Abstract
DNA polymerases are critical tools in biotechnology, enabling efficient and accurate amplification of DNA templates, yet many desired functions are not readily available in natural DNA polymerases. New or improved functions can be engineered in DNA polymerases by mutagenesis or through the creation of protein chimeras. Engineering often necessitates the development of new techniques, such as selections in water-in-oil emulsions that connect genotype to phenotype and allow more flexibility in engineering than phage display. Engineering efforts have led to DNA polymerases that can withstand extreme conditions or the presence of inhibitors, as well as polymerases with the ability to copy modified DNA templates. This review discusses polymerases for biotechnology that have been reported, along with tools to enable further development.
Keywords: Processivity, Fidelity, DNA modification, DNA damage, DNA polymerase
DNA polymerases
DNA polymerases copy DNA generally with high accuracy and efficiency; therefore, they are critical tools for molecular biology and biotechnology [1, 2]. These intrinsic characteristics are exploited for biotechnology tools that can be used in DNA sequencing and DNA replication and amplification, topics which have been recently reviewed [3–5]. DNA polymerases can also be engineered to address various biotechnology problems, such as amplifying DNA from crude environmental samples, copying modified DNA from patients, or sequencing DNA in ancient samples. The characterization of specialized Y-family damage-bypass (translesion synthesis (TLS), see Glossary) DNA polymerases greatly expanded the potential uses of DNA polymerases [6]. These DNA polymerases can copy DNA containing some types of damage accurately, inserting the complementary nucleotide opposite damaged bases, thus potentially allowing faithful amplification of damaged DNA. Due to these properties, TLS polymerases have been utilized for the study and identification of the repair of damaged nucleotides [7]. In addition, the mutagenic activity of Y-family DNA polymerases on undamaged DNA potentially provides additional applications in random mutagenesis and directed evolution experiments. The range of naturally occurring activities and specificities as well as the potential to introduce altered activities or improved efficiencies makes DNA polymerases especially attractive for manipulation. Engineering new functions in replicative and TLS polymerases to act on modified DNA or under challenging conditions, as well as the methods used for their development, will be a focus of this review.
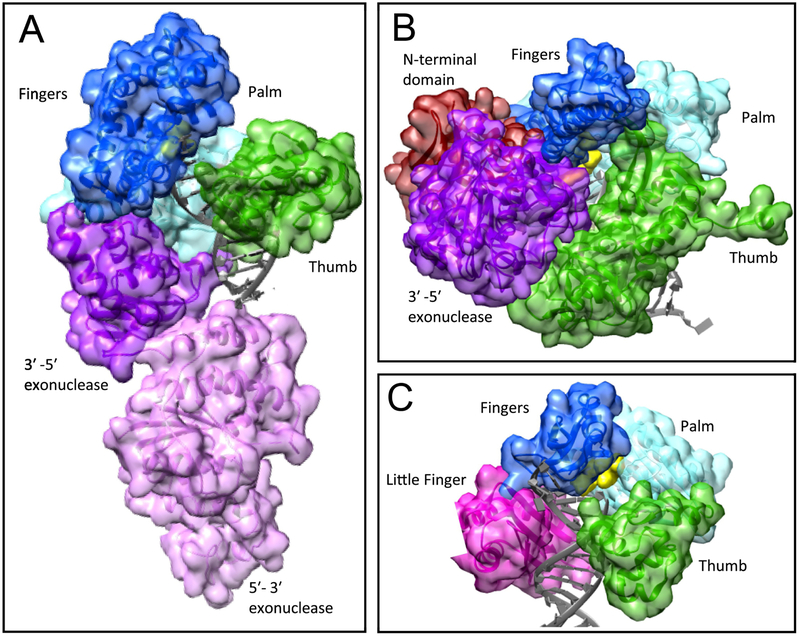
DNA polymerases are described as adopting a right-hand fold, in which the palm domain harbors the active site, the fingers domain binds the incoming nucleotide, and the thumb domain binds duplex DNA (Figure 1). Although there are eight different families of DNA polymerases (Table 1), they all catalyze DNA synthesis in a metal-dependent reaction in which the 3′-OH of the primer serves as the nucleophile that attacks the α-phosphate of the incoming nucleotide to form a new phosphodiester bond, releasing pyrophosphate (Figure 2A). DNA polymerases maintain fidelity via complementary base pairing of the incoming nucleotide with the nucleic acid template; most DNA polymerases exhibit high fidelity, although X- and Y-family DNA polymerases are notable exceptions. DNA processivity, which is the number of nucleotide additions to DNA in a single binding event of DNA polymerase, varies widely among DNA polymerases. Many polymerase engineering efforts have been directed toward developing polymerases that can accept modified DNA or incoming nucleotide substrates, have increased processivity, or have resistance to inhibitors. These types of modifications can extend the current capabilities of polymerases or expand them into new functional roles (summarized in Table 2).
FIGURE 1.
Structures of representative DNA polymerases with DNA in dark gray and the incoming nucleotide in yellow. Conserved polymerase domains are colored similarly in each structure: the thumb domain is in green, the palm domain is in cyan, and the fingers domain is in blue. A) A-family Thermus aquaticus polymerase I (PDB ID: 1TAQ), the 5′−3′ exonuclease domain is in light purple and the 3′−5′ exonuclease domain is in dark purple [79]. B) B-family Enterobacteria phage RB69 (PDB ID: 4FK4), the N-terminal domain is in maroon and the 3′−5′ exonuclease domain is in dark purple [80]. C) Y-family Escherichia coli polymerase IV (PDB ID: 4IRK), the little finger domain is in magenta [81].
Table 1.
Polymerase families and representativesa
| Family | E. coli | Human | Other |
|---|---|---|---|
| A | I | γ, ν, θ | Taq pol I (“Taq”), T7 DNA pol |
| B | II | α, δ, ε, ζ | RB69 |
| C | III | ||
| D | Archaeal replicative polymerase Dpol | ||
| X | β, λ, μ, TdT | ||
| Y | IV, V | ι, κ, η, Rev1 | |
| RT | Telomerase | ||
| AEP | PrimPol |
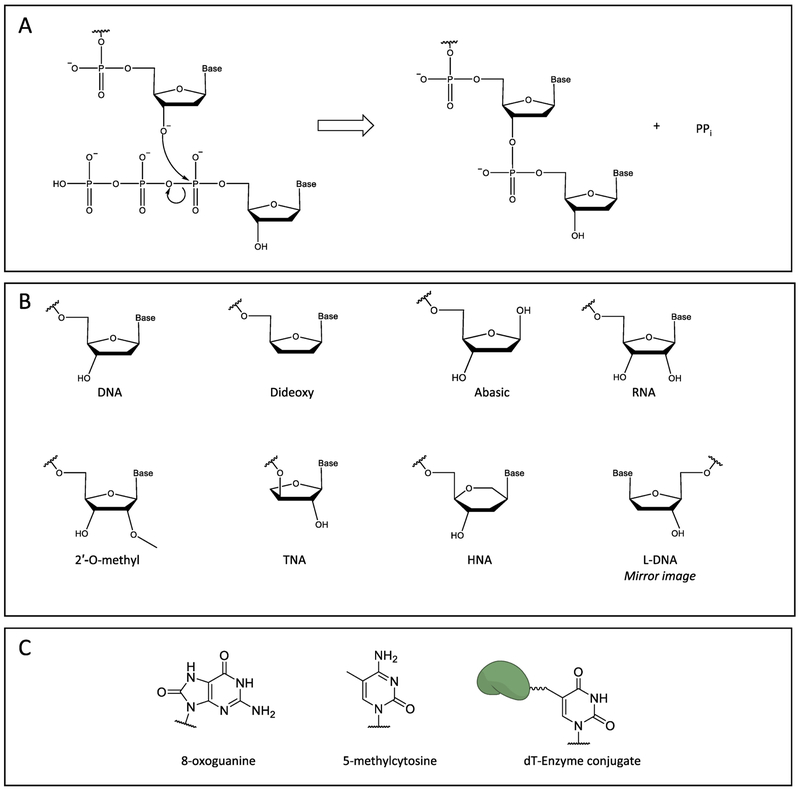
FIGURE 2:
Polymerization reaction and substrates. A) General polymerization reaction. B) Structures of nucleic acid sugars and modifications. C) Modified nucleobases.
Table 2.
Examples of engineered polymerases
| Polymerase | Application | Reference |
|---|---|---|
| Chimeras | ||
| BR3 Pol and KOD Pol chimeras | Activity in high salt and harsh environments | [8] |
| Taq Polymerase | Used to understand structure-function | [11] |
| Sulfolobus acidocaldarius Dbh | Increased processivity | [32] |
| RB69 and Human Cytomegalovirus Polymerase | Better model of viral polymerase for drug screening | [34] |
| Geobacillus sp. 777 Pol I | Use in harsh conditions | [37] |
| Taq and Pfu DNA polymerases | Use in harsh conditions | [38] |
| Thermophilic Y-fam pols | PCR of damage-containing samples | [46] |
| T7 RNA Polymerase | in vivo evolution | [74] |
| Chimeras combined with point mutations | ||
| Tth Pol I | Robust activity in CSR | [17] |
| A-fam Pols from Bst and Taq | Isothermal amplification; increased thermostability | [18] |
| Taq Pol I | Developing more efficient PCR | [26, 27] |
| Thermus sp. | Use in harsh conditions | [39] |
| A-Fam Pols from Taq, Tth, and Tfl | Amplification of ancient DNA samples | [43] |
| Tth and Taq | Epigenetic analysis | [44, 45] |
| Escherichia coli DNA polymerase I | Identifying novel mutations that confer antibiotic resistance | [75] |
| Terminal deoxynucleotidyl transferase | Template-independent oligonucleotide synthesis | [64] |
| Point mutation variants | ||
| Phi 29 DNA Polymerase | Robust whole genome amplification | [16] |
| T7 DNA Polymerase | DNA sequencing | [20, 21] |
| E. coli and Taq Pol I | DNA sequencing | [22] |
| KlenTaq | Hot start PCR | [24, 25] |
| Bst YoU | Early and inexpensive infection detection | [28] |
| T4 DNA Polymerase | Increased processivity | [29] |
| Dpo4 | Increased processivity | [30] |
| Dpo4 | Use in directed evolution to increase library diversity | [47] |
| Taq | SNP detection | [56] |
| Thermococcus sp. 9° N | Epigenetics | [57] |
| Thermococcus sp. 9° N | Synthetic biology | [67] |
| Tgo polymerase | Synthetic biology | [68] |
| Thermococcus sp. 9° N | Synthetic biology | [19] |
| KlenTaq | Forensic and ancient DNA amplification | [42] |
| Foot-and-mouth disease virus polymerase | Attenuated viruses for vaccines | [33] |
| Taq Polymerase | Amplification of crude DNA samples | [40, 41] |
| KOD exo- and KlenTaq | Epigenetics | [51] |
| Human PrimPol | Reverse transcription | [50] |
| KlenTaq LSIM variant | Identification of 2′-O-methylation sites | [52] |
| Tko exo- | Detecting 5-mC without mismatches | [55] |
| RT-KlenTaq | Epigenetics | [53] |
Polymerase engineering methods
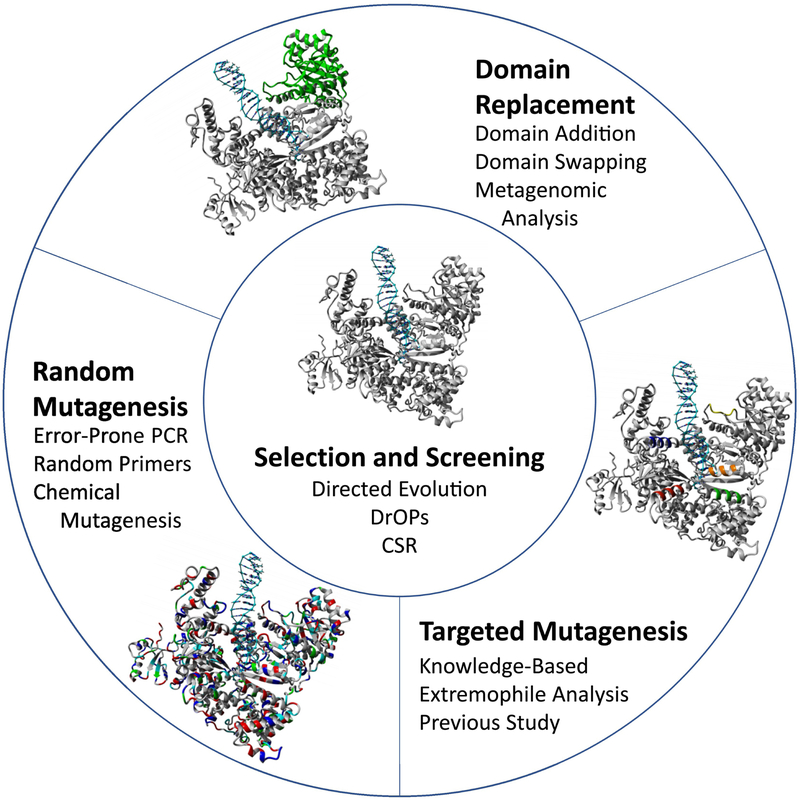
Engineering polymerases borrows many techniques used to engineer other enzymes, such as rational design based on information transfer or computational analysis, random mutagenesis and selection, or a combination of approaches (Figure 3). For example, analyzing a polymerase from an organism dwelling deep in the Red Sea allowed the creation of a halophilic polymerase [8]. Understanding the different structural and electrostatic features that allow viability in high temperatures and high salt made engineering salt tolerance possible. Understanding the roles of different residues in the bypass of abasic sites informed the study of incorporating unnatural amino acids to further dissect abasic site bypass [9]. The variant polymerases had reduced bypass activity due to variations in pKa of the modified tyrosine. Unnatural amino acids were used in this study to understand the bypass process, but they may be used in the future to engineer beneficial changes in polymerases [10]. Domain replacement is another route for engineering; for example, a chimeric Thermus aquaticus (Taq) polymerase with the proofreading domain from Escherichia coli pol I conferred exonuclease proofreading activity on Taq polymerase [11].
FIGURE 3:
Polymerase engineering strategies. In order to generate polymerases with new or altered activity, the addition of new residues or mutations at residues already present are usually made, candidates for which can be identified from a variety of sources. The replacement of full domains is one engineering route that relies on the swapping or addition of domains from other enzymes to incorporate new activity or function. Like domain swapping, many protein engineering methods involve rational design based on information-transfer methods from comparison to other proteins or computational analysis, random mutagenesis followed by selection, or a combination of the two approaches. Targeted mutations are often knowledge-based and rely on previous studies for an understanding on how to achieve a desired effect. Random mutagenesis relies on making mutations in a less targeted fashion, often through error-prone PCR or use of random primers. Due to the large sequence space of enzymes, often large numbers of variants must be screened or selected to identify the desired functionality. Methods such as directed evolution and compartmentalized-self replication have been developed and used due to their increased sampling abilities.
Directed evolution is a powerful technique in the development of new enzymes, including polymerases [12]. Phage display has produced several engineered polymerases, even though the sequence space accessible to sampling with this method may be somewhat limited [13, 14]. A method more specific to the evolution of polymerases is compartmentalized self-replication (CSR), whereby single polymerase clones are captured in emulsions [4, 15]. Polymerase chain reaction (PCR) occurs in the emulsion, which allows the next generation of clones to be created and expressed for another round of evolution. CSR has been utilized as a mechanism to produce polymerases with improved performance for further use in the CSR method itself [16–18].
Polymerase engineering methods are constantly evolving, in some cases becoming more targeted through increased knowledge and in others enabling screening of larger libraries through better technologies. One effort was the development of droplet-based optical polymerase sorting (DrOPS), in which single cells each carrying a single polymerase clone are encapsulated in droplets along with reagents to assay activity [19]. The system greatly reduces the amount of material used, which is especially important for reagents such as synthetic dTNPs that are challenging to produce in large quantities. The information from these large screening experiments may be utilized to inform further polymerase engineering, potentially aiding the rational design of new functions.
Engineering improved activity and robustness in DNA Polymerases
One of the earliest and most impactful examples of polymerase engineering was the development of T7 DNA polymerase as Sequenase and related enzymes for DNA sequencing. Engineering T7 DNA polymerase began with the recognition that eliminating its proofreading function allowed for more stable incorporation of chain-terminating dideoxy nucleotides (Figure 2B) [20] and the subsequent finding that pyrophosphorolysis, or addition of pyrophosphate to the last phosphodiester bond formed in the reverse of the polymerization reaction, reduces the efficiency of chain termination [21]. The latter problem was solved by adding pyrophosphatase to the sequencing reaction; manganese was also added to reduce discrimination between dideoxynucleotides and deoxynucleotides for more uniform sequencing results [21]. The single residue Y526 in T7 DNA pol is responsible for dNTP discrimination; introducing an analogous tyrosine in E. coli or Taq pol I decreased dNTP discrimination, allowing engineering of DNA pols, especially in DNA sequencing applications [22].
Although there are numerous thermostable DNA polymerases now in use in PCR applications, they are not without drawbacks, such as mispriming at nonspecific sites. This is particularly notable at ambient temperature, which can lead to the introduction of artifacts during reaction setup. Most commercial so-called hot start PCR products rely on reversible, thermolabile inhibition of the DNA polymerase, including by the addition of anti-polymerase antibodies or chemically-modified DNA primers or dNTPs [23]. Another solution to this problem is to identify cold-sensitive thermostable DNA polymerases that are inactive at ambient temperature. A screen of a library of KlenTaq variants identified mutations in the fingers domain that conferred cold sensitivity, with poor polymerase activity at 37 °C or 42 °C, while maintaining activity at 68 °C or 74 °C [24]. One variant identified from the screen was I707L, a mutation of an amino acid residue located ~20 Å from the active site. A crystal structure of the variant in complex with DNA and an incoming nucleotide reveals an overall similar structure as that of wild-type KlenTaq, with only minor local perturbations, indicating that the variant should retain activity [25]. However, the same variant in complex with only DNA shows an altered conformation of F749, leading to a reorganization of the fingers domain and opening of the active site. This opening allows two adenine residues from the template strand to block the active site and decrease activity; it is suggested that at low temperatures the polymerase can be trapped in this blocked state more easily than at higher temperatures [25].
Activity
Increasing the general performance of polymerases expands their use in areas such as PCR, CSR, and diagnostics. Metagenomic analysis of different soils has been a source of DNA sequences from unculturable organisms that may have useful, unusual activities [26, 27]. Chimeric proteins consisting of Taq Pol I and these gene fragments produced higher activity polymerases [26, 27]. Specific mutations were also created in the Taq fingers domain based on these chimeric proteins, producing variant Taq polymerases with better PCR performance [26]. Beyond PCR, activity of DNA polymerases can be altered for other clinical uses. Targeting conserved active site residues for mutation results in increased performance of Bst Pol I, which is used commercially for detection of the hand, foot, and mouth disease (HFMD) virus [28]. This variant has improved performance relative to commercial kits for the detection of HFMD and could be used in under-resourced areas at a lower cost than the commercial kit.
Processivity
To carry out their reactions, polymerases must translocate along a template strand and insert nucleotides sequentially, making processivity a major determinant of overall activity. Targeted mutagenesis can be used to increase DNA binding strength of the polymerase, in turn increasing processivity [29, 30]. The T4 DNA polymerase L412M motif A mutation produced a variant with increased processivity [29], which was extended to related polymerases yeast DNA pol δ and ε by making L-to-M mutations at analogous sites, showing the power of targeting conserved regions [29]. Targeting non-conserved residues can also lead to positive outcomes, as mutations in non-conserved regions of the thumb and fingers domains of Sulfolobus solfataricus Dpo4 increased processivity [30]. Besides mutagenesis, fusion of DNA binding domains can increase the processivity of polymerases [31, 32]. A multi-pronged approach, including fusion of a DNA-binding domain to Sulfolobus acidocaldarius Dbh, and bioinformatics and molecular simulation to identify mutations that increase DNA-binding affinity, led to increased processivity and DNA binding strength of this member of the typically poorly-processive Y-family [32].
Fidelity
The fidelity of a polymerase is an indication of the accuracy with which it inserts bases during polymerization. The fidelity of the Foot-and-Mouth Disease Virus polymerase was altered by mutation of Motif A residue W237, identified by crystal structure analysis [33]. Variants with both increased (W237F) and decreased (W237I and W237L) fidelity produced attenuated viruses suitable for vaccines. Altering the fidelity of a polymerase can also lead to better disease models, specifically human cytomegalovirus, by more accurately mimicking the behavior of a known viral polymerase [34]. Chimeras of RB69 and Human Cytomegalovirus polymerase were generated and their activity with known nucleotide analog inhibitors was determined, enabling the use of the engineered polymerase as a model of the viral polymerase that allows for screening of new drugs [34].
Robustness
DNA polymerases are commonly used in PCR and DNA sequencing of environmental samples, which can be mixed with a range of contaminants that inhibit DNA polymerases and other processing reactions. Thus, considerable effort has been invested in developing DNA polymerases that resist common inhibitors. Crime scene samples commonly contain contaminants like explosive substances, dyes, melanin, and other compounds that are inhibitors in PCR. These samples may be diluted or undergo DNA purification methods to counteract these inhibitory effects. Another option is the addition of DNA repair glycosylases and endonucleases, which excise some damage, allowing polymerases to act on the DNA. This workflow has been employed in challenging samples, such as in identifying the biological sex of a 4,000-year-old mummy [35]. In some cases, however, these methods result in DNA degradation or loss. Existing DNA polymerases were tested in a number of environments to simulate crime scene samples, including with ammonium nitrate or buried bone [36]. Other harsh conditions like high salt could interfere with polymerases binding DNA but can be overcome by fusing a sequence-nonspecific DNA binding motif such as a helix-hairpin-helix domain to polymerases or by creating chimeras of polymerases and DNA-binding domains of other proteins [37, 38]. Another method for engineering polymerases that can act under harsh conditions is to create an environment that has a specific selective pressure, such as using CSR to yield variant polymerases that can perform isothermal genome amplification, negating the need for thermophilic polymerases [16]. Engineering with a broad range of inhibitors led to the development of chimeras of different polymerases that resist humic acid, bone dust, and other contaminants [16, 39]. Variants of Taq polymerase have also been shown to perform amplification in crude DNA samples in the absence or presence of PCR enhancers, including detergents and osmoprotectants [40, 41].
DNA is susceptible to damage by natural processes and through processes involved in its analysis. This damage can lead to lesions that add bulk or obscure the identity of the DNA bases that then perturb polymerase activity. Variants of KlenTaq, obtained from error-prone PCR, were screened for high activity and selectivity, and one variant with the mutation M747K was found to bypass abasic sites, 8-oxoguanine, and UV-induced lesions (Figure 2C) more efficiently than the parental enzyme [42]. Chimeras were created through combinations of three different A-family polymerases from Thermus aquaticus, Thermus thermophilus, and Thermus flavus, and a mixture of the chimeras along with a parental DNA polymerase were more active on ancient DNA samples containing diverse DNA lesions than the parental polymerase alone [43]. Ancient DNA accrues many types of damage over time, so these polymerases were capable of amplifying DNA containing multiple mismatches, abasic sites, and other damage commonly found in these samples. Addition of bisulfite is a common treatment in epigenetics but can lead to sample loss and damage to DNA, prompting the development of a DNA polymerase that can replicate and amplify bisulfite-treated DNA [44, 45].
Harnessing the power of Y-family polymerases
Taq polymerase is commonly used in PCR methods because of its ability to perform at the high temperatures required during the reaction. However, Taq is not always able to amplify damaged DNA samples. One way to overcome this limitation is to engineer Taq or to use a polymerase that is not similarly limited, such as the Y-family polymerases. Y-family polymerases possess the ability to bypass non-canonical structures of DNA and are therefore attractive candidates to engineer for biotechnical applications. The Y-family polymerase Dpo4 is an appealing polymerase for engineering because of its ability to perform at high temperatures, like Taq. Unlike Taq, however, Dpo4 can act on DNA templates that contain certain types of damage. Seven novel thermostable Dpo4 orthologs, five from thermophilic organisms and two enhanced chimeras, were identified with properties similar to Taq that made them suitable for PCR, such as moderate processivity [46]. However, these Y-family polymerases can bypass damaged DNA that would block Taq polymerase. Combinations of Y-family polymerases and Taq were tested in PCR and had increased ability to amplify damaged DNA [46]. Directed evolution of Dpo4 identified a mutation, R336N, in the Y-family-specific little finger domain (Figure 1C) that increased the introduction of transversion mutations [47]. This variant can be utilized further in directed evolution attempts to increase library diversity [47]. Three residues in E. coli DNA polymerase V, another Y-family polymerase, were found to be important in DNA damage recognition [48]. The identification of such residues can be important in designing and tailoring these enzymes to accommodate other types of damage, even non-cognate damage, which could have applications in specific detection of DNA damage [46, 48].
Reporting naturally modified DNA
While DNA polymerases typically insert nucleotides using a DNA template, they can be modified to polymerize using RNA or modified DNA as templates. Natural reverse transcriptase polymerases (RT-Pols) can convert RNA to DNA, but they often have low stability or poor fidelity [49]. While human PrimPol can both initiate synthesis and elongate with dNTPs using DNA templates, an identified variant with a mutation in a conserved motif has improved activity with an RNA template [50].
A common naturally occurring modification of nucleotides is methylation as an epigenetic marker. Identifying and reporting these sites can be done by developing polymerases that can discriminate between natural nucleotides and their methylated counterparts [51–55]. One method to discriminate between modified and unmodified bases is to engineer polymerases that stall on the modification, leading to less product DNA or delays in replication. Coupled with qRT-PCR, the slower replication of DNA identifies the 2′-O-methylation site on RNA [52]. The ability of a polymerase to extend past mismatches can also be exploited for identifying methylation sites if extension capability is increased when the mismatch occurs at a methylation site [51].
This mismatch strategy was also used to detect specific single nucleotide polymorphisms (SNPs) [56, 57]. Variants of KlenTaq were generated with mutations at the basic residues of the active site that resulted in varying extension activities opposite 5-methylcytosine mismatches and also against specific allele sequences. Engineering efforts were made towards the detection of 5-methylcytosine by altering inherent activity with template 5mC relative to C, which negated the need for mismatched primers [55]. Another route is to alter the fidelity rather than activity at the methylated template base [53]. Mutations were made in the active site of the polymerase near the nascent base pair and screening in a parallel fluorescence assay identified a variant with increased misincorporation opposite methylation sites, leaving an identifiable pattern for next-gen sequencing. Similarly, differential polymerase activity was used to identify a mutation that is a biomarker for non-small cell lung cancer [58]. Identifying methylation sites and SNPs is clinically relevant and can be enabled by harnessing or modifying DNA polymerase activity.
Compatibility with unnatural or non-native substrates
While polymerases typically incorporate natural nucleotides, polymerase variants have been shown to incorporate non-natural nucleotides like modified dNTPs or even ribonucleotides using DNA templates [59, 60]. These variant polymerases can be used to build polymers of non-native nucleotides into macromolecules, such as aptamers that can bind to a specific target within biological systems [60]. The degradation by nucleases of these aptamers built with non-natural nucleotides is minimized due to their non-native structural features [61, 62]. These aptamers can act as riboswitches or markers, which can be used as therapeutic tools to modulate protein concentrations. Oligonucleotides containing non-natural nucleic acids that have high binding affinity to ochratoxin A showed stronger binding than natural DNA aptamers, making them suitable for poison treatment [63]. Variant polymerases have been used to synthesize oligonucleotides in a template-independent manner through the incorporation of a dNTP conjugated to a deoxynucleotidyl transferase [64]. Engineered polymerases have also been used to incorporate dNTPs linked to larger enzymes like glycoprotein horseradish peroxidase that has been used to visualize single-stranded DNA by the naked eye (Figure 2C) [65].
Synthetic biology
Synthetic biology creates artificial systems with some typical features of life, such as information transfer, a hallmark of DNA. Xeno-nucleic acids (XNA) offer such a possibility, but due to the different structural nature of the nucleotides, natural polymerases are often not capable of utilizing these substrates. Therefore, engineering efforts have been focused on the creation of polymerases with significant activity on a variety of XNAs [66]. New microfluidic technology allowed the high-throughput evaluation of a large number of variant polymerases for activity towards threose nucleic acids (TNA) [19]. While TNA utilizes the same bases as DNA, the sugars are linked to the extended chain at the 2′ and 3′ positions, rather than the natural 3′ and 5′ position (Figure 2B). These polymerases were able to extend XNA opposite DNA without the need for additional metals like manganese, which generally decrease fidelity, and thus allow synthetic nucleotide incorporation.
Another engineering strategy to increase polymerase activity with TNA was developed that relies on sampling scaffolds of homologous polymerases [67]. Gain-of-function mutations accumulated in one polymerase were predicted similarly to increase the activity of homologous polymerases. These positions were called “specificity determining residues,” and a number of these positions had been mutated in previous engineering efforts [67]. Targeting these positions in a number of polymerases resulted in variants with increased capacity to incorporate TNAs. Another significant need in synthetic biology is the ability to further replicate the synthetic nucleotide strands. Targeted mutagenesis to residues near the nascent strand and screening of Thermococcus gorgonarius polymerase variants identified variants with activity on 1,5-anhydrohexitol nucleic acids (HNAs) (Figure 2B) [68]. These polymerases were capable of transferring genetic information from DNA to HNA, as well as from HNA back to DNA, fulfilling the requirements for both storage and transfer of genetic information. Aside from their interaction with polymerases, nucleic acids interact with many other biomolecules. How xeno-nucleic acids interact with these other biomolecules will need to be studied, and possibly be modified, for further use in synthetic biology.
Designing mirror-image enzymes that produce mirror-image molecules offers a strategy to evade proteases and other defenses while maintaining biorthogonality [69]. In 2016, a small d-amino acid polymerase, African swine fever virus polymerase X, was chemically synthesized and was active. This synthesis demonstrated the feasibility of producing mirror-image enzymes capable of producing mirror-image DNA [70]. This group then chemically synthesized the larger polymerase Dpo4, which was also active [71], leading the way to chemically synthesize the mirror image of this polymerase [72, 73]. These mirror-image polymerases show specificity for L-nucleotides, which can be used to create mirror-image molecules like aptamers that offer more stability in biological systems because they can evade nucleases. However, design of these mirror-image biomolecules must balance binding to biological targets while evading other biological mechanisms such as clearance.
Engineering for Engineering
Compartmentalized self-replication (CSR) is a common technique employed in the engineering of polymerases with optimized activity under specific conditions. However, there are some limitations inherent in Taq DNA polymerase-based CSR, such as decreased activity in the emulsion compartments, limited product amount, and long run times. An engineered polymerase was able to provide higher product yields within fewer cycles of emulsion PCR [17]. A variety of techniques were combined to achieve this success, including a domain swap by deleting the Thermus thermophilus polymerase I exonuclease domain and adding a DNA binding domain. The polymerase sequence was codon optimized for E. coli expression and specific mutations were incorporated that resulted in increased activity in the homologous Taq polymerase. Chimeric polymerases resulting from gene shuffling of A-family polymerases from Bst and Taq also resulted in an improved polymerase for CSR [18]. This work produced a chimeric polymerase with increased strand-displacement activity that is capable of isothermal amplification, eliminating the need for thermal cycling equipment.
Polymerases are also essential in many directed evolution schemes for enzymes. Both random and targeted mutations are beneficial when trying to develop or alter enzymatic activity. A chimera consisting of the T7 RNA polymerase and a cytidine deaminase was able to target mutagenesis to DNA downstream of the T7 promoter [74]. This system is especially useful for in vivo evolution schemes as mutations can be made with limited effects on non-target DNA. Another chimera of DNA polymerases and a CRISPR-guided nickase enabled mutagenesis of all nucleotides within a tunable window at specific loci [75]. A plasmid-polymerase combination was also developed, such that the polymerase acts solely on the associated plasmid [76]. A second pol-plasmid pair was identified that is distinct from the first example and can operate in parallel [76]. Variants of the polymerase were then produced with reduced fidelity, which resulted in mutations in the target plasmid but not in the host yeast cells. The ability to modify and produce mutations in only a specific location with engineered polymerases is an enabling technology for further in vivo evolution.
Concluding Remarks
DNA polymerases represent incredibly useful tools for biotechnology, diagnostics, and protein engineering. One of the most widely-used tools in molecular biology is PCR, which relies on inherent characteristics of polymerases. Enhancing their natural properties can expand upon already developed methods, such as faster PCR runtimes for time-sensitive applications or for new applications. Engineering methods have been developed to construct new DNA polymerases with unusual properties often not present in natural DNA polymerases, including the ability to use XNAs as substrates to generate clinically relevant aptamers. Aside from building unnatural oligonucleotides, polymerases have also been engineered to report the presence of difficult-to-detect epigenetic markers. While new areas continue to emerge, more work is needed to develop improved polymerases and discover their application as useful tools. The feasibility of producing mirror-image polymerases or incorporating unnatural amino acids has been shown, but their full potential still needs to be realized. Further developments in polymerase engineering, in both methods and products, will help enable new biotechnological applications (see Outstanding Questions).
Outstanding questions:
How much can natural polymerases be changed; how different can their properties become?
How can the information gained from directed evolution and other techniques be used for rational engineering?
What do xeno-nucleic acids mean for the transfer of genetic material and other biomolecules they could interact with?
How can mirror-image biomolecules find balance between biological relevance and specific bioorthogonality?
Highlights:
New engineering methods enable DNA polymerases to carry out unnatural or enhanced functions
Methods have been developed to sample extended sequence space
The expanding polymerase universe is keeping pace with the growing chemical diversity of modified nucleotides
Polymerases can be made more resilient toward harsh conditions
Acknowledgments
Research in the Beuning laboratory is supported by the National Institutes of Health [R01GM123239 to P.J.B.] and the National Science Foundation [MCB-1615946 to P.J.B.]. T.A.C. was supported by a National Institute of Justice pre-doctoral fellowship [DOJ 2015-R2-CX-0011]. Molecular graphics and analyses performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Glossary
- Aptamer
Oligonucleotides that bind a specific target molecule
- Chimera
A protein composed of sequences and/or domains from two or more distinct proteins
- Compartmentalized self-replication (CSR)
Evolutionary technique characterized by encapsulation of single polymerase clones and DNA
- Epigenetics
Hereditary information that is not a result of changes in DNA sequences
- Exonuclease
Enzymes or domains that remove nucleotides from the end of a nucleic acid strand
- Fidelity
A property of polymerases describing their accuracy in inserting the correct nucleotide to form Watson-Crick base pairs
- Halophilic
Organisms that live and survive in high salt concentrations
- Metagenome
Genomic DNA that is often retrieved from soil samples, rather than specific organisms
- Processivity
A property of polymerases referring to the number of nucleotides added to the nucleic acid strand in a single polymerase binding event
- Reverse transcriptase (RT)
A polymerase that uses an RNA template to create a new strand of DNA
- Riboswitch
RNA molecule that can form different conformations, often by binding a ligand, that regulate gene expression
- Single nucleotide polymorphism (SNP)
a single change in the DNA sequence at a specific position in the genome
- Translesion synthesis (damage bypass)
Process by which a polymerase replicates a damaged DNA template
- Xeno-nucleic acids (XNAs)
orthogonal replication systems
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson KA (2010) The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta. 1804, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raper AT, et al. (2018) Kinetic Mechanism of DNA Polymerases: Contributions of Conformational Dynamics and a Third Divalent Metal Ion. Chem Rev. 118, 6000–6025. [DOI] [PubMed] [Google Scholar]
- 3.Aschenbrenner J and Marx A (2017) DNA polymerases and biotechnological applications. Curr Opin Biotechnol. 48, 187–195. [DOI] [PubMed] [Google Scholar]
- 4.Houlihan G, et al. (2017) Exploring the Chemistry of Genetic Information Storage and Propagation through Polymerase Engineering. Acc Chem Res. 50, 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner AF and Kelman Z (2014) DNA polymerases in biotechnology. Frontiers in Microbiology. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaisman A and Woodgate R (2017) Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol. 52, 274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, et al. (2017) Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene. Proc Natl Acad Sci U S A. 114, 6752–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi M, et al. (2018) Dynamic structure mediates halophilic adaptation of a DNA polymerase from the deep-sea brines of the Red Sea. FASEB J. fj201700862RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blatter N, et al. (2014) Modulating the pKa of a tyrosine in KlenTaq DNA polymerase that is crucial for abasic site bypass by in vivo incorporation of a non-canonical amino acid. Chembiochem. 15, 1735–1737. [DOI] [PubMed] [Google Scholar]
- 10.Zou H, et al. (2018) Biosynthesis and biotechnological application of non-canonical amino acids: Complex and unclear. Biotechnol Adv. 36, 1917–1927. [DOI] [PubMed] [Google Scholar]
- 11.Villbrandt B, et al. (2000) Domain exchange: chimeras of Thermus aquaticus DNA polymerase, Escherichia coli DNA polymerase I and Thermotoga neapolitana DNA polymerase. Protein Eng. 13, 645–654. [DOI] [PubMed] [Google Scholar]
- 12.Chen T and Romesberg FE (2014) Directed polymerase evolution. FEBS Letters. 588, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vichier-Guerre S, et al. (2006) A population of thermostable reverse transcriptases evolved from Thermus aquaticus DNA polymerase I by phage display. Angew Chem Int Ed Engl. 45, 6133–6137. [DOI] [PubMed] [Google Scholar]
- 14.Leconte AM, et al. (2005) Polymerase evolution: efforts toward expansion of the genetic code. J Am Chem Soc. 127, 12470–12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghadessy FJ, et al. (2001) Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci U S A. 98, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Povilaitis T, et al. (2016) In vitro evolution of phi29 DNA polymerase using isothermal compartmentalized self replication technique. Protein Eng Des Sel. 29, 617–628. [DOI] [PubMed] [Google Scholar]
- 17.Aye SL, et al. (2018) Engineering of DNA polymerase I from Thermus thermophilus using compartmentalized self-replication. Biochem Biophys Res Commun. 499, 170–176. [DOI] [PubMed] [Google Scholar]
- 18.Milligan JN, et al. (2018) Evolution of a Thermophilic Strand-Displacing Polymerase Using High-Temperature Isothermal Compartmentalized Self-Replication. Biochemistry. 57, 4607–4619. [DOI] [PubMed] [Google Scholar]
- 19.Larsen AC, et al. (2016) A general strategy for expanding polymerase function by droplet microfluidics. Nat Commun. 7, 11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabor S and Richardson CC (1989) Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 264, 6447–6458. [PubMed] [Google Scholar]
- 21.Tabor S and Richardson CC (1990) DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Effect of pyrophosphorolysis and metal ions. J Biol Chem. 265, 8322–8328. [PubMed] [Google Scholar]
- 22.Tabor S and Richardson CC (1995) A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc Natl Acad Sci U S A. 92, 6339–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul N, et al. (2010) Hot start PCR. Methods Mol Biol. 630, 301–318. [DOI] [PubMed] [Google Scholar]
- 24.Kermekchiev MB, et al. (2003) Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR. Nucleic Acids Res. 31, 6139–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu EY, et al. (2015) A conservative isoleucine to leucine mutation causes major rearrangements and cold sensitivity in KlenTaq1 DNA polymerase. Biochemistry. 54, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamagami T, et al. (2014) Mutant Taq DNA polymerases with improved elongation ability as a useful reagent for genetic engineering. Front Microbiol. 5, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamagami T, et al. (2016) A longer finger-subdomain of family A DNA polymerases found by metagenomic analysis strengthens DNA binding and primer extension abilities. Gene. 576, 690–695. [DOI] [PubMed] [Google Scholar]
- 28.Ma Y, et al. (2016) Enhancement of Polymerase Activity of the Large Fragment in DNA Polymerase I from Geobacillus stearothermophilus by Site-Directed Mutagenesis at the Active Site. Biomed Res Int. 2016, 2906484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reha-Krantz LJ, et al. (2014) Engineering processive DNA polymerases with maximum benefit at minimum cost. Frontiers in Microbiology. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, et al. (2017) Increased Processivity, Misincorporation, and Nucleotide Incorporation Efficiency in Sulfolobus solfataricus Dpo4 Thumb Domain Mutants. Appl Environ Microbiol. 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spibida M, et al. (2018) Fusion of DNA-binding domain of Pyrococcus furiosus ligase with TaqStoffel DNA polymerase as a useful tool in PCR with difficult targets. Appl Microbiol Biotechnol. 102, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, et al. (2017) DNA binding strength increases the processivity and activity of a Y-Family DNA polymerase. Sci Rep. 7, 4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai DK, et al. (2017) Attenuation of Foot-and-Mouth Disease Virus by Engineered Viral Polymerase Fidelity. J Virol. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tchesnokov EP, et al. (2009) Engineering of a chimeric RB69 DNA polymerase sensitive to drugs targeting the cytomegalovirus enzyme. J Biol Chem. 284, 26439–26446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loreille O, et al. (2018) Biological Sexing of a 4000-Year-Old Egyptian Mummy Head to Assess the Potential of Nuclear DNA Recovery from the Most Damaged and Limited Forensic Specimens. Genes (Basel). 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson M, et al. (2016) Comparison of DNA polymerases for improved forensic analysis of challenging samples. Forensic Sci Int Genet. 24, 55–59. [DOI] [PubMed] [Google Scholar]
- 37.Oscorbin IP, et al. (2017) Derivatives of Bst-like Gss-polymerase with improved processivity and inhibitor tolerance. Nucleic Acids Res. 45, 9595–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavlov AR, et al. (2002) Helix-hairpin-helix motifs confer salt resistance and processivity on chimeric DNA polymerases. Proc Natl Acad Sci U S A. 99, 13510–13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baar C, et al. (2011) Molecular breeding of polymerases for resistance to environmental inhibitors. Nucleic Acids Res. 39, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kermekchiev MB, et al. (2009) Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 37, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, et al. (2010) Direct DNA amplification from crude clinical samples using a PCR enhancer cocktail and novel mutants of Taq. J Mol Diagn. 12, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gloeckner C, et al. (2007) Evolving a thermostable DNA polymerase that amplifies from highly damaged templates. Angew Chem Int Ed Engl. 46, 3115–3117. [DOI] [PubMed] [Google Scholar]
- 43.d’Abbadie M, et al. (2007) Molecular breeding of polymerases for amplification of ancient DNA. Nat Biotechnol. 25, 939–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millar D, et al. (2015) A polymerase engineered for bisulfite sequencing. Nucleic Acids Res. 43, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loakes D, et al. (2009) Evolving a polymerase for hydrophobic base analogues. J Am Chem Soc. 131, 14827–14837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonald JP, et al. (2006) Novel thermostable Y-family polymerases: applications for the PCR amplification of damaged or ancient DNAs. Nucleic Acids Res. 34, 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kardashliev T, et al. (2014) A high-throughput screening method to reengineer DNA polymerases for random mutagenesis. Mol Biotechnol. 56, 274–283. [DOI] [PubMed] [Google Scholar]
- 48.Hawver LA, et al. (2015) Point mutations in Escherichia coli DNA pol V that confer resistance to non-cognate DNA damage also alter protein-protein interactions. Mutat Res. 780, 1–14. [DOI] [PubMed] [Google Scholar]
- 49.Sano S, et al. (2012) Mutations to create thermostable reverse transcriptase with bacterial family A DNA polymerase from Thermotoga petrophila K4. Journal of Bioscience and Bioengineering. 113, 315–321. [DOI] [PubMed] [Google Scholar]
- 50.Agudo R, et al. (2017) Engineering human PrimPol into an efficient RNA-dependent-DNA primase/polymerase. Nucleic Acids Res. 45, 9046–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aschenbrenner J, et al. (2014) Direct sensing of 5-methylcytosine by polymerase chain reaction. Angew Chem Int Ed Engl. 53, 8154–8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aschenbrenner J and Marx A (2016) Direct and site-specific quantification of RNA 2’-O-methylation by PCR with an engineered DNA polymerase. Nucleic Acids Res. 44, 3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aschenbrenner J, et al. (2018) Engineering of a DNA Polymerase for Direct m(6) A Sequencing. Angew Chem Int Ed Engl. 57, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drum M, et al. (2014) Variants of a Thermus aquaticus DNA polymerase with increased selectivity for applications in allele- and methylation-specific amplification. PLoS One. 9, e96640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huber C, et al. (2016) 5-methylcytosine-sensitive variants of Thermococcus kodakaraensis DNA polymerase. Nucleic Acids Res. 44, 9881–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen F, et al. (2010) Reconstructed evolutionary adaptive paths give polymerases accepting reversible terminators for sequencing and SNP detection. Proc Natl Acad Sci U S A. 107, 1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staiger N and Marx A (2010) A DNA polymerase with increased reactivity for ribonucleotides and C5-modified deoxyribonucleotides. Chembiochem. 11, 1963–1966. [DOI] [PubMed] [Google Scholar]
- 58.Cui M, et al. (2017) Detection of single nucleotide polymorphism by measuring extension kinetics with T7 exonuclease mediated isothermal amplification. Analyst. 143, 116–122. [DOI] [PubMed] [Google Scholar]
- 59.Lone S and Romano LJ (2003) Mechanistic insights into replication across from bulky DNA adducts: a mutant polymerase I allows an N-acetyl-2-aminofluorene adduct to be accommodated during DNA synthesis. Biochemistry. 42, 3826–3834. [DOI] [PubMed] [Google Scholar]
- 60.Randrianjatovo-Gbalou I, et al. (2018) Enzymatic synthesis of random sequences of RNA and RNA analogues by DNA polymerase theta mutants for the generation of aptamer libraries. Nucleic Acids Res. 46, 6271–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen T, et al. (2016) Evolution of Thermophilic DNA Polymerases for the Recognition and Amplification of C2’-Modified DNA. Nature chemistry. 8, 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Z, et al. (2017) Evolved polymerases facilitate selection of fully 2’-OMe-modified aptamers. Chem Sci. 8, 8179–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rangel AE, et al. (2018) In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 46, 8057–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palluk S, et al. (2018) De novo DNA synthesis using polymerase-nucleotide conjugates. Nat Biotechnol. 36, 645–650. [DOI] [PubMed] [Google Scholar]
- 65.Welter M, et al. (2016) Sequence-Specific Incorporation of Enzyme-Nucleotide Chimera by DNA Polymerases. Angew Chem Int Ed Engl. 55, 10131–10135. [DOI] [PubMed] [Google Scholar]
- 66.Pinheiro VB (2018) Engineering-driven biological insights into DNA polymerase mechanism. Curr Opin Biotechnol. 60, 9–16. [DOI] [PubMed] [Google Scholar]
- 67.Dunn MR, et al. (2016) Improving Polymerase Activity with Unnatural Substrates by Sampling Mutations in Homologous Protein Architectures. ACS Chemical Biology. 11, 1210–1219. [DOI] [PubMed] [Google Scholar]
- 68.Pinheiro VB, et al. (2012) Synthetic genetic polymers capable of heredity and evolution. Science. 336, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peplow M (2016) Mirror-image enzyme copies looking-glass DNA. Nature. 533, 303–304. [DOI] [PubMed] [Google Scholar]
- 70.Wang Z, et al. (2016) A synthetic molecular system capable of mirror-image genetic replication and transcription. Nat Chem. 8, 698–704. [DOI] [PubMed] [Google Scholar]
- 71.Xu W, et al. (2017) Total chemical synthesis of a thermostable enzyme capable of polymerase chain reaction. Cell Discov. 3, 17008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang W, et al. (2017) Mirror-image polymerase chain reaction. Cell Discov. 3, 17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pech A, et al. (2017) A thermostable D-polymerase for mirror-image PCR. Nucleic Acids Res. 45, 3997–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore CL, et al. (2018) A Processive Protein Chimera Introduces Mutations across Defined DNA Regions In Vivo. J Am Chem Soc. 140, 11560–11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Halperin SO, et al. (2018) CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature. 560, 248–252. [DOI] [PubMed] [Google Scholar]
- 76.Arzumanyan GA, et al. (2018) Mutually Orthogonal DNA Replication Systems In Vivo. ACS Synth Biol. 7, 1722–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ito J and Braithwaite DK (1991) Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19, 4045–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jain R, et al. (2018) Eukaryotic DNA polymerases. Curr Opin Struct Biol. 53, 77–87. [DOI] [PubMed] [Google Scholar]
- 79.Kim Y, et al. (1995) Crystal structure of Thermus aquaticus DNA polymerase. Nature. 376, 612–616. [DOI] [PubMed] [Google Scholar]
- 80.Xia S, et al. (2013) DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J Am Chem Soc. 135, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma A, et al. (2013) A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli. Nucleic Acids Res. 41, 5104–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]