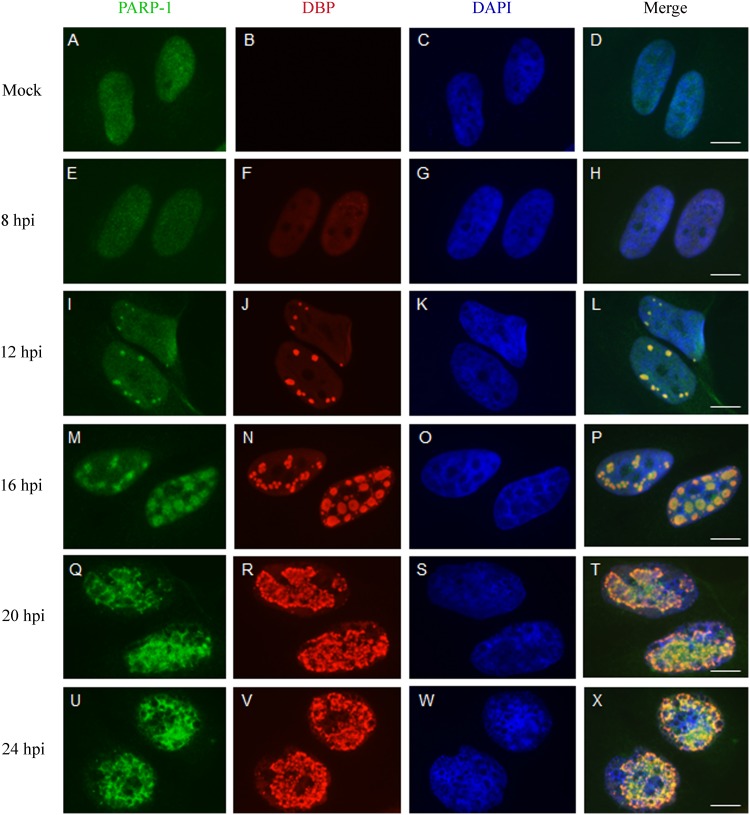
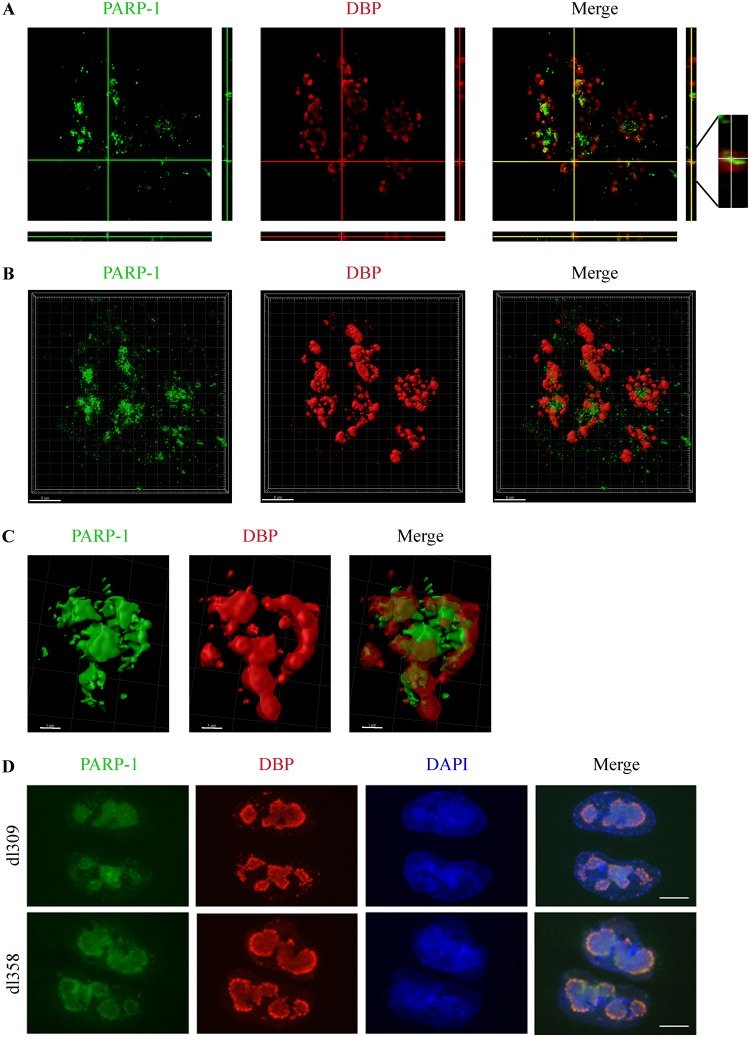
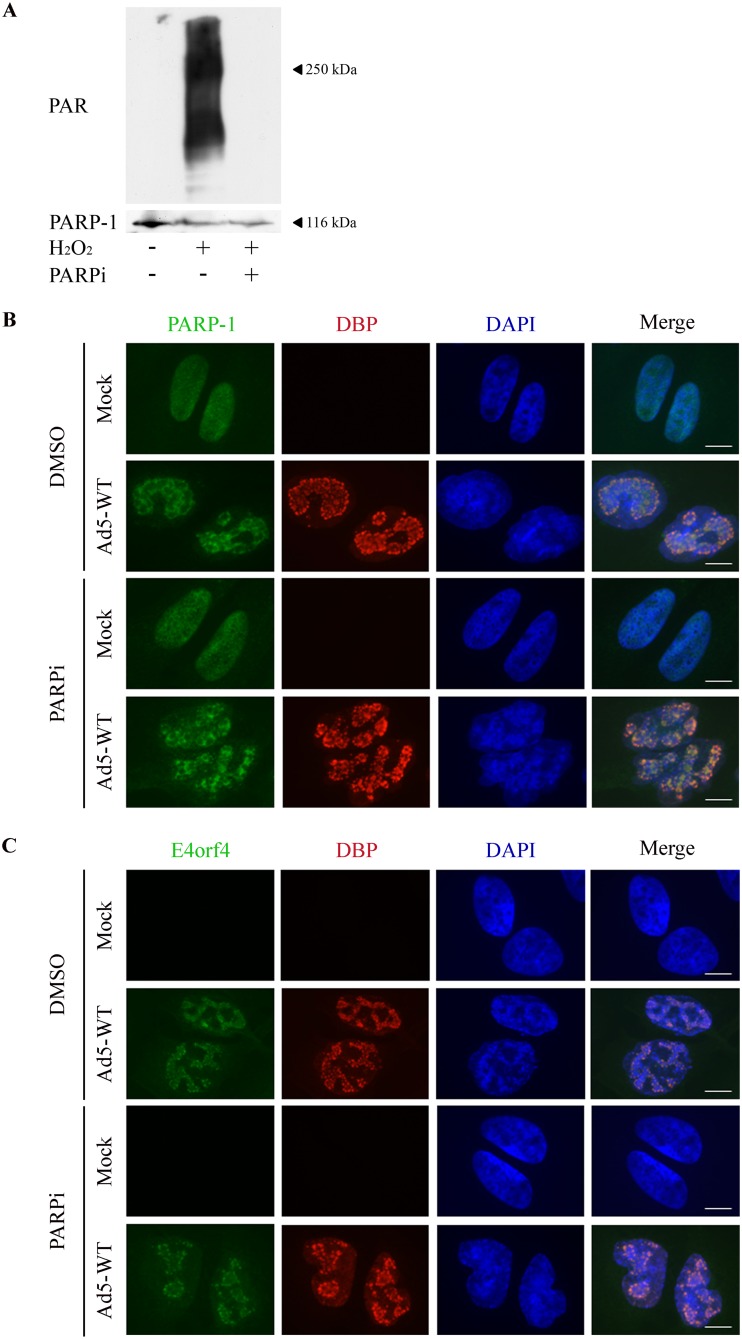
Several sensors and transducers of the DDR accumulate at Ad replication centers (RCs) and may be activated there by products of virus replication identified as DNA damage, unless inhibited by various Ad proteins (46, 47). Recent results from our laboratory also show that E4orf4 is found at Ad RCs as early as the DNA binding protein (DBP), an RC marker (30). We next investigated whether PARP-1 also accumulated in Ad RCs and examined the kinetics of its recruitment. HeLa cells were mock infected or infected with WT Ad5, and DBP as well as PARP-1 was immunostained at various times postinfection. As shown in Fig. 3, DBP started to accumulate in small replication foci in the infected cell nuclei at 8 h postinfection (p.i.), whereas more mature ring-like RC structures accumulated at 16 h p.i. and later. In parallel, PARP-1 also accumulated in the vicinity of Ad RCs. To better assess the relative localization of PARP-1 and DBP, both proteins were visualized by high-resolution microscopy. Figure 4A to C present a topological view and orthogonal projections of a representative infected cell nucleus as well as three-dimensional (3D) renderings of RCs within this nucleus. The results demonstrate that PARP-1 accumulated strongly inside some ring-like Ad RCs but could also be found at lower levels around other ring-like RCs. These results confirm findings shown in Fig. 3, demonstrating that PARP-1 is redistributed in the nucleus to the vicinity of Ad RCs. A comparison between PARP-1 recruitment to Ad RCs in the presence of E4orf4 (dl309 virus, a phenotypically WT Ad5) or in its absence (dl358) indicated that PARP-1 recruitment to Ad RCs did not require E4orf4 (Fig. 4D). Inhibition of PARP activity by addition of a PARP inhibitor to HeLa cells could reduce parylation efficiently (Fig. 5A) but did not prevent recruitment of both PARP-1 and E4orf4 to RCs (Fig. 5B and C). Thus, during Ad infection PARP-1 accumulates at viral RCs, where E4orf4 is also present, but the two proteins arrive there independently.
FIG 3.
PARP-1 is recruited to Ad replication centers. HeLa cells were infected with WT Ad5 and were fixed at various times postinfection (hpi, hours postinfection). The cells were stained with antibodies to PARP-1, to DBP, and with DAPI, as indicated at the top of the figure. Representative images obtained by confocal microscopy are shown for each time point. Scale bar, 10 μm.
FIG 4.
Characterization of the localization of PARP-1 at viral RCs. (A) HeLa cells were infected with WT Ad5 and were fixed and stained 24 h p.i .with antibodies to DBP and PARP-1. A high-resolution image of a representative infected cell nucleus is shown with horizontal and vertical orthogonal projections. (B) 3D renderings of a representative nucleus, prepared with the Imaris software, are shown. (C) Representative 3D renderings of one late replication center from the nucleus in panel B are shown. (D) HeLa cells were infected with dl309 (a phenotypically WT Ad) or with dl358 (a mutant Ad with dl309 backbone lacking E4orf4) virus. The cells were fixed at 24 h p.i. and stained with antibodies to DBP and PARP-1 and with DAPI. Representative images from confocal microscopy are shown. Scale bar, 10 μm.
FIG 5.
PARP inhibition does not affect recruitment of E4orf4 and PARP-1 to viral RCs. (A) HeLa cells were either left untreated or treated with H2O2 in the presence or absence of 4 μM PARP inhibitor (PARPi). Western blot analysis with PAR and PARP-1-specific antibodies is shown. (B and C) HeLa cells were either mock infected or infected for 24 h with WT Ad5 in the presence of dimethyl sulfoxide (DMSO) solvent or 4 μM PARP inhibitor (PARPi). The cells were stained with the specified antibodies and with DAPI, and representative images obtained by confocal microscopy are shown. Scale bar, 10 μm.
PARP-1 contribution to the ability of E4orf4 to reduce ATM and ATR signaling during virus infection.
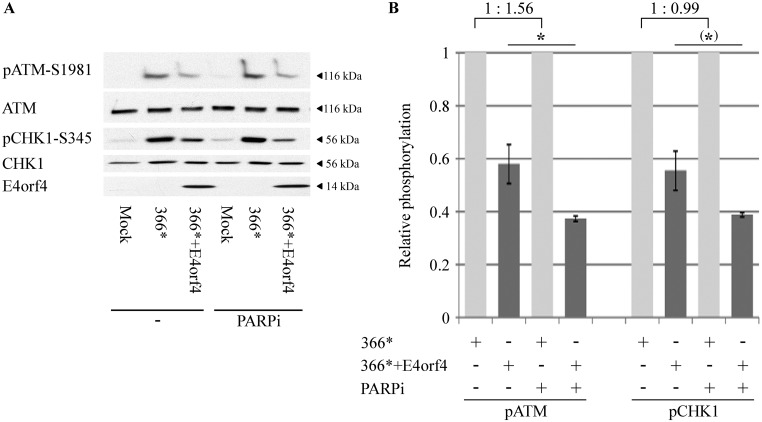
We have previously shown that E4orf4 diminished ATM and ATR signaling during Ad infection and following induction of DNA damage by drugs, but ATM and ATR were inhibited by E4orf4 independently of each other (28). These experiments were performed with the Ad mutants dl366*, lacking the E4 region, and dl366*+E4orf4, lacking all E4 open reading frames (ORFs) except E4orf4 (48). Those mutants were used to facilitate examination of E4orf4 effects without interference from other E4 gene products that inhibit the DDR. To determine whether PARP-1 contributed to the ability of E4orf4 to reduce ATM- or ATR-mediated DNA damage signaling, we explored whether inhibition of PARP impacted the effect of E4orf4 on phosphorylation of ATM and the ATR substrate Chk1 during mutant Ad infection. As shown in Fig. 6, addition of a PARP inhibitor facilitated a significantly larger decrease in ATM phosphorylation by E4orf4 (1.6-fold; P = 0.038) and a 1.4-fold-larger decrease of Chk1 phosphorylation, a difference of borderline significance (P = 0.06). Inhibition of PARP in the absence of E4orf4 did not reduce ATR signaling induced by dl366* infection and increased ATM signaling by 56% (Fig. 6B). These results demonstrate that inhibition of parylation contributed to the ability of E4orf4 to attenuate ATM and possibly ATR signaling during virus infection but was not sufficient by itself to attenuate these signaling pathways.
FIG 6.
Inhibition of PARP-1 aggravates the attenuation of ATM and ATR signaling by E4orf4 during Ad infection. (A) HeLa cells were either mock infected or infected with the Ad mutant dl366* lacking the whole E4 region or dl366*+E4orf4 expressing E4orf4 as the only E4 ORF. A PARP inhibitor (PARPi) was added to the infected cells for the duration of the infection starting at 2 h postinfection (p.i.), and another group of infected cells was left untreated. Proteins were harvested 24 h p.i., and Western blot analysis was carried out with the indicated antibodies for phosphorylated and nonphosphorylated proteins and for E4orf4. One representative blot is shown. (B) Blots from two independent experiments as described in panel A were subjected to densitometry. The levels of phosphorylated ATM and Chk1 as well as of the total proteins were calculated, and phospho-protein levels were normalized to levels of the total corresponding protein. Normalized phospho-protein levels in cells infected with dl366* (light gray bars) were defined as 1, and relative levels in dl366*+E4orf4-infected cells (dark gray bars) are shown. The relative ATM and Chk1 phosphorylation levels in dl366*-infected cells in the absence and presence of the PARP inhibitor are shown above the graph. Error bars represent standard error (n = 2). Statistical significance was determined by a Student's t test. *, P < 0.04; (*), P = 0.06.
E4orf4 reduces cellular parylation induced by DNA damage and by Ad infection.
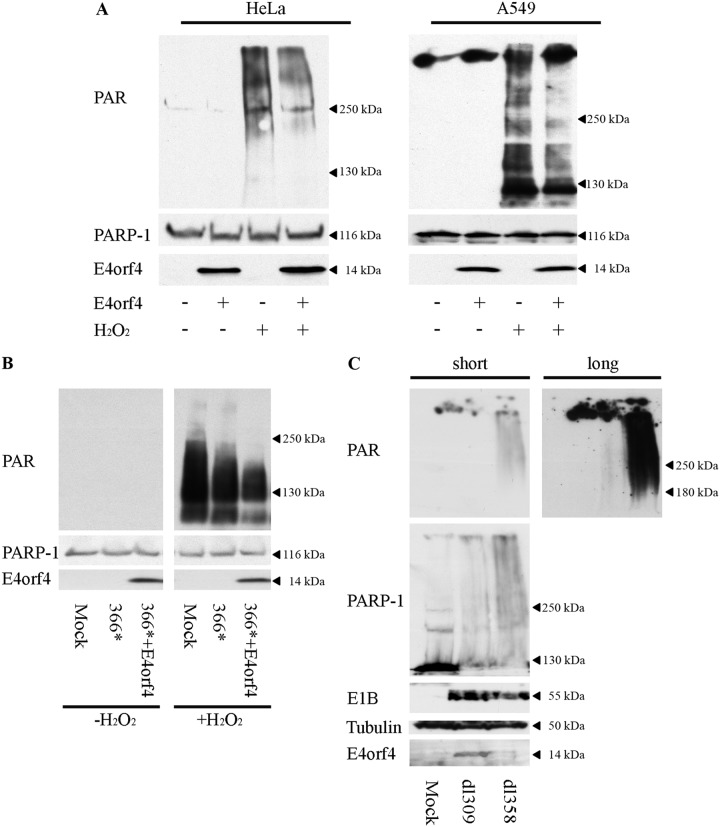
Because PARP inhibition assists E4orf4 in reducing DNA damage signaling (Fig. 6), we investigated whether E4orf4 naturally inhibits PARP-1 activity. We first tested the effect of E4orf4 on parylation stimulated by drug-induced DNA damage. E4orf4 was expressed in HeLa or A549 cells, and the cells were treated with H2O2 10 min prior to harvest or were left untreated. As seen in Fig. 7A, H2O2 treatment induced high levels of protein parylation in the cells in the absence of E4orf4, but lower parylation levels were observed in the presence of the viral protein. PARP-1 protein levels were not reduced by E4orf4. We next investigated whether E4orf4 diminished parylation upon virus infection. We could not detect parylation when HeLa cells were infected with the dl366* virus mutant (Fig. 7B, left), and we tested therefore whether E4orf4 could reduce drug-induced parylation in the context of viral infection. The results shown in Fig. 7B demonstrate that dl366* infection combined with a 10-min H2O2 treatment induced high levels of cellular parylation, and this parylation was reduced in the presence of virally expressed E4orf4 (dl366*+E4orf4). These results indicate that E4orf4 can reduce DNA damage-induced parylation during virus infection. It should be noted that we also consistently observed reduced parylation in H2O2-treated HeLa cells infected with the dl366* virus. However, E4orf4 made an additional pronounced contribution toward reducing H2O2-induced parylation. Because the E4-deficient virus dl366* did not induce detectable levels of parylation by itself, we also probed induction of parylation by a WT virus and examined whether deletion of E4orf4 alone (dl358) could enhance virus-induced parylation. As demonstrated in Fig. 7C, the dl309 Ad mutant, a phenotypically WT Ad5 with a backbone similar to that of dl358, induced relatively low parylation levels at 48 h p.i., but the dl358 virus lacking E4orf4 induced much higher parylation levels, indicating that E4orf4 reduced Ad-induced parylation. Thus, the results demonstrate that E4orf4 reduces parylation in the context of virus infection as well as when it is expressed alone in cells exposed to nonviral DNA damage.
FIG 7.
E4orf4 reduces drug- and virus-induced parylation. (A) HeLa and A549 cells were transfected with an empty vector or with a plasmid expressing Dox-inducible E4orf4. E4orf4 expression was induced for 4 h, and H2O2 was added to one set of cells at a concentration of 1 mM for 10 min prior to harvest. Whole-cell extracts were subjected to Western blot analysis with the indicated antibodies, and representative blots are shown. (B) HeLa cells were mock infected or infected with dl366* or dl366*+E4orf4 virus at 30 FFU/cell. H2O2 was added to one set of infected cells at 24 h p.i. at a concentration of 1 mM for 10 min prior to harvest. Whole-cell extracts were subjected to Western blot analysis with the indicated antibodies, and a representative blot is shown. The two parts of the picture are from the same exposed blot, but some lanes were removed from the middle. (C) HeLa cells were either mock-infected or infected with the Ad mutant virus dl309 (phenotypically WT Ad5) or dl358, which does not express E4orf4. Infections were performed at 10 FFU/cell. Cells were harvested 48 h p.i., and the levels of protein parylation in whole-cell extracts were analyzed by Western blotting. A representative blot is shown with two exposures (long and short) to facilitate visualization of very different levels of parylation.
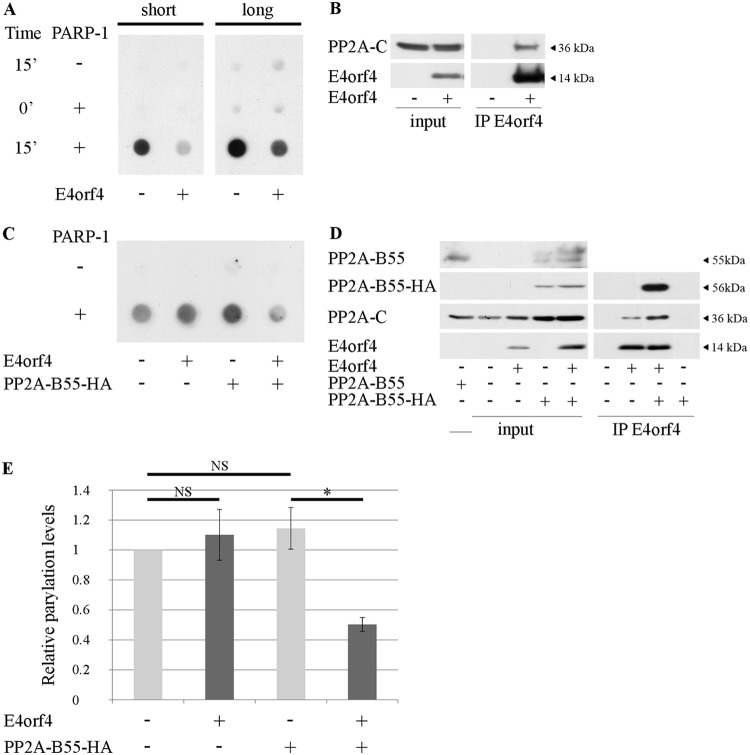
To test whether the decrease in parylation triggered by E4orf4 resulted from inhibition of PARP-1 enzymatic activity, in vitro parylation assays were performed (Fig. 8A). Reaction mixtures contained purified PARP-1 and immune complexes obtained from E4orf4-expressing or control cells by immunoprecipitation with an E4orf4-specific antibody (Fig. 8B). The mixtures were incubated in parylation buffer for 0 or 15 min and were then applied to nitrocellulose filters. Parylation activity was determined by staining the dot blots with PAR-specific antibodies. As shown in Fig. 8A, the immune complexes by themselves did not contain high PARP activity compared with that of the purified PARP-1. Addition of E4orf4 immune complexes reduced PARP-1 activity compared with the activity of this enzyme in the presence of control immune complexes. To probe the involvement of PP2A in inhibition of parylation by E4orf4, similar parylation reactions were performed using E4orf4 immune complexes from cells in which expression of the PP2A-B55 subunit was reduced by Dox-induced knockdown. This PP2A subunit mediates an interaction of E4orf4 with one of the PP2A heterotrimers, whereas PP2A-B56 subunits mediate E4orf4 interactions with other PP2A complexes (43, 44). Results shown in Fig. 8C to E demonstrate that when the PP2A-B55 levels were reduced, the levels of the PP2A-C subunit in the E4orf4-PP2A complexes were also decreased and parylation was no longer inhibited by E4orf4. However, when exogenous PP2A-B55 tagged with hemagglutinin (PP2A-B55-HA) was introduced into these cells, the ability of E4orf4 to associate with higher PP2A-C levels and to inhibit parylation was restored. The residual PP2A-C molecules in E4orf4 complexes from cells with reduced PP2A-B55 levels (Fig. 8D) could be part of the remaining B55-containing PP2A complexes as well as of B56-containing PP2A heterotrimers. The results described above support the conclusion that E4orf4, in collaboration with a PP2A heterotrimer containing the B55 subunit, inhibits PARP-1 activity.
FIG 8.
E4orf4 inhibits PARP-1 activity. (A) In vitro parylation assays were carried out. The reaction mixtures contained immune complexes isolated with E4orf4-specific antibodies from cell extracts containing an empty vector (−) or expressing E4orf4 (+), in parylation reaction buffer. Purified PARP-1 was added to two out of three sets of immune complexes, and reaction mixtures were incubated at 37°C for 15 min (15′) before termination. One set of reaction mixtures containing PARP-1 was kept on ice (time 0′). The terminated reactions were transferred to nitrocellulose, and dot blots were stained with PAR-specific antibodies. Two exposures of a representative blot are shown (short and long) to facilitate visualization of dots with very different intensities. (B) Ten percent of immune complexes used in the experiment shown in panel A as well as input lysates were subjected to Western blot analysis with the indicated antibodies. (C) L11 cells were induced with Dox for 3 days to reduce PP2A-B55 expression by shRNA-mediated knockdown. They were then transfected with plasmids expressing shRNA-resistant PP2A-B55-HA, E4orf4, or the corresponding empty vectors. The cells were harvested 24 h posttransfection, and cell extracts were subjected to immunoprecipitation with E4orf4-specific antibodies. Immune complexes were used in parylation reactions as described for panel A. (D) Ten percent of immune complexes used in the experiment shown in panel C as well as input lysates were subjected to Western blot analysis with the indicated antibodies. (E) Relative parylation levels in experiments described in panel C were determined by densitometry and normalization to the sample with reduced PP2A-B55 levels and no E4orf4 expression. The graph represents the average of three independent experiments. Error bars represent the standard error. *, P < 0.012; NS, not significant.
Investigation of candidate mechanisms underlying inhibition of PARP-1 by E4orf4.
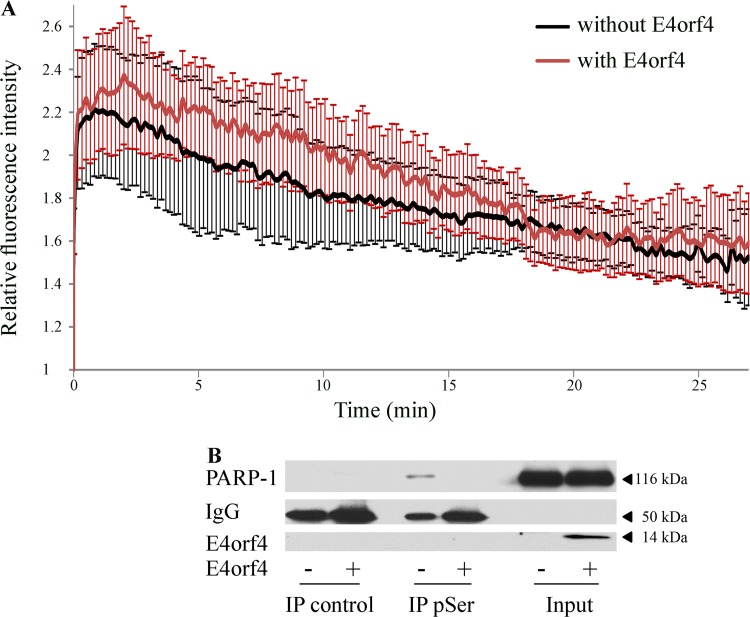
Various mechanisms could be considered to explain the inhibition of PARP-1 by E4orf4. One possible explanation involves premature removal of PARP-1 from DNA damage sites. To test this hypothesis, laser microirradiation experiments were performed using red fluorescent protein (RFP)-tagged PARP-1 and green fluorescent protein (GFP)-tagged E4orf4, and the kinetics of RFP-PARP-1 recruitment and removal from microirradiated damage sites in control and E4orf4-expressing cells was determined. The data in Fig. 9A demonstrate that E4orf4 had no significant effect on PARP-1 recruitment and release from damage sites.
FIG 9.
Investigation of mechanisms potentially responsible for inhibition of PARP-1 by E4orf4. (A) U2OS cells were transfected with a plasmid expressing WT E4orf4-GFP from a Dox-inducible promoter or a control plasmid expressing GFP, together with a plasmid expressing RFP-PARP-1. Following induction of E4orf4-GFP expression, cells with both RFP and GFP fluorescence were subjected to laser microirradiation at a preselected spot within the nucleus, and time-lapse microscopy was performed at 15-s time intervals to follow RFP-PARP-1 recruitment to the damage sites and its release. The fluorescence intensity of RFP signals at the laser-microirradiated sites was measured and normalized to the background level and to time zero. Relative fluorescence intensity at the damage site is shown. Error bars represent standard error (n = 10). A representative of two independent experiments is shown. (B) Clone 13 cells were either induced with Dox for 4 h to stimulate E4orf4 expression (+E4orf4) or were left uninduced (−E4orf4). Nuclear extracts were subjected to immunoprecipitation with a monoclonal antibody against phospho-serine (pSer) or with a Flag-tagged antibody, used as a negative control. The presence of PARP-1 in the immune complexes was determined by a Western blot analysis, and a representative blot is shown.
PARP-1 activity can be enhanced by various phosphorylation events (49–52). Because E4orf4 associates with PP2A and appears to reduce PARP-1 activity in a PP2A-dependent manner (Fig. 8), we examined whether PARP-1 phosphorylation was altered by E4orf4. Phospho-proteins were immunoprecipitated from cell extracts with a phospho-Ser (pSer)-specific antibody or with a Flag tag-specific control antibody, and the presence of PARP-1 in the immune complexes was determined by Western blot analysis. As shown in Fig. 9B, PARP-1 was precipitated from cell extracts lacking E4orf4 using the pSer antibodies but not with the control antibodies; however, PARP-1 was not detected in the pSer immune complexes from cells expressing E4orf4. Thus, E4orf4 expression is associated with reduced PARP-1 phosphorylation on Ser residues, which could potentially affect PARP-1 activity.
Inhibition of PARP activity enhances Ad replication.
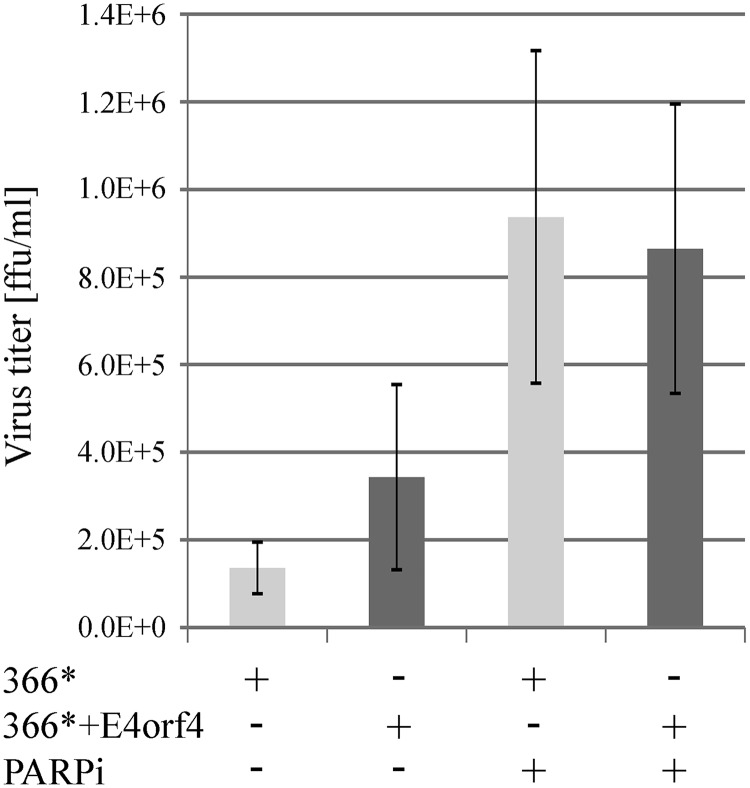
The finding that E4orf4 reduced parylation induced by Ad infection (Fig. 7) led us to investigate whether this E4orf4 function contributed to the efficiency of Ad replication. To address this possibility, we determined the effect of a PARP inhibitor on virus propagation. HeLa cells were infected with the dl366* and dl366*+E4orf4 viruses in the presence or absence of a PARP inhibitor, and the titer of progeny viruses obtained 24 h postinfection was determined. The data in Fig. 10 demonstrate that the PARP inhibitor increased the virus titer although its impact on dl366*+E4orf4 virus infection was weaker. Thus, the ability of E4orf4 to reduce parylation accounts, at least in part, for the previously reported contribution of E4orf4 to the efficiency of Ad infection (28).
FIG 10.
PARP inhibition enhances Ad replication. HeLa cells were infected with the Ad mutants dl366* and dl366*+E4orf4 at 30 FFU/cell. Cells were treated with a PARP inhibitor (PARPi) starting at 2 h p.i. or were left untreated. Viruses were collected from infected cells at 24 h postinfection, and their titers were determined. An average of two independent experiments is shown. Error bars represent standard error.
The role of PARP-1 in E4orf4-induced cell death.
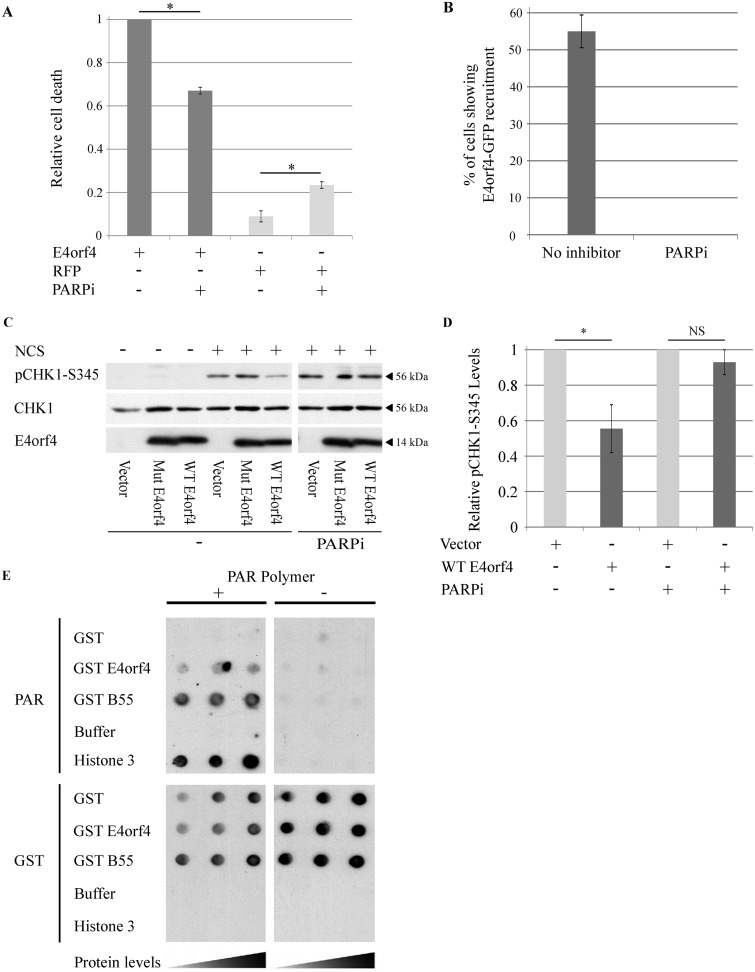
A second facet of E4orf4 biology is its ability to induce cancer cell death when it is expressed alone (40). We have previously demonstrated that E4orf4 expression resulted in accumulation of DNA damage after exposure to DNA-damaging drugs and in sensitization of cells to killing by sublethal concentrations of such drugs (28). DDR signaling is compromised in many cancers, causing cancer cells to be more sensitive to DDR inhibition. Thus, attenuation of DNA damage signaling may contribute to E4orf4-induced cancer cell death. Because we found that E4orf4 inhibited parylation by PARP-1, we set out to examine whether this E4orf4 function contributed to its ability to induce cell death. HeLa cells were transfected with a plasmid expressing E4orf4 or with a corresponding empty vector, with or without addition of a PARP inhibitor at a concentration that abolished parylation (Fig. 5A). Cell death levels were determined 24 h later using a 4′,6′-diamidino-2-phenylindole (DAPI) assay, as previously described (29, 34, 53). The results shown in Fig. 11A demonstrate that whereas the PARP inhibitor increased cell death in the absence of E4orf4, likely due to DDR inhibition leading to increased accumulation of damaged DNA, PARP inhibition significantly reduced the ability of E4orf4 to induce cell death. One possibility to explain these results is that E4orf4 must be recruited to DNA damage foci to inhibit the DDR and induce cancer cell death, but recruitment to these foci, unlike E4orf4 recruitment to viral RCs, requires PARP activity. We have previously shown that E4orf4 is recruited to sites of laser-microirradiated damage (30), and we now tested the effect of PARP inhibition on this recruitment. U2OS cells expressing E4orf4-GFP were subjected to laser microirradiation in the presence or absence of the PARP inhibitor, and recruitment of E4orf4 to irradiated sites was monitored by time-lapse confocal microscopy. The data shown in Fig. 11B demonstrate that whereas E4orf4-GFP recruitment to laser-microirradiated sites was evident in 55% of the tested cells, the PARP inhibitor completely abolished E4orf4 recruitment, which was no longer observed in any cells. These results demonstrate that protein parylation is required for E4orf4 recruitment to irradiation-induced DNA damage sites. Furthermore, the data shown in Fig. 11C and D demonstrate that PARP inhibition also prevented the previously reported PP2A-dependent ability of WT E4orf4 to reduce ATR signaling in drug-induced cells (28). These results further support the link between PARP-dependent E4orf4 recruitment to DNA damage sites and the ability of E4orf4 to inhibit damage signaling and to induce cell death when it is expressed alone.
FIG 11.
The role of PARP-1 in E4orf4-induced cell death. (A) HeLa cells were transfected with a plasmid expressing E4orf4 or with a control plasmid expressing the RFP protein. The cells were either left untreated or treated with a PARP inhibitor (4 μM; PARPi) at 3 h posttransfection, and another PARPi aliquot was added 12 h later. The cells were fixed at 24 h posttransfection and immunostained with an antibody against E4orf4 and counterstained with DAPI. Induction of cell death was measured by a DAPI assay. E4orf4-induced cell death in the absence of the PARP inhibitor was defined as 1, and relative cell death levels were determined for the other groups. The average of three independent experiments is shown. At least 150 transfected cells were analyzed in each experiment. Error bars represent standard error. Statistical significance was determined by a Student's t test (*, P < 0.015). (B) Recruitment of E4orf4-GFP to DNA damage sites and the effect of a specific PARP inhibitor (olaparib; PARPi) are shown as the percentage of cells in which E4orf4-GFP accumulated at laser-microirradiated sites. Error bars represent standard deviation (n > 25 from two independent experiments). (C) HeLa cells were transfected with an empty vector or with plasmids expressing WT E4orf4 or an E4orf4 mutant unable to bind PP2A (Mut E4orf4) from a Dox-inducible promoter. The cells were induced with Dox for 4 h and were treated with 0.5 ng/μl neocarzinostatin (NCS) or 4 μM PARP inhibitor (olaparib; PARPi) for 1 h prior to harvest. One set of cells was left untreated. Whole-cell extracts were subjected to Western blot analysis with the specified antibodies, and a representative blot is shown. The parts of this blot showing proteins in the presence or absence of a PARP inhibitor are from the same exposed blot, but some lanes were removed from the middle. The control lanes of this figure (without PARPi) were previously shown, combined with the missing lanes demonstrating the effect of a DNA-PK inhibitor on the ability of E4orf4 to reduce Chk1 phosphorylation (Fig. 2D in reference 30). (D) Blots as described in panel C from two independent experiments were subjected to densitometry. Relative phosphorylation levels of Chk1 were calculated as described for Fig. 6B and are shown in the graph. Error bars represent standard error (n = 2). Statistical significance was determined by a Student's t test. *, P < 0.05; NS, not significant. (E) GST and GST fused to E4orf4 or to the PP2A-B55 subunit, as well as histone H3, were spotted on two sets of nitrocellulose filters at three concentrations. One filter was incubated with PAR, and the other was left untreated. At the end of the incubation the filters were washed and probed with antibodies to PAR and then with GST-specific antibodies, which indicated that the levels of all GST proteins were similar.
Many proteins recruited to sites of DNA damage by PARP-1 do so by direct interaction with PAR chains (54). As our results showed that PARP-1 activity was required for recruitment of E4orf4 to laser microirradiated sites, we set out to determine whether E4orf4 can bind PAR chains. Indeed, a synthetic PAR polymer bound to dot-blotted recombinant GST-E4orf4 and to recombinant GST-PP2A-B55 as well as to purified histone H3, a known PAR-binding protein and positive control. The PAR polymer did not bind GST alone (Fig. 11E). These results indicate that when expressed outside the context of virus infection, E4orf4 is recruited to DNA damage sites most likely via direct interaction with PAR chains.
DISCUSSION
Many DNA viruses evolved ways to inhibit the DDR, which acts as an antiviral defense system. The importance of DDR inhibition to Ad infection is indicated by the variety of Ad proteins that participate in this process. E1B-55K, E4orf6, E4orf3, E4orf4, and core protein VII were all shown to contribute to inhibition of DDR branches regulated by ATM and ATR (28, 46, 47, 55, 56). Studies of the Ad E4orf4 protein revealed that this protein interferes with several different branches of the DDR. It was previously reported that E4orf4 reduced ATM and ATR signaling in virus-infected and in drug-treated cells but that neither kinase was required for inhibition of the other kinase by E4orf4 (28). It was then shown that DNA-PK was required for inhibition of ATM and ATR signaling by E4orf4 but that later in infection E4orf4 inhibited DNA-PK itself (30). In the current report we demonstrate that E4orf4 associates with PARP-1 and inhibits virus- and drug-induced parylation. The combined results indicate that E4orf4, which binds at least two DNA damage sensors (DNA-PK and PARP-1), has a multifaceted effect on the DDR, facilitating inhibition of various branches of this network.
It has been previously reported that Ad genome replication induced cellular parylation and that some late Ad proteins could be parylated (19). PAR polymer was suggested to act as a signaling molecule that induces cell death (57, 58), and its accumulation could possibly lead to premature death of Ad-infected cells. In addition, overstimulation of PARP activity leads to increased utilization of NAD+, the PARP substrate, resulting in NAD+ depletion as well as depletion of ATP, a precursor of NAD+. This depletion may cause necrotic cell death (59). Thus, inhibition of parylation would be beneficial for the virus as premature cell death would be attenuated, and ATP would not be dangerously depleted, relieving inhibition of ATP-dependent processes. Furthermore, inhibition of PARP contributed to the ability of E4orf4 to reduce ATM and possibly ATR signaling (Fig. 6), and inhibition of these DDR branches also increased the efficiency of Ad replication (28). Thus, inhibition of parylation by E4orf4 could increase virus replication via several mechanisms. We showed here that chemical inhibition of PARP moderately enhanced propagation of the Ad dl366* mutant virus lacking E4orf4 (Fig. 10). This moderate enhancement could be explained, at least in part, by the finding that the dl366* virus itself provided gene products that reduced parylation to some extent (Fig. 7B), and thus addition of another PARP inhibitor did not exhibit the full effect of PARP inhibition on virus infection. The observation that the impact of PARP inhibition on infection with the mutant virus expressing E4orf4 (dl366*+E4orf4) was weaker than its impact on dl366* infection (Fig. 10) is consistent with the conclusion that reduced parylation by E4orf4 contributed to enhanced Ad replication reported in the presence of E4orf4 (30).
Inhibition of PARP during Ad infection could be achieved by at least two mechanisms. One involves the E4orf4-dependent decrease in PARP-1 phosphorylation (Fig. 9B), which also contributes to PARP-1 inhibition following non-virus-induced DNA damage. The other could be linked to the sequestration of PARP inside RCs, as seen in Fig. 4. In addition, as reported previously (19), the Ad E1B-55K and E4orf3 proteins inhibit some consequences of PARP activity, and these results further confirm the importance of counteracting PARP signaling during Ad infection.
The nature of the contribution of PARP inhibition to E4orf4-reduced ATM and ATR signaling has not yet been defined. However, it was formerly shown that parylation of various substrates influenced ATM and ATR recruitment to chromatin and contributed to their activity (60, 61) or to the activity of their substrates (62). Therefore, inhibition of parylation could impact the E4orf4 effect on these pathways. Still, inhibition of ATM and ATR signaling cannot be due solely to diminished parylation because decreasing parylation in the context of a dl366* virus infection did not by itself reduce ATM and ATR signaling (Fig. 6). Chemical PARP inhibition replaced the requirement for E4orf4 binding to PARP-1, which leads to PARP inhibition. Under these conditions, the interaction of E4orf4 with DNA-PK could provide the additional contribution required for attenuation of ATM and ATR signaling (30). Based on the results described here and in Nebenzahl-Sharon et al. (30), we suggest that the combined functional interactions of E4orf4 with PARP-1 and DNA-PK are required to achieve a more complete effect of E4orf4 on the DDR.
The finding that E4orf4 is recruited to nonviral DNA damage sites in a PARP-dependent manner but inhibits PARP activity suggests that E4orf4 is first recruited to the damage sites, interacts with PARP-1, and then inhibits this enzyme. During Ad infection, E4orf4 and PARP-1 are both recruited to viral RCs as soon as RCs are visualized by DBP staining (Fig. 3) (30). The finding that E4orf4 and PARP-1 are recruited to Ad RCs independently suggests that they likely interact only after recruitment, leading to PARP inhibition by E4orf4. PARP-1 recruitment to RCs may occur similarly to its recruitment to DNA damage foci, and E4orf4 recruitment may require other early Ad proteins.
The discovery that PARP inhibition decreased the ability of E4orf4 to induce cell death is consistent with the requirement of PARP-1 activity for recruitment of E4orf4 to DNA damage sites where it can inhibit PARP-1 itself as well as other DDR proteins, leading to cancer cell death. As suggested previously, several types of cancer cells have deficiencies in components of DNA damage signaling, and this can make them more susceptible to inhibition of additional DDR pathways by E4orf4, leading to cancer-selective cell death. The finding that E4orf4 was recruited to damage sites in only 55% of the cells is not exceptional, and other examples include recruitment of BRCA1 to damage foci in only 50% of primary mammary epithelial (PME) cells (63). The degree of recruitment could be cell cycle dependent (64). The finding that PARP inhibition did not completely abolish E4orf4-induced cell death may result from the fact that E4orf4 interferes with additional cellular pathways, such as cell cycle regulation (36, 65), and their disruption can contribute to cancer cell death in a PARP-independent manner. It is also possible that the activity of the PARP inhibitor diminished with time during the 24-h incubation with the cells even though a second aliquot was added after 12 h.
In summary, this work describes a novel mechanism by which E4orf4 inhibits DNA damage signaling. Through an interaction with the DNA damage sensor PARP-1 and by reducing its phosphorylation, E4orf4 inhibits the ability of this enzyme to transmit a DNA damage signal by parylation. Inhibition of PARP assists E4orf4 in reducing ATM and ATR signaling and improves virus replication. Outside the context of virus infection, E4orf4 requires PARP activity to localize to damage sites and to inhibit damage signaling, an activity that is required for E4orf4-induced cancer-selective cell death.
MATERIALS AND METHODS
Cells, plasmids, transfection, virus mutants, and infections.
HeLa, A549, 293T, and U2OS cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). Cells used for transfection with tetracycline-inducible plasmids were cultured for 3 days prior to the start of the experiment in medium containing 10% tetracycline-free FCS (BD Bioscience or PAN-Biotech GmbH). Clone 13 and clone 3 cells containing Dox-inducible WT E4orf4 and the R81A/F84A E4orf4 mutant, respectively, were generated in our laboratory from T-Rex-293 cells (Invitrogen/Life Technologies) (28). L11 cells containing a Dox-inducible PP2A-B55alpha short hairpin RNA (shRNA) were constructed similarly (28).
GST and GST-E4orf4 as well as the PP2A-B55alpha mutant resistant to the PP2A-B55alpha shRNA have been previously described (20, 43). WT E4orf4 (20), WT E4orf4-GFP, and the R81A/F84A E4orf4 mutant that does not bind PP2A (45) were cloned into the tetracycline-inducible pcDNA4/TO vector (Invitrogen). The plasmids were transfected together with a plasmid expressing the tetracycline repressor (pcDNA6/TR; Invitrogen), using TurboFect (ThermoFisher Scientific), according to the manufacturer’s instructions. One day later the cells were induced with 1 μg/ml Dox for 3 to 4 h. When required by the experiment, H2O2 was added 10 min prior to harvest at a concentration of 1 mΜ, and the following reagents were added for the last hour of E4orf4 induction: a PARP inhibitor (4 μM olaparib; Selleck Chem), 50 nM okadaic acid (Santa Cruz Biotech), and 0.5 ng/μl neocarzinostatin (NCS).
Adenovirus mutants dl309 (phenotypically wild-type Ad5 in tissue culture), dl358 (dl309 backbone, lacking E4orf4) (66, 67), dl366*, lacking the complete E4 region, and dl366*+E4orf4, lacking all E4 open reading frames (ORFs) except E4orf4 (48), were propagated and titers were determined on W162 cells (T. Shenk, Princeton University [68]). dl366* and dl366*+E4orf4 virus infections were performed at a multiplicity of 30 fluorescence-forming units (FFU) per cell in HeLa cells. dl309 and dl358 infections were performed at 10 FFU per cell. The PARP inhibitor (olaparib; Selleck Chem) was added to infected cells at 2 h postinfection at a final concentration of 4 μM and was added again to the cells at 13 h postinfection using the same concentration. Untreated cells received equal quantities of the solvent. Infected cells were harvested for protein extraction, immunostaining, or virus collection at the times indicated in the figures and figure legends.
GST pulldown assays, cell extracts, immunoprecipitation, and Western blot analysis.
Purification of glutathione S-transferase (GST) fusion proteins from bacteria and GST pulldown assays were previously described (43). Whole-cell extracts were prepared in SDS sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 50 mM dithiothreitol [DTT], 10% glycerol), and viscosity was reduced by passage several times through a 27-gauge needle after a 3-min incubation at 95°C. Proteins were analyzed by Western blotting using the antibodies indicated in the figures. For immunoprecipitation assays, cell extracts were prepared by the Dignam method (53). Dignam buffer C (20 mM HEPES-KOH, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 25% [vol/vol] glycerol) was supplemented with 2× cOmplete protease inhibitor cocktail (Sigma), protease inhibitor cocktails for cells and yeast extracts (1:50 and 1:125, respectively; Sigma), and phosphatase inhibitor cocktails 2 and 3 (1:100; Sigma). Immunoprecipitation reactions for in vitro parylation assays were performed with cell extracts prepared in lysis buffer (50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.5% Nonidet P-40) supplemented with similar protease and phosphatase inhibitors.
Blot images were scanned with an Epson Photo 4990 scanner and were processed using Adobe Photoshop CS2. Band intensities were quantified using TotalLab software.
Antibodies were used as follows: E4orf4 (34); E1B-55K (clone 2A6) (69); Ad capsid proteins (a gift from T. Dobner); PP2A-C and PARP-1 (mouse) (BD Transduction Laboratories); PARP-1 (rabbit) (Cell signaling); PAR (Enzo Life Sciences); HA tag (Covance); pATM-S1981 and ATM (mouse) (Epitomics); pChk1-S345, ATM (rabbit), and Chk1 (Santa Cruz); Akt and pAkt (R&D Systems); pSer (Q5) (Qiagen); Flag tag and alpha-tubulin (Sigma); GST (Millipore).
Laser microirradiation.
U2OS cells were presensitized with 10 μM Hoechst 3334 dye (Sigma) for 15 min at 37°C. Laser microirradiation was performed using an inverted confocal microscope (LSM-700; Zeiss) equipped with a CO2 module and a 37°C heating chamber. A preselected spot within the nucleus was microirradiated with 10 iterations of a 405-nm laser with 100% power to generate localized DNA damage. Time-lapse images were acquired at 15-s time intervals. The fluorescence intensity of signals at laser microirradiated sites was measured using Zen 2009 software (Carl Zeiss). Data obtained were corrected for the loss of total fluorescence and normalized to the initial intensity.
Cell death assay: the DAPI assay.
DAPI assays, which determine nuclear aberrations, have been described previously (53, 70). The fraction of E4orf4-expressing cells with condensed, abnormal, or fragmented nuclei was determined in each experiment by counting at least 150 transfected nuclei. The average of two experiments was calculated. Error bars represent standard error, and statistical significance of the results was determined by a Student's t test.
Immunofluorescence microscopy.
HeLa cells were seeded on glass coverslips and infected as described above. At appropriate time points, the cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with PBS containing 0.25% Triton X-100 for 10 min at room temperature, and washed with PBS. The cells were blocked with PBS containing 10% goat serum and then incubated with primary antibodies diluted in blocking buffer. The antibodies used were anti-DBP rabbit polyclonal antibody (generously provided by Peter van der Vliet, University of Utrecht) and anti-PARP-1 mouse monoclonal antibody (Sigma-Aldrich). The cells were washed and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG (Invitrogen). The cells were washed a final time and mounted on slides with Immu-Mount (Thermo Shandon). Images were captured using a 63× objective with a Zeiss Axiovert 200 M digital deconvolution microscope and analyzed with AxioVision, version 4.8.2, SP3 software. For high-resolution microscopy, images were captured using a 100× objective with a Nikon N-SIM microscope. Image processing, reconstruction, and analysis were carried out using NIS-Elements Ar/NIS-Elements C software and NIS-A N-SIM Analysis. 3D renderings were generated using Imaris software.
PAR binding.
A PAR binding protocol was adapted from Rulten et al. (71). Briefly, dilutions of GST, GST-E4orf4, GST-PP2A-B55, and histone H3 were made in PBS and applied to nitrocellulose membrane by vacuum. The membrane was blocked in 5% nonfat milk in binding buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl) for 1 h. PAR (50 nM; Trevigen) was incubated with the membrane for 45 min in the same buffer and then rinsed three times with binding buffer for 5 min. The membrane was then incubated with anti-PAR antibody (10 H; Enzo Life Sciences) overnight and then washed again. PAR interactions were detected using anti-mouse horseradish peroxidase conjugate followed by chemiluminescence.
In vitro parylation.
Parylation reactions were performed by adding 25 μl of reaction buffer (50 mM Tris-HCl [pH 8.0], 20 mM NaCl, 4 mM MgCl2, 0.5 mM NAD+ [Merck Millipore], 0.25 mM DTT, 33.5 ng of sonicated salmon sperm DNA) to E4orf4 or control immune complexes bound to protein Sepharose A beads. Purified PARP-1 (2.94 ng, or 0.1 units; Trevigen) was added to each reaction mixture, and reaction mixtures were incubated for 15 min at 37°C with shaking at 1,000 rpm. One set of each immune complex (control and E4orf4) was incubated similarly in the absence of purified PARP-1. Reactions were terminated by addition of SDS sample buffer without glycerol, and products were boiled at 100°C for 3 min and loaded onto nitrocellulose filter using a dot blot apparatus. Parylation was detected by staining the membrane with anti‐PAR serum solution (10 H; Enzo Life Sciences).
ACKNOWLEDGMENTS
We are grateful to T. Dobner and T. Speiseder (Leibniz Institute for Experimental Virology) for gifts of cell lines, viruses, and antibodies and to Shani Bendori (Technion) for help with the Imaris software.
This work was supported by a grant (no. 261/15) to T.K. from the Israel Science Foundation and by a grant (no. 2013140) to T.K. and P.H. from the United States-Israel Binational Science Foundation.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Bartek J, Lukas J. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol 13:738–747. doi: 10.1016/S0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 2.Zhou BB, Elledge SJ. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 3.Sirbu BM, Cortez D. 2013. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 5:a012724. doi: 10.1101/cshperspect.a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. 2010. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 6.Wei H, Yu X. 2016. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics 14:131–139. doi: 10.1016/j.gpb.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkauskaite E, Jankevicius G, Ahel I. 2015. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol Cell 58:935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Dawicki-McKenna JM, Langelier MF, DeNizio JE, Riccio AA, Cao CD, Karch KR, McCauley M, Steffen JD, Black BE, Pascal JM. 2015. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol Cell 60:755–768. doi: 10.1016/j.molcel.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langelier MF, Zandarashvili L, Aguiar PM, Black BE, Pascal JM. 2018. NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun 9:844. doi: 10.1038/s41467-018-03234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. 2013. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell 52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. 2013. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J 280:3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, Chen Y, Li M, Yu X. 2014. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc Natl Acad Sci U S A 111:7278–7283. doi: 10.1073/pnas.1318367111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG. 1994. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim Biophys Acta 1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 14.Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I. 2011. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weitzman MD, Lilley CE, Chaurushiya MS. 2010. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol 64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 16.Hollingworth R, Grand RJ. 2015. Modulation of DNA damage and repair pathways by human tumour viruses. Viruses 7:2542–2591. doi: 10.3390/v7052542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnell AS, Grand RJ. 2012. DNA viruses and the cellular DNA-damage response. J Gen Virol 93:2076–2097. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- 18.Kuny CV, Sullivan CS. 2016. Virus-host interactions and the ARTD/PARP family of enzymes. PLoS Pathog 12:e1005453. doi: 10.1371/journal.ppat.1005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner RL, Wilkinson JC, Ornelles DA. 2014. E1B and E4 oncoproteins of adenovirus antagonize the effect of apoptosis inducing factor. Virology 456-457:205–219. doi: 10.1016/j.virol.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Israel H, Sharf R, Rechavi G, Kleinberger T. 2008. Adenovirus E4orf4 protein downregulates MYC expression through interaction with the PP2A-B55 subunit. J Virol 82:9381–9388. doi: 10.1128/JVI.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondesson M, Ohman K, Manervik M, Fan S, Akusjärvi G. 1996. Adenovirus E4 open reading 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J Virol 70:3844–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannervik M, Fan S, Strom AC, Helin K, Akusjarvi G. 1999. Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256:313–321. doi: 10.1006/viro.1999.9663. [DOI] [PubMed] [Google Scholar]
- 23.Muller U, Kleinberger T, Shenk T. 1992. Adenovirus E4orf4 protein reduces phosphorylation of c-fos and E1A proteins while simultaneously reducing the level of AP-1. J Virol 66:5867–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estmer Nilsson C, Petersen-Mahrt S, Durot C, Shtrichman R, Krainer AR, Kleinberger T, Akusjärvi G. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J 20:864–871. doi: 10.1093/emboj/20.4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanopka A, Mühlemann O, Petersen-Mahrt S, Estmer C, Öhrmalm C, Akusjärvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 26.O'Shea C, Klupsch K, Choi S, Bagus B, Soria C, Shen J, McCormick F, Stokoe D. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J 24:1211–1221. doi: 10.1038/sj.emboj.7600597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinberger T. 2014. Induction of cancer-specific cell death by the adenovirus e4orf4 protein. Adv Exp Med Biol 818:61–97. doi: 10.1007/978-1-4471-6458-6_4. [DOI] [PubMed] [Google Scholar]
- 28.Brestovitsky A, Nebenzahl-Sharon K, Kechker P, Sharf R, Kleinberger T. 2016. The adenovirus E4orf4 protein provides a novel mechanism for inhibition of the DNA damage response. PLoS Pathog 12:e1005420. doi: 10.1371/journal.ppat.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinberger T. 2015. Mechanisms of cancer cell killing by the adenovirus E4orf4 protein. Viruses 7:2334–2357. doi: 10.3390/v7052334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nebenzahl-Sharon K, Shalata H, Sharf R, Amer J, Khoury-Haddad H, Sohn SY, Ayoub N, Hearing P, Kleinberger T. 2019. Biphasic functional interaction between the adenovirus E4orf4 protein and DNA-PK. J Virol 93:e01365-18. doi: 10.1128/JVI.01365-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavoie JN, Nguyen M, Marcellus RC, Branton PE, Shore GC. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J Cell Biol 140:637–645. doi: 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livne A, Shtrichman R, Kleinberger T. 2001. Caspase activation by adenovirus E4orf4 protein is cell line-specific and is mediated by the death receptor pathway. J Virol 75:789–798. doi: 10.1128/JVI.75.2.789-798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcellus RC, Lavoie JN, Boivin D, Shore GC, Ketner G, Branton PE. 1998. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J Virol 72:7144–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shtrichman R, Kleinberger T. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J Virol 72:2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afifi R, Sharf R, Shtrichman R, Kleinberger T. 2001. Selection of apoptosis-deficient adenovirus E4orf4 mutants in S. cerevisiae. J Virol 75:4444–4447. doi: 10.1128/JVI.75.9.4444-4447.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornitzer D, Sharf R, Kleinberger T. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in S. cerevisiae and interacts with the anaphase promoting complex/cyclosome. J Cell Biol 154:331–344. doi: 10.1083/jcb.200104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maoz T, Koren R, Ben-Ari I, Kleinberger T. 2005. YND1 interacts with CDC55 and is a novel mediator of E4orf4-induced toxicity. J Biol Chem 280:41270–41277. doi: 10.1074/jbc.M507281200. [DOI] [PubMed] [Google Scholar]
- 38.Roopchand DE, Lee JM, Shahinian S, Paquette D, Bussey H, Branton PE. 2001. Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A. Oncogene 20:5279–5290. doi: 10.1038/sj.onc.1204693. [DOI] [PubMed] [Google Scholar]
- 39.Pechkovsky A, Lahav M, Bitman E, Salzberg A, Kleinberger T. 2013. E4orf4 induces PP2A- and Src-dependent cell death in Drosophila melanogaster and at the same time inhibits classic apoptosis pathways. Proc Natl Acad Sci U S A 110:E1724–33. doi: 10.1073/pnas.1220282110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shtrichman R, Sharf R, Barr H, Dobner T, Kleinberger T. 1999. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc Natl Acad Sci U S A 96:10080–10085. doi: 10.1073/pnas.96.18.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosen H, Sharf R, Pechkovsky A, Salzberg A, Kleinberger T. 2019. Selective elimination of cancer cells by the adenovirus E4orf4 protein in a Drosophila cancer model: a new paradigm for cancer therapy. Cell Death Dis 10:455–468. doi: 10.1038/s41419-019-1680-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sents W, Ivanova E, Lambrecht C, Haesen D, Janssens V. 2013. The biogenesis of active protein phosphatase 2A holoenzymes: a tightly regulated process creating phosphatase specificity. FEBS J 280:644–661. doi: 10.1111/j.1742-4658.2012.08579.x. [DOI] [PubMed] [Google Scholar]
- 43.Kleinberger T, Shenk T. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J Virol 67:7556–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shtrichman R, Sharf R, Kleinberger T. 2000. Adenovirus E4orf4 protein interacts with both Bα and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Bα. Oncogene 19:3757–3765. doi: 10.1038/sj.onc.1203705. [DOI] [PubMed] [Google Scholar]
- 45.Marcellus RC, Chan H, Paquette D, Thirlwell S, Boivin D, Branton PE. 2000. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Bα subunit of protein phosphatase 2A. J Virol 74:7869–7877. doi: 10.1128/jvi.74.17.7869-7877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carson CT, Orazio NI, Lee DV, Suh J, Bekker-Jensen S, Araujo FD, Lakdawala SS, Lilley CE, Bartek J, Lukas J, Weitzman MD. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J 28:652–662. doi: 10.1038/emboj.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J 22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang MM, Hearing P. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol 63:2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. 2006. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci U S A 103:7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright RH, Castellano G, Bonet J, Le Dily F, Font-Mateu J, Ballare C, Nacht AS, Soronellas D, Oliva B, Beato M. 2012. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev 26:1972–1983. doi: 10.1101/gad.193193.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong F, Soubeyrand S, Hache RJ. 2010. Activation of PARP-1 in response to bleomycin depends on the Ku antigen and protein phosphatase 5. Oncogene 29:2093–2103. doi: 10.1038/onc.2009.492. [DOI] [PubMed] [Google Scholar]
- 52.Cohen-Armon M. 2007. PARP-1 activation in the ERK signaling pathway. Trends Pharmacol Sci 28:556–560. doi: 10.1016/j.tips.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Brestovitsky A, Sharf R, Mittelman K, Kleinberger T. 2011. The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic Acids Res 39:6414–6427. doi: 10.1093/nar/gkr231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krietsch J, Rouleau M, Pic E, Ethier C, Dawson TM, Dawson VL, Masson JY, Poirier GG, Gagne JP. 2013. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med 34:1066–1087. doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karen KA, Hearing P. 2011. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J Virol 85:4135–4142. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah GA, O'Shea CC. 2015. Viral and cellular genomes activate distinct DNA damage responses. Cell 162:987–1002. doi: 10.1016/j.cell.2015.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. 2006. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A 103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.David KK, Andrabi SA, Dawson TM, Dawson VL. 2009. Parthanatos, a messenger of death. Front Biosci (Landmark ed) 14:1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fouquerel E, Sobol RW. 2014. ARTD1 (PARP1) activation and NAD+ in DNA repair and cell death. DNA Repair (Amst) 23:27–32. doi: 10.1016/j.dnarep.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khoury-Haddad H, Guttmann-Raviv N, Ipenberg I, Huggins D, Jeyasekharan AD, Ayoub N. 2014. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc Natl Acad Sci U S A 111:E728–37. doi: 10.1073/pnas.1317585111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kedar PS, Stefanick DF, Horton JK, Wilson SH. 2008. Interaction between PARP-1 and ATR in mouse fibroblasts is blocked by PARP inhibition. DNA Repair (Amst) 7:1787–1798. doi: 10.1016/j.dnarep.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min W, Bruhn C, Grigaravicius P, Zhou ZW, Li F, Kruger A, Siddeek B, Greulich KO, Popp O, Meisezahl C, Calkhoven CF, Burkle A, Xu X, Wang ZQ. 2013. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat Commun 4:2993. doi: 10.1038/ncomms3993. [DOI] [PubMed] [Google Scholar]
- 63.Tong WM, Yang YG, Cao WH, Galendo D, Frappart L, Shen Y, Wang ZQ. 2007. Poly(ADP-ribose) polymerase-1 plays a role in suppressing mammary tumourigenesis in mice. Oncogene 26:3857–3867. doi: 10.1038/sj.onc.1210156. [DOI] [PubMed] [Google Scholar]
- 64.Mosammaparast N, Kim H, Laurent B, Zhao Y, Lim HJ, Majid MC, Dango S, Luo Y, Hempel K, Sowa ME, Gygi SP, Steen H, Harper JW, Yankner B, Shi Y. 2013. The histone demethylase LSD1/KDM1A promotes the DNA damage response. J Cell Biol 203:457–470. doi: 10.1083/jcb.201302092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sriskandarajah N, Blanchette P, Kucharski TJ, Teodoro J, Branton PE. 2015. Analysis by live imaging of effects of the adenovirus E4orf4 protein on passage through mitosis of H1299 tumor cells. J Virol 89:4685–4689. doi: 10.1128/JVI.03437-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halbert DN, Cutt JR, Shenk T. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol 56:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones N, Shenk T. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 68.Weinberg DH, Ketner G. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J Virol 57:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sarnow P, Sullivan CA, Levine AJ. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510–517. doi: 10.1016/0042-6822(82)90054-X. [DOI] [PubMed] [Google Scholar]
- 70.Avital-Shacham M, Sharf R, Kleinberger T. 2014. NTPDASE4 gene products cooperate with the adenovirus E4orf4 protein through PP2A-dependent and -independent mechanisms and contribute to induction of cell death. J Virol 88:6318–6328. doi: 10.1128/JVI.00381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gomez-Herreros F, Hafezparast M, Caldecott KW. 2014. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res 42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]