Significance
Human cytomegalovirus (HCMV) is a major cause of birth defects and diseases in immune-compromised patients. HCMV is able to infect and establish latency in large populations by employing multiple strategies to evading the host immune response. Here, we report that HCMV suppresses cytokine-mediated antiviral responses by increasing the expression of cellular RNA-binding protein, Roquin. We show that Roquin inhibits cytokine production by directly binding to cytokine mRNAs and by repressing the expression of their transcription activator. This study highlights that cellular RNA metabolism, which is controlled by HCMV for immune evasion, can be the target for developing anti-HCMV therapeutics.
Keywords: human cytomegalovirus, RNA-binding protein, immune evasion, proinflammatory cytokine
Abstract
RNA represents a pivotal component of host–pathogen interactions. Human cytomegalovirus (HCMV) infection causes extensive alteration in host RNA metabolism, but the functional relationship between the virus and cellular RNA processing remains largely unknown. Through loss-of-function screening, we show that HCMV requires multiple RNA-processing machineries for efficient viral lytic production. In particular, the cellular RNA-binding protein Roquin, whose expression is actively stimulated by HCMV, plays an essential role in inhibiting the innate immune response. Transcriptome profiling revealed Roquin-dependent global down-regulation of proinflammatory cytokines and antiviral genes in HCMV-infected cells. Furthermore, using cross-linking immunoprecipitation (CLIP)-sequencing (seq), we identified IFN regulatory factor 1 (IRF1), a master transcriptional activator of immune responses, as a Roquin target gene. Roquin reduces IRF1 expression by directly binding to its mRNA, thereby enabling suppression of a variety of antiviral genes. This study demonstrates how HCMV exploits host RNA-binding protein to prevent a cellular antiviral response and offers mechanistic insight into the potential development of CMV therapeutics.
Human cytomegalovirus (HCMV) is a member of the β-herpesvirus family and a ubiquitous pathogen that establishes lifelong latency in large populations worldwide (1). During primary infection of HCMV, which rarely exhibits pathogenic symptoms in healthy humans, the virus overcomes innate and adaptive immune responses in the host and establishes permanent latency. Reactivation from latency can be dangerous in immune-compromised hosts, with HCMV congenital infection being a leading cause of neurological defects in infants (2–4). Currently, there is no therapeutic intervention capable of curing chronic HCMV infection, and no vaccines are available to prevent HCMV infection. This is largely because of the multiple strategies employed by HCMV to evade innate and adaptive immunity. Thus, having a thorough understanding of how the virus evades the host immune system is necessary for the development of anti-HCMV vaccines and therapeutics.
Primary HCMV infection provokes a robust innate immune response in terms of rapid induction of nuclear factor kappaB (NF-κB)- and IFN regulatory factor (IRF)3-signaling pathways in cells (5–7). Proinflammatory cytokines, type I interferons, and IFN-stimulated genes (ISGs) are produced and confer antiviral effects by targeting various steps of HCMV replication (8, 9). However, HCMV has evolved strategies of regulating cellular cytokines in order to counteract the host immune system. First, HCMV encodes cellular cytokine mimics and decoy receptors in order to modulate cytokine activity and restrict cell migration and differentiation (10–15). Second, HCMV inhibits production of cytokines by blocking the activation of transcription factors (16–19). Third, viral microRNAs (miRNAs) directly target the mRNAs of cellular chemokines and cytokines and posttranscriptionally repress their expression. We previously reported that the HCMV-encoded miRNA miR-UL148D down-regulates the cellular chemokine RANTES (20). Comprehensive studies of viral miRNAs revealed that they inhibit cytokine signaling, trafficking, and release, which strongly suppresses the host immune response (21, 22).
During HCMV lytic infection, host cells are influenced by certain unique features associated with RNA processing. In contrast to other viruses that induce shutdown of host mRNA translation, HCMV stimulates this activity by blocking protein kinase R activation (23) and modulates cellular RNA-processing machinery to accelerate viral gene translation (24–26). Additionally, viral mRNAs undergo extensive posttranscriptional modifications, such as alternative splicing and polyadenylation (27, 28), with elevated expressions of proteins regulating such modifications during HCMV infection (29, 30). Although these reports reveal that HCMV might require cellular RNA-processing machinery for efficient replication at multiple steps, how these RNA-binding proteins regulate host mRNAs and cellular functions remains largely unexplored.
In this study, we examined the role of cellular RNA-processing proteins in immune regulation during HCMV lytic production by employing loss-of-function screening. We found that 19 genes are required for HCMV lytic production, among which Roquin was critical for viral gene expression. HCMV-induced Roquin played a dominant role in the suppression of cytokine production. During viral infection, Roquin directly bound to and regulated expression of cellular RNA transcripts including those of cytokine genes and IRF1, which was crucial for increased cytokine suppression and enhanced viral replication. These results demonstrated areas of HCMV evolution to exploit host RNA-binding proteins in order to modulate the cellular environment in its favor.
Results
HCMV Requires Multiple Cellular RNA-Processing Activities for Its Efficient Lytic Production.
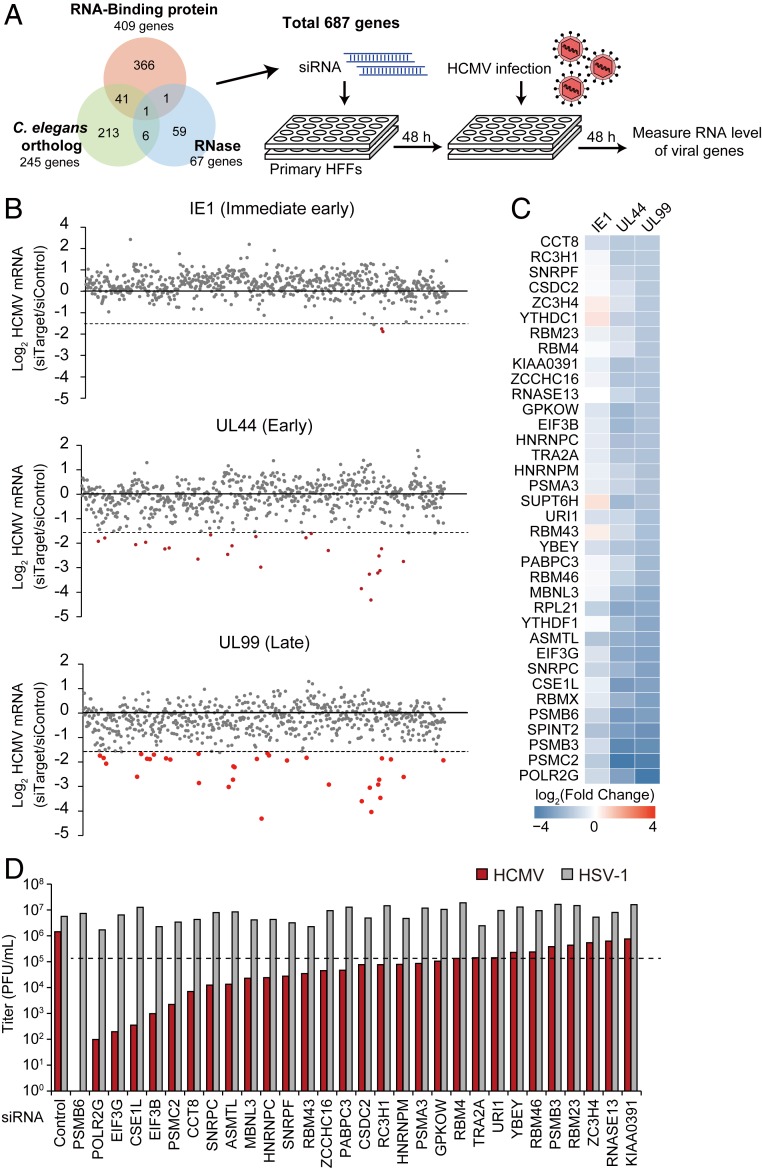
HCMV harbors multiple viral genes that modulate cellular mRNA processing, with some host cell genes capable of positively or negatively regulating viral infection. To identify genes essential for HCMV lytic production, an RNA interference (RNAi)-screening experiment was performed using 687 genes involved in cellular RNA processing. The small-interfering RNA (siRNA) library contained three gene categories: RNA-binding proteins, ribonucleases, and orthologs of Caenorhabditis elegans genes (Fig. 1A and Dataset S1). The gene information on RNA-binding proteins in humans was obtained from the RNA-binding Protein Database (31). The ribonuclease category included previously reported and predicted ribonucleases based on National Center for Biotechnology Information gene information. The orthologous genes from C. elegans were collected from candidate genes in the small RNA pathway (32) in order to include cellular genes containing neither an RNA-binding nor an RNase domain. We employed transient loss-of-function screening using siRNA to minimize cellular damage and artifacts potentially occurring as a result of genomic disruption. Additionally, we used primary human foreskin fibroblasts (HFFs) and HCMV low-passage strain Toledo to mimic the physiological environment of HCMV infection for screening.
Fig. 1.
Identification of RNA-binding proteins that regulate HCMV lytic infection through siRNA screening. (A) Experimental scheme of siRNA screening. RNases, RNA-binding proteins, and orthologous genes in the small-RNA pathway from C. elegans were collected for screening. HFF cells underwent two rounds of siRNA transfection, with infection with HCMV (MOI = 2). Viral gene expression was measured by qRT-PCR at 48 hpi and normalized against GAPDH mRNA levels. (B) Summarized results of RNAi screening. IE1, UL44, and UL99 mRNA levels were quantified. Genes showing a >threefold decrease are highlighted in red. (C) Genes showing a >threefold decrease in UL99 mRNA level are selected. Heatmap of fold changes [log2(siTarget/siCon)] of viral mRNA levels in cells depleted of each gene. (D) HCMV and HSV-1 viral replication as measured by plaque-forming assay. HFFs were infected with HCMV or HSV-1 (MOI = 0.1), and the cell-free supernatants were titrated at 7 dpi for HCMV and 2 dpi for HSV-1.
For RNAi screening, primary HFFs were transfected with mixed siRNAs comprising four kinds of specific siRNAs targeting the open reading frame (ORF) regions at a final concentration of 20 nM. At 2 d after transfection, the HFF cells were infected with virus at a multiplicity of infection (MOI) of 2, followed by treatment with the siRNA mixture again in order to maximize knockdown efficiency. The level of viral lytic production was quantified by detecting the mRNA levels of immediate early (IE1), early (UL44), and late (UL99) genes at 48 h postinfection (hpi) because most viral genes involved in lytic production are actively expressed at this time point. Therefore, it was expected that a functional gene influencing any phase of the HCMV life cycle would be identified through RNAi screening according to phase specificity. Screening returned 36 genes exhibiting a >threefold decrease in mRNA levels for viral late gene expression (Fig. 1B), with a majority of these genes also required for efficient viral early gene expression (Fig. 1C). To evaluate the effect on authentic viral production, secreted viruses were examined at a low-titer infection (0.1 MOI). We found that 19 genes exhibited a >10-fold decrease in viral production (Fig. 1D), with most of these genes not previously reported as having functions associated with viral production. Strikingly, replication of herpes simplex virus (HSV)-1, which belongs to the α-herpesvirus family, did not require these genes (Fig. 1D), suggesting an HCMV-specific role of cellular RNA-processing machinery. To identify the effect of the candidate genes on the HCMV lytic life cycle, viral protein expression was examined during a time-course infection accompanied by the transient knockdown of three candidate genes, MBNL3, ASMTL, and CSE1L (SI Appendix, Fig. S1), showing that viral gene expression was clearly repressed at the late stage of the viral life cycle. Therefore, HCMV requires multiple cellular genes involved in RNA processing, and the 19 newly identified cellular genes are involved in HCMV lytic production.
Roquin Is Essential for HCMV Lytic Production.
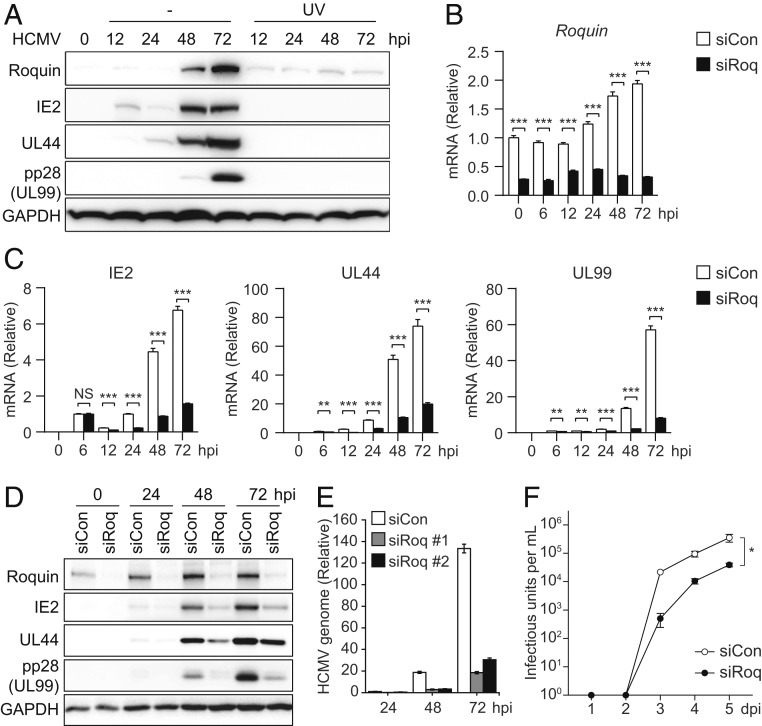
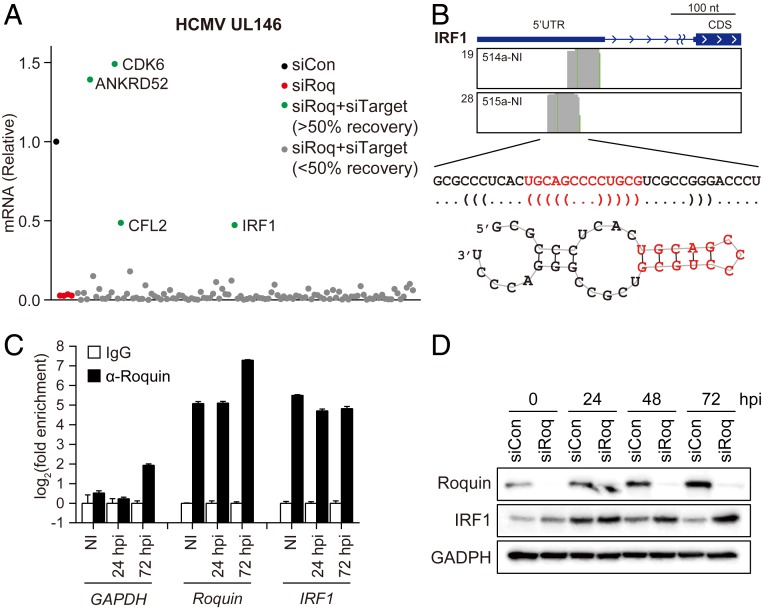
Among the 19 candidate genes, we investigated the RNA-binding protein Roquin (RC3H1) further. Human RC3H1 encodes the Roquin protein that contains a ROQ domain for RNA binding and serves an immunoregulatory role by targeting mRNA transcripts of several proinflammatory cytokines and signaling genes (33–36). Roquin-mediated cytokine control is crucial for T cell regulation (35), but its physiological roles regarding host–pathogen interactions have not been explored yet. We focused on Roquin based on its expression being significantly induced during HCMV infection of HFFs (Fig. 2A), as well as the absence of this induction during UV-irradiated HCMV infection, where immediate early (IE2), early (UL44), and late (UL99; pp28) viral gene expression was completely blocked. The involvement of viral genes in Roquin induction was further demonstrated by the absence of the increased Roquin expression in cells depleted of IE1, the major viral immediate early protein, which is necessary for the expression of most HCMV genes (SI Appendix, Fig. S2A). Treatment of ganciclovir, the viral DNA polymerase inhibitor repressing HCMV late gene expression, did not alter the increased expression of Roquin protein, implying that Roquin induction is mediated by HCMV immediate early or early genes (SI Appendix, Fig. S2B). We also observed that activation of cellular innate immune signaling by transfection of poly(dA:dT) or poly(I:C) had no impact on Roquin expression (SI Appendix, Fig. S2C). These results suggested that Roquin expression is actively induced by viral genes expressed during HCMV infection.
Fig. 2.
Pivotal role of Roquin during HCMV lytic production. (A) HFF cells were infected with HCMV Toledo (MOI = 2) and harvested at the indicated time points for immunoblot. HCMV was irradiated with UV-C (300 mJ/cm2) before infection. (B–F) HFF cells were treated with siCon or siRoq, infected with HCMV (MOI = 2), and harvested at the indicated time points. (B and C) mRNA levels of Roquin (B) and viral genes IE2, UL44, and UL99 (C) were measured by qRT-PCR. (D) Protein levels of various viral proteins and Roquin were measured by immunoblot. (E) Viral genomic DNA levels were measured in cells transfected with two different siRNAs targeting Roquin. (F) Cell-free virus in the supernatant was titrated by limiting dilution analysis. Data represent mean ± SEM, n = 3; *P < 0.05 according to two-way ANOVA with Dunnett’s multiple comparisons test. (B, C, and E) Data represent mean ± SEM, n = 3; **P < 0.01; ***P < 0.001 according to two-tailed Student’s t test; NS, not significant.
The impact of Roquin on viral replication was further examined during a detailed time-course infection of HFFs treated with control (siCon) or Roquin-targeting siRNA (siRoq) to induce transient knockdown of Roquin, after which the mRNA and protein levels of the viral genes were analyzed by qRT-PCR and immunoblot (Fig. 2 B–D). While Roquin expression was indeed repressed by siRoq (Fig. 2 B and D), mRNA and protein levels of viral IE2, UL44, and UL99 were also significantly down-regulated by Roquin silencing, with this down-regulation peaking at 72 hpi (Fig. 2 C and D). Reduction of several late transcripts also demonstrated that Roquin is necessary for efficient expression of viral genes (SI Appendix, Fig. S2D). We also examined the role of Regnase-1, which has redundant function with Roquin (37). Protein levels of Regnase-1 were also increased after 48 hpi, and Regnase-1 silencing resulted in reduction of viral gene expression (SI Appendix, Fig. S2E). However, the impact of Regnase-1 on viral genes was lower than that of Roquin, highlighting the pivotal role of Roquin during HCMV infection.
The expression of viral late gene UL83 (pp65) was measured in cells infected with two attenuated strains of HCMV (AD169 and Towne), revealing repressed UL83 expression in Roquin-knockdown cells and indicating that Roquin function during infection is conserved in multiple HCMV strains (SI Appendix, Fig. S2F). Additionally, we found that viral genomic DNA levels that had increased for up to 72 hpi were down-regulated by Roquin knockdown (Fig. 2E). Viral titers measured in cell culture supernatant also showed 10-fold decrease by Roquin silencing (Fig. 2F). These results suggested Roquin as a cellular factor with a critical role in HCMV lytic production, and that HCMV actively utilizes Roquin for efficient viral production.
Roquin Silencing Results in Increased Production of Proinflammatory Cytokines during HCMV Infection.
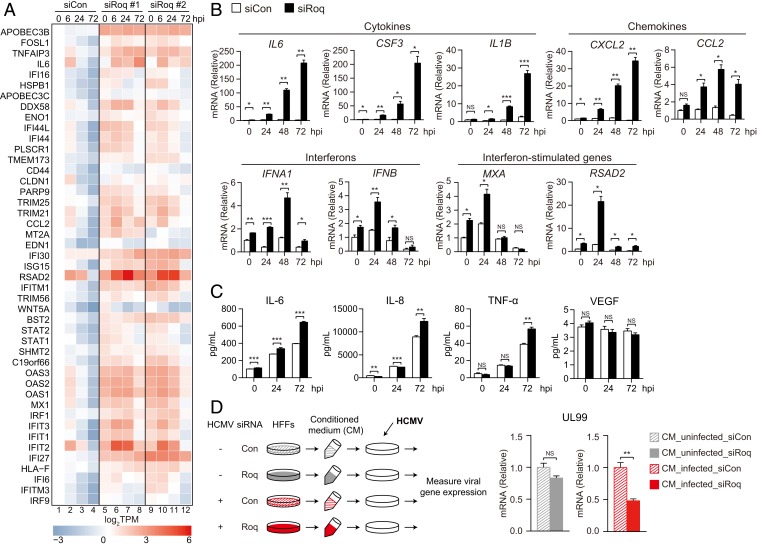
To investigate how cellular genes are regulated by Roquin during HCMV lytic replication, we performed RNA-sequencing (seq) in HFFs treated with control (siCon) or Roquin-targeting siRNA (siRoq) (38). The levels of RNA from cellular genes were measured in uninfected cells and HCMV-infected cells harvested at 6, 24, and 72 hpi, followed by comparison according to siCon or siRoq transfection and gene set enrichment analysis (GSEA) to examine the cellular pathways modulated by Roquin. The results showed that pathways related to the immune response and cytokine signaling were significantly enriched for genes up-regulated by Roquin knockdown (SI Appendix, Fig. S3 A and B and Dataset S2). We then evaluated the time-course expression patterns of genes from these pathways (Fig. 3A). In control cells, HCMV infection induced global decreases in the expression of these immune-related genes (Fig. 3A, lanes 1–4). Strikingly, upon Roquin silencing, this inhibition was completely blocked, and the expression of these genes was maintained at a high level (Fig. 3A, lanes 5–8 and 9–12). Moreover, the majority of these genes exhibited a higher fold induction by Roquin knockdown as viral replication proceeded, highlighting the role of Roquin throughout the infection process (SI Appendix, Fig. S3C). These results indicated that HCMV actively suppresses cellular immune responses during its lytic infection cycle by employing Roquin function.
Fig. 3.
Roquin knockdown results in increased expression of proinflammatory cytokines. (A) Heatmap of (log2TPM) immune-related gene expression at the indicated time points in siCon or siRoq cells. (B) HFF cells were treated with siCon or siRoq, infected with HCMV (MOI = 2), and harvested at the indicated time points. mRNA levels of immune-related genes were measured by qRT-PCR. (C) Cytokine levels from cell-culture supernatant were measured by multiplex cytokine bead assay. (D) After siRNA transfection and HCMV infection (MOI = 2), cell culture supernatant (conditioned media; CM) was harvested at 72 hpi and UV-irradiated to inactivate viral particles. Naïve HFFs were incubated with CM, infected with HCMV (MOI = 0.5), and harvested at 72 hpi. UL99 mRNA level was measured by qRT-PCR. (B–D) Data represent mean ± SEM, n = 3; *P < 0.05; **P < 0.01; ***P < 0.001 according to two-tailed Student’s t test; NS, not significant.
We confirmed the effect of Roquin on the expression of individual proinflammatory cytokine genes during HCMV infection. mRNA levels were measured from Roquin-silenced cells for genes encoding different cytokines (IL6, CSF3, and IL1B), chemokines (CXCL2 and CCL2), interferons (IFNB and IFNA1), and ISGs (MXA and RSAD2). mRNA expressions of these genes showed a marked increase by Roquin silencing in uninfected cells. However, this increase was further potentiated by HCMV infection, with cytokine levels showing >200-fold induction at 72 hpi (Fig. 3B). Notably, although the expression of cytokines and chemokines showed the highest increases at 72 hpi, IFN and ISG genes showed robust induction at 24 hpi. Thus, Roquin controls the expression of various antiviral genes at steady state to regulate immune homeostasis, and its function is maintained throughout the early and late phases of infection.
We then measured cytokine production in the supernatant of infected cells using a multiplex cytokine bead assay. Secretion of IL-6, IL-8, and tumor necrosis factor-α, which was increased upon HCMV infection, was elevated further in Roquin-knockdown cells and peaked at 72 hpi (Fig. 3C). Production of another proinflammatory cytokine, VEGF, showed no significant difference by Roquin knockdown, indicating that Roquin function is specific to a particular subset of immune-related genes.
To examine the impact of secreted cytokines on HCMV replication, we transferred the conditioned media harvested from Roquin-silenced HFFs to naïve cells and measured viral gene expression in the recipient cells (Fig. 3D). The media from Roquin-silenced but uninfected donor cells could not affect viral replication in the recipient cells (Fig. 3D, gray bars). Conversely, viral gene expression was significantly reduced when incubated with media from HCMV-infected, Roquin-silenced HFFs (Fig. 3D, red bars). Similar results were observed when recipient cells were infected at higher MOI (SI Appendix, Fig. S3D). Thus, although Roquin continually represses cytokine expression in uninfected cells, Roquin’s impact is further magnified during viral infection where antiviral cytokine levels are robustly induced.
Genome-Wide Identification of Roquin Target mRNAs during HCMV Infection.
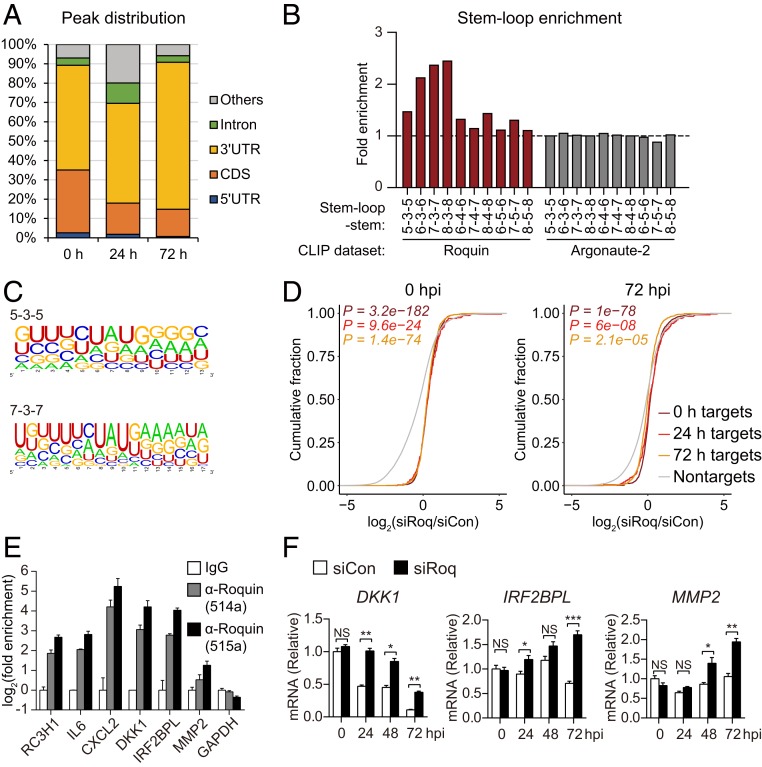
Roquin down-regulates gene expression by directly binding to the 3′ UTR of target mRNAs and controlling their stability (39). RNA-seq data showed that Roquin mediated the down-regulation of a number of genes during HCMV infection; therefore, we identified the RNAs directly bound by Roquin by cross-linking immunoprecipitation (CLIP)-seq analysis of uninfected or HCMV-infected HFFs at 24 and 72 hpi using two different antibodies (514a and 515a) (SI Appendix, Fig. S4A). Both antibodies successfully captured endogenous Roquin protein and Roquin-bound RNA (SI Appendix, Fig. S4B). CLIP-seq reads were mapped to the human or viral genome, which enabled identification of between ∼4,000 and 60,000 binding sites from each library (38). To define the Roquin target region, CLIP-seq clusters were built from each time point by overlapping (by one nucleotide) the libraries derived from the two different antibodies. We obtained 7,068, 462, and 1,907 clusters from 0, 24, and 72 hpi, respectively (SI Appendix, Fig. S4C and Dataset S3), with >50% of the clusters located in the 3′ UTR of cellular genes (Fig. 4A), consistent with previous reports (37–40). Although the majority of CLIP-seq clusters were mapped to the human genome, 23% and 9% of the clusters from 24 hpi and 72 hpi, respectively, were mapped to transcripts from the viral genome, indicating that Roquin can also bind to viral RNA molecules (SI Appendix, Fig. S4D).
Fig. 4.
Genome-wide identification of Roquin target genes. (A) Distribution of binding sites among RNA transcripts based on overlapped Roquin-binding clusters. (B) Enrichment of indicated stem-loop structures in the Roquin-binding clusters as compared with 100 dinucleotide shuffles of each sequence. (C) RNA sequences of Roquin-binding clusters comprising each indicated stem-loop structure. (D) Cumulative-distribution function plot of fold changes in CLIP target genes identified at each time point and following Roquin silencing. P values were calculated using a two-sided Mann–Whitney U test. (E) Roquin binding of the indicated mRNAs according to RNA IP. (F) RNA levels of CLIP target genes measured in Roquin-silenced cells by qRT-PCR. (E and F) Data represent mean ± SEM, n = 3; *P < 0.05; **P < 0 .01; ***P < 0 .001 according to two-tailed Student’s t test; NS, not significant.
We then analyzed the structural characteristics of the Roquin-binding sites. Secondary structure prediction using RNAfold (41) identified various stem-loop structures enriched in the Roquin CLIP-seq clusters from both noninfected or infected samples (Fig. 4B and SI Appendix, Fig. S4E). A total of 20%, 21%, and 22% of Roquin-binding clusters contained these stem-loop structures at 0, 24, and 72 hpi, respectively (SI Appendix, Fig. S4F). Stem loops with a three-nucleotide loop were the most common motif, and hairpins with longer stems were overrepresented. Given that no such enrichment of stem loops was observed from CLIP-seq data associated with the Argonaute-2 protein (22), enrichment of stem-loop structures likely represents a specific feature of Roquin-RNA binding. Sequences comprising each stem-loop were aligned using WebLogo (42) in order to identify sequence motifs (Fig. 4C). The results showed that a UAU motif was highly enriched at the loop position with a GC pair at the stem-initiation site. Although stem-loop structures involving a three-nucleotide loop and the GUAUC sequence represent a previously reported consensus binding motif of Roquin (37, 39, 43, 44), we also observed that Roquin is capable of binding stem loops of various lengths and sequences.
We next assessed the ability of Roquin to inhibit expression of target genes using RNA-seq data to measure fold changes in their expression between siCon- and siRoq-transfected cells. Compared with mRNAs not targeted by Roquin, mRNA levels of Roquin targets showed significant accumulation in Roquin-knockdown cells (Fig. 4D), indicating the degradative function of Roquin. Additionally, we verified that target genes identified from different CLIP-seq samples (noninfected, 24, or 72 hpi) showed elevated expression both in the presence and absence of viral infection (Fig. 4D). These results showed minimal changes in the Roquin targetome by HCMV infection, thereby suggesting that Roquin functions by down-regulating its target genes consistently throughout the infection cycle.
While CLIP-seq data confirmed previously reported Roquin targets such as IL6 and CXCL2, it also enabled identification of unrecognized Roquin-target genes involved in diverse cellular processes, including DKK1, IRF2BPL, and MMP2 (SI Appendix, Fig. S4 G and H). RNA IP verified Roquin binding to these target mRNAs (Fig. 4E), which were accumulated in Roquin-knockdown cells (Fig. 4F). Comparison of our Roquin targetome with previous studies (37, 40) revealed that 43% of the target genes identified in the present study overlapped with those reported previously (SI Appendix, Fig. S4I). Notably, both target genes that are unique to this study and that overlapped with previous reports were significantly up-regulated by Roquin silencing. These findings provided further evidence that Roquin selectively down-regulates target genes identified by CLIP-seq during viral replication.
IRF1 Is a Major Roquin Target.
To determine the specific Roquin target(s) involved in HCMV gene expression, we combined our Roquin CLIP-seq data with RNA-seq data from the Roquin-knockdown cells and performed another RNAi-screening experiment to evaluate the role(s) of Roquin targets (SI Appendix, Fig. S5A). We selected 109 genes associated with Roquin-binding peaks on their transcripts and increased expression upon Roquin silencing. Each gene was silenced along with Roquin in order to quantify changes in HCMV late gene (UL146) levels. Most of the candidate genes conferred no effect on the repressed viral gene expression induced by Roquin knockdown (Fig. 5A). However, respective silencing of four genes (ANKRD52, CDK6, CFL2, and IRF1) showed a >50% rescue of viral gene expression levels (Fig. 5A), implying that they are involved in reducing viral gene expression in Roquin-knockdown cells. Since IRF1 is a well-known transcriptional activator of various cytokines (45), we assumed that targeting IRF1 may be critical for Roquin-mediated control of cytokine and HCMV replication.
Fig. 5.
IRF1 is a primary target of Roquin. (A) Relative expression of viral late gene (UL146) measured in HFFs transfected with indicated siRNA, infected with HCMV (MOI = 2), and harvested at 72 hpi. Cells were transfected with siCon, siRoq, or cotransfected with siRoq and siRNAs targeting Roquin-binding targets (siTarget). (B) Roquin-specific CLIP reads mapped to IRF1 mRNA and its predicted secondary structure. (C) RNA IP results confirming Roquin binding to IRF1 mRNA (mean ± SEM, n = 3). (D) IRF1 protein levels measured by immunoblot from HFF cells treated with siCon or siRoq and infected with HCMV (MOI = 2).
CLIP-seq reads were mapped to the 5′ UTR region of IRF1 mRNA, and the binding sequences were predicted to contain a hairpin structure with a three-nucleotide loop, consistent with the consensus Roquin-binding site (Fig. 5B). This putative Roquin target site was unexpected, given that previous reports and our CLIP-seq data consistently showed that Roquin mainly binds to the 3′ UTR of target mRNA (39, 40). We observed that the levels of mRNAs harboring Roquin target sites in the 5′ UTR showed statistically significant up-regulation by Roquin knockdown (SI Appendix, Fig. S5B), implying that both 5′ and 3′ UTRs represent functional target sites of Roquin. Furthermore, we confirmed both that Roquin bound to IRF1 mRNA throughout the HCMV-infection process (Fig. 5C) as well the presence of up-regulated levels of IRF1 protein in Roquin-knockdown cells (Fig. 5D). IRF1 was increased both in the absence and presence of HCMV infection, indicating that Roquin constantly regulates IRF1 expression. Our findings demonstrated that IRF1 represents a target gene, the expression of which is suppressed by Roquin during HCMV infection.
We next examined the impact of Roquin on viral gene and IRF1 expression in THP-1–derived macrophages and glioblastoma U373MG cells. Similar to the results in HFFs, Roquin silencing reduced viral gene expression in both cell lines, where Roquin expression was increased by infection as well (SI Appendix, Fig. S5 C and D). However, Roquin silencing increased IRF1 level only in uninfected macrophages, implying that Roquin-mediated IRF1 regulation during HCMV infection might be specific to fibroblasts. These data indicated that while Roquin is necessary for lytic HCMV replication regardless of cell types, Roquin targetome might differ between cell types.
Roquin-Mediated Down-Regulation of IRF1 Is Critical to Mediating Cytokine Levels during HCMV Lytic Infection.
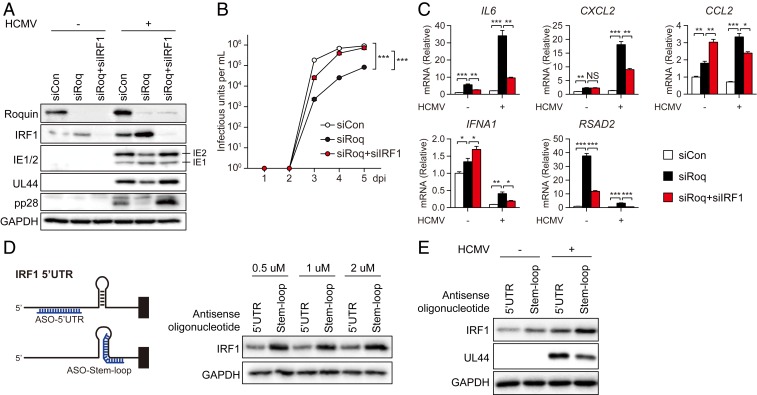
We then evaluated the role of Roquin-IRF1 axis in HCMV gene expression. HFF cells were transfected with either siRoq alone or siRoq and siRNA targeting IRF1 (siIRF1), followed by HCMV infection. We confirmed that IRF1 levels were elevated by Roquin knockdown and declined following siIRF1 transfection (Fig. 6A). Although levels of the viral genes IE2, UL44, and pp28 were all repressed by Roquin knockdown, they were rescued to control levels by IRF1 silencing. Viral production also showed significant increase when IRF1 was cosilenced with Roquin (Fig. 6B), indicating that IRF1 down-regulation by Roquin is critical for successful replication of HCMV.
Fig. 6.
IRF1 regulates cytokine production during HCMV replication. (A–C) HFF cells were treated with siCon, siRoq, or siRoq+siIRF1 and infected with HCMV (MOI = 2), followed by harvesting at 72 hpi. (A) Protein levels of Roquin, IRF1, and viral genes were measured by immunoblot. (B) Cell-free virus in the supernatant was titrated by limiting dilution analysis. Data represent mean ± SEM, n = 3; ***P < 0 .001 (siCon versus siRoq; siRoq versus siRoq+siIRF1) according to two-way ANOVA with Dunnett’s multiple comparisons test. (C) Cytokine mRNA levels were measured by qRT-PCR (mean ± SEM, n = 3; *P < 0.05; **P < 0 .01; ***P < 0 .001 according to two-tailed Student’s t test; NS, not significant). (D) HFF cells were transfected with ASOs (at the indicated concentrations) targeting the Roquin-binding site on IRF1 mRNA, and IRF1 levels were measured by immunoblot. (E) HFF cells were transfected with ASOs and infected with HCMV (MOI = 0.1). At 24 hpi, protein levels of IRF1 and viral gene (UL44) were measured by immunoblot.
IRF1 works as a transcription factor and regulates immune responses primarily through the induction of proinflammatory cytokines and promotion of antibacterial or antiviral immune responses (45). Additionally, genome-wide ChIP-seq analysis revealed putative IRF1-binding sites in the promoter regions of numerous genes associated with apoptosis and DNA-damage responses (46). To assess the role of Roquin and IRF1 on immune responses during HCMV infection, we measured cytokine production in cells with both genes silenced (Fig. 6C). We observed significant increases in the mRNA levels of various cytokines (IL6, CXCL2, and CCL2) and antiviral genes (IFNA1 and RSAD2) in Roquin-silenced cells, whereas these levels were attenuated to >50% by cosilencing of IRF1 and Roquin. This result suggested that cytokine regulation by Roquin during HCMV infection was, in part, dependent upon IRF1.
To further determine the importance of IRF1 targeting by Roquin, we blocked the interaction between Roquin and IRF1 mRNA specifically. We designed an antisense oligonucleotide (ASO) targeting the stem region in order to disrupt hairpin formation at Roquin-binding region, and a control ASO targeting a different region of the IRF1 5′ UTR in order to rule out the translation-inhibition effect of the 5′ UTR-targeting oligonucleotide (Fig. 6D). Transfection of cells with the stem loop-targeting ASO resulted in increased IRF1 protein levels (Fig. 6D), suggesting that Roquin recognizes and binds to the stem-loop region of the IRF1 5′ UTR to induce its down-regulation. Additionally, when ASO-transfected cells were infected with HCMV, the increased expression of IRF1 resulted in reduction of viral gene expression (Fig. 6E). Therefore, we conclude that Roquin represses IRF1 expression primarily to promote efficient replication of HCMV.
Discussion
In recent decades, the functional relationship between DNA viral infections and cellular RNA processing has been underestimated, and the associated mechanisms have remained largely unknown. Our report of an immune-regulatory function for the RNA-binding protein Roquin highlights the global requirement of cellular RNA processing machinery during HCMV infection. Loss-of-function screening using an siRNA library comprising 687 RNases or RNA-binding proteins identified 36 genes critical for viral gene expression and 19 genes indispensable for HCMV production. These genes showed no effect on HSV-1 replication, indicating their role is specifically required for HCMV. Because HCMV harbors a larger genome and a longer life cycle relative to HSV-1 (47), it is presumed to manipulate a more diverse set of cellular functions. Our screening results provide insight to promote further investigation into how diverse RNA-metabolic activity, such as mRNA splicing, export, and translation, is controlled by HCMV.
Cytokines are critical mediators of immune responses, and their expression must be tightly controlled in order to simultaneously inhibit pathogen replication and prevent host damage. HCMV actively induces massive alteration of IFN-responsive genes and cytokine mRNA levels during lytic infection; however, the mechanisms associated with this process remain unclear (48, 49). In the present study, we propose that Roquin is a key mediator of HCMV-induced immune regulation. We observed that protein and mRNA levels of proinflammatory cytokines and IFN-response molecules decreased over the duration of the infection, and that this down-regulation was completely reversed after blockage of Roquin induction (Fig. 3A). Expression levels of the corresponding genes were greatly increased and maintained at high level throughout the infection, which may prevent the successful replication of HCMV as shown in the conditioned media experiment (Fig. 3D).
Intriguingly, the expression of Roquin is markedly induced upon HCMV infection. Given the crucial role of Roquin involved in inflammatory diseases and immune cell homeostasis (33, 35), Roquin expression likely must be tightly regulated. Viruses modulate the expression patterns of cellular genes for their advantage, with HCMV reportedly inducing the altered expression of thousands of cellular genes (50). A previous study reported a threefold increase in peptides mapped to the Roquin protein following infection with HCMV, although its biological function was not addressed (51). Our data show that a specific viral gene is responsible for Roquin induction as virus could not induce Roquin after UV irradiation or IE1 knockdown. We assume that HCMV evolved to actively amplify Roquin to survive the proinflammatory cytokine storm during infection. Although Roquin is able to regulate its target genes in uninfected cells, immune-related genes are robustly increased by viral infection, so the virus may require Roquin induction to successfully control cytokines. Identification of this viral gene will help elucidate the detailed mechanism associated with Roquin exploitation by HCMV.
We identified genome-wide Roquin-binding sites at both the endogenous level and in virus-infected cells. The majority of Roquin-binding transcripts were derived from the human genome, and the targetome showed no significant difference upon HCMV infection. Roquin targets identified at a single time point in CLIP-seq data appeared to undergo continuous regulation by Roquin throughout the infection process (Fig. 4D). Our data suggest that target genes and their associated molecular functions are minimally altered by HCMV infection, but rather the virus enhances Roquin function by increasing its expression.
Roquin CLIP-seq showed that most target regions were mapped to the 3′ UTR of mRNAs, consistent with previous reports (39, 52). However, we also found Roquin-binding sites within 5′ UTRs or ORFs or introns of target genes, all of which were able to induce down-regulation of the transcript (SI Appendix, Fig. S5B). Moreover, ASO-mediated blocking of the Roquin-binding site on IRF1 5′ UTR could prevent down-regulation (Fig. 6D), indicating that the targeted region of Roquin is not limited to the 3′ UTR of mRNAs and that Roquin regulates target transcripts at multiple binding sites.
CLIP-seq and RNA-seq analyses identified IRF1 as a key Roquin target that plays a critical role in HCMV lytic infection. IRF1 acts as a transcription factor that promotes the expression of proinflammatory cytokines and IFNs. Our results showed that cytokine levels increased following Roquin silencing, and that this elevation was significantly reduced by cosilencing of IRF1 and Roquin. Moreover, we revealed that Roquin bound to and down-regulated the expression of other transcripts, such as those of STAT1, another important transcription factor in immune signaling, suggesting that these targets might be involved in amplifying the immune-regulatory function of Roquin. This suggests that in addition to posttranscriptional down-regulation of cellular genes, Roquin also targets transcripts of transcription factors, including IRF1, broadening its immune-regulatory function during HCMV infection.
Apart from IRF1, we also discovered that depletion of three Roquin target genes, ANKRD52, CDK6, and CFL2, could reverse Roquin-mediated reduction of viral late gene expression (Fig. 5A). Although further experiments are required to confirm Roquin’s impact on these genes, they may exert certain antiviral functions, which are prevented by Roquin induction during HCMV infection. ANKRD52 has been reported to promote miRNA-dependent regulation through Argonaute dephosphorylation (53), so Roquin may down-regulate its expression to inhibit cellular miRNAs involved in the innate antiviral responses (54). Furthermore, HCMV may induce the suppression of CDK6 and CFL2 to establish favorable cellular environment by controlling cell cycle (55) and cytoskeleton dynamics (56), respectively. Investigating the functions of the three genes would provide further insight on various antiviral responses affecting viral gene expression.
In summary, this study describes a HCMV-specific immune evasion strategy involving the RNA-binding protein Roquin. Our findings revealed the underlying mechanism associated with HCMV-induced global suppression of proinflammatory cytokines and antiviral genes, which had previously been observed but not clearly explained. The results showed that Roquin silencing resulted in activation of cytokine expression and delay of viral production, thereby suggesting cellular RNA-binding protein may be targeted for CMV therapeutics. Furthermore, this study emphasizes the importance of regulating RNA processing during the DNA virus infection cycle. Future investigations will target the role of other cellular RNA-binding proteins to elucidate other host–virus interactions involving RNA.
Materials and Methods
Cells and Viruses.
Primary HFF cells (American Type Culture Collection [ATCC]), HFFs immortalized with human telomerase, and glioblastoma U373MG cells were grown at 37 °C/5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM; HyClone) with 10% Fetal Bovine Serum (FBS; HyClone), GlutaMAX-I (Gibco), and penicillin-streptomycin (Gibco). THP-1 cells (ATCC) were cultured in Roswell Park Memorial Institute medium (RPMI 1640; HyClone) supplemented with 10% FBS and GlutaMAX-I. Differentiation of THP-1 was induced by treatment with 100 nM phorbol 12-myristate 13-acetate (PMA) for 24 h.
Infectious virus particles of HCMV were generated by transfecting HCMV Toledo-, AD169-, or Towne-BAC DNAs (gifts from T. Shenk; Princeton University, Princeton, NJ) into primary HFF cells via electroporation using Neon (Invitrogen). When a 100% cytopathic effect was observed, cell culture supernatants were harvested, centrifuged to remove cell debris, and stored at −−80 °C in 1-mL aliquots. To titrate the viral stocks, HFFs grown on cover glass were inoculated with diluted virus stocks for 1 h and fixed with 3.7% formaldehyde at 24 hpi. Cells were then permeabilized using 0.1% Triton X-100 and incubated in blocking buffer (2% BSA in PBS). Cells were stained with HCMV IE1 antibody (MAB810R; Millipore), followed by FITC-conjugated anti-mouse antibody (115-095-146; Jackson Laboratories), and mounted with DAPI-containing solution (H-1200; Vector Laboratory). The number of HCMV IE1-positive cells was counted, and the MOI was determined by calculating the ratio of IE1-positive cells to total cells.
For viral infection, HFFs or U373MG cells were incubated with virus diluted in serum-free DMEM for 1 h, washed with PBS, and incubated in complete media for further experiments. PMA-differentiated THP-1 cells were incubated with virus inoculum for 24 h, followed by PBS washing and incubation in fresh media. For UV-irradiated virus, virus stock was irradiated at 300 mJ/cm2 on ice using a Spectrolinker (Spectroline). Ganciclovir (G2536; Sigma) was treated at 5 µM after 1-h infection.
To measure viral production, naïve cells were incubated with serial dilutions of cell culture supernatant from 1 to 5 hpi, and infectious units were measured by counting IE1-positive cells using the same method explained above.
HSV-1 (a gift from J. H. Ahn; Sungkyunkwan University, Suwon, Republic of Korea) was prepared and used as previously described (57). For plaque assay, HFF cells were inoculated with serial dilutions of viruses and incubated with methyl cellulose overlay. At 3 dpi for HSV-1 and 10–14 dpi for HCMV, cells were stained with 1% crystal violet and plaques were counted.
DNA/RNA Transfection and RNAi Screening.
HFFs or U373MG cells were transfected with gene-specific siRNAs (siGenome; Dharmacon) or IE1 siRNA (GCGGGAGAUGUGGAUGGCUdTdT; synthesized by Bioneer) using Dharmafect-1 reagent (Dharmacon) at a final concentration of 20 nM. At 48 h posttransfection, another set of siRNAs was transfected to effectively knock down the target gene. The transfected cells were used for infection or subsequent analysis after 24 h. THP-1 cells were treated with 100 nM PMA for 24 h, then transfected with 40 nM siRNAs using Dharmafect-4 reagent (Dharmacon). Cells were treated again with siRNA mixture at 24 h after first transfection, then infected with HCMV after another 24 h. For stimulation of innate immune response, HFFs were transfected with 1 µg/mL poly(dA:dT) (Sigma) or poly(I:C) (Sigma) using Lipofectamine 3000 reagent (Invitrogen).
A total of 687 siRNAs for primary screening for RNA-processing proteins (gene information provided in Dataset S1) and 109 genes for secondary screening were obtained from the Dharmacon siGENOME library (Dharmacon). For each gene, four specific siRNAs were designed and transfected as a mixture. HFF cells were treated with siRNA for 48 h, infected with HCMV Toledo at 2 MOI, and treated with secondary siRNA. For primary siRNA screening, HFFs were transfected with 20 nM siRNAs and harvested at 48 hpi for analysis of IE1, UL44, and UL99 mRNA levels. For secondary screening, a total of 40 nM siRNAs was used for transfection (20 nM siRoq and 20 nM of siTarget), and UL146 mRNA levels were measured at 72 hpi. mRNA levels were quantified by qRT-PCR and normalized against GAPDH mRNA.
Other materials and methods used in this study are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Thomas Shenk (Princeton University) for providing HCMV bacterial artificial chromosome (BAC) clones and Prof. Jin-Hyun Ahn (Sungkyunkwan University) for providing HSV-1. This work was supported by Institute for Basic Science of the Ministry of Science Grant IBS-R008-D1 (to K.A.), Seoul National University (SNU)-Yonsei Research Cooperation Program through SNU, and funding from the BK21 plus fellowship and Global Ph.D. Fellowship Program 2013H1A2A1033331 (to J.S.) from a National Research Foundation grant funded by the Ministry of Education, Science, and Technology of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.A.B. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE132081).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909314116/-/DCSupplemental.
References
- 1.Manicklal S., Emery V. C., Lazzarotto T., Boppana S. B., Gupta R. K., The “silent” global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 26, 86–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wills M. R., Poole E., Lau B., Krishna B., Sinclair J. H., The immunology of human cytomegalovirus latency: CouldC latent infection be cleared by novel immunotherapeutic strategies? Cell. Mol. Immunol. 12, 128–138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanzieri T. M., Dollard S. C., Bialek S. R., Grosse S. D., Systematic review of the birth prevalence of congenital cytomegalovirus infection in developing countries. Int. J. Infect. Dis. 22, 44–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britt W. J., Congenital human cytomegalovirus infection and the enigma of maternal immunity. J. Virol. 91, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeFilippis V. R., Induction and evasion of the type I interferon response by cytomegaloviruses. Adv. Exp. Med. Biol. 598, 309–324 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Yurochko A. D., Mayo M. W., Poma E. E., Baldwin A. S. Jr, Huang E. S., Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-kappaB promoters. J. Virol. 71, 4638–4648 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle K. A., Pietropaolo R. L., Compton T., Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19, 3607–3613 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossini G., et al. , Interplay between human cytomegalovirus and intrinsic/innate host responses: AA complex bidirectional relationship. Mediators Inflamm. 2012, 607276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson S. E., Mason G. M., Wills M. R., Human cytomegalovirus immunity and immune evasion. Virus Res. 157, 151–160 (2011). [DOI] [PubMed] [Google Scholar]
- 10.McSharry B. P., Avdic S., Slobedman B., Human cytomegalovirus encoded homologs of cytokines, chemokines and their receptors: RolesR in immunomodulation. Viruses 4, 2448–2470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tadagaki K., et al. , Human cytomegalovirus-encoded UL33 and UL78 heteromerize with host CCR5 and CXCR4 impairing their HIV coreceptor activity. Blood 119, 4908–4918 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Randolph-Habecker J. R., et al. , The expression of the cytomegalovirus chemokine receptor homolog US28 sequesters biologically active CC chemokines and alters IL-8 production. Cytokine 19, 37–46 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Kotenko S. V., Saccani S., Izotova L. S., Mirochnitchenko O. V., Pestka S., Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. U.S.A. 97, 1695–1700 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang W. L. W., Baumgarth N., Yu D., Barry P. A., Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J. Virol. 78, 8720–8731 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lüttichau H. R., The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J. Biol. Chem. 285, 9137–9146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvis M. A., et al. , Human cytomegalovirus attenuates interleukin-1beta and tumor necrosis factor alpha proinflammatory signaling by inhibition of NF-kappaB activation. J. Virol. 80, 5588–5598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baillie J., Sahlender D. A., Sinclair J. H., Human cytomegalovirus infection inhibits tumor necrosis factor alpha (TNF-alpha) signaling by targeting the 55-kilodalton TNF-alpha receptor. J. Virol. 77, 7007–7016 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abate D. A., Watanabe S., Mocarski E. S., Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78, 10995–11006 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathers C., Schafer X., Martínez-Sobrido L., Munger J., The human cytomegalovirus UL26 protein antagonizes NF-κB activation. J. Virol. 88, 14289–14300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y., et al. , Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 8, e1002577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hook L. M., et al. , Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15, 363–373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S., et al. , Temporal landscape of microRNA-mediated host-virus crosstalk during productive human cytomegalovirus infection. Cell Host Microbe 17, 838–851 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Child S. J., Hakki M., De Niro K. L., Geballe A. P., Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78, 197–205 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoyagi M., Gaspar M., Shenk T. E., Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc. Natl. Acad. Sci. U.S.A. 107, 2640–2645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zielke B., Thomas M., Giede-Jeppe A., Müller R., Stamminger T., Characterization of the betaherpesviral pUL69 protein family reveals binding of the cellular mRNA export factor UAP56 as a prerequisite for stimulation of nuclear mRNA export and for efficient viral replication. J. Virol. 85, 1804–1819 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinney C., Perez C., Mohr I., Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 109, 5627–5632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatherer D., et al. , High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. U.S.A. 108, 19755–19760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern-Ginossar N., et al. , Decoding human cytomegalovirus. Science 338, 1088–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adair R., Liebisch G. W., Su Y., Colberg-Poley A. M., Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection. J. Gen. Virol. 85, 3541–3553 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Batra R., et al. , RNA-binding protein CPEB1 remodels host and viral RNA landscapes. Nat. Struct. Mol. Biol. 23, 1101–1110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook K. B., Kazan H., Zuberi K., Morris Q., Hughes T. R., RBPDB: AA database of RNA-binding specificities. Nucleic Acids Res. 39, D301–D308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabach Y., et al. , Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature 493, 694–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinuesa C. G., et al. , A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Glasmacher E., et al. , Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat. Immunol. 11, 725–733 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Athanasopoulos V., Ramiscal R. R., Vinuesa C. G., ROQUIN signalling pathways in innate and adaptive immunity. Eur. J. Immunol. 46, 1082–1090 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Schlundt A., Niessing D., Heissmeyer V., Sattler M., RNA recognition by Roquin in posttranscriptional gene regulation. Wiley Interdiscip. Rev. RNA 7, 455–469 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Mino T., et al. , Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161, 1058–1073 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Song J., Lee S., Cho D., Ahn K., Analysis of Roquin targetome during HCMV infection. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132081. Deposited 3 June 2019. [Google Scholar]
- 39.Leppek K., et al. , Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153, 869–881 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Murakawa Y., et al. , RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κB pathway. Nat. Commun. 6, 7367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenz R., et al. , ViennaRNA packagep 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: AA sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlundt A., et al. , Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation. Nat. Struct. Mol. Biol. 21, 671–678 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Tan D., Zhou M., Kiledjian M., Tong L., The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat. Struct. Mol. Biol. 21, 679–685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honda K., Taniguchi T., IRFs: MasterM regulators of signalling by tollt-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6, 644–658 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Rettino A., Clarke N. M.,Genome-wide identificationi of IRF1 binding sites reveals extensive occupancybsreo at cell death associated genes.cdag J Carcinog Mutagen. S6, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith J. D., De Harven E., Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. I. Sequence of viral replication. J. Virol. 12, 919–930 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browne E. P., Shenk T., Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. U.S.A. 100, 11439–11444 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gealy C., et al. , Posttranscriptional suppression of interleukin-6 production by human cytomegalovirus. J. Virol. 79, 472–485 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weekes M. P., et al. , Quantitative temporal viromics: AnA approach to investigate host-pathogen interaction. Cell 157, 1460–1472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nightingale K., et al. , High-definition analysis of host protein stability during human cytomegalovirus infection reveals antiviral factors and viral evasion mechanisms. Cell Host Microbe 24, 447–460.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Essig K., et al. , Roquin targets mRNAs in a 3′-UTR-specific manner by different modes of regulation. Nat. Commun. 9, 3810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Golden R. J., et al. , An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542, 197–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umbach J. L., Cullen B. R., The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23, 1151–1164 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez V., et al. , Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122-123 and UL37 immediate-early transcripts and at later times for virus production. J. Virol. 78, 11219–11232 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkie A. R., Lawler J. L., Coen D. M., A roler for nuclearn F-actin inductionai in human cytomegalovirus nuclear egresshcne. MBio 7, e01254-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seo M., et al. , MAP4-regulated dynein-dependent trafficking of BTN3A1 controls the TBK1-IRF3 signaling axis. Proc. Natl. Acad. Sci. U.S.A. 113, 14390–14395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.