Mechanisms of immunodeficiency related to STAT1 gain of function have not been previously studied in an animal model of viral pathogenesis. Using virological and immunological techniques, we examined the immune response to γHV68 in heterozygous mice that have an autosomal dominant mutation in the STAT1 coiled-coil domain (STAT1 R274W). We observed impaired control of infection, which was associated with diminished production of gamma interferon (IFN-γ), fewer effector CD4+ and CD8+ T cells, and a reduction in the number of antigen-specific CD8+ T cells. These findings indicate that a STAT1 gain-of-function mutation limits production of antiviral T cells, likely contributing to immunodeficiency against herpesviruses.
KEYWORDS: STAT transcription factors, T cells, herpesviruses, immune deficiency, innate immunity, gamma interferon
ABSTRACT
Autosomal dominant STAT1 mutations in humans have been associated with chronic mucocutaneous candidiasis (CMC), as well as with increased susceptibility to herpesvirus infections. Prior studies have focused on mucosal and Th17-mediated immunity against Candida, but mechanisms of impaired antiviral immunity have not previously been examined. To begin to explore the mechanisms of STAT1-associated immunodeficiency against herpesviruses, we generated heterozygous STAT1 R274W knock-in mice that have a frequently reported STAT1 mutation associated in humans with susceptibility to herpesvirus infections. In primary macrophages and fibroblasts, we found that STAT1 R274W had no appreciable effect on cell-intrinsic immunity against herpes simplex virus 1 (HSV-1) or gammaherpesvirus 68 (γHV68) infection. However, intraperitoneal inoculation of mice with γHV68 was associated with impaired control of infection at day 14 in STAT1 R274W mice compared with that in wild-type (WT) littermate control animals. Infection of STAT1 R274W mice was associated with paradoxically decreased expression of IFN-stimulated genes (ISGs) and gamma interferon (IFN-γ), likely secondary to defective CD4+ and CD8+ T cell responses, including diminished numbers of antigen-specific CD8+ T cells. Viral pathogenesis studies in WT and STAT1 R274W mixed bone marrow chimeric mice revealed that the presence of WT leukocytes was sufficient to limit infection and that antigen-specific STAT1 R274W CD8+ T cell responses were impaired even in the presence of WT leukocytes. Thus, in addition to regulating Th17 responses against Candida, a STAT1 gain-of-function mutant impedes antigen-specific T cell responses against a common gammaherpesvirus in mice.
IMPORTANCE Mechanisms of immunodeficiency related to STAT1 gain of function have not been previously studied in an animal model of viral pathogenesis. Using virological and immunological techniques, we examined the immune response to γHV68 in heterozygous mice that have an autosomal dominant mutation in the STAT1 coiled-coil domain (STAT1 R274W). We observed impaired control of infection, which was associated with diminished production of gamma interferon (IFN-γ), fewer effector CD4+ and CD8+ T cells, and a reduction in the number of antigen-specific CD8+ T cells. These findings indicate that a STAT1 gain-of-function mutation limits production of antiviral T cells, likely contributing to immunodeficiency against herpesviruses.
INTRODUCTION
Autosomal dominant STAT1 mutations in humans have been associated with increased susceptibility to infections with Candida as well as with DNA viruses, including alphaherpesviruses and gammaherpesviruses (1–5). Whereas aberrant mucosal interleukin-17A (IL-17A)-mediated immunity is thought to explain susceptibility to Candida (1, 2, 6), mechanisms of impaired antiviral immunity have not previously been explored, and no animal model of the disease has been described. Since activation of STAT1 by the type I (α/β), type II (γ), and type III (λ) interferon (IFN) receptors leads to upregulation of antiviral IFN-stimulated genes (ISGs) (7–10), a STAT1 gain-of-function mutation might be expected to upregulate ISGs and to enhance antiviral immunity. Thus, it is somewhat counterintuitive that STAT1 gain of function makes humans more susceptible to viruses.
STAT1 is comprised of N-terminal, coiled-coil, DNA-binding, SH2, and C-terminal transactivation domains (11). Human mutations in STAT1 have most frequently been reported in the highly conserved coiled-coil and DNA-binding domains (12–14). The most commonly reported mutations in the coiled-coil domain are in arginine 274 (R274Q and R274W) (1, 2, 15). Peripheral blood mononuclear cells (PBMCs) from patients with these mutations have diminished numbers of IL-17-producing T cells, which may explain susceptibility to Candida (1, 16). Furthermore, overexpression of R274 mutants in STAT1-deficient cell lines leads to upregulation of an IFN-γ luciferase reporter and a corresponding upregulation of ISGs (1, 17). Mutations in R274 also have been associated with increased phosphorylation of STAT1 upon stimulation with IFN-γ, IFN-α, or IL-27 (16–18).
In addition to upregulating expression of antiviral ISGs, STAT1 also regulates adaptive immune responses (19, 20). Therefore, we reasoned that STAT1-associated immunodeficiency may reflect a combination of innate and adaptive immune defects. To test this hypothesis, we generated heterozygous STAT1 R274W mice that have an autosomal dominant mutation in the highly conserved STAT1 coiled-coil domain. Here, we report our discoveries that heterozygous STAT1 R274W mice exhibit impaired antigen-specific CD8+ T cell responses during infection with gammaherpesvirus 68 (γHV68) and that this immunological defect is associated with increased viral burden at late but not early time points. The STAT1 R274W mutation had no impact on γHV68 replication in bone marrow-derived macrophages (BMDMs) or primary mouse embryonic fibroblasts (MEFs), suggesting that cell-intrinsic control of viral replication remains intact. We found that the dominant STAT1 R274W mutation impaired both CD4+ and CD8+ T cell responses, leading to diminished production of IFN-γ and a relative decrease in antiviral ISG expression during acute infection in vivo but not during infection of cultured cells. Studies in wild-type (WT) and STAT1 R274W mixed bone marrow chimeric mice revealed that WT leukocytes were sufficient to control infection and that antigen-specific STAT1 R274W CD8+ T cell responses are impaired even in the presence of WT leukocytes. Thus, the STAT1 R274W gain-of-function mutation impedes antigen-specific CD8+ T cell responses during gammaherpesvirus infection without significantly altering cell-intrinsic antiviral immunity.
RESULTS
Heterozygous STAT1 R274W mice exhibit impaired control of acute γHV68 infection.
We used CRISPR/Cas9 to generate knock-in mice with the STAT1 R274W mutation, which has been associated with increased susceptibility to herpesvirus and Candida infections in humans (1, 2, 15). The R274W mutation lies within the STAT1 coiled-coil domain, in a region that is highly conserved in mammals (Fig. 1A). Transgenic mice were produced on a C57BL/6J background, backcrossed for 3 generations prior to experiments, and then continuously backcrossed to wild-type (WT) animals. Mice were genotyped by Sanger sequencing (Fig. 1B). All of our experiments, including cell culture studies, were performed using WT littermate mice as controls.
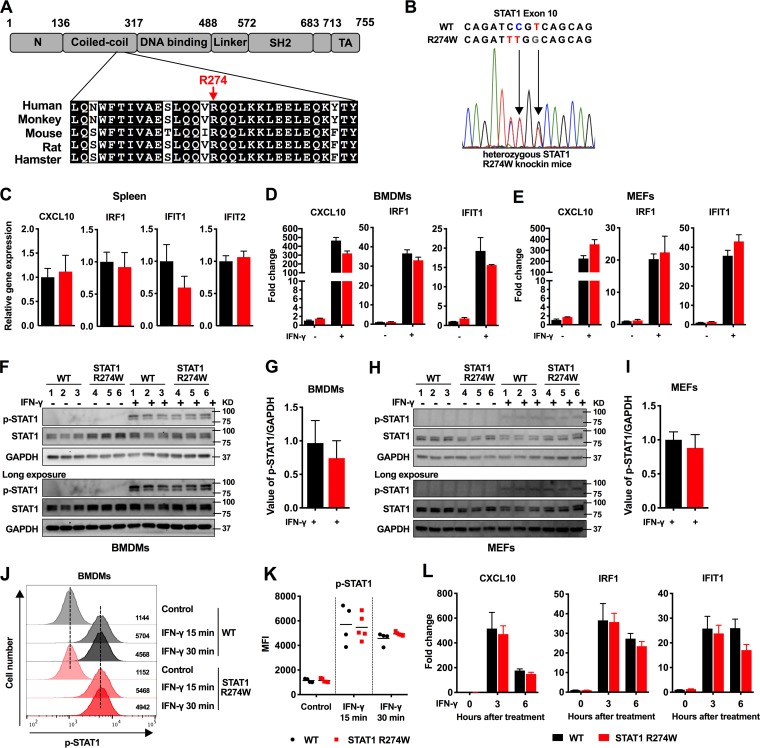
FIG 1.
Generation and initial characterization of heterozygous STAT1 R274W knock-in mice. (A) Schematic functional domain map of STAT1 with corresponding alignment of the highly conserved coiled-coil domain amino acid sequence, including arginine 274 (R274). (B) Electropherogram of WT and STAT1 R274W mutant alleles within exon 10. Three nucleotides and one amino acid (R274W) were altered in STAT1 exon 10. (C) Expression of indicated ISGs in the spleens of 5- to 7-week-old STAT1 R274W mice and WT littermates. Gene expression was measured by reverse transcription-quantitative PCR (qRT-PCR). Relative gene expression (or fold change) was calculated using the threshold cycle (ΔΔCT) method after normalizing to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data represent the mean of 10 to 12 spleens per group pooled from two independent experiments. (D and E) Expression levels of CXCL10, IRF1, and IFIT1 in WT and STAT1 R274W BMDMs (D) and MEFs (E) 6 h after stimulation with 100 ng/ml of recombinant murine IFN-γ. Data represent the mean of three independent experiments from separate WT and STAT1 R274W primary cell lines. BMDMs (F and G) and MEFs (H and I) were stimulated with 100 ng/ml IFN-γ for 30 min, and the phosphorylation of STAT1 (p-STAT1) was assessed by Western blotting with antibody to p-STAT1 (Tyr701), using STAT1 and GAPDH as loading controls. Relative p-STAT1 density was quantified using ImageJ software and was normalized to GAPDH (n = 3). (J) Representative flow cytometry histograms of p-STAT1 levels in BMDMs with and without 100 ng/ml IFN-γ treatment for 15 or 30 min. (K) Mean fluorescence intensity (MFI) of p-STAT1 in WT and STAT1 R274W BMDMs with or without 100 ng/ml IFN-γ treatment for 15 or 30 min. (L) Time course of ISG expression in WT or STAT1 R274W BMDMs at 0, 3, and 6 h after IFN-γ stimulation. Data in panels J through L represent the mean of n = 4 to 5 mice per group, pooled from two independent experiments. All data were analyzed by unpaired t test (P > 0.1 for all comparisons).
The STAT1 R274W mutation is considered a gain-of-function mutant that limits dephosphorylation of STAT1 in studies of transfected cells and transformed B cells (16, 17). Previous work indicated that overexpression of human R274 mutants in STAT1-deficient cells was associated with a slight hyperresponsiveness to IFN-γ, observed for both ISG upregulation and STAT1 phosphorylation (16, 17). Therefore, we reasoned that tissues and cells from heterozygous STAT1 R274W mice would exhibit ISG upregulation. However, when we examined naive STAT1 R274W mice and cells, we found no evidence of ISG upregulation in the spleen, in BMDMs, or in primary MEFs compared with WT littermate controls (Fig. 1C to E). We also did not observe increased phosphorylation of STAT1 by Western blot of knock-in BMDM or MEF lysates before or after treatment of cells with 100 ng/ml of IFN-γ for 30 min (Fig. 1F to I). We also observed no effect on STAT1 phosphorylation or ISG expression in BMDMs with an IFN-γ treatment dose response and time course (Fig. 1J to L). Thus, a heterozygous STAT1 R274W mutation in mice did not cause spontaneous or IFN-γ-inducible effects on STAT1 phosphorylation or ISG expression in MEFs or macrophages.
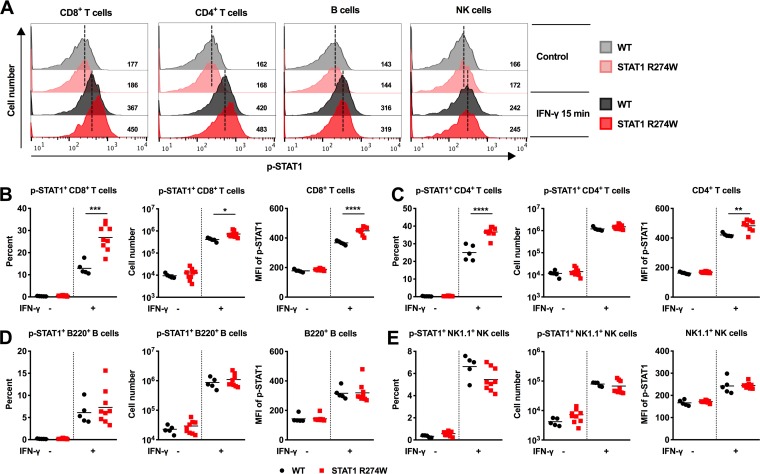
Since STAT1 gain-of-function mutations are thought to impact lymphocyte subsets, including Th17 cells in humans (4), we reasoned that hyperphosphorylation of STAT1 in response to IFN-γ may occur only in specific cell types. Indeed, treatment of cultured WT and STAT1 R274W splenocytes with IFN-γ resulted in hyperphosphorylation of STAT1 in mutant CD8+ and CD4+ T cells (Fig. 2A to C), but not in B cells or NK cells compared to WT littermate splenocytes (Fig. 2A, D to E). These results indicate that although the STAT1 R274W mutation does not alter T cell subsets at baseline in mice, the mutation may enhance signaling downstream of the IFN-γ receptor specifically in T cells.
FIG 2.
Hyperphosphorylation of p-STAT1 in IFN-γ-treated STAT1 R274W T cells. Splenocytes from 6- to 8-week-old STAT1 R274W and WT littermate control mice were treated with vehicle (medium) or 100 ng/ml IFN-γ for 15 min followed by flow cytometric analysis of p-STAT1 expression levels in CD8+ and CD4+ T cells, B cells, and NK cells. (A) Representative flow cytometry histograms of p-STAT1 in CD8+ T cells, CD4+ T cells, B cells, and NK cells with and without treatment with IFN-γ. (B through E) Percent p-STAT1+, number of p-STAT1+, and MFI of p-STAT1 in CD8+ T cells (B), CD4+ T cells (C), B220+ B cells (D), and NK1.1+ NK cells (E). All data represent the mean of n = 5 to 9 samples per genotype from three independent experiments and were analyzed by unpaired t test (****, P < 0.0001; ***, P < 0.0005; **, P < 0.005; *, P < 0.05).
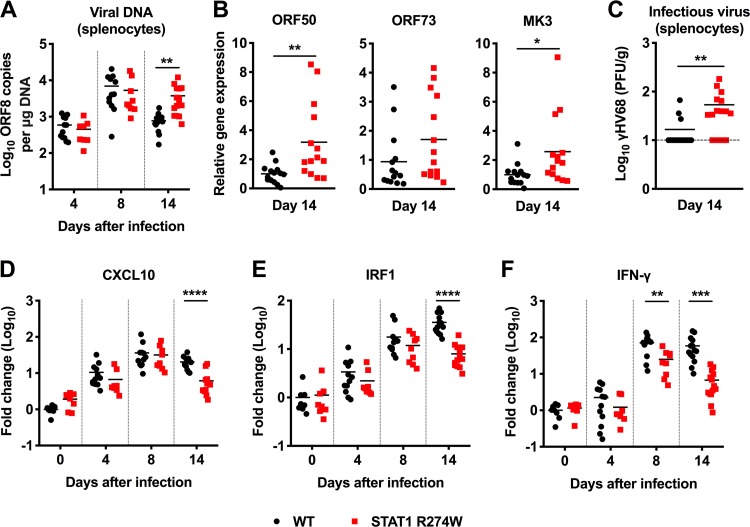
Humans with the STAT1 R274W mutation are more vulnerable to gammaherpesvirus infections, but the mechanism of increased susceptibility to virus infection has not previously been examined. To define the role of STAT1 R274W in gammaherpesvirus infection, we intraperitoneally inoculated 5- to 6-week-old STAT1 R274W and WT littermate control mice with 106 PFU of γHV68, and then measured viral genome copies by quantitative PCR (qPCR) on days 4, 8, and 14 after infection. On days 4 and 8, the numbers of γHV68 genome copies in the spleens of STAT1 R274W and WT littermate animals were equivalent (Fig. 3A). However, on day 14 we observed an approximately 5-fold increase in γHV68 genome copies in the spleens of STAT1 R274W mice (Fig. 3A). Lytic (e.g., open reading frame 50 [ORF50], MK3) but not latent (ORF73) genes were expressed at higher levels on day 14 in the heterozygous STAT1 R274W spleen (Fig. 3B), suggesting that the increased viral burden reflects either reactivation or diminished control of lytic infection. Indeed, plaque assays demonstrated higher levels of infectious virus in the spleens of STAT1 R274W mice than in WT control spleens on day 14 (Fig. 3C). More severe infection at late time points was unexpectedly associated with diminished expression of CXCL10, IRF1, and IFN-γ in the spleens of STAT1 R274W mice (CXCL10 ∼4.2-fold lower, IRF1 ∼4.5-fold lower, and IFN-γ ∼8.6-fold lower in STAT1 R274W; P < 0.005) (Fig. 3D to F).
FIG 3.
Heterozygous STAT1 R274W mice are more susceptible to γHV68 and have diminished expression of ISGs and IFN-γ during infection. WT and STAT1 R274W mice were intraperitoneally inoculated with 106 PFU of γHV68 or PBS control (mock, day 0). Mice were euthanized and the spleen harvested at days 0, 4, 8, and 14 after infection. (A) Viral genome copies in the splenocytes as measured by qPCR on days 4, 8, and 14 after infection. (B) Expression of open reading frame 50 (ORF50), ORF73, and MK3 in the spleen as measured by qPCR on day 14 after infection. (C) Levels of infectious virus in splenocytes as measured by plaque-forming assay on day 14 after infection. (D to F) Expression levels of CXCL10 (D), IRF1 (E), and IFN-γ (F) in the spleen as measured by qRT-PCR on days 0, 4, 8, and 14 after infection. All data represent the mean of n = 8 to 14 samples per genotype at each time point from at least two independent experiments and were analyzed by unpaired t test (****, P < 0.0001; ***, P < 0.0005; **, P < 0.005; *, P < 0.05).
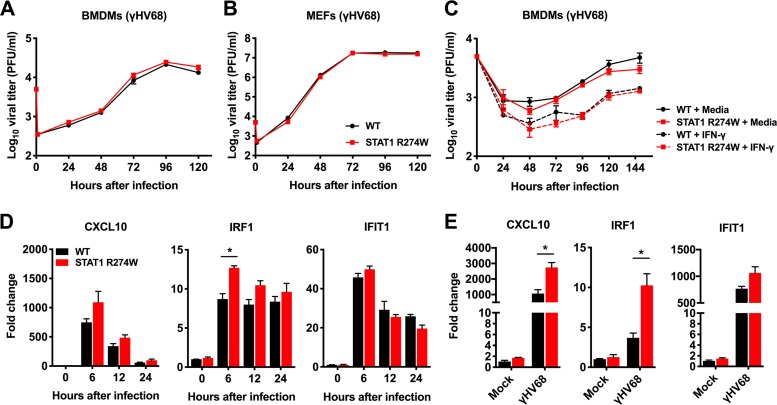
Since STAT1 upregulates expression of ISGs downstream of the IFN receptors, we reasoned that impaired cell-intrinsic antiviral immunity might create a more permissive environment for viral replication. However, multistep growth curve analysis in BMDMs and MEFs demonstrated similar γHV68 replication in WT and heterozygous STAT1 R274W mutant cells (Fig. 4A and B). Pretreatment of BMDMs with IFN-γ for 12 h resulted in similar levels of γHV68 replication in WT and STAT1 R274W BMDMs (Fig. 4C). However, consistent with the notion that STAT1 R274W does indeed have a subtle gain-of-function effect on ISG expression (17), we detected a slight increase in levels of Irf1 mRNA in STAT1 R274W cells compared to those in WT controls, as well as a 0.5-fold increase in CXCL10 expression in γHV68-infected MEFs (Fig. 4D and E). These results indicate that STAT1 R274W-associated effects on ISG expression do not appreciably impact γHV68 replication in our cell culture assays. Similar results were obtained by examining multistep growth curves and ISG expression after infection of BMDMs and MEFs with herpes simplex virus 1 (HSV-1) strain 17+ (data not shown), indicating consistent effects of STAT1 R274W for two herpesviruses of distinct subfamilies.
FIG 4.
Multistep γHV68 growth curve analysis and gene expression in primary BMDMs and MEFs generated from WT and heterozygous STAT1 R274W mice. (A and B) BMDMs and MEFs were infected with γHV68 at a multiplicity of infection (MOI) of 0.05. The medium was harvested for quantitation of infectious virus by plaque assay at the indicated time points. (C) BMDMs were pretreated with medium or IFN-γ at 100 ng/ml for 12 h before being infected with γHV68 at an MOI of 0.05. At the indicated time point after infection, the virus titer was determined by plaque assay. (D) Relative gene expression levels in BMDMs infected with γHV68 at an MOI of 10 for 0, 6, 12, and 24 h, followed by harvesting of cells for RNA isolation and qRT-PCR analysis. (E) Relative gene expression in MEFs infected with γHV68 at an MOI of 10 for 12 h, followed by harvesting of cells for RNA isolation and qRT-PCR analysis. Growth curve data in panels A through C represent the mean plus or minus standard error of the mean (SEM) of two independent experiments with 3 technical replicates for each growth curve experiment. Data in panel D represent the mean ±SEM of 4 or 5 biological replicates pooled from two independent experiments. Data in panel E represent the mean ±SEM of 4 biological replicates pooled from two independent experiments. Data in panels A, B, and C were analyzed by 2-way analysis of variance (ANOVA). Data in panels D and E were analyzed by unpaired t test. *, P < 0.05.
Impaired effector T cell responses in γHV68-infected STAT1 R274W mice.
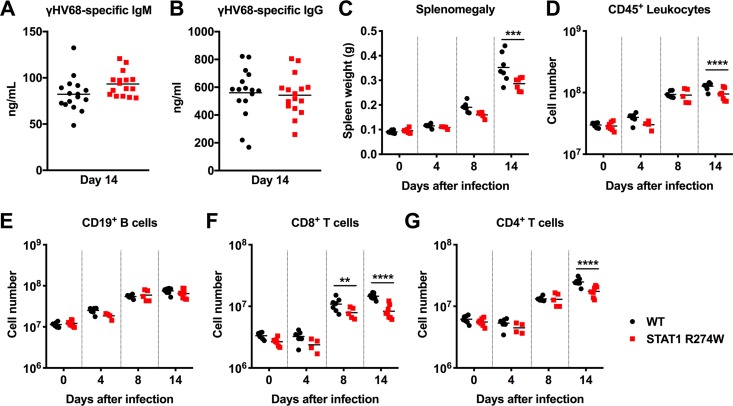
Since the STAT1 R274W mutation has no effect on γHV68 and HSV-1 replication in cell culture, we reasoned that a defect in adaptive immunity might explain increased levels of γHV68 in STAT1 R274W animals. First, we assessed levels of γHV68-specific IgM and IgG on day 14 after infection, but we observed no differences in levels of virus-specific IgM or IgG in STAT1 R274W mice and WT littermates (Fig. 5A and B), suggesting that the STAT1 gain-of-function mutation does not impact humoral immunity against γHV68. However, on day 14 after intraperitoneal inoculation with γHV68, but not at earlier time points, we found that the STAT1 R274W spleens were smaller than WT littermate control spleens (Fig. 5C). Flow cytometric analysis of splenocytes revealed that STAT1 R274W mice had a diminished total number of splenic CD45+ leukocytes on day 14 but not at earlier time points after infection (Fig. 5D). The total numbers of B cells, neutrophils, and monocytes were similar in WT and STAT1 R274W spleens, both before and after infection (Fig. 5E and data not shown). However, we observed an ∼2-fold decrease in the total number of CD8+ T cells on days 8 and 14 (Fig. 5F), as well as an ∼2-fold reduction in CD4+ T cells on day 14 (Fig. 5G). These results suggest that STAT1 R274W limits T cell responses during γHV68 infection.
FIG 5.
Virus-specific antibody responses and flow cytometric quantitation of splenic lymphocyte subsets in γHV68-infected WT and STAT1 R274W mice. At ages of 5 to 6 weeks, WT and STAT1 R274W mice were inoculated intraperitoneally with 106 PFU of γHV68 or PBS control (mock, day 0). (A and B) Serum was harvested on day 14 after infection. Virus-specific IgM (A) and IgG (B) levels were measured by ELISA. Data represent the mean of n = 16 biological replicates pooled from two independent experiments. P > 0.1 by unpaired t test. (C) Spleen weight prior to isolation of leukocytes. (D to G) Mice were euthanized and the spleens harvested at days 0, 4, 8, and 14 after infection for flow cytometric analysis. Numbers of CD45+ leukocytes (D), CD19+ B cells (E), CD8+ T cells (F), and CD4+ T cells (G). Data represent the mean of n = 4 to 8 samples from two independent experiments. Data were analyzed by unpaired t test. ****, P < 0.0001; ***, P < 0.0005; **, P < 0.005.
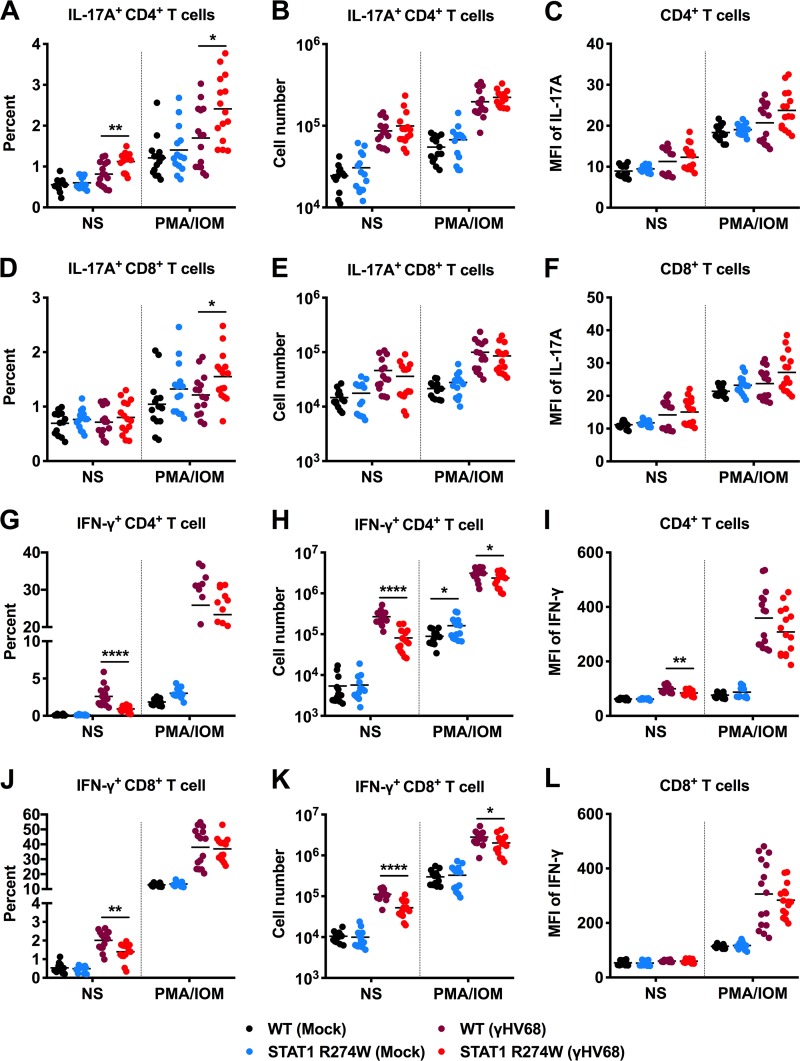
To evaluate CD4+ and CD8+ T cell subsets, we measured the percentage and number of T cells that produce IL-17A or IFN-γ in the spleens of mock- and γHV68-infected animals. Splenic T cells were also evaluated in the presence and absence of phorbol 12-myristate 13-acetate (PMA)-ionomycin under all conditions. We observed no difference prior to infection in the total number of IL-17A-producing CD4+ (Th17) or CD8+ (Tc17) cells (Fig. 6A to F) or in the amount of IL-17A produced by IL-17A+ CD4+ T cells based on mean fluorescence intensity (MFI) (Fig. 6C). In the presence or absence of PMA-ionomycin, we detected similar numbers of Th17 and Tc17 cells, whether we examined T cells from mock- or γHV68-infected WT and STAT1 R274W mice (Fig. 6A, B and D, and E). These results suggest that STAT1 R274W does not significantly impact the number of Th17 or Tc17 cells in uninfected mice.
FIG 6.
Flow cytometric analysis of CD4+ and CD8+ T cell subsets in the spleens of naive and γHV68-infected mice. At age 5 to 6 weeks, WT and STAT1 R274W mice were inoculated intraperitoneally with 106 PFU of γHV68 or PBS control (mock). Infected mice were euthanized and splenocytes harvested at day 14 for flow cytometric analysis. Splenocytes were cultured for 6 h with brefeldin A in the presence or absence (NS) of PMA-ionomycin (PMA/IOM). (A to C) Percentage (A), number (B), and IL-17A MFI (C) of IL-17A-producing CD4+ T cells from the spleens of mock- and γHV68-infected mice. (D to F) Percentage (D), number (E), and IL-17A MFI (F) of IL-17A-producing CD8+ T cells from the spleens of mock- and γHV68-infected mice. (G to I) Percentage (G), number (H), and IFN-γ MFI (I) of IFN-γ-producing CD4+ T cells from the spleens of mock- and γHV68-infected mice. (J to L) Percentage (J), number (K), and IFN-γ MFI (L) of IFN-γ-producing CD8+ T cells from the spleens of mock- and γHV68-infected mice. Data represent the mean of n = 14 to 16 samples from two independent experiments. Results were analyzed by unpaired t test. ****, P < 0.0001; **, P < 0.005; *, P < 0.05.
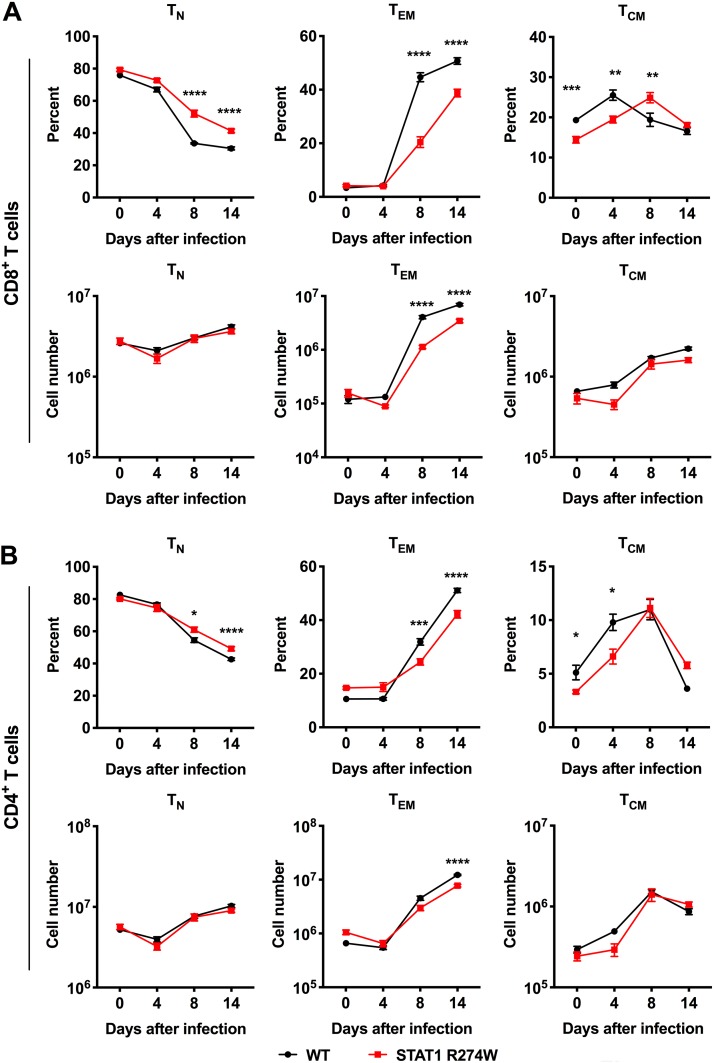
IFN-γ and T cells are critical for control of γHV68 infection (21–28). Upon examination of IFN-γ-expressing T cells, we observed a consistent ∼2- or 3-fold reduction in the total number of IFN-γ-producing CD4+ and CD8+ T cells in infected but not in naive STAT1 R274W animals compared to that in WT littermate controls (Fig. 6G, H, J, and K). There was also a subtle increase in the number of IFN-γ-producing naive CD4+ T cells upon stimulation with PMA-ionomycin. After γHV68 infection, there was a very small reduction in the amount of IFN-γ produced by unstimulated CD4+ T cells after γHV68 infection, but no difference in the amount of IFN-γ produced by CD8+ T cells in the presence or absence of stimulation or γHV68 infection (Fig. 6I and L). Diminished numbers of IFN-γ-producing CD4+ and CD8+ T cells could explain the lower levels of IFN-γ and ISG expression in the spleens of γHV68-infected STAT1 R274W mice on day 14 after infection (Fig. 3D to F). Additional evaluation of T cell subsets revealed a lower fraction and number of effector memory CD4+ and CD8+ T cells by day 8 after infection, further confirming an effect of STAT1 R274W on effector T cell populations (Fig. 7A and B).
FIG 7.
Flow cytometric quantitation of naive (TN), effector memory (TEM), and central memory (TCM) CD8+ and CD4+ T cell subsets in the spleens of naive and γHV68-infected mice. WT and STAT1 R274W mice were inoculated intraperitoneally at age 5 to 6 weeks with 106 PFU of γHV68 or PBS control (mock, day 0). Infected mice were euthanized and splenocytes harvested at days 0, 4, 8, and 14 for flow cytometric analysis of CD8+ and CD4+ naive (CD44loCD62Lhi) T cells, effector memory (CD44hiCD62Llo) T cells, and central memory (CD44hiCD62Lhi) T cells. (A and B) Percentage and numbers of naive (TN), effector memory (TEM), and central memory (TCM) CD8+ (A) and CD4+ (B) T cell subsets at the indicated time points. Data represent the mean ±SEM of n = 4 to 8 mice per genotype at each time point. Data were analyzed by 2-way ANOVA. ****, P < 0.0001; ***, P < 0.0005; **, P < 0.005; *, P < 0.05.
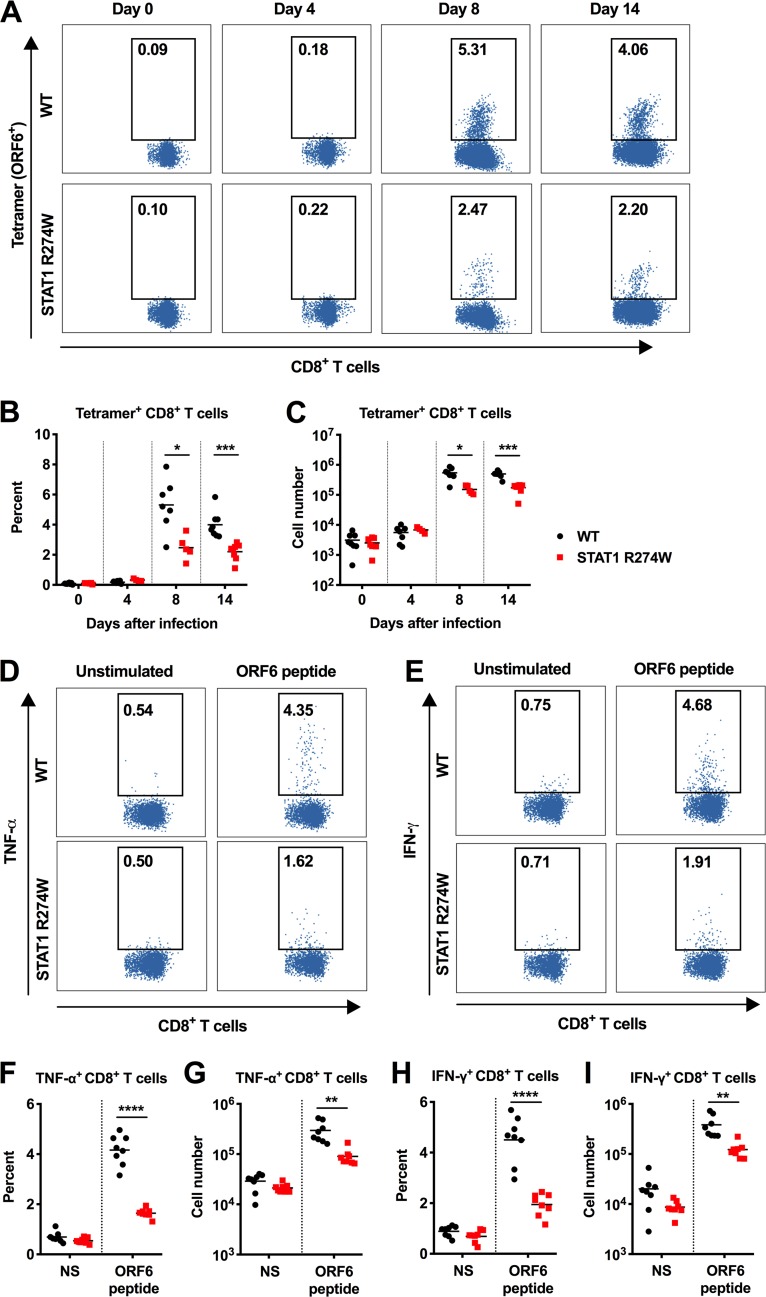
Next, we measured the number of antigen-specific CD8+ T cells binding the immunodominant ORF6 tetramer in WT and STAT1 R274W mice on days 0, 4, 8, and 14 after infection. ORF6 tetramer staining revealed that the spleens of STAT1 R274W mice have a reduced percentage and total number of antigen-specific CD8+ T cells on days 8 and 14 after infection (Fig. 8A to C). Consistent with this finding, ex vivo restimulation of splenic T cells with an immunodominant ORF6 peptide also revealed an ∼2-fold reduction in the percentage and number of tumor necrosis factor alpha (TNF-α)- and IFN-γ-positive CD8+ T cells (Fig. 8D to I).
FIG 8.
The STAT1 R274W mutation impairs antigen-specific CD8+ T cell responses during γHV68 infection. WT and STAT1 R274W mice were inoculated intraperitoneally at age 5 to 6 weeks with 106 PFU of γHV68 or PBS control (mock, day 0). Infected mice were euthanized and splenocytes harvested at days 0, 4, 8, and 14 for flow cytometric analysis. (A) Representative dot plots of antigen-specific CD8+ T cells quantitated by flow cytometric analysis following ORF6 tetramer staining. (B and C) Quantitation of the percentage (B) and number (C) of ORF6 tetramer+ CD8+ T cells by flow cytometry on days 0, 4, 8, and 14 after infection. (D and E) Representative dot plots of TNF-α- (D) and IFN-γ-expressing (E) CD8+ T cells at day 14 were quantitated by flow cytometric analysis after restimulation with or without (NS) the immunodominant ORF6 peptide. (F-G) Percentage (F) and number (G) of TNF-α-expressing CD8+ T cells. (H-I) Percentage (H) and number (I) of IFN-γ-expressing CD8+ T cells. Data represent the mean of n = 4 to 8 mice per genotype pooled from two independent experiments. Results were analyzed by unpaired t test. ****, P < 0.0001; ***, P < 0.0005; **, P < 0.005; *, P < 0.05.
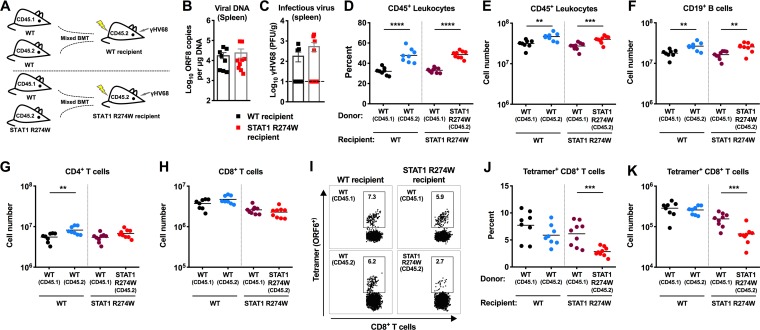
Because of the antigen-specific CD8+ T cell defects that we observed in STAT1 R274W animals, we reasoned that mixed bone marrow chimeric mice with both WT and STAT1 R274W leukocytes would allow us to more precisely define the relative phenotypes and functions of WT and STAT1 R274W CD8+ T cells during infection. We intravenously injected equal numbers of congenic CD45.1+ WT and CD45.2+ WT or STAT1 R274W bone marrow cells into lethally irradiated WT and STAT1 R274W recipient mice (Fig. 9A). This resulted in generation of two groups, WT/WT and WT/STAT1 R274W mixed bone marrow chimeras. Six weeks later, we confirmed reconstitution of peripheral blood leukocytes, including similar percentages of CD45.1+ and CD45.2+ cells in recipient animals (data not shown). Fourteen days after intraperitoneal inoculation with 106 PFU of γHV68, we observed no difference in levels of viral DNA or infectious virus between the spleens of WT/WT and WT/STAT1 R274W recipient mice (Fig. 9B and C). This suggests that the presence of WT leukocytes is sufficient to control γHV68 infection, even when STAT1 R274W leukocytes and radioresistant recipient cells are also present. Evaluation of leukocyte subsets revealed a slightly higher percentage and number of CD45.2 leukocytes, including B cells, in both groups of mixed bone marrow chimeras (Fig. 9D to F). This was anticipated, given the fact that radioresistant cells in recipient animals also express CD45.2+. Among CD4+ and CD8+ T cells, we observed only a slight increase in CD45.2+/CD4+ T cells in WT/WT chimeras, and no difference in the total number of CD8+ T cells in either group of chimeras (Fig. 9G and H). However, evaluation of antigen-specific CD8+ T cells on day 14 after infection revealed an ∼2.5-fold reduction in both the percentage and number of STAT1 R274W-expressing ORF6 tetramer+ CD8+ T cells (Fig. 9I to K). No difference was observed in the percentage or number of antigen-specific CD8+ T cells in WT/WT mixed chimeras or in WT CD45.1+ CD8+ cells in STAT1 R274W recipient animals (Fig. 9I to K). These results demonstrate that STAT1 R274W-associated impairment of antigen-specific CD8+ T cell responses can occur even in the presence of WT leukocytes. Impaired production of antigen-specific CD8+ T cells might be a consequence of cell-intrinsic antiproliferative effects of STAT1 (29, 30).
FIG 9.
Impaired STAT1 R274W antigen-specific CD8+ T cell responses after γHV68 infection of WT and STAT1 R274W mixed bone marrow chimeric mice. (A to K) STAT1 R274W and WT littermate animals were lethally irradiated followed by intravenous injection of a 1:1 mixture of CD45.1 WT bone marrow cells and either CD45.2 WT or STAT1 R274W bone marrow cells. This resulted in generation of WT/WT chimeras and WT/STAT1 R274W chimeras. Six weeks later, after reconstitution of similar numbers of circulating leukocytes, mice were intraperitoneally inoculated with 106 PFU of γHV68 and then euthanized 14 days after infection for virological and immunological analysis (A). (B) Viral DNA levels in the spleen as measured by qPCR. (C) Infectious virus levels in the spleens of mixed bone marrow chimeric mice as measured by plaque assay. The dashed line indicates the limit of detection of the assay at 10 PFU/g of tissue. (D and E) Flow cytometric analysis of the percentage (D) and number (E) of CD45.1+ and CD45.2+ leukocytes in the spleens of mixed bone marrow chimeric mice. (F to H) Numbers of CD19+ B cells, CD4+ T cells, and CD8+ T cells in the spleen after infection as measured by flow cytometry. (I) Representative dot plots of ORF6 tetramer+ CD8+ T cells in mixed bone marrow chimeric mice. (J and K) The percentage (J) and number (K) of ORF6 tetramer+ CD8+ T cells in mixed bone marrow chimeric mice. Data represent the mean of n = 8 to 9 chimeric mice per group pooled from two independent experiments. Results were analyzed by unpaired t test. ****, P < 0.0001; ***, P < 0.0005; **, P < 0.005.
Collectively, our findings demonstrate that STAT1 R274W impairs production of CD4+ and CD8+ effector T cells during γHV68 infection, without substantially affecting cell-intrinsic control of γHV68 replication in target cells, including BMDMs and fibroblasts. Because IFN-γ signaling, as well as antigen-specific T cells, plays a major role in controlling γHV68 infection in mice (24–27), the impaired production of antiviral T cells likely contributes to immunodeficiency caused by STAT1 gain of function.
DISCUSSION
Since STAT1 gain-of-function mutations cause chronic mucocutaneous candidiasis in humans, prior work in human cells has focused on Th17 responses that promote antifungal immunity. But STAT1 mutations are also associated with susceptibility to viruses (4), and the mechanism of antiviral immunodeficiency was not previously explored. Using a STAT1 R274W knock-in mouse model, we found that heterozygous STAT1 mutant mice exhibited impaired control of gammaherpesvirus infection in vivo. Although STAT1 R274W led to a slight upregulation of ISGs during infection with γHV68 in cell culture, viral replication was not impacted in vitro (Fig. 4). Furthermore, we observed lower IFN-γ production and ISG expression in vivo despite increased viral burden at late time points in STAT1 R274W mice (Fig. 3). These results corresponded with diminished numbers of infection-associated effector CD4+ and CD8+ T cells, including antigen-specific CD8+ T cells. We also found markedly diminished levels of IFN-γ in the spleen only at later time points (days 8 and 14), when antigen-specific T cell responses occur. Although we are not able to conclude definitively whether these adaptive immune defects are responsible for STAT1 R274W-associated susceptibility to viral infection, our studies of mixed bone marrow chimeric mice support the conclusion that STAT1 R274W impairs control of infection because of effects on antigen-specific CD8+ T cells. Furthermore, CD8+ T cells were previously demonstrated to play a major role in controlling γHV68 infection (21–28).
STAT1 integrates signals downstream of multiple antiviral cytokine receptors, including the type I, II, and III IFN receptors (10). Indeed, mice lacking IFN-γ, the IFN-γ receptor, or STAT1 are highly vulnerable to gammaherpesvirus infection (27). Although human STAT1 mutations have previously been studied extensively in cell culture (1), mechanisms of human STAT1 R274W-associated immunodeficiency have not previously been defined in an animal model of the disease. Cell culture and flow cytometric analysis of human PBMCs previously revealed that STAT1 mutations are associated with increased inducible expression of ISGs (1, 31). This prior work has provided key insight into mechanisms of STAT1-associated immunodeficiency in humans and transfected cells, but with some limitations, since the assays utilize overexpression systems, cells that lack endogenous STAT1 (1, 17), Epstein-Barr virus (EBV)-transformed B cells (1, 16, 17), or patient PBMCs that may have been latently infected (17, 31). Thus, although our work has substantial limitations related to potential species-specific effects of STAT1, the STAT1 R274W knock-in mouse model has allowed us to perform an immunological evaluation of STAT1 gain of function under isogenic conditions, without overexpression, and without EBV-mediated transformation of primary cells.
Since the STAT1 R274W mutation is regarded as gain of function (1, 2), we initially anticipated that we would detect increased basal and inducible upregulation of ISGs in STAT1 R274W BMDMs and MEFs. However, we found that STAT1 R274W was associated with only a mild upregulation of ISGs in cultured MEFs and BMDMs, and this was not enough to impact γHV68 replication (Fig. 4). However, treatment of cultured splenocytes with IFN-γ resulted in T cell-specific hyperphosphorylation of STAT1 (Fig. 2). This suggests that the STAT1 R274W mutation exerts T cell-specific effects on STAT1 signaling. Although we observed no impact of the mutation on γHV68 replication in primary MEFs or macrophages, we cannot exclude the possibility that γHV68 may replicate more efficiently in STAT1 R274W B cells. However, the results of our γHV68 pathogenesis studies in mixed bone marrow chimeric mice argue against this possibility.
In humans, STAT1 gain of function is thought to cause vulnerability to mucocutaneous candidiasis via its effects on Th17 cells (1, 2, 6), but it is not known whether defects in Th17-mediated immunity can create susceptibility to gammaherpesvirus infection. We discovered that STAT1 gain of function impairs CD4+ and CD8+ T cell responses (Fig. 5), including antigen-specific CD8+ T cell responses against γHV68 (Fig. 8). Antiviral CD4+ and CD8+ T cells are already known to play a major role in controlling γHV68 infection (21–28). However, it is not known whether a similar defect in antigen-specific T cell responses may occur during virus infection in humans, so this could be a topic of future study. Whether these animals will provide a suitable model for the study of mucocutaneous candidiasis also remains to be determined.
Our mixed bone marrow chimeric mouse studies support the conclusion that STAT1 R274W creates immunodeficiency because of its effects on antigen-specific CD8+ T cell responses. Impaired T cell responses may be a consequence of cell-intrinsic factors, since we observed an ∼2.5-fold reduction in antigen-specific STAT1 R274W CD8+ T cells compared to WT T cells in mixed bone marrow chimeric mice (Fig. 9I to K). However, our results do not exclude combined cell-intrinsic and extrinsic effects on T cell responses in STAT1 R274W mice. Prior studies of human cells revealed that STAT1 gain-of-function mutant macrophages may express higher levels of PD-L1 (32), which limits Th17 cell differentiation. Thus, one possibility is that upregulation of PD-L1 or other inhibitory proteins may dampen T cell responses against viruses, in addition to impairing Th17-mediated immunity against Candida. However, contributions of STAT1 R274W-associated cell-extrinsic factors are likely insufficient to impede antigen-specific CD8+ T cell responses, since numbers of tetramer-positive WT CD8+ T cells were not significantly different in WT and STAT1 R274W recipient animals in our studies of mixed bone marrow chimeric mice.
Although the majority of virus infections in STAT1 gain-of-function patients were herpesvirus infections (4), the reason STAT1 gain of function preferentially creates immunodeficiency against herpesviruses remains to be fully defined. In the future, cell type-specific STAT1 mutant mice will be useful to determine how STAT1 R274W impairs antiviral T cells. Finally, given our discovery that the STAT1 coiled-coil domain impacts T cells, our findings suggest that small molecular targeting of the STAT1 coiled-coil domain might be worth future exploration as a potential therapy, either to promote antiviral or antitumoral immunity with agonists or to limit T cell-mediated autoimmunity using antagonists.
MATERIALS AND METHODS
Design.
The goal of this project was to define mechanisms of immunodeficiency in STAT1 R274W mice during gammaherpesvirus infection. We conducted a power analysis for Institutional Animal Care and Use Committee-approved studies in order to predetermine the number of animals needed per experimental group. We used WT littermate mice as control animals in all studies of STAT1 R274W mice. No outliers were excluded from analyses. At least two or three independent in vivo experiments were performed to replicate findings. The number of biological replicates for each experiment is listed in the legends to Fig. 1 to 9.
Study approvals.
All protocols for animal studies were approved by the Institutional Animal Care and Use Committees at the Washington University School of Medicine (assurance no. A-3381-01).
Generation of STAT1 R274W knock-in mice.
Mice heterozygous for the STAT1 R274W mutation were generated by and obtained from Applied StemCell, Inc. Briefly, to generate STAT1 R274W knock-in mice, a single guide RNA (sgRNA; 5′-CTGCTGACGGATCTGCTGCA-3′) was designed to target STAT1 arginine 274 in exon 10. A single-stranded oligonucleotide donor (ssODN) encoding the amino acid substitution (R274W) with homology arms was synthesized (5′-GCATGTGGAGATTCTGTGACTCTCTCATCTAGTGTGTTTTGGGACTGTTGTTCATGGAGACCCCATGTCCTGTACCCTCCCAGGTTCACCATTGTTGCAGAGACaCTGCAGCAGATCtGgCAGCAGCTTAAAAAGCTGGAGGAGTTGGAACAGAAATTCACCTATGAGCCCGACCCTATTACAAAAAACAAGCAGGTGTT-3′; mutant nucleotides are indicated by underlined lowercase). C57BL/6J mice were superovulated and mated with C57BL/6J males. Single-cell embryos were isolated and injected with a combination of Cas9, guide RNA (gRNA), and ssODN. Modified embryos were transferred into pseudopregnant female recipient mice. One founder mouse was received and backcrossed to WT C57BL/6J animals for three generations and continued to be backcrossed in perpetuity since all studies involved heterozygous animals. WT littermate control mice were used for all experiments. A serendipitous synonymous mutation was identified in the third nucleotide of the codon for isoleucine 273. The synonymous mutation, which did not alter the amino acid sequence may have limited recutting of the mutant allele by Cas9.
Mice, viruses, and cells.
Mice were housed in specific pathogen-free mouse facilities at the Washington University School of Medicine. Experiments were performed on 5- to 6-week-old heterozygous STAT1 R274W mice, and WT littermates were used as controls. γHV68 strain WUMS (ATCC VR1465) was purchased from ATCC and used for all experiments. Virus was passaged and the titer was determined by infectious plaque assay in NIH 3T12 cells (ATCC CCL-164). HSV-1 strain 17+ (33), provided by the T. Margolis laboratory (Washington University, Saint Louis, MO), was passaged in Vero cells. 3T12 cells and mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 5% and 10% fetal bovine serum (FBS), respectively, along with 1% of l-glutamine, HEPES, and penicillin-streptomycin at 37°C in 5% CO2. MEFs from STAT1 R274W and WT littermate embryos on embryonic day 14.5 (E14.5) were obtained as previously described (34). Bone marrow-derived macrophages (BMDMs) from femurs and tibias of 7-week-old STAT1 R274W and WT littermate mice were generated by culturing bone marrow cells for 7 days in BMDM media (DMEM with 10% FBS, 1% penicillin-streptomycin, 1% l-glutamine, 1% HEPES, and 40 ng/ml of mouse macrophage colony-stimulating factor [M-CSF]).
Antibodies and other reagents.
Antibodies used for flow cytometry included CD45 (clone name 30-F11), CD45.1 (A20), CD45.2 (104), B220 (RA3-6B2), NK1.1 (PK136), CD11b (M1/70), F4/80 (BM8), CD19 (6D5), CD3 (145-2C11), CD4 (GK1.5), CD8α (53-6.7), CD8β (YTS156.7.7), CD44 (IM7), CD62L (MEL-14), IL-17A (TC11-18H10.1), and TNF-α (MP6-XT22) from BioLegend. Other antibodies utilized in flow cytometry included CD45 (30-F11; BD Biosciences), phospho-STAT1 (p-STAT1) (4a; BD), IFN-γ (XMG1.2; eBioscience), and FcBlock (2.4G2; BD). STAT1 antibody (no. 9172), phospho-STAT1 (Tyr701) rabbit monoclonal antibody (MAb) (no. 7649), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) rabbit MAb (no. 2118) were obtained from Cell Signaling Technology (CST). Other reagents were recombinant murine IFN-γ (315-05; PeproTech), mouse macrophage colony-stimulating factor (M-CSF) (300-25; PeproTech), phorbol 12-myristate 13-acetate (PMA) (P8139; Sigma), ionomycin (IOM) (I0634; Sigma), and brefeldin A (eBioscience).
Infections and tissue harvests.
Five- to 6-week-old STAT1 R274W heterozygous mice and WT littermates were inoculated intraperitoneally with 106 PFU of γHV68 in 100 μl of phosphate-buffered saline (PBS). Mice were euthanized under anesthesia at the indicated time points. Blood samples were collected via cheek pouch puncture into 1.5-ml tubes. For titration, mouse splenocytes or spleens were harvested and stored at −80°C prior to disruption. For flow cytometry experiments, harvested mouse spleens were stored in fluorescence-activated cell sorter (FACS) buffer (PBS with 4% FBS) or T cell medium (RPMI with 10% FBS and 0.1% 2-mercaptoethanol) for further processing.
Viral burden analysis.
Spleens were weighed and homogenized in 200 μl of PBS using zirconia beads and a MagNA Lyser (Roche Life Sciences). Total DNA was isolated from 15 μl of homogenate using the DNeasy blood and tissue kit (catalog no. 69504; Qiagen). γHV68 genome copies were quantitated using an established qPCR assay to detect ORF8, as described previously (28). A standard curve was used for absolute quantitation of genome copy numbers, also as previously described (28). Infectious γHV68 in spleens was analyzed by plaque assay as previously described (35, 36). In brief, 3T12 cells were seeded in 12-well plates 1 day prior to infection, and then four 10-fold serial dilutions of spleen homogenates were plated on 3T12 monolayers. After 2 h adsorption, cells were overlaid with 2 ml of 2% methylcellulose in minimum essential medium (MEM) supplemented with 5% FBS. Plates were fixed on day 6 with 4% paraformaldehyde (PFA) and stained with 0.5% crystal violet solution containing 20% methanol. Plates were air dried on paper towels for 24 h, followed by counting of PFU. The limit of detection was 10 PFU per gram of tissue.
Immunoblot analysis of STAT1 phosphorylation.
Mouse IFN-γ-treated BMDMs and MEFs were lysed in radioimmunoprecipitation assay (RIPA) buffer (catalog no. 9806S; CST) supplemented with a protease inhibitor (catalog no. 78430; Thermo Fisher) and phosphatase inhibitor (catalog no. 88667; Thermo Fisher). An equal amount of protein was loaded and separated on 10% SDS-PAGE gels (Bio-Rad), and then transferred to polyvinylidene fluoride membranes (EMD Millipore). Primary antibodies against phosphor-STAT1 (Tyr701), STAT1, and GAPDH were stained and detected by the use of horseradish-peroxidase-conjugated secondary anti-rabbit antibody (catalog no. 31460; Invitrogen). All blots were performed using Pierce ECL substrate (Thermo Fisher Scientific) and scanned with a ChemiDoc touch imaging system (Bio-Rad).
Viral growth curves.
BMDMs and MEFs were seeded in triplicate in 24-well plates, and cells were infected with γHV68 or HSV-1 at a multiplicity of infection (MOI) of 0.05 and 0.01, respectively. For γHV68 growth curves, supernatant was collected at 1, 24, 48, 72, 96, and 120 h after infection and then determined by plaque assay. In some experiments, cells were pretreated with IFN-γ as described in the legend to Fig. 4. For HSV-1 growth curve analysis, supernatant was collected at 1, 12, 24, 48, 72, and 96 h after infection, and Vero cells were infected with serial dilution of samples by incubation for 2 h and then overlaid with 2% methylcellulose in MEM supplemented with 1% FBS. After 3 to 4 days of incubation, monolayers were fixed in PFA and stained with 0.5% crystal violet solution, and then viral plaques were counted.
Measurement of gene expression by reverse transcription-quantitative PCR (qRT-PCR).
Total RNA from infected or uninfected spleen homogenates and from IFN-γ-stimulated, unstimulated, or infected BMDMs and MEFs was extracted using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. qRT-PCR was performed using a TaqMan RNA-to-CT 1-step kit (Applied Biosystems) according to the manufacturer’s instructions. Results were calculated using the threshold cycle (ΔΔCT) method, normalizing to GAPDH. Primers were obtained from Integrated Device Technology, Inc. (IDT).
Virus-specific antibody ELISAs.
γHV68-specific IgG and IgM enzyme-linked immunosorbent assays (ELISAs) were performed as described previously (28). Nunc MaxiSorp flat-bottom 96-well plates were coated with γHV68 for 12 h, followed by blocking with 1% bovine serum albumin for 1 h at room temperature. Wells were washed three times with wash buffer (PBS-0.1% Tween) before adding the serum samples. After 2 h of incubation, the wells were washed and incubated with horseradish peroxidase-conjugated IgG and IgM antibodies for 2 h before adding the tetramethylbenzidine (TMB) ELISA substrate (Thermo Fisher, USA). Reactions were stopped by adding 2 normal H2SO4. Plates were analyzed on a BioTek Synergy 2 multidetection microplate reader (BioTek) using Gen5 software (BioTek). Serially diluted mouse IgG (Sigma-Aldrich, USA) or IgM (EMD Millipore) were used for the standard curve.
Flow cytometry and intracellular cytokine stimulation.
Spleens were mashed and filtered through a 70μm cell strainer to make single cell suspensions. For analysis of surface markers, cells were stained ex vivo for 30 min on ice in FACS buffer using antibodies to CD45, CD19, CD3, CD4, CD8α, CD44, and CD62L. For intracellular cytokine quantification, splenocytes were stimulated for 6 h at 37°C with 50 ng/ml PMA and 1 μg/ml IOM in the presence of brefeldin A. Following the 6-h stimulation, cells were stained as above with CD45, CD19, CD4, and CD8α, then cells were washed and fixed, permeabilized (00-5123-43; eBioscience), and stained with IL-17A and IFN-γ antibodies. Samples were run on a BD FACSCanto instrument, and data were analyzed using Cytobank and FlowJo software. For phospho-STAT1 staining, BMDMs or splenocytes were stimulated for the indicated times with 100 ng/ml of IFN-γ. Subsequently, cells were fixed with 2% PFA for 10 min at room temperature, permeabilized with ice-cold methanol for 15 min on ice, and stained with p-STAT1, along with CD45, CD11b, and F4/80 in BMDMs or with CD45, B220, CD3, CD4, CD8, and NK1.1 in splenocytes for 1 h on ice. Sampling was performed on an Attune NxT flow cytometer and data analyzed using FlowJo software.
Measurement of antigen-specific CD8+ T cell responses.
We utilized tetramers binding to T cell receptors specific to the ORF6-derived peptide AGPHNDMEI of γHV68. Tetramers were generated by the Andrew and Jane M. Bursky Center for Human Immunology and Immunotherapy (CHiiPs) and subsequently validated by FACS and utilized for staining of antigen-specific T cells (28). Staining of intracellular IFN-γ and TNF-α was also performed as previously described (28), following 6 h of ex vivo restimulation with the ORF6 peptide in the presence of brefeldin A. Splenocytes were washed and stained to antibodies against CD45, CD19, CD4, CD8α, CD8β, TNF-α, and IFN-γ. Results were obtained by flow cytometry and analyzed using Cytobank.
Mixed bone marrow chimeric mouse studies.
Four- to 5-week-old and 5- to 6-week-old STAT1 R274W and WT littermate recipient mice were lethally irradiated, followed by intravenous injection with a 1:1 mixture of CD45.1 WT bone marrow cells and either CD45.2 WT or STAT1 R274W bone marrow cells in 100 μl of PBS. This resulted in generation of WT/WT chimeras and WT/STAT1 R274W chimeras. Six weeks later, after confirming reconstitution of circulating donor leukocytes by FACS, mice were intraperitoneally inoculated with 106 PFU of γHV68 and then euthanized 14 days after infection for virological and immunological analysis.
Statistical analysis.
All data were analyzed using GraphPad Prism software, and statistical analyses were performed as described in the legends to Fig. 1 to 9.
ACKNOWLEDGMENTS
We thank the Andrew M. and Jane M. Bursky Center for Human Immunology and Immunotherapy Programs Immunomonitoring Laboratory at Washington University for assistance with generation of the γHV68 ORF6 tetramer.
H.I. is supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (grant MI-F-2018-712). M.T.B. is supported by grants from the NIH (R01 AI141716 and R01 AI139314). The Miner laboratory is supported by grants from the NIH (K08 AR070918 and R01 AI143982) and by the Washington University Rheumatic Diseases Research Resource-based Center (grant WU RDRRC P30 AR073752).
Q.W., C.A.M., H.I., and D.J.P. performed experiments and analyzed data. Q.W. wrote portions of the initial manuscript and edited subsequent versions of the manuscript. M.T.B. guided experiments and edited the final version of the manuscript. J.J.M. conceived the project, guided experiments, analyzed data, wrote the majority of the initial manuscript, and edited subsequent and final versions of the manuscript.
We declare no competing interests.
REFERENCES
- 1.Liu L, Okada S, Kong X-F, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, Toth B, Flatot J, Migaud M, Chrabieh M, Kochetkov T, Bolze A, Borghesi A, Toulon A, Hiller J, Eyerich S, Eyerich K, Gulácsy V, Chernyshova L, Chernyshov V, Bondarenko A, Grimaldo RMC, Blancas-Galicia L, Beas IMM, Roesler J, Magdorf K, Engelhard D, Thumerelle C, Burgel P-R, Hoernes M, Drexel B, Seger R, Kusuma T, Jansson AF, Sawalle-Belohradsky J, Belohradsky B, Jouanguy E, Bustamante J, Bué M, Karin N, Wildbaum G, Bodemer C, Lortholary O, Fischer A, Blanche S, Al-Muhsen S, Reichenbach J, Kobayashi M, Rosales FE, Lozano CT, Kilic SS, Oleastro M, Etzioni A, Traidl-Hoffmann C, Renner ED, Abel L, Picard C, Maródi L, Boisson-Dupuis S, Puel A, Casanova J-L. 2011. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med 208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CA, Kullberg BJ, van der Meer JW, Lilic D, Veltman JA, Netea MG. 2011. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med 365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 3.Toth B, Mehes L, Tasko S, Szalai Z, Tulassay Z, Cypowyj S, Casanova JL, Puel A, Marodi L. 2012. Herpes in STAT1 gain-of-function mutation. Lancet 379:2500. doi: 10.1016/S0140-6736(12)60365-1. [DOI] [PubMed] [Google Scholar]
- 4.Toubiana J, Okada S, Hiller J, Oleastro M, Lagos Gomez M, Aldave Becerra JC, Ouachee-Chardin M, Fouyssac F, Girisha KM, Etzioni A, Van Montfrans J, Camcioglu Y, Kerns LA, Belohradsky B, Blanche S, Bousfiha A, Rodriguez-Gallego C, Meyts I, Kisand K, Reichenbach J, Renner ED, Rosenzweig S, Grimbacher B, van de Veerdonk FL, Traidl-Hoffmann C, Picard C, Marodi L, Morio T, Kobayashi M, Lilic D, Milner JD, Holland S, Casanova JL, Puel A. 2016. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood 127:3154–3164. doi: 10.1182/blood-2015-11-679902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee PP, Mao H, Yang W, Chan KW, Ho MH, Lee TL, Chan JF, Woo PC, Tu W, Lau YL. 2014. Penicillium marneffei infection and impaired IFN-gamma immunity in humans with autosomal-dominant gain-of-phosphorylation STAT1 mutations. J Allergy Clin Immunol 133:894–896.e895. doi: 10.1016/j.jaci.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 6.Puel A, Cypowyj S, Marodi L, Abel L, Picard C, Casanova JL. 2012. Inborn errors of human IL-17 immunity underlie chronic mucocutaneous candidiasis. Curr Opin Allergy Clin Immunol 12:616–622. doi: 10.1097/ACI.0b013e328358cc0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE Jr, 1992. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A 89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 9.Najjar I, Fagard R. 2010. STAT1 and pathogens, not a friendly relationship. Biochimie 92:425–444. doi: 10.1016/j.biochi.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann HH, Schneider WM, Rice CM. 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol 36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim CP, Cao X. 2006. Structure, function, and regulation of STAT proteins. Mol Biosyst 2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 12.Okada S, Puel A, Casanova JL, Kobayashi M. 2016. Chronic mucocutaneous candidiasis disease associated with inborn errors of IL-17 immunity. Clin Transl Immunology 5:e114. doi: 10.1038/cti.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smeekens SP, Plantinga TS, van de Veerdonk FL, Heinhuis B, Hoischen A, Joosten LA, Arkwright PD, Gennery A, Kullberg BJ, Veltman JA, Lilic D, van der Meer JW, Netea MG. 2011. STAT1 hyperphosphorylation and defective IL12R/IL23R signaling underlie defective immunity in autosomal dominant chronic mucocutaneous candidiasis. PLoS One 6:e29248. doi: 10.1371/journal.pone.0029248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takezaki S, Yamada M, Kato M, Park MJ, Maruyama K, Yamazaki Y, Chida N, Ohara O, Kobayashi I, Ariga T. 2012. Chronic mucocutaneous candidiasis caused by a gain-of-function mutation in the STAT1 DNA-binding domain. J Immunol 189:1521–1526. doi: 10.4049/jimmunol.1200926. [DOI] [PubMed] [Google Scholar]
- 15.Dhalla F, Fox H, Davenport EE, Sadler R, Anzilotti C, van Schouwenburg PA, Ferry B, Chapel H, Knight JC, Patel SY. 2016. Chronic mucocutaneous candidiasis: characterization of a family with STAT-1 gain-of-function and development of an ex-vivo assay for Th17 deficiency of diagnostic utility. Clin Exp Immunol 184:216–227. doi: 10.1111/cei.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soltesz B, Toth B, Shabashova N, Bondarenko A, Okada S, Cypowyj S, Abhyankar A, Csorba G, Tasko S, Sarkadi AK, Mehes L, Rozsival P, Neumann D, Chernyshova L, Tulassay Z, Puel A, Casanova JL, Sediva A, Litzman J, Marodi L. 2013. New and recurrent gain-of-function STAT1 mutations in patients with chronic mucocutaneous candidiasis from Eastern and Central Europe. J Med Genet 50:567–578. doi: 10.1136/jmedgenet-2013-101570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, Al Shehri T, Abinun M, Gennery AR, Mann J, Lendrem DW, Netea MG, Rowan AD, Lilic D. 2015. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC). Eur J Immunol 45:2834–2846. doi: 10.1002/eji.201445344. [DOI] [PubMed] [Google Scholar]
- 18.Vargas-Hernandez A, Mace EM, Zimmerman O, Zerbe CS, Freeman AF, Rosenzweig S, Leiding JW, Torgerson T, Altman MC, Schussler E, Cunningham-Rundles C, Chinn IK, Carisey AF, Hanson IC, Rider NL, Holland SM, Orange JS, Forbes LR. 2018. Ruxolitinib partially reverses functional natural killer cell deficiency in patients with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations. J Allergy Clin Immunol 141:2142–2155.e2145. doi: 10.1016/j.jaci.2017.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbert L, Pflanz S, De Waal Malefyt R, Kastelein RA. 2003. IL-27 and IFN-α signal via Stat1 and Stat3 and induce T-Bet and IL-12Rβ2 in naive T cells. J Interferon Cytokine Res 23:513–522. doi: 10.1089/10799900360708632. [DOI] [PubMed] [Google Scholar]
- 20.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. 2012. STATs shape the active enhancer landscape of T cell populations. Cell 151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usherwood EJ, Ross AJ, Allen DJ, Nash AA. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J Gen Virol 77:627–630. doi: 10.1099/0022-1317-77-4-627. [DOI] [PubMed] [Google Scholar]
- 22.Cardin RD, Brooks JW, Sarawar SR, Doherty PC. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med 184:863–871. doi: 10.1084/jem.184.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehtisham S, Sunil-Chandra NP, Nash AA. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J Virol 67:5247–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen JP, Cardin RD, Branum KC, Doherty PC. 1999. CD4+ T cell-mediated control of a gamma-herpesvirus in B cell-deficient mice is mediated by IFN-gamma. Proc Natl Acad Sci U S A 96:5135–5140. doi: 10.1073/pnas.96.9.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson PG, Cardin RD, Christensen JP, Doherty PC. 1999. Immunological control of a murine gammaherpesvirus independent of CD8+ T cells. J Gen Virol 80:477–483. doi: 10.1099/0022-1317-80-2-477. [DOI] [PubMed] [Google Scholar]
- 26.Tan CSE, Lawler C, Stevenson PG. 2017. CD8+ T cell evasion mandates CD4+ T cell control of chronic gamma-herpesvirus infection. PLoS Pathog 13:e1006311. doi: 10.1371/journal.ppat.1006311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weck KE, Dal Canto AJ, Gould JD, O’Guin AK, Roth KA, Saffitz JE, Speck SH, Virgin HW. 1997. Murine gamma-herpesvirus 68 causes severe large-vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus-induced vascular disease. Nat Med 3:1346–1353. doi: 10.1038/nm1297-1346. [DOI] [PubMed] [Google Scholar]
- 28.Bennion BG, Ingle H, Ai TL, Miner CA, Platt DJ, Smith AM, Baldridge MT, Miner JJ. 2019. A Human gain-of-function STING mutation causes immunodeficiency and gammaherpesvirus-induced pulmonary fibrosis in mice. J Virol 93. doi: 10.1128/JVI.01806-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durbin JE, Hackenmiller R, Simon MC, Levy DE. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 30.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 31.Mizoguchi Y, Tsumura M, Okada S, Hirata O, Minegishi S, Imai K, Hyakuna N, Muramatsu H, Kojima S, Ozaki Y, Imai T, Takeda S, Okazaki T, Ito T, Yasunaga S, Takihara Y, Bryant VL, Kong XF, Cypowyj S, Boisson-Dupuis S, Puel A, Casanova JL, Morio T, Kobayashi M. 2014. Simple diagnosis of STAT1 gain-of-function alleles in patients with chronic mucocutaneous candidiasis. J Leukoc Biol 95:667–676. doi: 10.1189/jlb.0513250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Ma CA, Lawrence MG, Break TJ, O’Connell MP, Lyons JJ, López DB, Barber JS, Zhao Y, Barber DL, Freeman AF, Holland SM, Lionakis MS, Milner JD. 2017. PD-L1 up-regulation restrains Th17 cell differentiation in STAT3 loss- and STAT1 gain-of-function patients. J Exp Med 214:2523–2533. doi: 10.1084/jem.20161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown SM, Ritchie DA, Subak-Sharpe JH. 1973. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol 18:329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- 34.Weck KE, Barkon ML, Yoo LI, Speck SH, Virgin HI. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol 70:6775–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cieniewicz B, Dong Q, Li G, Forrest JC, Mounce BC, Tarakanova VL, van der Velden A, Krug LT. 2015. Murine gammaherpesvirus 68 pathogenesis is independent of caspase-1 and caspase-11 in mice and impairs interleukin-1β production upon extrinsic stimulation in culture. J Virol 89:6562–6574. doi: 10.1128/JVI.00658-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krug LT, Collins CM, Gargano LM, Speck SH. 2009. NF-κB p50 plays distinct roles in the establishment and control of murine gammaherpesvirus 68 latency. J Virol 83:4732–4748. doi: 10.1128/JVI.00111-09. [DOI] [PMC free article] [PubMed] [Google Scholar]