Abstract
microRNAs (miRNAs) are recently identified small RNA molecules that regulate gene expression and significantly influence the essential cellular processes associated with CNS repair after trauma and neuropathological conditions including stroke and neurodegenerative disorders.
A number of specific miRNAs are implicated in regulating the development and propagation of CNS injury, as well as its subsequent regeneration. The review focuses on the functions of the miRNAs and their role in brain recovery following CNS damage. The article introduces a brief description of miRNA biogenesis and mechanisms of miRNA-induced gene suppression, followed by an overview of miRNAs involved in the processes associated with CNS repair, including neuroprotection, neuronal plasticity and axonal regeneration, vascular reorganization, neuroinflammation, and endogenous stem cell activation. Specific emphasis is placed on the role of multifunctional miRNA miR-155, as it appears to be involved in multiple neurorestorative processes during different CNS pathologies. In association with our own studies on miR-155, I introduce a new and unexplored approach to cerebral regeneration: regulation of brain tissue repair through a direct modulation of specific miRNA activity. The review concludes with discussion on the challenges and the future potential of miRNA-based therapeutic approaches to CNS repair.
Keywords: microRNA, miR-155, neurorestoration, post-stroke inflammation, cerebral blood flow, functional recovery
Introduction
CNS regeneration after injury is extremely limited, and spontaneous functional recovery is primarily depended upon the intrinsic mechanisms involving neuroprotection, neurogenesis, structural remodeling of spared axons and dendrites, and consolidation of compensatory neuronal circuits within the damaged tissue. These mechanisms involve significant changes in gene and protein expression and activation of complex molecular pathways recruited by CNS for survival. Among the actively involved molecules recently identified are short RNAs called MicroRNAs (miRNAs). miRNAs, constituting a substantial class of small non-coding RNAs, are characterized by short length (~22 nucleotides) and an extensive potential to suppress protein-coding genes in eukaryotes [1–3]. The discovery of miRNAs, in 1993, introduced a new role of short non-coding RNAs as regulatory molecules and considerably redefined the understanding of post-transcriptional gene regulation [4,5]. After more than 20 years of the extensive investigation, it is now postulated that miRNAs, assembled in the regulatory protein complexes, recognize and bind the complementary sequences of the messenger RNA (mRNA) and alter protein translation and synthesis, as well trigger mRNA destabilization and degradation. Most mammalian genes and more than 60% of human protein-coding genes are believed to be controlled by miRNAs [6]. New animal and plant microRNAs have been identified, and a stunning number and variety of these molecules demonstrate their widespread, prevalent, and important regulatory role. Recently, in addition to 1,900 of the previously known sequences, 3,707 novel mature miRNAs have been identified in 13 different human tissue types [7]. A detailed atlas of miRNA distribution in different human tissues presented the expression profiles of at least 1,300 widely distributed and tissue-specific human miRNAs [8].
microRNA Biogenesis
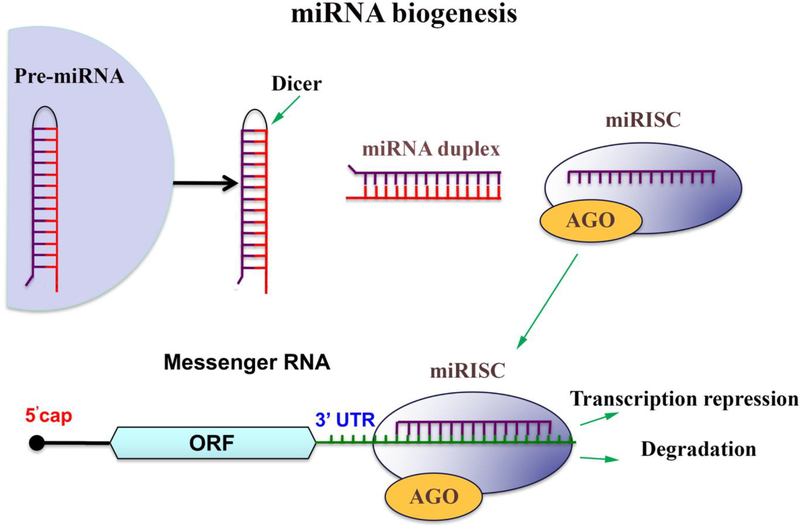
While initially discovered in the intergenic regions, miRNAs are also derived from intronic or exonic gene sequences. The microRNAs are transcribed in the nucleus, and their nuclear processing is regulated by RNAse III Drosha and its co-factor DGCR8/Pasha. The miRNA precursor is exported into the cytoplasm by Exportin 5, while further modification/maturation is mediated by an endoribonuclease Dicer. A small RNA duplex generated by Dicer associates with the Argonaut family proteins (AGO) which form a so-called RNA-induced silencing complex (RISC or miRISC) [9,10,2]. The unwinding of the AGO-loaded duplex and the generation of single-stranded mature miRNA are now believed to occur concomitantly with the RISC effector complex assembly [11,12]. The mechanism of RISC assembly and its exact composition are still being investigated. RISC is believed to be first assembled by AGO proteins, and to contain miRNA-loaded AGO bound to a glycine-tryptophan repeat-containing protein GW182. During the mRNA silencing, miRNA serves as a guide to target a complimentary mRNA, while miRNA-guided RISC effector protein complex mediates translational repression and/or mRNA degradation (Figure 1). The miRNA-mRNA targeting is governed by base pairing and occurs predominantly at the 3’ untranslated region (3’ UTR) of the target mRNA. A miRNA “seed region” (domain between the nucleotides 2 −7 at the 5ʹ end) is required for miRNA-mRNA interaction [1,13].
Figure 1: Canonical pathway of miRNA biogenesis.
The microRNAs are transcribed in the nucleus, the miRNA precursor is exported into the cytoplasm, and further modification/maturation is mediated by an endoribonuclease Dicer. A small RNA duplex generated by Dicer associates with the Argonaut family proteins (AGO) which form a so-called RNA-induced silencing complex (miRISC). RISC effector complex assembly involves the unwinding of the AGO-loaded duplex and generation of single-stranded mature miRNA. During the mRNA silencing, miRNA serves as a guide to target a complimentary mRNA, while miRNA-guided RISC effector protein complex mediates translational repression and/or mRNA degradation.
While the majority of miRNAs are generated via canonical pathway, several alternative biogenesis mechanisms are identified. Some unconventional miRNAs can be produced via non-canonical pathways through the nuclear microprocessing by Drosha or cytoplasmic processing by Dicer [14–16]. While the existence of non-canonical pathways exposes the complexity of miRNA biogenesis, the majority of functional miRNAs are generated via the conventional pathway. miRNA biogenesis and miRNA/RISC-induced silencing of the target mRNA is tightly regulated both on transcriptional and post-transcriptional levels. Among the regulators of miRNA functions are transcription factors such as MYC, p53, ZEB1 and ZEB2; epigenetic factors DNMT1 and DNMT2; RNA-binding proteins; SR proteins; heterogeneous nuclear ribonucleoproteins (hnRNPs); and other factors involved in RNA splicing [17].
miRNA biogenesis is described in detail in a number of reviews [1,9,18,13]. At present, miRNAs are regarded as “master regulators” of gene expression, and “grand managers” of numerous cellular processes, including cell growth, differentiation, maturation, proliferation, migration, interaction with other cells and extracellular components, metabolism, and apoptosis. Therefore, an increasing number of miRNAs are associated with different diseases and pathological conditions [19,1,20].
miRNAs and CNS damage
miRNAs have been found to play an important role during CNS development and they are abundantly expressed in adult human and rodent brain[21–23,7] and spinal cord [24,8]. CNS damage is accompanied by a significant dysregulation in miRNA expression profiles in the affected tissue, peripheral blood, and cerebrospinal fluid. Many research findings detected significant changes in microRNA profiles associated with TBI [25–28], stroke [29,30], epilepsy [31,32], Parkinson’s disease [33,34], Alzheimer’s disease [35,36], and spinal cord injury (SCI) [24,37]. These changes reflect concomitantly occurring injury and self-repair processes, which complicates the interpretation of the obtained data. Therefore, the most important and complicated task for understanding of the function of a specific miRNA is to distinguish whether these various and multiple changes in miRNA expression are harmful or beneficial for the regeneration and repair processes.
Recent findings have identified miRNAs’ novel role as the mediators of intracellular communication. Apart from direct intercellular interactions, crosstalk between neurons, astrocytes, microglia and endothelial cells involves indirect intercellular communication via small extracellular vesicles (EV) secreted by a cell and internalized by its neighboring cells. While various miRNAs can freely circulate in blood and cerebrospinal fluid, a large portion of them is transported as cargo within different types of lipid vesicles [38–40]. Along with proteins, lipids, and different RNA species, miRNAs represent a substantial component of the EV cargo molecules. Among the diverse secreted vesicle population are exosomes, small (30–200 nm) lipid bilayer enclosed vesicles, which recently have been implicated as the major mediators of intercellular microRNA delivery. Secretion of exosomes and their miRNA composition is significantly altered during CNS damage and repair processes [41,42].
miRNA functional analysis is performed to identify genes and processes regulated by specific miRNAs. These studies are based on gain- and loss-of-function experiments utilizing the inhibition or overexpression of miRNAs with specific synthetic inhibitor (antagomirs) or mimic (agomirs) oligonucleotides. While most of the miRNA research is based on in vitro experiments, systemic or local administration of the miRNA inhibitors and mimics are utilized to explore the miRNA function in various experimental animal models. In this review, we will focus on the microRNAs, which, based on the functional studies, are thought to have a profound positive or negative effect on regeneration in the animal models of CNS injury.
The processes and events accompanying CNS regeneration
The subacute phase of CNS injury is accompanied by the active spontaneous recovery process, triggering neuronal plasticity, axonal regeneration, post-injury angiogenesis and vasculogenesis, and neuroinflammation as an integral part of both damage and recovery processes. In addition, CNS damage results in a neurogenic response and a massive migration of neural progenitors into the lesion area, which may substantially contribute to recovery and repair processes [43,44]. All these events are associated with significant changes in gene and protein expression, and are thus broadly regulated by microRNAs. Some excellent reviews describe the involvement of miRNAs in acute and chronic CNS injury and neurodegenerative disorders, accompanied by neuronal and axonal injury, cell apoptosis, vascular damage, aberrant gliosis, and demyelination [45–47]. This review will focus mostly on regeneration, and will describe the miRNAs, which were experimentally proven to have a significant impact on each of the different components of recovery in the animal models of CNS injury.
Neuroprotection
Various microRNAs have been identified as critical regulators of cell survival following CNS injury. They control the levels of various target genes and signaling proteins, which are important for CNS tissue preservation and recovery. These microRNAs control signaling cascades involved in CNS cell survival and apoptosis; among them are components of hypoxia inducible factor −1α (HIF-1 α), mitogen activated protein (MAP) kinase, mammalian target of rapamycin (mTOR), transforming growth factor (TGF-β), Wnt, Notch, and p53 signaling pathways. Recently, the excellent reviews summarized the list of microRNAs, which are up- or down-regulated during the repair process and exerting either protective or damaging effects [48–50]. Based on the recent findings, Table 1 demonstrates some of the regulatory microRNAs and the associated experimentally verified direct target proteins underlying miRNA-based neuroprotection in animal models of CNS injury.
Table 1: In vivo inhibition or overexpression of specific microRNAs supports neuroprotection in the animal models of CNS injury.
Table 1 summarizes the list of regulatory microRNAs and their targets, which, after the experimental intervention using miRNA inhibitors and mimics, support neuronal survival and facilitate recovery in the animal models of CNS injury. The process is mediated via the experimentally verified direct target genes/proteins.
| miRNA | Animal model of CNS injury | Treatment | Involved direct miRNA targets |
|---|---|---|---|
| miR-210 | Stroke | Overexpression | mBDNF/proBDNF [51] |
| miR-124 | Stroke | Overexpression | Bcl-2 and Bcl-xl [52]; Usp14 [53] |
| miR-216a | Stroke | Overexpression | JAK2 [54] |
| miR-29b | Stroke | Overexpression | Aquaporin-4 [55] |
| miR-29c | Stroke | Overexpression | Birc2 [56] |
| miR-128–3p | Stroke | Overexpression | p38α [57] |
| miR-93 | Stroke | Overexpression | Nrf2 [58] |
| miR-223 | Stroke | Overexpression | GluR2 and NR2B [59] |
| miR-378 | Stroke | Overexpression | Caspase-3 [60] |
| miR-181 | Stroke | Inhibition | BCL2 and XIAP [61] |
| miR-181b | Stroke | Inhibition | HSPA5 and UCHL1 [62] |
| miR-155 | Stroke | Inhibition | Rheb [63] |
| miR-30a | Stroke | Inhibition | HSPA5 [64] |
| miR-27a | TBI | Overexpression | FoxO3a [50] |
| miR-144 | TBI | Inhibition | ADAM-10 [65] |
| miR-124 | Parkinson’s disease | Overexpression | Bim [66] |
| miR-7 | Parkinson’s disease | Overexpression | α-Synuclein [67] |
| miR-210 | CSI | Overexpression | PTP1B and ephrin-A3 [68] |
| miR-125b | CSI | Overexpression | Sema4D [69] |
| miR-486 | CSI | Inhibition | NeuroD6 [70] |
| miR-20a | CSI | Inhibition | Ngn1 [71] |
Neuronal plasticity and axonal regeneration
Neuronal plasticity, an intrinsic mechanism by which the CNS responds to the environment, is considerably activated after the injury and is directed toward post-damage survival. In both the brain and spinal cord, a reparative process based on neuronal plasticity includes the repair of damaged neuronal connections, and consists of different stages, such as clearance of debris, axonal sprouting, and formation of new functional synapses [72,73]. These events mainly take place in the peri-infarct or peri-impact area, however more distal (to the injury) portions of the CNS also undergo neuroanatomical changes, and thus contribute to the adaptation and compensation of impaired function. Axonal regeneration depends on the activity of different intrinsic and extrinsic factors that either inhibit or promote neuronal plasticity. The regeneration process is significantly impeded by multiple processes accompanying a sub-acute phase of injury, including generation of free radicals, ongoing demyelination, neuronal death, delayed cell death, activation of astrocytes and oligodendrocytes, formation of the barrier for the axonal sprouting, and a direct inhibition of neurite outgrowth [72,74]. Among the molecules, which inhibit post-injury plasticity are chondroitin sulphate proteoglycans and NogoA, while Inosine, Activin and other members of TGF-β family exert a neuroprotective effect and support neurite growth and axonal regeneration [72,75]. Many of these significant molecular pathways are believed to be also regulated by microRNAs [76–78]. In vivo experimental manipulation of the expression levels of several miRNAs has been proven beneficial for neurite outgrowth and axonal regeneration in the animal models of CNS injury. Among these microRNAs and their experimentally verified targets (shown in parenthesis), are: MicroRNA-431(Kremen1) [79]; miR-210 (EFNA3) [80]; miR-182 (BCAT2) [81]; miR-34a (synaptotagmin-1 SYT-1 and CTX1A) [82]; miR-127 (mitoNEET) [83]; miR-21 (PDCD4) [84]; and miR-320 (ARPP-1) [85].
Vascular reorganization
Significant vascular disfunction is associated with CNS trauma, stroke, and neurodegenerative disorders, including mild cognitive impairment and dementia, Alzheimer’s disease, arteriolosclerosis and cerebral amyloid angiopathy (small vessel disease), Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis, multiple sclerosis, and chronic traumatic encephalopathy [86,87]. Primary and secondary CNS injuries are associated with significant endothelial cell damage, which leads to destruction of microvasculature and development of hemorrhagic and ischemic states. Vascular injury and disfunction are followed by significant impairment and loss of the brain and spinal cord tissue [88,89]. Spontaneous regeneration processes involves post-injury angiogenesis and vasculogenesis [90,91]. The number of specific miRNAs is implicated in regulating endothelial morphogenesis and vascularization [92,93]. Vascular morphogenesis is a complex process that involves an intricate interplay between multiple cell signaling pathways. Formation of new capillaries includes endothelial cell morphogenesis, and subsequent maturation of intercellular junctions and the surrounding basement membrane. These stages are influenced by the number of signaling proteins, including VEGF, Notch, and TGF-β [94,95]. There are a number of miRNAs that are linked to the endothelial cell morphogenesis [96] in general, and particularly, in post-injury vascular remodeling [97,98]. In addition to the vascular reorganization, microRNAs regulate endothelial tight junctions, and thus, significantly influence the integrity and permeability of the blood brain barrier (BBB) and the blood-spinal cord barrier (BSCB). A significant number of microRNAs wave been implicated in regulating the endothelial barrier function through alteration of the expression levels and functions of the endothelial tight junction and the adherens junction proteins, including claudins, occludin, zonula occludens protein family (ZO-1,−2 and-3), VE-cadherin, and their regulatory signaling molecules [99–102]. Several regulatory microRNAs and their experimentally verified targets have been identified as the mediators of different molecular pathways involved in endothelial integrity and vascular remodeling in the animal models of CNS injury. Among them are miR-320a (AQP1) [102]; miR-363 (Timp-1 and THBS3) [103]; miR-150 (VEGF) [104]; and miR-155 (Annexin-2, claudin-1, DOCK-1, Rheb) [105,106].
Neuroinflammation
Post-injury inflammatory response represents an integral part of the injury, defense response, and recovery after CNS trauma, hypoxia, infection, and neurodegeneration. Neuroinflammation is associated with the elevation of cytokines; recruitment of neutrophils, lymphocytes, and monocytes; and activation of resident microglia, astrocytes, and endothelial cells. The cellular inflammatory response leads to the additional release of cytokines/chemokines and other pro-inflammatory factors [107,108]. While it is known that lower levels of pro-inflammatory cytokines and higher expression of anti-inflammatory cytokines are associated with a better clinical outcome, it is also accepted that inflammation-associated events play an active role in tissue remodeling and recovery. Among different factors triggered by the “injury” signals from the damaged CNS tissue are cytokines such as interleukins −1, −6, −4, −10 (IL-1α and IL-1β, IL-6, IL-4, IL-10) released within minutes after the insult, tumor necrosis factor-α (TNFα), interferon-γ (IFNγ), nitric oxide (NO), and cyclooxygenase-2 (COX-2) [109–111]. All these factors are context-dependent, and, at different times during the inflammation, exert either pro- or anti-inflammatory functions. Cytokine signaling is mediated via essential signaling pathways including JAK/STAT pathway, and negatively regulated by a number of molecules, such as SHIP-1, and SOCS family proteins. microRNAs are now known to be actively involved in every aspect of neuroinflammation, including the dynamic changes in cellular response and cytokine release/signaling, monocyte recruitment and infiltration, the interaction between different types of resident and infiltrated cells, and the de-activation and resolution of neuroinflammation. Based on these multiple regulatory functions, it is evident that miRNAs have an essential influence on both innate and adaptive immune responses. Increasing evidence supports the involvement of miRNAs as key regulators of neuroinflammation associated with various CNS pathologies [112–114]. Table 2 summarizes the list of regulatory microRNAs and their targets, which, after the interventional changes in their expression (inhibition or overexpression), modulate neuroinflammation and recovery in the animal models of CNS injury.
Table 2: In vivo inhibition or overexpression of specific microRNAs alters neuroinflammation in the animal models of CNS injury.
Table 2 summarizes the list of regulatory microRNAs and their targets, which, after the experimental intervention using miRNA inhibitors and mimics, modified the inflammation and supported recovery in the animal models of CNS injury. The process is mediated via the experimentally verified direct target genes/proteins.
| miRNA | Animal model of CNS injury | Treatment | Involved direct miRNA targets |
|---|---|---|---|
| miR-200b | TBI | Overexpression | c-Jun [115] |
| miR-3473b | Stroke | Inhibition | SOCS3 [116] |
| miR-155 | Stroke | Inhibition | SOCS-1, SHIP-1, and C/EBP-β [106] |
| miR-let-7c | TBI | Overexpression | Caspase-3 [117] |
| miR-27a | LPS-induced neuroinflammation (animal model of AD, PD, and ALS) | Overexpression | TLR4 and IRAK4 [118] |
| miR-367 | Intracerebral hemorrhage | Overexpression | IRAK4 [119] |
| miR-21 | EAE | Inhibition | SMAD-7 [120] |
Endogenous stem cell activation
In the adult mammalian brain, neural stem/progenitor cells (NSPCs) are primarily restricted to the subventricular zone of the lateral ventricles and subgranular zone of the dentate gyrus, and, in low numbers, in septum, striatum, and cortex. NSPCs have been identified in the spinal cord (the ependymal cell layer lining the central canal), optic nerve, and retina, where neurogenesis persists throughout adulthood [121–123]. The external signals and intracellular mechanisms that control NSPC generation, function and behavior following injury have been studied intensely, and various aspect of neuronal replacement and cell-based therapy have been introduced. In this review, we focus on the contribution of the endogenous neural stem cells to repair mechanisms, and the possible regulation of this process by microRNAs.
Neurogenesis
It is well established that oxygen is an important signal in all major aspects of stem cell biology. In vivo studies utilizing experimental models of ischemia showed that NSPCs strongly respond to hypoxia by massive proliferation and migration towards the stroke-induced brain lesion [43] indicating the importance of the NSPCs in the adaptation and possible recovery following acute brain damage or prolonged pathological conditions. Functional recovery from TBI is accompanied by active neurogenesis primarily in the hippocampal dentate gyrus [124–126]. In the spinal cord, neurogenesis occurs in response to specific kinds of spinal injury, including dorsal root lesion, compression, contusion, or dorsal funiculus incision [127–130].
Activation, proliferation, and differentiation of NPSCs in specific regions of the CNS are accompanied by significant changes in gene and protein profiles, which together lead to the upregulation of pro-neurogenic cell signaling cascades. Recent findings demonstrate that miRNAs specifically regulate neurogenesis, including the proliferation, migration, cell fate determination, and differentiation of various stem cell populations during CNS development and injury [131–134]. Most of these studies have been executed using neural stem/progenitor cells in vitro. Several studies utilizing the in vivo inhibition or overexpression approaches, identified the miRNAs and their direct targets that are actively involved in the endogenous stem cell activation in the animal models of CNS injury. It has been demonstrated that miR-7 overexpression resulting in suppression of its direct target NLRP3, supports SVZ neurogenesis in the animal model of Parkinson’s disease [135]. In the animal models of stroke, overexpression of miR-124a supports neurogenesis by directly targeting JAG1 protein [136], and miR-17–92 cluster mediates neural progenitor cell proliferation via its target protein PTEN [137].
Oligodendrogenesis
A small portion of SVZ neural progenitors produces oligodendrocyte progenitor cells (OPCs), which migrate into the white matter and cortex [138]. Loss of oligodendrocytes and demyelination associated with CNS damage are accompanied by active oligodendrogenesis, spontaneous remyelination, and myelination of new sprouting axons [139–141]. These processes are associated with the activation of parenchymal and SVZ-derived oligodendrocyte progenitor cells, and their migration toward the lesion [142–145]. It has been demonstrated that following CNS damage induced by stroke, OPCs migrate out of the SVZ and differentiate into myelin forming oligodendrocytes [146,140]. The number of OPCs significantly increases after TBI and spinal cord injury, which could contribute to the repair process by improving white matter function [147–150]. miRNAs have been identified as key regulators of oligodendrocyte functions, myelination, and OPCs generation and differentiation [151,140,152]. Functional in vivo studies have identified that increased expression of miR-146a promotes stroke-induced oligodendrogenesis [153], while overexpression of miR-219 after LPC-induced demyelination supports OPC maturation and regeneration processes in mice [154]. Transgenic mice with overexpressed miR-23a in oligodendrocytes demonstrated enhanced oligodendrocyte differentiation and myelination of CNS axons [155].
Multifunctional miRNA miR-155
Several miRNAs have been directly associated with different types of CNS pathologies. Among them is miR-155, a multifunctional and broadly conserved miRNA implicated in regulating various physiological and pathological processes. MiR-155 is processed from an exon of a noncoding RNA transcribed from the B-cell Integration Cluster (BIC) located on chromosome 21. BIC shows strong sequence homology among human, mouse, and chicken, implying an evolutionary conserved function [156–158]. miR-155 is specifically expressed in hematopoietic cells and cells involved in vascular remodeling [159,160]. In the brain tissue, the expression of miR-155 has been detected in the cerebrovascular endothelium, astrocytes, and microglia [158,161,162]. Apart from being regarded as a major pro-tumorigenic and pro-inflammatory miRNA, this miRNA is implicated in regulating hematopoietic lineage differentiation, endothelial and vascular function, and the progression of cardiovascular diseases [156,157,163]. Silencing of this miRNA is accompanied by reduced inflammation and improved regeneration processes [164–166].
Stroke-associated ischemic damage involves blood-brain barrier dysfunction, microvascular injury; post-ischemic inflammation; and, ultimately, the death of neurons, glia, and endothelial cells, which directly contributes to cerebral tissue damage and neuronal death [167,88,168]. miR-155 is involved in the progression of multiple CNS disorders and pathological conditions. Its increased expression is associated with poor prognosis in patients with amyotrophic lateral sclerosis [169], epilepsy [170], multiple sclerosis [171], and brain tumor [172,173]. miR-155 upregulation was detected in the animal model of stroke, while miR-155 inhibition supported post-stroke recovery [63,106]. Cerebral ischemia induces a cascade of biological events involving activation and upregulation of specific genes and cell signaling pathways, which is an essential component of post-stroke recovery. miR-155 and its target genes and proteins might play a significant role in the regeneration after stroke. Among predicted and experimentally identified miR-155 targets, with possible effect on stroke progression and outcome, are Rheb, Rictor, SMAD-5, SOCS-1, SHIP-1, and C/EBP-β [174,106,175,176]. In addition, it has been demonstrated that major TJ protein claudin-1 is a direct target of human miR-155 [105,177]. As the essential components of the mTOR, TGF-β/BMP, NO, PI3K/Akt, and JAK/STAT signaling pathways, these molecules broadly influence vascular function, neuroinflammation, and brain tissue remodeling.
Effect of systemic miR-155 inhibition after the experimental cerebral ischemia
Effect on cerebral vasculature
The in vitro studies identified miR-155 as a potential regulator of the endothelial morphogenesis: specific miR-155 antisense inhibitors supported capillary-like tube formation by the mouse brain endothelial cells [174]. Further experiments revealed that miR-155 inhibition improved HBMEC monolayer integrity and barrier function after oxygen glucose deprivation (OGD). In addition, miR-155 inhibition significantly increased the levels of major endothelial tight junction (TJ) proteins claudin-1 and ZO-1. Based on the detected association between ZO-1 and claudin-1 and their stabilization at the endothelial membrane, it was concluded that miR-155 inhibition strengthens the endothelial TJs after the OGD via stabilization of its direct target protein claudin-1.
Intravenous injections of a specific anti-miR-155 inhibitor, initiated at 48 hours after mouse distal middle cerebral artery occlusion (dMCAO), lead to a significant improvement of cerebrovascular functions, which reflected a significant enhancement of blood flow in the peri-infarct area, improvement of vascular integrity, and preservation of the capillary TJs.
Effect on brain damage and post-stroke neuroinflammation
An assessment of the brain tissue damage using MRI and electron microscopy (EM) demonstrated that at three weeks after stroke there was a significant (34%) reduction of the infarct size and a significant decrease in neuronal damage in miR-155 inhibitor-injected animals, as compared to the control group. miR-155 inhibition after dMCAO significantly altered the time course and the expression levels of the major cytokines (including IL-10, IL-4, IL-6, MIP-1α, IL-5, and IL-17) as well as considerably modified the microglia/macrophage phenotype in the peri-infarct area of stroke. Electron microscopy-based quantification detected a decreased number of phagocytically active peri-vascular microglia/macrophages in these animals [175].
Effect on the overall post-stroke recovery
Assessment of sensorimotor deficits (bilateral asymmetry/adhesive removal test) and gait/locomotion recovery (CatWalk system), as well as the weight-gain evaluation, indicated that the inhibitor-injected animals regained their sensorimotor deficits and recovered faster than controls [106]. Experiments utilizing miR-155 inhibition in the rat model of cerebral ischemia also demonstrated the efficacy of anti-miR-155 treatment in rats [63].
Molecular mechanisms of miR-155 inhibition-induced support of post-stroke recovery
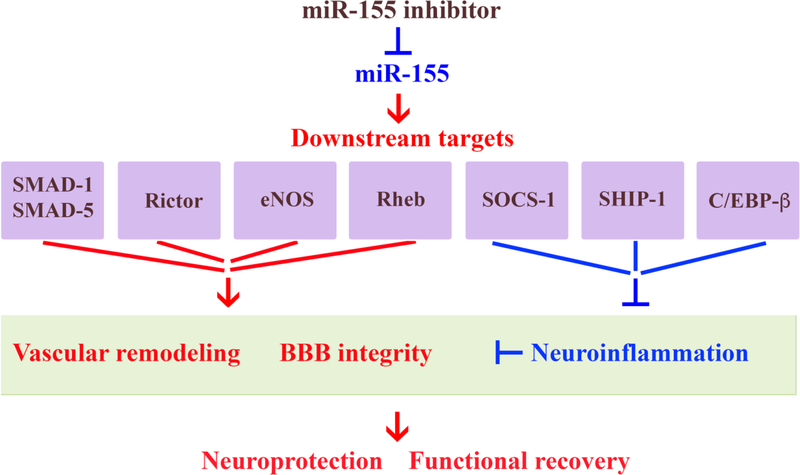
Based on all of the findings described above, it is possible to conclude that miR-155 inhibition has a beneficial effect on the regeneration after stroke. Systemic inhibition of miR-155 following the experimental cerebral ischemia supports cerebral microvasculature and improves cerebral blood supply to the peri-infarct area of stroke. These improvements are achieved via direct preservation of TJ integrity and suppression of early stage post-stroke inflammation. This recovery mechanism is facilitated by: 1) the initial preservation of vascular integrity, which prevents the propagation of the ischemic damage into the peri-infarct area; 2) the activation of IL-10-mediated neuroprotective mechanisms; and 3) transition from harmful phenotype toward the neuroprotective and reparative microglia/macrophage phenotype. All these support mechanisms could be mediated via the activation of miR-155 direct target proteins, including Rheb, SMAD-1, SMAD-2, SMAD-5, Rictor, eNOS, SOCS-1, SHIP-1, and C/EBP-β (Figure 2).
Figure 2: Possible molecular mechanisms mediating functional recovery supported by in vivo miR-155 inhibition after stroke.
miR-155 inhibition results in the increased expression of miR-155 target proteins SMAD-1 and SMAD5 (components of BMP signaling pathway), Rictor (mTOR pathway), eNOS (NO pathway), Rheb (activates Akt/ZO-1 pathway), which support microvascular function and strengthen the BBB integrity. Upregulation of other miR-155 targets, such as SOCS-1 and SHIP-1 (which suppress JAK/STAT-mediated cytokine signaling), and C/EBP (which activates anti-inflammatory IL-10), regulate post-stroke neuroinflammation and thus, support vascular integrity and neuronal survival. As a result, miR-155 inhibitor-induced support of BBB integrity leads to the reduction of brain edema, restoration blood flow in the peri-infarct area of stroke, and prevents delayed neuronal death in the peri-infarct area. This results in the reduced brain infarct, neuroprotection and improved functional recovery. Red font/arrow indicate upregulation/activation; blue - downregulation/inhibition.
miRNAs as the markers and therapeutic targets for CNS regeneration
The expression profiles of the miRNAs circulating in cerebrospinal fluid and blood reflect the molecular pathophysiology associated with CNS damage, thus making them promising biomarkers in diagnosis and prognosis of CNS trauma, diseases, and pathological conditions. There is an intensive search for the biomarkers of CNS damage focused on miRNA expression changes in blood, CSF, and saliva. The results are however characterized by significant variations, as they critically depend on methodology, timing of sample collection, and the disease form and stage (acute, subacute, chronic, etc.). To avoid these variable factors, the standardized protocols for circulating miRNA are needed for future studies [178]. Among the wide range of identified miRNAs, some are now considered as biomarkers for severity and prognosis for patients with Parkinson’s disease (miR-30b, miR-30c, miR-26a, miR-133b, and miR-126 [179,180]), acute ischemic stroke (let-7b, miR-16, miR-21, miR-106b, miR-320d, and miR-1246 [181]), Alzheimer’s disease (miR-29b-1, miR-29a, and miR-9 [182], spinal cord injury (miR-21, miR-133b, miR-9–3p, miR-219, miR-384–5p, mir-204–5p, mir-519d-3p, mir-20b-5p, and mir-6838–5p, [183,184]), and TBI (miR-1255b, miR-151–5p, miR-194, miR-195, miR-199a-3p, miR-20a, and miR-27a among others [185]). Please see the complete lists of miRNAs considered potentially to be used as biomarkers of CNS aberrations in previously published reviews [178],[186],[187].
As the changes in miRNA profiles strongly correlate with different CNS pathologies, a number of these small RNAs are considered to be used for targeted therapeutic approaches to enhance the regeneration processes in the future. Several miRNA-based therapies, focusing on targeted inhibition of specific miRNAs, have been recently introduced and supported by the leading RNA-therapeutic companies [188]. While several of these investigations remain in the pre-clinical stage, some of them, involving miR-122 inhibitor miravirsen or miR-34a inhibitor MRX34, have already entered into a phase II clinical trial for hepatitis C, and a phase I clinical study for cancer treatments, respectively. The studies demonstrate a substantial, prolonged, and highly specific decrease in plasma miR-122 levels in patients receiving subcutaneous injections of miravirsen [189]. MRX34 treatment demonstrated an acceptable safety and antitumor activity in patients with solid tumors [190]. Antagomir-based therapeutics have been potentially indicated for various human diseases and pathological conditions, including kidney disease (targeting miR-21), diabetes/obesity (using anti-miR-208), erythrocyte deficiency (anti-miR-451), and myocardial infarction (anti-miR-15) [188]. Despite the intense interest in the innovative miRNA targeting therapies, there are currently no trials related to CNS damage and repair. However, the emerging role of miRNAs as therapeutic agents for treatment of CNS pathologies has been proposed by numerous researchers and summarized in several recent reviews [191,178].
One of the major challenges of miRNA-based therapy is to achieve specific, safe, and efficient modulation of miRNAs. While the oral administration of miRNA inhibitors (or mimics) is inefficient, subcutaneous and intravenous delivery of oligonucleotides is also problematic because of their instability and limited bioavailability. Therefore, lipid-based delivery vehicles, viral vectors, nanoparticle-conjugated oligonucleotides, and biodegradable polymers are utilized for the introduction of synthetic miRNA inhibitors and mimics. Recently developed locked nucleic acid (LNA)-based technology greatly increased the pharmacokinetic properties of the miRNA inhibitors/mimics, improved their resistance to enzymatic degradation, and minimized off-target effects. As a future development, miRNA-based technology is expected to establish more sophisticated delivery systems targeting specific cells in human organisms. One of the novel approaches in miRNA-based therapy involves the exosome-mediated delivery of specific functional miRNAs. It has been proposed that administration of specific miRNA-enriched exosomes could be used as a therapy for stroke, TBI and other CNS pathologies [192–194]. Among the miRNAs involved in the beneficial effect of treatment with mesenchymal stromal cell-derived exosomes, are miR-133b and miR-17–92 cluster [195–197].
Besides the direct inhibition/overexpression of miRNAs using synthetic oligonucleotides or miRNA-enriched exosomes, a different approach could be pursued focusing on certain known drugs with a modulatory effect on certain miRNAs. In addition to the earlier described pharmaceutical compounds [178], several more drugs have demonstrated a strong and prolonged effect on specific miRNA expression in damaged CNS. Among them are: acetylbritannilactone (suppresses miR-155 expression), which reduces neuroinflammation after ischemia in mice [198]; resolvin D1 (targets miR-146b and miR-219a), which promotes recovery after the focal brain damage in rats [199]; dexmedetomidine (suppresses miR-124, −132, −134, and −155 [200]) and hesperidin (upregulates miR-132 [201]), which reduce neuroinflammation in LPS-treated rat brain; arctigenin (upregulates miR-16 and miR-199a), which provides neuroprotection against mechanical trauma injury in human neural cells [202]; amikacin (inhibits miR-497 maturation), which provides neuroprotection after the in vitro ischemia [203]; calycosin (upregulates miR-375) with a protective effect following ischemia/reperfusion in rats [204]; and finally, lithium, which was found to promote miR-124 expression and have a neuroprotective effect in the mouse model of stroke [205]. In addition, a number of natural agents have been found to have a strong modulatory effect on miRNAs. Interestingly, miR-155 is suppressed by turmeric, a polyphenolic compound derived from the dietary spice turmeric, known to have anti-inflammatory and anti-tumorigenic properties [206]. Natural compounds, including resveratrol, genistein, epigallocatechin-3-gallate, indole-3-carbinol, and other agents regulating miRNA expression are discussed in detail in recently published reviews [207,208].
Conclusion
Dysregulated miRNA functions following CNS damage have profound effects on their target genes, which are involved in both the progression of the CNS injury and the subsequent recovery process. Numerous studies have identified a number of miRNAs and their target genes, which are involved in the CNS repair process, promoting neuroprotection, neurogenesis, axon regeneration, neuronal plasticity, angiogenesis, and vasculogenesis. The associated changes in miRNA profiles provide evidence that many of them could be considered as markers for CNS damage and/or repair, and that their modulation could be beneficial for recovery. Thus, miRNAs represent an important class of molecules, which offer an understanding of CNS damage and repair, and represent the ideal biomarkers for diagnosis and prognosis. Most importantly, a targeted modulation of specific miRNA expression has promising potential as a strategy for treatment of CNS injuries. Despite the current challenges, miRNA-based therapy is expected to become an effective and innovative pharmaceutical approach in the future.
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke -NIH R01NS082225 grant.
Abbreviations:
- NogoA
neurite outgrowth inhibitor
- TGF-β
Transforming growth factor beta
- VEGF
vascular endothelial growth factor
- VE-cadherin
vascular endothelial cadherin
- TBI
traumatic brain injury
- Rheb
Ras homolog enriched in brain
- mTOR
mammalian target of rapamycin
- Rictor
Rapamycin-insensitive companion of mammalian target of rapamycin
- C/EBP-β
CCAAT/enhancer-binding protein beta
- BMP
Bone morphogenetic protein
- NO
nitric oxide
- JAK
Janus kinase
- STAT
signal transducers and activators of transcription
- SOCS
suppressor of cytokine signaling
- SHIP
Src homology 2 (SH2) domain-containing protein-tyrosine phosphatase
- LPS
Lipopolysaccharide
Footnotes
Declaration of Conflicting Interests
The author declares no competing financial interests.
References
- 1.Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S (2010) microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng 12:1–27. doi: 10.1146/annurev-bioeng-070909-105314 [DOI] [PubMed] [Google Scholar]
- 2.Fabian MR, Sonenberg N (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19 (6):586–593. doi: 10.1038/nsmb.2296 [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP (2018) Metazoan MicroRNAs. Cell 173 (1):20–51. doi: 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickens M, Takayama K (1994) RNA. Deviants--or emissaries. Nature 367 (6458):17–18. doi: 10.1038/367017a0 [DOI] [PubMed] [Google Scholar]
- 5.Ruvkun G, Wightman B, Ha I (2004) The 20 years it took to recognize the importance of tiny RNAs. Cell 116 (2 Suppl):S93–96, 92 p following S96 [DOI] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19 (1):92–105. doi: 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, Hatzimichael E, Kirino Y, Honda S, Lally M, Ramratnam B, Comstock CE, Knudsen KE, Gomella L, Spaeth GL, Hark L, Katz LJ, Witkiewicz A, Rostami A, Jimenez SA, Hollingsworth MA, Yeh JJ, Shaw CA, McKenzie SE, Bray P, Nelson PT, Zupo S, Van Roosbroeck K, Keating MJ, Calin GA, Yeo C, Jimbo M, Cozzitorto J, Brody JR, Delgrosso K, Mattick JS, Fortina P, Rigoutsos I (2015) Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci U S A 112 (10):E1106–1115. doi: 10.1073/pnas.1420955112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E, Keller A (2016) Distribution of miRNA expression across human tissues. Nucleic Acids Res 44 (8):3865–3877. doi: 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79:351–379. doi: 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 10.Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12 (2):99–110. doi: 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 11.Kawamata T, Seitz H, Tomari Y (2009) Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16 (9):953–960. doi: 10.1038/nsmb.1630 [DOI] [PubMed] [Google Scholar]
- 12.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y (2010) ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17 (1):17–23. doi: 10.1038/nsmb.1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15 (8):509–524. doi: 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448 (7149):83–86. doi: 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrera-Carrillo E, Berkhout B (2017) Dicer-independent processing of small RNA duplexes: mechanistic insights and applications. Nucleic Acids Res 45 (18):10369–10379. doi: 10.1093/nar/gkx779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22 (20):2773–2785. doi: 10.1101/gad.1705308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratnadiwakara M, Mohenska M, Anko ML (2017) Splicing factors as regulators of miRNA biogenesis - links to human disease. Semin Cell Dev Biol. doi: 10.1016/j.semcdb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 18.Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11 (9):597–610. doi: 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP, Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5 (5):396–400. doi: 10.1038/nrg1328 [DOI] [PubMed] [Google Scholar]
- 20.Bayraktar R, Van Roosbroeck K, Calin GA (2017) Cell-to-cell communication: microRNAs as hormones. Mol Oncol 11 (12):1673–1686. doi: 10.1002/1878-0261.12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohjoh H, Fukushima T (2007) Expression profile analysis of microRNA (miRNA) in mouse central nervous system using a new miRNA detection system that examines hybridization signals at every step of washing. Gene 391 (1–2):39–44. doi: 10.1016/j.gene.2006.11.018 [DOI] [PubMed] [Google Scholar]
- 22.Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T (2009) MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. Plos One 4 (10):e7225. doi: 10.1371/journal.pone.0007225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narayan A, Bommakanti A, Patel AA (2015) High-throughput RNA profiling via up-front sample parallelization. Nat Methods 12 (4):343–346. doi: 10.1038/nmeth.3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu NK, Wang XF, Lu QB, Xu XM (2009) Altered microRNA expression following traumatic spinal cord injury. Experimental neurology 219 (2):424–429. doi: 10.1016/j.expneurol.2009.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan YB, Sun ZL, Feng DF (2017) The Role of MicroRNA in Traumatic Brain Injury. Neuroscience 367:189–199. doi: 10.1016/j.neuroscience.2017.10.046 [DOI] [PubMed] [Google Scholar]
- 26.Martinez B, Peplow PV (2017) MicroRNAs as diagnostic markers and therapeutic targets for traumatic brain injury. Neural Regen Res 12 (11):1749–1761. doi: 10.4103/1673-5374.219025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei P, Li YH, Chen X, Yang SY, Zhang JN (2009) Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res 1284:191–201. doi:Doi 10.1016/J.Brainres.2009.05.074 [DOI] [PubMed] [Google Scholar]
- 28.Hu ZH, Yu DN, Almeida-Suhett C, Tu K, Marini AM, Eiden L, Braga MF, Zhu J, Li Z (2012) Expression of miRNAs and Their Cooperative Regulation of the Pathophysiology in Traumatic Brain Injury. Plos One 7 (6). doi: 10.1371/journal.pone.0039357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sepramaniam S, Tan JR, Tan KS, DeSilva DA, Tavintharan S, Woon FP, Wang CW, Yong FL, Karolina DS, Kaur P, Liu FJ, Lim KY, Armugam A, Jeyaseelan K (2014) Circulating microRNAs as biomarkers of acute stroke. Int J Mol Sci 15 (1):1418–1432. doi: 10.3390/ijms15011418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijayan M, Reddy PH (2016) Peripheral biomarkers of stroke: Focus on circulatory microRNAs. Biochim Biophys Acta 1862 (10):1984–1993. doi: 10.1016/j.bbadis.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henshall DC (2014) MicroRNA and epilepsy: profiling, functions and potential clinical applications. Curr Opin Neurol 27 (2):199–205. doi: 10.1097/WCO.0000000000000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raoof R, Jimenez-Mateos EM, Bauer S, Tackenberg B, Rosenow F, Lang J, Onugoren MD, Hamer H, Huchtemann T, Kortvelyessy P, Connolly NMC, Pfeiffer S, Prehn JHM, Farrell MA, O’Brien DF, Henshall DC, Mooney C (2017) Cerebrospinal fluid microRNAs are potential biomarkers of temporal lobe epilepsy and status epilepticus. Sci Rep 7 (1):3328. doi: 10.1038/s41598-017-02969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez B, Peplow PV (2017) MicroRNAs in Parkinson’s disease and emerging therapeutic targets. Neural Regen Res 12 (12):1945–1959. doi: 10.4103/1673-5374.221147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoss AG, Labadorf A, Beach TG, Latourelle JC, Myers RH (2016) microRNA Profiles in Parkinson’s Disease Prefrontal Cortex. Frontiers in aging neuroscience 8:36. doi: 10.3389/fnagi.2016.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bekris LM, Lutz F, Montine TJ, Yu CE, Tsuang D, Peskind ER, Leverenz JB (2013) MicroRNA in Alzheimer’s disease: an exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers 18 (5):455–466. doi: 10.3109/1354750X.2013.814073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, Bernier F, Yanagimachi M, Aoshima K, Oda Y (2013) Circulating miRNA biomarkers for Alzheimer’s disease. Plos One 8 (7):e69807. doi: 10.1371/journal.pone.0069807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieto-Diaz M, Esteban FJ, Reigada D, Munoz-Galdeano T, Yunta M, Caballero-Lopez M, Navarro-Ruiz R, Del Aguila A, Maza RM (2014) MicroRNA dysregulation in spinal cord injury: causes, consequences and therapeutics. Front Cell Neurosci 8:53. doi: 10.3389/fncel.2014.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 108 (12):5003–5008. doi: 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekstrom K, Valadi H, Sjostrand M, Malmhall C, Bossios A, Eldh M, Lotvall J (2012) Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J Extracell Vesicles 1. doi: 10.3402/jev.v1i0.18389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blandford SN, Galloway DA, Moore CS (2018) The roles of extracellular vesicle microRNAs in the central nervous system. Glia 66 (11):2267–2278. doi: 10.1002/glia.23445 [DOI] [PubMed] [Google Scholar]
- 41.Kanninen KM, Bister N, Koistinaho J, Malm T (2016) Exosomes as new diagnostic tools in CNS diseases. Biochim Biophys Acta 1862 (3):403–410. doi: 10.1016/j.bbadis.2015.09.020 [DOI] [PubMed] [Google Scholar]
- 42.Chen JJ, Zhao B, Zhao J, Li S (2017) Potential Roles of Exosomal MicroRNAs as Diagnostic Biomarkers and Therapeutic Application in Alzheimer’s Disease. Neural Plast 2017:7027380. doi: 10.1155/2017/7027380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arvidsson A, Collin T, Kirik D, Kokaia Z, Lindvall O (2002) Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med 8 (9):963–970. doi: 10.1038/nm747 nm747 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Kernie SG, Parent JM (2010) Forebrain neurogenesis after focal Ischemic and traumatic brain injury. Neurobiol Dis 37 (2):267–274. doi:S0969–9961(09)00313–1 [pii] 10.1016/j.nbd.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madathil SK, Nelson PT, Saatman KE, Wilfred BR (2011) MicroRNAs in CNS injury: potential roles and therapeutic implications. Bioessays 33 (1):21–26. doi: 10.1002/bies.201000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu NK, Xu XM (2011) MicroRNA in central nervous system trauma and degenerative disorders. Physiol Genomics 43 (10):571–580. doi: 10.1152/physiolgenomics.00168.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhalala OG, Srikanth M, Kessler JA (2013) The emerging roles of microRNAs in CNS injuries. Nat Rev Neurol 9 (6):328–339. doi: 10.1038/nrneurol.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Ji B, Cheng B, Chen J, Bai B (2014) Neuroprotection of microRNA in neurological disorders (Review). Biomed Rep 2 (5):611–619. doi: 10.3892/br.2014.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandran R, Mehta SL, Vemuganti R (2017) Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int 111:12–22. doi: 10.1016/j.neuint.2017.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun L, Zhao M, Wang Y, Liu A, Lv M, Li Y, Yang X, Wu Z (2017) Neuroprotective effects of miR-27a against traumatic brain injury via suppressing FoxO3a-mediated neuronal autophagy. Biochemical and biophysical research communications 482 (4):1141–1147. doi: 10.1016/j.bbrc.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 51.Zeng LL, He XS, Liu JR, Zheng CB, Wang YT, Yang GY (2016) Lentivirus-Mediated Overexpression of MicroRNA-210 Improves Long-Term Outcomes after Focal Cerebral Ischemia in Mice. CNS Neurosci Ther 22 (12):961–969. doi: 10.1111/cns.12589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y, Gui H, Li Q, Luo ZM, Zheng MJ, Duan JL, Liu X (2013) MicroRNA-124 protects neurons against apoptosis in cerebral ischemic stroke. CNS Neurosci Ther 19 (10):813–819. doi: 10.1111/cns.12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B, Koch JC, Bahr M, Hermann DM, Michel U (2013) MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol 126 (2):251–265. doi: 10.1007/s00401-013-1142-5 [DOI] [PubMed] [Google Scholar]
- 54.Tian YS, Zhong D, Liu QQ, Zhao XL, Sun HX, Jin J, Wang HN, Li GZ (2018) Upregulation of miR-216a exerts neuroprotective effects against ischemic injury through negatively regulating JAK2/STAT3-involved apoptosis and inflammatory pathways. J Neurosurg:1–12. doi: 10.3171/2017.5.JNS163165 [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Huang J, Ma Y, Tang G, Liu Y, Chen X, Zhang Z, Zeng L, Wang Y, Ouyang YB, Yang GY (2015) MicroRNA-29b is a therapeutic target in cerebral ischemia associated with aquaporin 4. J Cereb Blood Flow Metab 35 (12):1977–1984. doi: 10.1038/jcbfm.2015.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang LG, Li JP, Pang XM, Chen CY, Xiang HY, Feng LB, Su SY, Li SH, Zhang L, Liu JL (2015) MicroRNA-29c Correlates with Neuroprotection Induced by FNS by Targeting Both Birc2 and Bak1 in Rat Brain after Stroke. CNS Neurosci Ther 21 (6):496–503. doi: 10.1111/cns.12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao G, Ren P, Wang G, Yan F, Zhang Y (2017) MicroRNA-128–3p Protects Mouse Against Cerebral Ischemia Through Reducing p38alpha Mitogen-Activated Protein Kinase Activity. Journal of molecular neuroscience : MN 61 (2):152–158. doi: 10.1007/s12031-016-0871-z [DOI] [PubMed] [Google Scholar]
- 58.Wang P, Liang X, Lu Y, Zhao X, Liang J (2016) MicroRNA-93 Downregulation Ameliorates Cerebral Ischemic Injury Through the Nrf2/HO-1 Defense Pathway. Neurochem Res 41 (10):2627–2635. doi: 10.1007/s11064-016-1975-0 [DOI] [PubMed] [Google Scholar]
- 59.Harraz MM, Eacker SM, Wang X, Dawson TM, Dawson VL (2012) MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc Natl Acad Sci U S A 109 (46):18962–18967. doi: 10.1073/pnas.1121288109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang N, Zhong J, Han S, Li Y, Yin Y, Li J (2016) MicroRNA-378 Alleviates Cerebral Ischemic Injury by Negatively Regulating Apoptosis Executioner Caspase-3. Int J Mol Sci 17 (9). doi: 10.3390/ijms17091427 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Xu LJ, Ouyang YB, Xiong X, Stary CM, Giffard RG (2015) Post-stroke treatment with miR-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Experimental neurology 264:1–7. doi: 10.1016/j.expneurol.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng Z, Li J, Li Y, Yang X, Feng S, Han S, Li J (2013) Downregulation of miR-181b in mouse brain following ischemic stroke induces neuroprotection against ischemic injury through targeting heat shock protein A5 and ubiquitin carboxyl-terminal hydrolase isozyme L1. J Neurosci Res 91 (10):1349–1362. doi: 10.1002/jnr.23255 [DOI] [PubMed] [Google Scholar]
- 63.Xing G, Luo Z, Zhong C, Pan X, Xu X (2016) Influence of miR-155 on Cell Apoptosis in Rats with Ischemic Stroke: Role of the Ras Homolog Enriched in Brain (Rheb)/mTOR Pathway. Medical science monitor : international medical journal of experimental and clinical research 22:5141–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P, Zhang N, Liang J, Li J, Han S, Li J (2015) Micro-RNA-30a regulates ischemia-induced cell death by targeting heat shock protein HSPA5 in primary cultured cortical neurons and mouse brain after stroke. J Neurosci Res 93 (11):1756–1768. doi: 10.1002/jnr.23637 [DOI] [PubMed] [Google Scholar]
- 65.Sun L, Zhao M, Zhang J, Liu A, Ji W, Li Y, Yang X, Wu Z (2017) MiR-144 promotes beta-amyloid accumulation-induced cognitive impairments by targeting ADAM10 following traumatic brain injury. Oncotarget 8 (35):59181–59203. doi: 10.18632/oncotarget.19469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Ye Y, Zhu Z, Mo L, Lin C, Wang Q, Wang H, Gong X, He X, Lu G, Lu F, Zhang S (2016) MiR-124 Regulates Apoptosis and Autophagy Process in MPTP Model of Parkinson’s Disease by Targeting to Bim. Brain Pathol 26 (2):167–176. doi: 10.1111/bpa.12267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM (2009) Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A 106 (31):13052–13057. doi: 10.1073/pnas.0906277106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ujigo S, Kamei N, Hadoush H, Fujioka Y, Miyaki S, Nakasa T, Tanaka N, Nakanishi K, Eguchi A, Sunagawa T, Ochi M (2014) Administration of microRNA-210 promotes spinal cord regeneration in mice. Spine (Phila Pa 1976) 39 (14):1099–1107. doi: 10.1097/BRS.0000000000000356 [DOI] [PubMed] [Google Scholar]
- 69.Diaz Quiroz JF, Tsai E, Coyle M, Sehm T, Echeverri K (2014) Precise control of miR-125b levels is required to create a regeneration-permissive environment after spinal cord injury: a cross-species comparison between salamander and rat. Dis Model Mech 7 (6):601–611. doi: 10.1242/dmm.014837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jee MK, Jung JS, Choi JI, Jang JA, Kang KS, Im YB, Kang SK (2012) MicroRNA 486 is a potentially novel target for the treatment of spinal cord injury. Brain 135 (Pt 4):1237–1252. doi: 10.1093/brain/aws047 [DOI] [PubMed] [Google Scholar]
- 71.Jee MK, Jung JS, Im YB, Jung SJ, Kang SK (2012) Silencing of miR20a is crucial for Ngn1-mediated neuroprotection in injured spinal cord. Hum Gene Ther 23 (5):508–520. doi: 10.1089/hum.2011.121 [DOI] [PubMed] [Google Scholar]
- 72.Fawcett J (2009) Molecular control of brain plasticity and repair. Prog Brain Res 175:501–509. doi: 10.1016/S0079-6123(09)17534-9 [DOI] [PubMed] [Google Scholar]
- 73.Carmichael ST, Kathirvelu B, Schweppe CA, Nie EH (2017) Molecular, cellular and functional events in axonal sprouting after stroke. Experimental neurology 287 (Pt 3):384–394. doi: 10.1016/j.expneurol.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forostyak S, Jendelova P, Sykova E (2013) The role of mesenchymal stromal cells in spinal cord injury, regenerative medicine and possible clinical applications. Biochimie 95 (12):2257–2270. doi: 10.1016/j.biochi.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 75.Tedeschi A, Omura T, Costigan M (2017) CNS repair and axon regeneration: Using genetic variation to determine mechanisms. Experimental neurology 287 (Pt 3):409–422. doi: 10.1016/j.expneurol.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elramah S, Landry M, Favereaux A (2014) MicroRNAs regulate neuronal plasticity and are involved in pain mechanisms. Front Cell Neurosci 8:31. doi: 10.3389/fncel.2014.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghibaudi M, Boido M, Vercelli A (2017) Functional integration of complex miRNA networks in central and peripheral lesion and axonal regeneration. Prog Neurobiol 158:69–93. doi: 10.1016/j.pneurobio.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 78.Hu Z, Li Z (2017) miRNAs in synapse development and synaptic plasticity. Curr Opin Neurobiol 45:24–31. doi: 10.1016/j.conb.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu D, Murashov AK (2013) MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist Kremen1. Front Mol Neurosci 6:35. doi: 10.3389/fnmol.2013.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu YW, Jiang JJ, Yan G, Wang RY, Tu GJ (2016) MicroRNA-210 promotes sensory axon regeneration of adult mice in vivo and in vitro. Neurosci Lett 622:61–66. doi: 10.1016/j.neulet.2016.04.034 [DOI] [PubMed] [Google Scholar]
- 81.Wang WM, Lu G, Su XW, Lyu H, Poon WS (2017) MicroRNA-182 Regulates Neurite Outgrowth Involving the PTEN/AKT Pathway. Front Cell Neurosci 11:96. doi: 10.3389/fncel.2017.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G (2011) microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci U S A 108 (52):21099–21104. doi: 10.1073/pnas.1112063108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He QQ, Xiong LL, Liu F, He X, Feng GY, Shang FF, Xia QJ, Wang YC, Qiu DL, Luo CZ, Liu J, Wang TH (2016) MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection. Sci Rep 6:35205. doi: 10.1038/srep35205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang Y, Zhao S, Ding Y, Nong L, Li H, Gao G, Zhou D, Xu N (2017) MicroRNA21 promotes neurite outgrowth by regulating PDCD4 in a rat model of spinal cord injury. Mol Med Rep 16 (3):2522–2528. doi: 10.3892/mmr.2017.6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.White RE, Giffard RG (2012) MicroRNA-320 induces neurite outgrowth by targeting ARPP-19. Neuroreport 23 (10):590–595. doi: 10.1097/WNR.0b013e3283540394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider JA, Bennett DA (2010) Where vascular meets neurodegenerative disease. Stroke 41 (10 Suppl):S144–146. doi: 10.1161/STROKEAHA.110.598326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sweeney MD, Sagare AP, Zlokovic BV (2018) Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol 14 (3):133–150. doi: 10.1038/nrneurol.2017.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brouns R, De Deyn PP (2009) The complexity of neurobiological processes in acute ischemic stroke. Clin Neurol Neurosurg 111 (6):483–495. doi:S0303–8467(09)00082–1 [pii] 10.1016/j.clineuro.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 89.Nelson E, Gertz SD, Rennels ML, Ducker TB, Blaumanis OR (1977) Spinal cord injury. The role of vascular damage in the pathogenesis of central hemorrhagic necrosis. Arch Neurol 34 (6):332–333 [DOI] [PubMed] [Google Scholar]
- 90.Beck H, Plate KH (2009) Angiogenesis after cerebral ischemia. Acta Neuropathol 117 (5):481–496. doi: 10.1007/s00401-009-0483-6 [DOI] [PubMed] [Google Scholar]
- 91.Ng MT, Stammers AT, Kwon BK (2011) Vascular disruption and the role of angiogenic proteins after spinal cord injury. Transl Stroke Res 2 (4):474–491. doi: 10.1007/s12975-011-0109-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G (2006) MicroRNAs modulate the angiogenic properties of HUVECs. Blood 108 (9):3068–3071. doi:blood-2006-01-012369 [pii] 10.1182/blood-2006-01-012369 [DOI] [PubMed] [Google Scholar]
- 93.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S (2007) Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 101 (1):59–68. doi:CIRCRESAHA.107.153916 [pii] 10.1161/CIRCRESAHA.107.153916 [DOI] [PubMed] [Google Scholar]
- 94.Holderfield MT, Hughes CCW (2008) Crosstalk between vascular endothelial growth factor, notch, and transforming growth factor-beta in vascular morphogenesis. Circulation Research 102 (6):637–652. doi:Doi 10.1161/Circresaha.107.167171 [DOI] [PubMed] [Google Scholar]
- 95.Bobik A (2006) Transforming growth factor-betas and vascular disorders. Arterioscler Thromb Vasc Biol 26 (8):1712–1720. doi: 10.1161/01.ATV.0000225287.20034.2c [DOI] [PubMed] [Google Scholar]
- 96.Kuehbacher A, Urbich C, Dimmeler S (2008) Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci 29 (1):12–15. doi:S0165–6147(07)00277–5 [pii] 10.1016/j.tips.2007.10.014 [DOI] [PubMed] [Google Scholar]
- 97.Yin KJ, Hamblin M, Chen YE (2015) Angiogenesis-regulating microRNAs and Ischemic Stroke. Curr Vasc Pharmacol 13 (3):352–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Landskroner-Eiger S, Moneke I, Sessa WC (2013) miRNAs as modulators of angiogenesis. Cold Spring Harb Perspect Med 3 (2):a006643. doi: 10.1101/cshperspect.a006643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lopez-Ramirez MA, Reijerkerk A, de Vries HE, Romero IA (2016) Regulation of brain endothelial barrier function by microRNAs in health and neuroinflammation. FASEB J 30 (8):2662–2672. doi: 10.1096/fj.201600435RR [DOI] [PubMed] [Google Scholar]
- 100.Yin KJ, Hamblin M, Chen YE (2014) Non-coding RNAs in cerebral endothelial pathophysiology: emerging roles in stroke. Neurochem Int 77:9–16. doi: 10.1016/j.neuint.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cichon C, Sabharwal H, Ruter C, Schmidt MA (2014) MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers 2 (4):e944446. doi: 10.4161/21688362.2014.944446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li XQ, Fang B, Tan WF, Wang ZL, Sun XJ, Zhang ZL, Ma H (2016) miR-320a affects spinal cord edema through negatively regulating aquaporin-1 of blood-spinal cord barrier during bimodal stage after ischemia reperfusion injury in rats. BMC Neurosci 17:10. doi: 10.1186/s12868-016-0243-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Costa A, Afonso J, Osorio C, Gomes AL, Caiado F, Valente J, Aguiar SI, Pinto F, Ramirez M, Dias S (2013) miR-363–5p regulates endothelial cell properties and their communication with hematopoietic precursor cells. J Hematol Oncol 6 (1):87. doi: 10.1186/1756-8722-6-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He QW, Li Q, Jin HJ, Zhi F, Suraj B, Zhu YY, Xia YP, Mao L, Chen XL, Hu B (2016) MiR-150 Regulates Poststroke Cerebral Angiogenesis via Vascular Endothelial Growth Factor in Rats. CNS Neurosci Ther 22 (6):507–517. doi: 10.1111/cns.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lopez-Ramirez MA, Wu D, Pryce G, Simpson JE, Reijerkerk A, King-Robson J, Kay O, de Vries HE, Hirst MC, Sharrack B, Baker D, Male DK, Michael GJ, Romero IA (2014) MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J 28 (6):2551–2565. doi: 10.1096/fj.13-248880 [DOI] [PubMed] [Google Scholar]
- 106.Caballero-Garrido E, Pena-Philippides JC, Lordkipanidze T, Bragin D, Yang Y, Erhardt EB, Roitbak T (2015) In Vivo Inhibition of miR-155 Promotes Recovery after Experimental Mouse Stroke. J Neurosci 35 (36):12446–12464. doi: 10.1523/JNEUROSCI.1641-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cardoso AL, Guedes JR, de Lima MC (2016) Role of microRNAs in the regulation of innate immune cells under neuroinflammatory conditions. Curr Opin Pharmacol 26:1–9. doi:S1471–4892(15)00103–4 [pii] 10.1016/j.coph.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 108.Perera Nilupul M, Ma HK, Arakawa S, Howells DW, Markus R, Rowe CC, Donnan GA (2006) Inflammation following stroke. J Clin Neurosci 13 (1):1–8. doi: 10.1016/j.jocn.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 109.Lucas SM, Rothwell NJ, Gibson RM (2006) The role of inflammation in CNS injury and disease. Br J Pharmacol 147 Suppl 1:S232–240. doi: 10.1038/sj.bjp.0706400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lambertsen KL, Biber K, Finsen B (2012) Inflammatory cytokines in experimental and human stroke. J Cereb Blood Flow Metab 32 (9):1677–1698. doi: 10.1038/jcbfm.2012.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hausmann ON (2003) Post-traumatic inflammation following spinal cord injury. Spinal Cord 41 (7):369–378. doi: 10.1038/sj.sc.3101483 [DOI] [PubMed] [Google Scholar]
- 112.Su W, Aloi MS, Garden GA (2016) MicroRNAs mediating CNS inflammation: Small regulators with powerful potential. Brain Behav Immun 52:1–8. doi: 10.1016/j.bbi.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gaudet AD, Fonken LK, Watkins LR, Nelson RJ, Popovich PG (2017) MicroRNAs: Roles in Regulating Neuroinflammation. Neuroscientist:1073858417721150. doi: 10.1177/1073858417721150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thounaojam MC, Kaushik DK, Basu A (2013) MicroRNAs in the brain: it’s regulatory role in neuroinflammation. Mol Neurobiol 47 (3):1034–1044. doi: 10.1007/s12035-013-8400-3 [DOI] [PubMed] [Google Scholar]
- 115.Jadhav SP, Kamath SP, Choolani M, Lu J, Dheen ST (2014) microRNA-200b modulates microglia-mediated neuroinflammation via the cJun/MAPK pathway. J Neurochem 130 (3):388–401. doi: 10.1111/jnc.12731 [DOI] [PubMed] [Google Scholar]
- 116.Wang X, Chen S, Ni J, Cheng J, Jia J, Zhen X (2018) miRNA-3473b contributes to neuroinflammation following cerebral ischemia. Cell Death Dis 9 (1):11. doi: 10.1038/s41419-017-0014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lv J, Zeng Y, Qian Y, Dong J, Zhang Z, Zhang J (2018) MicroRNA let-7c-5p improves neurological outcomes in a murine model of traumatic brain injury by suppressing neuroinflammation and regulating microglial activation. Brain Res 1685:91–104. doi: 10.1016/j.brainres.2018.01.032 [DOI] [PubMed] [Google Scholar]
- 118.Lv YN, Ou-Yang AJ, Fu LS (2017) MicroRNA-27a Negatively Modulates the Inflammatory Response in Lipopolysaccharide-Stimulated Microglia by Targeting TLR4 and IRAK4. Cell Mol Neurobiol 37 (2):195–210. doi: 10.1007/s10571-016-0361-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yuan B, Shen H, Lin L, Su T, Zhong L, Yang Z (2015) MicroRNA367 negatively regulates the inflammatory response of microglia by targeting IRAK4 in intracerebral hemorrhage. J Neuroinflammation 12:206. doi: 10.1186/s12974-015-0424-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Murugaiyan G, da Cunha AP, Ajay AK, Joller N, Garo LP, Kumaradevan S, Yosef N, Vaidya VS, Weiner HL (2015) MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J Clin Invest 125 (3):1069–1080. doi: 10.1172/JCI74347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisen J (1999) Identification of a neural stem cell in the adult mammalian central nervous system. Cell 96 (1):25–34 [DOI] [PubMed] [Google Scholar]
- 122.Dietrich J, Kempermann G (2006) Role of endogenous neural stem cells in neurological disease and brain repair. Adv Exp Med Biol 557:191–220. doi: 10.1007/0-387-30128-3_12 [DOI] [PubMed] [Google Scholar]
- 123.Meletis K, Barnabe-Heider F, Carlen M, Evergren E, Tomilin N, Shupliakov O, Frisen J (2008) Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol 6 (7):e182. doi: 10.1371/journal.pbio.0060182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rice AC, Khaldi A, Harvey HB, Salman NJ, White F, Fillmore H, Bullock MR (2003) Proliferation and neuronal differentiation of mitotically active cells following traumatic brain injury. Experimental neurology 183 (2):406–417. doi:S0014488603002413 [pii] [DOI] [PubMed] [Google Scholar]
- 125.Richardson RM, Singh A, Sun D, Fillmore HL, Dietrich DW 3rd, Bullock MR (2010) Stem cell biology in traumatic brain injury: effects of injury and strategies for repair. J Neurosurg 112 (5):1125–1138. doi: 10.3171/2009.4.JNS081087 [DOI] [PubMed] [Google Scholar]
- 126.Sun D, Colello RJ, Daugherty WP, Kwon TH, McGinn MJ, Harvey HB, Bullock MR (2005) Cell proliferation and neuronal differentiation in the dentate gyrus in juvenile and adult rats following traumatic brain injury. J Neurotrauma 22 (1):95–105. doi: 10.1089/neu.2005.22.95 [DOI] [PubMed] [Google Scholar]
- 127.Darian-Smith C (2009) Synaptic plasticity, neurogenesis, and functional recovery after spinal cord injury. Neuroscientist 15 (2):149–165. doi: 10.1177/1073858408331372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wallace MC, Tator CH, Lewis AJ (1987) Chronic regenerative changes in the spinal cord after cord compression injury in rats. Surg Neurol 27 (3):209–219 [DOI] [PubMed] [Google Scholar]
- 129.Beattie MS, Bresnahan JC, Komon J, Tovar CA, Van Meter M, Anderson DK, Faden AI, Hsu CY, Noble LJ, Salzman S, Young W (1997) Endogenous repair after spinal cord contusion injuries in the rat. Experimental neurology 148 (2):453–463. doi: 10.1006/exnr.1997.6695 [DOI] [PubMed] [Google Scholar]
- 130.Lukovic D, Stojkovic M, Moreno-Manzano V, Jendelova P, Sykova E, Bhattacharya SS, Erceg S (2015) Concise review: reactive astrocytes and stem cells in spinal cord injury: good guys or bad guys? Stem Cells 33 (4):1036–1041. doi: 10.1002/stem.1959 [DOI] [PubMed] [Google Scholar]
- 131.Liu C, Zhao X (2009) MicroRNAs in adult and embryonic neurogenesis. Neuromolecular Med 11 (3):141–152. doi: 10.1007/s12017-009-8077-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi MA, Shi GP (2010) Intracellular delivery strategies for microRNAs and potential therapies for human cardiovascular diseases. Sci Signal 3 (146):pe40. doi:scisignal.3146pe40 [pii] 10.1126/scisignal.3146pe40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mathieu J, Ruohola-Baker H (2013) Regulation of stem cell populations by microRNAs. Adv Exp Med Biol 786:329–351. doi: 10.1007/978-94-007-6621-1_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bielefeld P, Mooney C, Henshall DC, Fitzsimons CP (2017) miRNA-Mediated Regulation of Adult Hippocampal Neurogenesis; Implications for Epilepsy. Brain Plast 3 (1):43–59. doi: 10.3233/BPL-160036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fan Z, Lu M, Qiao C, Zhou Y, Ding JH, Hu G (2016) MicroRNA-7 Enhances Subventricular Zone Neurogenesis by Inhibiting NLRP3/Caspase-1 Axis in Adult Neural Stem Cells. Mol Neurobiol 53 (10):7057–7069. doi: 10.1007/s12035-015-9620-5 [DOI] [PubMed] [Google Scholar]
- 136.Liu XS, Chopp M, Zhang RL, Tao T, Wang XL, Kassis H, Hozeska-Solgot A, Zhang L, Chen C, Zhang ZG (2011) MicroRNA profiling in subventricular zone after stroke: MiR-124a regulates proliferation of neural progenitor cells through Notch signaling pathway. Plos One 6 (8):e23461. doi: 10.1371/journal.pone.0023461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu XS, Chopp M, Wang XL, Zhang L, Hozeska-Solgot A, Tang T, Kassis H, Zhang RL, Chen C, Xu J, Zhang ZG (2013) MicroRNA-17–92 cluster mediates the proliferation and survival of neural progenitor cells after stroke. J Biol Chem 288 (18):12478–12488. doi: 10.1074/jbc.M112.449025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Suzuki SO, Goldman JE (2003) Multiple cell populations in the early postnatal subventricular zone take distinct migratory pathways: a dynamic study of glial and neuronal progenitor migration. J Neurosci 23 (10):4240–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang ZG, Chopp M (2009) Neurorestorative therapies for stroke: underlying mechanisms and translation to the clinic. Lancet Neurol 8 (5):491–500. doi: 10.1016/S1474-4422(09)70061-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang R, Chopp M, Zhang ZG (2013) Oligodendrogenesis after cerebral ischemia. Front Cell Neurosci 7:201. doi: 10.3389/fncel.2013.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.El Waly B, Macchi M, Cayre M, Durbec P (2014) Oligodendrogenesis in the normal and pathological central nervous system. Front Neurosci 8:145. doi: 10.3389/fnins.2014.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Flygt J, Djupsjo A, Lenne F, Marklund N (2013) Myelin loss and oligodendrocyte pathology in white matter tracts following traumatic brain injury in the rat. Eur J Neurosci 38 (1):2153–2165. doi: 10.1111/ejn.12179 [DOI] [PubMed] [Google Scholar]
- 143.Behrendt G, Baer K, Buffo A, Curtis MA, Faull RL, Rees MI, Gotz M, Dimou L (2013) Dynamic changes in myelin aberrations and oligodendrocyte generation in chronic amyloidosis in mice and men. Glia 61 (2):273–286. doi: 10.1002/glia.22432 [DOI] [PubMed] [Google Scholar]
- 144.Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A (2006) Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci 26 (30):7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Serwanski DR, Rasmussen AL, Brunquell CB, Perkins SS, Nishiyama A (2018) Sequential Contribution of Parenchymal and Neural Stem Cell-Derived Oligodendrocyte Precursor Cells toward Remyelination. Neuroglia 1 (1):91–105. doi: 10.3390/neuroglia1010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang RL, Chopp M, Roberts C, Jia L, Wei M, Lu M, Wang X, Pourabdollah S, Zhang ZG (2011) Ascl1 lineage cells contribute to ischemia-induced neurogenesis and oligodendrogenesis. J Cereb Blood Flow Metab 31 (2):614–625. doi: 10.1038/jcbfm.2010.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Flygt J, Gumucio A, Ingelsson M, Skoglund K, Holm J, Alafuzoff I, Marklund N (2016) Human Traumatic Brain Injury Results in Oligodendrocyte Death and Increases the Number of Oligodendrocyte Progenitor Cells. J Neuropathol Exp Neurol 75 (6):503–515. doi: 10.1093/jnen/nlw025 [DOI] [PubMed] [Google Scholar]
- 148.Flygt J, Clausen F, Marklund N (2017) Diffuse traumatic brain injury in the mouse induces a transient proliferation of oligodendrocyte progenitor cells in injured white matter tracts. Restor Neurol Neurosci 35 (2):251–263. doi: 10.3233/RNN-160675 [DOI] [PubMed] [Google Scholar]
- 149.Tripathi R, McTigue DM (2007) Prominent oligodendrocyte genesis along the border of spinal contusion lesions. Glia 55 (7):698–711. doi: 10.1002/glia.20491 [DOI] [PubMed] [Google Scholar]
- 150.Li N, Leung GK (2015) Oligodendrocyte Precursor Cells in Spinal Cord Injury: A Review and Update. Biomed Res Int 2015:235195. doi: 10.1155/2015/235195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Barca-Mayo O, Lu QR (2012) Fine-Tuning Oligodendrocyte Development by microRNAs. Front Neurosci 6:13. doi: 10.3389/fnins.2012.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Galloway DA, Moore CS (2016) miRNAs As Emerging Regulators of Oligodendrocyte Development and Differentiation. Front Cell Dev Biol 4:59. doi: 10.3389/fcell.2016.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu XS, Chopp M, Pan WL, Wang XL, Fan BY, Zhang Y, Kassis H, Zhang RL, Zhang XM, Zhang ZG (2017) MicroRNA-146a Promotes Oligodendrogenesis in Stroke. Mol Neurobiol 54 (1):227–237. doi: 10.1007/s12035-015-9655-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang H, Moyano AL, Ma Z, Deng Y, Lin Y, Zhao C, Zhang L, Jiang M, He X, Ma Z, Lu F, Xin M, Zhou W, Yoon SO, Bongarzone ER, Lu QR (2017) miR-219 Cooperates with miR-338 in Myelination and Promotes Myelin Repair in the CNS. Dev Cell 40 (6):566–582 e565. doi: 10.1016/j.devcel.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lin ST, Huang Y, Zhang L, Heng MY, Ptacek LJ, Fu YH (2013) MicroRNA-23a promotes myelination in the central nervous system. Proc Natl Acad Sci U S A 110 (43):17468–17473. doi: 10.1073/pnas.1317182110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Faraoni I, Antonetti FR, Cardone J, Bonmassar E (2009) miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 1792 (6):497–505. doi:S0925–4439(09)00042–8 [pii] 10.1016/j.bbadis.2009.02.013 [DOI] [PubMed] [Google Scholar]
- 157.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D (2010) MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33 (4):607–619. doi:S1074–7613(10)00351–1 [pii] 10.1016/j.immuni.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Elton TS, Selemon H, Elton SM, Parinandi NL (2013) Regulation of the MIR155 host gene in physiological and pathological processes. Gene 532 (1):1–12. doi: 10.1016/j.gene.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 159.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129 (7):1401–1414. doi: S0092-8674(07)00604-6 [pii] 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sun HX, Zeng DY, Li RT, Pang RP, Yang H, Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, Yan GJ, Zhou JG (2012) Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 60 (6):1407–1414. doi:HYPERTENSIONAHA.112.197301 [pii] 10.1161/HYPERTENSIONAHA.112.197301 [DOI] [PubMed] [Google Scholar]
- 161.Butovsky O, Jedrychowski MP, Cialic R, Krasemann S, Murugaiyan G, Fanek Z, Greco DJ, Wu PM, Doykan CE, Kiner O, Lawson RJ, Frosch MP, Pochet N, Fatimy RE, Krichevsky AM, Gygi SP, Lassmann H, Berry J, Cudkowicz ME, Weiner HL (2015) Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann Neurol 77 (1):75–99. doi: 10.1002/ana.24304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tarassishin L, Loudig O, Bauman A, Shafit-Zagardo B, Suh HS, Lee SC (2011) Interferon regulatory factor 3 inhibits astrocyte inflammatory gene expression through suppression of the proinflammatory miR-155 and miR-155*. Glia 59 (12):1911–1922. doi: 10.1002/glia.21233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weber M, Kim S, Patterson N, Rooney K, Searles CD (2014) MiRNA-155 targets myosin light chain kinase and modulates actin cytoskeleton organization in endothelial cells. Am J Physiol Heart Circ Physiol 306 (8):H1192–1203. doi:ajpheart.00521.2013 [pii] 10.1152/ajpheart.00521.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL (2011) Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol 187 (5):2213–2221. doi:jimmunol.1003952 [pii] 10.4049/jimmunol.1003952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S, Liew FY, McInnes IB (2011) MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A 108 (27):11193–11198. doi:1019536108 [pii] 10.1073/pnas.1019536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.van Solingen C, Araldi E, Chamorro-Jorganes A, Fernandez-Hernando C, Suarez Y (2014) Improved repair of dermal wounds in mice lacking microRNA-155. J Cell Mol Med. doi: 10.1111/jcmm.12255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Perera MN, Ma HK, Arakawa S, Howells DW, Markus R, Rowe CC, Donnan GA (2006) Inflammation following stroke. Journal of Clinical Neuroscience 13 (1):1–8. doi:Doi 10.1016/J.Jocn.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 168.Xiong X, Barreto GE, Xu L, Ouyang YB, Xie X, Giffard RG (2011) Increased brain injury and worsened neurological outcome in interleukin-4 knockout mice after transient focal cerebral ischemia. Stroke 42 (7):2026–2032. doi: 10.1161/STROKEAHA.110.593772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Koval ED, Shaner C, Zhang P, du Maine X, Fischer K, Tay J, Chau BN, Wu GF, Miller TM (2013) Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Human molecular genetics 22 (20):4127–4135. doi: 10.1093/hmg/ddt261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ashhab MU, Omran A, Kong H, Gan N, He F, Peng J, Yin F (2013) Expressions of tumor necrosis factor alpha and microRNA-155 in immature rat model of status epilepticus and children with mesial temporal lobe epilepsy. Journal of molecular neuroscience : MN 51 (3):950–958. doi: 10.1007/s12031-013-0013-9 [DOI] [PubMed] [Google Scholar]
- 171.Zhang J, Cheng Y, Cui W, Li M, Li B, Guo L (2014) MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. Journal of neuroimmunology 266 (1–2):56–63. doi: 10.1016/j.jneuroim.2013.09.019 [DOI] [PubMed] [Google Scholar]
- 172.Zhou J, Wang W, Gao Z, Peng X, Chen X, Chen W, Xu W, Xu H, Lin MC, Jiang S (2013) MicroRNA-155 promotes glioma cell proliferation via the regulation of MXI1. Plos One 8 (12):e83055. doi: 10.1371/journal.pone.0083055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yan Z, Che S, Wang J, Jiao Y, Wang C, Meng Q (2015) miR-155 contributes to the progression of glioma by enhancing Wnt/beta-catenin pathway. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 36 (7):5323–5331. doi: 10.1007/s13277-015-3193-9 [DOI] [PubMed] [Google Scholar]
- 174.Roitbak T, Bragina O, Padilla JL, Pickett GG (2011) The role of microRNAs in neural stem cell-supported endothelial morphogenesis. Vasc Cell 3:25. doi: 10.1186/2045-824X-3-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pena-Philippides JC, Caballero-Garrido E, Lordkipanidze T, Roitbak T (2016) In vivo inhibition of miR-155 significantly alters post-stroke inflammatory response. J Neuroinflammation 13 (1):287. doi: 10.1186/s12974-016-0753-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mashima R (2015) Physiological roles of miR-155. Immunology 145 (3):323–333. doi: 10.1111/imm.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pena-Philippides JC, Gardiner AS, Caballero-Garrido E, Pan R, Zhu Y, Roitbak T (2018) Inhibition of miR-155 Supports Endothelial Tight Junction Integrity Following Oxygen-Glucose Deprivation. J Am Heart Association 7 (13). doi: 10.1161/JAHA.118.009244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sun P, Liu DZ, Jickling GC, Sharp FR, Yin KJ (2018) MicroRNA-based therapeutics in central nervous system injuries. J Cereb Blood Flow Metab:271678X18773871. doi: 10.1177/0271678X18773871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Martins M, Rosa A, Guedes LC, Fonseca BV, Gotovac K, Violante S, Mestre T, Coelho M, Rosa MM, Martin ER, Vance JM, Outeiro TF, Wang L, Borovecki F, Ferreira JJ, Oliveira SA (2011) Convergence of miRNA expression profiling, alpha-synuclein interacton and GWAS in Parkinson’s disease. Plos One 6 (10):e25443. doi: 10.1371/journal.pone.0025443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kim W, Lee Y, McKenna ND, Yi M, Simunovic F, Wang Y, Kong B, Rooney RJ, Seo H, Stephens RM, Sonntag KC (2014) miR-126 contributes to Parkinson’s disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiol Aging 35 (7):1712–1721. doi: 10.1016/j.neurobiolaging.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dewdney B, Trollope A, Moxon J, Thomas Manapurathe D, Biros E, Golledge J (2018) Circulating MicroRNAs as Biomarkers for Acute Ischemic Stroke: A Systematic Review. J Stroke Cerebrovasc Dis 27 (3):522–530. doi: 10.1016/j.jstrokecerebrovasdis.2017.09.058 [DOI] [PubMed] [Google Scholar]
- 182.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ (2010) Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. Plos One 5 (2):e8898. doi: 10.1371/journal.pone.0008898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hachisuka S, Kamei N, Ujigo S, Miyaki S, Yasunaga Y, Ochi M (2014) Circulating microRNAs as biomarkers for evaluating the severity of acute spinal cord injury. Spinal Cord 52 (8):596–600. doi: 10.1038/sc.2014.86 [DOI] [PubMed] [Google Scholar]
- 184.Wang Y, Ye F, Huang C, Xue F, Li Y, Gao S, Qiu Z, Li S, Chen Q, Zhou H, Song Y, Huang W, Tan W, Wang Z (2018) Bioinformatic Analysis of Potential Biomarkers for Spinal Cord-injured Patients with Intractable Neuropathic Pain. Clin J Pain 34 (9):825–830. doi: 10.1097/AJP.0000000000000608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bhomia M, Balakathiresan NS, Wang KK, Papa L, Maheshwari RK (2016) A Panel of Serum MiRNA Biomarkers for the Diagnosis of Severe to Mild Traumatic Brain Injury in Humans. Sci Rep 6:28148. doi: 10.1038/srep28148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martirosyan NL, Carotenuto A, Patel AA, Kalani MY, Yagmurlu K, Lemole GM Jr., Preul MC, Theodore N (2016) The Role of microRNA Markers in the Diagnosis, Treatment, and Outcome Prediction of Spinal Cord Injury. Front Surg 3:56. doi: 10.3389/fsurg.2016.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Di Pietro V, Yakoub KM, Scarpa U, Di Pietro C, Belli A (2018) MicroRNA Signature of Traumatic Brain Injury: From the Biomarker Discovery to the Point-of-Care. Front Neurol 9:429. doi: 10.3389/fneur.2018.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS (2017) Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol Ther Nucleic Acids 8:132–143. doi: 10.1016/j.omtn.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.van der Ree MH, van der Meer AJ, van Nuenen AC, de Bruijne J, Ottosen S, Janssen HL, Kootstra NA, Reesink HW (2016) Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment Pharmacol Ther 43 (1):102–113. doi: 10.1111/apt.13432 [DOI] [PubMed] [Google Scholar]
- 190.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 35 (2):180–188. doi: 10.1007/s10637-016-0407-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Khoshnam SE, Winlow W, Farbood Y, Moghaddam HF, Farzaneh M (2017) Emerging Roles of microRNAs in Ischemic Stroke: As Possible Therapeutic Agents. J Stroke 19 (2):166–187. doi: 10.5853/jos.2016.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chopp M, Zhang ZG (2015) Emerging potential of exosomes and noncoding microRNAs for the treatment of neurological injury/diseases. Expert Opin Emerg Drugs 20 (4):523–526. doi: 10.1517/14728214.2015.1061993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xiong Y, Mahmood A, Chopp M (2017) Emerging potential of exosomes for treatment of traumatic brain injury. Neural Regen Res 12 (1):19–22. doi: 10.4103/1673-5374.198966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang ZG, Buller B, Chopp M (2019) Exosomes - beyond stem cells for restorative therapy in stroke and neurological injury. Nat Rev Neurol. doi: 10.1038/s41582-018-0126-4 [DOI] [PubMed] [Google Scholar]
- 195.Xin H, Li Y, Cui Y, Yang JJ, Zhang ZG, Chopp M (2013) Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J Cereb Blood Flow Metab 33 (11):1711–1715. doi: 10.1038/jcbfm.2013.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xin H, Li Y, Chopp M (2014) Exosomes/miRNAs as mediating cell-based therapy of stroke. Front Cell Neurosci 8:377. doi: 10.3389/fncel.2014.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang Y, Chopp M, Liu XS, Katakowski M, Wang X, Tian X, Wu D, Zhang ZG (2017) Exosomes Derived from Mesenchymal Stromal Cells Promote Axonal Growth of Cortical Neurons. Mol Neurobiol 54 (4):2659–2673. doi: 10.1007/s12035-016-9851-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wen Y, Zhang X, Dong L, Zhao J, Zhang C, Zhu C (2015) Acetylbritannilactone Modulates MicroRNA-155-Mediated Inflammatory Response in Ischemic Cerebral Tissues. Mol Med 21:197–209. doi: 10.2119/molmed.2014.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bisicchia E, Sasso V, Catanzaro G, Leuti A, Besharat ZM, Chiacchiarini M, Molinari M, Ferretti E, Viscomi MT, Chiurchiu V (2018) Resolvin D1 Halts Remote Neuroinflammation and Improves Functional Recovery after Focal Brain Damage Via ALX/FPR2 Receptor-Regulated MicroRNAs. Mol Neurobiol 55 (8):6894–6905. doi: 10.1007/s12035-018-0889-z [DOI] [PubMed] [Google Scholar]
- 200.Paeschke N, von Haefen C, Endesfelder S, Sifringer M, Spies CD (2017) Dexmedetomidine Prevents Lipopolysaccharide-Induced MicroRNA Expression in the Adult Rat Brain. Int J Mol Sci 18 (9). doi: 10.3390/ijms18091830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li M, Shao H, Zhang X, Qin B (2016) Hesperidin Alleviates Lipopolysaccharide-Induced Neuroinflammation in Mice by Promoting the miRNA-132 Pathway. Inflammation 39 (5):1681–1689. doi: 10.1007/s10753-016-0402-7 [DOI] [PubMed] [Google Scholar]
- 202.Song J, Li N, Xia Y, Gao Z, Zou SF, Yan YH, Li SH, Wang Y, Meng YK, Yang JX, Kang TG (2016) Arctigenin Confers Neuroprotection Against Mechanical Trauma Injury in Human Neuroblastoma SH-SY5Y Cells by Regulating miRNA-16 and miRNA-199a Expression to Alleviate Inflammation. Journal of molecular neuroscience : MN 60 (1):115–129. doi: 10.1007/s12031-016-0784-x [DOI] [PubMed] [Google Scholar]
- 203.Sinoy S, Fayaz SM, Charles KD, Suvanish VK, Kapfhammer JP, Rajanikant GK (2017) Amikacin Inhibits miR-497 Maturation and Exerts Post-ischemic Neuroprotection. Mol Neurobiol 54 (5):3683–3694. doi: 10.1007/s12035-016-9940-0 [DOI] [PubMed] [Google Scholar]
- 204.Wang Y, Dong X, Li Z, Wang W, Tian J, Chen J (2014) Downregulated RASD1 and upregulated miR-375 are involved in protective effects of calycosin on cerebral ischemia/reperfusion rats. J Neurol Sci 339 (1–2):144–148. doi: 10.1016/j.jns.2014.02.002 [DOI] [PubMed] [Google Scholar]
- 205.Doeppner TR, Kaltwasser B, Sanchez-Mendoza EH, Caglayan AB, Bahr M, Hermann DM (2017) Lithium-induced neuroprotection in stroke involves increased miR-124 expression, reduced RE1-silencing transcription factor abundance and decreased protein deubiquitination by GSK3beta inhibition-independent pathways. J Cereb Blood Flow Metab 37 (3):914–926. doi: 10.1177/0271678X16647738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ma F, Liu F, Ding L, You M, Yue H, Zhou Y, Hou Y (2017) Anti-inflammatory effects of curcumin are associated with down regulating microRNA-155 in LPS-treated macrophages and mice. Pharm Biol 55 (1):1263–1273. doi: 10.1080/13880209.2017.1297838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li Y, Kong D, Wang Z, Sarkar FH (2010) Regulation of microRNAs by natural agents: an emerging field in chemoprevention and chemotherapy research. Pharm Res 27 (6):1027–1041. doi: 10.1007/s11095-010-0105-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Phuah NH, Nagoor NH (2014) Regulation of microRNAs by natural agents: new strategies in cancer therapies. Biomed Res Int 2014:804510. doi: 10.1155/2014/804510 [DOI] [PMC free article] [PubMed] [Google Scholar]