Abstract
Group 2 innate lymphoid cells (ILC2s) are enriched in mucosal tissues (e.g. lung) and respond to epithelial cell-derived cytokines initiating type-2 inflammation. During inflammation, ILC2 numbers are increased in the lung. However, the mechanisms controlling ILC2 trafficking and motility within inflamed lungs remain unclear and are crucial for understanding ILC2 function in pulmonary immunity. Using several approaches, including lung intravital microscopy, we demonstrate that pulmonary ILC2s are highly-dynamic, exhibit amoeboid-like movement and aggregate in the lung peribronchial and perivascular spaces. They express distinct chemokine receptors, including CCR8, and actively home to CCL8 deposits located around the airway epithelium. Within lung tissue, ILC2s were particularly motile in extracellular matrix-enriched regions. We show that collagen-I drives ILC2 to dramatically change their morphology by remodeling their actin cytoskeleton to promote environmental exploration critical for regulating eosinophilic inflammation. Our study provides previously unappreciated insights into ILC2-migratory patterns during inflammation and highlights the importance of environmental guidance cues in the lung in controlling ILC2 dynamics.
Introduction
Innate lymphoid cells (ILC), are an emerging family of immune cells that originate from fetal liver and adult bone marrow (BM) progenitors (1, 2). Prenatally, ILC progenitors occupy peripheral sites from mid to late stages of fetal development (3), and are detectable in peripheral tissues by embryonic (E) day 15.5 in the mouse (4). Although ILC derive from common lymphoid progenitors, they lack specific antigen receptors and lymphoid cell lineage markers, but display diverse effector functions, analogous to T cells (5, 6). Based on distinct lineage-determining transcription factors, ILC segregate into three different subsets – ILC1, ILC2 and ILC3 (7–9). Each ILC subset mirrors a T helper cell subset via the production of T helper cell signature cytokines- IFN-γ (Th1/ILC1), IL-13 (Th2/ILC2) and IL-17 (Th17/ILC3) which act to combat infection by intracellular pathogens, helminths, and extracellular pathogens respectively (3). Anatomically, different ILC subsets are resident in particular barrier and non-barrier sites (3), including the BM, skin, secondary lymphoid organs, peripheral blood and non-lymphoid tissues. ILC3 are vital for the development of adaptive immune organs in utero, however after birth thymic ILC3 are replaced by ILC2 suggesting that these cells play an important role in the thymic microenivronment (10). Among non-lymphoid tissues, ILC2 are enriched in mucosal sites including the lung and small intestine, where they contribute to local tissue immunoregulation, repair and homeostasis (11, 12). In the mouse lung, ILC2 predominate (13), and rapidly expand during the first week of life (14, 15), populating collagen-rich structures associated with medium-sized blood vessels and airways (14). In a steady state, long term maintenance of ILC2 in peripheral tissues, including the lung, is mainly supported by self-renewal of proliferative local tissue resident progenitor populations (4, 15–18). Recent evidence suggests that ICAM-1 supports ILC2 development and function during lung inflammation (19). However, in T helper type-2 (Th2) cell induced lung inflammation, ILC2 exit the BM (19, 20) and concurrently numbers are increased in the blood and lung (21–24). Human and mouse ILC2 express β2 integrins and these have been proposed to be involved in cell trafficking since blocking β2 integrins results in reduced ILC2 numbers in the lung following allergen challenge (22). In addition, recent studies have identified a distinct pre-ILC2 population originating in the gut that migrates to the lung and other distal sites giving rise to inflammatory ILC2 (iILC2) that provide protection during worm infection (15). Within the lung, interaction of ILC2 with other immune cell populations is a critical factor for shaping type 2 inflammation (25).
Several factors, including the epithelial cell-derived cytokines IL-33, IL-25 and thymic stromal lymphopoietin (TSLP), contribute to ILC2 activation and function (26). Upon activation, ILC2 produce cytokines, including IL-4, IL-5, IL-13, colony stimulating factor 2 (CSF2; GM-CSF), and the epidermal growth factor family member amphiregulin (Areg) (13, 27–29). Lipid mediators, including the arachidonic acid metabolites leukotriene D4 (LTD4), prostaglandin D2 (PGD2) (30, 31) and sphingosine 1-phosphate (S1P) (15) serve as potent regulators of ILC2 activation, accumulation and function. In contrast PGE2 and PGI2 supress ILC2 function, inhibiting GATA-3, IL-5 and IL-13 expression and decreasing proliferation (32, 33)._We have previously demonstrated that epithelial cell-derived transforming growth factor (TGF)-β1 is critical for ILC2 activation and significantly enhances airway ILC2 chemoactivity and movement in vitro (34). Thus, ILC2 activation, behaviour and function are regulated by a wide variety of factors.
Tissue environments critically control optimal immune responses, coordinating timely and proportionate recruitment, motility, migration, chemotaxis, positioning, and cell–cell interaction of leukocytes within inflamed tissues (35). Factors, including lipids, cytokines and homing receptors (HRs) incorporating chemokine receptors (3) controlling migratory patterns of ILC2 in the bone marrow (BM), spleen, gut, mesenteric lymph nodes (36) and skin (37, 38) have been identified. Lung ILC2 express high levels of β1 and β2 integrins, and use β2 integrins selectively to migrate from the BM to the lung after intranasal (i.n.) allergen (Alternaria alternata) challenge (39). Organ specific imprinting confers differential gene expression patterns on tissue resident pulmonary ILC2 (40). However, ILC2 motility within the lung during inflammation remains unstudied, and little is known regarding the signals that might regulate it. Studies investigating T cell motility in the lung have described a combination of cell-intrinsic signals and physical guidance cues coupled with biochemical signals provided by the microenvironment in driving cell movement (41). However, the environmental guidance signals controlling ILC2 dynamic behaviour in the lung remain poorly defined. Here using IL13-eGFP mice (28), combined with several imaging approaches, including lung intravital microscopy, we have documented that ILC2 exhibit amoeboid-like movement in the peribronchial and perivascular space after IL-33 induced lung inflammation. We identified specific molecules that communicate between the inflamed pulmonary environment and ILC2, uncovering the major environmental factors from which ILC2 interpret locomotory cues within the lung.
Results
The number of ILC2 rapidly increases in the peribronchial / perivascular region after rIL-33 treatment
Under homeostatic conditions, ILC2 exist in relatively low numbers in the lung, but are rapidly enriched after recombinant IL-33 (rIL-33) or rIL-25 induced inflammation (20–24, 39), promoting tissue repair during inflammation and regulating immune homeostasis. In this respect, ILC2 are thought to occupy a specific niche in the lung in keeping with their putative role as tissue resident sentinel cells (14). However, whether this locational niche is shared with CD4+ T cells remains unknown. Using IL13-eGFP mice, we investigated ILC2 numbers in various sites in the lung both at homeostasis and after acute exposure to either the fungal allergen Alternaria alternata (Alt) or rIL-33. We first quantified GFP+CD45+ cells among live lymphocytes that were either lineage positive versus lineage negative cells (fig. S1A). ILC2 were further defined by pre-gating on live (determined using a fixable live dead dye), GFP+CD3-NKp46- cells that were lineage (TCRβ, TCRγδ, CD5, CD19, CD11b, CD11c, FCεR1, GR-1, F4/80, and TER-119) negative cells and co-expressed CD90.2, KLRG-1, CD127 and intracellular cytokine IL-13 (fig. S1B). As expected, in control mice, ILC2 were present in very low numbers in the airways, lung tissue and lung draining lymph nodes (mediastinal lymph node) (Fig. 1, A to C) as well as the blood, bone marrow, spleen, inguinal and mesenteric lymph nodes (fig. S2). However, during inflammation induced by i.n. administration of rIL-33 or Alt, ILC2 numbers significantly increased in the bronchoalveolar lavage fluid BAL (Fig. 1A), lung tissue (Fig. 1B) and the lung draining lymph nodes (Fig. 1C) as well as after rIL-33 treatment in the blood and bone marrow but not spleen, inguinal and mesenteric lymph nodes (fig. S2). To confirm that we were not excluding unstimulated ILC2 (IL-13-/GFP-) in our gating, we additionally re-evaluated ILC2 numbers by gating on CD45+lin- NKp46-CD3- cells and analysed the frequency of GATA-3+ (a faithful transcription factor to define all ILC2s) cells (fig. S3A). Here we used balb/c mice instead of IL-13 eGFP mice as the GFP signal was significantly lost following intra-nuclear staining for GATA-3. Balb/c mice were challenged with PBS, Alt or rIL-33 and lungs, BAL and blood ILC2s were evaluated by gating on GATA-3+ ILC2s. Our data suggests that ILC2s in mock treated mice are still very low in frequency compared to Alt and rIL-33 treatment (fig. S3B and C) and expressed significantly lower IL-13 and IL-5 (fig. S3D). Furthermore, by phenotyping GATA-3+ ILC2s for extracellular markers, our data suggests that each of the surface markers is altered as per the type of treatment as shown by the percent expression of each marker (fig. S3E-H) further highlighting the plasticity of ILC2s in inflammation.
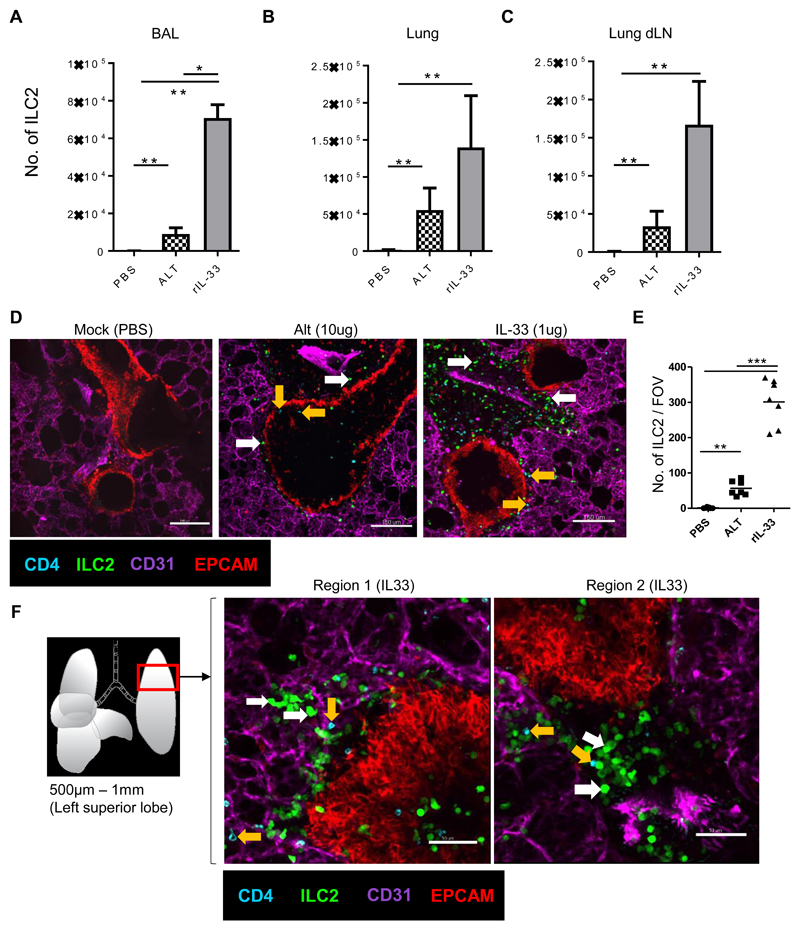
Fig 1. The number of ILC2 rapidly increase in the peribronchial / perivascular region after rIL-33 treatment.
IL13-eGFP mice were treated with 3 doses of rIL-33 (1μg per dose), Alt (10μg) or PBS (25μl) over 1 week and culled 24h after the final dose. The frequency of ILC2 (GFP+ CD45+ Linneg CD3-NKp46-CD127+CD90.2+KLRG1+CD25varIL-13+IL-5+) in the (A) airways (BAL fluid), (B) lung and (C) lung draining lymph nodes (mediastinal). Live viable precision cut lung slices of 200μm thickness were obtained and stained for CD31 (Magenta, the lung structure and blood vessels), CD4 (cyan, T cells, orange arrow), EpCAM (Red, to visualise bronchial epithelium) and GFP (ILC2, white arrow). Images of 1024μm x 1024μm field of view (FOV) were taken under a 20x objective using an inverted confocal microscope. (D) Images showing ILC2 (GFP+CD4-) CD4+ T cells (CD4+GFP-) location in PBS, rIL-33 and Alt treated mice, scale bar, 150 μm. (E) Number of ILC2 (GFP+CD4-) in lung sections per FOV taken under a 10x objective. (F) Schematic illustration of the lung depicting the anatomical location in the lung where precision cut lung slices were prepared. Representative images show two regions of the lung slice from a rIL-33 treated mouse showing distribution of ILC2 and CD4+ T cells, scale bar 150 μm. n = 4 mice per group (Mock(PBS)), n= 6 mice per group (Alt or rIL-33 treatment). Data representative of 4 experiments. *p < 0.05, **p < 0.01, ***p <0.001 and, **** p < 0.0001.
Similarly, imaging precision cut lung slices (PCLS) from rIL-33 treated IL13-eGFP mice, revealed substantialy more IL-13+GFP+ cells (fig. S4A) which largely accumulated around large airways and large blood vessels and to a lesser extent around alveolar capillaries (fig. S4B). To establish that the IL-13+GFP+ cells observed after rIL-33 treatment were pre-dominantly ILC2, we evaluated the proportion of GFP+ cells that were CD45+linnegCD4- and expressed ILC2 defining markers (CD90.2, CD127, KLRG1,CD25 and co-expressed intracellular IL-13). Our data showed that 98.86% of observed GFP+ cells were ILC2, and only 1.14% of GFP+ cells were CD4 T cells (fig. S4C). The source of IL-13 by ILC2 versus CD4+ T cells after 1 week of rIL-33 administration was further quantified (fig. S4D).
Since a minor proportion of CD4+ T cells produced IL-13 in our lung inflammation model, we included a CD4-T cell antibody to precisely evaluate the effects of rIL-33 administration on numbers and phenotype of CD4-GFP+ ILC2 in PCLS studies. Our PCLS imaging revealed that rIL-33 treatment induced a significant increase in GFP+ ILC2 numbers compared to Alt or mock treated mice (Fig. 1D). rIL-33 treatment induced a 6-fold greater ILC2 number compared to Alt treatment (Fig. 1,D and E), consistent with the number of ILC2 detected by flow cytometry (Fig. 1, A to C) and with a shared locational distribution in the inflamed lung tissue (fig. S4A). Interestingly GFP+ cells surrounding the bronchioles were significantly greater than the GFP+ cells in the alveolar tissue (fig. S4B). The majority of the GFP+ cells were IL-13 producing ILC2 (fig. S4, C and D) and hence represented IL-13+ activated ILC2, referred from here on as ILC2 for simplicity. Higher magnification views of the peribronchial space bordering the airways revealed an intimate association of some ILC2 with the airway epithelium and that ILC2 shared a locational niche with CD4+ T cells (Fig. 1F, depicting two representative images from a lung slice). Overall, we demonstrate that rIL-33 treatment significantly increased the number of lung ILC2 compared to Alt or PBS treatment and ILC2 accumulate around the airways of inflamed lungs.
IL-33 stimulation induces ILC2 motility around blood vessels and airways
Intranasal administration of either rIL-33 or Alt induces robust inflammation in the lung (42, 43). A hallmark of acute tissue inflammation is leukocyte recruitment. ILC2 are relatively rare in the lung compared to other leukocytes under homeostatic conditions but accumulate rapidly after rIL-33 induced inflammation (Fig. 1). We have previously demonstrated that the epithelial cell derived TGF-β induced after rIL-33 treatment promoted a high degree of ILC2 motility in vitro (34).
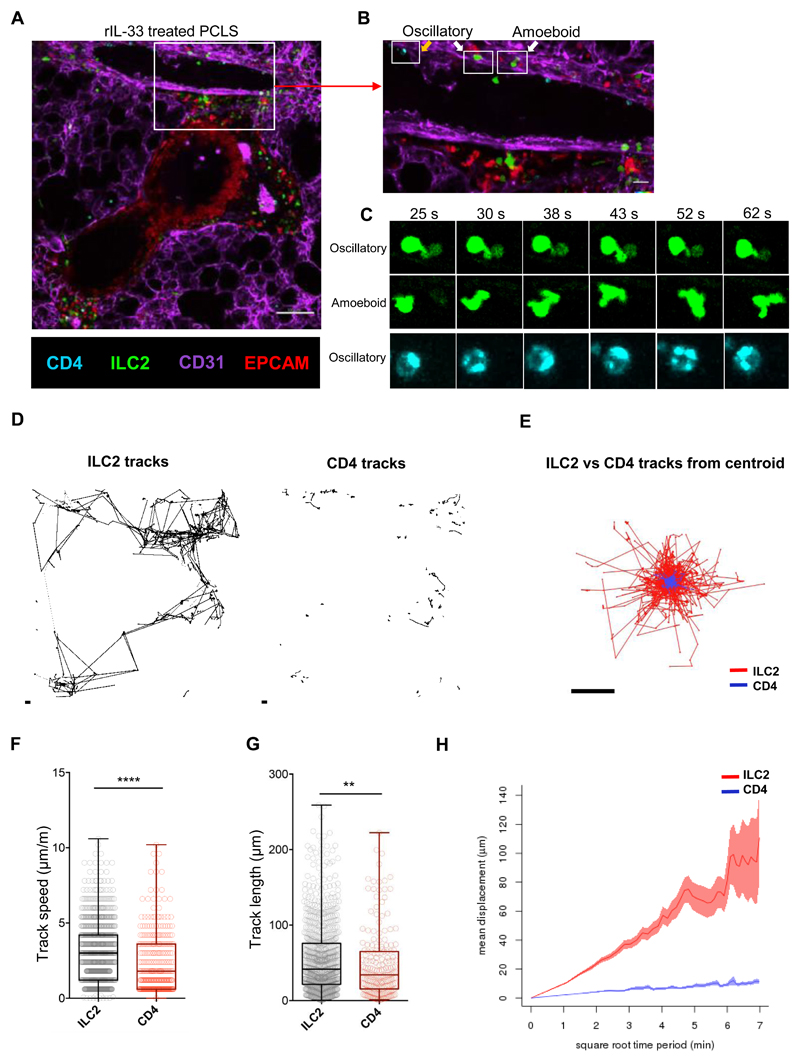
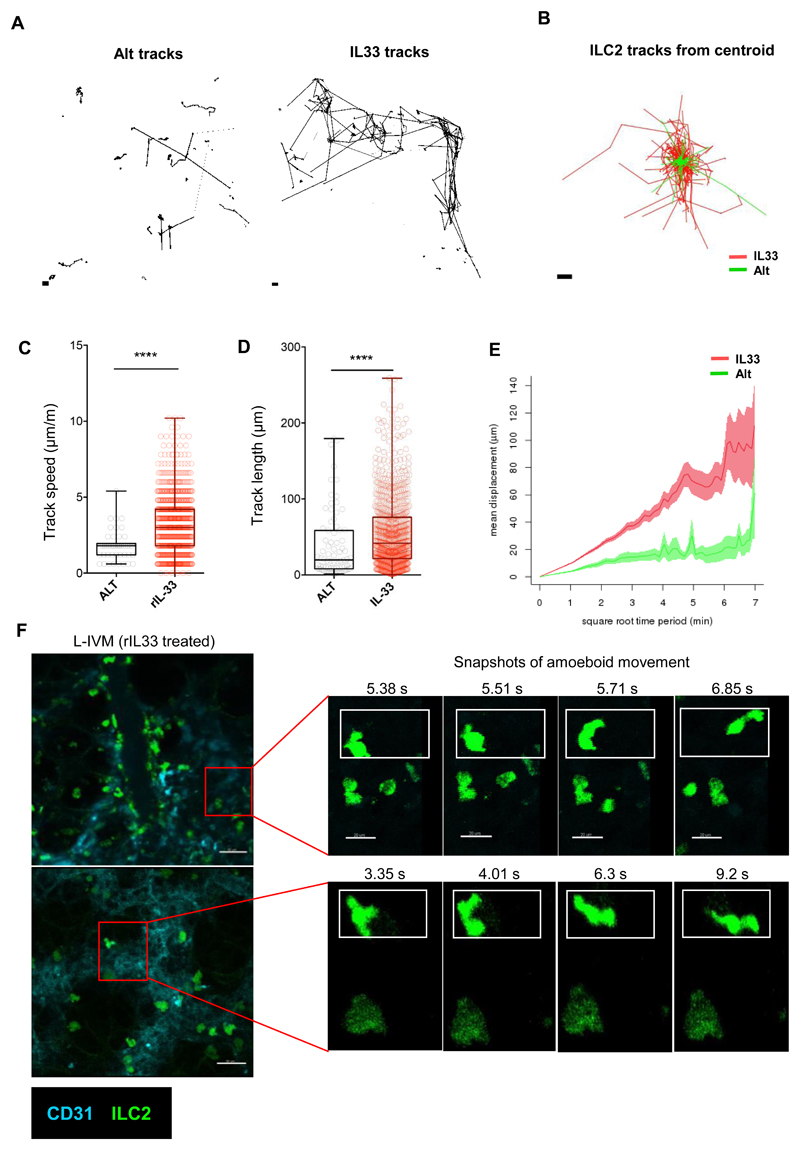
However, to date, dynamic movement of ILC2 has not been visualized in the lung. Since lung ILC2 are relatively rare at homeostasis (Fig. 1, B, D and E, fig. S3), we evaluated ILC2 movement in the inflamed lungs of IL13-eGFP mice. Imaging live PCLS from rIL-33 treated IL13-eGFP mice showed that ILC2 were motile in the peribronchial and perivascular space (Fig. 2A; movie S1). Closer examination of the lung tissue surrounding the blood vessel revealed that the majority of ILC2 exhibited ‘amoeboid-like’ exploratory movement (Fig. 2, B and C; movies S2 and S3A) with pseudopodia and bleb-like processes in the extravascular lung tissue surrounding the blood vessel, while a lesser proportion of ILC2 exhibited ‘oscillatory’ blebbing activity with little actual movement away from their start position (Fig. 2, B and C; movies S2 and S3B). Similar to oscillatory ILC2, CD4+ T cells (cyan) did not appear to displace from their point of origin in the lung tissue (Fig. 2B; movie S2). Cell tracking of ILC2 and CD4+ T cells (Fig. 2, D and E) revealed that in PCLS from rIL-33-treated mice, ILC2 track speed (Fig. 2F), track length (Fig. 2G) and track displacement (Fig. 2H) was significantly higher than that of CD4+ T cells. Interestingly, ILC2 from Alt treated mouse lungs moved more slowly, with shorter tracks, compared to rIL-33-treated mice (Fig. 3, A and B). In addition, rIL-33 treatment induced increased ILC2 track speed (Fig. 3C), track length (Fig. 3D) and track displacement (Fig. 3 E) compared to Alt treatment. Since rIL33 promoted greater ILC2 movement than Alt treatment, we focused on rIL-33 induced changes on ILC2 motility thereafter. Since PCLS do not recapitulate the forces exerted by blood-flow and breathing in vivo, we also performed lung intravital microscopy in live mice using a similar strategy to previous work (44). We found that consistent with our data in live PCLS (Fig. 1, D to F, Fig. 2, A and B), ILC2 were enriched in the extravascular tissue close to larger blood vessels and exhibited amoeboid-like movement within the lung tissue (Fig. 3F, movie S4A and 4B).
Fig 2. rIL-33 stimulation induces ILC2 motility around blood vessels and airways.
IL13-eGFP mice were treated with 3 doses of rIL-33 (1μg per dose), over 1 week and culled 24h after the final dose. Live viable precision cut lung slices (PCLS) of 200μm thickness were obtained and stained for CD31 (Magenta, the lung structure and blood vessels), CD4 (cyan, T cells, orange arrow), EpCAM (Red, to visualise bronchial epithelium) and GFP (ILC2, white arrow), and time-lapse video taken (1024μm x 1024μm field of view (FOV), 45 min duration under a 20x objective using an inverted confocal microscope) (A) Static image depicting the location of ILC2 and CD4+ T cells, scale bar 100 μm. (B) Zoomed in section of the blood vessel in figure 2A, scale bar 20 μm. (C) High power images of boxed cells in figure 2B showing differences in pattern of cell movement (oscillatory vs amoeboid movement). ILC2 and CD4+ T cells dynamics were tracked and plotted as (D) individual tracks or (E) tracks commencing from centroid and overlaid. (F) Track speed, (G) track length and (H) track displacement were quantified. Representative images shown in (A-C) are from rIL-33 treated mice, where n = 6 mice per treatment (3 slices per mouse were imaged). For (F-H) in box and whiskers plots, each dot represents an individual cell. Data are representative from 4 experiments where n = 6 mice per treatment. *p < 0.05, **p < 0.01, ***p < 0.001 and, **** p < 0.0001.
Fig 3. rIL-33 stimulation induces ILC2 motility around blood vessels and airways.
IL13-eGFP mice were treated with 3 doses of rIL-33 (1μg per dose) or Alt (10μg) over 1 week. Live precision cut lung slices were obtained and ILC2 dynamics were compared between the two and the differences were plotted as (A) individual tracks and (B) tracks commencing from centroid and overlaid. Differences in tracks between treatments were quantified as (C) track speed, (D) track length and (E) track displacement. Intravital microscopy (IVM) was performed in live IL-13eGFP mice after rIL-33 treatment (one 512μm x 512μm field of view (FOV) in a 1-hour-duration video). (F) Static images of different frames captured during the course of the video depicting amoeboid shape changes of ILC2 at separate time-points, scale bar 20 μm. n ≥ 4 mice per group. Data representative of 4 experiments. *p < 0.05, **p < 0.01, ***p < 0.001 and, **** p < 0.0001. Quantifications from (A-E) are representative of 4 experiments, where n = 6 mice per treatment (3 slices per mouse were imaged). For (F) IVM images are representative of 6 individual IL-33 treated mice. *p < 0.05, **p < 0.01, ***p < 0.001, and, **** p < 0.0001.
In summary, our results demonstrate that ILC2s are highly dynamic in the lung after treatment with either rIL-33 or allergen, where they exhibit amoeboid-like movement and travel faster over greater distances than CD4+ T cells.
ILC2s utilize distinct chemotactic pathways to home to inflammatory sites in the lung
We have shown that within the lung, ILC2s display a specific distribution pattern and movement behaviour distinct from T cells. However, the factors that regulate ILC2 dynamics are not well understood. Since ILC subsets have key features analogous to T cell populations, it has been suggested that molecular pathways which control motility, migration and tissue homing are common to ILC subsets and T cells, but this has not yet been proven. Therefore, we examined the pattern of chemokine receptor expression on ILC populations (defined as CD45+GFP+LinnegKLRG1+CD127+CD25var) isolated from the lungs of rIL-33 treated mice (or PBS treated controls) (fig. S5A and B). Surprisingly, despite the high degree of resemblance between ILCs and T cells, many of the prototypic chemokine receptors found on T cell subsets were absent on ILC, including CCR3, CCR4 (Th2), CCR6 (Th17), and CCR5 and CXCR3 (Th1). Notably, ILC did express both CCR1 and CCR8, which are associated with type 2 cytokine production by T cells in allergic contexts (45).
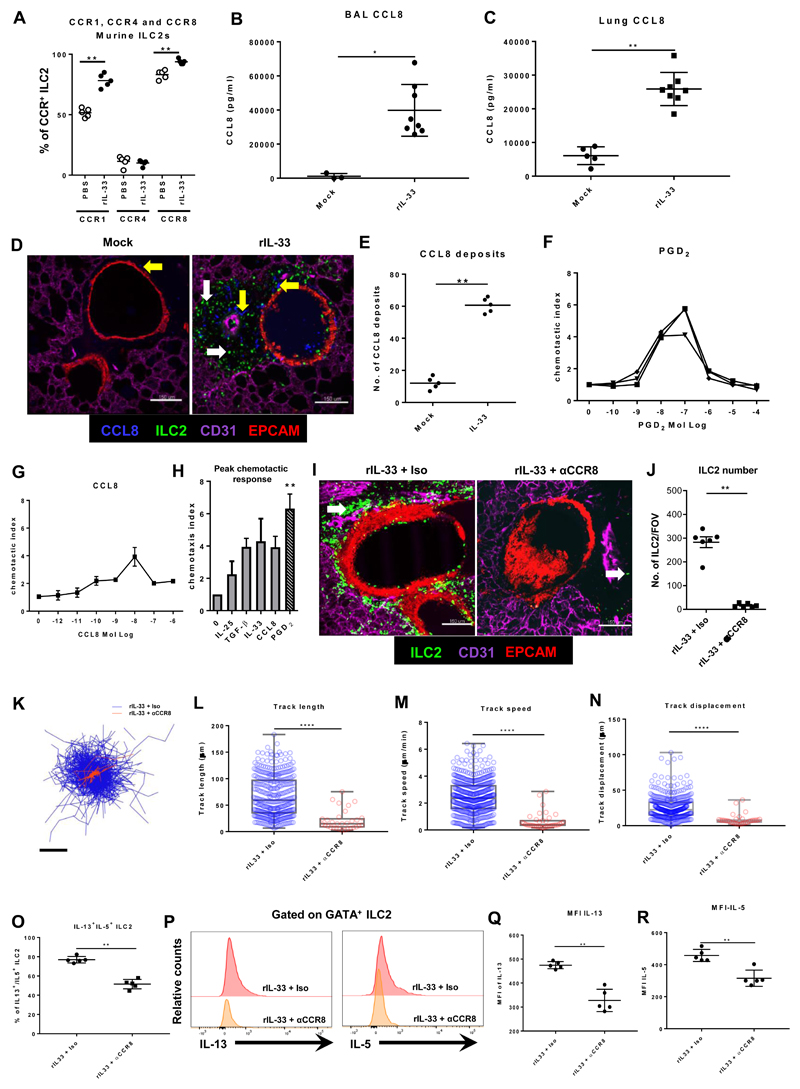
Therefore, we examined expression of CCR1, 4 and 8 specifically on IL-13+ ILC2, finding that the majority of IL-13+ ILC2 express CCR1 (>50%) and/or CCR8 (>90%) and that the expression of these chemokine receptors was higher in rIL-33 treated mice than in PBS treated controls (Fig. 4A). In contrast to T cells, we found that CCR4 expression was not associated with a type 2 phenotype in ILC with only 10% of IL-13+ ILC also expressing CCR4. Furthermore, in vivo activation of ILC2 with rIL-33 did not alter CCR4 expression (Fig. 4A). Interestingly, we also found significantly greater levels of CCL8 protein, a ligand for CCR8 in mice (46) in the airways (Fig. 4B) and lungs (Fig. 4C) of rIL-33 treated mice compared to PBS treated control mice. We additionally evaluated other CCR8 ligands (CCL1) and CCR1 ligands (RANTES) (fig. S5C and D) in lungs of mice exposed to rIL-33 compared to control treated mice. PCLS and intravital imaging demonstrated that ILC2 accumulate in the peribronchial and perivascular space and are highly motile in rIL-33 treated IL13-eGFP mice (Fig. 2, A to C and Fig. 3F). Hence, we determined whether we could identify CCL8 chemokine deposits in areas of ILC2 movement in live PCLS of rIL-33 treated IL13-eGFP mice. CCL8 was strongly expressed in the peribronchial space where ILC2 accumulate after rIL-33 treatment, at significantly greater levels than basal CCL8 expression in control mice (Fig. 4, D and E) with airway macrophages being the major producer for CCL8 in our model (fig. S5E).
Fig 4. ILC2 utilize distinct chemotactic pathways to home to inflammatory sites in the lung.
IL13-eGFP mice were treated with 3 doses of rIL-33 (1μg per dose) or PBS (25μl), over 1 week and culled 24h after the final dose. (A) The percentage of murine ILC2 (CD45+LinnegNKp46-CD3-) expressing CCR1, CCR4 and CCR8. CCL8 levels in murine (B) BAL and (C) lung. (D) Location of CCL8 expression and ILC2 and (E) quantified CCL8 deposits in PCLS stained for CD31 (Magenta, the lung structure and blood vessels), CCL8 (cyan, yellow arrow), EpCAM (Red, to visualise bronchial epithelium) and GFP (ILC2, white arrow), images of 1024μm x 1024μm FOV, scale bar 150 μm. Human ILC2 lines were generated and migration to varying concentrations of (F) PGD2 and (G) CCL8 were determined. (H) Peak migratory responses of a human ILC2 cell line to IL-25, TGF-β, rIL-33, CCL8 and PGD2. IL13-eGFP mice treated with rIL-33 were also treated with 5μg purified anti-mouse CCR8 antibody i.p., rCCL8 i.n. or an isotype control and PCLS obtained and stained. (I) Localisation of ILC2 in live PCLS. (J) Number of ILC2 per FOV under 10x objective. Time-lapse imaging of 45 min duration was performed and ILC2 (K) track from centroid, (L) track length and (M) track speed and (N) track displacement were quantified. In box and whiskers graphs each data point represents an individual cell. Balb/c mice treated with rIL-33 were further treated with rCCL8, αCCR8 or Isotype control antibody. (O) Percentage of IL-13+IL-5+ILC2 (CD45+lin-NKp46-CD3-GATA-3+). (P) Representation Histogram of MFI of IL-13 and IL-5 and quantification of MFI for (Q) IL-13 and (R) IL-5 from GATA+ ILC2. For panels A-E n ≥ 4 mice per group. Data representative of 4 experiments. For panels F-H n = 3 individual donors. Data representative of 3 experiments. For panels I-R, n = 5 mice per group. Data representative of 2 experiments *p < 0.05, **p < 0.01, ***p < 0.001 and,**** p < 0.0001.
To examine the migration of ILC2 to chemotactic agents we sorted human ILC2 from peripheral blood (CD45+, Linneg, CD161+, CD127+, CRTH2+ and C-Kit variable-) then cultured with recombinant IL-2, IL-7 and IL-33 for at least 4 weeks (resulting in >99% ILC2). These ILC2s expressed GATA-3 and IL-13 (fig. S6A) and produced IL-13 (fig. S6B). Among the three cytokines required for ILC2 maintenance, our results suggest that IL-7 signalling in ILC2 was critical for their survival, proliferation and cytokine production as demonstrated by viability (fig. S6C) GATA-3 expression (fig. S6D), Ki67 staining (fig. S6E) and IL-5 production (fig. S6F). We additionally compared differences in motility between ILC2 starved of IL-2 vs IL-33 compared to ILC2 that received all three cytokines. After 12h of live ex vivo imaging ILC2 track speed (fig. S6G) remained comparable (movie S5A, B and C). ILC2 that were starved of IL-7 could not be included in this set up as absence of IL-7 significantly reduced ILC2 viability (fig. S6C). Our findings are consistent with the literature and reemphasize the importance of IL-7 signalling being critical for the maintenance of ILC2 and other IL-7R+ lymphocyte populations (47).
The potential of in vitro cultured ILC2 to migrate to a known ILC2 chemotactic factor - the eicosanoid PGD2 (48), was examined using a modified Boyden chamber chemotaxis assay (Fig. 4F). As expected, all human ILC2 lines tested migrated to PGD2 and peak chemotaxis occurred at a concentration of 100nM PGD2 consistent with previous studies (48). Since we found CCL8 in the BAL and CCL8 deposits in the lung of rIL-33 treated mice (Fig. 4, D and E), we queried whether CCL8 could induce the migration of human ILC2. Using the same modified Boyden chamber assay, we found that CCL8 also induced human ILC2 chemotaxis in a dose-dependent manner, peaking at 10nM CCL8 (Fig. 4G). We next compared migration of ILC2 to factors thought to influence ILC2 movement. After determining the optimum concentrations for each factor, we found that although PGD2 was the most efficacious inducer of ILC2 migration, TGF-β, IL-33 and CCL8 all promoted comparable levels of migration (Fig. 4H). Taken together, these data identify specific chemokine receptor expression in ILC2 and show the corresponding ligand availability in areas of ILC2 accumulation. In order to confirm that CCL8-CCR8 axis plays an important functional role in ILC2 migration in vivo, we administered a blocking antibody against CCR8 or isotype Ab as a control and simultaneously treated mice with rIL-33. Our live PCLS data showed that blocking CCR8 significantly reduced ILC2 accumulation in the peribronchial region compared to control mice (Fig. 4, I and J) and ILC2 migration was impaired after CCR8 receptor blocking (movie S6, A and B) with shorter tracks depicted as track overlay between treatments (Fig. 4K) and reduced track length (Fig. 4L), track speed (Fig. 4M) and track displacement (Fig. 4N). Closer analysis of ILC2 from each of these treatments revealed that significantly less of the ILC2 in rIL-33 treated mice receiving the anti-CCR8 blocking antibody expressed IL-13+IL-5+ (Fig. 4O) and those cells that did express the type 2 cytokines had significantly less IL-13 and IL-5 as determined by the mean fluorescence intensity (MFI) (Fig. 4P-R). Overall, our data suggests that the CCL8/CCR8 pathway plays an essential role in inducing ILC2 accumulation, activation and movement within the lung during rIL-33 induced inflammation.
Extracellular matrix proteins, collagen-I, collagen-IV and fibronectin, promote increased ILC2 motility
In addition to chemokines, structural guidance cues such as extra cellular matrix (ECM) proteins have been shown to facilitate movement of immune cells, including T cells (41). In chronically inflamed lung tissue, as observed during asthma, aberrant ECM expression from tissue-remodelling can further influence immune cell activation and survival, thereby altering inflammatory immune responses (49). Therefore, we next examined whether ECM proteins influenced ILC2 movement. Fibronectin, collagen-I, III (fibrillar collagens) and collagen-IV (basement membrane collagen) are located around large airways and blood vessels and are altered during lung inflammation (50). Proteoglycans such as versican and tenascin C are changed during remodelling in various lung disorders, including asthma and chronic obstructive pulmonary disease (COPD) (51).
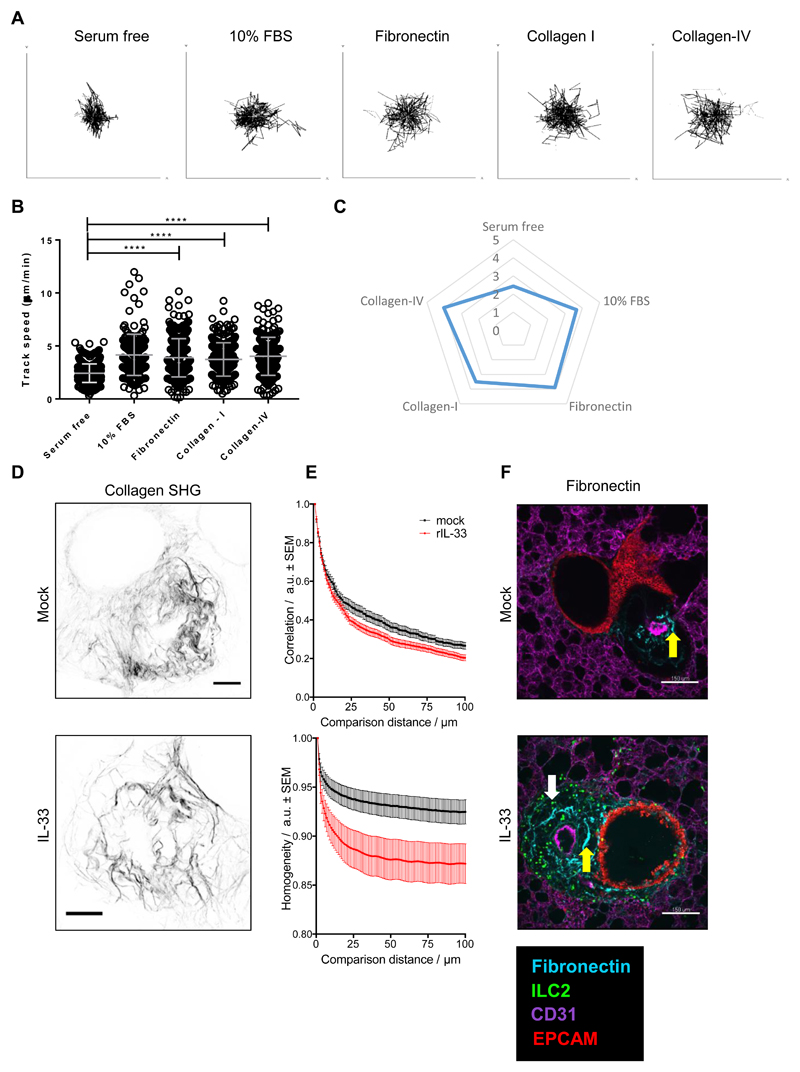
Therefore, the influence of collagens-I, -III and -IV, fibronectin, and proteoglycans, versican and tenascin C, on ILC2 movement was examined in a simplified 2D cell migration assay. Human ILC2 were cultured on these substrates and cell movement tracked over 12h in vitro by using an automated live cell imaging system using culture media with or without heat-inactivated fetal bovine serum (FBS) as controls. In contrast to serum-free media, collagen-I, fibronectin and collagen-IV significantly enhanced dynamics of ILC2, as shown in the cell tracks and speeds (Fig. 5, A to C, movie S7, A to E). As expected, FBS supplemented media significantly enhanced cell movement (Fig. 5, A to C and movie S7B). In contrast to collagen-I, fibronectin and collagen-IV, which showed strong effects on ILC2 dynamics, collagen-III, versican and tenascin C showed minimal influence on ILC2 movement (fig. S7A). Collagen fibres also produce contrast in two-photon microscopy by generating a strong second harmonic signal in tissue. Second harmonic generation (SHG) imaging suggested that the area around bronchioles and associated vessels had a strong fibrillar collagen component in both IL33-treated and control PCLS (Fig. 5D). We noticed a qualitative difference in the SHG signal with the collagen fibres in perivascular / peribroncheal areas of rIL-33 treated mice appearing ‘spikier’ than those in PBS treated control mice. Therefore, we analysed the geometry of the SHG images of fibrillar collagen in the perivascular / peribronchial area using a grey level co-occurrence matrix (GLCM) technique to quantify and compare the SHG signal (52, 53). Initially we plotted correlation versus comparison distance. This analysis compares the organisation of the extracellular matrix by considering how far it is possible to travel in a straight line away from a start point and observe the same intensity (grey level) between pairs of pixels. Images containing features that are generally organised in long straight lines will therefore have higher levels of intensity correlation over longer comparison distances and images that display structures more randomly orientated will lose this correlation at shorter comparison distances producing decays with steeper slopes. As can be seen from the data (Fig. 5E) GLCM SHG analysis of the perivascular / peribronchial area from IL-33 treated mice gave steeper decays than the controls, indicating a more disordered arrangement of shorter fibres. We also plotted homogeneity versus comparison distance. This parameter considers the distribution of grey levels at different comparison distances across the image. Therefore, images that have a texture less consistent with long fibrillar structures will have steeper decays in homogeneity with comparison distance. These data (Fig. 5, D and E) also highlighted contrast between the two conditions.
Fig 5. Extracellular matrix proteins, collagen-IV and fibronectin, promote increased ILC2 motility.
Human ILC2 lines were seeded on tissue culture plates coated with either 10% FBS, fibronectin, collagen-I, collagen-IV or serum free coating (control) for 24h. Cell movement was imaged via the JuLI imaging system and plotted as (A) individual tracks, (B) track speed dot plots and (C) track speed spider plot. IL13-eGFP mice were treated with 3 doses of rIL-33 (1μg per dose) or PBS (25μl), over 1 week and culled 24h after the final dose PCLS obtained. (D) SHG imaging of PCLS revealing collagen fibres, representative maximum intensity projections, scale bar 50μm. (E) GLCM analysis of SHG imaging. (F) Images of Fibronectin expression and localisation. PCLS stained for CD31 (Magenta, the lung structure and blood vessels), Fibronectin (cyan, yellow arrow), EpCAM (Red, to visualise bronchial epithelium) and GFP (ILC2, white arrow) and images of 1024μm x 1024μm field of view (FOV) were taken, scale bar 150μm. For panels A-C, n = 3 donors (in triplicate). Data representative of 3 experiments. For panels D-F n = 6 (in triplicate). *p < 0.05, **p < 0.01, ***p < 0.001 and,****, ****P < 0.0001.
In addition to collagen, fibronectin expression was also evaluated by staining for fibronectin in live PCLS from PBS or rIL-33 treated mice. Our results showed that fibronectin expression is enriched in the peribronchial region of rIL-33 treated mice (Fig. 5F). Collectively, our data indicate that collagen-I, -IV and fibronectin influence ILC2 speed in vitro. Furthermore, collagen and fibronectin are modified in areas where ILC2 accumulate during rIL-33 induced inflammation.
Collagen-I enhances ILC2 actin cytoskeletal remodelling and polarity
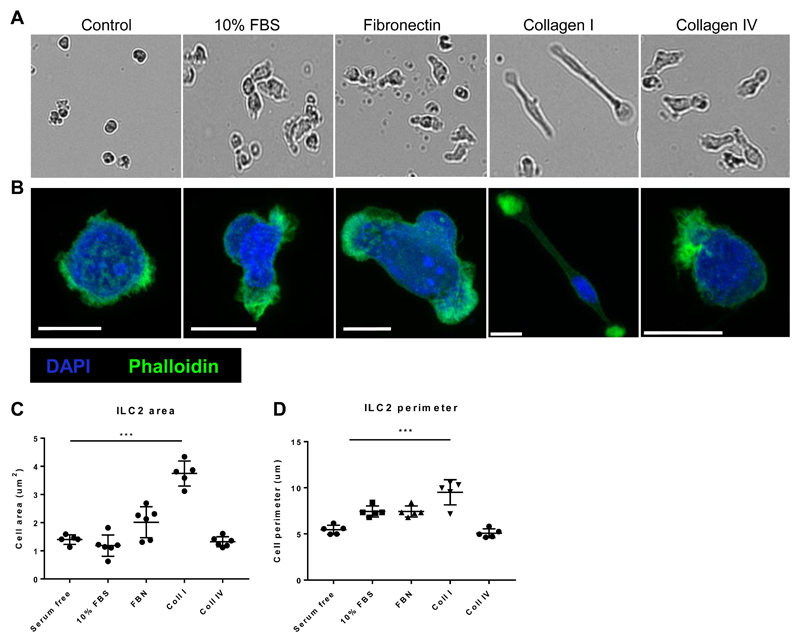
ECM protein ‘tracks’ can define the path of motile cells through the tissue (54). We observed that ILC2 exhibited amoeboid-like movement after rIL-33 induced inflammation (Fig. 2 and 3 F). Amoeboid cell movement involves induction of a simple polarized shape, dynamic pseudopod protrusion and retraction, flexible oscillatory shape changes, and rapid low-affinity crawling (55). Our results showed that ECM proteins collagen-I, fibronectin and collagen-IV promoted increased human ILC2 motility in a 2D migration assay. Therefore, we next tested whether each ECM protein directly influenced changes in ILC2 phenotype, cytokine production and shape change in the same 2D environment. We found that ILC2 GATA-3 expression (fig. S7B) and IL-5 production (Fig. S7C) remained comparable after incubation over collagen-I or collagen-IV. However collagen-I selectively induced the most pronounced ILC2 shape change with characteristically elongated ILC2 cell bodies visible (Fig. 6, A and B). In amoeboid movement, force generation (protrusion and contraction) and force transduction (adhesion), are controlled by actomyosin cytoskeletal remodelling (56). We imaged f-actin localisation and quantified ILC2 shape after 12h on the different ECM. ILC2 seeded on collagen-I were highly polarised with areas of f-actin accumulation (Fig. 6B) and had significantly greater cell area and perimeter (Fig. 6, C and D). Taken together these data indicate that collagen-I influences ILC2 shape towards the pro-migratory exploratory phenotype we described in vivo (Fig. 2 and 3 F).
Fig 6. Collagen-I enhances ILC2 actin cytoskeletal remodelling and polarity.
Human ILC2 lines were seeded on tissue culture plates coated with either 10% FBS, fibronectin, collagen-I, collagen-IV or serum free coating (control) and imaged after 12 hours. (A) Bright field images depicting change in shape. (B) Actin remodelling following staining with Phalloidin (green) and DAPI (cyan) and imaging using Airyscan detection (maximum intensity projections), scale bar 5μm. (C) Cell area. (D) Cell perimeter. n = 3 donors. Data representative of 2 experiments. *** P < 0.001.
Blocking collagen fibrillogenesis in vivo increases ILC2 dynamics and reduces eosinophil accumulation in the inflamed lung
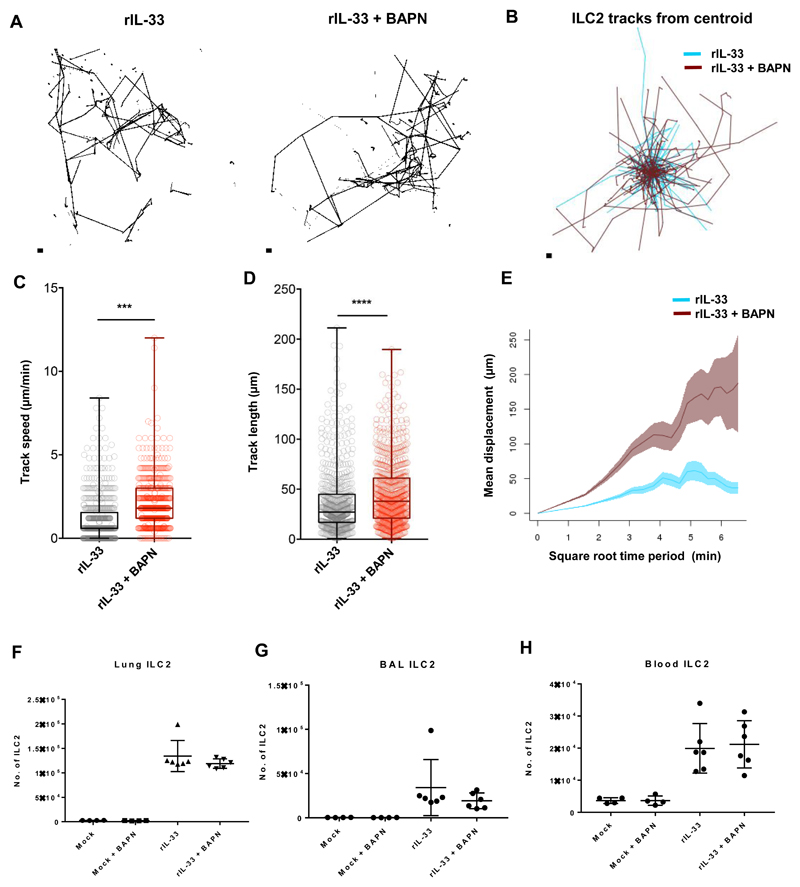
We have previously demonstrated that epithelial cell derived TGF-β1, induced after rIL-33 treatment, can drive ILC2 movement (34). Furthermore, our results show that collagen-I significantly alters ILC2 shape by elongating the cell body (Fig. 6). TGF-β has been shown to enhance collagen-I and III mRNA expression during remodeling and blocking collagen deposition restores baseline collagen levels (57). We observed changes to collagen geometry in the peribronchial space where ILC2 are highly dynamic after rIL-33 treatment (Fig. 5, D and E). Hence, we next investigated whether we could extend our findings from the 2D assays previously described in Fig 6 into in vivo 3D tissue by blocking newly synthesized collagen and measuring ILC2 movement after rIL-33 induced inflammation. The fibrillar structure of type 1 collagen morphology is stabilized by inter and intra molecular crosslinks initiated by lysyl oxidases whose expression can be inhibited by β-amino propionitrile (BAPN) (57). Following rIL-33 treatment, we administered BAPN i.p. Strikingly, mice that received rIL-33 along with the collagen crosslinking inhibitor showed longer tracks depicted as individual tracks (Fig. 7A) and track overlay between treatments (Fig. 7B). Cell track quantification revealed that ILC2 from lungs of mice that were rIL-33 treated along with BAPN displayed increased ILC2 track speed (Fig. 7C), track length (Fig. 7D) and track displacement (Fig. 7E) compared to rIL-33 treated mice suggestive of a reduced interaction with the ECM.
Fig 7. Blocking collagen fibrillogenesis in vivo increases ILC2 dynamics in the inflamed lung.
IL13-eGFP mice treated with rIL-33 were further treated with (BAPN) along with controls were culled 24 hours after the final dose. ILC2 dynamics from live PCLS were plotted as either (A) individual tracks or (B) tracks commencing from centroid and overlaid. Differences in tracks between treatments were quantified as (C) track speed, (D) track length and (E) track displacement. Total ILC2 () in (F) Lungs, (G) BAL and (H) Blood were enumerated. For panels A-E n ≥ 4 mice per group. Data representative of 4 experiments. For panels F-H n = 6 mice per group. Data representative of 2 experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
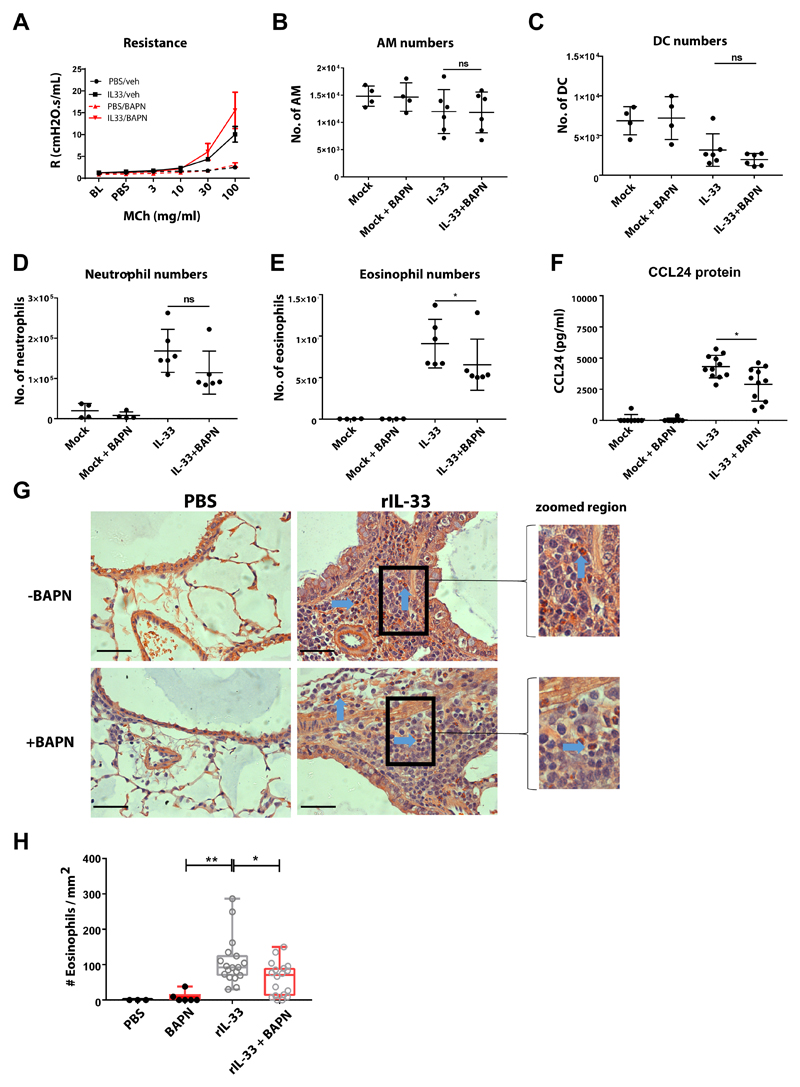
Although addition of BAPN altered ILC2 motility in rIL-33 treated mice, ILC numbers only slightly reduced in the lungs (Fig. 7F) with comparable ILC2 numbers in the BAL (Fig. 7G) and blood (Fig. 7H). We additionally tested if BAPN directly activated ILC2 by evaluating cytokine production by ex vivo sorted ILC2 from rIL-33 treated IL-13 eGFP mice. Our results showed that within the time frame of 36 h post incubation, IL-5 production by ILC2 remained unchanged after incubation with collagen-I + BAPN or collagen-I alone (fig. S8A). Interestingly addition, of rCCL8 to the cultures significantly increased IL-5 production (fig. S8A). We next evaluated if blocking collagen fibrillogenesis in rIL-33 treated mice impacted on the inflammatory infiltrate in the lungs and altered lung function. rIL-33 + BAPN treated mice had unchanged lung function (Fig. 8A) and comparable numbers of airway macrophages (Fig. 8B), dendritic cells (Fig. 8C), neutrophils (Fig. 8D) compared to mice treated with rIL-33 alone. Interestingly, blocking collagen fibrillogenesis in rIL-33 treated mice significantly reduced the frequency of eosinophils (Fig. 8E) and lung CCL24 levels (Fig. 8F) while eosinophil motility remained unaltered compared to mice treated with rIL33 treated alone (movie S8, A and B) and further quantified by track length (fig. S8B) and track speed (fig. S8C). We finally compared eosinophil accumulation in the peribronchial region of the lungs where ILC2 accumulate and found significantly lower number of eosinophils as demonstrated by congo red staining (Fig. 8G and H).
Fig 8. Blocking collagen fibrillogenesis reduces eosinophil accumulation in the inflamed lung.
Balb/c mice were treated with rIL-33 or PBS with or without BAPN. (A) Airway responsiveness to methacholine. Lung (B) alveolar macrophages (AM), (C) dendtitic cells (DC), (D) neutrophils and (E) were enumerated by flow cytometry. (F) CCL24 protein in lung tissue. (G) Congo red stained eosinophils (blue arrow) in lung histological samples. scale bar 50μm H Quantification of eosinophil from histological samples. For panels A-E n ≥ 6 mice per group. Data representative of 2 experiments. For panels G and H n ≥ 4 mice per group. Data representative of 2 experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Therefore, blocking newly synthesized collagen during rIL-33 treatment significantly changed ILC2 motility and the ensuing eosinophil accumulation. These data indicate that ILC2 interactions with the ECM may impact their function and contribution to eosinophilic inflammation.
Discussion
Immune cell recruitment in inflammation involves emigration of activated leukocytes from the vasculature across inflamed endothelium into the tissue space through activation of specific effector programs (58). The ensuing response must be robust enough to control infection or repair damage but sufficiently restrained so as to prevent excessive tissue damage and pathology. Mucosal surfaces, such as the lung, constantly encounter potentially harmful threats in the form of microbes, allergens and pollutants that can damage lung tissue and impair normal physiological lung function. Group 2 innate lymphoid cells (ILC2) are central in orchestrating type-2 immunity (28) and promoting tissue repair (13). We have previously shown that during lung inflammation, in vivo and in vitro, TGF-β expression influences ILC2 movement but the mechanisms which promote ILC2 migration are not well understood (34). Hence a more detailed spatio-temporal analysis of ILC2 induction and dynamics in the lung is of great importance to develop a thorough appreciation of ILC2 behaviour and function, and their interaction with other cells during type 2 inflammatory responses. Since ILC2 are a rare immune cell population in the lung at homeostasis (28), visualizing ILC2 by intravital imaging in the lungs of living animals has been challenging. Delivery of rIL-33 in mice drives type-2 immunity and ILC2 expand robustly and produce large amounts of IL-13 (28, 34) even in the absence of T and B cells (42). In this report, by utilizing a model of rIL-33 or Alt driven lung inflammation in IL-13eGFP knock-in mice, combined with sophisticated lung imaging, we demonstrate for the first time that ILC2 are highly dynamic in the lung in vivo. Following an acute inflammatory exposure, ILC2 numbers in the lung increased substantially by around 30 (Alt) - 100 (rIL-33) fold respectively. Simultaneously, our results show an incremental increase in the number of ILC2 in both the lung draining LN and circulation after Alt or rIL-33 induced inflammation. A recent study reported that administration of rIL-33 in mice promotes the egress of ILC2 progenitors (ILC2Ps) from the bone marrow to peripheral tissues while mice lacking IL-33 signalling had a significant accumulation of ILC2Ps in the bone marrow (59). This suggests that the rapid expansion in ILC2 numbers induced in the lung in our model of rIL-33 inflammation could result as a consequence of recruitment of ILC2Ps from the BM rather than in situ proliferation of a small steady state resident precursor. In addition, there was no increase in ILC2 numbers in the spleen or non-lung draining lymph nodes indicating that intranasal rIL-33 signals for ILC precursors in the bone marrow to traffic to the lungs. Our imaging studies also depict ILC2 accumulation in the extra-vascular tissue surrounding large blood vessels again indicating that these cells are perhaps likely to be recent emigrants from the BM.
In the lung, we found that ILC2 mainly accumulate in the peribronchial / perivascular area and to a lesser extent in the parenchyma. The location of ILC2 in the peribronchial area is consistent with their postulated role in promoting epithelial repair. In a previous study, we showed that ILC2 in the airway lumen are primed to respond to epithelial cell derived TGF-β induced after rIL-33 administration, as a result of high cell surface expression of TGF-βRII, and that epithelial cell derived TGF-β enhanced the migratory activity of ILC2 (34). Hence a critical molecular communication exists between ILC2 and bronchial epithelial cells to promote ILC2 movement during lung inflammation and could explain why ILC2 home to the peribronchial region of the lung tissue.
By using live lung slices, we demonstrated an amoeboid pattern of movement of lung ILC2 after rIL-33 treatment. This observation was recapitulated in the lungs of live intact mice using lung intravital microscopy of rIL-33 treated IL13eGFP mice. We found that similar to our data in live lung slices, ILC2 were enriched in the lung tissue surrounding larger blood vessels rather than the alveolar capillaries and characteristically move in an amoeboid manner during locomotion. However, despite their functional and phenotypic similarities to CD4+ T cells, ILC2 differed markedly in their migratory behaviour moving faster and to greater distances than CD4+ T cells in the inflamed lung. The physical proximity of motile ILC2 to epithelial cells further supports the notion that ILC2 may directly influence epithelial cell function and vice versa and may not be restricted to communication via secreted cytokines but could result from physical receptor-ligand interactions. It is known that motility patterns and distances travelled by various leukocyte subpopulations differ (60). In our study, we find that lung ILC2 exhibit amoeboid pattern of movement travelling distances in the range of 100-200μm perhaps displaying ’searching’ or ‘exploratory’ behaviour, consistent with their sentinel role at mucosal sites (61). Taken together, our data suggests that lung ILC2 may exhibit motility patterns similar to innate immune cells. This is consistent with findings in the skin, where dermal ILC2 patrol their environment with an average speed similar to that of dermal dendritic cells (38). This scanning behaviour of ILC2 may be suggestive of their need to detect local alarmins or danger signals by damaged epithelial cells in the lung after an inflammatory insult. Furthermore, the migration of lung ILC2 in close association with bronchial epithelial cells seems likely to ensure that factors secreted by the epithelium, such as TGF-β, can rapidly engage the ILC2. It presumably also ensures that signalling in the reverse direction, from the ILC2 to the epithelial cells, for promoting epithelial repair can take place efficiently.
Factors driving leukocyte motility include the distribution of adhesion molecules on leukocytes, as well as chemotactic signals. It has been well established that lung ILC2 express both β1 and β2 integrins at homeostasis and after Alt-induced inflammation (39). Furthermore, ILCs share many of the key features analogous to T cell populations, for example ILC2 and T cells both use the homing receptor CCR7 to actively migrate to lymphoid tissues (3). In addition, it has been shown recently that inflammatory iILC2, originating from precursors in the intestinal lamina propria express high levels of CCR9 and use sphingosine 1-phosphate receptors to exit the intestine into the lymph and move to distant sites like the lung where they display effector activity (15). Interestingly, progenitors to iILC2 do not exist in the lung and are not induced after inflammation induced via the intra nasal route (15). Hence, the chemokine migratory programs of ILC2 generated in the lung after intranasal instilled mediator driven inflammation have remained elusive until now and very little is understood about the process by which ILC2 are attracted to sites of immune insult in the lungs. At homeostasis, we found that CCR8 and CCR1 was expressed by the majority of murine lung ILC2, in contrast to other T cell subsets. However, following rIL-33 treatment, the percentage of CCR8 and CCR1 expressing ILC2 further increased.
In addition, TGF-β acts as an ILC2 chemo-activator enhancing murine ILC2 migration in a non-directional manner. Furthermore, both PGD2 and to a much lesser extent IL-33 have been shown to be chemotactic for human skin- and blood-derived ILC2 (48). Interestingly, we show for the first time that in addition to PGD2, CCL8 is chemotactic for ILC2 with nanomolar potency. We found that CCL8 deposits were strongly expressed in the peribronchial space where murine lung ILC2 accumulate. This could be attributed to the CCL8–CCR8 ligand-receptor axis which has been shown to be crucial in localizing pathogenic Th2 cells to the site of eosinophilic inflammation and in driving the allergic process in the skin (62). To further explore this hypothesis, we blocked CCR8 receptor in rIL-33 treated mice and analysed effects on ILC2 motility. Our data strongly suggest that signalling via CCR8 is critical for ILC2 cytokine (IL-13 and IL-5) production as well as motility of IL-13+ activated ILC2. A variety of resident pulmonary cells secrete CCL8, the CCR8 ligand in mice. We find that lung airway macrophages are the pre-dominant cellular source of CCL8 in rIL-33 treated mice. Our data suggest a previously undescribed model where macrophage derived CCL8 acts to increase ILC2 accumulation to areas proximal to epithelial cells.
Overall, our results provide insights into the independent and distinct migration pathways ILC2 utilize in contrast to T cells. Extracellular matrix proteins such as collagens are well-known to interact with leukocytes by directly binding to cell surface receptors (63). In this respect, changes to subepithelial collagen and extracellular matrix protein deposition are important pathophysiological components of airway remodeling in Th2 driven lung inflammation. We find strong fibrillar collagen deposition around bronchioles and associated vessels in both rIL33-treated and control PCLS where lung ILC2 accumulate. Interestingly rIL-33 treated mice showed quantitative differences in both correlation and homogeneity parameters of fibrillar collagen geometry compared to control mice. rIL-33 induced epithelial TGF-β is chemoactive for ILC2 and administration of TGF-β in mice increases collagen-I mRNA expression in mouse lungs (49, 57). This suggests that altered collagen distribution as a result of rIL-33 induced inflammation may influence ILC2 migratory programs. In line with this, our in vitro 2D migration assays indicate that collagen-I exclusively alters ILC2 morphology with enhanced cell spread and elongation of cell body. Hence in vivo, collagen-I fibres may support a more polarized ILC2 morphology, by increasing ILC2 traction, reducing ILC2 speed and allowing for longer dwell times in specific sites of inflammation in the lung. Cross-links are essential for the mechanical stability of collagen and have been shown to control T cell mobility (64). To test the overall contribution of newly synthesized collagen on ILC2 dynamics, we used beta-aminopropionitrile (BAPN) to block the formation of new collagen cross-links by inhibiting the enzyme lysyloxidase. Interestingly, blockade of collagen fibrillogenesis significantly increased ILC2 speed and travel distance after rIL-33 treatment. Hence suggesting that reduced collagen stability, resulting as a consequence of blocking new collagen fibrillogenesis may disrupt a controlled receptor-ligand interaction of ILC2 with collagen-I fibres, increasing overall dynamics of ILC2. Intriguingly, although collagen-I blocking increased ILC2 movement, rIL-33 induced eosinophilia was significantly reduced in the lungs of these mice. We have previously reported that after house dust mite treatment in mice, chemoactive epithelial derived TGF-β is essential for ILC2 activation and critically controls all hallmarks features of allergic responses including airway hyperresponsiveness and eosinophilia (34). Inferring from this knowledge our data suggests that although the initial cytokine driven instruction from structural cells initiates ILC2 movement, components of the inflamed lung environment namely, CCL8-CCR8 signaling and collagen-I are critical for regulating controlled ILC2 accumulation and dynamics imperative for ensuring robust type-2 inflammation. Hence although TGF-β serves as a chemokinetic mediator, CCL8 induced after rIL-33 treatment is an important chemotactic signal that mediates ILC2 motility and cytokine production via CCR8 receptor. Furthermore, the difference in interfibrillar geometry of collagen fibrils alters ILC2 migratory patterns and the dwell time of ILC2 with collagen-fibrils is a critical factor towards ILC2 behaviour and function in type-2 inflammation. Thus combined environmental signals from damaged lung tissue act in concert to mechanistically drive ILC2 migratory patterns in the inflamed lung.
ILC2 are defined by the combination of several extracellular and intracellular markers. Here we use multicolour flow cytometry of cell suspensions from dissociated lung tissue to identify and quantify these cells, but this approach loses spatiotemporal information by definition. Therefore, we used a combination of imaging using IL13-GFP reporter mice in combination with a handful of markers and dyes and detailed multicolour flow cytometry together to mitigate potential misidentification of the cells. However, in an ideal experiment we would be able to image tens of markers simultaneously. Newer approaches (for eg: imaging mass cytometry) are starting to address this, but none allow live organism / cell / tissue imaging to directly address the spatiotemporal dynamics of these populations, a significant challenge in the field.
Overall, using highly sophisticated imaging techniques, we reveal how ILC2 migrate within an inflamed tissue, interact with adjacent cells and lung environmental factors during a type-2 inflammatory response. Specifically, we show that lung ILC2 are highly motile in vivo and exhibit an exploratory amoeboid pattern of movement during inflammation. Additionally, we identify signature chemokine receptors employed by ILC2 to home to inflammatory sites in the lung. Our data provide novel insights into the mechanism by which diverse lung environmental factors combine to control ILC2 migration and provide direct evidence for collagen-I in exclusively regulating ILC2 shape and polarity, thereby influencing the outcome of an inflammatory response. Imaging these dynamic cellular events in real-time in the lungs of living animals has provided an important visual platform that could be employed for further interrogation of ILC2 and other immune cell functions in the context of allergen or pathogen driven Th2 inflammation.
Materials and Methods
Mice
Adult female IL13-eGFP (kind gift from A.N. McKenzie, MRC Laboratory of Molecular Biology, Cambridge) (28) were used for experiments between 7 and 10 weeks of age. Mice were housed in specific-pathogen-free conditions and given food and water ad libitum. All procedures were conducted in accordance to the institutional guidelines and under the approval of our Home Office project licence (granted under the Animals (Scientific Procedures) Act 1986). Mice were administered 10μg (Alternaria alternata) extract (Greer Laboratories) in 25μl of PBS intranasally 3 times a week for 1 week or PBS alone as controls. Alternatively, carrier-free recombinant murine rIL-33 (1μg per dose in 25μl PBS) (eBioscience) or PBS was administered 3 times a week for 1 week. Mice were culled 24hrs after the final cytokine or allergen dose.
Live Precision Cut Lung Slices
Ex vivo cultured precision-cut lung slices (PCLS) provided a 3D cell culture model to image ILC2 dynamics within the lung microenvironment and was adapted and modified from a previously described protocol (65). Briefly, prior to slicing, lungs were inflated with 0.8ml volume of 2% low melting agarose (Thermofisher) prepared in PBS. Following inflation lungs were carefully dissected out and placed in HBSS containing 2.5% HEPES (GIBCO, Life Technologies). The intact left superior lung lobe was excised and 200μm transverse sections were prepared from the upper 1mm region of the left superior lung lobe using a Compresstome VF-300 vibrating microtome (Precisionary Instruments). PCLS were incubated in complete RPMI (10% FCS, 2 mM L-glutamine, 100U/ml penicillin/streptomycin) (GIBCO, Life Technologies) for 15mins at 37°C to wash off excess agarose surrounding the tissue. Antibodies were prepared in complete DMEM solution and lung explants were incubated in staining solution for 1.5hrs at 37°C prior to imaging. Time lapse videos depicting ILC2 movement were generated over an hour using an inverted laser-scanning confocal microscope (SP5, Leica microsystems). Videos were quantified using Imaris version 8.1 (BitPlane, Oxford Instruments) and ILC2 track speed, length and displacement were quantified using Motilitylab software.
Intravital microscopy
Mouse lung intravital microscopy was performed as previously described (44, 66). Briefly, anaesthetised and mechanically ventilated mice were placed on a heat-mat in a right lateral position. A small custom-built circular suction chamber was surgically inserted using a small incision between the ribs of the mice for imaging on an upright laser-scanning confocal microscope with Hybrid detectors (Leica SP5). Gentle suction was applied via the vacuum port to bring the cover slip contained in the chamber into contact with the lung and stabilise the tissue. Time-lapse videos depicting ILC2 movement were generated using 488, 54 and 633nm laser lines through a 25x 0.95 N.A. long working distance water immersion objective.
Fixed Cell / PCLS imaging
Phalloidin stained ILC and fixed PCLS from IL-33 reported mice were imaged using a Zeiss LSM 880 Airyscan NLO system. DAPI was excited using a 405nm diode laser, Phalloidin AlexaFlour488 was excited at 488nm using an argon-ion laser and detected using a sub-Airy detector 32-element array (Airyscan) and processed using ‘auto’ 3D Airyscan processing options. Airyscan z-stacks were acquired and Imaris (Bitplane) was used to visualise each cell. SHG signals (produced at a central wavelength of 475nm) were collected from fixed PCLS using a Coherent Chameleon Discovery femtosecond pulsed laser tuned to 950 nm and collected using a non-descanned GaAsP detector with 450±50nm detection filter. A 25x 1.0 N.A. zeiss long working distance water immersion objective was used to collect all images.
SHG Analysis
50μm z-stacks were imaged over a region of 425.1μm by 425.1, with at least three stacks/mouse (n = 6 mice / condition). GLCM was performed similarly to previous studies (67, 68) Briefly, Image J (NIH) and the Norwegian University of Life Sciences (UMB) GLCM plugin was used for the texture analysis, modified to run automatically through the four directions, for each of the 100 comparison distances. A maximum projection image was automatically thresholded to produce a mask that was then applied to remove background noise bias in the GLCM analysis by selecting only the collagen SHG signal. The masked image was then passed to the modified GLCM texture plugin. The data files produced were processed using an ImageJ macro to output both the mean and individual values for each texture parameter for each image. These were then imported into Prism (GraphPad), where exponential decay models were fitted to the data.
Cell isolation (BAL, Lung and lung dLN)
The airways were washed three times with 400μl of PBS and bronchoalveolar lavage fluid was collected. Following centrifugation of the BAL fluid supernatants were stored at -80°C for further analysis and cells were resuspended in 500μl of complete RPMI media (10% FCS, 2mM L-glutamine, 100U/ml penicillin/streptomycin) (GIBCO, Life Technologies). For lung cell isolation, the left lung lobes were cut into small pieces and digested in complete media supplemented with 0.15mg/ml collagenase (Type D; Roche Diagnostics) and 25μg/ml DNase (Type 1; Roche Diagnostics) for 1h at 37°C. Lung homogenate obtained was then filtered through a 70μm sieve (BD Bioscience), washed, and resuspended in 1ml of complete media. Erylysis was performed on 200μl whole blood and leukocytes were washed twice and then then resuspended in 1ml of complete media.
Flow Cytometry and intracellular staining
For phenotyping ILC2 in IL-13 eGFP mice by flow cytometry, ILC2 were defined as live, singlet, lymphoid, GFP+CD45+ lineageneg (Lineage cocktail: TCRβ, TCRγδ, CD3e, CD5, CD19, CD11b, CD11c, FCεR1, GR-1, F4/80, NKp46 and TER-119) CD3- NKp46-, CD90.2+/CD127+/KLRG1+/CD25var cells with positive intracellular IL-13 and IL-5 staining. For intracellular cytokine staining, cells were stimulated with PMA (Sigma-Aldrich) and ionomycin (EMD Chemicals) in the presence of Brefeldin A (Sigma-Aldrich) and incubated for 4h at 37°C. Following stimulation, cells were washed and incubated for 20min with anti-CD16/32 (BD Pharmingen) prior to staining for extracellular antigens in 5% FCS/1% BSA in PBS for 30min at 4°C. All antibodies were purchased from eBioscience (table 2) with the exception of CD31, EpCAM, CD4 (Biolegend) also shown in table 1. Following staining for extracellular markers, cells were fixed with IC fixation buffer (E-Bioscience) for 10 mins at room temperature. Cells were then washed and stained for intracellular antigens using the Intracellular Fixation & Permeabilization Buffer Set (E Bioscience) or Foxp3 / Transcription Factor Staining Buffer Set (E-Bioscience) for GATA-3 staining.
Table S2. Antibodies used for flow cytometry.
| Antibody | Clone | Fluorochrome | Target species | Manufacturer |
|---|---|---|---|---|
| TCR-β | H57-597 | APC | Mouse | E Bioscience |
| TCR-γδ | ebio GL3 | APC | Mouse | E Bioscience |
| TER-119 | TER-119 | APC | Mouse | E Bioscience |
| CD19 | ebio ID3 | APC | Mouse | E Bioscience |
| CD11b | M1/70 | APC | Mouse | E Bioscience |
| CD5 | 53-7.3 | APC | Mouse | E Bioscience |
| F4/80 | BM8 | APC | Mouse | E Bioscience |
| FCεR1 | MAR1 | APC | Mouse | E Bioscience |
| GR-1 | RB6-8C5 | APC | Mouse | E Bioscience |
| CD11c | N418 | APC | Mouse | E Bioscience |
| NKp46 | 29A1.4 | PE-Dazzle | Mouse | E Bioscience |
| CD45 | 30-F11 | PerCP Cy5.5 | Mouse | E Bioscience |
| CD3e | 145-2C11 | APC-Cy7 | Mouse | E Bioscience |
| KLRG-1 | 2F1/KLRG1 | PE-Cy7 | Mouse | E Bioscience |
| CD90.2 | 53-2.1 | BV605 | Mouse | E Bioscience |
| CD25 | PC61 | BV510 | Mouse | E Bioscience |
| CD127 | A7R34 | BV711 | Mouse | E Bioscience |
| IL-13 | ebio 13A | PE | Mouse | E Bioscience |
| IL-5 | TRFK5 | BV421 | Mouse | E Bioscience |
| GATA-3 | TWAJ | PerCP-eFluor 710 | Mouse | E Bioscience |
Table S1. Antibodies used for imaging of PCLS.
| Antibody | Clone | Fluorochrome | Target species | Manufacturer |
|---|---|---|---|---|
| EPCAM | G8.8 | PE | Mouse | Biolegend |
| CD4 | RM4-5 | BV421 | Mouse | Biolegend |
| CD31 | 390 | Alexa-647 | Mouse | Biolegend |
Analysis was performed with LSRFortessa III and cell sorting on FACSAria III (BD Biosciences).
Lung histology and staining
Paraffin embedded lung sections (4-micron thickness) were stained with haematoxylin and Congo red-dyes. Images were taken under polarised light using an upright dry 40x objective. The number of eosinophils in the peribronchial space were quantified and corrected for the surface area scored.
Human ILC2 bulk cultures
ILC2 were enriched from whole blood using RosetteSepTM Human ILC2 Enrichment Kit (STEMCELL Technologies) and further sorted by FACs using CD45+ Lineageneg (CD1a, CD3, CD4, CD5, CD8, CD11c, CD14, CD16, CD19, CD20, CD34, FcγRI and CD123) (Biolegend), CD161+, CD127+, CRTH2+ and C-Kitvar (Biolegend). ILC2 were bulk cultured in IL-2, IL-7 (5ng/ml) and IL-33 (10ng/ml) (eBioscience) and cytokines and medium were replenished every 3-4 days. For investigations involving humans ILC2 isolation, an informed consent was obtained and the nature and possible consequences of the studies were explained to all donors.
CCR8 antibody blocking and rCCL8 treatment
CCR8 neutralisation studies were performed similar to previous reports (69). IL-13eGFP and balb/c mice were i.p. injected with 5μg purified anti-mouse CD198 (CCR8) antibody, (Clone: SA214G2) or purified Rat IgG2b, κ Isotype Ctrl Antibody two days prior to first dose of rIL-33 treatment and then combined with each of the three rIL-33 doses. Mice were analysed 24h post the third dose. Alternately 10μg Recombinant Mouse CCL8 (MCP-2) (Biolegend) was administered i.n. at the time of the three rIL-33 doses.
β-aminoproprionitrile (BAPN) treatment
BAPN treatment was carried out similar to previous reports (70). IL-13 eGFP or balb/c mice were i.p. injected with β-aminoproprionitrile (BAPN) (100 mg/kg, Sigma) dissolved in PBS and administered in 200 μl volume on day -1 and then daily for a week over the course of rIL-33 treatment.
Measurement of Airway hyperesponsiveness
Airway resistance in mice was evaluated adopting the flexiVent small animal ventilator (SciReq) as previously described (29) Prior to procedure mice were anesthetized with a combination of pentobarbital sodium (50 mg/kg, administered intraperitoneally) and ketamine (100 mg/kg, administered intramuscularly), tracheostomized, and connected to the flexiVent ventilator using a blunt-ended 19-gauge needle. The mice were ventilated by maintaining an average breathing frequency of 150 breaths/min, tidal volume of 10 ml/kg body weight and positive end-expiratory pressure of about 2 cm H2O. Differences in resistance to increasing concentrations of nebulized methacholine (3- 100 mg/ml) were analysed from the snap-shot perturbation measurements. The data was then plotted using multiple linear regression to the single-compartment model in the following form: pressure = resistance × flow + elastance × volume + fitting constant.
Chemotaxis Assay
Assays of chemotactic responsiveness were carried out as previously described using 96-well ChemoTx® plates with 5μm pores (Neuroprobe). Migrating cells were detected by the use of CellTiterGlo® Dye (Promega) and resulting luminescence measured using a TopCount scintillation counter (PerkinElmer). In all experiments, each data point was assayed in duplicate. Data are reported as Chemotactic Indices, defined as the migratory response to a particular stimulus divided by the migratory response to media alone.
Analysis of ILC2 dynamics on ECM proteins
Human ILC2 were obtained by sorting and differentiating ILC2 from peripheral blood. ILC2 lines were seeded on tissue culture plates coated with 10% FBS, 50μg/ml of fibronectin, collagen-I, and -IV. Human ILC2 dynamics was imaged on the JuLI imaging system for a period of 12h. ILC2 dynamics quantified as changes in speed and displacement were quantified using Image J software.
Statistical analysis
All data were analyzed using Graph Pad Prism 7. Time series videos were analysed and cell tracks were quantified by Imaris software. Tracks were imported to Prism for quantifying track length and track speed. Additionally tracks were imported into MotilityLab software to display tracks from centroid and differences in mean square displacement. All line graphs and bar charts are expressed as mean ±SD and data analyzed with non-parametric Mann-Whitney U test where significance was defined as *p < 0.05, **p < 0.01, ***p < 0.001 and,**** p < 0.0001.
Supplementary Material
One Sentence Summary.
Collagen-I and CCL8-CCR8 axis control the motility of ILC2 cells in the inflamed lung
Table S3. Antibodies used for sorting human ILC2.
| Antibody | Clone | Fluorochrome | Target species | Manufacturer |
|---|---|---|---|---|
| CD11c | 3.9 | FITC | Human | Biolegend |
| CD123 | 6H6 | FITC | Human | Biolegend |
| CD14 | 63D3 | FITC | Human | Biolegend |
| CD16 | B73.1 | FITC | Human | Biolegend |
| CD161 | HP-3G10 | PECy7 | Human | Biolegend |
| CD19 | HIB19 | FITC | Human | Biolegend |
| CD1a | HI149 | FITC | Human | Biolegend |
| CD20 | 2H7 | FITC | Human | Biolegend |
| CD3 | OKT3 | FITC | Human | Biolegend |
| CD34 | 581 | FITC | Human | Biolegend |
| CD4 | OKT4 | FITC | Human | Biolegend |
| CD5 | L17F12 | FITC | Human | Biolegend |
| CD127 | A019D5 | PerCP5.5 | Human | Biolegend |
| CRTH2 | BM16 | BV421 | Human | Biolegend |
| CD45 | H130 | BV605 | Human | Biolegend |
Acknowledgements
The authors thank Stephen Rothery, David Gaboriau and Andreas Bruckbauer from the FILM facility (in part funded by Wellcome Trust grant 104931/Z/14/Z), members of the Beatson Advanced Imaging Resource (BAIR) team for excellent technical and imaging assistance. We would additionally like to thank Jane Srivastava and Jessica Rowley, of the Imperial College Core Flow Cytometry facility for assistance with flow cytometry and Lorraine Lawrence for histological sectioning. We would additionally like to thank, Dhiren Patel, Helen Stoelting, Nuo En Chan and Megan Mc Fie for their expert technical assistance. We would additionally like to acknowledge Lucy Robson for thorough maintenance of the human ILC2 lines. Finally, we would like to acknowledge MotilityLab for providing the resource for carrying out our analysis on ILC2 dynamics.
Funding
L.M.C. thanks the MRC (MR/M01245X/1) and Cancer Research UK for funding (Institute group funding ref 23983). This study was funded by the Wellcome Trust grant 107059/Z/15/Z, awarded to C.M.L. who is a Wellcome Senior Fellow in Basic Biomedical Sciences.
Footnotes
Author Contributions
C.M.L. conceived the idea and directed the study. L.M.C. provided expertise in imaging techniques. J.E.P. provided expert advice on chemotaxis assays. M. B. H. and M.F.K provided expert advice on intravital imaging. F.P., L.D., L.G.G., L.M.C. and C.M.L. wrote the manuscript. F.P. L.D. designed the experiments and F.P., L.D., L.J.E., L.M.C., J.V., R.O. performed experiments. F.P., E.J.M., L.M.C and L.D. analysed the data. F.P., E.J.M., L.M.C. and L.D. generated figures. R.O. maintained and genotyped the mouse lines.
Competing Interests
The authors declare that they have no competing interest.
References
- 1.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, Di Santo JP. Gata3 drives development of RORgammat+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim CH, Hashimoto-Hill S, Kim M. Migration and Tissue Tropism of Innate Lymphoid Cells. Trends Immunol. 2016;37:68–79. doi: 10.1016/j.it.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bando JK, Liang HE, Locksley RM. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol. 2015;16:153–160. doi: 10.1038/ni.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- 6.Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. 2016;17:765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- 7.McKenzie ANJ, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 9.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones R, Cosway EJ, Willis C, White AJ, Jenkinson WE, Fehling HJ, Anderson G, Withers DR. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur J Immunol. 2018;48:1481–1491. doi: 10.1002/eji.201847511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 12.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, Kubota M, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang HE, Locksley RM. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, Usher N, Zhu J, Urban JF, Jr, Paul WE, Germain RN. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. 2018;359:114–119. doi: 10.1126/science.aam5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HK, Lund S, Baum R, Rosenthal P, Khorram N, Doherty TA. Innate type 2 response to Alternaria extract enhances ryegrass-induced lung inflammation. Int Arch Allergy Immunol. 2014;163:92–105. doi: 10.1159/000356341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, Fukunaga K, Asano K, Betsuyaku T, Koyasu S. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol. 2016;17:76–86. doi: 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- 19.Lei AH, Xiao Q, Liu GY, Shi K, Yang Q, Li X, Liu YF, Wang HK, Cai WP, Guan YJ, Gabrilovich DI, et al. ICAM-1 controls development and function of ILC2. J Exp Med. 2018;215:2157–2174. doi: 10.1084/jem.20172359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doherty TA, Scott D, Walford HH, Khorram N, Lund S, Baum R, Chang J, Rosenthal P, Beppu A, Miller M, Broide DH. Allergen challenge in allergic rhinitis rapidly induces increased peripheral blood type 2 innate lymphoid cells that express CD84. J Allergy Clin Immunol. 2014;133:1203–1205. doi: 10.1016/j.jaci.2013.12.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barlow JL, Bellosi A, Hardman CS, Drynan LF, Wong SH, Cruickshank JP, McKenzie AN. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J Allergy Clin Immunol. 2012;129:191–198 e191-194. doi: 10.1016/j.jaci.2011.09.041. [DOI] [PubMed] [Google Scholar]
- 22.Bartemes KR, Iijima K, Kobayashi T, Kephart GM, McKenzie AN, Kita H. IL-33-responsive lineage- CD25+ CD44(hi) lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J Immunol. 2012;188:1503–1513. doi: 10.4049/jimmunol.1102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol. 2014;134:671–678 e674. doi: 10.1016/j.jaci.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SG, Chen R, Kjarsgaard M, Huang C, Oliveria JP, O'Byrne PM, Gauvreau GM, Boulet LP, Lemiere C, Martin J, Nair P et al. Increased numbers of activated group 2 innate lymphoid cells in the airways of patients with severe asthma and persistent airway eosinophilia. J Allergy Clin Immunol. 2016;137:75–86 e78. doi: 10.1016/j.jaci.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 25.Ebbo M, Crinier A, Vely F, Vivier E. Innate lymphoid cells: major players in inflammatory diseases. Nat Rev Immunol. 2017;17:665–678. doi: 10.1038/nri.2017.86. [DOI] [PubMed] [Google Scholar]
- 26.Licona-Limon P, Kim LK, Palm NW, Flavell RA. TH2, allergy and group 2 innate lymphoid cells. Nat Immunol. 2013;14:536–542. doi: 10.1038/ni.2617. [DOI] [PubMed] [Google Scholar]
- 27.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. Innate production of T(H)2 cytokines by adipose tissue-associated c- Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 28.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, Jolin HE, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci U S A. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, Broide DH. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol. 2013;132:205–213. doi: 10.1016/j.jaci.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wojno ED, Monticelli LA, Tran SV, Alenghat T, Osborne LC, Thome JJ, Willis C, Budelsky A, Farber DL, Artis D. The prostaglandin D(2) receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 2015;8:1313–1323. doi: 10.1038/mi.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou W, Toki S, Zhang J, Goleniewksa K, Newcomb DC, Cephus JY, Dulek DE, Bloodworth MH, Stier MT, Polosuhkin V, Gangula RD, et al. Prostaglandin I2 Signaling and Inhibition of Group 2 Innate Lymphoid Cell Responses. Am J Respir Crit Care Med. 2016;193:31–42. doi: 10.1164/rccm.201410-1793OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maric J, Ravindran A, Mazzurana L, Bjorklund AK, Van Acker A, Rao A, Friberg D, Dahlen SE, Heinemann A, Konya V, Mjosberg J. Prostaglandin E2 suppresses human group 2 innate lymphoid cell function. J Allergy Clin Immunol. 2018;141:1761–1773 e1766. doi: 10.1016/j.jaci.2017.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denney L, Byrne AJ, Shea TJ, Buckley JS, Pease JE, Herledan GM, Walker SA, Gregory LG, Lloyd CM. Pulmonary Epithelial Cell-Derived Cytokine TGF-beta1 Is a Critical Cofactor for Enhanced Innate Lymphoid Cell Function. Immunity. 2015;43:945–958. doi: 10.1016/j.immuni.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lammermann T, Germain RN. The multiple faces of leukocyte interstitial migration. Semin Immunopathol. 2014;36:227–251. doi: 10.1007/s00281-014-0418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MH, Taparowsky EJ, Kim CH. Retinoic Acid Differentially Regulates the Migration of Innate Lymphoid Cell Subsets to the Gut. Immunity. 2015;43:107–119. doi: 10.1016/j.immuni.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, Huang LC, Johnson D, Scanlon ST, McKenzie AN, Fallon PG, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roediger B, Kyle R, Yip KH, Sumaria N, Guy TV, Kim BS, Mitchell AJ, Tay SS, Jain R, Forbes-Blom E, Chen X et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013;14:564–573. doi: 10.1038/ni.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karta MR, Rosenthal PS, Beppu A, Vuong CY, Miller M, Das S, Kurten RC, Doherty TA, Broide H. beta2 integrins rather than beta1 integrins mediate Alternaria-induced group 2 innate lymphoid cell trafficking to the lung. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, Locksley RM. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol. 2018 doi: 10.1038/s41590-018-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krummel MF, Bartumeus F, Gerard A. T cell migration, search strategies and mechanisms. Nat Rev Immunol. 2016;16:193–201. doi: 10.1038/nri.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo Y, Yoshimoto T, Yasuda K, Futatsugi-Yumikura S, Morimoto M, Hayashi N, Hoshino T, Fujimoto J, Nakanishi K. Administration of IL-33 induces airway hyperresponsiveness and goblet cell hyperplasia in the lungs in the absence of adaptive immune system. Int Immunol. 2008;20:791–800. doi: 10.1093/intimm/dxn037. [DOI] [PubMed] [Google Scholar]
- 43.Boitano S, Flynn AN, Sherwood CL, Schulz SM, Hoffman J, Gruzinova I, Daines MO. Alternaria alternata serine proteases induce lung inflammation and airway epithelial cell activation via PAR2. Am J Physiol Lung Cell Mol Physiol. 2011;300:L605–614. doi: 10.1152/ajplung.00359.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Headley MB, Bins A, Nip A, Roberts EW, Looney MR, Gerard A, Krummel MF. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016;531:513–517. doi: 10.1038/nature16985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaller MA, Kallal LE, Lukacs NW. A key role for CC chemokine receptor 1 in T-cell-mediated respiratory inflammation. Am J Pathol. 2008;172:386–394. doi: 10.2353/ajpath.2008.070537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Islam SA, Chang DS, Colvin RA, Byrne MH, McCully ML, Moser B, Lira SA, Charo IF, Luster D. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat Immunol. 2011;12:167–177. doi: 10.1038/ni.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W, Di Santo JP. Taming the beast within: regulation of innate lymphoid cell homeostasis and function. J Immunol. 2013;191:4489–4496. doi: 10.4049/jimmunol.1301759. [DOI] [PubMed] [Google Scholar]
- 48.Xue L, Salimi M, Panse I, Mjosberg JM, McKenzie AN, Spits H, Klenerman P, Ogg G. Prostaglandin D2 activates group 2 innate lymphoid cells through chemoattractant receptor-homologous molecule expressed on TH2 cells. J Allergy Clin Immunol. 2014;133:1184–1194. doi: 10.1016/j.jaci.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregory LG, Mathie SA, Walker SA, Pegorier S, Jones CP, Lloyd CM. Overexpression of Smad2 drives house dust mite-mediated airway remodeling and airway hyperresponsiveness via activin and IL-25. Am J Respir Crit Care Med. 2010;182:143–154. doi: 10.1164/rccm.200905-0725OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laurent GJ. Lung collagen: more than scaffolding. Thorax. 1986;41:418–428. doi: 10.1136/thx.41.6.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andersson-Sjoland A, Hallgren O, Rolandsson S, Weitoft M, Tykesson E, Larsson-Callerfelt AK, Rydell-Tormanen K, Bjermer L, Malmstrom A, Karlsson JC, Westergren-Thorsson G. Versican in inflammation and tissue remodeling: the impact on lung disorders. Glycobiology. 2015;25:243–251. doi: 10.1093/glycob/cwu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cicchi R, Kapsokalyvas D, De Giorgi V, Maio V, Van Wiechen A, Massi D, Lotti T, Pavone FS. Scoring of collagen organization in healthy and diseased human dermis by multiphoton microscopy. J Biophotonics. 2010;3:34–43. doi: 10.1002/jbio.200910062. [DOI] [PubMed] [Google Scholar]
- 53.Vennin C, Chin VT, Warren SC, Lucas MC, Herrmann D, Magenau A, Melenec P, Walters SN, Del Monte-Nieto G, Conway JR, Nobis M, et al. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leithner A, Eichner A, Muller J, Reversat A, Brown M, Schwarz J, Merrin J, de Gorter DJ, Schur F, Bayerl J, de Vries I, et al. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat Cell Biol. 2016;18:1253–1259. doi: 10.1038/ncb3426. [DOI] [PubMed] [Google Scholar]
- 55.Friedl P, Borgmann S, Brocker EB. Amoeboid leukocyte crawling through extracellular matrix: lessons from the Dictyostelium paradigm of cell movement. J Leukoc Biol. 2001;70:491–509. [PubMed] [Google Scholar]
- 56.Lammermann T, Sixt M. Mechanical modes of 'amoeboid' cell migration. Curr Opin Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Kenyon NJ, Ward RW, McGrew G, Last JA. TGF-beta1 causes airway fibrosis and increased collagen I and III mRNA in mice. Thorax. 2003;58:772–777. doi: 10.1136/thorax.58.9.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Honda T, Egen JG, Lammermann T, Kastenmuller W, Torabi-Parizi P, Germain RN. Tuning of antigen sensitivity by T cell receptor-dependent negative feedback controls T cell effector function in inflamed tissues. Immunity. 2014;40:235–247. doi: 10.1016/j.immuni.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stier MT, Zhang J, Goleniewska K, Cephus JY, Rusznak M, Wu L, Van Kaer L, Zhou B, Newcomb DC, Peebles RS., Jr IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J Exp Med. 2017 doi: 10.1084/jem.20170449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sumagin R, Prizant H, Lomakina E, Waugh RE, Sarelius IH. LFA-1 and Mac-1 define characteristically different intralumenal crawling and emigration patterns for monocytes and neutrophils in situ. J Immunol. 2010;185:7057–7066. doi: 10.4049/jimmunol.1001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Moltke J, Pepper M. Sentinels of the Type 2 Immune Response. Trends Immunol. 2017 doi: 10.1016/j.it.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Islam SA, Luster AD. T cell homing to epithelial barriers in allergic disease. Nat Med. 2012;18:705–715. doi: 10.1038/nm.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moreau JF, Pradeu T, Grignolio A, Nardini C, Castiglione F, Tieri P, Capri M, Salvioli S, Taupin JL, Garagnani P, Franceschi C. The emerging role of ECM crosslinking in T cell mobility as a hallmark of immunosenescence in humans. Ageing Res Rev. 2017;35:322–335. doi: 10.1016/j.arr.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Burgstaller G, Vierkotten S, Lindner M, Konigshoff M, Eickelberg O. Multidimensional immunolabeling and 4D time-lapse imaging of vital ex vivo lung tissue. Am J Physiol Lung Cell Mol Physiol. 2015;309:L323–332. doi: 10.1152/ajplung.00061.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods. 2011;8:91–96. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller BW, Morton JP, Pinese M, Saturno G, Jamieson NB, McGhee E, Timpson P, Leach J, McGarry L, Shanks E, Bailey P, et al. Targeting the LOX/hypoxia axis reverses many of the features that make pancreatic cancer deadly: inhibition of LOX abrogates metastasis and enhances drug efficacy. EMBO Mol Med. 2015;7:1063–1076. doi: 10.15252/emmm.201404827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rath N, Morton JP, Julian L, Helbig L, Kadir S, McGhee EJ, Anderson KI, Kalna G, Mullin M, Pinho AV, Rooman I, et al. ROCK signaling promotes collagen remodeling to facilitate invasive pancreatic ductal adenocarcinoma tumor cell growth. EMBO Mol Med. 2017;9:198–218. doi: 10.15252/emmm.201606743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Villarreal DO, L'Huillier A, Armington S, Mottershead C, Filippova EV, Coder BD, Petit RG, Princiotta MF. Targeting CCR8 Induces Protective Antitumor Immunity and Enhances Vaccine-Induced Responses in Colon Cancer. Cancer Res. 2018;78:5340–5348. doi: 10.1158/0008-5472.CAN-18-1119. [DOI] [PubMed] [Google Scholar]
- 70.Cheng T, Liu Q, Zhang R, Zhang Y, Chen J, Yu R, Ge G. Lysyl oxidase promotes bleomycin-induced lung fibrosis through modulating inflammation. J Mol Cell Biol. 2014;6:506–515. doi: 10.1093/jmcb/mju039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.