Significance
Many cellular structures are assembled via phase separation, forming liquid-like droplets in a manner analogous to oil in water. How can the cell control whether there is 1 droplet or many? Here, we provide insights into this question by studying the pyrenoid, an algal liquid-like organelle that mediates a substantial portion of global CO2 fixation. Whereas most algae have just 1 pyrenoid, we characterize a mutant that has approximately 10 pyrenoids per cell. Our results suggest a model where imposing an increased surface area on the liquid-like pyrenoid matrix favors the formation of multiple pyrenoids, with a concomitant loss of photosynthetic efficiency. Our findings advance our basic understanding of the biogenesis of the pyrenoid and of phase-separated organelles in general.
Keywords: carbon fixation, phase separation, starch, Rubisco, pyrenoid
Abstract
A phase-separated, liquid-like organelle called the pyrenoid mediates CO2 fixation in the chloroplasts of nearly all eukaryotic algae. While most algae have 1 pyrenoid per chloroplast, here we describe a mutant in the model alga Chlamydomonas that has on average 10 pyrenoids per chloroplast. Characterization of the mutant leads us to propose a model where multiple pyrenoids are favored by an increase in the surface area of the starch sheath that surrounds and binds to the liquid-like pyrenoid matrix. We find that the mutant’s phenotypes are due to disruption of a gene, which we call StArch Granules Abnormal 1 (SAGA1) because starch sheath granules, or plates, in mutants lacking SAGA1 are more elongated and thinner than those of wild type. SAGA1 contains a starch binding motif, suggesting that it may directly regulate starch sheath morphology. SAGA1 localizes to multiple puncta and streaks in the pyrenoid and physically interacts with the small and large subunits of the carbon-fixing enzyme Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase), a major component of the liquid-like pyrenoid matrix. Our findings suggest a biophysical mechanism by which starch sheath morphology affects pyrenoid number and CO2-concentrating mechanism function, advancing our understanding of the structure and function of this biogeochemically important organelle. More broadly, we propose that the number of phase-separated organelles can be regulated by imposing constraints on their surface area.
Compartmentalization is key to efficiency and specificity of cellular processes. Beyond the classic membrane-bound compartments widely observed in eukaryotic cells, many processes in both eukaryotic and prokaryotic cells are organized in membraneless cellular bodies (1–3). Our understanding of membraneless cellular bodies is undergoing a revolution, enabled by the recent discovery that many of these bodies can exhibit liquid-like behavior such as internal mixing and the ability to dynamically exchange components with the surrounding solution (1). The emerging paradigm is that many of these bodies assemble by phase separation of the constituent proteins and/or nucleic acids, mediated by weak multivalent interactions (3). Many fundamental questions about the principles that underlie the biogenesis and regulation of such phase-separated, liquid-like organelles remain to be addressed. A key outstanding question is how cells regulate whether they have 1 or many droplets of a particular phase-separated body.
The pyrenoid is a phase-separated, liquid-like organelle found in most eukaryotic algae that plays a significant role in global CO2 fixation (4, 5). The pyrenoid houses the CO2-fixing enzyme Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) and enhances the enzyme’s operating efficiency by supplying it with CO2 at a high concentration (5–7). This structure is an essential part of the CO2-concentrating mechanism (CCM) (5, 8).
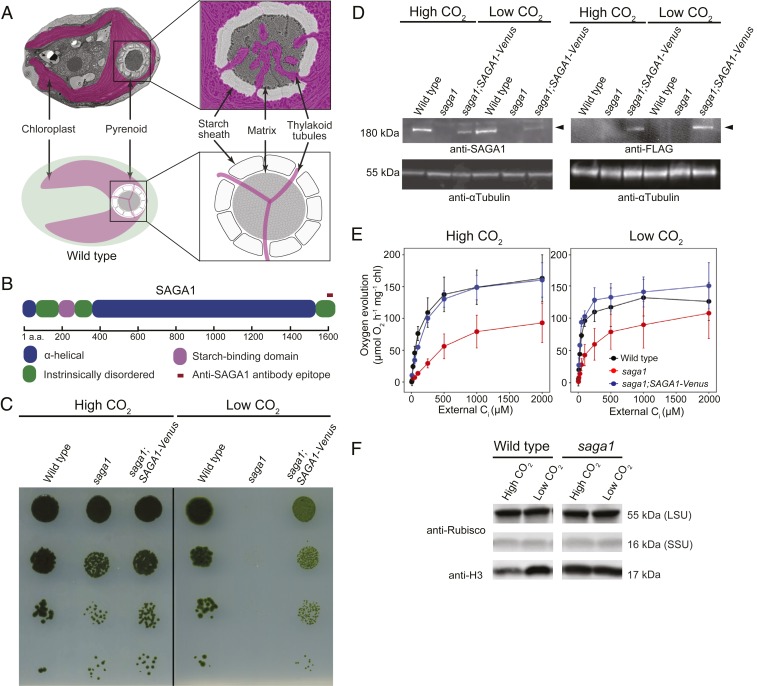
Most pyrenoids consist of 3 subcompartments discernible by electron microscopy (Fig. 1A) (9). At the core of the pyrenoid is the liquid-like pyrenoid matrix, where Rubisco is densely clustered (6, 10–13). Essential to this clustering is the recently characterized Rubisco linker protein EPYC1 (14, 15). In most species, the pyrenoid matrix is traversed by membrane tubules, which are continuous with the thylakoid membranes of the chloroplast. These thylakoid tubules have been postulated to deliver CO2 to Rubisco (16–18), at a concentration ∼100-fold the concentration outside the cell (19). In many species, the pyrenoid is surrounded by a starch sheath (20) made up of multiple starch granules, also known as starch plates. This starch has been suggested to act as a diffusion barrier that retains CO2 in the pyrenoid matrix (20). The molecular basis for the biogenesis of the thylakoid tubules and starch sheath, as well as their connections to the pyrenoid matrix, remains unknown in any species.
Fig. 1.
SAGA1, a gene with a predicted starch-binding domain and alpha helical stretch, is necessary for a functional CCM. (A) A Chlamydomonas cell is shown with the substructures of the pyrenoid labeled. (B) SAGA1 has a predicted starch binding domain followed by a long predicted alpha helical region. (C) Agar growth phenotypes of wild type, saga1, and saga1;SAGA1-Venus are shown. Serial 1:10 dilutions were spotted on TP minimal medium and grown at high (4%) and low (0.04%) CO2 under 500 μmol photons m−2⋅s−1 illumination. (D) SAGA1 protein levels in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low and high CO2 were probed with an anti-SAGA1 polyclonal antibody and with an anti-FLAG antibody. Anti-tubulin is shown as a loading control. (E) Oxygen evolution is shown as a function of external Ci for cells grown in high and low CO2. (F) Rubisco protein levels in wild type and saga1 grown at low and high CO2 were probed with a polyclonal antibody raised to Rubisco. Anti-histone H3 is shown as a loading control. LSU: large subunit; SSU: small subunit.
The model alga Chlamydomonas reinhardtii normally has only 1 pyrenoid per cell. Here we describe a pyrenoid-localized protein, SAGA1 (StArch Granules Abnormal 1), in whose absence cells have multiple pyrenoids and abnormally elongated starch sheaths. Our data lead us to propose a model where excessive starch surface area favors the formation of multiple pyrenoids, and where SAGA1 negatively regulates the surface area of the starch sheath to avoid the formation of multiple pyrenoids. Our findings provide a foundation for a molecular understanding of the interactions between the pyrenoid matrix and starch sheath and advance our knowledge of mechanisms that determine the number of phase-separated organelles.
Results
A Screen for CCM Mutants Identified SAGA1, a Gene with a Predicted Starch Binding Domain and Alpha Helical Stretch.
To identify Chlamydomonas mutants with defects in the CCM, we screened an insertional mutant library in search of mutants that require high CO2 to grow photosynthetically. The screen yielded multiple independent high-CO2-requiring mutants disrupted in the gene Cre11.g467712, which we call SAGA1 (StArch Granules Abnormal 1). The predicted gene product of SAGA1 is a protein of 1,626 amino acids (expasy.org; Fig. 1B). Amino acids 214–280 encode a predicted starch binding motif (CBM20; SI Appendix, Fig. S1), a feature conserved in amylolytic enzymes that bind granular starch (21–23). All 5 highly conserved residues necessary for starch binding are present in SAGA1, suggesting that SAGA1 can bind starch (SI Appendix, Fig. S1). Following the starch binding motif is a long (∼1,000 residues) predicted alpha helical region (Fig. 1B) that shares homology with proteins containing coiled-coil domains (SI Appendix, Fig. S2). SAGA1 also has a striking amino acid composition: ∼36% of SAGA1 consists of only 2 amino acids, alanine (25.3%) and glutamic acid (10.8%) (SI Appendix, Fig. S3). These marked features of SAGA1 are unique among characterized CCM-related proteins. SAGA1 homologs can be found in close Chlamydomonas relatives, including Volvox carteri and Tetrabaena socialis, and in more distant pyrenoid-containing eukaryotic algae, including Emiliana huxleyi (9) (SI Appendix, Tables S1 and S2).
SAGA1 Is Necessary for a Functional CCM.
To confirm the defective CCM growth phenotype, we performed spot assays on the saga1 mutant and the background strain. The saga1 mutant was unable to grow in low CO2 but grew under high CO2 (Fig. 1C). We observed similar growth phenotypes in 2 additional mutant alleles of SAGA1, saga1-2 and saga1-3 (SI Appendix, Fig. S4). We confirmed the presence of the insertional mutagenesis cassette in the SAGA1 locus by PCR (SI Appendix, Fig. S5). We carried out all further experiments using the saga1 mutant.
To test whether absence of SAGA1 causes the CCM mutant phenotype, we transformed the saga1 mutant using a construct encoding SAGA1 fused with a C-terminal Venus-3xFLAG tag and a selectable marker (SI Appendix, Fig. S6). Two of 9 transformants exhibiting both Venus fluorescence and antibiotic resistance also exhibited a substantial rescue of the low CO2 growth phenotypes of the saga1 mutant (Fig. 1C). The incomplete rescue of these strains could be due to their lower SAGA1 expression levels than observed in wild type (Fig. 1D), possibly resulting from use of an exogenous promoter and/or the removal of several introns in the transformed construct. Alternatively, the addition of a C-terminal Venus-3xFLAG tag could impair the function of SAGA1. We chose 1 of these rescued strains (denoted as saga1;SAGA1-Venus; Fig. 1C) for use in subsequent experiments. The presence of both the SAGA1-Venus cassette and the insertional mutagenesis cassette was confirmed using PCR (SI Appendix, Fig. S5).
Using a polyclonal antibody raised against the last 19 amino acids of SAGA1, we detected a ∼180-kDa band in the wild type and in the saga1;SAGA1-Venus-3xFLAG strain that was completely absent in the saga1 mutant (Fig. 1D and SI Appendix, Fig. S7). A band of similar size was detected in the protein sample of saga1;SAGA1-Venus-3xFLAG strain when probed with anti-FLAG antibody, confirming that the band of ∼180 kDa was the SAGA1 gene product (Fig. 1D). Unlike previously characterized components of the CCM such as EPYC1 (14) and LCIB (24), whose abundances increase under low CO2, SAGA1 was present at similar abundance per cell under both high and low CO2 (Fig. 1D). This observation suggests that SAGA1 may play a constitutive function under both high- and low-CO2 growth conditions.
We characterized CCM activity in the saga1 mutant and in the saga1;SAGA1-Venus-3xFLAG strain using photosynthetic O2 evolution as a proxy for whole-cell affinity for inorganic carbon (Ci; Fig. 1E). When acclimated to low-CO2 conditions, the saga1 mutant showed a decreased affinity for Ci relative to wild type, indicated by a high photosynthetic K0.5 value (∼160 µM Ci for saga1 vs. ∼40 µM Ci for wild type; Fig. 1E and SI Appendix, Table S3). In the saga1;SAGA1-Venus-3xFLAG strain, the affinity for Ci uptake was restored (∼42 µM Ci; Fig. 1E). These results indicate that SAGA1 is required for a functional CCM.
The saga1 mutant showed a decreased photosynthetic rate at saturating concentrations of Ci when acclimated to both low and high CO2 (Fig. 1E). The Rubisco protein content of saga1 was similar to wild type (Fig. 1F), suggesting that decreased photosynthetic efficiency of the saga1 mutant under both low- and high-CO2 growth conditions may be due to decreased availability of CO2 to Rubisco or decreased Rubisco activity. We conclude from these observations that SAGA1 is required for a functional CCM and for maximal CO2 uptake in cells acclimated to both low and high CO2.
saga1 Mutants Have Aberrant Starch Sheaths.
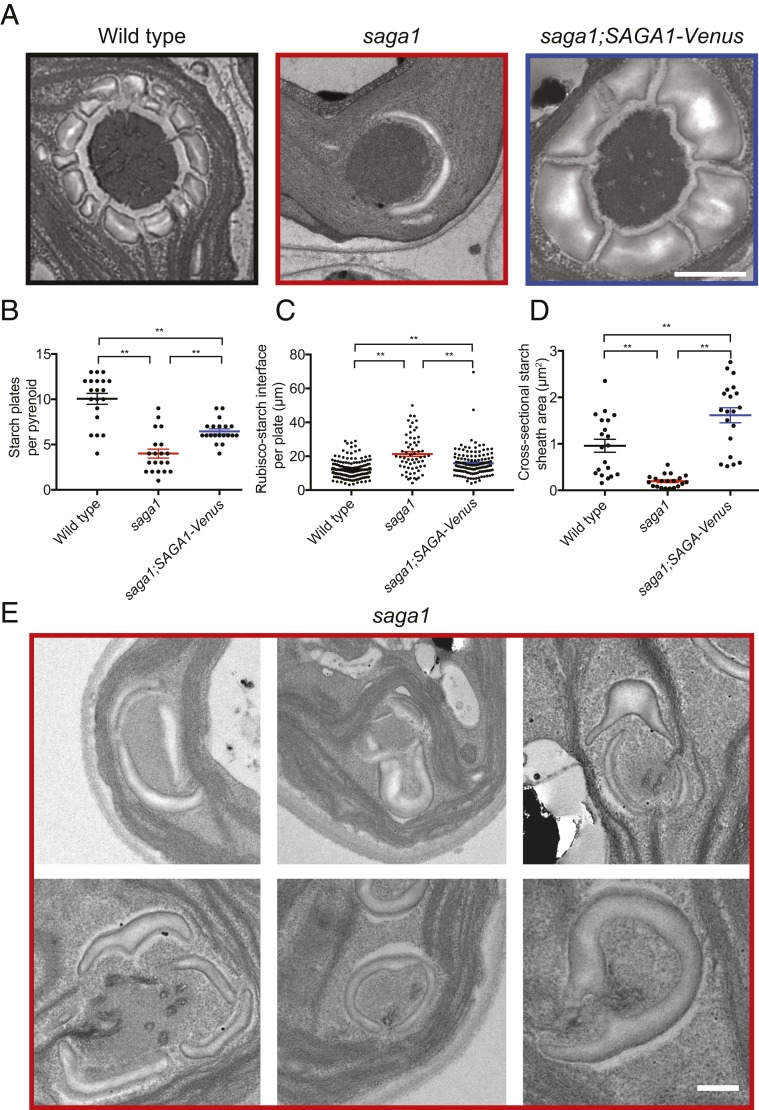
Because SAGA1 contains a starch binding motif, we sought to determine if the saga1 mutant had alterations in the starch sheath surrounding the pyrenoid matrix. Electron microscopy revealed that the saga1 mutant had an abnormal distribution of starch around the pyrenoid, with fewer starch plates observed per pyrenoid (Fig. 2 A and B). These abnormal starch plates were elongated: The length of each plate in the saga1 mutant was ∼30% longer that of wild-type starch plates (Fig. 2 A and C). Furthermore, the starch surrounding each saga1 pyrenoid appeared thinner, with a decreased total cross-sectional area (Fig. 2 A and D). We also occasionally observed starch plates that appeared to stack on each other, and single elongated starch plates that entirely or almost entirely surrounded regions of matrix (Fig. 2E).
Fig. 2.
The pyrenoids of the saga1 mutant have aberrant starch sheaths. (A) Representative TEMs showing pyrenoids of wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2, illustrating defects in the starch sheath. (B) Quantification of the number of starch granules per pyrenoid in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2 (n = 20 pyrenoids; Mann–Whitney U test). (C) Length of starch granules in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2 (n = 162, 114, 126 starch granules; Mann–Whitney U test). (D) The cross-sectional area of pyrenoid starch in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2 (n = 20 cells; Mann–Whitney U test). Error bars: SEM; **P < 0.001. (E) Example TEMs showing pyrenoids of saga1 cells with starch granules either “stacking” on top of each other or appearing to “pinch” away portions of the matrix. (Scale bars, 500 nm.)
The saga1;SAGA1-Venus strain showed partial complementation of the number of starch plates per pyrenoid and of the length of the starch plate–Rubisco interface (Fig. 2 A–C). The incomplete complementation could be due to the lower expression of SAGA1 relative to wild type (Fig. 1D). Interestingly, the pyrenoid starch cross-sectional area in the saga1;SAGA1-Venus strain was larger than that of wild type or that of the saga1 mutant (Fig. 2D), suggesting that pyrenoid starch plate biosynthesis was still abnormal in the saga1;SAGA1-Venus strain. Based on these results, we conclude that SAGA1 is required for normal starch sheath formation around the pyrenoid and that in the absence of SAGA1 pyrenoid plates are elongated and thinner.
saga1 Mutants Have Multiple Pyrenoids.
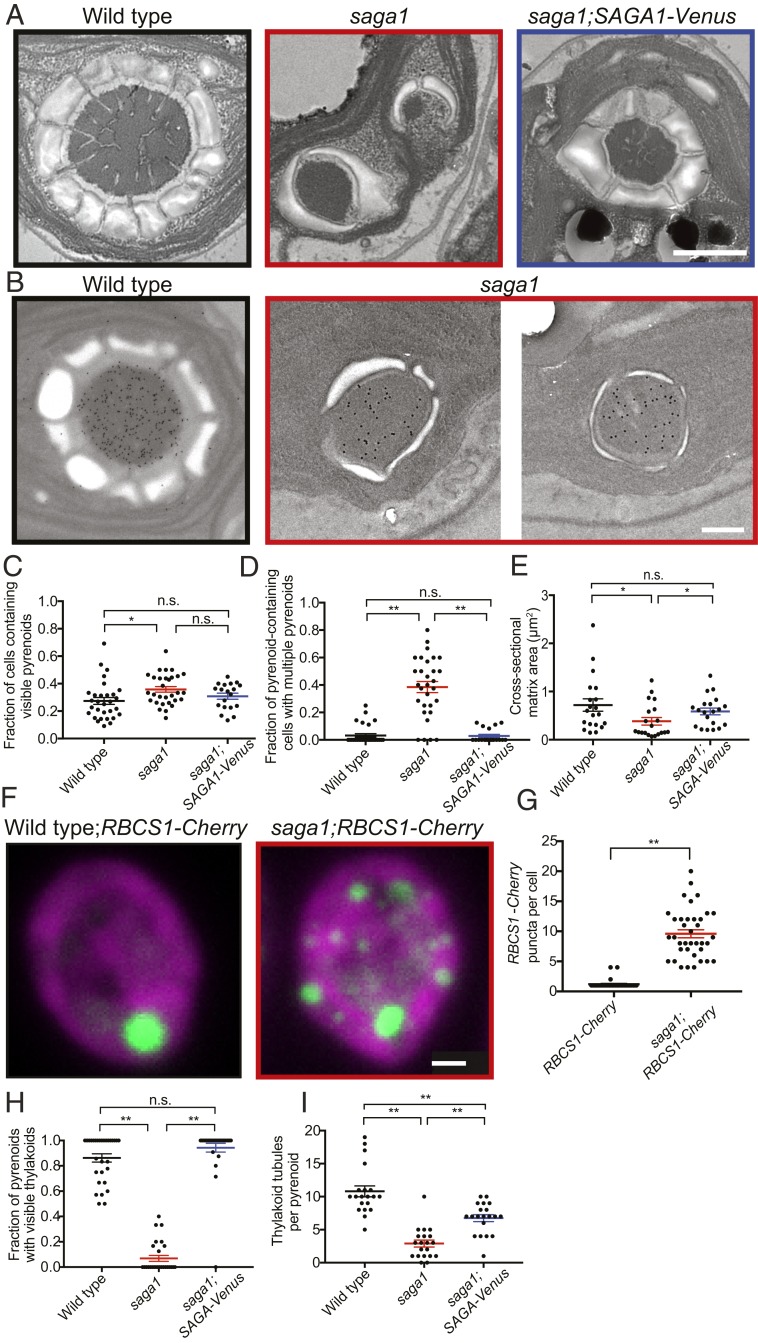
Surprisingly, the electron micrographs revealed that saga1 mutant cells have multiple pyrenoids, observed as electron-dense bodies identical in texture to the pyrenoid matrix found in wild-type cells (Fig. 3A and SI Appendix, Fig. S8). These multiple pyrenoids contained Rubisco, as detected by immunogold labeling and immunofluorescence (Fig. 3B and SI Appendix, Fig. S9 and Table S4). Whereas multiple pyrenoids were rare in wild-type cells, ∼40% of saga1 mutant cross-sections contained multiple pyrenoids (Fig. 3 C and D). Restoration of SAGA1 expression in the saga1;SAGA1-Venus strain eliminated the multiple pyrenoid phenotype, indicating that disruption of SAGA1 was responsible for this phenotype (Fig. 3D).
Fig. 3.
saga1 mutant cells have multiple pyrenoids and often lack thylakoid tubules. (A) Representative TEMs of wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2, illustrating the frequent observation of multiple pyrenoids in saga1. (Scale bar, 500 nm.) (B) Representative immunogold staining images of wild type and saga1 pyrenoids using an anti-Rubisco antibody. (Scale bar, 500 nm.) (C) The fraction of cells containing visible pyrenoids in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2 (n = 30 fields of view; Mann–Whitney U test). (D) The fraction of pyrenoid-containing cells that contain more than 1 pyrenoid in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2 (n = 30 fields of view; Mann–Whitney U test). (E) Pyrenoid matrix cross-sectional area in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2 (n = 20 pyrenoids). (F) Representative summed z-stacks of wild-type and saga1 cells constitutively expressing RBCS1-mCherry (green) grown in low CO2. Magenta is chlorophyll autofluorescence. (Scale bar, 1 μm.) (G) Quantification of the number of RBCS1-mCherry puncta per cell (wild type: 1.2 ± 0.7 puncta per cell; saga1: 9.6 ± 4.1; n = 35 cells, Mann–Whitney U test). (H) Quantification of the fraction of pyrenoids with visible thylakoid tubules (n = 30 fields of view; Mann–Whitney U test). (I) The number of thylakoid tubules per pyrenoid in wild-type, saga1, and saga1;SAGA1-Venus cells grown at low CO2 (n = 20 pyrenoids, Mann–Whitney U test). Error bars: SEM; n.s., not significant; P ≥ 0.05; *P < 0.05; **P < 0.001.
Given that saga1 mutant cells have the same amount of Rubisco per cell as wild type (Fig. 1F), and considering that this amount of Rubisco is distributed across multiple pyrenoids, one would expect the average pyrenoid matrix cross-sectional area to be smaller. Indeed, the average cross-sectional area of Rubisco matrix per pyrenoid in the saga1 mutant was lower than in wild type (Fig. 3E). Absolute saga1 cell size was similar to that of wild-type cells (SI Appendix, Fig. S10).
To count the number of pyrenoids per cell in the saga1 mutant and wild type, we expressed an mCherry-tagged Rubisco small subunit in each strain (14). In nearly all wild-type cells, the Rbcs1-mCherry signal was found in a single, large punctum at the base of the chloroplast, consistent with the presence of a single pyrenoid (Fig. 3F). Strikingly, in the saga1 mutant, we observed an average of ∼10 Rbcs1-mCherry puncta per cell (Fig. 3 F and G and SI Appendix, Fig. S11A), and no saga1 cell was observed to have a singular punctum. This remarkable number of pyrenoids per cell is consistent with the increased incidence of multiple pyrenoids we observed in the electron micrographs. The positions and sizes of these pyrenoids remained stable over the course of 1 h (SI Appendix, Fig. S11B). We conclude that the absence of SAGA1 leads to multiple pyrenoids.
Most Saga1 Mutant Pyrenoids Lack a Visible Pyrenoid Tubule Network.
In wild-type cells, thylakoid tubules traverse the pyrenoid matrix, forming a thylakoid tubule network (Fig. 3 A and H and SI Appendix, Fig. S12). However, nearly all pyrenoids of the saga1 mutant lacked such hallmark thylakoid tubules (Fig. 3 A and H).
Intriguingly, thylakoid tubules could still be observed in ∼8% of pyrenoids (9/113) in the saga1 mutant cell sections. In these pyrenoids, the number of observed tubules per pyrenoid was significantly decreased (Fig. 3I and SI Appendix, Fig. S12). Of the remaining tubuleless pyrenoids in the saga1 mutant, 48% had thylakoid membranes visible adjacent to the periphery of the pyrenoid matrix, failing to penetrate the matrix (SI Appendix, Fig. S12). Single pyrenoids with visible tubules were restored to wild-type levels in the SAGA1;SAGA1-Venus strain (Fig. 3H), indicating that the absence of pyrenoid tubules in most pyrenoids is due to the disruption of SAGA1.
SAGA1-Venus Localizes to the Pyrenoid.
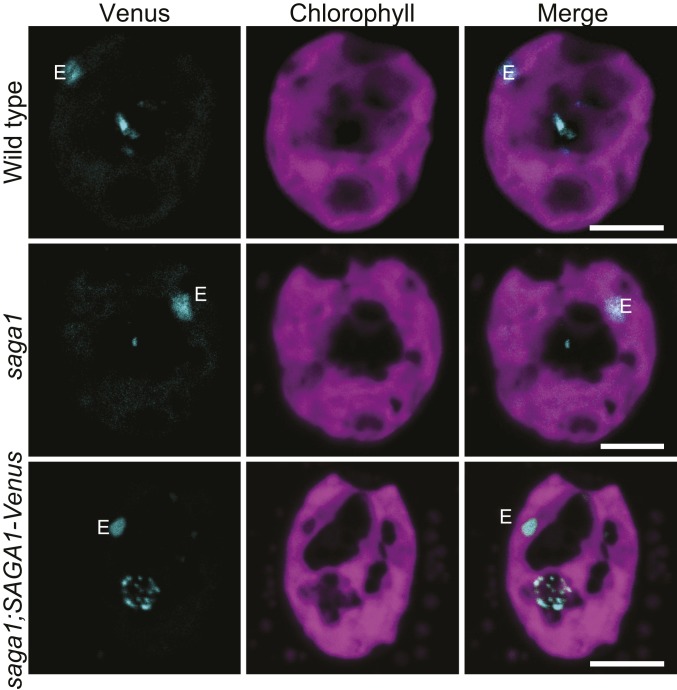
To better understand how absence of SAGA1 produces abnormal pyrenoids, we sought to determine the localization of SAGA1. In the saga1;SAGA1-Venus strain, we observed Venus fluorescence in a spherical region of low chlorophyll fluorescence at the base of the chloroplast, indicating that SAGA1 localizes to the restored pyrenoid in the complemented mutant (Fig. 4). Consistent with this finding, SAGA1 peptides have been detected in the pyrenoid proteome (25). Intriguingly, SAGA1-Venus fluorescence signal was not uniform but formed multiple small puncta and streaks in the pyrenoid. These streaks only partially resemble the peripheral mesh-like localization of LCI9, the star-like thylakoid localization of PSAH, and the punctate thylakoid localization of PSBP4 (26), suggesting that SAGA1 has a distinct localization from these proteins. Some of the SAGA1-Venus signal appeared to partially overlap with chlorophyll autofluorescence at the pyrenoid periphery (SI Appendix, Fig. S13). To determine if the overlap between the SAGA1-Venus fluorescence and chlorophyll autofluorescence was significant, we calculated the correlation between the pixel intensities of the 2 fluorescence channels, comparing to random expectation (see Methods for more details). We observed a subtle but significant overlap between the 2 fluorescence channels, suggesting that a portion of SAGA1 may colocalize with pyrenoid tubules (SI Appendix, Fig. S13 and Table S5). We conclude that SAGA1-Venus localizes to the pyrenoid.
Fig. 4.
SAGA1 was localized within the pyrenoid of the rescued mutant saga1;SAGA1-Venus after 24-h acclimation to ambient air (low CO2) to induce the CCM with wild type and saga1 as controls. All cells were grown at room temperature with illumination of about 50 µmol photons m−2⋅s−1. E: eyespot. White arrows indicated the location of the pyrenoid. (Scale bars, 5 µm.)
SAGA1 Binds to Rubisco Large and Small Subunits.
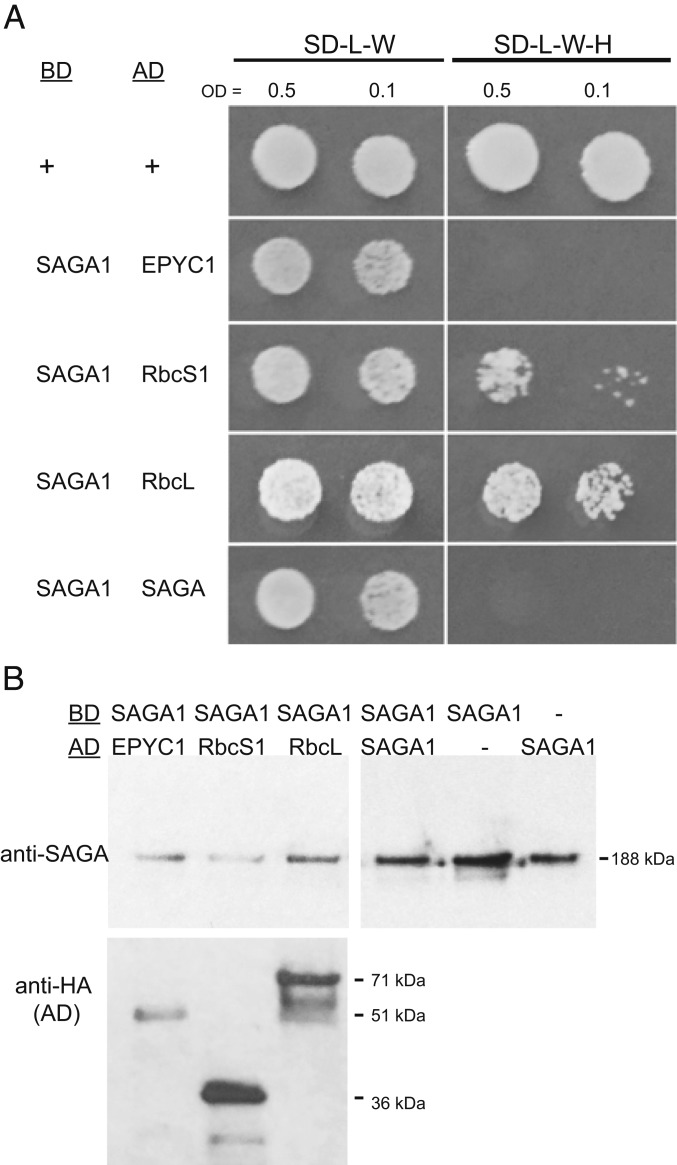
In our recent high-throughput immunoprecipitation–mass spectrometry study, we observed that SAGA1 coimmunoprecipitates with EPYC1, an abundant pyrenoid component that links Rubisco holoenzymes together to form the pyrenoid matrix (26). In order to validate this coimmunoprecipitation, we performed a yeast 2-hybrid assay to evaluate SAGA1’s binding interactions with EPYC1 as well as the large and small subunits of Rubisco. In this assay, SAGA1 interacted with the large and small subunits of Rubisco, but not with EPYC1 (Fig. 5A). These findings suggest that our previously observed coimmunoprecipitation of SAGA1 and EPYC1 was mediated by the binding of both proteins to Rubisco (Fig. 5A). The structural homology of SAGA1 to coiled-coil proteins raised the possibility that SAGA could dimerize (27, 28); however, SAGA1 did not appear to interact with itself in a yeast 2-hybrid assay (Fig. 5A). Western blots confirmed the presence and molecular weight of SAGA1 and the different preys (Fig. 5B). We conclude that SAGA1 binds to Rubisco large and small subunits.
Fig. 5.
SAGA1 binds Rubisco small and large subunits. (A) Yeast 2-hybrid assays of SAGA1 as the bait (BD; DNA-binding domain) and either EPYC1, Rbcs1, RbcL1, or SAGA1 as the prey (AD; activation domain). Saccharomyces cerevisiae expressing the bait were mated with cells expressing an AD and were grown to an OD of 0.5 and 0.1 and spotted onto either nonselective (SD-L-W) or selective (SD-LWH) media. (B) Protein levels of the bait and prey proteins in the mated strains were detected by Western blotting using either the anti-SAGA1 antibody or an anti-HA antibody, respectively.
Discussion
We Propose a Starch-Centric Model for SAGA1’s Function in Pyrenoid Biogenesis.
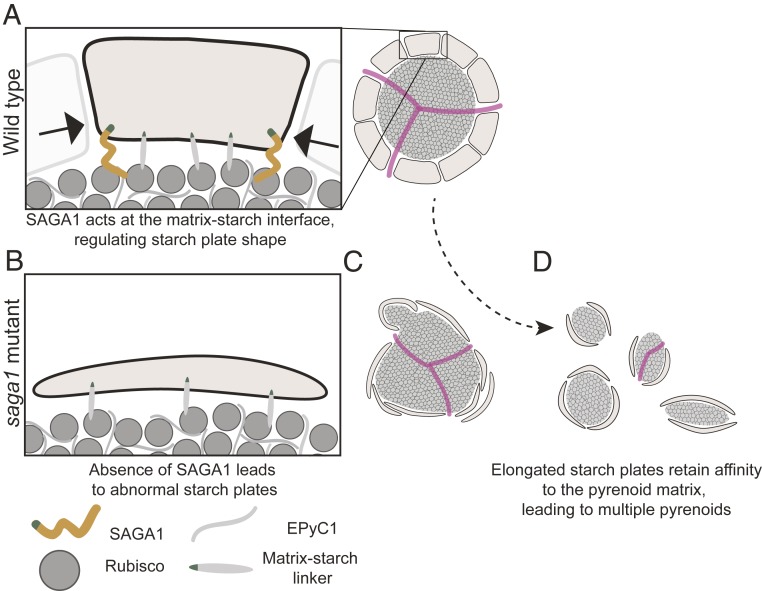
The saga1 mutant is puzzling because it has 3 prominent phenotypes: abnormal starch plates, multiple pyrenoids, and frequent lack of pyrenoid tubule networks. Occam’s razor suggests that SAGA1 likely has a single molecular function whose absence explains all 3 of these phenotypes. We propose a model where SAGA1 guides the formation of pyrenoid starch plates and, in the absence of SAGA1, defective pyrenoid starch plates lead to the formation of multiple pyrenoids, most of which lack tubules (Fig. 6).
Fig. 6.
A proposed model for the function of SAGA1. (A) Multiple proteins link starch and the Rubisco matrix. SAGA1 plays a role at the periphery of starch granules in restricting starch granule elongation. (B) Loss of SAGA1 results in elongated starch granules. (C) The elongated starch granules have increased surface area and their affinity for Rubisco favors a configuration of the Rubisco matrix that has increased surface area, resulting in multiple pyrenoids. (D) There is only 1 “knot” of thylakoid tubules per cell and, as a consequence, the extra pyrenoids in the saga1 mutant lack thylakoid tubules.
We were drawn to this model because the multiple pyrenoid phenotype is at odds with the recent description of the pyrenoid matrix as a phase-separated compartment (4). One would expect that the multiple pyrenoids of the saga1 mutant would rapidly resolve into a single pyrenoid due to Ostwald ripening, a physical phenomenon whereby large droplets grow by assimilating building blocks from smaller droplets (29). Indeed, Ostwald ripening appears to occur when the pyrenoid matrix is reconstituted in vitro by mixing the Rubisco-binding linker protein EPYC1 with Rubisco (15): Many small droplets initially form, but they rapidly resolve into a smaller number of larger droplets through growth of the large droplets and shrinkage of the small droplets. Some of the large droplets grow to over 5 µm in diameter, corresponding to a volume 2 orders of magnitude larger than that of a pyrenoid in a wild-type cell (Fig. 2A), suggesting that the natural tendency of a mixture of EPYC1 and Rubisco is to incorporate all available material into a single large pyrenoid. The presence of multiple stable pyrenoids thus suggests that in the saga1 mutant additional forces not present in the in vitro system act to counteract Ostwald ripening.
We propose that the excessive surface area of the abnormal starch plates in saga1 promotes the formation of multiple pyrenoids by counteracting Ostwald ripening. Central to our model is the assumption that starch plates have an affinity for pyrenoid matrix, such that the pyrenoid matrix material “wets” or adheres to 1 side of each starch plate. In wild-type cells, each cell has the appropriate amount of starch plate surface area to coat a single spherical pyrenoid. However, in the saga1 mutant, the starch plates elongate to reach a total surface area much greater than the surface area of a single pyrenoid. In our model, this mismatch leads to starch plates “pinching” portions of matrix off, which results in multiple smaller pyrenoids (Fig. 6). Indeed, some of the electron micrographs we have observed may represent intermediates in this process, including overlapping starch plates and “pinching” of matrix by single starch plates (Fig. 2E). Therefore, the multiple pyrenoids of the saga1 mutant represent a minimal-energy solution for putting a larger surface area of starch plates in contact with the same volume of liquid-like pyrenoid matrix.
The model raises the question of what mediates binding between starch plates and the matrix. We hypothesize that this binding is mediated by 1 or more proteins with starch-binding domains that also bind to Rubisco or EPYC1 (Fig. 6 and SI Appendix, Fig. S14). Furthermore, our model suggests that the starch sheath has an inherent asymmetric property that positions Rubisco to 1 side of the starch sheath plates. This asymmetry is unperturbed in the saga1 mutant; otherwise, one would expect abnormal starch plates to be surrounded by Rubisco matrix.
Previous studies have found that in the absence of pyrenoid starch cells contain a single pyrenoid and a functional CCM (30). This is in agreement with our proposed model where the multiple pyrenoid phenotype of the saga1 mutant is due to the presence of abnormal pyrenoid starch plates, as opposed to the absence of normal pyrenoid starch plates.
Excessive Starch Plate Surface Area:Matrix Volume Ratio Explains Multiple Pyrenoids in Other Mutants.
Our proposal that the formation of multiple pyrenoids is caused by an imbalance between the surface area and volume of the pyrenoid matrix is consistent with, and provides an explanation for, the previously unexplained increased incidence of multiple pyrenoids in other mutants, including epyc1 (14) and cia6 mutants (31). In these mutants, the pyrenoid matrix volume is decreased because much of the Rubisco is dissolved in the stroma. If pyrenoid starch biosynthesis is largely unchanged, one would expect that these mutants would have an excessive starch sheath surface area relative to pyrenoid matrix volume, which would favor multiple pyrenoids as the matrix adopts a configuration with a higher surface-to-volume ratio, just as we observe in the saga1 mutant.
The Lack of Tubules Observed in Most Mutant Pyrenoids May Be a Consequence of Cells Retaining a Single Pyrenoid Tubule Network.
While most pyrenoids of the saga1 mutant lacked the canonical thylakoid tubule network, a limited thylakoid tubule network was still observed in ∼10% of pyrenoids (Fig. 3H and SI Appendix, Fig. S12). We hypothesize that this observation is due to each saga1 mutant cell’s retaining a single, diminished pyrenoid tubule network, with only 1 of the ∼10 pyrenoids per cell containing the network. This proposed model explains the absence of thylakoid tubules in the majority of the pyrenoids of the saga1 mutant. This model is also consistent with previous work suggesting that the biogenesis of the pyrenoid tubule network is independent of matrix biogenesis, as mutants that entirely lack a pyrenoid matrix still contain a singular tubule network (14, 32).
The pyrenoid tubule networks observed in pyrenoids of the saga1 mutant have a reduced number of tubules and appear somewhat deformed (Fig. 3I). These abnormal pyrenoid tubule networks could be due to altered availability of spaces between starch plates where thylakoid tubules can enter the pyrenoid. Alternatively, the abnormal tubule networks could be explained by a regulatory response of the extent of the thylakoid tubule network to the size of the Rubisco matrix, which is decreased in saga1 mutants.
The absence of thylakoid tubule networks from most pyrenoids in the saga1 mutant indicates that most of these pyrenoids are not supplied with CO2 via the thylakoid network. Consequently, the majority of the Rubisco does not benefit from concentrated CO2, explaining the decreased affinity to CO2 and lower maximum photosynthetic rates we observed in the saga1 mutant (Fig. 1E). The absence of a singular pyrenoid and the diminished thylakoid tubule network of the saga1 mutant may also explain the decreased affinity of the mutant for Ci under high CO2 (SI Appendix, Table S3).
Recent work suggests that the pyrenoid tubules may serve as an “anchor” that localizes the pyrenoid to the base of the chloroplast, as the tubules still retain their canonical localization even in the absence of pyrenoid matrix (14). The scattering of pyrenoids across the stroma in the saga1 mutant (Fig. 3F) could thus be due to their lack of thylakoid tubules.
A Proposed Molecular Role for SAGA1 in Shaping Starch Plates.
While the mechanisms that control the shape of starch plates are poorly understood in any organism (33), a direct role for SAGA1 in regulation of starch plate shape is plausible given that the saga1 mutant exhibited abnormal starch plates, SAGA1 has a CBM20 starch binding domain, and other CBM20 motif-containing proteins have been demonstrated to influence starch biogenesis. Moreover, other coiled-coiled proteins are important for starch granule initiation in Arabidopsis (34) and pyrenoid starch synthesis in Chlamydomonas (35). The observation that SAGA1-Venus forms streaks near the periphery of the pyrenoid (Fig. 4) is consistent with a possible localization to the edges of starch plates, where SAGA1 could inhibit pyrenoid starch plate elongation (Fig. 6A). SAGA1 could influence starch-plate morphology by recruiting starch-remodeling enzymes, or by preventing access of starch-synthesis enzymes. Alternatively, SAGA1 itself could possess its own enzymatic activity; however, it shows no clear homology to any known starch-modifying enzymatic domain. The larger, but less numerous, starch granules we observed in the rescued strain suggest that SAGA1 may also play a role in creating the interfaces between starch plates for thylakoid tubules to enter and exit.
SAGA1’s binding interaction with Rubisco could serve to localize SAGA1 to the periphery of the pyrenoid matrix (Fig. 4). A size-exclusion effect may prevent the 180-kDa SAGA1 from entering inside the matrix. Indeed, we previously observed that most chloroplast proteins that can enter the pyrenoid matrix are smaller than ∼80 kDa (26). Other phase-separated bodies, such as P-granules, can act as size-exclusion barriers due to the effective mesh size of the condensed phase (36–38). SAGA1 contains a long alpha helical region (SI Appendix, Fig. S2); thus, if the Rubisco-binding region is on SAGA1’s C terminus, this alpha helical region could span the ∼30-nm gap between the starch sheath and matrix that has been observed in native electron microscopy studies (SI Appendix, Fig. S8) (14) and would allow the CBM20 domain to interact with the starch sheath. Our observation of a weak colocalization of SAGA1-Venus with chlorophyll autofluorescence (SI Appendix, Fig. S13) could indicate that excess SAGA1 may accumulate along the surface of the pyrenoid tubules, where it could still remain excluded from the matrix.
In addition to the model we presented above, alternative models can partially explain our data. SAGA1 could be implicated in thylakoid biogenesis or maintenance, for example by serving as a linker between the thylakoid tubules and the matrix. In this model, an absence of SAGA1 leads to fewer thylakoid tubules, which then liberates the matrix from its canonical location and leads to the formation of multiple pyrenoids. While also a compelling model, SAGA1’s starch binding domain, the elongated starch plates of the saga1 mutant, and the liquid-like nature of the pyrenoid matrix motivate our preference for a model centered around the ratio of starch plate surface area to matrix volume. We hope that our data and model will help guide future work on the molecular function of SAGA1.
Implications for Regulation of Phase-Separated Organelles.
Depending on the species of phase-separated organelle, the number of distinct droplets per cell can range from a single body, for example the Balbiani body (39), to dozens of bodies, for example stress granules (40). Moreover, many of these organelles change in number in response to environmental or developmental cues (4, 41). Illuminating work has demonstrated that the cytoskeleton (F-actin) regulates stress granule formation in Xenopus (42), while the control of the number of other phase separated bodies, such as P bodies and nucleoli, remains enigmatic (3).
Our model for how starch influences the number of pyrenoids suggests a basic principle for how cells could regulate the number of a species of phase-separated organelle, by constraining their surface-to-volume ratio through the interaction of a liquid-phase component with a peripheral component. More broadly, this principle could be useful for the regulation of number of phase-separated organelles in synthetic biology systems (43, 44).
Methods
Cloning.
The SAGA1 (Cre11.g467712) open reading frame (ORF) was assembled from 3 synthesized fragments, the first 2 containing introns and the third lacking introns. Twenty-one introns in total were included in the final sequence. Next, this assembled SAGA1 ORF was cloned into pRAM118 (GenBank accession no. MK357711). This cassette is identical to pLM005 (14), but the AphIII cassette for paromomycin resistance cassette is replaced with an AphII cassette for hygromycin resistance. This allows for transformation of the saga1 mutant.
Strains and Culture Conditions.
Wild-type C. reinhardtii cMJ030 was used for all experiments. The saga1 and saga1-2 mutants were isolated from a collection of high-CO2-requiring mutants generated by transformation of the pMJ016c mutagenesis cassette into cMJ030 (45). These mutants can be found at the Chlamydomonas Resource Center (saga1: CC5420; saga1-2: CC5421). The saga1-3 mutant was obtained from the CLiP collection (46) and can be found at the Chlamydomonas Resource Center (ID: LMJ.RY0402.060639). The saga1;SAGA1-Venus strain (CC5422) was generated by transformation of pRAM118 containing a Venus-tagged SAGA1 into the saga1 mutant. The saga1;RbcS1-mCherry strain (CC5423) was generated by transforming saga1 with a pLM006 construct containing Rbcs1 (14). Cells were grown to a concentration ∼2 × 106 cells per mL prior to experiments. All experiments were performed under photoautotrophic conditions in Tris-phosphate medium (47) with high CO2 (3% [vol/vol] or 5% [vol/vol] CO2-enriched air) or low CO2 (air, ∼0.04% [vol/vol] CO2).
Spot Tests.
Cells were pregrown in liquid Tris-acetate-phosphate medium (47). The cells were washed with Tris-phosphate medium and resuspended to a concentration of 6 × 105 cells per mL. Cells were then diluted by factors of 1, 10, 100, and 1,000. Fifteen microliters were spotted onto solid Tris-phosphate plates. The cells were grown under high- and low-CO2 concentrations for 7 d with 500 µmol photons m−2⋅s−1.
O2 Evolution.
The apparent affinity for Ci was determined using the oxygen evolution method as previously described in ref. 14 using an OXYVY1 Hansatech Oxyview System (Hansatech Instruments).
Microscopy.
Cell samples used for transmission electron microscopy (TEM) were fixed with 2% (vol/vol) glutaraldehyde and embedded in epoxy resin mix. TEM was performed using a Tecnai G2 80- to 200-kV transmission electron microscope (FEI Company) and imaged with AMT Image Capture Engine software (Advanced Microscopy Techniques). Measurement of the section area and perimeter of the cells, pyrenoids, and the surrounding starch sheath was performed using ImageJ2 (Fiji) (48–50).
Fluorescence microscopy on wild type, saga1, and saga1;SAGA1-Venus to observe the localization of SAGA1 was performed using Leica TCS SP5 confocal microscope (Leica Microsystems) and imaged with the Leica Application Suite Advanced Fluorescence software (Leica Microsystems). Fluorescence microscopy for saga1;RbcS1-mCherry was performed with a spinning-disk confocal microscope (3i custom-adapted Leica DMI6000). Samples were mounted on poly-l-lysine–coated plates (Ibidi) and covered with low-melting-temperature agarose (Invitrogen) to prevent desiccation during imaging.
The following excitation and emission settings were used: Venus, 514 nm excitation with 543/22 nm emission; mCherry, 561 nm excitation with 590/20 nm emission; and chlorophyll, 561 nm emission with 685/40 nm emission.
Analysis of Colocalization of SAGA1-Venus Fluorescence and Chlorophyll Autofluorescence.
Pyrenoids were visually demarcated using SAGA1-Venus fluorescence and intensity values of each pixel in the demarcated pyrenoid were extracted in the chlorophyll and Venus channel. The Spearman’s rank correlation coefficient was calculated between the chlorophyll and Venus channel for every pixel.
In order to compare to the expected Spearman’s rank coefficient that we would observe by chance, we independently scrambled the pixel intensity values of the chlorophyll and Venus channels in 100-pixel blocks and then calculated the Spearman’s rank coefficient. This was repeated 100 times to create a distribution of rho values specific to each image. The observed rho value was compared to this distribution to determine a P value. We performed the same analysis on a nonpyrenoid region of the chloroplast to control for bleed-through of the Venus channel into the chlorophyll channel. All rho values and P values are found in SI Appendix, Table S5.
Transformation of Chlamydomonas.
Constructs were transformed into the nuclear genome of saga1 mutant by electroporation as described previously (45). Transformants were first screened for hygromycin resistance then screened for Venus- and mCherry-expressing colonies using a Tecan Infinite M1000 PRO plate reader (Tecan Austria GmbH). Excitation and emission settings were as follows: Venus, 532-nm excitation with 555/20 emission; mCherry, 532-nm excitation with 610/30 nm emission; and chlorophyll, 633-nm excitation with 670/30 nm emission.
Western Blot Analysis.
Protein levels of SAGA1, Rubisco, alpha-tubulin, and histone H3 in wild type, saga1, and saga1;SAGA1-Venus were measured as described previously (51).
Analysis of Protein Interactions Using Yeast 2-Hybrid.
The 2-hybrid vectors pGBKT7 and pGADT7 were used to detect interactions between proteins of interest. Genes were amplified and cloned into each vector to create fusions with either the GAL4 DNA binding or activation domain, respectively. Yeast cells were then cotransformed with binding and activation domain vectors. Successful transformants were grown in liquid, harvested, diluted to an optical density at 600 nm (OD600) of 0.5 or 0.1, and plated onto SD-L-W and SD-L-W-H containing increasing concentrations of the HIS3 inhibitor triaminotriazole (3-AT) and incubated for 3 d before assessing growth. Protein extraction for Western blots was carried out by resuspending cells to an OD600 of 1 from an overnight liquid culture in a lysis buffer (50 mM Tris⋅HCl [pH 6], 4% [vol/vol] SDS, 8 M urea, 30% [vol/vol] glycerol, 0.1 M dithiothreitol, and 0.005% [wt/vol] bromophenol blue), incubating 65 °C for 30 min, and loading directly onto a 10% Bis-Tris protein gel (Expedeon). Proteins were transferred to a nitrocellulose membrane then probed with anti-SAGA (1:2,000) or anti-HA (1:5,000) primary antibody and HRP-linked goat anti-rabbit secondary antibody (1:10,000).
More detailed information on the materials and methods used in this study is provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank F. Fauser, R. Jinkerson, and J. Vilarrasa-Blasi for sharing phenotypic data for CLiP mutants; L. C. Mackinder and M. T. Meyer for advice and method support; J. N. Skepper, L. Carter, and the Cambridge Advanced Imaging Centre for advice and sample preparation for transmission electron microscopy; S. Ramundo for the pRAM118 vector; H. Cartwright, K. Geisler, and M. Szecówka for confocal microscopy support; and the H.G. and M.C.J. laboratories for helpful discussions. The project was funded by National Science Foundation (EF-1105617, IOS-1359682, and MCB-1146621); the National Institutes of Health (DP2-GM-119137); the Simons Foundation and Howard Hughes Medical Institute (55108535) grants to M.J.; and Biotechnology and Biological Sciences Research Council grants (BB/M007693/1) to H.G. and A.J.M. A.K.I. was supported by a National Institutes of Health Cell and Molecular Biology training grant (5T32GM 7276-42). K.X.C. was supported by the Cambridge-IDB International Scholarship, the Cambridge Philosophical Society, the Cambridge Trust, the Lundgren Fund, and the Houston Putnam Lowry Prize (Fitzwilliam College).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904587116/-/DCSupplemental.
References
- 1.Brangwynne C. P., et al. , Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Itakura A. K., Futia R. A., Jarosz D. F., It pays to be in phase. Biochemistry 57, 2520–2529 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeynaems S., et al. , Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman Rosenzweig E. S., et al. , The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer M. T., Whittaker C., Griffiths H., The algal pyrenoid: Key unanswered questions. J. Exp. Bot. 68, 3739–3749 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Kuchitsu K., Tsuzuki M., Miyachi S., Characterization of the pyrenoid isolated from unicellular green alga Chlamydomonas reinhardtii: Particulate form of RuBisCO protein. Protoplasma 144, 17–24 (1988). [Google Scholar]
- 7.Küken A., et al. , Effects of microcompartmentation on flux distribution and metabolic pools in Chlamydomonas reinhardtii chloroplasts. eLife 7, e37960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyachi S., Tsuzuki M., Maruyoma I., Gantar M., Miyachi S., Effects of CO2 concentration during growth on the intracellular structure of Chlorella and Scenedesmus (Chlorophyta). J. Phycol. 22, 313–320 (1986). [Google Scholar]
- 9.Meyer M., Griffiths H., Origins and diversity of eukaryotic CO2-concentrating mechanisms: Lessons for the future. J. Exp. Bot. 64, 769–786 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Borkhsenious O. N., Mason C. B., Moroney J. V., The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in chlamydomonas reinhardtii. Plant Physiol. 116, 1585–1591 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacoste-Royal G., Gibbs S. P., Immunocytochemical localization of ribulose-1,5-bisphosphate carboxylase in the pyrenoid and thylakoid region of the chloroplast of chlamydomonas reinhardtii. Plant Physiol. 83, 602–606 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morita E., Kuroiwa H., Kuoriwa T., Hisayoshi N., High localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in the pyrenoids of Chlamydomonas reinhardtii (Chlorophyta), as revealed by cryofixation and immunogold electron microscopy. J. Phycol. 33, 68–72 (1997). [Google Scholar]
- 13.Süss K.-H., Prokhorenko I., Adler K., In situ association of Calvin cycle enzymes, ribulose-1,5-bisphosphate carboxylase/oxygenase activase, ferredoxin-NADP+ reductase, and nitrite reductase with thylakoid and pyrenoid membranes of Chlamydomonas reinhardtii chloroplasts as revealed by immunoelectron microscopy.Plant Physiol. 107, 1387–1397 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackinder L. C. M., et al. , A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl. Acad. Sci. U.S.A. 113, 5958–5963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wunder T., Cheng S. L. H., Lai S.-K., Li H.-Y., Mueller-Cajar O., The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat. Commun. 9, 5076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raven J. A., CO2-concentrating mechanisms: A direct role for thylakoid lumen acidification? Plant Cell Environ. 20, 147–154 (1997). [Google Scholar]
- 17.Engel B. D., et al. , Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4, 1–29 (2015). Correction in: eLife4, e11383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pronina N. A., Avramova S., Georgiev D., Semenenko V. E., A pattern of carbonic anhydrase activity in Chlorella and Scenedesmus on cell adaptation to high light and low CO2 concentration. Sov. Plant Physiol. 28, 32–40 (1981). [Google Scholar]
- 19.Badger M. R., et al. , The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can. J. Bot. 76, 1052–1071 (1998). [Google Scholar]
- 20.Ramazanov Z., et al. , The induction of the CO2-concentrating mechanism is correlated with the formation of the starch sheath around the pyrenoid of Chlamydomonas reinhardtii. Planta 195, 210–216 (1994). [Google Scholar]
- 21.Goto M., Semimaru T., Furukawa K., Hayashida S., Analysis of the raw starch-binding domain by mutation of a glucoamylase from Aspergillus awamori var. kawachi expressed in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 60, 3926–3930 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Coutinho P. M., Nikolov Z., Ford C., Deletion analysis of the starch-binding domain of Aspergillus glucoamylase. Protein Eng. 8, 1049–1055 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Christiansen C., et al. , A CBM20 low-affinity starch-binding domain from glucan, water dikinase. FEBS Lett. 583, 1159–1163 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Yamano T., et al. , Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 51, 1453–1468 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Zhan Y., et al. , Pyrenoid functions revealed by proteomics in Chlamydomonas reinhardtii. PLoS One 13, e0185039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackinder L. C. M., et al. , A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlmann F., SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Matityahu A., Onn I., A new twist in the coil: Functions of the coiled-coil domain of structural maintenance of chromosome (SMC) proteins. Curr. Genet. 64, 109–116 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Ng J. D., et al. , The crystallization of biological macromolecules from precipitates: Evidence for Ostwald ripening. J. Cryst. Growth 168, 50–62 (1996). [Google Scholar]
- 30.Villarejo A., Martinez F., Plumed M. D. P., Ramazanov Z., The induction of the CO2 concentrating mechanism in a starch-less mutant of Chlamydomonas reinhardtii. Physiol. Plant. 98, 798–802 (1996). [Google Scholar]
- 31.Ma Y., Pollock S. V., Xiao Y., Cunnusamy K., Moroney J. V., Identification of a novel gene, CIA6, required for normal pyrenoid formation in Chlamydomonas reinhardtii. Plant Physiol. 156, 884–896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caspari O. D., et al. , Pyrenoid loss in Chlamydomonas reinhardtii causes limitations in CO2 supply, but not thylakoid operating efficiency. J. Exp. Bot. 68, 3903–3913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeeman S. C., Kossmann J., Smith A. M., Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Seung D., Schreier T. B., Bürgy L., Eicke S., Zeeman S. C., Two plastidial coiled-coil proteins are essential for normal starch granule initiation in Arabidopsis. Plant Cell 30, 1523–1542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Findinier J., et al. , Deletion of BSG1 in Chlamydomonas reinhardtii leads to abnormal starch granule size and morphology. Sci. Rep. 9, 1990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergeron-Sandoval L. P., Safaee N., Michnick S. W., Mechanisms and consequences of macromolecular phase separation. Cell 165, 1067–1079 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Updike D. L., Hachey S. J., Kreher J., Strome S., P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192, 939–948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei M.-T., et al. , Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 9, 1118–1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boke E., et al. , Amyloid-like self-assembly of a cellular compartment. Cell 166, 637–650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panas M. D., Ivanov P., Anderson P., Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lian X. J., Gallouzi I.-E., Oxidative stress increases the number of stress granules in senescent cells and triggers a rapid decrease in p21waf1/cip1 translation. J. Biol. Chem. 284, 8877–8887 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feric M., Brangwynne C. P., A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 15, 1253–1259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castellana M., et al. , Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol. 32, 1011–1018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuster B. S., et al. , Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 9, 2985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R., et al. , High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26, 1398–1409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., et al. , A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 51, 627–635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spreitzer R. J., Mets L., Photosynthesis-deficient mutants of chlamydomonas reinhardii with associated light-sensitive phenotypes. Plant Physiol. 67, 565–569 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rueden C. T., et al. , ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinnickel M. L., et al. , Novel thylakoid membrane greenCut protein CPLD38 impacts accumulation of the cytochrome b6f complex and associated regulatory processes. J. Biol. Chem. 288, 7024–7036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.